- 1The Sainsbury Laboratory, Norwich Research Park, Norwich, UK
- 2Commonwealth Scientific and Industrial Research Organisation, Agriculture Flagship, Canberra, ACT, Australia
The rust fungi (order: Pucciniales) are a group of widely distributed fungal plant pathogens, which can infect representatives of all vascular plant groups. Rust diseases significantly impact several crop species and considerable research focuses on understanding the basis of host specificity and nonhost resistance. Like many pathogens, rust fungi vary considerably in the number of hosts they can infect, such as wheat leaf rust (Puccinia triticina), which can only infect species in the genera Triticum and Aegilops, whereas Asian soybean rust (Phakopsora pachyrhizi) is known to infect over 95 species from over 42 genera. A greater understanding of the genetic basis determining host range has the potential to identify sources of durable resistance for agronomically important crops. Delimiting the boundary between host and nonhost has been complicated by the quantitative nature of phenotypes in the transition between these two states. Plant–pathogen interactions in this intermediate state are characterized either by (1) the majority of accessions of a species being resistant to the rust or (2) the rust only being able to partially complete key components of its life cycle. This leads to a continuum of disease phenotypes in the interaction with different plant species, observed as a range from compatibility (host) to complete immunity within a species (nonhost). In this review we will highlight how the quantitative nature of disease resistance in these intermediate interactions is caused by a continuum of defense barriers, which a pathogen needs to overcome for successfully establishing itself in the host. To illustrate continua as this underlying principle, we will discuss the advances that have been made in studying nonhost resistance towards rust pathogens, particularly cereal rust pathogens.
Introduction
A considerable body of research now exists at the genetic and molecular level on some of the mechanisms underlying nonhost resistance (NHR). This review will focus on what is known regarding the interaction of rust pathogens with their hosts and nonhosts and include key concepts in NHR derived from other plant–pathogen systems. Previous definitions have attempted to qualitatively distinguish between host and nonhost interactions, however, many plant–pathogen systems cannot be neatly classified into these two extremes. In reality a continuum of resistance outcomes is possible ranging from immunity to partial resistance with varying degrees of efficacy. As will be discussed, a variety of different nonhost outcomes occur for rust pathogens and thus, a definition of nonhost is proposed that incorporates the inherent quantitative nature of nonhost status.
The Rusts
Rusts (order: Pucciniales) are an order of obligate biotrophic fungal plant pathogens, of which many are agronomically important and affect major cereal crops such as wheat, barley, rye, and oat, as well as many other economically important plants ranging from legumes like soybean to trees like coffee (Agrios, 2005). The approximately 5,000 species of rust fungi tend to be specialized pathogens of specific host genera, but their life cycles can be very complex (Agrios, 2005). These life cycles range from the simple to the extreme, with the latter exemplified by macrocyclic, heteroecious rust fungi which have life cycles involving up to five different spore stages and two different hosts: a primary host allowing clonal reproduction and an alternate host to complete sexual reproduction (Agrios, 2005).
Puccinia graminis, causal agent of stem rust on wheat, barley, and oat, has a complex heteroecious life cycle with species in the Triticeae being telial/uredinial stage primary hosts (Leonard and Szabo, 2005). Asexual reproduction of the dikaryotic urediniospores can lead to epidemics on the primary hosts, with multiple infection cycles occurring during the growing season (Leonard and Szabo, 2005). Under appropriate developmental and unfavorable environmental conditions, the pathogen produces an alternative developmental morphology, telia, which erupt through the host tissue and produce diploid teliospores, which are considered survival structures. Haploid basidiospores are produced by the teliospore and infect an alternate spermagonial/aecial host, which in the case of Puccinia graminis are dicot barberry plants (Berberis spp.; Roelfs et al., 1992). Infection of barberry with basidiospores leads to the development of the spermagonial stage on the upper side of leaves. Haploid spermatia are formed and in the event that corresponding mating types are present, hyphae penetrate to form dikaryotic aecia on the lower side of leaves (Agrios, 2005). Aeciospores can infect the primary host to produce uredinia and thereby complete the sexual life cycle of the rust. Similar heteroecious life cycles occur for Puccinia striiformis and Puccinia triticina, the causal agents of stripe rust and wheat leaf rust disease, respectively (Bolton et al., 2008; Hovmøller et al., 2011).
Not all rust fungi share this five-stage (macrocyclic) life cycle, as some species only produce teliospores and basidiospores, i.e., they have a microcyclic life cycle (Agrios, 2005). In addition, not all macrocyclic rusts are heteroecious (i.e., need two different hosts to complete their life cycle). Autoecious rusts, such as the asparagus rust pathogen (Puccinia asparagi) and flax rust pathogen (Melampsora lini), complete their life cycle on a single host (Agrios, 2005).
The evolution of a heteroecious life cycle by some rusts, such as Puccinia graminis, is of interest, as it has clearly required a species jump by the progenitors of these modern pathogens. It is believed that the progenitors of cereal rusts parasitized dicot plants of the ancestral Berberidaceae in what is now northern Europe prior to the evolution of the Mahonia and Berberis genera in this family (Leppik, 1961; Wahl et al., 1984). The grasses subsequently evolved in the tropics and radiated out, speciating during this process. Eventually contact between rust infected Berberis plants and grasses occurred, enabling the evolution of modern heteroecious rust species that parasitize the Poaceae. One can only speculate if the initial parasitism of the grasses in this scenario required the pathogen to overcome an active NHR response or if these plants, which had never previously encountered a rust pathogen, were incapable of recognizing this pathogen. Rice being a tropical grass and a nonhost of all rust pathogens is unlikely to have been exposed to rust pathogens until relatively late, however, as described below, this plant species mounts an active defense response upon challenge with cereal rusts (Ayliffe et al., 2011). These data suggest that extended plant–rust coevolution is not a prerequisite for NHR recognition of rust pathogens.
Formae speciales – Evolution in Action
Many cereal rust species consist of subgroups, or formae speciales (ff. spp.), that have specialized to infect only certain plant species amongst the entire range of host plant species parasitized by the pathogen. The formae speciales concept was first introduced by Eriksson (1894). After inoculating a range of grass species with rust isolates obtained from different host plants, he was able to determine limitations on the host range of different rust isolates within the species. In the case of Puccinia striiformis, he identified five ff. spp., or specialized forms of rust in this species, these being Puccinia striiformis f. sp. tritici, Puccinia striiformis f. sp. hordei, Puccinia striiformis f. sp. secalis, Puccinia striiformis f. sp. elymi and Puccinia striiformis f. sp. agropyri, with each forma specialis largely restricted to infecting only plant hosts of the Triticeae, Hordeum spp., Secalis spp., Elymus spp., or Agropyron spp., respectively (Eriksson, 1894). In addition, Eriksson (1894) demonstrated that similar ff. spp. existed within the stem rust pathogen Puccinia graminis. Rust pathogen ff. spp. presumably represent the early stages of the evolution of new rust species and demarcation of the respective host and nonhost plant species of these rusts.
Rust Host Ranges
The host range of a pathogen is defined by the plant species it can infect and successfully complete its life cycle on (Thordal-Christensen, 2003; Schulze-Lefert and Panstruga, 2011). Some rust species such as Puccinia hordei (barley leaf rust), Puccinia sorghi (maize rust) and Puccinia kuehnii (sugarcane rust) have restricted host ranges confined to a few species in one or two genera (Wahl et al., 1984). A similar limited host range is apparent for Puccinia triticina, which infects hexaploid wheat and other wheat species from the genus Triticum (Roelfs et al., 1992), as well as several Aegilops species (Yehuda et al., 2004; Bolton et al., 2008), as its primary telial/uredinial hosts.
In contrast, the host range of Puccinia graminis is large and includes 365 species of plants in 54 genera, while that of Puccinia coronata (oat crown rust) includes 290 host species belonging to 72 genera (Wahl et al., 1984). Similarly, the natural host range of Phakopsora pachyrhizi (Asian soybean rust) is large and the pathogen can complete its life cycle on 31 species from 17 leguminous plant genera (Ono et al., 1992). Additional artificial inoculation studies confirmed that this rust species completes its life cycle on another 60 species from 26 genera, largely within the Papilionoideae, although care must be exercised when generalizing from artificial versus natural inoculations. More recent studies added a further 65 species from 25 genera, including 12 previously unreported genera, to this list of plants that allow infection or life cycle completion upon artificial inoculation with Phakopsora pachyrhizi (Slaminko et al., 2008). Further research will show whether those species that allow some degree of infection form part of the host range of Phakopsora pachyrhizi.
This large diversity in host range size is of significant interest. Why have some rust species evolved the ability to parasitize so many different hosts? Is this due to a series of aggressive host jumps or the development of a virulence arsenal more generically adapted for plant colonization? Conversely, is there extensive variation in plant species for resistance genes to nonhost rust pathogens? Alternatively, it may be the rapid evolution of a progenitor host species and the coevolution of the rust. Do rust species with a single host reflect the last remnants of a plant–pathogen interaction with both the plant and specialized pathogen headed for eventual extinction?
In the case of rust species the definition of host range is complicated by the elaborate life cycle of the pathogens. Host range in heteroecious rust species like cereal rusts are further expanded by an alternate host. As described above, the dicot hosts of cereal rusts are thought to be the original hosts. While many alternate hosts are known, our knowledge is still limited. Despite decades of searching for the alternate host of Puccinia striiformis, it was only recently that barberry was identified (Jin et al., 2010). A similar situation exists with Puccinia triticina, where the pathogen has been found to use different species of the genera Thalictrum and Isopyrum as alternate hosts (Roelfs et al., 1992; Bolton et al., 2008). These interactions between Puccinia triticina and the alternate hosts seem to be geographically restricted, complicating the understanding of host range in this context (Bolton et al., 2008). Regional adaptation to different alternate hosts was also reported for Puccinia graminis (Jin et al., 2014). North American Puccinia graminis f. sp. tritici populations east of the Rocky Mountains commonly use Berberis vulgaris as alternate host (Roelfs et al., 1992; Jin et al., 2014), yet it is only rarely found on this host in the northwestern United States (Jin et al., 2014). By isolating Puccinia graminis from aecia on Mahonia repens and Mahonia aquifolium and inoculating various wheat lines and Elymus glaucus, Jin et al. (2014) demonstrated how Puccinia graminis can maintain its virulence diversity in this region and also bypass agricultural selection pressure by completing its life cycle on Mahonia and Elymus. These two examples demonstrate our increasing understanding of the complete host range of these pathogens, especially concerning their potential host ranges on alternate hosts.
Species Jumps – New Hosts
In a study of 80 Pucciniaceae taxa, which collectively parasitize hosts in 33 angiosperm families, evidence for both coevolution of host and pathogen was observed in addition to numerous possible examples of host species jumps involving both telial and aecial forms of the pathogen (van der Merwe et al., 2008). These host jumps occurred on taxonomically unrelated plant species that were geographically associated with the pathogen (van der Merwe et al., 2008).
Similarly, de Vienne et al. (2013) conclude that cospeciation of pathogens and their hosts occurs over a short evolutionary time period, whereas the long-term evolution of plants and pathogens involves frequent host species jumps. From an extensive analysis of published literature they conclude that only 7% of cases represented convincing examples of cospeciation and host shifts constituted the most frequent form of pathogen speciation (de Vienne et al., 2013). This review included one rust study on three flower-mimic rusts (Puccinia monoica, Puccinia thlaspeos, and Puccinia consimilis) and their host genera (Roy, 2001). Flower-mimic rusts sexually reproduce by inducing the formation of pseudoflowers in their hosts, which facilitate fertilization of the rusts by attracted insects (Roy, 1993). Potentially influenced by localized rainfall differences, short distance dispersal of sexual spores by insects and proximity of potential hosts, geographic distance rather than phylogenetic distance predicted host jumps in this system (Roy, 2001).
