- 1Graduate School of Pharmaceutical Sciences, Chiba University, Chiba, Japan
- 2Research Planning Department, Wakunaga Pharmaceutical Company, Akitakata, Japan
- 3Central Research Institute, Wakunaga Pharmaceutical Company, Akitakata, Japan
- 4RIKEN Center for Sustainable Resource Science, Yokohama, Japan
S-Alk(en)yl-L-cysteine sulfoxides are pharmaceutically important secondary metabolites produced by plants that belong to the genus Allium. Biosynthesis of S-alk(en)yl-L-cysteine sulfoxides is initiated by S-alk(en)ylation of glutathione, which is followed by the removal of glycyl and γ-glutamyl groups and S-oxygenation. However, most of the enzymes involved in the biosynthesis of S-alk(en)yl-L-cysteine sulfoxides in Allium plants have not been identified. In this study, we identified three genes, AsGGT1, AsGGT2, and AsGGT3, from garlic (Allium sativum) that encode γ-glutamyl transpeptidases (GGTs) catalyzing the removal of the γ-glutamyl moiety from a putative biosynthetic intermediate of S-allyl-L-cysteine sulfoxide (alliin). The recombinant proteins of AsGGT1, AsGGT2, and AsGGT3 exhibited considerable deglutamylation activity toward a putative alliin biosynthetic intermediate, γ-glutamyl-S-allyl-L-cysteine, whereas these proteins showed very low deglutamylation activity toward another possible alliin biosynthetic intermediate, γ-glutamyl-S-allyl-L-cysteine sulfoxide. The deglutamylation activities of AsGGT1, AsGGT2, and AsGGT3 toward γ-glutamyl-S-allyl-L-cysteine were elevated in the presence of the dipeptide glycylglycine as a γ-glutamyl acceptor substrate, although these proteins can act as hydrolases in the absence of a proper acceptor substrate, except water. The apparent Km values of AsGGT1, AsGGT2, and AsGGT3 for γ-glutamyl-S-allyl-L-cysteine were 86 μM, 1.1 mM, and 9.4 mM, respectively. Subcellular distribution of GFP-fusion proteins transiently expressed in onion cells suggested that AsGGT2 localizes in the vacuole, whereas AsGGT1 and AsGGT3 possess no apparent transit peptide for localization to intracellular organelles. The different kinetic properties and subcellular localizations of AsGGT1, AsGGT2, and AsGGT3 suggest that these three GGTs may contribute differently to the biosynthesis of alliin in garlic.
Introduction
Production of cysteine-derived secondary metabolites, S-alk(en)yl-L-cysteine sulfoxides, is a pharmaceutically important characteristic of plants that belong to the genus Allium. These compounds are hydrolyzed by the endogenous vacuolar enzyme alliinase (EC. 4.4.1.4) upon tissue disruption to yield highly reactive alk(en)ylsulfenic acids that are spontaneously converted to various sulfur-containing compounds with diverse pharmacological activities, including antibacterial, antifungal, antivirus, immunostimulating, antioxidant, anticarcinogenic, antithrombotic, cholesterol- and triglyceride-lowering, and hypotensive effects (Jones et al., 2004; Rose et al., 2005; Iciek et al., 2009). To date, four major S-alk(en)yl-L-cysteine sulfoxides, S-allyl-L-cysteine sulfoxide (alliin), S-methyl-L-cysteine sulfoxide (methiin), S-trans-1-propenyl-L-cysteine sulfoxide (isoalliin), and S-propyl-L-cysteine sulfoxide (propiin), have been identified and isolated from Allium plants (Jones et al., 2004; Rose et al., 2005).
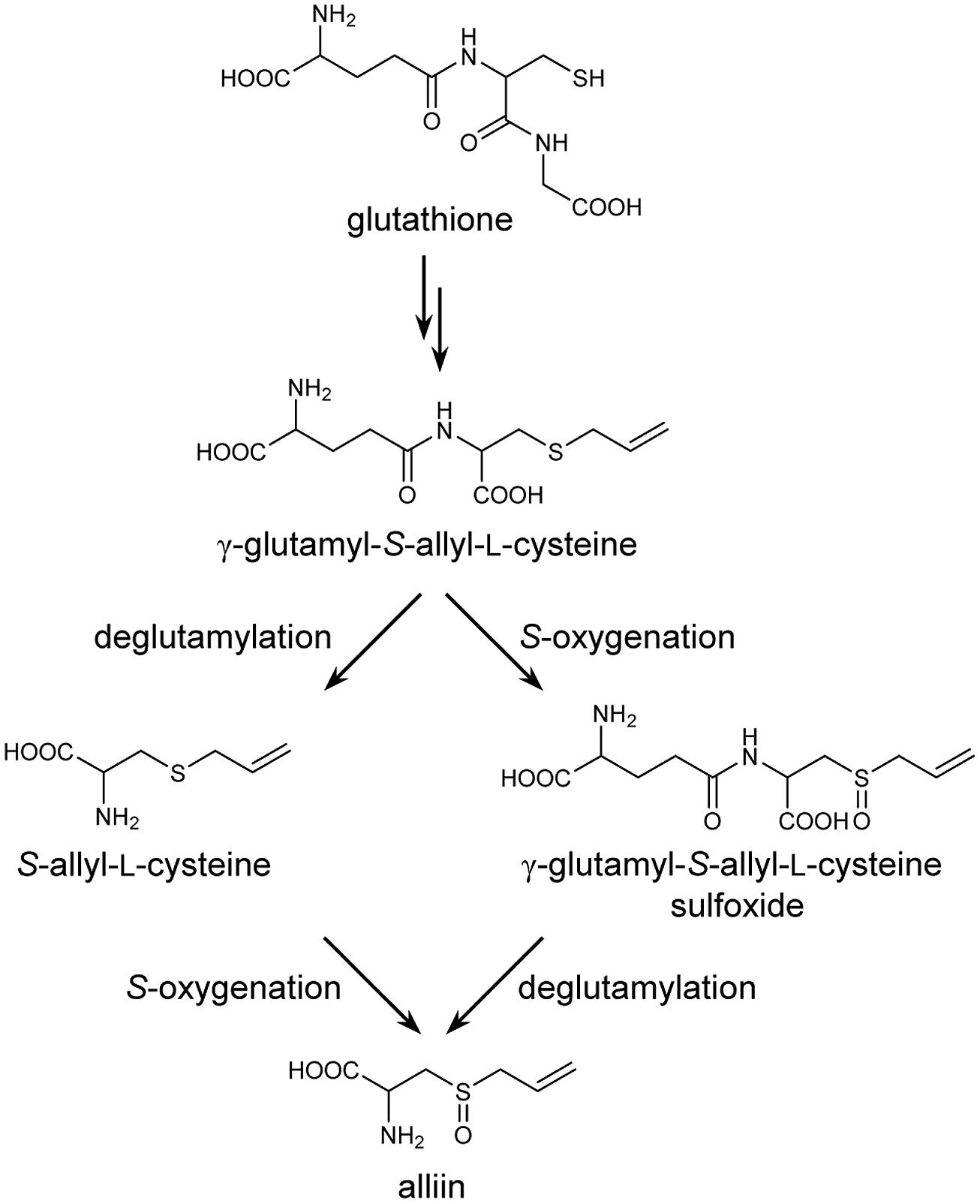
Biosynthesis of S-alk(en)yl-L-cysteine sulfoxides in Allium plants has previously been proposed to proceed via glutathione S-conjugates, according to the results of precursor feeding and pulse radiolabeling experiments (Suzuki et al., 1962; Turnbull et al., 1980; Lancaster and Shaw, 1989). In the proposed pathway, glutathione is S-alk(en)ylated at the cysteine residue, followed by the removal of a glycyl group to form a biosynthetic intermediate, γ-glutamyl-S-alk(en)yl-L-cysteine. This γ-glutamylated sulfide compound is further deglutamylated and S-oxygenated to yield S-alk(en)yl-L-cysteine sulfoxide (Figure 1). Although the results of pulse radiolabeling suggest that S-oxygenation may likely occur before deglutamylation in onion (Allium cepa; Lancaster and Shaw, 1989), the order of S-oxygenation and deglutamylation in other Allium plants remains unclear.
γ-Glutamyl transpeptidase (GGT; EC 2.3.2.2), also known as γ-glutamyl transferase, is the enzyme that catalyzes the transfer of the γ-glutamyl moiety of γ-glutamyl compounds to amino acids, short peptides (transpeptidation), or water (hydrolysis; Tate and Meister, 1981). The physiological role of GGT, commonly observed in bacteria, yeast, animals, and plants, is the catabolism of glutathione. Saccharomyces cerevisiae has a vacuolar GGT protein responsible for the degradation of glutathione in the vacuole during nitrogen starvation in order to supply the constituent amino acids of glutathione to the starved cell (Mehdi and Penninckx, 1997), whereas GGTs in Escherichia coli, mammals, and plants function in the breakdown of extracellular glutathione (Suzuki et al., 1999; Storozhenko et al., 2002; Dominici et al., 2005; Martin et al., 2007; Ohkama-Ohtsu et al., 2007a). GGT is also responsible for degrading glutathione-related compounds. For example, GGT is involved in the biosynthesis of the phytoalexin camalexin by removing a γ-glutamyl group from glutathione-indole-3-acetonitrile in Arabidopsis (Su et al., 2011), in the conversion of the endogenous glutathione S-conjugate leukotriene C4 to leukotriene D4 in rats (Anderson et al., 1982), and in the glutathione-mediated detoxification of xenobiotics in both animals and plants (Zhang et al., 2005; Grzam et al., 2007; Ohkama-Ohtsu et al., 2007b). Given that the removal of a γ-glutamyl group from the biosynthetic intermediate γ-glutamyl-S-alk(en)yl-L-cysteine is required for the biosynthesis of S-alk(en)yl-L-cysteine sulfoxides in the genus Allium, the involvement of GGTs in the biosynthesis of S-alk(en)yl-L-cysteine sulfoxides as deglutamylation enzymes has been proposed. The fact that the levels of biosynthetic intermediate γ-glutamyl peptides were decreased while GGT activity was increased during sprouting in onion bulbs also supports this idea (Lancaster and Shaw, 1991). To date, several efforts have been made to identify and characterize GGTs in Allium plants. A GGT partially purified from onion showed high substrate specificity toward γ-glutamyl compounds that are putative intermediates of S-alk(en)yl-L-cysteine sulfoxide biosynthesis, strongly suggesting the involvement of this GGT in the biosynthesis of S-alk(en)yl-L-cysteine sulfoxides (Lancaster and Shaw, 1994). Recently, a GGT protein was purified to homogeneity from sprouting onion bulbs, and a partial cDNA for this GGT, AcGGT, was cloned; however, in contrast to the previously partially purified onion GGT (Lancaster and Shaw, 1994), the purified AcGGT protein showed high affinity for glutathione and glutathione S-conjugates but could not utilize γ-glutamyl-trans-S-1-propenyl-L-cysteine sulfoxide as a good γ-glutamyl donor substrate, suggesting that AcGGT is not the major enzyme catalyzing deglutamylation in the biosynthesis of S-alk(en)yl-L-cysteine sulfoxides in onion (Shaw et al., 2005). A partial cDNA of AsGGT, which has high sequence homology to AcGGT, was isolated from garlic (Allium sativum), and its mRNA expression patterns suggested that AsGGT may play a role in synthesizing S-alk(en)yl-L-cysteine sulfoxides in garlic cloves during cold storage (Cho et al., 2012).
