- 1Department of Stress and Developmental Biology, Leibniz Institute of Plant Biochemistry, Halle/Saale, Germany
- 2National Institute of Plant Genome Research, New Delhi, India
Mitogen-activated protein kinase (MAPK) cascades are central signaling pathways activated in plants after sensing internal developmental and external stress cues. Knowledge about the downstream substrate proteins of MAPKs is still limited in plants. We screened Arabidopsis WRKY transcription factors as potential targets downstream of MAPKs, and concentrated on characterizing WRKY46 as a substrate of the MAPK, MPK3. Mass spectrometry revealed in vitro phosphorylation of WRKY46 at amino acid position S168 by MPK3. However, mutagenesis studies showed that a second phosphosite, S250, can also be phosphorylated. Elicitation with pathogen-associated molecular patterns (PAMPs), such as the bacterial flagellin-derived flg22 peptide led to in vivo destabilization of WRKY46 in Arabidopsis protoplasts. Mutation of either phosphorylation site reduced the PAMP-induced degradation of WRKY46. Furthermore, the protein for the double phosphosite mutant is expressed at higher levels compared to wild-type proteins or single phosphosite mutants. In line with its nuclear localization and predicted function as a transcriptional activator, overexpression of WRKY46 in protoplasts raised basal plant defense as reflected by the increase in promoter activity of the PAMP-responsive gene, NHL10, in a MAPK-dependent manner. Thus, MAPK-mediated regulation of WRKY46 is a mechanism to control plant defense.
Introduction
Upon pathogen encounter, plants respond through two layers of immunity: PTI (pattern-triggered immunity) and ETI (effector-triggered immunity; Jones and Dangl, 2006). PTI is the generalized defense mechanism in which conserved pathogen-associated molecular patterns (PAMPs) like bacterial flagellin, elongation factor Tu (EF-Tu) and fungal chitin are recognized at the cell surface by specialized pattern-recognition receptors (PRRs; Boller and Felix, 2009). One of the best studied plant PAMP–PRR interactions is that of the elicitor-active 22-amino acid peptide derived from bacterial flagellin (flg22) and the flg22 receptor, FLAGELLIN-SENSING 2 (FLS2), in Arabidopsis thaliana. FLS2 contains an extracellular leucine-rich repeat (LRR) domain for ligand binding, a transmembrane domain and a cytoplasmic serine/threonine kinase domain (Gómez-Gómez and Boller, 2002). After activation by flg22, FLS2 interacts with BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) to initiate PTI (Chinchilla et al., 2007). The general cellular responses in PTI include Ca2+ fluxes, reactive oxygen species (ROS) production, activation of mitogen-activated protein kinases (MAPKs), transcriptional reprogramming like PR-1 and WRKY gene expression and production of antimicrobial compounds like phytoalexins (Dodds and Rathjen, 2010). However, adapted pathogens are able to suppress PTI through effector molecules, which are mostly delivered into the plant cell. During plant-pathogen co-evolution, some plants have developed intracellular LRR-receptors to recognize, such effectors or the modifications they cause, and subsequently initiate the typically stronger ETI defense response (Chisholm et al., 2006). ETI usually results in localized cell death (hypersensitive response, HR) to restrict the growth of biotrophic pathogens.
Activation of MAPK signaling is vital for mounting an appropriate PTI response. A typical MAPK cascade consists of a modular complex comprising a MAPK kinase kinase (MAPKKK), phosphorylating a MAPK kinase (MAPKK), which phosphorylates a MAPK (Sinha et al., 2011; Meng and Zhang, 2013). Then, the activated MAPK specifically phosphorylates various substrate proteins, such as transcription factors, at conserved amino acid residues (S/T-P motif). After flg22 perception, two Arabidopsis MAPK pathways are activated. One pathway involves the MAPKKs, MKK4, and MKK5, which act upstream of the MAPKs, MPK3 and MPK6, leading to the activation of WRKY transcription factors that positively regulate defense gene expression (Asai et al., 2002). The second flg22-activated MAPK cascade is composed of MEKK1, MKK1/MKK2 (two redundant MAPKKs), and MPK4. Based on both genetic and biochemical studies, MPK4 exhibits negative control of salicylic acid (SA)-regulated plant defense (Petersen et al., 2000). A recent study identified a more complex scheme of MAPK-dependent defense gene expression. Here, MPK3 and MPK4 influenced subsets of defense-related genes both negatively and positively in their expression (Frei dit Frey et al., 2014). A fourth MAPK, MPK11, is also activated by flg22 treatment (Bethke et al., 2012) or other PAMPs (Eschen-Lippold et al., 2012).
Knowledge regarding the downstream targets of MAPKs is still fragmentary and high-throughput methods have been employed to identify the potential MAPK substrates. The various strategies used include yeast-two hybrid screens, protein microarrays and in vitro kinase assays. Microarray-based testing of 1690 Arabidopsis proteins identified 48 in vitro substrates for MPK3 and 39 for MPK6, with an overlapping set of 26 candidates (Feilner et al., 2005). High-density protein microarrays used to determine the phosphorylation targets of 10 different MAPKs identified 570 potential MAPK targets out of 2158 candidates (Popescu et al., 2009). Some of the well characterized MAPK targets relevant to pathogen response include the 1-aminocyclopropane-1-carboxylic acid synthase (ACS), ethylene response factor 104 (ERF104), tandem zinc finger 9 (TZF9), VirE1-interacting protein 1 (VIP1), WRKY33 transcription factor, MPK4 substrate 1 (MKS1), and several related proteins with so-called “VQ-motifs” (Meng and Zhang, 2013; Maldonado-Bonilla et al., 2014; Pecher et al., 2014).
WRKY transcription factors control transcriptional reprogramming to mediate cellular responses to diverse environmental cues (Eulgem and Somssich, 2007) and many WRKYs are targets of Arabidopsis MAPKs (Popescu et al., 2009). Besides regulating plant development, WRKYs are important positive and negative regulators of both PTI and ETI (van Verk et al., 2011). The defining feature of WRKYs is their DNA binding WRKY domain. The WRKY domain comprises the highly conserved WRKYGQK peptide sequence and a zinc finger motif (CX4-7 -CX22-23 -HXH/C, where X represents any amino acid and C/H are the conserved zinc-coordinating cysteine or histidine residues). This domain is responsible for binding to the W-box element with the consensus “(C/T)TGAC(T/C)” nucleotide sequence, although flanking DNA sequences appear to contribute to binding specificities (Ciolkowski et al., 2008). The 74 members of Arabidopsis WRKYs are divided into three groups based on the number of WRKY domains (two domains in group I, and one in groups II and III) and the arrangement of conserved cysteine/histidine residues of their zinc fingers (C–C–H–H in group I/II but C–C–H–C in group III; Rushton et al., 2010). In Arabidopsis, it has been shown that WRKY33 exists in nuclear complexes with MPK4 and MKS1. PAMP perception activates MPK4, leading to the nuclear dissociation of the MPK4–MKS1–WRKY33 complex and thereby releasing WRKY33 and MKS1. WRKY33 then activates expression of PAD3 (Phytoalexin Deficient 3), a key enzyme for the synthesis of antimicrobial camalexin (Qiu et al., 2008). Additionally, MPK3 and MPK6 are major contributors to fungus-induced camalexin accumulation by phosphorylating WRKY33 (Mao et al., 2011). Taken together, these two studies suggest transcriptional regulation of camalexin biosynthesis gene expression through a release of WRKY33 from an inhibitory MPK4-containing protein complex and a positive effect on WRKY33 function through MPK3/6-mediated WRKY33 phosphorylation.
