- 1College of Horticulture, Northwest A&F University, Yangling, China
- 2State Key Laboratory for Stress Biology of Arid Region Crop, Northwest A&F University, Yangling, China
- 3Institute of Vegetables, Hangzhou Academy of Agricultural Sciences, Hangzhou, China
The purple coloration of pepper leaves arises from the accumulation of anthocyanin. Three regulatory and 12 structural genes have been characterized for their involvement in the anthocyanin biosynthesis. Examination of the abundance of these genes in leaves showed that the majority of them differed between anthocyanin pigmented line Z1 and non-pigmented line A3. Silencing of the R2R3-MYB transcription factor CaMYB in pepper leaves of Z1 resulted in the loss of anthocyanin accumulation. Moreover, the expression of multiple genes was altered in the silenced leaves. The expression of MYC was significantly lower in CaMYB-silenced leaves, whereas WD40 showed the opposite pattern. Most structural genes including CHS, CHI, F3H, F3′5′H, DFR, ANS, UFGT, ANP, and GST were repressed in CaMYB-silenced foliage with the exception of PAL, C4H, and 4CL. These results indicated that MYB plays an important role in the regulation of anthocyanin biosynthetic related genes. Besides CaMYB silenced leaves rendered more sporulation of Phytophthora capsici Leonian indicating that CaMYB might be involved in the defense response to pathogens.
Introduction
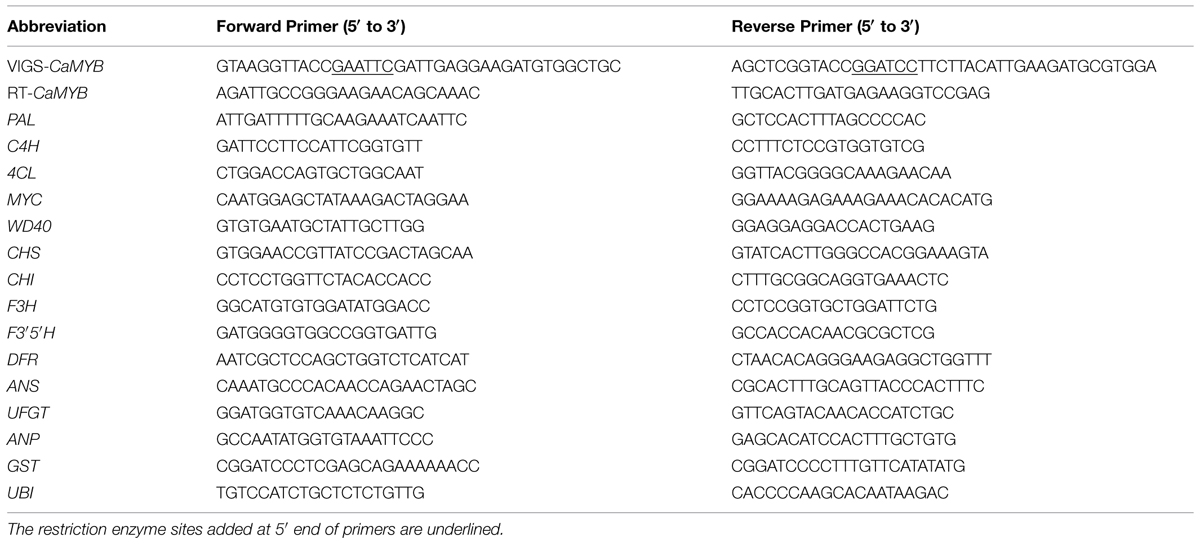
Flavonoids are secondary plant metabolites that fulfill numerous physiological functions such as pigmentation, health-promoting components, and protection against damage by ultraviolet light and phytopathogens (Lepiniec et al., 2006; Davies et al., 2012). Anthocyanins are soluble flavonoids pigments and its biosynthetic pathway has been elucidated. The enzymes involved in the pathway include phenylalanine ammonia-layse (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate: CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′,5′-hydroxylase (F3′5′H), dihydroflavonols 4-reductase (DFR), anthocyanin synthase (ANS), and UDP-glucose: flavonoid 3-glucosyltransferase (UFGT) (Aza-González et al., 2013; Figure 1). Also, an anthocyanin permease (ANP) and a glutathione S-transferase (GST) have been suggested to participate in sequestration of anthocyanin in the vacuole (Mathews et al., 2003).
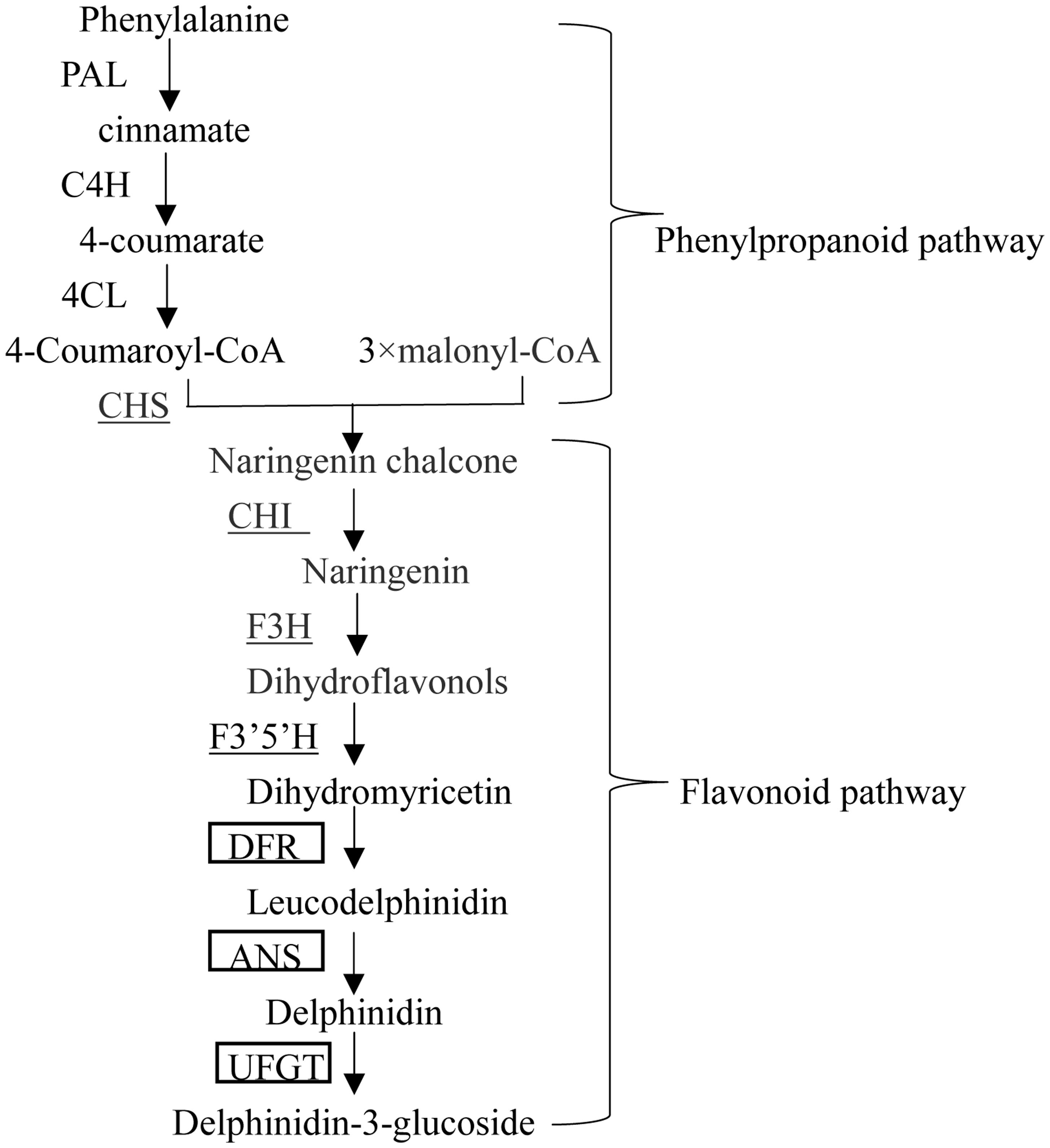
FIGURE 1. Anthocyanin biosynthesis pathway in pepper. Enzymes encoded by flavonoid early structural genes (EBGs) are underlined, and those boxed are encoded by flavonoid late structural genes (LBGs).
