- 1Plant Molecular Biology Laboratory, Department of Life Science, Sogang University, Seoul, South Korea
- 2College of Medicine, Seoul National University, Seoul, South Korea
- 3Stress Physiology and Molecular Biology Laboratory, School of Life Science, Jawaharlal Nehru University, New Delhi, India
Salinity is one of the important abiotic factors for any crop management in irrigated as well as rainfed areas, which leads to poor harvests. This yield reduction in salt affected soils can be overcome by improving salt tolerance in crops or by soil reclamation. Salty soils can be reclaimed by leaching the salt or by cultivation of salt tolerance crops. Salt tolerance is a quantitative trait controlled by several genes. Poor knowledge about mechanism of its inheritance makes slow progress in its introgression into target crops. Brassica is known to be a good reclamation crop. Inter and intra specific variation within Brassica species shows potential of molecular breeding to raise salinity tolerant genotypes. Among the various molecular markers, SSR markers are getting high attention, since they are randomly sparsed, highly variable and show co-dominant inheritance. Furthermore, as sequencing techniques are improving and softwares to find SSR markers are being developed, SSR markers technology is also evolving rapidly. Comparative SSR marker studies targeting Arabidopsis thaliana and Brassica species which lie in the same family will further aid in studying the salt tolerance related QTLs and subsequent identification of the “candidate genes” and finding out the origin of important QTLs. Although, there are a few reports on molecular breeding for improving salt tolerance using molecular markers in Brassica species, usage of SSR markers has a big potential to improve salt tolerance in Brassica crops. In order to obtain best harvests, role of SSR marker driven breeding approaches play important role and it has been discussed in this review especially for the introgression of salt tolerance traits in crops.
Introduction
Salinity in the soil is one of the serious obstacles for agriculture, due to which large areas of the agricultural lands are becoming unfertile. Three fourth of the total Earth surface is covered by saline water and hence significant proportion of this Earth is affected by saline conditions. Over 830 million hectares of land area in the entire earth are salt affected, either by saline water (403 million hectares) or by the conditions related with sodicity (434 million hectares; FAO, 2008) and it is more than six percent of the entire land area in the world. An excess amount of NaCl occurs as an abiotic environmental factor in many places such as salt deserts in the arid and semi-arid areas, coastal salt marshes and inland saline lakes (Kumar, 2013). During the last decades, apart from the natural salinity, salinization of soils due to intensive agriculture and irrigation has also been becoming a major problem in agriculture. When plants are exposed to salinity, it causes ion imbalance, ion toxicity and hyper osmotic stress (Yamaguchi and Blumwald, 2005; Kumar et al., 2013, 2014). It severely retards the crop growth and productivity. For most of the crops concentrations of 150 mM NaCl are highly toxic though, for a few crops, as low as 25 mM NaCl is lethal. Two main courses of actions were given special importance for providing the solution for salinity stress problem (Epstein, 1985; Ashraf, 1994; Flowers and Yeo, 1995; Grieve et al., 1999), which includes reclamation of saline soils by use of chemicals or by growing salt tolerant plants in the saline soils. Considering its low cost, feasible, and efficient approach, the latter strategy was being emphasized by many plant scientists during the past few decades. This includes cereals, legumes and other commercially important crops.
Apart from the cereals and legumes, oil seeds are very important for human food and are at the third position among the crops. At least forty different plant species are known to be grown for the oils production (Weiss, 1983). Among the oilseed crops, Brassicas which belongs to the family Brassicaceae are very important oilseed crops. The family Brassicaceae includes various crops, which are rich in nutritional and economic values. The members of the Brassica genus are sometimes collectively called as cabbages/mustards/-cole crops. Brassica contains more number of important horticultural and agricultural crops. The members of the Brassica genus also contains more number of weed species and wild relatives, making it a perfect platform for crop improvement practices, due to the presence of wide genetic base. Apart from the oilseeds (mustard seed, oilseed rape), almost every part of the plant of some species or the other are edible and grown for food, which includes the stems (kohlrali), root (Swedes, turnips), flower (Cualiflower, broccoli), and leaves (Cabbages, Brussels sprout). Brassica vegetables are most commonly regarded for their nutritional and medicinal properties (Beecher, 1994; Carvalhoa et al., 2006). They contain high amounts of soluble fiber and vitamin C (Divisi et al., 2006). Brassica contains different nutrients having potential anticancer properties like 3, 3′ diindolylmethane, sulforaphane, and selenium (Finley et al., 2005; Banerjee et al., 2012). Since Brassicas are of high agricultural importance, they are of much scientific interest.
Before discussing the markers and salinity tolerance in Brassica, it is important to know the relationship with in the Brassica species.
Relationships between Crop Brassicas
The relationship between six particular species in Brassica genus (B. carinata, B. juncea, B. napus, B. nigra, B. oleraceae, and B. rapa) is well described by the Triangle of U theory (Nagaharu, 1935; Figure 1). The Triangle of U theory explains the high chromosome number species [B. carinata (BBCC), 2n = 34; B. juncea (AABB), 2n = 36; and B. napus (AACC), 2n = 38] which are amphidiploids and possibly formed through the interspecific hybridization between the low chromosome number species in pairs [B. nigra (BB), 2n = 16; B. oleraceae (CC), 2n = 18; and B. rapa (AA), 2n = 20].
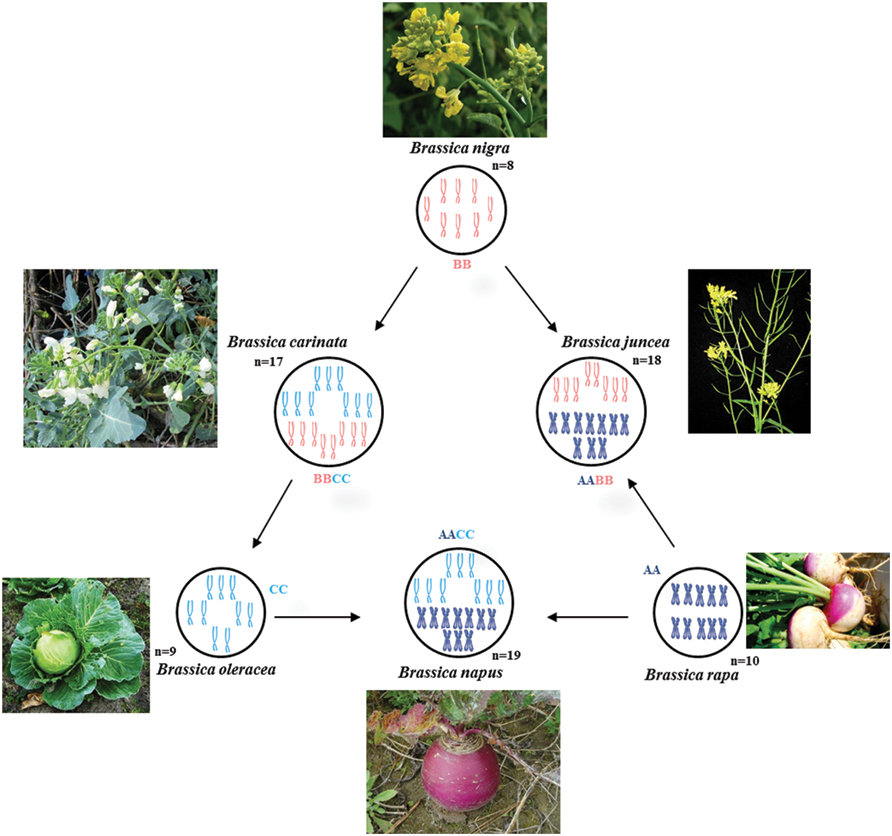
FIGURE 1. Triangle of U diagram illustrating the genetic relationships between six species of the genus Brassica, such as Chinese cabbage (AA), Cabbage (CC), Rapeseed (AACC), Indian mustard (AABB), and other common vegetables, showing the genetic relationships between Chromosomes from each of the genomes A, B, and C are represented by different colors.
Salinity Tolerance in Brassica
Salt stress tolerance is one of the highly complex processes in number of plant species. Many complex mechanisms are involved at different plant developmental levels. But some of these mechanisms are functional at a particular time in a given species. Apart from this, the effect of one process can exclude the effect of the other process at a particular time (Gorham et al., 1991; Ashraf, 1994; Yeo, 1998; Carvajal et al., 1999). Salt stress tolerance in plants is a developmentally regulated phenomenon and tolerance at one stage of development sometimes may not correlate with the stress tolerance at other stages. For example, in barely, corn, rice, tomato, and wheat, salt stress tolerance tends to increase as the plants become older. The situation becomes more complicated with polyploidy species in comparison with their respective diploid parents. Polyploid species can withstand the harmful environmental factors such as salt stress tolerance better than their own respective diploid parents. Different studies gives the indications that the amphidiploid species, Brassica carinata, B. juncea, and B. napus have the superiority over the diploid species, B. campestries, B. nigra, and B. oleraceae in terms of salinity tolerance (Ashraf et al., 2001). It was also found that the amphidiploid genotypes, B. carinata and B. napus were salt stress tolerant when compared with B. campestries. Another amphidiploid B. juncea is intermediate in salt stress tolerance (Ashraf and McNeilly, 1990). The continued survival of salt stress tolerant plants and the differences between the genotypes with salt sensitive plant species point out the presence of a genetic basis of salt stress tolerance.
