- 1Key Laboratory of Soil Environment and Pollution Remediation, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
- 2Key Laboratory of Coastal Zone Environmental Processes, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
Environmental pollutants have received considerable attention due to their serious effects on human health. There are physical, chemical, and biological means to remediate pollution; among them, bioremediation has become increasingly popular. The nitrogen-fixing rhizobia are widely distributed in the soil and root ecosystems and can increase legume growth and production by supplying nitrogen, resulting in the reduced need for fertilizer applications. Rhizobia also possess the biochemical and ecological capacity to degrade organic pollutants and are resistant to heavy metals, making them useful for rehabilitating contaminated soils. Moreover, rhizobia stimulate the survival and action of other biodegrading bacteria, thereby lowering the concentration of pollutants. The synergistic action of multiple rhizobial strains enhances both plant growth and the availability of pollutants ranging from heavy metals to persistent organic pollutants. Because phytoremediation has some restrictions, the beneficial interaction between plants and rhizobia provides a promising option for remediation. This review describes recent advances in the exploitation of rhizobia for the rehabilitation of contaminated soil and the biochemical and molecular mechanisms involved, thereby promoting further development of this novel bioremediation strategy into a widely accepted technique.
Introduction
Essential planetary functions such as primary production, the earth’s climate, biogeochemical and water cycling, and the maintenance of biodiversity have been severely undermined by anthropogenic activities (Teng et al., 2012; Alloway and Trevors, 2013; Valentín et al., 2013). Approximately 30% of the terrene environment is estimated to be degraded or contaminated, threatening agricultural production, and the environment (Alloway and Trevors, 2013; Valentín et al., 2013). In addition to contemporary pollutants such as heavy metals, hydrocarbons, and pesticides, a new generation of persistent organic pollutants (POPs) such as polybrominated diphenyl ethers (PBDEs), polychlorinated naphthalenes (PCNs), and perfluorooctanoic acid (PFOA) require urgent attention (Lohmann et al., 2007). Thus, there have been intensive studies investigating physico-chemical processes and bioaugmentation for their exploitation in multipurpose remediation technologies.
Although physico-chemical treatments (i.e., physical removal of contaminated soils, chemical extraction, and the application of chemical reagents) are still the most effective strategies to rapidly remediate heavily polluted sites, they are usually energy-intensive and intrusive for the environment (Segura and Ramos, 2013). In contrast, the less energy-demanding bioremediation utilizes living organisms and/or their bioproducts to clean up or stabilize inorganic/organic contaminants from the environment. Therefore, bioremediation is a promising alternative due to its relative low-level disturbance of contaminated sites, low cost and higher public acceptance compared with conventional remediation methods. Among the various types of bioremediation, phytoremediation is an environmentally friendly and cost effective approach that provides intangible benefits for the soil ecosystem, including soil carbon sequestration, soil quality improvement, biomass and biofuel production, and biodiversity maintenance (Rajkumar et al., 2012). Legumes are essential for nitrogen cycling in agriculture due to their symbiosis with the nitrogen-fixing rhizobia. Many reports have noted that some leguminous species are heavy-metal resistant and can significantly promote the dissipation of organic pollutants [i.e., polychlorinated biphenyls (PCBs), polycyclic aromatic hydrocarbons (PAHs), and amide herbicides; Fan et al., 2008; Hamdi et al., 2012; Carrasco-Gil et al., 2013; Li et al., 2013]. Intercropping of multiple leguminous plants may also become a promising in situ bioremediation strategy for contaminated sites (Sun et al., 2011a; Li et al., 2013).
The symbiosis between microorganisms and plants has been employed for the elimination of environmental contaminants to achieve high effectiveness and ecological sustainability. The effectiveness of phyto- or microbial-remediation is dependent on: (i) soil physio-chemical properties, such as pH, nutrient/organic matter content, soil surface properties, soil texture and bulk densities that influence plant–soil–water relationships and nutrient availability; (ii) toxicity or bioavailability of the targeted contaminants that reduce the productivity of the impacted soils, the biomass of plants and the degradative ability of microorganisms; (iii) plant species and traits; and (iv) the diversity and richness of the indigenous soil microbial communities or flora (Segura et al., 2009). However, these limitations can be addressed through the exploitation of the chemical interactions between the plants and the related rhizospheric microbes or endophytes (Abhilash et al., 2012). For the phytoremediation of heavy metals, heavy metal-resistant microbes can enhance plant growth, decrease metal phytotoxicity, and affect metal translocation and accumulation in plants (Li et al., 2012).
Rhizobiales, belonging to the alphaproteobacteria, are Gram-negative bacteria of agronomic importance because some species form nitrogen-fixing symbiotic relationships with leguminous plants (Sato et al., 2005). Rhizobia invade the roots of legumes and form nodules to fix atmospheric nitrogen into ammonia, which is then provided to the host plants. This activity allows the plants to grow in the absence of an external nitrogen source (Figure 1; Prell and Poole, 2006; Deakin and Broughton, 2009). Hydrogen (H2) is a by-product of the symbiotic nitrogen fixation process and has recently been revealed to be a common element with novel bioactive properties that enhances plant tolerance to abiotic factors (i.e., oxidative stress and heavy metal toxicity; Cui et al., 2013; Jin et al., 2013). Because nitrogen is of the utmost importance for agricultural productivity, rhizobia have attained a special position in the field of agriculture as a plant growth promoter.
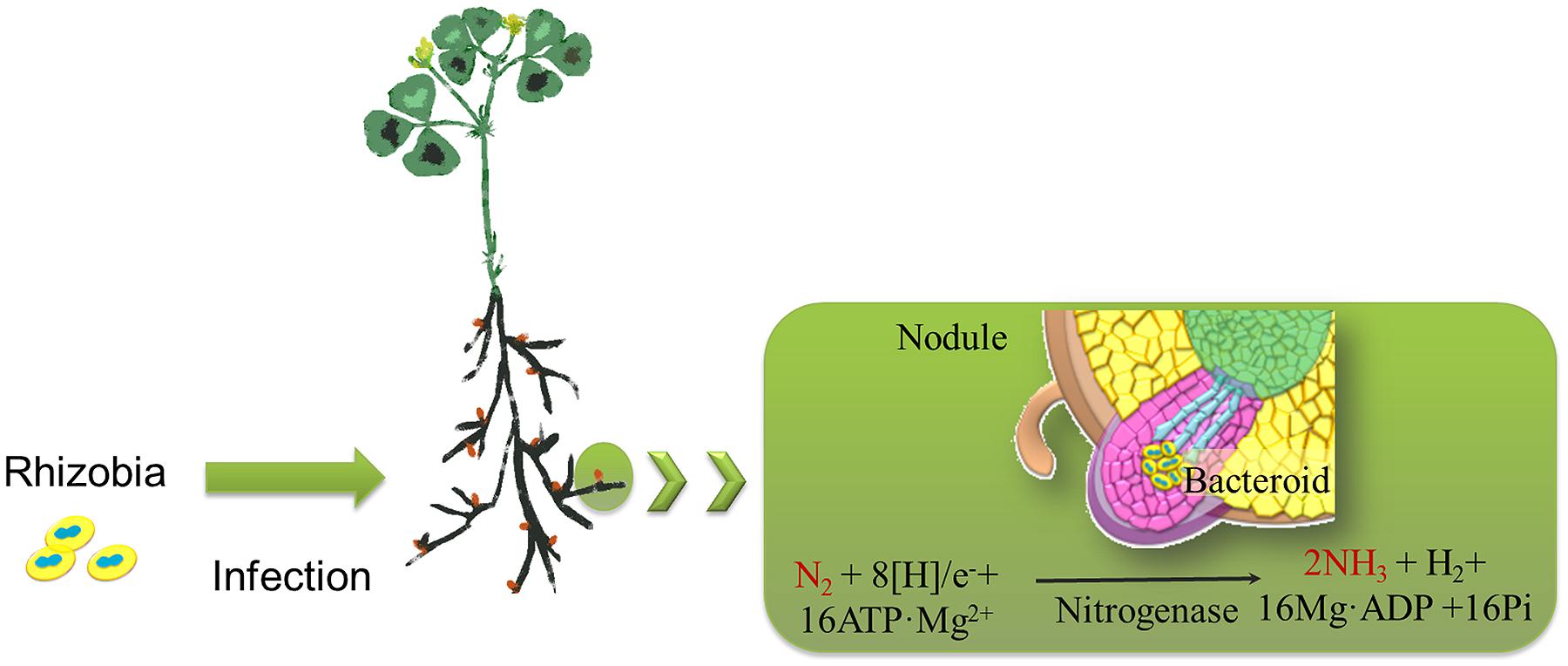
FIGURE 1. Schematic drawing representing the nitrogen-fixing process associated with rhizobia. Rhizobia invade the roots of legumes (i.e., alfalfa) and form nodules. During the process of biological nitrogen fixation in nodules, dinitrogen (N2) is reduced to two ammonia (NH3) molecules by the rhizobial nitrogenase. Hydrogen (H2) is a by-product of the symbiotic nitrogen fixation process.