In the current study, 48 Arabidopsis WRKY proteins were investigated as in vitro MPK3 and MPK6 phosphorylation targets; and one member, WRKY46 was selected for further characterisation since its importance in plant pathogen response and abiotic stresses is indicated in numerous studies (van Verk et al., 2011; Moreau et al., 2012; Ding et al., 2013, 2014; Gao et al., 2013). Our findings establish WRKY46 as an MAPK phospho-target mediating plant defense responses.
Materials and Methods
Plant Growth Conditions and Genotypes
All plants were grown in growth chambers with a photoperiod of 8 h light (120 μmol m-2 s-1; 22°C) and 16 h dark (20°C). Arabidopsis thaliana ecotype Col-0 was used as wild type. The mpk3 and mpk6 mutants (in Col-0 background) were described previously (Wang et al., 2008; Bethke et al., 2009). Nicotiana benthamiana was used for Agrobacterium tumefaciens-mediated transient expression experiments.
Site-Directed Mutagenesis
The WRKY46S168A, WRKY46S250A, and WRKY46S168A,S250A phosphorylation site variants were generated as described (Palm-Forster et al., 2012; Eschen-Lippold et al., 2014). Briefly, mutagenesis primers (as listed in Table 1) with a flanking BsaI site were used to amplify the entire pDONR220 vector harboring WRKY46. After DpnI digestion of the template plasmid, the amplified vector was gel-purified, digested with BsaI and ligated. The mutated WRKY46 variants were then transferred to destination vectors by standard GatewayTM LR- recombination-based cloning (Invitrogen). For the phosphomimetic WRKY46S168D,S250D variant, two fragments were amplified by PCR using the primer pairs, W46-S168D_3/W46-S250D_5 and W46-S168D_5/W46-S250D_3, and pUGW15-WRKY46 as a template. Both fragments were gel-purified and mixed and incubated in the presence of LguI and T4-DNA ligase in a PCR cycler machine (10 cycles of 10 min 37°C and 10 min 22°C).
Preparation of Recombinant Proteins and In Vitro Phosphorylation Assay
The 48 WRKY coding sequences were cloned in-frame into expression vector pDEST-N110 (Dyson et al., 2004) to generate N-terminally His10-tagged recombinant proteins. The vectors were transformed into KRX competent cells (Promega). Cells were grown in 2xYT media (16 g L-1 Bacto tryptone, 10 g L-1 Bacto yeast extract, 5 g L-1 NaCl, pH 7.2) to OD600 of 0.5–0.6 and protein expression was induced at 24°C overnight by adding 1 mM rhamnose. Denaturing purification was performed by resuspending the pellet from 100 mL culture in 20 mL of lysis buffer (100 mM sodium phosphate, 10 mM Tris, 6 M GuHCl, pH 8) and incubated at room temperature for 30 min. After centrifugation the supernatant was incubated for 1 h with 100 μL of equilibrated Ni-NTA-Agarose beads (Thermo Scientific). The beads were washed thrice with wash buffer (100 mM sodium phosphate, 10 mM Tris, 8 M Urea, pH 6.3). The proteins were refolded on the beads as previously described (Feilner et al., 2005). Briefly, 130 μL, 260 μL and 1 mL of native buffer (10 mM Tris pH 7.5, 1 mM PMSF) was consecutively added within a period of 30 min (on ice) to reduce urea concentration to <0.5 M. The supernatant after a brief centrifugation pulse was removed and 1 mL of native buffer added and refolding of the proteins allowed to proceed for another 2 h on ice.
In vitro kinase assay was performed as described (Feilner et al., 2005; Pecher et al., 2014) with slight modifications. Briefly, after visual estimation (by coomassie staining) of the amount of proteins purified, 5–10 μL of the Ni-NTA resin containing the bound protein was mixed with reaction buffer to give a final volume of 15 μL containing 25 mM Tris-Cl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 1 mM PMSF, 25 μM ATP, 1 μCi [γ-32P]ATP and ∼100 ng of active MAPKs. After 30 min incubation at 30°C, the reaction was stopped by addition of one volume of 2x SDS sample buffer. Samples were boiled at 95°C for 5 min and then separated on 10% SDS-PAGE gel and phosphorylation of the protein substrates visualized by phosphor-imaging (Typhoon Phosphorimaging System, GE Healthcare) after drying the gel.
ProQ Diamond Staining of Phosphoproteins
The MPK:WRKY proteins were incubated at approximately 1:10 (w/w) ratio for 30 min as described above (except for the lack of radioactive ATP). After separation by SDS-PAGE, the gel was fixed with 50% methanol and 10% acetic acid overnight. The gel was washed with water for 30 min and stained with 3x diluted Pro-Q diamond stain (Invitrogen) in the dark for 2 h. The gel was destained four times for 30 min each with 20 % acetonitrile, 50 mM sodium-acetate (pH 4.2). The gel was washed again with water for 10 min and was scanned at 400 V using a Typhoon Scanner (GE Healthcare).
Mass Spectrometric Analysis
Phosphorylated proteins were separated by 10% SDS-PAGE. The gel was stained with Colloidal Coomassie Blue Stain (Life Technologies) and washed with water. The corresponding gel bands were excised, destained by multiple washes with 30% acetonitrile, 100 mM NH4HCO3 and finally rinsed twice with water. After standard reduction and alkylation (Pecher et al., 2014), the proteins in the gel pieces were digested overnight at 37°C by incubation in 5% acetonitrile, 10 mM NH4HCO3 with 3 ng μL-1 modified trypsin (Promega). Peptides were extracted into 35% acetonitrile with 0.4% TFA, dried down in a Speed-vac and finally resuspended in 20 μL of 0.1% TFA. After 5 min of sonication in a sonifier water bath, the peptides were either frozen for storage or immediately analyzed on an LC-MS system consisting of a split-free nano-LC (Easy-nLC II, Proxeon, Thermo Scientific) coupled to a hybrid-FT-mass spectrometer (Orbitrap Velos Pro, Thermo Scientific). Additional targeted LC-MS measurements of selected phosphopeptides were performed using an inclusion list in a data dependent acquisition (DDA) scan method. Peptide fragmentation was achieved using CID with MSA (multi-stage activation). Proteins were identified and phosphorylation sites were identified using Mascot V.2.3.02 and the Phospho RS module in Proteome Discoverer v1.3.
In Silico Prediction of MAPK Targets
In silico functional prediction was performed with WRKY-family proteins. First, they were classified according to functional annotation clustering using DAVID 6.7 software1 (Huang et al., 2009). All known and predicted WRKY protein–protein interactions were retrieved from STRING 9.1 software2 (Franceschini et al., 2013). The data from both STRING 9.1 and DAVID 6.7 were integrated, analysed and visualized using CYTOSCAPE 3.0.2 software3 (Shannon et al., 2003). The WRKYs were broadly categorized into the functional groups of “general transcription” and “defense-regulated transcription factors.”
In Vivo Phosphorylation of WRKY46
For examining phosphorylation of WRKY46 in vivo, the WRKY46 coding sequence was cloned into pUGW15 (Nakagawa et al., 2007) to generate p35S-WRKY46 (which expresses an N-terminal HA-tagged WRKY46 under the control of the Cauliflower Mosaic Virus 35S promoter). Transformation of Arabidopsis mesophyll protoplasts was performed as described (Yoo et al., 2007; Ranf et al., 2011). After overnight incubation in the dark at 22°C, protoplasts were treated with either water or 100 nM flg22 for 15 min. Protoplasts were harvested by centrifugation and immediately frozen in liquid nitrogen. Dephosphorylation assays were performed as described (Maldonado-Bonilla et al., 2014). Briefly, lambda phosphatase (400 units; Upstate-Millipore) was added to the protoplast extracts for 1 h to dephosphorylate the proteins. After SDS-PAGE, the proteins were transferred to a nitrocellulose membrane (Macherey and Nagel) and immunoblotted against anti-HA.11 antibody (Eurogentec).
