- Food Biotechnology Division, National Food Research Institute, National Agriculture and Food Research Organization, Tsukuba, Japan
To shed unfertilized flowers or ripe fruits, many plant species develop a pedicel abscission zone (AZ), a specialized tissue that develops between the organ and the main body of the plant. Regulation of pedicel abscission is an important agricultural concern because pre-harvest abscission can reduce yields of fruit or grain crops, such as apples, rice, wheat, etc. Tomato has been studied as a model system for abscission, as tomato plants develop a distinct AZ at the midpoint of the pedicel and several tomato mutants, such as jointless, have pedicels that lack an AZ. This mini-review focuses on recent advances in research on the mechanisms regulating tomato pedicel abscission. Molecular genetic studies revealed that three MADS-box transcription factors interactively play a central role in pedicel AZ development. Transcriptome analyses identified activities involved in abscission and also found novel transcription factors that may regulate AZ activities. Another study identified transcription factors mediating abscission pathways from induction signals to activation of cell wall hydrolysis. These recent findings in tomato will enable significant advances in understanding the regulation of abscission in other key agronomic species.
Introduction
Similar to leaves, flowers and young fruits shed when the organs become unneeded or as a result of environmental stresses; for example, failure of pollination results in abscission of the unfertilized flowers. Also, in “June drop” in apple (Malus × domestica), some young fruitlets abscise at an early developmental stage (Bangerth, 2000). Just after flowering, apple trees often bear more fruits than they can support to maturity; thus the plants shed some fruits to limit fruit set. In addition, when fruits on a plant ripen, abscission of the fruits helps to disperse the seeds. The pedicel, a stem, or a stalk structure, connects at the base of the flower or fruit, attaching the organ to the plant body. In many species, an abscission zone (AZ) forms in the pedicel to enable regulated separation of the fruit or flower from the main plant body (Sexton and Roberts, 1982; Tabuchi et al., 2001; Roberts et al., 2002).
In agricultural applications, pedicel abscission is a critical trait directly affecting crop yields; thus the regulation of abscission has been important since ancient times. During the domestication of cereal crops such as rice (Oryza sativa), maize (Zea mays), or wheat (Triticum aestivum), early farmers selected for plants with reduced abscission (Doebley et al., 2006; Li et al., 2006; Lin et al., 2012). Cultivars carrying the trait conferring resistance to grain abscission retain the grain on the stalk, rather than dropping it on the ground.
Regulation of abscission also remains an important trait in modern breeding programs. Research in tomato has identified several mutations that block formation of the pedicel AZ, producing a “jointless” phenotype (Butler, 1936; Rick, 1967; Roberts et al., 2002), which has proven useful in tomato cultivars grown for industrial processing of tomato puree or juice. In these cultivars, fruits can be mechanically harvested without the pedicel and sepals because, in the absence of a breaking point in the pedicel AZ, the fruit detaches at the next breaking point, the calyx AZ at the proximal end of the fruit, and the green organs remains on the plant. This reduces the labor and time required to remove the pedicel and sepals during harvesting (Zahara and Scheuerman, 1988).
Fruit abscission is also an important trait for tree fruit production. In apple, abscission affects the fruit yield at several stages (Celton et al., 2014). The trees shed young fruitlets as “June drop,” as described above. Thinning of young fruits is an important practice to control fruit load and chemicals that induce partial abscission of fruit have been developed to reduce the labor required for thinning (Bangerth, 2000). After the early developmental stages, fruits at the expanding stage remain stably attached to the plant via the pedicels but the attachment gradually loosens during the initiation of ripening. However, severe weather can cause fruit to drop prematurely. For example, in Japan, the autumn fruit harvest coincides with the typhoon season and large numbers of fruits just before harvest time drop by the strong winds, which break a boundary between the plant body and pedicel, where the AZ is localized, resulting in severe damage to production (Yamamoto et al., 2012; Fujisawa et al., 2015).