Utilization of Brassica Genetic Diversity
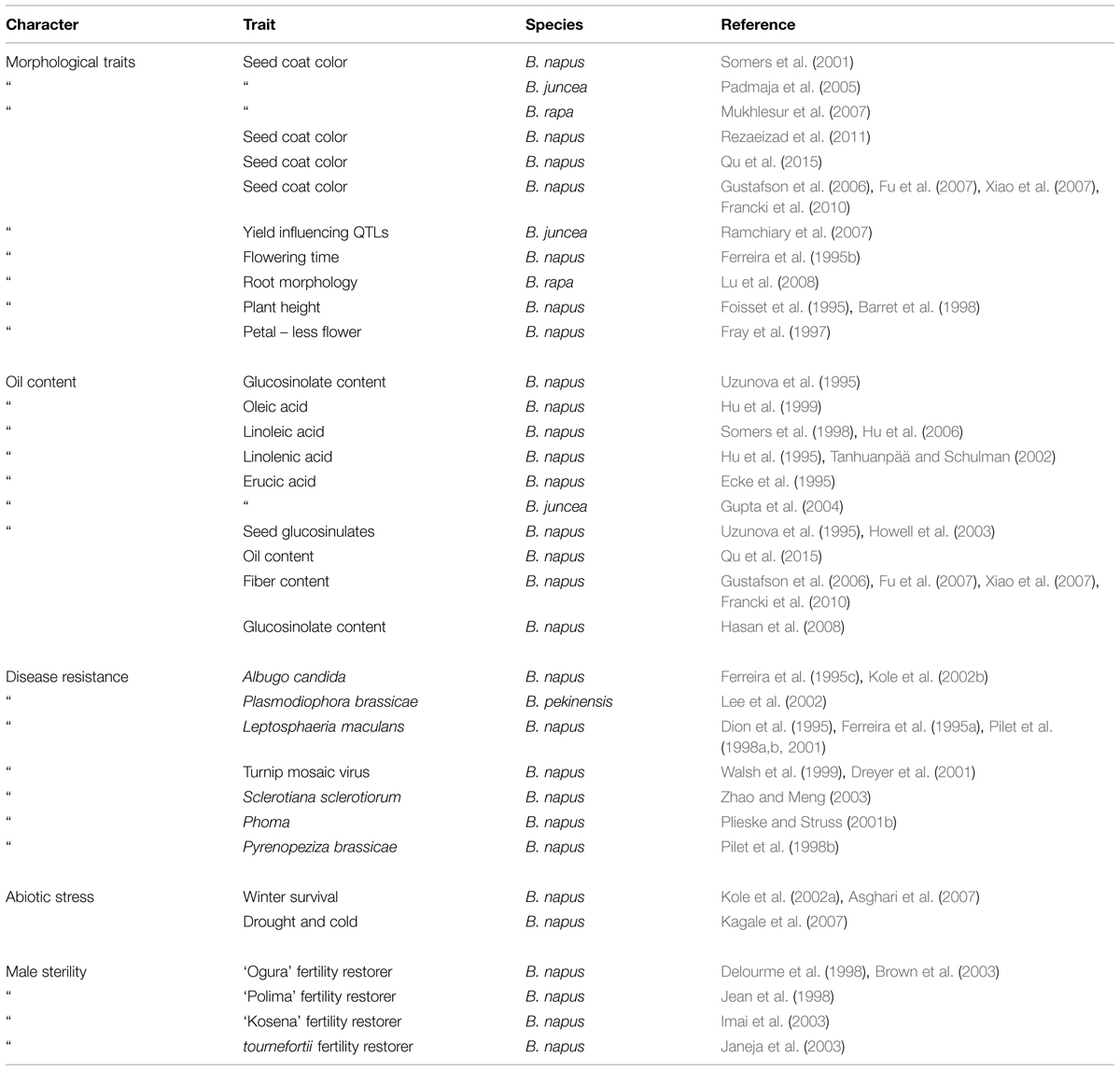
In the presence of environmental stresses, such as drought, salt, cold, nutrient deficiency, and water logging, growth of the Brassica plants, their oil production and reproduction capabilities are reduced. Hence, these Brassica species are normally grown on standard non-saline conditions in order to maximize the yield. If they are grown on salt affected soils, yield losses are severe. Therefore, improvement of their salt stress tolerance is of considerable economic value. For breeding program to be successful, presence of a significant heritable distinction with in the gene pool of these crops is a compulsory requirement (Becker et al., 1995; O’Neill et al., 2003; Kumar, 2015). Because of the close relationship and the presences of important inter and intra specific distinctions within Brassica species, the breeding programs for salinity stress tolerance have been highly benefited. It is also believed that support from other approaches such as mutagenesis, fusion of protoplast or recombinant technology can also be helpful in achieving the desired target (Diers et al., 1996; Riaz et al., 2001; Seyis et al., 2003). Even though there is a great inter and intra specific variation for salt stress tolerance within Brassica species, generation of new variation through induced mutation and utilization of those new variants gives more scope for enhancing salt stress tolerance. Due to the advancements of molecular techniques, the mutants can be identified and analyzed using DNA fingerprinting and mapping on PCR based markers such as SSR, RAPD, AFLP, and STMS (Diers and Osborn, 1994; Halldén et al., 1994; Thormann et al., 1994; Plieske and Struss, 2001a).
Molecular Markers in Breeding for Salinity Tolerance
Molecular genetics is one of the most important technologies in the today’s world. Stress tolerance and yield are difficult to breed using conventional methods because of their polygenic nature and are also largely influenced by environment and genotype. The complex quantitative feature of the most mechanisms involved in the salt stress tolerance is the main reason for the limited success of the modern salt tolerance breeding approaches (Yeo and Flowers, 1986). The association and application of the indirect selection markers which are genetically linked with the trait (s) of interest is a well-known approach for the betterment of the crop having difficult traits which includes salt stress tolerance (Im et al., 2014a). DNA marker technology has revolutionized the genome research and breeding in the recent decades. Implementation of various available markers and QTL mapping techniques have contributed for the good knowledge of genetic bases of various agriculturally significant traits such as resistance to biotic stresses, abiotic stresses tolerance, yield and nutritional quality in various crops (Xue et al., 2010; Ali et al., 2013).
Since, breeders use QTL linked markers to find the position of markers on the loci that controls the concerned traits; the number of methods to identify the phenotype is reduced. Therefore, the necessity for large-scale methods over time and space is significantly reduced. A few salt stress related QTL detected by SSR markers have been listed in the Table 1.
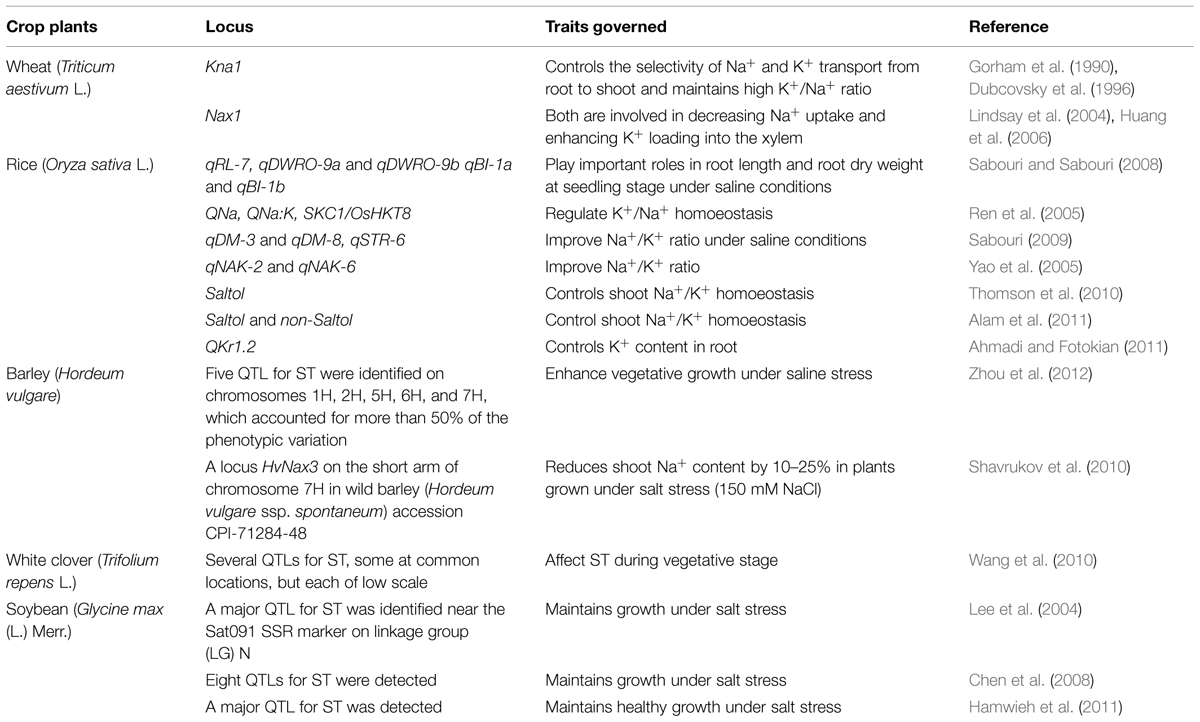
TABLE 1. Identification of quantitative trait loci (QTLs) by SSR markers for salt tolerance (ST) in different plant species.
In hexaploid bread wheat (Triticum aestivum), important locus (Kna1) has been reported that regulate the transport of Na+/K+ from root to shoot specifically, by containing a lower Na+/K+ ratio within the leaves (Gorham et al., 1987, 1990; Dubcovsky et al., 1996; Luo et al., 1996). Meanwhile, in durum Triticum turgidum L. ssp. durum Desf. (wheat) discharge process of Na+ is linked to Nax1 (Na+ exclusion 1; Huang et al., 2006, 2008), that might be related to the HKT8 (HKT1;5) and HKT7 (Na+ transporters HKT1;4). It has been reported that Nax1 loci efficiently reduce Na+ passage to shoot from root, by keeping Na+/K+ balance within the leaf of wheat by loading K+ into and excluding Na+ from, the xylem of the plant (James et al., 2006). Using F2 population of a hybrid within indica rice cultivar ‘IR36’ and japonica rice cultivar ‘Jiucaiqing,’ two QTLs identified for root Na+/K+ ratio, which were mapped to chromosomes 2 and 6 (Yao et al., 2005). For Salt tolerance traits different QTLs have been recognized in rice which include those at chromosome number 1- Saltol QTL, QNa, and SKC1/OsHKT8 along with, QNa:K on chromosome 4. Saltol describes many changes for the uptake of ion during salinity stress (Bonilla et al., 2002; Gregorio et al., 2002). For highest Na+ uptake QNa is QTL (Flowers et al., 2000). For Na+/K+, QNa:K is the corresponding QTL (Singh et al., 2001). For regulation of K+/Na+ ratio for homoeostasis in salt stress tolerant indica cultivar ‘Nona Bokra’ SKC1/OsHKT8 is the corresponding QTL (Lin et al., 2004; Ren et al., 2005). Also, many other QTLs are on the every chromosome except chromosome nine in the root for Na+/K+ ratio, and for exchange of ion three QTLs on chromosomes 10 and 3 (Sabouri and Sabouri, 2008), for tissue Na+/K+ ratio four QTLs and each for Na+ and K+ uptake on various chromosomes one QTL (Lang et al., 2001). Thereafter, 14 QTLs was identified for shoot and root Na+/K+ ratio and Na+ and K+ content on different rice chromosomes, recently (Ahmadi and Fotokian, 2011). Among these QTLs, on chromosome 1 for root K+ content, QKr1.2 was identified as one of very bright QTL as it explained around 30% of the variation of observed salt stress tolerance in rice. Furthermore, on rice chromosomes 8 and 10, two newly identified QTLs (SalTol8-1 and SalTol10-1) based on an F2 hybrid of a cross between a high salt stress tolerant line (IR61920-3B-22-2-1) and a medium salt stress tolerant line (BRRI-dhan40; Islam et al., 2011).
