- 1Biochemistry, Virginia Polytechnic and State University, Blacksburg, VA, USA
- 2Plant and Microbial Biology, North Carolina State University, Raleigh, NC, USA
Inositol phosphates (InsPs) are intricately tied to lipid signaling, as at least one portion of the inositol phosphate signaling pool is derived from hydrolysis of the lipid precursor, phosphatidyl inositol (4,5) bisphosphate. The focus of this review is on the inositol pyrophosphates, which are a novel group of InsP signaling molecules containing diphosphate or triphosphate chains (i.e., PPx) attached to the inositol ring. These PPx-InsPs are emerging as critical players in the integration of cellular metabolism and stress signaling in non-plant eukaryotes. Most eukaryotes synthesize the precursor molecule, myo-inositol (1,2,3,4,5,6)-hexakisphosphate (InsP6), which can serve as a signaling molecule or as storage compound of inositol, phosphorus, and minerals (referred to as phytic acid). Even though plants produce huge amounts of precursor InsP6 in seeds, almost no attention has been paid to whether PPx-InsPs exist in plants, and if so, what roles these molecules play. Recent work has delineated that Arabidopsis has two genes capable of PP-InsP5 synthesis, and PPx-InsPs have been detected across the plant kingdom. This review will detail the known roles of PPx-InsPs in yeast and animal systems, and provide a description of recent data on the synthesis and accumulation of these novel molecules in plants, and potential roles in signaling.
Myo-inositol (inositol) signaling is much like a language in that each molecular species used in the pathway, whether lipid or soluble in nature, can convey specific information to the cell, like a word. In this analogy, each combination of different numbers and positions of phosphates on the inositol ring and the presence of diacylglycerol linked via the C1 of inositol, also convey unique information (see Figure 1). Comprehensive analyses of both inositol and inositol phospholipids in signaling have been previously reviewed (Van Leeuwen et al., 2004; Gillaspy, 2011; Heilmann and Heilmann, 2014), so this review focuses on new inositol signaling molecules, the di-phospho (PP) and tri-phospho (PPP) inositol phosphates (PPx-InsPs), also known as inositol pyrophosphates. These high-energy molecules have been studied in non-plant eukaryotes, however, their existence and role in plants is newly emerging. The main questions regarding PPx-InsPs are: can these molecules be detected in plants, how are they synthesized and what type of information do they convey? Recent work addressing these questions will be discussed in the broader context of understanding how PPx-InsPs function in eukaryotes.
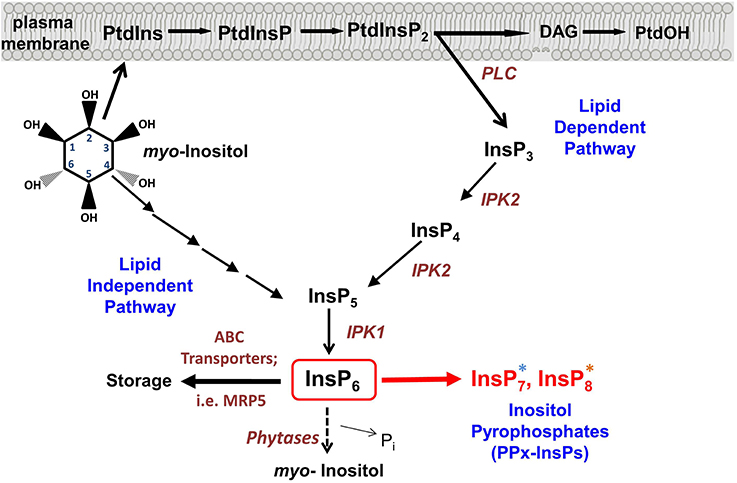
Figure 1. Synthesis of inositol pyrophosphate. Overview of the Inositol phosphate pathway, including both lipid dependent and lipid independent routes for synthesis of InsP6. Inositol Pyrophosphate (PPx-InsP) synthesis is indicated in red. Major lipid and inositol species are indicated in black and key enzymes are indicated in brown. A more detailed outline of PPx-InsP synthesis is depicted in Figure 2. The blue and orange asterisks correspond to the colored boxes in Figure 2.
History and Structure of PPx-InsPs
PPx-InsPs were first identified in Dictyostelium in 1993 (Glennon and Shears, 1993; Hawkins et al., 1993; Menniti et al., 1993; Stephens et al., 1993). They are similar to ATP and polyphosphates in that they contain a linear chain of two (PP) or three (PPP) phosphates separated by pyrophosphate bonds, linked to an InsP molecule (see Figure 2).
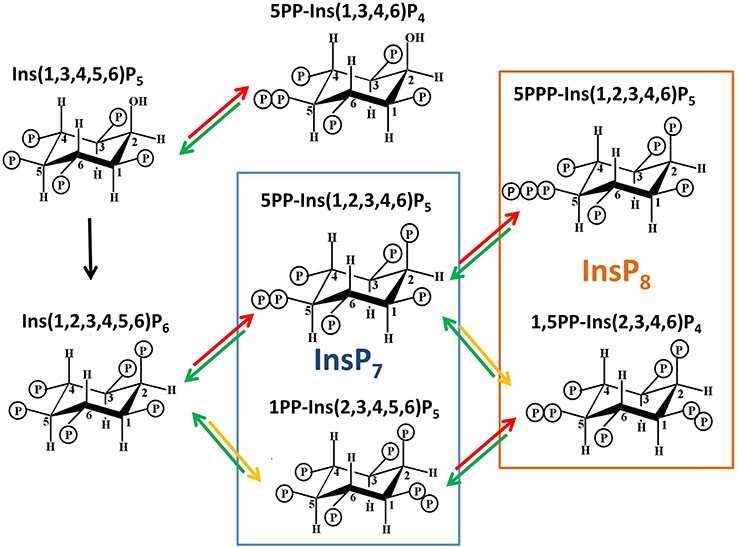
Figure 2. Structures of PPx-InsPs and pathway of proposed synthesis. The unboxed area is the last step in InsP6 synthesis, catalyzed by the IPK1 enzyme in plants. The boxed areas in blue and orange indicate InsP7 and InsP8 synthesis. The colored arrows indicate known enzymes in yeast. Red arrows indicate KCS1 activity, while yellow arrows indicate VIP activity. The green arrows indicate phosphatase activity by DDP1.
The PPx moieties at one or more positions on the inositol ring in PPx-InsPs are synthesized from InsP5 or InsP6 (Menniti et al., 1993; Shears et al., 2011), resulting in InsPs containing seven or eight phosphates (i.e., InsP7 and InsP8). The high energy pyrophosphate bonds present in PPx-InsPs may serve as a way to store energy in the cell, with the standard free energy of hydrolysis of InsP7 and InsP8 higher than that of ADP and ATP, respectively (Stephens et al., 1993). Indeed, the initial role proposed for PPx-InsPs was simply as a high energy molecules, as they can be broken down to generate ATP (Voglmaier et al., 1996; Huang et al., 1998). However, PPx-InsPs are present at very low amounts and they have high turnover rates, suggesting that they serve as more than energy storage molecules (Glennon and Shears, 1993; Menniti et al., 1993). Recently new physiological roles have been discovered for PPx-InsPs, supporting their role as dynamic and important signaling molecules.
Only a few of the theoretically possible PPx-InsP structures have been confirmed. The naming convention is to describe the position and number of the pyrophosphates first, followed by the name of the InsP. For example, Dictyostelium contains 6PP-InsP5 and 5PP-InsP5, and these contain a pyrophosphate on the 6th and 5th carbons of InsP5, respectively. Dictyostelium also contains 5,4/6(PP)2-InsP4 or 1/3,5(PP)2-InsP4, and in these cases the slash indicates one of the pyrophosphates present can occur at either of two carbons (i.e., at the C4 or C6 position, or at C1 or C3, respectively). The ratios of these different PPx-InsPs differ in various Dictyostelium species examined (Laussmann et al., 1997, 1998). Another member of the Amoebazoa kingdom, Entamoeba histolytica, has further diversity in that a PPx-InsP was identified containing neo-inositol, rather than myo-inositol (Martin et al., 2000). This difference could produce even more diversity in the language of InsPs, but it is not yet known if this occurs in other organisms.
