- Kihara Institute for Biological Research, Yokohama City University, Yokohama, Japan
The polycomb repressive complex 2 (PRC2) is an evolutionarily conserved multimeric protein complex in both plants and animals. In contrast to animals, plants have evolved a range of different components of PRC2 and form diverse complexes that act in the control of key regulatory genes at many stages of development during the life cycle. A number of studies, particularly in the model species Arabidopsis thaliana, have highlighted the role of PRC2 and of epigenetic controls via parent-of-origin specific gene expression for endosperm development. However, recent research in cereal plants has revealed that although some components of PRC2 show evolutionary conservation with respect to parent-of-origin specific gene expression patterns, the identity of the imprinted genes encoding PRC2 components is not conserved. This disparity may reflect the facts that cereal plant genomes have undergone different patterns of duplication during evolution compared to A. thaliana and that the endosperm development program is not identical in monocots and eudicots. In this context, we focus this review on the expression of imprinted PRC2 genes and their roles in endosperm development in cereals.
Introduction
The endosperm of plant seeds is the most important tissue in plants with regard to human life, because of its importance as a major source of dietary calories. Recent studies have highlighted the role played by polycomb repressive complex 2 (PRC2) as one of the controlling mechanisms of normal endosperm development (Kohler and Makarevich, 2006; Pien and Grossniklaus, 2007; Holec and Berger, 2012). PRC2 is an evolutionarily conserved, high molecular weight complex that was originally identified in Drosophila mutants because of its regulation of body-segmentation during embryogenesis (Pirrotta, 1995). Subsequently, PRC2 was shown to have methyltransferase activity for Lys27 of histone H3 (H3K27; Simon and Kingston, 2009). In Arabidopsis thaliana, the complex represses expression of target genes through epigenetic modification of the chromatin, and also controls parent-of-origin specific expression of downstream target genes and of the PRC2 component itself in the endosperm (Gehring, 2013). While most of our understanding of the role of PRC2 comes from studies in the model species A. thaliana, recent studies in cereal plants, such as maize, barley and rice, have also provided important insights.
In contrast to animal species, such as Drosophila, the components of the PRC2 complexes of plant species show considerable variation. Genome evolution in plants involved the generation of multi-gene families and also whole genome duplications, such as in A. thaliana, maize and rice (Spillane et al., 2007; Dickinson et al., 2012). It has been hypothesized that whole genome-duplication may reduce evolutionary forces on duplicated genes, resulting in the accumulation of nucleotide substitutions in genes or gain-of-function changes in expression patterns (Ohno, 1970). Additionally, the relaxation of evolutionary constraints might allow transposon insertion at various sites in genes, leading to their silencing (Lynch and Conery, 2000; Rodin and Riggs, 2003). The latter has been postulated to act as a novel epigenetic control through the process of neofunctionalization (Dickinson et al., 2012; Yoshida and Kawabe, 2013). In this intriguing scenario, genes that show specific expression patterns in the endosperm may be associated with targeted genome-wide DNA demethylation in the central cell of the female gametophyte (Dickinson et al., 2012). Mechanisms for imprinted gene expression have been described in many reports (Gehring, 2013); however, questions regarding the biological relevance of genomic imprinting still remain to be answered. The increased understanding of the role of PRC2 in different plant species should be of value to addressing many of the unanswered questions.
