- 1Department of Biology, Duke University, Durham, NC, USA
- 2University Herbarium and Department of Integrative Biology, University of California at Berkeley, Berkeley, CA, USA
- 3Botany Department, Cologne Biocenter, University of Cologne, Cologne, Germany
- 4Royal Botanic Gardens Edinburgh, Edinburgh, Scotland
- 5New York Botanical Garden, Bronx, NY, USA
- 6Department of Botany, University of British Columbia, Vancouver, BC, Canada
- 7Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada
- 8Department of Medicine, University of Alberta, Edmonton, AB, Canada
- 9BGI-Shenzhen, Shenzhen, China
- 10CSIRO, Centre for Australian National Biodiversity Research, Canberra, ACT, Australia
Plant phototropism, the ability to bend toward or away from light, is predominantly controlled by blue-light photoreceptors, the phototropins. Although phototropins have been well-characterized in Arabidopsis thaliana, their evolutionary history is largely unknown. In this study, we complete an in-depth survey of phototropin homologs across land plants and algae using newly available transcriptomic and genomic data. We show that phototropins originated in an ancestor of Viridiplantae (land plants + green algae). Phototropins repeatedly underwent independent duplications in most major land-plant lineages (mosses, lycophytes, ferns, and seed plants), but remained single-copy genes in liverworts and hornworts—an evolutionary pattern shared with another family of photoreceptors, the phytochromes. Following each major duplication event, the phototropins differentiated in parallel, resulting in two specialized, yet partially overlapping, functional forms that primarily mediate either low- or high-light responses. Our detailed phylogeny enables us to not only uncover new phototropin lineages, but also link our understanding of phototropin function in Arabidopsis with what is known in Adiantum and Physcomitrella (the major model organisms outside of flowering plants). We propose that the convergent functional divergences of phototropin paralogs likely contributed to the success of plants through time in adapting to habitats with diverse and heterogeneous light conditions.
Introduction
Light is the ultimate source of energy for almost all of life on earth, and a remarkable diversity of organisms uses photosynthesis to convert light into metabolic energy. Many of these organisms have also evolved phototropic/phototactic responses, and those in plants are particularly sophisticated—involving movement of shoots, leaves, and/or chloroplasts—in order to optimize their photosynthetic capacity. Charles Darwin pioneered modern research on phototropism by demonstrating that the coleoptile tip is the point of light perception (Darwin, 1880). Darwin proposed that a transmissible substance produced at the tip is responsible for inducing phototropic movements in plants. This insight led to the first discovery of a plant hormone, auxin, and later to the identification of the blue-light photoreceptors for phototropism—phototropins (Briggs et al., 2001; Christie and Murphy, 2013; Briggs, 2014).
Phototropins regulate key physiological responses that are under light control, including positive phototropism of shoots, negative phototropism of roots, chloroplast accumulation, and avoidance, stomatal opening, leaf expansion, and seedling elongation (Christie, 2007). Our current understanding of the function and biochemistry of phototropins originates from basic research on A. thaliana, and to a lesser extent on Adiantum capillus-veneris (a fern) and Physcomitrella patens (a moss). Only a few studies have attempted to address the origin and evolution of phototropins (Briggs et al., 2001; Lariguet and Dunand, 2005; Galván-Ampudia and Offringa, 2007) and all were based on limited sequence samples. The orthology of phototropin genes has therefore been ambiguous, confounding assignments of functional homology, and impeding our understanding of how phototropin evolution has allowed plants to adapt to light environments.
An extraordinary phototropin derivative is neochrome, which possesses supplementary red/far-red-sensing domains from phytochromes (Nozue et al., 1998). In ferns, neochrome can sense both blue and red/far-red light to modulate chloroplast movement and phototropism (Kanegae et al., 2006; Kanegae and Kimura, 2015). We previously reconstructed a phototropin phylogeny with an aim to elucidate the origin of neochromes (Li et al., 2014). However, that phylogeny had insufficient taxon sampling to accurately infer broad patterns of phototropin evolution, including the position of key phototropin duplications.
For this study, we greatly expanded our search for phototropins in genomes and transcriptomes from across land plants, green algae, red algae, glaucophytes, cryptophytes, haptophytes, and stramenopiles (Supplementary Tables S1 and S2). Using these data, we reconstructed a detailed phylogeny of phototropins and examined patterns of gene duplication. Our results suggest that phototropins likely originated in an ancestor of Viridiplantae (land plants + green algae). By reviewing published phototropin functional studies in light of our new gene phylogeny, we determined that phototropin paralogs repeatedly underwent functional divergences. These were likely to be important for adapting to diverse and heterogeneous light environments through time.
Materials and Methods
Mining Phototropin Homologs from Transcriptomes and Genomes
We searched a total of 194 transcriptomes and 26 genomes (Supplementary Table S1). To mine phototropin homologs, we used the BlueDevil python pipeline following Li et al. (2014) for transcriptomes, and for genomes we used BLASTp implemented in Phytozome (Goodstein et al., 2012) or individual genome portals (Supplementary Tables S1 and S2). A phototropin sequence from Anthoceros bharadwajii [voucher: Chantanaorrapint 229 (PSU)] was obtained by PCR and cloning (primers: photF1970 and photR4102; Li et al., 2014).
Sequence Alignment and Phylogenetic Reconstruction
We used MUSCLE (Edgar, 2004) with the default settings to align the amino acid sequences, and then back-translated these to nucleotides. The resulting alignment was manually improved based on known domain boundaries; unalignable regions were excluded prior to phylogenetic analyses. The final alignment length is 2,025 bp, within which most of the sequences are complete or near complete (Supplementary Figure S1).
We used PartitionFinder v1.1.1 (Lanfear et al., 2012) to obtain the optimal data partitioning scheme (by codon position) and the associated nucleotide substitution models (GTR + I+ Γ substitution model applied independently to the first, second, and third codon positions). Garli v2.0 (Zwickl, 2006) was employed to find the best maximum likelihood tree with “genthreshfortopoterm” set to 500,000 and eight independent runs from different random-addition starting trees. We carried out bootstrapping to assess branch support, using RAxML v8.1.11 (Stamatakis, 2006) with 1,000 replicates. The same partition scheme and models were used in MrBayes v3.2.3 (Ronquist et al., 2012) Bayesian inference. We carried out two independent MCMC runs, each with four chains and trees sampled every 1,000 generations (chain length: 6.451 × 109 generations). We unlinked substitution parameters and set the rate prior to vary among subsets. The resulting MCMC statistics were inspected in Tracer (Rambaut and Drummond, 2013) to ensure convergence and proper mixing; 25% of the total generations were discarded as burn-in before compiling the majority consensus tree. The alignment and tree files are deposited in Dryad1.
