- 1Key Laboratory of Molecular Epigenetics of Ministry of Education and Institute of Genetics and Cytology, Northeast Normal University, Changchun, China
- 2Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA
Ethylene regulates many aspects of plant growth and development including seed germination, leaf senescence, and fruit ripening, and of plant responses to environmental stimuli including both biotic and abiotic stresses. Ethylene response factors (ERFs) are plant-specific transcription factors and are a subfamily of the AP2 (APETALA2)/ERF transcription factor family. The function of many members in this large gene family remains largely unknown. ERF96, a member of the Group IX ERF family transcription factors, has recently been shown to be a transcriptional activator that is involved in plant defense response in Arabidopsis. Here we provide evidence that ERF96 is a positive regulator of abscisic acid (ABA) responses. Bioinformatics analysis indicated that there are a total four small ERFs in Arabidopsis including ERF95, ERF96, ERF97, and ERF98, and that ERF96 forms a cluster with ERF95 and ERF97. By using quantitative RT-PCR, we found that ERF96 is expressed in all tissues and organs examined except roots, with relatively high expression in flowers and seeds. Results from the protoplast transfection assay indicated that the EDLL motif-containing C-terminal domain is responsible for ERF96’s transcriptional activity. Although loss-of-function mutant of ERF96 was morphologically similar to wild type plants, transgenic plants overexpressing ERF96 had smaller rosette size and were delayed in flowering time. In ABA sensitivity assays, we found that ERF96 overexpression plants were hypersensitive to ABA in terms of ABA inhibition of seed germination, early seedling development and root elongation. Consistent with these observations, elevated transcript levels of some ABA-responsive genes including RD29A, ABI5, ABF3, ABF4, P5CS, and COR15A were observed in the transgenic plants in the presence of ABA. However, in the absence of ABA treatment, the transcript levels of these ABA-responsive genes remained largely unchanged. Our experiments also showed that water loss in ERF96 overexpression plants was slower than that in Col wild type plants. Stomatal closure assays indicated that ERF96 overexpression plants had reduced stomatal aperture in the presence of ABA. Taken together, our results suggest that ERF96 positively regulates ABA responses in Arabidopsis.
Introduction
Ethylene response factors (ERFs) are a subfamily of the AP2 (APETALA2)/ERF superfamily, one of the several plant-specific transcription factor families (Riechmann et al., 2000). According to the number and similarity of their DNA binding domains, AP2/ERF superfamily is classified into five subfamilies: ERF, AP2, dehydration-responsive element (DRE) binding protein, related to ABSCISIC ACID INSENSITIVE3 (ABI3)/VIVIPAROUS1 (VP1), and others (Sakuma et al., 2002). ERF proteins contain only one AP2/ERF domain (Riechmann et al., 2000; Sakuma et al., 2002; Licausi et al., 2013).
Ethylene response factors are involved in the regulation of plant growth and development, primary and secondary metabolism, and plant responses to environmental stimuli including biotic and abiotic stresses (Gutterson and Reuber, 2004; Nakano et al., 2006; Xu et al., 2011; Licausi et al., 2013; Mizoi et al., 2013; Müller and Munné-Bosch, 2015). To date, ERF transcription factors have been identified and characterized from a number of plant species such as Arabidopsis (Nakano et al., 2006), rice (Nakano et al., 2006; Sharoni et al., 2011; Rashid et al., 2012), cotton (Jin and Liu, 2008), poplar (Zhuang et al., 2008), soybean (Zhang et al., 2008), barley (Gil-Humanes et al., 2009), grape (Zhuang et al., 2009), maize (Zhuang et al., 2010), tomato (Sharma et al., 2010), apple (Zhuang et al., 2011), cucumber (Hu and Liu, 2011), wheat (Zhuang et al., 2011), kiwifruit (Yin et al., 2012), peach (Zhang et al., 2012a), plum (Du et al., 2012), castor bean (Xu et al., 2013), Chinese cabbage (Li et al., 2013; Song et al., 2013), Medicago truncatula (Zhang et al., 2013), sorghum (Yan et al., 2013), sweet orange (Ito et al., 2014), and potato (Charfeddine et al., 2015).
In Arabidopsis, there are a total of 147 genes encoding AP2/ERF transcription factors, and 122 of them encode ERF transcription factors (Nakano et al., 2006). Based on phylogenetic analysis using the AP2/ERF domains, ERF transcription factors in Arabidopsis can be further classified into 12 different groups, namely, groups I to X, VI-L and Xb-L (Nakano et al., 2006).
Some of the group I and V ERF transcription factors have been shown to be involved in the regulation of the expression of lipids and cell wall components biosynthesis genes, basic type defense-related genes, pathogenesis-related genes, and osmotin, chitinase and β-1,3-glucanase encoding genes (Licausi et al., 2013). Some of them have been shown to be involved in the regulation of plant responses to abiotic and biotic stresses by either activating or repressing abscisic acid (ABA)-responsive genes (Gutterson and Reuber, 2004; Nakano et al., 2006; Xu et al., 2008, 2011; Licausi et al., 2013; Mizoi et al., 2013). For example, AtERF4 over-expression plants were less sensitive to ABA inhibited root elongation which involves negative regulation of ethylene and ABA responses (Yang et al., 2005). AtERF7 binds to the GCC box and represses the expression of ABA-responsive genes (Zhang et al., 2007). ABR1 or ERF111 acts as a negative regulator of ABA responses during seed germination and ABA- and stress-regulated gene expression (Pandey et al., 2005) whereas transgenic plant overexpressing AtERF13 confers ABA hypersensitivity in Arabidopsis (Lee et al., 2010). AtERF15 was shown to act as a positive regulator of ABA responses (Lee et al., 2015). On the other hand, ABA can also induce the expression of some ERF genes. For example, the expression of cotton ERF gene GbERF, tobacco ERF gene NtCEF1 and tomato ERF gene JERF1/3 has been shown to be induced by ABA (Wang et al., 2004; Zhang et al., 2004; Lee et al., 2005).
Subgroup IXc in group IX ERF subfamily contains four small ERFs with amino acids ranged from 131 to 139. These four ERFs are ERF95, ERF96, ERF97, and ERF98. In addition to the AP2/ERF domain, these ERFs contain an unknown function motif named CMIX-1 (Nakano et al., 2006). Among them, ERF95, also named ESE1 (ETHYLENE AND SALT INDUCIBLE 1), and ERF98 has been shown to be involved in the regulation of salt tolerance (Zhang et al., 2011, 2012b). ERF97, previously named AtERF14, has been shown to regulate plant defense response (Oñate-Sánchez et al., 2007). Recently, ERF96 has also been shown to regulate plant defense response (Catinot et al., 2015). Here we provide evidence that ERF96 is involved in the regulation of ABA response in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotypic Columbia (Col-0) was used for protoplast isolation and plant transformation. The Arabidopsis mutant erf96-1, isolated from the transposon line GT_5_54244, is in Landsberg erecta (Ler) ecotypic background.
For seed germination, green seedlings, and root elongation assays, and for RNA isolation from seedlings, seeds were surface sterilized and sown on plates containing 0.6% (w/v) phytoagar solidified 1/2 Murashige and Skoog (MS) basal medium with vitamins (Plantmedia, Dublin, OH, USA) and 1% (w/v) sucrose. The plates were kept at 4°C in darkness for 2 days before being transferred to a growth room. For plant transformation and protoplasts isolation, seeds were sown directly into soil pots and grown in a growth room. The growth conditions in the growth room were set with temperature at 22°C, and photoperiod of 14 h light/10 h dark with light density of approximately 120 μmol m–2 s–1. For water loss and water-use efficiency assays, plants were grown in a growth chamber under a photoperiod of 10 h light/14 h dark photoperiod (short-day conditions).
Plasmid Construction
To generate the 35S:HA-ERF96 and 35S:GD-ERF96 constructs, the full-length open-reading frame (ORF) of ERF96 gene was amplified by RT-PCR using RNA isolated from 7-day-old Arabidopsis seedlings, and the PCR products were cloned in-frame with an N-terminal HA or GD tag into the pUC19 vector under the control of the double 35S enhancer promoter of CaMV (Wang et al., 2005). 35S:HA-ERF96 construct was digested with EcoR I restriction enzyme, and sub-cloned into the binary vector pPZP211 for plant transformation (Hajdukiewicz et al., 1994).
