- 1Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden of the Chinese Academy of Sciences, Wuhan, China
- 2Graduate University of Chinese Academy of Sciences, Beijing, China
- 3The New Zealand Institute for Plant & Food Research Ltd., Mt Albert Research Centre, Auckland, New Zealand
- 4School of Biological Sciences, University of Auckland, Auckland, New Zealand
Proanthocyanidins (PAs) are a group of natural phenolic compounds that have a great effect on both flavor and nutritious value of fruit. It has been shown that PA synthesis is regulated by R2R3-MYB transcription factors (TFs) via activation of PA-specific pathway genes encoding leucoanthocyanidin reductase and anthocyanidin reductase. Here, we report the isolation and characterization of a MYB gene designated PpMYB7 in peach. The peach PpMYB7 represents a new group of R2R3-MYB genes regulating PA synthesis in plants. It is able to activate transcription of PpLAR1 but not PpANR, and has a broader selection of potential bHLH partners compared with PpMYBPA1. Transcription of PpMYB7 can be activated by the peach basic leucine-zipper 5 TF (PpbZIP5) via response to ABA. Our study suggests a transcriptional network regulating PA synthesis in peach, with the results aiding the understanding of the functional divergence between R2R3-MYB TFs in plants.
Introduction
Proanthocyanidins (PAs), also named condensed tannins, are oligomers and polymers of flavan-3-ols, synthesized via the flavonoid pathway. PAs are widely distributed in the plant kingdom, appearing in flowers, fruits, stems, leaves, roots where they provide protection against predation (Mouradov and Spangenberg, 2014). PAs possess strong antioxidant properties (Dixon et al., 2005), and contribute to the prevention of age-related disorders such as cardiovascular diseases, obesity, and cancer (Chaturvedula and Prakash, 2011). However, PAs are also the main cause of astringency in fruits and beverages such as wine, fruit juice, tea, and beer. For example, persimmon accumulates a large amount of PAs in fresh fruits, which causes the bitter and astringent taste and negatively impacts the overall organoleptic quality. Many astringent fruits are not suitable for eating and need an artificial treatment to remove astringency by carbon dioxide and/or ethanol after harvest (Dixon et al., 2013). Fruits are the major dietary source of PAs. To facilitate the genetic improvement of nutritional value and organoleptic quality of fruits, it is necessary to elucidate the complicated network controlling PA biosynthesis.
The biosynthesis of PAs shares common steps with the anthocyanin pathway until the flavan-3,4-diol step (e.g., leucocyanidin in Figure 1). Leucocyanidin is converted into flavan-3-ols catechin and epicatechin through either via a single-step reaction catalyzed by leucoanthocyanidin reductase (LAR) or a two-step reaction catalyzed by leucoanthocyanidin dioxygenase (LDOX) and anthocyanidin reductase (ANR), respectively. However, little is known about the polymerization of flavan-3-ol monomers. To date, PA-specific pathway genes, LAR and ANR, have been characterized in a variety of fruit trees, such as grapevine (Bogs et al., 2005), persimmon (Ikegami et al., 2005, 2007; Akagi et al., 2009a,b), apple (Han et al., 2012; Henry-Kirk et al., 2012; Liao et al., 2015), strawberry (Schaart et al., 2013; Fischer et al., 2014), and peach (Ravaglia et al., 2013).
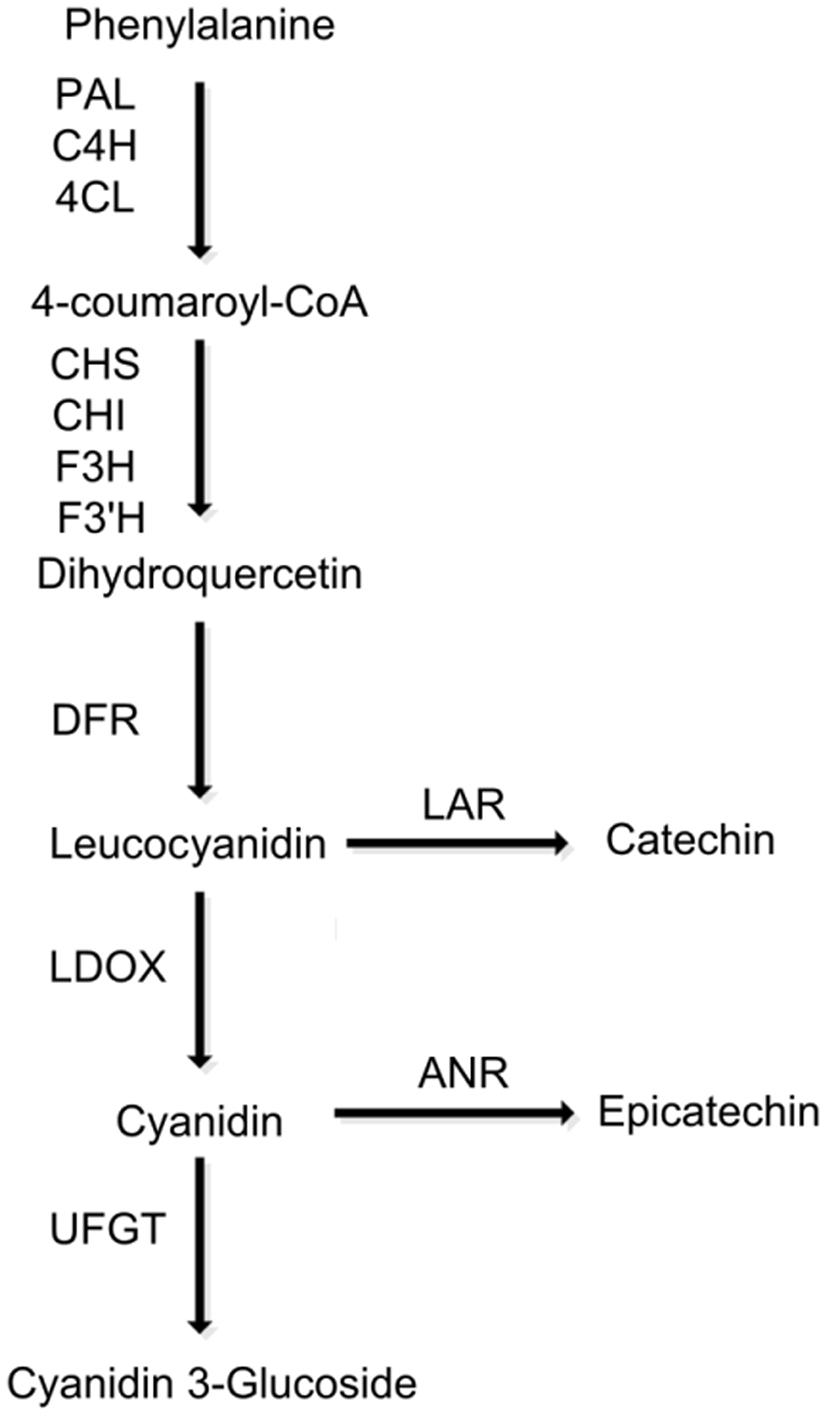
FIGURE 1. Schematic diagram of the flavonoid biosynthetic pathway in peach. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; DFR, dihydroflavonol reductase; LODX, leucoanthocyanidin dioxygenase; UFGT, UDP glucose, flavonoid 3-O-glucosyltransferase.
Proanthocyanidin accumulation is regulated at the transcriptional level by MYB transcription factors (TFs). To date, PA-related MYB activators have been identified in various plant species, such as AtMYB123 (AtTT2) and AtMYB5 in Arabidopsis (Nesi et al., 2001; Li et al., 2009), VvMYBPA1, VvMYBPA2, VvMYB5a, and VvMYB5b in grapevine (Deluc et al., 2006, 2008; Bogs et al., 2007; Terrier et al., 2009), MtPAR, MtMYB5, and MtMYB14 in Medicago (Verdier et al., 2012; Liu et al., 2014), LjTT2s in lotus (Yoshida et al., 2008), DkMYB2 and DkMYB4 in persimmon (Akagi et al., 2009b, 2010), PtMYB134 in poplar (Mellway et al., 2009), TaMYB14 from Trifolium arvense (Hancock et al., 2012), FtMYB1/FtMYB2 in tartary buckwheat (Bai et al., 2014), PpMYBPA1 in nectarine (Ravaglia et al., 2013) and MdMYB9/MdMYB11 in apple (Gesell et al., 2014; An et al., 2015). These PA-related MYB activators are divided into three phylogenetic groups (Akagi et al., 2010; Hancock et al., 2012).
Proanthocyanidin biosynthesis is affected by internal developmental cues such as phytohormones and external environmental stresses such as wounding (Zoratti et al., 2014; Liu et al., 2015). In fruit trees, the effects of developmental and environmental cues on PA synthesis have been investigated in persimmon as its fruit accumulates abundant PAs. DkbZIP5 can activate the PA-related MYB activator DkMYB4 via an ABA signal, resulting in the accumulation of PAs (Akagi et al., 2012). Whereas, ethylene responsive factors (ERFs) induce a decrease of PAs via the ethylene pathway (Min et al., 2012). Wounding induces PA synthesis through activating PA-related MYB regulator DkMYB2 (Akagi et al., 2010). More recently, jasmonate has been shown to activate PA synthesis in apple (An et al., 2015).