PRC2 in Cereal Plants
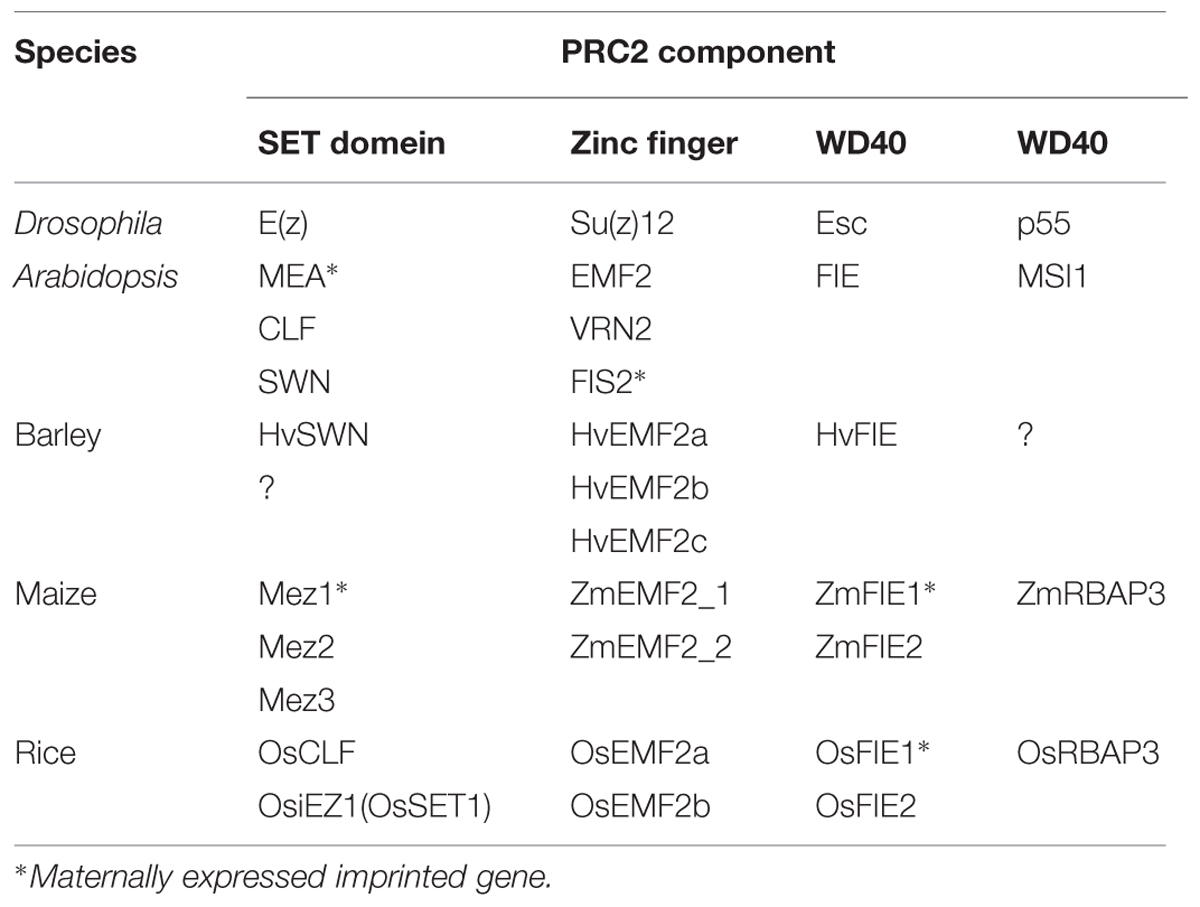
The PRC2 complex of animals has four major components: WD40 protein p55 (p55); Suppressor of Zeste 12 [Su(z)12]; Enhancer of Zeste [E(z)]; and extra sex combs (ESC; Schwartz and Pirrotta, 2013). These four components are conserved in A. thaliana and in cereal plants (Table 1). Although different combinations of the various subunits of PRC2 play distinct roles during development in A. thaliana, here we focus on the complex that determines endosperm fate. This complex has been termed FIS-class PRC2, and is encoded by the genes Multicopy Suppressors of IRA 1 (MSI1), Fertilization Independent Seed 2 (FIS2), MEDEA (MEA), and Fertilization Independent Endosperm (FIE), in A. thaliana (Kohler and Makarevich, 2006; Pien and Grossniklaus, 2007; Holec and Berger, 2012). To date, the characteristics of this complex have not been fully elucidated in cereal plants.
