- 1Laboratory of Plant Molecular Genetics, Graduate School of Biological Sciences, Nara Institute of Science and Technology, Ikoma, Japan
- 2Plant Genome Engineering Research Unit, Agrogenomics Research Center, National Institute of Agrobiological Sciences, Tsukuba, Japan
- 3Development of Agrobiological Resources, Faculty of Agriculture, Meijo University, Nagoya, Japan
Gene targeting (GT) refers to the designed modification of genomic sequence(s) through homologous recombination (HR). GT is a powerful tool both for the study of gene function and for molecular breeding. However, in transformation of higher plants, non-homologous end joining (NHEJ) occurs overwhelmingly in somatic cells, masking HR-mediated GT. Positive–negative selection (PNS) is an approach for finding HR-mediated GT events because it can eliminate NHEJ effectively by expression of a negative-selection marker gene. In rice—a major crop worldwide—reproducible PNS-mediated GT of endogenous genes has now been successfully achieved. The procedure is based on strong PNS using diphtheria toxin A-fragment as a negative marker, and has succeeded in the directed modification of several endogenous rice genes in various ways. In addition to gene knock-outs and knock-ins, a nucleotide substitution in a target gene was also achieved recently. This review presents a summary of the development of the rice PNS system, highlighting its advantages. Different types of gene modification and gene editing aimed at developing new plant breeding technology based on PNS are discussed.
Advantages of Developing a PNS System in Rice
In higher plants, the establishment of GT of endogenous natural genes through HR has been hampered by the overwhelming occurrence of NHEJ, i.e., random recombination, even when the transformed gene carries sequence(s) homologous to the target gene locus. Despite the clear demonstration of GT at an artificially generated selectable locus in tobacco somatic cells (Paszkowski et al., 1988), the frequency of GT was estimated to be 10-3 to 10-6 that of random integration. To overcome the low frequency of HR, various approaches for enhancement of HR and/or reduction of NHEJ have been attempted based on our existing knowledge of genome recombination and repair (Britt and May, 2003). In Arabidopsis, the yeast RAD54 gene—a member of the SWI2/SNF2 chromatin remodeling gene family—enhances GT frequency (Shaked et al., 2005); however, the procedure was still not efficient enough to detect GT of various endogenous genes. Induction of a DSB at the target site using an artificial endonuclease is now progressing as a means of establishing GT in several plant species (Shukla et al., 2009; Zhang et al., 2013; Endo and Toki, 2014; Puchta and Fauser, 2014), although most DSBs re-connected by NHEJ result in target gene disruption.
Positive–negative selection is a strategy for enriching transgenic cells carrying a targeted gene replacing an endogenous gene from among a large number of NHEJ-mediated random recombinants. PNS was first developed for gene knockouts in mice (Mansour et al., 1988). In the higher plant rice (Oryza sativa L.)—an important staple food crop—a reproducible PNS-mediated GT procedure applicable to endogenous genes was developed by Terada et al. (2002). In this latter study, the single copy Waxy locus (Os06g0133000) was targeted for knockout using a PNS vector carrying the hpt gene for positive selection followed by the effective transcriptional stop signal of the maize transposon En/Spm, positioned between the Waxy homologous sequences; two negative selection genes of DT-A (diphtheria toxin A-fragment from Corynebacterium diphtheriae) flanked both ends of the homologous sequence (Figures 1A,B). The DT-A acts as a counter-selection agent against NHEJ-mediated random and non-targeted recombinants, and is itself removed by HR between the target locus and the PNS vector (Figure 1C). DT-A induces ADP-ribosylation of elongation factor 2 in eukaryotic ribosomes and thus prevents protein synthesis (Pappenheimer, 1977; Iida and Terada, 2005). Because DT-A lacks the migration function, the negative selection is cell specific without any effect on neighboring cells (Day and Irish, 1997; Iida and Terada, 2004, 2005). To ensure strong selection against a large number of background recombinants, highly active promoters from the rice Actin1 gene (including its intron), cauliflower mosaic virus (CaMV35S with the caster bean catalase intron), and the maize Ubiquitin gene (also with its intron) were employed to express PNS markers in large-scale T-DNA-mediated rice transformation experiments (Terada et al., 2002, 2004). GT via HR was identified by PCR analysis of calli surviving PNS by detection of targeted-specific sequences reflecting insertion of the hpt-En/Spm into the Waxy locus (Figure 1D). Most survivors of PNS were derived from the random integration of the GT vector in which the DT-A genes have become non-functional due to rearrangements of the sequences (Terada et al., 2007). The GT frequency was calculated as 6.4 × 10-4 based on total transformants (six targeted lines per 9,300 calli), which lies within the range of 10-3 to 10-6 predicted in earlier GT experiments with an artificially generated selectable target gene locus (Paszkowski et al., 1988). We generally use the percentage of targeted lines obtained per number of surviving calli on PNS to define the efficiency of GT, in our case 0.94 % (six targeted lines per 638 calli). The heterozygosity of the Waxy locus in targeted T0 plants was confirmed by Southern blot and DNA sequence analysis at the waxy locus and by the Mendelian segregation of the Waxy-waxy phenotype in T1 plants (Terada et al., 2002).