In Arabidopsis, there are 125 R2R3-MYB TFs in the genome (Stracke et al., 2001), with many yet to be functionally characterized. In this work, we isolated and characterized an R2R3-MYB TF, designated PpMYB7, in peach. Phylogenetic analysis indicates that the PpMYB7 gene and MYB genes from other species form an outgroup of previously reported PA-MYB activators that affect transcription of both LAR and ANR genes. However, PpMYB7, unlike PpMYBPA1, only activates transcription of PpLAR1, but not for PpANR. Our study aids in the understanding of the functional divergence between R2R3-MYB TFs in plants, and in manipulating PA synthesis in peach.
Materials and Methods
Plant Materials
The variety ‘Baifeng’ of Peach (Prunus persica) is maintained at Wuhan Botanical Garden of the Chinese Academy of Sciences (Wuhan, Hubei Province, China). Fruit samples were collected at 30 (fruitlet), 65 (pre-ripening), 85 (early ripening stage), and 95 (full ripening stage) DAFB (days after full bloom) in 2012 and each stage consisted of 10 fruits. Fruits were mechanically peeled and cored, and the flesh was cut into small sections. Fruit samples for each stage were mixed and immediately frozen in liquid nitrogen, and then stored at -75°C until use.
Construction of Expression Vectors
A pair of primers, MYB7topoF and MYB7topoR, was designed to amplify the coding sequences of PpMYB7 using iProof High-Fidelity PCR kit (Bio-Rad, Hercules, CA, USA) and cDNA templates from fruit tissues of peach ‘Baifeng.’ PCR products were purified and inserted into pENTR/D-TOPO (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The positive clone was validated by direct sequencing and then inserted into binary vector pHEX2 using LR recombination cloning according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Similarly, the coding sequences of PpbHLH3 and PpbHLH33 were also cloned and inserted individually into pHEX2 vector using the same protocol as described for the PpMYB7 gene. In addition, a pair of primers, PpMYBPA1OEF (with a tail containing a HindIII site at the 5′ end) and PpMYBPA1OER (with a tail containing an XbaI site at the 5′ end), was designed to amplify the full coding region of PpMYBPA1. PCR products were digested with HindIII and XbaI and then inserted into pSAK277 vector. All the sequences of primers used for expression vector construction are listed in Supplementary Table S1.
Yeast Two Hybrid Assay
Yeast two-hybrid analysis was performed using Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech, Palo Alto, CA, USA). Firstly, the autoactivation activities of PpMYB7, PpbHLH3, and PpbHLH33 were tested. The full coding regions of the three genes were inserted into pGBKT7 vector. The primer sequences are listed in Supplementary Table S1. Empty pGBKT7 vector and the above three constructs were transformed individually into yeast strain Y2Hgold. The positive colonies were grown on SD-Trp and SD-Trp-Ade-His medium, and the results were observed after 3 days. The full coding region of PpMYB7 was inserted into pGADT7 vector, while partial coding sequences of PpbHLH3 (amino acid 1–235) and PpbHLH33 (amino acid 1–235) were inserted individually into pGBKT7 vector. The primer sequences are listed in Supplementary Table S1. Empty vector pGADT7 and its derivations were transformed individually into yeast strain Y187, whereas, empty vector pGBKT7 and its derivations were transformed individually into yeast strain Y2Hgold. After matting in 2 × YPDA medium, the positive colonies were grown on different types of medium, respectively, including DDO (SD-Trp-Leu), QDO/A (SD-Trp-Leu-Ade-His+AbA), and QDO/X/A (SD-Trp-Leu-Ade-His+X-α-Gal+AbA). Photographs were taken 3 days after incubation.
Dual Luciferase Reporter Assay
A dual luciferase reporter assay was conducted in Nicotiana benthamiana leaves according to a previous report (Espley et al., 2007). Upstream regions from the ATG start site of four peach genes, DFR (1.6 Kb), LAR1 (1.4 Kb), ANR (1.9 Kb), and UFGT (2.5 Kb), were isolated and inserted into multiple cloning site of vector pGreen 0800-LUC (Hellens et al., 2005). All the constructs were transformed into Agrobacterium tumefaciens GV3101, and incubated at 28°C for 2 days. The confluent bacteria was resuspended in infiltration buffer (10 mM MgCl2, 0.5 μM acetosyringone) and incubated at room temperature without shaking for 2 h before infiltration. Transient transformation was conducted by mixing 100 mL Agrobacterium strain GV3101 culture transformed with the reporter cassette with 450 μL Agrobacterium culture transformed with a cassette containing PpMYBPA1, PpMYB7, PpbHLH3, or PpbHLH33 fused to the 35S promoter. All analyses were repeated at least four times using biological replicates. The ratio of LUC to Ren activity was measured 3 days after infiltration using Dual-Glo® Luciferase Assay System (Promega Corporation, Madison, WI, USA).
Firefly Luciferase Complementation Assay
Firefly luciferase complementation assay was conducted using N. benthamiana young leaves according to a previous report (Chen et al., 2008). The full coding region of PpMYB7 was inserted into binary vector pCambia1300-NLuc, while the full coding regions of PpbHLH3 and PpbHLH33 were inserted individually into pCambia1300-CLuc. The primer sequences are listed in Supplementary Table S1. Preparation and infiltration of A. tumefaciens were performed using the same protocol as described for dual luciferase reporter assay. Luminescence units were measured using an Infinite M200 luminometer (Tecan, Mannerdorf, Switzerland). Leaf disks (exactly 2 cm in diameter) were punched adjacent to the infiltration site and firefly luciferase activity was assayed using Steady-Glo® Luciferase Assay System® (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions.
Quantification of Flavan-3-ols in Peach Fruits
Peach fruit sample was ground into fine powder, and 10 mg of powder was added to 1 mL of 70% (v/v) acetone containing 0.1% (w/v) ascorbic acid. The mixture was incubated at room temperature for 24 h in darkness. The extract was centrifuged and the supernatant was transferred to a 1.5 ml microcentrifuge tube. The extract was purified by adding equal amount of chloroform and the supernatant was collected. The solvent was evaporated, and the extract was resuspended in 500 μL of water/methanol (1:1, v/v). PAs were quantified using high-performance liquid chromatography (HPLC) according to our previously reported protocol (Liao et al., 2015). Briefly, the HPLC Separation was performed on a Hisep C18-T column (5 μm, 4.6 mm × 150 mm; Weltech, Co., Ltd., Wuhan, China). The PAs were observed under UV detector at 280 nm and determined according to retention time of standards, including catechin and epicatechin (Sigma). All analyses were repeated three times using biological replicates.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using Total RNA Rapid Extraction Kit (Zomanbio, Beijing, China). First strand cDNA was synthesized using PrimeScript@ Reverse Transcriptase (TaKaRa, Dalian, China). qRT-PCR was performed in a total volume of 20 μL reaction containing 100 ng of template cDNA, 0.2 μM of each primer, and 10 μL of 2 [SYBR premix Ex TaqTM (TaKaRa)]. The amplification program was as follows: one cycle of 30 s at 95°C, followed by 40 cycles of 5 s at 95°C, 34 s at 60°C. PpTEF2 (GDR accession no. ppa001368m) was selected as an internal control according to a previous report (Tong et al., 2009). The standard curve method was conducted to quantify the transcripts. All analyses were repeated three times. The sequences of primers used for real-time PCR analysis are listed in Supplementary Table S2.
Results
Identification of an R2R3-MYB Gene, PpMYB7, Which Potentially Regulates PA Synthesis in Peach
We have previously generated an RNA-Seq-based transcriptome database of peach (Wang et al., 2013). Screening the RNA-Seq data revealed two MYB TFs which showed high level of expression in fruits prior to ripening. One is PpMYBPA1, which is implicated in the regulation of PA accumulation in nectarine (Ravaglia et al., 2013). Another, designated PpMYB7 (NCBI accession no. KT159231), is a typical R2R3-MYB. The coding sequence of PpMYB7 in cv. Baifeng is identical to that of a predicted gene in cv. Lovell deposited in Genome Database for Rosaceae (GDR1) with an accession no. ppa016135m. It encodes a protein of 203 amino acid residues that is relatively shorter than most of R2R3-MYB proteins. The closest homolog of PpMYB7 is an MYB protein (GenBank accession no. XP_008238440) from Prunus mume of unknown function. PpMYB7 shows a low level (<40%) of identity in amino acid sequence with MYBs involved in the regulation of anthocyanins or PAs, such as AtPAP1 and AtMYB123 in Arabidopsis and VvMYBPA1 and VvMYBPA2 in grapevine.
Initially, we compared the PpMYB7 gene with all R2R3 MYB TFs in Arabidopsis and found it was closely related to TT2/AtMYB123 (Supplementary Figure S1). Subsequently, phylogenetic analysis was further conducted for PpMYB7 and MYB TFs in various plants. Surprisingly, the result showed that PpMYB7 does not fall into any groups of MYB genes with known function for flavonoid synthesis, but belongs to a new group (designated MYB7) with unknown function (Figure 2). Amino acid sequence alignment indicated that previously reported flavonoid-related MYB TFs contain one or more motifs in C-terminal region (Stracke et al., 2001), but PpMYB7 does not contain any motifs in C-terminal region (Figure 3). This result further confirms that PpMYB7 is different from previously reported flavonoid-related MYB TFs. In addition, a residue at positions 39 in the R2 domain and four residues at positions 90–93 in the R3 domain are key amino acid elements that control the specificity of MYB regulators for either the anthocyanin or PA pathway (Heppel et al., 2013). The key amino acid elements in PpMYB7 are the same as those in MYBs related to PAs but not anthocyanins (Figure 3), which suggests PpMYB7 is likely involved in the regulation of PA accumulation in peach.
