- 1National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China
- 2Institut de Biologie des Plantes, UMR8618, Université Paris-Sud 11, Orsay, France
Recent advances in rice flowering studies have shown that the accurate control of flowering by photoperiod is regulated by key mechanisms that involve the regulation of flowering genes including Heading date1 (Hd1), Early hd1 (Ehd1), Hd3a, and RFT1. The chromatin mechanism involved in the regulation of rice flowering genes is presently not well known. Here we show that the rice enhancer of zeste [E(z)] genes SDG711 and SDG718, which encode the polycomb repressive complex2 (PRC2) key subunit that is required for trimethylation of histone H3 lysine 27 (H3K27me3), are respectively, involved in long day (LD) and short day (SD) regulation of key flowering genes. The expression of SDG711 and SDG718 is induced by LD and SD, respectively. Over-expression and down-regulation of SDG711 respectively, repressed and promoted flowering in LD, but had no effect in SD. By contrast, down-regulation of SDG718 had no effect in LD but delayed flowering in SD. SDG711 and SDG718 repressed OsLF (a repressor of Hd1) respectively in LD and SD, leading to a higher expression of Hd1 thus late flowering in LD and early flowering in SD. SDG711 was also found to be involved in the repression of Ehd1 in LD. SDG711 was shown to directly target to OsLF and Ehd1 loci to mediate H3K27me3 and gene repression. The function of the rice E(z) genes in LD repression and SD promotion of flowering suggests that PRC2-mediated epigenetic repression of gene expression is involved in the accurate photoperiod control of rice flowering.
Introduction
The control of flowering time is a critical step for successful grain production in rice. Day length is a key factor controlling rice flowering. Most rice cultivars recognize 13.5 h light/10.5 h dark as a critical photoperiod to separate long day (LD) from short day (SD) periods, day length shorter than 13.5 h will greatly induce rice flowering (Itoh et al., 2010; Tsuji et al., 2013). Despite some genes are shared between Arabidopsis and rice flowering regulatory pathways, there are considerable differences between the regulation of flowering of both species such as absence of the vernalization pathway in rice (Shrestha et al., 2014). Heading date1 (Hd1), the rice ortholog of Arabidopsis CONSTANS (CO), has a dual role in flowering time control. It promotes flowering at SD and represses flowering in LD (Yano et al., 2000). (Hd3a is a rice ortholog of the Arabidopsis florigen gene FLOWERING LOCUS T (FT; Yano et al., 2000; Kojima et al., 2002; Tamaki et al., 2007). Hd1 activates Hd3a in SD, but in LD Hd1 is converted by phytochrome B to a repressor of Hd3a (Yano et al., 2000; Hayama and Coupland, 2004). Hd1 expression is controlled by Oryza sativa GIGANTEA (OsGI), a key factor of circadian rhythms in rice flowering control (Yano et al., 2000; Hayama et al., 2003). There is a second, Hd1-independent, photoperiod inductive pathway in rice. Early hd1 (Ehd1), a B-type response regulator that is activated by a SD flowering promoter (i.e., OsMADS51), activates the expression of Hd3a, RFT1 (another Rice FT gene), and OsMADS14, and mainly confers SD-dependent flowering promotion (Doi et al., 2004; Kim et al., 2007). In LD, Ehd1 is activated by Ehd2/OsId1/RID1 (Rice Indeterminate1) and OsMADS50, but repressed by Ghd7 (Grain number, plant height and heading date7; Lee et al., 2004; Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008; Xue et al., 2008; Itoh et al., 2010; Tsuji et al., 2013). Ehd1 activates the expression of Hd3a and RFT1 which activates MADS14 and MADS15 (Komiya et al., 2009). The tight control of expression of the flowering promoter Ehd1 and the flowering repressor Ghd7 allows to measure the slight differences in day lengths to control Hd3a and RFT1 transcription with a critical day length threshold (Itoh et al., 2010; Tsuji et al., 2013). Unlike Hd1 that is conserved with Arabidopsis CO, Ehd1, and Ghd7 are evolutionarily acquired rice-specific genes. Therefore, the control of florigen expression is regulated by key mechanisms that involve the regulation of Hd1 expression, the conversion of Hd1 function on Hd3a expression, and modulation of Ehd1/Ghd7 expression.
Recent studies have established a close relationship of epigenetic regulation with flowering. For instance, during the process of vernalization in Arabidopsis, polycomb repressive complex 2 (PRC2)-mediated trimethylation of histone H3 lysine 27 (H3K27me3) represses the expression of Flowering Locus C (FLC) to stimulate flowering (Sung and Amasino, 2004; De Lucia et al., 2008). In rice, SDG724 mediates H3K36me2/3 deposition at OsMADS50 and RFT1, promoting flowering and establishing a difference of functionality between paralogs RFT1 and Hd3a under LD or SD conditions (Sun et al., 2012). SDG725 also mediates H3K36me2/3 deposition at Ehd2, Ehd3, OsMADS50, Hd3a, and RFT1, promoting flowering under LD or SD conditions (Sui et al., 2013). The PRC2 complex was first discovered in Drosophila, which has four core proteins: ENHANCER OF ZESTE [E(z)], SUPPRESSOR OF ZESTE 12 [Su(z)12], EXTRA SEX COMBS (ESC) and P55 (Schuettengruber and Cavalli, 2009). The E(z) protein has the H3K27 methyltransferase activity (Cao et al., 2002; Czermin et al., 2002). Arabidopsis contains three E(z) genes [CURLY LEAF (CLF), SWINGER (SWN), and MEDEA (MEA)], three Su(z)12 genes [FERTILIZATION INDEPENDENT SEED 2 (FIS2), VERNALIZATION 2 (VRN2) and EMBRYONIC FLOWER 2 (EMF2)], and one ESC gene [FERTILIZATION INDEPENDENT ENDOSPERM (FIE)] (Luo et al., 1999; Ohad et al., 1999; Gendall et al., 2001; Yoshida et al., 2001; Hennig et al., 2003), which form three PRC2-like complexes. The FIS complex, which contains MEA/SWN, FIS2, FIE, and MSI1 (Multicopy Suppressor of IRA1), functions during gametogenesis and seed development. The others are involved in flowering control: the EMF complex that is comprised of CLF/SWN, EMF2, FIE, and MSI1 and is involved in the suppression of early flowering and the VRN complex that plays critical roles in the vernalization pathway by maintaining the high level of H3K27me3 on the FLC locus after vernalization (Hennig and Derkacheva, 2009). The VRN complex is associated with VERNALIZATION INSENSITIVE 3 (VIN3, a PHD-domain containing protein) and VIN3-like proteins to form PHD-VRN PRC2 complexes (Wood et al., 2006; De Lucia et al., 2008). The VIN3 protein enhances H3K27me3 throughout the target loci to a level sufficient for stable silencing.
The rice genome contains two genes for E(z) (OsiEZ1 and OsCLF), Su(Z)12 (OsEMF2a and OsEMF2b) and ESC (OsFIE1 and OsFIE2; Luo et al., 2009). No morphological changes are observed in osclf and osfie1 mutants, while osfie2 and osemf2b mutants display earlier flowering at LD and abnormal flower organs (Luo et al., 2009). Recent results have shown that rice VIN3-like proteins OsVIL2, OsVIL3, or RICE LEAF INCLINATION 2 (LC2, hereafter referred to as LC2) promote rice flowering through the photoperiod pathway (Wang et al., 2013; Yang et al., 2013). These results suggest that PRC2 and PRC2-associated genes are involved in photoperiod regulation of flowering in rice. However, how PRC2-mediated gene repression is involved in accurate photoperiod control of rice flowering is not clear.
