- 1Commonwealth Scientific and Industrial Research Organization Agriculture, Canberra, ACT, Australia
- 2State Key Laboratory of Cotton Biology, Institute of Cotton Research of Chinese Academy of Agricultural Sciences, Anyang, China
- 3Commonwealth Scientific and Industrial Research Organization Land and Water, Canberra, ACT, Australia
- 4Indian Council of Agricultural Research, Central Plantation Crops Research Institute, RC, Assam, India
- 5Commonwealth Scientific and Industrial Research Organization Food and Nutrition, Canberra, ACT, Australia
The safflower (Carthamus tinctorius L.) is considered a strongly domesticated species with a long history of cultivation. The hybridization of safflower with its wild relatives has played an important role in the evolution of cultivars and is of particular interest with regards to their production of high quality edible oils. Original safflower varieties were all rich in linoleic acid, while varieties rich in oleic acid have risen to prominence in recent decades. The high oleic acid trait is controlled by a partially recessive allele ol at a single locus OL. The ol allele was found to be a defective microsomal oleate desaturase FAD2-1. Here we present DNA sequence data and Southern blot analysis suggesting that there has been an ancient hybridization and introgression of the FAD2-1 gene into C. tinctorius from its wild relative C. palaestinus. It is from this gene that FAD2-1Δ was derived more recently. Identification and characterization of the genetic origin and diversity of FAD2-1 could aid safflower breeders in reducing population size and generations required for the development of new high oleic acid varieties by using perfect molecular marker-assisted selection.
Introduction
Hybridization between domestic crop plants and their wild relatives, and the potential for gene introgression, has received widespread interest in recent years (Ellstrand et al., 1999; Jarvis and Hodgkin, 1999; Felber et al., 2007; Arrigo et al., 2011). Spontaneous hybridization between domesticated and wild plants generally occurs wherever the crop plants remain in their place of origin. Genetic introgression has been defined as “the permanent incorporation of genes from one set of differentiated populations into another” (Stewart et al., 2003), and the gene introgression from wild relatives has long been considered important for the evolution of domestic crop species (Stebbins, 1959; Harlan, 1965; Slatkin, 1987; Prescott-Allen and Prescott-Allen, 1988; van Raamsdonk and van der Maesen, 1996). In these cases, the maintenance of new genetic combinations, which result in populations with new characteristics, depends on both natural and human selection (Jarvis and Hodgkin, 1999). There are many well-known breeding programmes involving deliberate introgression of desirable traits into crop plants as well as a growing number of studies documenting the natural hybridization and gene introgression between agricultural crops and their wild relatives (reviewed by Jarvis and Hodgkin, 1999). This is of particular interest in safflower (Carthamus tinctorius L.) with regards to high quality oil production.
C. tinctorius is considered a strongly domesticated species with a long history of cultivation and widespread distribution across the globe (Dempewolf et al., 2008). Hybridization with several sympatric wild species of Carthamus in the Mediterranean and Asia is suggested to have played an important role in the evolution of C. tinctorius (Ashri and Knowles, 1960; Schank and Knowles, 1964; Vilatersana et al., 2007). Indeed, hybridization between C. tinctorius and several wild relatives has been demonstrated to occur both artificially by hand pollination (Heaton and Klisiewicz, 1981), and naturally by open pollination (Ashri and Rudich, 1965). Notably, C. tinctorius can readily cross with both C. oxyacanthus and C. palaestinus to produce fertile hybrids (McPherson et al., 2004; Mayerhofer et al., 2011). Given that all three species coexist in the Old World and the range of C. oxyacanthus in the New World overlaps with the cultivated C. tinctorius, there is clear biological potential of an interspecific hybridization event should they be temporally sympatric. Each with 12 chromosomes, these species can be easily crossed, giving rise to fertile hybrids (Gill et al., 2011).
Safflower is grown mainly for its oil that can be classified into two types, one being characterized by high level of linoleic acid and the other being rich in oleic acid (HO varieties). While the oil with high level of linoleic acid is regarded as one of the most highly polyunsaturated vegetable oils (Velasco and Fernandez-Martinez, 2001), the oil with high level of oleic acid is considered particularly valuable because, in addition to its cholesterol-lowering effect, the high oleic oil also has high oxidative stability. These traits make it ideal for food applications without partial hydrogenation that can generate nutritionally undesirable trans fatty acids (Kinney and Clemente, 2005). The original safflower varieties were all high linoleic type. The high oleic trait that is controlled by a partially recessive allele ol at a single locus OL was first identified in an introduction from India (Knowles and Bill, 1964). The ol allele was subsequently incorporated into safflower breeding programs and the first HO safflower variety “UC-1” was released in 1966 in the US, which was followed by the releases of “Oleic leed” and the Saffola series including Saffola 317 (S-317), S-517, and S-518. The ol allele has also been used as the background genetic material for further enhancement of oleic acid content in safflower breeding programs worldwide (Weiske, 1997; Mundel and Bergman, 2009).
Oleic acid level in plant seed oil is primarily determined by the activity of microsomal Δ12 oleate desaturase FAD2 (Okuley et al., 1994). We isolated an unusually large FAD2 gene family with 11 members from safflower (Cao et al., 2013), among which FAD2-1 was demonstrated to be the key oleate desaturase gene specifically expressed in developing seeds and plays the major role in producing linoleic acid in safflower oil, while FAD2-2 appears to encode the house-keeping microsomal Δ12 oleate desaturase that has a generally constitutive expression throughout the plant. Other members of the FAD2 gene family were found to have divergent gene functions, with little role, if any, in the fatty acid composition of safflower seed oil (Cao et al., 2013). The ol allele in the high oleic variety was found to be a defective FAD2-1 with a single nucleotide deletion in the coding region that leads to premature termination of translation and subsequent nonsense-mediated mRNA decay (NMD) of FAD2-1, a process that typically degrades transcripts containing a premature termination codon (PTC) (Guan et al., 2012; Liu et al., 2013).