In this study, we cloned three genes encoding GGTs, AsGGT1, AsGGT2, and AsGGT3, that are suggested to be involved in the biosynthesis of alliin in garlic. The substrate preferences of AsGGT1, AsGGT2, and AsGGT3 suggest that a key biosynthetic intermediate, γ-glutamyl-S-allyl-L-cysteine, is deglutamylated by these GGTs prior to being S-oxygenated during alliin biosynthesis in garlic.
Materials and Methods
Plant Materials and Reagents
Total RNA was extracted from the bulbs of A. sativum L. ‘Fukuchi-howaito’. S-Allyl-L-cysteine was purchased from Tokyo Chemical Industry (Tokyo, Japan). Alliin [(RCSS)-S-allyl-L-cysteine sulfoxide] was synthesized and purified according to previously reported methods (Yu et al., 1994; Kubec et al., 1999; Kubec and Dadáková, 2008). γ-Glutamyl-S-allyl-L-cysteine was synthesized as follows. A mixture of 2.59 g of N-phthaloyl-L-glutamic anhydride and 1.93 g of S-allyl-L-cysteine in 10 mL of acetic acid was stirred at 60°C for 2 h. The solvent in the reaction mixture was removed in vacuo, and the residue was suspended in ethyl acetate and washed with brine. The organic layer was dried using sodium sulfate, and the solvent was removed in vacuo. After 1.5 g of residue was dissolved in 10 mL of methanol, 0.15 mL of hydrazine monohydrate was added, and this mixture was refluxed at 80°C for 1 h. Solvent was removed in vacuo, and the residue was washed with ethanol. The residue was recrystallized in a mixture of ethanol and water. The crystalline powder was applied to DowexTM 50Wx8 (The Dow Chemical Company, USA), and the column eluate and rinsing were combined and lyophilized. γ-Glutamyl-S-allyl-L-cysteine sulfoxide was synthesized as follows. γ-Glutamyl-S-allyl-L-cysteine was dissolved in water, and 1.1 equimolar of hydrogen peroxide was added. The mixture was stirred at room temperature, and the solvent was removed in vacuo. The residue was dried under reduced pressure with phosphorus (V) oxide at room temperature. Structures of synthesized γ-glutamyl-S-allyl-L-cysteine and γ-glutamyl-S-allyl-L-cysteine sulfoxide were confirmed using 13C-NMR and1H-NMR. All other chemicals were of analytical grade and were purchased from Sigma (St. Louis, MO, USA), Nacalai Tesque (Kyoto, Japan), or Wako Pure Chemical Industries (Osaka, Japan).
Cloning of AsGGT1, AsGGT2, and AsGGT3 from Garlic
Molecular biological experiments were performed according to the standard protocols (Sambrook et al., 1989), unless otherwise specified. Total RNA was extracted from garlic cloves by using the RNeasy plant mini kit (Qiagen, Valencia, CA, USA) and treated with DNase I (Life Technologies, Carlsbad, CA, USA). Reverse transcription (RT) was performed using SuperScript II reverse transcriptase (Life Technologies) and oligo-d(T)12-18. Partial cDNAs of AsGGT1 and AsGGT2 were amplified by PCR using ExTaq DNA polymerase (Takara, Tokyo, Japan) and oligonucleotide primers designed from the nucleotide sequences of two garlic EST clones, i.e., EPP005LLAA12S004013 and EPP005LLAA12S003688 in GarlicESTdb (Kim et al., 20091): AsGGT1-Core-F (5′-ATCGCCACTTCATATGAACC-3′) and AsGGT1-Core-R (5′-GATAATGCTAGATATGGCTC-3′) for AsGGT1; AsGGT2-Core-F (5′-CTCCTCCACATTAATGGAAC-3′) and AsGGT2-Core-R (5′-AAGTGGTCCCAAACATTTGTC-3′) for AsGGT2. For the amplification of a partial region of AsGGT3 cDNA, degenerate primers designed based on the sequences of conserved regions of known GGTs, GGT-degenerate-F (5′-ATHGTNYTNAAYAAYGARATG-3′) and GGT-degenerate-R (5′-CCNCCYTTNCKNGGRTC-3′), were used. Rapid amplification of cDNA ends (RACE) was performed using 5′-Full RACE Core Set (Takara) and 3′-Full RACE Core Set (TaKaRa), according to the manufacturer’s protocols. 5′-RACE was performed using the following primers: AsGGT1-5′-RACE-RT (5′-[Phos]TCTTCTGAACCG-3′), AsGGT1-5′-RACE-F1(5′-TGCTCTCACCACTCTGTTC-3′), AsGGT1-5′-RACE-F2 (5′-GACTCCATCTCTCATCAGTTC-3′), As GGT1-5′-RACE-R1 (5′-TCACGAACGATGAGCGATG-3′), and AsGGT1-5′-RACE-R2 (5′-CCAGTTTCTGATCAGAAGAAGC-3′) for AsGGT1; AsGGT2-5′-RACE-RT (5′-[Phos]TGAGCTCGTAA ACTC-3′), AsGGT2-5′-RACE-F1 (5′-TGTGCGACGGTATCCG ATCA-3′), AsGGT2-5′-RACE-F2 (5′-CTCAATCCAATTCAACCT AGAC-3′), AsGGT2-5′-RACE-R1 (5′-CATTGTGCAGCGGACGA TAG-3′), and AsGGT2-5′-RACE-R2 (5′-GGTTCCATTAATGTGG AGGAG-3′) for AsGGT2; AsGGT3-5′-RACE-RT (5′-[Phos]GTATC CATCGGGAAT-3′), AsGGT3-5′-RACE-F1 (5′-TGAAAAAGAAA GGGCAGCTC-3′), AsGGT3-5′-RACE-F2 (5′-GGTTTAGGGATT GCAAATGG-3′), AsGGT3-5′-RACE-R1 (5′-CCTCCACTTGCGC CTAGAG-3′), and AsGGT3-5′-RACE-R2 (5′-GGTGGCGGCATAT TGTTATT-3′) for AsGGT3. 3′-RACE was performed using 3 sites adaptor primer (5′-CTGATCTAGAGGTACCGGATCC-3′) and the following gene-specific primers: AsGGT1-3′-RACE-F1 (5′-AGCTGGTCTACATGCTGCATGG-3′) and AsGGT1-3′-RACE-F2 (5′-TCCCATGGAAGTCACTTTTCG-3′) for AsGGT1; AsGGT2-3′-RACE-F1 (5′-GCTTTTGATGCTAGAGAGACTGC-3′) and AsGGT2-3′-RACE-F2 (5′-ATCACTCCGACAAATGTTTG-3′) for AsGGT2; AsGGT3-3′-RACE-F (5′-TGAAAAAGAAAGGGCAGC TC-3′) for AsGGT3. cDNA clones of AsGGT1, AsGGT2, and AsGGT3 were re-isolated by RT-PCR using KOD plus DNA polymerase (Toyobo, Osaka, Japan) and the following primers: AsGGT1-F (5′-TCATATTCTGACGCAGATTCCACAG-3′) and AsGGT1-R (5′-TGTTCAATCATATTTTGTACAAATAGAC-3′) for AsGGT1; AsGGT2-F (5′-CGAGCAAATTAATTCATTTTGGCTC AC-3′) and AsGGT2-R (5′-GCATACCAATCGCCACAAACTC-3′) for AsGGT2; AsGGT3-F (5′-GTTAACAACAGGATTGGTCAATG CTC-3′) and AsGGT3-R (5′-CAGCAAACAACGCACTATTCAGT TTCTG-3′) for AsGGT3.
Heterologous Expression of AsGGT1, AsGGT2, and AsGGT3 in Yeast
The coding regions of AsGGT1, AsGGT2, and AsGGT3 were amplified by PCR using the cloned cDNA fragments described above, KOD plus DNA polymerase (Toyobo), and the following gene-specific primers: AsGGT1-FKpn3A (5′-GGTACCAAAATGAACCAAATGGCGCCGGCTTC-3′) and AsGGT1-stop-RXh (5′-CTCGAGCTATACACAAGCAGGACTTC CATC-3′) for AsGGT1; AsGGT2-FKpn3A (5′-GGTACCAAAATG GAACCGGCGCATGATGACTTAG-3′) and AsGGT2-stop-RXh (5′-CTCGAGTCACACACATGCAGGACTTCCATC-3′) for AsGG T2; AsGGT3-FKpn3A (5′-GGTACCAAAATGCTAATTAATTCATA CCCTGC-3′) and AsGGT3-stop-RXh (5′-CTCGAGTCAGTATCC ATCGGGAATACC-3′) for AsGGT3. The underlined sequences in the primers correspond to KpnI and XhoI restriction sites for subcloning. The amplified fragments were cloned into the pGEM-T easy vector (Promega, Madison, WI, USA). After their nucleotide sequences were confirmed, the coding regions of AsGGT1, AsGGT2, and AsGGT3 were cut out as KpnI-XhoI fragments and were inserted between the KpnI and XhoI sites in the yeast expression vector pYES2 (Life Technologies). The resulting plasmids, pYES2-AsGGT1, pYES2-AsGGT2, and pYES2-AsGGT3, and pYES2 empty vector were transformed into the Saccharomyces cerevisiae mutant strain BJ2168 (MATa, prb1-1122, prc1-407, pep4-3, ura3-52, leu2, trp1; Nippon Gene, Tokyo, Japan) by using the lithium acetate method (Gietz et al., 1992). The transformants were selected on SD minimal medium (Sherman, 1991) containing no uracil. For the induction of recombinant proteins, the yeast cells grown in SD minimal medium without uracil at 28°C for 1 days were transferred to 10 volumes of uracil-less SD medium containing 2% (w/v) galactose instead of glucose to activate the GAL1 promoter on pYES2, and cultured at 28°C for 1 days. The cells were harvested and disrupted at 4°C with 425–600-μm (diameter) glass beads in buffer G [10 mM Tris-HCl (pH 7.5), 300 mM sorbitol, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, and 1 μM pepstatin A]. The lysate was centrifuged at 10,000 × g for 5 min, and the supernatant was collected. Buffer G of the supernatant was subsequently replaced with 50 mM Tris-HCl (pH 8.0) by using the Sephadex column PD Mini Trap G-25 (GE Healthcare, Uppsala, Sweden), according to the manufacturer’s protocol. The eluted yeast crude proteins were used for the enzymatic activity assay described below. Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad, CA, USA) based on the Bradford method (Bradford, 1976), using bovine serum albumin as the standard.
Assays of GGT Enzyme Activities
Assays of GGT enzyme activities were performed by analyzing the amount of deglutamylated compounds produced from γ-glutamylated compounds by yeast crude proteins in 6 h at 37°C. The amount of deglutamylated compounds increased linearly over the 6-h incubation period.
Deglutamylation activities using γ-glutamyl-p-nitroanilide as the substrate were determined spectrophotometrically according to a previously described method (Orlowski and Meister, 1963), with slight modifications, as follows: the reaction mixture, which consisted of 0.0125 μg μl-1 yeast crude protein, 50 mM Tris-HCl (pH 8.0), 10 mM glycylglycine, and 1 mM γ-glutamyl-p-nitroanilide, was incubated for 6 h at 37°C, and p-nitroaniline released from γ-glutamyl-p-nitroanilide was monitored at 412 nm.
