- 1Ministry of Education Key Laboratory of Agriculture Biodiversity for Plant Disease Management, Yunnan Agricultural University, Kunming, China
- 2State Key Laboratory of Plant Genomics and Center for Plant Gene Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 3School of Life Sciences, Huaiyin Normal University, Huaian, China
SDS is a meiosis specific cyclin-like protein and required for DMC1 mediated double-strand break (DSB) repairing in Arabidopsis. Here, we found its rice homolog, OsSDS, is essential for meiotic DSB formation. The Ossds mutant is normal in vegetative growth but both male and female gametes are inviable. The Ossds meiocytes exhibit severe defects in homologous pairing and synapsis. No γH2AX immunosignals in Ossds meiocytes together with the suppression of chromosome fragmentation in Ossds-1 Osrad51c, both provide strong evidences that OsSDS is essential for meiotic DSB formation. Immunostaining investigations revealed that meiotic chromosome axes are normally formed but both SC installation and localization of recombination elements are failed in Ossds. We suspected that this cyclin protein has been differentiated pretty much between monocots and dicots on its function in meiosis.
Introduction
Meiosis is one of the key processes in sexual reproduction for all sexually propagating eukaryotic organisms. Meiosis includes one single round of DNA replication but followed by two successive rounds of nuclear segregations (meiosis I and II), and finally produces four haploid gametes with halved chromosomes. The meiotic prophase I is a complicated and prolonged stage, which can be divided into five substages, like leptotene, zygotene, pachytene, diplotene, and diakinesis, based on chromosome characterizations (Ashley and Plug, 1998; Dawe, 1998). During meiotic prophase I, pairing, synapsis and recombination of homologous chromosomes are coordinately accomplished. These events make sure the precise segregation of homologs, and generate both genetic conservation and diverse individuals in the future generations (Zickler and Kleckner, 1999; Page and Hawley, 2003).
In meiosis, DSBs are purposely produced to initiate homologous recombination. The formation of DSBs in meiosis is catalyzed by a type-II topoisomerase-like enzyme Spo11 (Bergerat et al., 1997; Keeney et al., 1997). Meanwhile, a series of cofactors are also required for this process. In budding yeast, the formation of meiotic DSBs requires at least nine other proteins (Rec102, Rec104, Rec114, Mei4, Mer2, Rad50, Mre11, Xrs2, and Ski8) for the cleavage mediated by SPO11 and further broken end resection (Paques and Haber, 1999; Keeney, 2001, 2008). The budding yeast Mre11-Rad50-Xrs2 (MRX) complex is homologous with the mammalian Mre11-Rad50-Nbs1 (MRN), which is required to incise the 5′ end of the break and then form 3′ single-strand tails (Symington, 2002; Mimitou and Symington, 2009). After that, one 3′ free single-strand DNA end recruits two RecA homologs, Rad51 and Dmc1, to mediate the single-end invasion (SEI) with its homologous duplex DNA (Bishop, 1994; Hunter and Kleckner, 2001), and the other 3′ free single-strand DNA end on the other side of the nick is captured simultaneously to form the Double Holliday Junction (DHJ). And then, the DHJ is exclusively processed into crossovers (COs), which represents the accomplishment of homologous recombination (Allers and Lichten, 2001; Bishop and Zickler, 2004; Borner et al., 2004). Consequently, meiotic DSBs are finally repaired during this process.
The function of SPO11 initiating meiotic recombination seems to be widely conserved within eukaryotes, as more and more homologs of SPO11 were identified in a wide range of organisms covering yeasts, flies, mice, humans, and plants (Dernburg et al., 1998; McKim and Hayashi-Hagihara, 1998; Celerin et al., 2000; Hartung and Puchta, 2000; Romanienko and Camerini-Otero, 2000; Grelon et al., 2001; Yu et al., 2010). Unlike animals and fungi where a single SPO11 is sufficient for meiotic DSBs formation, higher plants always possess multiple SPO11 homologs (Keeney et al., 1997; Grelon et al., 2001; Hartung et al., 2007; Shingu et al., 2012; Sprink and Hartung, 2014). But not every SPO11 homolog has the function to cleavage double-strand DNA and generate DSBs in plants. Arabidopsis owns three SPO11 homologs and they appear to function in two distinct processes, AtSPO11-1 and AtSPO11-2 in DSB formation, while AtSPO11-3 in DNA replication (Stacey et al., 2006; Shingu et al., 2012). While in monocot rice, there are five SPO11 homologs have been identified (Jain et al., 2006, 2008). Among them, only OsSPO11-4 has been proved to be with double-strand DNA cleavage activity (An et al., 2011). OsSPO11-1 is essential for homologous pairing, recombination and SC installation (Yu et al., 2010; Luo et al., 2014). So, it seems that the formation of meiotic DSBs is more complicated in plants.
Besides SPO11, several other DSB formation proteins have been identified recently in multicellular eukaryotes. Mei1 and Mei4 were shown to be required for DSB formation in mice (Libby et al., 2003; Kumar et al., 2010). Using a high throughput genetic screen, AtPRD1, AtPRD2, and AtPRD3 were identified to be essential for DSB formation in Arabidopsis (De Muyt et al., 2009). Nevertheless, AtDFO was also found to be necessary for DSB formation in Arabidopsis (Zhang et al., 2012). Studies in rice revealed that CRC1 works together with PAIR1 as a complex to regulate meiotic DSB formation (Miao et al., 2013).
Studies in budding yeast demonstrated that cyclin-dependent kinase Cdc7 and Cdc28 can directly regulate the meiotic DSB formation via the phosphorylation of Mer2 (Henderson et al., 2006; Sasanuma et al., 2008; Wan et al., 2008). However, in plants, only a few cyclins have been found involved in meiosis (Bulankova et al., 2013). The meiosis specific cyclin SDS was first found in A. thaliana, which play a specific role in regulating synapsis in prophase I (Azumi et al., 2002; Wang et al., 2004). In a recent study, SDS was found to be required for DMC1-mediated DSB repair (De Muyt et al., 2009). Although the rice SDS-RNAi plants showed the similar meiotic defects with those in Arabidopsis (Chang et al., 2009), the molecular mechanism of SDS in rice meiosis remains to be clear. Here, we identified the SDS homolog in rice by map-based cloning. Surprisingly, we found OsSDS is essential for DSB formation during rice meiosis, which is much different from that in Arabidopsis. We suspected this cyclin protein had been differentiated pretty much between monocots and dicots on its function in meiosis.