p55
The Drosophila p55 homolog in A. thaliana, MSI1, has been identified as a component of FIS-class PRC2 (Kohler et al., 2003; Guitton et al., 2004). MSI1 is a WD40 repeat protein; a loss-of-function mutant of MSI1 has been shown to display similar defects in cellularization and over-proliferation of endosperm as FIS-class PRC2 mutants. The MSI1 homologs of maize (Zea mays) and rice (Oryza sativa) have been identified (Table 1) but have yet to be studied in detail (Hennig et al., 2005).
Su(z)12
Three Su(z)12 homologs have been identified in the barley (Hordeum vulgare) genome, and are termed HvSu(z)12a, HvSu(z)12b, and HvSu(z)12c (Kapazoglou et al., 2010). All three genes are included in the Embryonic Flower 2 (EMF2) clade by phylogenetic analysis (Kapazoglou et al., 2010). HvSu(z)12b transcripts have been detected in all tested tissues and found to increase during seed development. Expression of HvSu(z)12c is limited to the young shoots and the developing seed; HvSu(z)12a has not been detected in any tested tissue (Kapazoglou et al., 2010). The rice genome has two homologs of Su(z)12, named OsEMF2a and OsEMF2b, that are expressed in a wide range of tissues (Luo et al., 2009). Interestingly, eudicots such as A. thaliana have a single copy of EMF2, while monocots have two or three EMF2-like genes. This suggests that the EMF2 gene family in the Poaceae (Gramineae) may have arisen from a recent duplication. No orthologs of VRN2 or FIS2 of A. thaliana have been identified in cereals (Luo et al., 2009).
E(z)
Analyses of the barley genome have identified one E(z) homolog, termed HvE(z), which is within the SWINGER (SWN) clade (Kapazoglou et al., 2010). Expression of HvE(z) occurs in both vegetative and reproductive tissues, and increases during seed development. The highest levels of HvE(z) expression have been found in young shoots (Kapazoglou et al., 2010). In maize, three E(z) homologs have been identified, namely, Mez1, Mez2, and Mez3 (Springer et al., 2002; Haun et al., 2007). The Mez1 sequence is similar to that of CLF, while Mez2 and Mez3 are more closely related to SWN. The Mez2 and Mez3 genes have high sequence identity, suggesting that they are duplicate genes formed during the paleotetraploid origin of maize (Springer et al., 2002). The three genes are widely expressed throughout the maize life cycle. Mez1 shows maternal-specific gene expression (imprinted) in the endosperm, but shows bi-allelic (non-imprinted) expression patterns in the embryo (Haun et al., 2007). Three splicing variants are transcribed from the Mez2 locus and show variations in their transcription among tissues (Springer et al., 2002). Analyses of sequence similarities indicate that the rice genome contains two homologs of E(z), namely, OsiEZ1(OsSET1) and OsCLF (Thakur et al., 2003; Luo et al., 2009). These two rice genes are widely expressed in a range of tissues (Luo et al., 2009). Homologs of E(z) in cereal plants fall into the CLF and SWN clades. The SWN clade is specific to flowering plants, while the CLF clade also contains homologs from spikemosses (Selaginella spp.; Luo et al., 2009). The maize homologs of E(z) are more diverse than those of other cereal plants; it seems that the multiplication of homologous genes provided diversity of PRC2 functions in maize. The MEA protein is a core component of FIS-class PRC2, which is related to seed development in A. thaliana. However, no MEA-like gene has been identified in cereals (Luo et al., 2009).