Target Enrichment for Confirming Phototropin Copy Number in Hornworts
The target enrichment data were from Li et al. (2015), whereby a hornwort (Anthoceros punctatus) DNA library was hybridized with 7,502 120mer RNA probes to enrich phototropin, phytochrome, and neochrome homologs. The probe sequences can be found in Dryad2. We used an enrichment protocol of Li et al. (2013), which can potentially capture sequences with similarity as low as 61%. The captured fragments were sequenced on one-tenth of a MiSeq (250 bp paired-end) run. The reads are deposited in NCBI SRA (SRP055877). We used Scythe v0.994 (Buffalo, 2014) to remove the adaptor sequences with the default prior contamination rate, and Sickle v1.33 (Joshi and Fass, 2011) to trim the low-quality bases with a quality threshold of 33. We assembled the processed reads using SOAPdenovo (Luo et al., 2012) with kmer of 33, 63, and 93, and used CAP3 (Huang, 1999) to merge the three assemblies from different kmer sizes. The phototropin contigs were identified by BLASTn (Camacho et al., 2009).
Results
The Origin of Phototropins
We show here that phototropins are present in all major land plant lineages (seed plants, ferns, lycophytes, mosses, liverworts, and hornworts), as well as in green algae (charophytes, chlorophytes, and prasinophytes; Figure 1A). In contrast, we did not recover phototropins from glaucophytes, red algae (rhodophytes), cryptophytes, haptophytes, or stramenopiles, indicating that the origin of phototropin most likely took place in a common ancestor of Viridiplantae (green algae + land plants; Figure 1A).
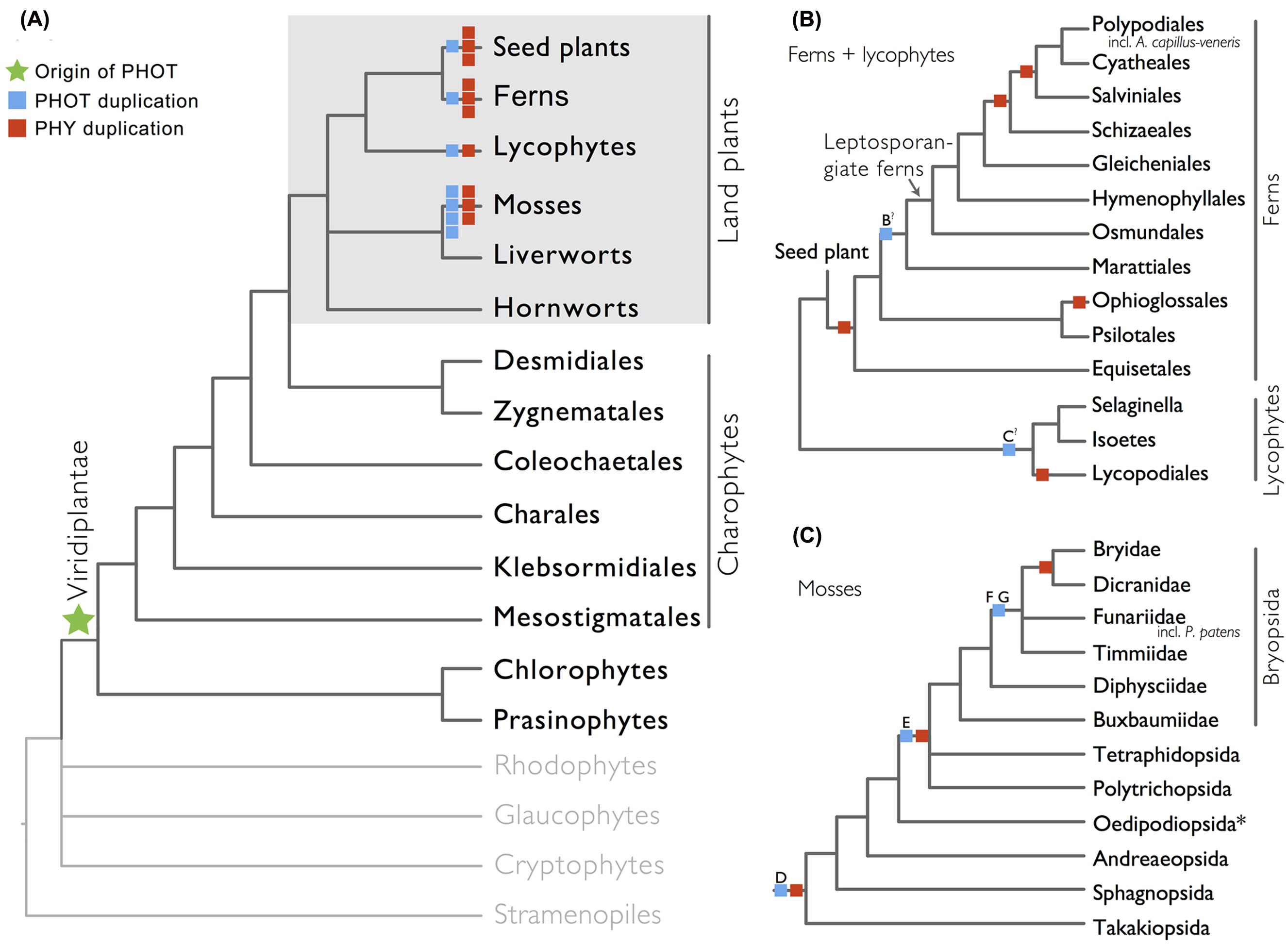
FIGURE 1. Organismal lineages screened for phototropin homologs. (A) Viridiplantae and algae. Lineages that lack phototropin are depicted in gray. Topology derived from Burki et al. (2012) and Wickett et al. (2014). Phototropin (PHOT) and phytochrome (PHY) duplications are only shown on land plant branches (within gray box). (B) Ferns and lycophytes; topology derived from Rothfels et al. (2015). (C) Mosses; topology derived from Cox et al. (2010). Capital letters above blue squares denote phototropin duplication events mentioned in the text and in Figures 2–4. “?” indicates that the exact phylogenetic position of the gene duplication event is ambiguous. “∗” indicates that the lineage was not sampled.
Phototropin Phylogeny
Our phototropin phylogeny is largely congruent with published organismal relationships (Forrest et al., 2006; Cox et al., 2010; Gontcharov and Melkonian, 2010; Villarreal and Renner, 2012; Wickett et al., 2014; Rothfels et al., 2015). Seed–plant phototropins form a monophyletic group that is sister to fern phototropins (Figure 2). Here we infer a single gene duplication event in seed plants, leading to A. thaliana AtPHOT1, and AtPHOT2. Because seed-plant PHOT1 and PHOT2 clades each include sequences from angiosperms and gymnosperms, the duplication event that gave rise to these two homologs predates the divergence of angiosperms from the ancestors of extant gymnosperms (“A” in Figure 2). We also find strong evidence for the monophyly of fern phototropins (Figure 2). Leptosporangiate ferns have two phototropin homologs that we designate fern PHOT1 and PHOT2, in reference to A. capillus-veneris AcPHOT1 and AcPHOT2 (Kagawa et al., 2004), respectively. The earliest-diverging fern lineages, Equisetales, Psilotales, and Ophioglossales, each have one phototropin gene, representing the pre-duplicated version of fern PHOT1 and PHOT2. The exact phylogenetic position of the split of fern PHOT1 and PHOT2 is ambiguous due to a lack of branch support, although it probably was prior to the most recent common ancestor of leptosporangiate ferns and Marattiales (“B” in Figures 1B and 2). We also infer a single duplication event in the lycophyte Selaginella, leading to Selaginella PHOT1 and PHOT2 based on the genome annotation of S. moellendorffii (Banks et al., 2011). The phylogenetic position of this duplication is unclear (“C” in Figure 3), but it must predate the common ancestor of extant Selaginella because the PHOT1 clade contains all known major Selaginella lineages (Korall and Kenrick, 2002). For Isoetales and Lycopodiales, we found only one phototropin homolog, but determining whether it is indeed a single-copy gene in these lineages will require confirmation with additional data.