Recently, rhizobia have been demonstrated to be available for the elimination of various types of organic pollutants from the environment, ranging from aromatic to linear hydrocarbons, chlorinated compounds, phenolic compounds, pesticides, and others (Table S1). Kaiya et al. (2012) reported that genus Rhizobium was one of the most abundant members of the degrading microcosm in dibenzofuran-contaminated soil. However, the bacterial catabolic enzymes and the pathways involved in the degradation of these compounds are only partially known (Figure 2). In addition to organic compounds, rhizobia have also been shown to have the potential to be a powerful tool for heavy metal bioremediation (Hao et al., 2014). Potential mechanisms involved are: (i) adsorption and accumulation of heavy metals; (ii) microbial secretion of enzymes and bioactive metabolites (i.e., extracellular polymeric substance, siderophores, and organic acids) to lessen their toxicity by altering the redox state of metals and increasing the complexation and bioavailability of metals; these actions can also indirectly aid phytoremediation (Hao et al., 2014); and (iii) microbial volatilization of heavy metals and their transformed products can also facilitate bioremediation, although this process has yet to be identified in rhizobia.
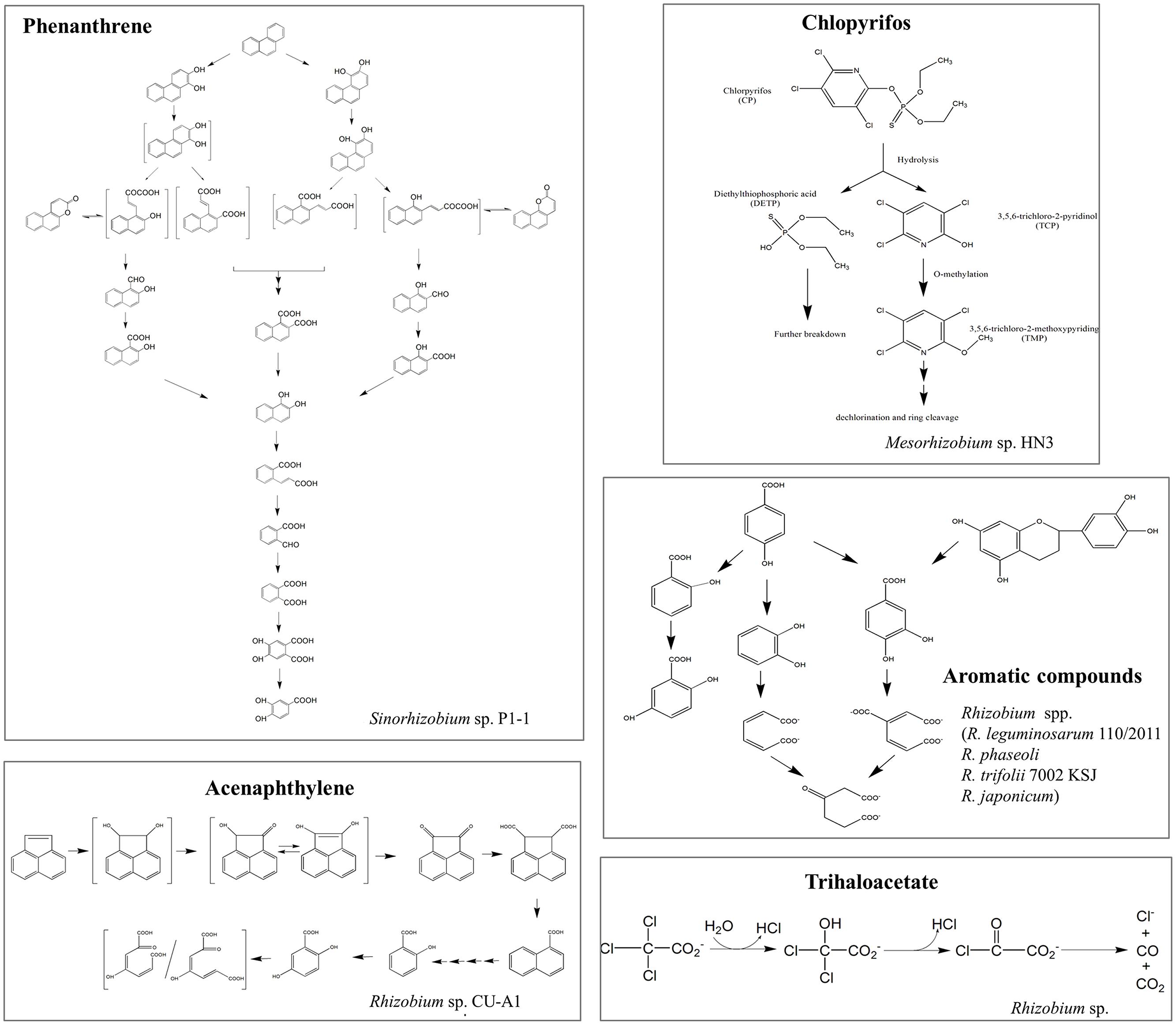
FIGURE 2. Schematic representations of the proposed degradation pathways of several organic pollutants in rhizobial strains. Organic pollutants: phenanthrene (Keum et al., 2006), chlorpyrifos (Jabeen et al., 2014), aromatic compounds (Muthukumar et al., 1982), acenaphthylene (Poonthrigpun et al., 2006), and trihaloacetate (Stringfellow et al., 1997). Structures in brackets represent the hypothetical metabolites.
Due to copious production of plant biomass in terrestrial ecosystems, microbial symbionts constitute the ‘unseen majority’ during phytoremediation (Fester et al., 2014). The nitrogen-fixing and plant growth-promoting traits of rhizobia directly improve plant biomass, soil fertility, bioavailability of contaminants, the uptake, and translocation of pollutants from soil to plant, and the ability to degrade organic pollutants and indirectly help phytostabilize metals. These traits could help rhizobia overcome the constraints associated with phytoremediation (see Assisted-phytoremediation) and achieve higher efficiency (Hao et al., 2014); therefore, the symbiotic relationship between rhizobia and legumes results in an enhanced removal rate for pollutants (Glick, 2010). In comparison with rhizospheric microorganisms (including non-symbiotic diazotrophs), the balanced and stable endophytic association between rhizobia and their host plants provides a sustainable way to improve the performance of host plants in the context of the plant life cycle (Li et al., 2012). Thus, rhizobial remediation may represent a low-input biotechnology with no need for repeated inoculations of microbial agents. Some rhizospheric microbes capable of removing pollutants cannot survive and achieve bioremediation in the soil environment because they cannot compete with indigenous organisms. Bacterial inoculation agents also have advantages compared with fungal elicitors, such as a short period for culture and elicitation of responses in host plants (Wang et al., 2015). Moreover, beneficial bacteria often trigger relatively weaker defense responses than fungal elicitors, which might facilitate the sustainable and balanced relationships between the bioremediation partners. This might be attributed to the smaller niches that bacteria occupied than fungi (Wang et al., 2015). Another advantage of using rhizobia in organic pollutant bioremediation is that rhizobial nitrogen-fixation removes the limitation caused by nitrogen deficiency in sites. The deposition of hydrocarbon skeletons significantly increases the accumulation of organic carbon in the soil and generates a very high C/N imbalance during bioremediation (Moreto et al., 2005; Dashti et al., 2010). The presence of rhizobia can also exert direct or indirect impacts on microbial-degrader communities in the soil, thereby comprehensively facilitating restoration (Li et al., 2013). The mechanisms involved in this process include: (i) improvement of environmental conditions (i.e., pH) and nutrient availability (i.e., nitrogen) and (ii) changes in the amounts and constituents of root exudates due to the enhancement of plant metabolic activities following inoculation with rhizobia (Johnson et al., 2005; Terrence et al., 2011). Nevertheless, because hyperaccumulator plants are often limited in their plant-metabolic capacities with toxic contaminants, researchers have proposed that in most cases it is the plant-associated microorganisms that are the real players mediating the plants’ impacts on the completed transformation of contaminant (Fester et al., 2014).
In this review, we will introduce the latest findings in the field of bioremediation by rhizobia and summarize the knowledge of the restoration mechanism and modified strategies for rhizobia involved in ecosystem revitalization in contaminated sites.
Rhizobia: Workers for Bioremediation
Degradation of Organic Pollutants
Many free-living rhizobial strains in the genera Agrobacterium, Rhizobium, Sinorhizobium, and Bradyrhizobium have a demonstrated capacity to thrive in or utilize PAHs, PCBs, aromatic heterocycles (i.e., pyridine), or other toxic organic compounds (Keum et al., 2006; Poonthrigpun et al., 2006; Tu et al., 2011). Hussien et al. (1974) first isolated 22 strains of Rhizobium capable of degrading phenolic compounds (i.e., catechol, protocatechuic acid, p-hydroxybenzoic acid, and salicylic acid). Among them, Rhizobium sp. and R. phaseoli 405 dissimilated p-hydroxybenzoate to salicylate and then to gentisic acid before oxidation. Catechol and protocatechuic acid were also directly cleaved by these species, whereas R. japonicum converted catechin to protocatechuic acid (Muthukumar et al., 1982; Table S1).
