- Department of Microbiology and Cell Science, University of Florida, Gainesville, FL, USA
Salicylic acid (SA) is a key defense signal molecule against biotrophic and hemibiotrophic pathogens in plants, but how SA is synthesized in plant cells still remains elusive. Identification of new components involved in pathogen-induced SA accumulation would help address this question. To this end, we performed a large-scale genetic screen for mutants with altered SA accumulation during pathogen infection in Arabidopsis using a bacterial biosensor Acinetobacter sp. ADPWH_lux-based SA quantification method. A total of 35,000 M2 plants in the npr1-3 mutant background have been individually analyzed for the bacterial pathogen Pseudomonas syringae pv. maculicola (Psm) ES4326-induced SA accumulation. Among the mutants isolated, 19 had SA levels lower than npr1 (sln) and two exhibited increased SA accumulation in npr1 (isn). Complementation tests revealed that seven of the sln mutants are new alleles of eds5/sid1, two are sid2/eds16 alleles, one is allelic to pad4, and the remaining seven sln and two isn mutants are new non-allelic SA accumulation mutants. Interestingly, a large group of mutants (in the npr1-3 background), in which Psm ES4326-induced SA levels were similar to those in the wild-type Columbia plants, were identified, suggesting that the signaling network fine-tuning pathogen-induced SA accumulation is complex. We further characterized the sln1 single mutant and found that Psm ES4326-induced defense responses were compromised in this mutant. These defense response defects could be rescued by exogenous SA, suggesting that SLN1 functions upstream of SA. The sln1 mutation was mapped to a region on the north arm of chromosome I, which contains no known genes regulating pathogen-induced SA accumulation, indicating that SLN1 likely encodes a new regulator of SA biosynthesis. Thus, the new sln and isn mutants identified in this genetic screen are valuable for dissecting the molecular mechanisms underlying pathogen-induced SA accumulation in plants.
Introduction
As sessile organisms, plants are under constant attack from diverse microbes including bacteria, fungi, oomycetes, and viruses. To ward off pathogens, plants activate their immune system to mount multiple defense responses, which are similar to animal innate immunity (Jones and Dangl, 2006). Recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors results in PAMP-triggered immunity (PTI). To achieve successful colonization, adapted pathogens can deliver effector molecules directly into the plant cells to suppress PTI, resulting in effector-triggered susceptibility (ETS) (Jones and Dangl, 2006). On the other hand, plants have evolved resistance (R) proteins to detect the presence of certain pathogen effector molecules, inducing effector-triggered immunity (ETI). Activation of PTI or ETI leads to generation of mobile signals, which induce a long-lasting broad-spectrum immune response known as systemic acquired resistance (SAR) (Durrant and Dong, 2004).
The phytohormone salicylic acid (SA) plays an essential role in these defense response pathways (Vlot et al., 2009). Exogenous application of SA or its analogs induces expression of defense genes including PR (pathogenesis-related) genes and disease resistance (White, 1979; Dong, 2004), whereas transgenic plants carrying the bacterial NahG gene, which encodes an SA hydroxylase, are hypersusceptible to pathogen infection and fail to develop SAR (Gaffney et al., 1993; Delaney et al., 1994; Lawton et al., 1995). Furthermore, Arabidopsis mutants with impaired SA biosynthesis during pathogen infection, such as sid2 (salicylic acid induction-deficient2) (Nawrath and Métraux, 1999; Wildermuth et al., 2001), eds5 (enhanced disease susceptibility5) (Nawrath and Métraux, 1999; Nawrath et al., 2002), and pad4 (phytoalexin deficient4) (Zhou et al., 1998; Jirage et al., 1999), show compromised defense responses. In contrast, mutants with elevated levels of SA, such as acd (accelerated cell death) (Greenberg et al., 1994; Rate et al., 1999), cpr (constitutive expressor of PR genes) (Bowling et al., 1997; Clarke et al., 1998), and ssi (suppressor of salicylate insensitivity of npr1-5) (Shah et al., 1999, 2001), display constitutive expression of PR genes and SAR.
Previous research has revealed that plants mainly utilize two distinct enzymatic pathways to synthesize SA, the phenylalanine ammonia-lyase (PAL) pathway and the isochorismate (IC) pathway (Vlot et al., 2009; Dempsey et al., 2011). Both pathways require the primary metabolite chorismate, which is derived from the shikimate pathway. Earlier studies using isotope feeding suggested that SA is synthesized from phenylalanine via either benzoate intermediates or coumaric acid catalyzed by a series of enzymes including PAL, benzoic acid 2-hydroxylase, and other unknown enzymes (León et al., 1995; Dempsey et al., 2011). SA can also be synthesized through isochorismate catalyzed by isochorismate synthase (ICS) and isochorismate pyruvate lyase (IPL). Two ICS enzymes, ICS1 and ICS2, exist in Arabidopsis, and ICS1 has been shown to play a major role in SA biosynthesis (Garcion et al., 2008). Intriguingly, no plant genes encoding IPL have been identified. In comparison to the PAL pathway, the IC pathway plays a more important role in synthesis of both basal and induced SA in Arabidopsis (Mauch-Mani and Slusarenko, 1996; Garcion et al., 2008). However, neither pathway has been fully defined so far.
Nawrath and Métraux (1999) conducted a forward genetic screen in Arabidopsis for mutants with altered levels of total SA after infection with the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 carrying the avirulence gene avrRpm1. Two mutants, sid1 and sid2, were identified, which did not accumulate SA during the infection (Nawrath and Métraux, 1999). The sid1 and sid2 mutants were shown to be allelic to eds5 and eds16, respectively, which were identified in another genetic screen for enhanced disease susceptibility (Rogers and Ausubel, 1997; Nawrath and Métraux, 1999). EDS5/SID1 encodes a chloroplast MATE (multidrug and toxin extrusion) transporter (Nawrath et al., 2002), and SID2/EDS16 encodes an SA biosynthetic enzyme ICS1 (Wildermuth et al., 2001). In this screen, an HPLC (high performance liquid chromatography)-based method was used to quantify SA levels in pathogen-infected leaf tissues from about 4500 individual M2 plants. Obviously, the genetic screen did not reach saturation.