To generate 35S:GD-EDLL and 35S:GD-ERF96ΔEDLL constructs, the nucleotide sequences encoding the EDLL motif containing C-terminal domain (amino acid 105–131) and ERF96ΔEDLL (amino acid 1–104) were amplified by RT-PCR using 35S:GD-ERF96 plasmids as template, and cloned in-frame with a GD tag into the pUC19 vector. The primers used to amplify the full-length ORF of ERF96 were 5′-ATGGATC-AAGGAGGTCGAGG-3′ and 5′-TCATTTCTTCTTGCCCTTG-3′. The primers used to amplify the EDLL motif were 5′-CATA-TGGAATTTGAGTACTTGGATG-3′ and 5′-TCATTTCTTCTT-GCCCTTG-3′. The primers used amplify the ERF96ΔEDLL were 5′-ATGGATCAAGGAGGTCGAGG-3′ and 5′-CTTAAGTC-AAAAAACTTGCCTAGAAG-3′.
Plant Transformation and Transgenic Plant Selection
Arabidopsis plants of ∼5-week-old with several mature flowers on the main inflorescence were used for transformation via Agrobacterium tumefaciens GV3101-mediated floral dip method (Clough and Bent, 1998). T1 seeds were planted on 1/2 MS medium containing 50 μg/ml Kanamycin and 100 μg/ml Carbenicillin for selecting transgenic plants. Phenotypes of transgenic plants were examined in the T1 generation and confirmed in T2 up to T4 generations. Overexpression of ERF96 in the transgenic plants was confirmed by RT-PCR. A minimum of five independent overexpression lines with similar phenotypes were obtained, and two homozygous lines were selected for further analysis.
Arabidopsis Leaf Mesophyll Protoplast Transfection Assay
The procedure for Arabidopsis leaf mesophyll protoplast isolation, transfection and GUS activity assay had been described previously (Wang et al., 2005, 2007, 2014; Zhou et al., 2014; Liu et al., 2015). Plasmid DNAs for reporter and effector genes were isolated using the GoldHi EndoFree Plasmid Maxi Kit (Kangwei) according to the manufacturer’s instructions. GUS activities were measured by using a Synergy™ HT micro-plate reader (BioTEK).
ABA Sensitivity Assays
For seed germination and green seedling assays, sterilized seeds were sown on 1/2 MS medium containing 0 (solvent alone), 1 or 2 μM ABA and grown in a growth room. Seed germination was scored 48 h after plates had been transferred into the growth room. Green seedlings were scored 10 days after transferring. The assays were repeated three times. For root elongation assay, 4-day-old seedlings grown on vertical plates were transferred to 1/2 MS medium plates containing 0 or 5.0 μM ABA and the plates were placed vertically. Root length was measured 6 days after seedling transferring. A minimum of 10 seedlings per line were used. For ABA-responsive gene expression assay, 7-day-old wild type and ERF96 transgenic seedlings grown on vertical plates were transferred into 1/2 MS liquid medium without phytoagar and incubated for 90 min, then treated with 50 μM ABA for 2 h before the seedlings were frozen in liquid N2.
RNA Isolation, RT-PCR, and Quantitative RT-PCR
Total RNA from Arabidopsis seedlings and different tissues and organs was isolated as described previously (Wang et al., 2014, 2015; Guo et al., 2015; Liu et al., 2015). cDNA was synthesized using 1 μg of total RNA by Oligo(dT)-primed reverse transcription using OMNISCRIPT RT Kit (QIAGEN). Quantitative RT-PCR (qRT-PCR) was used to examine the expression of ABA-responsive genes including RESPONSIVE DROUGHT 29A (RD29A), ABSCISIC ACID INSENSITIVE 5 (ABI5), ABSCISIC ACID RESPONSIVE ELEMENT-BINDING FACTOR 3 (ABF3), ABRE BINDING FACTOR 4 (ABF4), Δ1-PYRROLINE-5-CARBOXYLATE SYNTHETASE (P5CS) and COLD-RESPONSIVE 15A (COR15A). The expression of ACTIN2 (ACT2) was used as a control. qRT-PCR was performed on the Applied Biosystems 7500 real time PCR System using SYBR Green/ROX Master Mix (Thermo Scientific). The primers used for qRT-PCR examination of RD29A, ABF3, P5CS, COR15A, and ACT2 have been described previously (Strizhov et al., 1997; Nakashima et al., 2006; Liu et al., 2014, 2015; Lu et al., 2015; Wang et al., 2015). Other primers used for qRT-PCR were: ABI5, 5′-GGAGATTGCGGACATTG-ATGAG-3′ and 5′-GGGAACACTAGTAAAGCAGATC-3′, ABF4, 5′-ACTGGAAGCCGAAATTGAAAAGCTC-3′ and 5′-CACCATGGTCCGGTTAATGTCCT-3′, ERF95, 5′-CCAT-TCTCAATTTTCCTCAC-3′ and 5′-AACTCAATAACTTCC-CTCCC-3′, ERF96, 5′-GCGGCTAGAGCCTATG-3′ and 5′-GTACTTG GATGATAGTG-3′, ERF97, 5′-ACCGTGGAGT-AAGGAGAC-3′ and 5′-GAAGTTGAGAATGGCAGC-3′, ERF98, 5′-GGAGCAGCAACAACCAAT-3′ and 5′-AGCGAG-ATGACCCCTAAG-3′.
Water Loss and Stomatal Aperture Assays
Water loss was measured according to the method described by Tian et al. (2004). Briefly, rosettes of 5-week-old Col and ERF96 transgenic plants were cut and weighed at different time points after cutting. The experiments were performed at room temperature under dim light conditions with 50% relative humidity. Three plants per genotype were used, and water loss was calculated as the percentage of initial fresh weight at each time point.
Stomatal aperture bioassay was performed as described by Lee et al. (2013) with some modifications. Four rosette leaves were detached from 5-week-old plants and floated in stomatal opening solution (SOS: 50 mM KCl and 10 mM MES-KOH, pH 6.15, 10 μM CaCl2) in light for 2 h, then the solution was replaced with SOS containing 20 μM ABA. Leaves were incubated for another 2 h before stomata were observed. For each sample, 100 stomatas were randomly observed under a digital microscope (eclipse 80i, Nikon Instruments Inc., Shanghai, China) and the width of individual stomata was recorded using Image J1. The experiment was repeated three times.
Analysis of Water-Use Efficiency
About 5-week-old plants grown under short day conditions were sprayed with 75 μmol ABA or solvent containing distilled water as control. Instantaneous leaf water-use efficiency was measured 3 h after spraying by using a portable open-flow gas exchange system LI-6400 (LI-COR Biosciences, Lincoln, NE, USA), and calculated as described by Han et al. (2013).
Results
ERF96 is a Small ERF Transcription Factor
It has been reported that ERF96 belongs to the Group IX ERF family proteins (Nakano et al., 2006). In Arabidopsis, ERF95, ERF96, ERF97, and ERF98 are the only four small ERF proteins with amino acids ranged from 131 to 139 (Figure 1A). These four ERFs share 68–88% similarity and 47–74% identity at the amino acid level (Figure 1B). When the full-length amino acid sequence of ERF96 was used as a template to search for sequence homologues encoded by the Arabidopsis genome using the “Protein Homologs” search tool at Phytozome2, a total of 28 proteins were identified, and all of them are ERF proteins (Nakano et al., 2006). Phylogenetic analysis indicated that ERF95, ERF96, and ERF97 were closely clustered, whereas ERF98 was separated from this cluster (Figure 1C).