Resolving evolutionary questions within Carthamus has been challenging as low levels of genetic variation belie clear morphological differences between species (Vilatersana et al., 2005; Bowles et al., 2008). The development of molecular genetic techniques has greatly facilitated our ability to study low levels of introgression. In addition to the expressed-sequence tag (EST) and simple sequence repeat (SSR) markers used by Chapman et al. (2009), random amplified polymorphic DNA markers (Vilatersana et al., 2005), and conserved intron-spanning PCR markers (Chapman and Burke, 2007) have also been employed in studies addressing species relationships.
In this paper we present DNA sequence data and Southern blot analysis suggesting that there has been hybridization and introgression of the FAD2-1 gene from C. palaestinus into C. tinctorius, from which the ol allele (FAD2-1Δ) was derived more recently. We have mainly focused on the use of a large intron (~1.2 kb) present in the FAD2-1 5′ untranslated region (5′ UTR) which may evolve with less, if any, functional constrains and is therefore more suitable for elucidating evolutionary history than a coding region (Small et al., 2004). Such a 5′ UTR intron in FAD2-1 was previously used in evolutionary studies of Gossypium species (Liu et al., 2001). It has been suggested that FAD2-1 intron is large enough to be evolutionary meaningful, and it may be evolving at a quick enough rate for inferring evolutionary relationships among recently diverged lineages (Liu et al., 2001). In this regard it is particularly useful for elucidating the evolutionary pathways of Carthamus species.
Materials and Methods
Plant Materials and Genetic Extraction
In this study we used 16 C. tinctorius accessions (populations), 5 accessions of C. oxycanthus, and 1 accession of C. palaestinus, all of which were obtained from USDA Western Regional Plant Introduction Station (WRPIS; http://www.ars.usda.gov/main/site_main.htm?modecode=53481500; Table 1).
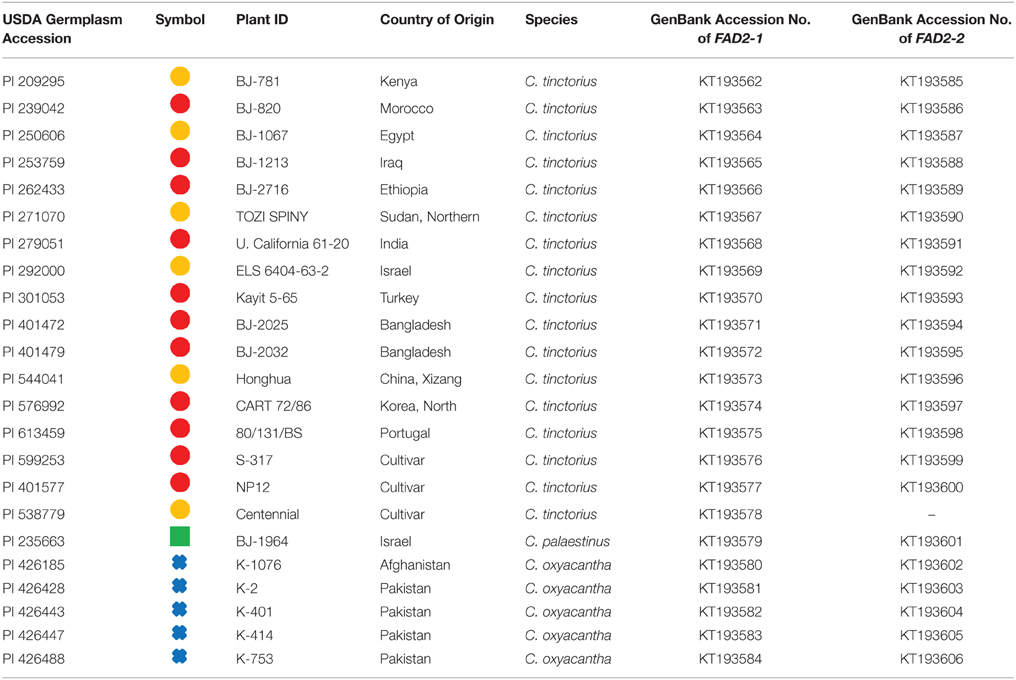
Table 1. Specimen details, their corresponding symbols, and GenBank numbers assigned to relevant DNA sequences.
Genetic Sequencing
The genomic DNA of Carthamus entries was isolated from fully expanded young leaves using CTAB buffer and further purified by CsCl gradient as described by Cao et al. (2013). The intron located in the 5′ UTR of FAD2-1 was amplified using the following oligonucleotides: 5′- GAGATTTTCAGAGAGCAAGCGCTT -3′ and 5′- CTTTGGTCTCGGAGGCAGACATA -3′ for FAD2-1 and 5′- CAAAAGGAGTTTCAGAAAGCCTCC -3′ and 5′- ACTCGTTGGATGCCTTCGAGTTC- 3′ for FAD2-2. All polymerase chain reaction (PCR) amplification reactions were performed in a final volume of 20 μL using 10 μL HotStart PCR mix (Qiagen, Hilden, Germany), 0.3 μM of each forward and reverse primers and 20 ng of Carthamus genomic DNA template. Reaction mixtures were denatured at 95°C for 15 min before undergoing 35 amplification cycles (94°C for 30 s, 57°C for 30 s and 72°C for 1 min), followed by a final extension at 72°C for 10 min. PCR products were subsequently purified with QIAquick® PCR Purification Kit (QIAGEN), and cloned into pGEM-T easy® (Promega, Madison, WI, USA). The entire inserts were sequenced bidirectionally using Big Dye Terminator (Applied Biosystems, Melbourne, Australia) reactions and reaction products separated on a capillary sequencer (Applied Biosystems). Sequences were assembled using CHROMAS PRO (Technelysium Ltd, Brisbane, Australia) and have been deposited in GenBank (Accession numbers are listed in Table 1).