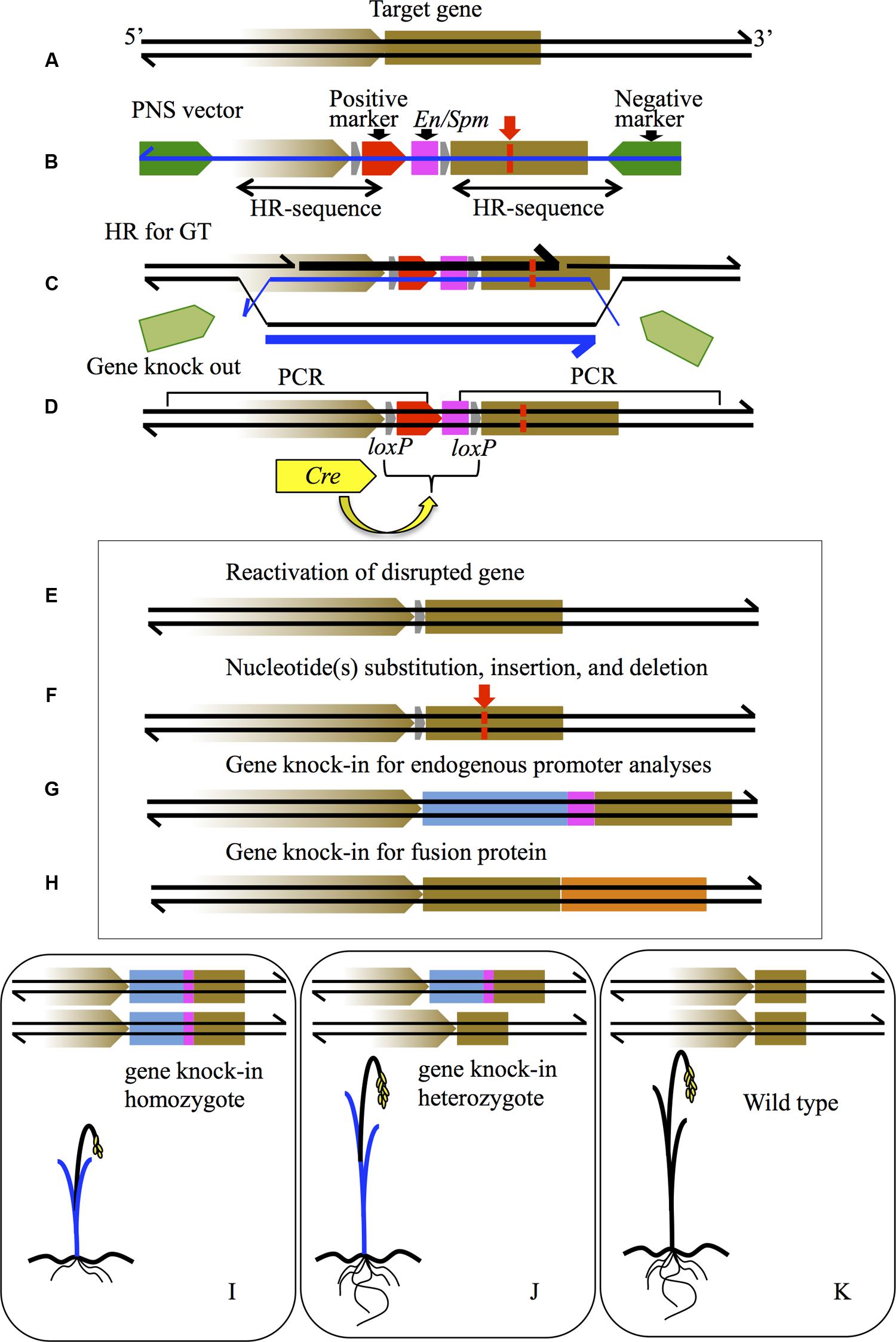
FIGURE 1. Schematic diagram of various gene modifications by PNS-mediated GT. (A) The brown box indicates the gene to be targeted on a genome sequence shown as black lines. The brown arrow represents the promoter of the gene. (B) PNS vector for GT. The green arrows are the negative markers; the red arrow is the positive marker. The pink box is the transcriptional stop sequence of En/Spm. The gray arrows are loxP sequences. Double-headed arrows under the vector indicate the homology regions for HR. The blue line is T-DNA sequence. (C) HR process for GT between the target gene and PNS vector. The thick lines of black and blue indicate newly synthesized DNA sequences in genome and T-DNA, respectively. (D) Gene knock-out of the target gene by insertion of a positive marker with En/Spm, which can be removed via subsequent Cre-loxP recombination caused by introduced Cre gene (yellow arrow). (E) Reactivation of knock-out gene in (D) by Cre-loxP recombination. (F) Nucleotide(s) substitution (red lines), insertion, and deletion in the target gene can be induced by designing a homology arm in the PNS vector in (B) and subsequent positive marker elimination by Cre-loxP recombination in (D). (G) Gene knock-in modification where the endogenous promoter sequence is connected to the GUS coding sequence (indicated as a blue box with En/Spm). (H) Gene knock-in modification where the mOrange coding sequence, indicated as an orange box, is connected precisely to the stop codon of the target gene; both endogenous promoter activity and protein localization of the target gene are detectable. (I–K) Diagrams of segregated plants from knock-in T0 into homozygote (I), heterozygote (J), and wild type (K). GUS expression image as blue leaves is shown in (I,J). Dwarf phenotype in (I) is a reflection of the disrupted target gene.