Materials and Methods
Plant Materials
The rice (Oryza sativa L.) spontaneous mutant Ossds-1 was isolated from an indica rice, Zhongxian 3037. The F2 and F3 mapping populations were generated by crossing the Ossds-1± heterozygous plants with a japonica cultivar, Zhonghua 11. The other two mutant alleles, Ossds-2 and Ossds-3, both were spontaneous mutants arose in tissue culture of Nipponbare. The meiotic mutant Osrad51c has been reported previously (Tang et al., 2014). The Ossds-1 Osrad51c double mutant was generated by crossing the two heterozygous Ossds-1± and Osrad51c±, and further identified from the F2 progeny. All plant materials were grown in paddy fields in the summer in Beijing or in the winter in Hainan.
Molecular Cloning of OsSDS
Total 861 sterile plants segregated from the F2 and F3 mapping populations were used for isolation the target gene. Sequence-tagged site (STS) markers were developed according to sequence differences between the japonica variety Nipponbare and the indica variety 9311, using the data published on the NCBI website (http://www.ncbi.nlm.nih.gov). All primers are listed in Supplemental Table 1.
RNAi Analysis
In the first exon, a 336-bp fragment of OsSDS cDNA sequence was chosen and amplified with the primers SDS-RNAi-F (adding a BamHI site) and SDS-RNAi-R (adding a SalI site) (Supplemental Table 1). RNAi vector construction and transformation were performed as described (Wang et al., 2009).
Complementation Test
The complementary plasmid was constructed by cloning the 10.8 kb OSJNBa0081P02 genomic DNA fragment containing the entire OsSDS coding region into the pCAMBIA-1300 vector. A control plasmid, containing 7.8 kb of the truncated OsSDS gene was also constructed. Both of these plasmids were transformed into EHA105 and then into embryonic calli of OsSDS± plants. The genotypes of the transgenic plants were further identify using the Primers SDS-JD (Supplemental Table 1).
Real-Time PCR for Transcript Expression Assay
Total RNA was extracted from the root, internode, leaf and panicle of Zhongxian 3037. Real-time PCR analysis was performed using the Bio-Rad CFX96 real-time PCR instrument (Bio-Rad, http://www.bio-rad.com/) and EvaGreen (Biotium, http://www.biotium.com/). The RT-PCR was carried out using the gene-specific primer pairs SDS-RT-F and SDS-RT-R for OsSDS. The primers Ubi-RT-F and Ubi-RT-R for ubiquitin were used as an internal control for the normalization of RNA sample. The results were analyzed using OPTICON MONITOR 3.1 (Bio-Rad). Each experiment had three replicates.
Cloning the Full-Length OsSDS cDNA
Total RNA was extracted from the panicle of Zhongxian 3037. The 3′ RACE and 5′ RACE were performed according to the protocol of the kit (3′-Full RACE Core Set and 5′ -Full RACE Core Set; Takara, http://www.takara-bio.com/). 3′ RACE was carried out using primers 3R-1F, 3R-2F, 3R-3F, and adaptor primer (P-ada). During 5′RACE, the RNA was reverse transcribed with 5′ (P)-labeled primer (SDS-4Rb); the first and second PCRs were performed using two sets of OsSDS specific primers (5R-1 and 5R-2). The 3′ RACE-PCR and 5′ RACE-PCR products were cloned and sequenced. OsSDS amino acid sequence translation and alignment were completed with the Vector NTI 11.5 (Invitrogen). OsSDS gene structure diagram was generated from GSDS (http://gsds.cbi.pku.edu.cn/index.php).
Meiotic Chromosome Preparation
Young panicles with appropriate size of both Ossds mutants and wild type were collected, fixed in Carnoy's solution (ethanol:glacial acetic, 3:1) and stored at −20°C. Microsporocytes at the appropriate meiotic stage were squashed and stained with acetocarmine. After washing the chromosome preparations with 45% acetic acid and freezing them in liquid nitrogen, the coverslips were quickly removed with a razor blade and the slides harbored with samples were dehydrated through an ethanol series (70, 90, and 100%) for 5 min each and finally air-dried. Chromosomes on slides were counterstained with 4, 6-diamidinophenylindole (DAPI) in an anti-fade solution (Vector Laboratories, Burlingame, CA, USA). Finally, images were captured under the ZEISS A2 fluorescence microscope with a micro CCD camera (Zeiss, http://www.zeiss.de/en).
Fluorescence Immunolocalization
Fresh young panicles (40–60 mm) were fixed with 4% (w/v) paraformaldehyde for 30 min at room temperature. Anthers with appropriate stage were squashed into one drop of 1×PBS solution added on a slide. Then, covering the slide with a coverslip and pressing it with appropriate strength, the slide together with the coverslip was frozen thoroughly in liquid nitrogen. After quickly prizing up the coverslip, the slide was dehydrated through an ethanol series (70, 90, and 100%). The following immunolocalization procedure was performed as described (Tang et al., 2014).
The polyclonal antibodies against γ-H2AX, OsMSH5, OsREC8, PAIR2, PAIR3, ZEP1, OsMER3, and OsZIP4 used in this study have been described previously (Wang et al., 2009, 2010; Shao et al., 2011; Shen et al., 2012; Zhang et al., 2012; Luo et al., 2013; Miao et al., 2013).
Results
Characterization of a Sterile Mutant
We identified a spontaneous mutant exhibiting complete sterility in a rice field of Zhongxian 3037. From the heterozygous plant progeny related to this mutation, the normal fertile plants and the sterile plants were segregated in a 3:1 ratio, indicating that it was a single recessive mutation (χ2 = 0.57; P > 0.05). The mutant plant was normal during vegetative growth and could not be distinguished from the wild type based on plant morphology (Supplemental Figure 1A). However, when come into reproductive stage, its spikelets exhibited complete sterility (Supplemental Figure 1B). So we further examined the mature pollen viability of the mutant by staining with 1% iodine potassium iodide solution (I2-KI) (Supplemental Figures 1C,D). Only empty and shrunken pollen grains were observed in the mutant plant, indicating that microspores of the mutant are all abnormal and inviable. Moreover, when pollinated with wild-type pollens, the mutant spikelets still did not set any seeds, indicating that its female gametes were also affected.
