- Biotechnology Division, Defence Institute of Bio-Energy Research, Haldwani, India
NAC genes contribute to enhance survivability of plants under conditions of environmental stress and in secondary growth of the plants, thereby building biomass. Thus, genetic transformation of plants using NAC genes provides a possibility to tailor biofuel plants. Over-expression studies have indicated that NAC family genes can provide tolerance to various biotic and abiotic stresses, either by physiological or biochemical changes at the cellular level, or by affecting visible morphological and anatomical changes, for example, by development of lateral roots in a number of plants. Over-expression of these genes also work as triggers for development of secondary cell walls. In our laboratory, we have observed a NAC gene from Lepidium latifolium contributing to both enhanced biomass as well as cold stress tolerance of model plants tobacco. Thus, we have reviewed all the developments of genetic engineering using NAC genes which could enhance the traits required for biofuel plants, either by enhancing the stress tolerance or by enhancing the biomass of the plants.
Introduction
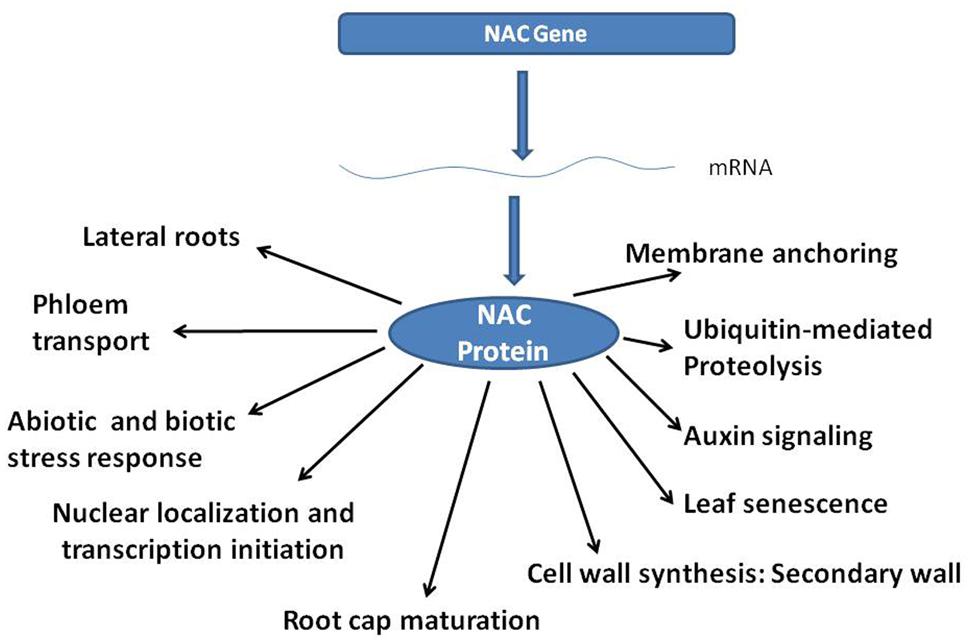
NAC (NAM, ATAF, CUC) genes containing the NAC domain, constitute one of the largest plant-specific transcription factor (TF) families. NAC family TFs are characterized by a highly conserved N-terminal DNA binding domain (Olsen et al., 2005) and a diversified C-terminal domain that generally regulates the transcriptional activation (Yamaguchi et al., 2008). Functional characterization of the NAC family genes using over-expression studies (He et al., 2005) have helped understanding various biological roles of NAC proteins as indicated in Figure 1 Interestingly, NAC genes contribute to enhanced survivability of plants under stress (Puranik et al., 2012), and in secondary growth of the plants (Nakano et al., 2015), thereby building biomass.
Biofuel plants of the future, so-called fourth generation biofuels are perceived to be the ones that survive under harshest of conditions owing to their abilities to withstand stress, and produce large amounts of biomass that can be converted to green diesel, via biomass-to-liquid technologies.
Here, we review the literature on functional characterization of NAC genes through over-expression to establish their roles in stress tolerance and biomass production, the two important parameters for biofuel plants.