For the analysis of deglutamylation activities toward γ-glutamyl-S-allyl-L-cysteine and γ-glutamyl-S-allyl-L-cysteine sulfoxide, the enzyme assay reaction mixture consisted of 0.67 μg μl-1 yeast crude protein, 50 mM Tris-HCl (pH 8.0), 10 mM glycylglycine, and 1 mM γ-glutamyl-S-allyl-L-cysteine or γ-glutamyl-S-allyl-L-cysteine sulfoxide was incubated for 6 h at 37°C. For the determination of the enzyme activity in a pH range of 6.0–7.0, the reaction mixture containing 50 mM 2-(N-morpholino)ethanesulfonic acid buffer, instead of 50 mM Tris-HCl buffer, was used. For the determination of the enzyme activity in a pH range of 7.0–9.0, 50 mM Tris-HCl buffer at pH values from 7.0 to 9.0, instead of 50 mM Tris-HCl (pH 8.0), was used. For the analysis of the effects of glycylglycine as a γ-glutamyl acceptor, deglutamylation activity was determined in the reaction mixture with or without 10 mM glycylglycine. The reaction was initiated by the addition of yeast crude proteins. After incubation at 37°C, proteins in the reaction mixture were removed using a centrifugal ultrafiltration device (molecular weight cut-off, 10 kD; Kurabo, Osaka, Japan). S-Allyl-L-cysteine and alliin in the ultrafiltrated solution were quantified using high-performance liquid chromatography (HPLC). For the kinetic analysis, assays were carried out with γ-glutamyl-S-allyl-L-cysteine concentrations ranging from 12.5 to 1000 μM for AsGGT1, 0.5–8 mM for AsGGT2, and 1–25 mM for AsGGT3. Km values were calculated from triplicate date sets according to the Michaelis-Menten equation.
Analysis of Sulfur-Containing Metabolites by Using HPLC
The enzymatic products were analyzed quantitatively by using HPLC (Hitachi, Tokyo, Japan) with the cation-exchange column (TSKgel Aminopak, Tosoh, Tokyo, Japan). For the determination of S-allyl-L-cysteine, a mobile phase composed of 67 mM sodium citrate, 8% (v/v) ethanol, and 0.01% (v/v) octanoic acid (pH 3.26) was used for an isocratic elution. The column temperature was 35°C. For the determination of alliin, the mobile phase consisted of 22 mM trisodium citrate and 80 mM citric acid and the column temperature was 40°C. After separation, S-allyl-L-cysteine and alliin were fluorescently derivatized using o-phthalaldehyde and were detected using a fluorescence detector (excitation 340 nm, emission 455 nm). Identification of S-allyl-L-cysteine and alliin in the enzymatic reaction mixture was based on comparisons of retention times of the synthesized standards.
Subcellular Localization Analysis
For the construction of the fusion gene constructs of 35Spro:As GGT1N100:GFP, 35Spro:AsGGT2N100:GFP, and 35Spro:AsGGT3N100 :GFP, partial coding regions of AsGGT1, AsGGT2, and AsGGT3 that encode the N-terminal 100 amino acid residues were amplified by PCR using KOD plus DNA polymerase (Toyobo) and the following gene-specific primers: AsGGT1-FSal (5′-GTCGACATGAACCAAATGGCGCCGGCTTCTTC-3′) and AsGGT1-N100-RNco (5′-CCATGGAACCACCACCACCACC ACCTTTTCTCAGAACTGAAGCTCC-3′) for AsGGT1; AsGGT2-FSal (5′-GTCGACATGGAACCGGCGCATGATGACTTAG-3′) and AsGGT2-N100-RNco (5′-CCATGGAACCACCACCACCACCACC TAAAGCGTCCACAGCATGACC-3′) for AsGGT2; AsGGT3-FSal (5′-GTCGACATGCTAATTAATTCATACCCTGCATATC-3′) and AsGGT3-N100-RNco (5′-CCATGGAACCACCACCACCACCACC ACTAACAACTCCTAAACAAAATG-3′) for AsGGT3. The sequence encoding hexa-Gly residues was generated downstream of the sequence of AsGGT1, AsGGT2, and AsGGT3 by the PCR. The underlined sequences in the primers correspond to SalI and NcoI restriction sites for subcloning. The amplified DNA fragments were cloned into the pGEM-T easy vector (Promega) to confirm the nucleotide sequence. Partial AsGGT1, AsGGT2, and AsGGT3, fused with the sequence encoding hexa-Gly residues, were cut out as SalI-NcoI fragments and were inserted between the SalI and NcoI sites in pTH2 (Chiu et al., 1996). Each of the resulting plasmids was co-introduced with pDsRed plasmid (Kitajima et al., 2009) into onion epidermal cells by particle bombardment at 150 psi, using a Helios gene gun (Bio-Rad). After bombardment, onion peels were incubated for 26–47 h on B5 medium (Gamborg et al., 1968) in the dark at 25°C. GFP and DsRed fluorescence in the onion cells were observed using a LSM710 confocal laser-scanning microscope (Carl Zeiss, Oberkochen, Germany).
Phylogenetic Analysis
Phylogenetic analysis was performed using MEGA version 6 software (Tamura et al., 2013) based on the ClustalW multiple alignment. A phylogenetic tree was generated using the neighbor-joining method.
Results
Identification of Three Genes Encoding γ-Glutamyl Transpeptidases in Garlic
We found two garlic EST clones, EPP005LLAA12S004013 and EPP005LLAA12S003688, that show high sequence homology with known GGTs in GarlicESTdb (Kim et al., 20092). Utilizing the sequence information on these EST clones, we obtained two different full-length cDNA clones by 5′- and 3′-RACE and RT-PCR from the RNA of garlic cloves and designated them as AsGGT1 (GenBank Accession No. LC008010) and AsGGT2 (GenBank Accession No. LC008011). In addition, we amplified one garlic cDNA fragment using degenerate primers designed based on the conserved regions of known plant GGTs. A full-length cDNA clone was obtained by RACE and RT-PCR, and was designated as AsGGT3 (GenBank Accession No. LC008012). The cDNAs of AsGGT1, AsGGT2, and AsGGT3 coded for polypeptides of 627, 622, and 605 amino acids, respectively. The deduced amino acid sequences of AsGGT1 and AsGGT2 shared 69% identity, whereas the amino acid sequence identity of AsGGT3 with AsGGT1 and AsGGT2 was 46 and 43%, respectively. The amino acid sequence of AsGGT3 showed 99% sequence identity with that of a partial sequence of garlic AsGGT (Cho et al., 2012) in their 158 aa overlapped region and showed 92% sequence identity with that of a partial sequence of onion AcGGT (Shaw et al., 2005) in their 543 aa overlapped region. Arabidopsis thaliana has three functional GGTs, AtGGT1, AtGGT2, and AtGGT4. Among them, AtGGT4 is known to have a long N-terminal sequence that determines vacuolar localization, compared to AtGGT1 and AtGGT2 that localize in the extracellular space (Grzam et al., 2007; Ohkama-Ohtsu et al., 2007b). As in Arabidopsis AtGGT4, the deduced amino acid sequences of AsGGT1, AsGGT2, and AsGGT3 had longer N-terminal sequences than those of Arabidopsis AtGGT1 and AtGGT2, suggesting the presence of the N-terminal signal sequences for targeting to cellular organelles in AsGGT1, AsGGT2, and AsGGT3. N-terminal regions of the deduced amino acid sequences of AsGGT1, AsGGT2, and AsGGT3 were not highly similar to each other or to that of AtGGT4, despite the high sequence similarities among the rest of their regions.
A phylogenic tree was generated by the neighbor-joining method, using the amino acid sequences of known GGTs from plants, yeast, bacteria, and humans (Figure 2). All plant GGTs were classified into the same branch, which was further divided into two distinct subgroups. AsGGT1 and AsGGT2 belonged to the subgroup containing Arabidopsis AtGGT4 that functions in the degradation of glutathione S-conjugates in the vacuole (Grzam et al., 2007; Ohkama-Ohtsu et al., 2007b), whereas AsGGT3 belonged to the subgroup containing Arabidopsis AtGGT1 and AtGGT2 that function in the breakdown of extracellular glutathione (Martin et al., 2007; Ohkama-Ohtsu et al., 2007a) together with onion AcGGT (Shaw et al., 2005).
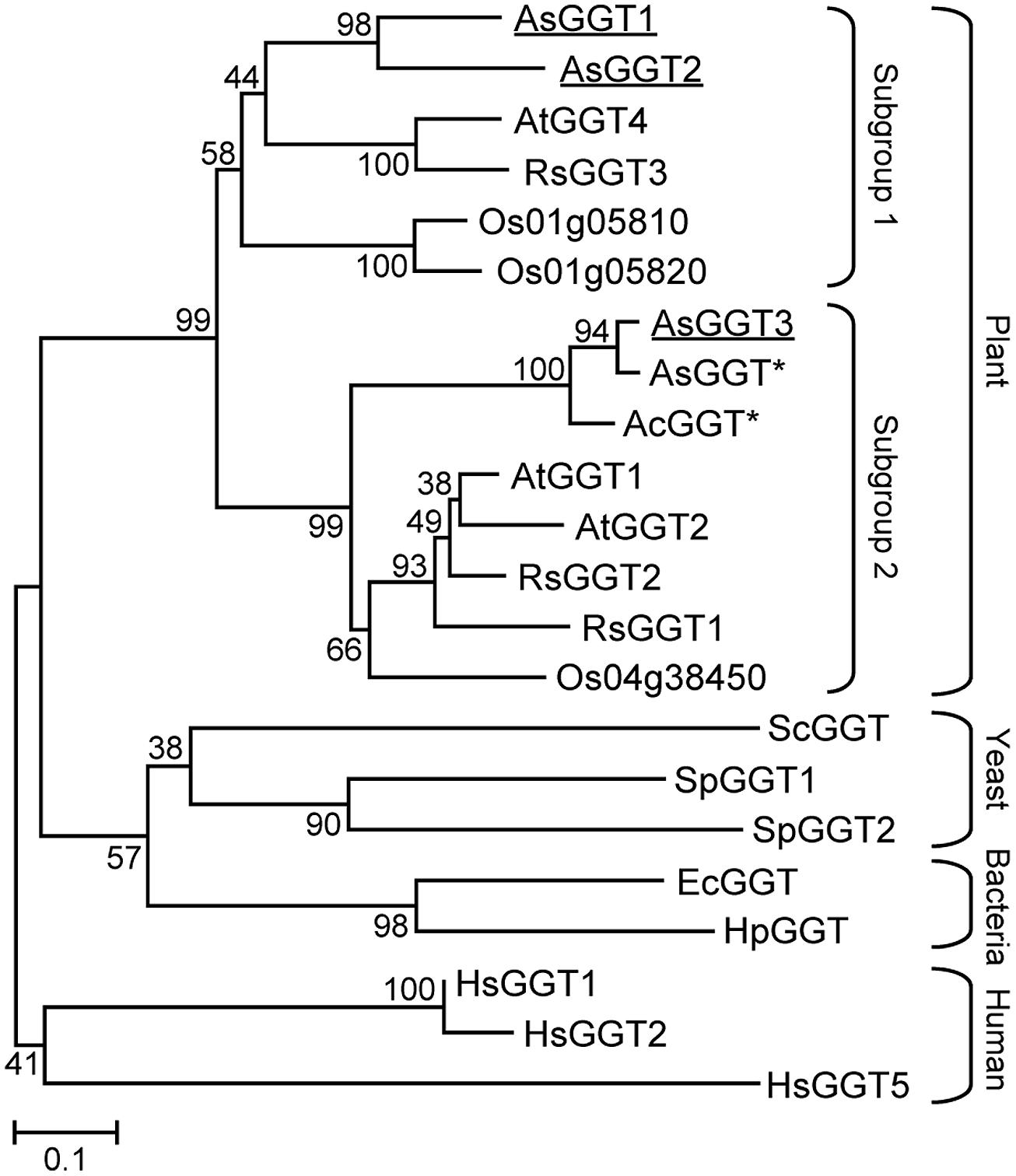
FIGURE 2. Phylogenetic tree for the γ-glutamyl transpeptidases. An unrooted tree was constructed using MEGA version 6 software based on the ClustalW multiple alignment. Bootstrap values (1000 replicates) are shown next to the branches. Plant GGTs are classified into two subgroups. Garlic AsGGT1, AsGGT2, and AsGGT3 analyzed in this study are underlined. Asterisks indicate partial amino acid sequences. Abbreviations for species are: Ac, Allium cepa; As, Allium sativum; At, Arabidopsis thaliana; Ec, Escherichia coli; Hp, Helicobacter pylori; Hs, Homo sapiens; Os, Oryza sativa; Rs, Raphanus sativus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe. The GenBank accession numbers for the sequences are shown in parentheses: AcGGT (AAL61611); AsGGT1 (LC008010); AsGGT2 (LC008011); AsGGT3 (LC008012); AtGGT1 (AEE87097); AtGGT2 (AEE87099); AtGGT4 (AEE85602); EcGGT (AAA23869); HpGGT (AAD08162); HsGGT1 (AAH25927); HsGGT2 (XP_006724458); HsGGT5 (AAH73999); Os01g05810 (BAD61112); Os01g05820 (BAD61113); Os04g38450 (CAD40892); RsGGT1 (BAC45233); RsGGT2 (BAC56855); RsGGT3 (BAD22536); ScGGT (DAA09609); SpGGT1 (AAN01227); SpGGT2 (AAQ57121).
