- Department of Life Sciences, University of Parma, Parma, Italy
Hyperaccumulators are plants that can extract heavy metal ions from the soil and translocate those ions to the shoots, where they are sequestered and detoxified. Hyperaccumulation depends not only on the availability of mobilized metal ions in the soil, but also on the enhanced activity of metal transporters and metal chelators which may be provided by the plant or its associated microbes. The rhizobiome is captured by plant root exudates from the complex microbial community in the soil, and may colonize the root surface or infiltrate the root cortex. This community can increase the root surface area by inducing hairy root proliferation. It may also increase the solubility of metals in the rhizosphere and promote the uptake of soluble metals by the plant. The bacterial rhizobiome, a subset of specialized microorganisms that colonize the plant rhizosphere and endosphere, makes an important contribution to the hyperaccumulator phenotype. In this review, we discuss classic and more recent tools that are used to study the interactions between hyperaccumulators and the bacterial rhizobiome, and consider future perspectives based on the use of omics analysis and microscopy to study plant metabolism in the context of metal accumulation. Recent data suggest that metal-resistant bacteria isolated from the hyperaccumulator rhizosphere and endosphere could be useful in applications such as phytoextraction and phytoremediation, although more research is required to determine whether such properties can be transferred successfully to non-accumulator species.
Hyperaccumulators and the rhizobiome
Hyperaccumulators are plants that accumulate metals and/or metalloids in their leaves at concentrations several orders of magnitude higher than the levels tolerated by other species. The hyperaccumulator phenotype has evolved in environments where restrictive growth conditions allow the adaptation of only a few plant species (Baker et al., 2000; Pollard et al., 2014). The root environment is a dynamic microsystem in which microbes, roots, and the soil interact, and the roots can gain access to soil nutrients and metals (Alford et al., 2010). Root system development, root morphology and chemotropism are all recognized as equally important for establishing the hyperaccumulator phenotype, although the mechanisms involved are still not fully understood (Moradi et al., 2009). Bacteria and fungi colonizing the rhizosphere (immediately surrounding the root) and the endosphere (compartments within the root) play an important role in the establishment of interactions between hyperaccumulators and the soil. These microbes tend to be metal tolerant and can promote plant growth in contaminated soils by several mechanisms: inducing the formation of hairy roots (thus increasing the root surface area), enhancing the solubility and uptake of metal ions and producing phytormons and metabolites (Rajkumar et al., 2012). The beneficial effects of metal-tolerant microbes have attracted attention because of their biotechnological applications in plant-based remediation strategies (Salt et al., 1995). The characterization of the hyperaccumulator rhizobiome is therefore needed to facilitate such applications (Mastretta et al., 2006; Rajkumar et al., 2009; Ma et al., 2011a; Sessitsch et al., 2013). In this review, we discuss classical and more recent omics-based methods that are used, and can be used, to study the interaction between hyperaccumulators and the bacterial rhizobiome, combined with advanced microscopy techniques for the visualization of microbe–host systems, emphasizing the potential applications of these microorganisms in phytotechnology.
Culture-Dependent vs. Culture-Independent Methods
The structure and diversity of microbial communities in the rhizosphere and endosphere of several plants (the rhizobiome) has been analyzed in detail at the molecular level in order to characterize the interaction between microbes and plants (Sørensen et al., 2009). However, less than 10% of hyperaccumulator species have been investigated (Alford et al., 2010). The principal approach to study bacteria in the hyperaccumulator rhizobiome was mainly based on traditional culture-dependent techniques (Table 1). These comprise fractionation protocols, in which roots are shaken in high ionic solutions to remove bacteria from soil particles and the rhizobacteria are collected by washing. Roots surfaces are then sterilized and sonicated to remove the epidermal cells and macerated for the extraction of endophytes. Serial dilutions are then prepared from the wash fraction (rhizobacteria) and the crushed roots (endophytes) and these are inoculated on rich agar medium supplemented with heavy metals. This allows bacterial strains showing high resistance to metals to be selected and tested for the production of metabolites such as siderophores, organic acids and phytohormones, which may be responsible for promoting root growth and solubilization of metals (Figures 1A,B). If the isolated bacterial strains are amenable to laboratory cultivation, they can be used to inoculate hyperaccumulator plants so as to optimize plant growth and metal extraction capacity. Alternatively, they can be used to inoculate higher-biomass non-accumulator plants to determine the transferability of metal tolerance traits, which would make those species more suitable for phytoremediation applications (Table 1).
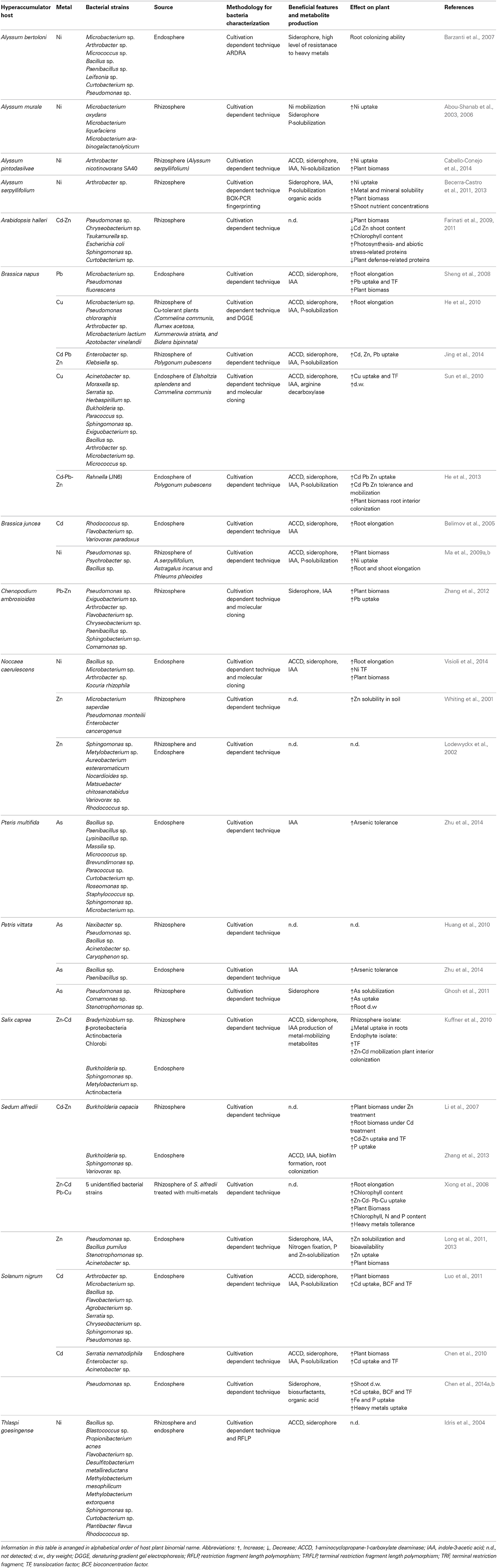
Table 1. Studies resuming recent literature about culture-dependent hyperaccumulators endophytes and rhizobacteria and their effects on plant growth and metal accumulation.