The HPLC-based method used by Nawrath and Métraux (1999) is extremely costly and time-consuming, which would not be practical for a large-scale genetic screen. Recently, an SA biosensor, named Acinetobacter sp. ADPWH_lux, was developed (Huang et al., 2005). This bacterial strain was derived from Acinetobacter sp. ADP1 and contains a chromosomal integration of an SA-inducible lux-CDABE operon, which encodes a luciferase (LuxA and LuxB) and the enzymes that produce its substrate (LuxC, LuxD, and LuxE). In the presence of SA, methylsalicylic acid, and acetylsalicylic acid, the operon is activated, resulting in emission of 490-nm light (Huang et al., 2005). Measurement of SA from tobacco mosaic virus-infected tobacco leaves with the biosensor and gas chromatography/mass spectrometry (GC/MS) yielded similar results, demonstrating that this strain is suitable for quantification of SA in plants (Huang et al., 2006). DeFraia et al. developed an improved methodology for Acinetobacter sp. ADPWH_lux-based SA quantification for both free SA and SA O-β-glucoside (SAG) in crude plant extracts (Defraia et al., 2008). Based on this, Marek et al. (2010) established a further simplified protocol for estimation of free SA levels in crude plant extracts in a high-throughput format (Marek et al., 2010). The efficacy and effectiveness of the newly developed SA biosensor-based method were confirmed by HPLC and verified in a small-scale mutant screen.
To better understand SA biology, we conducted a large-scale forward genetic screen aimed at isolating more Arabidopsis mutants with altered SA accumulation upon pathogen infection. We expected that mutants accumulating significantly altered levels of SA during pathogen infection will help study how SA is synthesized in plant cells and uncover important regulators of plant immunity. This screen allowed us to identify nine new mutants with significantly altered levels of pathogen-induced SA in the npr1-3 genetic background. Among them, seven produced SA levels lower than npr1 (sln) and two displayed increased SA accumulation in npr1 (isn). Enhanced disease resistance tests demonstrated that the seven new sln npr1-3 mutants are more susceptible to bacterial pathogen infection, while both isn npr1-3 mutants are more resistant than npr1-3. We further characterized the sln1 single mutant and found that the sln1 mutation compromised the bacterial pathogen P. syringae pv. maculicola (Psm) ES4326-induced defense responses. Moreover, exogenous SA induced both PR gene expression and disease resistance in sln1, indicating that SLN1 functions upstream of SA. Finally, the sln1 mutation was mapped to a region on the north arm of chromosome I, which contains no known genes involved in regulating pathogen-induced SA accumulation, suggesting that SLN1 encodes a new SA pathway component.
Materials and Methods
Plant Materials and Growth Conditions
The wild type used was the Arabidopsis thaliana (L.) Heynh. Columbia (Col-0) ecotype, and the mutant alleles used were npr1-3 (Glazebrook et al., 1996), npr1-L (GT_5_89558), eds5-1 (Nawrath et al., 2002), sid2-1 (Nawrath and Métraux, 1999; Wildermuth et al., 2001), pad4-1 (Glazebrook et al., 1996; Jirage et al., 1999), eps1-1 (Zheng et al., 2009), and pbs3-1 (Nobuta et al., 2007). The eds5-1 npr1-3, sid2-1 npr1-3, and pad4-1 npr1-3 double mutants were created by crossing npr1-3 with eds5-1, sid2-1, and pad4-1, respectively. Homozygous plants were identified by genotyping (Tables S1 and S2). Arabidopsis seeds were sown on autoclaved soil (Sunshine MVP, Sun Gro Horticulture, http://www.sungro.com) and cold-treated at 4°C for 3 days. Plants were grown at approximately 22°C under a 16-h light/8-h dark regime.
Pathogen Infection
The bacterial strains Psm ES4326 and Pst DC3000/avrRpt2 were grown overnight in liquid King's B medium. Bacterial cells were collected by centrifugation and diluted in 10 mM MgCl2. Inoculation of plants was performed by pressure infiltration with a 1 mL needleless syringe (Clarke et al., 1998). For SA measurement, Psm ES4326 and Pst DC3000/avrRpt2 suspensions with an OD600 of 0.001 were used for inoculation. The susceptibility phenotype was tested using a low-titer inoculum (OD600 = 0.0001) of Psm ES4326. In planta growth of Psm ES4326 was assayed 3 days after inoculation as previously described (Clarke et al., 1998). For SA-induced resistance assay, SA-treated plants were inoculated with a Psm ES4326 suspension (OD600 = 0.001) and the bacterial growth was determined 3 days post-inoculation.
SA Measurement
Free SA measurement using the SA biosensor was conducted as described by Marek et al. (2010). SA measurement with HPLC was performed as described by Verberne et al. (2002).
RNA Extraction and Quantitative PCR
RNA extraction was carried out as described previously (Cao et al., 1997). For reverse transcription (RT), ~10 μg of total RNA was treated with DNase I (Ambion) at 37°C for 30 min for digestion of contaminating DNA. After inactivation of the DNase, ~2 μg of total RNA was used as a template for first-strand cDNA synthesis using the M-MLV Reverse Transcriptase first-strand synthesis system (Promega). The resulting cDNA products were diluted 20-fold with autoclaved distilled water, and 2.5 μL of the diluted solution was used for quantitative PCR (qPCR). qPCR was performed in an Mx3005P qPCR system (Stratagene). All qPCR reactions were performed with a 12.5 μL reaction volume using the SYBR Green protocol under the following conditions: denaturation program (95°C for 10 min), amplification and quantification program repeated for 40 cycles (95°C for 30 s, 55°C for 1 min, 72°C for 1 min), and melting curve program (95°C for 1 min, 55°C for 30 s, and 95°C for 30 s). The primers used for qPCR in this study are listed in Table S2.
Statistical Methods
Statistical analyses were performed with Prism 5 (GraphPad Software, Inc., La Jolla, CA). One-Way analysis of variance (ANOVA) was used to determine statistical significance among genotypes or treatments. In addition, two-way analysis of variance was used to examine the effects of genotypes, treatments, and the interaction of these two factors on disease resistance. Post-hoc comparison was performed using Fisher's least significant difference LSD test and represented by different letters. Alternatively, statistical analyses were performed using Student's t-test for comparison of two data sets (Assuming Unequal Variances).