NAC Engineered Plants for Growth in Degraded Lands
Stress Tolerance
Members of NAC family genes from different plants have been shown to provide tolerance to biotic and abiotic stresses (Jiang et al., 2014; Mao et al., 2014; Shah et al., 2014). ATAF sub-family in particular is involved in response to environmental stimulus, and the mechanism of action of NAC may be ABA dependent or independent. Ohnishi et al. (2005) provided comprehensive evidence that OsNAC6 gene in rice was induced by cold, salt, drought, abscisic acid (ABA), jasmonic acid (JA), and wounding. Subsequently, similar results were also reported by Hu et al. (2006, 2008) for SNAC1 and SNAC2 genes in rice, which too were induced by cold, drought and salinity stresses. Similarly, GmNAC11 and GmNAC20 genes from soybean were shown to be differentially expressed in response to multiple abiotic stress and plant hormones as a transcriptional activator of other stress responsive genes like DREB1A, ERD11, cor15A, ERF5, RAB18, and KAT2 genes (Hao et al., 2011). A well-characterized NAC gene is AtNAC2 from Arabidopsis thaliana, whose elevated expression levels have been reported in ethylene and auxin overproducer mutants, when exposed to salt stress. The gene also gets induced by ABA and drought stresses (He et al., 2005). In our laboratory, we have identified a cold-inducible NAC gene from a Brassicaceae family member Lepidium latifolium (LlaNAC), which was subsequently validated by over-expressing the gene in tobacco plants (Grover et al., 2014). The popularity of the NAC gene as a tool to induce stress tolerance in plants by genetic engineering can well be assessed by overlooking the number of such reports within the calendar year 2015 (Table 1).

TABLE 1. Recent reports on demonstration of acquiring stress tolerance in NAC over-expressor plants.
From human perspective, a good biofuel crop would be the one which would also be resistant to the abiotic stresses. Interestingly, a small number of NAC proteins have also been reported to respond to the abiotic stresses (reviewed by Nuruzzaman et al., 2013). Examples include OsNAC111 in rice, belonging to the TERN subgroup, and has been shown to provide tolerance to over-expressor plants against Magnaporthe oryzae (Yokotani et al., 2014).
Physiological and Morphological Adaptations
NAC family genes are also involved in a number of growth and development processes, as well as in tissue formation, which in turn help a plant to survive stress. For example, membrane-associated NAC TFs, up-regulated by stress conditions, have been found associated to a variety of morphological features like delayed flowering, reduced growth, curled leaves, etc., under stress conditions (Kim et al., 2007a,b). Arabidopsis lines over-expressing RhNAC3 gene from rose displayed hypersensitivity during seed germination and leaf closure on ABA or drought stresses (Jiang et al., 2014). A number of NAC genes have also been reported downstream to ETHYLENE-INSENSITIVE2 or similar genes, thereby participating in leaf senescence, fruit ripening, etc. (Ay et al., 2014; Kim et al., 2014; Nieuwenhuizen et al., 2015). Zhao et al. (2015) reported delayed leaf senescence and higher nitrogen concentrations in grain by over-expressing wheat TaNAC-S.
Another well known effect of NAC over-expression in plants is lateral root formation, which is generally observed as a response or phenotypic adaptation to water scarcity. The effect has best been studied for AtNAC2 from Arabidopsis thaliana, being associated to lateral root development under salt stress. Further, model plants over-expressing AtNAC2 gene were also shown to have extensive lateral root development (He et al., 2005).
NAC Engineered Plants for Biomass Production
Biomass, i.e., deposition of photosynthetic free energy, is an important source of biofuels today. Vegetative tissues of specialized crops like switch grass, miscanthus and poplar are primarily used for this purpose. Therefore, any engineering event in plants that prolongs or enhances vegetative meristematic activity is desirable from biomass production point of view. In our laboratory, we raised LlaNAC over-expressor lines that could accumulate 2–3 times more biomass and chlorophyll pigments than the wild-types. In addition, these plants matured early, had shorter life cycles (Grover et al., 2014), and could capture 3–5 times more carbon dioxide. It is pertinent to mention that the nearest homolog, ANAC056 from Arabidopsis thaliana, of the gene that we cloned (i.e., LlaNAC) clustered with VND subfamily of genes (Zhong et al., 2010a). Expression of ANAC056 is predominant in cork, xylem, silique, hypocotyls, and stamen. VND genes clearly participate in secondary growth in perennials, their overall effects in annuals and herbaceous plants like Arabidopsis and Lepidium, shall be more thoroughly evaluated. VND proteins function in formation of various tissues other than xylem vessels too (Bennett et al., 2010).