A protein complex consisting of a MYB transcription factor, a basic helix-loop-helix (bHLH) protein and WD40 protein (MBW complex) has been proposed to regulate the expression of structural genes (Broun, 2005; Hatlestad et al., 2015), among which MYB appears to be the major determinant of anthocyanin accumulation (Hichri et al., 2011). By interacting with the promoters of late structural genes (LBGs), the MBW complex conveys significant modulation on the expression of these genes, while its effect on the early structural genes (EBGs) is marginal (Nesi et al., 2001).
Pepper (Capsicum annuum L.) is an important horticultural crop cultivated both as an ornamental or vegetable crop. The purple skin color of pepper is preferred by consumers and has become a quality marker for pepper cultivar breeding. The appearance of purple color is due to the abundance of anthocyanin. The anthocyanin biosynthetic pathway has been investigated extensively in pepper fruits, while little research has been done in pepper leaves. Borovsky et al. (2004) isolated A (CaMYB) from pepper fruits and it was uniquely expressed in various organs of purple pepper as opposed to green pepper. Only the late genes in the pathway are dependent on the expression of A (CaMYB) in pepper, similar to the results which have been previously investigated in Petunia (Quattrocchio et al., 1999). Stommel et al. (2009) evaluated the expression of anthocyanin structural (CHS, DFR, and ANS) and regulatory (MYC, MYBA, and WD) genes in foliar tissue from pigmented and non-pigmented C. annuum genotypes, showing that there was no significant difference in the expression of regulatory genes, but pronounced difference in the transcription of structural genes between anthocyanin-pigmented versus and green-colored leaves. These two studies showed inconsistent results regarding the regulation of structural genes in Capsicum and also provided incomplete data on the expression of rest genes in anthocyanin biosynthetic pathway in chili pepper, especially in leaves.
Virus-induced gene silencing (VIGS) is a well-established technology in many species to characterize the function of genes, but it has not been applied in studying the anthocyanin biosynthetic pathway in pepper foliage. In this study, we explored the VIGS application in studying the regulation of anthocyanin biosynthetic pathway in pepper leaves. CaMYB was knocked down in purple pepper line Z1 using the tobacco rattle virus (TRV) based gene silencing technique (Wang et al., 2013). We examined the anthocyanin content and expression of anthocyanin regulatory gene (CaMYB, MYC, and WD40) and 12 structural genes in silenced leaves. Based on these results, we propose that CaMYB is a key factor in anthocyanin biosynthesis pathway in pepper leaves. In addition, CaMYB may be involved in the defense response to pathogens in the purple pepper line.
Materials and Methods
Plant Materials and Growth Conditions
The purple pepper line Z1 and green pepper line A3 were provided by College of Horticulture, Northwest A&F University. Z1 is an anthocyanin pigmented cultivar having purple stem and immature fruit under regular growth conditions. Its leaf is purple under the sixth position, and interspersed with green portions above the sixth position. Pepper seeds were germinated on damp sponge at 28°C in the dark for 2 days. Sprouted seedlings were sown into plastic trays containing steam-sterilized growing media in the illumination incubator at day/night temperatures of 23/18°C with relative humidity of 60% and a 16-h light and 8-h dark photoperiod.
VIGS of CaMYB in Pepper
The pTRV vector and Agrobacterium tumefaciens strain GV3101 were prepared for VIGS (Wang et al., 2013; Ji et al., 2014). We cloned CaMYB gene in purple pepper Z1 and found that the sequence in the coding region of CaMYB is the same as that deposited in the GenBank (AJ608992). Multiple sequence alignment was performed using MEGA6 to reveal the homology among genes involved in anthocyanin biosynthesis from different species. Online software siRNA-scan1 was used to avoid off-target silencing (Xu et al., 2006). A 332-bp fragment harboring a conserved and a non-conserved region of CaMYB was cloned into the pTRV2 vector by One Step Cloning Kit to yield the pTRV2: CaMYB construct. Empty vector (pTRV: 00) was served as a negative control (NC). The pTRV2: PDS, which contains the phytoene desaturase sequence to induce a photo-bleaching phenotype, was used as a positive control (Figure 2). The primers were shown in Table 1. A 10-mL culture of each strain was incubated for 24–36 h at 28°C in LB broth containing 50 mL-1 kanamycin, 50 mL-1 Gentamicin, and 50 mL-1 rifampicin. The primary culture was resuspended into Induction Medium containing 50 mL-1 kanamycin, 20 mg mL-1rifampicin, 50 mL-1 Gentamicin, 200 μM acetosyringone, and was shacked at 28°C for 20–24 h according to Velásquez et al. (2009). The cells were precipitated by centrifugation for 10 min at 3,000 × g and resuspended in the same volume of Agrobacterium infiltration buffer containing 10 mM MgCl2 and 10 mM MES at pH 5.7 till the OD600 reaching 0.5. A. tumefaciens GV3101 containing pTRV1 was mixed with GV3101 containing either pTRV2:00 or pTRV2: CaMYB in a 1:1 ratio. After mixing them, 400 μM acetosyringone was added.