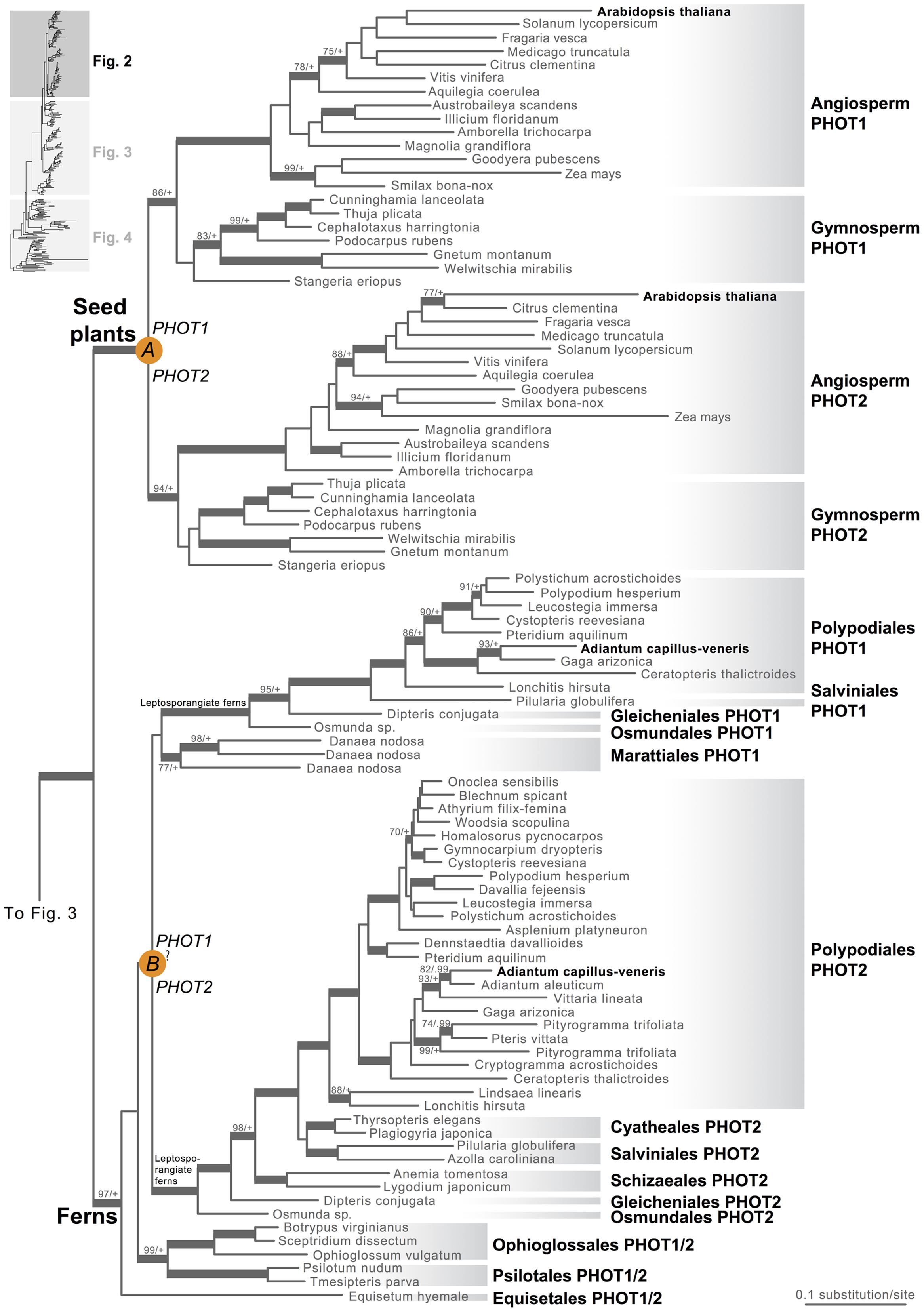
FIGURE 2. Phylogenetic relationships of seed plant and fern phototropins. The phylogeny tree continues to Figures 3 and 4. Orange circles indicate inferred phototropin (PHOT) duplication events. The italicized capital letter within each circle corresponds to the duplication event mentioned in the text, and the numbers/letters adjacent to each orange circle are the names of the gene duplicates. Support values associated with branches are maximum likelihood bootstrap values (BS)/Bayesian posterior probabilities (PP); these are only displayed (along with thickened branches) if BS > 70 and PP > 0.95. “+” denotes BS = 100 or PP = 1.00. Thickened branches without numbers are 100/1.0. “?” indicates that the exact phylogenetic position of the gene duplication event is ambiguous.
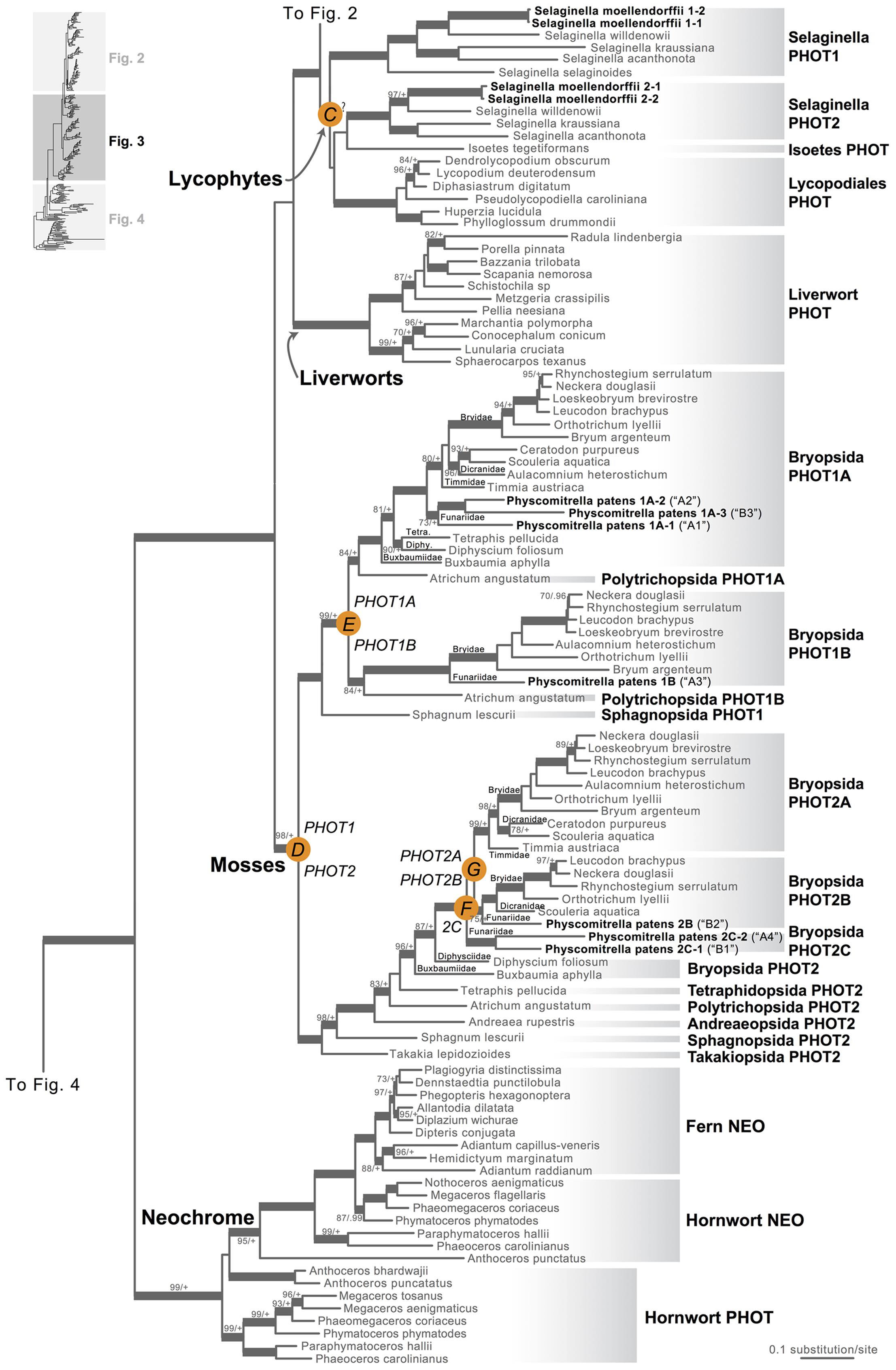
FIGURE 3. Phylogenetic relationships of lycophyte and bryophyte phototropins. The phylogeny tree is continued from Figure 2. Previous gene annotations for Physcomitrella patens are in parentheses. Orange circles indicate inferred phototropin (PHOT) duplication events. The italicized capital letter within each circle corresponds to the duplication event mentioned in the text, and the numbers/letters adjacent to each orange circle are the names of the gene duplicates. Support values associated with branches are maximum likelihood bootstrap values (BS)/Bayesian posterior probabilities (PP); these are only displayed (along with thickened branches) if BS > 70 and PP > 0.95. “+” denotes BS = 100 or PP = 1.00. Thickened branches without numbers are 100/1.0. “?” indicates that the exact phylogenetic position of the gene duplication event is ambiguous.