Polycyclic aromatic hydrocarbons, a class of hazardous chemicals consisting of two or more fused benzene rings in various structural configurations, are listed as priority toxic pollutants by the U.S. Environmental Protection Agency due to their carcinogenicity, mutagenicity, and toxicity (Poonthrigpun et al., 2006). Ahmad et al. (1997) first isolated and characterized a variety of strains of Rhizobium meliloti in soils contaminated with aromatic/chloroaromatic hydrocarbons. They also found that the rhizobial population was composed of several phenotypically and genetically distinct strains and that all were effective in symbiotic N2-fixation. Acenaphthylene and phenanthrene are ubiquitous PAHs in the environment. Acenaphthylene (600 mg liter-1) can be totally degraded by Rhizobium sp. strain CU-A1 within three days through the naphthalene-1, 8-dicarboxylic acid metabolism pathway (Poonthrigpun et al., 2006). Sinorhizobium sp. C4 can utilize phenanthrene as a sole carbon source, and 16 intermediate metabolites involved in this degradation pathway have been identified (Keum et al., 2006).
Polychlorinated biphenyls are a class of POPs differing in the number of chlorine atoms (1–10) attached to their biphenyl rings (Passatore et al., 2014). Damaj and Ahmad (1996) revealed that rhizobia could act as a promising candidate for PCB degradation. Tu et al. (2011) demonstrated that Sinorhizobium meliloti ACCC17519 degraded more than 70% of 2,4,4′-TCB (PCB28), which was more efficient than the reported performance of other rhizobial strains. In experiments under aerobic conditions, 2-hydroxy-6-oxo-6-phenylhex-2,4-dienoic acid (HOPDA), the meta cleavage product in the classic PCBs-degradative pathway, was identified using GC-MS as the principal intermediate during the biotransformation of 2,4,4′-TCB by S. meliloti.
Some toxic aromatic acids and their hydroaromatic biosynthetic intermediates (i.e., quinate and shikimate) commonly distributed in plants and the rhizosphere have also been found to support the growth of rhizobia (Parke et al., 1985). Mimosine [β-N-(3-hydroxy-4-pyrid-one)-aminopropionic acid], an aromatic toxin produced by the roots of Leucaena sp., is toxic to both bacteria and eukaryotic cells (Awaya et al., 2005). Some Leucaena-nodulating Rhizobium strains have been reported to be able to utilize mimosine as a source of carbon and nitrogen (Soedarjo et al., 1995; Soedarjo and Borthakur, 1998), highlighting the catabolic capacity of aromatic compounds in rhizobia.
Degradative Mechanisms Involved in Organic Pollutants
Because rhizobial strains are regularly exposed and adapted to recalcitrant pollutants, they are presumed to have developed enzymatic profiles essential for the degradation of these toxins (Figure 2; Zinjarde et al., 2014). Some aromatic acids and hydroaromatic biosynthetic intermediates (i.e., quinate and shikimate) commonly found within plants and in the rhizosphere could also support the growth of diverse rhizobial species (Parke et al., 1985). Indeed, genes for the catabolism of these compounds have been widely found in the genomes of rhizobiales and are conserved among different members. Linking rhizobial degradative gene expression to the decontamination and recycling of pollutants is crucial for illustrating the role played by these rhizobia in environment revitalization.
The microbial PCB-degradation system includes two major metabolic steps: (i) anaerobic reductive dechlorination, where PCBs are transformed into less chlorinated congeners; and (ii) aerobic breakdown of the biphenyl structure in lower-halogenated congeners (containing less than five chlorines), resulting in chloro-HOPDA (2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate), and the production of chlorobenzoic acid, ring opening, and potentially complete mineralization (Passatore et al., 2014). The oxidative pathways of PCB degradation in rhizobia have been documented in several studies. Aerobic rhizobial degradation of PCBs typically proceeds via the oxidative biphenyl pathway encoded by the bph genes that include a multi-component dioxygenase (bph A, E, F, and G), a dehydrogenase (bph B), a second dioxygenase (bph C), and a hydrolase (bph D) in other bacteria. Researchers first found that genomic DNAs from Rhizobium and Bradyrhizobium hybridized strongly with the Comamonas testosteroni-derived bphABC gene probe, indicating the presence of a similar oxidative degradation system in rhizobia (Damaj and Ahmad, 1996; Ahmad et al., 1997). Xu et al. (2010) and Tu et al. (2011) showed that R. meliloti and S. meliloti could utilize 2,4,4′-TCB (PCB 28) as a sole carbon and energy source under aerobic conditions, and HOPDA has been identified as the main intermediate during the biotransformation of 2,4,4′-TCB by S. meliloti.
However, little is known about the dehalogenases responsible for the reductive dechlorination of PCBs in the aerobic rhizobia strains (Aken et al., 2010). Most dehalogenase-producing bacteria are anaerobic species and contain more than one dehalogenase. Leigh et al. (1988) reported that a fast-growing Rhizobium sp. utilized 2, 2-dichloropropionate (2, 2-DCP) and D, L-2-chloropropionate (DL-2CP) as sole sources of carbon and energy and contained three forms of inducible dehalogenases.
The degradation pathways of different rhizobial species vary. Muthukumar et al. (1982) reported that p-hydroxybenzoate was metabolized to protocatechuate that in turn was cleaved by protocatechuate 3, 4-dioxygenase via the ortho pathway in R. leguminosarum, R. phaseoli, and R. trifolii, whereas R. japonicum degraded p-hydroxybenzoate to catechol that was cleaved by catechol 1,2-dioxygenase. Moreover, the regulatory mechanisms for rhizobial bioremediation are related to various rhizobial catabolism pathways, thereby indicating that further investigations are needed. For example, the presence of glutamate favored the degradation of p-hydroxybenzoic and salicylic acids but had little effect on catechol during the rhizobial degradation of aromatic compounds (Hussien et al., 1974).
Researchers have also proposed that although some rhizobial species carry DNA sequences that are homologous to degradative genes, they may not be involved in the degradation process and may instead function in some other capacity (Ahmad et al., 1997). Sato et al. (2005) showed that M. loti MAFF303099 and Bradyrhizobium japonicum USDA110 possessed dehalogenase-like opening reading frame fragments (ORFs) in their genomes and produced functional haloalkane dehalogenases, but they did not function as halogenated compound degraders. Other researchers have reported that some strains of rhizobia (i.e., S. meliloti) did not possess genes for the bioremediation of pollutants (Chen et al., 2005). These outcomes could be due to the inefficient induction of degradative genes limited by plant exudates, O2-tension, cell density signals, and other environmental factors, the genetic background of the rhizobia, the interdependent regulation of the genes, the flux of metabolic intermediates, or the presence of their end products and non-specific transformation products (Damaj and Ahmad, 1996).
Remediation of Metallic Contaminants
Metals and metalloids are persistent toxins in organisms at high concentrations due to the fact that they are non-degradable and irreversibly immobilized in the environment. The selective pressure exerted by metals on microorganisms results in microbial populations with a high tolerance to metals as well as some tolerant metal hyper-accumulators (Pereira et al., 2006). There has been increasing concern over heavy metal resistance (i.e., arsenic, cadmium, zinc, copper, and lead) in free-living or symbiotic rhizobia and its effects on their potential for bioremediation (Hao et al., 2014). Heavy metal-resistant strains are commonly isolated from nodules of the metallicolous legume (i.e., Robinia pseudoacacia, Anthyllis vulneraria, and Glycine max) from mining tailings or contaminated sites. Smith and Giller (1992) first isolated strains of heavy metal-resistant R. leguminosarum from contaminated sewage sludge and mine spoils. Plant-growth promoting Rhizobium sp. and Bradyrhizobium sp. are the most common rhizobial strains identified at heavy metal-contaminated sites (Sriprang et al., 2002).
The mechanisms involved in the metal-resistance system have been studied at the nucleic acid and protein level in many rhizobial species. The results of Hao et al. (2012) revealed that the Mesorhizobium amorphae CCNWGS0123 genome carried multiple genes potentially involved in copper resistance, including cusAB (encoding a resistance-nodulation-cell division protein family-type copper efflux system), genes encoding P1B-type ATPases involved in heavy metal transition and translocation, and operons encoding copper resistance determinants. S. meliloti has been reported to detoxify arsenic through an aquaglyceroporin (AqpS) that has a physiological function in arsenic detoxification and resistance (ARS) due to the presence of the ars genes (Yang et al., 2005). By using the comparative RNAseq-based approach, Maynaud et al. (2013) identified genes that specifically responded to zinc and cadmium and demonstrated that several genes encoding metal efflux and sequestration systems were significantly up-regulated; thus, these genes could be considered to be involved in the most widely represented mechanisms of rhizobial metal tolerance. Pereira et al. (2006) reported that metals (zinc, lead, cadmium, chromium, and nickel) negatively influenced bacterial protein profiles, especially for polypeptide expression, whereas in tolerant Rhizobium strains these alterations mostly increased correspondingly.