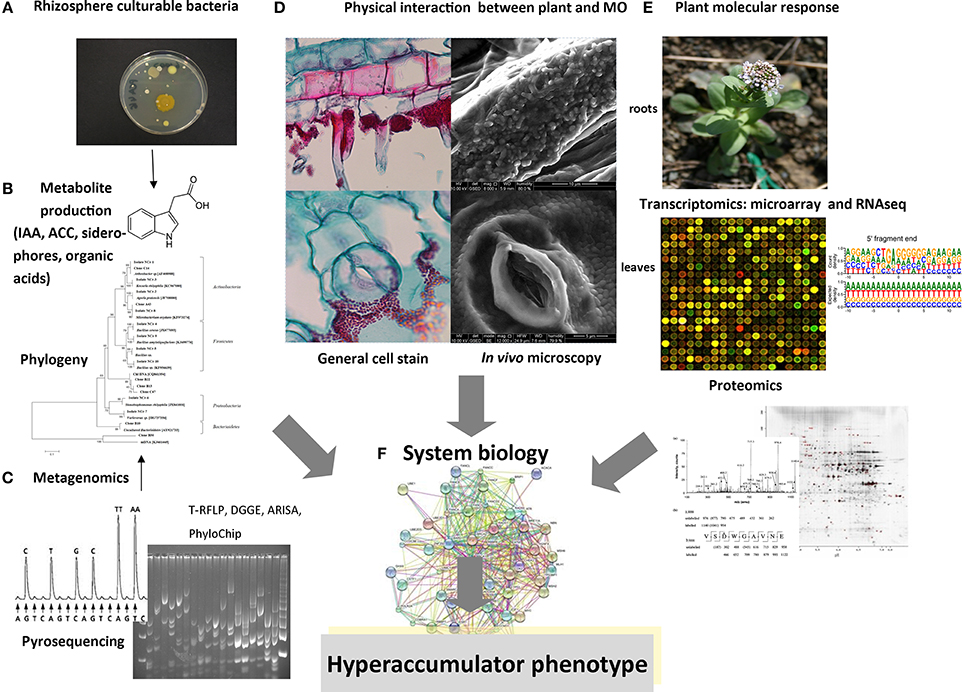
Figure 1. Schematic representation of multidisciplinary approaches for the analysis of rhizobiome–hyperaccumulator interactions. (A) Bacteria in the rhizosphere and endosphere that are amenable to laboratory cultivation can be analyzed for metal resistance and (B) for the production of metabolites. (C) Metagenomics involves the extraction of DNA from roots and rhizosphere compartments, followed by techniques such as 16S rDNA amplification, cloning, and sequencing, fingerprinting (RFLP analysis, DGGE, ARISA, PhyloChip), or direct next-generation sequencing. (D) Physical interactions between bacteria and plants can be visualized using advanced optical and electron microscopy methods following the inoculation of seeds or roots with bacteria. (E) Microarray and RNA-Seq technologies can be used to analyze bacterial gene expression following exposure to root exudates, or to compare root/shoot gene expression between plants cultivated in the presence and absence of bacteria. Proteomic analysis can also identify plant proteins that are modulated by the rhizobiome and characterize the roles of bacteria that promote the hyperaccumulator phenotype. (F) The goal of future studies will be to collect and correlate data from all these methods in a system biology approach to define the molecular basis of metal accumulation in plants.
Thus far, bacteria found to be associated with hyperaccumulators using cultivation-dependent methods mainly comprise the Gram-positive genera Arthrobacter, Microbacterium, Bacillus and Curtobacterium, and the Gram-negative genera Pseudomonas, Sphingomonas, and Variovorax (Table 1). These genera have been found in both the rhizosphere and the endosphere of hyperaccumulators regardless the specific metal composition of the soil. Other genera appear to be more prevalent in soils containing specific metals, e.g., Chrysobacterium and Burkholderia tend to be associated with Cd/Zn hyperaccumulators. The data presented in Table 1, summarizing recent literature about endophytes and rhizobacteria of hyperaccumulators, show that the transfer of such bacteria to non-accumulator plants has in many cases boosted the accumulation of plant biomass, root proliferation, metal uptake and metal translocation to aerial parts of the plant, although some species benefit from only a subset of these effects. A complete absence of beneficial effects was observed in only two cases. These results highlight both the general positive impact of the bacteria rhizobiome on plant growth and its specific influence on metal accumulation.
Metal resistance is a common feature of the rhizobiome in metalliferous soils but not among the bacteria inhabiting normal soils. Furthermore, endophytic bacteria from the inner tissues of hyperaccumulators may have adapted to withstand higher metal concentrations than the rhizobacteria (Idris et al., 2004). The presence of endophytic populations in different organs and tissues (e.g., roots, stems, and leaves) may also explain their variable levels of metal tolerance (Barzanti et al., 2007; Long et al., 2011; Ma et al., 2011b; He et al., 2013).