Because PNS-mediated GT occurs via HR between homologous sequences present on both the vector and a corresponding sequence at the targeted locus (Figures 1B–D), the procedure could be used to introduce desired mutations of various types into any gene of interest (Figures 1E–H). After the first successful GT of the Waxy locus (Terada et al., 2002), many endogenous rice genes (more than 10 loci) have been targeted and altered to desired forms (Terada et al., 2007; Yamauchi et al., 2009, 2014; Moritoh et al., 2012; Ono et al., 2012; Ozawa et al., 2012; Dang et al., 2013; Osakabe et al., 2014). At the early stage of PNS development for higher plants, the codA gene was employed for negative selection rather than DT-A because the toxic effect of DT-A was not only non-conditional but also very strong, so that even transient expression of DT-A would kill any cell receiving the PNS vector. On the other hand, the toxic effect of DT-A expressed transiently can be suppressed by the following T-DNA-mediated transformation process, compared with results obtained in direct transformation methods delivering a double-stranded DNA vector in GT experiments in mice and yeast. The applicability of T-DNA for HR-mediated GT has been confirmed for an artificially generated selectable target locus in tobacco (Offringa and Hooykaas, 1995). T-DNA-mediated transformation of the monocot plant rice—a non-host of Agrobacterium—was developed by Hiei et al. (1994). A single-stranded T-DNA carrying the GT vector for PNS is transformed into the plant nucleus, where it is then thought to be converted into double-stranded DNA (dsDNA), and integrated into the host genome (Loyter et al., 2005). Transient expression of DT-A could be delayed due to the time required for ssDNA to dsDNA conversion of T-DNA.
The codA gene, which encodes cytosine deaminase [catalyzes conversion of 5-fluorocytosine (5-FC) to the toxic 5-fluorouracil (5-FU)], was used as a conditional negative marker for establishment of PNS-mediated GT in Lotus japonicas and Arabidopsis thaliana; however, codA was found to be insufficient for negative selection of GT events (Thykjaer et al., 1997; Gallego et al., 1999; Wang et al., 2001; Iida and Terada, 2005). The codA gene can be improved by introducing a single amino acid substitution: D314A (Mahan et al., 2004), and negative selection using this modified codA (D314A) was recently found to be functionally comparable to that using DT-A (Osakabe et al., 2014). The rice CAOMT (caffeic acid O-methyltransferase) gene was targeted successfully by PNS using modified codA (D314A). Development of suitable negative selection markers is important to improve PNS-mediated GT and to make it more publicly acceptable, especially as a procedure for molecular breeding. The embryonic rice calli used for PNS-mediated GT, which maintain totipotency for regeneration, are postulated to be as HR-reactive as mouse embryonic stem cells (where HR is common, occurring with a frequency of more than 10-2 among transformation events; Jasin et al., 1996). Such calli consist of small, compact, and vigorously proliferating cells that have the additional advantage of being easy to handle for large scale-transformation (Terada et al., 2004).
Applications of PNS and the Variety of Possible Gene Modifications
As shown in Figures 1D–H, genome sequences can be modified to various forms by PNS-mediated GT, i.e., not only gene knockouts but also gene knock-ins have been established, as well as nucleotide insertions, deletions, and substitutions. In addition to the Waxy knockout, the Alcohol dehydrogenase2 (Adh2) gene, Os11g0210500 (Terada et al., 2007) and Adh1 (Os11g0210300) on chromosome 11 were targeted independently by the same PNS strategy, despite both genes being surrounded by redundant sequences of repetitive Copia-like and Gypsy-like retroelements (Tarchini et al., 2000; Iida and Terada, 2005). Recently, it was shown that disruption of the single copy rice gene Xyl, encoding β1,2-gxylosyl-transferase, resulted in the absence of xylose residues in targeted homozygotes (Ozawa et al., 2012). To date, more than ten gene loci distributed in different positions on rice chromosomes have been targeted and altered to different forms. GT efficiency ranges from about 1.0–10 % among PNS survivors, and is assumed to depend on characteristics of the DNA sequence required for HR, such as sequence repeat(s) and palindromic elements, as well as other genomic processes such as DNA replication and/or transcription, and epigenetic modifications of DNA and chromosome(s).