In this work we show that the two rice E(z) genes, OsCLF (or SDG711, Os06g16390, here after referred to as SDG711) and OsiEZ1 (or SDG718, Os03g19480, here after referred to as SDG718), displayed distinct function in photoperiod regulation of flowering in rice. SDG718 is induced in SD and represses OsLF, a repressor of Hd1 (Zhao et al., 2011), leading to a higher expression of Hd1 (that activates Hd3a in SD) and thus early flowering. SDG711 is induced in LD and represses OsLF, Ehd1, and other flowering-promoting genes leading to late flowering in LD. The data suggested that the two E(z) genes are involved respectively, in LD and SD signals to differentially control key flowering genes expression and contribute to the accurate photoperiod control of flowering time in rice.
Materials and Methods
Plant Materials and Growing Conditions
Rice cultivar (Oryza sativa spp. japonica) “Zhonghua 11” (ZH11) and “DongJin” (DJ) were used for genetic transformation in this study. T-DNA insertion line of SDG711 (3A-60654.R) was obtained from the Postech rice mutant database (http://signal.salk.edu/cgi-bin/RiceGE). Insertion was confirmed by PCR using the specific primers F and R and a T-DNA left side primer RB2. The primers used for genotyping and real-time PCR analysis are listed in Table S1. The rice plants were grown either in a paddy field in summer in Wuhan (day length >13.5 h) or in controlled growth chambers for 6 week-old under SD (10 h light at 30°C/14 h dark at 25°C) or 8 week-old LDs (14 h light at 30°C/10 h dark at 25°C) conditions as described previously (Yang et al., 2013).
Expression Analysis by Northern Blots and Rt-PCR
Total RNA was isolated from rice callus, stems, leaves, flag leaves, shoots, panicles, endosperm, and roots using TRIzol reagent (Invitrogen). Three μg of total RNA were reverse-transcribed in a reaction of 20 μl by using DNase I and SuperScript III (Invitrogen) according to the manufacturer’s instruction to obtain cDNA. For northern blotting analysis, fifteen micrograms of total RNA extracted from field-grown rice leaves was separated in 1.2% (w/v) formamide-denaturing agarose gels, then transferred to nylon membranes. Gene-specific probes were labeled with 32P-dCTP using the Random Primer kit (Invitrogen) and hybridized to the RNA blots. The probe of SDG711 was amplified from SDG711 cDNA using primers Insitu-SDG711-F and Insitu-SDG711-R (Table S1), resulting in a fragment of 505 bp of the cDNA.
Real-time PCR was performed in an optical 96-well plate that included SYBR Premix EX Taq and 0.5 μl of Rox Reference Dye II (Takara), 1 μl of the reverse transcription reaction, and 0.25 μM of each gene-specific primer in a final volume of 25 μl on a PRISM 7500 PCR instrument (Applied Biosystems). The reactions were performed at 95°C for 10 s, 45 cycles of 95°C for 5 s, and 60°C for 40 s. Disassociation curve analysis was performed as follows: 95°C for 15 s, 60°C for 20 s, and 95°C for 15 s. Data were collected using the ABI PRISM 7500 sequence detection system following the instruction manual. The relative expression levels were analyzed using the 2–ΔΔCT method (Livak and Schmittgen, 2001). The rice ACTIN1 gene was used as the internal control. Accession numbers of genes analyzed in this study: SDG711: LOC_Os06g16390; SDG718: LOC_Os03g19480; OsLF: LOC_Os05g46370; Ehd1: LOC_Os10g32600; RFT1: LOC_Os06g06300; Hd1: LOC_Os06g16370; Hd3a: LOC_Os06g06320; RID1: LOC_Os10g28330; OsGI: LOC_Os01g08700; Ghd7: LOC_Os07g15770; OsMADS14: LOC_Os03g54160; OsMADS15: LOC_Os07g01820; OsMADS50: LOC_Os03g03070; OsMADS51: LOC_Os01g69850. The primers for real-time PCR are listed in Table S1.
Vector Construction and Plant Transformation
For over-expression (OX) vector, the SDG711 full-length cDNA was amplified from DJ leaf mRNA using primer set OXSDG711-F and OXSDG711-R, then inserted into the OX vector pU1301 under the control of the maize ubiquitin gene promoter within KpnI sites (Sun and Zhou, 2008). For RNAi vectors, gene-specific sequences of SDG711 and SDG718, spanning from 2352 bp to 2860 bp relative to the translation start site and from 918 bp to 1599 bp relative to the translation start site, respectively, were amplified from cDNA using primer sets RiSDG711-F/RiSDG711-R and RiSDG718-F/RiSDG718-R, then inserted into the RNAi vector pDS1301 (Huang et al., 2007). Sequence amplified using the primers sets are listed in Table S1. The constructs were transformed into DJ (SDG711 OX and RNAi) and ZH11 (SDG718 RNAi) plants by Agrobacterium tumefaciens (strain EHA105) – mediated transformation as previously described (Huang et al., 2007).
Western Blotting Analysis
For Western blot analysis, histone-enriched fractions were extracted from wild type (WT), mutant, and transgenic leaves as described previously (Huang et al., 2007). Antibodies used in Western blot are: anti-H3K27me3 (07-449, Millipore), anti-H3K27me2 (ab24684, Abcam), anti-H3K27me1 (ab113671, Abcam), anti-H3 (ab1791, Abcam), anti-H3K4me3 (07-473, Millipore), anti-H3K4me2 (07-430, Millipore), anti-H3K4me1 (07-436, Millipore), anti-H3K9me3 (ab8898, Abcam), anti-H3K9me2 (07-441, Millipore), anti-H3K9me1 (ab9045, Abcam) H3K36me1 (ab9048, Abcam), anti-H3K36me2 (ab9049, Abcam), and anti-H3K36me3 (ab9050, Abcam). Anti-SDG711 was prepared by immunizing rabbits with SDG711 protein produced in Escherichia coli (in pET-28a vector) and purified with His-tag protein purification beads (V8550, GE Healthcare). The anti-serum was affinity-purified with protein-A agarose beads purchased from Millipore (16-157).
Chromatin Immunoprecipitation (ChIP)
The chromatin immunoprecipitation (ChIP) experiment was performed as described (Huang et al., 2007). Rice leaves (0.8–1.0 g fresh weight) were harvested at the end of the dark period of 8 week-old LD grown and 6 week-old SD grown plants and crosslinked in 1% formaldehyde under vacuum. Chromatin was extracted and fragmented to 200–750 bp by sonication, and ChIP was performed using the following antibodies: H3K27me3 (07-449, Millipore), H3K4me3 (07-473, Millipore), and Anti-SDG711. The precipitated and input DNA samples were then analyzed by real-time PCR with gene-specific primers listed in Table S1. All assays were performed at least three times from two biological replicates.
Results
The Rice E(z) Homologous Genes Display Distinct Photoperiodic Expression Patterns
Phylogenetic analysis has previously shown that rice E(z) homologous genes, SDG711 and SDG718, are closely related to Arabidopsis CLF and SWN, respectively (Luo et al., 2009; Figure S1). The two rice genes share 51% amino acid sequence identity, with much higher conservations in the catalytic SET domain and the protein interaction domains (SAND, Cys-rich). Analysis of expression of the genes by real-time PCR indicated that SDG711 was widely expressed in different tissues/organs, whereas SDG718 was more expressed in leaves than other tested tissues/organs (Figure 1A). Because OsEMF2b and PRC2-interacting PHD domain protein genes are involved in photoperiod regulation of flowering time in rice (Luo et al., 2009; Wang et al., 2013; Yang et al., 2013), we studied whether the rice E(z) genes were photoperiod-responsive. We analyzed mRNA isolated from leaves of 6 week-old SD (10 h light/14 h dark)- and 8 week-old LD (14 h light/10 h dark)- grown plant leaves harvested at intervals of 4 h during a 36 h period. The analysis revealed that SDG711 mRNA levels were higher in LD than in SD, whereas that of SDG718 were higher in SD than LD (Figure 1B).