In Dictyostelium and the animal kingdom, PPx-INsPs synthesized from InsP5 exist (Laussmann et al., 1997, 1998; Draskovic et al., 2008), however at present there is no data indicating they are found in plants. This review will focus on the PPx-InsP species synthesized from InsP6. The most abundant InsP7 isomer has been confirmed through NMR as 5PP-InsP5 (Mulugu et al., 2007; Draskovic et al., 2008). A second InsP7 was first speculated to be pyrophosphorylated at C4 or C6 (i.e., 4/6), but was later conclusively identified as 1/3PP-InsP5 (Lin et al., 2009). Recent work in animals has shown that the 1- rather than 3- position is phosphorylated, thus 1PP-InsP5 is likely to be the second type of InsP7 present in animals (Wang et al., 2012). Given this, we use 1PP-InsP5 as the updated nomenclature for this second molecular species of InsP7. Studies in yeast and humans identified that both of these InsP7 molecules are present. The InsP8 species confirmed are 1,5(PP)2-InsP4 (in vivo) and 5PPP-InsP5(in vitro) (Draskovic et al., 2008; Lin et al., 2009).
Methods Used to Detect PPx-InsPs
Several methods have been used to detect PPx-InsPs, each having distinct strengths and limitations. The most common method is to introduce a radiolabeled precursor, often 3H-myo-inositol or 3H-InsP6, followed by HPLC separation of InsP species produced after a given time (Azevedo and Saiardi, 2006). This method is very sensitive, but labor-intensive, and while it can resolve isomers of the lower InsPs, currently it is not possible to separate different PPx-InsP isomers. As well, one is limited to analysis of cells/tissues that can take up the radiolabelled precursor. However, this method has an advantage in that one can be fairly certain of the identity of the resulting labeled peaks on the HPLC chromatograms. A nonradioactive high-performance anion-exchange chromatographic method based on metal dye detection (MDD)-HPLC can also be used to detect PPx-InsPs. The advantage of this method is that it can separate different isomers of InsP7, however this method requires a 3 pump HPLC unit which increases the complexity of the system and limits its use (Mayr, 1988). Another method of separation of InsPs is thin layer chromatography, which utilizes either radiolabeling or dye for detection of InsP species (Otto et al., 2007). This method, in general, does not have great sensitivity, and is most often used with purified PPx-InsPs.
Since acidic conditions can cause the degradation of PPx-InsPs, HPLC analyses may underestimate the amount of PPx-InsPs present (Losito et al., 2009). A new method developed to limit exposure of extracted PPx-InsPs to acid buffers is polyacrylamide gel separation by electrophoresis (PAGE), and subsequent staining with either DAPI or toluidine blue to detect InsPs (Losito et al., 2009). The PAGE technique is sensitive enough to visualize PPx-InsPs from cell/tissue extracts, and it can separate different InsP7 and InsP8 isomers. However, its distinct advantage is that it may allow for a better estimation of PPx-InsP abundance because of the speed of analysis. The disadvantage of using PAGE is that conclusive identification of stained “bands” as PPx-InsPs is best verified with a separate technology, as other molecules could be present and give rise to bands. It is important to note that co-migration with InsP and PPx-InsP standards is required for all of these methods, and follow-up NMR is needed to conclusively identify the specific structure of PPx-InsP species.
Plants Contain PPx-InsPs
Plants have large amounts of one of the precursors to PPx-InsPs, InsP6, which is well studied as a phosphorous storage molecule (Raboy, 2003). Given this, it seems likely that plants also synthesize the PPx-InsPs. Previous studies had noted InsP molecules more polar than InsP6 in barley, duckweed, and potato (Brearley and Hanke, 1996; Flores and Smart, 2000; Lemtiri-Chlieh et al., 2000; Dorsch et al., 2003). Acting on this information, we recently utilized both HPLC separation of radiolabelled plant tissues and PAGE to delineate the presence of InsP7 and InsP8 in higher plants including Arabidopsis, Camelina sativa, cotton, and maize (Desai et al., 2014). These two methods provided a complementary analysis of higher phosphorylated InsPs, including the PPx-InsPs. Since PPx-InsPs are low abundance molecules, it is not surprising that Arabidopsis seeds were found to contain less than 2% of the total inositol pool as inositol pyrophosphates (1.33% InsP7, 0.24% InsP8). Vegetative tissue from Arabidopsis was also analyzed and PPx-InsPs were found in both seedlings (0.64% InsP7, 0.14% InsP8) and mature leaves (1.00% InsP7). InsP7 was detected in other plant species as well, including another member of the Brassica family, Camelina sativa (1.40% in seedlings), and an unrelated dicot, cotton (Gossypium hirsutum) in the leaves and in shoots and roots of seedlings. PAGE analysis was used to detect PPx-InsPs in both Arabidopsis and maize seed in this same work (Desai et al., 2014). These findings indicate that PPx-InsPs may play a role during the plant life cycle throughout the plant kingdom, both in monocots and dicots.
Critical to our detection of the PPx-InsPs in plants was the use of a mutant containing elevated InsP7 and InsP8. The Multidrug Resistance associated Protein 5 (MRP5) is a high affinity ABC-binding cassette transporter that specifically binds to InsP6 (Nagy et al., 2009) (Figure 1). Studies on MRP5 have indicated the likely role of this transporter is in moving InsP6 into the storage vacuole (Nagy et al., 2009). The subcellular localization of MRP5 has been reported as both the plasma membrane (Suh et al., 2007) and the vacuolar membrane (Nagy et al., 2009), however it has been suggested that the plasma membrane localization is an artifact resulting from ectopic expression (Nagy et al., 2009). MRP5 was first identified as an important player in stomatal responses, since guard cells of the loss-of-function mrp5 mutant are insensitive to ABA and Ca2+ (Klein et al., 2003). This alteration in guard cell function results in reduction of water loss and use, allowing mrp5 mutants some resistance to drought (Klein et al., 2003). The maize paralogue of MRP5 (called MRP4), results in decreased levels of InsP6 in seeds when mutated (Shi et al., 2007; Nagy et al., 2009). Our recent study showed that in addition to decreased levels of InsP6, mrp5 mutants have elevated levels of InsP7 and InsP8 in seeds (Desai et al., 2014). These changes are less striking in vegetative tissue, perhaps as a result of overall lower levels of InsP6 (Desai et al., 2014), or reduced MRP5 expression (Nagy et al., 2009) in vegetative tissues. The guard cell phenotype of mrp5 mutants has been attributed to an increase in cytosolic InsP6, which could mobilize Ca2+, leading to inhibition of inward rectifying K+ channels, and changes in turgor pressure resulting in a decreased stomatal aperture (Lemtiri-Chlieh et al., 2000, 2003; Nagy et al., 2009). With the identification of elevated PPx-InsPs in mrp5 mutants, an alternative hypothesis for alterations in mrp5 guard cell signaling is that changes in InsP7 and InsP8 may be involved.
It should be noted that the recent study identifying PPx-InsPs in plants was not able to discern the enantiomers present (Desai et al., 2014). Thus, it is not known whether plants contain 5PP-InsP7 or 1PP-InsP7 similar to yeast and animals, or (4/6)PP-InsP7, similar to Dictyostelium. Efforts were made to purify the plant PPx-InsPs, however no informative NMR data was obtained (Desai et al., 2014). The identity of the plant isomers is key, and in itself may yield insights into the pathway, as different types of enzymes in other organisms give rise to specific PPx-InsP isomers. The following section describes this relationship between PPx-InsP synthesis and isomers in detail.
Synthesis of PPx-InsPs by KCS1/IP6K Enzymes
There are two classes of genes shown to encode enzymes required for synthesis of PPx-InsPs. Figure 3 shows the presence and names of these genes in species relevant to this review. These two classes of genes encode distinct enzymes that catalyze the addition of pyrophosphates at specific positions on the inositol ring (Figure 2). The first class is named the InsP6 kinases (IP6Ks), and the kinase activity of these enzymes phosphorylates the 5-position of InsP5, InsP6, and InsP7, yielding 5PP-InsP4 or 5PP-InsP5 and two possible forms of InsP8: 1/3,5PP-InsP5 and 5PPP-InsP5 (Draskovic et al., 2008). In yeast, this class of enzymes is named KCS1, and was first identified in a suppressor screen of the yeast Protein Kinase C (pkc1) mutant (Huang and Symington, 1995). Kcs1 encodes a protein closely related to the bZIP family of transcription factors, although analysis of its two potential leucine zipper motifs indicates the secondary alpha-helical structure for DNA binding is not formed (Huang and Symington, 1995). Instead, the altered structure of this alpha helix in addition to a two-turn 310 helix, forms a pocket for InsP6 binding (Wang et al., 2014).