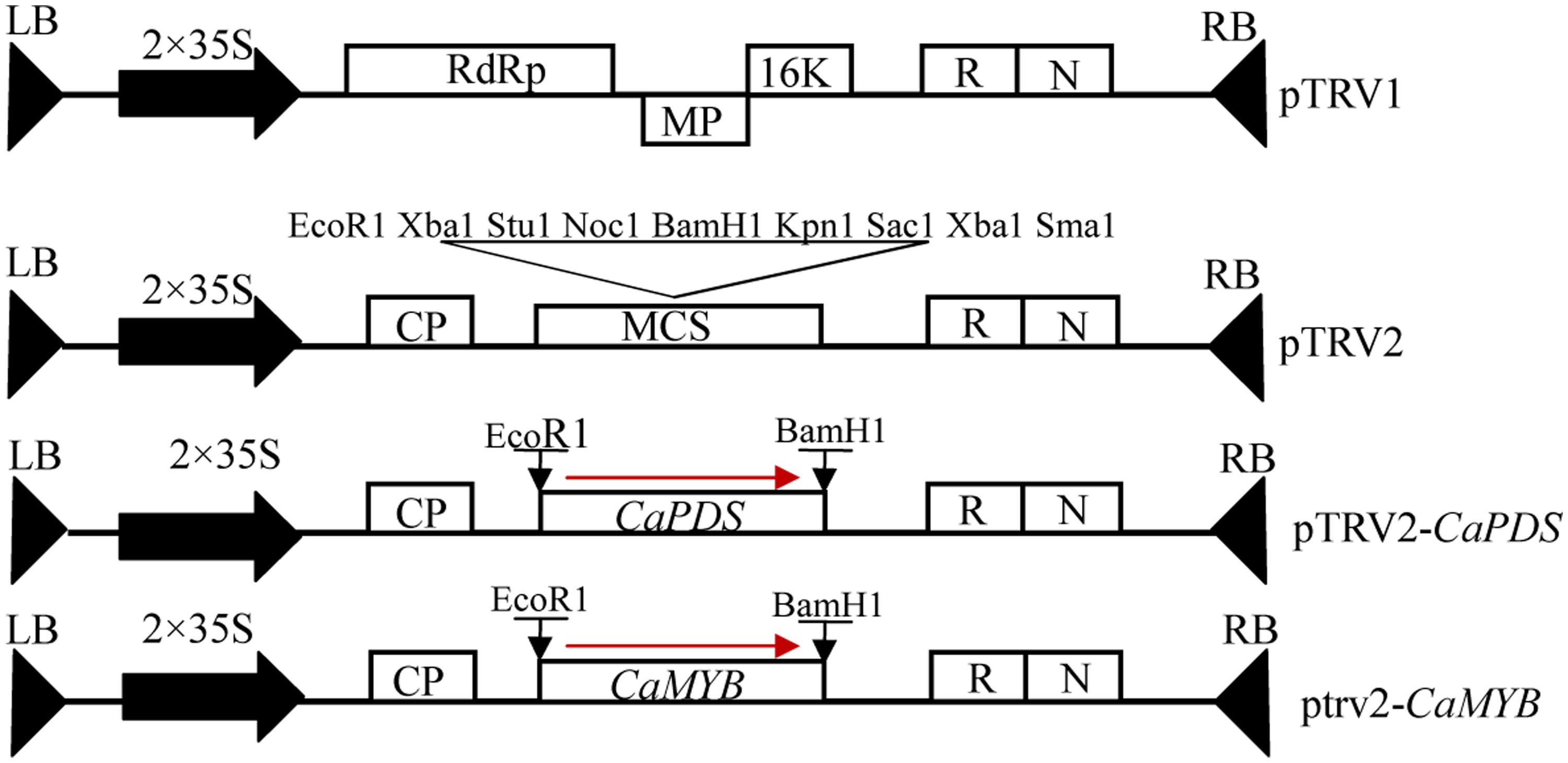
FIGURE 2. Schematic representation of the silencing vector. LB, left borders of the T-DNA; RB, right borders of the T-DNA; 2 × 35 S, two copies of the cauliflower mosaic virus 35 S promoter; CP, coat protein; RdRp, RNA-dependent RNA polymerase; MP, movement protein; 16K, 16 KDa protein; R, ribozyme; N, nos-terminator; MCS, multiple cloning sites.
The plants with the fourth leaf fully expanded were used for VIGS. We punched holes with a needle on both sides of main veins. Then the leaves were infiltrated with a 1 ml syringe without needle. The inoculated plants were grown at 18°C for 48 h in 60% relative humidity under dark and then placed in a growth room at 25°C with a 16-h light/8-h dark photoperiod.
Pathogen Preparation
Preparation and identification of Phytophthora capsici inocula was conducted according to Zhang et al. (2013b). The strain PC of P. capsici was grown on potato dextrose agar (PDA) medium in darkness at 28°C. Twenty-one days after the induction of TRV-mediated gene silencing, detaching leaves from non-infiltrated, pTRV2: 00 and pTRV2: CaMYB plants were exposed to an 8 mm diameter mycelium plug from P. capsici. Then the leaves were placed in a petri dish containing moist filter paper followed by incubation in the dark at 28°C for 3 days.
RNA Analysis
The total RNA was extracted from the leaves of the inoculated plants of Z1 at the same position as the bleaching leaves of pTRV2: PDS plants using the RNA simple kits (Tiangen Biotech, Beijing, China). RNA samples (1 μg) were reverse transcript into complementary DNA (cDNA) using the High Fidelity PrimeScript RT-PCR kit (TaKaRa) according to the manufacturer protocol. The primers of CaMYB designed from the specific sequences outside of the silencing region were used for detecting the gene silencing efficiency. Real-time PCR was used to compare the expression of anthocyanin biosynthetic genes including CaMYB, MYC, WD40, PAL, C4H, 4CL, CHS, CHI, F3H, F3′5′H, DFR, ANS, UFGT, ANP, GST, UBI between anthocyanin-pigmented and non-pigmented leaves. Sequences related to these genes were identified by comparison with sequences in the GenBank database and the pepper whole-genome sequences database2 (Kim et al., 2014). Some of the pepper specific primer sets were adopted from Lightbourn et al. (2007) and Aza-González et al. (2013; Primer sequences were listed in Table 1). The relative fold difference in mRNA levels was calculated using the 2-ΔΔCt formula with CaUbi as the internal control.
Measurement of Anthocyanin Production
For extraction of anthocyanin, 5 g leaves of Z1 line were agitated gently in the dark for 24 h at 4°C in 2 mL of 3 M mixture of HCl : water : MeOH, 1:3:16 (v/v/v; Gould et al., 2000). The extracts were centrifuged. High-performance liquid chromatography (HPLC) analysis of anthocyanin was carried out with a HP1200 Liquid Chromatograph equipped with a diode array detector (Agilent Technology, Palo Alto, CA, USA) using the method as described by Stommel et al. (2009). The anthocyanin was identified by comparing both the retention time and the absorption spectrum with those of a commercial standard (delphinidin-3-p-coumaroyl-rutinoside-5-glucoside).
Results
Phylogenetic Analyses
CaMYB is clustered with the R2R3 MYB transcription factors that are involved in the regulation of anthocyanin biosynthesis in other plant species, such as Arabidopsis, maize, petunia, tobacco, and eggplant (Figure 3). CaMYB is closely related to the subgroup 6 MYBs in Arabidopsis (Dubos et al., 2010), and the nearest MYB is SmMYB2 in eggplant which belongs to the Solanaceae. In pepper genome, there are six genes showing high similarity to CaMYB, and the similarity is heterogeneous ranging from 49 to 67% at the protein level.