NAC proteins target a number of genes in the genome including those which are involved in stress responses, growth, and secondary wall synthesis (discussed below) or cambial activities. Previous estimated have suggested that as many as 72 genes are the target genes to NAC proteins (Shamimuzzaman and Vodkin, 2013). Such versatility in their action is partly due to their ability to form homo- and hetero-dimers, which act as transcriptional switches (Olsen et al., 2005), and partly because many NAC TFs are downstream to each other (Grover et al., 2014).
Secondary Cell Wall Synthesis
Secondary cell walls are the most abundant biomass and renewable source of energy. Interestingly, secondary cell wall biosynthesis is regulated by a subset of closely related NAC domain proteins, i.e., NST1/ANAC043, NST2/ANAC066, and NST3/SND1 (SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1)/ANAC012 as master transcriptional switches (Zhong et al., 2008). These proteins bind to a triggering expression of other genes downstream by binding at 19-bp consensus sequence, (T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T) called as secondary wall NAC binding element (SNBE; Zhong et al., 2010b). Downstream genes, SND2 and SND3 (Zhong et al., 2008) up-regulate genes associated with cellulose, xylan, mannan, and lignin biosynthesis and polymerization.
Other NAC TFs belonging to this sub-family include Vascular NAC related domain proteins (VND6 and VND7) which act as regulators of secondary cell wall biosynthesis specifically in vascular vessels (Yamaguchi et al., 2008). These genes are expressed in fiber cells of inflorescence stems, hypocotyls, valve endocarp layer and in the cells surrounding vascular vessels in replum of siliques (Shah et al., 2014). Examples include BdSWN5 in Brachypodium distachyon, OsSWN1 in grasses (Chai et al., 2015), and wood associated NAC domain proteins (WND2B and WND6B) in poplar (Zhong et al., 2010a; Zhao et al., 2014). The role of alternative splicing too has been suggested in function of these genes as regulators of secondary cell wall biosynthesis (Zhao et al., 2014). Interestingly, a number of other NAC genes too lie downstream of VND subfamily genes (Shamimuzzaman and Vodkin, 2013). Wood-associated NAC domain TF (PtrWNDs), are alternatively spliced (Zhao et al., 2014) occurs exclusively in secondary xylem fiber cells. The two PtrWND1B isoforms play antagonistic roles in regulating cell wall thickening during fiber cell differentiation in Populus sp.
Cellular maturation of root cap is also regulated by NAC TF family members, viz., SMB (SOMBRERO), BRN1 (BEARSKIN1), and BRN2, along with VND and NST genes (Bennett et al., 2010). Interestingly, SMB, BRN1, and BRN2 over-expression show similar phenotypic patterns to VND/NST genes (Bennett et al., 2010).
Perspectives
NAC gene family is not only one of the largest gene families in plants, but is also one of the best characterized gene families in plants. A random search on google scholar using the key words “Characterization of NAC genes” returns more than 100 relevant result links. Inclusion of information from all of these papers would have required publication of a monograph on NAC genes, and is thus out of the scope of this mini-review. However, there have been few or even negligible efforts on realization of potential of the NAC genes by preparing genetically engineered crops for food or fuel purposes. Thus, while NAC genes remain a favorite among Plant Molecular Biologists, they are yet to be adopted by Plant Biotechnologists.
Sufficient evidence is available on, how NAC over-expressing plants negotiate stress better than the wild-type plants. A transgenic crop over-expressing NAC gene would thus have much wider applicability and adaptability to a conventional breed or variety. Conversely, the agrochemicals can judiciously be directed to food crops. Further, a number of reports implicating role of NAC proteins in secondary wall synthesis and thereby biomass accumulation are available, thereby opening a lucrative opportunity for designing second or fourth generation biofuel plants. An obvious objection would be that stress tolerance and biomass accumulation are different properties, and are not accounted by a single NAC gene. Panorama of traits are generally observed in transgenic plants designed with an objective to characterize these genes. For example, Xia et al. (2010) observed biotic and abiotic stress tolerance due to the effect of a single gene, Uauy et al. (2006) reported senescence controlling NAC gene improving grain protein, zinc, and iron content in wheat, NAC gene from Lepidium latifolium improved both stress tolerance as well as biomass characters in over-expressing tobacco lines (Grover et al., 2014). Much recently, Chen et al. (2015) found OsNAC2 over-expression affects height of the plant, insensitivity to gibberellic acid and delays flowering.