Also, in Hordeum vulgare L. (barley, Table 1), many studies have discovered QTLs for salt stress tolerance related phenotypes. Recently, 30 QTLs were identified for 10 different traits, such as K+ and shoot Na+ content, yield-related traits, several growth and Na+/K+ ratio, in populations grown on normal soil and salt affected soil. In the three species of Helianthus sp. (sunflower) and Helianthus paradoxus, ion-uptake traits related QTL analysis from highly salt affected habitat and its relative ancestor H. petiolaris and H. annuus which are both relatively salt sensitive, identified 14 ion uptake QTLs (Lexer et al., 2003). Additional studies are required to decide the benefits of unreported QTLs within crop breeding to improve salt stress tolerance. Since molecular marker techniques for breeding is economical and rapid, this technique is a very powerful method to enhance breeding programs to improve plant tolerance toward salinity. Especially, DNA markers are very important in plant breeding for the selection of polygenic traits, because of the absence of genotype X environment interaction, epistatic effect, and also ease in the picking up of homozygous plants and the homozygous lines can be greatly distinguished from the others at an early generation. Before the crosses of parental lines, molecular characterization of germplasm can help the genetic variations among the parental genotypes increase. Genetic diversity present in the breeding population is maximized and the labor time that is required for either direct selection in traditional breeding or in direct selection through QTLs minimizes. Even though this kind of procedure remains encouraging, its implementation to the complex traits such as salt stress tolerance may be limited due to the close genetic relationship between, wide confidence intervals and, big sample size requirement for screening of the segregating populations, parental population, and possible interactions between genotype and environment for QTL study.
Available Marker Systems in Brassica
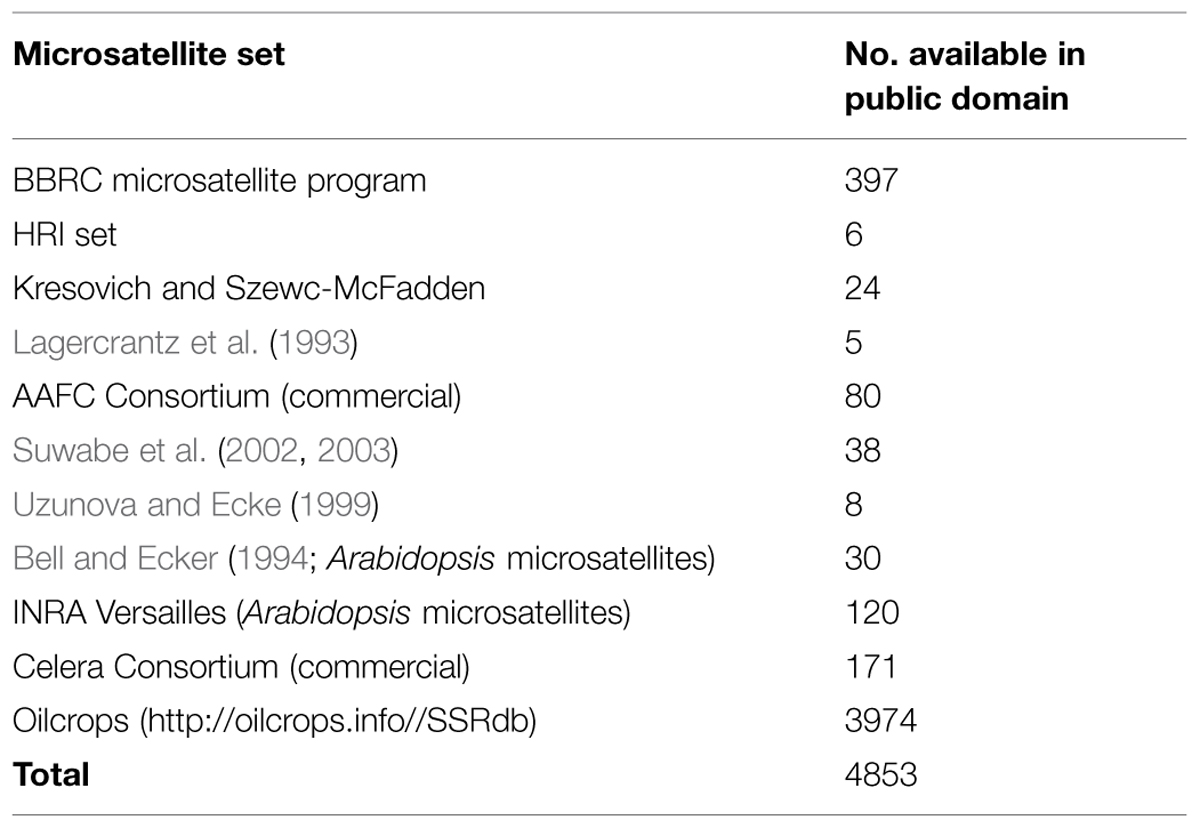
In Brassica, genome research with the application of marker assisted program began to emerge in the late 1980s when the first RFLP linkage map for B. oleraceae (Slocum et al., 1990), B. napus (Landry et al., 1991), and B. rapa (Song et al., 1991) was developed. For phylogenetic studies and genetic mapping in Brassica, RFLPs and RAPDs have been extensively used (Williams et al., 1990). However, the discovery of the PCR (Mullis and Faloona, 1987) leads the potential to increase the variety and density of marker in the already existing genetic maps with ISSR, AFLP and with the microsatellites (Grist et al., 1993), also called as SSR. SSRs are highly important resource of map-based alignment among distinct crosses, because of their robust, simple, and relatively inexpensive analysis and highly polymorphic nature. The number of available Brassica SSRs (microsatellite) primers is increasing (http://www.brassica.info/ssr/SSRinfo.htm) the list is given in the Table 2. Brassica genome integration greatly assisted the release of highly polymorphic mapped based, robust SSR markers of the entire B. nigra, B. rapa, B. napus, and B. oleracea genome into public domain. A large number of SSRs (microsatellite) markers have been developed among the cultivated Brassica species such as B. oleracea (AACC) and the diploids B. rapa (AA), B. nigra (BB), and B. oleracea (CC) which have been shown to be applicable within and between different Brassica species. One of the main limit to develop SSR markers in some Brassica crops is the lack of finished genome sequence. However, thanks to development of sequencing technology, B. rapa, B. oleracea, and B. napus are sequenced, recently (Wang et al., 2011; Yu et al., 2013; Shi et al., 2014; Sharma et al., 2015). From this sequence, 140998, 229389, 420991 mono- to hexanucleotide repeat microsatellites are identified using PERL5 script MIcroSAtellite (Thiel et al., 2003). From these identified microsatellites, 115869, 185662, and 356522 SSR markers were developed using in silico method, respectively (Shi et al., 2014; Table 3). In the past few years, the research work has clearly proven the power of candidate gene studies and genetic maps of high density for the location of molecular markers that are closely linked with the useful trait (s) within Brassica have been developed and most of them have been successfully integrated into the Brassica oilseed breeding programs (Table 4).
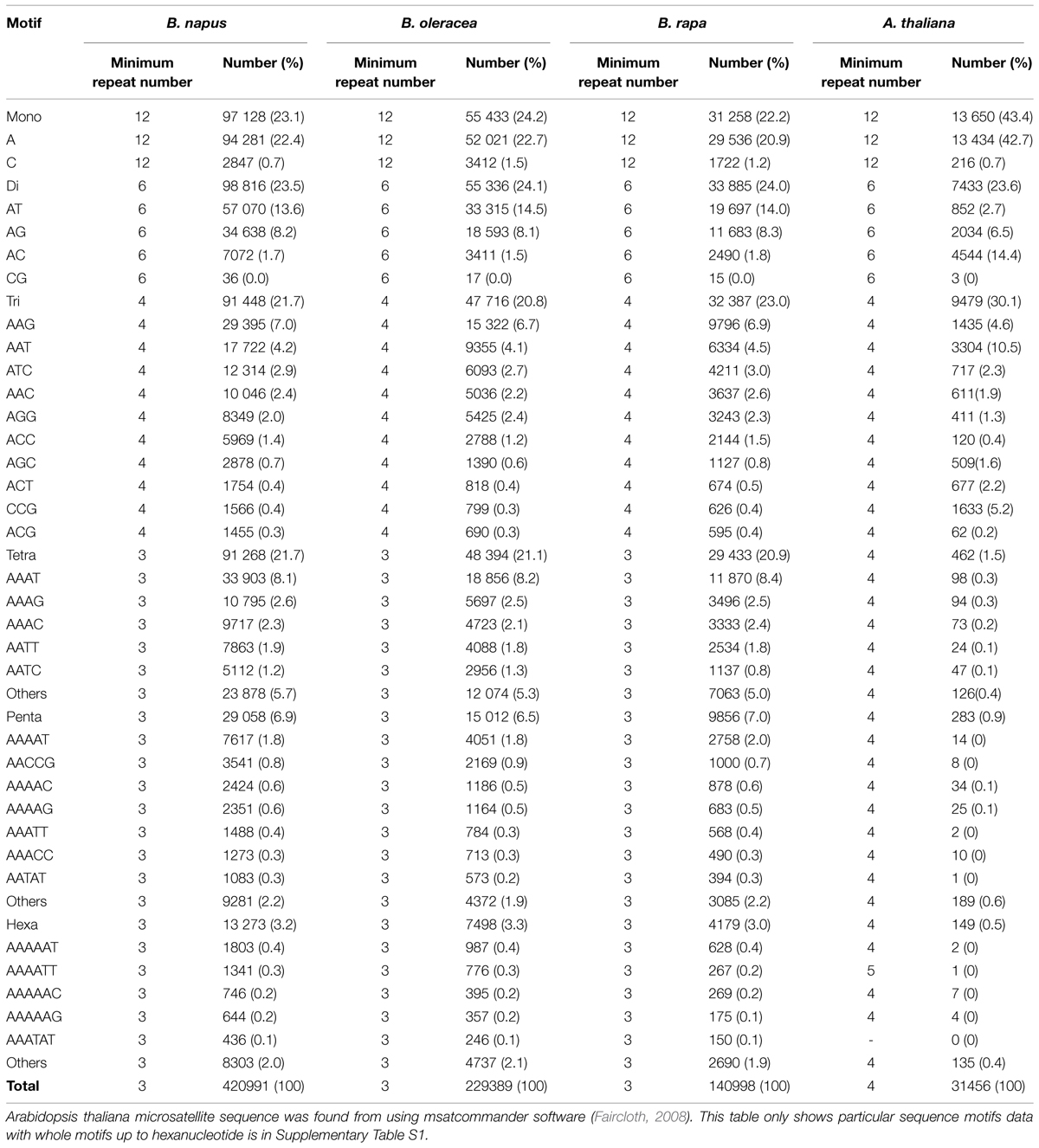
TABLE 3. Microsatellite sequence from the genome of Brassica rapa, B. oleracea, B. napus (Shi et al., 2014), and Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000).