Accession Number
The locus numbers for the genes discussed in this study are as follows: NPR1 (At1g64280), EDS5 (At4g39030), ICS1 (At1g74710), PAD4 (At3g52430), EPS1 (At5g67160), PBS3 (At5g13320), PR1 (At2g14610), PR2 (At3g57260), PR5 (At1g75040), UBQ5 (At3g62250).
Results
Isolation of SA Accumulation Mutants
In order to identify new components involved in pathogen-induced SA accumulation, we took advantage of the SA biosensor-based method to screen for mutants with altered levels of pathogen-induced SA in Arabidopsis. Approximately 35,000 M2 plants from an ethyl methanesulfonate-mutagenized population (20 pools, each from ~500 M1 plants) in the npr1-3 mutant background were individually analyzed for free SA levels after infection with the bacterial pathogen Psm ES4326. The npr1-3 mutant was used as the starting material for the genetic screen, because it accumulates significantly higher levels of SA than wild type upon bacterial pathogen infection (Figures 1A,B; Cao et al., 1997; Ryals et al., 1997; Shah et al., 1997; Zhang et al., 2010). Plants that accumulated significantly higher or lower levels of pathogen-induced SA than npr1-3 were considered to be putative SA accumulation mutants. Approximately 350 such mutants were identified in the primary screen. To confirm these putative mutants, eight plants of each mutant line were tested for Psm ES4326-induced SA accumulation using the SA biosensor in the M3 generation (Marek et al., 2010). Nineteen mutants with drastically altered levels of pathogen-induced SA, including 17 sln npr1-3 and two isn npr1-3 mutants, were chosen for further analysis (Figure 1A). SA levels accumulated in the remaining mutants were significantly lower than those in npr1-3, but slightly higher than those in the wild-type plants (data not shown). Contamination from other mutants in the lab was excluded by checking the mutant plants under ultraviolet (UV) illumination, since the npr1-3 mutant carries a fuhl-2 allele, which lacks sinapoyl malate in the leaf epidermis and appears red under UV light (Chapple et al., 1992; Glazebrook et al., 1996). In addition, the presence of the npr1-3 mutation in the identified mutants was confirmed with a derived cleaved amplification polymorphism sequence (dCAPS) marker (Table S1).
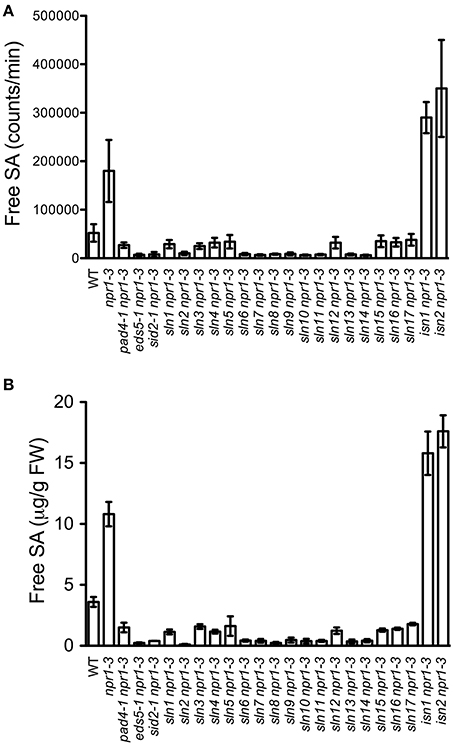
Figure 1. Pathogen-induced free SA levels in the SA accumulation mutants. (A) Luminescence from crude extracts of Psm ES4326-infected wild-type, npr1-3, pad4-1 npr1-3, eds5-1 npr1-3, sid2-1 npr1-3, and 19 putative mutant leaf tissues measured with the SA biosensor. (B) Free SA levels in Psm ES4326-infected wild-type, npr1-3, pad4-1 npr1-3, eds5-1 npr1-3, sid2-1 npr1-3, and 19 putative mutant plants detected by the HPLC-based method. Values are the mean of eight (A) or three (B) samples with standard deviation (SD). The experiments were repeated three times with similar results.
To confirm that the 19 mutants accumulate altered levels of SA after pathogen infection, we measured free SA levels accumulated in these mutants after Psm ES4326 infection using HPLC. Similarly to the results obtained using the SA biosensor, upon Psm ES4326 infection, the 17 sln npr1-3 mutants accumulated dramatically lower levels of free SA and the two isn npr1-3 mutants produced higher levels of free SA than the npr-3 mutant (Figure 1B). These results suggest that the sln mutations may reside in genes that are required for pathogen-induced SA biosynthesis, whereas the isn mutations may be located in suppressors of SA accumulation.
Pathogen Resistance of the SA Accumulation Mutants
SA accumulation is generally associated with resistance to biotrophic and hemibiotrophic bacterial pathogens (An and Mou, 2011). To investigate whether susceptibility or resistance to bacterial pathogens in the 19 SA accumulation mutants described above is also affected, we inoculated 4-week-old plants with a low-titer inoculum (OD600 = 0.0001) of the virulent bacterial pathogen Psm ES4326. Interestingly, all sln npr1-3 mutants developed enhanced disease symptoms (data not shown) and supported more bacterial growth (2- to 7-fold) compared with the npr1-3 mutant (Figure 2), suggesting that the SLN genes are required for resistance to the bacterial pathogen. In contrast, the two isn npr1-3 mutants supported less Psm ES4326 growth than npr1-3, although the bacteria still grew to a slightly higher titer in the isn npr1-3 mutants than in the wild-type plants (Figure 2), indicating that the increased levels of SA in the isn npr1-3 mutants may activate NPR1-independent disease resistance.