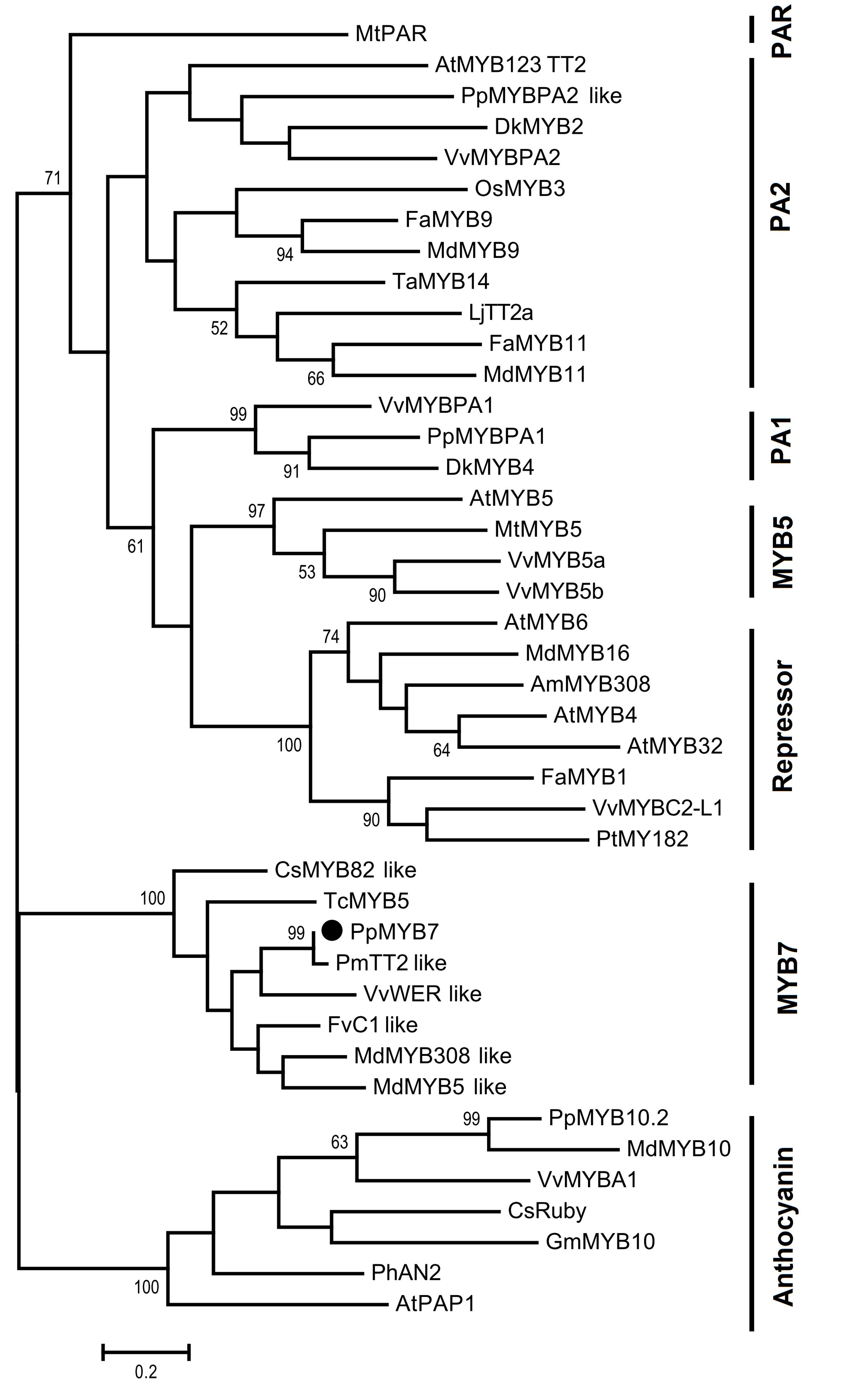
FIGURE 2. Phylogenetic tree derived from amino acid sequences of genes encoding R2R3-MYB transcription factors. The PpMYB7 gene isolated in this study is highlighted with a black circle. The full length amino acid sequences were aligned using Muscle software and phylogenetic tree was conducted with MEGA version 6.0 using the maximum likelihood method. Numbers on branches represent bootstrap estimates for 1,000 replicate analyses and values <50% are not indicated. The scale bar represents 0.2 substitutions per site. The GenBank or TAIR accession numbers are as follows: Arabidopsis thaliana AtMYB4 (AT4G38620), AtMYB5 (AT3G13540), AtMYB6 (AT4G09460), AtPAP1 (AT1G56650), AtMYB123 (AT5G35550), AtMYB32 (AT4G34990), Vitis vinifera VvMYBA1 (AB097923), VvMYBPA2 (EU919682), VvMYBPA1 (AM259485), VvMYB5a (AAS68190), VvMYB5b (AAX51291), VvMYBC2-L1 (JX050227), VvWER_like (XP_010646852); Prunus persica PpMYB10.2 (EU155160), PpMYB7 (KT159231), PpMYBPA1 (CV047374), and PpMYBPA2 like (XM_007203070); Malus domestica MdMYB308_like (XP_008369485), MdMYB5_like (XP_008356551), MdMYB10 (DQ267897), MdMYB16 (HM122617), MdMYB9 (ABB84757), MdMYB11 (AAZ20431); Fragaria x ananassa FaMYB9 (JQ989281), FaMYB11 (JQ989282), FaMYB1 (AF401220); Oryza sativa OsMYB3 (D88619); Diospyros kaki DkMYB2 (AB503699), DkMYB4 (AB503701); Trifolium arvense TaMYB14 (JN049641); Petunia x hybrid PhAN2 (ABO21074); Citrus sinensis CsRuby (NM_001288889), CsMYB82_like (XP_006477150); Garcinia mangostana GmMYB10 (FJ197137); Lotus japonicas LjTT2a (AB300033); Fagopyrum tataricum FtMYB1 (AEC32973); Antirrhinum majus AmMYB308 (P81393); Prunus mume PmTT2_like (XP_008238440); Fragaria vesca FvC1_like (XP_004299414); Theobroma cacao TcMYB5 (XP_007039783); Populus tremula x Populus tremuloides PtMYB182 (KP723392), Medicago truncatula MtMYB5 (XP_003601609), MtPAR (HQ337434).
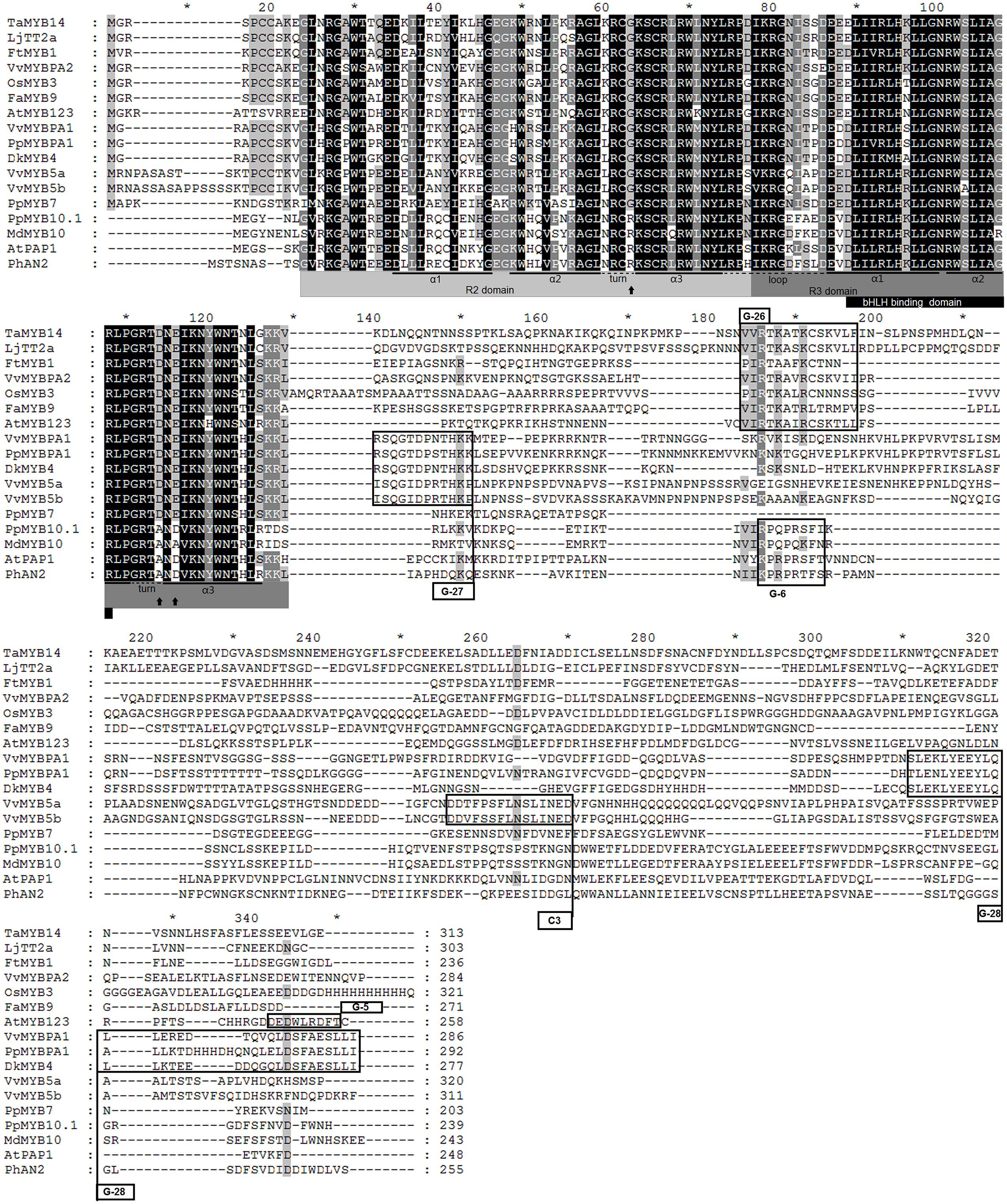
FIGURE 3. Amino acid sequence alignment of the full length amino acid sequences of PpMYB7 and other known proanthocyanidin and anthocyanin MYB regulators in plants. Conserved residues are highlighted in black, and partial conservation is indicated in gray. The alpha helices of R2 and R3 repeats are indicated with black lines. Black arrows indicate key residues that mediate specificity for either the anthocyanin or PA pathway (Heppel et al., 2013). Conserved motifs in the C-terminal region (G-5, G-6, G-26, G-27, and G-28) were numbered according to Stracke et al. (2001) and Heppel et al. (2013), and C3 motif according to Deluc et al. (2008).