Sequence Analyses
Sequences were aligned using ClustalW in MEGA4 (Tamura et al., 2007) and general sequence statistics were also calculated in MEGA4 (Tamura et al., 2007). jModelTest was used to statistically select the best fit model of nucleotide substitution for data analysis (Guindon and Gascuel, 2003; Felsenstein, 2005; Posada, 2008). The HKY (Hasegawa et al., 1985) model of nucleotide substitution gave the best fit for the data. A phylogenetic maximum-likelihood tree was constructed using RAxML (Stamatakis, 2006). Support values for inferred relationships between taxa were estimated using 1000 bootstrap (BP) replicates. We conducted Bayesian phylogenetic analysis in MrBayes (Huelsenbeck and Ronquist, 2001) using the HKY model (Hasegawa et al., 1985) with inverse gamma-distributed rate variation across sites and a proportion of invariable sites. The analysis was run for 200,000 generations with the sampling frequency set to every 100th generation. The analysis was continued until the standard deviation of split frequencies was below 0.01 after which the parameter values were summarized with a burn-in value corresponding to 25% of the samples used in each analysis. To ensure that the analyses were not trapped in local optima, the data set was run independently five times. Consensus trees were viewed in TREEVIEW (Page, 1996).
Southern Blot Analyses
Ten micrograms of genomic DNA prepared from the Carthamus leaves was digested with one of the three restriction enzymes, including BglII, or HindIII or EcoRV overnight and electrophoresed through 1% agarose gel prior to blotting onto a Hybond-N+ nylon membrane (Amersham, UK). The filters were probed with α-P32 dCTP-labeled safflower FAD2-1 or FAD2-2 gene specific DNA fragment. Hybridization was performed in 6 × SSPE, 10% Denhardt's solution, 0.5% SDS, 100 μg/mL denatured salmon sperm DNA overnight at 65°C. After a brief wash in 2 × SSC/0.1% SDS at 50°C, the filter was washed three times in 0.2 × SSC/0.1% SDS at 50°C for 20 min each prior to autoradiography.
Fatty Acid Analyses
The oil from mature safflower seeds was expressed onto a filter paper disc and directly methylated in 2 mL of 0.02 M sodium methoxide for 1 h at 90°C, followed by addition of 1.5 mL of hexane and 2 mL of water. After vortexing and phase separation, the upper hexane layer containing the fatty acid methylesters (FAME) was transferred to a new microvial and was analyzed by Agilent 6890N gas chromatography with a 30 m BPX70 column as described by Cao et al. (2013).
Results
DNA Sequence Analyses of FAD2-1 and FAD2-2
The entire 5′ UTR intron of FAD2-1 was amplified (1199 bp) from all the 22 Carthamus accessions, including 17 C. tinctorius, 1 C. palaestinus, and 4 C. oxycanthus accessions, using the primers based on the its conserved flanking regions in the 5′ UTR (Table 1). All of the introns started with GT and ended with AG, consistent with the plant consensus exon/intron boundaries (Simpson and Filipowicz, 1996).
FAD2-1 intron sequences were found to be highly conserved with a total of 144 variable sites and 4 haplotypes identified across all species. Haplotype 1 was present in C. palaestinus and C. tinctorius (including PI 401577, the high oleic acid accession); haplotype 2 was unique to the C. tinctorius PI 599253, the high oleic acid cultivar S-317; haplotype 3 was exclusive to C. oxyacantha; and haplotype 4 was identified in C. tinctorius, including the PI 538779, the high linoleic cultivar Centennial.
Maximum likelihood analysis of FAD2-1 sequences shows C. oxyacantha is clearly monophyletic (bootstrap = 100%), however, C. tinctorius appears to be paraphyletic (Figure 1). Eleven C. tinctorius FAD2-1 sequences form a strongly supported clade with C. palaestinus. The remaining 5 C. tinctorius FAD2-1 sequences create a separate clade. All of the C. oxyacantha group together. Two different FAD2-1 intron sequences, belonging to the two distinct clades mentioned above, likely with a heterozygous FAD2-1 locus, were isolated from PI 271070 and it was therefore subsequently removed from the analysis. The two high oleic acid mutants PI 401577 (S-317) and PI 401479, harboring the ol allele, were closely associated with C. palaestinus, along with 9 other C. tinctorius accessions.
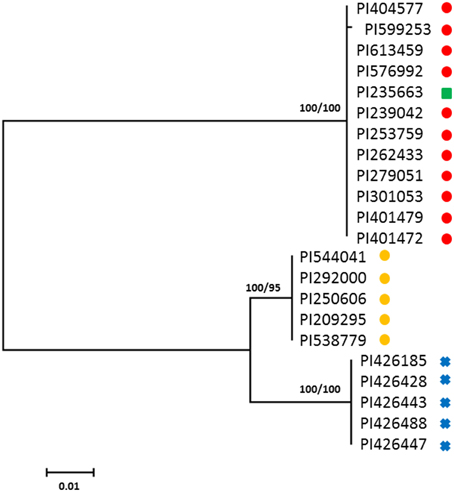
Figure 1. Phylogenetic maximum-likelihood trees obtained from the analysis of FAD2-1 datasets for three species: C. oxyacantha, C. palaestinus, and C. tinctorius. Bootstrap values are reported for each node unless less than 70 (RAxML/MrBayes). The scale bar represents the branch length as a measure of substitutions per site.  , C. tinctorius group A;
, C. tinctorius group A;  , C. tinctorius group B;
, C. tinctorius group B;  , C. palaestinus;
, C. palaestinus;  , C. oxyacantha.
, C. oxyacantha.
Similar to FAD2-1, the entire 5′ UTR intron of FAD2-2 (3178 bp) was amplified from all the 22 Carthamus accessions, using the primers based on its conserved flanking regions in the 5′ UTR (Table 1). Again, all of the introns started with GT and ended with AG.
A total of 18 haplotypes of FAD2-2 were identified with 178 variable sites across all species, including 4 haplotypes for C. oxycanthus (n = 5); 1 haplotype for C. palaestinus (n = 1); and 13 haplotypes for C. tinctorius (n = 16). In phylogenetic analyses all the three species were genetically distinct from one another. In contrast to FAD2-1, C. oxyacantha and G. palaestinus FAD2-2 appeared to be more closely related to each other than to C. tinctorius (Figure 2). We have tracked the origin of samples and found that all clades appear to be widespread across the globe, without a distinct distribution pattern (Figure 3).