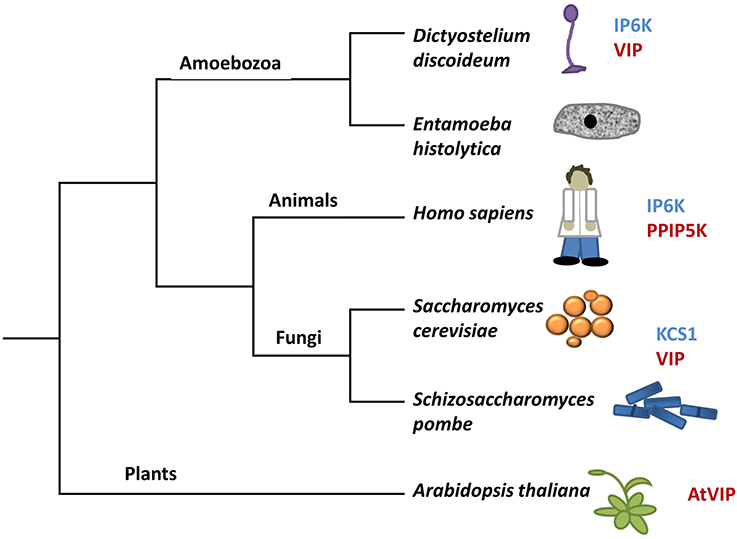
Figure 3. A modified tree of life indicating the composition of genes in different species that encode kinases capable of phosphorylating InsP6. Genes in blue have sequence identity with KCS1, whereas genes in red have sequence identity with VIP. The tree depicts evolutionary relationships between groups discussed in the review.
Under low energy conditions, KCS1 can generate ATP from InsP6 (Wundenberg et al., 2014). This dual function of KCS1 to both degrade InsP6 and generate InsP7 presents the possibility of KCS1 acting as an ATP/ADP ratio sensor (Wundenberg et al., 2014). Given the considerable amount of InsP6 in plants, if present, a KCS1-like enzyme could generate a significant source of ATP under low energy conditions. However, sequence homology searches using the yeast KCS1 and human IP6K proteins indicate that there are no KCS1/IP6K homologs in plants (Bennett et al., 2006; Desai et al., 2014).
In the absence of a plant KCS1/IP6K, one might expect that 5PP-InsP5 could not be synthesized. However, there is the possibility of another InsP kinase in plants acting as a KCS1/IP6K in the synthesis of 5PP-InsP5. The larger InsP kinase family (Pfam PF03770) has a PxxxDxKxG (“PDKG”) catalytic motif and includes the InsP3 kinases (IP3Ks), IP6K, and inositol polyphosphate multikinases (IPMKs). Recent phylogenetic studies on these enzymes has suggested that the InsP kinase family common ancestor is an IP6K precursor (Bennett et al., 2006). The rationale is that the substrate-binding pocket for InsP6 is larger, and from this common ancestor, the binding pocket would shrink to become more specific for other InsPs (Wang et al., 2014). Not all extant species have developed kinases solely acting on InsP3: Entamoeba histolytica still has an IP3K which retains IP6K activity (Wang et al., 2014), presenting the possibility that if plants have a KCS1/IP6K, it may be distinct from that of yeast and mammals.
Synthesis of PPx-InsPs by VIP/PPIP5K Enzymes
The second class of enzymes capable of synthesizing PPx-InsPs are the VIPs, which are also known as diphosphoinositol pentakisphosphate kinases (PPIP5Ks) in animals (Shears et al., 2013) (Figures 2, 3). In quick succession, two groups identified VIPs in yeast and mammalian cells (Choi et al., 2007; Fridy et al., 2007; Mulugu et al., 2007). The name PPIP5K originated as scientists were looking for an enzyme capable of phosphorylating PP-InsP5 to produce the InsP8, which had been observed in mammalian cell extracts (Stephens et al., 1993). The PPIP5K that was identified has a higher affinity for InsP7 than InsP6 (Choi et al., 2007). These enzymes produce a structurally distinct InsP7 with recent NMR work delineating 1PP-InsP7 as the product (Wang et al., 2012). These enzymes can also phosphorylate 5PP-InsP5 to produce 1,5(PP)2-InsP4 (Figure 2) or speculatively, even 1PPP-InsP5.
The Vip genes are conserved across eukaryotes, including plants (Mulugu et al., 2007; Desai et al., 2014). They have a dual domain structure consisting of an N-terminal ATP grasp domain with kinase activity and a C-terminal histidine acid phosphatase domain or “phytase” domain (Mulugu et al., 2007) (see Figure 4). The human PPIP5K1 phosphatase domain is not active with InsP5, InsP6, PP-InsP4, or PP-InsP5 as the substrate or even p-nitrophenyl phosphate, a generic substrate for acid phosphatases (Gokhale et al., 2011). This is probably due to the fact that PPIP5Ks lack a conserved histidine essential for phosphatase activity. In addition, the phosphatase catalytic region is interrupted by a Pleckstrin Homology (PH) domain (Gokhale et al., 2011). The PH domain is found in signaling proteins and is responsible for binding phospholipids or molecules derived from their head group (Scheffzek and Welti, 2012). The hybrid PH domain in PPIP5K1 preferentially binds PtdIns(3,4,5)P3 and can also bind InsP6 allowing PPIP5K1 to translocate from the cytoplasm to the plasma membrane when the PtdIns(3,4,5)P3 signaling pathway is activated (Gokhale et al., 2011). Critical to ligand binding is arginine 417 in the PH domain of PPIP5K1 (Gokhale et al., 2011).
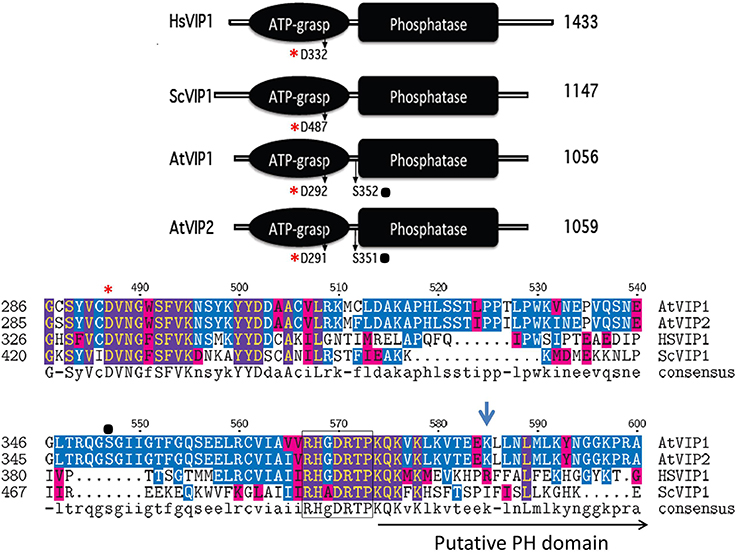
Figure 4. Schematic alignment of the kinase and phosphatase domains of VIPs. The ATP grasp/RimK/kinase (ATP-grasp) and the histidine acid Phosphatase (Phosphatase) domains within the VIP proteins from Homo sapiens (Hs, Genbank AAH57395), Saccharomyces cerevisiae (Sc, NP_013514) and Arabidopsis thaliana (AtVip1 Gene ID, 821297; AtVip2 Gene ID, 831359). The red asterisks denotes in both panels the conserved aspartic acid residue (D) required for kinase activity, and the black dot, the known phosphorylated serine residues within the Arabidopsis VIPs. The lower panel contains the amino acid alignment of the boundary region between the ATP-Grasp and Phosphatase domains. The beginning of the Histidine Acid phosphatase domain is boxed, followed by the PH domain and Arg417 is marked by the blue arrow.
All plant species searched through BLAST contained multiple expressed Vip homologs (Desai et al., 2014). Arabidopsis contains two conserved Vip genes, AtVip1 (At3g01310) and AtVip2 (At5g15070) and the encoded proteins have 94% similarity to each other, but only 50% and 59% similarity to yeast ScVIP1 and human HsVIP1 respectively (Desai et al., 2014). As with yeast and human VIPs, Arabidopsis VIP1 and VIP2 contain a RimK/ATP-Grasp domain (kinase domain) and a histidine acid-phosphatase domain (Figure 4). A BLAST analysis identifies potential Vip genes in several other plant species indicating that PPx-InsP synthesis is conserved across the plant kingdom. The kinase domain of the Arabidopsis VIPs contains a conserved aspartic acid (D), which as in yeast and humans, is necessary for activity (Choi et al., 2007; Mulugu et al., 2007; Desai et al., 2014). This residue is also conserved in mouse and Drosophila VIPs, however its role in activity has yet to be confirmed (Fridy et al., 2007). The AtVIP1 and AtVIP2 phosphatase domains are also interrupted with a putative PH domain (Gokhale et al., 2011) (Figure 4), however binding to PtdInsPs has not been tested to date. The arginine residue required for PtdInsP-binding of the human PPIP5K is not conserved in the AtVIPs, however, the substituted lysine at this position provides a similar charge as arginine and there are other arginine residues located nearby (Figure 4). Phosphoproteomics has shown that AtVIPs have a serine adjacent to the phosphatase domain that is phosphorylated. This is not conserved in ScVIP and HsVIP (http://phosphat.mpimp-golm.mpg.de) and may represent a unique mechanism for regulation of VIP activity specific to plants (Desai et al., 2014).
