- 1Plant Genome Engineering Research Unit, Agrogenomics Research Center, National Institute of Agrobiological Sciences, Tsukuba, Japan
- 2Kihara Institute for Biological Research, Yokohama City University, Yokohama, Japan
Stable transformation with T-DNA needs the coordinated activities of many proteins derived from both host plant cells and Agrobacterium. In dicot plants, including Arabidopsis, it has been suggested that non-homologous end-joining (NHEJ)—one of the main DNA double-strand break repair pathways—is involved in the T-DNA integration step that is crucial to stable transformation. However, how this pathway is involved remains unclear as results with NHEJ mutants in Arabidopsis have given inconsistent results. Recently, a system for visualization of stable expression of genes located on T-DNA has been established in rice callus. Stable expression was shown to be reduced significantly in NHEJ knock-down rice calli, suggesting strongly that NHEJ is involved in Agrobacterium-mediated stable transformation in rice. Since rice transformation is now efficient and reproducible, rice is a good model plant in which to elucidate the molecular mechanisms of T-DNA integration.
Introduction
Agrobacterium tumefaciens enables genetic transformation of many plant species via the movement of transferred-DNA (T-DNA) of Ti plasmids into the plant nucleus. Despite its use in both basic research and molecular breeding in several crops, many species and varieties are still recalcitrant to Agrobacterium-mediated transformation. To overcome this limitation requires not only optimization of cell and tissue culture conditions but also elucidation of the molecular mechanisms of all the events that occur during Agrobacterium-mediated transformation.
In the conventional Agrobacterium-mediated transformation system in rice, callus derived from the scutellum of mature seeds or immature embryos is generally used for Agrobacterium inoculation (Nishimura et al., 2006; Hiei and Komari, 2008). Since these tissues have a stereo architecture consisting of many cells, Agrobacterium can only infect cells on the surface, and, unlike protoplasts, it is quite difficult to isolate single cells from these tissues without any selection pressure. Indeed, there are far fewer transformed than non-transformed cells when Agrobacterium-inoculated primary calli derived from mature seeds are cultured without selection pressure in a conventional transformation system (Saika and Toki, 2009; Saika et al., 2011b). Therefore, transformed cells in which antibiotic- and herbicide-resistance genes located on T-DNA are present, but not expressed to a sufficient level, cannot be distinguished from non-transformed cells.
Agrobacterium-mediated transformation has many steps (Gelvin, 2010; Pitzschke and Hirt, 2010; Magori and Citovsky, 2011; Shiboleth and Tzfira, 2012). The last step, i.e., stable transgene expression that results from T-DNA integration into the host genome (and the avoidance of the gene silencing system) is crucial to the clonal propagation of transformed cells of rice as referred to above. Meanwhile, transient expression of transgenes is often observed at an earlier stage when transgenes are expressed stably and constantly from T-DNA that has not integrated into the rice genome. After import into the nucleus of the T-complex, which consists of single stranded T-DNA (ssT-DNA) and proteins such as virD2 and virE2, ssT-DNA is replicated to double-strand T-DNA (dsT-DNA). Hypothetical models of dsT-DNA formation have been proposed as shown in Figure 1 (Liang and Tzfira, 2013). Subsequently, transgenes located on the dsT-DNA are expressed. However, this expression is not continuous because naked T-DNA that has not integrated into the host genome is susceptible to degradation (Gelvin, 2010). The transient expression of a cytokinin biosynthesis gene, which leaves no selection marker or vector backbone in the host genome, has been exploited in a transformation system for Solanaceous plants (Richael et al., 2008). However, it is thought that transient transgene expression is not crucial to the production of clonally propagated transformed cells in the case of rice.