Pedicel AZ Structure and Development in Tomato
The AZ, a specialized tissue for organ abscission, forms at a predetermined site on the organ that will abscise. Anatomical studies revealed that an AZ includes several layers of small, densely cytoplasmic cells that forms at an early stage of pedicel development and proliferation of the cells is observed during fruit development (Addicott, 1982; Sexton and Roberts, 1982; Tabuchi and Arai, 2000; Patterson, 2001). These properties suggest that these cells may be arrested in an undifferentiated state (van Nocker, 2009). In tomato, initial differentiation of the pedicel AZ occurs when the flower sepal differentiates from the primordium. AZ cells first form in the inner region of the young pedicel and then the AZ structure gradually extends to the outer tissues (Tabuchi, 1999; Liu et al., 2014). The innermost cell layer has a critical role in AZ development, as examination of chimeric plants consisting of layers of jointless mutant cells and wild-type cells showed that the genotype of the inner layer (L3) determines cell fates of overlaying layers L1 (outer layer) and L2 (middle layer) and whether they differentiate into AZ tissue (Szymkowiak and Irish, 1999). At the flower anthesis stage, pedicel AZ tissues have developed into six to eight cell layers that extend across the pedicel. The AZ cells around the vascular tissue and cortex can still divide (Tabuchi and Arai, 2000), suggesting that the AZs in flower pedicels maintain meristem-like activity.
Normally, pedicel abscission is induced if flower fertilization fails or the fruit ripens fully. Pedicel abscission can also be induced artificially by flower removal (Roberts et al., 1984; Meir et al., 2010; Nakano et al., 2013) or ethylene treatment (Roberts et al., 1984; Wang et al., 2013); several studies have used these treatments to analyze abscission. Roberts et al. (1984) observed that cell separation for abscission first took place at the cortex within the distal side of the AZs if the pedicel was treated with ethylene. Also, Tabuchi et al. (2001) reported that pedicel abscission occurred first at the epidermis of the AZ if abscission was induced by emasculation. Dissolution of the middle lamella commonly occurred in response to either treatment, and cell wall hydrolysis enzymes and remodeling proteins, such as polygalacturonase (tomato abscission-related polygalacturonase; TAPG), endo-β-1,4-glucanase (also referred as cellulase; Cel), xyloglucan endotransglucosylase/hydrolase (XTH), and expansin, play a critical role in abscission (Roberts et al., 2002; Tucker et al., 2007; Cai and Lashbrook, 2008). The abscission-inducing treatment also caused enlargement of the epidermal cells in tomato (Tabuchi et al., 2001). Cell enlargement during abscission also occurs in other plant systems such as bean leaves (McManus et al., 1998) and Arabidopsis flower organs (Shi et al., 2011), and this enlargement may confer mechanical force to facilitate abscission (Shi et al., 2011). The abscised surface of the proximal side formed thickened and lignified cell walls, implying that a protective layer forms to prevent pathogen invasion (Tabuchi et al., 2001).
MADS-Box Family Transcription Factors Regulate Pedicel AZ Development in Tomato
The most important breakthrough in abscission research was the identification of the jointless (j) mutant locus (Mao et al., 2000), which causes the plant to fail to develop pedicel AZs. The j locus was isolated by map-based cloning and the wild-type gene encodes a MADS-box transcription factor. In the same year, independent work on an early-flowering mutant identified Arabidopsis SHORT VEGETATIVE PHASE (SVP), which encodes a MADS-box protein with high similarity to J (Hartmann et al., 2000). Although J and SVP have high amino acid sequence similarity, they have distinct functions, with SVP acting as a repressor of the floral transition. Moreover, Arabidopsis plants do not shed fruits from the pedicels. Also, in several tree fruit species, SVP homologs may play roles in bud dormancy (Li et al., 2009; Yamane et al., 2011; Wu et al., 2012).