Based on studies in other bacteria, the metal resistance of rhizobia might be attributed to (Figure 3): (i) changes in the metal efflux of microbial cell membranes; (ii) intracellular chelation due to the production of metallothionein proteins (Nies, 1995); and (iii) the transformation of heavy metals to their less toxic oxidated forms through microbial metabolism (Nies, 2003). For example, the increased contents of reductive agents (i.e., glutathione concentrations) in microbial cells might reduce the toxicity of cadmium, thereby contributing to cadmium-resistance (Bright and Bulgheresi, 2010). Moreover, the metabolism of rhizobia also increases metal bioavailability in the soil through alterations in the soil pH, resulting in the release of chelators (i.e., siderophores) and organic acids capable of enhancing the complexation of metals and their mobility (Schalk et al., 2011). Microbial volatilization is another preferred method of metal bioremoval (i.e., selenium and mercury) in many rhizosphere bacteria (Souza et al., 2001; Zhang et al., 2012), although the mechanisms for the volatilization of metals in rhizobia have yet to be identified. Studies have suggested that engineering rhizobia for the volatilization of heavy metals could be a valuable avenue for tackling soil pollution. For example, Chen et al. (2014) demonstrated that Pseudomonas putida KT2440 endowed with the arsM gene encoding the As(III) S-adenosylmethionine (SAM) methyltransferase from Rhodopseudomonas palustris could remove arsenic from contaminated soil through microbial arsenic methylation and volatilization.
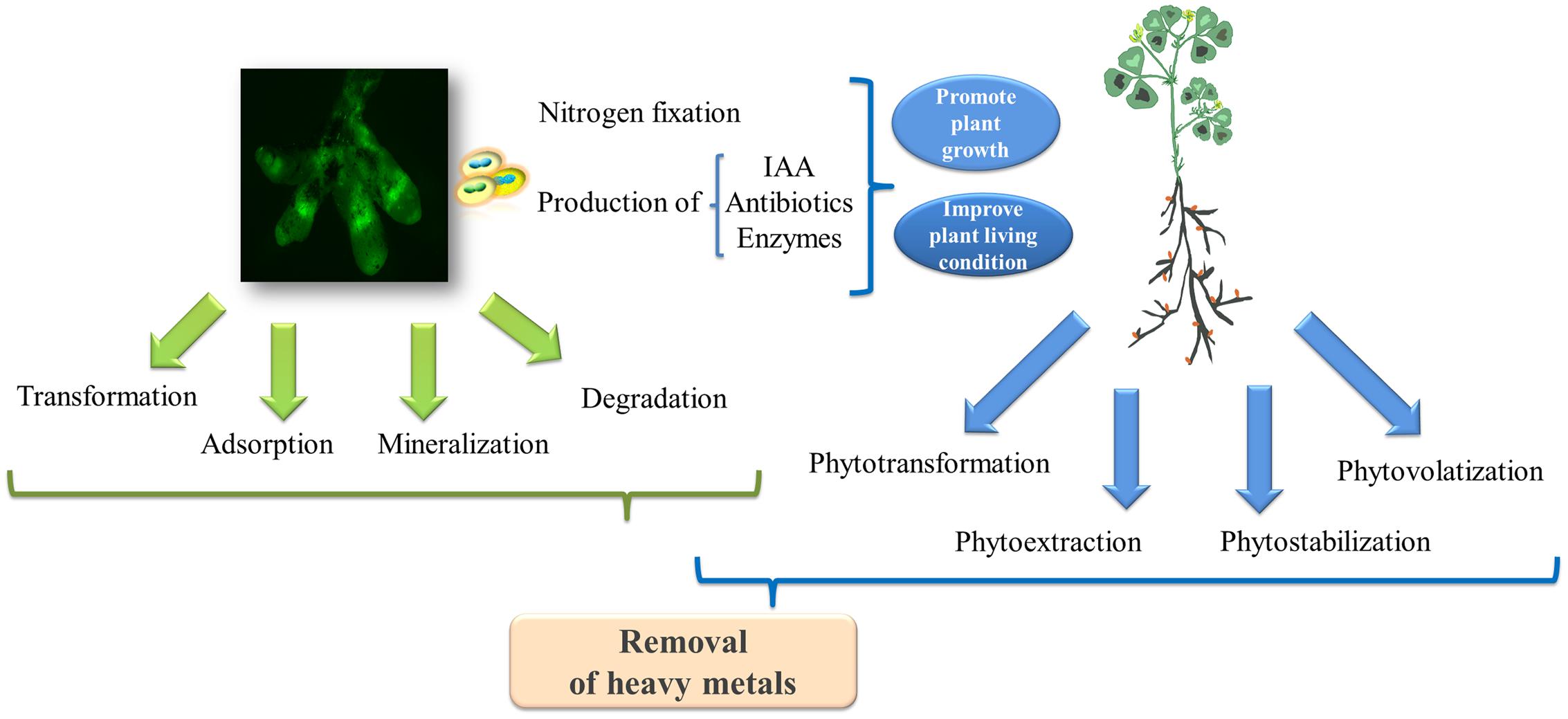
FIGURE 3. The biodegradation mechanisms involved in the legume-rhizobia symbiosis for the removal of heavy metals. The green fluorescent protein (GFP)-labeled rhizobia in the nodules was photographed by Chen Tu at the Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences.
Assisted-Phytoremediation
Taking advantage of microbe-plant cross-talk offers a low-input biotechnology for ecosystem revitalization in toxic and nutrient-limited environments (bio-augmentation; Abhilash et al., 2012). Bacterial bioremediation in the field is often affected by a variety of factors, including microbial competition, fluctuating environmental conditions, and limited nutrients (Chen et al., 2005). Thus, the selection of effective contaminant-degrading bacteria alone is not enough to ensure optimal remediation. Moreover, phytoremediation is often regarded as slow and incomplete due to limitations in plant-metabolic capacities, rooting depths, and the seasonality of plant growth (Abhilash et al., 2012). It has also been proposed that the storage and accumulation of organic pollutants (or their metabolites) and toxic metals in plant tissues reduces plant survival and results in atmospheric contamination via volatilization through the leaves (Abhilash et al., 2012). Therefore, much work is still required for phytoremediation to achieve an effective performance within a reasonable time frame (Glick, 2010; Fester et al., 2014). The inclusion of microorganisms interacting with plants will address the weaknesses of the two individual systems. Legumes are considered to be pioneer plants for phytoremediation (Hao et al., 2014). The huge variety in the metabolic pathways employed by microbes makes them valuable tools to assist phytoremediation. The endophytic helper rhizobia, acting as ‘microbial logistics,’ break down contaminants that have accumulated in nodules, greatly reducing phytovolatilization, and facilitating phytoremediation in the rhizosphere and other environments. Sun et al. (2011b) showed that the accumulation of PCBs was higher in nodules than any other part of the alfalfa plant, indicating that high concentration of PCBs accumulated in alfalfa nodules. Furthermore, Li et al. (2013) demonstrated that Rhizobium was effective in removing PCBs when inoculated with alfalfa in pot experiments.
Legumes can enhance or stabilize rhizobial degradation and the biotransformation of various pollutants, which offers many advantages (de-Bashan et al., 2011): (i) the stimulation of legume growth conferred by the plant growth-promoting traits of rhizobia, including nitrogen fixation, phosphorus solubilization, phytohormone synthesis, siderophore release, and the production of ACC deaminase and volatile compounds (i.e., acetoin and 2, 3-butanediol); (ii) the phytostabilization and phytoextraction of heavy metals by the combined actions of the plants and microbes; (iii) the immobilization of contaminants and increasing soil organic content via root exudates; (iv) the provision of additional nitrogenous compounds to the soil, thereby improving soil fertility and supporting biological growth; and v) the modification of the structure and diversity of microflora [i.e., rhizobacteria and arbuscular mycorrhiza fungi (AMF)], which help immobilize metals, promote microbial degradation, and enhance the growth and phyto-stabilization of the legumes.
Various species of rhizobia are appropriate candidates for the phytoremediation of organic contaminants. The phytoremediation of organic contaminants mainly occurs by three mechanisms: phytoextraction, phytodegradation, and phytovolatilization. These three mechanisms demand a high biomass of plants and high availability of pollutants, both of which can be achieved by the N-fixation process and the secretions of rhizobia (Johnson et al., 2004). For example, the collaboration between Medicago sativa (alfalfa) and S. meliloti for the removal of aromatic pollutants has been extensively studied. Mehmannavaza et al. (2002) reported that a combined treatment (S. meliloti A-025 and alfalfa grown together) was the most effective for PCB biotransformation after 44 days. However, after 270 days alfalfa grown alone became the most effective treatment, whereas S. meliloti alone was the least effective. Xu et al. (2008) conducted further field experiments and found that PCB removal from rhizosphere soil was clearly enhanced in alfalfa simultaneously inoculated with R. meliloti at 90 days after planting. Similarly, in a pot study Teng et al. (2011) found that planting alfalfa inoculated with R. meliloti significantly lowered the initial soil PAH concentrations by 51.4% compared with unplanted control soil.