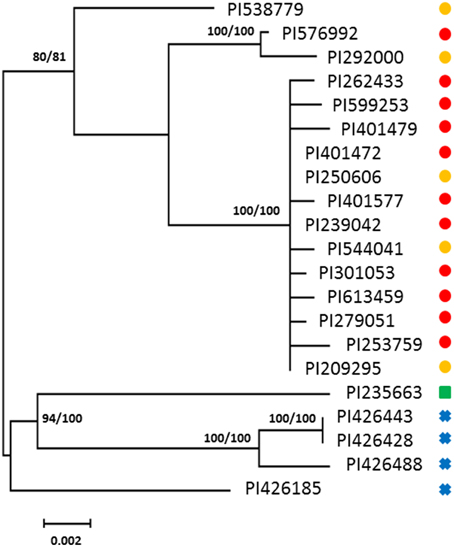
Figure 2. Phylogenetic maximum-likelihood trees obtained from the analysis of FAD2-2 datasets for three species: C. oxyacantha, C. palaestinus, and C. tinctorius. Bootstrap values are reported for each node unless less than 70 (RAxML/MrBayes). The scale bar represents the branch length as a measure of substitutions per site.  , C. tinctorius group A;
, C. tinctorius group A;  , C. tinctorius group B;
, C. tinctorius group B;  , C. palaestinus;
, C. palaestinus;  , C. oxyacantha.
, C. oxyacantha.
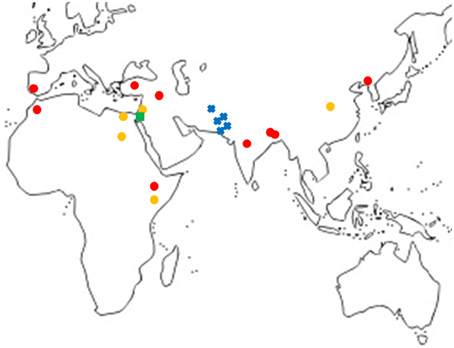
Figure 3. Map showing the locations of samples grouped according to FAD2-1 sequences excluding cultivars.  , C. tinctorius group A;
, C. tinctorius group A;  , C. tinctorius group B;
, C. tinctorius group B;  , C. palaestinus;
, C. palaestinus;  , C. oxyacantha.
, C. oxyacantha.
Southern Blot Analyses
DNA blot analysis of FAD2-1 using enzymes BglII and EcoRV consistently showed three distinct restriction fragment length polymorphism (RFLP) patterns in C. tinctorius. Group 1 consisting of 5 accessions, as labeled by a yellow dot in Figure 4, showed a single band of approximately 6.0 kb by BglII digestion, and 2.5 kb by EcoRV digestion. Group 2 consisting of 11 other accessions, as labeled by Red dots, showed a single band of approximately 4.0 kb in length by BglII digestion, and 7.0 kb by EcoRV digestion. Group three is a single accession, PI 271070 labeled by a yellow dot, showed two bands in either restriction enzyme digestion, combining the RFLP pattern of both group 1 and group 2. This particular plant was likely a heterozygous hybrid between groups 1 and 2 of C. tinctorius. Of particular interest, in both BglII and EcoRV digestions, C. palaestinus (labeled by a green square) showed the exactly same RFLP pattern as that of the Group 2 in C. tinctorius. This is consistent with the sequence analysis of FAD2-1 intron, which showed higher sequence similarity between group 2 C. tinctorius and C. palaestinus, distinct from group 1 C. tinctorius. In both restriction enzyme digestions, the four C. oxycanthus accessions showed distinct patterns from either C. tinctorius or C. palaestinus.
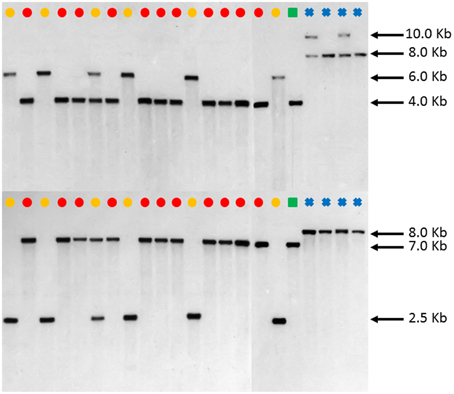
Figure 4. Southern blot FAD2-1 digestion radiographs (top using enzyme BglII, bottom using enzyme EcoRV). Samples are in order according to their listing in Table 1 starting with PI 209295 and ending with PI 426488.  , C. tinctorius group A;
, C. tinctorius group A;  , C. tinctorius group B;
, C. tinctorius group B;  , C. palaestinus;
, C. palaestinus;  , C. oxyacantha.
, C. oxyacantha.
However, such an observation was not made in the FAD2-2 RFLP patterns as illustrated in Figure 5. Except PI 292000, the other 16 C. tinctorius accessions (labeled with yellow or red dots) shared the same RFLP pattern (a band of approximately 3.0 kb) with C. palaestinus (labeled with a green square) and two C. oxycanthus (labeled with blue crosses) in the BglII digestion, whereas the C. palaestius showed a clear distinction from all the 17 C. tinctorius accessions, but shared a band of approximately 12.0 kb with all four C. oxycanthus in the HindIII digestion.
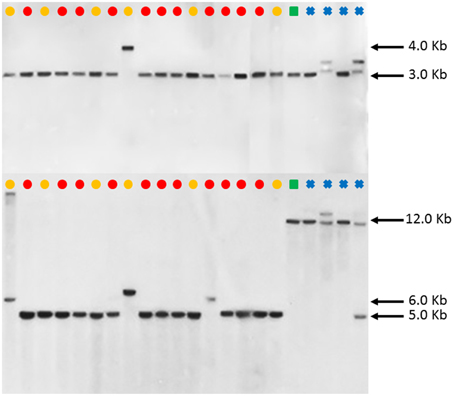
Figure 5. Southern blot FAD2-2 digestion radiographs (top using enzyme BglII, bottom using enzyme HindIII). Samples are in order according to their listing in Table 1 starting with PI 209295 and ending with PI 426488.  , C. tinctorius group A;
, C. tinctorius group A;  , C. tinctorius group B;
, C. tinctorius group B;  , C. palaestinus;
, C. palaestinus;  , C. oxyacantha.