FIGURE 3. Phylogenetic analysis of CaMYB and other MYBs. Protein sequences used for alignment are as follow: SmMYB2, KF727477; PhAN2, EF423868.1; PHZ, HQ116170; NtAn2, FJ472647; AtMYB113, NM_105308; AtMYB114, NM_105309; AtPAP1, NM_104541; AtPAP2, NM_105310; ZmC1, P10290.1. Ca, Capsicum annuum L; Sm, Solanum melongena; Ph, Petunia hybrid; Nt, Nicotiana tabacum; At, Arabidopsis thaliana; Zm, Zea mays.
Expression of Biosynthetic Genes in both Anthocyanin-Pigmented and Non-Pigmented Pepper
The expression of genes involved in the anthocyanin biosynthetic pathway was monitored in leaves of pepper line A3 (non-anthocyanin pigmented) and Z1 (anthocyanin pigmented; Figure 4). These genes included phenylpropanold pathway genes (PAL, C4H, 4CL), flavonoid pathway genes (CHS, CHI, F3H, F3′5′H, DFR, ANS, UFGT, ANP, and GST) and regulatory genes (CaMYB, MYC, and WD40). The transcription level of CaMYB was undetectable in A3, but maintained a high level in Z1. The expression of MYC was extremely low in A3 as opposed to Z1. On the contrary, the expression of WD40 was lower in Z1 than in A3. Structural genes could be classified into three groups on the basis of expression pattern. Nine genes including PAL, C4H, F3H, F3′5′H, DFR, ANS, UFGT, ANP, and GST showed higher expression in Z1 than in A3, whereas CHI showed higher expression in A3 than in Z1. The transcript level of 4CL and CHS was similar between A3 and Z1.
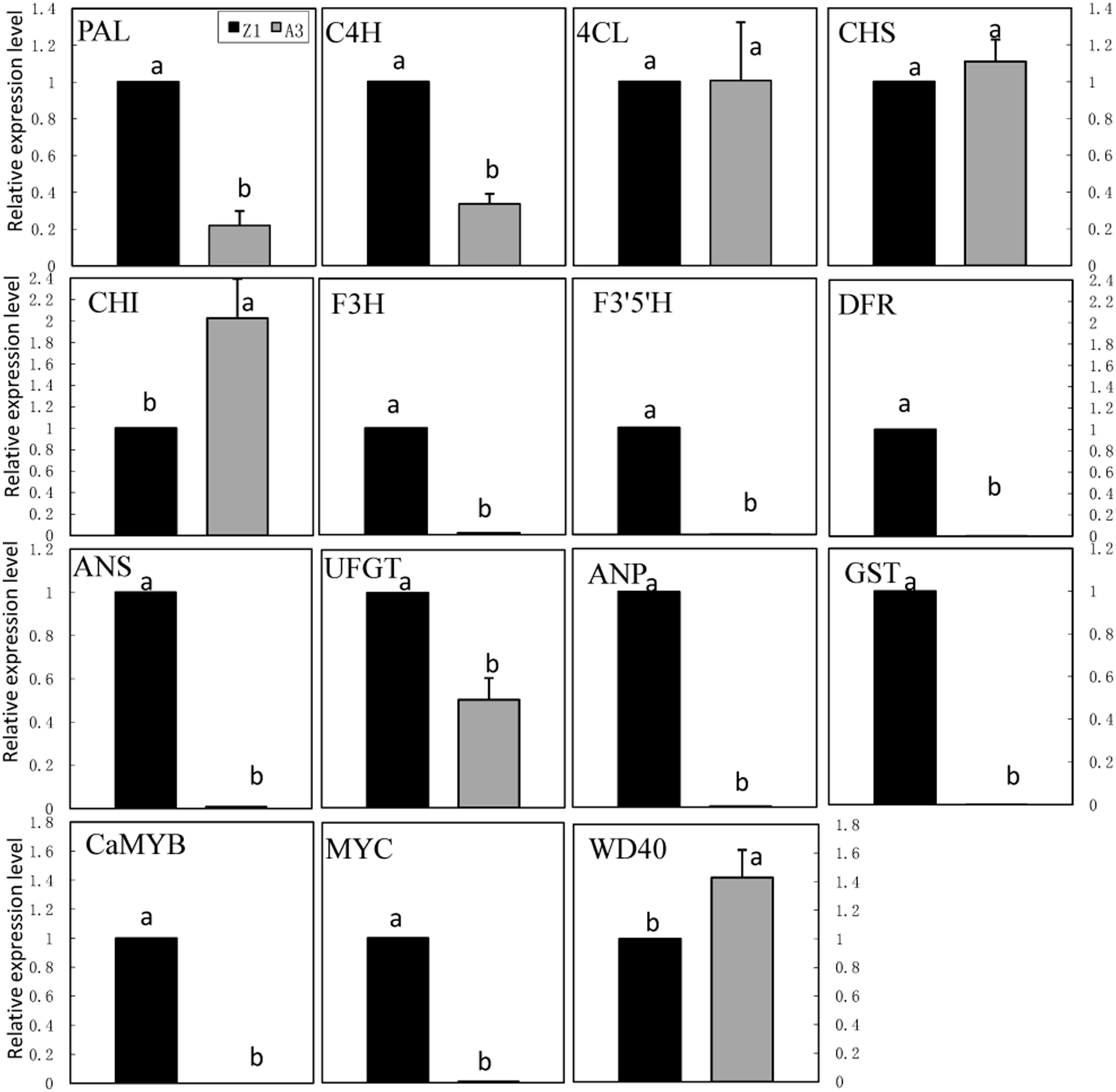
FIGURE 4. Anthocyanin biosynthesis and regulatory genes expression in foliage of line A3 and Z1. Error bars represent the mean ± SD of three independent biological replicates. Bars with different lower case letters in each group indicate significant differences using Duncan’s multiple range test at p < 0.05.
Silencing of CaMYB Reduced the Accumulation of Anthocyanins
As an indication of successful silencing, CaPDS-silenced plants showed photo bleaching (Figure 5A). Z1 leaves infiltrated with pTRV2: 00 (NC) appeared to be less purple colored compared with non-infiltrated Z1 leaves (CK). Z1 leaves infiltrated with pTRV2: CaMYB exhibited green coloration, and CaMYB abundance was significantly decreased in CaMYB-silenced plants compared to the NC (Figure 5B).