The phytoremediation of heavy metals might be impeded by their low bio-availability due to their insolubility and soil-bound properties (DalCorso et al., 2013). A collaborative system consisting of metal(loid) resistant rhizobia and legumes holds promising potential for the removal of metal and metalloid ions (e.g., the Mimosa pudica-Cupriavidus taiwanensis symbionts system; Chen et al., 2008). Hao et al. (2014) reviewed the phytoremediation of heavy and transition metals aided by legume-rhizobia symbiosis and concluded that rhizobial metabolism increased: (i) the uptake and translocation of metals from soils to plants due to their increased bioavailability; (ii) microbial extracellular polymeric substance production and enzyme activities that could immobilize and/or change the redox state of metals to lessen their toxicity to plants (Hao et al., 2014; e.g., Rhizobium sp. RP5 enhancement of Ni and Zn bioavailability to leguminous plants through the secretion of siderophores (Wani et al., 2007)); and (iii) the promotion of the leguminous biomass and bioaccumulation of metals in plants (Carrasco et al., 2005). A variety of metal(loid) resistance genes as well as genes reported to promote host plant growth were identified in a draft genome sequence, indicating their potential for aiding phytoremediation (Hao et al., 2012). For example, inoculation with Bradyrhizobium sp. ameliorated cadmium, zinc, and nickel stress in green gram (Vigna radiata) or annual ryegrass (Lolium multiflorum) by increasing the bioavailability of cadmium and the reduction of the metal concentrations in the plant organs and was accompanied by an increase in the plant biomass (Wani et al., 2007; Guo and Chi, 2014).
Mixed systems with multiple plant species or microbial symbionts have also been used for restoration, such as in PAH-spiked (chrysene-amended) agricultural soil (Johnson et al., 2004). Moreover, different legume species respond distinctly to the inoculation of metal-resistant rhizobia. Cadmium concentrations in the roots of L. multiflorum increased following inoculation with Bradyrhizobium sp. YL-6, but significantly decreased in the roots and shoots of Glycine max (Guo and Chi, 2014). Moreover, Camargo et al. (2011) found that inoculation with Rhizobium sp. produced differences in the growth of Stizolobium deeringianum Bort in atrazine-contaminated soil, whereas no differences were found in the growth of S. aterrimum Piper & Tracy.
Other Strategies to Address Recalcitrant Pollutants
Optimization of Pollutant-Degrading Microbial Consortia
The augmentation of the diversity and richness of degrading microbial consortia in contaminated sites has been regarded as one of the key reasons that rhizobia enhance the biodegradation of organic pollutants. By manipulating sterile and non-sterile soils, Johnson et al. (2004) found that the removal of chrysene was not caused by the direct degradation or uptake by R. leguminosarum bv. trifolii itself, but as a result of the stimulated plant growth and rhizospheric microflora promoted by the symbiotic association between white clover/ryegrass and this rhizobial strain. Many studies have suggested that the symbiotic association between alfalfa and rhizobia (i.e., S. meliloti) increases the counts of culturable PAH-degrading bacteria, soil microbial activity and the carbon utilization ability of the soil microbial community (Teng et al., 2011).
Rhizobia have the potential to directly modify rhizosphere microflora by improving environmental conditions and nutrient availability. Nitrogen is a major limiting factor in bioremediation and is often added to contaminated soils to stimulate the existing microbial communities (Terrence et al., 2011). Thus, the organic nitrogen resource fixed by rhizobia is one of the key factors that facilitates the growth and activity of other soil biodegraders. That being said, changes in the amounts and constituents of root exudates (i.e., organic acids, vitamins, and hormones) and secondary metabolites of legumes due to the colonization by rhizobia could be an additional promoting element for the optimization of rhizospheric microflora (Johnson et al., 2005).
Synergistic Interactions With Other Microbes
The collaboration between multiple beneficial microbes has been exploited for more comprehensive and sustainable rehabilitation. Simultaneous inoculation of multiple beneficial microbes often provides complementary and additive benefits to plants, revealing the compatibility, and synergy between distinct mutualists (Larimer et al., 2012). The synergistic promotion of plant biomass and activities of indigenous microbial species caused by dual colonization of rhizobia and other microbes could be a potent tool to further intensify bioremediation efficiency. For example, arbuscular mycorrhizal fungus (AMF) could form extended mycelial networks that not only provide organic nitrogen to host plants but also possess the catabolic capacity to remove organic pollutants (Harms et al., 2011). Teng et al. (2010) reported that the co-inoculation of Rhizobium sp. and AMF enhanced the removal rate of PCBs. This could be due to their combined contribution to plant growth and development by improving nutrient conditions (Saini et al., 2004) and the additive degradative ability of contaminants conferred by the two cleaners (Harms et al., 2011). The positive synergism of these interactions could be limited by plant species and soil conditions. Additionally, combined approaches using both biostimulation (with exogenous carbon sources) and bioaugmentation may be necessary to sustain the timely and effective in situ microbial biodegradation of pollutants (Andeer et al., 2013).
However, questions remain due to the considerable complexity of the soil environment. For example, the determination of the best method to establish appropriate numbers of foreign pollutant-degrading bacteria in the contaminated sites and to ensure that multiple degraders co-work during the entire rehabilitation process requires further studies (Segura and Ramos, 2013).
Transgenic Rhizobia in Bioremediation
Recent advances in ‘omics’ technologies have provided opportunities to exploit genomic, transcriptomic, proteomic, and metabolomic means to modify the traits of ‘biological designers’ in order to maximize their phytoremediation efficiency (Abhilash et al., 2012). Bioengineering could potentially be used to manipulate the tolerance, accumulation, and degradation potentials of plants and microbes against pollutants.
Rhizobial transgenics can be harnessed for the accumulation of inorganic contaminants and detoxification of organic pollutants. Ike et al. (2007) transferred two resistance genes [synthetic tetrameric metallothionein (MTL4), and a cDNA encoding the phytochelatin synthase from Arabidopsis thaliana (AtPCS)] into M. huakuii subsp. rengei B3. The results from this study showed that the two recombinant strains accumulated more cadmium compared with the free-living cells. When Astragalus sinicus was inoculated with the two recombinant strains, the increased cadmium accumulation in nodules was observed. However, the accumulation of copper was not promoted by the expression of MTL4 in M. huakuii B3 in nodules of A. sinicus, indicating that the functioning of the MTL4 gene might vary in response to different metal stresses (Sriprang et al., 2002). Genetic horizontal transfer of plasmid pJP4 that encodes genes for mercury resistance and 2,4-D degradation into Bradyrhizobium in non-sterile soil (Kinkle et al., 1993) reportedly resulted in the co-metabolism of herbicide 2,4-D (Feng et al., 1994) and the enhancement of microbial PCB degradation in the soil (Donelly et al., 1994). The work of Chen et al. (2005) showed that when corresponding degradative genes were introduced into S. meliloti, the degradation rate of PCBs increased in the transformed strain. The dechlorination of PCBs by alfalfa inoculated with the S. meliloti transformants was more than twice that of alfalfa grown with wild-type S. meliloti. Therefore, the effectiveness of phytostabilization could be improved by developing such downstream functional strategies.
Studies have shown that the simultaneous expression of multiple genes related to degradation or metal accumulation exerts additive effects on the removal of pollutants (Ike et al., 2007). However, it is difficult to control the expression levels of the transferred genes in the recipient cells and the effect of certain individual genes may be limited to a narrow scope of impact as a consequence, thereby restricting its application. Further exploitation of the genomic databases of these rhizobia and the identification of the various functions of different genes are required in order to identify the most effective genes for bioremediation.
Conclusion
Rhizobia have been recognized as a potential strategy to simultaneously enhance soil nitrogen content, reduce the use of fertilizers, and increase H2 concentration (hydrogen fertilizers) in the rhizosphere through symbiotic nitrogen fixation. Rhizobia also possess the biochemical and ecological capacity to degrade environmental organic chemicals and to decrease the risk associated with metals and metalloids in contaminated sites. Rhizobia-assisted phytoremediation provides further environmental and economic benefits for bioremediation. The exploitation of microbe–microbe or plant–microbe interactions between intra-species and inter-species communication in the rhizosphere could represent more integrative approaches to further facilitate bioremediation. Researchers have proposed that the wide adoption of these biological adaptation strategies would result in the development of environmentally friendly management techniques (i.e., biological carbon sequestration, bioenergy, and bioremediation: the “3B” technique) to further enhance biodiversity and relieve environmental stressors (Teng et al., 2012).
Symbiotic nitrogen fixation and the reductive dechlorination of organic pollutants are both oxygen-sensitive and energetically costly for rhizobia (Morris and Schmidt, 2013). The presence of leghemoglobin maintains the low O2 concentrations in the root nodules (within the nanomolar range) and protects nitrogenase from inhibition by O2 (Becker et al., 2004). Therefore, researchers have proposed that the micro-oxic environment formed in nodules might also provide the proper conditions for the reductive degradation of organic pollutants. Moreover, Rhizobium sp. have a demonstrated capacity for partial denitrification in soils; some strains have been associated with nitrate reductase (NAR) activities, especially under micro-oxic conditions (Streeter and DeVine, 1983). Some reports have provided evidence of NAR-mediated dechlorination of PCB153 in free-living cells of Phanerochaete chrysosporium and crude enzyme extracts from Medicago sativa leaves, even under aerobic conditions (De et al., 2006; Magee et al., 2008). O’Hara et al. (1983) reported that S. meliloti possessed denitrifying activities in both its free-living and symbiotic forms. However, Becker et al. (2004) only found two genes (nirV and norB) that were induced in the S. meliloti bacteroids (Becker et al., 2004). Therefore, the involvement of metabolic enzymes in the potential degradation of organic compounds in rhizobia requires further study.