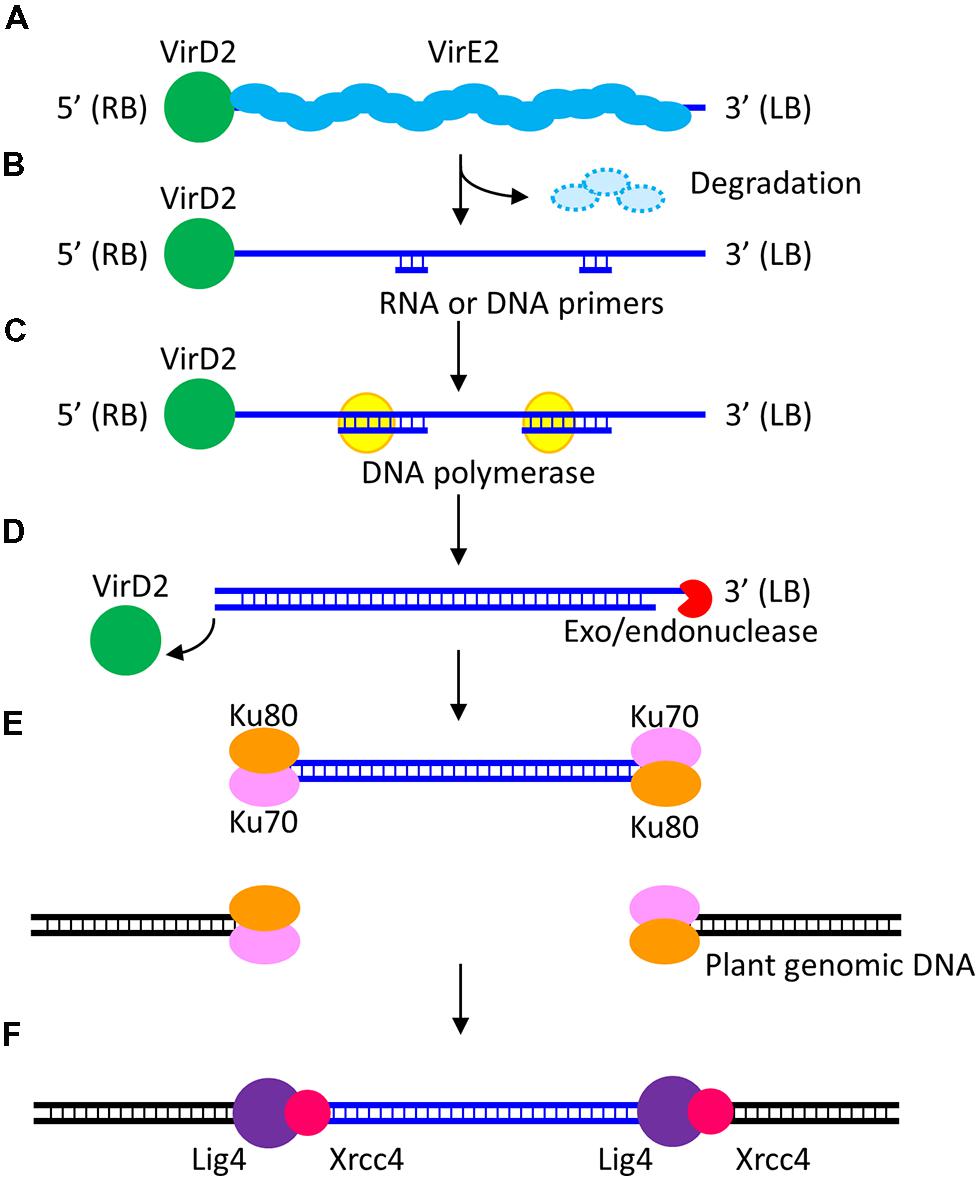
FIGURE 1. A model of T-DNA integration into the host genome via the non-homologous end-joining (NHEJ) pathway. A T-complex composed of ssT-DNA and virulence proteins VirD2 and VirE2 is transported to the host cell nucleus (A). In the nucleus, VirE2 proteins, which bind the ssT-DNA, are removed by proteasomal degradation. Short DNA or RNA primers anneal randomly to the ssT-DNA (B) and then serve as primers for polymerization of the nascent T-DNA strand (C). The 3′ end of the T-DNA is thought to be degraded by nucleases (D). dsT-DNA molecules integrate into plant genomic double strand breaks (DSBs) via the NHEJ pathway. The Ku70/80 heterodimer binds and protects the DSB ends (E). Lig4 is a ligase that joins the DSB ends. Xrcc4 interacts with Lig4 and enhances its activity (F).