Expression Profiles of MYB Regulators and Structural Genes of PA Synthesis in Peach Fruits
We screened our RNA-Seq database of peach fruit transcriptome (Wang et al., 2013) and three candidate genes encoding biosynthetic steps of PA synthesis were identified, including PpANR gene (ppa008295m), PpLAR1 (ppa009439m), and PpLAR2 (ppa016135m). Expression profiles of these three genes and two MYB regulators, PpMYB7 and PpMYBPA1, were investigated in peach fruits using qRT-PCR analysis and two biological replicates was conducted in two successive years 2012 and 2013. Since the result in 2013 (Supplementary Figure S2) is similar to that in 2012, only the result in 2012 was described here.
Both PpMYB7 and PpMYBPA1 showed a decreasing expression trend throughout fruit development, and their transcript levels were extremely low or almost undetectable after 85 DAFB (days after full bloom; Figure 4). Similarly, both PpLAR1 and PpANR had the highest level of expression in fruits at 30 DAFB, and showed a decrease in expression throughout fruit development. The transcript levels of PpLAR1 and PpANR decreased significantly after 65 DAFB. In addition, the transcript of PpLAR2 was undetectable at all tested stages of fruit development (data not shown). Therefore, the expression level of PpMYB7 and PpMYBPA1 is correlated with that of PpLAR1 and PpANR in peach fruits.
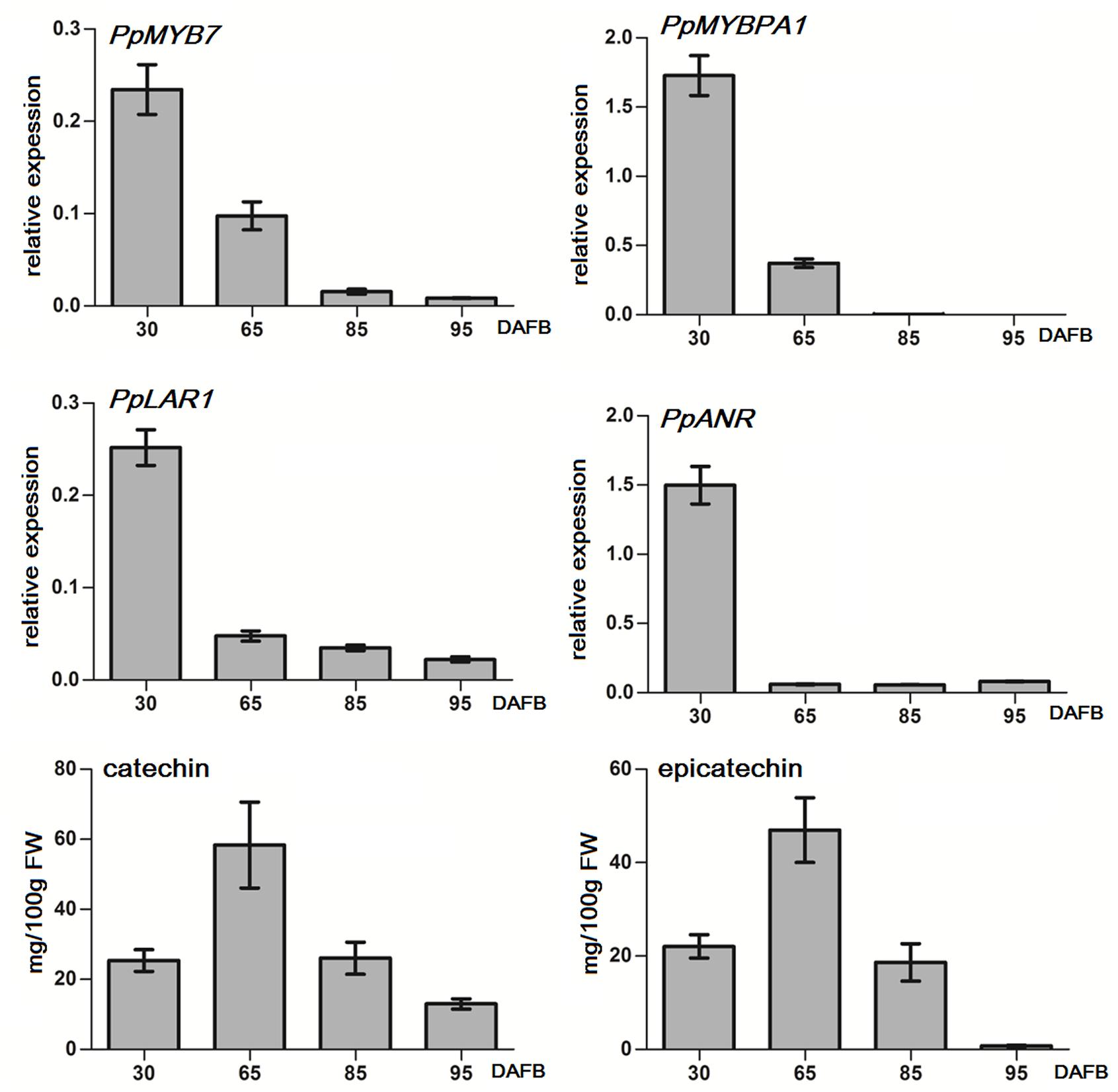
FIGURE 4. Quantitative real-time PCR (qRT-PCR) analysis of the expression profiles of genes related to PA synthesis and flavan-3-ol content in fruits of peach cv. Baifeng that were at different stages of development in 2012. qRT-PCR was done for three technical replicates. Catechin and epicatechin contents represent the means of three biological repeats. Error bars show SE of the mean. DAFB stands for days after full bloom. A biological replicate was conducted for fruit samples collected in 2013, and the results (Supplementary Figure S2) are similar to those in 2012.
In addition, we also investigated the accumulation of flavan-3-ols in flesh throughout fruit development (Figure 4). The accumulation of both catechin and epicatechin showed an increase during early fruit developmental stages, with a peak at 65 DAFB at the S3 development stage, and then decreased during ripening. The accumulation of epicatechin was almost undetectable at the full ripening stage (95 DAFB). These results suggest that the expression of MYB regulators and the genes encoding biosynthetic steps of PA synthesis precedes the accumulation of flavan-3-ols in peach fruits.
PpMYB7 Specifically Activates PpDFR and PpLAR1
To elucidate the role of PpMYB7 in the regulation of PA accumulation, dual luciferase reporter assays of promoter activity were conducted. As both peach MYB regulators had residues that predicted interaction with bHLH TFs to activate the flavonoid pathway genes (Zimmermann et al., 2004; Hichri et al., 2011), two peach bHLHs (PpbHLH3 and PpbHLH33) that are expressed in fruits (Zhou et al., 2015) were selected as candidate partners of PpMYB7. PpbHLH3 belongs to bHLH2/AN1/TT8 clade and PpbHLH33 belongs to bHLH1/JAF13/EGL3 clade. Our previous study showed that PpbHLH3 rather than PpbHLH33 participates in regulation of anthocyanin synthesis in fruits (Zhou et al., 2015). Four genes of the phenylpropanoid biosynthetic pathway, PpDFR, PpLAR1, PpANR, and PpUFGT, were selected to test the interaction of their promoters with PpMYB7. Infiltration of PpMYB7, bHLH3, or bHLH33 alone resulted in very low activity against the promoters of all the tested genes (Figure 5). Co-infiltration of PpMYB7 with PpbHLH3 or PpbHLH33 showed a significant increase in PpDFR and PpLAR1 promoter activity, with ratios of LUC to REN ranging from 1.6 to 2.0. In contrast, co-infiltration of PpMYB7 with PpbHLH3 or PpbHLH33 showed little activity against both PpUFGT and PpANR promoters. These results suggest that the PpMYB7 gene regulates the biosynthesis of catechin but not epicatechin in peach.