However, very recent examples of changes in rust host range do exist, which are likely to be a direct consequence of agricultural practices. The cultivation of Australian eucalyptus species in South America has enabled these plants to be parasitized by a rust species endemic to this continent, Puccinia psiidi, which was not present in Australia. This rust has now unfortunately arrived in Australia with many of the native plants susceptible to this pathogen, with growth occurring on 107 species in 30 genera (Carnegie and Lidbetter, 2012). Apart from Australian flora, this pathogen attacks more than 129 species in 33 genera of the Myrtaceae (Carnegie and Lidbetter, 2012). The parasitism of eucalyptus species by this rust probably does not constitute a host jump in the true sense, but rather a host expansion via an opportunistic introduction to susceptible species not previously exposed to this pathogen.
In some rare cases somatic hybridization of two rusts species or ff. spp. has produced a hybrid pathogen with an expanded host range (Park and Wellings, 2012). For example, a Puccinia graminis hybrid lineage was formed by hybridization of Puccinia graminis f. sp. tritici and Puccinia graminis f. sp. secalis to produce a hybrid rust with new virulence specificities (Burdon et al., 1981, 1982). Similarly, a hybrid rust between Melampsora medusae and Melampsora larici-populina was reported in New Zealand that had a virulence spectrum distinct to that of either presumptive parent (Spiers and Hopcroft, 1994).
Sexual recombination events have not been conclusively recorded between rust taxa apart from two potential cases. In one case a potential hybrid between Melampsora medusae and Melampsora occidentalis, called Melampsora x columbiana, was suggested to have arisen by this process, although somatic hybridization followed by some degree of parasexuality could not be excluded (Newcombe et al., 2000, 2001). Similarly, some limited evidence exists for sexual recombination occurring between North American pine blister rusts, specifically between Cronartium comandrae, an endemic species, and Cronartium ribicola, an exotic species (Joly et al., 2006).
Interestingly, the parasitism of new hosts is not always dependent upon major changes in pathogen biology such as somatic hybridization. For example, the specialization of Phytophthora into the species Phytophthora infestans and Phytophthora mirabilis, infecting Solanum spp. and Mirabilis jalapa, respectively, is directly reflected in a mutation of a single fungal gene (an effector gene – see below) that is associated with a host jump 1,300 years ago (Dong et al., 2014).
Rust – Host Interactions: Levels of Resistance
A rust that is capable of parasitizing a plant species is said to be an adapted pathogen of that species, i.e., it can form all the necessary cellular components for colonization and successful reproduction. This same rust species will be incapable of parasitizing the vast majority of plant species for which it is a nonadapted pathogen. Infection of a host plant by urediniospores from an adapted rust pathogen in many cases involves germination of the spore on the leaf surface and growth of a germ tube across the leaf surface, whereupon it identifies a plant stoma by a thigmotropic response, leading to the production of an appressorium over the stoma (Figures 1D and 2F). (Note: although Figures 1 and 2 depict NHR outcomes the same fungal structures are produced during host infection.) From the appressorium an infection peg is inserted between the stomatal guard cells and a substomatal vesicle is produced within the leaf apoplast. Some rust species (e.g., Phakopsora pachyrizi) enter the host plant by germinated urediniospores forming an appressorium on the leaf surface and directly penetrating through the plant epidermis with an appressorium, and subsequently the hypha infect intercellular space. Infection of the alternate hosts of cereal rusts is also performed in this latter manner.
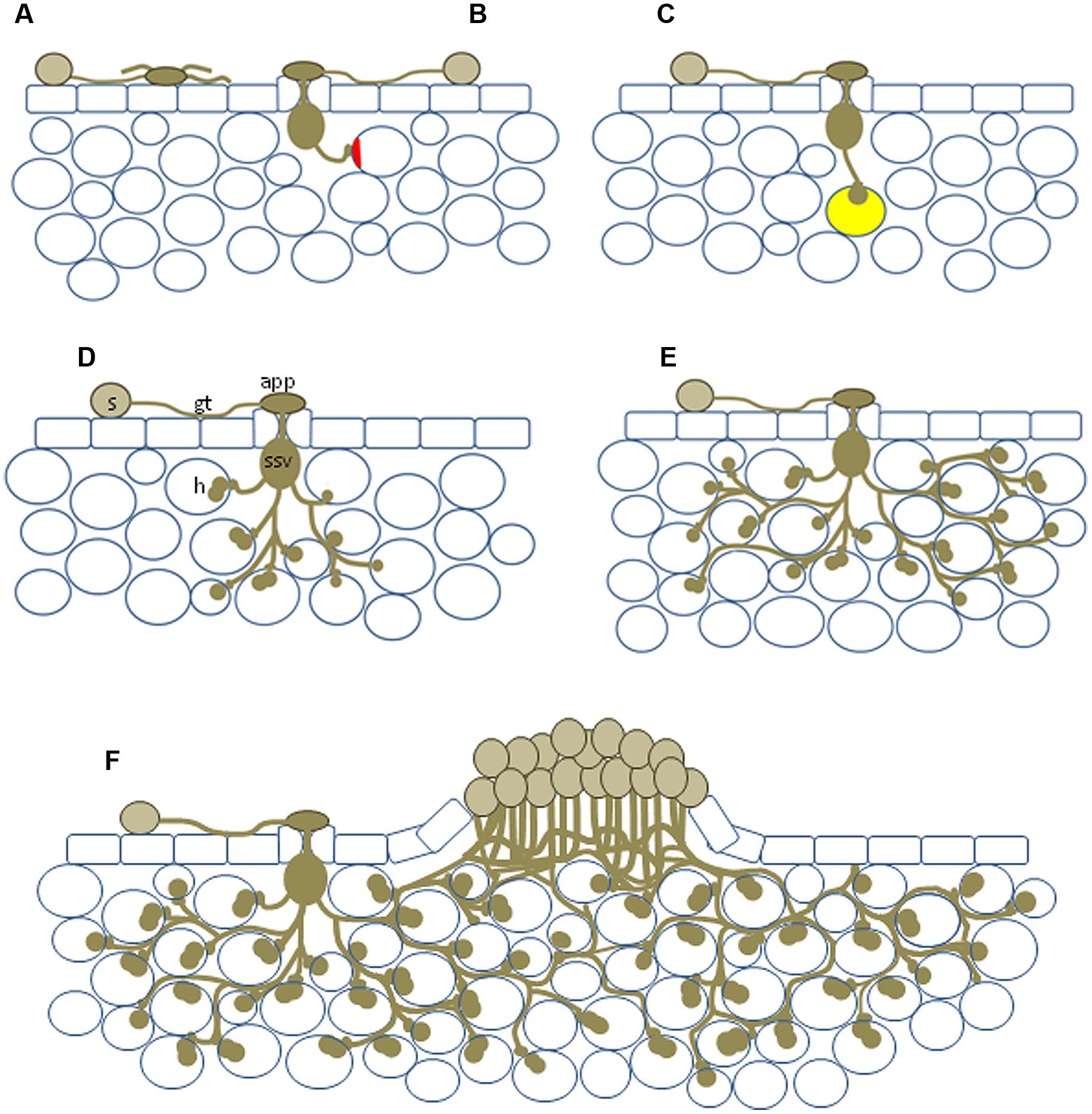
FIGURE 1. Diagram showing the range of potential NHR outcomes. (A) Basic incompatibility in which a spore germinates to produce an aberrant appressorium-like structure in the absence of a stoma; (B) pre-haustorial resistance in which a germination event enters the leaf but is unable to penetrate the cell wall, plant cell wall appositions (shown in red) can occur; (C) infection resulting in the formation of a single haustorium, autofluorescence (yellow) can be associated with these events; (D) haustoria produced in multiple plant cells; (E) relatively large infection site encompassing numerous mesophyll cells but sporulation never observed; (F) formation of a sporulating uredinia, usually much smaller than those observed on susceptible host plants. s, urediniospore; gt, germ tube; app, appressorium; ssv, substomatal vesicle; h, haustorium.
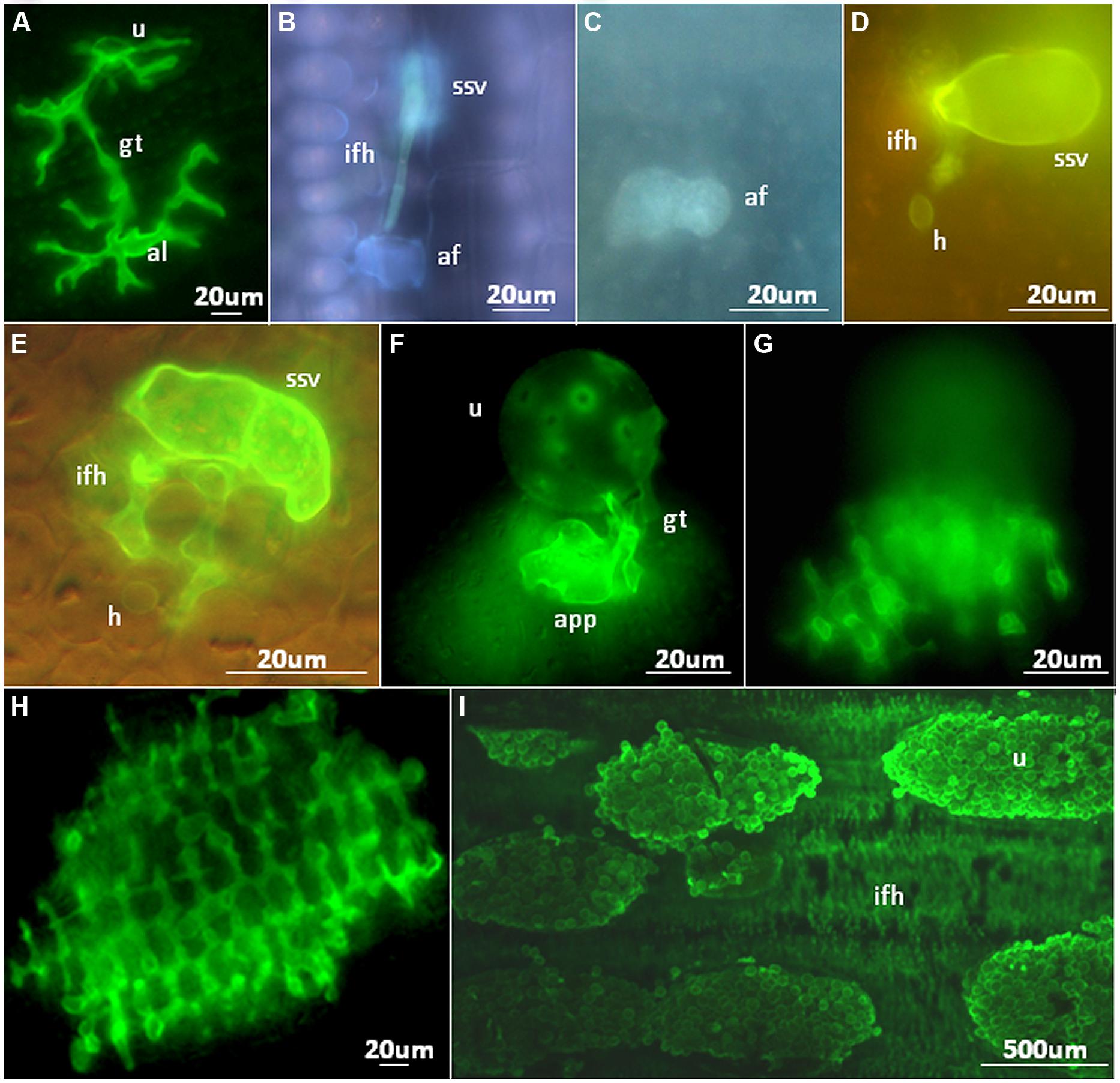
FIGURE 2. Microscopic analyses of NHR outcomes to rust pathogens. (A) Growth of a Melampsora lini (flax rust) germ tube on the surface of a rice leaf. An aberrant appressorium-like structure (al) has been produced. (B) Pre-haustorial resistance against a Puccinia graminis f. sp. tritici infection site on Setaria italica. Contact of a fungal infection hyphae with a single mesophyll cell results in autofluorescence. (C,D) An autofluorescent Brachypodium distachyon mesophyll cell (C) containing a Puccinia striiformis f. sp. tritici haustorium (D). (E) A Puccinia hordei infection site on rice with a single, non-autofluorescent mesophyll cell containing a haustorium. (F,G) A Puccinia striiformis f. sp. tritici urediniospore on the surface of a rice leaf that has produced an appressorium (F) and underlying infection hyphae (G) that encompass multiple mesophyll cells. (H) A Puccinia graminis f. sp. tritici infection site on a rice leaf producing infection hyphae that encompass numerous mesophyll cells. Each dark, circular structure surrounded by green stained fungal infection hyphae is a single mesophyll cell. (I) Multiple Puccinia striiformis f. sp. tritici uredinia on a Brachypodium distachyon leaf producing urediniospores with underlying infection hyphae also apparent. af, autofluorescent plant cell; gt, spore germ tube; ifh, infection hyphae; h, haustoria; ssv, substomatal vesicle; u, urediniospore. All microscopic images were produced as described by Ayliffe et al. (2011).