Further studies identified two additional tomato MADS-box genes regulating pedicel AZ development, Macrocalyx (MC) and SlMBP21. MC was originally identified in a study of rin (ripening inhibitor), which regulates fruit ripening. The rin mutation produces non-ripening fruits with large sepals (Vrebalov et al., 2002). The cloning study identified two nearby genes, RIN and MC, both of which encode MADS-box genes. RIN regulates ripening and MC regulates sepal size (Vrebalov et al., 2002). The rin mutation also shows a weak effect on pedicel AZ development and antisense-mediated knockdown revealed that MC also plays a role in pedicel AZ development (Nakano et al., 2012). A comprehensive interaction study of tomato MADS-box proteins using yeast two-hybrid system initially identified SlMBP21 as a MADS-box protein interacting with J (Leseberg et al., 2008). A gene knockdown study revealed that SlMBP21 also participates in pedicel AZ development (Liu et al., 2014). These studies showed physical interactions among J, MC, and SlMBP21, suggesting that these three MADS-box proteins form a complex. At an early stage of AZ initiation, these MADS-box genes are co-expressed in vascular tissue derived from the L3 layer required for AZ development (Szymkowiak and Irish, 1999; Liu et al., 2014). In Arabidopsis, the J homolog SVP and the MC homolog AP1 likely form a dimer as an active form to regulate floral identity (Gregis et al., 2009). SEP family proteins, including SlMBP21, play an important role in forming multimers of MADS-box proteins by acting as a glue (Immink et al., 2009). Thus, multimer formation of J, MC, and SlMBP21 may be a conserved activity among plant species, although the targets of biological regulation by homologous MADS-box proteins may differ in each plant.
Is the regulation of pedicel AZ development by the MADS-box transcription factors conserved in other plant species, or is it specific to tomato? Ectopic expression of the apple SVP family MADS-box gene MdJb in a tomato j mutant restored the formation of pedicel AZ structure in the j mutant (Nakano et al., 2015). The restored AZs showed abscission-associated expression of cell wall hydrolysis enzyme genes and complete pedicel abscission, as in wild-type tomato plants. The results suggest that the regulation of pedicel AZ development in plants by the MADS-box transcription factors may be conserved, but other plant systems remain to be examined. Further investigation will be required to understand the mechanism of AZ development in other plant species.
Genes Expressed in Tomato Pedicel AZs
Before abscission, pedicel AZs attach the flowers firmly to the plant body, but when the AZ cells perceive an abscission-stimulating signal, the adhesion immediately starts to loosen. During abscission, the gene expression pattern in the AZ changes drastically; genes for cell wall hydrolysis enzymes, such TAPG and Cel, and for factors regulating programmed cell-death increase intensely and specifically at the AZ (Roberts et al., 2002; Cai and Lashbrook, 2008; Meir et al., 2010; Bar-Dror et al., 2011). In addition to these genes, a transcriptome study during initiation of abscission found many genes possibly responsible for regulatory roles in abscission, such as genes for transcription factor families of ARF, Aux/IAA, KNOX, HAT, bHLH, AP2, NAC, AGL, and WRKY, genes for components of signal transduction pathways such as a LRR-RLK and a Ser/Thr protein kinase, and a gene for a component of a RNA-induced silencing complex, AGO1 (Meir et al., 2010). The analyses also provided specific expression patterns of phytohormone-related genes, which confirmed and improved a conventional abscission-inducing model with the substantial evidence (Patterson, 2001; Roberts et al., 2002; Meir et al., 2010); a decrease in auxin provides the first signal for abscission, and reactions to the decrease in auxin, including down-regulation of genes induced by auxin (such as Aux/IAA genes and other transcription factor genes) and up-regulation of genes repressed by auxin, confers ethylene-sensitivity and abscission competence to the AZ. Then increased ethylene production, due to the up-regulation of genes for ethylene biosynthesis (such as ACS, encoding 1-aminocyclopropane-1-carboxylate (ACC) synthase), leads to AZ-specific up-regulation of the genes for abscission, such as genes encoding cell wall-modifying proteins and pathogenesis-related proteins, development of a protective layer on the surface of the abscised tissue, and so on.