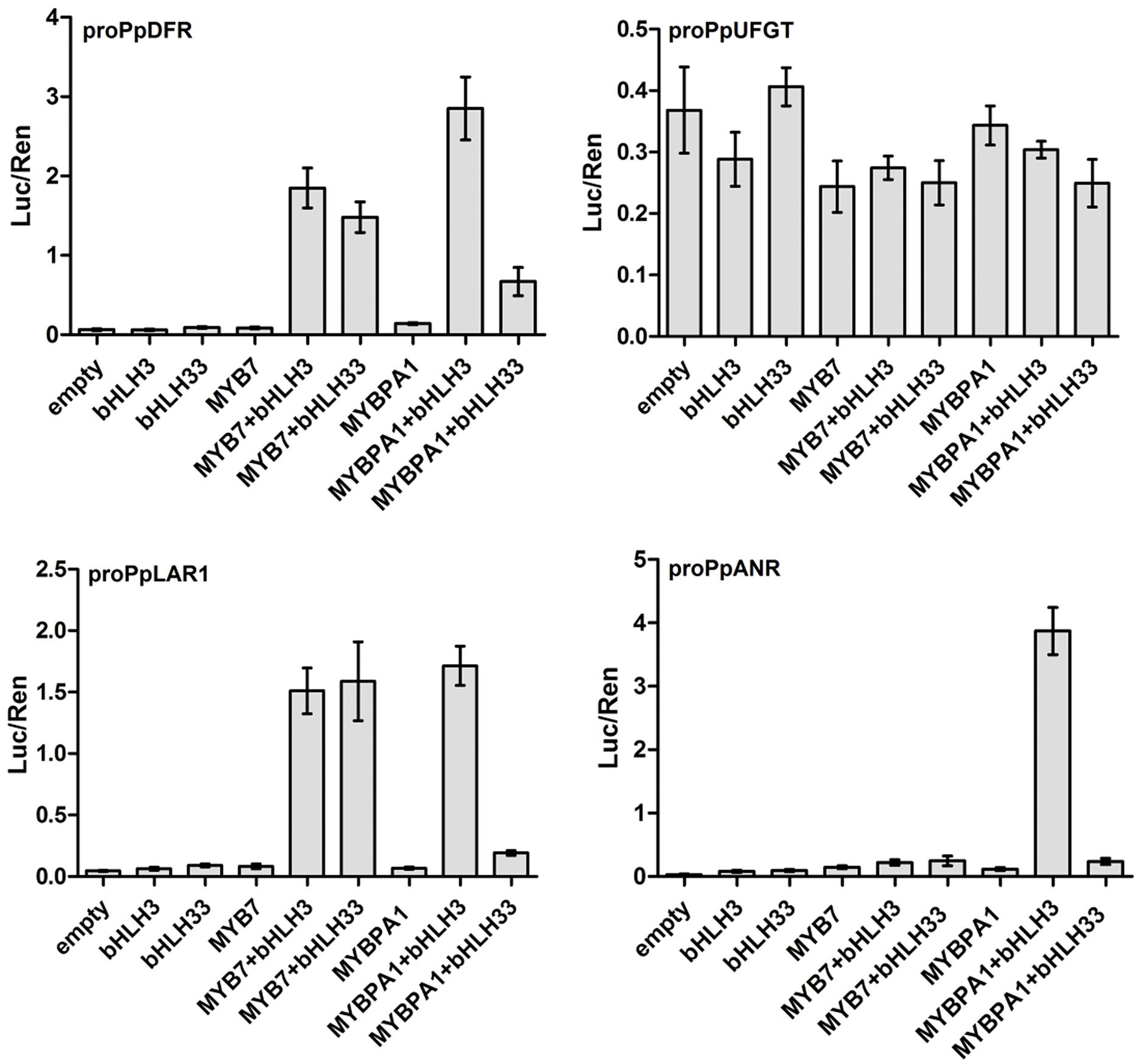
FIGURE 5. Analysis of the interaction of PpMYB7 and PpMYBPA1 with the promoters of PpDFR, PpLDOX, PpLAR1, and PpANR in transiently transfected Nicotiana benthamiana leaves. Luc and Ren were measured 3 days after infiltration, and error bars show SE of four biological replicates.
Unlike PpMYB7, PpMYBPA1 had good activity against both PpLAR1 and PpANR promoters and this activity required PpbHLH3 but not PpbHLH33. Similarly, a high activity was observed for the PpDFR promoter when PpMYBPA1 was co-transformed with PpbHLH3, whereas, co-transformation of PpMYBPA1 and PpbHLH33 failed to activate the promoter as much as that of PpMYBPA1 and PpbHLH3. In addition, MYBPA1, like PpMYB7, also showed very low activity against the PpUFGT promoter.
PpMYB7 Binding to PpbHLH3 and PpbHLH33 In vivo
Interaction of PpMYB7 with PpbHLH3 and PpbHLH33, which was observed in dual luciferase reporter assay as described above, was further tested using both yeast two-hybrid (Y2H) and firefly luciferase complementation assays. Firstly, the autoactivation was tested for PpMYB7, PpbHLH3, and PpbHLH33. The full coding regions of the three TFs were amplified and inserted into pGBKT7 vector. Autoactivation test results showed that PpMYB7-pGBKT7 and PpbHLH33-pGBKT7 were able to grow in SD-Trp-Ade-His medium, but not for the empty pGBKT7 or PpbHLH3-pGBKT7 (Supplementary Figure S3). This indicates that PpMYB7 and PpbHH33 have autoactivation ability. To avoid any false positive results and to keep the consistency of the two bHLHs, both PpbHLH3 and PpbHLH33 were truncated and their N-terminal domain (amino acid residues 1–235) was amplified and subjected to Y2H assay according to previously reported method (Hernandez et al., 2004). Empty-AD and empty-BD vectors were used as negative controls. Transformants with the PpMYB7 and the N-terminal domain of PpbHLH3 or PpbHLH33 showed growth on both DDO and QDO/A media, and their color turned to blue when grown on QDO/X/A medium (Figure 6A). In contrast, the negative controls could grow on DDO medium, but not on QDO/A medium. Moreover, R2R3 domain of PpMYB7 (amino acid 1–118) was also inserted into pGBKT7 vector to test its interaction with PpbHLH1-235 and PpbHLH331-235, and the result showed that PpMYB71-118 was able to interact with both PpbHLH1-235 and PpbHLH331-235 (Figure 6B).
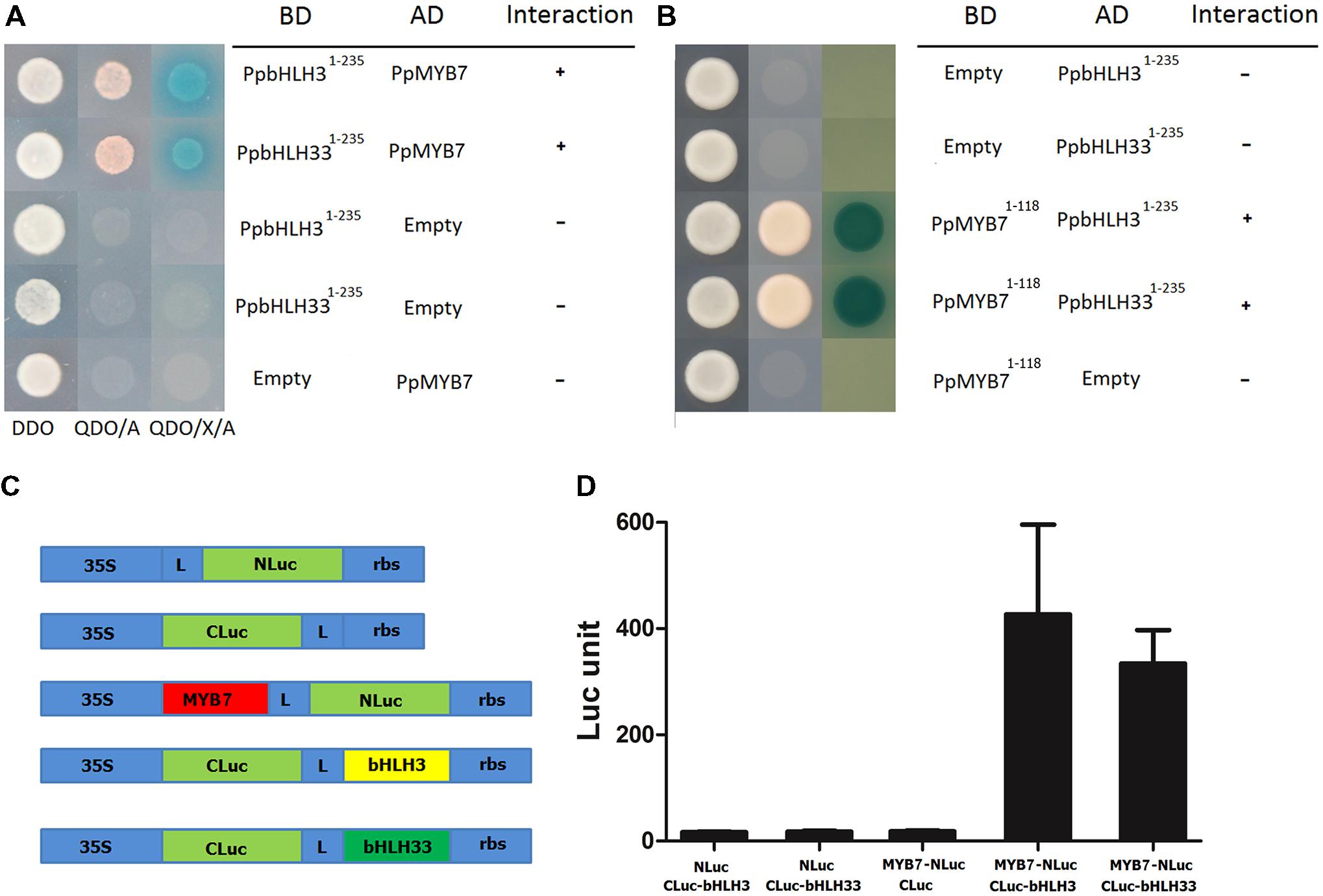
FIGURE 6. Interaction of PpMYB7 with PpbHLH3 or PpbHLH33 in vivo. (A,B) Yeast two hybrid assay. DDO, SD-Trp-Leu medium; QDO/A, SD-Trp-Leu-Ade-His+AbA medium; QDO/X/A, SD-Trp-Leu-Ade-His+X-α-Gal+AbA medium. (C) Schematic diagram of the NLuc, CLuc, and NLuc/CLuc constructs. L, Gly/Ser linker; rbs, Transcription terminator derived from the Rubisco small subunit gene. (D) Firefly luciferase complementation assay in young N. Benthamiana leaves. The error bars stand for SE of six biological replicates.