After substomatal vesicle formation, infection hyphae emerge from this vesicle and ramify through the apoplastic space, inserting haustoria into adjacent plant cells as they extend outward (Figures 1D and 2G,H). Haustoria are specialized fungal feeding structures that arise from haustorium mother cells and penetrate the plant cell wall and invaginate, but do not penetrate, the plant cell membrane (Figures 2D,E). Nutrient transport and molecular trafficking occurs across this plant–pathogen interface called the extrahaustorial matrix (Staples, 2001; Kemen et al., 2005; Garnica et al., 2014). The infection site established from the urediniospore continues to expand and colonize additional plant cells. New urediniospores develop from fungal pedicels, called uredinia, which emerge from the center of the infection site. The uredinia erupt through the leaf surface and thousands of urediniospores are wind dispersed to repeat this asexual infection cycle (Figures 1F and 2I). Given the relative simplicity of the asexual cycle compared with the sexual cycle and that asexual colonization is the causal route of disease on cereals this stage is the most extensively characterized plant–rust interaction.
Successful infection of a plant host by rust requires the pathogen to overcome numerous defense barriers. The first barrier, while not a plant defense mechanism per se, is nonetheless a real obstacle to nonadapted rust pathogen infection and that is a requirement of basic compatibility. A consequence of host–pathogen coevolution is that pathogens can specialize to such a degree they have difficulty in effectively recognizing other plant species as potential hosts. In this case the majority of germinated spores are incapable of identifying a stoma on nonhost plant leaves, a key requirement for appressorium production (see below). It is noteworthy that numerous rust pathogens can be induced to produce appressoria on simple membranes with suitable sized ridges, suggesting leaf topography is a major determinant in this process (Dickinson, 1949; Heath, 1977; Hoch et al., 1987; Allen et al., 1991). For those rusts like Phakopsora pachyrizi that enter the leaf by directly penetrating the plant epidermis hydrophobicity or wax signals play a role in inducing pre-penetration structures, rather than thigmotropism (Uppalapati et al., 2012; Ishiga et al., 2013).
The plant epidermis constitutes a formidable defense to potential pathogens. In the case of mildew pathogens, which also directly penetrate the epidermis, it is a significant barrier to nonadapted mildews. Three genes in Arabidopsis thaliana, PEN1, PEN2, and PEN3 (PENETRATION), which encode a syntaxin vesicle targeting protein, glucosyltransferase, and ATP-binding cassette (ABC) transporter, respectively, are essential components of this epidermal cell-based defense mechanism, which when perturbed enables nonadapted mildew pathogen species to penetrate the leaf (Collins et al., 2003; Lipka et al., 2005, 2010; Stein et al., 2006; Underwood and Somerville, 2013). Underlying penetration resistance is a second layer of defense mediated by components of salicylic acid (SA) based responses. Arabidopsis thaliana plants with mutations in both penetration resistance genes and genes in this second layer of defense can become hosts for nonadapted mildew pathogens such as the pea mildew pathogen Erysiphe pisi (Stein et al., 2006). As will be discussed in depth below, these PEN genes play a role in penetration resistance to the rust pathogen Phakopsora pachyrhizi on nonhost plants, confirming common epidermal defense mechanisms against these diverse pathogen species.
In some pathosystems preformed chemicals play a role in defining host range with nonadapted pathogens unable to overcome these chemical defenses. For example, oats produce antimicrobial triterpene glycosides (avenacins) which effectively make it a nonhost to Gaeumannomyces graminis var. tritici, the causal agent of take-all disease in wheat and barley (Bowyer et al., 1995; Papadopoulou et al., 1999; Qi et al., 2004). As yet preformed chemical barriers have not been demonstrated as a major constraint on rust parasitism.
Active plant defense mechanisms must also be overcome for successful rust parasitism to occur. Extensive research on host resistance in numerous plant pathosystems has given rise to a two-layered paradigm of the active plant defense systems, consisting of pathogen-associated molecular pattern (PAMP) or damage associated molecular pattern (DAMP) triggered immunity (PTI/DTI) and effector-triggered immunity (ETI; Jones and Dangl, 2006). PAMPs are highly conserved, indispensible pathogen molecules which include chitin and xylanases in the case of fungal pathogens (Zipfel, 2008), while DAMPs are endogenous plant molecules released during interactions with pathogens (Boller and Felix, 2009). Recognition of PAMPs or DAMPs via membrane localized pattern recognition receptors (PRRs) leads to a signaling cascade that alerts the plant to the presence of a pathogen and induces PTI (Ishiga et al., 2013). This PTI response frequently involves ion fluxes, the production of reactive oxygen species, protein phosphorylation, ethylene biosynthesis, and callose deposition (Boller and Felix, 2009).
The successful colonization of a plant host requires that the rust pathogen must suppress PTI which is achieved, as for many other pathogens, by the deployment of hundreds of small, secreted effector proteins into plant tissues (Giraldo and Valent, 2013). Rust pathogen effector proteins are produced by haustoria and secreted into the extrahaustorial matrix (i.e., the space between the haustorium cell wall and the plant cytoplasmic membrane) from where many are thought to move into the plant cytoplasm. These effectors likely suppress PTI and alter plant cell homeostasis for the pathogens’ benefit, as observed for other pathogenic fungi (Rafiqi et al., 2010; Giraldo and Valent, 2013). Predicted effector complements of rust pathogens are diverse and presumably play a large role in defining their host range. Amongst different rust species common predicted effector proteins can be identified, although their sequence divergence is high, with more related rust pathogens having more similar effector complements, although each rust species also contains a complement of unique effector proteins (Duplessis et al., 2011; Nemri et al., 2014).
However, not all plants within a given species are susceptible to all isolates of a rust pathogen. As for other pathosystems, plants have evolved resistance proteins, typified by nucleotide binding site–leucine rich repeat (NBS–LRR) proteins, that each recognize a specific rust effector, called an avirulence (Avr) protein. Upon recognition, a more extreme defense response termed ETI is activated, which frequently involves hypersensitive cell death. NBS–LRR proteins that recognize adapted rust pathogens have been isolated from flax (Lawrence et al., 1995, 2010; Anderson et al., 1997; Dodds et al., 2001a,b), maize (Collins et al., 1999; Webb et al., 2002), wheat (Feuillet et al., 2003; Huang et al., 2003; Cloutier et al., 2007; Loutre et al., 2009; Periyannan et al., 2013; Saintenac et al., 2013), and barley (Wang et al., 2013). The best characterized interactions between rust effectors and plant NBS–LRR resistance proteins are from the flax rust pathogen, Melampsora lini, and the flax plant, Linum usitatissimum, where some NBS–LRR proteins recognize flax rust effector proteins by direct protein–protein interaction (Dodds et al., 2006; Catanzariti et al., 2010). In most pathosystems effector recognition by R (resistance) proteins is indirect and involves detection of specific modifications of host proteins mediated by the effector (i.e., guard hypothesis). As individual effectors are often dispensable, different isolates of a rust pathogen species have different effector complements, generally consisting of allelic variants rather than novel genes. In addition, different members of a host plant species contain different R gene complements. Rust infections can therefore lead to either a resistant or a susceptible outcome, depending upon the plant and pathogen genotypes involved. These molecular interactions form the basis of the gene-for-gene hypothesis first formulated by Flor (1942) using the flax-flax rust system.
In addition to NBS–LRR encoding rust resistance genes, several adult plant resistance (APR) genes have been cloned which were originally identified as quantitative trait loci (QTLs) with moderate rust resistance effects at the adult plant stage. One example is the Lr34/Yr18/Sr57/Pm38 gene of wheat which confers resistance to a range of pathogens, including Puccinia triticina, Puccinia striiformis f. sp. tritici, Puccinia graminis f. sp. tritici, and Blumeria graminis f. sp. tritici (wheat powdery mildew; Dyck et al., 1966; Krattinger et al., 2009; Spielmeyer et al., 2013). Lr34 is remarkable in that it provides resistance to all tested isolates of each pathogen and has not been overcome during decades of deployment. This APR gene encodes an ABC transporter protein, although the substrate of this protein is unknown (Krattinger et al., 2009). A second APR gene to be cloned is the Yr36 gene that confers broad spectrum resistance to Puccinia striiformis f. sp. tritici (Fu et al., 2009). This gene encodes a protein with an N-terminal kinase domain fused to a C-terminal steroidogenic acute regulatory protein-related lipid transfer domain (START domain). The mechanism of resistance mediated by Yr36 is largely unknown. In some cases combining of APR genes can lead to increased resistance, i.e., these genes have additive effects, which appears to be the case for the Lr34 and Yr36 genes (Uauy et al., 2005).
Nonhost Resistance to Rust Pathogens
Despite the omnipresence of potential pathogens in the environment and the constant threat of infection, disease is the exception, not the rule, and generally the vast majority of plants are healthy. This is due to the fact that plants can only be infected by a very limited number of the potential pathogens present in the environment. It therefore becomes apparent that all plants are nonhosts to the vast majority of pathogens, which highlights the effectiveness of NHR in the vast majority of cases (Heath, 1991).
Ideally a species would be classified as a nonhost, if all accessions of the species were resistant to all isolates of the pathogen and uniform levels of resistance were observed in all interactions between both species. In practice, delimiting the host range of a pathogen is complicated by the quantitative nature of phenotypes in the transition from host to nonhost. Interactions in this transitional phase may involve only a few accessions of a species being infected by a pathogen or only some isolates of a pathogen being able to infect a plant species. In addition, a continuum of disease phenotypes exists in the interaction with different plant species, which range from full susceptibility (host) to complete immunity (nonhost). The following examples demonstrate some of the phenotypic outcomes that can occur between nonadapted rust pathogens and nonhost plant species.
Arabidopsis thaliana and Nonadapted Rust Pathogens
Several groups have focused their attention on the interaction of the model plant Arabidopsis thaliana with several nonhost pathogens. Germination of Puccinia triticina urediniospores on Arabidopsis thaliana was similar to that on wheat (91% on Arabidopsis thaliana compared to 95% and 93% on susceptible and resistant wheat varieties, respectively), but identification of stomata and appressoria formation were significantly reduced (12% on Arabidopsis thaliana compared to 85% and 83% on susceptible and resistant wheat varieties, respectively; Shafiei et al., 2007). As 62% of pathogen penetration attempts on Arabidopsis thaliana lead to guard cell death that prevented further fungal growth, only 0.2% of the urediniospores managed to form haustoria within leaf mesophyll cells (compared to 72% and 4% on susceptible and resistant wheat varieties, respectively). These numbers did not increase when testing Arabidopsis thaliana mutants defective in defense-related pathways. Using natural variation between Arabidopsis thaliana accessions Col-0 and Ler, Shafiei et al. (2007) identified three QTLs that controlled 41% of sub-stomatal vesicle frequency and two QTLs controlled 21% of guard cell death. Several of these loci co-segregated with genes encoding NBS–LRR or receptor-like kinase (RLK) proteins (Shafiei et al., 2007).