Molecular Mechanism of Stable Transformation via Agrobacterium in Arabidopsis
The molecular mechanisms of T-DNA integration into the plant genome in Agrobacterium-mediated transformation remain unclear. The sequences of T-DNA integration sites were determined in Arabidopsis and tobacco plants over 20 years ago (Gheysen et al., 1991; Mayerhofer et al., 1991). Based on these pioneer reports, two major models—the strand-invasion model and the double strand break (DSB) repair model—were proposed (Gelvin, 2010; Pitzschke and Hirt, 2010; Magori and Citovsky, 2011; Shiboleth and Tzfira, 2012). In the strand-invasion model, T-DNA integration is thought to be caused by microhomology-mediated repair between ssT-DNA and the plant genome. On the other hand, in the DSB repair model, dsT-DNA is thought to integrate via non-homologous end-joining (NHEJ)—one of the main DSB repair pathways—into the DSB sites that occur randomly in the plant genome (Figure 1).
Many proteins involved in the NHEJ pathway, such as Ku70, Ku80, Ligase 4 (Lig4), and Xrcc4, have been characterized in model plants such as Arabidopsis and rice (West et al., 2000; Riha et al., 2002; Kimura and Sakaguchi, 2006; Hong et al., 2010; Nishizawa-Yokoi et al., 2012). To summarize previous reports in Arabidopsis, on the whole, the frequency of stable transformation tends to decrease in NHEJ mutants except for xrcc4, although this point remains controversial. It was reported initially that the frequency of stable transformation increased twofold in in planta floral dip transformation experiments in Arabidopsis ku80 mutants (Gallego et al., 2003), while another report demonstrated a two to threefold decrease (Friesner and Britt, 2003). However, a root tumorigenesis assay in Arabidopsis ku80 mutants confirmed that the stable transformation frequency was decreased severely, although the frequency of transient transformation remained comparable to controls (Li et al., 2005b). In addition, overexpression of Ku80 enhanced stable but not transient transformation frequency (Li et al., 2005b). Moreover, recent reports have shown that stable transformation frequency was decreased two to fourfold in Arabidopsis ku80 mutants in both root tumorigenesis assay and in in planta floral dip assay (Jia et al., 2012; Mestiri et al., 2014), and stable transformation frequency was also decreased fourfold in Arabidopsis ku70 mutants in the in planta floral dip assay (Jia et al., 2012). Thus, Arabidopsis Ku70/80 plays a positive role in stable transformation. It was proposed that Ku80 proteins interact with dsT-DNA molecules and direct them to DSB sites (Li et al., 2005b). On the other hand, there are contrary reports in Arabidopsis lig4 mutants: some authors found that stable transformation frequency was decreased to 40–70% in in planta floral dip assay (Friesner and Britt, 2003), but others found comparable rates to wild-type in both root tumorigenesis assay and in planta floral dip assay (van Attikum et al., 2003). These inconsistencies are probably due to differences in cell type (somatic cells vs germ cells) and/or experimental procedures (Gelvin, 2010; Nishizawa-Yokoi et al., 2012; Lacroix and Citovsky, 2013). Interestingly, the possibility that Xrcc4 plays a negative role in stable transformation via interaction with virE2 has been raised (Vaghchhipawala et al., 2012). In this latter theory, active Xrcc4 proteins are inhibited by virE2 proteins and T-DNA is integrated into DSB sites that are either not repaired or that occur de novo by suppression of the NHEJ pathway at a higher frequency. Agrobacterium might bring stable transformation to a successful conclusion by the good use of host proteins involved in the NHEJ pathway, among others Ku80 and Xrcc4. Just recently, Mestiri et al. (2014) reported that DSB repair pathways including NHEJ are redundantly involved in stable transformation. Stable transformation frequencies were decreased in the single mutants, ku80, xrcc1, xrcc2, and xpf (Mestiri et al., 2014), which are deficient in NHEJ, Ku-independent NHEJ (known as alternative NHEJ to distinguish it from classical Ku-dependent NHEJ), microhomology-mediated end joining/single strand annealing and homologous recombination, respectively. Interestingly, stable transformation was markedly but not completely suppressed in a quadruple mutant (Mestiri et al., 2014), possibly because the quadruple mutant did not lose DSB repair activity completely (Charbonnel et al., 2011).