ESC
Barley genome sequencing identified a single homolog of ESC (Kapazoglou et al., 2010); however, two duplicated genes for FIE-like proteins are present in both maize and rice genomes (Springer et al., 2002; Luo et al., 2009). In barley, HvFIE is widely expressed in vegetative and reproductive tissues. Similarly, ZmFIE2 is expressed in a range of tissues in maize (Springer et al., 2002; Danilevskaya et al., 2003). These various genes are therefore the likely functional orthologs in cereals of FIE in A. thaliana. ZmFIE1 in maize and OsFIE1 in rice are predominantly expressed in the endosperm, and both display maternal-specific expression patterns (Danilevskaya et al., 2003; Gutierrez-Marcos et al., 2006). In maize, analysis using methylation sensitive restriction enzymes and PCR has shown that genome-wide DNA hypomethylation of the maternally derived genome occurs in the endosperm (Lauria et al., 2004). Related to this finding, differentially methylated regions (DMRs) have been identified that involve hypomethylation of the maternal allele of the ZmFIE1 and ZmFIE2 genes (Gutierrez-Marcos et al., 2006). The promoter region of ZmFIE1 is demethylated in the central cell but not in the sperm cells; this asymmetric pattern of DNA methylation is inherited to the endosperm, where the maternally derived ZmFIE1 is expressed while the paternally derived allele is silenced. The 5′ region of ZmFIE2 is hypomethylated in many tissues, but subjected to de novo DNA methylation only on the paternally derived allele in the endosperm after fertilization. These DMRs may be a mechanism for maternal specific gene expression during early endosperm development (Gutierrez-Marcos et al., 2006). Similarly, transcription of the paternal OsFIE1 allele during early endosperm development is likely silenced by DNA methylation (Luo et al., 2009; Ishikawa et al., 2011; Zhang et al., 2012). The sequences and expression patterns of maize ZmFIE1 and rice OsFIE1 are very similar suggesting an orthologous relationship between these genes. In maize, ZmFIE1 and ZmFIE2 are located on different chromosomes (Springer et al., 2002), whereas rice OsFIE1 and OsFIE2 are located in the same genomic region on chromosome 8. Phylogenetic analysis of these maize and rice genes suggest that the two maize genomic regions arose from reciprocal deletion of one of the ancestral paralogs during maize genome evolution (Swigonova et al., 2004). The fact that rice OsFIE1 and OsFIE2 are closely positioned on the same chromosome suggests they arose through an intraspecies gene duplication event (Luo et al., 2009).
Roles for PRC2 Complexes in Cereal Endosperm
In a comparison of gene expression patterns in two barley cultivars that have seeds of different sizes, differential expression of HvFIE and HvE(z) was shown to occur during seed development (Kapazoglou et al., 2010). HvFIE expression was found to increase immediately after fertilization in both cultivars, and then to decline in the cultivar producing larger seeds, but to increase in the cultivar with smaller seeds. The expression patterns of HvFIE are consistent with the predicted role of PRC2 in cereal plants, namely, the repression of endosperm development. HvFIE and HvE(z) expression can also be induced by the plant hormone abscisic acid (ABA), which is known to be involved in seed maturation, dormancy, and germination (Kapazoglou et al., 2010). These findings suggest that genes for PRC2 components can act at both earlier and later stages of endosperm development in barley; this may reflect the developmental program of endosperm of cereal species. Although the syncytial phase during early endosperm development is conserved in A. thaliana and cereal species, embryonic growth in A. thaliana later results in the consumption of the endosperm; by contrast, the endosperm persists in cereals (Sabelli and Larkins, 2009; Dante et al., 2014).