In view of the above, it is justified to conclude that an open minded screening of NAC over-expressor plants is required to evaluate the potential for biofuel plants. Implementation of NAC transgenic plants for economic benefits in biofuel production may be dictated by the local requirements, and necessary strategy can be drafted based on the huge literature available in public domain.
Author Contributions
SS conceived the idea. SS and AG built the article together, and MN approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research in our lab is funded by Defence Research and Development Organization (DRDO). Director DIBER is acknowledged for approving the manuscript for publication.
References
Ay, N., Raum, U., Balazadeh, S., Seidensticker, T., Fischer, A., Reuter, S., et al. (2014). Regulatory factors of leaf senescence are affected in Arabidopsis plants over-expressing the histone methyltransferase SUVH2. J. Plant Growth Regul. 33, 119–136. doi: 10.1007/s00344-013-9384-y
Bennett, T., van den Toorn, A., Sanchez-Perez, G. F., Campiho, A., Willemsen, V., Snel, B., et al. (2010). SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22, 640–654. doi: 10.1105/tpc.109.072272
Chai, M., Bellizzi, M., Wan, C., Cui, Z., Li, Y., and Wang, G.-L. (2015). The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 58, 44–51. doi: 10.1007/s12374-014-0400-y
Chen, X., Lu, S., Wang, Y., Zhang, X., Lv, B., Luo, L., et al. (2015). OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberllic acid pathway in rice. Plant J. 82, 302–314. doi: 10.1111/tpj.12819
Fang, Y., Liao, K., Du, H., Xu, Y., Song, H., Li, X., et al. (2015). A stress-repsonsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 25, 3472–3490. doi: 10.1093/jxb/erv386
Grover, A., Singh, S., Pandey, P., Patade, V. Y., Gupta, S. M., and Nasim, M. (2014). Overexpression of NAC gene from Lepidium latifolium enhances biomass, shortens life cycle and induces cold stress tolerance in tobacco: potential for engineering fourth generation biofuel crops. Mol. Biol. Rep. 11, 7479–7489. doi: 10.1007/s11033-014-3638-z
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313. doi: 10.1111/j.1365313X.2011.04687.x
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365313X.2005.02575.x
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Hu, H., You, J., Fang, Y., Zhu, X., Qi, Z., and Xiong, L. (2008). Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 67, 169–181. doi: 10.1007/s11103-008-9309-5
Huang, Q., Wang, Y., Li, B., Chang, J., Chen, M., Li, K., et al. (2015). TaNAC29, a NAC transcription factor in wheat, enahces salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15:268. doi: 10.1186/s12870-015-0644-9
Jiang, G., Jiang, X., Lu, P., Liu, J., Gao, J., and Zhang, C. (2014). The rose (Rosa hybrida) NAC transcription factor 3 gene, RhNAC3, involved in ABA signaling pathway both in rose and Arabidopsis. PLoS ONE 9:e109415. doi: 10.1371/journal.pone.0109415
Kim, H. J., Hong, S. H., Kim, Y. W., Lee, I. H., Jun, J. H., Phee, B. K., et al. (2014). Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2 mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036. doi: 10.1093/jxb/eru112
Kim, S. G., Kim, S. Y., and Park, C. M. (2007a). A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226, 647–654. doi: 10.1007/s00425.007.513.3
Kim, S. Y., Kim, S. G., Kim, Y. S., Seo, P. J., Bae, M., Yoon, H. K., et al. (2007b). Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 35, 203–213. doi: 10.1093/nar/gkl1068
Mao, X., Chen, S., Li, A., Zhai, C., and Jing, R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 9:e84359. doi: 10.1371/journal.pone.0084359
Movahedi, A., Zhang, J., Yin, T., and Zhuge, Q. (2015). Functional analysis of two orthologous NAC genes, CarNAC3, and CarNAC6 from Cicer arietinum, involved in abiotic stresses in poplar. Plant Mol. Biol. Rep. 33, 1539–1551. doi: 10.1007/s11105-015-0855-0
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Nieuwenhuizen, N. J., Chen, X., Wang, M. Y., Matich, A. J., Perez, R. L., Allan, A. C., et al. (2015). Natural variation in Monoterpene synthesis in Kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3 like transcription factors. Plant Physiol. 167, 1243–1258. doi: 10.1104/pp.114.254367