Complementary to the results with Puccinia triticina, stomatal penetration frequencies of Arabidopsis thaliana by three Puccinia striiformis f. sp. tritici isolates was also found to be significantly lower than those observed for wheat, however, no haustoria were formed following penetration (Cheng et al., 2013). The authors attribute this to an active defense response involving callose deposition and accumulation of antimicrobial phenolic compounds, but found no signs of reactive oxygen species release. However, unlike the Puccinia triticina study (Shafiei et al., 2007), increased hyphal growth of the stripe rust pathogen lead to the occasional formation of haustoria in npr1-1 (non-expressor of PR genes) mutant Arabidopsis thaliana plants and in plants depleted in SA, an important plant hormone involved in defense responses (Cheng et al., 2013). Additionally, transcription of the SA-response genes PR1b and PR5 and the jasmonic acid (JA) response gene PDF1.2 was up-regulated during infection of wild type Col-0, although at different time points during infection.
Nonhost resistance of Arabidopsis thaliana has also been examined with other rusts, such as Phakopsora pachyrhizi (described below), Hemileia vastatrix, causal agent of coffee leaf rust, and Uromyces vignae, causal agent of cowpea rust (Mellersh and Heath, 2003; Azinheira et al., 2010). Hemileia vastatrix penetrated Arabidopsis thaliana via stomata, but was unable to form haustoria for nutrient uptake (Azinheira et al., 2010). Similar to the other nonadapted rust infections of Arabidopsis thaliana, penetration of Col-0 by Hemileia vastatrix also lead to a hypersensitive response of the guard cells, callose deposition in epidermal and mesophyll cells, as well as a build-up of phenolic compounds. Moreover, expression of PR1b peaked at 18 h post-inoculation, whereas expression of PDF1.2 peaked at 42 h post-inoculation (Azinheira et al., 2010). Mellersh and Heath (2003) infected 17 Arabidopsis thaliana accessions with Uromyces vignae and all but one accession displayed pre-haustorial NHR. Haustoria formation, however, increased in mutants defective in the SA pathway, which lead to callose deposition around haustoria. The authors conclude that initial defense gene expression limits fungal growth, with an additional SA-dependent layer preventing haustoria formation (Mellersh and Heath, 2003).
Wheat Rust on Beans and Bean Rust on Wheat
Similar observations to those observed in Arabidopsis thaliana upon cereal rust infection were made in broad bean (Vicia faba) upon infection with Puccinia striiformis (Cheng et al., 2012). Only 1.2% of germinated urediniospores were able to locate a stoma on the leaf surface. However, having located a stoma, the majority (96%) of these infection attempts then entered the leaf. Many of these infection sites within the leaf showed aberrant substomatal vesicle morphology and only 2% of infection hyphae produced haustoria. Infection sites were associated with hydrogen peroxide production, callose deposition, and upregulation of SA responsive genes like PR1. Plant cells containing haustoria were autofluorescent, consistent with a hypersensitive cell death response (Cheng et al., 2012).
A similar response was observed when the bean rust pathogen, Uromyces fabae, was inoculated on wheat (Zhang et al., 2011). Again, germ tubes from very few (2%) germinated spores were able to identify a stoma and produce an appressorium. Of the few infection sites that successfully entered the leaf, only 4% managed to produce haustoria and these infected mesophyll cells became callose encased, although cell death was not observed. Most infection hyphae that entered the leaf apoplast were blocked when contact was made with mesophyll cells and cell wall appositions were formed. Reactive oxygen species were present at infection sites.
Limited haustoria formation was also observed on nonhost plants in a study involving Vigna sinensis (cowpea), Phaseolus vulgaris (French bean), Phaseolus lunatus (Lima bean), Pisum sativum (garden pea), Vicia faba (broad bean), Brassica oleracea (cabbage), Helianthus annus (sunflower), and Zea mays (maize) with three rust pathogens, Uromyces phaseoli var. vignae, Puccinia helianthi (sunflower rust) and Puccinia sorghi (Heath, 1977). In most interactions the rust pathogen frequently germinated to identify a stoma and produce an appressorium, with the majority of infections terminating following formation of a haustorial mother cell within the apoplast. In most interactions haustoria were not observed and in those rare cases were they did occur they were usually associated with plant cell death. A distribution of infection site outcomes was observed on single leaves that ranged from germinated spores unable to locate a stoma to those that produced haustoria (Heath, 1977).
Nonhost Resistance to the Asian Soybean Rust Pathogen
As described above, Phakopsora pachyrhizi urediniospores germinate to produce appressoria that directly penetrate the plant epidermis. Phakopsora pachyrhizi urediniospores also germinate on the leaf surface of nonhost plants such as Arabidopsis thaliana and barley and produce appressoria (Loehrer et al., 2008; Hoefle et al., 2009). Penetration is often unsuccessful with the underlying epidermal cells producing callose enriched cell wall appositions at attempted infection sites in both species. However, numerous infection hyphae did successfully penetrate the epidermis of both species and reached underlying mesophyll cells, but the fungus did not successfully invade the mesophyll of either species (Loehrer et al., 2008; Hoefle et al., 2009). Similar to mildew pathogens these observations imply dual defense mechanisms, including both pre-invasive and post-invasive, that act against this soybean rust pathogen in nonhost interactions.
Barley plants with mutations in the Ror1 gene, a homolog of the Arabidopsis thaliana PEN1 gene, have much higher rates of epidermal penetration by the nonadapted soybean rust pathogen, consistent with PEN-mediated pre-invasive defenses also being effective in this NHR response (Hoefle et al., 2009). Similarly, infection of Arabidopsis thaliana plants defective in any of three PEN genes enabled the fungus to grow within the mesophyll intercellular spaces and occasionally produce haustoria, with this effect being most pronounced in pen3 mutants (Loehrer et al., 2008; Langenbach et al., 2013). Pathogen growth was further enhanced in pen plants that were also deficient in either SA (pen3/sid2; pen2/pad4/sag101) or JA (pen3/jar1) signaling pathways, with an increase in haustoria production (Loehrer et al., 2008; Langenbach et al., 2013). An additional gene, BRT1, involved in phenylpropanoid metabolism was also shown to contribute to post-invasive defense in a pen2 mutant background, again demonstrating inducible defense mechanisms (Langenbach et al., 2013).
Nonhost Interactions of Rice
Five cereal rust species (Puccinia graminis f. sp. tritici, Puccinia hordei, Puccinia triticina, Puccinia striiformis f. sp. tritici, and Puccinia sorghi) were shown to infect rice and produce all the infection structures necessary for colonization, including haustoria (Figure 2E). In some instances infection sites were large and encompassed hundreds of mesophyll cells (Figures 2F–H), however, sporulation was never observed (Ayliffe et al., 2011). Nonhost rice plants responded with active defense responses (callose deposition, reactive oxygen species production and rarely cell death) that likely play a role in limiting colonization and consequently prevent sporulation (Ayliffe et al., 2011). However, it appeared that cereal rusts were able to take up nutrients from rice to develop the relatively large infection sites observed in some cases, as the limited energy stored in rust spores is unlikely to support the observed degree of growth. Interestingly, for Melampsora lini, the flax rust pathogen, only 37% of spores that germinated on a rice leaf developed appressoria (compared to 92% on the host). These were often morphologically deformed or not positioned at stomata (Figure 1A), the normal route of penetration. The authors conclude that NHR has a component determined by basic compatibility, reflected by the phylogenetic distance between the host and the nonhost. A subsequent study (Yang et al., 2014) looked for differences in nonhost interactions between Puccinia striiformis f. sp. tritici and the two rice subspecies japonica (11 varieties) and indica (12 varieties). Germination rates were similar for all varieties, but most of the germ tubes did not recognize stomata on the japonica varieties, whereas successful penetration and substomatal vesicle formation was more common in the indica varieties. This coincided with reactive oxygen species release and hypersensitive response at attempted sites of infection in japonica varieties, which were not observed in indica varieties.
Nonhost Rust Interactions of Brachypodium spp.
Brachypodium distachyon is a model grass for temperate cereals such as wheat and barley due to its small genome, small stature, rapid life cycle, and relatively recent divergence from the Triticeae 35–40 million years ago (Draper et al., 2001; Bossolini et al., 2007). As Brachypodium distachyon (sensu lato) has recently been divided into the three species Brachypodium distachyon, Brachypodium stacei, and Brachypodium hybridum (López-Alvarez et al., 2012), many studies include representatives from more than one of these species. Brachypodium distachyon is host to a rust pathogen, Puccinia brachypodii, and genetic studies indicate that resistance to this pathogen is polygenically inherited (Barbieri et al., 2011, 2012). Infection of Brachypodium species with nonadapted rust species and ff. spp. results in macroscopic symptoms including immunity, small necrotic lesions, or the production of small sporulating pustules, depending on the rust species, Brachypodium species, or accession and growth conditions (Draper et al., 2001; Barbieri et al., 2012; Ayliffe et al., 2013; Figueroa et al., 2013). Lesions and pustule development have been observed on some Brachypodium spp. upon infection with the Puccinia graminis ff. spp. tritici, lolii, phlei-pratensis, aveneae, and phalaridi and the Puccinia striiformis ff. spp. tritici, hordei, and bromi, but less so with Puccinia triticina. Interestingly, the majority of stem rusts of the Aveneae/Poeae (Puccinia graminis ff. spp. lolii, phlei-pratensis, aveneae, and phalaridi) produced small sporulating pustules on most Brachypodium spp. accessions tested, arguing these pathosystems are closer to an intermediate host response than NHR (Ayliffe et al., 2013; Figueroa et al., 2013). In contrast, the majority of Brachypodium spp. accessions were immune to rust pathogens of the Triticeae (Puccinia graminis f. sp. tritici and Puccinia striiformis f. sp. tritici), which is consistent with a true nonhost relationship. The increased susceptibility of Brachypodium spp. to rust pathogens of the Poeae is not consistent with the proposed equidistant phylogenetic relationship between the Brachypodieae, Poeae, and Triticeae (Soreng et al., 2007).
Microscopic analyses elucidated differences in the resistance response of Brachypodium distachyon to Puccinia graminis rusts. In one study the response was prehaustorial (Figueroa et al., 2013), whereas in a second study a range of infection site sizes was observed on single leaves. These varied from substomatal vesicles to larger sites that encompassed many mesophyll cells with frequent haustoria production and occasional sporulation (Ayliffe et al., 2013). Presumably this difference is due to a combination of the different environmental conditions, different rust isolates, and different Brachypodium genotypes used in each study. Frequent callose deposition and H2O2 production around infection sites was associated with resistance in the latter study. Cell death was relatively rare in most cases suggesting that hypersensitive cell death is not a major component of this defense response. Additionally, Ayliffe et al. (2013) reported similar NHR responses to Puccinia striiformis f. sp. tritici (Figures 2C,D,I) and that immunity segregated as a single dominant gene in one Brachypodium distachyon mapping family (BdTR10h × TEK4) and potentially as two dominant, linked resistance genes in a second family (BdTR13k × Bd21).
Barley and Nonadapted Rusts
In some cases, a vast majority of a given plant species may be immune to a pathogen species apart from a few isolated lines. This so called “near NHR” has been demonstrated in barley (Hordeum vulgare) with Puccinia triticina, Puccinia hordei-murini, Puccinia hordei-secalini, and Puccinia persistens (Neu et al., 2003; Jafary et al., 2008; Niks, 2014). Based on a screen of 56 Puccinia triticina isolates on the barley accession Bowman, Neu et al. (2003) identified a highly virulent isolate that was subsequently tested on a panel of 18 barley lines. Phenotypes ranged from immunity, to intermediate resistance (small uredinia), and one line was very susceptible (i.e., producing many pustules), demonstrating the intermediate position of this interaction on the scale from full compatibility to complete incompatibility. Even in the immune accession Cebada Capa 20% of infection units produced large infection sites and in some cases even initiated uredinia formation. Interestingly, expression of HvNR-F6, an ortholog of the rice PAMP receptor Xa21 (Song et al., 1995), was upregulated after infection with the nonadapted pathogen Puccinia triticina and the adapted pathogen Puccinia hordei. The authors discuss “a continuum of resistance paradigms,” which may involve similar mechanisms, especially in cases where the pathogens or host and nonhost plants are closely related, as demonstrated in the case studied.