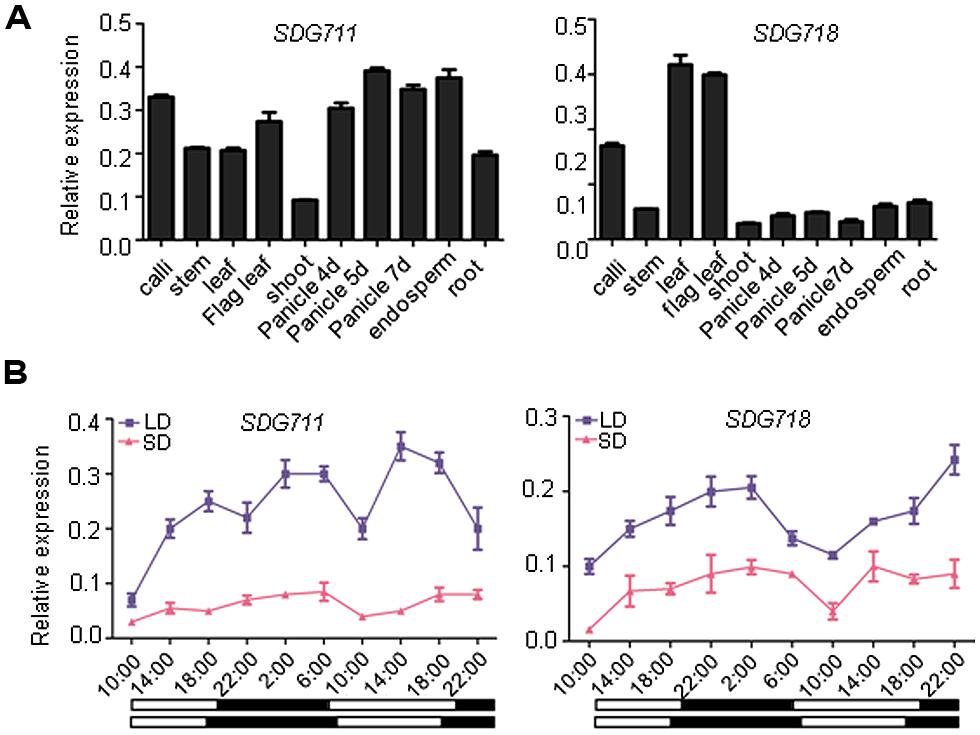
FIGURE 1. Expression profiles of SDG711 and SDG718. (A) Real-time PCR detection of transcript levels of the genes in indicated organs or developmental stages. (B) transcript levels of the two genes in young leaves harvested at intervals of 4 h during a 36 h period from plants grown under long day (LD) or short day (SD) conditions. Dark periods are indicated by black bars. Bars = mean ± SD from two biological repeats. Values are shown as relative to ACTIN transcript levels.
SDG711 is Involved In LD Repression of Rice Flowering
To study the function of SDG711, we produced transgenic rice plants in DJ which contains functional Hd1 (Naranjo et al., 2014) to knockdown the gene by RNAi and to over-express the gene by using the maize ubiquitin gene promoter (Sun and Zhou, 2008). Analysis of the transgenic plants revealed several lines with reduced expression and six lines with increased expression of the gene (Figure 2A). To check the protein level of SDG711, we performed Western blot analysis of protein extracts from the leaves of the WT and transgenic plants grown in LD by using antibodies generated against E. coli-produced SDG711 protein. The analysis confirmed the OX and RNAi of the gene in the transgenic lines (Figure 2B). Three single copy T3 homozygous lines (offspring of single insertion T2 lines that did not segregate transgene-negative individuals) per transgene were selected for phenotypic analysis. During vegetative growth the transgenic plants did not display any visible morphological defects but exhibited altered HD (or flowering time) compared to the WT plants. In LD (14 h light/10 h dark), the HD of the OX plants was largely delayed (20–25 days, p < 0.001, student’s t-test), while that of the RNAi plants was significantly earlier (10–16 days, p < 0.001, student’s t-test) than WT (Figure 3). However, in SD (10 h light/14 h dark) the HD of SDG711 OX and RNAi plants was not significantly different from WT (Figure 3). These data suggested that SDG711 may have a function to repress flowering in LD. In addition to the effect on flowering time, the OX plants produced a higher number of stamens (Figure S2). The pollen viability was reduced in both the OX and RNAi plants of SDG711 (Figure S2). These observations suggested that SDG711 might play a role in fertility. To confirm the transgenic results, we characterized a T-DNA mutant that had the insertion located in the 5′-UTR of the gene (Figures S3A,B). One single insertion was identified by Southern blotting (Figure S3C). Real-time PCR revealed a higher level of transcripts of the gene (Figure S3D). Northern blotting experiments revealed that the increased transcripts were about the same size as in WT (Figure S3E). The mutant displayed the same phenotype as that of the OX plants (Figure 3). After three backcrosses the phenotype co-segregated with the presence of the insertion, indicating that the mutation was a gain-of-function mutation.
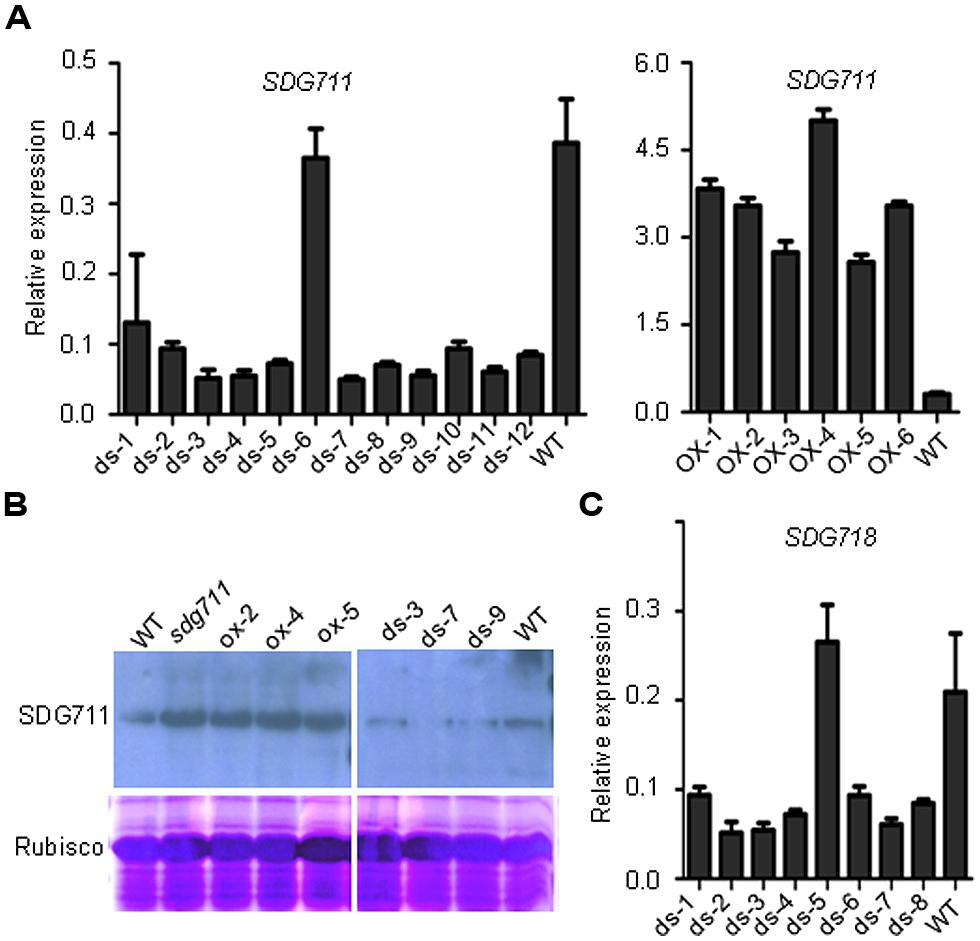
FIGURE 2. Characterization of SDG711 and SDG718 transgenic plants. (A) Detection of SDG711 mRNA levels in wild type (WT) and 12 independent RNAi (ds; left), and six over-expression (OX) lines (right).(B) Detection of SDG711 protein levels in WT, three OX and three RNAi lines, and the gain-of-function mutant (sdg711). (C) Detection of SDG718 mRNA levels in WT and eight independent RNAi lines. Y-axis: relative expression levels to ACTIN transcripts. Bars = means ± SD from three technical repeats.