Our understanding of the genetic and molecular influences of bioremediation effects is not complete, and the goal of transforming this strategy into practice has not yet been fully achieved. The suitable selection of rhizobial strains or consortia in combination with plant hosts, indicators of successful bioremediation under field conditions and the mechanisms involved constitute future work that should be pursued for the initiation of successful efforts in this area. The successful execution of this versatile bioremediation strategy also requires a thorough understanding of the factors regulating the growth, metabolism, and functions of degradative rhizobia and indigenous microbial communities at contaminated sites. Furthermore, to date the major work of the “Rhizobial bioremediation” field has been mostly conducted under controlled laboratory conditions and not in the field, where further practical investigations and testing are required before bioremediation can become a widely accepted technique. The selection of suitable rhizobial strains will be necessary for the remediation of certain polluted sites.
In conclusion, this review provides a comprehensive framework for applying the versatile rhizobia to revitalize contaminated soils. The selective introduction of degradative rhizobia into hyperaccumulator plants could facilitate the accelerated removal of mixed pollutants from soils. Using this approach, the exploitation of these degradative, nitrogen-fixing and endophytic pollutant-cleaners could become a highly efficient, eco-friendly and low-input bioremediation technology for the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was funded by grants from the support of the National Natural Science Foundation of China (No. 41371309) and K. C. Wong Education Foundation of Hong Kong.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00032/abstract
References
Abhilash, P. C., Powell, J. R., Singh, H. B., and Singh, B. K. (2012). Plant–microbe interactions: novel applications for exploitation in multipurpose remediation technologies. Trends Biotechnol. 30, 416–420. doi: 10.1016/j.tibtech.2012.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ahmad, D., Mehmannavaz, R., and Damaj, M. (1997). Isolation and characterization of symbiotic N2-fixing Rhizobium meliloti from soils contaminated with aromatic and chloroaromatic hydrocarbons: PAHs and PCBs. Int. Biodeter. Biodegr. 39, 33–43. doi: 10.1016/S0964-8305(96)00065-0
Aken, B., Van Correa, P. A., and Schnoor, J. L. (2010). Phytoremediation of polychlorinated biphenyls: new trends and promises. Environ. Sci. Technol. 44, 2767–2776. doi: 10.1021/es902514d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Alloway, B. J., and Trevors, J. T. (2013). Heavy Metals in Soils-Trace Metals and Metalloids in Soils and their Bioavailability. New York: Springer Dordrecht Heidelberg.
Andeer, P., Stahl, D. A., Lillis, L., and Strand, S. E. (2013). Identification of microbial populations assimilating nitrogen from RDX in munitions contaminated military training range soils by high sensitivity stable isotope probing. Environ. Sci. Technol. 47, 10356–10363. doi: 10.1021/es401729c
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Awaya, J. D., Fox, P. M., and Borthakur, D. (2005). pyd Genes of Rhizobium sp. strain TAL1145 are required for degradation of 3-hydroxy-4-pyridone, an aromatic intermediate in mimosine metabolism. J. Bacteriol. 187, 4480–4487. doi: 10.1128/JB.187.13.4480-4487.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Becker, A., Bergès, H., Krol, E., Bruand, C., Rüberg, S., Capela, D.,et al. (2004). Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant Microbe Interact. 17, 292–303. doi: 10.1094/MPMI.2004.17.3.292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bright, M., and Bulgheresi, S. (2010). A complex journey: transmission of microbial symbionts. Appl. Soil Ecol. 8, 218–230. doi: 10.1038/nrmicro2262
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Camargo, D., Bispo, K. L., and Sene, L. (2011). Association of Rhizobium sp. with two legumes on atrazine tolerance. Rev. Ceres. 58, 425–431. doi: 10.1590/S0034-737X2011000400004
Carrasco, J. A., Armario, P., Pajuelo, E., Burgos, A., Caviedes, M. A., López, R.,et al. (2005). Isolation and characterization of symbiotically effective Rhizobium resistant to arsenic and heavy metals after the toxic spill at the Aznalcollar pyrite mine. Soil Biol. Biochem. 37, 1131–1140. doi: 10.1016/j.soilbio.2004.11.015
Carrasco-Gil, S., Siebner, H., LeDuc, D. L., Webb, S. M., Millaìn, R., Andrews, J. C.,et al. (2013). Mercury localization and speciation in plants grown hydroponically or in a natural environment. Environ. Sci. Technol. 47, 3082–3090. doi: 10.1021/es303310t
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, J., Sun, G. X., Wang, X. X., de Lorenzo, V., Rosen, B. P., and Zhu, Y. G. (2014). Volatilization of arsenic from polluted soil by Pseudomonas putida engineered for expression of the arsM arsenic(III) S-adenosine methyltransferase gene. Environ. Sci. Technol. 48, 10337-10344. doi: 10.1021/es502230b
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, W. M., Wu, C. H., James, E. K., and Chang, J. S. (2008). Biosorption capability of Cupriavidus taiwanensis and its effects on heavy metal removal by nodulated Mimosa pudica. J. Hazard. Mater. 151, 364–371. doi: 10.1016/j.jhazmat.2007.05.082
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Y. Q., Adam, A., Toure, O., and Dutta, S. K. (2005). Molecular evidence of genetic modification of Sinorhizobium meliloti: enhanced PCB bioremediation. J. Ind. Microbiol. Biotechnol. 32, 561–566. doi: 10.1007/s10295-005-0039-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cui, W. T., Gao, C. Y., Fang, P., Lin, G. Q., and Shen, W. B. (2013). Alleviation of cadmium toxicity in Medicago sativa by hydrogen-rich water. J. Hazard. Mater. 260, 715–724. doi: 10.1016/j.jhazmat.2013.06.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
DalCorso, G., Fasani, E., and Furini, A. (2013). Recent advances in the analysis of metal hyperaccumulation and hypertolerance in plants using proteomics. Front. Plant Sci. 4:280. doi: 10.3389/fpls.2013.00280
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Damaj, M., and Ahmad, D. (1996). Biodegradation of polychlorinated biphenyls by rhizobia: a novel finding. Biochem. Bioph. Res. Commun. 218, 908–915. doi: 10.1006/bbrc.1996.0161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dashti, N., Khanafer, M., El-Nemr, I., Sorkhoh, N., Ali, N., and Radwan, S. (2010). The potential of oil-utilizing bacterial consortia associated with legume root nodules for cleaning oily soils. Chemosphere 74, 1354–1359. doi: 10.1016/j.Chemosphere.2008.11.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De, S., Perkins, M., and Dutta, S. K. (2006). Nitrate reductase gene involvement in hexachlorobiphenyl dechlorination by Phanerochaete chrysosporium. J. Hazard. Mater. 135, 350–354. doi: 10.1016/j.jhazmat.2005.11.073
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deakin, W. J., and Broughton, W. J. (2009). Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Appl. Soil Ecol. 7, 312–320. doi: 10.1038/nrmicro2091
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
de-Bashan, L. E., Hernandez, J. P., and Bashan, Y. (2011). The potential contribution of plant growth-promoting bacteria to reduce environmental degradation–a comprehensive evaluation. Appl. Soil Ecol. 61, 171–189. doi: 10.1016/j.apsoil.2011.09.003
Donelly, P. K., Hedge, R. S., and Fletcher, J. S. (1994). Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere 28, 981–988. doi: 10.1016/0045-6535(94)90014-0