Many bacteria present in the rhizosphere and endosphere cannot be cultivated under laboratory conditions but are nevertheless important mediators of plant–soil interactions. Approximately 99% of microorganisms in the biosphere have never been recovered by standard cultivation techniques and it is necessary to study these species using cultivation-independent approaches. Methods based on direct isolation and analysis of proteins, lipids and nucleic acids from environmental samples have been developed to reveal structure diversity, functions and dynamics of microbial communities without cultivation (Kirk et al., 2004; Rastogi and Sani, 2011) but, so far, only few of these methods have been applied to study bacterial communities associated with hyperaccumulators, which are not suitable for laboratory cultivation.
The extraction of total genomic DNA from the rhizosphere and/or endosphere of hyperaccumulators followed by the preparation of 16S rDNA clone libraries provides sufficient material for an initial survey of microbial diversity and facilitates the identification of novel taxa. This method has been used to study bacteria associated to Solanum nigrum roots and copper and lead tolerant/resistant plants (Sun et al., 2010; Zhang et al., 2012; Chen et al., 2014a,b). The sequencing of clone libraries based on amplified 16S rRNA genes offers the highest phylogenetic resolution but microbial diversity can be underestimated because many clones are required to document the richness present in rhizosphere samples. One additional bottleneck is the co-amplification of 16S rDNA from plant organelles, although this can be overcome using primers specific for bacterial rDNA (Chelius and Triplett, 2001). These primers exclude chloroplast DNA and give a larger mitochondrial PCR product (Idris et al., 2004). Genetic fingerprinting techniques (Figure 1C) have been used as an alternative to clone libraries and these are more sensitive because they are applied directly to the extracted genomic DNA. Denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphism (T-RFLP) analysis and automated ribosomal intergenic spacer analysis (ARISA) have been applied to hyperaccumulators (He et al., 2010; Gupta et al., 2014). Although they do not always allow the immediate taxonomic identification of all species in the community, they can track the dominant members in a complex environment and allow the comparison of bacterial communities in different settings (Rastogi and Sani, 2011).
The PhyloChip16S rDNA microarray could also provide a high-throughput and comprehensive overview of microbial communities in environmental samples, but cross-hybridization is a major limitation and this method cannot detect novel taxa because only sequences represented on the chip are interrogated (Sanguin et al., 2006; Rastogi and Sani, 2011).
More recently, the emergence of next-generation sequencing (NGS) technologies such as the Roche/454, Illumina/Solexa, Life/APG, and HeliScope/Helicos BioSciences platforms (Figure 1C) has revolutionized environmental microbiology and made it possible to resolve complex microbiomes with greater accuracy and associate the diversity of microbial communities with their niche functions (Knief, 2014). Notably, short-read methods such as pyrosequencing have dramatically reduced the time and cost of microbial whole-genome sequencing projects, and have also facilitated the ultra-high-throughput sequencing of hypervariable regions of 16S rRNA genes with 2–3 orders of magnitude greater coverage than Sanger sequencing. Even short hypervariable sequences (100–350 bp) provide sufficient phylogenetic information for taxonomic profiling, so multiple environmental samples can be combined in a single run, and the reads can be parsed using their assigned nucleotide barcode, which is added to the templates by PCR (Metzker, 2010). NGS technologies have been used to study the rhizobiome of Arabidopsis thaliana (Bulgarelli et al., 2012; Lundberg et al., 2012), Populus deltoides (Gottel et al., 2011), Lactuca sativa (Schreiter et al., 2014), and Zea mays (Peiffer et al., 2013) but this approach has not yet been used to characterize the microbial community associated with hyperaccumulators (Knief, 2014). However, NGS has been used to analyze the bacterial community in soils polluted with heavy metals and to determine the impact of heavy metal contamination on the composition of the community (Berg et al., 2012; Gołębiewski et al., 2014). It has also been used to determine the impact of genotype and soil type on the microbiome (Peiffer et al., 2013; Ge et al., 2014; Zachow et al., 2014). Metagenomics therefore appears to be an ideal approach to investigate the diversity and ecology of the hyperaccumulator rhizobiome.
In situ Analysis of Plant–Rhizobiome Interactions
Over the last two decades there have been significant developments in the methods used for the localization and in situ visualization of microbes inside and around plant roots, matching the advances in molecular microbiology described above (Figure 1D). Specific bacterial populations can be detected and localized using fluorescence microscopy or electron microscopy combined with tagging techniques (Sørensen et al., 2009). There have been few studies of the physical plant–microbe interactions around the roots of hyperaccumulators, but the role of specific bacterial strains can be investigated by monitoring the colonization and survival of inoculums under real environmental conditions using in situ microscopy (Figure 1D). In this context, environmental scanning electron microscopy (ESEM) is a powerful system that allows the observation of biological specimens in situ without sample preparation (Stabentheiner et al., 2010). The ESEM specimen chamber operates slightly above the saturation vapor pressure of water. Under such conditions, water remains in the liquid phase and hydrated biological samples can be observed without fixation and dehydration, which is normally required for conventional scanning electron microscopy. As an example, this method has been used to investigate the physical association between the roots and shoots of the Ni hyperaccumulator Noccaea caerulescens and Ni-resistant endophytic bacteria and rhizobacteria of the genera Microbacterium and Arthrobacter (Visioli et al., 2014).
In the model plant species A. thaliana, conventional epifluorescence microscopy and confocal laser scanning microscopy has been used to localize bacteria in the rhizosphere and within plant tissues (Compant et al., 2005; Bulgarelli et al., 2012; Cardinale, 2014). The abundance and composition of the bacterial community can be investigated using strain-specific fluorescent antibodies, fluorescence in situ hybridization (FISH) against rRNA or mRNA targets, and more recent methods such as catalyzed reporter deposition (CARD)-FISH (Pernthaler et al., 2002; Bulgarelli et al., 2012; Lundberg et al., 2012).