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Ohnishi, T., Sugahara, S., Yamada, T., Kikuchi, K., Yoshiba, Y., Hirano, H. Y., et al. (2005). OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 80, 135–139. doi: 10.1266/ggs.80.135
Olsen, A. N., Ernst, H. A., Lo, L. L., and Skriver, K. (2005). DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169, 785–797. doi: 10.1016/j.plantsci.2005.05.035
Pandurangaiah, M., Lokanadha Rao, G., Sudhakarbabu, O., Nareshkumar, A., Kiranmai, K., Lokesh, U., et al. (2014). Overexpression of horsegram (Macrotyloma uniflorum Lam. Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Mol. Biotechnol. 56, 758–769. doi: 10.1007/s12033-014-9754-0
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Shah, S. T., Pang, C., Hussain, A., Fan, S., Song, M., Zamir, R., et al. (2014). Molecular cloning and functional analysis of NAC family genes associated with leaf senescence and stresses in Gossypium hirsutum L. Plant Cell. Tiss. Organ. Cult. 117, 167–186. doi: 10.1007/s11240-014-0430-7
Shamimuzzaman, M., and Vodkin, L. (2013). Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genomics 14:477. doi: 10.1186/1471-2164-14-477
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., and Dubcovsky, J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. doi: 10.1126/science.1133649
Wang, B., Guo, X., Wang, C., Ma, J., Niu, F., Zhang, H., et al. (2015). Identification and characterization of plant-specific NAC gene family in canola (Brassica napus L.) reveal novel members involved in cell death. Plant Mol. Biol. 87, 395–411. doi: 10.1007/s11103-015-0286-1
Xia, N., Zhang, G., Liu, X. Y., Deng, L., Cai, G. L., Zhang, Y., et al. (2010). Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 37, 3703–3712. doi: 10.1007/s11033-010-0023-4
Yamaguchi, M., Kubo, M., Fukuda, H., and Demura, T. (2008). Vascular related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55, 652–664. doi: 10.1111/j.1365-313X.2008.03533.x
Yang, X., Wang, X., Ji, L., Yi, Z., Fu, C., Ran, J., et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 34, 943–958. doi: 10.1007/s00299-015-1756-2
Yokotani, N., Tsuchida-Mayama, T., Ichikawa, H., Ohme-Takagi, M., and Kaku, H. (2014). OsNAC111, a blast disease-responsive transcription factor in rice, positively regulates the expression of defense-related genes. Mol. Plant Microbe Interact. 27, 1027–1034. doi: 10.1094/MPMI-03-14-0065-R
Zhao, D., Derkx, A. P., Liu, D.-C., Buchner, P., and Hawkesford, M. J. (2015). Overexpression of a NAC transcription factor delays leaf senescence and increase grain nitrogen concentration in wheat. Plant Biol. 17, 904–913. doi: 10.1111/plb.12296
Zhao, Y., Sun, J., Xu, P., Zhang, R., and Li, L. (2014). Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 164, 765–776. doi: 10.1104/pp.113.231134
Zhong, R., Lee, C., and Ye, Z. H. (2010a). Functional characterization of poplar wood associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055. doi: 10.1104/pp.109.148270
Zhong, R., Lee, C., and Ye, Z. H. (2010b). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant. 3, 1087–1103. doi: 10.1093/mp/ssq062
Keywords: NAC, genetically engineered plants, abiotic stress tolerance, secondary growth, cell wall synthesis, biomass
Citation: Singh S, Grover A and Nasim M (2016) Biofuel Potential of Plants Transformed Genetically with NAC Family Genes. Front. Plant Sci. 7:22. doi: 10.3389/fpls.2016.00022
Received: 29 October 2015; Accepted: 08 January 2016;
Published: 26 January 2016.
Edited by:
Camila Caldana, Brazilian Bioethanol Science and Technology Laboratory – National Center for Research in Energy and Materials/Brazilian Association for Synchrotron Light Technology, BrazilReviewed by:
Paulo Arruda, Universidade Estadual de Campinas, BrazilGong-yin Ye, Zhejiang University, China
Copyright © 2016 Singh, Grover and Nasim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sadhana Singh, sadhnasingh1986@gmail.com
†These authors have contributed equally to this work.
 Sadhana Singh
Sadhana Singh Atul Grover†
Atul Grover† M. Nasim
M. Nasim