- 1Bioscience Centre, International Institute of Tropical Agriculture, Nairobi, Kenya
- 2Department of Biochemistry and Biotechnology, Kenyatta University, Nairobi, Kenya
Banana (Musa spp.) is an important staple food as well as cash crop in tropical and subtropical countries. Various bacterial, fungal, and viral diseases and pests such as nematodes are major constraints in its production and are currently destabilizing the banana production in sub-Saharan Africa. Genetic engineering is a complementary option used for incorporating useful traits in banana to bypass the long generation time, polyploidy, and sterility of most of the cultivated varieties. A robust transformation protocol for farmer preferred varieties is crucial for banana genomics and improvement. A robust and reproducible system for genetic transformation of banana using embryogenic cell suspensions (ECS) has been developed in this study. Two different types of explants (immature male flowers and multiple buds) were tested for their ability to develop ECS in several varieties of banana locally grown in Africa. ECS of banana varieties “Cavendish Williams” and “Gros Michel” were developed using multiple buds, whereas ECS of “Sukali Ndiizi” was developed using immature male flowers. Regeneration efficiency of ECS was about 20,000–50,000 plantlets per ml of settled cell volume (SCV) depending on variety. ECS of three different varieties were transformed through Agrobacterium-mediated transformation using gusA reporter gene and 20–70 independent transgenic events per ml SCV of ECS were regenerated on selective medium. The presence and integration of gusA gene in transgenic plants was confirmed by PCR, dot blot, and Southern blot analysis and expression by histochemical GUS assays. The robust transformation platform was successfully used to generate hundreds of transgenic lines with disease resistance. Such a platform will facilitate the transfer of technologies to national agricultural research systems (NARS) in Africa.
Introduction
Banana and plantain (Musa spp.) are the eighth most important staple food and cash crops in tropical and subtropical countries (FAOSTAT, 2013; Tripathi et al., 2014a). They are produced in more than 140 countries and territories across the globe with an annual production of about 144 million tones (FAOSTAT, 2013). The crop is mainly grown by smallholder farmers for food and domestic markets. Uganda is the largest banana producer in Africa with about 10 million tones harvested from over 1.8 million ha (FAOSTAT, 2012). Furthermore, Uganda is known for having the highest consumption rate of 1.6 kg for an individual per day (FAOSTAT, 2001).
Banana production is constrained by various biotic stresses, such as fungal, bacterial, and viral diseases and pests such as weevils and nematodes (Jones, 2000; Tushemereirwe et al., 2004). Currently, banana production in east and central Africa is devastated by the banana Xanthomonas wilt (BXW) disease caused by Xanthomonas campestris pv. musacearum. BXW disease affects the livelihoods of millions of farmers (Tripathi et al., 2009). About 30–60% of yield losses in banana production are also due to abiotic stresses such as drought and soil fertility (Kalyebara et al., 2007). Because banana is an important staple food crop, there is a need to develop varieties for disease resistance and enhanced yield. The improved varieties can be developed through conventional breeding or/and transgenic technologies. However, genetic improvement of banana for disease and pest resistance has not been adequately supported by a successful conventional breeding strategy. The recalcitrant nature of banana in conventional breeding has been attributed to the long generation time, limited genetic variability, absence of disease, or pest resistance in banana germplasm, sterility, and various levels of ploidy (Lorenzen et al., 2010). Due to many restrictions using conventional breeding, the advancement of effective regeneration and genetic transformation provides a substitute for banana improvement. The regeneration system is a critical factor for the production of any genetically modified plant (Hansen and Wright, 1999).
Genetic transformation of banana can be achieved using different methods, such as Agrobacterium-mediated transformation, electroporation, and micro-projectile bombardment. However, Agrobacterium-mediated transformation is the most preferred method due to its advantages, such as integration of low copy number of transgenes into the host plant genome and transfer of relatively large segments of DNA with minimal rearrangements (Lindsey, 1992; Gelvin, 2003). Several protocols for Agrobacterium-mediated transformation are available using different explants such as embryogenic cell suspension (ECS) cultures (Ganapathi et al., 2001; Khanna et al., 2004; Kosky et al., 2010; Tripathi et al., 2012) and apical meristematic tissues for various varieties of banana and plantain (May et al., 1995; Tripathi et al., 2005, 2008). The most commonly used target tissue for transformation is ECS; however, production of such cell suspensions is laborious, time consuming, and extremely variety dependent. Production of ECS and the optimization of a transformation system for each specific variety therefore becomes a prerequisite for genetic improvement. Cell suspensions of various varieties have been developed using basal leaf sheaths and corm section (Novak et al., 1989), highly proliferating multiple meristems (Dheda et al., 1991; Strosse et al., 2006), zygotic embryos (Marroquin et al., 1993), and immature male flowers (Escalant et al., 1994; Cote et al., 1996; Navarro et al., 1997; Becker et al., 2000; Grapin et al., 2000).
The availability of a robust transformation protocol for any variety is a must to produce large numbers of independent transgenic events on a regular basis. We report here a robust and reproducible system for genetic transformation of banana using ECS of “Gros Michel,” “Cavendish Williams,” and “Sukali Ndiizi.” An efficient system for developing ECS using either immature male flowers or highly proliferating multiple buds was established. This is the first comparative study where both immature flowers as well as multiple buds were used to develop embryogenic cells. The amenability of these ECS for transformation and regeneration was investigated to provide a complete system to develop large numbers of transgenic plants with economically useful traits.
Materials and Methods
Plant Materials
Immature male buds of banana varieties “Sukali Ndiizi” (AAB genome), “Cavendish Williams,” “Gros Michel,” (AAA genome), and “Ngombe” (AAA-EAHB) were collected from banana growing fields at Kenya Agricultural and Livestock Research Organization (KALRO), Kenya. Varieties “Cavendish Williams,” “Gros Michel,” and “Mpologoma” (AAA-EAHB) were obtained from the IITA collection as in vitro plantlets and used to develop highly proliferating multiple buds.
Preparation of Multiple Buds and Immature Male Flowers
The in vitro plantlets of varieties “Cavendish Williams,” “Gros Michel,” and “Mpologoma” were micropropagated as described by Tripathi et al. (2012). Small buds produced at the base of the shoot were transferred to multiple bud induction medium (MBI, Supplementary Table 1) and cultured in the dark at 26 ± 2°C. The multiple buds were sub-cultured on MBI medium at 4-week intervals until groups of tiny buds were obtained. About 3–5 monthly subcultures were done to acquire good quality multiple buds.
For immature male flowers, male inflorescences of varieties “Sukali Ndiizi,” “Cavendish Williams,” “Gros Michel,” and “Ngombe” were collected from the field within a month after bunch appearance. The outermost part of the inflorescence was removed and floral apices were surface sterilized with 70% ethanol for 2 min. The floral apices were then washed in sterile distilled water thrice. The floral buds were reduced in size (about 2 cm in length) by removing bracts under sterile conditions.
Development of Embryogenic Callus
Multiple buds were isolated on Callus Induction Medium (CIM1, Supplementary Table 1) for initiation of friable embryogenic calli as described by Tripathi et al. (2012). Three hundred explants for each variety were cultured in each experiment. A total of 900 explants were used in three experiments. The cultures were kept in the dark until callus initiated without changing any medium. The cultures were inspected consistently for development of friable embryogenic calli.