Before the onset of pedicel abscission, the plant maintains firm cell-to-cell adhesion at the AZs to allow continuous growth from the flower to the mature fruit. To maintain the adhesion and the competence to react to an abscission-inducing signal, the AZ cells might undergo specific regulation. A transcriptome analysis comparing gene expression between pedicel AZs and the flanking pedicel regions at anthesis (Nakano et al., 2013) identified about 90 genes specifically expressed in AZ cells, including genes for transcription factors, phytohormone-related proteins, cell wall modification enzymes, lipid metabolism, and others. Most interestingly, the AZ-specific gene set included transcription factor genes that encode key regulators of meristem-associated functions, including a tomato homolog of WUSCHEL (LeWUS), GOBLET (GOB), LATERAL SUPPRESSOR (Ls), and Blind (Bl). WUS expressed in Arabidopsis shoot apex is required for maintenance of stem cells in an undifferentiated state (Mayer et al., 1998). GOB is a member of the NAC family transcription factor genes and its Arabidopsis homolog genes, CUP-SHAPED COTYLEDONs (CUCs), are involved in shoot meristem formation and specification of organ boundaries (Aida et al., 1997; Blein et al., 2008; Berger et al., 2009). Ls and its Arabidopsis homolog are known to regulate axillary meristem initiation (Schumacher et al., 1999; Greb et al., 2003). Bl and its Arabidopsis homolog of REGULATOR OF AXILLARY MERISTEM (RAX) also involved in axillary meristem formation (Schmitz et al., 2002; Keller et al., 2006). These transcription factors were suppressed in the pedicels of AZ-deficient plants, the j mutant, and MC- and SlMBP21-suppressed plants (Nakano et al., 2012; Liu et al., 2014). Also, LeWUS, GOB, and Ls were down-regulated immediately after an abscission-inducing treatment while Bl was up-regulated (Nakano et al., 2013). These characteristic expression patterns suggest that these transcription factor genes play important roles in the AZs. Similar to meristems, AZs include small cells that likely exist in an undifferentiated state (van Nocker, 2009); thus, these transcription factors may regulate the maintenance of these undifferentiated cells in both tissues. In rice flower pedicels, homologs of Bl, GOB, and Ls are expressed specifically in the AZs, indicating that the mechanism of regulation by these transcription factors may be conserved in monocots and dicots (Nakano and Ito, 2013).
A recent study showed an intriguing result on the undifferentiated properties of the AZ cells. Constitutive expression of a miRNA-resistant form of a tomato homolog of the REVOLUTA gene, encoding a Class III homeodomain-leucine zipper (HD-ZIP III) transcription factor, caused the transgenic plants to produce ectopic flowers from the pedicel AZs (Hu et al., 2014). In the AZs at anthesis, the transgenic plants expressed Bl and GOB at significantly higher levels than the wild-type plants. The results imply that pedicel AZs include undifferentiated cells that have the potential to develop into flower primordia, and the transcription factors expressed in the AZs may coordinately regulate the maintenance and proliferation of the undifferentiated AZ cells.
These transcriptome analyses identified genes specifically expressed in tomato pedicel AZs, and of them, two transcription factor genes, SlERF52 and KD1, were further analyzed for their effect on AZ functions, as described in the next section.
Transcription Factors Connecting Abscission-Inducing Signals and Abscission Processes
Of the transcription factor genes expressed in tomato pedicel AZs, the ERF family transcription factor gene SlERF52 was further investigated by RNAi-mediated knockdown assays (Nakano et al., 2014). The SlERF52-knockdown plants developed pedicel AZ structures similarly to wild-type plants; however, the responses to an abscission-inducing treatment differed in the knockdown and wild-type plants. In wild-type plants, removing the anthesis-stage flower from the pedicel usually induces pedicel abscission within 2 days; in the SlERF52-knockdown plants, pedicel abscission took significantly longer. The knockdown disturbed the abscission-specific up-regulation of the genes for hydrolytic enzymes, such as TAPG and Cel, indicating that the suppression of the hydrolytic enzymes caused the delay in abscission. The result suggests that the SlERF52 ERF transcription factor functions as a component of a signaling pathway for pedicel abscission and plays a key role in the induction of expression of genes involved in cell wall hydrolysis. The induction of the hydrolytic enzyme genes during abscission, however, may require an additional factor to activate SlERF52. The expression levels of SlERF52 did not differ before and after the abscission-inducing treatment; thus, the expression level of SlERF52 cannot explain the activation of abscission. On the other hand, before the onset of abscission, the AZ-specific expression of LeWUS, Ls, and GOB requires SlERF52, implying that SlERF52 acts before and during abscission, but the transcriptional targets of SlERF52 apparently differ in the two stages. Explaining the functional switching of SlERF52 may require additional factors, such as stage-specific co-factors of SlERF52 or repressor proteins at SlERF52-binding sites. The identification of the switching mechanism will provide further insights into the regulation of pedicel abscission.