Molecular Mechanism of Stable Transformation via Agrobacterium in Rice
Unlike Arabidopsis, rice is not a natural host plant of Agrobacterium; however, the combination of a sophisticated tissue culture system and activation of Agrobacterium using the phenolic compound acetosyringon, allows transgenic rice plants to be produced via this method (Hiei et al., 1994). Human cells and yeast—also not natural hosts of Agrobacterium—are also susceptible to Agrobacterium-mediated transformation (Bundock et al., 1995; Kunik et al., 2001). Moreover, the NHEJ pathway is involved in T-DNA integration in yeast (van Attikum et al., 2001; van Attikum and Hooykaas, 2003). These findings suggest that Agrobacterium uses a fundamental DNA repair system that is common to both host and non-host organisms, including rice, in the step leading to stable transformation. Since no conventional root transformation system or in planta floral dip transformation system has been established in rice, we are currently restricted to rice callus for analyzing stable transformation frequencies. Tumorigenesis assays are not applicable to rice callus because this tissue has already dedifferentiated. In Arabidopsis and Nicotiana, stable transformation frequencies are estimated by visualization of transformed cells (Mysore et al., 1998; Anand et al., 2007). In rice, transient and stable transgene expression can also be observed using green fluorescent protein—a non-destructive and visible selection marker (Toki et al., 2006; Saika et al., 2012). Visualization of transformed cells can quantify stable transformation events rapidly and easily compared to clonal propagation of transformed cells using antibiotics. We have established a sequential monitoring system for stable transformation events in rice callus that uses the enhancer trap of the click beetle luciferase gene, which is 30 times more sensitive than firefly luciferase (Saika et al., 2012). Using this system, we showed that the stable transformation frequency is decreased in knock-down lines of OsKu70, OsKu80, and OsLig4 genes and the knock-out line of the OsKu70 gene (Nishizawa-Yokoi et al., 2012; Saika et al., 2012), suggesting that the NHEJ pathway is involved in the stable transformation process also in rice.
Prospects
According to the DSB repair model, DSBs could occur in cells successfully transformed with T-DNA. In Arabidopsis, DSBs induce cell death and endocycle, which halts cell division (Adachi et al., 2011). Cells in which these events occur cannot propagate clonally if stable transformation is successful. Thus, transformation frequency in Arabidopsis might be underestimated in mutants in which DSBs occurs at higher frequency, such as NHEJ deficient mutants. However, in rice callus, endocycle does not occur even under genotoxic conditions inducing DSBs (Endo et al., 2012). Thus, transformation frequency may be estimated more accurately in rice callus than in Arabidopsis.