For immature male flowers, tiny flowers were isolated under stereomicroscope and cultured on Callus Induction Medium (CIM2). About 6–9 tiny flowers were incubated per 90-mm petri dish, and a total of 300 explants were cultured for callus induction in three experiments for each variety. The cultures were kept in the dark at 26 ± 2°C until callus was obtained without sub-culturing. The cultures were checked biweekly for development of friable embryogenic calli.
Development of Embryogenic Cell Suspension
Creamish yellow, translucent friable embryogenic calli of each variety were identified under the microscope and transferred into a 25 ml conical flask containing liquid Callus Induction Medium (LCIM1 or LCIM2 depending upon the explants). Initially a 5 ml medium was used in each 25 ml conical flask up to 1 week; gradually the medium was increased to 10 ml in 3 weeks. On the fourth week, fine granular cells were transferred into a new 25 ml conical flask. After 8 weeks of culture, fine cells were transferred into 250 ml conical flasks containing 30–40 ml medium for further proliferation and maturation. These embryogenic cells were washed and replenished with a new medium every 10–14 days (Tripathi et al., 2012).
Testing of Regeneration Capacity of ECS of Various Varieties
The concentration of fast dividing embryogenic cells was adjusted to 3–5% SCV with either LCIM1 or LCIM2. About 1 ml of SCV of the diluted ECS of each variety (“Sukali Ndiizi,” “Cavendish Williams,” and “Gros Michel”) was spread on nylon mesh and cultured on semisolid Embryo Development Medium (EDM, Supplementary Table 1) in 90 mm Petri dish for 1–2 months. The embryos developed on EDM were regenerated into complete plantlets as described by Tripathi et al. (2012). The regeneration efficiency of each variety was calculated as number of plantlets regenerated per ml SCV. The experiments were repeated three times.
Agrobacterium Strain and Plasmid
The binary vector pCAMBIA2301 containing the neomycin phosphotransferase (nptII) gene as the selectable marker and β-glucuronidase (gusA) gene as a reporter was used for all the transformation experiments (Supplementary Figure 1). The culture of Agrobacterium tumefaciens strain EHA 105 harboring pCAMBIA2301 was prepared for transformation as described by Tripathi et al. (2012).
Transformation, Selection, and Regeneration
Embryogenic cell suspensions of the varieties “Cavendish Williams,” “Gros Michel,” and “Sukali Ndiizi” were transformed through Agrobacterium-mediated transformation followed by selection and regeneration of transgenic lines using a modified protocol of Tripathi et al. (2012). The modification was that after co-cultivation of ECS with Agrobacterium, Agro-infected embryogenic cells were transferred on EDM without antibiotic selection for 1 week; after that these cells were shifted to selective medium supplemented with 100 mg/l kanamycin. Shoots obtained on selective medium were shifted to proliferation medium (PM, Supplementary Table 1) for multiplication and maintenance. Transformation efficiency was calculated as number of PCR positive transgenic lines regenerated on kanamycin selective medium per ml SCV of ECS of each cultivar. Transformation experiments were repeated three times. The putative transgenic shoots were then transferred to rooting medium (RM, Supplementary Table 1). Rooted plantlets of transgenic and non-transgenic control lines of “Cavendish Williams,” “Gros Michel,” and “Sukali Ndiizi” were transferred to sterile soil in pots as reported by Tripathi et al. (2014a). Plant morphology was compared between transgenic and non-transgenic control plants.
Histochemical GUS Assay
Transient and stable GUS assay was performed with embryogenic cells and with different parts of transgenic plant tissues as described (Jefferson, 1987; Tripathi et al., 2012). Transient expression of gusA gene was examined in Agro-infected embryogenic cell suspension after 3 days of co-cultivation, while stable expression of the reporter gene was investigated in leaves, petioles, pseudostem, and roots isolated from putative transgenic plants regenerated on selective medium. Non-transformed ECS, leaves, and roots of non-transgenic control plants were also included in the experiment. Photographs were taken using stereomicroscope SMZ1500 attached to a Nikon camera and computer.
PCR Analysis of Transgenic Plants
Plant genomic DNA was isolated from regenerated putative transgenic fresh leaves using a DNeasy kit (Qiagen, GmbH, Germany). PCR was performed using gusA gene specific primers [forward primer 5′- TTTAACTATGCCGGGATCCATCGC -3′ and reverse primer 5′- CCAGTCGAGCATCTCTTCAGCGTA -3′] (Nyaboga et al., 2014). PCR was performed in a total volume of 20 μl, containing 100 ng genomic DNA, 2 μl 10X buffer, 0.5 μl of 10 mM dNTP, 0.5 μl of 10 μM of each primers, and 1 unit of Hot star Taq DNA polymerase (Qiagen, Germany). The PCR conditions were: initial denaturation at 95°C for 10 min, 35 cycles of denaturation at 94°C for 15 s, annealing at 62°C for 45 s and extension at 72°C for 50 s followed by final extension at 72°C for 7 min and holding at 4°C. The amplified PCR products were separated by electrophoresis on 0.8% (w/v) agarose gel. Both positive (plasmid DNA) and negative (gDNA of non-transgenic control plant) were included in each experiment.
Dot Blot and Southern Blot Analysis
The integration of gusA gene into the genome of banana was analyzed using dot blot and Southern hybridization. Dot blot was performed using gDNA extracted from 24 PCR positive transgenic lines as described by Nyaboga et al. (2014).
For Southern blot analysis, gDNA was isolated from fresh leaves of in vitro grown transgenic plants using cetyltrimethylammonium bromide (CTAB) method developed by Gawel and Jarret (1991). About 25 μg of gDNA of transgenic lines and non-transgenic control plant were restricted using enzyme HindIII (New England Biolabs, USA). Southern blot analysis was performed as described by Tripathi et al. (2014a).
Statistical Analysis
All the experiments were repeated three times and data were analyzed using the statistical program Minitab 14. The means and standard error presented were for three experiments. One-way analysis of variance (ANOVA) was performed and interaction between means were separated by least significant difference (LSD) at P = 0.05.
Results and Discussion
Genetic improvement of bananas is crucial to create new varieties possessing traits of agronomic importance, such as high yield, combined with resistance or tolerance to biotic and abiotic stresses. Other desirable features include better fruit quality, early maturity, short stature, photosynthetic efficiency, strong roots, and uniform fruits (Pillay et al., 2002; Bakry et al., 2009). Conventional plant breeding in banana is challenging due to long generation period, polyploidy, parthenocarpy, low genetic variability, and sterility. Genetic transformation can complement conventional breeding for developing improved varieties of banana. It is also a useful tool for functional genomics (Peraza-Echeverria et al., 2007; Roux et al., 2008; Santos-Rosa et al., 2009). Therefore, it is essential to establish a high throughput regeneration and genetic transformation system, which can be applicable for many farmer-preferred varieties of banana and plantain.
Even though Agrobacterium tumefaciens-mediated genetic transformation of banana and plantain has been developed (Ganapathi et al., 2001; Khanna et al., 2004; Tripathi et al., 2012), the transformation efficiency of several farmer-preferred varieties, is still inadequate, and only a limited number of varieties has been genetically modified. Several factors such as variety, age of the explants, Agrobacterium strains, plasmid vector, the selectable marker genes, selection agent, and regeneration conditions affect the efficiency of genetic transformation (Hiei et al., 1997; Cheng et al., 2004). Most of the transformation protocols are variety dependent and it is difficult to overcome this by using only highly virulent strains of Agrobacterium (Hansen et al., 1994; Liu and Nester, 2006; Nyaboga et al., 2014) or optimizing plant regeneration conditions (Lee et al., 2002; Zuo et al., 2002). Therefore, there is a need to optimize the protocol for each economically important variety.