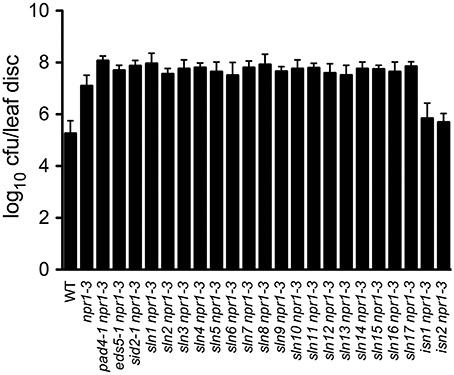
Figure 2. Pathogen growth in the SA accumulation mutants. Leaves of 4-week-old plants were inoculated with a Psm ES4326 suspension (OD600 = 0.0001). The in planta bacterial titers were determined 3 days post-inoculation. Data represent the mean of eight independent samples with SD. cfu, colony-forming units. The experiment was repeated three times with similar results.
Allelism Test
Analyses of the F1 plants from crosses between the 19 SA accumulation mutants and npr1-3 indicated that all sln and isn mutations are recessive. Several recessive mutations, including eds5 (Nawrath and Métraux, 1999; Nawrath et al., 2002), sid2 (Nawrath and Métraux, 1999; Wildermuth et al., 2001), pad4 (Glazebrook et al., 1996; Zhou et al., 1998; Jirage et al., 1999), eds1 (Parker et al., 1996; Falk et al., 1999), eps1 (Zheng et al., 2009), and pbs3/win3/gdg1 (Jagadeeswaran et al., 2007; Lee et al., 2007; Nobuta et al., 2007), have been shown to compromise pathogen-induced SA accumulation. We reasoned that the sln mutants are unlikely alleles of eps1, pbs3, and eds1, since no difference in pathogen-induced free SA levels was detected between eps1-1 or pbs3-1 and the wild type using the SA biosensor (Figure S1), and two EDS1 genes are present in the Arabidopsis ecotype Col-0 (Feys et al., 2005). We therefore tested for allelism between the sln mutants and eds5, sid2, or pad4. Pathogen-induced SA levels in F1 plants were measured using the SA biosensor and compared with those in their parents. These allelism tests revealed that seven sln mutants are alleles of eds5, two are sid2 alleles, and one is allelic to pad4 (Table 1).
We also performed complementation tests for allelism among the remaining seven sln mutants. They were crossed to each other and the resulting F1 plants were tested for the ability to accumulate SA after Psm ES4326 infection using the SA biosensor. We found that the sln mutations complemented each other, suggesting that they are located in different genes required for pathogen-induced SA accumulation (Table 1). Moreover, complementation test indicated that the two isn mutations reside in two different genes, which are likely involved in suppressing pathogen-induced SA accumulation (Table 1).
Characterization of the sln1 npr1-3 Mutant
To have a better understanding of the sln mutations, we further characterized one of the newly identified SA accumulation mutants, sln1 npr1-3. The sln1 npr1-3 mutant was morphologically similar to npr1-3 (Figure 3A). F1 plants from a backcross of sln1 npr1-3 and npr1-3 accumulated similar levels of free SA as npr1-3, suggesting that sln1 is recessive. SA analysis of F2 progeny showed that sln1 segregated as a single Mendelian locus (high SA:low SA, 33:8; χ2 = 0.6585, 0.25 < P < 1).
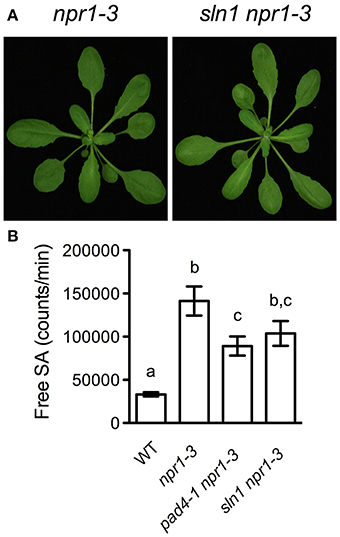
Figure 3. Further characterization of sln1 npr1-3. (A) Photos of 4-week-old soil-grown npr1-3 and sln1 npr1-3 plants. (B) Luminescence from crude extracts of Pst DC3000/avrRpt2-infected wild-type, npr1-3, pad4-1 npr1-3, sln1 npr1-3 leaf tissues measured with the SA biosensor. Values are the mean of eight independent samples with SD. Different letters above the bars indicate significant differences (P < 0.05, One-Way ANOVA). The experiment was repeated three times with similar results.
It was reported that the pad4 mutation does not affect free SA accumulation in response to the avirulent bacterial pathogen Pst DC3000/avrRpt2 (Zhou et al., 1998). To test whether the sln1 mutation influences the avirulent pathogen-induced SA accumulation, we challenged sln1 npr1-3 plants with Pst DC3000/avrRpt2. As shown in Figure 3B, Pst DC3000/avrRpt2 induced significant SA accumulation in both sln1 npr1-3 and pad4-1 npr1-3 plants. Although free SA levels accumulated in the sln1 npr1-3 plants were still slightly lower than those in the npr1-3 plants, the difference was not as dramatic as that detected in the Psm ES4326-infected plants (Figure 1). These results indicate that the avirulent pathogen Pst DC3000/avrRpt2-triggered SA accumulation is largely independent of SLN1.
SA Accumulation in the sln1 Single Mutant
Since the sln1 mutation is able to reduce SA accumulation in npr1-3, it may affect SA accumulation in the presence of NPR1. To test this, we isolated sln1 single mutant in the F2 progeny of a cross between sln1 npr1-3 and the wild-type Col-0 using the npr1-3 dCAPS marker (Table S1) and based on SA levels accumulated in the plants upon Psm ES4326 infection. As shown in Figures 4A,B, both free SA and total SA levels accumulated in the sln1 single mutant plants after Psm ES4326 infection were significantly lower than those in the wild type. We also found that Psm ES4326-induced expression of ICS1, which is responsible for pathogen-induced SA accumulation (Wildermuth et al., 2001), was significantly reduced in the sln1 single mutant compared with that in the wild type (Figure 4C), indicating that SLN1 may regulate SA accumulation through ICS1.