In general, gene promoter activities can be studied by analyzing transgenic plants carrying chimeric genes with the promoter of interest fused to the coding sequence of a visual marker such as GUS or GFP, although expression of visual markers can be unstable depending on positional effects and multicopy integrations of the chimeric gene (Yamauchi et al., 2009). In addition, promoters in chimeric genes do not always reflect their original functionality because of the length limitation of promoter regions that can be applied for gene transformation. In a knock-in GT experiment, the GUS coding sequence attached to hpt-En/Spm was connected to the promoter of the target gene (Figure 1G; Yamauchi et al., 2009, 2014). Because almost all PNS-mediated GT in rice occurs in a heterozygous manner without any additional insertion of the GT vector (Terada et al., 2002, 2007), endogenous promoter activity in the original gene locus can be detected in the targeted heterozygote (Figure 1J). Simultaneously, the phenotypic alteration derived from the targeted gene function in addition to endogenous promoter activity is detectable in the targeted homozygote (Figure 1I) when compared to the segregated wild type homozygote as a control plant (Figure 1K).
Genes functioning in genome methylation were studied by knock-in (plus simultaneous knock-out) GT experiments. The genes for maintenance of CG methylation, methyltransferase OsMet1a, Os03g0798300 (Yamauchi et al., 2009) and OsMet1b, Os07g0182900 (Yamauchi et al., 2014) were selected based on DNA sequence characteristics and encoded protein motifs, and then targeted precisely. A functional ATG in the target gene was detected by 5′ RACE analyzes and adjusted to become the initiation codon of GUS in the knock-in vector. Strong GUS expression was detected in tissues with active cell division, such as meristems in shoot and root, in addition to callus tissue in knock-in plants of OsMet1a and OsMet1b, respectively. Due to the knock-in of this single locus, dose-dependent GUS expression reflecting targeted-homozygote and -heterozygote was detected clearly in a OsMet1a knock-in mutant (Yamauchi et al., 2009). In addition, promoter activities of OsMet1a and OsMet1b were detected as GUS expression in shoot and root in knock-in hetero- or homozygotes. The original promoter activity of OsMet1a and OsMet1b was precisely compared through GUS expression in shoots of GT- derived heterozygotes (Yamauchi et al., 2014).
Domains rearranged methylase 2, OsDRM2, Os03g0110800 (Moritoh et al., 2012), encoding both de novo and non-CG methyltransferase, and Repressor of Silencing, OsROS1a, Os01g11900 encoding DNA demethylase (Ono et al., 2012) were altered by a knock-in approach by PNS-mediated GT. Whereas no morphological phenotype was detected in Arabidopsis drm1 drm2 mutants, in rice, the OsDRM2 knock-in homozygote exhibited drastic growth delay, dwarfism, and sterility, indicating the unique function of OsDRM2 (Moritoh et al., 2012). Osros1a-GUS was detected in pollen and unfertilized ovules; concomitantly, an arrest of endosperm growth was observed in heterozygous knock-in rice (Ono et al., 2012). All these results show that the PNS-mediated GT procedure is able to generate novel mutant rice plants based on information gleaned from DNA sequence(s) and encoded protein motif(s), and that the functions of both endogenous promoters and genes can be studied effectively with plants segregated for the targeted locus.
Recently, visual markers such as GFP, mOrange, and AsRed2 have been developed, expression of which is detectable in living plant tissues without chemical treatment(s). Connecting sequences encoding these visual markers to the 3′-end of the target gene results in expression of chimeric fusion proteins that allow the spatiotemporal localization of the protein of interest to be visualized (Figure 1H).
Positive-Marker Free Gene Editing by PNS-Mediated GT Induced by Site-Specific Recombination
Positive–negative selection-mediated GT can be used to introduce nucleotide substitution(s) at a targeted locus. Indeed, several nucleotides of the Adh2 locus have been substituted successfully by a modified GT vector through HR (Johzuka-Hisatomi et al., 2008; Figure 1F). Furthermore, because the positive marker of the hpt-En/Spm is placed between the two loxPs in the same orientation, the positive marker can be removed by Cre-loxP-mediated site-specific recombination after GT (Figures 1D–F). In Waxy GT, the hpt-En/Spm between two loxPs was indeed eliminated by transient expression of the Cre gene, which was transformed into calli derived from the targeted-waxy homozygote, resulting in Waxy reactivation in pollen (Figure 1E; Terada et al., 2010). These results indicate that nucleotide(s) in rice genome sequence can be substituted precisely by PNS-mediated GT followed by Cre-loxP recombination to excise the positive marker.
The OsRac1 gene (Os01g0229400) was edited by introducing a single nucleotide substitution of G56T, which results in a constitutively active enzyme, by GT-mediated single nucleotide substitution and subsequent positive marker elimination. OsRac1 belongs to the Rac/Rop small GTPase family and acts as a molecular switch in rice immunity. The substitution of guanine (G) with thymine (T) at the 56th nucleotide in exon1 of OsRac1 alters the 19th glycine (G) to valine (V). The mutated OsRac1(G19V) is constitutively active and increases resistance to rice blast fungus (Magnaporthe oryzae) when expressed from the CaMV35S promoter, although rice fertility was seriously reduced (Ono et al., 2001). To see whether the mutated OsRac1(G19V) driven by endogenous promoter in the original locus would generate blast-fungus-resistant rice, the G56T nucleotide substitution was introduced in OsRac1 through PNS-mediated GT and subsequent removal of the positive marker by Cre-loxP recombination (Dang et al., 2013). In the first step, OsRac1-GT occurred at a high frequency of 5.3% among the 94 calli surviving PNS; all five callus lines obtained carried the G56T substitution. Then, in the second step, β-estradiol-inducible Cre was transformed into each targeted callus line, and plants were regenerated from calli after induction of Cre expression. In total, seven fertile, hpt-free, rice plants with the G56T substitution in OsRac1 were obtained from a single GT line. All plants expressed OsRac1(G19V) in the leaves with a blast fungus resistance phenotype; however, the level of the OsRac1(G19V) expression was unexpectedly low and the mutation was associated with a dwarf phenotype.