In addition, the localization and functional analysis of specific bacteria within plant tissues can be achieved by expressing reporter genes encoding fluorescent marker proteins such as green fluorescent protein (GFP), which can be integrated directly into the bacterial chromosome or into a plasmid that is subsequently introduced into the bacteria (Sørensen et al., 2009). One of the advantages of the reporter gene strategy is that the marker protein can be expressed constitutively or induced by external factors such as the presence/concentration of specific chemicals, including metals (Ramos et al., 2002; Rothballer et al., 2005). Gram-positive Actinobacteria such as the genera Arthrobacter and Microbacterium are present in the hyperaccumulator rhizobiome and have been shown to promote plant growth and metal absorption. Although these bacteria are usually recalcitrant to transformation, a Microbacterium strain was recently transformed with a GFP probe before inoculation onto sugarcane plants to study plant–microbe interactions (Lin et al., 2012). The success of this approach suggests that a similar strategy could be used to investigate microbial interactions with hyperaccumulators.
The Plant Growth-Promoting Capacity of the rhizobiome
As shown in Table 1, the rhizobacteria and endophytes associated with hyperaccumulators often promote the growth of their host plants and increase their capacity for metal accumulation. This could be achieved by the production of siderophores and carboxylic acids, or the solubilization of phosphates to increase the mobility of metals in the rhizosphere (Li et al., 2007; Ma et al., 2009b; Cabello-Conejo et al., 2014), thus enhancing the accumulation of metals by roots and shoots (Sheng et al., 2008; Sun et al., 2010; Luo et al., 2011; He et al., 2013). Plant-associated microbes can also promote growth indirectly by protecting their hosts against pathogens, or directly by producing phytohormones (such as indole acetic acid, abscisic acid and gibberellic acid) or by secreting enzymes such as 1-aminocyclopropane-1-carboxylic acid deaminase which, reducing ethylene levels, allows plant growth and resistance to environmental stresses. Classical culture-based methods are often used to test bacteria for the production of siderophores and other secondary metabolites (Sheng et al., 2008). However, more sensitive approaches based on gas chromatography and mass spectrometry can be used to detect metabolites produced by bacteria in situ without sample preparation, e.g., nanospray desorption ionization (nano-DESI) (Traxler and Kolter, 2012; Watrous et al., 2012). These are highly sensitive techniques that rapidly determine the metabolites present in a sample and they help to identify and quantify new compounds produced by bacteria which can be beneficial for plant growth and metal accumulation in contaminated soils.
Transcriptomics and Proteomics
The composition and genetic capabilities of the hyperaccumulator rhizobiome can be characterized by metagenomics analysis as described above, but this does not reveal the ability of microbes to respond to particular stimuli, and the functions of the majority of microbial species that inhabit intercellular spaces in the root are still poorly understood (Hirsch and Mauchline, 2012). Transcriptomics and proteomics can be useful in this regard because both approaches show how the complementary functions of plants and their associated microbial communities are expressed (Figure 1E). The study of such interactions not only contributes to our understanding of plant–microbe relationships but also facilitates the development of novel strategies to promote phytoremediation.
Microarrays provide a useful platform to analyze the transcriptomes of plants and the microbes that inhabit the rhizosphere, although they can only monitor the expression of genes that are represented on the array (Mark et al., 2005; Fan et al., 2012; Kwak et al., 2012). In contrast, NGS can be used to sequence entire transcriptomes with unparalleled accuracy, resolution and throughput, and with no limitations in terms of sequence coverage. This approach is known as RNA-Seq, and has already been used to study of abiotic stress responses (including exposure to metals) in bacteria isolated from the rhizosphere or contaminated soils, and for comparative studies of gene expression in the roots of hyperaccumulators adapted to grow in different metalliferous soils (Maynaud et al., 2013; Halimaa et al., 2014; López-Leal et al., 2014). This is an ideal approach for the analysis of plant–rhizobiome interactions at the transcriptomic level. RNA-Seq and microarray analysis can also be complemented with comparative proteomics to determine the proteins that are synthesized or modified during such interactions (DalCorso et al., 2013; Visioli and Marmiroli, 2013). For example, comparative proteomics was used to look for proteins expressed by the Cd/Zn accumulator Arabidopsis halleri in the presence or absence of specific Cd-resistant microbes or the entire autochthonous rhizobiome. The presence of the rhizobiome correlated with the accumulation of both Cd and Zn in the shoots, and this involved the upregulation of proteins involved in photosynthesis and the Calvin cycle, whereas defense proteins and antioxidant enzymes were down-regulated (Farinati et al., 2009, 2011). The A. halleri proteome responded differently to the presence of the total rhizobiome compared to selected bacterial strains, indicating that the rhizobiome as a community is required for the most efficient hyperaccumulation phenotype (Farinati et al., 2011).
In the same manner that metagenomics can be used to sample the genetic potential of the microbial community, metaproteomics can be used to sample the proteins present among complex environmental consortia in extreme environments such as metalliferous soils. However, several empirical, technical, computational, and experimental design challenges remain to be addressed, including the development of efficient techniques for protein extraction from soils and subsequent sample preparation (Leary et al., 2013). Several organic compounds in the soil (e.g., humic acids) can interfere with protein identification, and the samples are prone to degradation so the amount of available metaproteomic data is currently limited (Leary et al., 2013). Even so, the presence (or absence) of specific microbial proteins will eventually be useful as an indicator for positive interactions between the plant root and soil microbes, allowing the prediction of hyperaccumulator phenotypes.