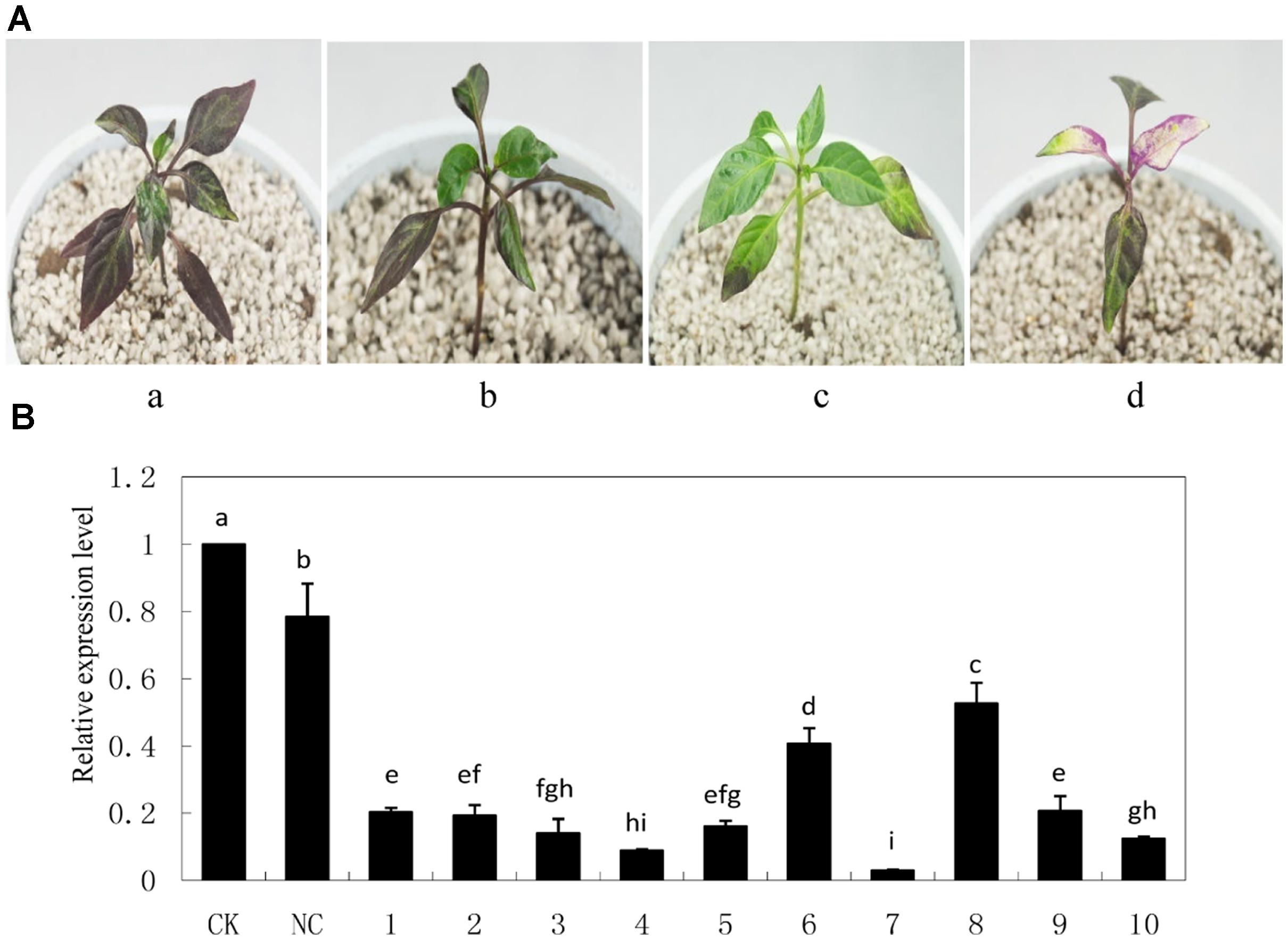
FIGURE 5. Silencing efficiency of CaMYB in pepper plants using a tobacco rattle virus (TRV)-based Virus-induced gene silencing (VIGS) system. (A) Phenotypes of pepper plants 21 days after infiltration of different vectors. a: non-infiltrated control (CK); b: negative control (NC) plants (PTRV2:00); c: CaMYB-silenced plant (PTRV2:CaMYB); d: PDS-silenced plant (PTRV2:PDS). (B) Quantitative real time-PCR analysis of CaMYB expression levels in leaves of CK, NC plants and CaMYB-silenced plants. Ten silencing plants were analyzed (numbered 1–10). Error bars represent mean ± SD for three technical replicates for each plant. Bars with different lower case letters in each group indicate significant differences using Duncan’s multiple range test at p < 0.05.
Anthocyanin from leaves of CK, NC plants, CaMYB-silenced plants and CaPDS-silenced plant were extracted and measured by HPLC (Figure 6). The color intensity of the extracted solutions was different among four samplings (Figure 6B). CaMYB-silenced foliage with a green phenotype accumulated far less anthocyanin; a 31- and 37-fold increase of anthocyanin accumulation was detected in NC and in CK, respectively, compared to that in CaMYB-silenced leaves (Figure 6C).
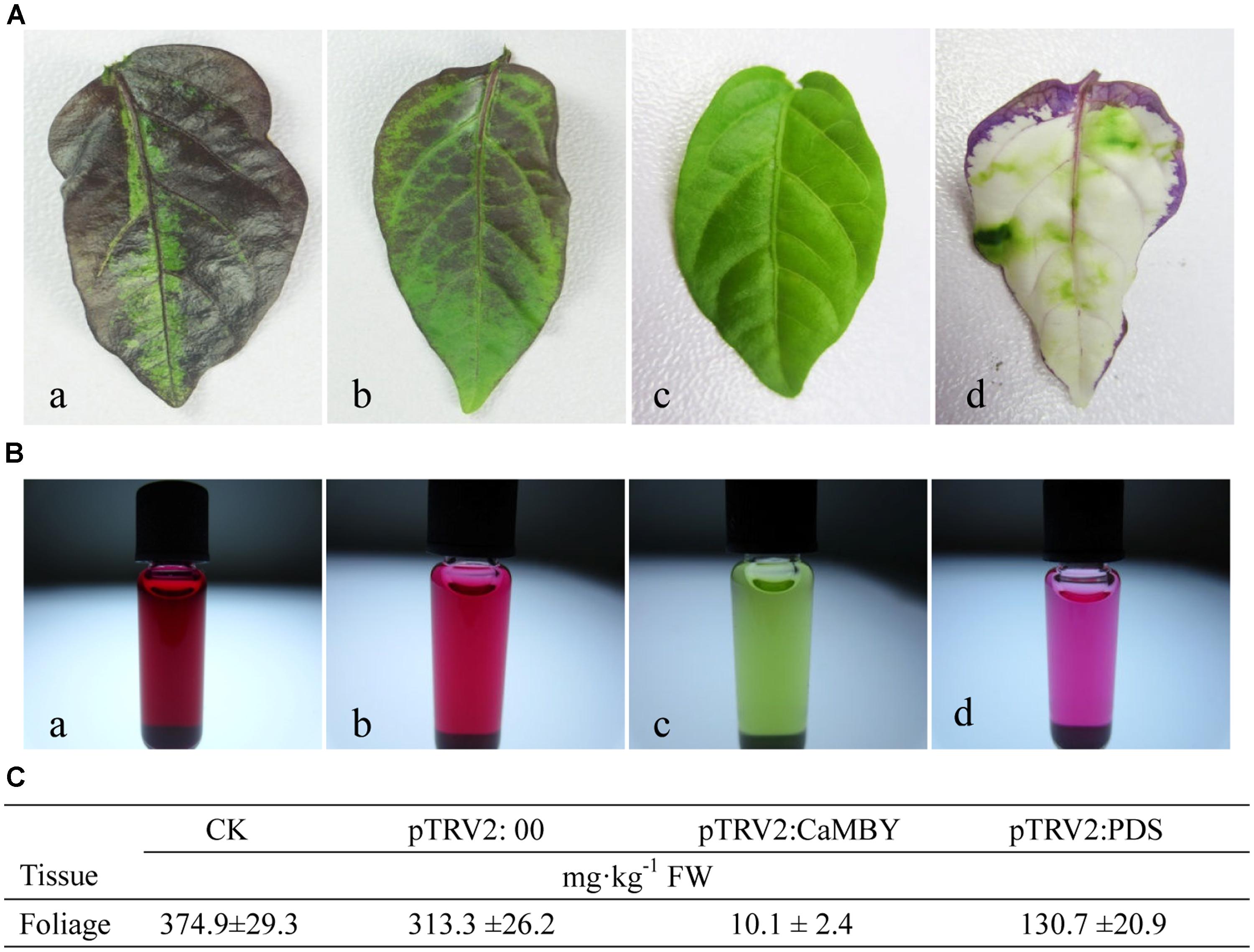
FIGURE 6. Variation of anthocyanin accumulation in leaves infiltrated with different vectors. (A) Leaves were detached from: a: non-infiltrated control (CK); b: NC plants (PTRV2:00); c: CaMYB-silenced plant (PTRV2: CaMYB); d: PDS-silenced plant (PTRV2: PDS). (B) Extracted solutions from the corresponding four samplings. (C) Anthocyanin content of the four samplings by High-performance liquid chromatography (HPLC). Means ± SD (n = 3).