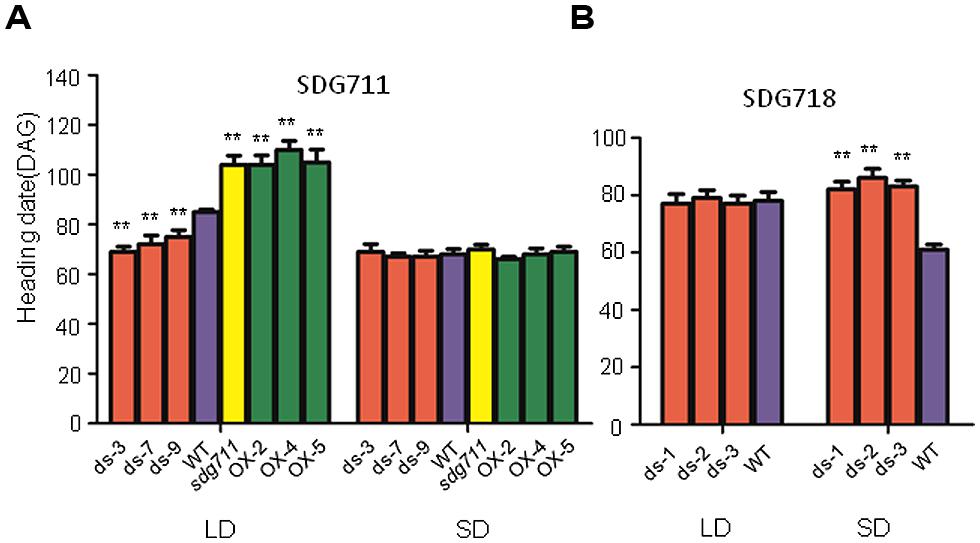
FIGURE 3. Flowering time of SDG711 and SDG718 transgenic plants. (A) Heading date (DAG: days after germination) of 3 SDG711 RNAi (ds) lines, three OX lines and the gain-of-function mutant (sdg711) compared to WT in LD and SD conditions. (B) Heading date of three SDG718 RNAi (ds) lines compared to WT in LD and SD conditions. Twenty plants per genotype were surveyed. Significance of differences between WT and transgenic plants are indicated by (∗∗p < 0.01, Student’s t-test).
To study the effect of SDG711 OX or RNAi on histone methylation, we performed Western blotting analysis of histones isolated from leaves of the gain-of-function mutant, an RNAi and an OX line. The analysis revealed that compared to WT the levels of H3K27me3 were lower in the RNAi line, but higher in the gain-of-function mutant and OX plants (Figure S4), suggesting that SDG711 was required for the overall H3K27me2/3 in rice.
The flowering time phenotype suggested that SDG711 had a function to suppress flowering in LD while without a clear effect in SD. We therefore analyzed the expression of flowering regulatory genes in both pathways. mRNA were isolated from 8 week-old LD or 6 week-old SD plant leaves at intervals of 4 h during a 24 h period. In LD, the mRNA levels of LD flowering activators including Ehd1, Hd3a, MADS14, and MADS15 were clearly decreased in the OX, but increased in the RNAi plants compared to WT (Figure 4A). By contrast, the transcript levels of the LD flowering repressor Ghd7 were not clearly altered in the transgenic plants (Figure S5). The expression of RID1 and OsMADS50 also appeared unchanged in the transgenic plants (Figure 4A; Figure S5). Interestingly, Hd1 that acts as a repressor of Hd3a in LD was highly induced by SDG711 OX and repressed by SDG711 RNAi (Figure 4A), while the expression level of OsGI that activates Hd1 was not changed (Figure S5). Because E(z) proteins are supposed to be transcriptional repressors, the activation of Hd1 in SDG711 OX plants might be due to an indirect effect. Recent data have shown that OsLF, a bHLH protein that directly represses Hd1 (Wang et al., 2013). Examination of OsLF transcripts in the transgenic plants indicated that OsLF was repressed by SDG711 OX, but activated by SDG711 RNAi in LD (Figure 4A). The expression of flowering regulatory genes supported the flowering time phenotype observed in the transgenic plants (Figure 3). Analysis of the gain-of-function mutant confirmed the above observed effects of SDG711 OX on flowering time gene expression (Figure S6). In SD, the expression levels of the flowering genes were not clearly affected by SDG711 OX or RNAi, except the peak expression of OsLF in the RNAi plants was about 4 h earlier than the WT (Figure 4B). These results suggested that SDG711 is mostly involved in LD repression of rice flowering time.
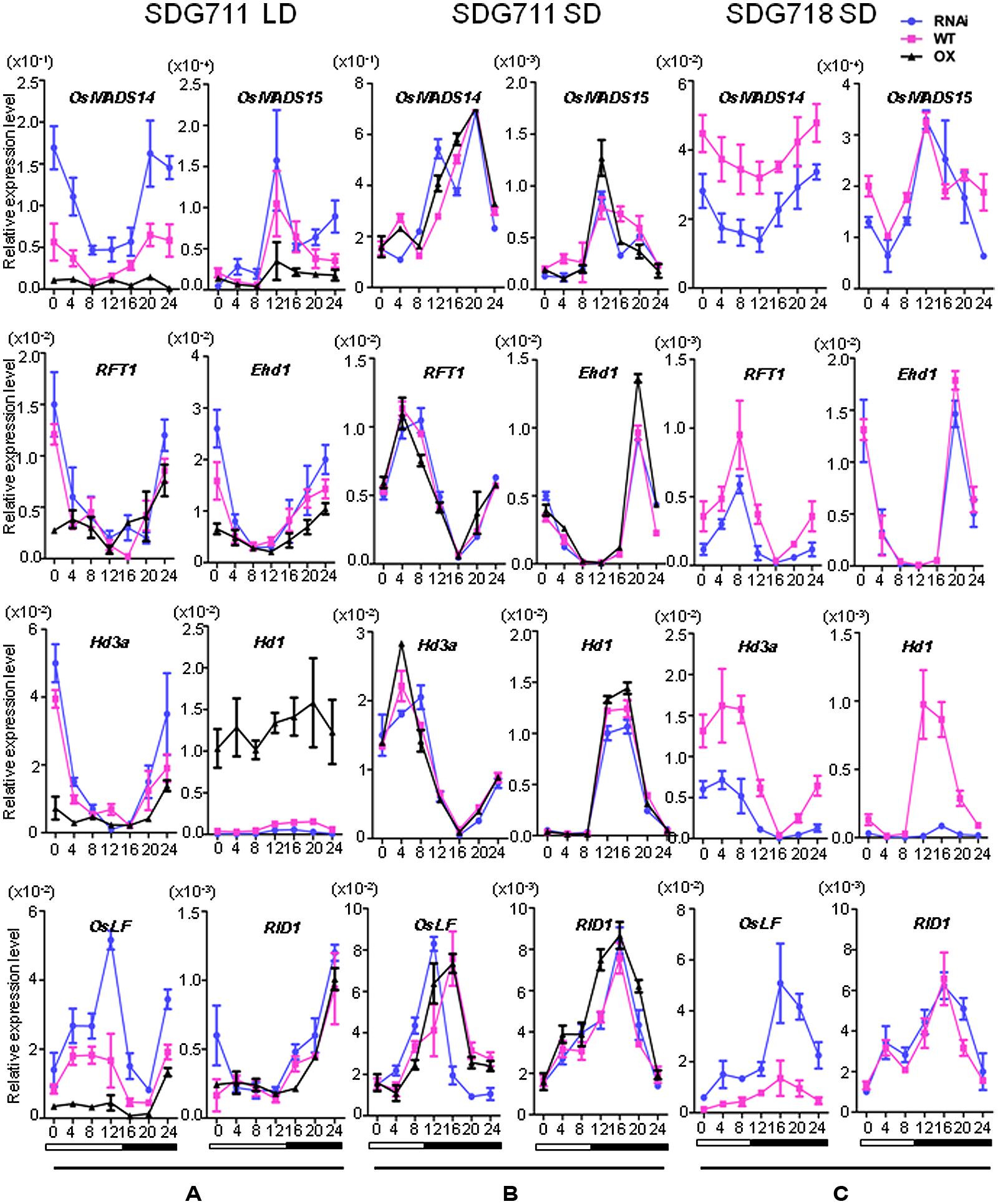
FIGURE 4. Expression levels of key flowering regulatory genes in mature leaves of SDG711 and SDG718 transgenic plants compared to WT. (A) Key flowering genes expression in SDG711 transgenic plants compared to WT in LD. (B) Key flowering genes expression in SDG711 transgenic plants compared to WT in SD. (C) Key flowering genes expression in SDG718 RNAi plants compared to WT in SD. Leaves of transgenic plants were harvested from plants of 3 T3 lines grown for 8 week-old under LDs and 6 week-old under SDs. Bars = mean ± SD from three biological repeats.