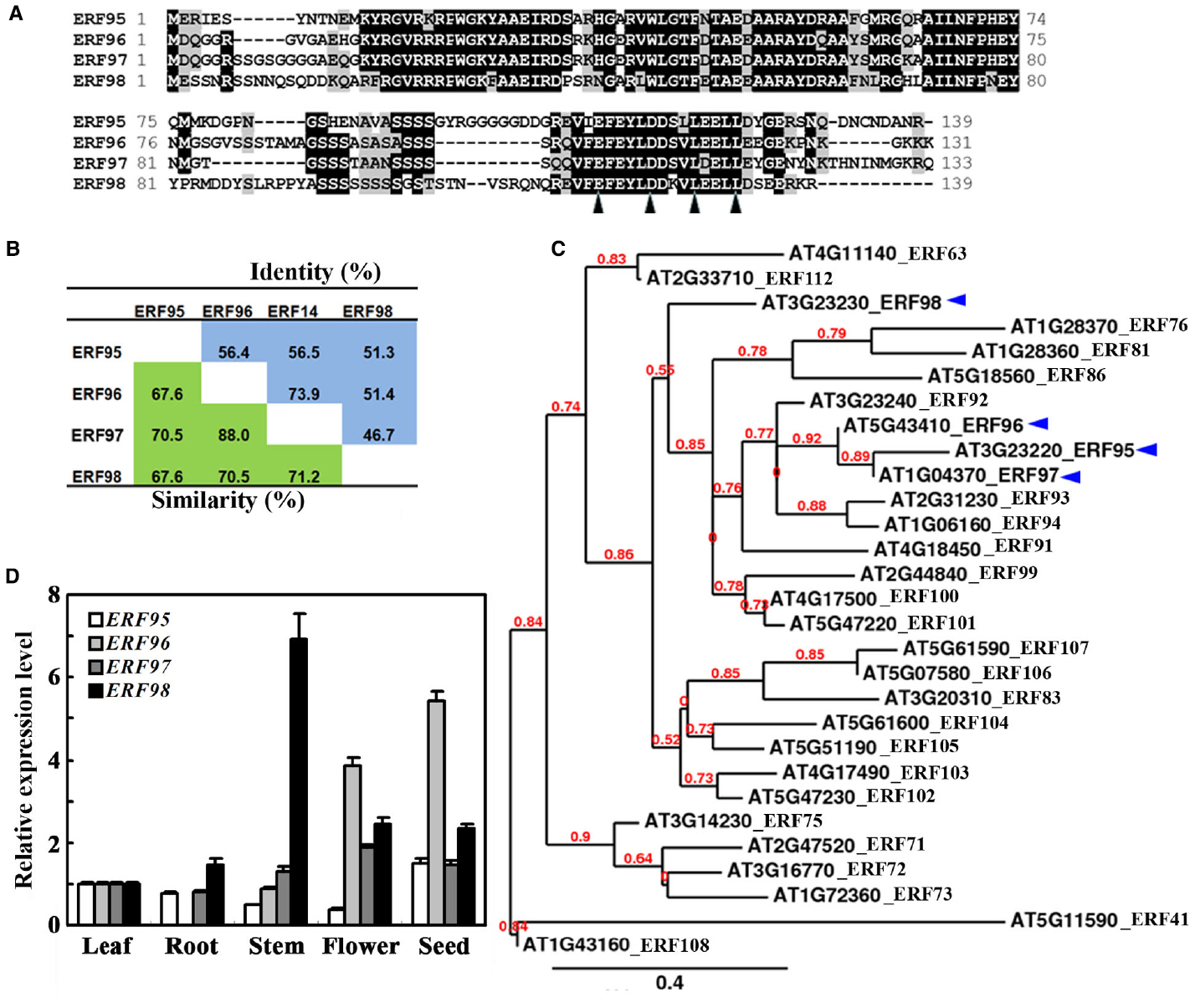
Figure 1. Small ERFs in Arabidopsis. (A) Amino acid sequence alignment of small ERF proteins in Arabidopsis. Identical amino acids were shaded in black, and similar amino acids were in grey. Arrowheads indicate the conserved amino acid residues in the EDLL motif. (B) Amino acid similarity and identity of small ERF proteins in Arabidopsis. (C) Phylogenetic analysis of ERF96 and its homologues in Arabidopsis. (D) Expression pattern of ERF95, ERF96, ERF97, and ERF98. RNA was isolated from different tissues and organs of Col wide-type plants. Leaves and roots were from 4-week-old plants. qRT-PCR was used to examine the expression of the small ERF genes. The expression of ACT2 was used as a control. The transcript level of the ERF genes in leaves was set at 1. Data represent mean ± SD of three biological replicates.
Expression Patterns of ERF96
Previous experiments have shown that ERF95 and ERF98 are involved in the regulation of plant response to abiotic stresses (Zhang et al., 2012b), whereas ERF96 and ERF97 are involved in the regulation of plant response to biotic stresses (Oñate-Sánchez et al., 2007; Catinot et al., 2015). Because some of the ERFs have been shown to regulate plant responses to abiotic and biotic stresses by regulating ABA-responsive genes (Gutterson and Reuber, 2004; Nakano et al., 2006; Xu et al., 2008, 2011; Licausi et al., 2013; Mizoi et al., 2013), we wanted to examine whether small ERFs may involve in the regulation of ABA signaling in Arabidopsis by taking ERF96 as an example. To do that, we first examined the expression pattern of ERF96 in Arabidopsis by using qRT-PCR. As shown in Figure 1D, relatively high expression of ERF96 was observed in seeds and flowers whereas the transcript of ERF96 in roots was undetectable. For comparison, we also examined the expression pattern of the other three small ERF genes. We found that none of them had an expression pattern similar to that of ERF96 (Figure 1D).
EDLL Motif is Responsible for ERF96’s Transcriptional Activity
Based on amino acid sequence analysis, small ERFs including ERF96 do not contain an obvious activation or repression domain. However, ERF96 functions as a transcription activator in protoplasts transfection assays (Catinot et al., 2015), and it contains an EDLL motif, a motif that has been shown to be presented in several ERF transcriptional activators (Tiwari et al., 2012), at its C-terminus (Figure 1A). We thus wanted to examine whether the EDLL motif is responsible for ERF96’s transcriptional activity by using the Arabidopsis mesophyll protoplast transient expression system (Wang et al., 2007). Plasmids of GAL4 DNA binding domain (GD) fused with ERF96, EDLL containing C-terminal domain (EDLL) or ERF096ΔEDLL were cotransfected with GAL4:GUS reporter plasmids into protoplasts. Cotransfection of GD plasmids was used as a control. As shown in Figure 2, both ERF96 and EDLL activated the expression of the reporter gene whereas ERF96ΔEDLL failed to do so, suggesting that the EDLL motif is responsible for ERF96’s transcriptional activity.
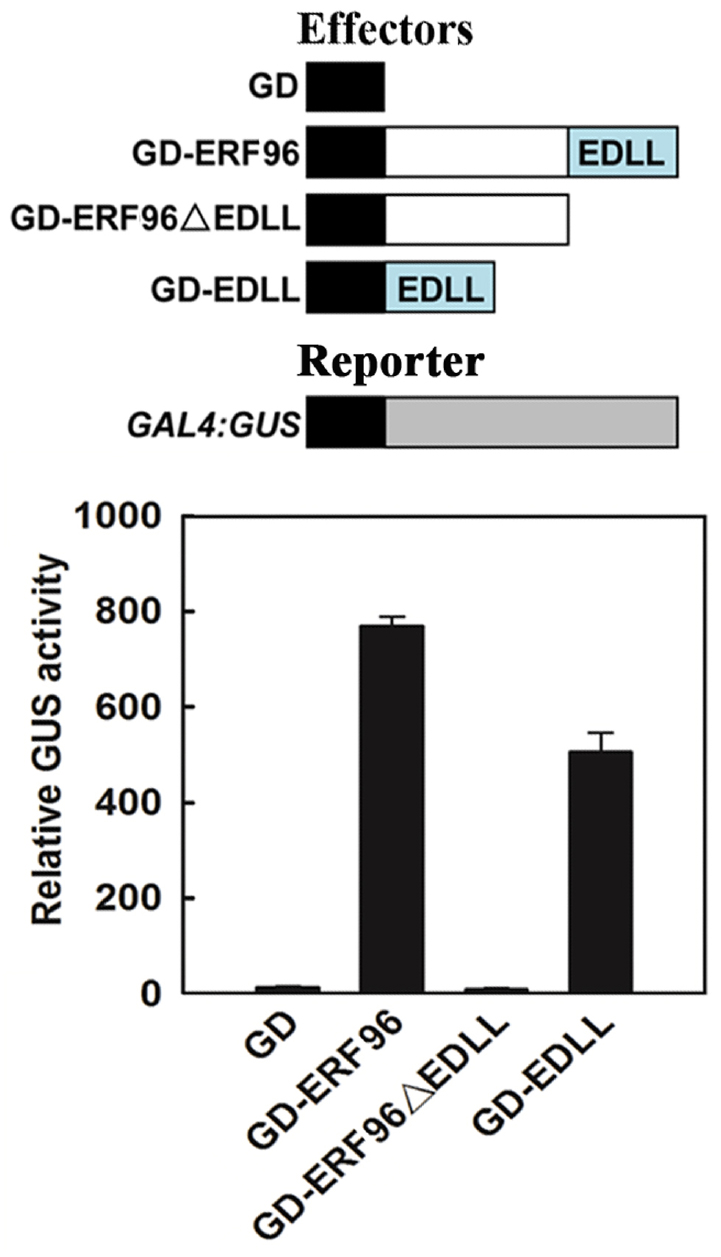
Figure 2. The EDLL motif is responsible for ERF96’s transcriptional activity. Plasmids of GD-ERF96, GD-ERF96ΔEDLL and GD-EDLL or GD alone (as a control) were co-transfected with a GAL4:GUS reporter into protoplasts isolated from rosette leaves of Col wild type plants. Transfected protoplasts were incubated in the darkness for 20–22 h before GUS activities were measured. Data represent mean ± SD of three replicates. Effectors and reporter used were diagrammed on the top of the figure.