Importantly, both AtVIPs encode catalytically active proteins that allow specific yeast mutants to synthesize InsP7 (Desai et al., 2014). There appears to be an intriguing difference in the AtVIPs as compared to the human and yeast VIPs. In the human PPIP5K and yeast VIPs the kinase domain alone is more active than the full-length protein (Fridy et al., 2007; Mulugu et al., 2007), while both full-length AtVIPs are more active than their kinase domains alone (Desai et al., 2014). One explanation for this difference is that the phosphatase domain in the yeast VIP and human PPIP5K may auto-inhibit the kinase activity, and this control may be missing in the AtVIPs. Data from yeast supports this idea of auto-inhibition (Fridy et al., 2007). A second explanation is that the unique presence of a phosphorylated serine in the AtVIPs might provide regulatory control.
Although NMR data on the plant PPx-InsPs is not yet available, it seems reasonable to speculate that only the 1PP-PPx-InsPs should be synthesized in plants, since VIP enzymes are known to phosphorylate at this position. Indeed, the architecture of the VIP catalytic site is what determines the position of phosphorylation (Shears et al., 2013), and as discussed above, 5PP-InsPs may not be synthesized. Intriguing however, both InsP7 and InsP8 have been found in plants (Desai et al., 2014). This argues for the presence of a plant enzyme capable of 5PP-InsP synthetic ability, since the only structurally verified isomers of InsP8 in any organism, 1,5(PP)2-InsP4 and 5PPP-InsP5, both require phosphorylation at a C5 position (Laussmann et al., 1997, 1998; Draskovic et al., 2008; Lin et al., 2009). Further, in yeast, KCS1 and VIP are known to act sequentially to phosphorylate each other's InsP7 product, resulting in 1,5(PP)2-InsP4 synthesis (Figure 2). Alternatively, the plant InsP8 molecule may be unique in nature and may not require phosphorylation at the 5-position. Thus, either the plant VIPs are different enough that they can phosphorylate a different position, or there are other enzymes in the plant that can phosphorylate InsP6 or InsP7. It is important to note one final structural implication of PPx-InsPs: InsP7 produced by either IP6K or VIP may not be equivalent since the charge from phosphate is distributed differently in each, and the shapes are not superimposable. As a result, different InsP7 enantiomers may interact with different proteins and act in different signaling conditions or pathways.
The similarity of AtVIP1 and AtVIP2 proteins (94% similarity), and the ability of each gene product to restore InsP7 synthesis in yeast, suggests that these two genes function similarly at the biochemical level. Thus, the expression patterns of each gene may provide information on where and when InsP7 is synthesized. Recent studies showed that AtVip1 expression is high in vegetative tissues, including shoot of seedlings as well as mature leaf and stem. In contrast, AtVip2 is most abundantly expressed in roots and reproductive tissues (Desai et al., 2014) suggesting differential spatial regulation. Additionally, since both AtVip1 and AtVip2 are expressed together in some vegetative tissues, they may be differentially regulated at a subcellular level. Using subcellular prediction tools, we found compelling predictions for AtVIP1 nuclear localization and an AtVIP2 cytosolic location within the plant cell. Discerning whether the two AtVIPs really do function in these compartments will require experimental validation.
Meaning of PPx-InsPs: How are They Likely to Function in Plants?
If we continue the analogy of InsP as words, our next challenge will be to understand what these words mean and what information they convey. We will describe how InsP7 is known to modify or interact with proteins in other model systems, and how these actions lead to changes in known signaling functions in yeast and animals. Although little is known about PPx-InsPs function in plants and mechanisms regulating inositol signaling differ between plants and animals, it is likely that InsP7 conveys plant signaling information by virtue of modifying or interacting with proteins. Drawing parallels from the known and hypothesized roles of InsP7 in other organisms, we speculate that PPx-InsPs function in several plant signaling pathways, including, but not limited to energy homeostasis, phosphate (Pi) sensing, and immune responses. In the following sections, we will elaborate on published data from other model systems that indicates a role for PPx-InsPs in these pathways.
Energy Homeostasis
PPx-InsPs are involved in energy homeostasis both at the cellular and organismal level. Maintaining energy homeostasis, or the balance of intake/production, storage and use of energy often in the form of ATP or sugar, is essential for all living organism. In animals, the AMP Kinase (AMPK) is often named as an energy sensor. Under low energy conditions, AMP is bound, activating the AMP kinase and reprograming the cell to maximize energy acquisition and minimize energy use (Hardie, 2011). In opposition is mammalian Target Of Rapamycin (mTOR), which under high energy conditions promotes growth and cell division (Dunlop and Tee, 2009). In plants, these two enzymes also form the base for maintaining energy homeostasis.
At the cellular level, PPx-InsPs regulate ATP levels through what has been referred to as the “glycolic/mitochondrial metabolic ratio” in yeast (Szijgyarto et al., 2011). Yeast mutants lacking KCS1 have up to five fold higher level of ATP than their wild type controls while overexpression of KCS1 results in a decrease of ATP (Szijgyarto et al., 2011). A similar result is seen with kcs1 mutant mouse embryonic fibroblast (MEF) cells, where levels of ADP and AMP are low. Further studies showed that the yeast kcs1 and kcs1/vip1 mutants as well as MEF ip6k mutants have reduction or loss of functional mitochondria. This loss of mitochondrial function with high ATP levels can be explained by an increase in glycolysis and a reduction of ATP used in metabolic pathways. Thus, the lack of InsP7 synthesis in these mutants leads to changes in ATP synthesis and utilization. InsP7 in this system most likely affects ATP levels by altering transcription of genes that control glycolysis. Specifically, in yeast promoters of glycolytic regulatory genes have a CT-box that binds to the General Control Response 1 (GCR1) transcription factor. InsP7 may function to regulate GCR1 directly by a non-catalytic transfer of the β-phosphate from InsP7 to an already phosphorylated serine residue in GCR1, resulting in a pyrophosphorylated serine. This modification likely causes a conformational change in GCR1, allowing it to bind the CT-box, thereby regulating the expression of glycolytic regulatory genes (Szijgyarto et al., 2011).
This addition of a new pyrophosphate bond on an already phosphorylated serine residue in a target substrate protein is a proposed mechanism unique to InsP7. First demonstrated in yeast, InsP7was shown to modify proteins important for ribosomal biogenesis and endosomal trafficking (Saiardi et al., 2004). The pyrophosphorylated serine in target proteins is surrounded by acidic residues, possibly enhancing the recruitment of Mg2+ as a cofactor. This modification would be more permanent than phosphorylation by ATP (Bhandari et al., 2007), as no known enzymes exist to remove the pyrophosphate. One limitation to acceptance of this mechanism is that it has been difficult to verify whether such pyrophosphorylated serines occur in vivo.
At the whole organism level, InsP7 functions in sugar homeostasis through regulating insulin release and glucose uptake in animals. In mammals, InsP7 acts as an inhibitor of the Protein Kinase B (also known as Akt) pathway, reducing glucose uptake, insulin sensitivity and protein translation. In response to growth factors, Akt is normally phosphorylated by a protein kinase named PDK1 (3-phosphoinositide-dependent protein kinase 1), which activates the GSK3β (Glycogen synthase kinase 3) and mTOR signaling pathways. Activation of Akt requires binding of its PH domain to PtdIns(3,4,5)P3, associated with the plasma membrane. When bound to PtdIns(3,4,5)P3, Akt undergoes a conformational change which exposes its activation domain, allowing Akt to be phosphorylated and activated by PDK1 (Calleja et al., 2007). InsP7, produced by IP6K1, acts as an inhibitor of Akt by competing for binding to the PH domain within Akt. This effectively prevents the phosphorylation of T308 of Akt (Chakraborty et al., 2010), and dampens Akt signaling. ip6k1 loss-of-function mutant mice are smaller than their wild type littermates and have lower circulating levels of insulin, but are not diabetic and they have normal blood glucose levels (Bhandari et al., 2008). These genetic data underscore the role of InsP7 in global metabolic control.