Numerous QTLs were found to confer resistance to Puccinia triticina, Puccinia hordei-murini, Puccinia hordei-secalini, and Puccinia persistens when inoculated on three barley doubled haploid populations (Cebada Capa × SusPtrit, Vada × SusPtrit, and Oregon Wolfe Barley Dominant × Recessive). The authors mirror this observation with the diversity of QTLs governing resistance in host systems (Jafary et al., 2008) and compare their results with the location of the major hypersensitive response gene (Rph7) and several partial resistance (Rphq) genes to the host pathogen Puccinia hordei. As in previous studies (Neu et al., 2003), Rph7 was not found to be involved in resistance against these nonadapted rusts, but the Rphq QTLs colocalized with the QTLs identified against these pathogens. The authors conclude that qualitative R genes are not commonly involved in this “near NHR,” but genes conferring partial resistance to adapted pathogens may play a role in NHR (Jafary et al., 2008; Niks, 2014).
Similar polygenic interactions were observed between Puccinia graminis f. sp. avenea and barley, where transgressive segregants that allowed pathogen sporulation, albeit associated with a mesothetic response, were observed in a mapping family derived from two immune parents (Dracatos et al., 2014). In both parents clear prehaustorial resistance was identified, which involved infection hyphae touching the plant cell surface, but not penetrating the plant cell wall. Host cells responded with localized deposition of autofluorescent material at contact sites, however, no cell death was apparent (Dracatos et al., 2014). The barley prehaustorial resistance to the oat stem rust pathogen argues that either effector production and recognition is not limited to the plant–haustoria interface, such as the case of Rpg1 in the barley-Puccinia graminis f. sp. tritici interaction (Nirmala et al., 2010, 2011), or alternatively this is an extreme PTI response. This contrasts the ability of Puccinia graminis f. sp. avenea to frequently sporulate on Brachypodium spp. accessions and Puccinia graminis f. sp. tritici to infect rice cells to produce large infections sites that encompass numerous mesophyll cells and contain haustoria.
The Continuum of Infection Outcomes
These examples demonstrate the continuum of rust infection outcomes in what is collectively categorized as NHR and range from:
-
A basic incompatibility where the pathogen is physically incapable of efficiently infecting the host (e.g., flax rust on rice; Figures 1A and 2A).
-
Leaf entry but the rust is unable, or rarely able, to form haustoria (cereal rusts on Arabidopsis thaliana and bean, bean rust on wheat etc.; Figures 1B and 2B). This is a very typical outcome for many nonadapted rust pathogens.
-
Infection where all the fungal structures necessary for parasitism are produced, but sporulation never occurs (cereal rusts on rice; Figures 1C–E and 2D–H).
-
The formation of occasional tiny sporulating pustules (rusts with Triticeae hosts on Brachypodium spp.; Figures 1F and 2I).
-
The frequent formation of very small pustules (rusts with Aveneae/Poeae hosts on Brachypodium spp.).
-
“Near NHR” (wheat leaf rust on barley).
The above observations are generalizations in that rare infection sites do develop further in each case. These observations are largely consistent with the proposed layered model of NHR mechanisms (Thordal-Christensen, 2003). It is also apparent that within each nonadapted rust pathosystem a range of infection outcomes can occur on a single leaf with a proportion of infections sites not entering the apoplast, a proportion entering but not forming haustoria, a proportion forming haustoria, and so on. Each NHR interaction is therefore a distribution of outcomes with the median outcome lying between extreme resistance and partial susceptibility depending upon the NHR pathosystem.
Unsurprisingly, in the above examples it is generally observed that the more related the host and nonhost plant species, the greater the rust pathogen colonization on the nonhost. In the case of wheat rusts, for example, the extent of infection on Arabidopsis thaliana was very limited, but increased on rice and even more so on Brachypodium spp. Presumably, elements of basic incompatibility and inappropriate plant signals decrease as host and nonhost relatedness increase. In addition, the efficacy of pathogen virulence molecules, like effectors, may also increase with this increasing relatedness, as the plant molecules targeted by these effectors either increase in similarity or a greater proportion become identical between the host and nonhost (Dong et al., 2014).
The increasing colonization of rusts on nonhosts that are more related to the pathogens’ host species seems to hold up to a point, but then terminates somewhat spectacularly with minimal pathogen growth accompanied by a hypersensitive cell death response occurring in cells containing haustoria, e.g., wheat and Puccinia coronata f. sp. avenea (Moerschbacher et al., 1990). These observations generally fit the molecular evolutionary model proposed by Schulze-Lefert and Panstruga (2011). According to this model, PTI immunity plays a key role in NHR when pathogens attempt to infect more distantly related nonhost species, but a point is reached when host and nonhost plants are each infected by similar pathogen species (Schulze-Lefert and Panstruga, 2011). So similar in fact, that these two pathogen species share common effector molecules. Hence, the nonhost plant may contain pre-existing R proteins capable of directly recognizing effectors or effector activities deployed by the nonadapted pathogen.
As illustrated by the different plant–pathogen interactions discussed, it is important to consider the status of an interaction based on initial differentiation of the rust, colonization, and life cycle completion. The continuum of outcomes listed above and visualized in Figures 1 and 2 highlight that a pathogen’s ability to complete its life cycle on plant species will decrease faster than its ability to penetrate the plant and produce infection structures (Figure 3A). In an extreme host interaction, a panel of accessions would be severely colonized and forming pustules. Conversely, a panel of accessions from a true nonhost species would show no signs of colonization or life cycle completion by a nonadapted pathogen. We propose the terms “intermediate host” and “intermediate nonhost” to classify the continuum of rust infection outcomes observed in the transition from host to nonhost. The pathogens’ inability to penetrate the leaf or form infection structures such as haustoria (Figures 1A,B and 2A,B) would be requirements for describing a plant as nonhost to a pathogen. In an intermediate nonhost, the pathogen can overcome the plants initial layers of defense and produce infection structures typically observed on hosts, but is unable to complete its life cycle (Figures 1C–E and 2D–H) or sporulation is a very rare exception (Figure 1F). Frequent formation of small pustules (Figures 1F and 2I) or the “near NHR” of wheat leaf rust on barley constitutes an intermediate host system. Figure 3 illustrates the expected frequencies for pustule formation and colonization based on a representative panel of the genetic diversity of plant and pathogens in host, intermediate host, intermediate nonhost, and nonhost systems, respectively.
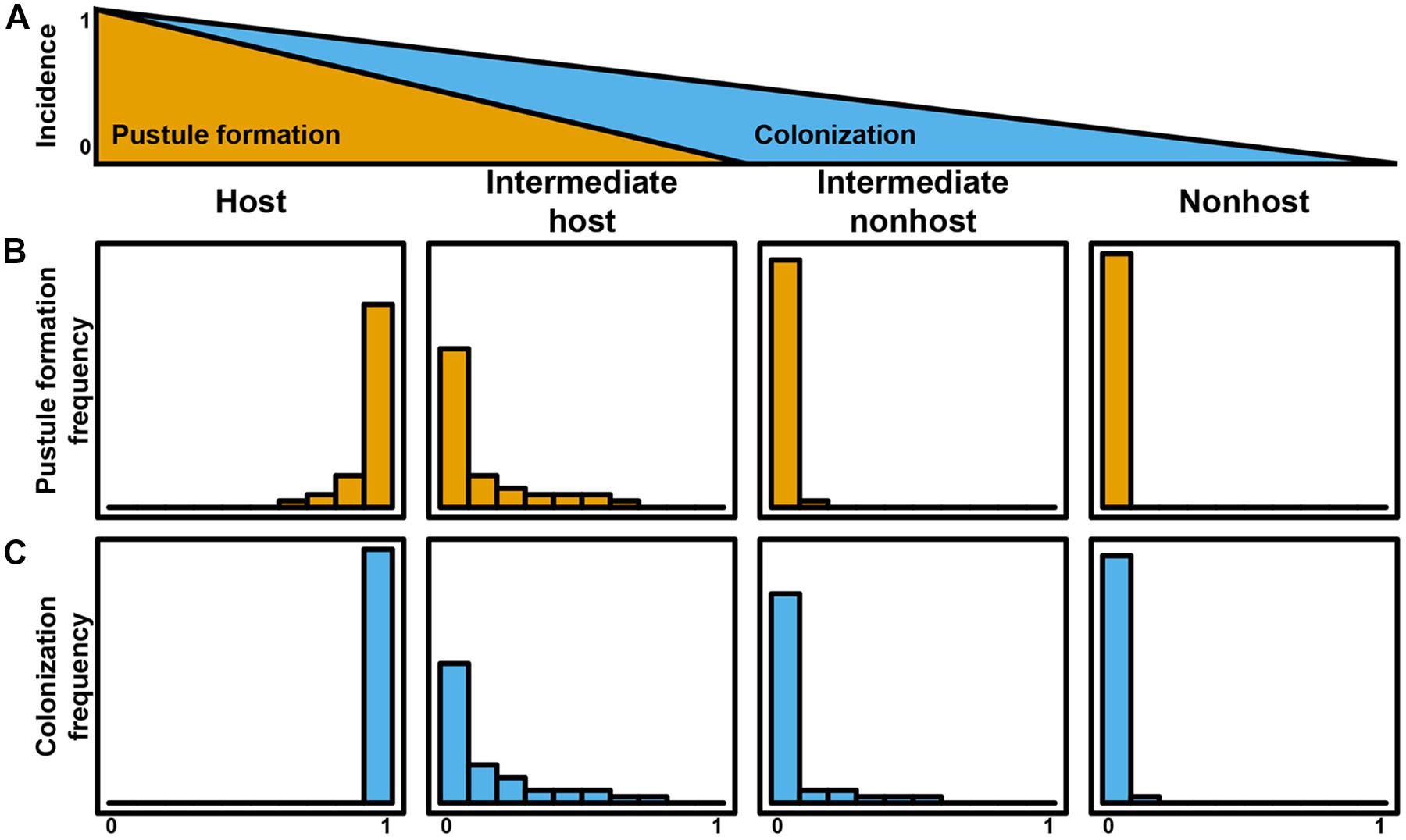
FIGURE 3. Quantitative plant–rust interaction model for the transition from host to nonhost. The incidence of life cycle completion of a nonadapted pathogen decreases faster than incidences of colonization (A). Pustule formation frequency (B) and colonization frequency (C) define host systems, intermediate host systems, intermediate nonhost systems, and nonhost systems.
But it’s More Complicated than that – of Course
The above examples reasonably fit current evolutionary models regarding NHR. However, this is not always the case, particularly as host and nonhost species become more closely related. Histological studies of six species of the Poaceae (Lolium perenne, Avena sativa, Hordeum vulgare, Triticum aestivum, Triticosecale, and Secale cereale) inoculated with Puccinia coronata f. sp. lolii, Puccinia coronata f. sp. avenea, Puccinia hordei, Puccinia triticina, and Puccinia melanocephala (sugarcane rust) were undertaken and in all interactions fungi developed haustorial mother cells (Luke et al., 1987). Haustoria were never formed in any nonhost interaction with Puccinia melanocephala, consistent with the sugarcane host of this pathogen being the most divergent compared with the other plant species. However, Puccinia hordei also never formed haustoria, even though its host, barley, is relatively closely related to the other plant hosts of these rust species (Niks, 1983; Luke et al., 1987). In addition, Puccinia melanocephala showed poor appressorium development on barley and ryegrass (Lolium), but frequent appressorium development on oats, approximately equivalent to that observed on the host.
Moreover, in this study Puccinia coronata f. sp. lolii and Puccinia triticina produced haustoria in all nonhost species, but Puccinia coronata f. sp. avenea frequently produced haustoria on barley and rye but not wheat (Luke et al., 1987). Different infection sites on the same leaf led to variable outcomes ranging from no observable effect to hypersensitive cell death. In some cases cell death (collapsed, fluorescent cells) was not apparent in haustoria infected nonhost cells, which is not consistent with ETI induced by haustoria derived effectors providing resistance. Alternatively, it is possible that resistance is determined by recognition of secreted apoplastic effectors or conserved PAMPs. These data indicate that infection outcomes of nonadapted rust pathogens are often not entirely predictable and do not always readily fit current molecular models of NHR. A caveat is that microscopic phenotypes do not necessarily provide information on the underlying resistance mechanism. For example, the inability to produce haustoria in one pathosystem could be due to a phytoalexin while in a second pathosystem it may be due to PTI. Hypersensitive cell death based resistance is not always a hallmark of NBS–LRR mediated rust resistance (e.g., Sr33) and some rust resistance genes can elicit both cell death and non-cell death responses on the same infected leaf (e.g., Sr45; Periyannan et al., 2013). PTI may also result in cell death in some cases, for example by necrosis inducing elicitors from Phytophthora pathogens (Khatib et al., 2004; Larroque et al., 2013). Therefore, phenotypic similarities do not always necessarily confirm common resistance mechanisms.