, C. oxyacantha.
Fatty Acid Analyses
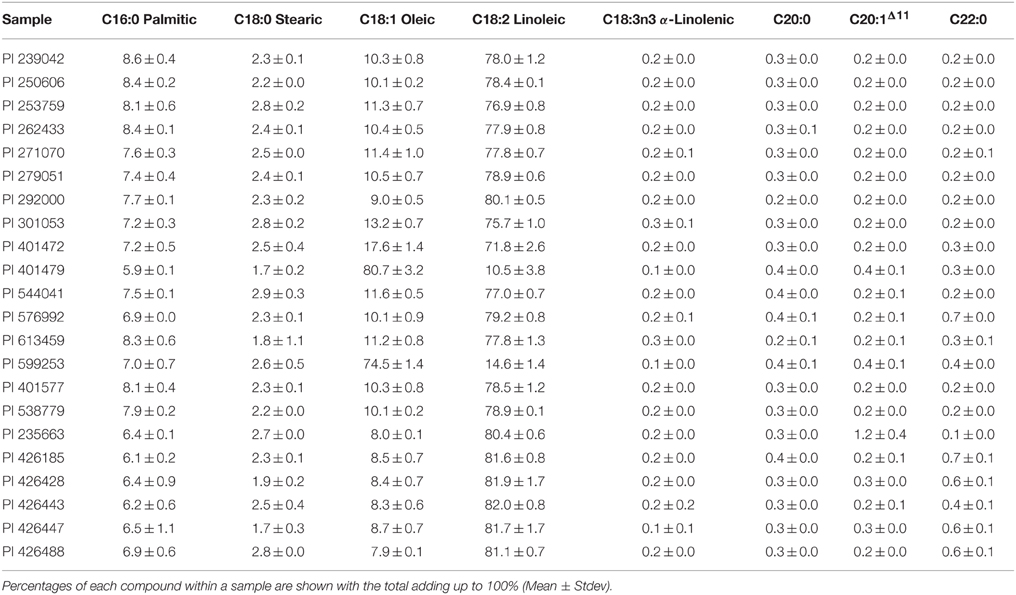
The fatty acid composition of the seed oil from each of the 22 accessions of Carthamus spp. was analyzed. The majority of samples, except two, contained high levels of linoleic acid, ranging between 71 and 82%, and an oleic acid content of between 8 and 17% (Table 2). The two high oleic acid C. tinctorius accessions were PI 599253 containing 74.5 ± 1.4 and PI 401479 containing 80.7 ± 3.2% oleic acid, respectively. Both high oleic accessions also contained low concentrations of linoleic acid, showing a clear precursor/product relationship. PI 599253 is a commercial cultivar developed in the California, USA, by incorporating the ol allele originally found in an Indian accession. PI 401479 contains significantly higher level of oleic acid than S-317, indicating that a different mechanism may operate between these two high oleic acid mutants.
Discussion
In this study we have found that among FAD2-1 intron sequences of C. tinctorius accessions sampled across the world, there is a group aligned closely with that of C. palaestinus, while the remaining sequences clearly support the conclusions of Chapman et al. (2009), showing C. oxyacantha, C. palaestinus, and C. tinctorius are all genetically distinct from one another. Such a division within C. tinctorius was not observed in FAD2-2 intron. We have also presented corroborative evidence showing three distinct RFLP patterns for FAD2-1 that differed to the RFLP patterns for FAD2-2. From these analyses we propose that there was a natural outcrossing event that led to FAD2-1 gene introgression from C. palaestinus into C. tinctorius.
Genetic introgression is unidirectional (Martinsen et al., 2001) and occurs as a result of repeated backcrossing of hybrids to the parental species (Stewart et al., 2003). Interspecific gene flow between cultivated and weedy Carthamus species (Berville et al., 2005), and other Compositae crops, such as sunflower (Burke et al., 2002), chicory (Kiaer et al., 2007), and lettuce (Hayes and Ryder, 2007) have been reported, fuelling debates about transgene escape in the evolution of super weeds. In this study, we have found clear evidence for the introgression of the FAD2-1 gene from C. palaestinus into more than half of the C. tinctorius samples examined. This finding is not unexpected as hybrids between these two species can be readily acquired under field conditions (Ashri and Rudich, 1965; Heaton and Klisiewicz, 1981).
The cultivated safflower is believed to have had a single origin of domestication in the Fertile Crescent region approximately 4000 years ago, and reproductive barriers between crops and wild progenitors appears to be weak (Weiss, 2000). The ability of safflower to intercross and produce fertile offspring with C. palaestinus or C. oxyacanthus has been well documented (Ashri and Knowles, 1960; Ashri and Rudich, 1965; McPherson et al., 2004). All three species have the same number of chromosomes and the distribution of C. palaestinus and C. oxyacanthus in the Near East is consistent with safflower originating in this region. In addition, C. palaestinus is considered to be the progenitor species of safflower (Chapman and Burke, 2007). It is therefore plausible to suggest that the hybridization between C. palaestinus and C. tinctorius has occurred. Furthermore, given that C. palaestinus is only found in desert areas of Western Iraq, Jordan, and Southern Israel (Knowles, 1976; Smith, 1996), this particular hybridization event is most like to have occurred within this area.
Since its initial domestication from the Fertile Crescent, safflower cultivation has spread to other parts of the world. Initially safflower was classified into phenotypically distinguished seven “centers of similarity” including the Far East, India-Pakistan, the Middle East, Egypt, Sudan, Ethiopia, and Europe (Knowles, 1969; Knowles and Ashri, 1995). The combination of poor reproductive barriers and the opportunity for secondary contact via migration may have allowed the introgression of FAD2-1 from C. palaestinus into C. tinctorius. In addition, the long-term retention of ancestral polymorphism observed in FAD2-1 in C. tinctorius could be attributable to migrant alleles from occasional interspecific hybridization with C. palaestinus. These migrant alleles, represented by FAD2-1 as described here, may increase the diversity within populations and slow the process of allelic coalescence. A substantial increase in the sample size for C. palaestinus is clearly needed to more accurately understand the introgression event of FAD2-1. Additional information on the conditions, such as climatic variation, which may have enabled spatial and temporal sympatry of the two species and subsequently their hybridization in the Fertile Crescent, would also improve our understanding of this event.