In Vivo Stability of WRKY46
To check the stability of WRKY46 after PAMP elicitation, protoplasts were transfected with p35S-WRKY46 constructs as described above. Protoplasts were treated with 2.5 μM cycloheximide to stop the translation and immediately elicited with 100 nM flg22. Samples were harvested by centrifugation at the indicated time points and were subjected to immunoblotting with anti-HA11 antibody to check the levels of WRKY46 proteins.
Promoter-Luciferase (LUC) Assay
The promoter-LUC assay was performed as described earlier (Ranf et al., 2011). Briefly, protoplasts were isolated and co-transformed with three vectors: an effector construct for WRKY46 overexpression, pNHL10-LUC (reporter construct) and pUBQ10-GUS (normalization construct). After overnight incubation in the dark at 22°C, 200 μM luciferin were added and mixed gently by inverting the tubes. Then, 100 μL of protoplasts were transferred into 96-well microtiter plates suitable for luminescence measurements (Greiner) and were elicited with 100 nM flg22/elf18, 200 μg/mL chitin or water as control. The luciferase activity kinetics were measured for 3 h with a 96-well plate luminescence reader (Luminoscan, Thermo Scientific). After the measurement, protoplast samples were directly lysed by adding 10 μL of 10-fold concentrated GUS extraction buffer (final concentration: 50 mM NaPO4 pH 7.0, 1 mM EDTA, 0.1% Triton X-100, 10 mM beta-mercaptoethanol) and vortexing. GUS-activity was measured upon incubation with 4-methylumbelliferyl glucuronide (4-MUG; 15 min at 37°C), based on the fluorometric detection of the reaction product 4-methyl umbelliferone (4-MU). Values are expressed as LUC/GUS ratios relative to the CFP-transfected controls (at timepoint 0 min).
Localization of WRKY46
WRKY46 was cloned into pEXSG-YFP vector (Feys et al., 2005) to express WRKY46 with a C-terminal YFP-tag. ERF104-CFP (Bethke et al., 2009) was used as a nuclear marker. Protoplasts were isolated, transfected with WRKY46-YFP with or without ERF104-CFP constructs, incubated overnight and observed under a laser scanning confocal microscope (LSM 710 Laser Scanning System; Carl Zeiss) as described Reddy et al. (2014). For YFP detection, excitation wavelength, and emission filters were 514 nm/band-pass 520–540 nm. For CFP detection, excitation wavelength and emission filters were 458 nm/band-pass 465–500 nm. Chloroplast auto-fluorescence was recorded using band-pass 650–710 nm settings.
Statistical Analyses
All statistical analyses were performed using GraphPad Prism 54.
Results
Arabidopsis MPK3 Phosphorylates WRKY46 In Vitro
We expressed 48 members out of the 74 WRKYs, covering all three WRKY groups, as recombinant proteins. MPK3 and MPK6 were also prepared and activated by incubating with a constitutively active MAPKK (abbreviated as MKK5DD; Lee et al., 2004). In vitro kinase assays were performed using the 48 purified WRKY proteins as substrates for MPK3 or MPK6 (Figure 1). Most of the WRKYs (or their truncated products) were phosphorylated by both kinases. The WRKYs showed differential phosphorylation intensities, possibly reflecting either the strength of their interaction with MPK3/MPK6 or the number of putative MAPK-targeted (SP/TP) sites present in them.
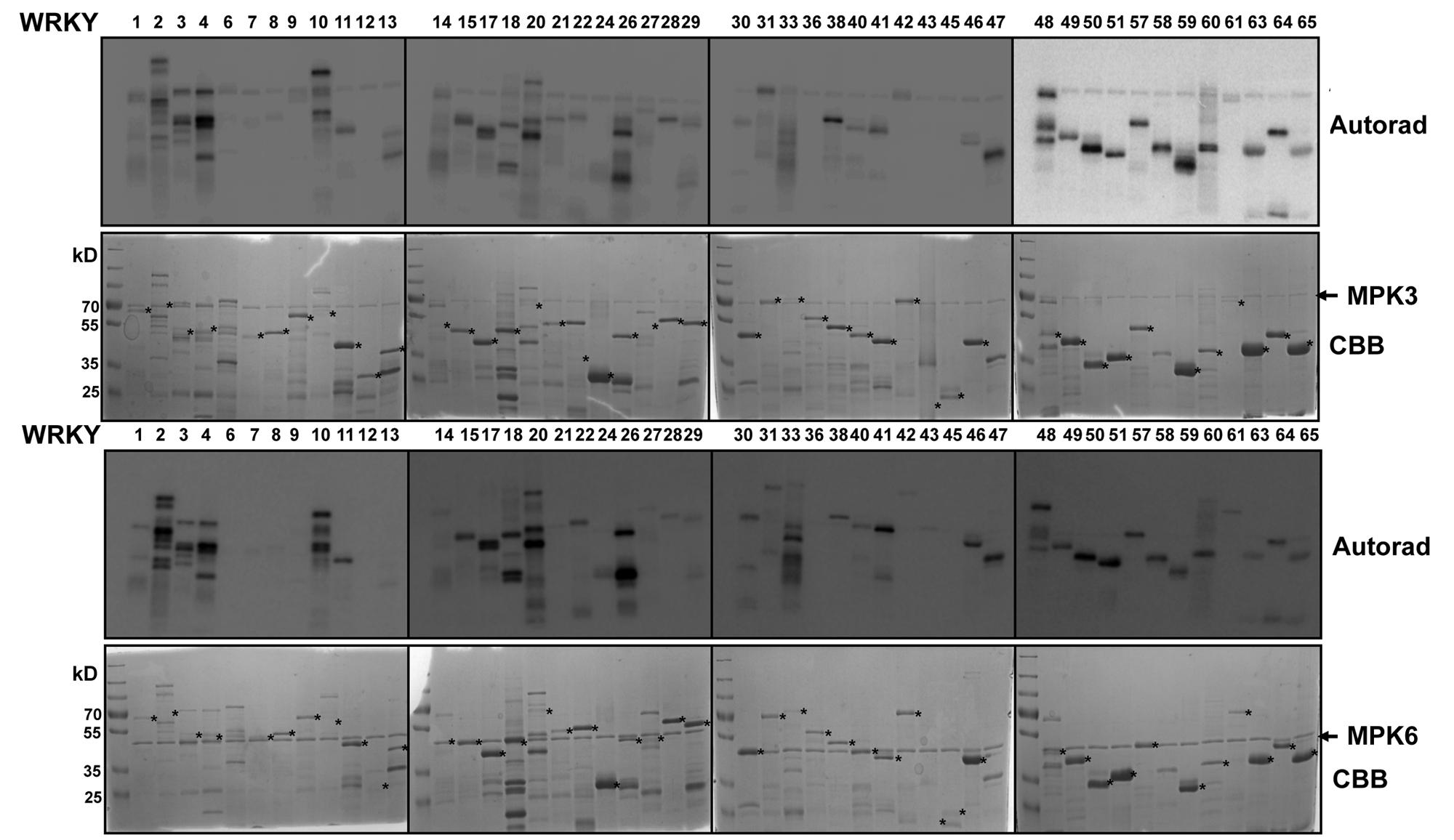
FIGURE 1. In vitro phosphorylation of Arabidopsis WRKYs by MPK3 and MPK6. Forty-eight Arabidopsis WRKY transcription factors were expressed in bacteria as His-tagged recombinant proteins, purified and used as substrates for in vitro phosphorylation by GST-tagged MPK3 (Upper) or untagged MPK6 proteins (Lower). Expected sizes of the WRKYs are marked with asterisks. Note that truncated products or aberrant migration patterns in SDS-PAGE, as seen in the Coomassie Brilliant Blue staining (CBB), are seen for several WRKYs. The experiment was repeated twice with similar results.