In addition, PpMYB7 was fused to N-terminus of luciferase (NLuc), while PpbHLH3 or PpbHLH33 was fused to C-terminus of luciferase (CLuc; Figure 6C). Empty-NLuc and empty-CLuc vectors were used as controls. Co-expression of PpMYB7-NLuc and bHLH3-CLuc or bHLH33-CLuc was able to rescue intense luciferase activity, which did not occur in any controls, bHLH3 or bHLH33-CLuc with NLuc and PpMYB7-NLuc with CLuc (Figure 6D). Taken together, these results indicate that PpMYB7 can bind to both PpbHLH3 and PpbHLH33 in vivo.
Cis-regulatory Elements in the Promoter Region of PpMYB7 and PpMYBPA1
A 1.9-kb fragment upstream of the ATG start codon of PpMYB7 and PpMYBPA1 was examined to identify cis-regulatory elements using the PLACE software2. As a result, three abscisic acid response elements (ABREs) were found at positions -220, -970, and -1003 bp upstream of the start codon of PpMYBPA1, whereas, a drought-responsive element 2 (DRE2) was found at position -112 from the start codon of PpMYB7 (Figure 7A). ABA-dependent DkbZIP5 can recognize ABREs to induce the expression of PA-specific regulator DkMYB4 in persimmon (Akagi et al., 2012). The coding sequence of the DkbZIP5 gene was compared against the peach reference genome (Verde et al., 2013), and eight homologs were identified. Phylogenetic analysis indicated that an ABF-, AREB-, and ABI5-Like bZIP showed the closest relationship to DkbZIP5 (Supplementary Figure S4) and was designated PpbZIP5 (NCBI accession no. KT223015). The coding sequence of PpbZIP5 was isolated to conduct a dual luciferase assay. As expected, PpbZIP5 was able to activate the promoters of both PpMYB7 and PpMYBPA1 in the presence of exogenously supplied ABA (Figures 7B,C).
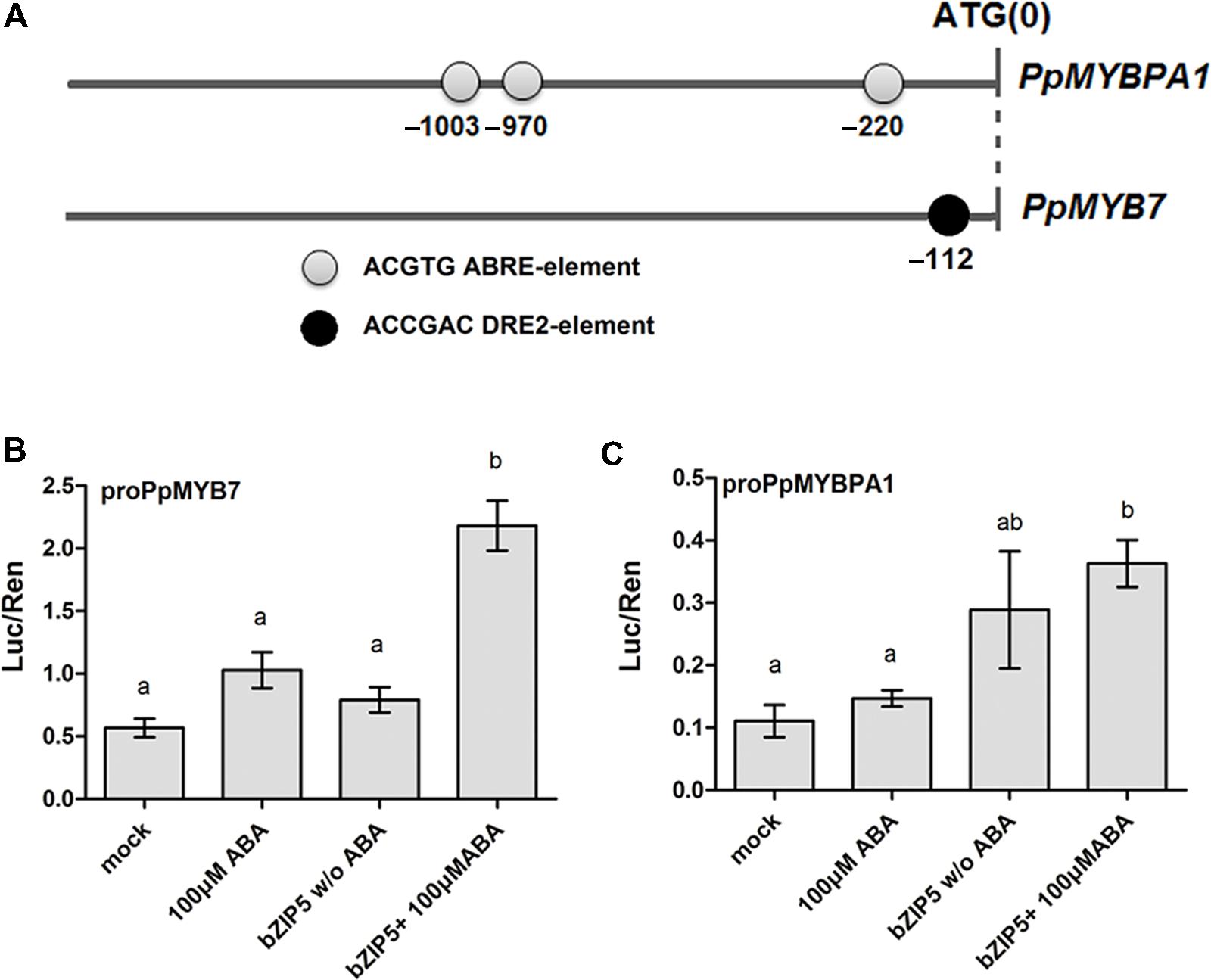
FIGURE 7. Functional analysis of the promoter sequences of PpMYB7 and PpMYBPA1. (A) Cis-regulatory elements in the promoter sequences of PpMYBPA1 and PpMYB7. (B,C) Analysis of the interaction of PpbZIP5 and ABA with the promoters of PpMYB7 and PpMYBPA1 in transiently transfected N. benthamiana leaves. Mock represents the control soaked in sterile distilled water. Different lowercase letters indicate the significant differences among treatments by Tukey’s test at P < 0.05. Error bars show SE of the mean (four biological replicates).
Discussion
Peach PpMYB7 Represents a New Group of R2R3-MYB Genes Regulating PA Synthesis in Plants
In plants, PA biosynthesis is regulated by the MBW complex formed by interaction of three types of TFs: R2R3-MYBs, basic helix-loop-helix (bHLH) proteins and WD40 repeats. In this complex, MYB TFs play a critical role in recognizing cis-elements in the promoters of PA pathway genes (Schaart et al., 2013). Here, phylogenetic analysis reveals that the previously reported PA-related R2R3-MYB activators are divided into four groups (Figure 2). Three groups, PA1 (Bogs et al., 2007; Akagi et al., 2009b), PA2 (Baudry et al., 2004; Terrier et al., 2009; Hancock et al., 2012; Liu et al., 2014), and MYB5 (Deluc et al., 2008; Xu et al., 2014), contain members from various plant species, while one group PAR only contains one member MtPAR from Medicago truncatula (Verdier et al., 2012). In addition, one group which is characterized as repressors closely clusters with PA1 and MYB5 groups (Cavallini et al., 2015). It is worth noting that some MYB repressors, such as FaMYB1 (Aharoni et al., 2001), PtMYB182 (Yoshida et al., 2015), and VvMYBC2-L1 (Huang et al., 2014) are related to the biosynthesis of anthocyanins and PAs.
Our study reveals functionally a new MYB7 group, which consists of the peach PpMYB7 and its homologs in species as diverse as citrus, grape, strawberry, and apple (Figure 2). The MYB genes within this group, like the PA2 group MYBs, consist of three exons and two introns, and are more closely related to the PA-related MYBs than to the anthocyanin-related MYBs. Dual luciferase assays demonstrate that PpMYB7 can activate transcription of PA-specific pathway gene PpLAR, but not for the anthocyanin-specific gene UFGT PpUFGT. To date, all the characterized anthocyanin-related MYBs, such as AtMYB75/AtPAP1 (Borevitz et al., 2000), VvMYBA1 (Walker et al., 2007), MdMYB10 (Espley et al., 2007), and PpMYB10.1 (Zhou et al., 2015), are able to activate transcription of UFGT. Thus, the dual luciferase assay clearly suggests that the PpMYB7 gene is a PA-related MYB regulator.