Map-based Cloning of OsSDS
To isolate the mutated gene, we constructed a population by crossing heterozygous plants with a japonica cultivar Zhonghua 11. A total of 861 sterile plants segregated from the F2 and F3 populations were used for mapping the target gene. Linkage analysis initially mapped the target gene onto the long arm of chromosome 3, which subsequently further delimited to a 130 kb region. Within this region, we found one candidate gene (Os03g12414) annotated as a putative cyclin with high similarity with SOLO DANCERS (SDS) gene in Arabidopsis. Thus, this candidate gene was chosen to be amplified and sequenced. Sequencing analysis revealed that there was a transversion happened from base C to T in the first exon, which produces a premature termination codon (TGA) and causes early termination (Figure 1A). We named the mutant here Ossds-1 and suspected the mutation of Os03g12414 leading to the sterile phenotype. We also obtained two other alleles arose from tissue culture of Nipponbare, named Ossds-2 and Ossds-3, respectively. They all showed the same defects as Ossds-1. Sequencing analysis showed that there were a transversion from base G to C at the third exon causing corresponding amino acid A replaced by P in Ossds-2 and two bases (GC) deletion at the fifth exon resulting in frame-shift mutation causing a premature termination codon (TGA) in Ossds-3 (Figure 1A). As earlier termination close to the start codon in Ossds-1 compared with the other mutants, it was selected for the subsequent experiments described blow. To further confirm the mutant phenotype was resulted by the mutation of OsSDS gene, RNA interference (RNAi) approach was carried out to down-regulate SDS in rice variety Yandao 8. We got 27 transgenic plants with 19 plants exhibited complete sterility. Additionally, the transformation of plasmid pCAMBIA-1300 containing the whole OsSDS gene was successful in rescuing the sterility of the mutant plants, just as respected. These results strongly confirmed that the mutation of OsSDS gene led to the sterile phenotype as described above.
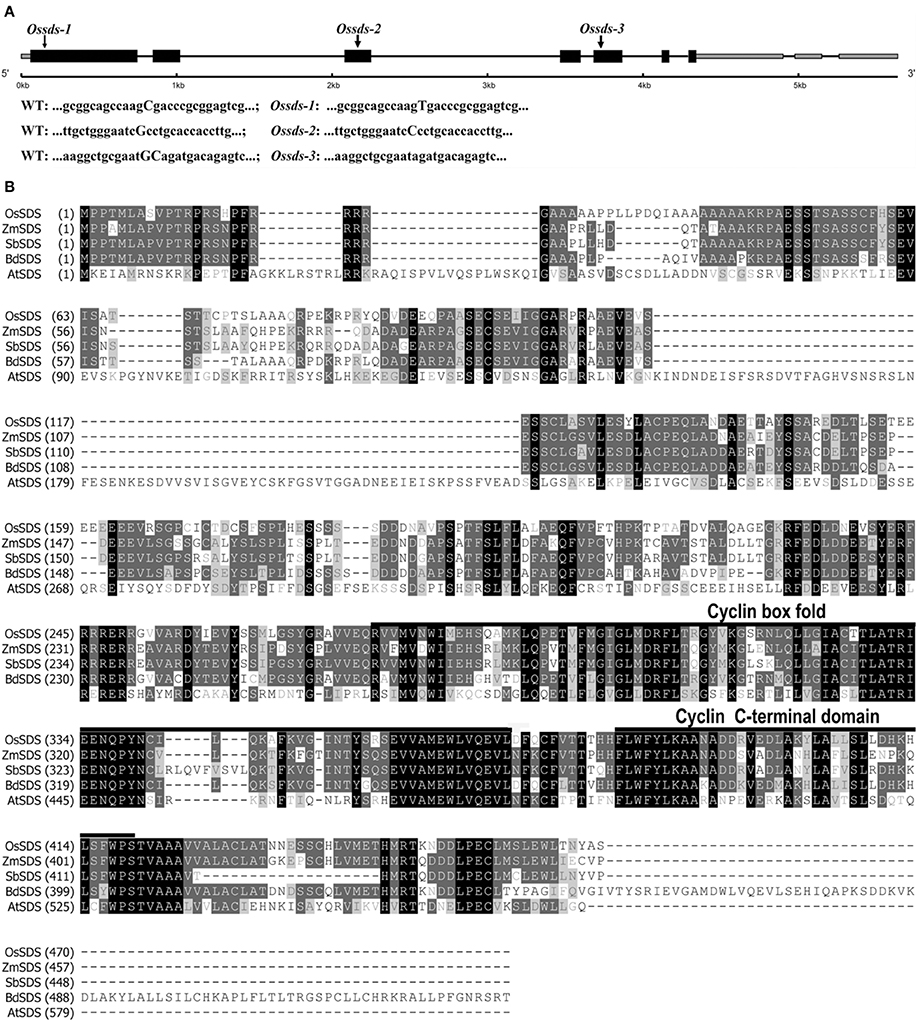
Figure 1. Gene structure of OsSDS and sequence alignment of SDS proteins. (A) Schematic of representation of gene structure and three mutation sites in OsSDS. Exons are represented by black boxes, intones are represented by black lines, 5′UTR and 3′UTR are represented by gray boxes. The capitalized bases in sequences of wild type and mutants represent accurate mutation sites of the three allelic mutants. (B) Multiple alignment of SDS protein sequences from different organisms. The numbers at the left of the sequences are the amino acid numbers. The black boxes represent identical sequences; the dark gray boxes represent conservative sequences; the light gray boxes represent weakly similar sequences. A predicted cyclin box fold domain (276–366 amino acids) and a predicted cyclin C-terminal domain (378–419 amino acids) are underlined.
Expression of OsSDS was also analyzed by real-time PCR (RT-PCR). As shown in Supplemental Figure 2, transcripts were all detected at root, internode, leaf and panicle, indicating that OsSDS is not a meiosis-specific gene.