To narrow down the candidate WRKYs possibly involved in plant defense responses, in silico data-mining for protein-protein interaction (STRING 9.1, Franceschini et al., 2013) and functional annotation of roles in defense or transcriptional regulation (DAVID 6.7, Huang et al., 2009) was carried out with the WRKYs and MAPKs used in this study. A network of at least eight WRKYs, including WRKY46, was linked to MPK3 and may be involved in plant defense responses (Figure 2A).
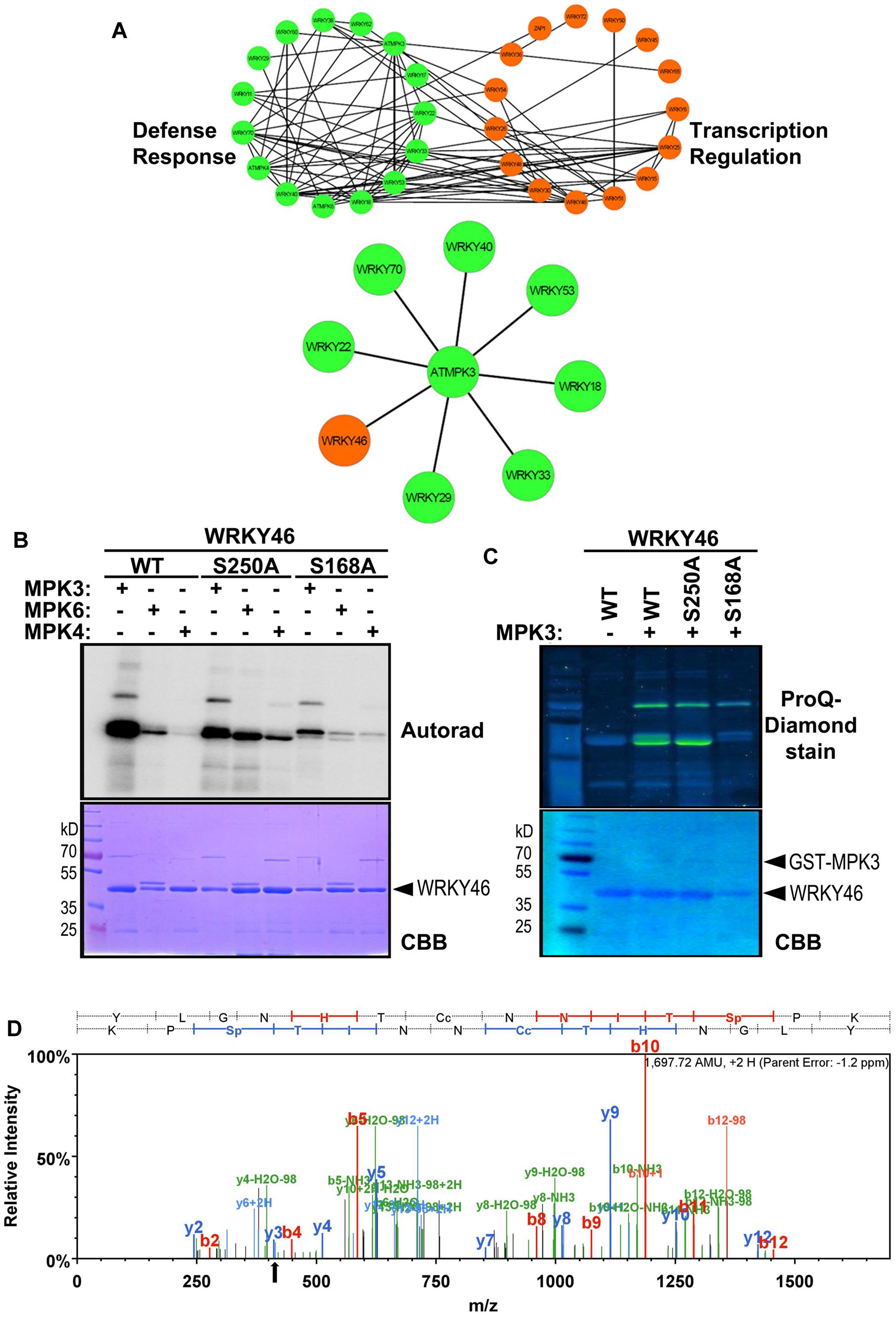
FIGURE 2. WRKY46 is a potential defense-related phosphorylation target of MPK3. (A) The functional annotation of all WRKYs was performed using DAVID 6.7 tool dividing all WRKYs into two groups (defense response, DR, or transcriptional regulation, TR; top network). The known and the predicted protein-protein interactions were analyzed by STRING 9.1 tool. Finally, all the information was integrated, analyzed and visualized using CYTOSCAPE 3.0.2 program. The (bottom) network shows WRKY46 as a potential defense-related MPK3 target along with many known MPK3 interaction partners. (B) In vitro phosphorylation of WRKY46 by MAPKs and phospho-site mapping. His-tagged WRKY46 and its phospho-site variants (S168A and S250A) were expressed using bacterial expression systems and used as substrates for phosphorylation by GST-MPK3, GST-MPK4, and tag-free MPK6 proteins, which were pre-activated with constitutively active MKK5DD. The proteins from the in vitro kinase assays were separated by SDS-PAGE and phosphorylation visualized by autoradiography (Autorad). Note that the MAPK bands can be seen above the WRKY46 proteins (GST-MPK3 or -MPK4 = ∼70 kD; MPK6 = ∼48 kD) The experiment was repeated twice with similar results. (C) Phosphorylation of WRKY46 and its variants by MPK3 was performed as described in (B; but using non-radioactive ATP). ProQ diamond® staining was used to visualize phosphorylated proteins. CBB of the gels was performed as loading control. (D) Mass spectrometry (MS) analysis of WRKY46. The phosphorylated WRKY46 bands (from C) were excised from the gels, trypsin digested and analyzed by LC-MS/MS. A phospho-peptide demonstrating phosphorylation of WRKY46 at position S168 (as reflected by y3 peak at 411 m/z) is shown.
Arabidopsis WRKY46 is a group III WRKY of 295 amino acids (molecular weight of 33.6 kDa) and contains two consensus MAPK phosphorylation sites at positions S168 and S250. We performed site-directed mutagenesis to change these two sites from Ser to Ala. To determine which phosphorylation site of WRKY46 can be phosphorylated by MAPKs in vitro, His-tagged recombinant WRKY46 protein variants were purified and used for MAPK phosphorylation assays. Using MPK3, MPK4 and MPK6 with comparable kinase activities (estimated by kinase assays with artificial myelin-basic protein as substrate), MPK3 showed the strongest phosphorylation of WRKY46 (Figure 2B). The observation was also confirmed by using a non-radioactive kinase assay with MPK3 and subsequent Pro-Q diamond phosphoprotein staining (Figure 2C). The MPK3-phosphorylated WRKY46 bands were excised from the non-radioactive gel, trypsin-digested and subjected to mass spectrometry (MS) analysis. A phospho-peptide containing phosphorylated S168 was detected (Figure 2D). Phosphorylation at S250, however, cannot be excluded since the peptide coverage of the MS analysis did not include this region of the protein. Nevertheless, MPK3-mediated phosphorylation of the WRKY46S168A variant was strongly reduced compared to WRKY46S250A (Figures 2B,C), so that S168 may be the preferred in vitro phospho-site. The recombinant WRKY46S168A,S250A double mutant was, unfortunately, poorly expressed in bacteria and it was difficult to purify sufficient amounts for comparison in the in vitro kinase assay. Curiously, the phosphorylated WRKY46 band(s) often show a double band, except for the WRKY46S250A variant; this may mean that phosphorylation at S250 results in a mobility shift in SDS-PAGE (Figures 2B,C).