Transformation frequency in Arabidopsis and rice depends on the variety (Nam et al., 1997; Saika and Toki, 2010). Furthermore, optimal conditions for cell and tissue culture differ among rice varieties (Hiei and Komari, 2008). For example, Koshihikari—an elite variety from Japan—shows lower regeneration frequency due to lower activity of nitrite reductase (Nishimura et al., 2005; Ozawa and Kawahigashi, 2006). This makes it difficult to compare transformation frequency precisely among rice varieties since different callus culture conditions can affect Agrobacterium activity and growth. Thus, in order to compare transformation frequency precisely, experiments must be performed under reliable, reproducible and identical conditions using mutants with the same genetic background. Recent technological advances have made it much easier to produce plants with knockouts of targeted genes: successful gene knockouts using artificial nucleases such as transcription activator-like effector nucleases (TALENs) and CRISPR/Cas have already been reported in rice (Li et al., 2012; Feng et al., 2013; Jiang et al., 2013; Miao et al., 2013; Shan et al., 2013; Xu et al., 2014). A series of mutants in which genes involved in the NHEJ pathway are disrupted can now be produced easily in the same variety. Moreover, targeted mutagenesis using artificial nucleases enables “null” and “truncated” mutants to be produced as necessary, unlike conventional mutagenesis approaches and T-DNA tagging lines. These new technologies could help resolve some of the conflicting results in this field. For example, the involvement in stable transformation of the Arabidopsis lig4 mutant described above remains controversial, and conflicting results have also been reported in mutants in pathways other than NHEJ. For example, the plant protein VirE2 binding protein 1 (VIP1) has long been considered important in transient and/or stable transformation (Tzfira et al., 2001, 2002; Li et al., 2005a), but results that dispute this view have been reported recently (Shi et al., 2014). A study using knockout mutants of Lig4 and Vip1 in rice produced by artificial nucleases could lead to an answer.
Gene targeting (GT) is a transformation technology that can modify a targeted gene in a predicted manner. Successful examples of gene modification via GT have been reported in several higher plants including rice and Arabidopsis. However, rice is currently the only flowering plant in which target modification via GT can be performed routinely (Endo et al., 2007; Saika et al., 2011a; Nishizawa-Yokoi et al., 2014; Osakabe et al., 2014). In particular, GT using positive-negative selection enables the introduction of desirable mutations that cause amino acid substitutions in the targeted rice gene (Wakasa et al., 2012; Dang et al., 2013; Nishizawa-Yokoi et al., 2014). Molecular analyses and structure-based protein engineering can reveal the essential amino acids involved in protein–protein interactions. For example, as mentioned above, Agrobacterium virE2 protein interacts with Xrcc4 proteins (Vaghchhipawala et al., 2012) It is easy to produce mutant rice plants in which mutated Xrcc4 proteins are expressed. The comparison of transient/stable transformation frequencies in xrcc4 mutant rice lines deficient in the interaction with virE2 protein or the enhancement of Lig4 activity will be able to provide clues as to the exact roles of Xrcc4 protein in stable transformation. Similarly, a set of rice mutants expressing mutated proteins that have lost the ability to interact with proteins derived from Agrobacterium will be useful in analyzing transformation frequency. Such studies will offer new insights into the stable transformation process. Moreover, further analysis will enable the discovery of chemicals effective in strictly activating or suppressing stable transformation, and these could be applied not only in experimental procedures in plants that are recalcitrant to Agrobacterium-mediated transformation but also in cases where T-DNA integration is not desired, such as site-directed mutagenesis using artificial nucleases and GT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Rothnie for English editing. We also thank Dr. Endo, Ms. Ohtsuki, Ms. Mori, and Mr. Mikami for discussion. Our work is supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan to Hiroaki Saika and Seiichi Toki (Genomics-based Technology for Agricultural Improvement, PGE-1001, 1002), and the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry to Hiroaki Saika.
References
Adachi, S., Minamisawa, K., Okushima, Y., Inagaki, S., Yoshiyama, K., Kondou, Y.,et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 10004–10009. doi: 10.1073/pnas.1103584108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anand, A., Krichevsky, A., Schomack, S., Lahaye, T., Tzfira, T., Tang, Y. H.,et al. (2007). Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants. Plant Cell 19, 1695–1708. doi: 10.1105/tpc.106.042903
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bundock, P., Den Dulk-Ras, A., Beijersbergen, A., and Hooykaas, P. J. (1995). Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214.