Overexpression of ERF96 Affects Plant Growth and Development
To functionally characterize ERF96 in Arabidopsis, we first tried to identify loss-of-function mutant of ERF96. We found one transposon insertion line (GT_5_54244) through T-DNA Express: Arabidopsis Gene Mapping Tool3. In this line, the transposon was inserted at the first exon of ERF96 gene (Figure 3A). RT-PCR analysis indicated that the full-length ERF96 transcript was absent in this line (Figure 3B), suggesting that it represents a loss-of-function allele. This allele was designated as erf96-1. The erf96-1 mutant displayed wild-type morphology at both vegetative and reproductive stages (Figures 3C,D). We then generated transgenic lines overexpressing ERF96 in Col-0 ecotypic background. Morphologically, ERF96 overexpression plants had smaller rosette size when compared with Col wild-type (Figures 4A,C). ERF96 overexpression plants also showed late flowering phenotypes (Figures 4A,D).

Figure 3. Identification of the erf96-1 mutant. (A) Transposon insertion site in the erf96-1 mutant. (B) RT-PCR analysis of ERF96 transcript. RNA was isolated from 7-day-old seedlings. Expression of ACT2 was used as a control. (C) The erf96-1 mutant at vegetative stage. (D) The erf96-1 mutant at reproductive stage. Bar in (C,D), 1 cm.
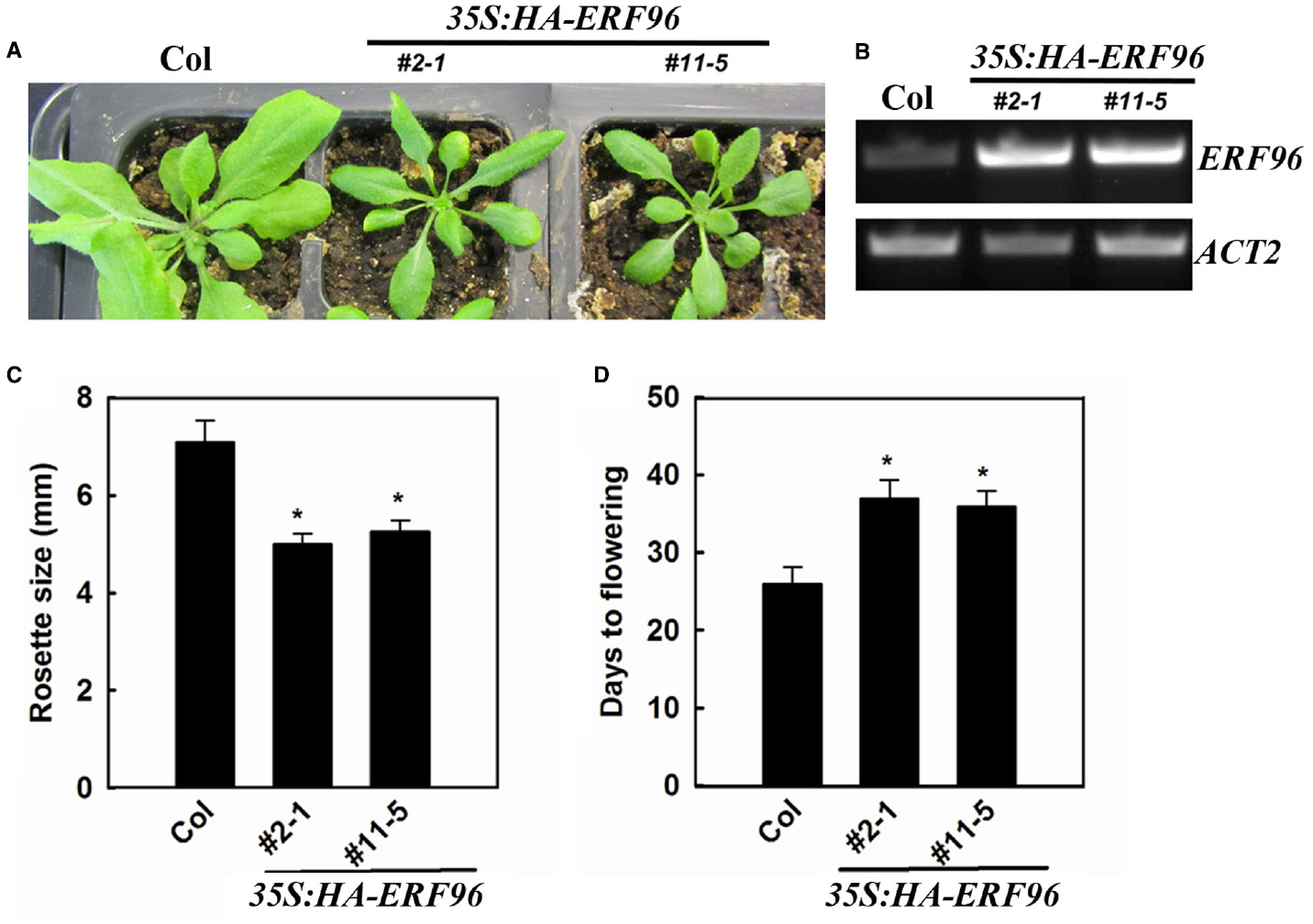
Figure 4. Phenotypes of transgenic plants overexpressing ERF96. (A) Phenotypes of 4-week old ERF96 overexpression plants. (B) Expression of ERF96 in the transgenic plants. RNA was isolated from 7-day-old seedlings, and RT-PCR was used to examine the expression of ERF96. Expression of ACT2 was used as a control. (C) Rosette size of the ERF96 overexpression plants. (D) Flowering time of the ERF96 overexpression plants. Data in (C,D) represent the mean ± SD of 10 individual plants. Asterisk (*) indicates significantly different from Col wild-type (p < 0.05).
Transgenic Plants Overexpressing ERF96 are Hypersensitive to ABA
Some ERF proteins are involved in plant response to abiotic and biotic stresses (Gutterson and Reuber, 2004; Nakano et al., 2006; Xu et al., 2011; Licausi et al., 2013; Mizoi et al., 2013; Müller and Munné-Bosch, 2015). Available evidence suggested that this is also true for the small ERF proteins (Oñate-Sánchez et al., 2007; Zhang et al., 2011, 2012b; Catinot et al., 2015). Because ABA acts as one of the most important stress hormones and some ERFs such as AtERF4, AtERF7, and AtERF111 have been shown to be involved in ABA responses (Pandey et al., 2005; Yang et al., 2005; Zhang et al., 2007), we examined the ABA sensitivity of ERF96 overexpression lines and the erf96-1 mutant. We used the three different assays that have been commonly used to assess ABA sensitivities, including seed germination, early seedling development and root elongation. We found that in each of these assays, ERF96 overexpression plants were hypersensitive to ABA (Figure 5), suggesting that ERF96 positively regulates ABA responses. However, erf96-1 mutant had near wild-type responses in each of these assays (data not shown).