In Vivo Phosphorylation of WRKY46
To determine whether WRKY46 is phosphorylated in vivo, WRKY46 was cloned into a vector expressing HA-tagged proteins under control of the Cauliflower Mosaic Virus 35S promoter and transfected into Arabidopsis protoplasts. After an overnight incubation (∼15 h) to enable protein expression, protoplasts were treated with flg22 (100 nM) or an equivalent volume of water (as control) for 15 min. Protoplasts were harvested, proteins extracted, and immunoblotted against an anti-HA antibody. An aliquot of proteins from the flg22-treated sample was incubated with lambda phosphatase to dephosphorylate the proteins. As reflected by a mobility shift in SDS-PAGE, which is abolished by the phosphatase treatment, WRKY46 is already phosphorylated before any PAMP elicitation (Figure 3A). Since the MAPKs are typically not activated prior to PAMP treatment, it appears that there are other kinase(s) that target WRKY46. As there is no additional phospho-shift after PAMP treatment, it is also not possible to use this assay as an indicator for in vivo phosphorylation by PAMP-activated MAPKs, such as MPK3 or MPK6 (see below).
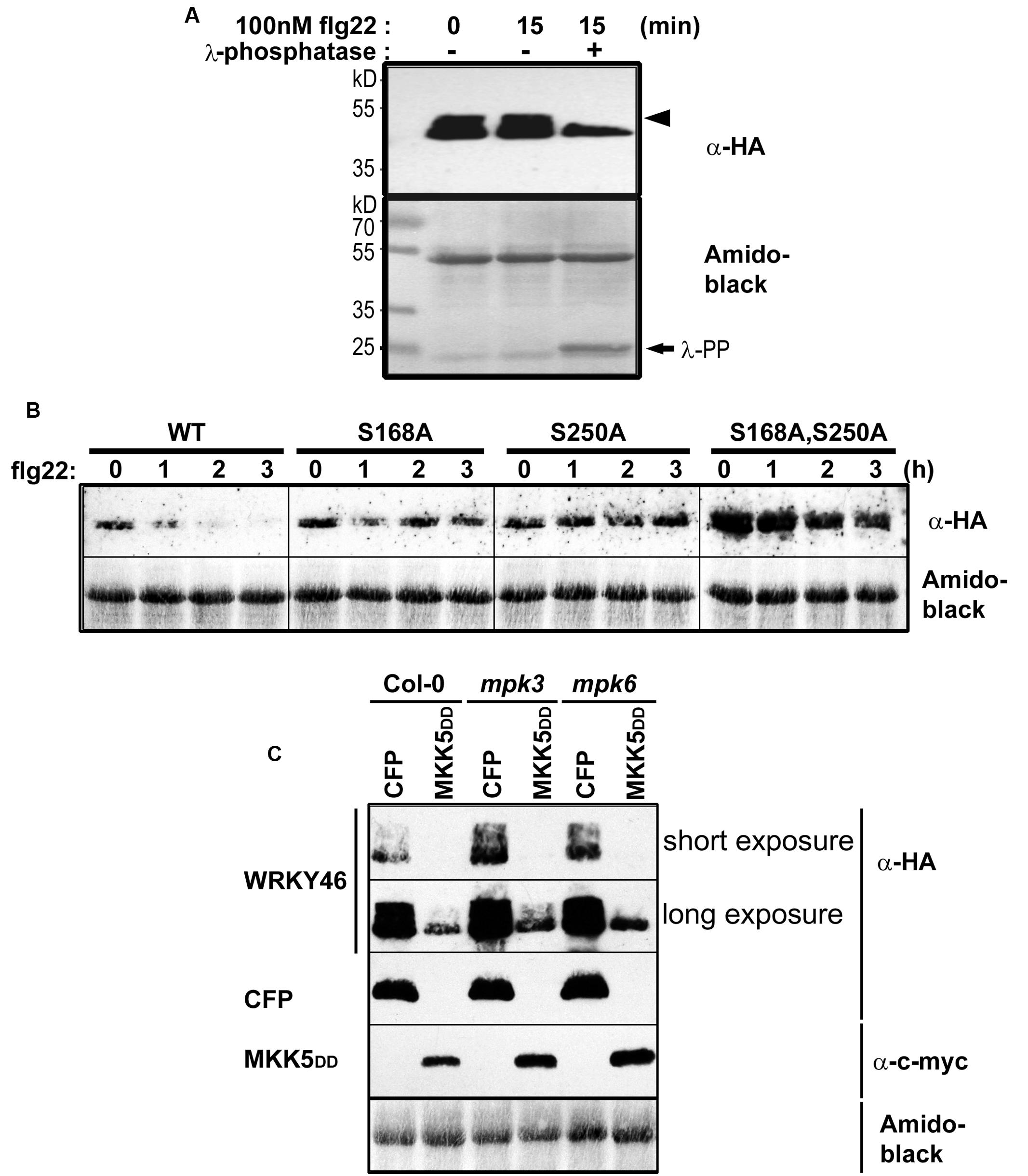
FIGURE 3. In vivo phosphorylation and alteration in WRKY46 stability. (A) Arabidopsis mesophyll protoplasts were transfected with p35S-WRKY46 (expressing an HA-tagged protein). After overnight incubation, protoplasts were elicited with 100 nM flg22 for 15 min. Total protein extracts were incubated with or without lambda phosphatase (λ-PP) for 1 h (37°C), separated by SDS-PAGE and western blot analysis was performed using anti-HA antibodies. Amido black staining was used to monitor equal loading of the proteins. The experiment was performed twice. Note a mobility shift of WRKY46 (indicative of phosphorylation) is marked by an arrowhead. (B) Arabidopsis mesophyll protoplasts expressing different WRKY46 variants were simultaneously treated with 100 nM flg22 and 2.5 μM cycloheximide (to block translation) and harvested at the time points indicated. The protein levels of WRKY46 variants were estimated by immunoblotting using anti-HA antibody. Amido black staining (showing the Rubisco large subunit band) was performed to monitor equal loading of proteins. The experiment was repeated at least five times. Note that the gels were blotted on the same membrane to allow direct comparison of band intensities. (C) Arabidopsis mesophyll protoplasts derived from the indicated genotype were co-transfected with p35S-WRKY46 and either p35S-mycMKK5DD (expressing a constitutively active MAPK kinase that activates MPK3 and MPK6) or a as control, a CFP-expressing plasmid. Immunoblot with the indicated antibodies and loading control monitoring by amido black staining were performed as described above.
Phosphorylation of WRKY46 Alters Its In Vivo Stability
The half-life of proteins is critical for the regulation of various signaling pathways and stability of several MAPK substrates has been shown to be regulated through phosphorylation after PAMP treatment (Liu and Zhang, 2004; Maldonado-Bonilla et al., 2014; Pecher et al., 2014). Therefore, as an alternative to using phospho-shift in gels, we investigated WRKY46 stability after elicitation of transiently transformed protoplasts with flg22. To facilitate visualization of the reduced protein levels after PAMP addition, cycloheximide was also added to block protein translation. Protoplasts were harvested at different time points after elicitation and subjected to immunoblotting. Reduction in wild type WRKY46 levels was seen 1 h after flg22 treatment, with little-to-no WRKY46 detectable after 2 h (Figure 3B). By contrast, flg22-induced reduction in protein levels was less pronounced for the WRKY46S168A, WRKY46S250A and WRKY46S168A,S250A mutant variants over the tested period of 3 h. In general, the highest level of protein expression was observed for the WRKY46S168A,S250A double phosphorylation site mutant (Figure 3B). Taken together, phosphorylation of WRKY46 (at S168 and/or S250) is required for the flg22-induced degradation in vivo and the non-phosphorylatable WRKY46 is more stable.