All liverwort transcriptomes we examined contained a single phototropin (Figure 3), a result consistent with the recent demonstration that phototropin in Marchantia polymorpha is a single-copy gene (Komatsu et al., 2014). Hornwort phototropins also appear to be single-copy based on our screening of hornwort transcriptomes and a low-coverage genome draft of Anthoceros punctatus (Li et al., 2014). To further investigate gene copy number in hornworts, we used a target-enrichment strategy to sequence all phototropin-, phytochrome- and neochrome-like genomic fragments in Anthoceros punctatus. We found no additional divergent phototropin copies, and recovered one phytochrome and one neochrome gene copy.
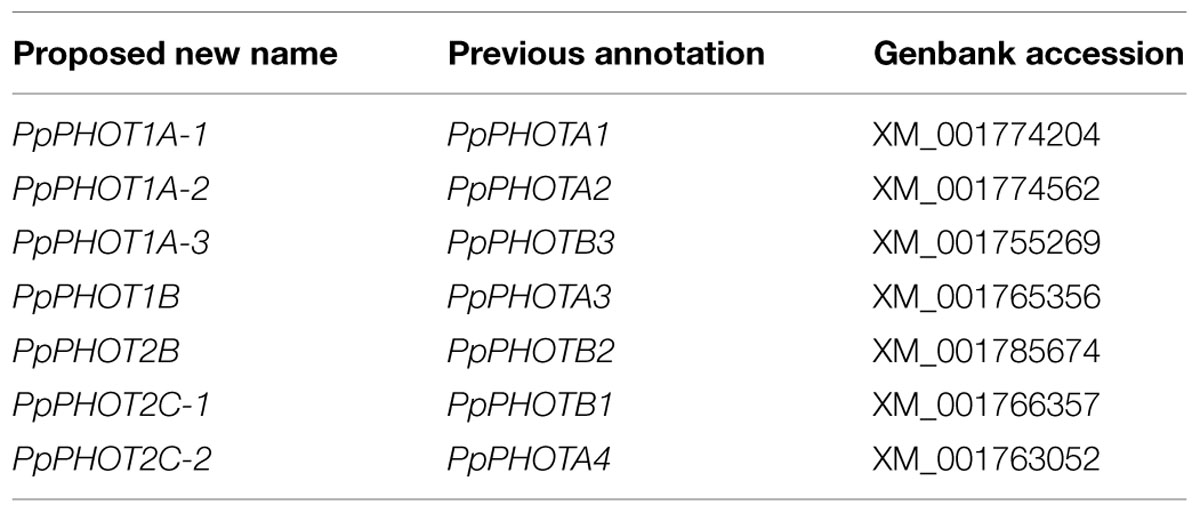
Moss phototropins, on the other hand, have a significantly more complex evolutionary history (Figures 1C and 3). We discovered that the published phototropin annotations from the moss P. patens genome (Rensing et al., 2008) do not correctly reflect gene orthology. Because “PHOTAs” and “PHOTBs” are intermingled, we reclassified the moss phototropins based on the phylogenetic relationships inferred here (Table 1, Figure 3). Prior to the initial divergences among extant mosses, a gene duplication (“D” in Figures 1C and 3) gave rise to moss PHOT1 and PHOT2. In moss PHOT1, a second duplication occurred in a common ancestor of Bryopsida and Polytrichopsida (“E” in Figures 1C and 3) that split moss PHOT1 into moss PHOT1A and PHOT1B. In moss PHOT2, two additional duplications occurred (“F” and “G” in Figures 1C and 3) subsequent to the divergence of Diphysciidae (Bryopsida), resulting in moss PHOT2A-C. Both moss PHOT2A and PHOT2B are present in Dicranidae and Bryidae, whereas moss PHOT2C is only known in P. patens (Funariidae). P. patens may also have lost the PHOT2A homolog. Alternatively, because the placement of PHY2C is not supported, these P. patens sequences could belong to PHY2A, requiring only one gene duplication that resulted in moss PHY2A and PHY2B. Most green algal transcriptomes and genomes revealed a single phototropin gene (Figure 4), with the exception of those from Zygnematales, where two phototropin homologs are present (PHOTA and PHOTB).