To study whether SDG711 protein levels were differentially regulated in LD and SD, Western blotting analysis of protein extracts from WT, RNAi and OX plants grown in LD and SD harvested at 12:00 (mid day, after 7 h in light for LD condition and 5 h in light for SD condition) and 24:00 am (mid night, after 5 h in dark for LD condition and 7 h in dark for SD condition) was performed by using anti-SDG711. The results shown in Figure 5 revealed that SDG711 protein levels were lower in SD compared in LD and that the high SDG711 level of the OX line was reduced in SD and in the dark, suggesting that the stability of SDG711 protein may be regulated by day length and light/dark conditions.
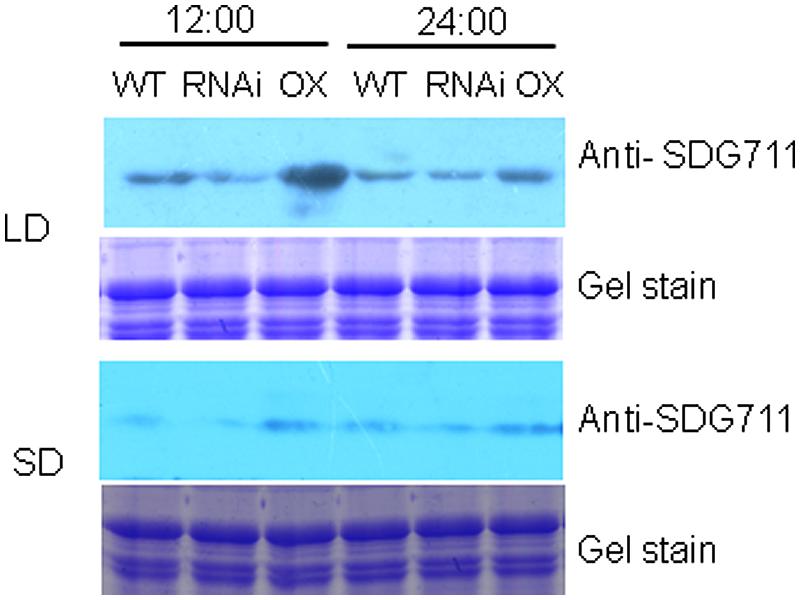
FIGURE 5. SDG711 protein accumulation in LD - and SD- grown WT, SDG711 RNAi and OX mature leaves of plants from 3 T3 lines harvested at noon (12:00) and mid night (24:00). Protein extracts were resolved by SDS-PAGE and Coomassie stained before Western blotting with anti-SDG711.
SDG718 Promotes Flowering in SD
To study whether SDG718 had a function in flowering time control, we obtained several RNAi lines in ZH11 variety (Figure 2C), which also has a functional Hd1 (Luan et al., 2009; Naranjo et al., 2014). These plants showed a similar HD as the WT in LD, but flowered later (>20 days) than WT in SD (Figure 3B). Analysis of flowering time gene expression revealed that SDG718 RNAi clearly induced OsLF, but repressed the SD activators including Hd1, Hd3a, OsMADS14, and RFT1 (Figure 4C), suggesting that SDG718 played a role in promoting flowering in SD. However, the SDG718 RNAi plants did not show any clear defect in flower organ and pollen viability (Figure S2).
SDG711 and SDG718 Regulate H3K27me3 On Flowering Genes
To study whether knockdown and OX of SDG711 altered chromatin modification of flowering regulatory genes, we analyzed H3K27me3 on RID1, Ehd1, RFT1, Hd3a, MADS14, MADS15, Hd1, and OsLF in WT, OX and RNAi plants grown under LD conditions by ChIP assays. The 8 week-old plant leaves were harvested at the end of the darkness period under LD conditions. Because H3K27me3 is located in gene body with enrichment on the 5′ end of the gene in rice (Hu et al., 2012; Li et al., 2013), we analyzed the ChIP by real-time PCR using two primer sets, one corresponding to the 5′ region, the other to the gene body (Figure 6). Among these genes, Hd1 appeared to be unmodified by H3K27me3 in either the 5′ region or the gene body, further supporting an indirect effect of SDG711 on its expression. RID displayed a very low level of H3K27me3 (Figure 6A), consistent with the observation that the expression of the gene was not affected in the SDG711 transgenic and the gain-of-function mutant plants (Figure 4; Figures S5 and S6). The remaining genes displayed H3K27me3 in the 5′ region and/or gene body, suggesting that regulation of these genes might involve PRC2 function. H3K27me3 on these genes (except the gene body of RFT1 which displayed a relatively weak level of H3K27me3) was clearly reduced in the RNAi but increased in the OX plants (Figure 6A), which reversely correlated with their expression changes in the transgenic plants. These data suggested that SDG711-mediated H3K27me3 was involved in the regulation of the genes.