Development of ECS for Various Banana Varieties
ECS are the best explant for genetic transformation of banana because chimerism can be avoided in this system as transgenic events are obtained from a single cell (Roux, 2004). Immature male flowers from cultivars “Cavendish Williams,” “Gros Michel,” “Sukali Ndiizi,” “Ngombe” and multiple buds from cultivars “Cavendish Williams,” “Gros Michel,” and “Mpologoma” were tested as explants to obtain friable embryogenic callus and ECS (Figure 1 and Table 1). Yellow creamish friable calli were developed using immature male flowers from banana cultivars “Cavendish Williams,” “Gros Michel,” “Ngombe,” and “Sukali Ndiizi” in 4–6 months (Figure 1). These friable calli were transferred in LCIM2 for development of ECS. Yellowish and creamish granular embryogenic cells were observed after 1–2 months in “Sukali Ndiizi” (Figure 2). These fine cells increased and uniform cell suspension was obtained in 3–4 months following initiation (Figure 2). However, all calli for “Cavendish Williams,” “Gros Michel,” and “Ngombe” turned black after transferring to liquid medium and no embryogenic cells were observed. This could be due to the quality of calli, which was not good for the production of embryogenic cells.
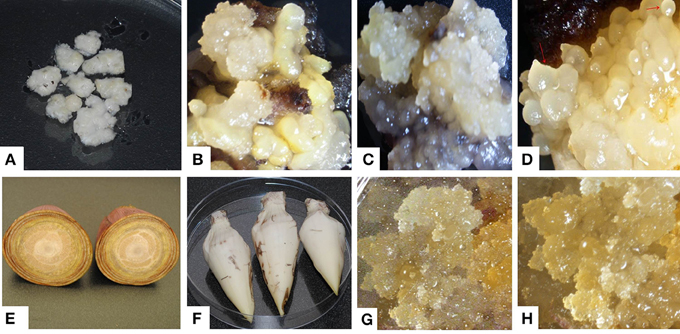
Figure 1. Induction of embryogenic calli using multiple buds and immature male flowers of various banana varieties. (A) Multiple buds, (B) embryogenic calli of “Cavendish Williams,” (C) embryogenic calli of “Gros Michel,” (D) embryogenic calli of “Mpologoma,” (E,F) banana inflorescences, (G) embryogenic calli of “Sukali Ndiizi” and (H) embryogenic calli of “Ngombe.”
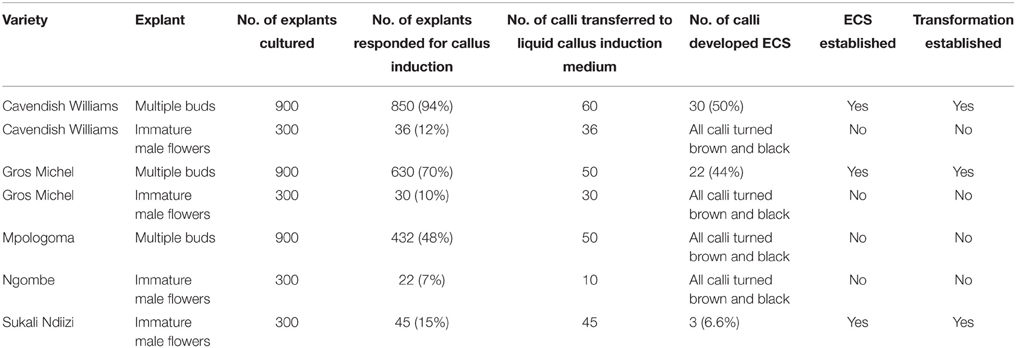
Table 1. Summary of development of embryogenic cell suspension (ECS) and transformation for various banana varieties.
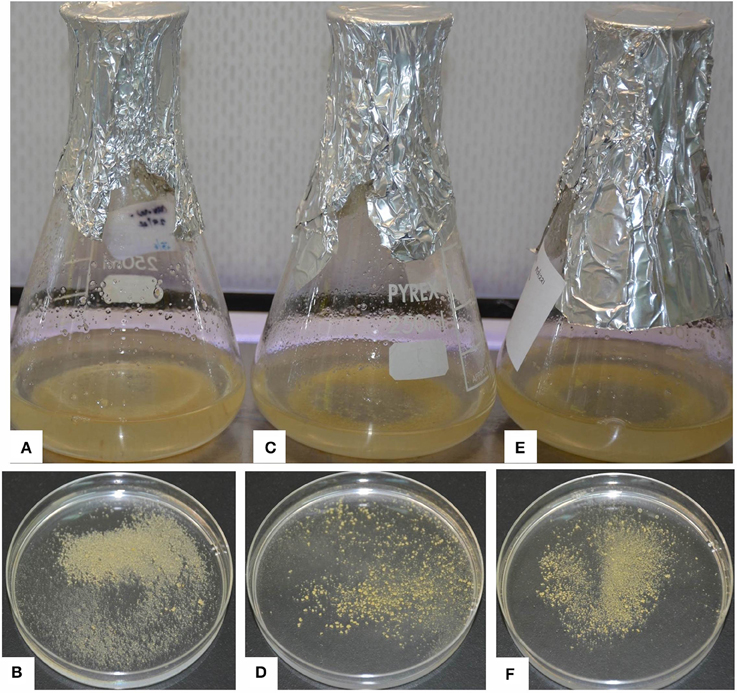
Figure 2. Embryogenic cell suspensions (ECS) of various varieties of banana. (A) ECS of “Cavendish Williams” in liquid, (B) ECS of “Cavendish Williams” on plate, (C) ECS of “Gros Michel” in liquid, (D) ECS of “Gros Michel” on plate, (E) ECS of “Sukali Ndiizi” in liquid, (F) ECS of “Sukali Ndiizi” on plate.
Friable calli were developed using multiple buds of “Cavendish Williams,” “Gros Michel,” and “Mpologoma” after 3–5 months of culture of explants in the dark (Figure 1). Fine embryogenic cells were obtained from creamish yellowish calli in about 1–2 months after transferring them to LCIM1. Embroygenic cells of “Cavendish Williams” and “Gros Michel” were multiplied and the uniform cell suspensions were established in 3–4 months after initiation (Figure 2). However, the calli of “Mpologoma” turned brownish black and did not produce any embryogenic cells.
The ECS developed for different cultivars using either of the explants were maintained for 1–1.5 years by subculturing every 10–14 days. The quantity of embryogenic cells was doubled every 2 weeks, but the regeneration capacity of ECS was observed to decrease with time. In this study, we noticed that both the explants were amenable for development of embryogenic cells depending upon the cultivar (Table 1). However, multiple buds are preferred explants as they can be easily produced using tissue culture plantlets under laboratory conditions. To get immature male flowers, researchers need to have access to field-grown plants, which is sometimes not possible. Previous reports demonstrated the development of embryogenic cell suspension of “Cavendish Williams” using immature male flowers (Xu et al., 2005; Ghosh et al., 2009; Youssef et al., 2010; Chong-Pe'rez et al., 2012). However, we could not obtain ECS of “Cavendish Williams” from immature male flowers, but we successfully obtained ECS from multiple buds. In this study, we compared two types of explants, multiple buds and immature male flowers, for development of ECS with various cultivars. ECS were obtained from both multiple buds and immature male flowers depending upon cultivar (Table 1).