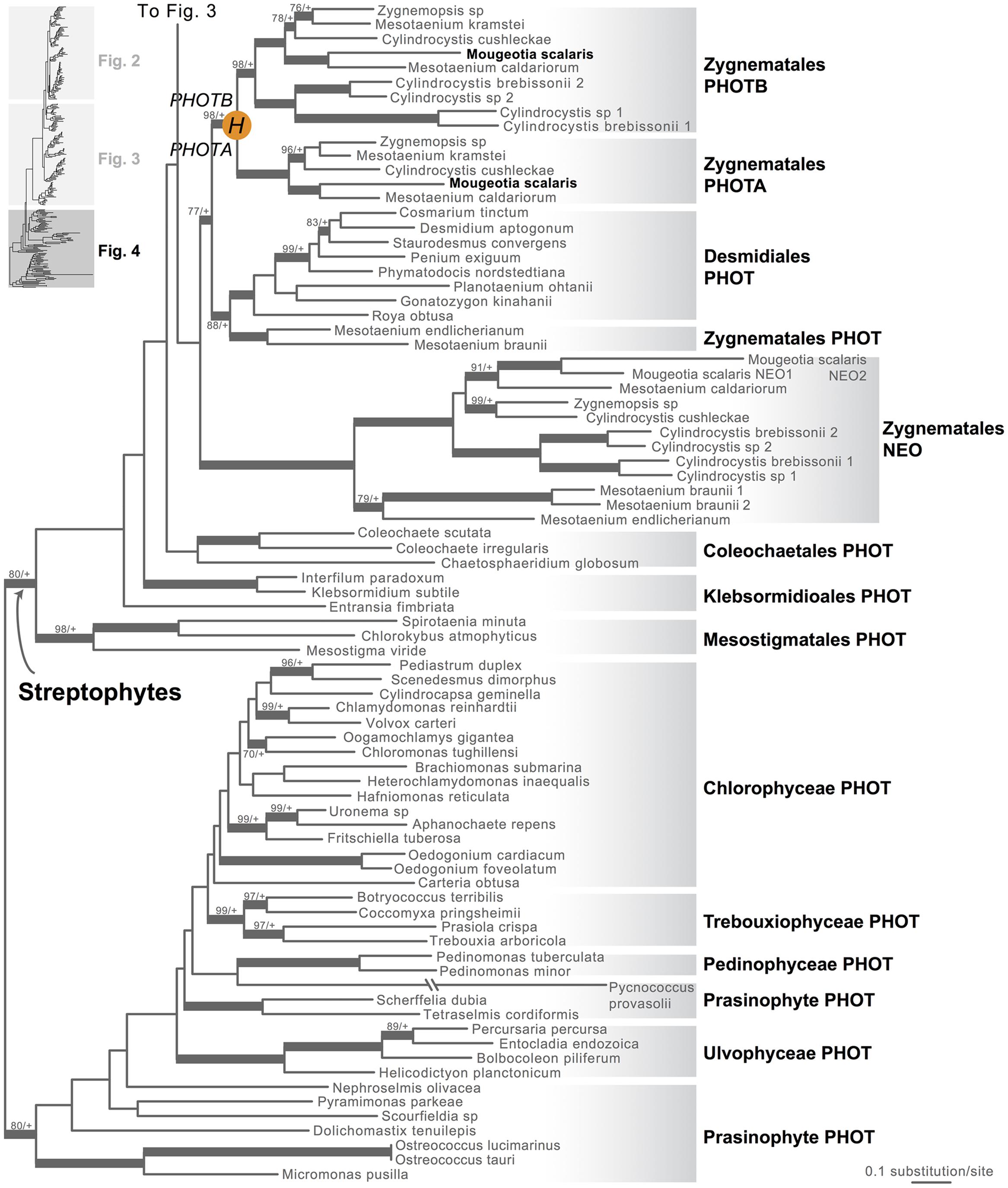
FIGURE 4. Phylogenetic relationships of algal phototropins. The phylogenetic tree is continued from Figure 3. Orange circles indicate inferred phototropin (PHOT) duplication events. The italicized capital letter within each circle corresponds to the duplication event mentioned in the text, and the numbers/letters adjacent to each orange circle are the names of the gene duplicates. Support values associated with branches are maximum likelihood bootstrap values (BS)/Bayesian posterior probabilities (PP); these are only displayed (along with thickened branches) if BS > 70 and PP > 0.95. “+” denotes BS = 100 or PP = 1.00. Thickened branches without numbers are 100/1.0.
All Algal Neochromes Lack the Conserved Cysteine Residue at the LOV2 Domain
Neochrome (neo, Figures 3 and 4) is a unique chimeric phototropin variant that possesses supplementary red/far-red-sensing domains from phytochromes (Nozue et al., 1998). Recent studies have revealed two independent origins of neochromes, one in zygnematalean algae and the other in hornworts (Suetsugu et al., 2005; Li et al., 2014). Interestingly, the neochromes found in ferns were determined to be derived from hornworts via horizontal gene transfer (Li et al., 2014). Neochrome perceives both blue and red/far-red light to mediate phototropic responses in ferns (Kawai et al., 2003; Kanegae et al., 2006; Kanegae and Kimura, 2015), and it appears to have played a significant role in their diversification (Schneider et al., 2004; Schuettpelz and Pryer, 2009). The function of neochrome in hornworts and zygnematalean algae, however, remains unclear. Because some zygnematalean algae have plate-like chloroplasts that rotate in response to both blue and red/far-red light irradiation (Haupt and Scheuerlein, 1990), it was hypothesized that algal neochrome, originally discovered in Mougeotia scalaris, is the candidate gene responsible for this movement (Suetsugu et al., 2005). However, neochrome in M. scalaris responds only to red/far-red light and not to blue light (Suetsugu et al., 2005; Kagawa and Suetsugu, 2007).
To explore whether M. scalaris might be anomalous among zygnematalean algae in having a neochrome that is not responsive to blue light, we examined all the algal neochromes that we recovered. As is the case with the neochrome of M. scalaris, none has the conserved cysteine residue in the LOV2 domain (Figure 5) that is essential for the formation of flavin mononucleotide (FMN) chromophore adduct and blue-light signal transduction (Christie, 2007; but see Kanegae and Kimura, 2015). Furthermore, many of the FMN-interacting residues (Crosson and Moffat, 2001) are also not conserved in zygnematalean neochromes (Figure 5). It is thus possible that all zygnematalean algae use neochromes only for sensing red/far-red light, and use other blue-light photoreceptors (most likely phototropins; Kagawa and Suetsugu, 2007; Banas et al., 2012) to maneuver chloroplast rotations. However, Kanegae and Kimura (2015) recently discovered that in fern neochromes, substitution of the cysteine residue did not completely abolish blue-light-induced phototropism. They further suggested that the phytochrome chromophore, phytochromobilin, could have the capacity to perceive blue light and then relay the signals. Therefore, we cannot rule out the possibility that some zygnematalean neochromes can sense blue light through a FMN-independent mechanism.