Application and Translation of Nonhost Resistance
As described above, an often-cited advantage of NHR is that it is considered durable and broad spectrum (Thordal-Christensen, 2003; Mysore and Ryu, 2004; Schulze-Lefert and Panstruga, 2011). These two qualities of disease resistance are strongly sought after in agricultural crop plants. The possibility of transferring NHR to an agricultural host plant is therefore an attractive proposition. There are a number of examples of successful transfer of resistance from a nonhost species to a host (Wulff et al., 2011). The first example involved the transfer of the maize NBS–LRR encoding gene Rxo1 to rice (Zhao et al., 2005). This gene was identified by the detection of maize lines that showed a strong hypersensitive response when infiltrated with the rice bacterial pathogen Xanthomonas oryzae pv. oryzicola (Zhao et al., 2004b) or by expression of a single type III effector protein from this bacterium in maize (Zhao et al., 2004a). Xanthomonas oryzae pv. oryzicola is the causal agent of bacterial leaf streak on rice, but is a non-pathogen of maize. A single maize gene that conferred the strong hypersensitive response phenotype induced in some lines was isolated by positional cloning and the NBS–LRR protein encoded by this gene provided resistance to Xanthomonas oryzae pv. oryzicola when transferred to rice. Intriguingly, Rxo1 also provides protection in maize against an adapted maize bacterial pathogen, Burkholderia andropogonis (Zhao et al., 2005).
In the Brassicaceae species Arabidopsis thaliana, a leucine rich repeat receptor-kinase, EFR, recognizes a conserved, abundant bacterial translation initiation factor protein, Ef-Tu, to activate PTI (Zipfel et al., 2006). No equivalent receptor is present in solanaceous plants and transfer of this gene to tomato and Nicotiana benthamiana provided increased resistance against bacterial pathogens (Agrobacterium tumefaciens, Pseudomonas syringae, Ralstonia solanacearum, Xanthomonas perforans), some of which are nonadapted pathogens of Arabidopsis thaliana (Lacombe et al., 2010). It is of interest that a fully functional signaling pathway enabling EFR to function in these solanaceous species was pre-existing.
As described above, the wheat Lr34/Yr18/Sr57/Pm38 gene encodes an ABC transporter-like plasma membrane protein that provides broad spectrum resistance to Puccinia triticina, Puccinia striiformis f. sp. tritici, Puccinia graminis f. sp. tritici, and Blumeria graminis f. sp. tritici. However, this resistance is both partial and only effective in adult plants (Krattinger et al., 2009). Transfer of this gene to barley provides resistance to pathogens adapted to both wheat and barley (e.g., Puccinia graminis f. sp. tritici) and pathogens adapted for colonization of barley alone (Puccinia hordei and Blumeria graminis f. sp. hordei; Risk et al., 2013). Associated with this resistance, however, is a necrotic response in barley seedlings and adult plants that is consistent with an accelerated senescence response making this resistance a non-viable option in barley in its current form.
These are all examples of resistance genes from one species conferring resistance in a second species against pathogens that do not parasitize the gene donor plant. Potentially, these genes may also play a role in protecting the donor plants against these nonadapted pathogen species. This conclusion, however, is entirely presumptive, as the absence of functional Rxo1 and Lr34 alleles in maize and wheat, respectively, do not increase their susceptibility to nonadapted pathogens (Zhao et al., 2005). Yet, redundancy in NHR mechanisms is likely to mask any individual contributions of these genes and adds difficulty in confirming their roles in NHR.
An important question is whether identifying single genes potentially involved in NHR and deploying them individually into host plants will result in durable resistance. Given the well-documented transient efficacy of these genes it seems unlikely that a single NBS–LRR encoding gene will be able to provide durable resistance, unless those members involved in NHR recognize an indispensible subset of either pathogen effector proteins or effector modifications of plant proteins. Conversely, a PRR would seem to offer a greater chance of durability given the conservation of PAMP molecules, suggesting biological constraints act upon these molecules that prevent mutation or deletion for resistance avoidance. However, at least on an evolutionary time scale, PTI pathways are ultimately suppressed by the deployment of novel effectors (Cui et al., 2009). Moreover, the durability of an APR gene like Lr34 in providing resistance to pathogens of a different species after heterologous transfer would be very difficult to predict, as important factors such as the substrate of this transporter and the resulting mechanism of resistance are currently unknown.
An alarming possibility is that single gene deployment of NHR genes in new species may result in loss of their efficacy due to pathogen adaptation, which in turn may make the gene donor species susceptible to a previously nonadapted pathogen. Is this scenario likely? A case in point is the Yr9 resistance gene introgressed into wheat from rye (Secale cereale) and widely deployed to provide resistance against Puccinia striiformis f. sp. tritici. After the introduction of Puccinia striiformis f. sp. tritici to Australia, triticale (an artificial hybrid of wheat and rye) retained its resistance to Puccinia striiformis f. sp. tritici for almost a quarter of a century until a race virulent for Yr9 emerged. Due to the breakdown of two additional resistance genes (YrJ and Yr27), Puccinia striiformis f. sp. tritici is now affecting the triticale industry in Australia (Wellings, 2012). The interaction between Puccinia striiformis f. sp. tritici and triticale illustrates the influence that humans can have on pathogen evolution. Introgression of resistance into a major crop grown over a large area places a strong selective pressure on the pathogen and one could imagine that if a sufficient number of rye resistance genes get defeated in wheat or triticale, this could even lead to greater susceptibility of rye to Puccinia striiformis f. sp. tritici.
While in some instances very few genes confer apparent immunity to a pathogen species, in other instances it appears that NHR is polygenic. The loss of a single NHR gene in this latter case is unlikely to result in susceptibility of the donor gene species. Redundancy in NHR mechanisms to mildew pathogens has been well-established in Arabidopsis thaliana with penetration resistance mechanisms, contributed to by the PEN proteins, preceding a second layer of resistance dependent upon SA signaling, as described above (Fan and Doerner, 2012). Other forms of polygenic NHR do not involve multiple independent layers of resistance, but rather multiple loci, each conferring minor additive affects, as observed in the interaction between barley and Puccinia triticina (Neu et al., 2003; Jafary et al., 2008; Niks, 2014). This latter resistance, however, is unlikely to be of practical use due to the genetic complexity required to achieve useful levels of resistance.
The significance of the potential erosion of NHR is fundamentally linked to the agronomic status of the gene donor species. For example, the potential erosion of NHR by the deployment of genes from Brachypodium spp. to wheat seems of minimal concern given this species is of no practical agricultural significance. This process would be akin to the already well-established deployment of host resistance genes from wild grasses such as Aegilops tauschii and Triticum monococcum into wheat (McIntosh et al., 1995), with little concern for the probable future breakdown of host resistance in the wild wheat relatives. In contrast, the potential erosion of rice NHR to rust is potentially of far more significance, although the biological likelihood of such an event is entirely unknown, particularly given the likely polygenic nature of this resistance (Ayliffe et al., 2011). An obvious approach to alleviate some of these concerns would be to treat cloned NHR genes like any other R gene and avoid their single gene deployment.
Summary
Rust evolution has involved numerous host species jumps that involve plants with wide taxonomic divides and which in some cases have been essential for the formation of heteroecious pathogen life cycles. These host jumps demonstrate that plant NHR mechanisms to rusts can be overcome by the pathogen – but generally over long evolutionary periods of time. Modern ff. spp. offer an insight into the process of coevolution and speciation of host and nonhost plant species with rust pathogens. Phenotypically, the NHR response to rusts covers a range of interaction outcomes from basic incompatibility to active defense responses, presumably based upon the same surveillance mechanisms well-established in other pathosystems. However, phenotypic outcomes of rust infections by nonadapted pathogens can be difficult to predict and do not necessarily follow phylogenetic relationships of host and nonhost species. These phenotypic observations have been known for some time, yet, progress in understanding the molecular mechanisms of NHR in a number of plant–rust pathosystems has been made. These interactions tend toward partial resistance rather than true NHR immunity due to the more quantifiable and differential infection outcomes observed. Nonetheless, the genes underlying this resistance will be of great interest, as will be the determination of their potential application in agriculture.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Andrew Dawson and Jeremy Burdon for critical review of the manuscript. Jan Bettgenhaeuser is supported by a Doctoral Training Fellowship from the Biotechnology and Biological Sciences Research Council. Matthew J. Moscou acknowledges funding from the Human Frontier Science Program (LT000218/2011) and the Gatsby Charitable Foundation. Brian Gilbert is supported by an OCE Postdoctoral Research Fellowship and Michael Ayliffe acknowledges support from the Two Blades Foundation.
References
Allen, E. A., Hazen, B. E., Hoch, H. C., Kwon, Y., Leinhos, G. M. E., Staples, R. C.,et al. (1991). Appressorium formation in response to topographical signals by 27 rusts species. Phytopathology 81, 323–331. doi: 10.1094/Phyto-81-323
Anderson, P. A., Lawrence, G. J., Morrish, B. C., Ayliffe, M. A., Finnegan, E. J., and Ellis, J. G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. doi: 10.1105/tpc.9.4.641
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ayliffe, M., Devilla, R., Mago, R., White, R., Talbot, M., Pryor, A.,et al. (2011). Nonhost resistance of rice to rust pathogens. Mol. Plant Microbe Interact. 24, 1143–1155. doi: 10.1094/MPMI-04-11-0100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ayliffe, M., Singh, D., Park, R., Moscou, M., and Pryor, T. (2013). Infection of Brachypodium distachyon with selected grass rust pathogens. Mol. Plant Microbe Interact. 26, 946–957. doi: 10.1094/MPMI-01-13-0017-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Azinheira, H. G., Silva, M. C., Talhinhas, P., Medeira, C., Maia, I., Petitot, A.,et al. (2010). Non-host resistance responses of Arabidopsis thaliana to the coffee leaf rust fungus (Hemileia vastatrix). Botany 88, 621–629. doi: 10.1139/B10-039
Barbieri, M., Marcel, T. C., and Niks, R. E. (2011). Host status of false brome grass to the leaf rust fungus Puccinia brachypodii and the stripe rust fungus P. striiformis. Plant Dis. 95, 1339–1345. doi: 10.1094/PDIS-11-10-0825
Barbieri, M., Marcel, T. C., Niks, R. E., Francia, E., Pasquariello, M., Mazzamurro, V.,et al. (2012). QTLs for resistance to the false brome rust Puccinia brachypodii in the model grass Brachypodium distachyon L. Genome 55, 152–163. doi: 10.1139/G2012-001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bolton, M. D., Kolmer, J. A., and Garvin, D. F. (2008). Wheat leaf rust caused by Puccinia triticina. Mol. Plant Pathol. 9, 563–575. doi: 10.1111/j.1364-3703.2008.00487.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bossolini, E., Wicker, T., Knobel, P. A., and Keller, B. (2007). Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J. 49, 704–717. doi: 10.1111/j.1365-313X.2006.02991.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bowyer, P., Clarke, B. R., Lunness, P., Daniels, M. J., and Osbourn, A. E. (1995). Host range of a plant pathogenic fungus determined by a saponin detoxifying enzyme. Science 267, 371–374.
Burdon, J. J., Marshall, D. R., and Luig, N. H. (1981). Isoenzyme analysis indicates that a virulent cereal rust pathogen is a somatic hybrid. Nature 293, 565–566. doi: 10.1038/293565a0
Burdon, J. J., Marshall, D. R., Luig, N. H., and Gow, D. J. S. (1982). Isozyme studies on the origin and evolution of Puccinia graminis f. sp. tritici in Australia. Aust. J. Biol. Sci. 35, 231–238.