Regeneration Capacity of ECS of Different Varieties
Three-month old ECS of “Cavendish Williams,” “Gros Michel,” and “Sukali Ndiizi” were tested for regeneration and their regeneration efficiencies were compared (Figures 3, 4). White globular embryos appeared in 1–2 months on EDM, which were then transferred to Embryo Maturation Medium (EMM, Supplementary Table 1) for 1 month. The mature white embryos were cultured on Embryo Germination Medium (EGM, Supplementary Table 1), where they started germinating in 1–2 months and produced whitish shoots which were subsequently transferred onto proliferation medium for shoot development (Figure 3). We observed that once cells suspensions were established, regeneration of complete plantlets were similar irrespective of explants from which cell suspensions were originated.

Figure 3. Regeneration of embryogenic cell suspensions. (A) Embryogenic cell suspension, (B) embryogenic cells plated on nylon mesh cultured on embryo development medium, (C) regenerating white embryos on embryo development medium, (D) embryos on embryo maturation medium, (E) embryos on germination medium, (F) regenerated shoots on proliferation medium.
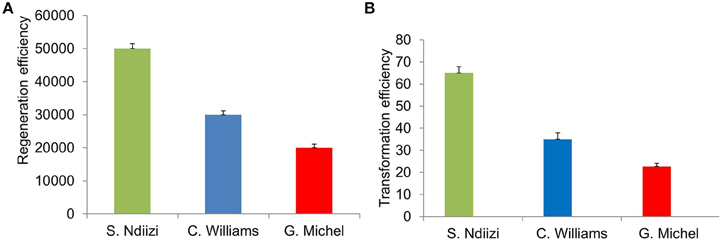
Figure 4. Graphs showing regeneration and transformation efficiency of various banana varieties using embryogenic cell suspension (ECS). (A) Regeneration efficiency as number of plantlets regenerated from 1 ml of settled cell volume of ECS and (B) transformation efficiency as number of transgenic lines generated from 1 ml of settled cell volume of ECS transformed. Values were presented mean ± S.E.
The regenerated shoots were transferred to light with photoperiod of 16 h where they turned green and subsequently transferred onto rooting medium for 2–4 weeks. About 99% of well-rooted plants were successfully hardened and established in pots in the glasshouse. The plants exhibited identical morphology compared with other plants developed using suckers.
Approximately 20,000–50,000 plantlets were developed from 1 ml SCV of ECS (Figure 4A). Maximum numbers of plantlets (about 50,000 plantlets per ml SCV) were regenerated from ECS of “Sukali Ndiizi,” whereas “Gros Michel” was found to be least responsive, developing about 20,000 plantlets per ml SCV. Approximately 30,000–40,000 plantlets per ml SCV were regenerated for “Cavendish Williams.” It was noted that regeneration efficiency is cultivar dependent, which was also observed in previous reports. Regeneration of 25,000–30,000 plants per 1 ml of SCV was reported for plantain cultivars (Strosse et al., 2006; Tripathi et al., 2012).
In this study, we obtained complete plantlets from ECS in 13–15 months for different banana varieties, similar to our previous report (Tripathi et al., 2012). We have observed somaclonal variations such as retarded growth, and thinner and variegated leaves in about 3–5% regenerated plants in the glasshouse. To minimize somaclonal variation, we use ECS for transformation and regeneration only for 1–1.5 years after their establishment. Production of embryogenic cell suspension cultures and regeneration is time intensive and variety dependent. It has been reported previously that regeneration of whole plantlets takes 14–42 months for banana and 18–27 months for plantain starting from callus induction (Strosse et al., 2006).
Transformation, Selection, and Regeneration of Transgenic Plant
ECS of three varieties “Gros Michel,” “Cavendish Williams,” and “Sukali Ndiizi” were transformed with Agrobacterium tumefaciens strain EHA 105 containing the pCAMBIA2301 plasmid (Supplementary Figure 1). Agro-infected cells multiplied and proliferated on EDM supplemented with kanamycin, whereas the non-transformed control cells turned black. Creamish white colored transformed embryos developed on selective medium in 1–2 months (Figure 5A). These embryos matured in 1 month after culturing them on EMM (Figure 5B) and later on the matured embryos were transferred to EGM (Figure 5C). The germination of putative transgenic plants started 1–2 months after transferring the mature embryos on EGM. The fully developed shoots with few leaves were transferred on proliferation medium (PM) and cultured in a light and dark 16/8 h cycle (Figures 5D–E). The shoots were proliferated and individual shoots were cultured on rooting medium without selection. Roots were developed in 2–4 weeks in all the shoots. Proliferation and elongation of shoots for “Gros Michel” was found to be challenging. Shoots cultured on proliferation medium turned into nodular structures instead of multiplying and elongating. To rectify this challenge, the shoots germinated on EGM were transferred into hormone-free medium for 1 month and then transferred to proliferation medium. This modification provided complete shoots from “Gros Michel.” Kanamycin was noted to have an inhibitory effect on regeneration of transformed shoots in previous studies (Yao et al., 1995; Tripathi et al., 2012), thus after regeneration of complete shoots antibiotic was withdrawn from the proliferation and rooting medium.

Figure 5. Agrobacterium-mediated transformations of embryogenic cell suspensions. (A) Agro-infected cells proliferating on selection medium, (B) embryos on embryo development medium, (C) embryos on embryo maturation medium, (D) shoot germination on embryo germination medium, (E) complete shoots in proliferation medium, (F) potted transgenic plants in glasshouse.
Approximately 20–70 transgenic lines per ml SCV were generated depending on the banana variety used in about 7–9 months after Agro-inoculation of embryogenic cells. Maximum number of transgenic lines (60–70) was obtained from ECS of “Sukali Ndiizi” and minimum number of lines (20–30) was obtained for “Gros Michel.” About 30–40 lines were developed from 1 ml SCV of embryogenic cells of “Cavendish Williams” (Figure 4B). The transformation efficiency was well-correlated to regeneration efficiency of embryogenic cells of various varieties. Higher regeneration efficiency can provide more independent transgenic shoots. Regeneration efficiency of transgenic lines depends on quality and age of embryogenic cells used for transformation. Newly developed fine cells provide more transgenic plants. Bigger clumps of cells are not good for transformation as they just turned brown and black. The transformation efficiencies obtained in this study were similar to previous reports. About 25–65 plants per 50 mg of SCV of embryogenic suspension cells were reported for variety “Cavendish” and “Lady Finger” (Khanna et al., 2004; Ghosh et al., 2009). Similarly, Ganapathi et al. (2001) reported approximately 40 transgenic plants per 0.5 ml packed cell volume of ECS of variety “Rasthali” (AAB). However, Chong-Pe'rez et al. (2012) recently reported more than 600 independent transgenic lines from 50 mg of settled cells of Dwarf Cavendish.