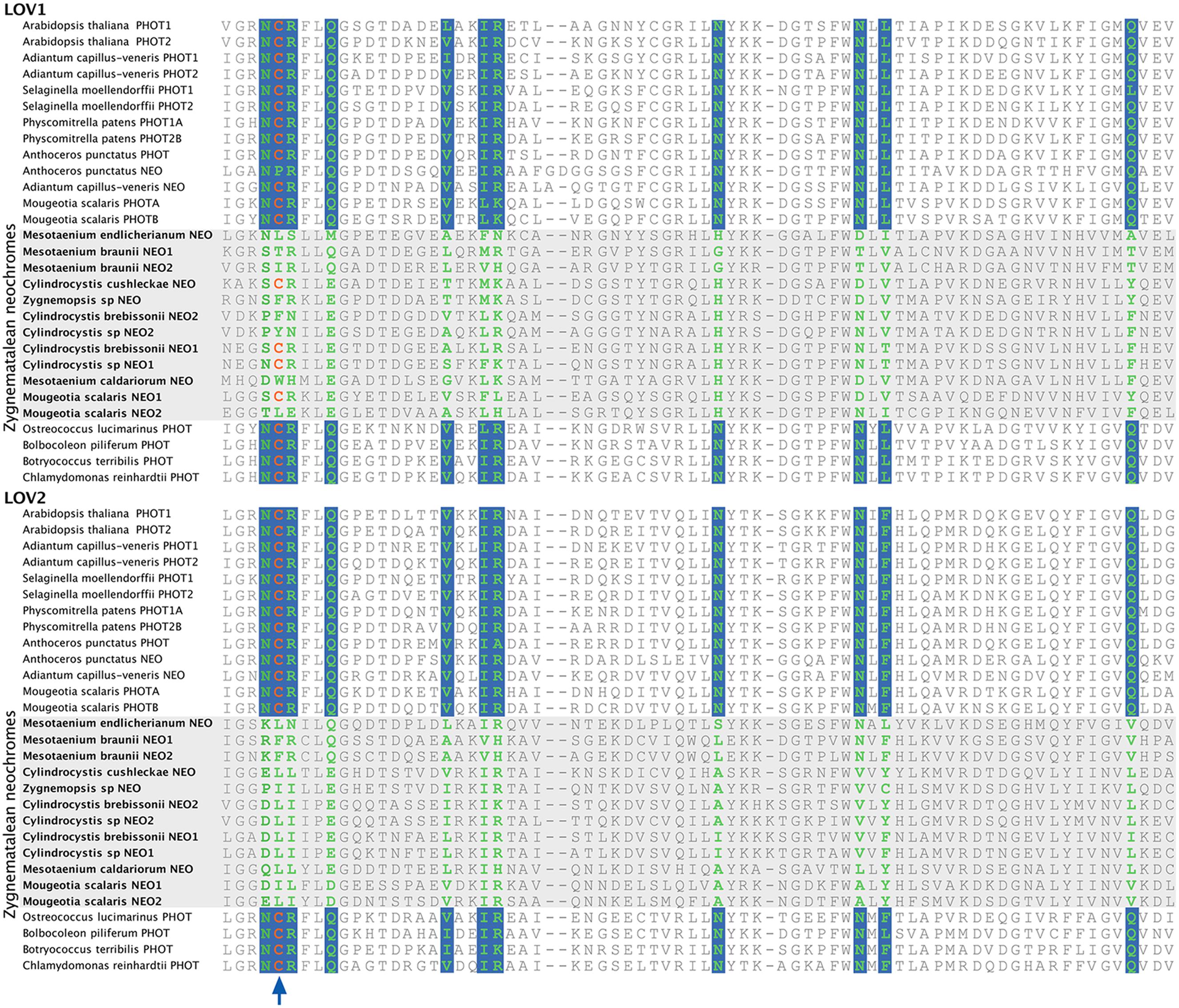
FIGURE 5. Alignment of a portion of LOV1 and LOV2 domains in selected phototropins and neochromes. The site for flavin mononucleotide (FMN) adduct formation is marked by an arrow, and the FMN-interacting sites are shown in green with a blue background. All zygnematalean neochromes (highlighted in a gray box) lack the conserved cysteine residue in the LOV2 domain, and several residues that interact with FMN are also not conserved.
Discussions
A New Phototropin Gene Orthology
With a detailed phototropin phylogeny that encompasses all of green plant representatives, we were able to discover new phototropin lineages and pinpoint the timing of gene duplications (Figures 1–4). This new understanding of phototropin gene orthology refutes the previous assertion that the “PHOT2” ortholog is the ancestral phototropin and that “PHOT1” evolved later in seed plants (Galván-Ampudia and Offringa, 2007); the ancestral phototropin is neither “PHOT1” nor “PHOT2.” Rather, a single PHOT ancestral sequence gave rise to the multiple gene copies found in seed plants, lycophytes, ferns, mosses, and zygnematalean algae (Figures 1–4). Independent duplications occurred subsequently, such that PHOT1 and PHOT2 in each of these plant lineages are more closely related to one another than they are to other PHOT1s or PHOT2s.
Convergent Sub-Functionalization of Phototropins
Our findings on gene orthology also have important implications for understanding the functional evolution of phototropins. Plants often respond differently under low- and high-light levels; chloroplasts, in particular, accumulate on the periclinal walls under weak light, but retreat to anticlinal walls when the light intensity is too high. Our phylogenetic reconstruction suggests that phototropins repeatedly duplicated and diverged, and that after doing so, they subsequently specialized in mediating either low– or high-light responses, although functional redundancies do exist (Christie, 2007). Of the two phototropins known in A. thaliana, Atphot1 mediates phototropism under low light intensity, and is more sensitive than Atphot2 in triggering chloroplast accumulation (Sakai et al., 2001). Atphot2, in contrast, responds predominantly to high-light intensity, and is primarily responsible for chloroplast avoidance under strong light (Kagawa et al., 2001; Luesse et al., 2010). A similar functional differentiation can also be seen in the fern A. capillus-veneris Acphot1 and Acphot2. Acphot2 controls chloroplast avoidance under high-light intensity, whereas Acphot1 has little or no role in this response (Kagawa et al., 2004). Similarly, in the moss P. patens, Ppphot1A-2 (see Table 1) is the primary mediator for the chloroplast avoidance response, and plays a redundant role in the accumulation behavior (Kasahara et al., 2004).
The single phototropin in the liverwort M. polymorpha can respond to a wide range of light intensities and triggers both chloroplast avoidance and accumulation responses (Komatsu et al., 2014). Because liverworts represent one of the deepest splits in land-plant phylogeny (Wickett et al., 2014), their unspecialized phototropins suggest that the ancestral land plant phototropin was probably a “general-purpose” photoreceptor. The subsequent and parallel specializations of phototropin into forms that are responsive to low–, or high-light intensities may have played an important role in the adaptation of early land plants to Earth’s changing landscapes. From the formation of the earliest forests by cladoxylopsid ferns about 385 million years ago (Stein et al., 2007) through to today’s angiosperm-dominated terrestrial ecosystems, light environments have become increasingly complex, and deeply shaded habitats have expanded. Possessing duplicated phototropin genes dedicated to functioning under different light intensities would have been advantageous (Galen et al., 2004) compared with having a single, general-purpose phototropin. The fact that land plant lineages with duplicated phototropins (seed plants, leptosporangiate ferns and mosses) are more species-rich than those without (liverworts, hornworts, and non-leptosporangiate ferns) is consistent with such an advantage, although many other traits could also have contributed to this disparity in diversity.
Compared to land-plant phototropins, much less is known about the function of algal phototropins, where most of the research done on Chlamydomonas reinhardtii shows that phototropins regulate sexual processes (Huang and Beck, 2003), eyespot size and phototactic behavior (Trippens et al., 2012). Interestingly, the phototropin gene from C. reinhardtii can partially rescue A. thaliana phot1 phot2 double mutant phenotypes, suggesting that the phototropin signal transduction pathway is deeply conserved from green algae to seed plants (Onodera et al., 2005). Future studies focusing on phototropins across more algal lineages (Figure 4) should help to clarify early phototropin evolution in unicellular organisms, and the genetic basis of its functional differentiation in land plants.