Nevertheless, the PpMYB7 gene is functionally different from the PpMYBPA1 gene, a member in the PA1 group. The PpMYB7 gene activates transcription of PpLAR but not PpANR, whereas, the PpMYBPA1 gene can activate transcription of both PpLAR and PpANR. Similar to PpMYBPA1, all the previously reported PA-related MYB activators except VvMYB5a can also induce the transcription of both PpLAR and PpANR. The VvMYB5a gene shares a similar activation pattern with the PpMYB7 gene although they do not have an orthologous relationship. In grapevine, the VvMYB5a gene can only activate VvLAR, but its ectopic expression also activates transcription of AtBAN encoding ANR in Arabidopsis and induces the accumulation of epicatechin but not catechin in tobacco (Deluc et al., 2006, 2008). It is known that the α-helices of the R2R3 domain play a critical role in directing MYBs to bind the promoters of flavonoid structural genes (Ogata et al., 1994; Williams and Grotewold, 1997; Jia et al., 2004). The third α-helix of the R2 domain and all the three α-helices of the R3 domain are conserved between PpMYB7 and the PA-related MYB activators, but they show divergence in the first and second α-helices of the R2 domain. The first two flexible α-helices of the R2 domain are likely important in the recognition of a specific gene target as they contribute to secondary structure and ternary folding of the R2R3 domain (Jia et al., 2004). More research is needed to address if the divergence in activation pattern between PpMYB7 and other PA-related MYB activators has arisen from their genetic variation in the first and second α-helices of the R2 domain.
Proanthocyanidin-related MYBs require a bHLH partner for the trans-activation of PA pathway genes (Bogs et al., 2007; Deluc et al., 2008). For example, MdMYB9 requires MdbHLH3 as a partner to regulate the biosynthesis of PAs in apple fruit (Gesell et al., 2014; An et al., 2015). bHLH TFs involved in the regulation of the flavonoid pathway from a variety of species are divided into two clades, bHLH2/AN1/TT8 and bHLH1/JAF13/EGL3 (Montefiori et al., 2015).
A previous study indicates that PpMYBPA1 requires a partner PpbHLH3 to activate transcription of PpLAR (Ravaglia et al., 2013). Here, our study further demonstrates that PpMYBPA1 can activate both PpLAR and PpANR, and requires a partner of PpbHLH3 but not PpbHLH33. Similar phenomenon was also observed in anthocyanin accumulation of fruits such as apple (Espley et al., 2007) and peach (Zhou et al., 2015), where MYB regulators MdMYB10 and PpMYB10.1 uses bHLH3 rather than bHLH33 as a partner to activate transcription of anthocyanin pathway genes. However, PpMYB7, unlike PpMYBPA1, can use either PpbHLH3 or PpbHLH33 as a partner. A residue at the position 72 in the bHLH binding domain shows a great variation. This residue is identical between PpMYB7 and PpMYB10.1, which differ in the bHLH selection. PpMYB10.1 is able to physically interact with both PpbHLH3 and PpbHLH33 (unpublished data). Thus, it seems that PA- or anthocyanin-related MYBs can interact with either bHLH3 or bHLH33 to form a complex, but some of the MYB-bHLH33 complexes could not directly activate flavonoid biosynthesis. In Solanaceae, MYB regulators act with JAF13 to activate the expression of AN1, resulting in the generation of a MYB/AN1 complex that induces the transcription of anthocyanin structural genes (Montefiori et al., 2015). This hierarchy of bHLHs has also been reported in other dicots such as Arabidopsis and Petunia (Albert et al., 2014). In peach, there are two flavonoid-related bHLH regulators, PpbHLH3 and PpbHLH33, which belong to bHLH2/AN1/TT8 clade and bHLH1/JAF13/EGL3 clade, respectively. Our recent study indicates that the expression levels of PpbHLH3 and PpbHLH33 are similar between white-flesh and blood-flesh peaches, whereas, the expression of PpMYB10.1 is significantly higher in blood-flesh than in white-flesh peaches (Zhou et al., 2015). Thus, it appears that the bHLH heirarchy aspect is not conserved in peach because the bHLH3 expression is not activated in blood peach.
Taken together, the PpMYB7 gene represents a novel group of PA-related MYBs, which may have diverged in function from the MYBPA1, MYBPA2, MYB5, and PAR genes in plants. In peach, at least two MYB genes, PpMYBPA1 and PpMYB7, are involved in the regulation of PA accumulation in fruits. Multiple MYBs such as VvMYBPA1, VvMYBPA2, VvMYB5a, and VvMYB5b are also reported to regulate PA accumulation in grapevine. Similarly, three MYB regulators, MtPAR, MtMYB5, and MtMYB14 are involved in the regulation of PA accumulation in Medicago truncatula (Verdier et al., 2012; Liu et al., 2014). Thus, it seems that PA biosynthesis is controlled by a network of MYBs, as demonstrated by the recent finding that the MBW complex accommodates two different R2R3-MYBs to regulate PA synthesis (Albert et al., 2014; Liu et al., 2014). In addition, we checked the publicly available PLAZA database3 and found that the MYB7 gene is lost in some plant species, such as Arabidopsis, Brassica rapa, potato, and tomato. This suggests that the complex transcriptional regulatory networks of PA biosynthesis have diverged among different plant species.
Abscisic Acid may be Involved in the Regulation of PA Biosynthesis in Peach Fruit
Proanthocyanidins accumulate most abundantly in immature fruits such as apple (Henry-Kirk et al., 2012), bilberry (Jaakola et al., 2002), banana (Barnell and Barnell, 1945), and strawberry (Symons et al., 2012; Schaart et al., 2013), and levels decrease toward ripening In this study, a decreasing trend was also observed for flavan-3-ols in peach fruit, consistent with a previous report in nectarine (Ravaglia et al., 2013). However, few studies have been reported any mechanism underlying dynamic accumulation of PAs in fruits.
Several studies show that phytohormones such as abscisic acid (ABA) participate in the regulation of PA synthesis in fruits such as persimmon (Akagi et al., 2012) and grapevine (Lacampagne et al., 2009). In persimmon, DkbZIP5 can bind ABRE cis-elements in the promoter of PA-related MYB activator DkMYB4 via an ABA signal, resulting in the accumulation of PAs (Akagi et al., 2012). In this study, three ABRE cis-elements were identified in the promoter of PpMYBPA1. However, the promoter of PpMYB7 does not contain any ABRE cis-elements, but has an ABA-dependent DRE2 element (Busk et al., 1997). Both PpMYBPA1 and PpMYB7 can be activated by PpbZIP5 via the ABA signaling. ABA biosynthesis decreases in peach fruits at late stages of development (Zhang et al., 2009). Thus, it seems that ABA may play an important role in the regulation of PA biosynthesis in peach fruit.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was supported by funds received from the National 863 program of China (No. 2011AA100206) and the Chinese Academy of Sciences Visiting Professorship for Distinguished Scientists (No. 2015DB009).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00908
Footnotes
- ^ http://www.rosaceae.org/species/prunus_persica/genome_v1.0
- ^ http://www.dna.affrc.go.jp/PLACE/
- ^ http://bioinformatics.psb.ugent.be/plaza/
References
Aharoni, A., De Vos, C. H., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Akagi, T., Ikegami, A., Suzuki, Y., Yoshida, J., Yamada, M., Sato, A., et al. (2009a). Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kaki Thunb.) fruit. Planta 230, 899–915. doi: 10.1007/s00425-009-0991-6
Akagi, T., Ikegami, A., Tsujimoto, T., Kobayashi, S., Sato, A., Kono, A., et al. (2009b). DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 151, 2028–2045. doi: 10.1104/pp.109.146985
Akagi, T., Ikegami, A., and Yonemori, K. (2010). DkMyb2 wound-induced transcription factor of persimmon (Diospyros kaki Thunb.), contributes to proanthocyanidin regulation. Planta 232, 1045–1059. doi: 10.1007/s00425-010-1241-7
Akagi, T., Katayama-Ikegami, A., Kobayashi, S., Sato, A., Kono, A., and Yonemori, K. (2012). Seasonal abscisic acid signal and a basic leucine zipper transcription factor, dkbzip5, regulate proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 158, 1089–1102. doi: 10.1104/pp.111.191205
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
An, X. H., Tian, Y., Chen, K. Q., Liu, X. J., Liu, D. D., Xie, X. B., et al. (2015). MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. doi: 10.1093/pcp/pcu205
Bai, Y. C., Li, C. L., Zhang, J. W., Li, S. J., Luo, X. P., Yao, H. P., et al. (2014). Characterization of two tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis. Physiol. Plant. 152, 31–40. doi: 10.1111/ppl.12199
Barnell, H., and Barnell, E. (1945). Studies in tropical fruits XVI. The distribution of tannins within the banana and the changes in their condition and amount during ripening. Ann. Bot. 9, 77–99.