While insulin is not made by plants, gene homologs functioning in the Akt, GSK3β, and mTOR pathways exist in plants, and have been implicated in growth control pathways. Plants contain homologs of both Akt (i.e., Adi3: AvrPto-dependent Pto-interacting protein 3) and the kinase that activates Akt, PDK1. Most studies indicate that plants do not synthesize PtdIns(3,4,5)P3, however plant PDK1 is known to bind phosphatidic acid via its PH domain, allowing membrane localization (Anthony et al., 2004) and the subsequent activation of Adi3 (Devarenne and Martin, 2007). Adi3 can suppress the activity of a major regulator of plant metabolism and AMPK homolog, SnRK1 (Sucrose non-fermenting Related Kinase 1), through phosphorylation of a SnRK1 multiple subunit complex (Avila et al., 2012). GSK-3 kinases are negative regulators of signal transduction pathways controlling metabolism and developmental events across the animal kingdom. In plants, GSK3 homologs are involved in brassinosteroid (BR) signaling pathways. Specifically, the brassinosteroid insensitive 2 (BIN2) protein is a GSK-3 that functions as a negative regulator of BR signal transduction (Yan et al., 2009; Clouse, 2011). As with the animal GSK-3 signaling, BR signal transduction is required for proper metabolic and developmental control throughout the life of a plant. In the case of mTOR, the Arabidopsis gene homologs are known to be important regulators of metabolic changes in response to glucose. Arabidopsis TOR signaling has been linked to transcriptional reprogramming of central and secondary metabolism and other processes within plants (Xiong and Sheen, 2014).
We do not yet know whether InsP7 in plants regulates transcription via pyrophosphorylation or whether InsP7 can compete with binding to PH domains of plant proteins, however, both are potential mechanisms by which InsP7 could act. Determining whether plants use InsP7 to regulate metabolism or growth, and the mechanistic details of such regulation will benefit from the development of genetic resources to examine Atvip loss-of-function mutants. In addition, we need to know whether PPx-InsPs levels are altered by changes in energy or metabolic status. Answering these questions is now possible as the basis for detecting and measuring PPx-InsPs in plants has been established, and the AtVip genes have been cloned and shown to encode active proteins.
Pi Sensing
PPx-InsPs are also involved in perceiving and maintaining Pi levels and numerous studies link PPx-InsPs to low Pi responses in other organisms. Plant Pi homeostasis is a highly regulated process (Zhang et al., 2014) and it is important to consider whether PPx-InsPs play a role in this process in plants. Pi sensing involves the perception of Pi present in the environment, followed by acquisition, remobilization and recycling of Pi to maintain Pi homeostasis. In yeast, the response to Pi starvation is regulated by the Pi-responsive (PHO) signaling pathway, including the Pho80-Pho85 cyclin-CDK (cyclin dependent kinase) complex (Lenburg and O'shea, 1996; Carroll and O'shea, 2002). When Pi levels are low, the Pho80-Pho85 protein complex is inactive. As a result, the Pho4 transcription factor is not phosphorylated and remains in the nucleus where it acts to activate PHO genes (Kaffman et al., 1994; O'neill et al., 1996).
Though there is some lack of consensus for the exact mechanism by which PPx-InsPs control Pi sensing, it is clear that they play a role in Pi homeostasis. One group found that low Pi elevates InsP7, and genetic evidence suggested that it was 1PP-InsP5, although this was not experimentally confirmed (Lee et al., 2007). This group showed that InsP7 physically interacts with Pho81, inactivating the Pho80-Pho85 complex, and ultimately leading to changes in gene expression required to maintain metabolic homeostasis under low Pi conditions. In addition, this pathway was dependent on the activity of the yeast VIP genes (Lee et al., 2007). The finding that InsP7 is elevated in yeast in response to low Pi has been contested by other investigators. Exposure of wild type yeast to Pi-free medium for 20 min resulted in a decrease of intracellular PPx-InsPs levels by 80%, without affecting InsP6 levels (Lonetti et al., 2011).
The change in InsP7 levels is not the only phenotype that suggests KCS1 is involved in the yeast low Pi response. An intriguing connection between KCS1 and Pi sensing comes from recent studies that found that Pho4 binds to intragenic regions of the Kcs1 gene, promoting the transcription of intragenic and antisense RNA. The authors suggested that the truncated KCS1 protein produced could down-regulate KCS1 function (Nishizawa et al., 2008). An alternative hypothesis is that this truncated KCS1 protein has an altered enzymatic property yet to be determined (Saiardi, 2012). Further work is needed to examine these possibilities and determine the function of intragenic and antisense Kcs1 RNA. In addition, recent work examining the lipidome of numerous yeast mutants found similarities in changes in sphingolipids of pho85 and kcs1, but not vip1 mutants, suggesting that similar metabolic changes take place in pho85 and kcs1 mutants (Da Silveira Dos Santos et al., 2014). Together these data suggest that either Vip or Kcs1 genes, or possibly both, are linked to Pi sensing and sphingolipid homeostasis in yeast.
In animal cells, IP6K has been identified as a stimulator of Pi uptake in response to low nutrients. A study found that the mRNAs expressed in rabbit duodenum from a rabbit fed a low Pi diet can stimulate Na+-dependent Pi uptake when injected into Xenopus oocytes (Yagci et al., 1992). From this pool of mRNAs, the Pi uptake stimulator (PiUS) gene was isolated and confirmed to increase Pi uptake when expressed in Xenopus oocytes (Norbis et al., 1997). The PiUS gene was later found to encode an IP6K, and the gene is now known as Ip6k2 (Saiardi et al., 1999).
In plants, InsPs are essential for Pi response and homeostasis. Arabidopsis mutants in ipk1, which catalyzes the addition of a phosphate at the 2-position to select substrates, have a 83% reduction in InsP6 levels compared to wild type, and are hypersensitive to Pi (Stevenson-Paulik et al., 2005). These mutants have increased uptake of Pi and root to shoot translocation of Pi (Kuo et al., 2014). Many plant responses to Pi starvation (PSR) are regulated at a transcriptional and post-transcriptional level. A recent study has shown that a sub set of PSR genes involved in Pi uptake, translocation and remobilization are up regulated in the ipk1 mutant under Pi sufficient conditions (Kuo et al., 2014). Additionally, increased expression of a subset of PSR genes was shown to correlate with a reduction of histone H2A.Z occupancy (Smith et al., 2010) and interestingly, H2A.Z occupancy at chromatin sites associated with several PSR genes was found to be significantly reduced in ipk1 (under both sufficient and low Pi) compared to wild type (Kuo et al., 2014). However, Arabidopsis mutants with lower InsP6 levels including mips1 (myo-inositol 1-phosphate synthase), do not show an accumulation of Pi, indicating that InsP6 per se is probably not the molecule utilized for sensing Pi (Kuo et al., 2014). This implicates other InsPs or the PPx-InsPs as controllers of Pi sensing. In particular, since PPx-InsPs are synthesized from InsP6 substrates, these molecules might serve as critical players in sensing Pi. We note that the conversion between InsP6 and the PPx-InsPs might be important as InsP6 serves an important function in phosphorous storage (Raboy, 2003).
Studies on yeast and animal mutant responses to low Pi were among the first to highlight a specific property of InsPs that may be especially important for understanding PPx-InsP function. Response to environmental stress, including Pi starvation, requires the fine tuning of TOR and the TORC1 complex activity. This results in the down regulation of ribosomal and protein synthesis regulatory genes, as well as the up regulation of stress response genes (Loewith et al., 2002). Working in parallel with the inactivation of TORC1, the histone deacetylase (HDAC) enzyme is recruited to turn off expression of ribosomal and protein synthesis regulatory genes (Alejandro-Osorio et al., 2009). This HDAC activity is dependent on InsP4, which is known to act as a “molecular glue” allowing the HDAC Rpd3L complex to interact with its co-repressor, SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) (Watson et al., 2012). InsP7 has been hypothesized to interact with this same complex (Worley et al., 2013), suggesting that PPx-InsPs may play a role in chromatin remodeling thereby regulating gene expression.
There are other known cases of InsPs serving a role as a type of molecular glue, and these bear mentioning. InsP6 and InsP5 have been found in the auxin (TIR1; Transport Inhibitor Response 1) (Tan et al., 2007) and jasmonic acid receptor, COI1 (Coronatine Insensitive 1) (Sheard et al., 2010; Mosblech et al., 2011), respectively. In both of these examples, InsPs are lodged between the F-Box and the repressor protein target in the E3 ubiquitin ligase complex. When the hormone is present, the repressors for auxin and jasmonic acid, Aux/IAA (Auxin inducible) and JAZ (Jasmonate Zim-domain protein) respectively, are degraded, allowing for transcription of hormone responsive genes. TIR1 is a member of a family of F-box proteins whose five members differ slightly in expression, biochemical activity or function (Parry et al., 2009) and Aux/IAA belongs to an even larger family of 29 proteins (Remington et al., 2004). Like TIR1, COI1 is a member of the F-Box family while JAZ is a 12 member subgroup of TIFY (named for the shared TIF[F/Y]XG motif) (Chini et al., 2007; Vanholme et al., 2007). With all the potential combinations of hormone receptors and repressor proteins, it is interesting to speculate whether other InsPs, including PPx-InsPs, may function as cofactors in hormone signaling to regulate transcription.