Generally, in bacteria, yeast, plants, and mammals, GGT is a heteromeric protein consisting of large and small subunits, both of which are generated from a common inactive precursor polypeptide by autoprocessing (Penninckx and Jaspers, 1985; Storozhenko et al., 2002; Suzuki and Kumagai, 2002; Ikeda and Taniguchi, 2005; Shaw et al., 2005; Nakano et al., 2006; Boanca et al., 2007). Some plants, such as tomato, onion, and radish, are suggested to have GGT proteins consisting of a single polypeptide, although their sequence information remains unknown (Lancaster and Shaw, 1994; Martin and Slovin, 2000; Nakano et al., 2006). The deduced amino acid sequences of AsGGT1, AsGGT2, and AsGGT3 possessed the conserved threonine residue required for autocatalytic processing and the amino acid residues necessary for GGT activity, which were previously identified by biochemical and structural analyses of GGTs from humans and E. coli (Ikeda et al., 1993, 1995a,b; Okada et al., 2006).
In vitro Characterization of Recombinant AsGGT1, AsGGT2, and AsGGT3
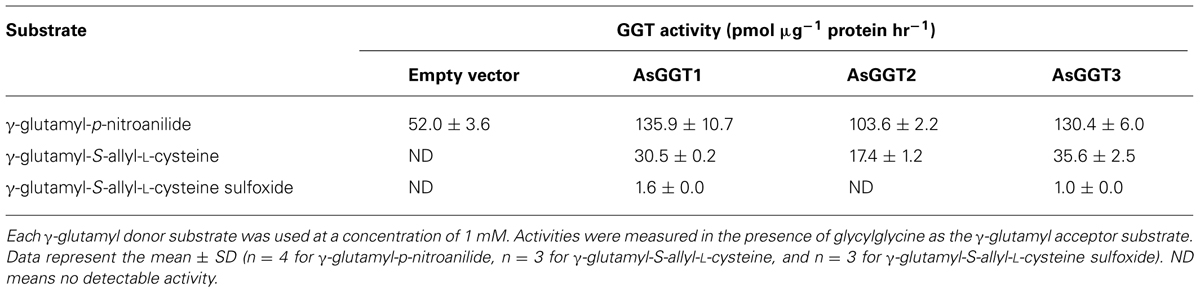
Recombinant proteins of AsGGT1, AsGGT2, and AsGGT3 were independently expressed in budding yeast, and the crude protein extracts were used for the in vitro enzymatic activity assays. To confirm whether recombinant proteins of AsGGT1, AsGGT2, and AsGGT3 were expressed as mature GGT enzymes in yeast cells, we first examined deglutamylation activities of these recombinant proteins by using the standard procedure that utilizes γ-glutamyl-p-nitroanilide, a common synthetic γ-glutamyl donor substrate for known GGTs (Orlowski and Meister, 1963). For a γ-glutamyl acceptor substrate, we used dipeptide glycylglycine. Crude protein extracts from control yeast carrying the empty vector converted γ-glutamyl-p-nitroanilide to p-nitroaniline (Table 1), showing that yeast endogenous GGT could utilize γ-glutamyl-p-nitroanilide as a γ-glutamyl donor substrate, as reported previously (Payne and Payne, 1984). The amounts of p-nitroaniline released from γ-glutamyl-p-nitroanilide in assays using crude protein extracts from yeast expressing AsGGT1, AsGGT2, and AsGGT3, respectively, were significantly higher than that in assays using crude protein extracts from control yeast (Table 1), indicating that the recombinant proteins of AsGGT1, AsGGT2, and AsGGT3 were successfully expressed and folded to form mature functional GGT proteins that can utilize γ-glutamyl-p-nitroanilide as a γ-glutamyl donor substrate in yeast cells.
Next, we examined the enzymatic activities of AsGGT1, AsGGT2, and AsGGT3 toward γ-glutamyl-S-allyl-L-cysteine and γ-glutamyl-S-allyl-L-cysteine sulfoxide, which are two possible biosynthetic intermediates in alliin biosynthesis, as potential γ-glutamyl donor substrates (Figures 3 and 4). Activities were measured in the presence of glycylglycine as a γ-glutamyl acceptor substrate. When γ-glutamyl-S-allyl-L-cysteine was used as a γ-glutamyl donor substrate, S-allyl-L-cysteine was not formed at a detectable level in assays using crude protein extracts from control yeast (Figure 3; Table 1), indicating that yeast endogenous GGT could not use γ-glutamyl-S-allyl-L-cysteine as a γ-glutamyl donor substrate. By contrast, considerable amounts of S-allyl-L-cysteine were detected in assays using crude protein extracts prepared from yeast cells expressing AsGGT1, AsGGT2, and AsGGT3 (Figure 3; Table 1), demonstrating that the recombinant proteins of AsGGT1, AsGGT2, and AsGGT3 can convert γ-glutamyl-S-allyl-L-cysteine to S-allyl-L-cysteine. The deglutamylation activities of AsGGT1, AsGGT2, and AsGGT3 toward γ-glutamyl-S-allyl-L-cysteine were decreased in the absence of glycylglycine (Figure 5), indicating that these garlic GGTs can catalyze transpeptidation more effectively than hydrolysis. In the presence of glycylglycine, the activities of AsGGT2 were higher with lower pH (Figure 5). Kinetic characterization of the recombinant AsGGT1, AsGGT2, and AsGGT3 exhibited typical Michaelis-Menten behavior, and the apparent Km values of AsGGT1, AsGGT2, and AsGGT3 for γ-glutamyl-S-allyl-L-cysteine were 86 μM, 1.1 mM, and 9.4 mM, respectively, in the presence of glycylglycine (Figure 6).
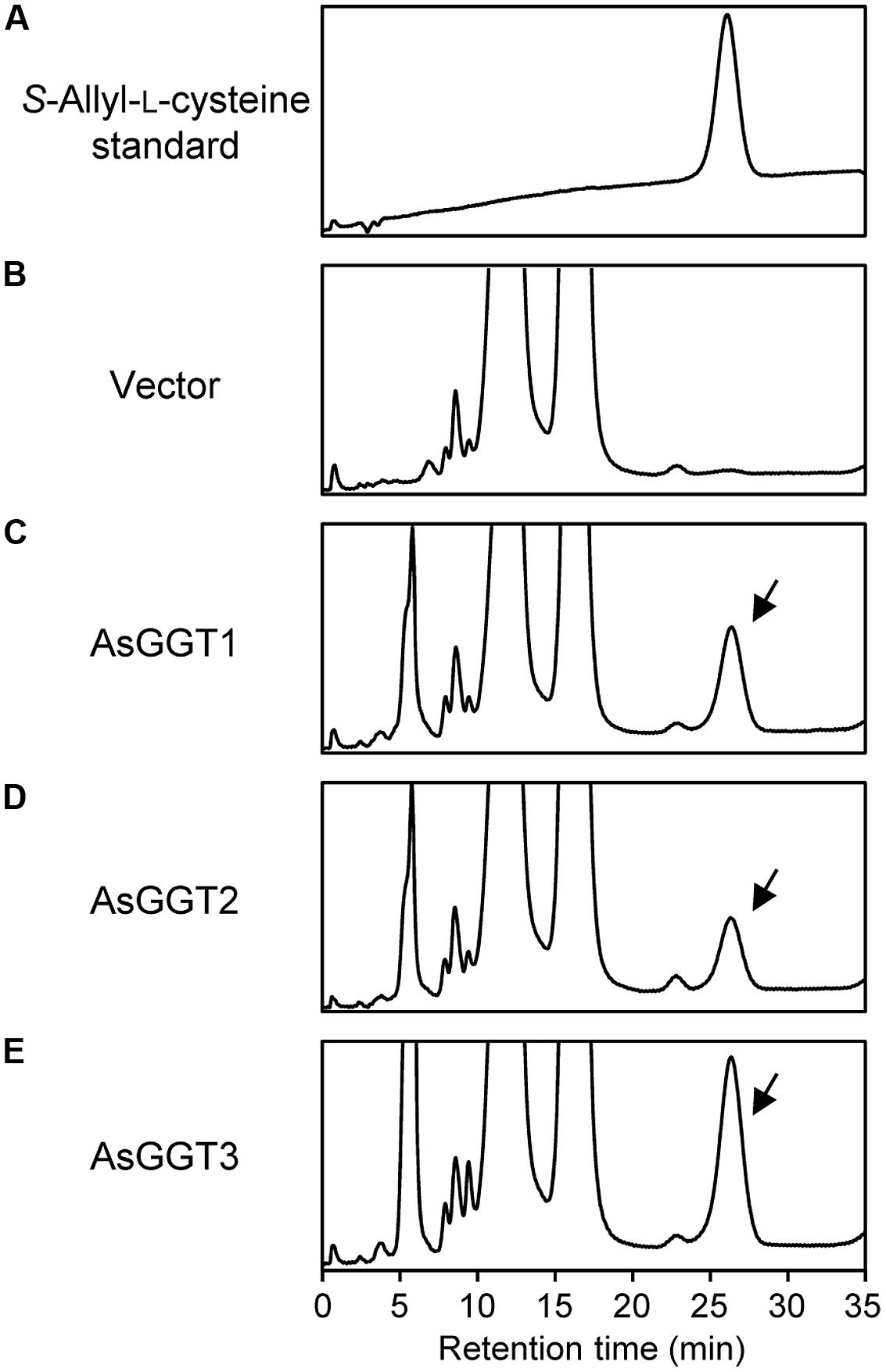
FIGURE 3. Deglutamylation activities of recombinant AsGGT1, AsGGT2, and AsGGT3 toward γ-glutamyl-S-allyl-L-cysteine. High-performance liquid chromatography (HPLC) elution profiles of the S-allyl-L-cysteine standard (A) and the reaction products from γ-glutamyl-S-allyl-L-cysteine by the crude protein extracts of yeast carrying empty vector (B) or yeast expressing AsGGT1 (C), AsGGT2 (D), and AsGGT3 (E) are shown. Arrows indicate peaks of S-allyl-L-cysteine in the reaction products.