Another transcription factor gene expressed specifically in the tomato pedicel AZ, KD1, a KNOTTED1-LIKE HOMEOBOX (KNOX) family gene, was investigated for function in pedicel and petiole AZs (Ma et al., 2015). Down-regulation of KD1 significantly delayed abscission of the pedicel and even petiole and up-regulation of KD1 promoted abscission. The investigation suggested that KD1 controls abscission by regulating genes that modulate auxin levels (Ma et al., 2015). Identification of the regulator of abscission in both pedicels and petioles provides substantial evidence that abscission in these tissues involves identical regulatory mechanisms, in contrast to their distinct mechanisms regulating AZ development (Szymkowiak and Irish, 1999). Further investigation of the relationship between SlERF52 and KD1 may reveal their activities in abscission processes more clearly.
Conclusion
These recent advances in our understanding of the regulation of pedicel abscission revealed key factors involved in AZ development and signal transduction in the initiation of abscission. Figure 1 shows a current model of development of AZs and induction of abscission in tomato pedicels. The MADS-box transcription factor complex regulates pedicel AZ development. The developed AZ contains undifferentiated cells, probably maintained by a mechanism similar to that found in meristems. The signals of decreased auxin and increased ethylene induce abscission and SlERF52 and KD1 possibly connect the phytohormone signaling pathway and abscission processes. A remaining mystery is another tomato jointless mutation, jointless-2 (j-2), which is the best used mutation in practical breeding programs of processing tomatoes. A candidate gene for the mutation was reported but it has not been fully identified yet (Yang et al., 2005).
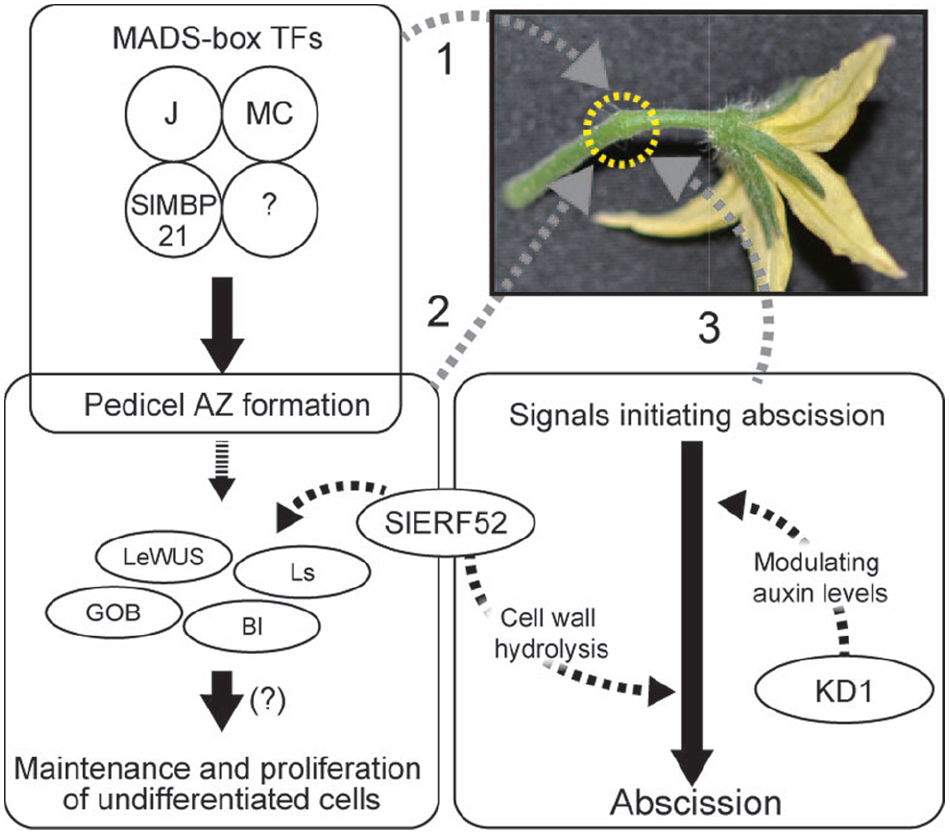
Figure 1. Regulation of pedicel AZ functions. 1. MADS-box proteins form tetramers and regulate pedicel AZ formation. 2. Undifferentiated cells are maintained in the pedicel AZ. AZ-specific transcription factors may be involved in the maintenance and proliferation of the undifferentiated cells. Expression of these transcription factor genes requires the activity of SlERF52. 3. In response to abscission-initiating signals, KD1 and SlERF52 activate abscission by modulating auxin levels and up-regulating genes encoding cell wall hydrolysis enzymes, respectively.