Immune Response
The innate immune system is the first line of defense in both plants and animals. The first step in the innate immune response pathway involves the recognition of pathogen-associated molecular patterns (PAMPs) by the host pattern recognition receptors (PRRs) on the cell surface or cytoplasm. In plants, pathogen detection, signaling, and immune response takes place in most cells, while animal immune systems have evolved specialized mobile immune cells. Detection of PAMPs by PRRs initiate signaling cascades which can result in Ca2+ release, activation of kinases, and transcription factors, production of reactive oxygen species and alterations in other signaling pathways within the organism (Jones and Dangl, 2006). In animals, RIG-1 (retinoic acid-inducible gene 1) is a PRR in the cytoplasm, which detects double stranded viral DNA and activates a signaling cascade in which the transcription factor IRF3 (Interferon regulatory factor 3) is phosphorylated. IFR3 then moves into the nucleus and promotes the expression of type-1 interferon genes. The interferon proteins then stimulate anti-viral or anti-bacterial activity in leukocytes (Trinchieri, 2010). Recently, this innate response pathway was linked to PPx-InsPs. An in vitro study found both InsP7 and InsP8 are capable of inducing an interferon response through the RIG-1 signaling pathway. The authors of this study speculated that 1PP-InsP5 is the physiologically relevant molecule and acts a co-factor for protein interactions or by β-phosphorylation of a serine residue on IRF3, a type of PPx-InsP-driven protein pyrophosphorylation event that we have previously discussed (Pulloor et al., 2014).
The innate immune system in plants and animals share many similarities in the use of PAMPs and PRR as a method of detecting pathogens. Plants have a large diversity of PRR and Resistance (R) genes, however homologs of the RIG-1/IRF3 pathway have not been found in plants. Therefore, while it is interesting to speculate that PPx-InsPs may regulate specific defense transcription factors in plants, the lack of RIG-1 and IRF-3 homologs suggests that this pathway may be unique to animal innate immunity signaling.
A second role of PPx-InsPs in the animal innate immune response involves the afore-mentioned PDK1/Akt signaling pathway. This complex pathway regulates multiple central biological processes including cell survival, proliferation, growth, and metabolism (Hemmings and Restuccia, 2012). In the immune system, neutrophil activation is tightly controlled, with 5PP-InsP5 acting as a negative regulator. As described previously, 5PP-InsP5 competes for binding with Akt through the PH domain. Upon infection, 5PP-InsP5 levels drop allowing Akt to translocate to the membrane and allow for the induction of PtdIns(3,4,5)P3 signaling, leading to neutrophil activation and superoxide production. ip6k mutant neutrophils have increased bactericidal activity and ROS production (Prasad et al., 2011). Akt triggers reactive oxygen species and nitric oxide production, and is not limited to neutrophils, it can also regulate programmed cell death in other cell types (Lam, 2004). The inhibition of Akt signaling by InsP7 is a general phenomenon, however, the mechanism of regulation and the biological outcome may differ depending on tissue and signaling context (Prasad et al., 2011).
A common characteristic of the innate immune response is the programed cell death of infected cells to reduce the spread of disease. In plants, localized programed cell death stimulated by the hypersensitive response occurs rapidly in response to pathogen infection (Morel and Dangl, 1997). As mentioned previously, plants have a homologous pathway to the PDK1/Akt pathway in mammals. In plants, the homolog to Akt is Adi3, which acts in the immune response as a negative regulator of cell death through the MAPK kinase cascade. Akt and Adi3 kinases may be a target for pathogen manipulation of the host In the case of Pseudomonas infection of tomato, the bacterial effector protein AvrPto interacts with the Adi3 presumably to manipulate cell death (Devarenne et al., 2006). It is intriguing to speculate whether PPx-InsPs may serve as innate immunity signaling molecules in plants, perhaps by acting to antagonize Adi3 signaling. However it should be noted that although functionally similar, Akt and Adi3 share only 21.4% amino acid identity, suggesting differences in regulation and possibly function (Devarenne et al., 2006).
A final connection between PPx-InsPs and plant innate immune signaling involves plant mutants defective in the synthesis or metabolism of InsPs. Transgenic plants constitutively expressing the human type 1 inositol polyphosphate 5-phosphatase (InsP 5-ptase, the enzyme which dephosphorylates InsP3), showed a compromised defense response, including decreased expression of defense genes and a reduction in the systemic acquired immunity in response to a bacterial pathogen (Hung et al., 2014). Furthermore, plants defective in synthesis of myo-inositol and InsP6 were also more susceptible to disease, including viral, bacterial and fungal pathogen infection (Murphy et al., 2008). It was concluded that InsP6 and not its precursors is the critical InsP for this phenotype, however, the authors of this study could not rule out a role for PPx-InsPs in this process due to the difficulty in detection (Murphy et al., 2008). Crops with altered InsP profiles, specifically low InsP6, have been developed to combat issues of nutrition and Pi pollution (Raboy, 2007). If InsP6 or PPx-InsPs play a role in pathogen resistance and immune response, it could negatively impact the performance of these so-called low phytate crops.
Concluding Remarks
PPx-InsPs have recently been identified in higher plants, adding new molecular players in the plant inositol signaling pathway. Both InsP7 and InsP8 have been detected in a handful of plant species. With two Vip/PPIP5K gene homologs as the only identified kinase to synthesize PPx-InsPs, the predicted species are 1PP-InsP5 and either 1,3(PP)2-InsP4 or 1PPP-InsP5. Further work is needed to identify the stereochemistry of plant PPx-InsPs and to clarify the regulatory components involved in their synthesis and metabolism. Drawing parallels to known roles of PPx-InsPs in other eukaryotes, plant PPx-InsPs may have a role in energy, Pi sensing, and innate immunity signaling pathways. Thus, identification of PPx-InsPs in plants presents a new avenue and tool that may be useful for improving crop yield, reduced fertilizer demand, and improved growth under stress.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a NSF collaborative grant (MCB1051646 to GG and MCB 1052034 to IYP) and a NIFA award (2013-02277 to GG and IYP).
References
Alejandro-Osorio, A. L., Huebert, D. J., Porcaro, D. T., Sonntag, M. E., Nillasithanukroh, S., Will, J. L., et al. (2009). The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 10:R57. doi: 10.1186/gb-2009-10-5-r57
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anthony, R. G., Henriques, R., Helfer, A., Meszaros, T., Rios, G., Testerink, C., et al. (2004). A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581. doi: 10.1038/sj.emboj.7600068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Avila, J., Gregory, O. G., Su, D., Deeter, T. A., Chen, S., Silva-Sanchez, C., et al. (2012). The beta-subunit of the SnRK1 complex is phosphorylated by the plant cell death suppressor Adi3. Plant Physiol. 159, 1277–1290. doi: 10.1104/pp.112.198432
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Azevedo, C., and Saiardi, A. (2006). Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 1, 2416–2422. doi: 10.1038/nprot.2006.337
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bennett, M., Onnebo, S. M., Azevedo, C., and Saiardi, A. (2006). Inositol pyrophosphates: metabolism and signaling. Cell. Mol. Life Sci. 63, 552–564. doi: 10.1007/s00018-005-5446-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhandari, R., Juluri, K. R., Resnick, A. C., and Snyder, S. H. (2008). Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 2349–2353. doi: 10.1073/pnas.0712227105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhandari, R., Saiardi, A., Ahmadibeni, Y., Snowman, A. M., Resnick, A. C., Kristiansen, T. Z., et al. (2007). Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. U.S.A. 104, 15305–15310. doi: 10.1073/pnas.0707338104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brearley, C. A., and Hanke, D. E. (1996). Inositol phosphates in barley (Hordeum vulgare L.) aleurone tissue are stereochemically similar to the products of breakdown of InsP6 in vitro by wheat-bran phytase. Biochem. J. 318(Pt 1), 279–286.