To directly link this destabilization effect to MAPK activities, we co-expressed MKK5-DD, a constitutively active MAPK kinase that activates MPK3 and MPK6 (Lee et al., 2004, 2015; Lassowskat et al., 2014) together with WRKY46. We also performed these experiments in the mpk3 or mpk6 mutant backgrounds to specifically activate only MPK6 or MPK3, respectively. As seen in Figure 3C, specific activation of MPK3 and/or MPK6 led to reduced WRKY46 protein levels – thus pointing to a direct effect of MPK3/MPK6-mediated phosphorylation on WRKY46 stability. As mpk3mpk6 double mutants are embryo-lethal and even the so-called “rescued” mutants have severe growth defects (Wang et al., 2007, 2008), it is difficult to test the effect of knocking-out both MAPKs on WRKY46 stability. The data therefore also suggests functional redundancy between MPK3 and MPK6 in regulating WRKY46 destabilization.
WRKY46 is a Nuclear Protein and Elevates the Basal Plant Defense Status
To examine WRKY46 subcellular localization, we transiently expressed WRKY46-YFP in Arabidopsis protoplasts. To visualize the nucleus, protoplasts were co-transfected with a CFP-tagged ERF104 that was previously shown to be nuclear-localized (Bethke et al., 2009). The resulting CFP and YFP fluorescence signals were both in the nuclei (Figure 4A). To exclude that WRKY46 might have been tethered to the ERF104 nuclear protein, WRKY46 was expressed alone where the WRKY46-YFP signals were also in the nucleus (Supplementary Figure S1). Western blotting using an anti-GFP monoclonal antibody revealed predominantly protein bands with the expected molecular weights for intact WRKY46-YFP and ERF104-CFP fusion proteins (Figure 4A), so that the nuclear fluorescence signals detected is not due to free CFP/YFP cleavage products. In addition, we also tested the non-phosphorylatable WRKY46S168A,S250A variant, which is also localized in the nucleus (Supplementary Figure S1). These findings are in agreement with WRKY46 functioning as a putative transcription factor in the nucleus (Ding et al., 2014, 2015) and also the increased protein stability of WRKY46S168A,S250A (Figure 3A) is not due to its mis-localization.
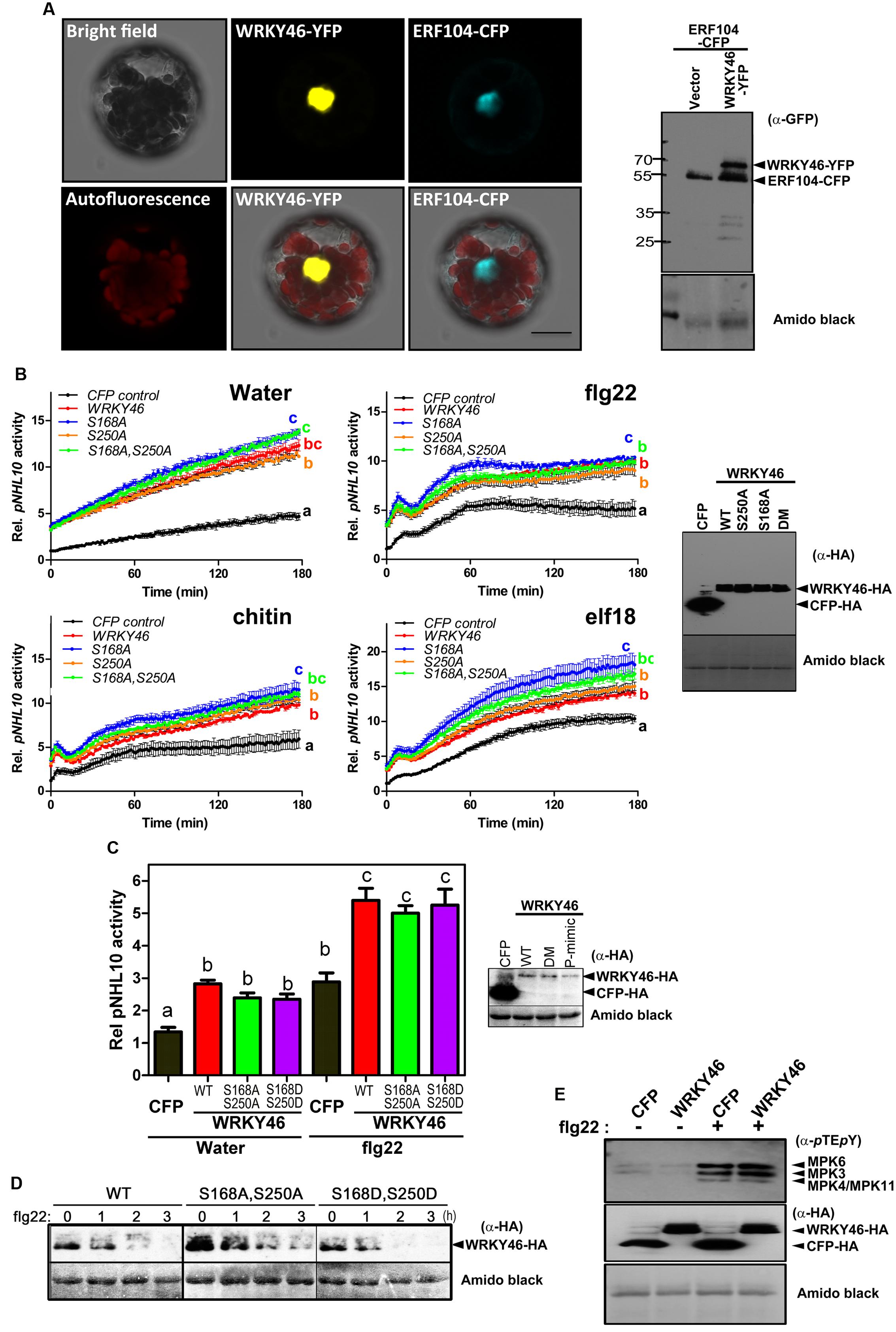
FIGURE 4. WRKY46 is nuclear localized and boosts defense-related promoter activity. (A) Arabidopsis Col-0 mesophyll protoplasts were co-transfected with WRKY46-YFP and ERF104-CFP constructs, with ERF104 serving as a nuclear marker. Confocal laser scanning micrographs of the protoplasts show the nuclear localization of WRKY46. Chlorophyll autofluorescence is also included to visualize chloroplasts. Scale Bar = 20 μm. (B) p35S-WRKY46 wild type and mutant variants were transfected into Arabidopsis mesophyll protoplasts along with pNHL10-LUC and pUBQ10-GUS reporter constructs. After overnight incubation, protoplasts were elicited with water, 100 nM flg22/elf18 or 200 μg mL-1 crab shell chitin. Luciferase activity was measured for 3 h. After the measurements, GUS-assays were performed with total protein extracts. The data are presented as LUC/GUS ratios relative to the untreated CFP-expressing control (at timepoint 0 min) with error bars indicating standard deviation (SD) of the mean. Different letters indicate statistically significant differences at time point 60 min [two-way repeated measures (RM) ANOVA with Bonferroni post tests, p < 0.01]. The experiment was performed three times with similar results (DM = S168A, S250A double phospho-site mutant). (C) Experiments after transfection with the indicated WRKY46 variants or CFP as a control were conducted as described in (B) above and the peak pNHL10 activities (typically at ∼40 min post elicitation with flg22) plotted. The different alphabets mark statistically distinct groups after one-way ANOVA analysis with Bonferroni’s multiple comparison test. Right panel is the immunoblot analysis of HA-tagged proteins. (P-mimic = S168D, S250D double phospho-mimic mutant). (D) WRKY46 protein stability after flg22 treatment was analyzed as described in Figure 3B. (E) Overexpression of WRKY46 does not alter MAPK activation. Arabidopsis mesophyll protoplasts were either transfected with constructs for expressing CFP (as a control) or WRKY46. After overnight incubation, protoplasts were treated with either water or 100 nM flg22 for 10 min. The SDS-PAGE-separated proteins were immunoblotted with anti-pTEpY antibody to check for MAPK activation. The experiment was performed twice with similar results. In all cases, equivalent expression of (intact) proteins in the protoplasts was tested by immunoblotting with the indicated antibodies. Amido black staining of the nitrocellulose membranes was used to estimate equal loading (The staining of the Rubisco large subunit is shown).