The outline of the current model constructed in tomato will facilitate further detailed studies on pedicel functions in tomato and other plants and these studies will provide new applications for fruit crops to improve their productivities.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by Science Technique Research Promotion program for Agriculture, Forestry, Fisheries and Food industry (grant number 25005A).
References
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Bangerth, F. (2000). Abscission and thinning of young fruit and their regulation by plant hormones and bioregulators. Plant Growth Regul. 31, 43–59. doi: 10.1023/A:1006398513703
Bar-Dror, T., Dermastia, M., Kladnik, A., Znidaric, M. T., Novak, M. P., Meir, S., et al. (2011). Programmed cell death occurs asymmetrically during abscission in tomato. Plant Cell 23, 4146–4163. doi: 10.1105/tpc.111.092494
Berger, Y., Harpaz-Saad, S., Brand, A., Melnik, H., Sirding, N., Alvarez, J. P., et al. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136, 823–832. doi: 10.1242/dev.031625
Blein, T., Pulido, A., Vialette-Guiraud, A., Nikovics, K., Morin, H., Hay, A., et al. (2008). A conserved molecular framework for compound leaf development. Science 322, 1835–1839. doi: 10.1126/science.1166168
Cai, S., and Lashbrook, C. C. (2008). Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 146, 1305–1321. doi: 10.1104/pp.107.110908
Celton, J. M., Kelner, J. J., Martinez, S., Bechti, A., Khelifi Touhami, A., James, M. J., et al. (2014). Fruit self-thinning: a trait to consider for genetic improvement of apple tree. PLoS ONE 9:e91016. doi: 10.1371/journal.pone.0091016
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Fujisawa, M., Kobayashi, K., Johnston, P., and New, M. (2015). What drives farmers to make top-down or bottom-up adaptation to climate change and fluctuations? A comparative study on 3 cases of apple farming in Japan and South Africa. PLoS ONE 10:e0120563. doi: 10.1371/journal.pone.0120563
Greb, T., Clarenz, O., Schafer, E., Muller, D., Herrero, R., Schmitz, G., et al. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17, 1175–1187. doi: 10.1101/gad.260703
Gregis, V., Sessa, A., Dorca-Fornell, C., and Kater, M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60, 626–637. doi: 10.1111/j.1365-313X.2009.03985.x
Hartmann, U., Höhmann, S., Nettesheim, K., Wisman, E., Saedler, H., and Huijser, P. (2000). Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21, 351–360. doi: 10.1046/j.1365-313x.2000.00682.x
Hu, G., Fan, J., Xian, Z., Huang, W., Lin, D., and Li, Z. (2014). Overexpression of SlREV alters the development of the flower pedicel abscission zone and fruit formation in tomato. Plant Sci. 229, 86–95. doi: 10.1016/j.plantsci.2014.08.010
Immink, R. G., Tonaco, I. A., De Folter, S., Shchennikova, A., Van Dijk, A. D., Busscher-Lange, J., et al. (2009). SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 10, R24. doi: 10.1186/gb-2009-10-2-r24
Keller, T., Abbott, J., Moritz, T., and Doerner, P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18, 598–611. doi: 10.1105/tpc.105.038588
Leseberg, C. H., Eissler, C. L., Wang, X., Johns, M. A., Duvall, M. R., and Mao, L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59, 2253–2265. doi: 10.1093/jxb/ern094
Li, C., Zhou, A., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311, 1936–1939. doi: 10.1126/science.1123604