Importance of Microsatellite (SSR) Marker
Over the past few years, various new PCR based marker such as AFLPs, RAPDs, and microsatellites have been developed and applied in crop improvement program. Microsatellites markers have great deal of potential among all the markers. Litt and Luty (1989) coined the term microsatellite. Microsatellites markers are also defined as simple sequence repeats (SSRs) which are based on unique DNA sequences that are flanking short repetitive traits of simple sequence motifs, for example – di or tri nucleotides. They are randomly distributed within the eukaryotic genomes (Smith and Devey, 1994). They are variable with respect to the number of repeats, pedigree analysis and are highly efficient in the fingerprinting and show co-dominant inheritance of different crops. The extra ordinary level of instructive polymorphism at SSR locus originated from the apparent tendency of development or replication or unequal crossing-over event at the time of meiosis. The strengths of SSR markers include their high numbers in eukaryotes, the codominance of alleles, and their arbitrary dispensation throughout the genome with special consortium with in low-copy regions (Morgante et al., 2002). Also, low quantities of template DNA (10–50 ng/reaction) are required, because of the PCR based technique. Reproducibility of SSR markers is high due to the use of lengthy PCR primers (Im et al., 2014b), and its use does not even require high quality DNA. Even though, the RFLP was one of the first markers used for genome analysis, RFLP technique is laborious and RFLP is less polymorphic than SSR marker. Also, improved technique that is more simple and efficient to find polymorphism in SSR marker makes SSR marker more useful (Kumar et al., 2015). Furthermore, since conventional microsatellite generating method using genomic libraries (Weising et al., 2005; Smykal et al., 2006) was replaced by in silico microsatellite generating method, many software generating microsatellite was made, such as MISA, MicrosatDesign, msatminer, msatcommander, IMEx, WebSat (Thiel et al., 2003; Singan and Colbourne, 2005; Thurston and Field, 2005; Mudunuri and Nagarajaram, 2007; Faircloth, 2008; Martins et al., 2009). These programs and genome sequence information make SSR marker generation procedure convenient. Therefore, the number of available SSR markers increases rapidly and available SSR marker in the genome becomes more dense. Therefore, the amount of SSR marker database is increasing. Even though, there are not so many reports about QTL for salt stress tolerance, many QTL has been identified and QTL information was enhanced by association mapping using SSR markers in Brassica crops. In Brassica napus, 53 SSR markers were found to be significantly associated to three phenolic fractions and 11 markers found to be associated to total phenolic acid contents. Among these markers, four SSR markers are derived from QTL for seed color (Rezaeizad et al., 2011). Twenty five and 11 SSR markers were found to be associated with seed coat color and oil content. Among these markers six SSR markers are associated with both of coat color and oil content (Qu et al., 2015). Also, association study between known major QTL and SSR marker is useful to find candidate gene because of high density of SSR marker in genome. Main QTL for seed color and fiber content on one of the homoeolog chromosomes A9 or C8 in B. napus has been described by many studies (Gustafson et al., 2006; Fu et al., 2007; Xiao et al., 2007). SSR markers bridge the sequence contig overlaying this QTL was identified and four of these SSR markers from small genomic region less than 50 kb which are strongly associated with seed color and fiber content traits were identified (Francki et al., 2010). These SSR markers in known major QTL or near these QTL which are strongly associated with the trait are useful to find candidate gene. SSR markers can be analyzed interspecifically. After mapping SSR markers in several species, comparative mapping of SSR markers can be made using alignment of orthologous loci (Yu et al., 2004; Zhang et al., 2007; Yu and Li, 2008). This will help in identifying the origin of QTL, find candidate gene as well as structural collinearity in genome between two species can also be dissected out.
The Candidate Gene Approach
Many traits of agricultural importance, which includes salinity tolerant traits, exhibits quantitative inheritance, which is mostly the result of multiple genes influenced by the environment. Due to the their imprecise localization on the genetic maps and multiplicity of genes defining a quantitative trait and their incomplete effects on phenotypic differentiation, the candidate gene approach is more adapted to the QTL characterization. The candidate gene approach has been emphasized as an encouraging method for combining QTL analysis with the large-scale data available on the cloning and genes characterization (Pflieger et al., 2001). Genes likely to be involved in the biochemical pathways, in this technique, that lead to a trait articulation, are engaged as molecular markers for QTL analysis. The Brassica species are the closest crop relative of the model crucifer Arabidopsis thaliana and the complete sequencing of this model crop has also created the way to relative examination into the complicated structure of Brassica genomes (Chalhoub et al., 2003). The comparative studies of flanked genome regions of known genes shows the extensive co-linearity between Arabidopsis and Brassica genome segments on a small scale level (Fourmann et al., 1998; Babula et al., 2003). Over the long chromosome stretches, the large scale synteny makes way to use the sequence data from the markers bound QTLs or genes of interest in Brassica to determine candidate genes from the chromosome segment in Arabidopsis. For example, different homeologs regions in B. napus and B. rapa that have different QTLs that regulate flowering time, each show useful similarity to the Arabidopsis chromosomal part containing a particular number of genes that influence the flowering time (Ferreira et al., 1995b; Lagercrantz et al., 1996).
Furthermore, comparative mapping between Brassica and Arabidopsis with SSR marker is helpful to identify candidate gene. Comparative mapping based on SSR marker between B. rapa and A. thaliana shows corresponding regions in A. thaliana for Crr1 and Crr2 which are QTL for club root resistance in B. rapa are in a small region of A. thaliana chromosome four where one of the region of disease resistance gene cluster has been identified. Therefore, it seems that the gene for club root resistance in B. rapa is related with disease resistance gene cluster in A. thaliana (Suwabe et al., 2006).
While comparing the salt stress tolerance of a particular Brassica species at the early growth stages, B. carinata, B. juncea, and B. napus, had better salinity tolerance than B. campestris (Ashraf and McNeilly, 1990). The reactions of four Brassica species, B. campestris, B. carinata, B. juncea, and B. napus to four different salts, CaCl2, MgCl2, NaCl, and Na2SO4, was tested at the seedling and germination stage using sand culture and solution (Ashraf et al., 1989). Effect of NaCl was more significantly within the effect of four salts and it inhibits the germination rate of all four species. There was no uniform connection between results for seedling growth and germination rate, exceptions in B. napus, which showed more seedling growth and better germination rate under the salinity stress as compared to the other three species.
Huge and still growing Arabidopsis EST database and the amalgamation into the comparative Brassica genome study helps fine mapping of the genomic re-arrangements and the recognition of regions that contains genes crop plants. Also recognition of the association of given traits in Brassica crops with the candidate genes of Arabidopsis and the generation of the molecular markers that have association with the corresponding genes (Panjabi et al., 2008). For example the Co (constans) gene isolated from Arabidopsis, which is involved in late flowering is a putative candidate gene (genes isolated in model species establish the putative CGs for the agronomic species) for two QTLs, which control the flowering time in B. napus (Putterill et al., 1995; Robert et al., 1998). Putative CGs involved in fatty acid metabolism, were mapped on the rapeseed genome. In B. rapa, a important QTL for flowering time was found in the region homologous to the Arabidopsis chromosome five top, where many genes that regulate flowering are located (Kole et al., 2001). In B. napus, many cold and drought induced genes were isolated and characterized and there is a huge correlation between the development of freezing tolerance and the expression of some of these genes, which seems to be up-regulated by cold stress (Kole et al., 2002a; Asghari et al., 2007; Kagale et al., 2007).
In B. oleracea, fifteen QTLs regulating the flowering time were situated to the Arabidopsis genomic segments that contain flowering time genes that affect flowering (Okazaki et al., 2007). Similarly large sets of Arabidopsis QTLs were found for Brassica QTLs that were influencing leaves and whole plant structure (Lan and Paterson, 2001). Also, based on synteny, SSR markers near the QTL for glucosinolate content in B. napus whose orthologs in A.thaliana is linked to candidate genes were identified and four putative candidate genes for glucosinolate biosynthesis were identified (Hasan et al., 2008.). Furthermore, Using in silico analysis about B. oleracea genome and synteny with A. thaliana, putative seven candidate major loci for regulating glucosinolate content were proposed (Sotelo et al., 2014.). Due to this Arabidopsis genome sequence is a very informative resource for identifying and further assessment of candidate genes that may account for the control of complicated traits in Brassica at the genetic level. But main difficulty in the application of genetic information of Arabidopsis for the map-based cloning, candidate gene identification and marker development in Brassica crop species is hindered by the complicated structure of the polyploidy Brassica genome.