Calleja, V., Alcor, D., Laguerre, M., Park, J., Vojnovic, B., Hemmings, B. A., et al. (2007). Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 5:e95. doi: 10.1371/journal.pbio.0050095
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carroll, A. S., and O'shea, E. K. (2002). Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27, 87–93. doi: 10.1016/S0968-0004(01)02040-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chakraborty, A., Koldobskiy, M. A., Bello, N. T., Maxwell, M., Potter, J. J., Juluri, K. R., et al. (2010). Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 143, 897–910. doi: 10.1016/j.cell.2010.11.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Choi, J. H., Williams, J., Cho, J., Falck, J. R., and Shears, S. B. (2007). Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 282, 30763–30775. doi: 10.1074/jbc.M704655200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clouse, S. D. (2011). Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23, 1219–1230. doi: 10.1105/tpc.111.084475
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Da Silveira Dos Santos, A. X., Riezman, I., Aguilera-Romero, M. A., David, F., Piccolis, M., Loewith, R., et al. (2014). Systematic lipidomic analysis of yeast protein kinase and phosphatase mutants reveals novel insights into regulation of lipid homeostasis. Mol. Biol. Cell 25, 3234–3246. doi: 10.1091/mbc.E14-03-0851
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Desai, M., Rangarajan, P., Donahue, J. L., Williams, S. P., Land, E. S., Mandal, M. K., et al. (2014). Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J. 80, 642–653. doi: 10.1111/tpj.12669
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Devarenne, T. P., Ekengren, S. K., Pedley, K. F., and Martin, G. B. (2006). Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J. 25, 255–265. doi: 10.1038/sj.emboj.7600910
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Devarenne, T. P., and Martin, G. B. (2007). Manipulation of plant programmed cell death pathways during plant-pathogen interactions. Plant Signal. Behav. 2, 188–189. doi: 10.4161/psb.2.3.4150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dorsch, J. A., Cook, A., Young, K. A., Anderson, J. M., Bauman, A. T., Volkmann, C. J., et al. (2003). Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry 62, 691–706. doi: 10.1016/S0031-9422(02)00610-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Draskovic, P., Saiardi, A., Bhandari, R., Burton, A., Ilc, G., Kovacevic, M., et al. (2008). Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 15, 274–286. doi: 10.1016/j.chembiol.2008.01.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dunlop, E. A., and Tee, A. R. (2009). Mammalian target of rapamycin complex 1: signalling inputs, substrates and feedback mechanisms. Cell. Signal. 21, 827–835. doi: 10.1016/j.cellsig.2009.01.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Flores, S., and Smart, C. C. (2000). Abscisic acid-induced changes in inositol metabolism in Spirodela polyrrhiza. Planta 211, 823–832. doi: 10.1007/s004250000348
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fridy, P. C., Otto, J. C., Dollins, D. E., and York, J. D. (2007). Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 282, 30754–30762. doi: 10.1074/jbc.M704656200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gillaspy, G. E. (2011). The cellular language of myo-inositol signaling. New Phytol. 192, 823–839. doi: 10.1111/j.1469-8137.2011.03939.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glennon, M. C., and Shears, S. B. (1993). Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem. J. 293(Pt 2), 583–590.
Gokhale, N. A., Zaremba, A., and Shears, S. B. (2011). Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem. J. 434, 415–426. doi: 10.1042/BJ20101437
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hardie, D. G. (2011). AMPK and autophagy get connected. EMBO J. 30, 634–635. doi: 10.1038/emboj.2011.12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hawkins, P. T., Stephens, L. R., and Piggott, J. R. (1993). Analysis of inositol metabolites produced by Saccharomyces cerevisiae in response to glucose stimulation. J. Biol. Chem. 268, 3374–3383.
Heilmann, M., and Heilmann, I. (2014). Plant phosphoinositides-complex networks controlling growth and adaptation. Biochim. Biophys. Acta doi: 10.1016/j.bbalip.2014.09.018. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hemmings, B. A., and Restuccia, D. F. (2012). PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 4:a011189. doi: 10.1101/cshperspect.a011189
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, C. F., Voglmaier, S. M., Bembenek, M. E., Saiardi, A., and Snyder, S. H. (1998). Identification and purification of diphosphoinositol pentakisphosphate kinase, which synthesizes the inositol pyrophosphate bis(diphospho)inositol tetrakisphosphate. Biochemistry 37, 14998–15004. doi: 10.1021/bi981920l
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, K. N., and Symington, L. S. (1995). Suppressors of a Saccharomyces cerevisiae pkc1 mutation identify alleles of the phosphatase gene PTC1 and of a novel gene encoding a putative basic leucine zipper protein. Genetics 141, 1275–1285.
Hung, C. Y., Aspesi, P. Jr., Hunter, M. R., Lomax, A. W., and Perera, I. Y. (2014). Phosphoinositide-signaling is one component of a robust plant defense response. Front. Plant Sci. 5:267. doi: 10.3389/fpls.2014.00267
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaffman, A., Herskowitz, I., Tjian, R., and O'shea, E. K. (1994). Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science 263, 1153–1156. doi: 10.1126/science.8108735
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Klein, M., Perfus-Barbeoch, L., Frelet, A., Gaedeke, N., Reinhardt, D., Mueller-Roeber, B., et al. (2003). The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J. 33, 119–129. doi: 10.1046/j.1365-313X.2003.016012.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuo, H. F., Chang, T. Y., Chiang, S. F., Wang, W. D., Charng, Y. Y., and Chiou, T. J. (2014). Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J. 80, 503–515. doi: 10.1111/tpj.12650
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lam, E. (2004). Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5, 305–315. doi: 10.1038/nrm1358
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Laussmann, T., Hansen, A., Reddy, K. M., Reddy, K. K., Falck, J. R., and Vogel, G. (1998). Diphospho-myo-inositol phosphates in Dictyostelium and Polysphondylium: identification of a new bisdiphospho-myo-inositol tetrakisphosphate. FEBS Lett. 426, 145–150. doi: 10.1016/S0014-5793(98)00329-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Laussmann, T., Reddy, K. M., Reddy, K. K., Falck, J. R., and Vogel, G. (1997). Diphospho-myo-inositol phosphates from Dictyostelium identified as D-6-diphospho-myo-inositol pentakisphosphate and D-5,6-bisdiphospho-myo-inositol tetrakisphosphate. Biochem. J. 322(Pt 1), 31–33.
Lee, Y. S., Mulugu, S., York, J. D., and O'shea, E. K. (2007). Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316, 109–112. doi: 10.1126/science.1139080
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lemtiri-Chlieh, F., Macrobbie, E. A., and Brearley, C. A. (2000). Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. U.S.A. 97, 8687–8692. doi: 10.1073/pnas.140217497
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lemtiri-Chlieh, F., Macrobbie, E. A., Webb, A. A., Manison, N. F., Brownlee, C., Skepper, J. N., et al. (2003). Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc. Natl. Acad. Sci. U.S.A. 100, 10091–10095. doi: 10.1073/pnas.1133289100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lenburg, M. E., and O'shea, E. K. (1996). Signaling phosphate starvation. Trends Biochem. Sci. 21, 383–387. doi: 10.1016/0968-0004(96)10048-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, H., Fridy, P. C., Ribeiro, A. A., Choi, J. H., Barma, D. K., Vogel, G., et al. (2009). Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 284, 1863–1872. doi: 10.1074/jbc.M805686200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., et al. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468. doi: 10.1016/S1097-2765(02)00636-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lonetti, A., Szijgyarto, Z., Bosch, D., Loss, O., Azevedo, C., and Saiardi, A. (2011). Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974. doi: 10.1074/jbc.M111.266320
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Losito, O., Szijgyarto, Z., Resnick, A. C., and Saiardi, A. (2009). Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS ONE 4:e5580. doi: 10.1371/journal.pone.0005580
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin, J. B., Laussmann, T., Bakker-Grunwald, T., Vogel, G., and Klein, G. (2000). Neo-inositol polyphosphates in the amoeba Entamoeba histolytica. J. Biol. Chem. 275, 10134–10140. doi: 10.1074/jbc.275.14.10134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mayr, G. W. (1988). A novel metal-dye detection system permits picomolar-range h.p.l.c. analysis of inositol polyphosphates from non-radioactively labelled cell or tissue specimens. Biochem. J. 254, 585–591.
Menniti, F. S., Miller, R. N., Putney, J. W. Jr., and Shears, S. B. (1993). Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 268, 3850–3856.