Fully developed rooted transgenic plants were transferred to pots in the glasshouse and about 96–100% plants were successfully established (Figure 5F). Morphologically all the transgenic plants were similar to control non-transgenic control plants. Transformation experiments were repeated three times using several lines of cells, confirming the reproducibility of the developed transformation system.
In this study we used the same protocol to transform ECS and regenerate complete plants, regardless of explants from where ECS was derived. The detailed protocol for transformation and regeneration is presented in Figure 6.
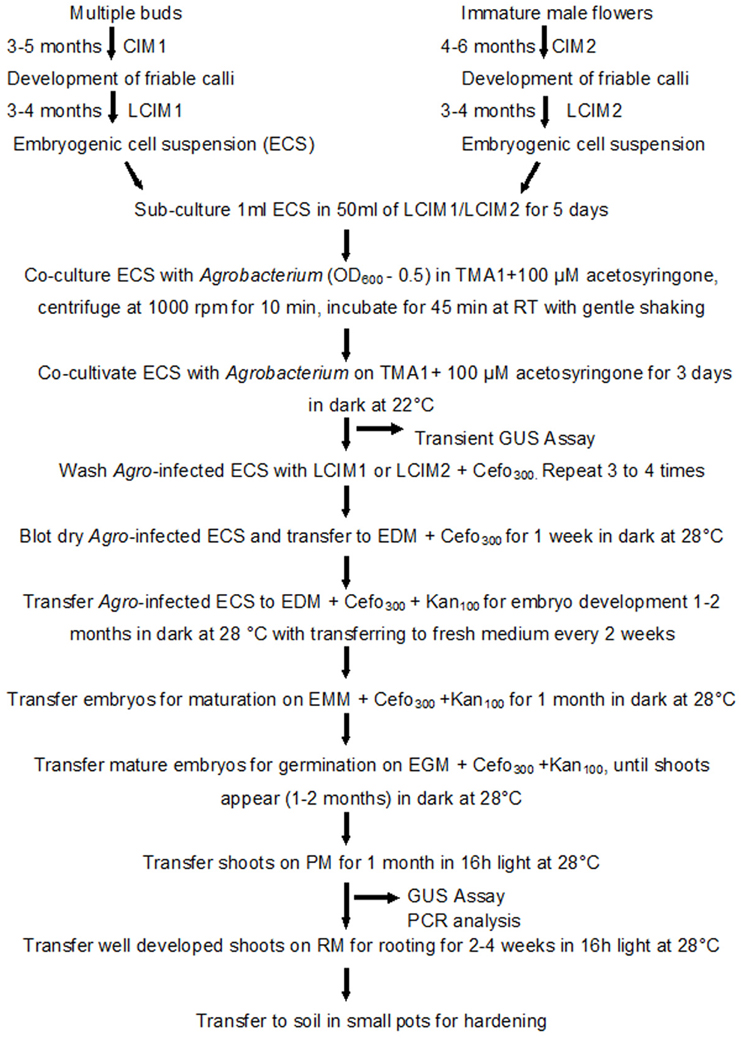
Figure 6. Schematic diagram showing various steps of genetic transformation of banana using embryogenic cell suspensions (ECS) derived from multiple buds or immature male flowers. CIM, callus induction medium; LCIM, liquid callus induction medium; TMA1, Agrobacterium resuspension medium; EDM, embryo development medium; EMM, embryo maturation medium; EGM, embryo germination medium; RM, rooting medium; Cefo300, 300 mg/l cefotaxime; Kan100, 100 mg/l Kanamycin.
Histochemical GUS Assay
GUS assay of Agro-infected embryogenic cells showed blue coloration confirming transient expression of the reporter gusA gene after 3 days of co-cultivation (Figure 7A). A blue coloration was seen in germinated embryos, leaves, petioles, pseudostems, and roots isolated from transgenic plants regenerated on kanamycin selection medium, confirming stable expression of gusA reporter gene (Figures 7B–F). The blue coloration in various tissues of transgenic plants demonstrated uniform transformation. No blue coloration was observed in control non-transformed embryogenic cells (Figure 7G), leaves, and roots of non-transgenic control plants (Figures 7H,I).
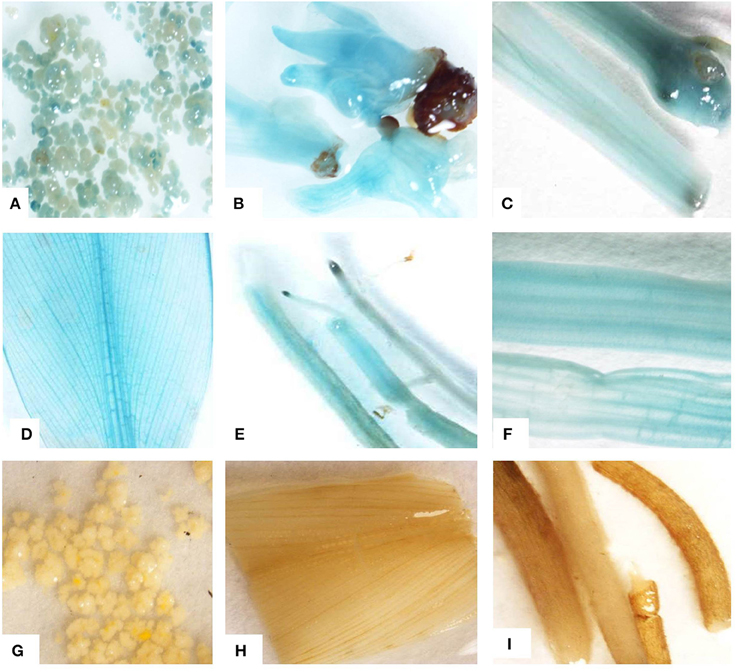
Figure 7. Histochemical GUS assay. (A) Transient expression of gusA gene in embryogenic cells after 72 h of Agro-infection, (B) stable expression of gusA gene in regenerating transgenic embryos, (C) pseudostem, (D) leaf, (E) roots, and (F) leaf petioles from transgenic plant, (G) non-transformed embryogenic cells, (H) leaf, and (I) roots from non-transgenic control plant. All the photographs were taken by Nikon SMZ 1500 microscopic camera attached with computer.
Molecular Characterization of Transgenic Lines
The presence of the transgene in randomly selected transgenic lines of “Sukali Ndiizi,” “Cavendish Williams,” and “Gros Michel” was confirmed by PCR analysis using gusA specific primers. The amplicon of expected size of 528 bp was obtained in all the transgenic lines tested validating the presence of gusA reporter gene in the genome of transgenic lines (Figure 8). No amplified PCR product was detected in non-transgenic control plants.
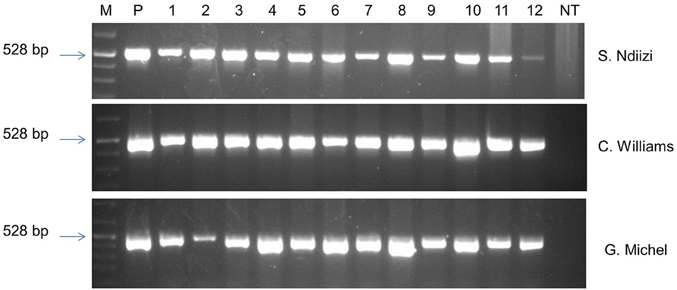
Figure 8. PCR analysis of genomic DNA of putative transgenic and non-transgenic control plants of various banana varieties using gusA specific primers. M, marker; P, plasmid DNA; NT, non-transgenic control plant.