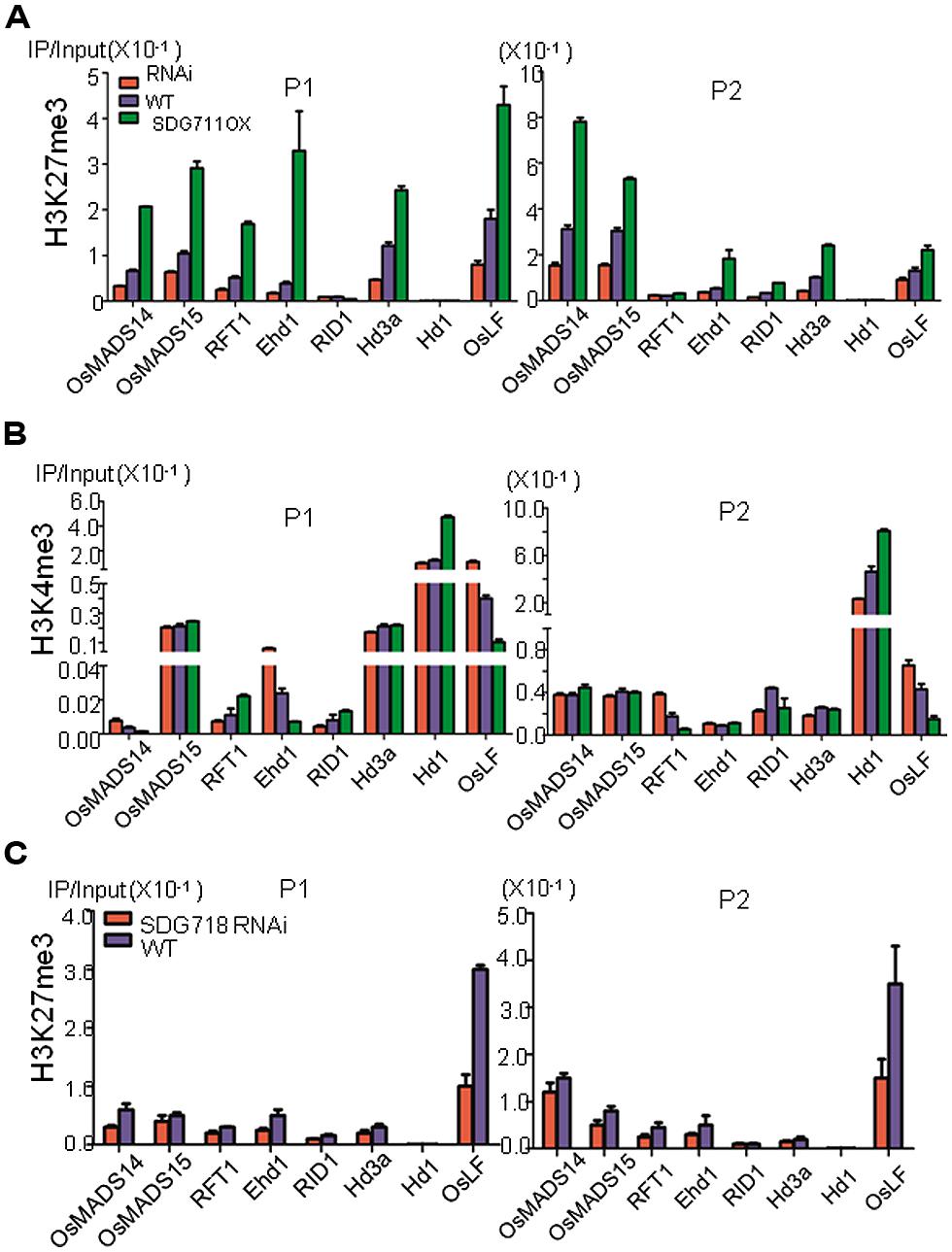
FIGURE 6. SDG711 and SDG718 function in H3K27me3 and H3K4me3 on key flowering genes. (A) chromatin immunoprecipitation (ChIP) analysis of H3K27me3 on the indicated flowering genes in WT, SDG711 RNAi and OX plants (mature leaves pooled from 3 T3 lines). (B) ChIP analysis of H3K4me3 on the indicated flowering genes in WT, SDG711 RNAi and OX plants (pooled from 3 T3 lines). H3K27me3 and H3K4me3 enrichments on the 5′ end (P1) and the coding region (P2) of the flowering genes were detected by quantitative PCR. Three biological repeats were performed, one repetition is shown. One other biological replicate is shown in Figure S7. Bars = mean ± SD from three technical repeats. (C) ChIP analysis of H3K27me3 on the indicated flowering genes in WT and SDG718 RNAi plants pooled from 3 T3 lines. Leaves of SDG711 transgenic plants were harvested from 8 week-old LD plants grown, and leaves of SDG718 transgenic plants were harvested from 6 week-old SD plants grown.
Because H3K27me3 is antagonistic to H3K4me3 on gene activity, we therefore analyzed whether alteration of H3K27me3 affected H3K4me3 on the flowering genes in the transgenic plants. The analysis revealed relatively higher levels of H3K4me3 in Hd1, Hd3a, MADS15, Ehd1, and OsLF than in MADS14, RFT1, and RID in WT plants (Figure 6B). H3K4me3 levels in Hd1, MADS15, and Hd3a were not affected by the SDG711 transgenes except some increases on Hd1 in the OX plants (Figure 6B). The increased H3K4me3 on Hd1 may be due to increased expression of the gene in the OX plants, as H3K4me3 is thought to be associated with active genes (Hu et al., 2011). However, H3K4me3 on Ehd1 and OsLF was increased in SDG711 RNAi but decreased in SDG711 OX plants (Figure 6B), which conversely correlated to that of H3K27me3 and suggested that SDG711-mediated H3K27me3 might affect H3K4me3 in the two loci, which may be linked to the repression of the genes. Analysis of SDG718 RNAi plants also revealed a clear decrease of H3K27me3 in OsLF in SD (Figure 6C).
SDG711 Binds to Ehd1 and OsLF Loci
To further assess the function of SDG711 on the flowering gene regulation, we performed anti-SDG711 ChIP assays and analyzed by real-time PCR using the same primer sets as for the histone methylation ChIP. Non-immunized serum was used as control. The analysis revealed that in LD-grown WT plants SDG711 binding was clearly enriched in the 5′ end and the gene body of Ehd1 and OsLF compared to the other tested genes (Figure 7A). However, some enrichment was also observed in the gene body of RID1. The SDG711 enrichment on the three genes was sensibly reduced in RNAi and clearly enhanced in the OX plants (Figure 7A), suggesting that SDG711 may directly target to the genes in LD. In SD-grown plants, the enrichment of SDG711 on the three genes was much weaker than that in SD (Figure 7B), which was consistent with lower accumulation of SDG711 in SD (Figure 5) and suggested that SDG711 enrichment in the flowering genes was regulated by day length.
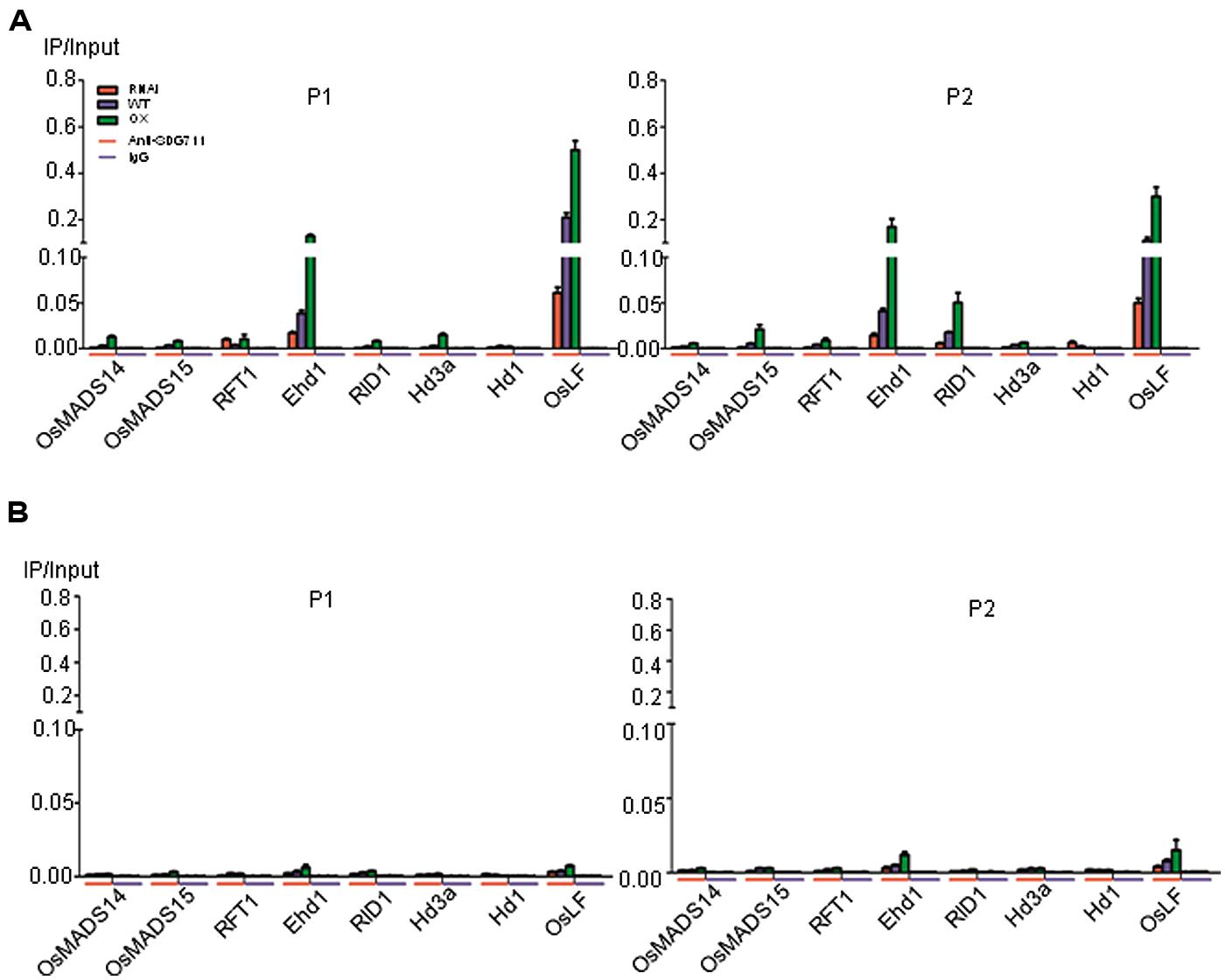
FIGURE 7. Direct association of SDG711 protein with flowering genes. (A) SDG711 protein enrichment on the indicated flowering genes in LD-grown WT, SDG711 RNAi and OX seedlings tested by ChIP with anti-SDG711. Non-immunized rabbit serum (IgG) was used as control. Three biological repeats were performed, one repetition is shown. The two other repeats are shown in Figure S8. Bars = means ± SD from three technical repeats. (B) SDG711 protein enrichment on the indicated flowering genes in SD-grown plants. Leaves were harvested from plants grown for 8 week-old LD and 6 week-old SD plants grown.