Carnegie, A. J., and Lidbetter, J. R. (2012). Rapidly expanding host range for Puccinia psidii sensu lato in Australia. Aust. Plant Pathol. 41, 13–29. doi: 10.1007/s13313-011-0082-6
Catanzariti, A. M., Dodds, P. N., Ellis, J. G., and Staskawicz, B. J. (2010). The interaction of avirulence and resistance gene products in flax rust disease – providing advances in rust research. Can. J. Plant Pathol. 32, 11–19. doi: 10.1080/07060661003621068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, Y., Zhang, H., Yao, J., Han, Q., Wang, X., Huang, L.,et al. (2013). Cytological and molecular characterization of non-host resistance in Arabidopsis thaliana against wheat stripe rust. Plant Physiol. Biochem. 62, 11–18. doi: 10.1016/j.plaphy.2012.10.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, Y., Zhang, H., Yao, J., Wang, X., Xu, J., Han, Q.,et al. (2012). Characterization of non-host resistance in broad bean to the wheat stripe rust pathogen. BMC Plant Biol. 12:96. doi: 10.1186/1471-2229-12-96
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cloutier, S., McCallum, B. D., Loutre, C., Banks, T. W., Wicker, T., Feuillet, C.,et al. (2007). Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 65, 93–106. doi: 10.1007/s11103-007-9201-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collins, N., Drake, J., Ayliffe, M., Sun, Q., Ellis, J., Hulbert, S.,et al. (1999). Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11, 1365–1376. doi: 10.1105/tpc.11.7.1365
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collins, N. C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J. L.,et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. doi: 10.1038/nature02076
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cui, H., Xiang, T., and Zhou, J. M. (2009). Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell. Microbiol. 11, 1453–1461. doi: 10.1111/j.1462-5822.2009.01359.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
de Vienne, D. M., Refrégier, G., López-Villavicencio, M., Tellier, A., Hood, M. E., and Giraud, T. (2013). Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 198, 347–385. doi: 10.1111/nph.12150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dickinson, S. (1949). Studies in the physiology of obligate parasitism: IV. The formation on membranes of haustoria by rust hyphae and powdery mildew germ-tubes. Ann. Bot. 13, 345–353.
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Teh, T., Wang, C. I., Ayliffe, M. A.,et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 8888–8893. doi: 10.1073/pnas.0602577103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dodds, P. N., Lawrence, G. J., and Ellis, J. G. (2001a). Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27, 439–453. doi: 10.1046/j.1365-313X.2001.01114.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dodds, P. N., Lawrence, G. J., and Ellis, J. G. (2001b). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178.
Dong, S., Stam, R., Cano, L. M., Song, J., Sklenar, J., Yoshida, K.,et al. (2014). Effector specialization in a lineage of the Irish potato famine pathogen. Science 343, 552–555. doi: 10.1126/science.1246300
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dracatos, P. M., Ayliffe, M., Khatkar, M., Fetch, T., Singh, D., and Park, R. (2014). Inheritance of pre-haustorial resistance to Puccinia graminis f. sp. avenae in barley (Hordeum vulgare L.). Mol. Plant Microbe Interact. 27, 1253–1262. doi: 10.1094/MPMI-05-14-0140-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Draper, J., Mur, L. A., Jenkins, G., Ghosh-Biswas, G. C., Bablak, P., Hasterok, R.,et al. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555. doi: 10.1104/pp.010196
Duplessis, S., Cuomo, C. A., Lin, Y. C., Aerts, A., Tisserant, E., Veneault-Fourrey, C.,et al. (2011). Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. U.S.A. 108, 9166–9171. doi: 10.1073/pnas.1019315108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dyck, P. L., Samborski, D. J., and Anderson, R. G. (1966). Inheritance of adult-plant leaf rust resistance derived from the common wheat varieties Exchange and Frontana. Can. J. Genet. Cytol. 8, 665–671.
Eriksson, J. (1894). Ueber die specialisirung des parasitismus bei den getreiderostpilzen. Ber. Dtsch. Bot. Ges. 12, 292–331.
Fan, J., and Doerner, P. (2012). Genetic and molecular basis of nonhost disease resistance: complex, yes; silver bullet, no. Curr. Opin. Plant Biol. 15, 400–406. doi: 10.1016/j.pbi.2012.03.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., and Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. U.S.A. 100, 15253–15258. doi: 10.1073/pnas.2435133100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Figueroa, M., Alderman, S., Garvin, D. F., and Pfender, W. F. (2013). Infection of Brachypodium distachyon by formae speciales of Puccinia graminis: early infection events and host-pathogen incompatibility. PLoS ONE 8:e56857. doi: 10.1371/journal.pone.0056857
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X.,et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garnica, D. P., Nemri, A., Upadhyaya, N. M., Rathjen, J. P., and Dodds, P. N. (2014). The ins and outs of rust haustoria. PLoS Pathog. 10:e1004329. doi: 10.1371/journal.ppat.1004329
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giraldo, M. C., and Valent, B. (2013). Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. doi: 10.1038/nrmicro3119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heath, M. C. (1977). A comparative study of non-host interactions with rust fungi. Physiol. Plant Pathol. 10, 73–76. doi: 10.1016/0048-4059(77)90009-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heath, M. C. (1991). The role of gene-for-gene interactions in the determination of host species specificity. Phytopathology 81, 127–130.
Hoch, H. C., Staples, R. C., Whitehead, B., Comeau, J., and Wolf, E. D. (1987). Signaling for growth orientation and cell differentiation by surface topography in Uromyces. Science 235, 1659–1662. doi: 10.1126/science.235.4796.1659
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoefle, C., Loehrer, M., Schaffrath, U., Frank, M., Schultheiss, H., and Hückelhoven, R. (2009). Transgenic suppression of cell death limits penetration success of the soybean rust fungus Phakopsora pachyrhizi into epidermal cells of barley. Phytopathology 99, 220–226. doi: 10.1094/PHYTO-99-3-0220
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hovmøller, M. S., Sørensen, C. K., Walter, S., and Justesen, A. F. (2011). Diversity of Puccinia striiformis on cereals and grasses. Annu. Rev. Phytopathol. 49, 197–217. doi: 10.1146/annurev-phyto-072910-095230
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, L., Brooks, S. A., Li, W., Fellers, J. P., Trick, H. N., and Gill, B. S. (2003). Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164, 655–664.
Ishiga, Y., Upplapapti, S. R., and Mysore, K. S. (2013). Expression analysis reveals a role for hydrophobic or epicuticular wax signals in pre-penetration structure formation of Phakopsora pachyrhizi. Plant Signal. Behav. 8:e26959. doi: 10.4161/psb.26959
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jafary, H., Albertazzi, G., Marcel, T. C., and Niks, R. E. (2008). High diversity of genes for nonhost resistance of barley to heterologous rust fungi. Genetics 178, 2327–2339. doi: 10.1534/genetics.107.077552
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jin, Y., Rouse, M., and Groth, J. (2014). Population diversity of Puccinia graminis is sustained through sexual cycle on alternate hosts. J. Integr. Agric. 13, 262–264. doi: 10.1016/S2095-3119(13)60647-4
Jin, Y., Szabo, L. J., and Carson, M. (2010). Century-old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology 100, 432–435. doi: 10.1094/PHYTO-100-5-0432
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joly, D. L., Langor, D. W., and Hamelin, R. C. (2006). Molecular and morphological evidence for interspecific hybridization between Cronartium ribicola and C. comandrae on Pinus flexilis in Southwestern Alberta. Plant Dis. 90:1552. doi: 10.1094/PD-90-1552A
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kemen, E., Kemen, A. C., Rafiqi, M., Hempel, U., Mendgen, K., Hahn, M.,et al. (2005). Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant Microbe Interact. 18, 1130–1139. doi: 10.1094/MPMI-18-1130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khatib, M., Lafitte, C., Esquerreì-Tugayeì, M.-T., Bottin, A., and Rickauer, M. (2004). The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162, 501–510. doi: 10.1111/j.1469-8137.2004.01043.x
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H.,et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lacombe, S., Rougon-Cardoso, A., Sherwood, E., Peeters, N., Dahlbeck, D., van Esse, H. P.,et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. doi: 10.1038/nbt.1613
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Langenbach, C., Campe, R., Schaffrath, U., Goellner, K., and Conrath, U. (2013). UDP-glucosyltransferase UGT84A2/BRT1 is required for Arabidopsis nonhost resistance to the Asian soybean rust pathogen Phakopsora pachyrhizi. New Phytol. 198, 536–545. doi: 10.1111/nph.12155
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Larroque, M., Belmas, E., Martinez, T., Vergnes, S., Ladouce, N., Lafitte, C.,et al. (2013). Pathogen-associated molecular pattern-triggered immunity and resistance to the root pathogen Phytophthora parasitica in Arabidopsis. J. Exp. Bot. 64, 3615–3625. doi: 10.1093/jxb/ert195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lawrence, G. J., Anderson, P. A., Dodds, P. N., and Ellis, J. G. (2010). Relationships between rust resistance genes at the M locus in flax. Mol. Plant Pathol. 11, 19–32. doi: 10.1111/j.1364-3703.2009.00563.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lawrence, G. J., Finnegan, E. J., Ayliffe, M. A., and Ellis, J. G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7, 1195–1206. doi: 10.1105/tpc.7.8.1195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leonard, K. J., and Szabo, L. J. (2005). Stem rust of small grains and grasses caused by Puccinia graminis. Mol. Plant Pathol. 6, 99–111. doi: 10.1111/j.1364-3703.2005.00273.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leppik, E. E. (1961). Some viewpoints on the phylogeny of rust fungi. IV. Stem rust genealogy. Mycologia 53, 378–405. doi: 10.2307/3756583
Lipka, U., Fuchs, R., Kuhns, C., Petutschnig, E., and Lipka, V. (2010). Live and let die – Arabidopsis nonhost resistance to powdery mildews. Eur. J. Cell Biol. 89, 194–199. doi: 10.1016/j.ejcb.2009.11.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lipka, V., Dittgen, J., Bednarek, P., Bhat, R., Wiermer, M., Stein, M.,et al. (2005). Pre-and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183. doi: 10.1126/science.1119409
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loehrer, M., Langenbach, C., Goellner, K., Conrath, U., and Schaffrath, U. (2008). Characterization of nonhost resistance of Arabidopsis to the Asian soybean rust. Mol. Plant Microbe Interact. 21, 1421–1430. doi: 10.1094/MPMI-21-11-1421
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
López-Alvarez, D., López-Herranz, M. L., Betekhtin, A., and Catalán, P. (2012). A DNA barcoding method to discriminate between the model plant Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). PLoS ONE 7:e51058. doi: 10.1371/journal.pone.0051058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loutre, C., Wicker, T., Travella, S., Galli, P., Scofield, S., Fahima, T.,et al. (2009). Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 60, 1043–1054. doi: 10.1111/j.1365-313X.2009.04024.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luke, H. H., Barnett, R. D., and Pfahler, P. L. (1987). Xenoparasite-nonhost reactions in Puccinia-Gramineae pathosystems. Phytopathology 77, 1488–1491. doi: 10.1094/Phyto-77-1488
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts. An Atlas of Resistance Genes. East Melbourne: CSIRO Publications. doi: 10.1007/978-94-011-0083-0
Mellersh, D. G., and Heath, M. C. (2003). An investigation into the involvement of defense signaling pathways in components of the nonhost resistance of Arabidopsis thaliana to rust fungi also reveals a model system for studying rust fungal compatibility. Mol. Plant Microbe Interact. 16, 398–404. doi: 10.1094/MPMI.2003.16.5.398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moerschbacher, B. M., Noll, U., Ocampo, C. A., Flott, B. E., Gotthardt, U., Wüslefeld, A.,et al. (1990). Hypersensitive lignification response as the mechanism of non-host resistance of wheat against oat crown rust. Physiol. Plant. 78, 609–615. doi: 10.1111/j.1399-3054.1990.tb05249.x