Full-Length cDNA Cloning and Deduced Protein Sequence of OsSDS
The full-length cDNA of OsSDS gene was obtained by performing RT-PCR combined with 5′ and 3′ rapid amplification of cDNA ends PCR (RACE-PCR) using specific primers. We found that the OsSDS cDNA is comprised of 2786 bp containing an open reading frame (ORF) of 1410 bp and 1376 bp 5′ and 3′ untranslated regions (UTRs). The OsSDS cDNA sequence obtained is consistent with one mRNA (GenBank: AK065907.1) from the public network database (http://www.ncbi.nlm.nih.gov/). As shown in Figure 1A, the OsSDS gene contains seven exons and six introns. The predicted 469 amino acid protein of OsSDS shares as low as 30.6% identity with SDS in Arabidopsis, but with high similarity at the C-terminal (Figure 1B). Compared with the dicots Arabidopsis, OsSDS shares more than 60% identity with those in monocots, such as Zea mays, Sorghum bicolor, and Brachypodium. Conserved domain searching in NCBI revealed there are two conserved domains in SDS, namely, cyclin box fold domain (residues 276–366) and cyclin C-terminal domain (residues 378–419) (Figure 1B). Blast searching in NCBI revealed that SDS is a plant specific protein and owns one single copy in the plant kingdom.
Meiotic Defects in Ossds
To clarify whether the sterile phenotype of Ossds is caused by meiosis defects, chromosome behaviors at different stages of pollen mother cells (PMCs) from both wild type and Ossds-1 mutants were investigated. In wild type, chromosomes began to condense and became visible as thin thread-like structures at leptotene. After that, homologous chromosomes started to pair at zygotene. Fully synapsis between homologs formed at pachytene (Figure 2A). After further chromosome condensation, 12 bivalents were clearly observed at diakinesis (Figure 2B). Accompanying with the spindle installation, these bivalents aligned on the equatorial plate at metaphase I (Figure 2C). Then, homologous chromosomes separated and moved to the two opposite poles from anaphase I to telophase I (Figures 2D,E). During meiosis II, sister chromatids of each chromosome separated from each other and finally tetrads formed (Figure 2F).
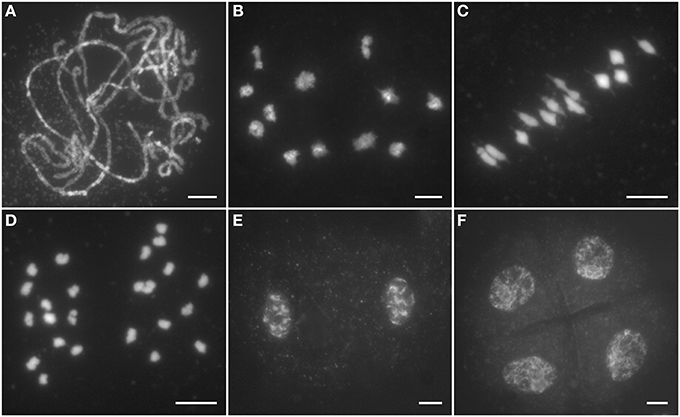
Figure 2. Male meiosis of the wild type. (A) Pachytene; (B) Diakinesis; (C) Metaphase I; (D) Anaphase I; (E) Dyad; (F) Tetrad. Chromosomes stained with 4,6-diamidino-2-phenylindole (DAPI). Bars = 5 μm.
In Ossds-1 PMCs, chromosome behaviors were similar to the wild type from leptotene to zygotene. However, obvious abnormities were first observed at pachytene stage, where the Ossds-1 mutant shows severely defects in homologous chromosome pairing and synapsis, and fully synapsed homologs were never observed (Figure 3A). Due to the lack of chromosome pairing, only univalents were observed at diakinesis (Figure 3B). During metaphase I, the univalents were unable to align on the equator plate (Figure 3C). From anaphase I to telophase I, they randomly segregated into two daughter cells (Figure 3D). In meiosis II, dyads and tetrads always exhibited different sizes caused by uneven chromosome segregation (Figures 3E,F). Cytological observation of meiocytes from the other two alleles, Ossds-2 and Ossds-3, as well as OsSDS RNAi plants showed the same meiotic defects as described in Ossds-1 (Supplemental Figure 3). Therefore, we proposed that the sterility of Ossds was caused by uneven segregation of homologous chromosomes without pairing and recombination during propose I.
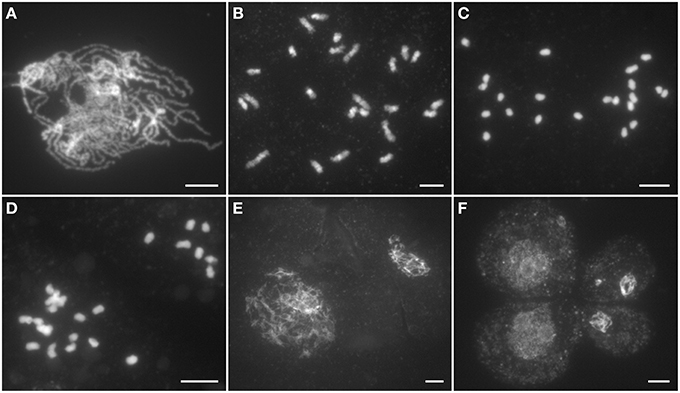
Figure 3. Male meiosis of the Ossds-1 mutant. (A) Pachytene; (B) Diakinesis; (C) Metaphase I; (D) Anaphase I; (E) Dyad; (F) Tetrad. Chromosomes stained with DAPI. Bars = 5 μm.