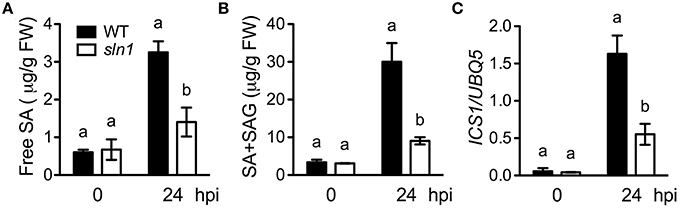
Figure 4. Pathogen-induced SA levels and ICS1 expression in the sln1 single mutant. Leaves of 4-week-old soil-grown wild-type and sln1 plants were infiltrated with a suspension of Psm ES4326 (OD600 = 0. 001). The inoculated leaves were harvested 24 h post-inoculation (hpi) for SA measurement using HPLC or ICS1 expression analysis using qPCR. (A) Free SA levels in Psm ES4326-infected wild-type and sln1 plants. (B) Total SA levels in Psm ES4326-infected wild-type and sln1 plants. (C) ICS1 expression levels in Psm ES4326-infected wild-type and sln1 plants. Values are the mean of three independent samples with SD. Different letters above the bars indicate significant differences (P < 0.05, Student's t-test). The comparison was made separately for each time point. Expression of ICS1 in (C) was normalized against constitutively expressed UBQ5. The experiments were repeated three times with similar results.
Pathogen Resistance of the sln1 Single Mutant
We then investigated pathogen growth in the sln1 single mutant. After infected with a low-titer inoculum (OD600 = 0.0001) of Psm ES4326, the sln1 single mutant plants developed enhanced disease symptoms (Figure 5A), and supported ~15-fold more bacterial growth than the wild type (Figure 5B). We also tested pathogen-induced PR gene expression in the sln1 single mutant. As shown in Figures 5C–E, Psm ES4326-induced PR1 expression was significantly reduced in the sln1 single mutant, but the induction of PR2 and PR5 in sln1 was comparable to that in the wild type. Taken together, these results indicate that SLN1 is required for defense responses against the bacterial pathogen Psm ES4326.
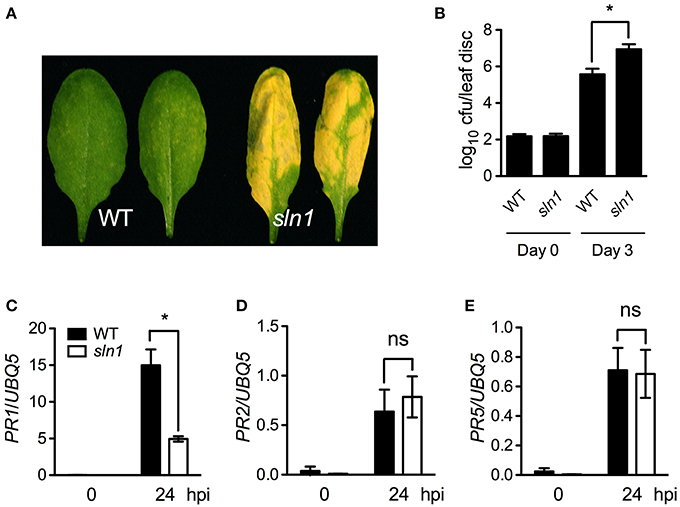
Figure 5. Defense responses in the sln1 single mutant. (A) Disease symptoms of Psm ES4326-infected wild-type and sln1 leaves. Four-week-old soil-grown plants were inoculated with a suspension of Psm ES4326 (OD600 = 0.0001). Photos were taken 3 days post-inoculation. (B) Growth of Psm ES4326 in wild-type and sln1 plants. Four-week-old soil-grown plants were inoculated with a suspension of Psm ES4326 (OD600 = 0.0001). The in planta bacterial titers were determined immediately and 3 days post-inoculation. Data represent the mean of eight independent samples with SD. (C–E) Psm ES4326-induced PR1 (C), PR2 (D), and PR5 (E) gene expression in wild-type and sln1 plants. Four-week-old soil-grown plants were inoculated with a suspension of Psm ES4326 (OD600 = 0.001). Total RNA was extracted from leaf tissues collected at 24 hpi and subjected to qPCR analysis. Data represent the mean of three independent samples with SD. An asterisk (*) above the bars indicates significant differences (P < 0.05, Student's t-test). ns, not significant. All experiments were repeated three times with similar results.
Since the sln1 mutation inhibits pathogen-induced SA accumulation, exogenous SA may restore defense responses in sln1 plants. Indeed, SA treatment induced similar levels of PR1 gene expression and resistance to Psm ES4326 in the sln1 single mutant and the wild-type plants (Figures 6A,B). Based on these results, we concluded that the signaling pathway downstream of SA in sln1 is intact. Thus, SLN1 most likely functions in a signal amplification loop upstream of SA.
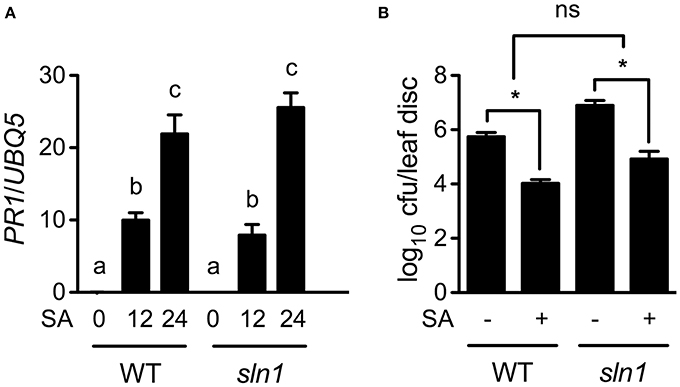
Figure 6. Exogenous SA-induced PR gene expression and resistance in sln1. (A) Exogenous SA-induced PR1 expression in wild-type and sln1 plants. Four-week-old soil-grown wild-type and sln1 plants were soaked with an SA water solution (1 mM). Total RNA was extracted from leaf tissues collected at the indicated time points and analyzed for PR1 expression using qPCR. Values are the mean of three independent samples with SD. Different letters above the bars indicate significant differences (P < 0.05, One-Way ANOVA). The comparison was made separately for each genotype. (B) Exogenous SA-induced resistance to Psm ES4326 in wild-type and sln1 plants. Plants were treated as in (A). Twelve hours later, the plants were inoculated with a suspension of Psm ES4326 (OD600 = 0.001). The in planta bacterial titers were determined 3 days post-inoculation. Values are the mean of eight independent samples with SD. An asterisk (*) above the bars indicates significant differences (P < 0.05, Two-Way ANOVA). ns, not significant. These experiments were repeated three times with similar results.