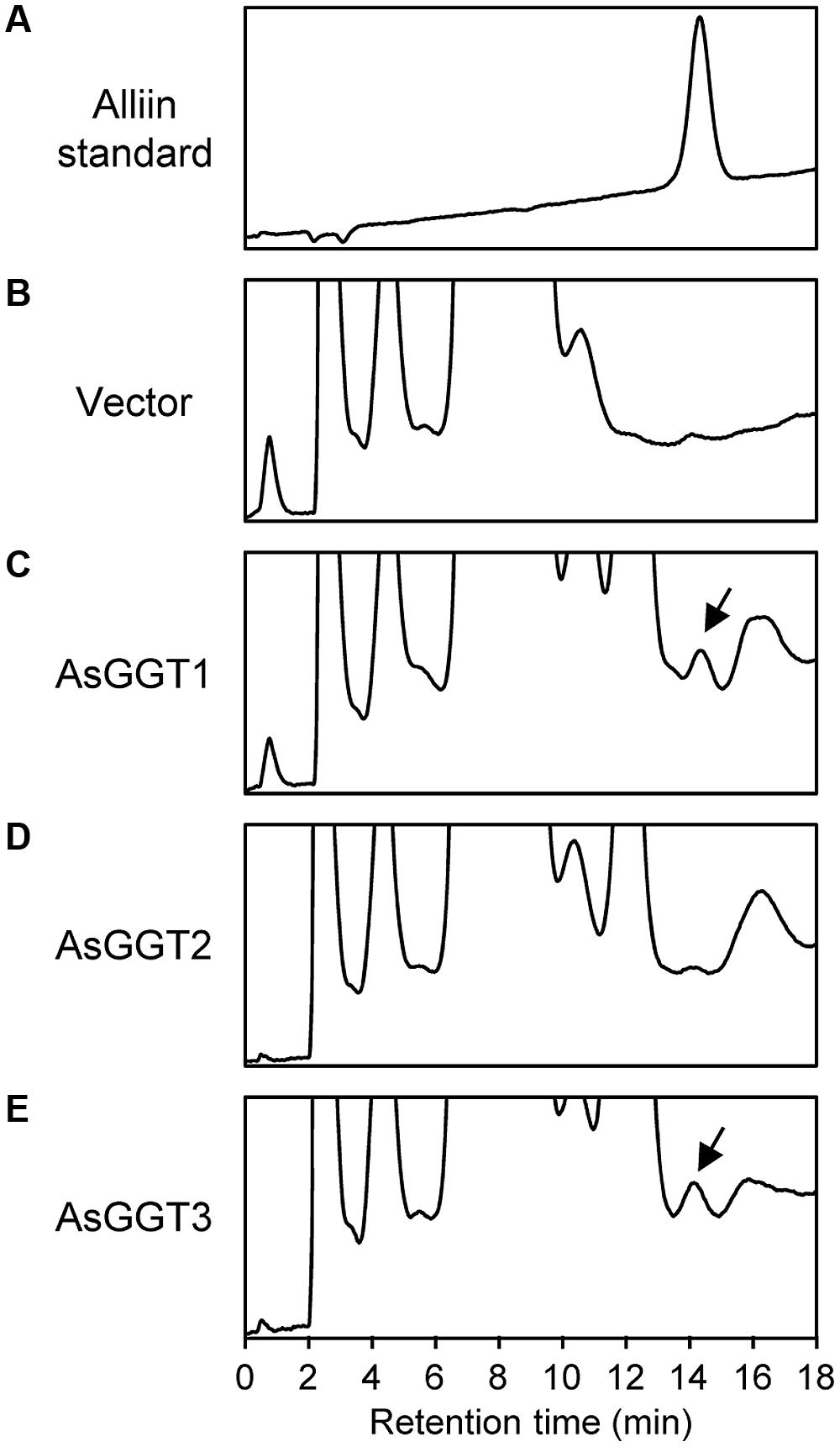
FIGURE 4. Deglutamylation activities of recombinant AsGGT1, AsGGT2, and AsGGT3 toward γ-glutamyl-S-allyl-L-cysteine sulfoxide. HPLC elution profiles of the alliin standard (A) and the reaction products from γ-glutamyl-S-allyl-L-cysteine sulfoxide by the crude protein extracts of yeast carrying empty vector (B) or yeast expressing AsGGT1 (C), AsGGT2 (D), and AsGGT3 (E) are shown. Arrows indicate peaks of alliin in the reaction products.
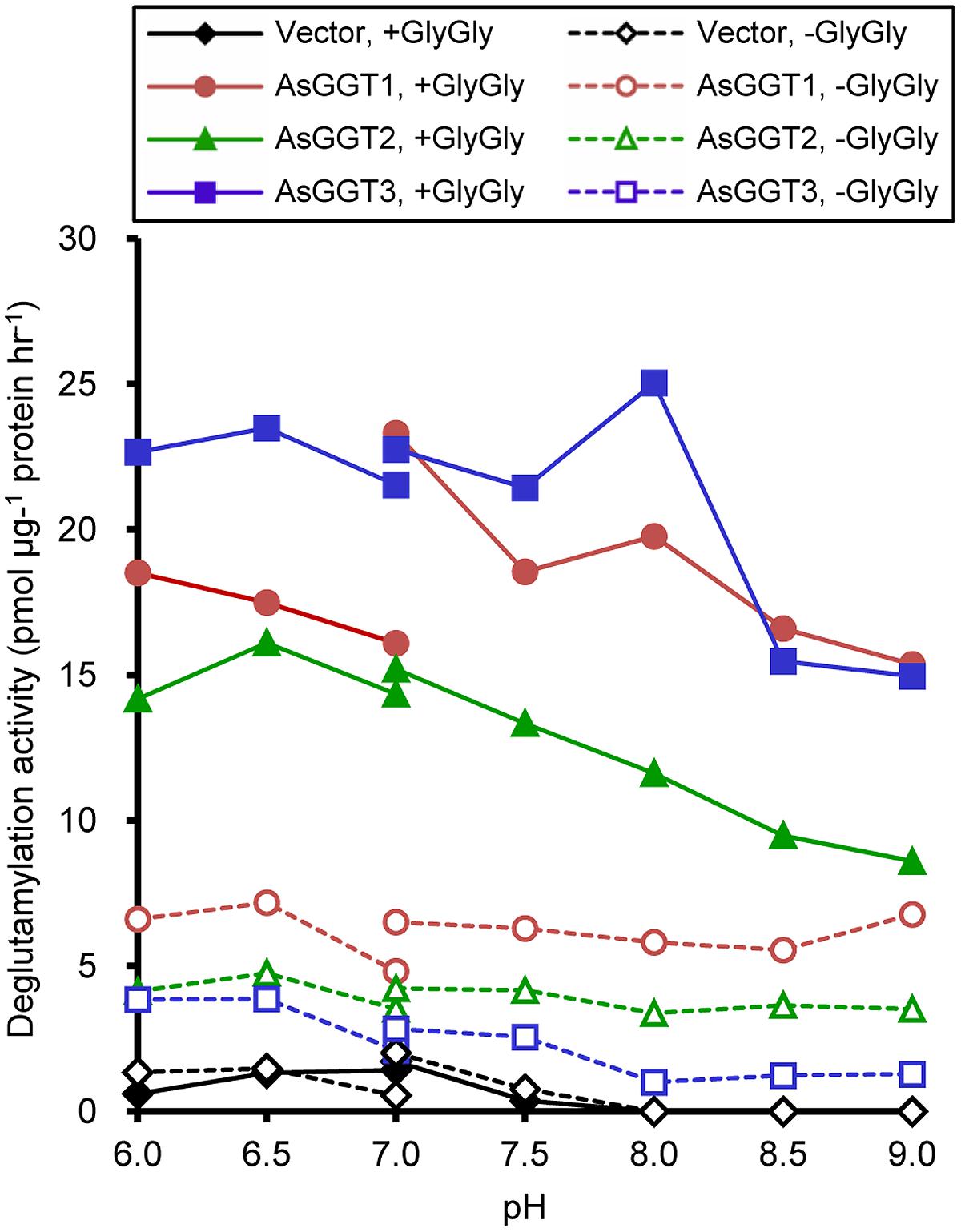
FIGURE 5. pH dependence of transpeptidation and hydrolysis by AsGGT1, AsGGT2, and AsGGT3. Production of S-allyl-L-cysteine from γ-glutamyl-S-allyl-L-cysteine was analyzed in the presence or absence of glycylglycine as the γ-glutamyl acceptor substrate. 2-(N-morpholino)ethanesulfonic acid buffer was used to cover the pH range from 6.0 to 7.0, and Tris-HCl buffer was used to cover the pH range from 7.0 to 9.0.