OsSDS is Essential for Meiotic DSB Formation
The above cytological observation indicated Ossds showed similar defects with the loss of function of CRC1, a meiotic DSB formation protein in rice (Miao et al., 2013). We wondered whether OsSDS is also required for meiotic DSB formation. Phosphorylation of histone H2AX occurs within a few minutes after a DSB initiated in mitosis (Banath et al., 2010), and the same kind phosphorylation takes place during the meiotic DSB formation (Dickey et al., 2009). To verify this hypothesis, a dual-immunostaining experiment was carried out utilizing antibodies specific for γH2AX and OsREC8 raised from rabbit and mouse, respectively, in the meiocytes both from wild type and Ossds-1. OsREC8, one of the key cohesion proteins in rice meiosis, is localized onto meiotic chromosomes from leptotene to metaphase I (Wang et al., 2009). Here, we used it as a biomarker to indicate the rice meiotic chromosomes in the prophase I specifically. Results showed that numerous dot and patchy immunosignals of γH2AX were detected at zygotene in wild type (Figure 4A). However, no γH2AX immunosignals was detected in Ossds-1 meiocytes at the corresponding stage (Figure 4B), showing that OsSDS is essential for meiotic DSB formation.
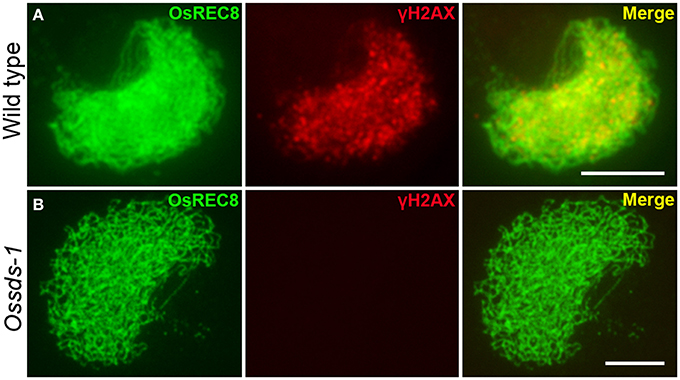
Figure 4. Immunostaining of γ-H2AX at zygotene in the wild type and Ossds-1 mutant. OsREC8 signals were used to indicate the chromosome axes. Bars = 5 μm.
During meiosis, all DSBs will be repaired by the repair system for maintaining genome stability. The loss function of repair proteins always results in chromosome fragmentation. OsRAD51C has been proved to be involved in meiotic DSB repair in rice (Tang et al., 2014). To verify this speculation, the Ossds-1 Osrad51c double mutant was generated by crossing the two heterozygous Ossds-1± and Osrad51c±, and further identified from their F2 progeny. In the Osrad51c mutant meiocytes, cytological observation shows 24 irregularly univalents appeared at diakinesis (Figure 5A). These univalents did not align well along the equatorial plate, and several chromosome fragments started to be shown at metaphase I (Figure 5B). Numerous chromosome fragments detained at the equatorial plate, while those massive chromosome bodies with centromeres moved into the two opposite poles at anaphase I (Figure 5C). While in the Ossds-1 Osrad51c meiocytes, chromosome behaviors were very much similar to Ossds-1 at the corresponding stages (Figures 5D–F). Together with the above γH2AX immunostaining data, we demonstrated very strong evidence that OsSDS is essential for DSB formation during rice meiosis.
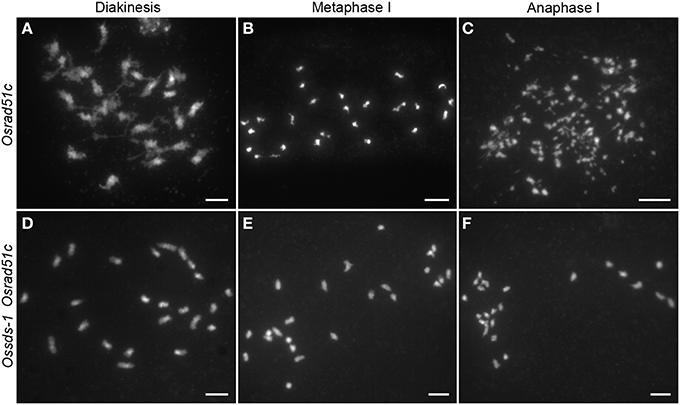
Figure 5. Comparison of chromosome behaviors between Osrad51c and the Ossds-1 Osrad51c double mutant. Chromosomes were stained with DAPI. Bars = 5 μm.
Meiotic Chromosome Axes Normally Formed but SC Installation Failed in Ossds
In rice, there are three axis-associated proteins OsREC8, PAIR2 and PAIR3 have been reported playing important roles in SC assembly (Nonomura et al., 2006; Shao et al., 2011; Wang et al., 2011). Except for those axial elements, ZEP1, the central element of SC, has also been identified (Wang et al., 2010). To investigate what kind of SC installation defects happened due to the loss function of OsSDS, we conducted immunodetection experiments using antibodies against PAIR2, PAIR3, and ZEP1 in Ossds-1 microsporocytes. The results showed that OsREC8 was normally localized onto chromosomes in Ossds-1 meiocytes. Moreover, both PAIR2 and PAIR3 were overlapped very well with OsREC8 at zygotene, indicating their normal localization along the chromosome axis (Figures 6A,B). However, ZEP1 signals always appeared as short dots or discontinuous lines at early prophase I (Figure 6C), showing the deficient central element installation of SC in the mutant. Thus, the meiotic chromosome axes normally formed but SC installation failed in Ossds.
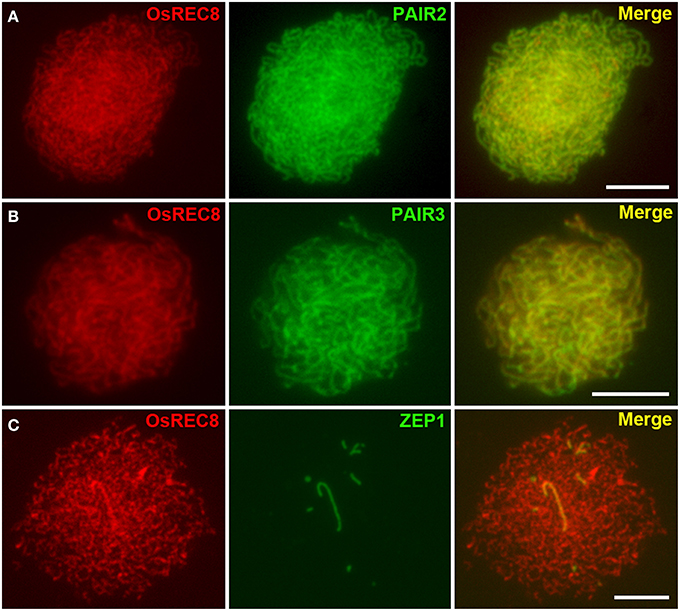
Figure 6. Dual immunostaining detection of several meiotic proteins in the Ossds-1. (A) OsREC8 (red) and PAIR2 (green) signals at late zygotene; (B) OsREC8 (red) and PAIR3 (green) signals at pachytene; (C) OsREC8 (red) and ZEP1 (green) signals at pachytene. Bars = 5 μm.