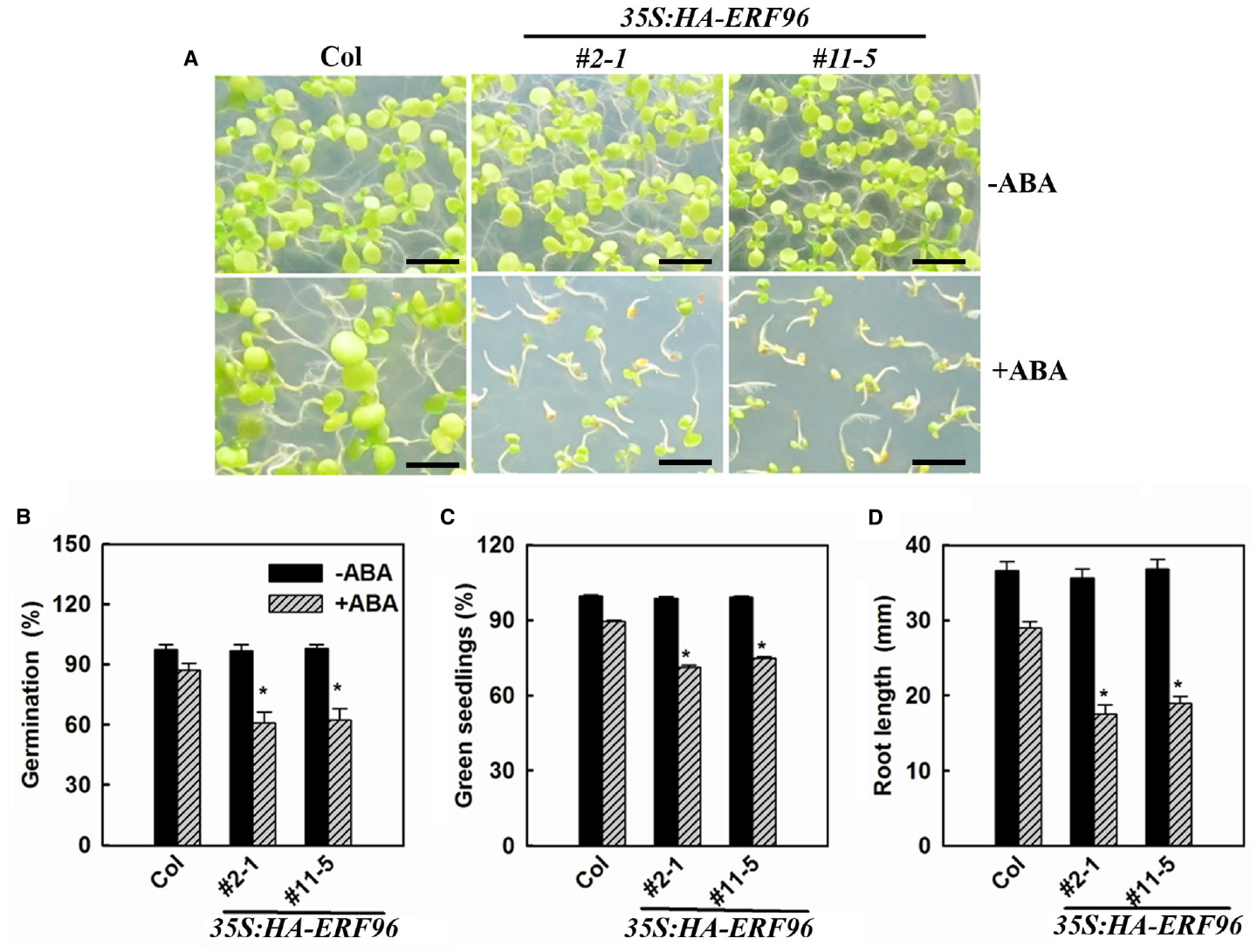
Figure 5. ERF96 overexpression plants are hypersensitive to ABA. (A) Ten-day-old Col wild type and ERF96 overexpression seedlings grown on plates in the presence or absence of 2.0 μM ABA. Bar, 5 mm. (B) Effects of ABA on seed germination. Seeds were sown on plates in the presence or absence of 1.0 μM ABA, and percentage of seed germination was scored 48 h after the plates had been transferred into a growth room. (C) Effects of ABA on seedling greening. Seeds were sown on plates in the presence or absence 2.0 μM ABA. The percentage of green seedlings was scored 10 days after the plates had been transferred into a growth room. (D) Effects of ABA root elongation. The primary root length was measured 4 days after seedlings had been transferred to 1/2 MS plates containing 5.0 μM ABA. Data in (B,C) represent the mean ± SD of three replicates. Data in (D) represent the mean ± SD of 10 individual seedlings. Asterisk (*) indicates significantly different from Col in the presence of ABA (p < 0.05).
Expression of Some ABA-Responsive Genes is Increased in Transgenic Plants Overexpressing ERF96 Upon ABA Treatment
Because the expression of ERF96 was not induced by ABA (Hoth et al., 2002), and ERF96 overexpression plants displayed ABA hypersensitivity in each of those three different assays (Figure 5), we wanted to further examine whether ERF96 may regulate the expression of ABA-responsive genes. Because erf96-1 mutant was morphologically similar to wild type (Figure 3), had a near wild type response to ABA, and ERF96 RNAi plants displayed a wild type defense response (Catinot et al., 2015), our assays hereon focused on ERF96 overexpression plants. qRT-PCR was used to examine the expression of several selected ABA-responsive genes including RD29A, ABI5, ABF3, ABF4, P5CS, and COR15A (Baker et al., 1994; Strizhov et al., 1997; Kang et al., 2002; Nakashima et al., 2006; Liu and Stone, 2010). As shown in Figure 6, when compared with that in Col wild type seedlings, the expression of all the genes examined, but RD29A, remained largely unchanged in the transgenic plant seedlings in the absence of ABA. ABA treatment induced the expression of all of these genes in Col wild type seedlings, and the expression of these ABA-responsive genes was further elevated in ERF96 overexpression plant seedlings when compared with that in the Col wild-type seedlings (Figure 6). These results indicate that ERF96 positively regulates ABA-induced gene expression, suggesting that ERF96 positively regulates ABA responses.
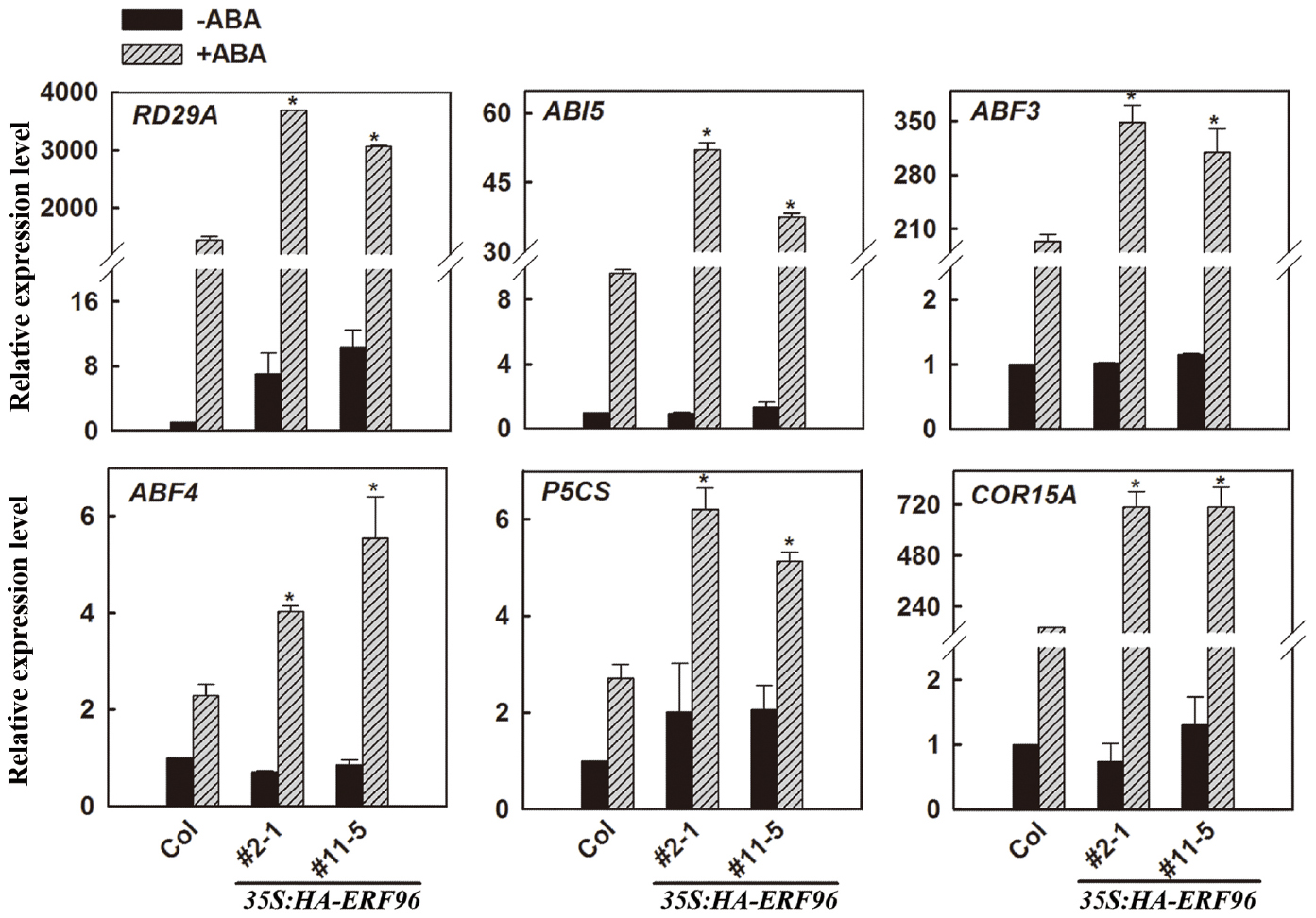
Figure 6. Expression of ABA-responsive genes in ERF96 overexpression plants. RNA was isolated from 7-day-old seedlings with or without ABA treatment. qRT-PCR was used to examine the expression of RD29A, ABI5, ABF3, ABF4, P5CS, and COR15A. The expression of ACT2 was used as a control. The transcript level of the corresponding gene in Col without ABA treatment was set at 1.0. Data represent the means ± SD of three biological replicates. Asterisk (*) indicates significantly different from that in ABA-treated Col wild type seedlings (p < 0.05).