Conclusions
The composition of the bacterial rhizobiome coupled with the genomic, transcriptomic, and proteomic analysis of plant–microbe interactions may help us to understand in more detail the associations between hyperaccumulators and the surrounding bacterial communities of the endosphere and rhizosphere. It will be interesting to compare the rhizobiome of different facultative metallophytes, such as N. caerulescens adapted to grow in different metalliferous and non-metalliferous soils (Pollard et al., 2014), because this will help to isolate the bacteria that contribute to the hyperaccumulator phenotype. However, the rhizosphere is a dynamic environment with the community undergoing rapid spatiotemporal changes in response to external factors. The metabolic profiling of microbial colonies by in situ mass spectrometry (Traxler and Kolter, 2012) should therefore be integrated with omics-based profiling methods in a systems biology approach (Figure 1F) to facilitate the investigation of interactions between the rhizobiome and hyperaccumulator plants, thus providing an advanced toolkit for phytotechnology applications.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abou-Shanab, R. A., Angle, J. S., and Chaney, R. L. (2006). Bacterial inoculants affecting nickel uptake by Alyssum murale from low, moderate and high Ni soils. Soil Biol. Biochem. 38, 2882–2889. doi: 10.1016/j.soilbio.2006.04.045
Abou-Shanab, R. A., Angle, J. S., Delorme, T. A., Chaney, R. L., van Berkum, P., Moawad, H., et al. (2003). Rhizobacterial effects on nickel extraction from soil and uptake by Alyssum murale. New Phytol. 158, 219–224. doi: 10.1046/j.1469-8137.2003.00721.x
Alford, E. M., Pilon-Smits, E. A. H., and Paschke, M. W. (2010). Metallophytes—a view from the rhizosphere. Plant Soil 337, 33–50. doi: 10.1007/s11104-010-0482-3
Baker, A. J. M., McGrath, S. P., Reeves, R. D., and Smith, J. A. C. (2000). “Chapter 5. Metal hyper accumulator plants: a review of the ecology and physiology of a biological resource for phytoremediation of metal-polluted soils,” in Phytoremediation of Contaminated Soil and Water, eds N. Terry and G. Bañuelos (BocaRaton, FL: CRC Press), 85–107.
Barzanti, R., Ozino, F., Bazzicalupo, M., Gabrielli, R., Galardi, F., Gonnelli, C., et al. (2007). Isolation and characterization of endophytic bacteria from the nickel hyperaccumulator plant Alyssum bertolonii. Microb. Ecol. 53, 306–316. doi: 10.1007/s00248-006-9164-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Becerra-Castro, C., Kidd, P. S., Kuffner, M., Prieto-Fernandez, A., Hann, S., Monterroso, C., et al. (2013). Bacterially induced weathering of ultramafic rock and its implications for phytoextraction. Appl. Environ. Microbiol. 79, 5094–5103. doi: 10.1128/AEM.00402-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Becerra-Castro, C., Prieto-Fernández, Á., Álvarez-López, V., Cabello-Conejo, M. I., Acea, M. J., Kidd, P. S., et al. (2011). Nickel solubilizing capacity and characterization of rhizobacteria isolated from hyperaccumulating and non-hyperaccumulating subspecies of Alyssum serpyllifolium. Int. J. Phytoremediation 13, 229–244. doi: 10.1080/15226514.2011.568545
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Belimov, A. A., Hontzeas, N., Safronova, V. I., Demchinskaya, S. V., Piluzza, G., Bullitta, S., et al. (2005). Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 37, 241–250. doi: 10.1016/j.soilbio.2004.07.033
Berg, J., Brandt, K. K., Al-Soud, W. A., Holm, P. E., Hansen, L. H., Sørensen, S. J., et al. (2012). Selection for Cu-tolerant bacterial communities with altered composition, but unaltered richness, via long-term Cu exposure. Appl. Environ. Microbiol. 78, 7438–7446. doi: 10.1128/AEM.01071-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bulgarelli, D., Rott, M., Schlaeppi, K., van Themaat, E. V. L., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cabello-Conejo, M., Becerra-Castro, C., Prieto-Fernández, A., Monterroso, C., Saavedra-Ferro, A., Mench, M., et al. (2014). Rhizobacterial inoculants can improve nickel phytoextraction by the hyperaccumulator Alyssum pintodasilvae. Plant Soil 379, 35–50. doi: 10.1007/s11104-014-2043-7
Cardinale, M. (2014). Scanning a microhabitat: plant-microbe interactions revealed by confocal laser microscopy. Front. Microbiol. 5:94. doi: 10.3389/fmicb.2014.00094
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chelius, M. K., and Triplett, E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41, 252–263. doi: 10.1007/S002480000087
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, L., Luo, S., Chen, J., Wan, Y., Li, X., Liu, C., et al. (2014b). Comparative analysis of endophytic bacterial communities associated with hyperaccumulators growig in mine soils. Environ. Sci. Pollut. Res. Int. 21, 7538–7547. doi: 10.1007/s11356-014-2670-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, L., Luo, S. L., Li, X. J., Wan, Y., Chen, J. L., and Liu, C. B. (2014a). Interaction of Cd-hyperaccumulator Solanum nigrum L. and functional endophyte Pseudomonas sp. Lk9 on soil heavy metals uptake. Soil Biol. Biochem. 68, 300–308. doi: 10.1016/j.soilbio.2013.10.021
Chen, L., Luo, S. L., Xiao, X., Guo, H. J., Chen, J. L., Wan, Y., et al. (2010). Application of plant growth-promoting endophytes (PGPE) isolated from Solanum nigrum L. for phytoextraction of Cd-polluted soils. Appl. Soil Ecol. 46, 383–389. doi: 10.1016/j.apsoil.2010.10.003
Compant, S., Duffy, B., Nowak, J., Clément, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
DalCorso, G., Fasani, E., and Furini, A. (2013). Recent advances in the analysis of metal hyperaccumulation and hypertolerance in plants using proteomics. Front. Plant Sci. 4:280. doi: 10.3389/fpls.2013.00280
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fan, B., Cravalhais, L. C., Becker, A., Fedoseyenko, D., Von Wiren, N., and Borriss, R. (2012). Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol. 12:116. doi: 10.1186/1471-2180-12-116