OsSDS is Critical for Faithful Localization of Recombination Elements onto Meiotic Chromosomes
Meiotic homologous recombination finally falls into two recombination pathways by forming two type crossovers, interference-sensitive COs (class I) and interference-insensitive COs (class II). ZMM proteins are closely associated with class I crossovers formation (Chen et al., 2008; Shinohara et al., 2008). In rice, OsMSH5, OsMER3 and OsZEP4 are three ZMM proteins participating in the class I COs formation (Wang et al., 2009; Shen et al., 2012; Luo et al., 2013). To determine whether the defective OsSDS also affects the localization of OsMSH5, OsMER3, and OsZEP4, dual immunolocalization were carried out using antibodies against OsMSH5, OsMER3, and OsZEP4. In the wild-type microsporocytes, immunostaining experiments showed that OsMSH5, OsMER3, and OsZEP4 were observed as punctuate foci on chromosomes at zygotene (Figures 7A–C), and these foci persisted until late pachytene stage (Wang et al., 2009; Shen et al., 2012; Luo et al., 2013). However, no obvious signal foci of OsMSH5, OsMER3, and OsZEP4 were observed on chromosomes in Ossds-1 microsporocytes at the corresponding stage (Figures 7D–F), indicating that OsSDS is critical for the localization of OsMSH5, OsMER3 and OsZEP4.
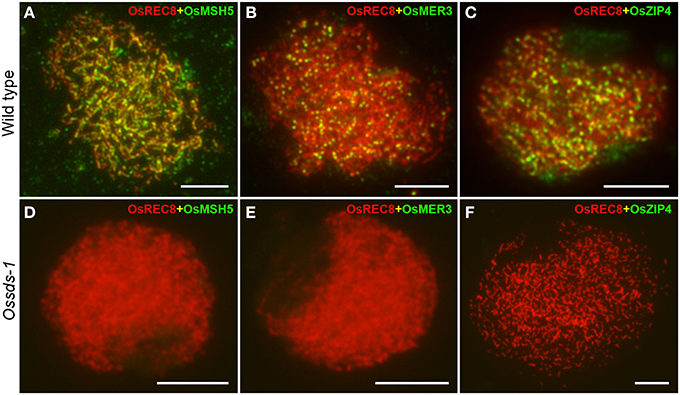
Figure 7. Immunostaining detection of three ZMM proteins in the wild type and Ossds-1. (A–C) Immunostaining for OsMSH5, OsMER3, and OsZIP4 at zygotene in the wild type. (D–F) Immunostaining for OsMSH5, OsMER3, and OsZIP4 at zygotene in Ossds-1. OsREC8 was used indicating chromosome axes. Bars = 5 μm.
Discussion
During meiosis, Spo11-catalyzed DSB formation is a highly conserved biological process among eukaryotes (Li and Ma, 2006). As increased data on plant meiosis research, several DSB formation proteins have been identified in higher plants, such as PRD1, PRD2, PRD3, and AtDFO in Arabidopsis (De Muyt et al., 2009; Zhang et al., 2012). And in rice, CRC1 was also reported to be involved in meiotic DSB formation besides those OsSPO11 homologs (Miao et al., 2013; Luo et al., 2014). Deficiency of these proteins always cause severe defects in homologous chromosome pairing, synapsis and nondisjunction.
MRE11 is known as an important DSB processing protein. The Atmre11 sds double mutant exhibited similar phenotype with the Atmre11 single mutant (De Muyt et al., 2009), proposing that SDS may be not required for meiotic DSB formation in Arabidopsis. Moreover, in sds mutant, the localization of DMC1 was abnormal while RAD51 was normally loaded. Thus, the role of SDS in Arabidopsis was thought to be necessary for DMC1-mediated DSB repair utilizing the homologous chromosome (De Muyt et al., 2009). While in rice, we provided evidences that OsSDS is essential for meiotic DSB formation. SDS is a plant specific cyclin protein. Amino acid sequences alignment revealed that SDS orthologs between dicots and monocots showed very low identities, Arabidopsis SDS sharing 30.6% identity with OsSDS, and as low as 26.1% with BdSDS. While among monocots, they share high identities. For example, ZmSDS shares 69.1% with OsSDS, and 83.1% with SbSDS. The diverged sequences between dicots and monocots suggest that SDS function in meiosis has been differentiated a lot during its evolution.
To date, the precise mechanisms of DSB formation are still unclear. It has been reported that the NBS1 protein is recruited to the end of the DSB soon after a DSB formed, which then initiates the formation of the NBS1/MRE11/RAD50 complex (Kobayashi, 2004). After that, the ATM protein phosphorylates the serine 139 residue of H2AX through its auto-phosphorlation and leading to γH2AX formation (Kinner et al., 2008). Studies revealed that γH2AX plays dual role in the DSB triggered signaling pathway: one is recruiting more MRN complex to the DSB site thus enhancing the signalization by a positive feedback loop; the other one is binding the DNA damage repair proteins (Paull et al., 2000; Minter-Dykhouse et al., 2008). The process of H2AX phosphorlation takes place just within a few minutes after a DSB occurrence (Banath et al., 2010). We did not detect any γH2AX immunosignals in Ossds, indicating OsSDS is required for DSB formation during rice meiosis.