Patterns of Phototropin Copy Expansion and Stasis Resemble that of Phytochromes
The evolutionary pattern that we observe here for phototropins shows a striking resemblance to that for phytochromes. Both photoreceptors (phytochromes and phototropins) duplicated repeatedly in seed plants, ferns, lycophytes, and mosses, while remaining single-copy in liverworts and hornworts (Figure 1; Li et al., 2015). Although this pattern of concerted gene family expansion and stasis could be due to whole genome duplications (WGD), the exact evolutionary positions of gene duplication events in these two photoreceptors differ—they did not all happen along the same phylogenetic branches (Figure 1), suggesting that WGD is not solely responsible. Recent studies have shown that phototropins and phytochromes not only share cross-talk in their signal transduction pathways (Lariguet et al., 2006; de Carbonnel et al., 2010; Demarsy et al., 2012), but also can physically interact (Jaedicke et al., 2012). The extent to which phototropins and phytochromes are co-evolving would be an interesting topic for future research.
Conclusion
In summary, we have leveraged recent genomic and transcriptomic data to discover phototropins from across a broad sample of photosynthetic eukaryotes. Our study reveals that phototropins are unique to Viridiplantae, and that gene family expansion and stasis have operated uniquely within each of the various land plant lineages, a pattern similar to that of phytochromes (Li et al., 2015). Existing functional data for phototropins, interpreted in light of our gene phylogeny, suggest a history of repeated gene duplications followed by parallel functional divergences. Our broad phylogenetic approach is an important complement to ongoing photobiological research focused on a small number of plant model organisms, and will enable new research linking ecology, evolution, and photochemistry to understanding how plants adapt (and have adapted) to variable light environments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Science Foundation Doctoral Dissertation Improvement Grant DEB-1407158 (to KMP and F-WL), and National Science Foundation Graduate Research Fellowship (to F-WL). We thank Pryer lab members and two reviewers for suggestions and comments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00637
Footnotes
References
Banas, A. K., Aggarwal, C., Labuz, J., Sztatelman, O., and Gabrys, H. (2012). Blue light signalling in chloroplast movements. J. Exp. Bot. 63, 1559–1574. doi: 10.1093/jxb/err429
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., Depamphilis, C., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963. doi: 10.1126/science.1203810
Briggs, W. R. (2014). Phototropism: some history, some puzzles, and a look ahead. Plant Physiol. 164, 13–23. doi: 10.1104/pp.113.230573
Briggs, W. R., Beck, C. F., Cashmore, A. R., Christie, J. M., Hughes, J., Jarillo, J. A., et al. (2001). The phototropin family of photoreceptors. Plant Cell 13, 993–997. doi: 10.1105/tpc.13.5.993
Buffalo, V. (2014). Scythe – A Bayesian Adapter Trimmer (Version 0.994 BETA). Available at: https://github.com/vsbuffalo/scythe
Burki, F., Okamoto, N., Pombert, J. F., and Keeling, P. J. (2012). The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Biol. Sic. 279, 2246–2254. doi: 10.1098/rsbp.2011.2301
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Christie, J. M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant. Biol. 58, 21–45. doi: 10.1146/annurev.arplant.58.032806.103951
Christie, J. M., and Murphy, A. S. (2013). Shoot phototropism in higher plants: new light through old concepts. Am. J. Bot. 100, 35–46. doi: 10.3732/ajb.1200340
Cox, C. J., Goffinet, B., Wickett, N. J., Boles, S. B., and Shaw, A. J. (2010). Moss diversity: a molecular phylogenetic analysis of genera. Phytotaxa 9, 175–195. doi: 10.11646/phytotaxa.9.1.10
Crosson, S., and Moffat, K. (2001). Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2995–3000. doi: 10.1073/pnas.051520298
Darwin, C. (1880). The Power of Movement in Plants. London: John Murray. doi: 10.5962/bhl.title.102319
de Carbonnel, M., Davis, P., Roelfsema, M. R. G., Inoue, S.-I., Schepens, I., Lariguet, P., et al. (2010). The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 152, 1391–1405. doi: 10.1104/pp.109.150441
Demarsy, E., Schepens, I., Okajima, K., Hersch, M., Bergmann, S., Christie, J. M., et al. (2012). Phytochrome Kinase Substrate 4 is phosphorylated by the phototropin 1 photoreceptor. EMBO J. 31, 3457–3467. doi: 10.1038/emboj.2012.186
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Forrest, L. L., Davis, E. C., Long, D. G., Crandall-Stotler, B. J., Clark, A., and Hollingsworth, M. L. (2006). Unraveling the evolutionary history of the liverworts (Marchantiophyta): multiple taxa, genomes and analyses. Bryologist 109, 303–334. doi: 10.1639/0007-2745(2006)109[303:UTEHOT]2.0.CO;2
Galen, C., Huddle, J., and Liscum, E. (2004). An experimental test of the adaptive evolution of phototropins: blue-light photoreceptors controlling phototropism in Arabidopsis thaliana. Evolution 58, 515–523. doi: 10.1111/j.0014-3820.2004.tb01675.x
Galván-Ampudia, C. S., and Offringa, R. (2007). Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 12, 541–547. doi: 10.1016/j.tplants.2007.10.004
Gontcharov, A. A., and Melkonian, M. (2010). Molecular phylogeny and revision of the genus Netrium (Zygnematophyceae, Streptophyta): Nucleotaenium gen. nov. J. Phycol. 46, 346–362. doi: 10.1111/j.1529-8817.2010.00814.x
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Haupt, W., and Scheuerlein, R. (1990). Chloroplast movement. Plant Cell Environ. 13, 595–614. doi: 10.1111/j.1365-3040.1990.tb01078.x
Huang, K., and Beck, C. (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 100, 6269–6274. doi: 10.1073/pnas.0931459100
Huang, X. (1999). CAP3: a DNA sequence assembly program. Genome Res. 9, 868–877. doi: 10.1101/gr.9.9.868
Jaedicke, K., Lichtenthaler, A. L., Meyberg, R., Zeidler, M., and Hughes, J. (2012). A phytochrome-phototropin light signaling complex at the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 109, 12231–12236. doi: 10.1073/pnas.1120203109
Joshi, N. A., and Fass, J. N. (2011). Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33). Available at: https://github.com/najoshi/sickle
Kagawa, T., Kasahara, M., Abe, T., Yoshida, S., and Wada, M. (2004). Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol. 45, 416–426. doi: 10.1093/pcp/pch045
Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., et al. (2001). Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141. doi: 10.1126/science.291.5511.2138
Kagawa, T., and Suetsugu, N. (2007). Photometrical analysis with photosensory domains of photoreceptors in green algae. FEBS Lett. 581, 368–374. doi: 10.1016/j.febslet.2006.12.041
Kanegae, T., Hayashida, E., Kuramoto, C., and Wada, M. (2006). A single chromoprotein with triple chromophores acts as both a phytochrome and a phototropin. Proc. Natl. Acad. Sci. U.S.A. 103, 17997–18001. doi: 10.1073/pnas.0603569103
Kanegae, T., and Kimura, I. (2015). A phytochrome/phototropin chimeric photoreceptor of fern functions as a blue/far-red light-dependent photoreceptor for phototropism in Arabidopsis. Plant J. 83, 480–488. doi: 10.1111/tpj.12903
Kasahara, M., Kagawa, T., Sato, Y., Kiyosue, T., and Wada, M. (2004). Phototropins mediate blue and red light-induced chloroplast movements in Physcomitrella patens. Plant Physiol. 135, 1388–1397. doi: 10.1104/pp.104.042705
Kawai, H., Kanegae, T., Christensen, S., Kiyosue, T., Sato, Y., Imaizumi, T., et al. (2003). Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature 421, 287–290. doi: 10.1038/nature01310
Komatsu, A., Terai, M., Ishizaki, K., Suetsugu, N., Tsuboi, H., Nishihama, R., et al. (2014). Phototropin encoded by a single-copy gene mediates chloroplast photorelocation movements in the liverwort Marchantia polymorpha. Plant Physiol. 166, 411–427. doi: 10.1104/pp.114.245100
Korall, P., and Kenrick, P. (2002). Phylogenetic relationships in Selaginellaceae based on rbcL sequences. Am. J. Bot. 89, 506–517. doi: 10.3732/ajb.89.3.506
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Lariguet, P., and Dunand, C. (2005). Plant photoreceptors: phylogenetic overview. J. Mol. Evol. 61, 559–569. doi: 10.1007/s00239-004-0294-2
Lariguet, P., Schepens, I., Hodgson, D., Pedmale, U., Trevisan, M., Kami, C., et al. (2006). PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. U.S.A. 103, 10134–10139. doi: 10.1073/pnas.0603799103
Li, C., Hofreiter, M., Straube, N., Corrigan, S., and Naylor, G. J. P. (2013). Capturing protein-coding genes across highly divergent species. Biotechniques 54, 321–326. doi: 10.2144/000114039
Li, F.-W., Melkonian, M., Rothfels, C. J., Villarreal, J. C., Stevenson, D. W., Graham, S. W., et al. (2015). Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat. Commun. 6:7852. doi: 10.1038/ncomms8852
Li, F.-W., Villarreal, J. C., Kelly, S., Rothfels, C. J., Melkonian, M., Frangedakis, E., et al. (2014). Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc. Natl. Acad. Sci. U.S.A. 111, 6672–6677. doi: 10.1073/pnas.1319929111
Luesse, D. R., DeBlasio, S. L., and Hangarter, R. P. (2010). Integration of Phot1, Phot2, and PhyB signalling in light-induced chloroplast movements. J. Exp. Bot. 61, 4387–4397. doi: 10.1093/jxb/erq242
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18. doi: 10.1186/2047-217X-1-18
Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H., Yeh, K., et al. (1998). A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. U.S.A. 95, 15826–15830. doi: 10.1073/pnas.95.26.15826
Onodera, A., Kong, S.-G., Doi, M., Shimazaki, K.-I., Christie, J. M., Mochizuki, N., et al. (2005). Phototropin from Chlamydomonas reinhardtii is functional in Arabidopsis thaliana. Plant Cell Physiol. 46, 367–374. doi: 10.1093/pcp/pci037
Rambaut, A., and Drummond, A. J. (2013). Tracer v1.6. Available at: http://tree.bio.ed.ac.uk/software/tracer/
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rothfels, C. J., Li, F.-W., Sigel, E. M., Huiet, L., Larsson, A., Burge, D. O., et al. (2015). The evolutionary history of ferns inferred from 25 low-copy nuclear genes. Am. J. Bot. 102, 1089–1107. doi: 10.3732/ajb.1500089
Sakai, T., Kagawa, T., Kasahara, M., Swartz, T. E., Christie, J. M., Briggs, W. R., et al. (2001). Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. U.S.A. 98, 6969–6974. doi: 10.1073/pnas.101137598
Schneider, H., Schuettpelz, E., Pryer, K. M., Cranfill, R., Magallón, S., and Lupia, R. (2004). Ferns diversified in the shadow of angiosperms. Nature 428, 553–557. doi: 10.1038/nature02361
Schuettpelz, E., and Pryer, K. M. (2009). Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc. Natl. Acad. Sci. U.S.A. 106, 11200–11205. doi: 10.1073/pnas.0811136106
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stein, W. E., Mannolini, F., Hernick, L. V., Landing, E., and Berry, C. M. (2007). Giant cladoxylopsid trees resolve the enigma of the Earth’s earliest forest stumps at Gilboa. Nature 446, 904–907. doi: 10.1038/nature05705
Suetsugu, N., Mittmann, F., Wagner, G., Hughes, J., and Wada, M. (2005). A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc. Natl. Acad. Sci. U.S.A. 102, 13705–13709. doi: 10.1073/pnas.0504734102
Trippens, J., Greiner, A., Schellwat, J., Neukam, M., Rottmann, T., Lu, Y., et al. (2012). Phototropin influence on eyespot development and regulation of phototactic behavior in Chlamydomonas reinhardtii. Plant Cell 24, 4687–4702. doi: 10.1105/tpc.112.103523
Villarreal, J. C., and Renner, S. S. (2012). Hornwort pyrenoids, carbon-concentrating structures, evolved and were lost at least five times during the last 100 million years. Proc. Natl. Acad. Sci. U.S.A. 109, 18873–18878. doi: 10.1073/pnas.1213498109
Wickett, N. J., Mirarab, S., Nguyen, N., Warnow, T., Carpenter, E., Matasci, N., et al. (2014). Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111, E4859–E4868. doi: 10.1073/pnas.1323926111
Keywords: blue-light, convergent evolution, land plants, photoreceptors, phototropism
Citation: Li F-W, Rothfels CJ, Melkonian M, Villarreal JC, Stevenson DW, Graham SW, Wong GK-S, Mathews S and Pryer KM (2015) The origin and evolution of phototropins. Front. Plant Sci. 6:637. doi: 10.3389/fpls.2015.00637
Received: 11 July 2015; Accepted: 31 July 2015;
Published: 12 August 2015.
Edited by:
Hirokazu Tsukaya, The University of Tokyo, JapanReviewed by:
Noriyuki Suetsugu, Kyoto University, JapanJon Hughes, Justus Liebig University Giessen, Germany
Copyright © 2015 Li, Rothfels, Melkonian, Villarreal, Stevenson, Graham, Wong, Mathews and Pryer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fay-Wei Li, Department of Biology, Duke University, Biological Sciences Building, 130 Science Drive, Durham, NC 27708, USA, fay.wei.li@duke.edu
 Fay-Wei Li
Fay-Wei Li Carl J. Rothfels
Carl J. Rothfels Michael Melkonian3
Michael Melkonian3 Juan C. Villarreal
Juan C. Villarreal Dennis W. Stevenson
Dennis W. Stevenson Sean W. Graham
Sean W. Graham Gane K.-S. Wong
Gane K.-S. Wong Kathleen M. Pryer
Kathleen M. Pryer