In A. thaliana, the imprinted genes MEA and FIS2 encode PRC2 components and are involved in endosperm development through repression of the AGL62 gene expression that controls the timing of cellularization (Kang et al., 2008; Hehenberger et al., 2012). In contrast to A. thaliana, MEA and FIS2 orthologs have not been identified in barley, maize, or rice genomes. In rice, with the exception of OsFIE1, genes encoding PRC2 components are widely expressed in a range of tissues. OsFIE1 shows specific expression in the endosperm and is the only imprinted PRC2 gene in rice endosperm (Luo et al., 2009); the gene is expected to be involved in multiple processes during endosperm development including cellularization. Plants homozygous for the Osfie1 mutation do not display an obvious endosperm phenotype compared to wild type plants (Luo et al., 2009); by contrast, RNAi transgenic plant lines showed autonomous endosperm development (Li et al., 2014). This outcome may be due to off-target effects of the OsFIE2 RNAi construct which silenced both OsFIE1 and OsFIE2 in the endosperm of the transgenic rice (Li et al., 2014). By contrast, the specific down-regulation of OsFIE2 by RNAi results in the production of small seeds, which contain shrunken and defective endosperm and a relatively large embryo (Nallamilli et al., 2013). Although a sporophytic effect of the knock-down mutation, due to the dominant nature of RNAi construct, cannot be discounted in the latter experiment, this result suggests OsFIE2 has a positive regulatory role in either early or late development of rice endosperm, in contrast to the role of FIS-class PRC2 in the endosperm of A. thaliana. It should be possible to more clearly determine the role of OsFIE2 through use of the appropriate mutant alleles in combination with TALLEN or CRISPER/Cas technology (Kim and Kim, 2014). Such analyses would elucidate the role of OsFIE2 in endosperm development, especially in relation to the timing of cellularization. There is evidence from interspecific and interploidy crosses in rice that the timing of cellularization and the eventual size of the endosperm are related (Ishikawa et al., 2011; Sekine et al., 2013). Therefore, investigation of cellularization in PRC2 mutants will be an essential approach to understanding the action of PRC2 in cereal endosperm.
Recently, an epigenetic allele of Epi-df was identified; this allele is a gain-of-function variant that likely resulted from hypomethylation of the 5′ region of OsFIE1 without any change in nucleotide sequence (Zhang et al., 2012). On the Epi-df mutant background, OsFIE1 is ectopically expressed in vegetative tissues and the normally silent paternally derived allele is active in the endosperm (Zhang et al., 2012). The Epi-df plants show dwarfism and floral organ defects in a dominant fashion; the latter prevented investigation of the endosperm phenotype. By contrast, a recent study showed that expression of OsFIE1 is correlated with the timing of cellularization (Folsom et al., 2014). Under moderately high temperature conditions, OsFIE1 expression increases, and this elevated level of expression is correlated with precocious endosperm cellularization (Folsom et al., 2014). Similarly, overexpression of OsFIE1 causes decreased seed sizes and weights (Folsom et al., 2014). This is in contrast with the outcome of OsFIE2 overexpression, which does not result in phenotypic changes in plants (Nallamilli et al., 2013). Overall, these findings suggest the possibility that OsFIE1 and OsFIE2 may have non-equivalent roles in endosperm development (Figure 1). Further analyses will be required to clarify precisely the roles of PRC2 in the cereal endosperm development.