Li, Z., Reighard, G. L., Abbott, A. G., and Bielenberg, D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60, 3521–3530. doi: 10.1093/jxb/erp195
Lin, Z., Li, X., Shannon, L. M., Yeh, C. T., Wang, M. L., Bai, G., et al. (2012). Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44, 720–724. doi: 10.1038/ng.2281
Liu, D., Wang, D., Qin, Z., Zhang, D., Yin, L., Wu, L., et al. (2014). The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J. 77, 284–296. doi: 10.1111/tpj.12387
Ma, C., Meir, S., Xiao, L., Tong, J., Liu, Q., Reid, M. S., et al. (2015). A KNOTTED1-LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 167, 844–853. doi: 10.1104/pp.114.253815
Mao, L., Begum, D., Chuang, H. W., Budiman, M. A., Szymkowiak, E. J., Irish, E. E., et al. (2000). JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406, 910–913. doi: 10.1038/35022611
Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815.
McManus, M. T., Thompson, D. S., Merriman, C., Lyne, L., and Osborne, D. J. (1998). Transdifferentiation of mature cortical cells to functional abscission cells in bean. Plant Physiol. 116, 891–899.
Meir, S., Philosoph-Hadas, S., Sundaresan, S., Selvaraj, K. S., Burd, S., Ophir, R., et al. (2010). Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 154, 1929–1956. doi: 10.1104/pp.110.160697
Nakano, T., Fujisawa, M., Shima, Y., and Ito, Y. (2013). Expression profiling of tomato pre-abscission pedicels provides insights into abscission zone properties including competence to respond to abscission signals. BMC Plant Biol. 13:40. doi: 10.1186/1471-2229-13-40
Nakano, T., Fujisawa, M., Shima, Y., and Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65, 3111–3119. doi: 10.1093/jxb/eru154
Nakano, T., and Ito, Y. (2013). Molecular mechanisms controlling plant organ abscission. Plant Biotechnol. 30, 209–216. doi: 10.5511/plantbiotechnology.13.0318a
Nakano, T., Kato, H., Shima, Y., and Ito, Y. (2015). Apple SVP family MADS-box proteins and the tomato pedicel abscission zone regulator JOINTLESS have similar molecular activities. Plant Cell Physiol. doi: 10.1093/pcp/pcv034 [Epub ahead of print].
Nakano, T., Kimbara, J., Fujisawa, M., Kitagawa, M., Ihashi, N., Maeda, H., et al. (2012). MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 158, 439–450. doi: 10.1104/pp.111.183731
Patterson, S. E. (2001). Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 126, 494–500. doi: 10.1104/pp.126.2.494
Rick, C. M. (1967). Fruit and pedicel characters derived from galapagos tomatoes. Econ. Bot. 21, 171–184.
Roberts, J. A., Elliott, K. A., and Gonzalez-Carranza, Z. H. (2002). Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53, 131–158. doi: 10.1146/annurev.arplant.53.092701.180236
Roberts, J. A., Schindler, C. B., and Tucker, G. A. (1984). Ethylene-promoted tomato flower abscission and the possible involvement of an inhibitor. Planta 160, 159–163.
Schmitz, G., Tillmann, E., Carriero, F., Fiore, C., Cellini, F., and Theres, K. (2002). The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. U.S.A. 99, 1064–1069. doi: 10.1073/pnas.022516199
Schumacher, K., Schmitt, T., Rossberg, M., Schmitz, G., and Theres, K. (1999). The lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. U.S.A. 96, 290–295.
Sexton, R., and Roberts, J. A. (1982). Cell biology of abscission. Annu. Rev. Plant Physiol. Plant Mol. Biol. 33, 133–162.