Conclusion
Today’s agriculture certainly requires salt tolerant Brassicas for the very commercial purpose of the crop. Recent in-depth investigations at physiological and moleuclar levels have identified many ways by which wild type plants cope with salinity stress. Thanks to close relationship and the significant inter and intra specific variation within Brassica species which shows huge potential for breeding for salt stress tolerance in Brassica crops. Nonetheless, it is clear that to link the salt tolerance trait with QTL position on the chromosome, a proper breeding program assisted with the markers is a prerequisite.
Among the various molecular markers available for this purpose, SSRs are gaining huge attention. SSR markers have many advantages such as high polymorphism, relatively simple methods for identification and most importantly, small amounts of plant material is required for the PCR-based experiment. Other advantages of SSR markars are their random distribution, moderate genome coverage and co-dominant inheritance.
Since Brassica and Arabidopsis belongs to the same family, many studies and database of Arabidopsis are helpful to do breeding in order to improve salt tolerance. Although genome of B. Rapa (485 Mb) and B. Oleracea (630 Mb) is bigger than the genome of A. thaliana (Wang et al., 2011; Yu et al., 2013) and A. thaliana genome segments are dispersed and rearranged in many Brassica crops (Kowalski et al., 1994; Lagercrantz, 1998; Parkin et al., 2005), it has many genomic segments which are highly conserved. Therefore, comparative molecular marker mapping is quite informative. There are 140998, 229389, 420991 mono to hexanucleotide repeat SSRs from the genome of B. rapa, B. oleracea, and B. napus, respectively, out of which 31456 are microsatellite sequence which are candidates of SSR markers from the genome of A. thaliana (Table 3, Supplementary Data S1). There is tendancy that major motif sequences of candidates of SSR marker in both Brassica crops and A. thaliana are A/T rich and the motif sequences that are abundant in the genome of Brassica crops are also abundant in A. thaliana. Even microsatellite sequence in A. thaliana genome is much less than that in Brassica crops, the number of SSR marker is enough to show potential of comparative study of salt tolerance QTL using SSR marker in Brassica crops and A. thaliana. Furthermore, since EST-SSR marker developed from EST-database is more useful to find the candidate genes, the available huge EST-database is helpful for identifying the candidate gene from the QTL. Also, other physiological and agronomical traits can be studied with SSR markers to make a robust and healthy plants with high yield.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Mrs. Nisha Kumari for proof reading and editing of this paper. I would also like to thanks Jawaharlal Nehru University New Delhi, and Sogang University Seoul for providing logistics support and space to work. This work was supported by a grant from IAEA, Vienna and Sogang University, Republic of Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00688
References
Ahmadi, J., and Fotokian, M. H. (2011). Identification and mapping of quantitative trait loci associated with salinity tolerance in rice (Oryza sativa) using SSR markers. Iran. J. Biotechnol. 9, 21–30.
Alam, R., Sazzadur, R. M., Seraj, Z. I., Thomson, M. J., Ismail, A. M., Tumimbang-Raiz, E., et al. (2011). Investigation of seedling-stage salinity tolerance QTLs using backcross lines derived from Oryza sativa L. Pokkali. Plant Breeding 130, 430–437. doi: 10.1111/j.1439-0523.2010.01837.x
Ali, A., Yang, E. M., Lee, S. Y., and Chung, S. M. (2013). Evaluation of chloroplast genotypes of Korean cucumber cultivars (Cucumis sativus L.) using sdCAPS markers related to chilling tolerance. Korean J. Hortic. Sci. Technol. 31, 219–223. doi: 10.7235/hort.2013.12161
Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Asghari, A., Mohammadi, S. A., Mohgaddam, M., and Mohammaddoost, H. R. (2007). Identification of QTLS controlling winter survival in Brassica napus using RAPD markers. Biotechnol. Biotechnol. Equip. 21, 413–416. doi: 10.1080/13102818.2007.10817485
Ashraf, M. (1994). Breeding for salinity tolerance in plants. Crit. Rev. Plant Sci. 13, 17–42. doi: 10.1080/713608051
Ashraf, M., Bokhari, M. H., and Mehmood, S. (1989). Effects of four different salts on germination and seedling growth of four Brassica species. Biologia 35, 173–187.
Ashraf, M., and McNeilly, T. (1990). Responses of four Brassica species to sodium chloride. Environ. Exp. Bot. 30, 475–487. doi: 10.1016/0098-8472(90)90013-T
Ashraf, M., McNeilly, T., and Nazir, M. (2001). Comparative salt tolerance of amphidiploid and diploid Brassica species. Plant Sci. 160, 683–689. doi: 10.1016/S0168-9452(00)00449-0
Babula, D., Kaczmarek, M., Barakat, A., Delseny, M., Quiros, C. F., and Sadowski, J. (2003). Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol. Genet. Genomics 268, 656–665. doi: 10.1007/s00438-002-0782-2
Banerjee, S., Parasramka, M. A., and Sarkar, F. H. (2012). “Cellular, molecular and biological insight into chemopreventive and therapeutic potential of 3,3’-diindolylmethane (DIM),” in Nutraceuticals and Cancer, ed. F. H. Sarkar (New York, NY: Springer Netherlands), 111–133. doi: 10.1007/978-94-007-2630-7_6
Barret, P., Delourme, R., Foisset, N., and Renard, M. (1998). Development of a SCAR (sequence characterized amplified region) marker for molecular tagging of the dwarf BREIZH (Bzh) gene in Brassica napus L. Theor. Appl. Genet. 97, 828–833. doi: 10.1007/s001220050962
Becker, H. C., Engqvist, G. M., and Karlssom, B. (1995). Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theor. Appl. Genet. 91, 62–67. doi: 10.1007/BF00220859
Beecher, C. (1994). Cancer preventive properties of varieties of Brassica Oleaceae: a review. Am. J. Clin. Nutr. 59, 1166S–1170S.
Bell, C. J., and Ecker, J. R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. doi: 10.1006/geno.1994.1023
Bonilla, P., Dvorak, J., Mackill, D., Deal, K., and Gregorio, G. (2002). RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp. Agric. Sci. 85, 68–76.
Brown, G. G., Formanova, N., Jin, H., Wargachuk, R., Dendy, C., Patil, P., et al. (2003). The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35, 262–272. doi: 10.1046/j.1365-313X.2003.01799.x
Carvajal, M., Martinez, M., and Alcaraz, C. F. (1999). Physiological function of water channels as affected by salinity in roots of paprika pepper. Physiol. Plant. 105, 95–101. doi: 10.1034/j.1399-3054.1999.105115.x
Carvalhoa, I. S., Mirandab, I., and Pereira, H. (2006). Evaluation of oil composition of some crops suitable for human nutrition. Ind. Crops Prod. 24, 75–78. doi: 10.1016/j.indcrop.2006.03.005
Chalhoub, B., Budin, K., Aubourg, S., Belcram, H., Laffont, P., Delourme, R., et al. (2003). “Comparative genomic approaches for high-throughput physical and genetic mapping and gene cloning in oilseed rape (Brassica napus) and application to breeding,” in Proceedings of the 11th International Rapeseed Congress, Vol. 1, Copenhagen, 78.
Chen, H., Cui, S., Fu, S., Gai, J., and Yu, D. (2008). Identification of quantitative trait loci associated with salt tolerance during seedling growth in soybean (Glycine max L.). Aust. J. Agric. Res. 59, 1086–1091. doi: 10.1071/AR08104
Delourme, R., Foisset, N., Horvais, R., Barret, P., Champagne, G., Cheung, W. Y., et al. (1998). Characterisation of the radish introgression carrying the Rfo restorer gene for the Ogu-INRA cytoplasmic male sterility in rapeseed (Brassica napus L.). Theor. Appl. Genet. 97, 129–134. doi: 10.1007/s001220050876
Diers, B. W., McVetty, P. B. E., and Osborn, T. C. (1996). Relationship between heterosis and genetic distance based on restriction fragment length polymorphism markers in oilseed rape (Brassica napus L.). Crop Sci. 36, 79–83. doi: 10.2135/cropsci1996.0011183X003600010014x
Diers, B. W., and Osborn, T. C. (1994). Genetic diversity of oilseed Brassica napus germplasm based on restriction fragment length polymorphisms. Theor. Appl. Genet. 88, 662–668. doi: 10.1007/BF01253968
Dion, Y., Gugel, R. K., Rakow, G. F. W., Seguin-Swartz, G., and Landry, B. S. (1995). RFLP mapping of resistance to the blackleg disease [causal agent, Leptosphaeria maculans (Desm.) Ces. et de Not. In canola (Brassica napus L). Theor. Appl. Genet. 91, 1190–1194. doi: 10.1007/BF00220928
Divisi, D., Tommaso, D. S., Salvemini, S., Garramone, M., and Crisci, R. (2006). Diet and cancer. Acta. Biomed. 77, 118–123.
Dreyer, F., Graichen, K., and Jung, C. (2001). A major quantitative trait locus for resistance to turnip yellows virus (TuYV, syn. beet western yellows virus, BWYV) in rapeseed. Plant Breed. 120, 457–462. doi: 10.1046/j.1439-0523.2001.00646.x
Dubcovsky, J., Maria, G. S., Epstein, E., Luo, M. C., and Dvorak, J. (1996). Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor. Appl. Genet. 92, 448–454. doi: 10.1007/BF00223692
Ecke, W., Uzunova, M., and Weißleder, K. (1995). Mapping the genome of rapeseed (Brassica napus L.). II. Localization of genes controlling erucic acid systhesis and seed oil content. Theor. Appl. Genet. 91, 972–977. doi: 10.1007/BF00223908
Epstein, R. (1985). Extinction-induced resurgence: preliminary investigations and possible applications. Psychol. Rec. 35, 143–153.