Excision of the selectable marker gene via the Cre-loxP or Flp-FRT system leaves recognition sequences for Cre and Flp recombinases—loxP and FRT sites, respectively—at the excised sites. For GT applications in the field, it is considered preferable to use marker excision systems that do not leave such sequences, to be more equivalent to spontaneous mutagenesis. The piggyBac transposon derived from the lepidopteran cabbage looper moth integrates into the host genome at TTAA elements and excises without leaving a footprint at the excised site (Cary et al., 1989). Using an assay system that allows transposition of piggyBac transposon to be visualized as luminescence derived from reconstituted luciferase expression cassettes, we demonstrated that the piggyBac transposon is capable of accurate and effective transposase-mediated transposition in plant cells (Nishizawa-Yokoi et al., 2014a).
To generate marker-free plants harboring only the desired mutation in the target locus, we attempted to introduce two point mutations accompanied by two amino acid changes—tryptophan (TGG) to leucine (TTG) at amino acid 548 (W548L), and serine (AGT) to isoleucine (ATT) at amino acid 627 (S627I)—into the rice acetolactate synthase (ALS) gene via PNS-mediated GT and subsequent marker excision by piggyBac transposition (Figure 2). Mutation of W548L/S627I in the ALS gene confers increased tolerance to the herbicide bispyribac (BS) on rice plants (Endo et al., 2007). Four-week-old rice calli were infected with Agrobacterium harboring a GT vector containing hpt and DT-A genes as PNS markers (Figure 2A). Four independent GT callus lines were identified by PCR analysis with the primer sets shown in Figure 2B and, among them, two lines were inoculated with Agrobacterium harboring a hyperactive piggyBac transposase (hyPBase, Yusa et al., 2011) expression vector driven by a constitutive promoter. Plants regenerated from hyPBase-expressing GT calli were subjected to marker excision analysis by cleaved amplified polymorphic sequence (CAPS) analysis. More than 90% of regenerated plants contained two point mutations in the ALS gene and lacked the piggyBac transposon carrying the hpt gene, suggesting that these regenerated plants indeed represented marker-free rice plants containing the desired mutations at the target locus (Nishizawa-Yokoi et al., 2014b). Our approach, i.e., GT with PNS and subsequent marker excision, provides a general strategy for targeted modification of endogenous genes in plants.
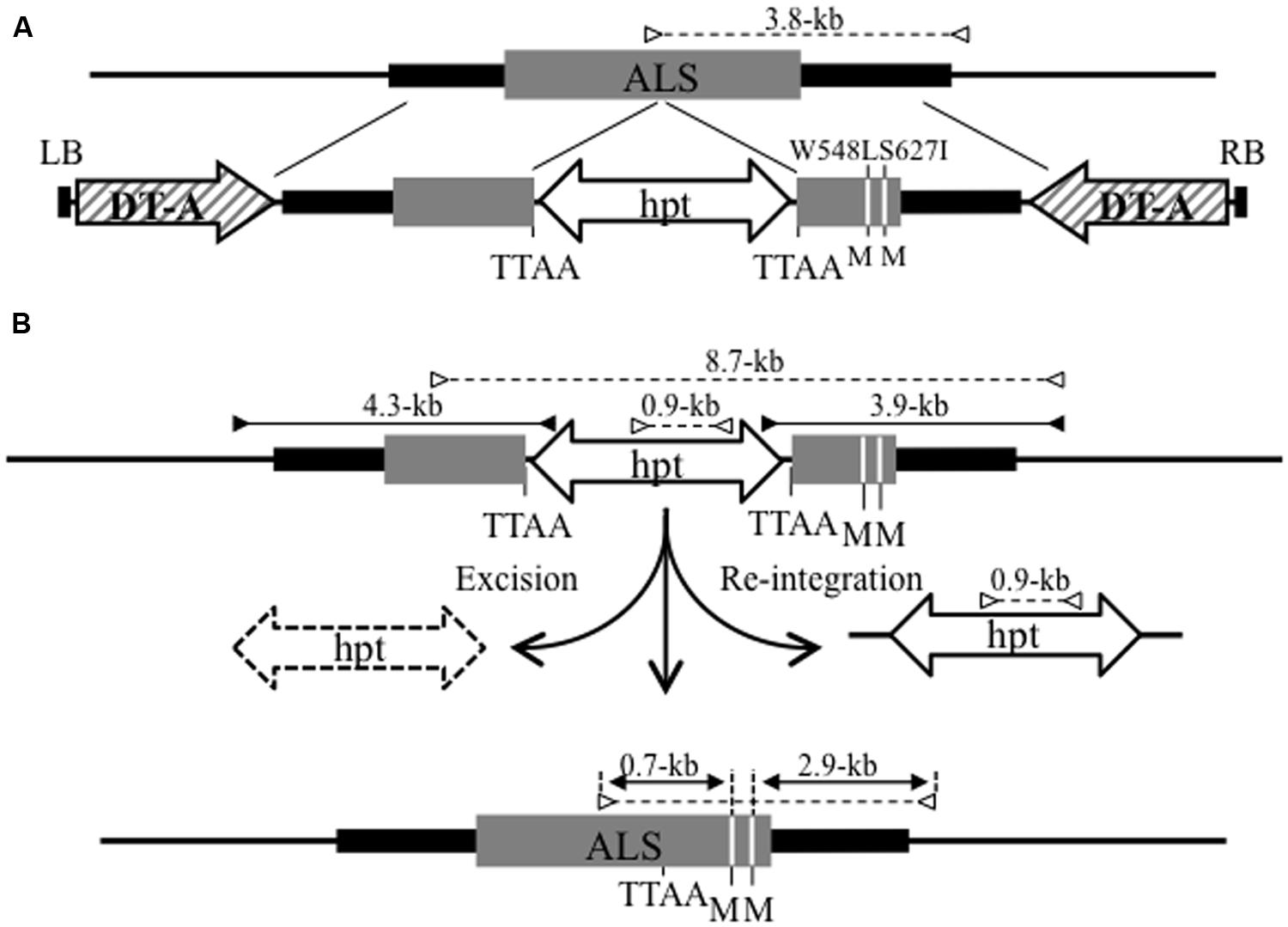
FIGURE 2. Strategy for the introduction of point mutations into the ALS locus via GT and subsequent marker excision from the GT locus using the piggyBac transposon. (A) Schematic diagram of GT at the ALS locus. The top line indicates the genomic structure of the wild-type ALS gene region. The bottom line shows the T-DNA region of the GT vector carrying DT-A as a negative selection marker and a 6.4-kb fragment containing an ALS coding region (gray box) with W548L and S627I mutations (white lines) and silent mutations (TTAA site added 301-bp upstream of W548L; GCTGAC to GAATTC) for the insertion of piggyBac transposon harboring hpt gene as a positive selection marker. The W548 L and S627I mutations create novel Mfe I restriction sites (M). LB, left