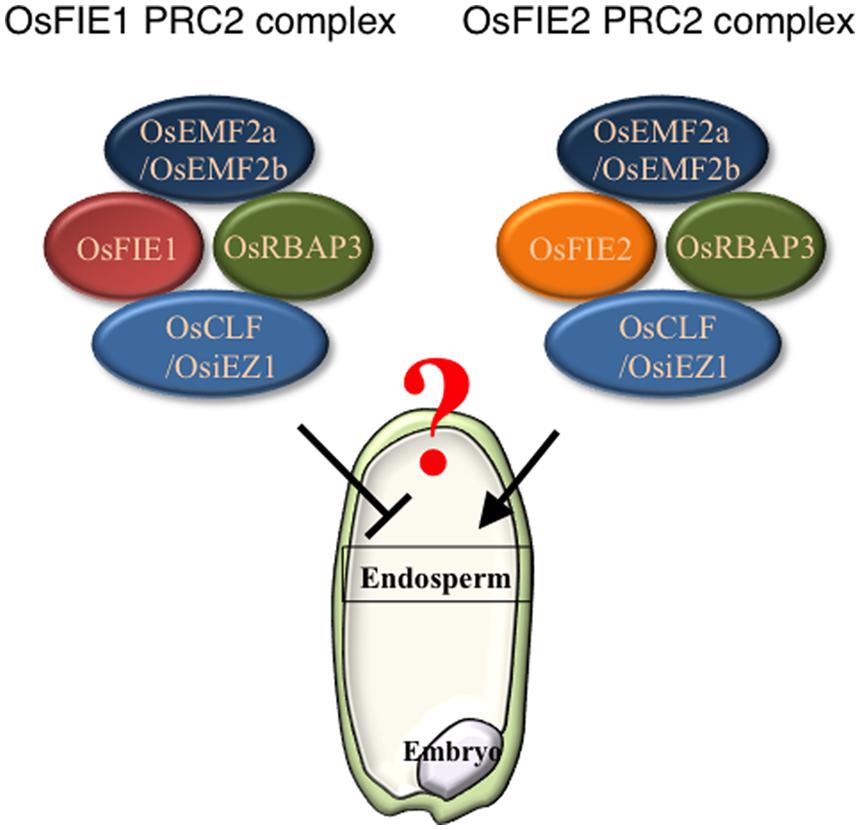
FIGURE 1. Polycomb repressive complex 2 (PRC2) components of OsFIE1 and OsFIE2 may have distinct roles in rice endosperm. Based on recent findings, PRC2 complexes that contain OsFIE1 and OsFIE2 are likely to have distinct roles. In the endosperm, the OsFIE1 protein is produced from the maternally derived allele and contributes to the FIE1-containing PRC2 (left), By contrast, OsFIE2 protein derived from both maternal and paternal alleles is used to form FIE2-containing PRC2 (right).
Conclusion
The data generated by cereal genome sequencing initiatives have enabled the identification of PRC2 genes in crop plant species. Detailed analyses of the expression of these genes have revealed remarkable differences in their behavior compared to orthologs in A. thaliana. Endosperm specific variants of the Su(Z)12 homolog and E(z) homolog have been found, namely, MEA and FIS2; however, no variants of the ESC homolog are known in A. thaliana. By contrast, two ESC homologs FIE1 and FIE2 are present in maize and rice genomes. Although FIE is not consistently imprinted in A. thaliana (Yadegari et al., 2000), its homologs in maize and rice show maternal specific expression (Gutierrez-Marcos et al., 2006; Luo et al., 2009). In general, ESC and its homologs are WD40 repeat scaffolding proteins and do not seem to have any enzymatic activity. However, their animal counterparts have been shown to have binding activity for the N-terminal histone tail of H3 and to cause allosteric effects on the histone methyltransferase activity of EZH2; binding to chromatin residues associated with a repressive state of gene expression, such as H3K9me3, induces histone methyltransferase activity, while binding to chromatin residues associated with active transcription reduces its activity. Therefore, the protein–protein interactions of each PRC2 component are important determinants of the activity of the PRC2 complex. Further study of cereal PRC2 complexes will undoubtedly provide greater insights into their roles in endosperm development.
Acknowledgments
Funding on this topic is provided by Grant-in-Aid for Scientific Research on Innovative Areas (23113001 and 23113003 to TK) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by a fellowship from the Japan Society for the Promotion of Science to KT.