Faircloth, B. C. (2008). msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol. Ecol. Resour. 8, 92–94. doi: 10.1111/j.1471-8286.2007.01884.x
FAO. (2008). FAO Land and Plant Nutrition Management Service. Available at: http://www.fao.org/ag/agl/agll/spush
Ferreira, M. E., Rimmer, S. R., Williams, P. H., and Osborn, T. C. (1995a). Mapping loci controlling Brassica napus resistance to Leptosphaeria maculans under different screening conditions. Phytopathology 85, 213–217. doi: 10.1094/Phyto-85-213
Ferreira, M. E., Satagopan, J., Yandell, B. S., Williams, P. H., and Osborn, T. C. (1995b). Mapping loci controlling vernalization requirement and flowering time in Brassica napus. Theor. Appl. Genet. 90, 727–732. doi: 10.1007/BF00222140
Ferreira, M. E., Williams, P. H., and Osborn, T. C. (1995c). Mapping of a locus controlling resistance to Albugo candida in Brassica napus using molecular markers. Phytopathology 85, 218–220.
Finley, J. W., Sigrid-Keck, A., Robbins, R. J., and Hintze, K. J. (2005). Selenium enrichment of Broccoli: interactions between selenium and secondary plant compounds. J. Nutr. 135, 1236–1238.
Flowers, T. J., Koyama, M. L., Flowers, S. A., Sudhakar, C., Singh, K. P., and Yeo, A. R. (2000). QTL: their place in engineering tolerance of rice to salinity. J. Exp. Bot. 51, 99–106. doi: 10.1093/jexbot/51.342.99
Flowers, T. J., and Yeo, A. R. (1995). Breeding for salinity resistance in crop plants. Where next? Aust. J. Plant Phys. 22, 875–884. doi: 10.1071/PP9950875
Foisset, N., Delourme, R., Barret, P., and Renard, M. (1995). Molecular tagging of the dwarf Breizh (Bzh) gene in Brassica napus. Theor. Appl. Genet. 91, 756–761. doi: 10.1007/BF00220955
Fourmann, M., Barret, P., Renard, M., Pelletier, G., Delourme, R., and Brunel, D. (1998). The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor. Appl. Genet. 96, 852–858. doi: 10.1007/s001220050812
Francki, M., Snowdon, R. J., Wittkop, B., Rezaidad, A., Hasan, M., Lipsa, F., et al. (2010). Regional association analysis delineates a sequenced chromosome region influencing antinutritive seed meal compounds in oilseed rape. Genome 53, 917–928. doi: 10.1139/g10-052
Fray, M. J., Puangsomlee, P., Goodrich, J., Coupland, G., Evans, E. J., Arthur, A. E., et al. (1997). The genetics of stamenoid petal production in oilseed rape (Brassica napus) and equivalent variation in Arabidopsis thaliana. Theor. Appl. Genet. 94, 731–736. doi: 10.1007/s001220050472
Fu, F. Y., Liu, L. Z., Chai, Y. R., Chen, L., Yang, T., Jin, M. Y., et al. (2007). Localization of QTLs for seed color using recombinant inbred lines of Brassica napus in different environments. Genome 50, 840–854. doi: 10.1139/G07-068
Gorham, J., Britol, A., Young, E. M., and Wyn Jones, R. G. (1991). The presence of the enhanced K+/Na+ discrimination trait in diploid Triticum species. Theor. Appl. Genet. 82, 729–736. doi: 10.1007/BF00227318
Gorham, J., Hardy, C., Jones, R. G. W., Joppa, L. R., and Law, C. N. (1987). Chromosomal l location of a K/Na discrimination character in the D-genome of wheat. Theor. Appl. Genet. 74, 584–588. doi: 10.1007/BF00288856
Gorham, J., Jones, R. G. W., and Bristol, A. (1990). Partial characterization of the trait for enhanced K+-Na+ discrimination in the D-genome of wheat. Planta 180, 590–597. doi: 10.1007/BF02411458
Gregorio, G. B., Senadhira, D., Mendoza, R. D., Manigbas, N. L., Roxas, J. P., and Guerta, C. Q. (2002). Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 76, 91–101. doi: 10.1016/S0378-4290(02)00031-X
Grieve, C. M., Suarez, D. L., and Shannon, M. C. (1999). Effect of saline irrigation water composition on selenium accumulation by wheat. J. Plant Nutr. 22, 1443–1450. doi: 10.1080/01904169909365725
Grist, S. A., Firgaira, F. A., and Morley, A. A. (1993). Dinucleotide repeat polymorphisms isolated by the polymerase chain reaction. Biotechniques 15, 304–309.
Gupta, V., Mukhopadhyay, A., Arumugam, N., Sodhi, Y. S., Pental, D., and Pradhan, A. K. (2004). Molecular tagging of erucic acid trait in oilseed mustard (Brassica juncea) by QTL mapping and single nucleotide polymorphisms in FAE1 gene. Theor. Appl. Genet. 108, 743–749. doi: 10.1007/s00122-003-1481-z
Gustafson, J. P., Badani, A. G., Snowdon, R. J., Wittkop, B., Lipsa, F. D., Baetzel, R., et al. (2006). Colocalization of a partially dominant gene for yellow seed colour with a major QTL influencing acid detergent fibre (ADF) content in different crosses of oilseed rape (Brassica napus). Genome 49, 1499–1509. doi: 10.1139/g06-091
Halldén, C., Nilsson, N. O., Rading, I. M., and Säll, T. (1994). Evaluation of RFLP and RAPD markers in comparison of Brassica napus breeding lines. Theor. Appl. Genet. 88, 123–128. doi: 10.1007/BF00222404
Hamwieh, A., Tuyen, D., Cong, H., Benitez, E., Takahashi, R., and Xu, D. (2011). Identification and validation of a major QTL for salt tolerance in soybean. Euphytica 179, 451–459. doi: 10.1007/s10681-011-0347-8
Hasan, M., Friedt, W., Pons-Kühnemann, J., Freitag, N. M., Link, K., and Snowdon, R. J. (2008). Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus). Theor. Appl. Genet. 116, 1035–1049. doi: 10.1007/s00122-008-0733-3
Howell, P. M., Sharpe, A. G., and Lydiate, D. J. (2003). Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus). Genome 46, 454–460. doi: 10.1139/g03-028
Hu, J., Li, G., Struss, D., and Quiros, C. F. (1999). SCAR amd RAPD markers associated with 18-carbon fatty acid rapeseed, Brassica napus. Plant Breed. 118, 145–150. doi: 10.1046/j.1439-0523.1999.118002145.x
Hu, J., Quiros, C., Arus, P., Struss, D., and Robbelen, G. (1995). Mapping of a gene determining linolenic acid concentration in rapeseed with DNA-based markers. Theor. Appl. Genet. 90, 258–262. doi: 10.1007/BF00222211
Hu, X., Sullivan-Gilbert, M., Gupta, M., and Thompson, S. A. (2006). Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus L.). Theor. Appl. Genet. 113, 497–507. doi: 10.1007/s00122-006-0315-1
Huang, S. B., Spielmeyer, W., Lagudah, E. S., James, R. A., Platten, J. D., Dennis, E. S., et al. (2006). A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 142, 1718–1727.
Huang, S., Spielmeyer, W., Lagudah, E. S., and Munns, R. (2008). Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J. Exp. Bot. 59, 927–937. doi: 10.1093/jxb/ern033
Im, C. H., Kim, M. K., Kim, K. H., Cho, S. J., Lee, J. J., Joung, W. K., et al. (2014a). Breeding of Pleurotus eryngii with a high temperature tolerance trait. J. Mushrooms 12, 187–192. doi: 10.4489/KJM.2013.41.2.91
Im, C. H., Kim, K. H., Je, H. J., Ali, A., Kim, M. K., Joung, W. K., et al. (2014b). Multiplex simple sequence repeat (SSR) markers discriminating Pleurotus eryngii cultivar. Korean J. Mycol. 42, 159–164. doi: 10.4489/KJM.2014.42.2.159
Imai, R., Koizuka, N., Fujimoto, H., Hayakawa, T., Sakai, T., and Imamura, J. (2003). Delimitation of the fertility restorer locus Rfk1 to a 43-kb contig in Kosena radish (Raphanus sativus L.). Mol. Genet. Genomics 269, 388–394. doi: 10.1007/s00438-003-0847-x
Islam, M. R., Salam, M. A., Hassan, L., Collard, B. C. Y., Singh, R. K., and Gregorio, G. B. (2011). QTL mapping for salinity tolerance in at seedling stage in rice. Emirates J. Food Agric. 23, 137–146. doi: 10.9755/ejfa.v23i2.6348
James, R. A., Davenport, R. J., and Munns, R. (2006). Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 142, 1537–1547. doi: 10.1104/pp.106.086538
Janeja, H. S., Banga, S. S., and Lakshmikumaran, M. (2003). Identification of AFLP markers linked to fertility restorer genes for tournefortii cytoplasmic male-sterility system in Brassica napus. Theor. Appl. Genet. 107, 148–154. doi: 10.1007/s00122-003-1225-0
Jean, M., Brown, G. G., and Landry, B. S. (1998). Targeted mapping approaches to identify DNA markers linked to the Rfp1 restorer gene for the ‘Polima’ CMS of canola (Brassica napus L.). Theor. Appl. Genet. 97, 431–438. doi: 10.1007/s001220050913
Kagale, S., Divi, U. K., Krochko, J. E., Keller, W. A., and Krishna, P. (2007). Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225, 353–364. doi: 10.1007/s00425-006-0361-6
Kole, C., Quijada, P., Michael, S. D., Amasino, R., and Osborn, T. C. (2001). Evidence for homology of flowering-time genes VFR2 from Brassica rapa and FLC from Arabidopsis thaliana. Theor. Appl. Genet. 102, 425–430. doi: 10.1007/s001220051663
Kole, C., Thormann, C. E., Karlsson, B. H., Palta, J. P., Gaffney, P., Yandell, B., et al. (2002a). Comparative mapping of loci controlling winter survival and related traits in oilseed Brassica rapa and B. napus. Mol. Breed. 9, 201–210. doi: 10.1023/A:1019759512347
Kole, C., Williams, P. H., Rimmer, S. R., and Osborn, T. C. (2002b). Linkage mapping of genes controlling resistance to white rust (Albugo candida) in Brassica rapa (syn. campestris) and comparative mapping to Brassica napus and Arabidopsis thaliana. Genome 45, 22–27. doi: 10.1139/g01-123
Kowalski, S. P., Lan, T. H., Feldmann, K. A., and Paterson, A. H. (1994). Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 138, 499–510.