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Farinati, S., DalCorso, G., Bona, E., Corbella, M., Lampis, S., Cecconi, D., et al. (2009). Proteomic analysis of Arabidopsis halleri shoots in response to the heavy metals cadmium and zinc and rhizosphere microorganisms. Proteomics 9, 4837–4850. doi: 10.1002/pmic.200900036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Farinati, S., DalCorso, G., Panigati, M., and Furini, A. (2011). Interaction between selected bacterial strains and Arabidopsis halleri modulates shoot proteome and cadmium and zinc accumulation. J. Exp. Bot. 62, 3433–3447. doi: 10.1093/jxb/err015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ge, Y., Schimel, J. P., and Holden, P. A. (2014). Analysis of run-to run variation of bar-coded pyrosequencing for evaluationg bacterial community shifts and individual taxa dynamics. PLoS ONE 9:e99414. doi: 10.1371/journal.pone.0099414
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ghosh, P., Rathinasabapathi, B., and Ma, L. Q. (2011). Arsenic-resistant bacteria solubilized arsenic in the growth media and increased growth of arsenic hyperaccumulator Pteris vittata L. Bioresour. Technol. 102, 8756–8761. doi: 10.1016/j.biortech.2011.07.064
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gołębiewski, M., Deja-Sikora, E., Cichosz, M., Tretyn, A., and Wróbel, B. (2014). 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb. Ecol. 67, 635–647. doi: 10.1007/s00248-013-0344-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gottel, N. R., Castro, H. F., Kerley, M., Yang, Z., Podar, M., Karpinets, T., et al. (2011). Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 77, 5934–5944. doi: 10.1128/AEM.05255-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gupta, R., Mathimaran, R., Wiemken, A., Boller, T., Bisaria, V. S., and Sharma, S. (2014). Non-target effects of bioinoculants on rhizospheric microbial communities of Cajanus cajan, Appl. Soil Ecol. 76, 26–33. doi: 10.1016/j.apsoil.2013.12.001
Halimaa, P., Blande, D., Aarts, M. G., Tuomainen, M., Tervahauta, A., and Kärenlampi, S. (2014). Comparative transcriptome analysis of the metal hyperaccumulator Noccaea caerulescens. Front. Plant Sci. 5:213. doi: 10.3389/fpls.2014.00213
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, H., Ye, Z., Yang, D., Yan, J., Xiao, L., Zhong, T., et al. (2013). Characterization of endophytic Rahnella sp. JN6 from Polygonum pubescens and its potential in promoting growth and Cd, Pb, Zn uptake by Brassica napus. Chemosphere 90, 1960–1965. doi: 10.1016/j.chemosphere.2012.10.05
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, L. Y., Zhang, Y. F., Ma, H. Y., Su, L. N., Chen, Z. J., Wang, Q. Y., et al. (2010). Characterization of copper-resistant bacteria and assessment of bacterial communities in rhizosphere soils of copper-tolerant plants. Appl. Soil Ecol. 44, 49–55. doi: 10.1016/j.apsoil.2009.09.004
Hirsch, P., and Mauchline, T. H. (2012). Who's who in the plant root microbiome? Nat. Biotechnol. 30, 961–962. doi: 10.1038/nbt.2387
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, A., Teplitski, M., Rathinasabapathi, B., and Ma, L. Q. (2010). Characterization of arsenic-resistant bacteria from the rhizosphere of arsenic hyperaccumulator Pteris vittata. Can. J. Microbiol. 56, 236–246. doi: 10.1139/W10-005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Idris, R., Trifonova, R., Puschenreiter, M., Wenzel, W. W., and Sessitsch, A. (2004). Bacteria communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microbiol. 70, 2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jing, Y. X., Yan, J. L., He, H. D., Yang, D. J., Xiao, L., Zhong, T., et al. (2014). Characterization of bacteria in the rhizosphere soils of Polygonum pubescens and their potential in promoting growth and Cd, Pb, Zn uptake by Brassica napus. Int. J. Phytoremediation 16, 321–333. doi: 10.1080/15226514.2013.773283
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kirk, J. L., Beaudette, L. A., Hart, M., Moutoglis, P., Klironomos, J. N., Lee, H., et al. (2004). Methods of studying soil microbial diversity. J. Microbiol. Methods 58, 169–188. doi: 10.1016/j.mimet.2004.04.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Knief, C. (2014). Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 5:216. doi: 10.3389/fpls.2014.00216
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuffner, M., De Maria, S., Puschenreiter, M., Fallmann, K., Wieshammer, G., Gorfer, M., et al. (2010). Culturable bacteria from Zn- and Cd-accumulating Salix caprea with differential effects on plant growth and heavy metal availability. J. Appl. Microbiol. 108, 1471–1484. doi: 10.1111/j.1365-2672.2010.04670.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kwak, Y. S., Bonsall, R. F., Okubara, P. A., Paulitz, T. C., Thomashow, L. S., and Weller, D. M. (2012). Factors impacting the activity of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens against take-all of wheat. Soil Biol. Biochem. 54, 48–56. doi: 10.1016/j.soilbio.2012.05.012
Leary, D. H., Hervey, W. J. IV., Deschamps, J. R., Kusterbeck, A. W., and Vora, G. J. (2013). Which metaproteome? The impact of protein extraction bias on metaproteomic analyses. Mol. Cell. Probes 27, 193–199. doi: 10.1016/j.mcp.2013.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, W. C., Ye, Z. H., and Wong, M. H. (2007). Effects of bacteria an enhanced metal uptake of the Cd/Zn-hyperaccumulating plant, Sedum alfredii. J. Exp. Bot. 58, 4173–4182. doi: 10.1093/jxb/erm274