Preliminary Mapping of the sln1 Mutation
To map the sln1 mutation, sln1 npr1-3 (in the Col-0 genetic background) was crossed with npr1-L (an npr1 T-DNA insertion mutant in the polymorphic ecotype Landsberg erecta) to generate a segregating population. Preliminary mapping using 74 F2 plants, which accumulated extremely low levels of SA after Psm ES4326 infection, revealed that sln1 is located between gene At1g01448 and the molecular marker PAI1.2 (Figure 7). To our knowledge, this region does not contain any known genes regulating pathogen-induced SA accumulation. Therefore, SLN1 likely encodes a new regulator of SA biosynthesis. Further fine-mapping and/or whole genome sequencing will help identify the sln1 mutation.
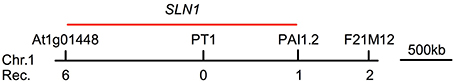
Figure 7. Preliminary mapping of the sln1 mutation. A total of 74 F2 progeny homozygous for sln1 were used to determine the approximate position of the sln1 mutation using bulked segregant analysis. The sln1 mutation was linked to the molecular marker PT1 on chromosome 1. Out of the 74 F2 plants, six were heterozygous at gene At1g01448, and one was heterozygous at the molecular marker PAI1.2. The heterozygotes found by these two markers were mutually exclusive. No heterozygotes were found at PT1. The SLN1 gene is likely located in the vicinity of PT1, as indicated by the red bar. Rec., recombinant.
Discussion
In this study, we performed a forward genetic screen for Arabidopsis mutants with altered SA accumulation during pathogen infection using the newly developed SA biosensor method (Marek et al., 2010). Compared with the HPLC and GC/MS approaches, the SA biosensor method is much faster and less expensive (Malamy et al., 1992; Verberne et al., 2002; Marek et al., 2010). Using this method, we screened a large population (35,000) of M2 plants in less than 1 year. Approximately 350 putative SA accumulation mutants in the npr1-3 genetic background were identified. Among them, 17 are sln npr1-3 mutants, producing significantly lower levels of SA than npr1-3 after Psm ES4326 infection, and two are isn npr1-3 mutants, accumulating higher levels of SA than npr1-3 (Figures 1A,B). Interestingly, upon Psm ES4326 infection, SA levels accumulated in the remaining putative mutants (in the npr1-3 background) were significantly lower than those in npr1-3, but slightly higher than those in the wild-type plants, suggesting the existence of a larger number of regulatory components involved in pathogen-induced SA accumulation. Indeed, genetic studies have uncovered a complicated signaling network that regulates SA accumulation. This consists of upstream SA signaling components (such as EDS1, PAD4, and NDR1), downstream SA signaling components (such as NPR1), transcription factors (such as CBP60g and SARD1), metabolic enzymes (such as EPS1 and PBS3), and various positive and negative feedback loops (Cao et al., 1997; Ryals et al., 1997; Shah et al., 1997; Zhou et al., 1998; Jirage et al., 1999; Shapiro and Zhang, 2001; Wildermuth et al., 2001; Jagadeeswaran et al., 2007; Lee et al., 2007; Nobuta et al., 2007; Zheng et al., 2009; Zhang et al., 2010; Wang et al., 2011).
The SA accumulation phenotype of the sln mutants is similar to that of eds5, sid2, and pad4 mutants (Zhou et al., 1998; Jirage et al., 1999; Nawrath and Métraux, 1999; Wildermuth et al., 2001; Nawrath et al., 2002). EDS5 and SID2 encode a chloroplast MATE transporter and an SA biosynthetic enzyme ICS1, respectively, which are two important components in the SA biosynthesis pathway. PAD4 is a lipase-like protein involved in an SA positive signal-amplification loop required for activation of defense responses (Jirage et al., 1999). Complementation tests indicated that seven out of the 17 sln mutants are new alleles of eds5, two are alleles of sid2, and one is allelic to pad4, and the other seven sln and two isn mutants are new non-allelic mutants (Table 1). Although this is a large-scale genetic screen, the low frequency of alleles for the new sln and isn mutants indicates that our genetic screen has not been saturated.
Several other recessive mutations have also been reported to impair pathogen-induced SA accumulation. In the eps1-1 mutant, pathogen-induced accumulation of SAG was greatly reduced, but free SA levels were comparable to those in the wild type. EPS1 encodes a novel member of the BAHD acyltransferase superfamily, which is predicted to be directly involved in the synthesis of a precursor or regulatory molecule for SA biosynthesis (Zheng et al., 2009). Similarly, the pbs3-1 mutant displayed decreased pathogen-induced accumulation of SAG, but varied in free SA accumulation between studies (Jagadeeswaran et al., 2007; Lee et al., 2007; Nobuta et al., 2007). PBS3 belongs to the acyl adenylate/thioesterforming enzyme superfamily. The exact functions of both EPS1 and PBS3 in SA biosynthesis, however, have not been clearly defined. Consistent with these studies, we found that free SA levels in the eps1-1 and pbs3-1 mutants were comparable to those in the wild type when assayed with the SA biosensor (Figure S1). Thus, the sln mutations are unlikely located in either EPS1 or PBS3, since these mutations greatly influenced Psm ES4326-induced free SA accumulation (Figures 1A,B). Additionally, although the eds1 mutation significantly affects pathogen-induced accumulation of both free SA and SAG (Falk et al., 1999), the sln mutants are unlikely alleles of eds1, because there are two EDS1 genes lying in tandem on chromosome 3 of the Arabidopsis ecotype Col-0 (Feys et al., 2005). Therefore, the SLN genes may encode new signaling components downstream of recognition of pathogen infection, or new enzymes directly involved in the synthesis of a precursor and/or regulatory molecule for SA biosynthesis.