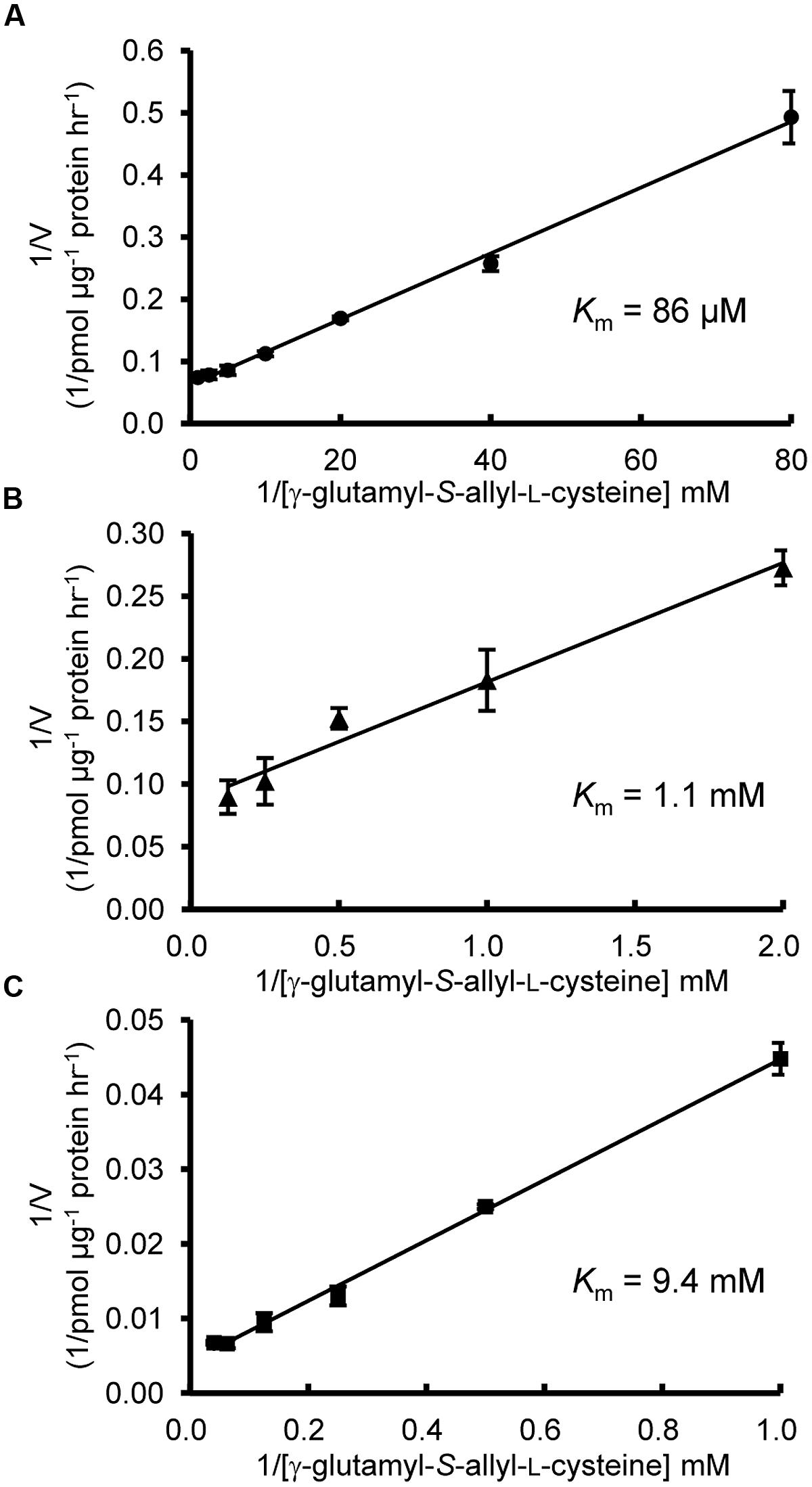
FIGURE 6. Kinetic characterization of AsGGT1, AsGGT2, and AsGGT3. Lineweaver-Burk plots between deglutamylation activity and γ-glutamyl-S-allyl-L-cysteine concentrations. Assays were carried out with γ-glutamyl-S-allyl-L-cysteine concentrations ranging from 12.5 to 1000 μM for AsGGT1 (A), 0.5–8 mM for AsGGT2 (B), and 1–25 mM for AsGGT3 (C). Each data point represents the mean ± SD (n = 3).
When γ-glutamyl-S-allyl-L-cysteine sulfoxide was used as a γ-glutamyl donor substrate, only small amounts of alliin were detected in assays using crude protein extracts from yeast expressing AsGGT1 or AsGGT3 (Figure 4; Table 1), suggesting the recombinant AsGGT1 and AsGGT3 exhibit a weak activity to deglutamylate γ-glutamyl-S-allyl-L-cysteine sulfoxide. However, under the conditions we examined, the activities of AsGGT1 and AsGGT3 to deglutamylate γ-glutamyl-S-allyl-L-cysteine sulfoxide were too weak to perform further enzymatic characterization. AsGGT2 exhibited no detectable deglutamylation activity toward γ-glutamyl-S-allyl-L-cysteine sulfoxide (Figure 4; Table 1). These results indicate that AsGGT1, AsGGT2, and AsGGT3 are the functional GGT proteins with a preference for γ-glutamyl-S-allyl-L-cysteine as a γ-glutamyl donor substrate over γ-glutamyl-S-allyl-L-cysteine sulfoxide.
Subcellular Localization of AsGGT1, AsGGT2, and AsGGT3
The probable subcellular localization of AsGGT1, AsGGT2, and AsGGT3 was computationally analyzed using the program TargetP v1.13 and WoLF PSORT4. Both programs predicted that these three garlic GGT proteins lack signal peptides for secretion or localization to cellular organelles.
To determine subcellular localization of AsGGT1, AsGGT2, and AsGGT3, the green fluorescent protein (GFP)-fusion constructs of AsGGT1, AsGGT2, and AsGGT3 were transiently expressed in onion epidermal cells under the control of the cauliflower mosaic virus 35S RNA promoter by using the particle bombardment method. Since N-terminal sequences of AsGGT1, AsGGT2, and AsGGT3 are long and may contain the signal sequence for secretion or targeting to cellular organelles, three types of fusion proteins, GFP C-terminally fused to the N-terminal 100-amino acid residues of GGT (AsGGT1N100-GFP, AsGGT2N100-GFP, and AsGGT3N100-GFP), GFP C-terminally fused to the N-terminal 300-amino acid residues of GGT (AsGGT1N300-GFP, AsGGT2N300-GFP, and AsGGT3N300-GFP), and GFP C-terminally fused to the full-length GGT protein (AsGGT1Full-GFP, AsGGT2Full-GFP, and AsGGT3Full-GFP), were analyzed. As a control of cytosolic localization, DsRed protein was simultaneously expressed with each GFP-fusion protein. The green fluorescent signals derived from AsGGT1N100-GFP and AsGGT3N100-GFP overlapped with the red fluorescence of DsRed (Figures 7A,C), suggesting that N-terminal regions of AsGGT1 and AsGGT3 have no signal sequence for secretion or targeting to cellular organelles. Similarly, the green fluorescent signals from AsGGT1N300-GFP, AsGGT1Full-GFP, AsGGT3N300-GFP, and AsGGT3Full-GFP were observed in the cytosol, although the fluorescent signal intensities were much weaker (data not shown). By contrast, the green fluorescence from AsGGT2N100-GFP was observed predominantly in the vacuole (Figure 7B), indicating that AsGGT2 has a signal sequence for targeting to the vacuole within its N-terminal 100 amino acid residues. When AsGGT2N300-GFP and AsGGT2Full-GFP were expressed, weak fluorescent signals were detected both in the vacuole and cytosol (data not shown).
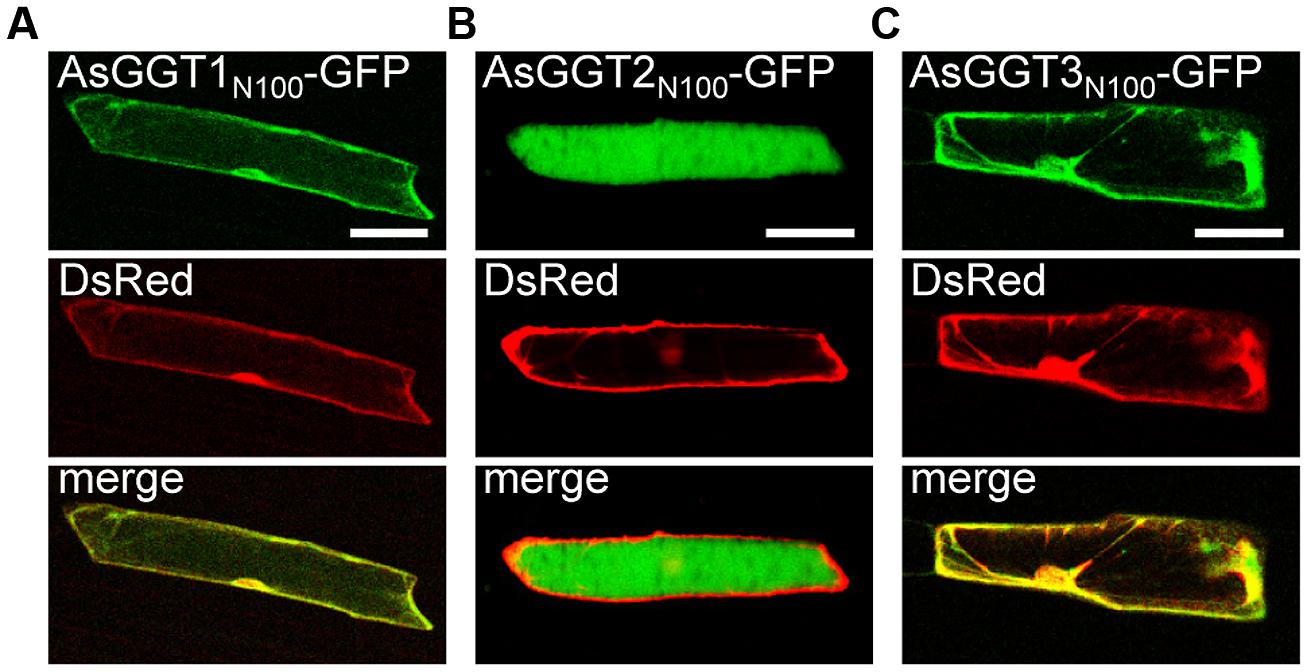
FIGURE 7. Transient expression of GFP fusion proteins of AsGGT1, AsGGT2, and AsGGT3 in onion epidermal cells. GFP fusion proteins, AsGGT1N100-GFP (A), AsGGT2N100-GFP (B), or AsGGT3N100-GFP (C), were transiently co-expressed with DsRed in onion epidermal cells. DsRed was used as a control for cytosolic localization. Bars = 100 μm.
Discussion
In this study, we identified three novel genes encoding GGTs, AsGGT1, AsGGT2, and AsGGT3, from garlic by utilizing their partial sequence information found in a publicly available EST database or by utilizing sequence information of conserved regions of known plant GGTs. The deduced amino acid sequences of AsGGT1, AsGGT2, and AsGGT3 contained threonine residues required for autoprocessing and the residues required for GGT activity (Ikeda et al., 1993, 1995a,b; Okada et al., 2006), suggesting that AsGGT1, AsGGT2, and AsGGT3 all encode functional GGT proteins. The cDNA sequence of AsGGT3 and the previously identified partial cDNA sequence of garlic AsGGT (Cho et al., 2012) were almost identical in their overlapping region, suggesting that these two cDNAs were derived from a single gene encoding GGT. The high sequence homology between garlic AsGGT3 and onion AcGGT (Shaw et al., 2005) may indicate that they are orthologs. By contrast, AsGGT1 and AsGGT2 showed relatively low sequence similarity with garlic AsGGT (Cho et al., 2012) and onion AcGGT (Shaw et al., 2005). Phylogenetic analysis revealed that AsGGT3 belongs to a subgroup different from that containing AsGGT1 and AsGGT2 (Figure 2), suggesting that the biochemical characteristics of AsGGT3 might be somewhat different from those of AsGGT1 and AsGGT2.