Expression of Anthocyanin Biosynthetic Genes in CaMYB-Silenced Plants
Real-time quantitative PCR was carried out to examine the transcription of anthocyanin biosynthetic genes in leaves of CK, NC and CaMYB-silenced plants (Figure 7). The expression of MYC was significantly lower in CaMYB-silenced leaves than in CK and NC. In contrast, WD40 showed higher expression in CaMYB-silenced leaves than in CK and NC. The transcription of nine structural genes including CHS, CHI, F3H, F3′5′H, DFR, ANS, UFGT, ANP, and GST were repressed in CaMYB-silenced foliage, while PAL, C4H, and 4CL maintained stable and even higher expression in silenced foliage.
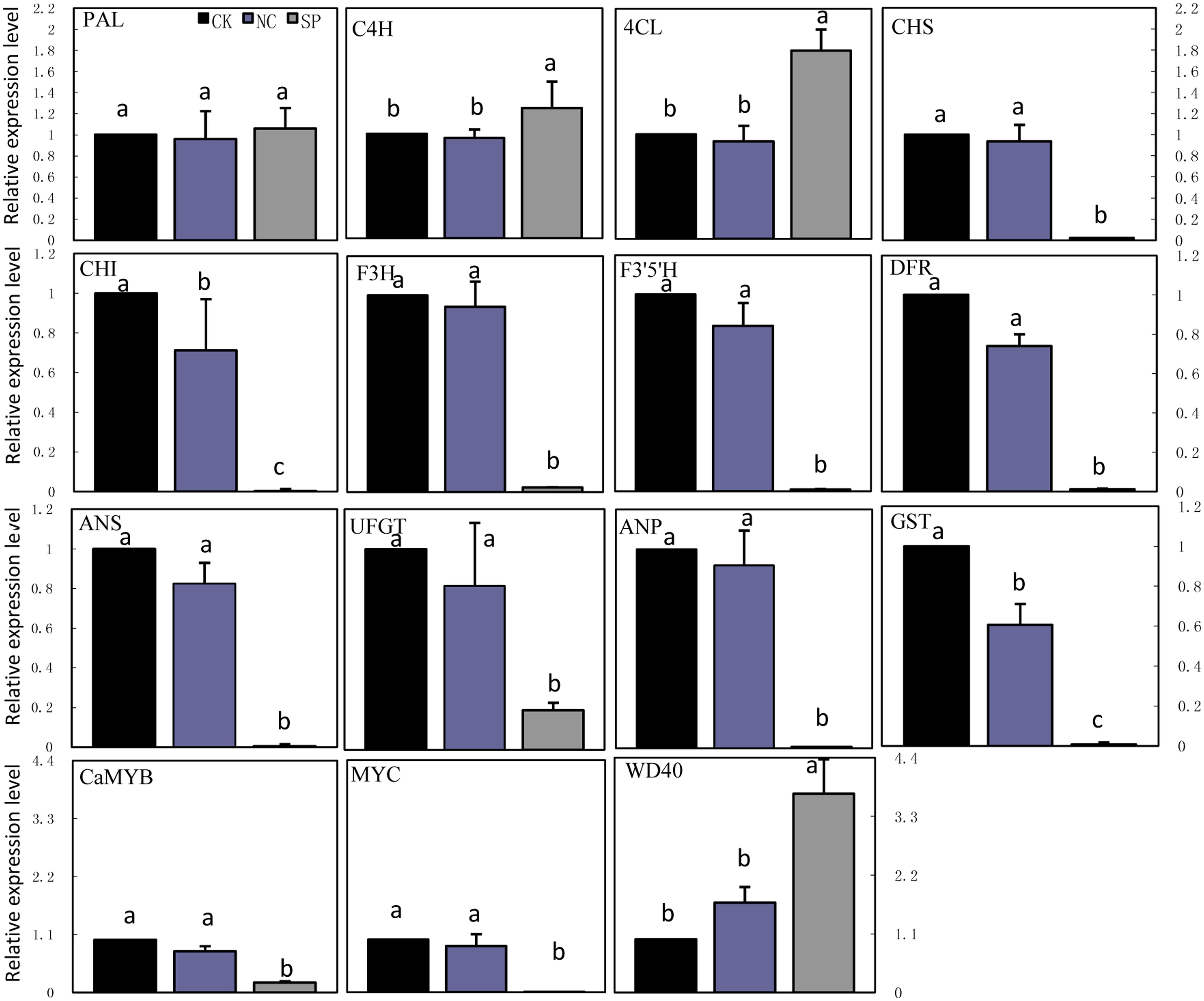
FIGURE 7. Expression of anthocyanin biosynthetic genes in foliage infiltrated with different vectors. CK, non-infiltrated control leaf; NC, negative control leaf (PTRV2:00); SP, CaMYB-silenced plants. Error bars represent the mean ± SD of three independent biological replicates. Bars with different lower case letters in each group indicate significant differences using Duncan’s multiple range test at p < 0.05.
Detached Leaves Inoculation Assays
To determine the role of CaMYB in the basal defense, detached leaves from CK, NC, and pTRV2: CaMYB plants were exposed to an 8 mm diameter mycelium plug from P. capsici 21 days after the performance of the VIGS. Water-soaked lesions occurred 40 h after inoculation on the silenced Z1 leaves, and 8 h later the whole leaves became water-soaked (Figure 8). In contrast, water-soaked lesions were visible on CK and the NC plants at 72 h after inoculation and the symptoms developed slowly.