In addition to components upstream of SA biosynthesis, the downstream component, NPR1 (nonexpressor of PR genes1), which has been shown to be an important regulator of defense responses (Cao et al., 1997; Dong, 2004), also regulates SA levels. Mutations in the NPR1 gene enhance SA accumulation during pathogen infection, suggesting that NPR1 is a feedback inhibitor of SA biosynthesis (Figures 1A,B; Clarke et al., 2000; Wildermuth et al., 2001; Zhang et al., 2010). Here we found that eds5 npr1-3, sid2 npr1-3, pad4 npr1-3, and sln npr1-3 double mutants accumulated significantly lower levels of SA than npr1-3 (Figures 1A,B), suggesting that these mutations (eds5, sid2, pad4, and sln) suppress npr1-mediated SA hyperaccumulation. On the other hand, these double mutants were more susceptible to Psm ES4326 than npr1-3 (Figure 2), indicating that EDS5, SID2, PAD4, and the SLN genes may contribute to NPR1-independent defense responses (Glazebrook, 2001). NPR1-independent defense signaling pathways have been shown to be activated in various Arabidopsis mutants, including sni1 (Li et al., 1999), snc1 (Li et al., 2001), ssi (Shah et al., 1999, 2001), and cpr (Bowling et al., 1997; Clarke et al., 1998). The two isn mutations appear to also activate NPR1-independent disease resistance (Figure 2).
We further isolated and characterized the sln1 single mutant. The sln1 plants exhibited significantly reduced levels of Psm ES4326-induced SA and supported more Psm ES4326 growth than the wild-type plants (Figures 4A,B, 5B), suggesting that SLN1 plays an important role in activation of defense responses against this pathogen. Interestingly, the sln1 mutation appears to differentially influence pathogen-induced PR1, PR2, and PR5 expression. Psm ES4326-induced PR1 expression was greatly reduced in sln1 plants, but induction of PR2 and PR5 was nearly unaffected (Figures 5C–E). In this regard, sln1 is also similar to eds5, sid2, and pad4, which cause reduced induction of PR1, but have no effect on the expression of PR2 and PR5 (Rogers and Ausubel, 1997; Zhou et al., 1998; Nawrath and Métraux, 1999). On the other hand, pathogen-induced expression of PR1, PR2, and PR5 is strongly reduced in NahG transgenic plants (Nawrath and Métraux, 1999), which argues against the idea that an SA-independent pathway exists to control PR2 and PR5 expression. It is possible that the low levels of SA accumulated in the SA biosynthesis mutants are sufficient for induction of PR2 and PR5, but not for PR1.
In summary, we identified a group of new SA accumulation mutants, including seven sln mutants and two isn mutants, in a genetic screen using the newly developed SA biosensor-based method. Further characterization of these sln and isn mutants and cloning of the SLN and ISN genes will shed new light on the molecular mechanisms underlying pathogen-induced SA accumulation and SA-mediated defense signaling in plants.
Author Contributions
Yezhang Ding and Zhonglin Mou designed research. Yezhang Ding and Danjela Shaholli performed mutant screen. Yezhang Ding characterized mutants and analyzed data. Yezhang Ding and Zhonglin Mou wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Hui Wang (NERC/Centre for Ecology and Hydrology-Oxford, Oxford, UK) for the SA biosensor strain Acinetobacter sp. ADPWH_lux and Dr. Sixue Chen (University of Florida) for access to the HPLC equipment. We are grateful to Dr. Roger Innes (Indiana University) for the pbs3-1 mutant and Dr. Zhixiang Chen (Purdue University) for the eps1-1 mutant. We thank the Arabidopsis Biological Resource Center at Ohio State University for providing the T-DNA insertion line GT_5_89558 (npr1-L). This work was supported by a grant from the National Science Foundation (IOS-0842716) awarded to Zhonglin Mou.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00763/abstract
Figure S1. SA accumulation in eps1 and pbs3. Luminescence from crude extracts of Psm ES4326-infected wild-type, npr1-3, eps1-1, and pbs3-1 leaf tissues measured with the SA biosensor. Values are the mean of six samples with standard deviation (SD). The experiment was repeated with similar results.
References
An, C., and Mou, Z. (2011). Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428. doi: 10.1111/j.1744-7909.2011.01043.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bowling, S. A., Clarke, J. D., Liu, Y., Klessig, D. F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. doi: 10.1105/tpc.9.9.1573
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, H., Glazebrook, J., Clark, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chapple, C. C., Vogt, T., Ellis, B. E., and Somerville, C. R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4, 1413–1424. doi: 10.1105/tpc.4.11.1413
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clarke, J. D., Liu, Y., Klessig, D. F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10, 557–569.
Clarke, J. D., Volko, S. M., Ledford, H., Ausubel, F. M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. doi: 10.1105/tpc.12.11.2175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Defraia, C. T., Schmelz, E. A., and Mou, Z. (2008). A rapid biosensor-based method for quantification of free and glucose-conjugated salicylic acid. Plant Methods 4, 28. doi: 10.1186/1746-4811-4-28
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., et al. (1994). Central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. doi: 10.1126/science.266.5188.1247
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dempsey, D. A., Vlot, A. C., Wildermuth, M. C., and Klessig, D. F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156. doi: 10.1199/tab.0156
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–752. doi: 10.1016/j.pbi.2004.07.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Falk, A., Feys, B. J., Frost, L. N., Jones, J. D., Daniels, M. J., and Parker, J. E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U.S.A. 96, 3292–3297. doi: 10.1073/pnas.96.6.3292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feys, B. J., Wiermer, M., Bhat, R. A., Moisan, L. J., Medina-Escobar, N., Neu, C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stablizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613. doi: 10.1105/tpc.105.033910
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garcion, C., Lohmann, A., Lamodière, E., Catinot, J., Buchala, A., Doermann, P., et al. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. doi: 10.1104/pp.108.119420
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis–2001 status. Curr. Opin. Plant Biol. 4, 301–308. doi: 10.1016/S1369-5266(00)00177-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glazebrook, J., Rogers, E. E., and Ausubel, F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982.