The deglutamylation activities of AsGGT1, AsGGT2, and AsGGT3 toward alliin biosynthetic intermediates were demonstrated by in vitro biochemical assays, using recombinant proteins expressed in yeast. In the hypothetical alliin biosynthetic pathway, two different routes from the intermediate γ-glutamyl-S-allyl-L-cysteine to alliin are possible, according to differences in the order of deglutamylation and S-oxygenation reactions (Figure 1): a potential route via deglutamylation of γ-glutamyl-S-allyl-L-cysteine to yield S-allyl-L-cysteine that is further S-oxygenated to alliin, and an alternative route via S-oxygenation of γ-glutamyl-S-allyl-L-cysteine to form γ-glutamyl-S-allyl-L-cysteine sulfoxide that is further deglutamylated to yield alliin. Our results demonstrated that AsGGT1, AsGGT2, and AsGGT3 actively deglutamylate γ-glutamyl-S-allyl-L-cysteine, whereas these GGTs have almost no deglutamylation activity toward γ-glutamyl-S-allyl-L-cysteine sulfoxide (Figures 3 and 4; Table 1). It can be speculated that the intermediate γ-glutamyl-S-allyl-L-cysteine is mainly deglutamylated prior to being S-oxygenated in alliin biosynthesis in garlic. This hypothesis is also supported by our recent study on flavin-dependent S-oxygenase, which preferably utilizes S-allyl-L-cysteine, rather than γ-glutamyl-S-allyl-L-cysteine, as the substrate (unpublished results). The presence of dipeptide glycylglycine as a γ-glutamyl acceptor increased the deglutamylation activities of AsGGT1, AsGGT2, and AsGGT3 (Figure 5), showing that these GGTs catalyze transpeptiation more efficiently than hydrolysis, as in GGTs from Arabidopsis and onion (Storozhenko et al., 2002; Shaw et al., 2005).
There are two characteristic differences among the AsGGT1, AsGGT2, and AsGGT3 proteins. One is in their affinity for γ-glutamyl-S-allyl-L-cysteine (Figure 6). The apparent Km values of AsGGT1 and AsGGT2 for γ-glutamyl-S-allyl-L-cysteine determined in this study (86 μM and 1.1 mM, respectively) are lower than or comparable to those of partially purified onion GGT for γ-glutamyl-S-propenyl-L-cysteine (Km = 1.68 mM) and for γ-glutamyl-S-methyl-L-cysteine (Km = 0.55 mM; Lancaster and Shaw, 1994). By contrast, AsGGT3 exhibited a relatively low affinity for γ-glutamyl-S-allyl-L-cysteine (Km = 9.4 mM). This is in agreement with the results of a previous study that onion AcGGT, which shares high sequence homology with AsGGT3, could not utilize γ-glutamyl-trans-S-1-propenyl-L-cysteine sulfoxide (a major γ-glutamylated biosynthetic intermediate in onion) as a good γ-glutamyl donor substrate (Shaw et al., 2005). In garlic, the content of alliin is increased dramatically before and during the maturation of bulbs (Ueda et al., 1991). Alliin is largely found in leaves before the formation of bulbs and in the initial stage of bulb maturation, whereas it is found predominantly in bulbs in the later stage of bulb formation (Ueda et al., 1991; Koch and Lawson, 1996). It is suggested that leaves of garlic actively biosynthesize alliin before the formation of bulbs and in the initial stage of bulb maturation. However, in leaves at the same stages, γ-glutamyl-S-allyl-L-cysteine is present in trace levels (0.01 mg g-1 fresh weight; Matsuura et al., 1996). The concentration of γ-glutamyl-S-allyl-L-cysteine in cells of these tissues is calculated to be approximately 38 μM, when the content of water in tissues is estimated to be 90%. By contrast, the content of γ-glutamyl-S-allyl-L-cysteine in mature bulbs is ∼5 mg g-1 fresh weight (Matsuura et al., 1996; Ichikawa et al., 2006a,b), and the concentration of γ-glutamyl-S-allyl-L-cysteine in cells of bulbs is calculated to be 26 mM, when the content of water in tissues is estimated to be 65%. The highly accumulated γ-glutamyl-S-allyl-L-cysteine in bulbs is stored during dormancy of bulbs at -3°C, while it is rapidly converted to alliin when bulb dormancy is broken at 4°C (Ichikawa et al., 2006a). Based on these observations, we hypothesize that AsGGT1 and AsGGT2, which exhibit high-affinity for γ-glutamyl-S-allyl-L-cysteine, would contribute to the biosynthesis of alliin in leaves during the formation and maturation of bulbs, while AsGGT3 may contribute to alliin biosynthesis in bulbs during dormancy-breaking or, alternatively, the main in vivo function of AsGGT3 may not be the deglutamylation of γ-glutamyl-S-allyl-L-cysteine. The other major difference observed among AsGGT1, AsGGT2, and AsGGT3 is in their subcellular localization. Transient expression analyses of GFP-fused AsGGT2 proteins in onion cells suggested that AsGGT2 is predominantly localized in the vacuole in vivo. In contrast to the almost exclusive localization of AsGGT2N100-GFP in the vacuole (Figure 7B), the green fluorescence signals from AsGGT2N300-GFP and AsGGT2Full-GFP were detected both in the vacuole and cytosol (data not shown), suggesting that a part of AsGGT2N300-GFP and AsGGT2Full-GFP polypeptides was not properly processed and/or assembled and thus was not sorted to the vacuole in the heterologous expression system we used. Alternatively, AsGGT2 may localize both in the vacuole and cytosol in garlic cells. The signal sequence for targeting to the vacuole of AsGGT2 is located within its N-terminal 100 amino acids, as in Arabidopsis AtGGT4 (Grzam et al., 2007; Ohkama-Ohtsu et al., 2007b). To date, several sequence motifs for vacuolar targeting have been identified from plants (Xiang et al., 2013). However, we could not identify the potential motif for vacuolar targeting that is conserved between AsGGT2 and AtGGT4. Future studies are needed to determine the sequence motif and the mechanism for their targeting to the vacuole. Consistent with vacuolar localization of AsGGT2, the deglutamylation activities of AsGGT2 toward γ-glutamyl-S-allyl-L-cysteine were increased under weakly acidic conditions (Figure 5). AsGGT2 is suggested to contribute alliin biosynthesis mainly in the vacuole. In addition, AsGGT2 may function in the breakdown of glutathione S-conjugates in the vacuole in a similar manner as Arabidopsis AtGGT4 (Grzam et al., 2007; Ohkama-Ohtsu et al., 2007b). In contrast, GFP-fusion proteins of AsGGT1 and AsGGT3 were retained in the cytosol (Figures 7A,C), suggesting that AsGGT1 and AsGGT3 have no apparent signal sequence for targeting to the cellular organelles in their N-terminal peptides. To the best of our knowledge, there have been no reports of GGT proteins localizing in the cytosol. Future investigations will reveal whether AsGGT1 and AsGGT3 are new types of GGT proteins that localize and function in the cytosol in vivo or not. To date, the subcellular distribution of alliin biosynthetic intermediates and enzymes in garlic remains largely unclear, although the previous cell fractionation experiment suggested that γ-glutamyl peptides and S-alk(en)yl-L-cysteine sulfoxides are mainly located in the cytosol in onion (Lancaster et al., 1989). Our results suggest that AsGGT1, AsGGT2, and AsGGT3 contribute differently to alliin biosynthesis, according to differences in their kinetic properties and localization patterns. Recently, five γ-glutamyl peptidases (GGPs), which have similar catalytic functions but no sequence homology with GGTs, were identified from Arabidopsis. Among these, GGP1 and GGP3 were shown to be cytosolic proteins that play major roles in the removal of γ-glutamyl groups from glutathione S-conjugates in the biosynthesis of glucosinolates and camalexins (Geu-Flores et al., 2009, 2011). Although identification of GGPs from garlic has not been reported to date, it is likely that GGPs exist and function in the deglutamylation reaction in alliin biosynthesis, perhaps together with GGTs.
In the present study, we succeeded in identifying three garlic GGTs, AsGGT1, AsGGT2, and AsGGT3, that can deglutamylate an alliin biosynthetic intermediate, γ-glutamyl-S-allyl-L-cysteine. Future investigations of the in vivo functions of AsGGT1, AsGGT2, and AsGGT3 will provide a better understanding of the molecular mechanisms underlying the biosynthesis of alliin in garlic, which can be applied to future metabolic engineering of plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Toshiaki Mitsui (Niigata University) for providing the pDsRed vector. This work was supported in part by JSPS KAKENHI Grant Number 23770036 and 26460118 (to Naoko Yoshimoto), and Grant Number 22108008 (to Kazuki Saito) of Japan.
Footnotes
- ^ http://garlicdb.kribb.re.kr
- ^ http://garlicdb.kribb.re.kr
- ^ http://www.cbs.dtu.dk/services/TargetP/
- ^ http://wolfpsort.org/
References
Anderson, M. E., Allison, R. D., and Meister, A. (1982). Interconversion of leukotrienes catalyzed by purified γ-glutamyl transpeptidase: concomitant formation of leukotriene D4 and γ-glutamyl amino acids. Proc. Natl. Acad. Sci. U.S.A. 79, 1088–1091. doi: 10.1073/pnas.79.4.1088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boanca, G., Sand, A., Okada, T., Suzuki, H., Kumagai, H., Fukuyama, K.,et al. (2007). Autoprocessing of Helicobacter pylori γ-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad. J. Biol. Chem. 282, 534–541. doi: 10.1074/jbc.M607694200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiu, W. L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. doi: 10.1016/S0960-9822(02)00483-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cho, J., Park, M., Choi, D., and Lee, S. K. (2012). Cloning and expression of γ-glutamyl transpeptidase and its relationship to greening in crushed garlic (Allium sativum) cloves. J. Sci. Food Agric. 92, 253–257. doi: 10.1002/jsfa.4610
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dominici, S., Paolicchi, A., Corti, A., Maellaro, E., and Pompella, A. (2005). Prooxidant reactions promoted by soluble and cell-bound γ-glutamyltransferase activity. Methods Enzymol. 401, 484–501. doi: 10.1016/S0076-6879(05)01029-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gamborg, O. L., Miller, R. A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geu-Flores, F., Møldrup, M. E., Böttcher, C., Olsen, C. E., Scheel, D., and Halkier, B. A. (2011). Cytosolic γ-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23, 2456–2469. doi: 10.1105/tpc.111.083998
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geu-Flores, F., Nielsen, M. T., Nafisi, M., Møldrup, M. E., Olsen, C. E., Motawia, M. S.,et al. (2009). Glucosinolate engineering identifies a γ-glutamyl peptidase. Nat. Chem. Biol. 5, 575–577. doi: 10.1038/nchembio.185
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gietz, D., Jean, A. S., Woods, D. A., and Schiestl, R. H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. doi: 10.1093/nar/20.6.1425
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grzam, A., Martin, M. N., Hell, R., and Meyer, A. J. (2007). γ-Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S-conjugates in Arabidopsis. FEBS Lett. 581, 3131–3138. doi: 10.1016/j.febslet.2007.05.071
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ichikawa, M., Ide, N., and Ono, K. (2006a). Changes in organosulfur compounds in garlic cloves during storage. J. Agric. Food Chem. 54, 4849–4854. doi: 10.1021/jf060083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ichikawa, M., Ide, N., Yoshida, J., Yamaguchi, H., and Ono, K. (2006b). Determination of seven organosulfur compounds in garlic by high-performance liquid chromatography. J. Agric. Food Chem. 54, 1535–1540. doi: 10.1021/jf051742k
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iciek, M., Kwiecień, I., and Włodek, L. (2009). Biological properties of garlic and garlic-derived organosulfur compounds. Environ. Mol. Mutagen. 50, 247–265. doi: 10.1002/em.20474
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ikeda, Y., Fujii, J., Anderson, M. E., Taniguchi, N., and Meister, A. (1995a). Involvement of Ser-451 and Ser-452 in the catalysis of human γ-glutamyl transpeptidase. J. Biol. Chem. 270, 22223–22228. doi: 10.1074/jbc.270.38.22223
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ikeda, Y., Fujii, J., Taniguchi, N., and Meister, A. (1995b). Human γ-glutamyl transpeptidase mutants involving conserved aspartate residues and the unique cysteine residue of the light subunit. J. Biol. Chem. 270, 12471–12475. doi: 10.1074/jbc.270.21.12471
Ikeda, Y., Fujii, J., and Taniguchi, N. (1993). Significance of Arg-107 and Glu-108 in the catalytic mechanism of human γ-glutamyl transpeptidase. J. Biol. Chem. 268, 3980–3985.