Charbonnel, C., Allain, E., Gallego, M. E., and White, C. I. (2011). Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair 10, 611–619. doi: 10.1016/j.dnarep.2011.04.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dang, T. T., Shimatani, Z., Kawano, Y., Terada, R., and Shimamoto, K. (2013). Gene editing a constitutively active OsRac1 by homologous recombination-based gene targeting induces immune responses in rice. Plant Cell Physiol. 54, 2058–2070. doi: 10.1093/pcp/pct147
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Endo, M., Nakayama, S., Umeda-Hara, C., Ohtsuki, N., Saika, H., Umeda, M.,et al. (2012). CDKB2 is involved in mitosis and DNA damage response in rice. Plant J. 69, 967–977. doi: 10.1111/j.1365-313X.2011.04847.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Endo, M., Osakabe, K., Ono, K., Handa, H., Shimizu, T., and Toki, S. (2007). Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J. 52, 157–166. doi: 10.1111/j.1365-313X.2007.03230.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D.-L., Wei, P.,et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Friesner, J., and Britt, A. B. (2003). Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 34, 427–440. doi: 10.1046/j.1365-313X.2003.01738.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gallego, M. E., Bleuyard, J. Y., Daoudal-Cotterell, S., Jallut, N., and White, C. I. (2003). Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 35, 557–565. doi: 10.1046/j.1365-313X.2003.01827.x
Gelvin, S. B. (2010). Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu. Rev. Phytopathol. 48, 45–68. doi: 10.1146/annurev-phyto-080508-081852
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gheysen, G., Villarroel, R., and Van Montagu, M. (1991). Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev. 5, 287–297. doi: 10.1101/gad.5.2.287
Hiei, Y., and Komari, T. (2008). Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834. doi: 10.1038/nprot.2008.46
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hong, J.-P., Byun, M. Y., An, K., Yang, S.-J., An, G., and Kim, W. T. (2010). OsKu70 is associated with developmental growth and genome stability in rice. Plant Physiol. 152, 374–387. doi: 10.1104/pp.109.150391
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jia, Q., Bundock, P., Hooykaas, P. J. J., and Pater, S. D. (2012). Agrobacterium tumefaciens T-DNA integration and gene targeting in Arabidopsis thaliana non-homologous end-joining mutants. J. Bot. 2012, 13. doi: 10.1155/2012/989272
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. doi: 10.1093/nar/gkt780
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kimura, S., and Sakaguchi, K. (2006). DNA repair in plants. Chem. Rev. 106, 753–766. doi: 10.1021/cr040482n
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kunik, T., Tzfira, T., Kapulnik, Y., Gafni, Y., Dingwall, C., and Citovsky, V. (2001). Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 98, 1871–1876. doi: 10.1073/pnas.041327598
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lacroix, B., and Citovsky, V. (2013). The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int. J. Dev. Biol. 57, 467–481. doi: 10.1387/ijdb.130199b1
Li, J. X., Krichevsky, A., Vaidya, M., Tzfira, T., and Citovsky, V. (2005a). Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 5733–5738. doi: 10.1073/pnas.0404118102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, J. X., Vaidya, M., White, C., Vainstein, A., Citovsky, V., and Tzfira, T. (2005b). Involvement of KU80 in T-DNA integration in plant cells. Proc. Natl. Acad. Sci. U.S.A. 102, 19231–19236. doi: 10.1073/pnas.0506437103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liang, Z. B., and Tzfira, T. (2013). In vivo formation of double-stranded T-DNA molecules by T-strand priming. Nat. Commun. 4, 2253. doi: 10.1038/ncomms3253
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Magori, S., and Citovsky, V. (2011). Epigenetic control of Agrobacterium T-DNA integration. Biochim. Biophys. Acta Gene 1809, 388–394. doi: 10.1016/j.bbagrm.2011.01.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Bakkeren, G., Crameri, A., Angelis, K.,et al. (1991). T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 10, 697–704.