It is likely that the introgression FAD2-1 from C. palaestinus to C. tinctorius was a single event that spread to more than half of the safflower samples chosen randomly from around the world. If the introgression had been extensive and dating further back in time, we expect that the C. palaestinus FAD2-1 gene would have been more evenly distributed among all the C. tinctorius individuals and harder to recognize as an introgressed gene. The mutant allele conferring the high oleic acid trait in C. tinctorius appears to have a more recent origin than the introgression event, by deriving from a single nucleotide deletion in the coding region of FAD2-1 following its introgression into C. tinctorius.
A typical oleic acid profile conferred by the ol allele is about 74% as shown here in PI 599253 (Table 2), a cultivar derived from the original high oleic germplasm identified by Knowles and Hill (1964). In this study we have included another high oleic acid C. tinctorius accession, PI 401479, which accumulates more than 80% oleic acid in its total fatty acids, clearly above the upper-margin of ol mutant (Table 2). This very high oleic acid germplasm was identified by Fernández-Martinez et al. (1993) and genetically characterized by Hamdan et al. (2009, 2012) who demonstrated that the additional oleic acid content (compared to that of PI 599253) was a result of the combined effects of the recessive ol allele and a number of other modifying genes with a positive effect on oleic acid content, as previously suggested by Knowles (1972). In confirmation of this hypothesis, we found that the FAD2-1 sequences of both PI 599253 and PI 401479 are identical and contain the same genetic mutation of a single nucleotide deletion linked to the high oleic acid trait (data not presented).
Genetic resources remain critically important for the development and improvement of safflower cultivars and germplasm. The identification and characterization of the genetic diversity of FAD2-1 enables the exploitation of the high oleic acid trait by safflower breeders in an efficient manner. Further, the development of molecular markers from expressed sequences from genes of importance, such as FAD2-1 in this case, should aid plant breeders in reducing the population size and number of generations required for the development of new high oleic acid varieties by using marker-assisted selection. In this context it is pivotal to understand the genetic origin and variation of FAD2-1 alleles in safflower population.
The relative level of oleic/linoleic acid in Carthamus seed oil controlled by the expression of FAD2-1 is known as a neutral trait that does not offer any evolutionary advantages. Nevertheless, the case study reported here will contribute to a growing body of literature showing that crop origins are genetically more complex than once thought. Beyond the current study, the whole genome sequencing approach may provide detailed information on genetic content and the origins of the introgressed regions through comparison to genomes of C. tinctorius and C. palaestinus. For instance, in a recent study with next-generation sequencing, significant introgression of genes from wild species to cultivated crops, and interspecific gene flow has left its imprint in the genome of several Compositae crops and their wild relatives (Hodgins et al., 2014). Through the current case study with FAD2-1, it is tempting to speculate that the diversity of safflower germplasm is not only complex in genetic structure but also dynamic with constantly evolving entities. Reticulate evolution, facilitated by introgression, may have also influenced the observed complexities among Carthamus species.
Furthermore, wild progenitor species and feral forms of domesticated species have been reservoirs of genetic variations that could be beneficial to crops, such as disease resistance or increased stress tolerance (Tanksley and McCouch, 1997; McCouch, 2004; Fernie et al., 2006). For example, bacterial resistance gene Pto, which was introgressed from the wild tomato species, S. pimpinellifolium, in the 1930's, and later positionally cloned (Martin et al., 1993; Pedley and Martin, 2003). Despite the current availability of transgenic approaches allowing tapping of tertiary gene pools from distant taxa, primary and secondary gene pools from the same and related species are still the most important sources of genetic variation for plant breeders.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the technical assistance from Luch Hac, Annette Kasprzak, and Lijun Tian. This research was supported by Crop Biofactory Initiatives (CBI) and the Australian Grain Research and Development Corporation (GRDC).
References
Arrigo, N., Guadagnuolo, R., Lappe, S., Pasche, S., Parisod, C., and Felber, F. (2011). Gene flow between wheat and wild relatives: empirical evidence from Aegilops geniculata, Ae. neglecta and Ae. Triuncialis. Evol. Appl. 4, 685–695. doi: 10.1111/j.1752-4571.2011.00191.x
Ashri, A., and Knowles, P. F. (1960). Cytogenetics of safflower (Carthamus L.) species and their hybrids. Agron. J. 52, 11–17. doi: 10.2134/agronj1960.00021962005200010004x
Ashri, A., and Rudich, J. (1965). Unequal reciprocal natural hybridization rates between two Carthamus L. species. Crop Sci. 5, 190–191. doi: 10.2135/cropsci1965.0011183X000500020032x
Berville, A., Breton, C., Cinliffe, K., Darmency, H., Good, A. G., Gressel, J., et al. (2005). “Issues of ferality or potential for ferality in oats, olives, the Vigna group, ryegrass species, safflower and sugarcane,” in Crop Ferality and Volunteerism, ed J. Gressel (Baton Rouge, LA: CRC Press), 231–255.
Bowles, V., Hall, J., and Good, A. G. (2008). “Creation of microsatellite markers for investigation of relationships among closely related Carthamusspecies,” in Proceedings of the 7th International Safflower Conference (Wagga Wagga, NSW).
Burke, J. M., Tang, S., Knapp, S. J., and Rieseberg, L. H. (2002). Genetic analysis of sunflower domestication. Genetics 161, 1257–1267.