Ikeda, Y., and Taniguchi, N. (2005). Gene expression of γ-glutamyltranspeptidase. Methods Enzymol. 401, 408-425. doi: 10.1016/S0076-6879(05)01025-6
Jones, M. G., Hughes, J., Tregova, A., Milne, J., Tomsett, A. B., and Collin, H. A. (2004). Biosynthesis of the flavor precursors of onion and garlic. J. Exp. Bot. 55, 1903–1918. doi: 10.1093/jxb/erh138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, D.-W., Jung, T.-S., Nam, S.-H., Kwon, H.-R., Kim, A., Chae, S.-H.,et al. (2009). GarlicESTdb: an online database and mining tool for garlic EST sequences. BMC Plant Biol. 9:61. doi: 10.1186/1471-2229-9-61
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kitajima, A., Asatsuma, S., Okada, H., Hamada, Y., Kaneko, K., Nanjo, Y.,et al. (2009). The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21, 2844–2858. doi: 10.1105/tpc.109.068288
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koch, H. P., and Lawson, L. D. (1996). Garlic: The Science and Therapeutic Application of Allium sativum L. and Related Species, 2nd Edn. Baltimore, MD: Williams & Wilkins.
Kubec, R., and Dadáková, E. (2008). Quantitative determination of S-alk(en)ylcysteine-S-oxides by micellar electrokinetic capillary chromatography. J. Chromatogr. A 1212, 154–157. doi: 10.1016/j.chroma.2008.10.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kubec, R., Svobodová, M., and Velíšek, J. (1999). Gas chromatographic determination of S-alk(en)ylcysteine sulfoxides. J. Chromatogr. A 862, 85–94. doi: 10.1016/S0021-9673(99)00902-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lancaster, J. E., Reynolds, P. H. S., Shaw, M. L., Dommisse, E. M., and Munro, J. (1989). Intra-cellular localization of the biosynthetic pathway to flavor precursors in onion. Phytochemistry 28, 461–464. doi: 10.1016/0031-9422(89) 80032-9
Lancaster, J. E., and Shaw, M. L. (1989). γ-Glutamyl peptides in the biosynthesis of S-alk(en)yl-L-cysteine sulphoxides (flavor precursors) in Allium. Phytochemistry 28, 455–460. doi: 10.1016/0031-9422(89)80031-7
Lancaster, J. E., and Shaw, M. L. (1991). Metabolism of γ-glutamyl peptides during development, storage and sprouting of onion bulbs. Phytochemistry 30, 2857–2859. doi: 10.1016/S0031-9422(00)98212-8
Lancaster, J. E., and Shaw, M. L. (1994). Characterization of purified γ-glutamyl transpeptidase in onions: evidence for in vivo role as a peptidase. Phytochemistry 36, 1351–1358. doi: 10.1016/S0031-9422(00)89723-X
Martin, M. N., Saladores, P. H., Lambert, E., Hudson, A. O., and Leustek, T. (2007). Localization of members of the γ-glutamyl transpeptidase family identified sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol. 144, 1715–1732. doi: 10.1104/pp.106.094409
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin, M. N., and Slovin, J. P. (2000). Purified γ-glutamyl transpeptidases from tomato exhibit high affinity for glutathione and glutathione S-conjugates. Plant Physiol. 122, 1417–1426. doi: 10.1104/pp.122.4.1417
Matsuura, H., Inagaki, M., Maeshige, K., Ide, N., Kajimura, Y., and Itakura, Y. (1996). Changes in contents of γ-glutamyl peptides and fructan during growth of Allium sativum. Plant Med. 62, 70–71. doi: 10.1055/s-2006-957805
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mehdi, K., and Penninckx, M. J. (1997). An important role for glutathione and γ-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology 143, 1885–1889. doi: 10.1099/00221287-143-6-1885
Nakano, Y., Okawa, S., Yamauchi, T., Koizumi, Y., and Sekiya, J. (2006). Purification and properties of soluble and bound γ-glutamyltransferases from radish cotyledon. Biosci. Biotechnol. Biochem. 70, 369–376. doi: 10.1271/bbb.70.369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ohkama-Ohtsu, N., Radwan, S., Peterson, A., Zhao, P., Badr, A. F., Xiang, C.,et al. (2007a). Characterization of the extracellular γ-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J. 49, 865–877. doi: 10.1111/j.1365-313X.2006.03004.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ohkama-Ohtsu, N., Zhao, P., Xiang, C., and Oliver, D. J. (2007b). Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 49, 878–888. doi: 10.1111/j.1365-313X.2006.03005.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okada, T., Suzuki, H., Wada, K., Kumagai, H., and Fukuyama, K. (2006). Crystal structures of γ-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc. Natl. Acad. Sci. U.S.A. 103, 6471–6476. doi: 10.1073/pnas.0511020103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orlowski, M., and Meister, A. (1963). γ-Glutamyl-p-nitroanilide: a new convenient substrate for determination and study of L- and D-γ-glutamyltranspeptidase activities. Biochim. Biophys. Acta 73, 679–681. doi: 10.1016/0926-6569(63)90197-4
Payne, G. M., and Payne, J. W. (1984). γ-Glutamyltransferase is not involved in the bulk uptake of amino acids, peptides or γ-glutamyl-amino acids in yeast (Saccharomyces cerevisiae). Biochem. J. 218, 147–155.
Penninckx, M. J., and Jaspers, C. J. (1985). Molecular and kinetic properties of purified γ-glutamyl transpeptidase from yeast (Saccharomyces cerevisiae). Phytochemistry 24, 1913–1918. doi: 10.1016/S0031-9422(00)83091-5
Rose, P., Whiteman, M., Moore, P. K., and Zhu, Y. Z. (2005). Bioactive S-alk(en)yl cysteine sulfoxide metabolites in the genus Allium: the chemistry of potential therapeutic agents. Nat. Prod. Rep. 22, 351–368. doi: 10.1039/B417639C
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd Edn. New York: Cold Spring Harbor Laboratory Press.
Shaw, M. L., Pither-Joyce, M. D., and McCallum, J. A. (2005). Purification and cloning of a γ-glutamyl transpeptidase from onion (Allium cepa). Phytochemistry 66, 515–522. doi: 10.1016/j.phytochem.2005.01.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 4–21. doi: 10.1016/0076-6879(91)94004-V
Storozhenko, S., Belles-Boix, E., Babiychuk, E., Hérouart, D., Davey, M. W.,Slooten, L.,et al. (2002). γ-Glutamyl transpeptidase in transgenic tobacco plants. Cellular localization, processing, and biochemical properties. Plant Physiol. 128, 1109–1119. doi: 10.1104/pp.010887
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Su, T., Xu, J., Li, Y., Lei, L., Zhao, L., Yang, H.,et al. (2011). Glutathione-indole-3-acetonitrile in required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23, 364–380. doi: 10.1105/tpc.110.079145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suzuki, H., Hashimoto, W., and Kumagai, H. (1999). Glutathione metabolism in Escherichia coli. J. Mol. Catal. B Enzymat. 6, 175–184. doi: 10.1016/S1381-1177(98)00116-7
Suzuki, H., and Kumagai, H. (2002). Autocatalytic processing of γ-glutamyltranspeptidase. J. Biol. Chem. 277, 43536–43543. doi: 10.1074/jbc.M207680200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suzuki, T., Sugii, M., and Kakimoto, T. (1962). Incorporation of L-valine-[14C] into S-(2-carboxypropyl)glutathione and S-(2-carboxypropyl)cysteine in garlic. Chem. Pharm. Bull. 10, 328–331. doi: 10.1248/cpb.10.328
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tate, S. S., and Meister, A. (1981). γ-Glutamyl transpeptidase: catalytic, structural and functional aspects. Mol. Cell. Biochem. 39, 357–368. doi: 10.1007/BF00232585
Turnbull, A., Galpin, I. J., and Collin, H. A. (1980). Comparison of the onion plant (Allium cepa) and onion tissue culture. III. Feeding of 14C labeled precursors of the flavor precursor compounds. New Phytol. 85, 483–487. doi: 10.1111/j.1469-8137.1980.tb00762.x
Ueda, Y., Kawajiri, H., Miyamura, N., and Miyajima, R. (1991). Content of some sulfur-containing components and free amino acids in various strains of garlic. Nippon Shokuhin Kogyo Gakkaishi 38, 429–434. doi: 10.3136/nskkk1962. 38.429
Xiang, L., Etxeberria, E., and Van den Ende, W. (2013). Vacuolar protein sorting mechanisms in plants. FEBS J. 280, 979–993. doi: 10.1111/febs.12092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, T.-H., Wu, C.-M., Rosen, R. T., Hartman, T. G., and Ho, C.-T. (1994). Volatile compounds generated from thermal degradation of alliin and deoxyalliin in an aqueous solution. J. Agric. Food Chem. 42, 146–153. doi: 10.1021/jf0003 7a026
Keywords: γ-glutamyl transpeptidase, deglutamylation, alliin, secondary metabolism, garlic
Citation: Yoshimoto N, Yabe A, Sugino Y, Murakami S, Sai-ngam N, Sumi S-i, Tsuneyoshi T and Saito K (2015) Garlic γ-glutamyl transpeptidases that catalyze deglutamylation of biosynthetic intermediate of alliin. Front. Plant Sci. 5:758. doi: 10.3389/fpls.2014.00758
Received: 31 October 2014; Accepted: 09 December 2014;
Published online: 08 January 2015.
Edited by:
Stanislav Kopriva, University of Cologne, GermanyReviewed by:
Naoko Ohtsu, Tokyo University of Agriculture and Technology, JapanPete Rose, University of Lincoln, UK
Copyright © 2015 Yoshimoto, Yabe, Sugino, Murakami, Sai-ngam, Sumi, Tsuneyoshi and Saito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuki Saito, Graduate School of Pharmaceutical Sciences, Chiba University, Inohana 1-8-1, Chuo-ku, Chiba 260-8675, Japan e-mail: ksaito@faculty.chiba-u.jp
 Naoko Yoshimoto
Naoko Yoshimoto Ayami Yabe1
Ayami Yabe1 Tadamitsu Tsuneyoshi
Tadamitsu Tsuneyoshi Kazuki Saito
Kazuki Saito