border; RB, right border. (B) Strategy for precise marker excision from the GT locus using piggyBac transposon. The top line reveals the structure of the modified ALS locus resulting from HR between the GT vector and wild-type locus. The bottom line represents the ALS locus modified by GT and subsequent precise marker excision via piggyBac transposition. The primer sets used for PCR that identify transgenic calli in which a GT event occurred at the ALS locus are shown as black arrows. White arrows indicate the primer sets used for CAPS analysis to evaluate the frequency of marker excision via piggyBac transposition. The numbers on each arrow reveal the length of the PCR fragments.
In a related genome editing strategy, DSBs were induced in the target gene using the zinc-finger nuclease (Shukla et al., 2009), TALENs (transcription activator-like effectors nuclease) from Xanthomonas, and CRISPR (clustered regularly interspaced short palindromic repeats)-associated (Cas9) systems in Arabidopsis, tobacco, maize, and rice (Zhang et al., 2013). DSBs are expected to enhance HR; indeed, effective HR induction was detected in an artificially targeted site (Puchta, 1999); however, for endogenous genes, most DSBs are repaired immediately by NHEJ and become associated with nucleotide deletions, substitutions, and insertions, resulting in gene-disruption-mediated mutants that could be screened for plant improvements (Shukla et al., 2009; Zhang et al., 2013; Puchta and Fauser, 2014). Precise nucleotide sequence design of a target gene by HR is still difficult even using induced DSB at a known target locus. Although PNS-mediated GT does not enhance HR, it can be combined with DSB induction in various plants with agricultural value, as well as in rice, in the search for new plant breeding technologies (NPBT).
Outlook
Positive–negative selection-mediated GT, which retains the unique competence for T-DNA mediated HR, has been developed in rice. In addition to gene knock-out, visualization of endogenous gene expression has been detected by gene knock-in. Further precise connection of a visual marker to the gene of interest will provide novel information about behavior of the protein in developing rice plants. Precise modification of target genes will be applicable to detailed functional analysis as well as rice breeding. Combination of PNS-mediated GT and genome editing strategy is expected to expand the availability of GT procedure and its application to various plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. Shimamoto for great support for our research; S. Moritoh, H. Asao, M. Shimamoto, M. Matsumoto, and C. Namba for excellent technical assistance; S. Ignacimuthu for manuscript preparation; M. Nishimura for generous support. This work was supported by The Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project grant); The Ministry of Education, Culture Sports, Science and Technology, “Mext” Grant-in-Aid for Science Research; Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
Abbreviations
DSBs, double strand break; DT-A, diphtheria toxin A-fragment; GT, gene targeting; hpt, hygromycin phosphotransferase; HR, homologous recombination; NHEJ, non-homologous end joining; PCR, polymerase chain reaction; PNS, positive–negative selection; T-DNA, transfer DNA.