Mestiri, I., Norre, F., Gallego, M. E., and White, C. I. (2014). Multiple host-cell recombination pathways act in Agrobacterium-mediated transformation of plant cells. Plant J. 77, 511–520. doi: 10.1111/tpj.12398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X.,et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mysore, K. S., Bassuner, B., Deng, X. B., Darbinian, N. S., Motchoulski, A., Ream, W.,et al. (1998). Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 11, 668–683. doi: 10.1094/mpmi.1998.11.7.668
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nam, J., Matthysse, A. G., and Gelvin, S. B. (1997). Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9, 317–333. doi: 10.1105/tpc.9.3.317
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishimura, A., Aichi, I., and Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
Nishimura, A., Ashikari, M., Lin, S., Takashi, T., Angeles, E. R., Yamamoto, T.,et al. (2005). Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proc. Natl. Acad. Sci. U.S.A. 102, 11940–11944. doi: 10.1073/pnas.0504220102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa-Yokoi, A., Endo, M., Ohtsuki, N., Saika, H., and Toki, S. (2014). Precision genome editing in plants via gene targeting and piggyBac-mediated marker excision. Plant J. doi: 10.1111/tpj.12693 [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa-Yokoi, A., Nonaka, S., Saika, H., Kwon, Y. I., Osakabe, K., and Toki, S. (2012). Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol. 196, 1048–1059. doi: 10.1111/j.1469-8137.2012.04350.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Osakabe, K., Nishizawa-Yokoi, A., Ohtsuki, N., Osakabe, Y., and Toki, S. (2014). A mutated cytosine deaminase gene, codA (D314A), as an efficient negative selection marker for gene targeting in rice. Plant Cell Physiol. 55, 658–665. doi: 10.1093/pcp/pct183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ozawa, K., and Kawahigashi, H. (2006). Positional cloning of the nitrite reductase gene associated with good growth and regeneration ability of calli and establishment of a new selection system for Agrobacterium-mediated transformation in rice (Oryza sativa L.). Plant Sci. 170, 384–393. doi: 10.1016/j.plantsci.2005.09.015
Pitzschke, A., and Hirt, H. (2010). New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 29, 1021–1032. doi: 10.1038/emboj.2010.8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Richael, C. M., Kalyaeva, M., Chretien, R. C., Yan, H., Adimulam, S., Stivison, A.,et al. (2008). Cytokinin vectors mediate marker-free and backbone-free plant transformation. Transgenic Res. 17, 905–917. doi: 10.1007/s11248-008-9175-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Riha, K., Watson, J. M., Parkey, J., and Shippen, D. E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826. doi: 10.1093/emboj/21.11.2819
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saika, H., Nonaka, S., Osakabe, K., and Toki, S. (2012). Sequential monitoring of transgene expression following Agrobacterium-mediated transformation of rice. Plant Cell Physiol. 53, 1974–1983. doi: 10.1093/pcp/pcs135
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saika, H., Oikawa, A., Matsuda, F., Onodera, H., Saito, K., and Toki, S. (2011a). Application of gene targeting to designed mutation breeding of high-tryptophan rice. Plant Physiol. 156, 1269–1277. doi: 10.1104/pp.111.175778
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saika, H., Sakamoto, W., Maekawa, M., and Toki, S. (2011b). Highly efficient visual selection of transgenic rice plants using green fluorescent protein or anthocyanin synthetic genes. Plant Biotechnol. 28, 107–110. doi: 10.5511/plantbiotechnology.10.1104a