Fan, S. X., Li, P. J., Gong, Z. Q., Ren, W. X., and He, N. (2008). Promotion of pyrene degradation in rhizosphere of alfalfa (Medicago sativa L.). Chemosphere 71, 1593–1598. doi: 10.1016/j.Chemosphere.2007.10.068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feng, L., Van Zwieten, I., Kennedy, I. R., Rolfe, B. G., and Gartner, E. (1994). Expression of the 2,4-D degrading plasmid pJP4 of Alcaligenes eutrophus in Rhizobium trifolii. Acta Biotechnol. 14, 119–129. doi: 10.1002/abio.370140202
Fester, T., Giebler, J., Wick, L. Y., Schlosser, D., and Kästner, M. (2014). Plant–microbe interactions as drivers of ecosystem functions relevant for the biodegradation of organic contaminants. Curr. Opin. Biotech. 27, 168–175. doi: 10.1016/j.copbio.2014.01.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glick, B. R. (2010). Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 28, 367–374. doi: 10.1016/j.biotechadv.2010.02.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guo, J. K., and Chi, J. (2014). Effect of Cd-tolerant plant growth-promoting rhizobium on plant growth and Cd uptake by Lolium multiflorum Lam. and Glycine max (L.) Merr. in Cd-contaminated soil. Plant Soil 375, 205–214. doi: 10.1007/s11104-013-1952-1
Hamdi, H., Benzarti, S., Aoyama, I., and Jedidi, N. (2012). Rehabilitation of degraded soils containing aged PAHs based on phytoremediation with alfalfa (Medicago sativa L.). Int. Biodeter. Biodegr. 67, 40–47. doi: 10.1016/j.ibiod.2011.10.009
Hao, X. L., Lin, Y. B., Johnstone, L., Baltrus, D. A., Miller, S. J., Wei, G. W.,et al. (2012). Draft genome sequence of plant growth-promoting rhizobium Mesorhizobium amorphae, isolated from zinc-lead mine tailings. J. Bacteriol. 194, 736–737. doi: 10.1128/JB.06475-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hao, X., Taghavi, S., Xie, P., Orbach, M. J., Alwathnani, H. A., Rensing, C.,et al. (2014). Phytoremediation of heavy and transition metals aided by legume-rhizobia symbiosis. Int. J. Phytoremediat. 16, 179–202. doi: 10.1080/15226514.2013.773273
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Harms, H., Schlosser, D., and Wick, L. Y. (2011). Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. 9, 177–192. doi: 10.1038/nrmicro2519
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hussien, Y. A., Tewfik, M. S., and Hamdi, Y. A. (1974). Degradation of certain aromatic compounds by rhizobia. Soil Biol. Biochem. 6, 377–381. doi: 10.1016/0038-0717(74)90047-9
Ike, A., Sriprang, R., Ono, H., Murooka, Y., and Yamashita, M. (2007). Bioremediation of cadmium contaminated soil using symbiosis between leguminous plant and recombinant rhizobia with the MTL4 and the PCS genes. Chemosphere 66, 1670–1676. doi: 10.1016/j.Chemosphere.2006.07.058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jabeen, H., Iqbal, S., and Anwar, S. (2014). Biodegradation of chlorpyrifos and 3, 5, 6-trichloro-2-pyridinol by a novel rhizobial strain Mesorhizobium sp. HN3. Water Environ. J. doi: 10.1111/wej.12081
Jin, Q. J., Zhu, K. K., Cui, W. T., Xie, Y. J., Han, B., and Shen, W. B. (2013). Hydrogen gas acts as a novel bioactive molecule in enhancing plant tolerance to paraquat-induced oxidative stress via the modulation of heme oxygenase-1 signalling system. Plant Cell Environ. 36, 956–969. doi: 10.1111/pce.12029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Johnson, D., Anderson, D., and McGrath, S. (2005). Soil microbial response during the phytoremediation of a PAH contaminated soil. Soil Biol. Biochem. 37, 2334–2336. doi: 10.1016/j.soilbio.2005.04.001
Johnson, D. L., Maguire, K. L., Anderson, D. R., and McGrath, S. P. (2004). Enhanced dissipation of chrysene in planted soil: the impact of a rhizobial inoculum. Soil Biol. Biochem. 36, 33–38. doi: 10.1016/j.soilbio.2003.07.004
Kaiya, S., Utsunomiya, S., Suzuki, S., Yoshida, N., Futamata, H., Yamada, T.,et al. (2012). Isolation and functional gene analyses of aromatic-hydrocarbon-degrading bacteria from a polychlorinated-dioxin-dechlorinating process. Microbes Environ. 27, 127–135. doi: 10.1264/jsme2.ME11283
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keum, Y. S., Seo, J. S., Hu, Y. T., and Li, Q. X. (2006). Degradation pathways of phenanthrene by Sinorhizobium sp. C4. Appl. Microbiol. Biotechnol. 71, 935–941. doi: 10.1007/s00253-005-0219-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kinkle, B. K., Sadowsky, M. J., Schmidt, E. L., and Koskinen, W. C. (1993). Plasmids pJP4 and r68.45 can be transferred between populations of bradyrhizobia in nonsterile soil. Appl. Environ. Microbiol. 59, 1762–1766.
Larimer, A. L., Bever, J. D., and Clay, K. (2012). Consequences of simultaneous interactions of fungal endophytes and arbuscular mycorrhizal fungi with a shared host grass. Oikos 121, 2090–2096. doi: 10.1111/j.1600-0706.2012.20153.x
Leigh, J. A., Skinner, A. J., and Cooper, R. A. (1988). Partial purification, stereospecificity and stoichiometry of three dehalogenases from a Rhizobium species. FEMS Microbiol. Lett. 49, 353–356. doi: 10.1111/j.1574-6968.1988.tb02756.x
Li, H. Y., Wei, D. Q., Shen, M., and Zhou, Z. P. (2012). Endophytes and their role in phytoremediation. Fungal Divers. 54, 11–18. doi: 10.1007/s13225-012-0165-x
Li, Y., Liang, F., Zhu, Y. F., and Wang, F. P. (2013). Phytoremediation of a PCB-contaminated soil by alfalfa and tall fescue single and mixed plants cultivation. J. Soil. Sediment. 13, 925–931. doi: 10.1007/s11368-012-0618-6
Lohmann, R., Breivik, K., Dachs, J., and Muir, D. (2007). Global fate of POPs: current and future esearch directions. Environ. Pollut. 150, 150–165. doi: 10.1016/j.envpol.2007.06.051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Magee, K. D., Michael, A., Ullah, H., and Dutta, S. K. (2008). Dechlorination of PCB in the presence of plant nitrate reductase. Environ. Toxicol. Pharmacol. 25, 144–147. doi: 10.1016/j.etap.2007.10.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maynaud, G., Brunel, B., Mornico, D., Durot, M., Severac, D., Dubois, E.,et al. (2013). Genome-wide transcriptional responses of two metal-tolerant symbiotic Mesorhizobium isolates to zinc and cadmium exposure. BMC Genomics 14:292. doi: 10.1186/1471-2164-14-292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mehmannavaza, R., Prasher, S. O., and Ahmad, D. (2002). Rhizospheric effects of alfalfa on biotransformation of polychlorinated biphenyls in a contaminated soil augmented with Sinorhizobium meliloti. Process Biochem. 37, 955–963. doi: 10.1016/S0032-9592(01)00305-3
Moreto, M., Silvestri, S., Ugo, P., Zorzi, G., Abbodanzi, F., Baiocchi, C.,et al. (2005). Polycyclic aromatic hydrocarbons degradation by composting in a soot contaminated alkaline soil. J. Hazard. Mater. 26, 141–148. doi: 10.1016/j.jhazmat.2005.06.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morris, R. L., and Schmidt, T. M. (2013). Shallow breathing: bacterial life at low O2. Appl. Soil Ecol. 2013, 11, 205–212. doi: 10.1038/nrmicro2970
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muthukumar, G., Arunakumari, A., and Mahadevan, A. (1982). Degradation of aromatic compounds by Rhizobium spp. Plant Soil 69, 163–169. doi: 10.1007/BF02374511
Nies, D. H. (1995). The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 177, 2707–2712.
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/S0168-6445(03)00048-2
O’Hara, G. W., Daniel, R. M., and Steele, K. W. (1983). Effect of oxygen on the synthesis, activity and breakdown of the Rhizobium denitrification system. J. Gen. Microbiol. 129, 2405–2412. doi: 10.1099/00221287-129-8-2405
Parke, D., Rivelli, M., and Ornston, L. N. (1985). Chemotaxis to aromatic and hydroaromatic acids: comparison of Bradyrhizobium japonicum and Rhizobium trifolii. J. Bacteriol. 163, 417–422.