References
Britt, A. B., and May, G. D. (2003). Re-engineering plant gene targeting. Trends Plant Sci. 8, 90–95. doi: 10.1016/S1360-1385(03)00002-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cary, L. C., Goebel, M., Corsaro, B. G., Wang, H. G., Rosen, E., and Fraser, M. J. (1989). Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172, 156–169. doi: 10.1016/0042-6822(89)90117-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dang, T. T., Shimatani, Z., Kawano, Y., Terada, R., and Shimamoto, K. (2013). Gene editing; a constitutively active OsRac1 by homologous recombination based gene targeting induces immune responses in rice. Plant Cell Physiol. 54, 2058–2070. doi: 10.1093/pcp/pct147
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Day, C. D., and Irish, V. F. (1997). Cell ablation and the analysis of plant development. Trends Plant Sci. 2, 106–111. doi: 10.1016/S1360-1385(97)01004-2
Endo, M., Osakabe, K., Ono, K., Handa, H., Shimizu, T., and Toki, S. (2007). Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J. 52, 157–166. doi: 10.1111/j.1365-313X.2007.03230.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Endo, M., and Toki, S. (2014). Toward establishing an efficient and versatile gene targeting system in higher plants. Biocatal. Agric. Biotechnol. 3, 2–6. doi: 10.1016/j.bcab.2013.10.002
Gallego, M. E., Sirand-Pugnet, P., and White, C. I. (1999). Positive-negative selection and T-DNA stability in Arabidopsis transformation. Plant Mol. Biol. 39, 83–93. doi: 10.1023/A:1006192225464
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iida, S., and Terada, R. (2004). A tale of two integrations, transgene and T-DNA:gene targeting by homologous recombination in rice. Curr. Opin. Biotechnol. 15, 132–138. doi: 10.1016/j.copbio.2004.02.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iida, S., and Terada, R. (2005). Modification of endogenous natural genes by gene targeting in rice and other higher plants. Plant Mol. Biol. 59, 205–219. doi: 10.1007/s11103-005-2162-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jasin, M., Moynahan, M. E., and Richardson, C. (1996). Targeted transgenesis. Proc. Natl. Acad. Sci. U.S.A. 93, 8804–8808. doi: 10.1073/pnas.93.17.8804
Johzuka-Hisatomi, Y., Terada, R., and Iida, S. (2008). Efficient transfer of base changes from a vector to the rice genome by homologous recombination: involvement of heteroduplex formation and mismatch correction. Nucleic Acids Res. 36, 4727–4735. doi: 10.1093/nar/gkn451
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loyter, A., Rosenbluh, J., Zakai, N., Li, J., Kozlovsky, S. V., Tzfira, T.,et al. (2005). The plant VirE2 interacting protein 1. A molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiol. 138, 1318–1321. doi: 10.1104/pp.105.062547
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mahan, S. D., Ireton, G. C., Stoddard, B. L., and Black, M. E. (2004). Alanine-scanning mutagenesis reveals a cytosine deaminase mutant with alteredsubstrate preference. Biochemistry 43, 8957–8964. doi: 10.1021/bi049720z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mansour, S. L., Thomas, K. R., and Capecchi, M. R. (1988). Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336, 348–352. doi: 10.1038/336348a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moritoh, S., Eun, C. H., Ono, A., Asao, H., Okano, Y., Yamaguchi, K.,et al. (2012). Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 71, 85–98. doi: 10.1111/j.1365-313X.2012.04974.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa-Yokoi, A., Endo, M., Osakabe, K., Saika, H., and Toki, S. (2014a). Precise marker excision system using an animal-derived piggyBac transposon in plants. Plant J. 77, 454–463. doi: 10.1111/tpj.12367
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa-Yokoi, A., Endo, M., Ohtsuki, N., Saika, H., and Toki, S. (2014b). Precision genome editing in plants via gene targeting and piggyBac-mediated marker excision. Plant J. doi: 10.1111/tpj.12693 [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Offringa, R., and Hooykaas, P. (1995) “Gene targeting in plants,” in Gene Targeting, ed. M. A. Vega (Boca Raton, FL: CRC Press), 84–121.
Ono, A., Yamaguchi, K., Fukada-Tanaka, S., Terada, R., Mitsui, T., and Iida, S. (2012). A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 71, 564–574. doi: 10.1111/j.1365-313X.2012.05009.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ono, E., Wong, H. L., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Natl. Acad. Sci. U.S.A. 98, 759–764. doi: 10.1073/pnas.98.2.759
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Osakabe, K., Nishizawa-Yokoi, A., Ohtsuki, N., Osakabe, Y., and Toki, S. (2014). A mutated cytosine deaminase gene, codA (d314a), as an efficient negative selection marker for gene targeting in rice. Plant Cell Physiol. 55, 658–665. doi: 10.1093/pcp/pct183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ozawa, K., Wakasa, Y., Ogo, Y., Matsuo, K., Kawahigashi, H., and Takaiwa, F. (2012). Development of an efficient Agrobacterium-mediated gene targeting system for rice and analysis of rice knockouts laking Granule-bound starch synthase (waxy) and β1,2-Xylosyltransferase. Plant Cell Physiol. 53, 755–761. doi: 10.1093/pcp/pcs016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pappenheimer, A. M. (1977). Diphtheria toxin. Annu. Rev. Biochem. 46, 69–94. doi: 10.1146/annurev.bi.46.070177.000441
Paszkowski, J., Baur, M., Bogucki, A., and Potrykus, I. (1988). Gene targeting in plants. EMBO J. 7, 4021–4026. doi: 10.1387/ijdb.130194hp
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Puchta, H. (1999). Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152, 1173–1181.