Dot and Southern blot analysis were performed with genomic DNA isolated from PCR positive transgenic lines. Twenty four transgenic lines along with the positive control and non-transgenic control were tested in three replicates by dot blot analysis. All the transgenic lines tested showed a positive signal except for the control non-transgenic control plant (Figure 9). Dot blot is a rapid technique, which can be used for quick testing of large numbers of transgenic lines for transgene integration.
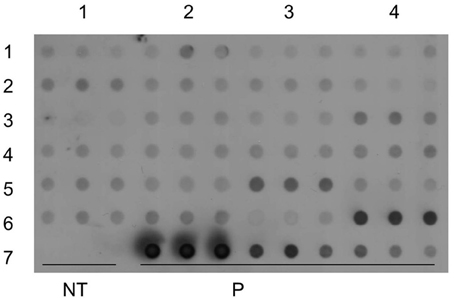
Figure 9. Molecular analysis of transgenic lines to confirm integration of transgene. Dot blot analysis of transgenic lines in triplicates, pCAMBIA2301 as a positive control in triplet (10, 5, 1 ng) and control non-transgenic plant.
The gDNA of randomly selected transgenic and non-transgenic control plants were restricted with HindIII enzyme. Southern blot analysis was performed and to confirm the transgene integration. HindIII has a single restriction site in the pCAMBIA2301 construct used for plant transformation. All 14 independent transgenic lines tested showed positive bands; however, no band was observed for the control non-transgenic control plant (Supplementary Figure 2). The copy number of integrated gusA gene was 1–4 in all the tested transgenic lines.
Testing of Robustness of the Regeneration and Genetic Transformation Protocol
A robust high throughput regeneration and genetic transformation system is required for rapid testing of large numbers of gene constructs in any crop. The main focus of research in our laboratory is to control the deadly banana Xanthomonas wilt (BXW) disease, which is the most important biotic constraint to banana production in east and central Africa (Tripathi et al., 2009, 2014b). In the absence of resistant banana germplasm and considering the difficulties associated with conventional breeding, we are applying transgenic technologies to develop BXW resistant bananas using farmer-preferred cultivars. This necessitates the development of a robust regeneration and transformation protocol capable of producing large numbers of independent transgenic lines on a routine basis. The regeneration and transformation platform established in this study has opened the door for genetic advancement of banana by inserting desirable traits such as disease and pest resistance. To validate the reproducibility of this system, various Agrobacterium-mediated transformation experiments were performed to develop BXW resistant banana using different gene constructs. In total, 867 transgenic lines were generated from ECS of three cultivars (“Cavendish Williams,” “Gros Michel,” and “Sukali Ndiizi”) with various gene constructs (Table 2). These transgenic lines were tested by PCR analysis using primers specific to target gene and selection marker gene to confirm the presence of all the transgenes. The amplicons of expected sizes provide strong evidence of a complete T-DNA insertion. PCR positive transgenic lines were further evaluated for BXW disease resistance under glasshouse conditions. As hundreds of transgenic lines are produced for testing the efficacy of each gene construct for disease resistance, only lines showing enhanced disease resistance are advanced further for detailed molecular analysis such as Southern blotting, RT-PCR, qRT-PCR, and northern blotting. The transgenic lines showing the presence of transgene/s, enhanced disease resistance, low copy integration, and expression of the transgene were selected for the confined field trial. The various steps of production of transgenic banana, molecular analysis, and evaluation for disease resistance are summarized in Figure 10. Currently, we are also developing large numbers of transgenic banana plantlets using gene constructs with other traits such as virus and fungus resistance for glasshouse screening and confined field trials.
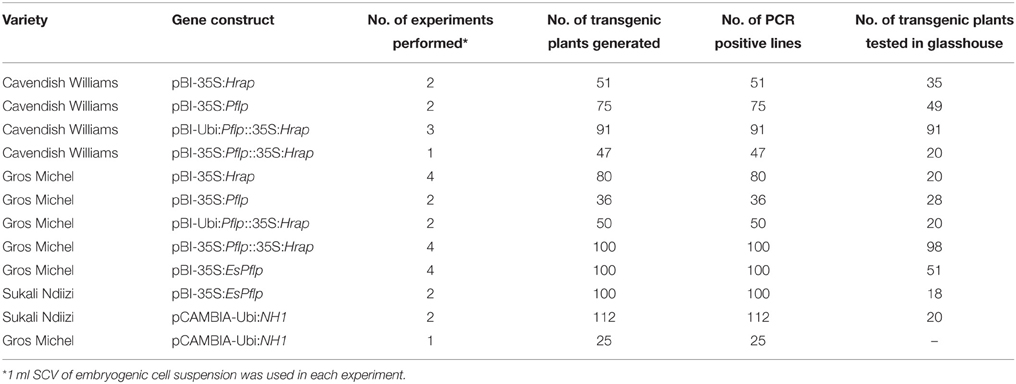
Table 2. Transgenic plants generated using various gene constructs and embryogenic cell suspensions of “Cavendish Williams,” “Gros Michel,” and “Sukali Ndiizi,” in order to develop banana Xanthomonas wilt resistant transgenic banana.
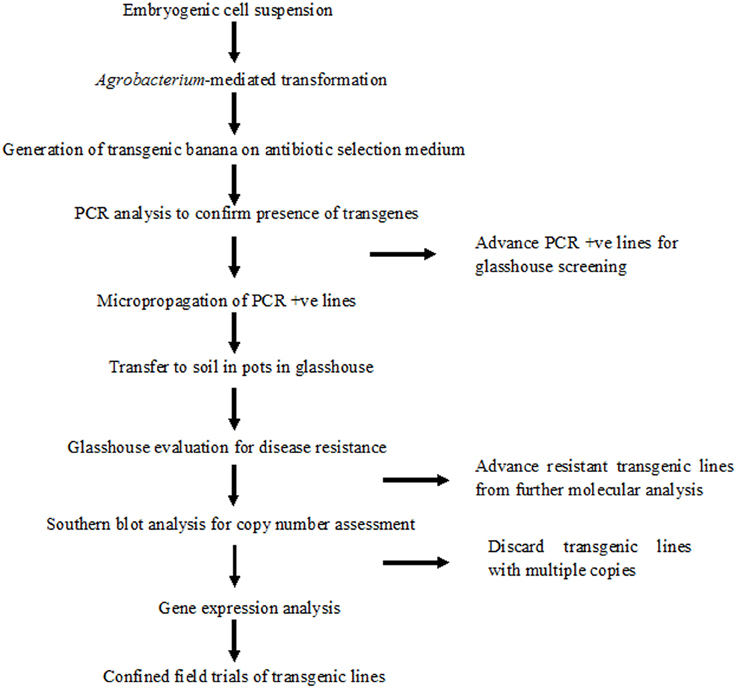
Figure 10. Schematic representation of production of banana transgenic lines, molecular analysis and evaluation for disease resistance.