Morel, J. B., and Dangl, J. L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. doi: 10.1038/sj.cdd.4400309
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mosblech, A., Thurow, C., Gatz, C., Feussner, I., and Heilmann, I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65, 949–957. doi: 10.1111/j.1365-313X.2011.04480.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mulugu, S., Bai, W., Fridy, P. C., Bastidas, R. J., Otto, J. C., Dollins, D. E., et al. (2007). A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109. doi: 10.1126/science.1139099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murphy, A. M., Otto, B., Brearley, C. A., Carr, J. P., and Hanke, D. E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56, 638–652. doi: 10.1111/j.1365-313X.2008.03629.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nagy, R., Grob, H., Weder, B., Green, P., Klein, M., Frelet-Barrand, A., et al. (2009). The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 284, 33614–33622. doi: 10.1074/jbc.M109.030247
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, M., Komai, T., Morohashi, N., Shimizu, M., and Toh-e, A. (2008). Transcriptional repression by the Pho4 transcription factor controls the timing of SNZ1 expression. Eukaryot. Cell 7, 949–957. doi: 10.1128/EC.00366-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Norbis, F., Boll, M., Stange, G., Markovich, D., Verrey, F., Biber, J., et al. (1997). Identification of a cDNA/protein leading to an increased Pi-uptake in Xenopus laevis oocytes. J. Membr. Biol. 156, 19–24. doi: 10.1007/s002329900183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O'neill, E. M., Kaffman, A., Jolly, E. R., and O'shea, E. K. (1996). Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271, 209–212. doi: 10.1126/science.271.5246.209
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Otto, J. C., Mulugu, S., Fridy, P. C., Chiou, S. T., Armbruster, B. N., Ribeiro, A. A., et al. (2007). Biochemical analysis of inositol phosphate kinases. Meth. Enzymol. 434, 171–185. doi: 10.1016/S0076-6879(07)34010-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parry, G., Calderon-Villalobos, L. I., Prigge, M., Peret, B., Dharmasiri, S., Itoh, H., et al. (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. U.S.A. 106, 22540–22545. doi: 10.1073/pnas.0911967106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Prasad, A., Jia, Y., Chakraborty, A., Li, Y., Jain, S. K., Zhong, J., et al. (2011). Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat. Immunol. 12, 752–760. doi: 10.1038/ni.2052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pulloor, N. K., Nair, S., Kostic, A. D., Bist, P., Weaver, J. D., Riley, A. M., et al. (2014). Human genome-wide RNAi screen identifies an essential role for inositol pyrophosphates in Type-I interferon response. PLoS Pathog. 10:e1003981. doi: 10.1371/journal.ppat.1003981
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raboy, V. (2003). myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64, 1033–1043. doi: 10.1016/S0031-9422(03)00446-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raboy, V. (2007). The ABCs of low-phytate crops. Nat. Biotechnol. 25, 874–875. doi: 10.1038/nbt0807-874
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Remington, D. L., Vision, T. J., Guilfoyle, T. J., and Reed, J. W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752. doi: 10.1104/pp.104.039669
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saiardi, A. (2012). How inositol pyrophosphates control cellular phosphate homeostasis? Adv. Biol. Regul. 52, 351–359. doi: 10.1016/j.jbior.2012.03.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saiardi, A., Bhandari, R., Resnick, A. C., Snowman, A. M., and Snyder, S. H. (2004). Phosphorylation of proteins by inositol pyrophosphates. Science 306, 2101–2105. doi: 10.1126/science.1103344
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saiardi, A., Erdjument-Bromage, H., Snowman, A. M., Tempst, P., and Snyder, S. H. (1999). Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9, 1323–1326. doi: 10.1016/S0960-9822(00)80055-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scheffzek, K., and Welti, S. (2012). Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 586, 2662–2673. doi: 10.1016/j.febslet.2012.06.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shears, S. B., Gokhale, N. A., Wang, H., and Zaremba, A. (2011). Diphosphoinositol polyphosphates: what are the mechanisms? Adv. Enzyme Regul. 51, 13–25. doi: 10.1016/j.advenzreg.2010.09.008
Shears, S. B., Weaver, J. D., and Wang, H. (2013). Structural insight into inositol pyrophosphate turnover. Adv. Biol. Regul. 53, 19–27. doi: 10.1016/j.jbior.2012.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shi, J., Wang, H., Schellin, K., Li, B., Faller, M., Stoop, J. M., et al. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25, 930–937. doi: 10.1038/nbt1322
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, A. P., Jain, A., Deal, R. B., Nagarajan, V. K., Poling, M. D., Raghothama, K. G., et al. (2010). Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 152, 217–225. doi: 10.1104/pp.109.145532
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stephens, L., Radenberg, T., Thiel, U., Vogel, G., Khoo, K. H., Dell, A., et al. (1993). The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s). J. Biol. Chem. 268, 4009–4015.
Stevenson-Paulik, J., Bastidas, R. J., Chiou, S. T., Frye, R. A., and York, J. D. (2005). Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 102, 12612–12617. doi: 10.1073/pnas.0504172102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suh, S. J., Wang, Y. F., Frelet, A., Leonhardt, N., Klein, M., Forestier, C., et al. (2007). The ATP binding cassette transporter AtMRP5 modulates anion and calcium channel activities in Arabidopsis guard cells. J. Biol. Chem. 282, 1916–1924. doi: 10.1074/jbc.M607926200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Szijgyarto, Z., Garedew, A., Azevedo, C., and Saiardi, A. (2011). Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805. doi: 10.1126/science.1211908
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tan, X., Calderon-Villalobos, L. I., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Trinchieri, G. (2010). Type I interferon: friend or foe? J. Exp. Med. 207, 2053–2063. doi: 10.1084/jem.20101664
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Leeuwen, W., Okresz, L., Bogre, L., and Munnik, T. (2004). Learning the lipid language of plant signalling. Trends Plant Sci. 9, 378–384. doi: 10.1016/j.tplants.2004.06.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Voglmaier, S. M., Bembenek, M. E., Kaplin, A. I., Dorman, G., Olszewski, J. D., Prestwich, G. D., et al. (1996). Purified inositol hexakisphosphate kinase is an ATP synthase: diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc. Natl. Acad. Sci. U.S.A. 93, 4305–4310. doi: 10.1073/pnas.93.9.4305
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., Derose, E. F., London, R. E., and Shears, S. B. (2014). IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family. Nat. Commun. 5:4178. doi: 10.1038/ncomms5178
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., Falck, J. R., Hall, T. M., and Shears, S. B. (2012). Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 8, 111–116. doi: 10.1038/nchembio.733
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watson, P. J., Fairall, L., Santos, G. M., and Schwabe, J. W. (2012). Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481, 335–340. doi: 10.1038/nature10728
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Worley, J., Luo, X., and Capaldi, A. P. (2013). Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Rep. 3, 1476–1482. doi: 10.1016/j.celrep.2013.03.043
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wundenberg, T., Grabinski, N., Lin, H., and Mayr, G. W. (2014). Discovery of InsP6-kinases as InsP6-dephosphorylating enzymes provides a new mechanism of cytosolic InsP6 degradation driven by the cellular ATP/ADP ratio. Biochem. J. 462, 173–184. doi: 10.1042/BJ20130992
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xiong, Y., and Sheen, J. (2014). The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 164, 499–512. doi: 10.1104/pp.113.229948
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yagci, A., Werner, A., Murer, H., and Biber, J. (1992). Effect of rabbit duodenal mRNA on phosphate transport in Xenopus laevis oocytes: dependence on 1,25-dihydroxy-vitamin-D3. Pflugers Arch. 422, 211–216. doi: 10.1007/BF00376204
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yan, Z., Zhao, J., Peng, P., Chihara, R. K., and Li, J. (2009). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150, 710–721. doi: 10.1104/pp.109.138099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Z., Liao, H., and Lucas, W. J. (2014). Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 56, 192–220. doi: 10.1111/jipb.12163
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: plant inositol signaling, inositol hexakisphosphate, VIP, inositol pyrophosphate, energy metabolism
Citation: Williams SP, Gillaspy GE and Perera IY (2015) Biosynthesis and possible functions of inositol pyrophosphates in plants. Front. Plant Sci. 6:67. doi: 10.3389/fpls.2015.00067
Received: 24 November 2014; Accepted: 26 January 2015;
Published online: 12 February 2015.
Edited by:
Eric Ruelland, Centre National de la Recherche Scientifique, FranceReviewed by:
Tzyy-Jen Chiou, Academia Sinica, TaiwanIngo Heilmann, Martin-Luther-University Halle-Wittenberg, Germany
Copyright © 2015 Williams, Gillaspy and Perera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Imara Y. Perera, Plant and Microbial Biology, North Carolina State University, Campus Box 7612, Raleigh, NC, USA e-mail: imara_perera@ncsu.edu
 Sarah P. Williams
Sarah P. Williams Glenda E. Gillaspy
Glenda E. Gillaspy Imara Y. Perera
Imara Y. Perera