Greenberg, J. T., Guo, A., Klessig, D. F., and Ausubel, F. M. (1994). Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 7, 551–563. doi: 10.1016/0092-8674(94)90217-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, W. E., Huang, L., Preston, G. M., Martin, N., Carr, J. P., Li, Y. H., et al. (2006). Quantitative in situ assay of salicylic acid in tobacco leaves using a genetically modified biosensor strain of Acinetobacter sp. ADP1. Plant J. 46, 1073–1083. doi: 10.1111/j.1365-313X.2006.02758.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, W. E., Wang, H., Zheng, H., Huang, L., Singer, A. C., Thompson, I., et al. (2005). Chromosomally located gene fusions constructed in Acinetobacter sp. ADP1 for the detection of salicylate. Environ. Microbiol. 7, 1339–1348. doi: 10.1111/j.1462-5822.2005.00821.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jagadeeswaran, G., Raina, S., Acharya, B. R., Maqbool, S. B., Mosher, S. L., Appel, H. M., et al. (2007). Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 51, 234–246. doi: 10.1111/j.1365-313X.2007.03130.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jirage, D., Tootle, T. L., Reuber, T. L., Frost, L. N., Feys, B. J., Parker, J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96, 13583–13588. doi: 10.1073/pnas.96.23.13583
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lawton, K. A., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., and Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 8, 863–870. doi: 10.1094/MPMI-8-0863
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, M. W., Lu, H., Jung, H. W., and Greenberg, J. T. (2007). A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol. Plant Microbe Interact. 20, 1192–1200. doi: 10.1094/MPMI-20-10-1192
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
León, J., Shulaev, V., Yalpani, N., Lawton, M. A., and Raskin, I. (1995). Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 92, 10413–10417 doi: 10.1073/pnas.92.22.10413
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, X., Clarke, J. D., Zhang, Y., and Dong, X. (2001). Activation of an EDS1- mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14, 1131–1139. doi: 10.1094/MPMI.2001.14.10.1131
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, X., Zhang, Y., Clarke, J. D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. doi: 10.1016/S0092-8674(00)81962-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malamy, J., Hennig, J., and Klessig, D. F. (1992). Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell 4, 359–366. doi: 10.1105/tpc.4.3.359
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marek, G., Carver, R., Ding, Y., Sathyanarayan, D., Zhang, X., and Mou, Z. (2010). A high-throughput method for isolation of salicylic acid metabolic mutants. Plant Methods 6, 21. doi: 10.1186/1746-4811-6-21
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mauch-Mani, B., and Slusarenko, A. J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212. doi: 10.1105/tpc.8.2.203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nawrath, C., Heck, S., Parinthawong, N., and Métraux, J. P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. doi: 10.1105/tpc.010376
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nawrath, C., and Métraux, J. P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404.
Nobuta, K., Okrent, R. A., Stoutemyer, M., Rodibaugh, N., Kempema, L., Wildermuth, M. C., et al. (2007). The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 144, 1144–1156. doi: 10.1104/pp.107.097691
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parker, J. E., Frost, L. N., Falk, A., Gunn, N. D., and Daniels, M. J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. doi: 10.1105/tpc.8.11.2033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rate, D. N., Cuenca, J. V., Bowman, G. R., Guttman, D. S., and Greenberg, J. T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defense, and cell growth. Plant Cell 11, 1695–1708. doi: 10.1105/tpc.11.9.1695
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rogers, E. E., and Ausubel, F. M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316. doi: 10.1105/tpc.9.3.305
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H. Y., et al. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IkB. Plant Cell 9, 425–439.
Shah, J., Kachroo, P., and Klessig, D. F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders Defensin gene expression SA dependent. Plant Cell 11, 191–206. doi: 10.1105/tpc.11.2.191
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shah, J., Kachroo, P., Nandi, A., and Klessig, D. F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574. doi: 10.1046/j.1365-313x.2001.00992.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10, 69–78. doi: 10.1094/MPMI.1997.10.1.69
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shapiro, A. D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. doi: 10.1104/pp.010096
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Verberne, M. C., Brouwer, N., Delbianco, F., Linthorst, H. J., Bol, J. F., and Verpoorte, R. (2002). Method for the extraction of the volatile compound salicylic acid from tobacco leaf material. Phytochem. Anal. 13, 45–50. doi: 10.1002/pca.615
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, L., Tsuda, K., Truman, W., Sato, M., Nguyen le, V., Katagiri, F., et al. (2011). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. doi: 10.1111/j.1365-313X.2011.04655.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
White, R. F. (1979). Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99, 410–412. doi: 10.1016/0042-6822(79)90019-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Chen, S., and Mou, Z. (2010). Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J. Plant Physiol. 167, 144–148. doi: 10.1016/j.jplph.2009.08.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zheng, Z., Qualley, A., Fan, B., Dudareva, N., and Chen, Z. (2009). An important role of a BAHD acyl transferase-like protein in plant innate immunity. Plant J. 57, 1040–1053. doi: 10.1111/j.1365-313X.2008.03747.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, N., Tootle, T. L., Tsui, F., Klessig, D. F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. doi: 10.1105/tpc.10.6.1021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: salicylic acid, genetic screen, NPR1, Arabidopsis thaliana, disease resistance, sln mutant, isn mutant
Citation: Ding Y, Shaholli D and Mou Z (2015) A large-scale genetic screen for mutants with altered salicylic acid accumulation in Arabidopsis. Front. Plant Sci. 5:763. doi: 10.3389/fpls.2014.00763
Received: 16 October 2014; Paper pending published: 04 November 2014;
Accepted: 10 December 2014; Published online: 07 January 2015.
Edited by:
Hua Lu, University of Maryland Baltimore County, USACopyright © 2015 Ding, Shaholli and Mou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonglin Mou, Department of Microbiology and Cell Science, University of Florida, Museum Road, Building 981, Gainesville, FL 32611, USA e-mail: zhlmou@ufl.edu
 Yezhang Ding
Yezhang Ding Danjela Shaholli
Danjela Shaholli Zhonglin Mou
Zhonglin Mou