Discussion
Our data suggest that SDG711 and SDG718 repress OsLF in LD and SD, respectively, leading to the activation of Hd1 that inhibits Hd3a and flowering in LD but activates Hd3a and RFT1 and flowering in SD (Figure 8). Therefore, the two E(z) genes likely contribute to the accurate photoperiod control of flowering in rice. The increased level of SDG711 expression in LD and that of SDG718 in SD may be critical for OsLF repression. The SD-induced SDG718 expression is consistent with the results showing that OsVIL genes that promote flowering in SD are also induced in SD (Wang et al., 2013). The SDG711-mediated LD repression of flowering is supported by previous results showing that mutants of other PRC2 genes such as osfie2 and osemf2b also display an early flowering phenotype in LD (Luo et al., 2009). The observations that the SDG711 OX only enhanced LD repression of flowering but did not affect flowering in SD (Figure 3), suggest that SDG711 may be also regulated at posttranscriptional levels by day length. This hypothesis is supported by the observation that accumulation of SDG711 protein in the OX lines in LD was reduced in SD and that SDG711 binding to key flowering genes was largely reduced in SD (Figure 7B). This day length-dependent stability of the protein may allow SDG711 to regulate flowering only in LD. The changes of H3K27me3 levels on the OsLF locus in SDG711 transgenic plants imply that PRC2-mediated repression may involve the deposition of H3K27me3 on the gene. As SDG711 was shown to be directly associated with the OsLF locus in LD (Figure 7), OsLF may be a primary target for rice PRC2-mediated LD-repression of flowering. The results are in agreement with recent data showing directly repression of OsLF by LC2 that interacts with the OsEMF2b protein (Wang et al., 2013; Yang et al., 2013). However, it is not excluded that SDG711 or SDG718 represses other genes that mediate Hd1 repression.
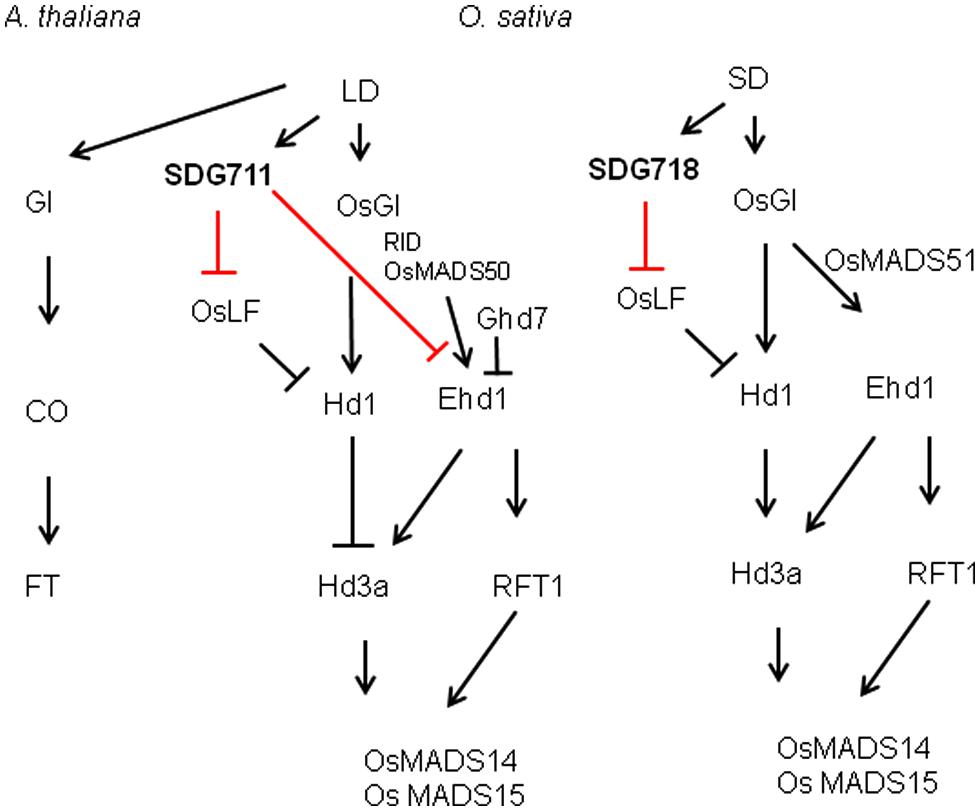
FIGURE 8. Summary of SDG711 and SDG718 function in LD and SD pathways of rice flowering control compared to LD activated flowering in Arabidopsis.
Previous studies have shown that the flowering promoter Ehd1 and the flowering repressor Ghd7 could enable manipulation of slight differences in day lengths to control Hd3a transcription with a critical day length threshold (Itoh et al., 2010). Ehd1 and Ghd7 provide a gating mechanism to set critical day length for Hd3a expression in SD, in which Ehd1 is repressed by the morning activation of Ghd7 in LD (Itoh et al., 2010; Tsuji et al., 2013). The observations that SDG711 transgenic and gain-of-function mutant plants affected the expression of Ehd1 but not that of its upstream regulators (i.e., Ghd7, MADS50 or RID1) and that SDG711 was associated with the Ehd1 locus (Figure 6; Figures S5 and S6), indicate that SDG711 is a direct repressor of Ehd1 in LD. In addition, H3K27me3 on the Ehd1 locus was modulated by the expression level of SDG711 (Figure 6A). Therefore, besides Ghd7 and OsMADS50, SDG711 represents an additional LD repressor of Ehd1 (Figure 8). The observations that Ehd1 expression was not clearly affected by down-regulation of SDG718 and SDG711 or up-regulation of SDG711 in SD (Figures 4B,C), suggest that PRC2 may mainly target to the OsLF-Hd1 pathway instead of that of Ehd1 in SD. Alternatively, the SD-induced expression of Ehd1 (mainly due to the repression of Ghd7 in SD) may be overwhelming, which may mask the effect of SDG718 RNAi. In addition, the chromatin analysis data indicated that key flowering genes displayed different histone modification patterns (Figure 6). The changes of H3K27me3 levels on the marked genes caused by SDG711 RNAi and OX and SDG718 RNAi, which are correlated with their expression change in the transgenic plants in LD or SD, suggest that SDG711/SDG718 may be also involved in the deposition of the mark on these loci. Collectively, the data demonstrating that the SDG711 and SDG718 are involved respectively, in the LD and SD signaling to promote LD repression and SD activation of flowering, suggest the involvement of PRC2 in the accurate photoperiod control of flowering in rice.