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, L., Guo, W., Xing, Y., Zhang, X., Li, Z., Hu, C., et al. (2012). The actinobacterium Microbacterium sp. 16SH accepts pBBR1-based pPROBE vectors, forms biofilms, invades roots, and fixes N2 associated with micropropagated sugarcane plants. Appl. Microbiol. Biotechnol. 93, 1185–1195. doi: 10.1007/s00253-011-3618-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lodewyckx, C., Mergeay, M., Vangronsveld, J., Clijsters, H., and Van Der Lelie, D. (2002). Isolation, characterization, and identification of bacteria associated with the zinc hyperaccumulator Thlaspi caerulescens, subsp. Calaminaria. Int. J. Phytoremediation 4, 105–115. doi: 10.1080/15226510208500076
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Long, X. X., Chen, X. M., Wong-Chung, J. W. C., Wei, Z. B., and Wu, Q. T. (2011). Isolation and characterization endophytic bacteria from hyperaccumulator Sedum alfredii Hance and their potential to promote phytoextraction of zinc polluted soil. World J. Microbiol. Biotechnol. 27, 1197–1207. doi: 10.1007/s11274-010-0568-3
Long, X. X., Chen, X. M., Wong-Chung, J. W. C., Wei, Z. B., and Wu, Q. T. (2013). Feasibility of enhanced phytoextraction of Zn contaminated soil with Zn mobilizing and plant growth promoting endophytic bacteria. Trans. Nonferrous Met. Soc. China 23, 2389-2396. doi: 10.1016/S1003-6326(13)62746-6
López-Leal, G., Tabche, M. L., Castillo-Ramírez, S., Mendoza-Vargas, A., Ramírez-Romero, M. A., and Dávila, G. (2014). RNA-Seq analysis of the multipartite genome of Rhizobium etli CE3 shows different replicon contributions under heat and saline shock. BMC Genomics 15:770. doi: 10.1186/1471-2164-15-770
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, S. L., Chen, L., Chen, J. L., Xiao, X., Xu, T. Y., Wan, Y., et al. (2011). Analysis and characterisation of cultivabe heavy metal-resistant bacterial endophytes isolated from Cd-hyperaccumulator Solanum nigrum L. and their potential use for phytoremediation. Chemosphere 85, 1130–1138. doi: 10.1016/j.chemosphere.2011.07.053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, Y., Prasad, M. N. V., Rajkumar, M., and Freitas, H. (2011a). Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 29, 248–258. doi: 10.1016/j.biotechadv.2010.12.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, Y., Rajkumar, M., and Freitas, H. (2009a). Improvement of plant growth and nickel uptake by nickel resistant-plant growth promoting bacteria. J. Hazard. Mater. 166, 1154–1161. doi: 10.1016/j.jhazmat.2008.12.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, Y., Rajkumar, M., and Freitas, H. (2009b). Isolation and characterization of Ni mobilizing PGPB from serpentine soils and their potential in promoting plant growth and Ni accumulation by Brassica spp. Chemosphere 75, 719–725. doi: 10.1016/j.chemosphere.2009.01.056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, Y., Rajkumar, M., Luo, Y., and Freitas, H. (2011b). Inoculation of endophytic bacteria on host and non-host plants-effects on plant growth and Ni uptake. J. Hazard. Mater. 195, 230–237. doi: 10.1016/j.jhazmat.2011.08.034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mark, G. L., Dow, J. M., Kiely, P. D., Higgings, H., Haynes, J., Baysse, C., et al. (2005). Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 17454–17459. doi: 10.1073/pnas.0506407102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mastretta, C., Barac, T., Vangronsveld, J., Newman, L., Taghavi, S., and van der Lelie, D. (2006). Endophytic bacteria and their potential application to improve the phytoremediation of contaminated environments. Biotechnol. Genet. Eng. Rev. 23, 175–207. doi: 10.1080/02648725.2006.10648084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maynaud, G., Brunel, B., Mornico, D., Durot, M., Severac, D., Dubois, E., et al. (2013). Genome-wide transcriptional responses of two metal-tolerant symbiotic Mesorhizobium isolates to zinc and cadmium exposure. BMC Genomics 14:292. doi: 10.1186/1471-2164-14-292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Metzker, M. L. (2010). Sequencing technologies – the next generation. Nat. Rev. Genet. 11, 31–46. doi: 10.1038/nrg2626
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moradi, A. B., Conesa, H. M., Robinson, B. H., Lehmann, E., Kaestner, A., and Schulin, R. (2009). Root responses to soil Ni heterogeneity in a hyperaccumulator and a non-accumulator species. Environ. Pollut. 157, 2189–2196. doi: 10.1016/j.envpol.2009.04.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peiffer, J., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553 doi: 10.1073/pnas.1302837110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pernthaler, A., Pernthaler, J., and Amann, R. (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pollard, A. J., Reeves, R. D., and Baker, A. J. M. (2014). Facultative hyperaccumulation of heavy metals and metalloids. Plant Sci. 218, 8–17. doi: 10.1016/j.plantsci.2013.11.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rajkumar, M., Ae, N., and Freitas, H. (2009). Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 77, 153–160. doi: 10.1016/j.chemosphere.2009.06.047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rajkumar, M., Sandhya, S., Prasad, M. N. V., and Freitas, H. (2012). Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 30, 1562–1574. doi: 10.1016/j.biotechadv.2012.04.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ramos, H. J. O., Roncato-Maccari, L. D. B., Souza, E. M., Soares-Ramos, J. R. L., Hungria, M., and Pedrosa, F. O. (2002). Monitoring Azospirillum-wheat interactions using the gfp and gusA genes constitutively expressed from a new broad-host range vector. J. Biotechnol. 97, 243–252. doi: 10.1016/S0168-1656(02)00108-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rastogi, G., and Sani, R. K. (2011). “Chapter two molecular techniques to assess microbial community structure, function and dynamics in the environment” in Microbes and Microbial Technology: Agricultural and Environmental Applications, eds I. Ahmad, F. Ahmad, and J. Pichtel (New York, NY: Springer Science+Business Media LLC), 29–57.