Kumar, K., Kumar, M., Kim, S. R., Ryu, H., and Cho, Y. G. (2013). Insights into genomics of salt stress response in rice. Rice (N. Y.) 6, 27. doi: 10.1186/1939-8433-6-27
Kumar, M. (2013). Crop plants and abiotic stresses. J. Biomol. Res. Ther. 3, e125. doi: 10.4172/2167-7956.1000e125
Kumar, M. (2015). Detection of genetic variation in crop plants. J. Biomol. Res. Ther. 4, e140. doi: 10.4172/2167-7956.1000e140
Kumar, M., Kim, S. R., Sharma, P. C., and Pareek, A. (2015). Simple and efficient way to detect small polymorphic bands in plants. Genomics Data 5, 218–222. doi: 10.1016/j.gdata.2015.06.006
Kumar, M., Lee, S. C., Kim, J. Y., Kim, S. J., Aye, S. S., and Kim, S. R. (2014). Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 57, 383–393. doi: 10.1007/s12374-014-0487-1
Lagercrantz, U. (1998). Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150, 1217–1228.
Lagercrantz, U., Ellegren, H., and Andersson, L. (1993). The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 21, 1111–1115. doi: 10.1093/nar/21.5.1111
Lagercrantz, U., Putterill, J., Coupland, G., and Lydiate, D. (1996). Comparative mapping in Arabidopsis and Brassica, fine scale genome collinearity and congruence of genes controlling flowering time. Plant J. 9, 13–20. doi: 10.1046/j.1365-313X.1996.09010013.x
Lan, T. H., and Paterson, A. H. (2001). Comparative mapping of QTLs determining the plant size of Brassica oleracea. Theor. Appl. Genet. 103, 383–397. doi: 10.1007/s001220100615
Landry, B. S., Hubert, N., Etoh, T., Harada, J. J., and Lincoln, S. E. (1991). A genetic map for Brassica napus based on restriction fragment length polymorphisms detected with expressed DNA sequences. Genome 34, 543–552. doi: 10.1139/g91-084
Lang, N. T., Yanagihara, S. S., and Buu, B. C. (2001). A microsatellite marker for a gene conferring salt tolerance on rice at the vegetative and reproductive stages. Sabrao J. Breed. Genet. 33, 1–10.
Lee, G. P., Baek, N. K., and Park, K. W. (2002). Genetic mapping of resistant genes in Brassica pekinensis against Plasmodiophora brassicae race 6. Plant Pathol. J. 18, 266–270. doi: 10.5423/PPJ.2002.18.5.266
Lee, G. J., Carter, T. E. Jr., Villagarcia, M. R., Li, Z., Zhou, X., Gibbs, M. O., et al. (2004). A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor. Appl. Genet. 109, 1610–1619. doi: 10.1007/s00122-004-1783-9
Lexer, C., Welch, M. E., Durphy, J. L., and Rieseberg, L. H. (2003). Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol. Ecol. 12, 1225–1235. doi: 10.1046/j.1365-294X.2003.01803.x
Lin, H. X., Zhu, M. Z., Yano, M., Gao, J. P., Liang, Z. W., Su, W. A., et al. (2004). QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 108, 253–260. doi: 10.1007/s00122-003-1421-y
Lindsay, M. P., Lagudah, E. S., Hare, R. A., and Munns, R. (2004). A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct. Plant Biol. 31, 1105–1114. doi: 10.1071/FP04111
Litt, M., and Luty, J. A. (1989). A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Amr. J. Hum. Genet. 44, 397–401.
Lu, G., Cao, J., Yu, X., Xiang, X., and Chen, H. (2008). Mapping QTLs for root morphological traits in Brassica rapa L. based on AFLP and RAPD markers. J. Appl. Genet. 49, 23–31. doi: 10.1007/BF03195245
Luo, M. C., Dubcovsky, J., Goyal, S., and Dvorak, J. (1996). Engineering of interstitial foreign chromosome segments containing the K+/Na+ selectivity gene Kna1 by sequential homoeologous recombination in durum wheat. Theor. Appl. Genet. 93, 1180–1184. doi: 10.1007/BF00230144
Martins, W. S., Lucas, D. C. S., Neves, K. F. S., and Bertioli, D. J. (2009). WebSat - A Web Software for MicroSatellite marker development. Bioinformation 3, 282–283. doi: 10.6026/97320630003282
Morgante, M., Hanafey, H., and Powell, W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genome. Nature Genet. 30, 194–200. doi: 10.1038/ng822
Mudunuri, S. B., and Nagarajaram, H. A. (2007). IMEx: imperfect microsatellite extractor. Bioinformatics 23, 1181–1187. doi: 10.1093/bioinformatics/btm097
Mukhlesur, R., Mcvetty, P. B. E., and Li, G. (2007). Development of SRAP, SNP and Multiplexed SCAR molecular markers for the major seed coat color gene in Brassica rapa L. Theor. Appl. Genet. 115, 1101–1107. doi: 10.1007/s00122-007-0636-8
Mullis, K., and Faloona, F. (1987). Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 44, 224–249.
Nagaharu, U. (1935). Genome analysis in Brassica with special reference to the experimental formation of Brassica napus and peculiar mode of fertilization. Japan J. Bot. 7, 389–452.
Okazaki, K., Sakamoto, K., Kikuchi, R., Saito, A., Togashi, E., Kuginuki, Y., et al. (2007). Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 114, 595–608. doi: 10.1007/s00122-006-0460-6
O’Neill, R., Snowdon, R. J., and Köhler, W. (2003). Population genetics aspects of biodiversity. Prog. Bot. 64, 115–137. doi: 10.1007/978-3-642-55819-1_7
Padmaja, K. L., Arumugam, N., Gupta, V., Mukhopadhyay, A., Sodhi, Y. S., Pental, D., et al. (2005). Mapping and tagging of seed coat colour and the identifcation of markers for marker-assisted manipulation of the trait in Brassica juncea microsatellite. Theor. Appl. Genet. 111, 8–14. doi: 10.1007/s00122-005-1933-8
Panjabi, P., Jagannath, A., Bisht, N. C., Padmaja, K. L., Sharma, S., Gupta, V., et al. (2008). Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9:113. doi: 10.1186/1471-2164-9-113
Parkin, I. A. P., Gulden, S. M., Sharpe, A. G., Lukens, L., Trick, M., Osborn, T. C., et al. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171, 765–781. doi: 10.1534/genetics.105.042093
Pflieger, S., Lefebvre, V., and Causse, M. (2001). The candidate gene approach in plant genetics: a review. Mol. Breed. 7, 275–291. doi: 10.1023/A:1011605013259
Pilet, M. L., Delourme, R., Foisset, N., and Renard, M. (1998a). Identification of loci contributing to quantitative resistance to blackleg disease, causal agent Leptsosphaeria maculans (Desm.) Ces. et de Not., in winter rapseed (Brassica napus L.). Theor. Appl. Genet. 96, 23–30. doi: 10.1007/s001220050704
Pilet, M. L., Delourme, R., Foisset, N., and Renard, M. (1998b). Identification of QTL involved in field resistance to light leaf spot (Pyrenopeziza brassicae) and blackleg resistance (Leptosphaeria maculans) in winter rapeseed (Brassica napus L.). Theor. Appl. Genet. 97, 398–406. doi: 10.1007/s001220050909
Pilet, M. L., Duplan, G., Archipiano, H., Barret, P., Baron, C., Horvais, R., et al. (2001). Stability of QTL for field resistance to blackleg across two genetic backgrounds in oilseed rape. Crop Sci. 41, 197–205. doi: 10.2135/cropsci2001.411197x
Plieske, J., and Struss, D. (2001a). Microsatellite markers for genome analysis in Brassica. I. development in Brassica napus and abundance in Brassicaceae species. Theor. Appl. Genet. 102, 689–694. doi: 10.1007/s001220051698
Plieske, J., and Struss, D. (2001b). STS markers linked to Phoma resistance genes of the Brassica B-genome revealed sequence homology between Brassica nigra and Brassica napus. Theor. Appl. Genet. 102, 483–488. doi: 10.1007/s001220051671
Putterill, J., Robson, F., Lee, K., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Qu, C., Hasan, M., Lu, K., Liu, L., Zhang, K., Fu, F., et al. (2015). Identification of QTL for seed coat colour and oil content in Brassica napus by association mapping using SSR markers. Can. J. Plant Sci. 95, 387–395. doi: 10.4141/cjps2013-411
Ramchiary, N., Padmaja, K. L., Sharma, S., Gupta, V., Sodhi, Y. S., Mukhopadhyay, A., et al. (2007). Mapping of yield influencing QTL in Brassica juncea: implications for breeding of a major oilseed crop of dryland areas. Theor. Appl. Genet. 115, 807–817. doi: 10.1007/s00122-007-0610-5
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Rezaeizad, A., Wittkop, B., Snowdon, R., Hasan, M., Mohammadi, V., Zali, A., et al. (2011). Identification of QTLs for phenolic compounds in oilseed rape (Brassica napus L.) by association mapping using SSR markers. Euphytica 177, 335–342. doi: 10.1007/s10681-010-0231-y
Riaz, A., Li, G., Quresh, Z., Swati, M. S., and Quiros, C. F. (2001). Genetic diversity of oilseed Brassica napus inbred lines based on sequence-related amplified polymorphism and its relation to hybrid performance. Plant Breed. 120, 411–415. doi: 10.1046/j.1439-0523.2001.00636.x
Robert, L. S., Robson, F., Sharpe, A., Lydiate, D., and Coupland, G. (1998). Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus. Plant Mol. Biol. 37, 763–772. doi: 10.1023/A:1006064514311
Sabouri, H. (2009). QTL detection of rice grain quality traits by microsatellite markers using an indica rice (Oryza sativa L.) combination. J. Genet. 88, 81–85. doi: 10.1007/s12041-009-0011-4
Sabouri, H., and Sabouri, A. (2008). New evidence of QTLs attributed to salinity tolerance in rice. Afr. J. Biotechnol. 7, 4376–4383. doi: 10.1007/s11033-013-2561-z
Seyis, F., Snowdon, R. J., Lühs, W., and Friedt, W. (2003). Molecular characterization of novel resynthesized rapeseed (Brassica napus) lines and analysis of their genetic diversity in comparison with spring rapeseed cultivars. Plant Breed. 122, 473–478. doi: 10.1111/j.1439-0523.2003.00859.x
Sharma, R., Mishra, M., Gupta, B., Parsania, C., Singla-Pareek, S. L., and Pareek, A. (2015). De novo assembly and characterization of stress transcriptome in a salinity tolerant variety CS52 of Brassica juncea. PLoS ONE 10:e0126783. doi: 10.1371/journal.pone.0126783
Shavrukov, Y., Gupta, N. K., Miyazaki, J., Baho, M. N., Chalmers, K. J., Tester, M., et al. (2010). HvNax3-a locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum). Funct. Integr. Genomics 10, 277–291. doi: 10.1007/s10142-009-0153-8
Shi, J., Huang, S., Zhan, J., Yu, J., Wang, X., Hua, W., et al. (2014). Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res. 21, 53–68. doi: 10.1093/dnares/dst040
Singan, V., and Colbourne, J. K. (2005). MicrosatDesign is a Pipeline for Transforming Sequencer Trace Files into DNA Markers. CGB Technical Report 2005-01. Bloomington, IN: The Center for Genomics and Bioinformatics, Indiana University.