References
Danilevskaya, O. N., Hermon, P., Hantke, S., Muszynski, M. G., Kollipara, K., and Ananiev, E. V. (2003). Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15, 425–438. doi: 10.1105/tpc.006759
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dante, R. A., Larkins, B. A., and Sabelli, P. A. (2014). Cell cycle control and seed development. Front. Plant Sci. 5:493. doi: 10.3389/fpls.2014.00493
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dickinson, H., Costa, L., and Gutierrez-Marcos, J. (2012). Epigenetic neofunctionalisation and regulatory gene evolution in grasses. Trends Plant Sci. 17, 389–394. doi: 10.1016/j.tplants.2012.04.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Folsom, J. J., Begcy, K., Hao, X., Wang, D., and Walia, H. (2014). Rice fertilization-independent endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165, 238–248. doi: 10.1104/pp.113.232413
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gehring, M. (2013). Genomic imprinting: insights from plants. Annu. Rev. Genet. 47, 187–208. doi: 10.1146/annurev-genet-110711-155527
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guitton, A. E., Page, D. R., Chambrier, P., Lionnet, C., Faure, J. E., Grossniklaus, U.,et al. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131, 2971–2981. doi: 10.1242/dev.01168
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gutierrez-Marcos, J. F., Costa, L. M., Dal Pra, M., Scholten, S., Kranz, E., Perez, P.,et al. (2006). Epigenetic asymmetry of imprinted genes in plant gametes. Nat. Genet. 38, 876–878. doi: 10.1038/ng1828
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Haun, W. J., Laoueille-Duprat, S., O’Connell, M. J., Spillane, C., Grossniklaus, U., Phillips, A. R.,et al. (2007). Genomic imprinting, methylation and molecular evolution of maize enhancer of zeste (Mez) homologs. Plant J. 49, 325–337. doi: 10.1111/j.1365-313X.2006.02965.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hehenberger, E., Kradolfer, D., and Kohler, C. (2012). Endosperm cellularization defines an important developmental transition for embryo development. Development 139, 2031–2039. doi: 10.1242/dev.077057
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hennig, L., Bouveret, R., and Gruissem, W. (2005). MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 15, 295–302. doi: 10.1016/j.tcb.2005.04.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Holec, S., and Berger, F. (2012). Polycomb group complexes mediate developmental transitions in plants. Plant Physiol. 158, 35–43. doi: 10.1104/pp.111.186445
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ishikawa, R., Ohnishi, T., Kinoshita, Y., Eiguchi, M., Kurata, N., and Kinoshita, T. (2011). Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J. 65, 798–806. doi: 10.1111/j.1365-313X.2010.04466.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kang, I. H., Steffen, J. G., Portereiko, M. F., Lloyd, A., and Drews, G. N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20, 635–647. doi: 10.1105/tpc.107.055137
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kapazoglou, A., Tondelli, A., Papaefthimiou, D., Ampatzidou, H., Francia, E., Stanca, M. A.,et al. (2010). Epigenetic chromatin modifiers in barley: IV. The study of barley polycomb group (PcG) genes during seed development and in response to external ABA. BMC Plant Biol. 10:73. doi: 10.1186/1471-2229-10-73
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, H., and Kim, J. S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334. doi: 10.1038/nrg3686
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kohler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE polycomb group complex and required for seed development. EMBO J. 22, 4804–4814. doi: 10.1093/emboj/cdg444
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kohler, C., and Makarevich, G. (2006). Epigenetic mechanisms governing seed development in plants. EMBO Rep. 7, 1223–1227. doi: 10.1038/sj.embor.7400854
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lauria, M., Rupe, M., Guo, M., Kranz, E., Pirona, R., Viotti, A.,et al. (2004). Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell 16, 510–522. doi: 10.1105/tpc.017780
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, S., Zhou, B., Peng, X., Kuang, Q., Huang, X., Yao, J.,et al. (2014). OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol. 201, 66–79. doi: 10.1111/nph.12472