It has been reported that cyclin-dependent kinase Cdc7 and Cdc28 can directly regulate the meiotic DSB formation via the phosphorylation of Mer2 in budding yeast (Henderson et al., 2006; Sasanuma et al., 2008; Wan et al., 2008). However, we still do not know the molecular mechanism of how rice cyclin OsSDS being involved in meiotic DSB formation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (U1302261, 31360260, and 31400169), and the Natural Science Foundation of Jiangsu Province (BK20140454).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00021/abstract
References
Allers, T., and Lichten, M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. doi: 10.1016/S0092-8674(01)00416-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
An, X. J., Deng, Z. Y., and Wang, T. (2011). OsSpo11-4, a rice homologue of the archaeal TopVIA protein, mediates double-strand DNA cleavage and interacts with OsTopVIB. PLoS ONE 6:e20327. doi: 10.1371/journal.pone.0020327
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ashley, T., and Plug, A. (1998). Caught in the act: deducing meiotic function from protein immunolocalization. Curr. Top. Dev. Biol. 37, 201–239. doi: 10.1016/S0070-2153(08)60175-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Azumi, Y., Liu, D., Zhao, D., Li, W., Wang, G., Hu, Y., et al. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21, 3081–3095. doi: 10.1093/emboj/cdf285
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Banath, J. P., Klokov, D., Macphail, S. H., Banuelos, C. A., and Olive, P. L. (2010). Residual gamma H2AX foci as an indication of lethal DNA lesions. BMC Cancer 10:4. doi: 10.1186/1471-2407-10-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergerat, A., De Massy, B., Gadelle, D., Varoutas, P. C., Nicolas, A., and Forterre, P. (1997). An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417. doi: 10.1038/386414a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bishop, D. K. (1994). RecA homologs Dmc1 and Rad51 interact to form multiple nuclear-complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092. doi: 10.1016/0092-8674(94)90038-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bishop, D. K., and Zickler, D. (2004). Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15. doi: 10.1016/S0092-8674(04)00297-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Borner, G. V., Kleckner, N., and Hunter, N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45. doi: 10.1016/S0092-8674(04)00292-2
Bulankova, P., Akimcheva, S., Fellner, N., and Riha, K. (2013). Identification of Arabidopsis meiotic cyclins reveals functional diversification among plant cyclin genes. PLoS Genet. 9:e1003508. doi: 10.1371/journal.pgen.1003508
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Celerin, M., Merino, S. T., Stone, J. E., Menzie, A. M., and Zolan, M. E. (2000). Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J. 19, 2739–2750. doi: 10.1093/emboj/19.11.2739
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, L., Ma, H., and Xue, H. W. (2009). Functional conservation of the meiotic genes SDS and RCK in male meiosis in the monocot rice. Cell Res. 19, 768–782. doi: 10.1038/cr.2009.52
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, S. Y., Tsubouchi, T., Rockmill, B., Sandler, J. S., Richards, D. R., Vader, G., et al. (2008). Global analysis of the meiotic crossover landscape. Dev. Cell 15, 401–415. doi: 10.1016/j.devcel.2008.07.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dawe, R. K. (1998). Meiotic chromosome organization and segregation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 371–395. doi: 10.1146/annurev.arplant.49.1.371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Muyt, A., Pereira, L., Vezon, D., Chelysheva, L., Gendrot, G., Chambon, A., et al. (2009). A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 5:e1000654. doi: 10.1371/journal.pgen.1000654
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dernburg, A. F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A. M. (1998). Meiotic recombination in C-elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398. doi: 10.1016/S0092-8674(00)81481-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dickey, J. S., Redon, C. E., Nakamura, A. J., Baird, B. J., Sedelnikova, O. A., and Bonner, W. M. (2009). H2AX: functional roles and potential applications. Chromosoma 118, 683–692. doi: 10.1007/s00412-009-0234-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600. doi: 10.1093/emboj/20.3.589
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hartung, F., and Puchta, H. (2000). Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 28, 1548–1554. doi: 10.1093/nar/28.7.1548
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hartung, F., Wurz-Wildersinn, R., Fuchs, J., Schubert, I., Suer, S., and Puchta, H. (2007). The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19, 3090–3099. doi: 10.1105/tpc.107.054817
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henderson, K. A., Kee, K., Maleki, S., Santini, P. A., and Keeney, S. (2006). Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125, 1321–1332. doi: 10.1016/j.cell.2006.04.039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hunter, N., and Kleckner, N. (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70. doi: 10.1016/S0092-8674(01)00430-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jain, M., Tyagi, A. K., and Khurana, J. P. (2006). Overexpression of putative topoisomerase 6 genes from rice confers stress tolerance in transgenic Arabidopsis plants. FEBS J. 273, 5245–5260. doi: 10.1111/j.1742-4658.2006.05518.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jain, M., Tyagi, A. K., and Khurana, J. P. (2008). Constitutive expression of a meiotic recombination protein gene homolog, OsTOP6A1, from rice confers abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Rep. 27, 767–778. doi: 10.1007/s00299-007-0491-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keeney, S. (2001). Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53. doi: 10.1016/S0070-2153(01)52008-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keeney, S. (2008). Spo11 and the formation of DNA double-strand breaks in Meiosis. Genome Dyn. Stab. 2, 81–123. doi: 10.1007/7050_2007_026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keeney, S., Giroux, C. N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. doi: 10.1016/S0092-8674(00)81876-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kinner, A., Wu, W. Q., Staudt, C., and Iliakis, G. (2008). gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36, 5678–5694. doi: 10.1093/nar/gkn550
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kobayashi, J. (2004). Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J. Radiat. Res. 45, 473–478. doi: 10.1269/jrr.45.473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, R., Bourbon, H. M., and De Massy, B. (2010). Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24, 1266–1280. doi: 10.1101/gad.571710
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, W. X., and Ma, H. (2006). Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 16, 402–412. doi: 10.1038/sj.cr.7310052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Libby, B. J., Reinholdt, L. G., and Schimenti, J. C. (2003). Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 15706–15711. doi: 10.1073/pnas.2432067100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, Q., Li, Y., Shen, Y., and Cheng, Z. (2014). Ten years of gene discovery for meiotic event control in rice. J. Genet. Genomics 41, 125–137. doi: 10.1016/j.jgg.2014.02.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, Q., Tang, D., Wang, M., Luo, W. X., Zhang, L., Qin, B. X., et al. (2013). The role of OsMSH5 in crossover formation during rice meiosis. Mol. Plant 6, 729–742. doi: 10.1093/mp/sss145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McKim, K. S., and Hayashi-Hagihara, A. (1998). mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12, 2932–2942. doi: 10.1101/gad.12.18.2932
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miao, C. B., Tang, D., Zhang, H. G., Wang, M., Li, Y. F., Tang, S. Z., et al. (2013). CENTRAL REGION COMPONENT1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25, 2998–3009. doi: 10.1105/tpc.113.113175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mimitou, E. P., and Symington, L. S. (2009). DNA end resection: many nucleases make light work. DNA Repair 8, 983–995. doi: 10.1016/j.dnarep.2009.04.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Minter-Dykhouse, K., Ward, I., Huen, M. S. Y., Chen, J., and Lou, Z. K. (2008). Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. J. Cell Biol. 181, 727–735. doi: 10.1083/jcb.200801083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nonomura, K. I., Nakano, M., Eiguchi, M., Suzuki, T., and Kurata, N. (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119, 217–225. doi: 10.1242/jcs.02736
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Page, S. L., and Hawley, R. S. (2003). Chromosome choreography: the meiotic ballet. Science 301, 785–789. doi: 10.1126/science.1086605
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Paques, F., and Haber, J. E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404.