Singh, R. K., Mishra, B., and Vandna, J. (2001). Segregations for alkalinity tolerance in three rice crosses. SABRAO J. 33, 31–34.
Slocum, M. K., Figdore, S. S., Kennard, W. C., Suzuki, J. Y., and Osborn, T. C. (1990). Linkage arrangement of restriction fragment length polymorphisms in Brassica oleracea. Theor. Appl. Genet. 80, 57–64. doi: 10.1007/BF00224016
Smith, D. N., and Devey, M. E. (1994). Occurrence and inheritance of microsatellites in Pinus radia. Genome 37, 977–983. doi: 10.1139/g94-138
Smykal, P., Weising, K., Nybom, H., Wolf, K., and Kahl, G. (2006). DNA fingerprinting in plants. principles, methods and applications, 2nd edition. Biol. Plant. 50, 799. doi: 10.1007/s10535-006-0138-6
Somers, D. J., Friesen, K. R. D., and Rakow, G. (1998). Identifcation of molecular markers associated with linoleic acid desaturation in Brassica napus. Theor. Appl. Genet. 96, 897–903. doi: 10.1007/s001220050817
Somers, D. J., Rakow, G., Prabhu, V. K., and Friesen, K. R. D. (2001). Identification of a major gene and RAPD markers for yellow seed coat color in B. napus. Genome 44, 1077–1082. doi: 10.1139/gen-44-6-1077
Song, K. M., Suzuki, J. Y., Slocum, M. K., Williams, P. H., and Osborn, T. C. (1991). A linkage map of Brassica rapa (syn. campestris) based on restriction fragment length polymorphisms. Theor. Appl. Genet. 82, 296–304. doi: 10.1007/BF02190615
Sotelo, T., Soengas, P., Velasco, P., Rodríguez, V. M., and Cartea, M. E. (2014). Identification of metabolic qtls and candidate genes for glucosinolate synthesis in Brassica oleracea leaves, seeds and flower buds. PLoS ONE 9:e91428. doi: 10.1371/journal.pone.0091428
Suwabe, K., Iketani, H., Nunome, T., Kage, T., and Hirai, M. (2002). Isolation and characterization of microsatellites in Brassica rapa L. Theor. Appl. Genet. 104, 1092–1098. doi: 10.1007/s00122-002-0875-7
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Fujimura, M., Nunome, T., et al. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 107, 997–1002. doi: 10.1007/s00122-003-1309-x
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Kondo, M., Fujimura, M., et al. (2006). Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173, 309–319. doi: 10.1534/genetics.104.038968
Tanhuanpää, P., and Schulman, A. (2002). Mapping of genes affecting linolenic acid content in Brassica rapa ssp. oleifera. Mol. Breed. 10, 51–62. doi: 10.1023/A:1020357211089
Thiel, T., Michalek, W., Varshney, R. K., and Graner, A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422. doi: 10.1007/s00122-002-1031-0
Thomson, M. J., De, O. M., Egdane, J., Rahman, M. A., Sajise, A. G., Adorada, D. L., et al. (2010). Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3, 148–160. doi: 10.1007/s12284-010-9053-8
Thormann, C. E., Ferreira, M. E., Camargo, L. E. A., Tivang, J. G., and Osborn, T. C. (1994). Comparison of RFLP and RAPD markers to estimating genetic relationships within and among cruciferous species. Theor. Appl. Genet. 88, 973–980. doi: 10.1007/BF00220804
Thurston, M., and Field, D. (2005). Msatminer: Detection and Characterization of Microsatellites. Oxford: Centre for Ecology & Hydrology.
Uzunova, M. I., and Ecke, W. (1999). Abundance, polymorphism and genetic mapping of microsatellites in oilseed rape (Brassica napus L.). Plant Breeding 118, 323–326. doi: 10.1046/j.1439-0523.1999.00371.x
Uzunova, M., Ecke, W., Weissleder, K., and Robbelen, G. (1995). Mapping the genome of rapeseed (Brassica napus L.). I. Construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor. Appl. Genet. 90, 194–204. doi: 10.1007/BF00222202
Walsh, J. A., Sharpe, A. G., Jenner, C. E., and Lydiate, D. J. (1999). Characterisation of resistance to turnip mosaic virus in oilseed rape (Brassica napus) and genetic mapping of TuRB01. Theor. Appl. Genet. 99, 1149–1154. doi: 10.1007/s001220051319
Wang, J., Drayton, M. C., George, J., Cogan, N. O., Baillie, R. C., Hand, M. L., et al. (2010). Identification of genetic factors influencing salt stress tolerance in white clover (Trifolium repens L.) by QTL analysis. Theor. Appl. Genet. 120, 607–619. doi: 10.1007/s00122-009-1179-y
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Weising, K., Nybom, H., Wolff, K., and Kahl, G. (2005). DNA Fingerprinting in Plants: Principles, Methods, and Applications, 2nd Edn. Boca Raton, FL: CRC Press Taylor & Francis Group.
Weiss, E. (1983). Oilseed Crops, Tropical Agriculture Series. London: Longman Scientific and Technical, 530–564.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A., and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18, 6531–6535. doi: 10.1093/nar/18.22.6531
Xiao, S., Xu, J., Li, Y., Zhang, L., Shi, S., Shi, S., et al. (2007). Generation and mapping of SCAR and CAPS markers linked to the seed coat color gene in Brassica napus using a genome-walking technique. Genome 50, 611–618. doi: 10.1139/G07-044
Xue, D. W., Zhou, M. X., Zhang, X. Q., Chen, S., Wei, K., Zeng, F. R., et al. (2010). Identification of QTLs for yield and yield components of barley under different growth conditions. J. Zhejiang Univ. Sci. B 11, 169–176. doi: 10.1631/jzus.B0900332
Yamaguchi, T., and Blumwald, E. (2005). Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci. 10, 615–620. doi: 10.1016/j.tplants.2005.10.002
Yao, M. Z., Wang, J. F., Chen, H. Y., Zhai, H. Q., and Zhang, H. S. (2005). Inheritance and QTL mapping of salt tolerance in rice. Rice Sci. 12, 25–32.
Yeo, A. R. (1998). Molecular biology of salt tolerance in the context of whole-plant physiology. J. Exp. Bot. 49, 915–929. doi: 10.1093/jxb/49.323.915
Yeo, A. R., and Flowers, T. J. (1986). “Salinity resistance in rice and a pyramiding approach to breeding varieties for saline soils,” in Plant Growth, Drought, and Salinity, eds N. C. Turner and J. B. Passioura (Melbourne, VIC: CSIRO), 161–173.
Yu, H., and Li, Q. (2008). Exploiting EST databases for the development and characterization of EST-SSRs in the Pacific Oyster (Crassostrea gigas). J. Hered. 99, 208–214. doi: 10.1093/jhered/esm124
Yu, J. K., La Rota, M., Kantety, R. V., and Sorrells, M. E. (2004). EST derived SSR markers for comparative mapping in wheat and rice. Mol. Genet. Genomics 271, 742–751. doi: 10.1007/s00438-004-1027-3
Yu, J., Zhao, M., Wang, X., Tong, C., Huang, S., Tehrim, S., et al. (2013). Bolbase: a comprehensive genomics database for Brassica oleracea. BMC Genomics 14:664. doi: 10.1186/1471-2164-14-664
Zhang, Y., Sledge, M. K., and Bouton, J. H. (2007). Genome mapping of white clover (Trifolium repens L.) and comparative analysis within the Trifolieae using cross-species SSR markers. Theor. Appl. Genet. 114, 1367–1378. doi: 10.1007/s00122-007-0523-3
Zhao, J., and Meng, J. (2003). Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor. Appl. Genet. 106, 759–764. doi: 10.1007/s00122-002-1171-2
Keywords: Brassica, salt stress, abiotic stress, SSR markers, QTL
Citation: Kumar M, Choi J-Y, Kumari N, Pareek A and Kim S-R (2015) Molecular breeding in Brassica for salt tolerance: importance of microsatellite (SSR) markers for molecular breeding in Brassica. Front. Plant Sci. 6:688. doi: 10.3389/fpls.2015.00688
Received: 22 April 2015; Accepted: 20 August 2015;
Published: 04 September 2015.
Edited by:
Sarvajeet Singh Gill, Maharshi Dayanand University, IndiaReviewed by:
Juan Francisco Jimenez Bremont, Instituto Potosino de Investigacion Cientifica y Tecnologica, MexicoNarottam Dey, Visva-Bharati University, India
Copyright © 2015 Kumar, Choi, Kumari, Pareek and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manu Kumar and Seong-Ryong Kim, Plant Molecular Biology Laboratory, Department of Life Science, Sogang University, Seoul 121-742, South Korea, manukumar007@gmail.com; manukumar7@sogang.ac.kr; sungkim@sogang.ac.kr
 Manu Kumar
Manu Kumar Ju-Young Choi
Ju-Young Choi Nisha Kumari
Nisha Kumari Ashwani Pareek
Ashwani Pareek Seong-Ryong Kim
Seong-Ryong Kim