ERF96 Regulates Water Loss
Abscisic acid is a critical regulator of stomatal movements that are associated with water loss in plants. Having shown that ERF96 functions as a positive regulator of ABA responses, we wanted to further examine whether ERF96 overexpression plants display altered water loss. The water loss from the detached whole rosette of Col wild-type and ERF96 transgenic plants grown in short-day conditions was measured. As shown in Figure 7A, ERF96 overexpression plants lost water significantly slower than the Col wild-type plants. Consistent with these results, stomatal closure assays indicated that stomatal aperture in ERF96 overexpression plants was small than wild type in the presence of ABA (Figures 7B,C). We also examined water-use efficiency in the ERF96 transgenic plants. We found that, in the absence of ABA treatment, instantaneous leaf water-use efficiency in the ERF96 transgenic plants was similar to that in the Col wild-type plants (Figure 7D). In the presence of ABA treatment, however, the instantaneous leaf water-use efficiency in the ERF96 transgenic plants was higher when compared to that in the Col wild-type plants (Figure 7D).
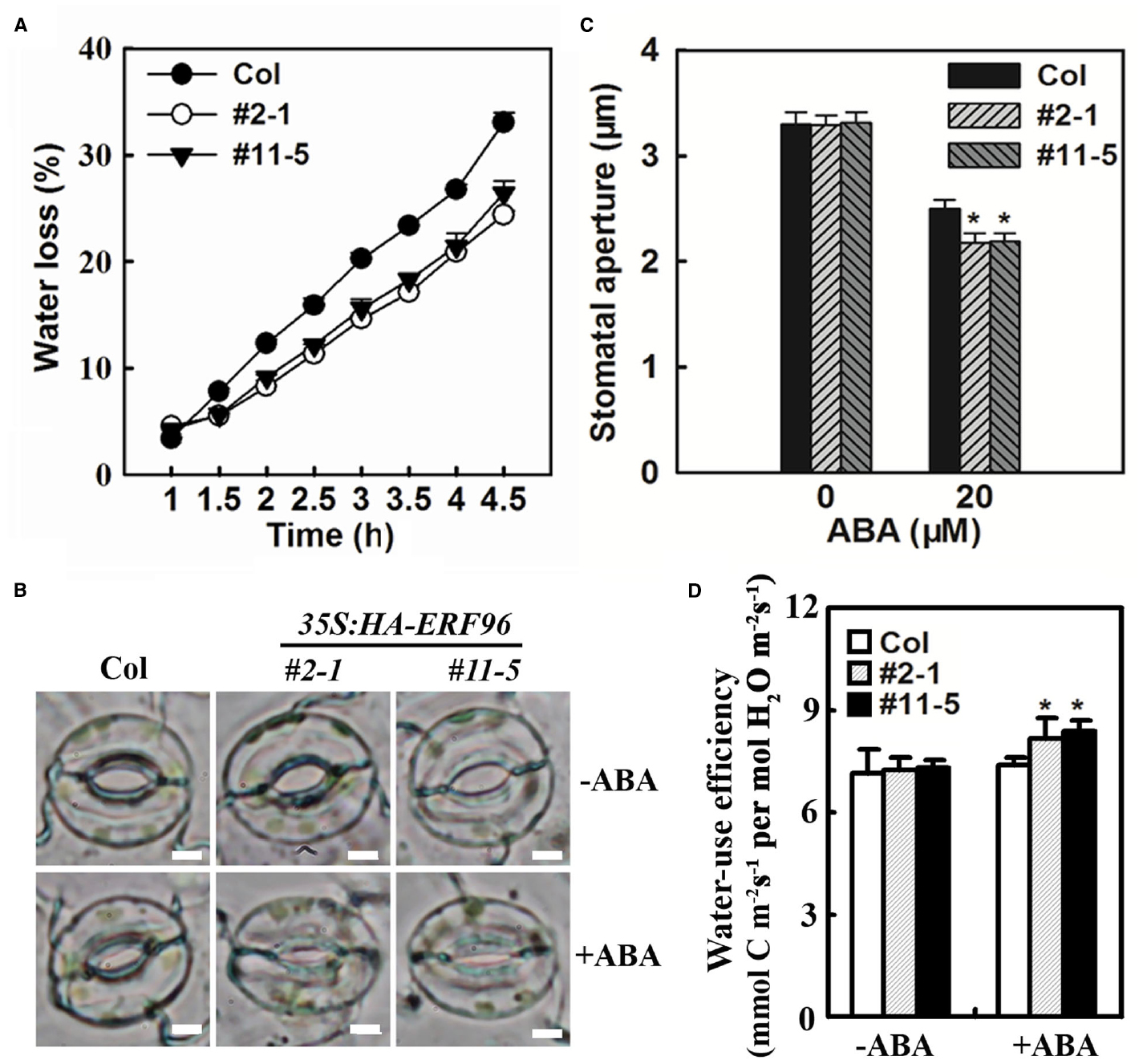
Figure 7. Water loss and stomatal aperture in ERF96 overexpression plants. (A) Water loss assay in Col wild type plants and ERF96 overexpression plants. Whole rosettes of 5-week-old plants grown under short-day conditions were cut off from the base and used for water loss assay. Data represent the mean ± SD of three replicates. (B) Stomatal aperture in Col wild and ERF96 overexpression plants. Shown are representative images of stomata before or after ABA treatment. Bar, 4 μm. (C) Measurement of stomatal apertures in Col wild and ERF96 overexpression plants. Data represent the means ± SD of 100 stomata. Asterisk (*) indicates significantly different from that in Col before ABA treatment (p < 0.05). (D) Instantaneous leaf water-use efficiency in Col wild and ERF96 overexpression plants. Data represent the means ± SD of three replicates. Asterisk (*) indicates significantly different from that in Col before ABA treatment (p < 0.05).
Discussion
ERF96 has recently been shown to regulate plant defense response (Catinot et al., 2015). In this study, we provide molecular and genetic evidence that ERF96 is a positive regulator of ABA responses.
ERF96 is Involved in the Regulation of ABA Signaling
Among the small ERFs, ERF95 enhances transcript levels of salt-related genes, such as COR15A, RD29A, P5CS2, and HOOKLESS1 (HLS1, Zhang et al., 2011). ERF97 is able to activate the transcription of some ERF-target genes including PLANT DEFENSIN1.2 (PDF1.2) and BASIC CHITINASE (CHIB, Oñate-Sánchez et al., 2007), possibly through binding of the GCC box, which has been shown to be a binding site for several other Arabidopsis ERFs (Fujimoto et al., 2000). ERF98 promotes the expression of genes related to the AsA-GSH cycle, such as ASCORBATE PEROXIDASE 3 (APX3), APX6, CHLOROPLASTIC DHAR (ChlDHAR), CYTOSOLIC DHAR (CytDHAR), and GLUTATHIONE REDUCTASE 1 (GR1, Zhang et al., 2012b). ERF96 functions as a transcriptional activator, and it can directly activate some of the jasmonic acid/ethylene-response defense genes including PDF1.2a, PATHOGENESIS-RELATED 3 (PR-3), PR-4, and VEGETATIVE STORAGE PROTEIN 2 (VSP2; Catinot et al., 2015).