Paull, T. T., Rogakou, E. P., Yamazaki, V., Kirchgessner, C. U., Gellert, M., and Bonner, W. M. (2000). A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895. doi: 10.1016/S0960-9822(00)00610-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Romanienko, P. J., and Camerini-Otero, R. D. (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987. doi: 10.1016/S1097-2765(00)00097-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sasanuma, H., Hirota, K., Fukuda, T., Kakusho, N., Kugou, K., Kawasaki, Y., et al. (2008). Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 22, 398–410. doi: 10.1101/gad.1626608
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shao, T., Tang, D., Wang, K. J., Wang, M., Che, L. X., Qin, B. X., et al. (2011). OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 156, 1386–1396. doi: 10.1104/pp.111.177428
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shen, Y., Tang, D., Wang, K. J., Wang, M., Huang, J., Luo, W. X., et al. (2012). ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 125, 2581–2591. doi: 10.1242/jcs.090993
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shingu, Y., Tokai, T., Agawa, Y., Toyota, K., Ahamed, S., Kawagishi-Kobayashi, M., et al. (2012). The double-stranded break-forming activity of plant SPO11s and a novel rice SPO11 revealed by a Drosophila bioassay. BMC Mol. Biol. 13:1. doi: 10.1186/1471-2199-13-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shinohara, M., Oh, S. D., Hunter, N., and Shinohara, A. (2008). Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40, 299–309. doi: 10.1038/ng.83
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sprink, T., and Hartung, F. (2014). The splicing fate of plant SPO11 genes. Front. Plant Sci. 5:214. doi: 10.3389/fpls.2014.00214
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stacey, N. J., Kuromori, T., Azumi, Y., Roberts, G., Breuer, C., Wada, T., et al. (2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48, 206–216. doi: 10.1111/j.1365-313X.2006.02867.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Symington, L. S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670. doi: 10.1128/MMBR.66.4.630-670.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tang, D., Miao, C. B., Li, Y. F., Wang, H. J., Liu, X. F., Yu, H. X., et al. (2014). OsRAD51C is essential for double-strand break repair in rice meiosis. Front. Plant Sci. 5:167. doi: 10.3389/fpls.2014.00167
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wan, L., Niu, H., Futcher, B., Zhang, C., Shokat, K. M., Boulton, S. J., et al. (2008). Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 22, 386–397. doi: 10.1101/gad.1626408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, G. F., Kong, H. Z., Sun, Y. J., Zhang, X. H., Zhang, W., Altman, N., et al. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135, 1084–1099. doi: 10.1104/pp.104.040436
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, K. J., Tang, D., Wang, M., Lu, J. F., Yu, H. X., Liu, J. F., et al. (2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 122, 2055–2063. doi: 10.1242/jcs.049080
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, K. J., Wang, M., Tang, D., Shen, Y., Qin, B. X., Li, M., et al. (2011). PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 22, 12–19. doi: 10.1091/mbc.E10-08-0667
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, M., Wang, K. J., Tang, D., Wei, C. X., Li, M., Shen, Y., et al. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430. doi: 10.1105/tpc.109.070789
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, H. X., Wang, M., Tang, D., Wang, K. J., Chen, F. L., Gong, Z. Y., et al. (2010). OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 119, 625–636. doi: 10.1007/s00412-010-0284-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, C., Song, Y., Cheng, Z. H., Wang, Y. X., Zhu, J., Ma, H., et al. (2012). The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant J. 72, 271–281. doi: 10.1111/j.1365-313X.2012.05075.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zickler, D., and Kleckner, N. (1999). Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33, 603–754. doi: 10.1146/annurev.genet.33.1.603
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: rice, OsSDS, meiosis, DSB formation
Citation: Wu Z, Ji J, Tang D, Wang H, Shen Y, Shi W, Li Y, Tan X, Cheng Z and Luo Q (2015) OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 6:21. doi: 10.3389/fpls.2015.00021
Received: 28 October 2014; Paper pending published: 13 December 2014;
Accepted: 10 January 2015; Published online: 03 February 2015.
Edited by:
Kang Chong, Chinese Academy of Sciences, ChinaCopyright © 2015 Wu, Ji, Tang, Wang, Shen, Shi, Li, Tan, Cheng and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhukuan Cheng, State Key Laboratory of Plant Genomics and Center for Plant Gene Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, No.1 West Beichen Road, Chaoyang District, Beijing 100101, China e-mail: zkcheng@genetics.ac.cn;
Qiong Luo, Ministry of Education Key Laboratory of Agriculture Biodiversity for Plant Disease Management, Yunnan Agricultural University, Heilongtan, Guandu District, Kunming 650201, China e-mail: qiongbf@aliyun.com
†These authors have contributed equally to this work.
 Zhigang Wu
Zhigang Wu Jianhui Ji
Jianhui Ji Ding Tang
Ding Tang Hongjun Wang
Hongjun Wang Yi Shen2
Yi Shen2 Yafei Li
Yafei Li Zhukuan Cheng
Zhukuan Cheng Qiong Luo
Qiong Luo