To investigate the role of WRKY46 in plant defense, we analyzed expression of a luciferase (LUC) reporter driven by a routinely used W-box-containing defense-related promoter, NDR1/HIN1-like 10 (NHL10), in Arabidopsis protoplasts co-expressing either different variants of WRKY46 or CFP (as a control for expression of a defense-unrelated protein). Protoplasts were treated with either water, bacterial (flg22/elf18) or fungal (chitin) elicitors and luciferase activity was measured. In protoplasts expressing any WRKY46 variant, an elevated basal promoter activity (time point 0 min) was observed for the water and also the PAMP-treated samples (Figure 4B). Also at later time points, all treatments showed an overall elevation of NHL10 promoter activity in the WRKY46-expressing protoplasts. This suggests that WRKY46 is a positive regulator of the defense-related NHL10 gene. Interestingly, the S168A variant consistently showed a higher boost on the NHL10 promoter activity. Since western blotting showed comparable expression of the respective WRKY46 variants (Figure 4B, right), the enhanced activity of WRKY46S168A is not simply due to enhanced protein stability/expression (see Figure 3B). To further test if phosphorylation of WRKY46 influences its effect on transcription controlled by the NHL10 promoter, we created a WRKY46S168D,S250D phospho-mimic. For simplicity, we tested only flg22 and concentrated only on the peak NHL10 promoter activity. As seen in Figure 4C, there was no difference in activities of the phospho-mimic (WRKY46S168D,S250D) or the non-phosphorylatable (WRKY46S168A,S250A) variants compared to wild type WRKY46. However, in line with the role of phosphorylation in regulating WRKY46 stability (compare Figure 3B), the phospho-mimic variant is expressed at lower levels (Figure 4C). We therefore compared the stability of WRKY46 phosphosite-mutated variants. While trace levels of wild type WRKY46 are expressed 2 h post flg22 treatment, the phospho-mimetic WRKY46 is hardly detectable (Figure 4D). Thus, this serves as indirect evidence that mutated residues are functionally mimicking the phosphorylated serines. Taken together, transcriptional activity of WRKY46 does not seem to be directly controlled by its phosphorylation status.
Since the NHL10 reporter is often used as a reporter for MAPK impact on defense gene expression (Boudsocq et al., 2010), it is possible that there is a feedback regulation on MAPK activities after WRKY46 overexpression. This has been shown for the MAPK substrate, MYC2, which exerts feedback regulation on its phosphorylating MAPK, MPK6, during blue light-induced signaling (Sethi et al., 2014). To investigate whether also WRKY46 affects MAPK activities in planta, protoplasts expressing either WRKY46 (or CFP as a control) were treated with water or flg22 for 10 min and MAPK activation was analyzed. Treatment with flg22 leads to strong activation of MPK6, MPK3, and MPK4/11 in both WRKY46 and CFP expressing samples without obvious differences (Figure 4E). Thus, WRKY46 does not seem to have feedback regulatory activity on the MAPK pathway; and the enhanced NHL10 promoter activity is likely due to direct action of the overexpressed WRKY46.
Transgenic WRKY46-overexpressing Arabidopsis plants have been shown to be more resistant to bacterial pathogens (Hu et al., 2012). To validate this, we transiently overexpressed Arabidopsis WRKY46 in Nicotiana benthamiana leaves by agro-infiltration. After 24 h, the leaves were challenged with Pseudomonas syringae pv. tabaci and harvested 2 days later. Significantly less bacteria were counted in the leaves overexpressing WRKY46 compared to leaves expressing CFP as a control (Figure 5A). Taken together, these observations suggest that WRKY46 is a positive regulator of plant defense.
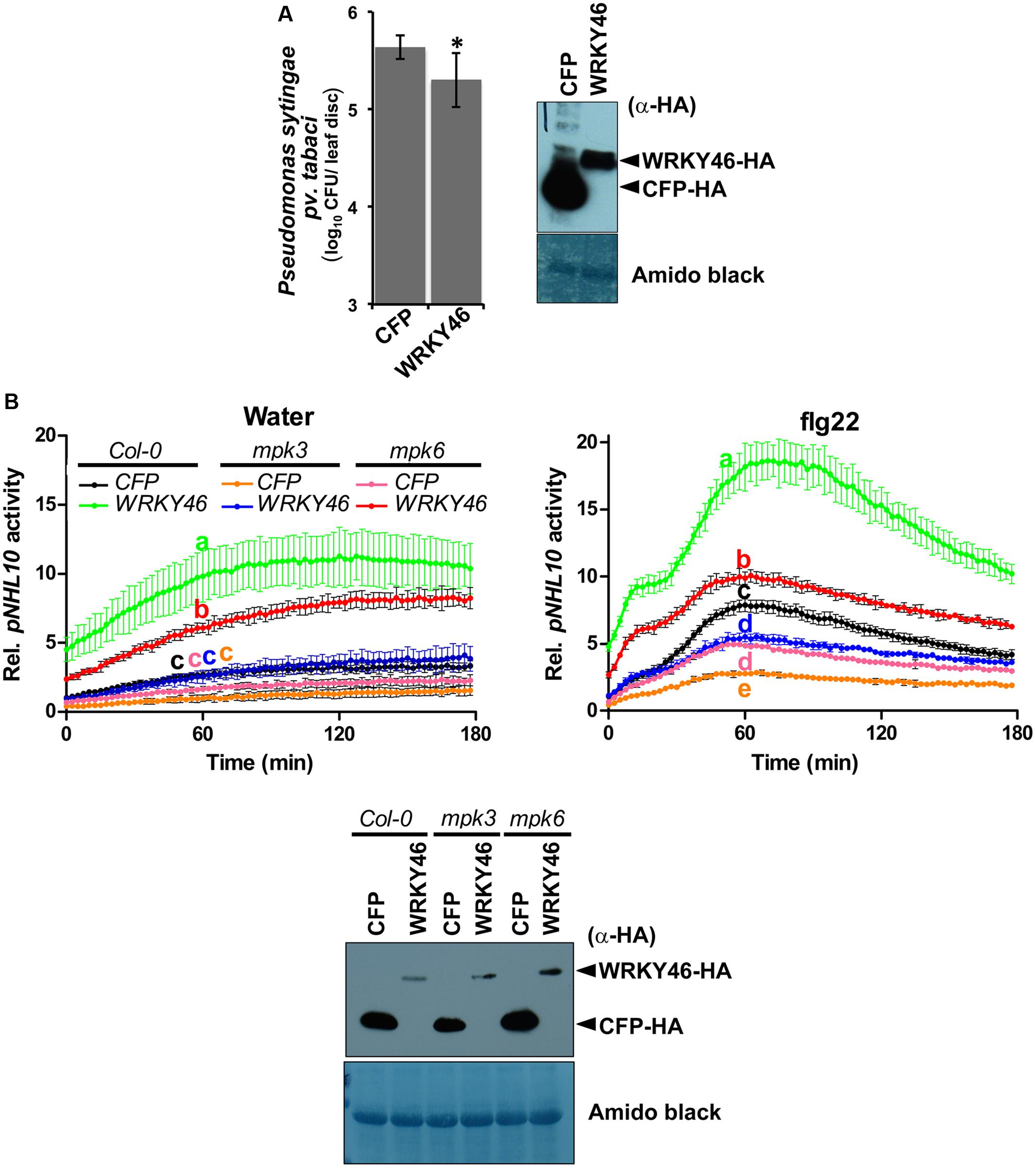
FIGURE 5. WRKY46 enhances bacterial resistance and MPK3/6 are required for the WRKY46-mediated boost of NHL10-promoter activity. (A) WRKY46 overexpression slightly enhances resistance against P. syringae in tobacco. WRKY46 was transiently expressed in N. benthamiana leaves by Agrobacterium-mediated delivery. After 48 h, leaves were challenged with P. syringae pv. tabaci. Colony forming units (CFU) were calculated for samples harvested 2 days post infection. Data shown is the average of three triplicates with error bars indicating standard deviation (∗p < 0.05; Student’s t-test). (B) p35S-WRKY46 was transfected together with pNHL10-LUC and pUBQ10-GUS constructs into protoplasts derived from Arabidopsis Col-0, mpk3 and mpk6, and analyzed as described above (B). Error bars indicate standard deviation. Different letters indicate statistically significant differences at time point 60 min (two-way RM ANOVA with Bonferroni post tests, p < 0.01). The experiment was performed three times with similar results. In all cases, protein expression is validated by western blotting as described above.