We provided evidence that ERF96 functions as a positive regulator of ABA responses (Figure 5). Expression of some ABA-responsive genes including RD29A, ABI5, ABF3, ABF4, P5CS, and COR15A was elevated upon ABA treatment (Figure 6). However, considering the fact that the expression of some ABA-responsive genes remained largely unchanged in the ERF96 overexpression plants in the absence of ABA (Figure 6), it is unlikely that ERF96 directly activates the expression of these ABA-responsive genes. Further studies will be required to pinpoint the precise action site of ERF96 in the ABA signaling network.
ERF96 May Function Redundantly with Other ERFs to Regulate Plant Growth and ABA Responses
Functional redundancy has been a common theme for members of ERF transcription factors. So far, phenotypes have only been observed in loss-of-function alleles of a very limited number of ERFs, such as the loss-of-function mutants of AtERF4 and NICOTIANA BENTHAMIANA CELL DEATH (NbCD1, McGrath et al., 2005; Nasir et al., 2005). The absence of scorable phenotypes in null alleles of most ERFs has made it difficult to assess the function of ERFs through loss-of-function studies. Although overexpression study has its limitation in defining gene function, it has been helpful for the characterization of some ERFs. For example, overexpression of AtERF7 reduced ABA responses in guard cells and decreased drought tolerance (Song et al., 2005), overexpression of CYTOKININ RESPONSE FACTOR 5 (CRF5) increased pathogen resistance and activated the expression of a large number of GCC-box pathogenesis-related genes (Liang et al., 2010), and overexpression of TRANSLUCENT GREEN (TG) in transgenic plants conferred enhanced drought tolerance (Zhu et al., 2014). Plants overexpressing ERF96 showed enhanced defense response, however, ERF96 RNAi plants had wild type response (Catinot et al., 2015). We found that loss-of-function allele of ERF96 did not show any morphological phenotypes and had a near wild-type ABA sensitivity, whereas ERF96 overexpression plants displayed morphological phenotypes and showed hypersensitivity to ABA (Figures 3–5). Thus it is likely that ERF96 may function redundantly with other ERFs including small ERFs. Combination of double, triple and higher order mutations in ERFs may help address the functional redundancy of ERFs.
In summary, we provide molecular, biochemical and genetic evidence that ERF96 functions as a positive regulator of ABA responses. Overexpression of ERF96 also conferred reduction in water loss from leaf surface likely through more sensitive stomatal closure. Manipulating the expression level of ERF96 may help improve crops’ water use efficiency in agriculture.
Author Contributions
SW and JC conceived the study. XW, SW, and JC designed the experiments. XW, SL, and HT performed the experiments. XW, SW, and JC analyzed the data. XW drafted the manuscript, and all authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Key Laboratory of Molecular Epigenetics of MOE (130014542), a startup fund from Northeast Normal University (www.nenu.edu.cn), the Programme for Introducing Talents to Universities (B07017), and by the Plant–Microbe Interfaces Scientific Focus Area in the Genomic Science Program, United States Department of Energy, Office of Science, Biological and Environmental Research. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the United States Department of Energy under contract DE-AC05-00OR22725. XW was partially supported by a visiting scholarship from the China Scholarship Council.
Footnotes
- ^ http://imagej.nih.gov/ij
- ^ http://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://signal.salk.edu/cgi-bin/tdnaexpress
References
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2015). ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate- and ethylene-responsive defence genes. Plant Cell Environ. doi: 10.1111/pce.12583 [Epub ahead of print].
Charfeddine, M., Saïdi, M. N., Charfeddine, S., Hammami, A., and Gargouri Bouzid, R. (2015). Genome-wide analysis and expression profiling of the ERF transcription factor family in Potato (Solanum tuberosum L.). Mol. Biotechnol. 57, 348–358. doi: 10.1007/s12033-014-9828-z
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Du, D., Hao, R., Cheng, T., Pan, H., Yang, W., Wang, J., et al. (2012). Genome-wide analysis of the AP2/ERF gene family in Prunus mume. Plant Mol. Biol. Rep. 31, 741–750. doi: 10.1007/s11105-012-0531-6
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Gil-Humanes, J., Piston, F., Martin, A., and Barro, F. (2009). Comparative genomic analysis and expression of the APETALA2-like genes from barley, wheat, and barley-wheat amphiploids. BMC Plant Biol. 9:66. doi: 10.1186/1471-2229-9-66
Guo, H., Zhang, W., Tian, H., Zheng, K., Dai, X., Liu, S., et al. (2015). An auxin responsive CLE gene regulates shoot apical meristem development in Arabidopsis. Front. Plant Sci. 6:295. doi: 10.3389/fpls.2015.00295
Gutterson, N., and Reuber, T. L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7, 465–471. doi: 10.1016/j.pbi.2004.04.007
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. doi: 10.1007/BF00014672
Han, X., Tang, S., An, Y., Zheng, D., Xia, X., and Yin, W. (2013). Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 64, 4589–4601. doi: 10.1093/jxb/ert262
Hoth, S., Morgante, M., Sanchez, J. P., Hanafey, M. K., Tingey, S. V., and Chua, N. H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115, 4891–4900. doi: 10.1242/jcs.00175
Hu, L., and Liu, S. (2011). Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 34, 624–633. doi: 10.1590/S1415-47572011005000054
Ito, T. M., Polido, P. B., Rampim, M. C., Kaschuk, G., and Souza, S. G. (2014). Genome-wide identification and phylogenetic analysis of the AP2/ERF gene superfamily in sweet orange (Citrus sinensis). Genet. Mol. Biol. 13, 7839–7851. doi: 10.4238/2014.September.26.22
Jin, L. G., and Liu, J. Y. (2008). Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton (Gossypium hirsutum). Plant Physiol. Biochem. 46, 46–53. doi: 10.1016/j.plaphy.2007.10.004
Kang, J. Y., Choi, H. I., Im, M. Y., and Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. doi: 10.1105/tpc.010362
Lee, J. H., Kim, D. M., Lee, J. H., Kim, J., Bang, J. W., Kim, W. T., et al. (2005). Functional characterization of NtCEF1, an AP2/EREBP-type transcriptional activator highly expressed in tobacco callus. Planta 222, 211–224. doi: 10.1007/s00425-005-1525-5
Lee, S. B., Lee, S. J., and Kim, S. Y. (2015). AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 34, 71–81. doi: 10.1007/s00299-014-1688-2
Lee, S. C., Lim, C. W., Lan, W., He, K., and Luan, S. (2013). ABA signaling in guard cells entails a dynamic protein–protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 6, 528–538. doi: 10.1093/mp/sss078
Lee, S. J., Park, J. H., Lee, M. H., Yu, J. H., and Kim, S. Y. (2010). Isolation and functional characterization of CE1 binding proteins. BMC Plant Biol. 10:277. doi: 10.1186/1471-2229-10-277
Li, M. Y., Wang, F., Jiang, Q., Li, R., Ma, J., and Xiong, A. S. (2013). Genome-wide analysis of the distribution of AP2/ERF transcription factors reveals duplication and elucidates their potential function in Chinese Cabbage (Brassica rapa ssp. pekinensis). Plant Mol. Biol. Rep. 31, 1002–1011. doi: 10.1007/s11105-013-0570-7
Liang, Y. S., Ermawati, N., Cha, J. Y., Jung, M. H., Su’udi, M., Kim, M. G., et al. (2010). Overexpression of an AP2/ERF-type transcription factor CRF5 confers pathogen resistance to Arabidopsis plants. J. Korean Soc. Appl. Biol. Chem. 53, 142–148. doi: 10.3839/jksabc.2010.024
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, D., Li, W., Cheng, J., and Hou, L. (2014). Expression analysis and functional characterization of a cold-responsive gene COR15A from Arabidopsis thaliana. Acta Physiol. Plant 36, 2421–2432. doi: 10.1007/s11738-014-1615-8
Liu, H., and Stone, S. L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22, 2630–2641. doi: 10.1105/tpc.110.076075