Rothballer, M., Schmid, M., Fekete, A., and Hartmann, A. (2005). Comparative in situ analysis of ipdC-gfpmut3 promoter fusions of Azospirillum brasilense strains Sp7 and Sp245. Environ. Microbiol. 7, 1839–1846. doi: 10.1111/j.1462-2920.2005.00848.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Salt, D. E., Blaylock, M., Kumar, N. P. B. A., Dushenkov, V., Ensley, B. D., Ilan Chet, I., et al. (1995). Phytoremediation: a novel strategy for the removal of toxic metals from the environment using plants. Nat. Biotechnol. 13, 468–474.
Sanguin, H., Remenant, B., Dechesne, A., Thioulouse, J., Vogel, T. M., Nesme, X., et al. (2006). Potential of a 16rRNA-based taxonomic microarray for analysing the rhizosphere effects of maize on Agrobacterium spp. and bacterial communities. Appl. Environ. Microbiol. 72, 4302–4312. doi: 10.1128/AEM.02686-05
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schreiter, S., Ding, G. C., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 8:144. doi: 10.3389/fmicb.2014.00144
Sessitsch, A., Kuffner, M., Kidd, P., Vangronsveld, J., Wenzel, W. W., Fallman, K., et al. (2013). The role of plant-associate bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 60, 182–194. doi: 10.1016/j.soilbio.2013.01.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sheng, X. F., Xia, J. J., Jiang, C. Y., He, L. Y., and Qian, M. (2008). Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 156, 1164–1170. doi: 10.1016/j.envpol.2008.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sørensen, J., Nicoliasen, M. H., Ron, E., and Simonet, P. (2009). Molecular tools in rhizosphere microbiology—from single-cell to whole-community analysis. Plant Soil 321, 483–512 doi: 10.1007/s11104-009-9946-8
Stabentheiner, E., Zankel, A., and Pölt, P. (2010). Environmental scanning electron microscopy (ESEM) a versatile tool in studying plants. Protoplasma 246, 89–99. doi: 10.1007/s00709-010-0155-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, L. N., Zhang, Y. F., He, L. Y., Chen, Z. J., Wang, Q. Y., Qian, M., et al. (2010). Genetic diversity and characterization of heavy metal-resistant-endophytic bacteria from two copper-tolerant plant species on copper mine wasteland. Bioresour. Technol. 101, 501–509. doi: 10.1016/j.biortech.2009.08.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Traxler, M. F., and Kolter, R. (2012). A massively spectacular view of the chemical lives of microbes. Proc. Natl. Acad. Sci. U.S.A. 109, 10128–10129. doi: 10.1073/pnas.1207725109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Visioli, G., D'Egidio, S., Vamerali, T., Mattarozzi, M., and Sanangelantoni, A. M. (2014). Culturable endophytic bacteria enhance Ni translocation in the hyperaccumulator Noccaea caerulescens. Chemosphere 117, 538–544. doi: 10.1016/j.chemosphere.2014.09.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Visioli, G., and Marmiroli, N. (2013). The proteomics of heavy metal hyperaccumulation by plants. J. Proteomics 79, 133–145. doi: 10.1016/j.jprot.2012.12.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watrous, J., Roach, P., Alexandrov, T., Heath, B. S., Yang, J. Y., Kersten, R. D., et al. (2012). Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U.S.A. 109, 1743–1752. doi: 10.1073/pnas.1203689109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Whiting, S. N., de Souza, M. P., and Terry, N. (2001). Rhizosphere bacteria mobilize Zn for hyperaccumulation by Thlaspi caerulescens. Environ. Sci. Technol. 35, 3144–3150. doi: 10.1021/es001938v
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xiong, J., He, Z., Liu, D., Mahmood, Q., and Yang, X. (2008). The role of bacteria in the heavy metals removal and growth of Sedum alfredii Hance in an aqueous medium. Chemosphere 70, 489–494. doi: 10.1016/j.chemosphere.2007.06.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zachow, C., Müller, H., Tilcher, R., and Berg, G. (2014). Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima-ancestor of all beet crop- and modern sugar beets. Front. Microbiol. 5:415. doi: 10.3389/fmicb.2014.00415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, W. H., Huang, Z., He, L. Y., and Sheng, X. F. (2012). Assessment of bacterial communities and characterization of lead-resistant bacteria in the rhizosphere soils of metal-tolerant Chenopodium ambrosioides g rown on lead-zinc mine tailings. Chemosphere 87, 1171–1178. doi: 10.1016/j.chemosphere.2012.02.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Li, L., Zhu, Z., Yang, X., Wang, Y., and An, Q. (2013). Colonization and modulation of host growth and metal uptake by endophytic bacteria of Sedum alfredii. Int. J. Phytoremediation 15, 51–64. doi: 10.1080/15226514.2012.670315
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, L. J., Guan, D. X., Luo, J., Rathinasabapathi, B., and Ma, L. Q. (2014). Characterization of arsenic-resistant endophytic bacteria from hyperaccumulators Pteris vittata and Pteris multifida. Chemosphere 113, 9–16. doi: 10.1016/j.chemosphere.2014.03.081
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: hyperaccumulators, rhizosphere, endosphere, metals, omics, microscopy, phytoremediation
Citation: Visioli G, D'Egidio S and Sanangelantoni AM (2015) The bacterial rhizobiome of hyperaccumulators: future perspectives based on omics analysis and advanced microscopy. Front. Plant Sci. 5:752. doi: 10.3389/fpls.2014.00752
Received: 10 October 2014; Accepted: 08 December 2014;
Published online: 07 January 2015.
Edited by:
Giovanni DalCorso, University of Verona, ItalyReviewed by:
Petra Kidd, Consejo Superior de Investigaciones Científicas, SpainGiovanni Vallini, University of Verona, Italy
Copyright © 2015 Visioli, D'Egidio and Sanangelantoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanna Visioli, Department of Life Sciences, University of Parma, Parco Area delle Scienze 33/A, 43124 Parma, Italy e-mail: giovanna.visioli@unipr.it
 Giovanna Visioli
Giovanna Visioli Sara D'Egidio
Sara D'Egidio Anna M. Sanangelantoni
Anna M. Sanangelantoni