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bogs, J., Downey, M. O., Harvey, J. S., Ashton, A. R., Tanner, G. J., and Robinson, S. P. (2005). Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 139, 652–663. doi: 10.1104/pp.105.064238
Bogs, J., Jaffe, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361. doi: 10.1104/pp.106.093203
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. doi: 10.2307/3871236
Busk, P. K., Jensen, A. B., and Pagès, M. (1997). Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J. 11, 1285–1295. doi: 10.1046/j.1365-313X.1997.11061285.x
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Chaturvedula, V. S. P., and Prakash, I. (2011). The aroma, taste, color and bioactive constituents of tea. J. Med. Plants Res. 5, 2110–2124.
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Deluc, L., Barrieu, F., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., et al. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140, 499–511. doi: 10.1104/pp.105.067231
Deluc, L., Bogs, J., Walker, A. R., Ferrier, T., Decendit, A., Merillon, J. M., et al. (2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147, 2041–2053. doi: 10.1104/pp.108.118919
Dixon, R. A., Liu, C., and Jun, J. H. (2013). Metabolic engineering of anthocyanins and condensed tannins in plants. Curr. Opin. Biotechnol. 24, 329–335. doi: 10.1016/j.copbio.2012.07.004
Dixon, R. A., Xie, D. Y., and Sharma, S. B. (2005). Proanthocyanidins–a final frontier in flavonoid research? New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Fischer, T. C., Mirbeth, B., Rentsch, J., Sutter, C., Ring, L., Flachowsky, H., et al. (2014). Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria x ananassa). New Phytol. 201, 440–451. doi: 10.1111/nph.12528
Gesell, A., Yoshida, K., Tran, L. T., and Constabel, C. P. (2014). Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 240, 497–511. doi: 10.1007/s00425-014-2098-y
Han, Y., Vimolmangkang, S., Soria-Guerra, R. E., and Korban, S. S. (2012). Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 63, 2437–2447. doi: 10.1093/jxb/err415
Hancock, K. R., Collette, V., Fraser, K., Greig, M., Xue, H., Richardson, K., et al. (2012). Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol. 159, 1204–1220. doi: 10.1104/pp.112.195420
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. doi: 10.1186/1746-4811-1-13
Henry-Kirk, R. A., Mcghie, T. K., Andre, C. M., Hellens, R. P., and Allan, A. C. (2012). Transcriptional analysis of apple fruit proanthocyanidin biosynthesis. J. Exp. Bot. 63, 5437–5450. doi: 10.1093/jxb/ers193
Heppel, S. C., Jaffé, F. W., Takos, A. M., Schellmann, S., Rausch, T., Walker, A. R., et al. (2013). Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 82, 457–471. doi: 10.1007/s11103-013-0074-8
Hernandez, J. M., Heine, G. F., Irani, N. G., Feller, A., Kim, M. G., Matulnik, T., et al. (2004). Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 279, 48205–48213. doi: 10.1074/jbc.M407845200
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201, 795–809. doi: 10.1111/nph.12557
Ikegami, A., Eguchi, S., Kitajima, A., Inoue, K., and Yonemori, K. (2007). Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci. 172, 1037–1047. doi: 10.1016/j.plantsci.2007.02.010
Ikegami, A., Yonemori, K., Kitajima, A., Sato, A., and Yamada, M. (2005). Expression of genes involved in proanthocyanidin biosynthesis during fruit development in a Chinese pollination-constant, nonastringent (PCNA) persimmon, ‘Luo Tian Tian Shi.’ J. Am. Soc. Hortic. Sci. 130, 830–835. doi: 10.1186/1471-2164-15-112
Jaakola, L., Maatta, K., Pirttila, A. M., Torronen, R., Karenlampi, S., and Hohtola, A. (2002). Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130, 729–739. doi: 10.1104/pp.006957
Jia, L., Clegg, M. T., and Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies Indica and Japonica genomes. Plant Physiol. 134, 575–585. doi: 10.1104/pp.103.027201
Lacampagne, S., Gagné, S., and Gény, L. (2009). Involvement of abscisic acid in controlling the proanthocyanidin biosynthesis pathway in grape skin: new elements regarding the regulation of tannin composition and leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) activities and expression. J. Plant Growth Regul. 29, 81–90.
Li, S. F., Milliken, O. N., Pham, H., Seyit, R., Napoli, R., Preston, J., et al. (2009). The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21, 72–89. doi: 10.1105/tpc.108.063503
Liao, L., Vimolmangkang, S., Wei, G., Zhou, H., Korban, S. S., and Han, Y. (2015). Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 6:243. doi: 10.3389/fpls.2015.00243
Liu, C., Jun, J. H., and Dixon, R. A. (2014). MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol. 165, 1424–1439. doi: 10.1104/pp.114.241877
Liu, M., Tian, H., Wu, J., Cang, R., Wang, R., Qi, X., et al. (2015). Relationship between gene expression and the accumulation of catechin during spring and autumn in tea plants (Camellia sinensis L.). Hortic. Res. 2, 15011. doi: 10.1038/hortres.2015.11
Mellway, R. D., Tran, L. T., Prouse, M. B., Campbell, M. M., and Constabel, C. P. (2009). The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 150, 924–941. doi: 10.1104/pp.109.139071
Min, T., Yin, X. R., Shi, Y. N., Luo, Z. R., Yao, Y. C., Grierson, D., et al. (2012). Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. J. Exp. Bot. 63, 6393–6405. doi: 10.1093/jxb/ers296
Montefiori, M., Brendolise, C., Dare, A. P., Lin-Wang, K., Davies, K. M., Hellens, R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Mouradov, A., and Spangenberg, G. (2014). Flavonoids: a metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 5:620. doi: 10.3389/fpls.2014.00620
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. doi: 10.2307/3871430
Ogata, K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., et al. (1994). Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648. doi: 10.1016/0092-8674(94)90549-5
Ravaglia, D., Espley, R. V., Henry-Kirk, R. A., Andreotti, C., Ziosi, V., Hellens, R. P., et al. (2013). Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13:68. doi: 10.1186/1471-2229-13-68
Schaart, J. G., Dubos, C., Romero De La Fuente, I., van Houwelingen, A. M., de Vos, R. C., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Symons, G., Chua, Y.-J., Ross, J., Quittenden, L., Davies, N., and Reid, J. (2012). Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 63, 4741–4750. doi: 10.1093/jxb/ers147
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verriès, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Tong, Z., Gao, Z., Wang, F., Zhou, J., and Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10:71. doi: 10.1186/1471-2199-10-71
Verde, I., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S., and Marroni, F. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–494. doi: 10.1038/ng.2586
Verdier, J., Zhao, J., Torres-Jerez, I., Ge, S., Liu, C., He, X., et al. (2012). MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 109, 1766–1771. doi: 10.1073/pnas.1120916109
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wang, L., Zhao, S., Gu, C., Zhou, Y., Zhou, H., Ma, J., et al. (2013). Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol. Biol. 83, 365–377. doi: 10.1007/s11103-013-0093-5
Williams, C., and Grotewold, E. (1997). Differences between plant and animal Myb domains are fundamental for DNA binding activity, and chimeric Myb domains have novel DNA binding specificities. J. Biol. Chem. 272, 563–571. doi: 10.1074/jbc.272.1.563
Xu, W., Grain, D., Bobet, S., Le Gourrierec, J., Thévenin, J., Kelemen, Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. N. Phytol. 202, 132–144. doi: 10.1111/nph.12620
Yoshida, K., Iwasaka, R., Kaneko, T., Sato, S., Tabata, S., and Sakuta, M. (2008). Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol. 49, 157–169. doi: 10.1093/pcp/pcn009
Yoshida, K., Ma, D., and Constabel, C. P. (2015). The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 167, 693–710. doi: 10.1104/pp.114.253674
Zhang, M., Leng, P., Zhang, G., and Li, X. (2009). Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J. Plant Physiol. 166, 1241–1252. doi: 10.1016/j.jplph.2009.01.013
Zhou, H., Lin-Wang, K., Wang, H., Gu, C., Dare, A. P., Espley, R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. doi: 10.1111/tpj.12792
Zimmermann, I. M., Heim, M. A., Weisshaar, B., and Uhrig, J. F. (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40, 22–34. doi: 10.1111/j.1365-313X.2004.02183.x
Keywords: peach, PpMYB7, leucoanthocyanidin reductase, anthocyanidin reductase, proanthocyanidins
Citation: Zhou H, Lin-Wang K, Liao L, Gu C, Lu Z, Allan AC and Han Y (2015) Peach MYB7 activates transcription of the proanthocyanidin pathway gene encoding leucoanthocyanidin reductase, but not anthocyanidin reductase. Front. Plant Sci. 6:908. doi: 10.3389/fpls.2015.00908
Received: 18 July 2015; Accepted: 12 October 2015;
Published: 26 October 2015.
Edited by:
David Caparros-Ruiz, Centre for Research in Agricultural Genomics, Consortium CSIC-IRTA-UAB-UB, SpainReviewed by:
Keiko Yonekura-Sakakibara, RIKEN, JapanNick Albert, Plant & Food Research, New Zealand
Copyright © 2015 Zhou, Lin-Wang, Liao, Gu, Lu, Allan and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuepeng Han, yphan@wbgcas.cn
 Hui Zhou
Hui Zhou Kui Lin-Wang
Kui Lin-Wang Liao Liao
Liao Liao Chao Gu
Chao Gu Ziqi Lu
Ziqi Lu Andrew C. Allan
Andrew C. Allan Yuepeng Han
Yuepeng Han