Liu, S., Hu, Q., Luo, S., Li, Q., Yang, X., Wang, X., et al. (2015). Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 6:388. doi: 10.3389/fpls.2015.00388
Lu, Y., Sasaki, Y., Li, X., Mori, I. C., Matsuura, T., Hirayama, T., et al. (2015). ABI1 regulates carbon/nitrogen-nutrient signal transduction independent of ABA biosynthesis and canonical ABA signalling pathways in Arabidopsis. J. Exp. Bot. 66, 2763–2771. doi: 10.1093/jxb/erv086
McGrath, K. C., Dombrecht, B., Manners, J. M., Schenk, P. M., Edgar, C. I., Maclean, D. J., et al. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. doi: 10.1104/pp.105.068544
Mizoi, J., Ohori, T., Moriwaki, T., Kidokoro, S., Todaka, D., Maruyama, K., et al. (2013). GmDREB2A;2, a canonical Dehydration-Responsive Element-Binding Protein2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161, 346–361. doi: 10.1104/pp.112.204875
Müller, M., and Munné-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Nasir, K. H. B., Takahashi, Y., Ito, A., Saitoh, H., Matsumura, H., Kanzaki, H., et al. (2005). High-throughput in planta expression screening identifies a class II ethylene-responsive element binding factor-like protein that regulates plant cell death and non-host resistance. Plant J. 43, 491–505. doi: 10.1111/j.1365-313X.2005.02472.x
Oñate-Sánchez, L., Anderson, J. P., Young, J., and Singh, K. B. (2007). AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 43, 400–409. doi: 10.1104/pp.106.086637
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L., and Luan, S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139, 1185–1193. doi: 10.1104/pp.105.066324
Rashid, M., He, G. Y., Yang, G. X., Hussain, J., and Yan, X. (2012). AP2/ERF transcription factor in rice: genome-wide canvas and syntenic relationships between monocots and eudicots. Evol. Bioinform. 8, 321–355. doi: 10.4137/EBO.S9369
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sharma, M. K., Kumar, R., Solanke, A. U., Sharma, R., Tyagi, A. K., and Sharma, A. K. (2010). Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol. Genet. Genomics 284, 455–475. doi: 10.1007/s00438-010-0580-1
Sharoni, A. M., Nuruzzaman, M., Satoh, K., Shimizu, T., Kondoh, H., Sasaya, T., et al. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52, 344–360. doi: 10.1093/pcp/pcq196
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Song, X., Li, Y., and Hou, X. (2013). Genome-wide analysis of the AP2/ERF transcription factor superfamily in chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics. 14, 573. doi: 10.1186/1471-2164-14-573
Strizhov, N., Abrahám, E., Okrész, L., Blickling, S., Zilberstein, A., Schell, J., et al. (1997). Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 12, 557–569. doi: 10.1046/j.1365-313X.1997.00537.x
Tian, L., DellaPenna, D., and Zeevaart, J. A. D. (2004). Effect of hydroxylated carotenoid deficiency on ABA accumulation in Arabidopsis. Physiol. Plant. 122, 314–320. doi: 10.1111/j.1399-3054.2004.00409.x
Tiwari, S. B., Belachew, A., Ma, S. F., Young, M., Ade, J., Shen, Y., et al. (2012). The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. doi: 10.1111/j.1365-313X.2012.04935.x
Wang, H., Huang, Z., Chen, Q., Zhang, Z., Zhang, H., Wu, Y., et al. (2004). Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol. Biol. 55, 183–192. doi: 10.1007/s11103-004-0113-6
Wang, S., Kwak, S. H., Zeng, Q., Ellis, B. E., Chen, X. Y., Schiefelbein, J., et al. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134, 3873–3882. doi: 10.1242/dev.009597
Wang, S., Li, E., Porth, I., Chen, J. G., Mansfield, S. D., and Douglas, C. J. (2014). Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 4, 5054. doi: 10.1038/srep05054
Wang, S., Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2005). AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17, 1979–1993. doi: 10.1105/tpc.105.031096
Wang, X., Wang, X., Hu, Q., Dai, X., Tian, H., Zheng, K., et al. (2015). Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 83, 300–311. doi: 10.1111/tpj.12887
Xu, W., Li, F., Ling, L., and Liu, A. (2013). Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genomics 14:785. doi: 10.1186/1471-2164-14-785
Xu, Z., Chen, M., Li, L. C., and Ma, Y. (2008). Functions of the ERF transcription factor family in plants. Botany 86, 969–977. doi: 10.1139/B08-041
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yan, H. W., Hong, L., Zhou, Y. Q., Jiang, H. Y., Zhu, S. W., Fan, J., et al. (2013). A genome-wide analysis of the ERF gene family in sorghum. Genet. Mol. Res. 12, 2038–2055. doi: 10.4238/2013.May.13.1
Yang, Z., Tian, L., Latoszek-Green, M., Brown, D., and Wu, K. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596. doi: 10.1007/s11103-005-7294-5
Yin, X. R., Allan, A. C., Xu, Q., Burdon, J., Dejnoprat, S., Chen, K. S., et al. (2012). Differential expression of kiwifruit ERF genes in response to postharvest abiotic stress. Postharvest Biol. Technol. 66, 1–7. doi: 10.1016/j.postharvbio.2011.11.009
Zhang, G., Chen, M., Chen, X., Xu, Z., Guan, S., Li, L.C., et al. (2008). Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 59, 4095–4107. doi: 10.1093/jxb/ern248
Zhang, C. H., Shangguan, L. F., Ma, R. J., Sun, X., Tao, R., Guo, L., et al. (2012a). Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica). Genet. Mol. Res. 11, 4789–4809. doi: 10.4238/2012.October.17.6
Zhang, Z., Wang, J., Zhang, R., and Huang, R. (2012b). The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 71, 273–287. doi: 10.1111/j.1365-313X.2012.04996.x
Zhang, H., Li, W., Chen, J., Yang, Y., Zhang, Z., Zhang, H., et al. (2007). Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 63, 63–71. doi: 10.1007/s11103-006-9072-4
Zhang, H., Lu, X., Huang, D., and Huang, R. (2004). The ethylene-, jasmonate-, abscisic acid- and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220, 262–270. doi: 10.1007/s00425-004-1347-x
Zhang, L., Li, Z., Quan, R., Li, G., Wang, R., and Huang, R. (2011). An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865. doi: 10.1104/pp.111.179028
Zhang, Z., Zhou, Q., Yang, Z., and Jiang, J. (2013). Discovery AP2/ERF family genes in silico in Medicago truncatula. Afr. J. Biotechnol. 12, 3636–3642.
Zhou, L., Zheng, K., Wang, X., Tian, H., Wang, X., and Wang, S. (2014). Control of trichome formation in Arabidopsis by poplar single-repeat R3 MYB transcription factors. Front. Plant Sci. 5:262. doi: 10.3389/fpls.2014.00262
Zhu, D., Wu, Z., Cao, G., Li, J., Wei, J., Tsuge, T., et al. (2014). TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol. Plant 7, 601–615. doi: 10.1093/mp/sst152
Zhuang, J., Cai, B., Peng, R. H., Zhu, B., Jin, X. F., Xue, Y., et al. (2008). Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Bioph. Res. Commun. 371, 468–474. doi: 10.1016/j.bbrc.2008.04.087
Zhuang, J., Chen, J. M., Yao, Q. H., Xiong, F., Sun, C. C., Zhou, X. R., et al. (2011). Discovery and expression profile analysis of AP2/ERF family genes from Triticum aestivum. Mol. Biol. Rep. 38, 745–753. doi: 10.1007/s11033-010-0162-7
Zhuang, J., Deng, D. X., Yao, Q. H., Zhang, J. A., Xiong, F., Chen, J. M., et al. (2010). Discovery, phylogeny and expression patterns of AP2-like genes in maize. Plant Growth Regul. 62, 51–58. doi: 10.1007/s10725-010-9484-7
Keywords: ERF96, ethylene response factor, transcription factor, ethylene, ABA, Arabidopsis
Citation: Wang X, Liu S, Tian H, Wang S and Chen J-G (2015) The Small Ethylene Response Factor ERF96 is Involved in the Regulation of the Abscisic Acid Response in Arabidopsis. Front. Plant Sci. 6:1064. doi: 10.3389/fpls.2015.01064
Received: 19 September 2015; Accepted: 16 November 2015;
Published: 26 November 2015.
Edited by:
David J. Burritt, University of Otago, New ZealandReviewed by:
Biswapriya B. Misra, University of Florida, USAAntonio Ferrante, Università degli Studi di Milano, Italy
Copyright © 2015 Wang, Liu, Tian, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shucai Wang, wangsc550@nenu.edu.cn
 Xiaoping Wang1,2
Xiaoping Wang1,2