Passatore, L., Rossetti, S., Juwarkar, A. A., and Massacci, A. (2014). Phytoremediation and bioremediation of polychlorinated biphenyls (PCBs): state of knowledge and research perspectives. J. Hazard. Mater. 278, 189–202. doi: 10.1016/j.jhazmat.2014.05.051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pereira, S. I. A., Lima, A. I. G., Figueira, E. M., and de Almeida, P. F. (2006). Heavy metal toxicity in Rhizobium leguminosarum biovar viciae isolated from soils subjected to different sources of heavy-metal contamination: effects on protein expression. Appl. Soil Ecol. 33, 286–293. doi: 10.1016/j.apsoil.2005.10.002
Poonthrigpun, S., Pattaragulwanit, K., Paengthai, S., Kriangkripipat, T., Juntongjin, K., Thaniyavarn, S.,et al. (2006). Novel intermediates of acenaphthylene degradation by Rhizobium sp. strain CU-A1: evidence for naphthalene-1,8-dicarboxylic acid metabolism. Appl. Environ. Microbiol. 72, 6034–6039. doi: 10.1128/AEM.00897-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Prell, J., and Poole, P. (2006). Metabolic changes of rhizobia in legume nodules. Trends Biotechnol. 14, 161–168. doi: 10.1016/j.tim.2006.02.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rajkumar, M., Sandhya, S., Prasad, M. N. V., and Freitas, H. (2012). Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 30, 1562–1574. doi: 10.1016/j.biotechadv.2012.04.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saini, V. K., Bhandari, S. C., and Tarafdar, J. C. (2004). Comparison of crop yield, soil microbial C, N and P, N-fixation, nodulation and mycorrhizal infection in inoculated and non-inoculated sorghum and chickpea crops. Field Crop. Res. 89, 39–47. doi: 10.1016/j.fcr.2004.01.013
Sato, Y., Monincová, M., Chaloupková, R., Prokop, Z., Ohtsubo, Y., Minamisawa, K.,et al. (2005). Two rhizobial strains, Mesorhizobium loti MAFF303099 and Bradyrhizobium japonicum USDA110, encode haloalkane dehalogenases with novel structures and substrate specificities. Appl. Environ. Microb. 71, 4372–4379. doi: 10.1128/AEM.71.8.4372-4379.2005
Schalk, I. J., Hannauer, M., and Braud, A. (2011). New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 13, 2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Segura, A., and Ramos, J. L. (2013). Plant–bacteria interactions in the removal of pollutants. Curr. Opin. Biotech. 24, 467–473. doi: 10.1016/j.copbio.2012.09.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Segura, A., Rodríguez-Conde, S., Ramos, C., and Ramos, J. L. (2009). Bacterial responses and interactions with plants during rhizoremediation. Microb. Biotechnol. 2, 452–464. doi: 10.1111/j.1751-7915.2009.00113.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, S. R., and Giller, K. E. (1992). Effective Rhizobium leguminosarum biovar trifolii present in five soils contaminated with heavy metals from long-term applications of sewage sludge or metal mine spoil. Soil Biol. Biochem. 24, 781–788. doi: 10.1016/0038-0717(92)90253-T
Soedarjo, M., and Borthakur, D. (1998). Mimosine, a toxin produced by the tree-legume Leucaena provides a nodulation competition advantage to mimosine-degrading Rhizobium strains. Soil Biol. Biochem. 30, 1605–1613. doi: 10.1016/S0038-0717(97)00180-6
Soedarjo, M., Hemscheidt, T. K., and Borthakur, D. (1995). Mimosine, a toxin present in leguminous trees (Leucaena spp.), induces a mimosine-degrading enzyme activity in some strains of Rhizobium. Appl. Environ. Microbiol. 60, 4268–4272.
Souza, M. P., Amini, A., Dojka, M. A., Pickering, I. J., Dawson, S. C., Pace, N. R.,et al. (2001). Identification and characterization of bacteria in a selenium-contaminated hypersaline evaporation pond. Appl. Environ. Microbiol. 67, 3785–3794. doi: 10.1128/AEM.67.9.3785-3794.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sriprang, R., Hayashi, M., Yamashita, M., Ono, H., Saeki, K., and Murooka, Y. (2002). A novel bioremediation system for heavy metals using the symbiosis between leguminous plant and genetically engineered rhizobia. J. Biotechnol. 99, 279–293. doi: 10.1016/S0168-1656(02)00219-5
Streeter, J. G., and DeVine, P. J. (1983). Evaluation of nitrate reductase activity in Rhizobium japonicum. Appl. Environ. Microbiol. 46, 521–524. doi: 10.1038/nrmicro2970
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stringfellow, J. M., Cairns, S. S., Cornish, A., and Cooper, R. A. (1997). Haloalkanoate dehalogenase I1 (DehE) of a Rhizobium sp. molecular analysis of the gene and formation of carbon monoxide from trihaloacetate by the enzyme. Eur. J. Biochem. 250, 789–793. doi: 10.1111/j.1432-1033.1997.00789.x
Sun, M. M., Fu, D. Q., Teng, T., Shen, Y. Y., Luo, Y. M., Li, Z. G.,et al. (2011a). In situ phytoremediation of PAH-contaminated soil by intercropping alfalfa (Medicago sativa L.) with tall fescue (Festuca arundinacea Schreb.) and associated soil microbial activity. J. Soil. Sediment. 11, 980–989. doi: 10.1007/s11368-011-0382-z
Sun, X. H., Teng, Y., Luo, Y. M., Tu, C., and Li, Z. G. (2011b). Accumulation, distribution and chemical speciation of PCBs in different parts of alfalfa. Soils 43, 595–599.
Teng, Y., Luo, Y., Sun, X., Tu, C., Xu, L., Liu, W.,et al. (2010). Influence of arbuscular mycorrhiza and Rhizobium on phytoremediation by alfalfa of an agricultural soil contaminated with weathered PCBs: a field study. Int. J. Phytoremediation 12, 516–533. doi: 10.1080/15226510903353120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Teng, Y., Shen, Y. Y., Luo, Y. M., Sun, X. H., Sun, M. M., Fu, D. Q.,et al. (2011). Influence of Rhizobium meliloti on phytoremediation of polycyclic aromatic hydrocarbons by alfalfa in an aged contaminated soil. J. Hazard. Mater. 186, 1271–1276. doi: 10.1016/j.jhazmat.2010.11.126
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Teng, Y., Xu, Z. H., Luo, Y. M., and Reverchon, F. (2012). How do persistent organic pollutants be coupled with biogeochemical cycles of carbon and nutrients in terrestrial ecosystems under global climate change? J. Soil. Sediment. 12, 411–419. doi: 10.1007/s11368-011-0462-0
Terrence, H. B., Etienne, Y., Christine, M., David, J., Lyle, G. W., and Charles, W. G. (2011). Identification of nitrogen incorporating bacteria in petroleum-contaminated arctic soils by using [15N] DNA-based stable isotope probing and pyrosequencing. Appl. Environ. Microbiol. 77, 4163–4171. doi: 10.1128/AEM.00172-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tu, C., Teng, Y., Luo, Y. M., Li, X. H., Sun, X. H., Li, Z. G., et al., (2011). Potential for biodegradation of polychlorinated biphenyls (PCBs) by Sinorhizobium meliloti. J. Hazard. Mater. 186, 1438–1444. doi: 10.1016/j.jhazmat.2010.12.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Valentín, L., Nousiainen, A., and Mikkonen, A. (2013). “Introduction to organic contaminants in soil: concepts and risks,” in Emerging Organic Contaminants in Sludges: The Handbook of Environmental Chemistry, eds A. G. Kostianoy and D. Barceló (Berlin, Heidelberg: Springer-Verlag), 1–29.
Wang, X. M., Yang, B., Ren, C, G., Wang, H. W., Wang, J. Y., and Dai, C. C. (2015). Involvement of abscisic acid and salicylic acid in signal cascade regulating bacterial endophyte-induced volatile oil biosynthesis in plantlets of Atractylodes lancea. Physiol. Plant. 153, 30–42. doi: 10.1111/ppl.12236
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wani, P. A., Khan, M. S., and Zaidi, A. (2007). Effect of metal tolerant plant growth promoting Bradyrhizobium sp.(vigna) on growth, symbiosis, seed yield, and metal uptake by greengram plants. Chemosphere 70, 36–45. doi: 10.1007/s00244-007-9097-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, L., Teng, T., Luo, Y. M., and Li, Z. G. (2010). Effects of Rhizobium meliloti on PCBs degradation and transformation in solution culture. Environ. Sci. 31, 255–259.
Xu, L., Teng, Y., Li, Z. G., Norton, J. M., and Luo, Y. M. (2008). Enhanced removal of polychlorinated biphenyls from alfalfa rhizosphere soil in a field study: the impact of a rhizobial inoculum. Sci. Total Environ. 408, 1007–1013. doi: 10.1016/j.scitotenv.2009.11.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, H. C., Cheng, J. J., Finan, T. M., Rosen, B. P., and Bhattacharjee, H. (2005). Novel pathway for arsenic detoxification in the legume symbiont Sinorhizobium meliloti. J. Bacteriol. 187, 6991–6997. doi: 10.1128/JB.187.20.6991-6997.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, W. W., Chen, L. X., and Liu, D. Y. (2012). Characterization of a marine-isolated mercury-resistant Pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Appl. Microbiol. Biotechnol. 93, 1305–1314. doi: 10.1007/s00253-011-3454-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zinjarde, S., Apte, M., Mohite, P., and Kumar, A. R. (2014). Yarrowia lipolytica and pollutants: interactions and applications. Biotechnol. Adv. 32, 920–933. doi: 10.1016/j.biotechadv.2014.04.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: rhizobia, nitrogen-fixation, bioremediation, organic pollutants, heavy metals, rhizospheric degraders, transgenic rhizobia
Citation: Teng Y, Wang X, Li L, Li Z and Luo Y (2015) Rhizobia and their bio-partners as novel drivers for functional remediation in contaminated soils. Front. Plant Sci. 6:32. doi: 10.3389/fpls.2015.00032
Received: 15 September 2014; Accepted: 13 January 2015;
Published online: 05 February 2015.
Edited by:
Anna Manara, University of Verona, ItalyReviewed by:
Richard S. Winder, Natural Resources Canada, CanadaTiziana Pandolfini, University of Verona, Italy
Copyright © 2015 Teng, Wang, Li, Li and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Teng, Key Laboratory of Soil Environment and Pollution Remediation, Institute of Soil Science, Chinese Academy of Sciences, East Beijing Road No. 71, Nanjing, Jiangsu 210008, China e-mail: yteng@issas.ac.cn
 Ying Teng
Ying Teng Xiaomi Wang1
Xiaomi Wang1 Yongming Luo
Yongming Luo