SDG711 and SDG718 are closely related to Arabidopsis CLF and SWN, respectively. It is suggested that CLF and SWN act redundantly to regulate vegetative growth. No vegetative phenotype observed in SDG711 and SDG718 transgenic plants support the idea that the two rice genes may also have a redundant function in the vegetative growth. Arabidopsis genome contains a third E(z) gene, MEA, which is mainly involved in the regulation of gene imprinting and embryo and endosperm development. The counterpart of MEA is not found in rice. Therefore the question arises as that whether either SDG711 or SDG718 plays a role in reproductive development. Our data showing that besides flowering time, SDG711 RNAi and OX affects stamen number, pollen viability, and fertility (Figure S2) suggests that this E(z) gene may be involved in reproductive development in addition to flowering time control in rice.
Author Contributions
Xiaoyun Liu and Dao-Xiu Zhou designed the experiments and wrote the manuscript. Xiaoyun Liu, Chao Zhou, and Yu Zhao performed the experiments. Shaoli Zhou performed some images processing. Wentao Wang performed some materials cultivation. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Caiguo Xu and Prof. Xianghua Li for help in field experiments and management. This work was supported by grants from National Science Foundation of China (31371241 and 31371468), the 863 Key Project “Rice Functional Genomics” of the Chinese Ministry of Science grants (2012AA10A303), the special transgenic program of the Chinese Ministry of Agriculture (2014ZX0800902B), and the Melinda and Bill Gates Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00591/abstract
References
Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P.,et al. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043. doi: 10.1126/science.1076997
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Czermin, B., Melfi, R., Mccabe, D., Seitz, V., Imhof, A., and Pirrotta, V. (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196. doi: 10.1016/S0092-8674(02)00975-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Lucia, F., Crevillen, P., Jones, A. M., Greb, T., and Dean, C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 105, 16831–16836. doi: 10.1073/pnas.0808687105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z.,et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gendall, A. R., Levy, Y. Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. doi: 10.1016/S0092-8674(01)00573-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135, 677–684. doi: 10.1104/pp.104.042614
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hennig, L., and Derkacheva, M. (2009). Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 25, 414–423. doi: 10.1016/j.tig.2009.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hennig, L., Taranto, P., Walser, M., Schonrock, N., and Gruissem, W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130, 2555–2565. doi: 10.1242/dev.00470
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, Y., Liu, D., Zhong, X., Zhang, C., Zhang, Q., and Zhou, D. X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. U.S.A. 109, 5773–5778. doi: 10.1073/pnas.1203148109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, Y., Shen, Y., Conde, E. S. N., and Zhou, D. X. (2011). The role of histone methylation and H2A.Z occupancy during rapid activation of ethylene responsive genes. PLoS ONE 6:e28224. doi: 10.1371/journal.pone.0028224
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, L., Sun, Q., Qin, F., Li, C., Zhao, Y., and Zhou, D. X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144, 1508–1519. doi: 10.1104/pp.107.099473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Itoh, H., Nonoue, Y., Yano, M., and Izawa, T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42, 635–638. doi: 10.1038/ng.606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T.,et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Komiya, R., Yokoi, S., and Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. doi: 10.1242/dev.040170
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, S., Kim, J., Han, J. J., Han, M. J., and An, G. (2004). Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 38, 754–764. doi: 10.1111/j.1365-313X.2004.02082.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, T., Chen, X., Zhong, X., Zhao, Y., Liu, X., Zhou, S.,et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736. doi: 10.1105/tpc.113.118802
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luan, W., Chen, H., Fu, Y., Si, H., Peng, W., Song, S.,et al. (2009). The effect of the crosstalk between photoperiod and temperature on the heading-date in rice. PLoS ONE 4:e5891. doi: 10.1371/journal.pone.0005891
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, M., Bilodeau, P., Koltunow, A., Dennis, E. S., Peacock, W. J., and Chaudhury, A. M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96, 296–301. doi: 10.1073/pnas.96.1.296
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, M., Platten, D., Chaudhury, A., Peacock, W. J., and Dennis, E. S. (2009). Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant 2, 711–723. doi: 10.1093/mp/ssp036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsubara, K., Yamanouchi, U., Wang, Z. X., Minobe, Y., Izawa, T., and Yano, M. (2008). Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 148, 1425–1435. doi: 10.1104/pp.108.125542
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naranjo, L., Talon, M., and Domingo, C. (2014). Diversity of floral regulatory genes of japonica rice. BMC Genomics 15:101–121. doi: 10.1186/1471-2164-15-101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J. J.,et al. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416. doi: 10.1105/tpc.11.3.407
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, S. J., Kim, S. L., Lee, S., Je, B. I., Piao, H. L., Park, S. H.,et al. (2008). Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 56, 1018–1029. doi: 10.1111/j.1365-313X.2008.03667.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schuettengruber, B., and Cavalli, G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542. doi: 10.1242/dev.033902
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shrestha, R., Gomez-Ariza, J., Brambilla, V., and Fornara, F. (2014). Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann. Bot. doi: 10.1093/aob/mcu032 [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sui, P., Shi, J., Gao, X., Shen, W. H., and Dong, A. (2013). H3K36 methylation is involved in promoting rice flowering. Mol. Plant 6, 975–977. doi: 10.1093/mp/sss152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, C., Fang, J., Zhao, T., Xu, B., Zhang, F., Liu, L.,et al. (2012). The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24, 3235–3247. doi: 10.1105/tpc.112.101436
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, Q., and Zhou, D. X. (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U.S.A. 105, 13679–13684. doi: 10.1073/pnas.0805901105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sung, S., and Amasino, R. M. (2004). Vernalization and epigenetics: how plants remember winter. Curr. Opin. Plant Biol. 7, 4–10. doi: 10.1016/j.pbi.2003.11.010
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsuji, H., Taoka, K., and Shimamoto, K. (2013). Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr. Opin. Plant Biol. 16, 228–235. doi: 10.1016/j.pbi.2013.01.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., Hu, J., Qian, Q., and Xue, H. W. (2013). LC2 and OsVIL2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF. Mol. Plant 6, 514–527. doi: 10.1093/mp/sss096
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wood, C. C., Robertson, M., Tanner, G., Peacock, W. J., Dennis, E. S., and Helliwell, C. A. (2006). The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. U.S.A. 103, 14631–14636. doi: 10.1073/pnas.0606385103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, C., You, C., Li, C., Long, T., Chen, G., Byrne, M. E.,et al. (2008). RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 12915–12920. doi: 10.1073/pnas.0806019105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L.,et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, J., Lee, S., Hang, R., Kim, S. R., Lee, Y. S., Cao, X.,et al. (2013). OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 73, 566–578. doi: 10.1111/tpj.12057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T.,et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. doi: 10.1105/tpc.12.12.2473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yoshida, N., Yanai, Y., Chen, L., Kato, Y., Hiratsuka, J., Miwa, T.,et al. (2001). EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13, 2471–2481. doi: 10.1105/tpc.010227
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, X. L., Shi, Z. Y., Peng, L. T., Shen, G. Z., and Zhang, J. L. (2011). An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15. N. Biotechnol. 28, 788–797. doi: 10.1016/j.nbt.2011.04.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Oryza sativa, heading date, epigenetics, epigenomics, histone methylation, PRC2, OsCLF, OsiEZ1
Citation: Liu X, Zhou C, Zhao Y, Zhou S, Wang W and Zhou D-X (2014) The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 5:591. doi: 10.3389/fpls.2014.00591
Received: 02 September 2014; Accepted: 10 October 2014;
Published online: 31 October 2014.
Edited by:
Raúl Alvarez-Venegas, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoReviewed by:
Hairong Wei, Michigan Technological University, USADonna Glassop, Commonwealth Scientific and Industrial Research Organisation, Australia
Copyright © 2014 Liu, Zhou, Zhao, Zhou, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dao-Xiu Zhou, Institut de Biologie des Plantes, UMR8618, Université Paris-Sud 11, 90405 Orsay, France e-mail: dao-xiu.zhou@u-psud.fr
 Xiaoyun Liu
Xiaoyun Liu Chao Zhou
Chao Zhou Yu Zhao1
Yu Zhao1 Wentao Wang
Wentao Wang