Mysore, K. S., and Ryu, C. M. (2004). Nonhost resistance: how much do we know? Trends Plant Sci. 9, 97–104. doi: 10.1016/j.tplants.2003.12.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nemri, A., Saunders, D. G., Anderson, C., Upadhyaya, N. M., Win, J., Lawrence, G. J.,et al. (2014). The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front. Plant Sci. 5:98. doi: 10.3389/fpls.2014.00098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Neu, C., Keller, B., and Feuillet, C. (2003). Cytological and molecular analysis of the Hordeum vulgare–Puccinia triticina nonhost interaction. Mol. Plant Microbe Interact. 16, 626–633. doi: 10.1094/MPMI.2003.16.7.626
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Newcombe, G., Stirling, B., and Bradshaw, H. D. Jr. (2001). Abundant pathogenic variation in the new hybrid rust Melampsora × columbiana on hybrid poplar. Phytopathology 91, 981–985. doi: 10.1094/PHYTO.2001.91.10.981
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Newcombe, G., Stirling, B., McDonald, S., and Bradshaw, H. D. Jr. (2000). Melampsora × columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycol. Res. 104, 261–274. doi: 10.1017/S0953756299001665
Niks, R. E. (1983). Comparative histology of partial resistance and the nonhost reaction to leaf rust pathogens in barley and wheat seedlings. Phytopathology 73, 60–64. doi: 10.1094/Phyto-73-60
Niks, R. E. (2014). How specific is non-hypersensitive host and nonhost resistance of barley to rust and mildew fungi? J. Integr. Agric. 13, 244–254. doi: 10.1016/S2095-3119(13)60648-6
Nirmala, J., Drader, T., Chen, X., Steffenson, B., and Kleinhofs, A. (2010). Stem rust spores elicit rapid RPG1 phosphorylation. Mol. Plant Microbe Interact. 23, 1635–1642. doi: 10.1094/MPMI-06-10-0136
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nirmala, J., Drader, T., Lawrence, P. K., Yin, C., Hulbert, S., Steber, C. M.,et al. (2011). Concerted action of two avirulent spore effectors activates reaction to Puccinia graminis 1 (Rpg1)-mediated cereal stem rust resistance. Proc. Natl. Acad. Sci. U.S.A. 108, 14676–14681. doi: 10.1073/pnas.1111771108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ono, Y., Buriticá, P., and Hennen, J. F. (1992). Delimitation of Phakopsora, Physopella, and Cerotelium and their species on Leguminosae. Mycol. Res. 96, 825–850. doi: 10.1016/S0953-7562(09)81029-0
Papadopoulou, K., Melton, R. E., Leggett, M., Daniels, M. J., and Osbourn, A. E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. U.S.A. 96, 12923–12928. doi: 10.1073/pnas.96.22.12923
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, R. F., and Wellings, C. R. (2012). Somatic hybridization in the Uredinales. Annu. Rev. Phytopathol. 50, 219–239. doi: 10.1146/annurev-phyto-072910-095405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L.,et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. doi: 10.1126/science.1239028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qi, X., Bakht, S., Leggett, M., Maxwell, C., Melton, R., and Osbourn, A. (2004). A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. U.S.A. 101, 8233–8238. doi: 10.1073/pnas.0401301101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rafiqi, M., Gan, P. H., Ravensdale, M., Lawrence, G. J., Ellis, J. G., Jones, D. A.,et al. (2010). Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22, 2017–2032. doi: 10.1105/tpc.109.072983
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Risk, J. M., Selter, L. L., Chauhan, H., Krattinger, S. G., Kumlehn, J., Hensel, G.,et al. (2013). The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol. J. 11, 847–854. doi: 10.1111/pbi.12077
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roelfs, A. P., Singh, R. P., and Saari, E. E. (1992). Rust Disease of Wheat: Concepts and Methods of Disease Management. Mexico, DF: CIMMYT. 81.
Roy, B. A. (1993). Floral mimicry by a plant pathogen. Nature 362, 56–58. doi: 10.1038/362056a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roy, B. A. (2001). Patterns of association between crucifers and their flower-mimic pathogens: host jumps are more common than coevolution or cospeciation. Evolution (N. Y.) 55, 41–53.
Saintenac, C., Zhang, W., Salcedo, A., Rouse, M. N., Trick, H. N., Akhunov, E.,et al. (2013). Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341, 783–786. doi: 10.1126/science.1239022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schulze-Lefert, P., and Panstruga, R. (2011). A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16, 117–125. doi: 10.1016/j.tplants.2011.01.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shafiei, R., Hang, C., Kang, J. G., and Loake, G. J. (2007). Identification of loci controlling non-host disease resistance in Arabidopsis against the leaf rust pathogen Puccinia triticina. Mol. Plant Pathol. 8, 773–784. doi: 10.1111/j.1364-3703.2007.00431.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Slaminko, T. L., Miles, M. R., Frederick, R. D., Bonde, M. R., and Hartman, G. L. (2008). New legume hosts of Phakopsora pachyrhizi based on greenhouse evaluations. Plant Dis. 92, 767–771. doi: 10.1094/PDIS-92-5-0767
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T.,et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Soreng, R. J., Davis, J. I., and Voionmaa, M. A. (2007). A phylogenetic analysis of Poaceae tribe Poeae sensu lato based on morphological characters and sequence data from three plastid-encoded genes: evidence for reticulation, and a new classification for the tribe. Kew Bull. 62, 425–454.
Spielmeyer, W., Mago, R., Wellings, C., and Ayliffe, M. (2013). Lr67 and Lr34 rust resistance genes have much in common – they confer broad spectrum resistance to multiple pathogens in wheat. BMC Plant Biol. 13:96. doi: 10.1186/1471-2229-13-96
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spiers, A. G., and Hopcroft, D. H. (1994). Comparative studies of the poplar rusts Melampsora medusae, M. larici-populina and their interspecific hybrid M. medusae-populina. Mycol. Res. 98, 889–903. doi: 10.1016/S0953-7562(09)80260-8
Staples, R. C. (2001). Nutrients for a rust fungus: the role of haustoria. Trends Plant Sci. 6, 496–498. doi: 10.1016/S1360-1385(01)02126-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stein, M., Dittgen, J., Sánchez-Rodríguez, C., Hou, B. H., Molina, A., Schulze-Lefert, P.,et al. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18, 731–746. doi: 10.1105/tpc.105.038372
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thordal-Christensen, H. (2003). Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 6, 351–357. doi: 10.1016/S1369-5266(03)00063-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Uauy, C., Brevis, J. C., Chen, X., Khan, I., Jackson, L., Chicaiza, O.,et al. (2005). High-temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theor. Appl. Genet. 112, 97–105. doi: 10.1007/s00122-005-0109-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Underwood, W., and Somerville, S. C. (2013). Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc. Natl. Acad. Sci. U.S.A. 110, 12492–12497. doi: 10.1073/pnas.1218701110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Uppalapati, S. R., Ishiga, Y., Doraiswamy, V., Bedair, M., Mittal, S., Chen, J.,et al. (2012). Loss of abaxial leaf epicuticular wax in Medicago truncatula irg1/palm1 mutants results in reduced spore differentiation of anthracnose and nonhost rust pathogens. Plant Cell 24, 353–370. doi: 10.1105/tpc.111.093104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van der Merwe, M. M., Walker, J., Ericson, L., and Burdon, J. J. (2008). Coevolution with higher taxonomic host groups within the Puccinia/Uromyces rust lineage obscured by host jumps. Mycol. Res. 112, 1387–1408. doi: 10.1016/j.mycres.2008.06.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wahl, I., Anikster, Y., Manisterski, J., and Segal, A. (1984). “Evolution at the center of origin,” in The Cereal Rusts, eds W. R. Bushnell and A. P. Roelfs (Orlando, FL: Academic Press), 39–77. doi: 10.1016/B978-0-12-148401-9.50008-X
Wang, X., Richards, J., Gross, T., Druka, A., Kleinhofs, A., Steffenson, B.,et al. (2013). The rpg4-mediated resistance to wheat stem rust (Puccinia graminis) in barley (Hordeum vulgare) requires Rpg5, a second NBS-LRR gene, and an actin depolymerization factor. Mol. Plant Microbe Interact. 26, 407–418. doi: 10.1094/MPMI-06-12-0146-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Webb, C. A., Richter, T. E., Collins, N. C., Nicolas, M., Trick, H. N., Pryor, T.,et al. (2002). Genetic and molecular characterization of the maize rp3 rust resistance locus. Genetics 162, 381–394.
Wellings, C. (2012). “Stripe rust resistance in triticale in Australia: pathogen change exposes genetic vulnerability,” in Proceedings of the 13th International Cereal Rusts and Powdery Mildews Conference, ed. W.-Q. Chen (Beijing: China Agricultural Sciences and Technology Press), 38–39.
Wulff, B. B. H., Horvath, D. M., and Ward, E. R. (2011). Improving immunity in crops: new tactics in an old game. Curr. Opin. Plant Biol. 14, 468–476. doi: 10.1016/j.pbi.2011.04.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, Y., Zhao, J., Xing, H., Wang, J., Zhou, K., Zhan, G.,et al. (2014). Different non-host resistance responses of two rice subspecies, japonica and indica, to Puccinia striiformis f. sp. tritici. Plant Cell Rep. 33, 423–433. doi: 10.1007/s00299-013-1542-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yehuda, P. B., Eilam, T., Manisterski, J., Shimoni, A., and Anikster, Y. (2004). Leaf rust on Aegilops speltoides caused by a new forma specialis of Puccinia triticina. Phytopathology 94, 94–101. doi: 10.1094/PHYTO.2004.94.1.94
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, H., Wang, C., Cheng, Y., Wang, X., Li, F., Han, Q.,et al. (2011). Histological and molecular studies of the non-host interaction between wheat and Uromyces fabae. Planta 234, 979–991. doi: 10.1007/s00425-011-1453-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, B., Ardales, E. Y., Raymundo, A., Bai, J., Trick, H. N., Leach, J. E.,et al. (2004a). The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol. Plant Microbe Interact. 17, 771–779. doi: 10.1094/MPMI.2004.17.7.771
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, B. Y., Ardales, E., Brasset, E., Claflin, L. E., Leach, J. E., and Hulbert, S. H. (2004b). The Rxo1/ Rba1 locus of maize controls resistance reactions to pathogenic and non-host bacteria. Theor. Appl. Genet. 109, 71–79. doi: 10.1007/s00122-004-1623-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, B., Lin, X., Poland, J., Trick, H., Leach, J., and Hulbert, S. (2005). A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl. Acad. Sci. U.S.A. 102, 15383–15388. doi: 10.1073/pnas.0503023102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zipfel, C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. doi: 10.1016/j.coi.2007.11.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D., Boller, T.,et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. doi: 10.1016/j.cell.2006.03.037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: host range, disease resistance, nonadapted pathogen, NHR, formae speciales, Pucciniales
Citation: Bettgenhaeuser J, Gilbert B, Ayliffe M and Moscou MJ (2014) Nonhost resistance to rust pathogens – a continuation of continua. Front. Plant Sci. 5:664. doi: 10.3389/fpls.2014.00664
Received: 29 September 2014; Accepted: 07 November 2014;
Published online: 11 December 2014.
Edited by:
Gitta Coaker, University of California, Davis, USAReviewed by:
Ralph Panstruga, RWTH Aachen University, GermanyGuus Bakkeren, Agriculture and Agri-Food Canada, Canada
Copyright © 2014 Bettgenhaeuser, Gilbert, Ayliffe and Moscou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Ayliffe, Commonwealth Scientific and Industrial Research Organisation, Agriculture Flagship, P.O. Box 1600, Clunies Ross Street, Acton, Canberra, ACT 2601, Australia e-mail: michael.ayliffe@csiro.au; Matthew J. Moscou, The Sainsbury Laboratory, Norwich Research Park, Norfolk, Norwich NR4 7UH, UK e-mail: matthew.moscou@tsl.ac.uk
 Jan Bettgenhaeuser
Jan Bettgenhaeuser Brian Gilbert2
Brian Gilbert2 Michael Ayliffe
Michael Ayliffe Matthew J. Moscou
Matthew J. Moscou