Conclusion
In conclusion, we developed a high-throughput technique for Agrobacterium-mediated transformation and regeneration of economically significant banana varieties grown in east African countries and globally. The ECS of three banana varieties—“Cavendish Williams,” “Gros Michel,” and “Sukali Ndiizi”—which are highly prone to fungal as well as bacterial diseases, were successfully transformed using Agrobacterium-mediated system. Transformed ECS have the potential to generate transgenic plants with high efficiency, therefore, only a few milliliters of cells can provide a sufficient number of transformed plants with the desired traits. This high-throughput protocol can be used for functional genomics and improvement of banana varieties. Using this protocol, hundreds of transgenic banana lines with bacterial wilt resistance were generated (Table 2). This study showed potential for genetic manipulation of banana varieties for developing resistance to biotic and abiotic stresses. It is possible that with further optimization, this protocol can be applicable to other varieties of banana in future. This type of high-throughput system also serves as an important platform for transferring technologies to NARS in sub-Saharan Africa on banana improvement.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the United States Agency for International Development (USAID) for funding and Biosciences eastern and central Africa (BecA) located at the International Livestock Research Institute in Nairobi, Kenya for providing the laboratory facilities. We acknowledge Sarah Macharia, Kenneth Mburu, Kenneth Kogo, and Mark Adero for providing technical assistance. We would like to thank Evans Nyaboga for critically reviewing the manuscript. We acknowledge Academia Sinica, Taiwan, and the University of California, Davis for providing gene constructs. We also thank African Agricultural Technology Foundation (AATF) for negotiating license of Pflp and Hrap genes.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01025
References
Bakry, F., Carreel, F., Jenny, C., and Horry, J. P. (2009). “Genetic improvement of banana,” in Breeding Plantation Tree Crops: Tropical Species, eds S. M. Jain and P. M.Priyadarshan (Berlin: Springer), 3–51. doi: 10.1007/978-0-387-71201-7_1
Becker, D. K., Dugdale, B., Smith, M. K., Harding, R. M., and Dale, J. (2000). Genetic transformation of Cavendish banana (Musa spp. AAA group) cv. Grand Nain via microprojectile bombardment. Plant Cell Rep. 19, 229–234. doi: 10.1007/s002990050004
Cheng, M., Lowe, B. A., Spencer, T. M., Ye, X. D., and Armstrong, C. L. (2004). Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cell. Dev. Biol. Plant 40, 31–45. doi: 10.1079/IVP2003501
Chong-Pe'rez, B., Reyes, M., Rojas, L., Ocaña, B., Pe'rez, B., Kosky, R. G., et al. (2012). Establishment of embryogenic cell suspension cultures and Agrobacterium-mediated transformation in banana cv. “Dwarf Cavendish” (Musa AAA): effect of spermidine on transformation efficiency. Plant Cell Tiss. Org. Cult. 111, 79–90. doi: 10.1007/s11240-012-0174-1
Cote, F. X., Domergue, R., Monmarson, S., Schwendiman, J., Teisson, C., and Escalant, J. V. (1996). Embryogenic cell suspensions from the male flower of Musa AAA cv. Grand Nain. Physiol. Plant 97, 285–290.
Dheda, D., Dumortier, F., Panis, B., Vuylsteke, D., and De Langhe, E. (1991). Plant regeneration in cell suspension cultures of the cooking banana cv. Bluggoe (Musa spp, ABB group). Fruits 46, 125–135.
Escalant, J. V., Teisson, C., and Cote, F. X. (1994). Amplified somatic embryogenesis from male flowers of triploid banana and plantain varieties (Musa sp.). In Vitro Cell. Dev. Biol. Plant 30, 181–186. doi: 10.1007/BF02823029
FAOSTAT Agriculture Data (2001). Available online at: http://www.fao.org/ag
FAOSTAT Agriculture Data (2012). Available online at: http://www.fao.org/ag
FAOSTAT Agriculture Data (2013). Available online at: http://www.fao.org/ag
Ganapathi, T. R., Higgs, N. S., Balint-Kurti, P. J., Arntzen, C. J., May, G. D., and van Eck, J. M. (2001). Agrobacterium-mediated transformation of the embryogenic cell suspensions of the banana cultivar “Rasthali” (AAB). Plant Cell Rep. 20, 157–162. doi: 10.1007/s002990000287
Gawel, N. J., and Jarret, R. L. (1991). A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol. Biol. Rep. 9, 262–266. doi: 10.1007/BF02672076
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Ghosh, A., Ganapathi, T. R., Nath, P., and Bapat, V. A. (2009). Establishment of embryogenic cell suspension cultures and Agrobacterium-mediated transformation in an important Cavendish banana cv. Robusta (AAA). Plant Cell Tiss. Org. Cult. 97, 131–139. doi: 10.1007/s11240-009-9507-0
Grapin, A., Ortíz, J. L., Lescot, T., Ferrière, N., and Cote, F. X. (2000). Recovery and regeneration of embryogenic cultures from female flowers of False Horn Plantain. Plant Cell Tiss. Org. Cult. 61, 237–244. doi: 10.1023/A:1006423304033
Hansen, G., Das, A., and Chilton, M. D. (1994). Constitutive expression of the virulence genes improves the efficiency of plant transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 91, 7603–7607. doi: 10.1073/pnas.91.16.7603
Hansen, G., and Wright, M. S. (1999). Recent advances in transformation of plants. Trends Plant Sci. 4, 226–231. doi: 10.1016/S1360-1385(99)01412-0
Hiei, Y., Komari, T., and Kubo, T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35, 205–218. doi: 10.1023/A:1005847615493
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405.
Kalyebara, R., Wood, S., and Abodi, P. (2007). “Assessing the potential impact of selected technologies on the banana industry in Uganda,” in An Economic Assessment of Banana Genetic Improvement and Innovations in the Lake Victoria Region of Uganda and Tanzania, eds M. Smale and W. K. Tushemereirwe (Washington, DC: International Food Policy Research Institute), 141–156.
Khanna, H., Becker, D., Kleidon, J., and Dale, J. (2004). Centrifugation assisted Agrobacterium tumefaciens mediated transformation (CAA) of embryogenic cell suspensions of banana (Musa spp.) Cavendish AAA and Lady finger AAB. Mol. Breed. 14, 239–252. doi: 10.1023/B:MOLB.0000047771.34186.e8
Kosky, R. G., Chong-Pérez, B., López-Torres, J., Reyes, M., Bermúdez-Caraballoso, I., Martín, N. M., et al. (2010). Plantain (Musa spp. cv. Navolean AAB) transgenic plants from Agrobacterium tumefaciens-mediated transformation of embryogenic cell suspensions. Biotecnol. Veget. 10, 209–218. Available online at: http://132.248.9.34/hevila/Biotecnologiavegetal/2010/vol10/no4/2.pdf
Lee, K. S., Jeon, H. S., and Kim, M. Y. (2002). Optimization of a mature embryo-based in vitro culture system for high-frequency somatic embryogenic callus induction and plant regeneration from japonica rice cultivars. Plant Cell Tiss. Org. Cult. 71, 237–244. doi: 10.1023/A:1020305432315
Lindsey, K. (1992). Genetic manipulation of crop plants. J. Biotechnol. 26, 1–28. doi: 10.1016/0168-1656(92)90067-J
Liu, P., and Nester, E. W. (2006). Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc. Natl. Acad. Sci. U.S.A. 103, 4658–4662. doi: 10.1073/pnas.0600366103
Lorenzen, J., Tenkouano, A., Bandyopadhyay, R., Vroh, B., Coyne, D., and Tripathi, L. (2010). Over view of banana and plantain (Musa spp.) improvement in Africa: past and future. Acta Hortic. 879, 595–603. doi: 10.17660/ActaHortic.2010.879.66
Marroquin, C. G., Paduscheck, C., Escalant, J. V., and Teisson, C. (1993). Somatic embryogenesis and plant regeneration through cell suspension in Musa acuminata. In Vitro Cell. Dev. Biol. Plant 29, 43–46. doi: 10.1007/BF02632238
May, G., Afza, R., Mason, H., Wiecko, A., Novak, F., and Arntzen, C. (1995). Generation of transgenic banana (Musa acuminata) plants via Agrobacterium-mediated transformation. Biotechnology 13, 486–492. doi: 10.1038/nbt0595-486
Morel, G., and Wetmore, R. H. (1951). Tissue culture of monocotyledons. Am. J. Bot. 38, 138–140. doi: 10.2307/2437836
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Navarro, C., Escobedo, R. M., and Mayo, A. (1997). In vitro plant regeneration from embryogenic cultures of a diploid and a triploid, Cavendish banana. Plant Cell Tiss. Org. Cult. 5, 17–25. doi: 10.1023/A:1005965030075
Novak, F. J., Afza, R., van Duren, M., Perea-Dallos, M., Conger, B. V., and Xiaolang, T. (1989). Somatic embryogenesis and plant regeneration in suspension cultures of dessert (AA and AAA) and cooking (ABB) bananas (Musa spp.). Bio. Technol. 7, 147–158.