Cao, S., Zhou, X. R., Wood, C. C., Green, A. G., Singh, S. P., Liu, L., et al. (2013). A large and functionally diverse family of Fad2 genes in safflower (Carthamus tinctorius L.). BMC Plant Biol. 13:5. doi: 10.1186/1471-2229-13-5
Chapman, M. A., and Burke, J. M. (2007). DNA sequence diversity and the origin of cultivated safflower (Carthamus tinctorius L.; Asteraceae). BMC Plant Biol. 7:60. doi: 10.1186/1471-2229-7-60
Chapman, M. A., Hvala, J., Strever, J., Matvienko, M., Kozik, A., Michelmore, R. W., et al. (2009). Development, polymorphism and cross-taxon utility of EST-SSR markers from safflower (Carthamus tinctorius L.). Theor. Appl. Genet. 120, 85–91. doi: 10.1007/s00122-009-1161-8
Dempewolf, H., Rieseberg, L. H., and Cronk, Q. C. (2008). Crop domestication in the Compositae: a family-wide trait assessment. Genet. Resour. Crop Evol. 55, 1141–1157. doi: 10.1007/s10722-008-9315-0
Ellstrand, N. C., Prentice, H. C., and Hancock, J. F. (1999). Gene flow and introgression from domesticated plants into their wild relatives. Annu. Rev. Ecol. Syst. 30, 539–563. doi: 10.1146/annurev.ecolsys.30.1.539
Felber, F., Kozlowski, G., Arrigo, N., and Guadagnuolo, R. (2007). “Genetic and ecological consequences of transgene flow to the wild flora,” in Advances in Biochemical Engineering/Biotechnology, eds A. Fiechter and C. Suatter (Berlin: Springer-Verlag), 173–205.
Felsenstein, J. (2005). PHYLIP (Phylogeny Inference Package). Seattle, WA: Department of Genome Sciences, University of Washington.
Fernández-Martinez, J., del Rio, M., and de Haro, A. (1993). Survey of safflower (Carthamus tinctorius L.) germplasm for variants in fatty acid composition and other seed characters. Euphytica 69, 115–122. doi: 10.1007/BF00021734
Fernie, A. R., Tadmor, Y., and Zamir, D. (2006). Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 9, 196–202. doi: 10.1016/j.pbi.2006.01.010
Gill, B. S., Friebe, B. R., and White, F. F. (2011). Alien introgressions represent a rich source of genes for crop improvement. Proc. Natl. Acad. Sci. U.S.A. 108, 7657–7658. doi: 10.1073/pnas.1104845108
Guan, L. L., Xu, Y. W., Wang, Y. B., Chen, L., Shao, J. F., and Wu, W. (2012). Isolation and characterization of a temperature-regulated microsomal oleate desaturase gene (ctFAD2-1) from safflower (Carthamus tinctorius L.). Plant Mol. Biol. Rep. 30, 391–402. doi: 10.1007/s11105-011-0349-7
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hamdan, Y. S., García-Moreno, M., Fernández-Martínez, J., Velasco, L., and Pérez-Vich, B. (2012). Mapping of major and modifying genes for high oleic acid content in safflower. Mol. Breed. 30, 1279–1293. doi: 10.1007/s11032-012-9714-y
Hamdan, Y. S., Pérez-Vich, B., Velasco, L., and Fernández-Martínez, J. (2009). Inheritance of high oleic acid content in safflower. Euphytica 168, 61–69. doi: 10.1007/s10681-008-9879-y
Harlan, J. R. (1965). The possible role of weedy races in the evolution of cultivated plants. Euphytica 14, 173–176. doi: 10.1007/BF00038984
Hasegawa, M., Kishino, H., and Yano, T. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 21, 160–174. doi: 10.1007/BF02101694
Hayes, R. J., and Ryder, E. J. (2007). Introgression of novel alleles for partial resistance to big vein disease from Lactuca virosa into cultivar lettuce. Hortscience 42, 35–39.
Heaton, T. C., and Klisiewicz, J. M. (1981). A disease-resistant safflower alloploid from Carthamus tinctorius L. Can. J. Plant. Sci. 61, 219–224. doi: 10.4141/cjps81-033
Hodgins, K. A., Lai, Z., Oliveira, L. O., Still, D. W., Scascitelli, M., Barker, M. S., et al. (2014). Genomics of Compositae crops: reference transcriptome assemblies and evidence of hybridization with wild relatives. Mol. Ecol. Resour. 14, 166–177. doi: 10.1111/1755-0998.12163
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogeny. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jarvis, D. I., and Hodgkin, T. (1999). Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agroecosystems. Mol. Ecol. 8, S159–S173. doi: 10.1046/j.1365-294x.1999.00799.x
Kiaer, L. P., Philipp, M., Jørgensen, R. B., and Hauser, T. P. (2007). Genealogy, morphology and fitness of spontaneous hybrids between wild and cultivated chicory (Cichorium intybus). Heredity 99, 112–120. doi: 10.1038/sj.hdy.6800973
Kinney, A. J., and Clemente, T. E. (2005). Modifying soybean oil for enhanced performance in biodiesel blends. Fuel Process. Technol. 86, 1137–1147. doi: 10.1016/j.fuproc.2004.11.008
Knowles, P. F. (1969). Centers of plant diversity and conservation of crop germ plasm: Safflower. Econ. Bot. 23, 324–329. doi: 10.1007/BF02860678
Knowles, P. F. (1972). The plant geneticist' contribution toward changing lipid and amino acid composition of safflower. J. Am. Oil Chem. Soc. 49, 27–29. doi: 10.1007/BF02545133
Knowles, P. F. (1976). “Safflower,” in Evolution of Crop Plants, ed N. W. Simmonds (New York, NY: Longman), 31–33.
Knowles, P. F., and Ashri, A. (1995). “Safflower: Carthamus tinctorius (Compositae),” in Evolution of Crop Plants, eds J. Smartt and N. W. Simmonds (Harlow: Longman), 47–50.