Saika, H., and Toki, S. (2009). Visual selection allows immediate identification of transgenic rice calli efficiently accumulating transgene products. Plant Cell Rep. 28, 619–626. doi: 10.1007/s00299-009-0671-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saika, H., and Toki, S. (2010). Mature seed-derived callus of the model indica rice variety Kasalath is highly competent in Agrobacterium-mediated transformation. Plant Cell Rep. 29, 1351–1364. doi: 10.1007/s00299-010-0921-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z.,et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shi, Y., Lee, L. Y., and Gelvin, S. B. (2014). Is VIP1 important for Agrobacterium-mediated transformation? Plant J. 9, 848–860. doi: 10.1111/tpj.12596
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shiboleth, Y., and Tzfira, T. (2012). “Agrobacterium-mediated plant genetic transformation,” in Plant Biotechnology and Agriculture: Prospects for the 21st Century, eds A. Altman and P. M. Hasegawa (Waltham, MA: Academic Press), 99–116. doi: 10.1016/B978-0-12-381466-1.00007-9
Toki, S., Hara, N., Ono, K., Onodera, H., Tagiri, A., Oka, S.,et al. (2006). Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976. doi: 10.1111/j.1365-313X.2006.02836.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tzfira, T., Vaidya, M., and Citovsky, V. (2001). VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20, 3596–3607. doi: 10.1093/emboj/20.13.3596
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tzfira, T., Vaidya, M., and Citovsky, V. (2002). Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc. Natl. Acad. Sci. U.S.A. 99, 10435–10440. doi: 10.1073/pnas.162304099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vaghchhipawala, Z. E., Vasudevan, B., Lee, S., Morsy, M. R., and Mysore, K. S. (2012). Agrobacterium may delay plant nonhomologous end-joining DNA repair via XRCC4 to favor T-DNA integration. Plant Cell 24, 4110–4123. doi: 10.1105/tpc.112.100495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Attikum, H., Bundock, P., and Hooykaas, P. J. J. (2001). Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 20, 6550–6558. doi: 10.1093/emboj/20.22.6550
van Attikum, H., Bundock, P., Overmeer, R. M., Lee, L. Y., Gelvin, S. B., and Hooykaas, P. J. J. (2003). The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 31, 4247–4255. doi: 10.1093/nar/gkg458
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Attikum, H., and Hooykaas, P. J. J. (2003). Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 31, 826–832. doi: 10.1093/nar/gkg183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wakasa, Y., Hayashi, S., Ozawa, K., and Takaiwa, F. (2012). Multiple roles of the ER stress sensor IRE1 demonstrated by gene targeting in rice. Sci. Rep. 2, 944. doi: 10.1038/srep00944
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
West, C. E., Waterworth, W. M., Jiang, Q., and Bray, C. M. (2000). Arabidopsis DNA ligase IV is induced by γ-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 24, 67–78. doi: 10.1046/j.1365-313x.2000.00856.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, R., Li, H., Qin, R., Wang, L., Li, L., Wei, P.,et al. (2014). Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 7, 5. doi: 10.1186/s12284-014-0005-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Agrobacterium, non-homologous end-joining, rice, stable transformation, T-DNA
Citation: Saika H, Nishizawa-Yokoi A and Toki S (2014) The non-homologous end-joining pathway is involved in stable transformation in rice. Front. Plant Sci. 5:560. doi: 10.3389/fpls.2014.00560
Received: 29 August 2014; Accepted: 29 September 2014;
Published online: 17 October 2014.
Edited by:
Toshihiko Komari, Japan Tobacco Inc., JapanReviewed by:
Sylvia De Pater, Leiden University, NetherlandsKinya Toriyama, Tohoku University, Japan
Copyright © 2014 Saika, Nishizawa-Yokoi and Toki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroaki Saika, Plant Genome Engineering Research Unit, Agrogenomics Research Center, National Institute of Agrobiological Sciences, 2-1-2 Kannondai, Tsukuba, Ibaraki 305-8602, Japan e-mail: saika@affrc.go.jp
 Hiroaki Saika
Hiroaki Saika Ayako Nishizawa-Yokoi
Ayako Nishizawa-Yokoi Seiichi Toki
Seiichi Toki