FIGURE 8. Disease symptoms developed on the non-infiltrated (CK), empty vector control (pTRV2: 00) and silenced (TRV2: CaMYB) leaves infected by Phytophthora capsici. (A) Non-infiltrated control; (B) leaves were infiltrated with TRV empty vector; (C) leaves were infiltrated with TRV2: CaMYB vector. Pictures were taken at 40 and 48 h after the inoculation.
Discussion
It has been reported that purple pepper leaves can enhance the photosynthesis and alleviate the oxidative stress. The anthocyanin epidermal layer may limit photo-inhibitory effects on PSII, the formation of reactive oxygen species and oxidative cellular damages of Tradescantia pallida leaves, supporting the role of anthocyanin pigments in the regulation of photosynthesis for excess absorbed light irradiance (Dewez and Perreault, 2013). Also, pepper seedlings can be consumed fresh and the purple pigments adds nutritive value. Thus it is worthwhile to expand the understanding of anthocyanin biosynthetic pathway in leaves.
Phylogenetic analysis indicates that CaMYB may execute similar functions as that in other plant species (Figure 2). In Arabidopsis, plants harboring a silencing construct targeting MYBs showed decreased MYB gene expression and visible depletion of pigmentation (Gonzalez et al., 2008). In this study, silencing of CaMYB resulted in considerable loss of anthocyanin accumulation in pepper leaves (Figures 5 and 6). Likewise, disabling of CaMYB in pepper fruits reduced the anthocyanin content with lower efficiency (Aguilar-Barragán and Ochoa-Alejo, 2014) in comparison with the results from leaves in this study. These evidence supports that MYBs are essential for anthocyanin production in various organs, including pepper leaves.
The transcription levels of CaMYB and MYC were undetectable in non-pigmented foliage (A3), but an increase in expression of WD40 was observed in A3 compared with Z1 (Figure 4), which is inconsistent with the results from Stommel et al. (2009) probably owing to different lines used in the two studies. The expression of MYC was significantly lower in CaMYB-silenced leaves, whereas WD40 showed the opposite pattern (Figure 7). It has been noted that R2R3-MYB gene alone is capable of regulating the expression of the bHLH (MYC) partner (Baudry et al., 2006; Albert et al., 2014). MYB may induce the expression of MYC, co-regulating the accumulation of anthocyanin. WD40 transcription factor serves as a scaffold that allows the interactions between different MYC and MYB proteins, such as Transparent Testa Glabra1 (TTG1) in Arabidopsis thaliana (Gonzalez et al., 2008). In Petunia sp., AN11 (WD40) protein activated AN2 and AN4 (MYB domain proteins; Quattrocchio et al., 1993; Spelt et al., 2000). More direct evidence is needed to clarify the mechanisms of MBW complex in the anthocyanin biosynthesis in pepper foliage.
Based on the different expression mode of A (CaMYB)and four anthocyanin biosynthetic related genes in both anthocyanin-pigmented and non-pigmented pepper lines, Borovsky et al. (2004) concluded that the early genes (CHS, CHI) in the flavonoid pathway are regulated independently of A, while expression of the late genes (DFR, ANS) is A-dependent. In this study, the expression modes of the anthocyanin biosynthesis related genes in Z1 and A3 indicated that the transcription of LBGs and EBGs except CHS and CHI may be dependent of CaMYB (Figures 1 and 4). This observation was further confirmed by VIGS showing that the expression of flavonoid pathway genes were altered in CaMYB-silenced plants (Figure 7). The expressions of CHS, CHI, F3′5′H, DFR, and 3GT (UFGT) genes were decreased in pepper fruits transformed with the TRV2- MYB construct (Aguilar-Barragán and Ochoa-Alejo, 2014), which also indicated that CHS, CHI were dependent on the expression of MYB (CaMYB). The transcription of F3H did not differ between fruits treated with either the empty TRV2 or TRV2- MYB and the normal fruits, indicating that F3H might be modulated in a different manner in pepper leaf from that in pepper fruits.
It was noted that the flavonoid pathway genes were effectively suppressed, while phenylproanold pathway genes showed a stable (PAL) or an even higher (C4H, 4CL) expression level in CaMYB-silenced leaf (Figure 7). Presumably CaMYB directly activates transcription of CHS, which is the first gene with lower expression in the anthocyanin biosynthetic pathway in CaMYB-silenced plants. Espley et al. (2007) identified the interaction signature motif in MdMYB10, showing that the protein interacts with bHLH proteins to activate transcription of DFR in apple (Espley et al., 2007). In A. thaliana, ternary complex (MYB–MYC–WD40) is important in stimulating anthocyanin formation by activating transcription of DFR and ANS along with trichome formation (Nemie-Feyissa et al., 2014). Here, VIGS provides a useful tool to preliminary screen the target sites of transcription factors, however, sophisticated experiments are needed to identify the authentic targets of transcription factors.
CaMYB-silencing plants rendered susceptibility to P. capsici (Figure 8). It has been observed that anthocyanins often served as the first line of defense against pathogen attacks (Wang et al., 2011). Disruption of Del/Ros1 partially reduced the accumulation of anthocyanins on purple tomato fruits, and areas with less anthocyanin were more susceptible to Botrytis cinerea than purple areas (Zhang et al., 2013a). In potato, common scab resistance was correlated with the expression of MYB and three bHLH genes, indicating that they might be involved in the regulation of the defense response against the common scab pathogen (Tai et al., 2013). Presumably CaMYB gene plays a role, at least partially, in resistance against P. capsici. Therefore effective deployment of CaMYB may aid to develop pepper cultivars with favorable anthocyanin production and resistance level.
Conclusion
This study provides a more comprehensive study on the regulation of anthocyanin biosynthetic genes in pepper leaves. Differential gene expression between leaf of A3 and Z1 and in pepper leaves by VIGS suggests that CaMYB in C. annuum. L regulates the expression of all the flavonoid pathway genes. The gene expression analysis in CaMYB-silenced leaves demonstrates that the transcription factor of CaMYB plays a vital role in the MBW activation complex for anthocyanin accumulation, and MYC might be positively regulated by CaMYB, while WD40 might be negatively regulated by CaMYB in pepper leaves. Furthermore, detached leaves inoculation assay from VIGS system revealed that CaMYB may play an important role in resistance to Phytophthora capsici Leonian in purple pepper. The evidence presented in this study in conjunction with results acquired from further research will help clarify the mechanisms of MBW complex in the anthocyanin biosynthesis and defense response in pepper and other plants.