Shi, C. L., Stenvik, G. E., Vie, A. K., Bones, A. M., Pautot, V., Proveniers, M., et al. (2011). Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23, 2553–2567. doi: 10.1105/tpc.111.084608
Szymkowiak, E. J., and Irish, E. E. (1999). Interactions between jointless and wild-type tomato tissues during development of the pedicel abscission zone and the inflorescence meristem. Plant Cell 11, 159–175.
Tabuchi, T. (1999). Comparison on the development of abscission zones in the pedicels between two tomato cultivars. J. Japan Soc. Hort. Sci. 68, 939–999.
Tabuchi, T., and Arai, N. (2000). Formation of the secondary cell division zone in tomato pedicels at different fruit growing stages. J. Japan Soc. Hort. Sci. 69, 156–160. doi: 10.2503/jjshs.69.156
Tabuchi, T., Ito, S., and Arai, N. (2001). Anatomical studies of the abscission process in the tomato pedicels at flowering stage. J. Japan Soc. Hort. Sci. 70, 63–65. doi: 10.2503/jjshs.70.63
Tucker, M. L., Burke, A., Murphy, C. A., Thai, V. K., and Ehrenfried, M. L. (2007). Gene expression profiles for cell wall-modifying proteins associated with soybean cyst nematode infection, petiole abscission, root tips, flowers, apical buds, and leaves. J. Exp. Bot. 58, 3395–3406. doi: 10.1093/jxb/erm188
van Nocker, S. (2009). Development of the abscission zone. Stewart Postharvest Rev. 1, 1–6. doi: 10.2212/spr.2009.1.5
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Wang, X., Liu, D., Li, A., Sun, X., Zhang, R., Wu, L., et al. (2013). Transcriptome analysis of tomato flower pedicel tissues reveals abscission zone-specific modulation of key meristem activity genes. PLoS ONE 8:e55238. doi: 10.1371/journal.pone.0055238
Wu, R. M., Walton, E. F., Richardson, A. C., Wood, M., Hellens, R. P., and Varkonyi-Gasic, E. (2012). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63, 797–807. doi: 10.1093/jxb/err304
Yamamoto, T., Ito, M., and Harako, T. (2012). Mechanism of wind fruit drop and its prevention by physical reinforcement near fruit stalk in apple and pear (in Japanese). Bull Yamagata Univ. Agr. Sci. 16, 133–143.
Yamane, H., Ooka, T., Jotatsu, H., Hosaka, Y., Sasaki, R., and Tao, R. (2011). Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 62, 3481–3488. doi: 10.1093/jxb/err028
Yang, T. J., Lee, S., Chang, S. B., Yu, Y., De Jong, H., and Wing, R. A. (2005). In-depth sequence analysis of the tomato chromosome 12 centromeric region: identification of a large CAA block and characterization of pericentromere retrotranposons. Chromosoma 114, 103–117. doi: 10.1007/s00412-005-0342-8
Keywords: abscission, pedicel, MADS-box, ERF, tomato
Citation: Ito Y and Nakano T (2015) Development and regulation of pedicel abscission in tomato. Front. Plant Sci. 6:442. doi: 10.3389/fpls.2015.00442
Received: 30 March 2015; Accepted: 29 May 2015;
Published: 11 June 2015.
Edited by:
Shimon Meir, Agriculture Research Organization, IsraelReviewed by:
Cai-Zhong Jiang, United States Department of Agriculture - Agricultural Research Service, USAAmnon Lers, Agriculture Research Organization, Israel
Copyright © 2015 Ito and Nakano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuhiro Ito, Food Biotechnology Division, National Food Research Institute, National Agriculture and Food Research Organization, 2-1-12 Kannondai, Tsukuba, Ibaraki 305-8642, Japan, yasuito@affrc.go.jp
†Present address: Toshitsugu Nakano, Institute of Crops Research and Development, Vietnam National University of Agriculture, Trau Quy, Gia Lam, Hanoi, Vietnam
 Yasuhiro Ito
Yasuhiro Ito