Knowles, P. F., and Bill, A. B. (1964). Inheritance of fatty acid content in the seed oil of a safflower introduction from Iran. Crop Sci. 4, 406–409. doi: 10.2135/cropsci1964.0011183X000400040023x
Knowles, P. F., and Hill, A. B. (1964). Inheritance of fatty acid content in the seed oil of a safflower introduction from Iran. Crop Sci. 4, 406–409. doi: 10.2135/cropsci1964.0011183X000400040023x
Liu, Q., Brubaker, C. L., Green, A. G., Marshall, D. R., Sharp, P. J., and Singh, S. P. (2001). Evolution of the FAD2-1 fatty acid desaturase 5′ UTR intron and the molecular systematics of Gossypium (Malvaceae). Am. J. Bot. 88, 92–102. doi: 10.2307/2657130
Liu, Q., Cao, S., Zhou, X., Wood, C., Green, A., and Singh, S. (2013). Nonsense-mediated mRNA degradation of CtFAD2-1 and development of a perfect molecular marker for ol mutation in high oleic safflower (Carthamus tinctorius L.). Theor. Appl. Genet. 126, 2219–2231. doi: 10.1007/s00122-013-2129-2
Martin, G. B., Brommonschenkel, S. H., Chunwongse, J., Frary, A., Ganal, M. W., Spivey, R., et al. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436. doi: 10.1126/science.7902614
Martinsen, G. D., Whitham, T. G., Turek, R. J., and Keim, P. (2001). Hybrid populations selectively filter gene introgression between species. Evolution 55, 1325–1335. doi: 10.1111/j.0014-3820.2001.tb00655.x
Mayerhofer, M., Mayerhofer, R., Topinka, D., Christianson, J., and Good, A. G. (2011). Introgression potential between safflower (Carthamus tinctorius) and wild relatives of the genus Carthamus. BMC Plant Biol. 11:47. doi: 10.1186/1471-2229-11-47
McCouch, S. (2004). Diversifying selection in plant breeding. PLoS Biol. 2:e347. doi: 10.1371/journal.pbio.0020347
McPherson, M. A., Good, A. G., Topinka, K. C., and Hall, L. M. (2004). Theoretical hybridization potential of transgenic safflower (Carthamus tinctorius L.) with weedy relatives in the New World. Can. J. Plant Sci. 84, 923–934. doi: 10.4141/P03-150
Mundel, H. H., and Bergman, J. W. (2009). “Safflower,” in Oil Crops, Handbook of Plant Breeding 4, eds J. Vollmann and I. Rajcan (Berlin: Springer-Verlag), 423–447.
Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E., and Browse, J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158. doi: 10.1105/tpc.6.1.147
Page, R. D. M. (1996). TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358.
Pedley, K. F., and Martin, G. B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243. doi: 10.1146/annurev.phyto.41.121602.143032
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Prescott-Allen, R., and Prescott-Allen, C. (1988). Genes from the Wild using Wild Genetic Resources for Food and Raw Materials. London: Earthscan Publications Limited.
Schank, S. C., and Knowles, P. F. (1964). Cytogenetics of pairs of hybrids of Carthamus species (Compositae). Am. J. Bot. 51, 1093–1102. doi: 10.2307/2439957
Simpson, G. G., and Filipowicz, W. (1996). Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32, 1–41. doi: 10.1007/BF00039375
Slatkin, M. (1987). Gene flow and the geographic structure of natural populations. Science 236, 787–792. doi: 10.1126/science.3576198
Small, R. L., Cronn, R. C., and Wendel, J. F. (2004). Use of nuclear genes for phylogeny reconstruction in plants. L.A.S. Johnson Review No. 2. Aust. Syst. Bot. 17, 145–170. doi: 10.1071/SB03015
Stamatakis, A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stewart, C. N., Halfhill, M. D., and Warwick, S. I. (2003). Transgene introgression from genetically modified crops to their wild relatives. Nat. Rev. Gen. 4, 806–817. doi: 10.1038/nrg1179
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tanksley, S. D., and McCouch, S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066. doi: 10.1126/science.277.5329.1063
van Raamsdonk, L. W. D., and van der Maesen, L. D. G. (1996). Crop-weed complexes: the complex relationship between crop plants and their wild relatives. Acta Bot. Neerl. 45, 135–155. doi: 10.1111/j.1438-8677.1996.tb00505.x
Velasco, L., and Fernandez-Martinez, J. (2001). “Breeding for oil quality in Safflower,” in Proceedings of the 5th International Safflower Conference, eds J. W. Bergman and H. H. Mundel (Sidney, MO), 133–137.
Vilatersana, R., Brysting, A. K., and Brochmann, C. (2007). Molecular evidence for hybrid origins of the invasive polyploids Carthamus creticus and C. turkestanicus (Cardueae, Asteraceae). Mol. Phylogenet. Evol. 44, 610–621. doi: 10.1016/j.ympev.2007.05.008
Vilatersana, R., Garnatje, T., Susanna, A., and Garcia-Jacas, N. (2005). Taxonomic problems in Carthamus (Asteraceae): RAPD markers and sectional classification. Bot. J. Linn. Soc. 147, 375–383. doi: 10.1111/j.1095-8339.2005.00375.x
Weiske, A. C. (1997). Safflower Products with Very High Level of Unsaturated Fatty Acids. Patent No 5,912,416. U.S. Patent. Richmond, CA: California Oils Corporation.
Keywords: hybridization, integration, phylogenetics, safflower, FAD2-1
Citation: Rapson S, Wu M, Okada S, Das A, Shrestha P, Zhou X-R, Wood C, Green A, Singh S and Liu Q (2015) A case study on the genetic origin of the high oleic acid trait through FAD2-1 DNA sequence variation in safflower (Carthamus tinctorius L.). Front. Plant Sci. 6:691. doi: 10.3389/fpls.2015.00691
Received: 15 June 2015; Accepted: 21 August 2015;
Published: 09 September 2015.
Edited by:
Guo-Bo Chen, University of Queensland, AustraliaCopyright © 2015 Rapson, Wu, Okada, Das, Shrestha, Zhou, Wood, Green, Singh and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Liu, Commonwealth Scientific and Industrial Research Organization Agriculture, PO Box 1600, Canberra, ACT 2601, Australia, qing.liu@csiro.com
 Sara Rapson1
Sara Rapson1 Man Wu
Man Wu Shoko Okada
Shoko Okada Alpana Das
Alpana Das Pushkar Shrestha
Pushkar Shrestha Xue-Rong Zhou
Xue-Rong Zhou Craig Wood
Craig Wood Surinder Singh
Surinder Singh Qing Liu
Qing Liu