Author Contributions
ZZ, D-WL, and Z-HG conceived the research. ZZ, D-WL, J-HJ, H-XZ performed the research. ZZ, D-WL, YL performed interpretation of data. ZZ, D-WL, and H-XZ took the photos. ZZ, D-WL and Y-XY performed statistical analyses. ZZ, D-WL wrote the paper. ZZ, D-WL, and Z-HG revised the paper. D-WL and Z-HG provided the materials and resources for the research. ZZ, D-WL, W-GC, and Z-HG performed the integrity of the work. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The following organizations are acknowledged for their financial support: National Natural Science Foundation of China (No.31401879, 31272163), Northwest A&F University Basic Fund for Scientific Research (QN2013016), and the Hangzhou Municipal Science and Technology Commission Project (20140932H01).
Footnotes
References
Aguilar-Barragán, A., and Ochoa-Alejo, N. (2014). Virus-induced silencing of MYB and WD40 transcription factor genes affects the accumulation of anthocyanins in chilli pepper fruit. Biol. Plant. 58, 567–574. doi: 10.1007/s10535-014-0427-4
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). Conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in Eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Aza-González, C., Herrera-Isidrón, L., Núñez-Palenius, H. G., De La Vega, O. M., and Ochoa-Alejo, N. (2013). Anthocyanin accumulation and expression analysis of biosynthesis-related genes during chili pepper fruit development. Biol. Plant. 57, 49–55. doi: 10.1007/s10535-012-0265-1
Baudry, A., Caboche, M., and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779. doi: 10.1111/j.1365-313X.2006.02733.x
Borovsky, Y., Oren-Shamir, M., Ovadia, R., De Jong, W., and Paran, I. (2004). The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. Theor. Appl. Genet. 109, 23–29. doi: 10.1007/s00122-004-1625-9
Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8, 272–279. doi: 10.1016/j.pbi.2005.03.006
Davies, K. M., Albert, N. W., and Schwinn, K. E. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638. doi: 10.1071/FP12195
Dewez, D., and Perreault, F. (2013). Effect of the anthocyanic epidermal layer on Photosystem II and I energy dissipation processes in Tradescantia pallida (Rose) Hunt. Acta Physiol. Plant. 35, 463–472. doi: 10.1007/s11738-012-1089-5
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Gould, K. S., Markham, K. R., Smith, R. H., and Goris, J. J. (2000). Functional role of anthocyanins in the leaves of Quintinia serrata A. Cunn. J. Exp. Bot. 51, 1107–1115. doi: 10.1093/jexbot/51.347.1107
Hatlestad, G. J., Akhavan, N. A., Sunnadeniya, R. M., Elam, L., Cargile, S., Hembd, A., et al. (2015). The beet Y locus encodes an anthocyanin MYB-like protein that activates the betalain red pigment pathway. Nat. Genet. 47, 92–96. doi: 10.1038/ng.3163
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Ji, J. J., Huang, W., Li, D. W., Yin, Y. X., Chai, W. G., and Gong, Z. H. (2014). A CMS-related gene, Ψatp6-2, causes increased ATP hydrolysis activity of the mitochondrial F1Fo-ATP synthase and induces male sterility in pepper (Capsicum annuum L.). Plant Mol. Biol. Rep. 32, 1–12. doi: 10.1007/s11105-014-0702-8
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Lepiniec, L., Debeaujon, I., Routaboul, J. M., Baudry, A., Pourcel, L., Nesi, N., et al. (2006). Genetics and biochemistry of seed flavonoids. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252
Lightbourn, G. J., Stommel, J. R., and Griesbach, R. J. (2007). Epistatic interactions influencing anthocyanin gene expression in Capsicum annuum. J. Am. Soc. Hortic. Sci. 132, 824–829.
Mathews, H., Clendennen, S. K., Caldwell, C. G., Liu, X. L., Connors, K., Matheis, N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15, 1689–1703. doi: 10.1105/tpc.012963
Nemie-Feyissa, D., Olafsdottir, S. M., Heidari, B., and Lillo, C. (2014). Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves. Phytochemistry 98, 34–40. doi: 10.1016/j.phytochem.2013.12.006
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114.
Quattrocchio, F., Wing, J. F., Leppen, H. T., Mol, J. N., and Koes, R. E. (1993). Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5, 1497–1512. doi: 10.1105/tpc.5.11.1497
Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., et al. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. doi: 10.1105/tpc.11.8.1433
Spelt, C., Quattrocchio, F., Mol, J. N., and Koes, R. (2000). Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1631. doi: 10.1105/tpc.12.9.1619
Stommel, J. R., Lightbourn, G. J., Winkel, B. S., and Griesbach, R. J. (2009). Transcription factor families regulate the anthocyanin biosynthetic pathway in Capsicum annuum. J. Am. Soc. Hortic. Sci. 134, 244–251.
Tai, H. H., Goyer, C., and Murphy, A. M. (2013). Potato MYB and bHLH transcription factors associated with anthocyanin intensity and common scab resistance. Botany 91, 722–730. doi: 10.1139/cjb-2012-0025
Velásquez, A. C., Chakravarthy, S., and Martin, G. B. (2009). Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J. Vis. Exp. 28, 1292.
Wang, J. E., Li, D. W., Zhang, Y. L., Zhao, Q., He, Y. M., and Gong, Z. H. (2013). Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. Eur. J. Plant Pathol. 136, 625–638. doi: 10.1007/s10658-013-0193-8
Wang, Y., Chen, S., and Yu, O. (2011). Metabolic engineering of flavonoids in plants and microorganisms. Appl. Microbiol. Biotechnol. 91, 949–956. doi: 10.1007/s00253-011-3449-2
Xu, P., Zhang, Y., Kang, L., Roossinck, M. J., and Mysore, K. S. (2006). Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 142, 429–440. doi: 10.1104/pp.106.083295
Zhang, Y., Butelli, E., Stefano, R. D., Schoonbeek, H., Magusin, A., Pagliarani, C., et al. (2013a). Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility. Curr. Biol. 23 , 1094–1100. doi: 10.1016/j.cub.2013.04.072
Keywords: Capsicum annuum L., purple leaves, CaMYB, virus-induced gene silencing, anthocyanin biosynthesis, P. capsici resistance
Citation: Zhang Z, Li D-W, Jin J-H, Yin Y-X, Zhang H-X, Chai W-G and Gong Z-H (2015) VIGS approach reveals the modulation of anthocyanin biosynthetic genes by CaMYB in chili pepper leaves. Front. Plant Sci. 6:500. doi: 10.3389/fpls.2015.00500
Received: 12 April 2015; Accepted: 22 June 2015;
Published: 07 July 2015.
Edited by:
Hinanit Koltai, Agricultural Research Organization, Volcani Center, IsraelReviewed by:
Michael H. Walter, Leibniz Institute of Plant Biochemistry, GermanyZhaoqi Zhang, South China Agricultural University, China
Copyright © 2015 Zhang, Li, Jin, Yin, Zhang, Chai and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Hui Gong, College of Horticulture, Northwest A&F University, Number 3 Taicheng Road, Yangling, Shaanxi 712100, China, zhgong@nwsuaf.edu.cn
†These authors have contributed equally to this work.
 Zhen Zhang1†
Zhen Zhang1† Wei-Guo Chai
Wei-Guo Chai Zhen-Hui Gong
Zhen-Hui Gong