Puchta, H., and Fauser, F. (2014). Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 78, 727–741. doi: 10.1111/tpj.12338
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shaked, H., Melamed-Bessudo, C., and Levy, A. A. (2005). High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc. Natl. Acad. Sci. U.S.A. 102, 12265–12269. doi: 10.1073/pnas.0502601102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shukla, V. K., Doyon, Y., Miller, J. C., DeKelver, R. C., Moehle, E. A., Worden, S. E.,et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459, 437–441. doi: 10.1038/nature07992
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. (2000). The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted co linearity with maize chromosome 4. Plant Cell 12, 381–391. doi: 10.1105/tpc.12.3.381
Terada, R., Asao, H., and Iida, S. (2004). A large-scale Agrobacterium-mediated transformation procedure with a strong positive-negative selection for gene targeting in rice (Oryza sativa L.). Plant Cell Rep. 22, 653–659. doi: 10.1007/s00299-003-0752-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Terada, R., Johzuka-Hisatomi, Y., Saitoh, M., Asao, H., and Iida, S. (2007) Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 144, 846–856. doi: 10.1104/pp.107.095992
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Terada, R., Nagahara, M., Furukawa, K., Shimamoto, M., Yamaguchi, K., and Iida, S. (2010). Cre-lox mediated marker elimination and gene reactivation at waxy locus created in the rice genome based on strong positive-negative selection. Plant Biotechnol. 27, 29–37. doi: 10.5511/plantbiotechnology.27.29
Terada, R., Urawa, H., Inagaki, Y., Tsugane, K., and Iida, S. (2002). Efficient gene targeting by homologous recombination in rice. Nat. Biotechnol. 20, 1030–1034. doi: 10.1038/nbt737
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thykjaer, T., Finnemann, J., Schauser, L., Christensen, L., Poulsen, C., and Stougaard, J. (1997). Gene targeting approaches using positive- negative selection and large flanking regions. Plant Mol. Biol. 35, 523–530. doi: 10.1023/A:1005865600319
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H. X., Viret, J.-F., Eldridge, A., Perera, R., Signer, E. R., and Chiurazzi, M. (2001). Positive–Negative selection for homologous recombination in Arabidopsis. Gene 272, 249–255. doi: 10.1016/S0378-1119(01)00532-7
Yamauchi, T., Johzuka-Hisatomi, Y., Fukada-Tanaka, S., Terada, R., Nakamura, I., and Iida, S. (2009). Homologous recombination-mediated knock-in targeting of the MET1a gene for a maintenance DNA methyltransferase reproducibly reveals dosage-dependent spatiotemporal gene expression in rice. Plant J. 60, 386–396. doi: 10.1111/j.1365-313X.2009.03947.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamauchi, T., Johzuka-Hisatomi, Y., Terada, R., Nakamura, I., and Iida, S. (2014). The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol. Biol. 85, 219–232. doi: 10.1007/s11103-014-0178-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yusa, K., Zhou, L., Li, M., Bradley, A., and Craig, N. (2011). A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 108, 1531–1536. doi: 10.1073/pnas.1008322108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z.,et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 687–688. doi: 10.1038/nbt.2650
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: gene targeting, homologous recombination, positive–negative selection, rice, knock-in, marker-free, site specific recombination, gene editing
Citation: Shimatani Z, Nishizawa-Yokoi A, Endo M, Toki S and Terada R (2015) Positive–negative-selection-mediated gene targeting in rice. Front. Plant Sci. 5:748. doi: 10.3389/fpls.2014.00748
Received: 31 August 2014; Accepted: 08 December 2014;
Published online: 05 January 2015.
Edited by:
Toshihiko Komari, Japan Tobacco Inc., JapanReviewed by:
Barbara Hohn, Friedrich Miescher Institute for Biomedical Research, SwitzerlandHolger Puchta, Karlsruhe Institute of Technology, Germany
Copyright © 2015 Shimatani, Nishizawa-Yokoi, Endo, Toki and Terada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rie Terada, Development of Agrobiological Resources, Faculty of Agriculture, Meijo University, 1-501 Shiogamaguchi, Tempaku-ku, Nagoya 468-8502, Aichi, Japan e-mail: teradar@meijo-u.ac.jp
 Zenpei Shimatani
Zenpei Shimatani Ayako Nishizawa-Yokoi
Ayako Nishizawa-Yokoi Masaki Endo
Masaki Endo Seiichi Toki
Seiichi Toki Rie Terada
Rie Terada