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Luo, M., Platten, D., Chaudhury, A., Peacock, W. J., and Dennis, E. S. (2009). Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant 2, 711–723. doi: 10.1093/mp/ssp036
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Nallamilli, B. R., Zhang, J., Mujahid, H., Malone, B. M., Bridges, S. M., and Peng, Z. (2013). Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet. 9:e1003322. doi: 10.1371/journal.pgen.1003322
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pien, S., and Grossniklaus, U. (2007). Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta 1769, 375–382. doi: 10.1016/j.bbaexp.2007.01.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pirrotta, V. (1995). Chromatin complexes regulating gene expression in Drosophila. Curr. Opin. Genet. Dev. 5, 466–472. doi: 10.1016/0959-437X(95)90050-Q
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rodin, S. N., and Riggs, A. D. (2003). Epigenetic silencing may aid evolution by gene duplication. J. Mol. Evol. 56, 718–729. doi: 10.1007/s00239-002-2446-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sabelli, P. A., and Larkins, B. A. (2009). The development of endosperm in grasses. Plant Physiol. 149, 14–26. doi: 10.1104/pp.108.129437
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwartz, Y. B., and Pirrotta, V. (2013). A new world of Polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 14, 853–864. doi: 10.1038/nrg3603
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sekine, D., Ohnishi, T., Furuumi, H., Ono, A., Yamada, T., Kurata, N.,et al. (2013). Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J. 76, 792–799. doi: 10.1111/tpj.12333
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Simon, J. A., and Kingston, R. E. (2009). Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708. doi: 10.1038/nrm2763
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spillane, C., Schmid, K. J., Laoueille-Duprat, S., Pien, S., Escobar-Restrepo, J. M., Baroux, C.,et al. (2007). Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448, 349–352. doi: 10.1038/nature05984
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Springer, N. M., Danilevskaya, O. N., Hermon, P., Helentjaris, T. G., Phillips, R. L., Kaeppler, H. F.,et al. (2002). Sequence relationships, conserved domains, and expression patterns for maize homologs of the polycomb group genes E(z), esc, and E(Pc). Plant Physiol. 128, 1332–1345. doi: 10.1104/pp.010742
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Swigonova, Z., Lai, J., Ma, J., Ramakrishna, W., Llaca, V., Bennetzen, J. L.,et al. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14, 1916–1923. doi: 10.1101/gr.2332504
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thakur, J. K., Malik, M. R., Bhatt, V., Reddy, M. K., Sopory, S. K., Tyagi, A. K.,et al. (2003). A POLYCOMB group gene of rice (Oryza sativa L. subspecies indica), OsiEZ1, codes for a nuclear-localized protein expressed preferentially in young seedlings and during reproductive development. Gene 314, 1–13. doi: 10.1016/s0378-1119(03)00723-6
Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y.,et al. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12, 2367–2382. doi: 10.1105/tpc.12.12.2367
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yoshida, T., and Kawabe, A. (2013). Importance of gene duplication in the evolution of genomic imprinting revealed by molecular evolutionary analysis of the type I MADS-box gene family in Arabidopsis species. PLoS ONE 8:e73588. doi: 10.1371/journal.pone.0073588
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, L., Cheng, Z., Qin, R., Qiu, Y., Wang, J. L., Cui, X.,et al. (2012). Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24, 4407–4421. doi: 10.1105/tpc.112.102269
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: endosperm, epigenetics, cereal plants, polycomb, imprinting
citation: Tonosaki K and Kinoshita T (2015) Possible roles for polycomb repressive complex 2 in cereal endosperm. Front. Plant Sci. 6:144. doi: 10.3389/fpls.2015.00144
Received: 31 October 2014; Accepted: 23 February 2015;
Published online: 12 March 2015.
Edited by:
Ravishankar Palanivelu, University of Arizona, USACopyright © 2015 Tonosaki and Kinoshita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaoru Tonosaki and Tetsu Kinoshita, Kihara Institute for Biological Research, Yokohama City University, 641-12 Maioka, Totsuka, Yokohama, Kanagawa 244-0813, Japan tonosaki@yokohama-cu.ac.jp; tkinoshi@yokohama-cu.ac.jp
 Kaoru Tonosaki
Kaoru Tonosaki Tetsu Kinoshita
Tetsu Kinoshita