Nyaboga, E., Tripathi, J. N., Manoharan, R., and Tripathi, L. (2014). Agrobacterium-mediated genetic transformation of yam (Dioscorea rotundata): an important tool for functional study of genes and crop improvement. Front. Plant Sci. 5:463. doi: 10.3389/fpls.2014.00463
Peraza-Echeverria, S., James-Kay, A., Canto-Canché, B., and Castillo-Castro, E. (2007). Structural and phylogenetic analysis of Pto-type disease resistance gene candidates in banana. Mol. Genet. Genomics 278, 443–453. doi: 10.1007/s00438-007-0262-9
Pillay, M., Tenkouano, A., and Hartman, J. (2002). “Bananas and plantains: future challenges in Musa breeding,” in Crop Improvement, Challenges in the Twenty-First Century, ed M. S. Kang (New York, NY: Food Products Press), 223–252.
Roux, N. (2004). “Mutation induction in Musa,” in Banana Improvement: Cellular, Molecular Biology, and Induced Mutations, eds S. Mohan Jain and R Swennen (Enfield, NH: Science Publishers), 23–32.
Roux, N., Baurens, F. C., Dolezel, J., Hribova, E., Heslop-Harrison, P., Town, C., et al. (2008). “Genomics of banana and plantain (Musa spp.), major staple crops in the tropics,” in Genomics of Tropical Crop Plants, Plant Genetics and Genomics: Crops and Models, eds P. H. Moore and R. Ming (New York, NY: Springer), 83–111.
Santos-Rosa, H., Kirmizis, A., Nelson, C., Bartke, T., Saksouk, N., Cote, J., et al. (2009). Histone H3 tail clipping regulates gene expression. Nat. Struct. Mol. Biol. 16, 17–22. doi: 10.1038/nsmb.1534
Schenk, R. U., and Hildebrandt, A. C. (1972). Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can. J. Bot. 50, 199–204. doi: 10.1139/b72-026
Strosse, H., Schoofs, H., Panis, B., Andre, E., Reyniers, K., and Swennen, R. (2006). Development of embryogenic cell suspensions from shoot meristematic tissue bananas and plantains (Musa spp.). Plant Sci. 170, 104–112. doi: 10.1016/j.plantsci.2005.08.007
Tripathi, J., Muwonge, A., and Tripathi, L. (2012). Highly efficient regeneration and transformation protocol for plantain cv. Gonja manjaya (Musa spp. AAB) using embryogenic cell suspension. In Vitro Cell. Dev. Biol. Plant 48, 216–224. doi: 10.1007/s11627-011-9422-z
Tripathi, J. N., Lorenzen, J., Bahar, O., Ronald, P., and Tripathi, L. (2014a). Transgenic expression of the rice Xa21 pattern-recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum. Plant Biotech. J. 12, 663–673. doi: 10.1111/pbi.12170
Tripathi, L., Mwangi, M., Abele, S., Aritua, V., Tushemereirwe, W. K., and Bandyopadhyay, R. (2009). A threat to banana production in East and Central Africa. Plant Dis. 93, 440–451. doi: 10.1094/PDIS-93-5-0440
Tripathi, L., Tripathi, J. N., and Hughes, J., d A. (2005). Agrobacterium-mediated transformation of plantain (Musa spp.) cultivar Agbagba. Afr. J. Biotechnol. 4, 1378–1383. doi: 10.4314/ajb.v4i12.71498
Tripathi, L., Tripathi, J. N., Kiggundu, A., Korie, S., Shotkoski, F., and Tushemereirwe, W. K. (2014b). Field trial of Xanthomonas wilt disease-resistant bananas in East Africa. Nat. Biotechnol. 32, 868–870. doi: 10.1038/nbt.3007
Tripathi, L., Tripathi, J. N., and Tushemereirwe, W. K. (2008). Rapid and efficient production of transgenic East African Highland Banana (Musa spp.) using intercalary meristematic tissues. Afri. J. Biotechnol. 7, 1438–1445.
Tushemereirwe, W. K., Kangire, A., and Ssekiwoko, F. (2004). First report of Xanthomonas campestris pv. musacearum on banana in Uganda. Plant Path. 53, 802. doi: 10.1111/j.1365-3059.2004.01090.x
Xu, C. X., Pains, B., Strosse, H., Li, H. P., Xiao, H. G., Fan, H., et al. (2005). Establishment of embryogenic cell suspensions and plant regeneration of the dessert banana “Williams” (Musa AAA group). J. Hortic. Sci. Biotechnol. 80, 523–528.
Yao, J. L., Cohen, D., Atkinson, R., Richardson, K., and Morris, B. (1995). Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep. 14, 407–412. doi: 10.1007/BF00234044
Youssef, M. A., James, C. A., Mosqueda, A. M., Ku-Cauich, J. R., Grijalva-Arango, R., and Escobedo-GM, R. M. (2010). Influence of genotype and age of explant source on the capacity for somatic embryogenesis of two “Cavendish” banana cultivars (Musa acuminata Colla, AAA). Afr. J. Biotechnol. 9, 2216–2223.
Keywords: banana, Agrobacterium-mediated transformation, embryogenic cell suspension, immature male flowers, multiple buds
Citation: Tripathi JN, Oduor RO and Tripathi L (2015) A High-Throughput Regeneration and Transformation Platform for Production of Genetically Modified Banana. Front. Plant Sci. 6:1025. doi: 10.3389/fpls.2015.01025
Received: 16 July 2015; Accepted: 05 November 2015;
Published: 26 November 2015.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Martha Magaretha O'Kennedy, CSIR, South AfricaBeena M. Ravindran, Leibniz Institute-DSMZ, Germany
Copyright © 2015 Tripathi, Oduor and Tripathi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leena Tripathi, l.tripathi@cgiar.org
 Jaindra N. Tripathi
Jaindra N. Tripathi Richard O. Oduor
Richard O. Oduor Leena Tripathi
Leena Tripathi