MAPKs, and Particularly MPK3, are Required for the Transcriptional Activity of WRKY46
As mentioned above, we could not use the protein mobility phospho-shift in SDS-PAGE to demonstrate an in vivo PAMP-activated MAPK phosphorylation of WRKY46, which we routinely use for several other MAPK substrates (Bethke et al., 2009; Maldonado-Bonilla et al., 2014; Pecher et al., 2014). To alternatively address the role of MPK3 and MPK6 in the function of WRKY46, we investigated NHL10 promoter activities in the respective mpk mutants. In protoplasts derived from mpk3 or mpk6 plants, a statistically significant reduction of the WRKY46-mediated boost of the NHL10 promoter activity was observed in comparison to the wild type for both the water (control) or flg22 treatments (Figure 5B), indicating that both kinases are important for full WRKY46 activity. Notably, mpk3 showed a more dramatic effect than the mpk6 mutant, where the WRKY46-mediated boost on the basal NHL10 promoter activity was completely abolished (See Figure 5B left, compare the green trace to the blue trace). Furthermore, the flg22-activation of NHL10 promoter was also compromised compared to the CFP control (Figure 5B, right, blue and black traces, respectively) in the mpk3 background. Thus, despite functional redundancy between MPK3 and MPK6 (Figure 3C), MPK3 may play a stronger in vivo role in regulating the transcriptional activity of WRKY46 (on the NHL10 promoter). This observation is in agreement to a preferential in vitro phosphorylation of WRKY46 by MPK3 than MPK6 (Figure 2B).
Discussion
In order to adapt to new environmental conditions, plants have to reprogram transcription of the appropriate genes. Transcription factors play major roles in the control of gene expression. The WRKY family is among the ten largest families of transcription factors in higher plants and is found throughout the green lineage (green algae and land plants; Rushton et al., 2010). WRKY proteins are involved in growth development, such as embryogenesis (Lagace and Matton, 2004), trichome development (Johnson et al., 2002), senescence (Robatzek and Somssich, 2001), and plant responses to various biotic and abiotic stresses (Cai et al., 2008; Wu et al., 2009). Only a few components of signaling pathways interacting with WRKY transcription factors have been identified. In addition to histone deacetylases and calmodulin (CaM)-binding domain proteins, MAP kinases are one of the major protein classes interacting with WRKY transcription factors (Kim and Zhang, 2004). The ability of WRKY proteins to interact with many other proteins makes it challenging to decipher the entire pathways regulated by them. WRKY proteins can interact among themselves, with VQ-domain proteins, calmodulin-like proteins, chromatin remodeling proteins and most importantly with MAP kinases (Chi et al., 2013). MAP kinase pathways are involved in regulating the activity of WRKY33 and WRKY34 to control the plant defense response and pollen development, respectively (Mao et al., 2011; Guan et al., 2014). It is plausible that the MPK3/6 cascade acts as a molecular hub to integrate different signaling networks of WRKY transcription factors with different upstream cues. This is further substantiated by the fact that almost 70% of the WRKYs are differentially regulated by bacterial pathogens or SA treatment, which also activates the MPK3/6 pathway (Dong et al., 2003).
In the current study, 48 out of 74 WRKY members were identified as in vitro phosphorylation substrates of MPK3 and MPK6. In silico analysis revealed that WRKY46 may be a putative target of MPK3, which is supported by in vitro kinase assays showing a stronger phosphorylation by MPK3 compared to MPK6. MS analysis identified S168 of WRKY46 as a phosphorylation site targeted by MPK3. However, subsequent experiments suggest that S250, the other putative phosphorylation site, is also phosphorylated. Direct in vivo MPK3 or MPK6 activation by expressing a constitutively active MKK5 led to destabilization of WRKY46 protein. Thus, MPK3 and/or MPK6 (or perhaps also other unknown kinases activated by these MAPKs) phosphorylate S168, S250 and other positions to regulate its stability. In this respect, a WRKY46 promoter-based reporter assay revealed transcription activation of WRKY46 by the calcium-dependent protein kinases, CPK-3, -4, -5, -6, -10, -11, and -30 and several WRKYs were directly phosphorylated by CPKs (Gao et al., 2013).
WRKY46 regulates responses to several abiotic stresses in Arabidopsis. For instance, it controls the expression of several genes involved in osmoprotection and redox homeostasis under dehydration stress (Ding et al., 2015). It is also involved in controlling the light-dependent stomatal opening in guard cells (Ding et al., 2014), which may serve as potential entry points for pathogens and therefore contribute to responses to pathogens. Indeed, WRKY46 is also involved in biotic stresses. Similar to the NHL10 gene shown in this work, over-expression of WRKY46 in protoplasts was found to increase the expression of endogenous AVRPPHB SUSCEPTIBLE 3 (PBS3) gene, which plays an important role in SA metabolism (van Verk et al., 2011). WRKY46 acts in concert with other WRKYs, including WRKY54 as a positive regulator and WRKY70 as a negative defense regulator, to resist infection by the necrotrophic Erwinia amylovora (Moreau et al., 2012). It also coordinates with WRKY53 and WRKY70 to control defense response against the hemi-biotrophic pathogen, Pseudomonas syringae (Hu et al., 2012). In general, expression of many WRKY genes is induced by pathogen-related stimuli, possibly in a feedback amplification loop. For instance, the expression of WRKY46 was highly induced within hours after P. syringae and SA treatments (Dong et al., 2003). WRKY46 transcripts accumulated in protoplasts expressing avrRpm1, avrB, or avrRpt2 in an RPM1- or RPS2- dependent manner, thereby making it an early marker gene in ETI signaling and candidate ETI regulator (Gao et al., 2013). The induction of its expression by PAMPs (Figure 4B) also makes it a PTI marker gene. Since WRKY46 can contribute to both PTI and ETI signaling, it is plausible that it mediates interplay between PTI and ETI and this will be an interesting question to address in the future.
The MAPK phosphorylation could control the function of transcription factors or their associated proteins by regulating their in planta stability (Pecher et al., 2014; Weyhe et al., 2014). Recently, phosphorylation of LIP5 (LYST-INTERACTING PROTEIN5), a positive regulator of multivesicular body (MVB) biogenesis by MPK6 was found to increase its stability (Wang et al., 2015). Similarly, phosphorylation of Arabidopsis ERF6, ACS2, and ACS6 by MPK3/MPK6 increase the stability of these substrates (Li et al., 2012). In case of WRKY46, our data provides evidence that MAPK-mediated phosphorylation upon flg22-treatment controls its stability in vivo. The phosphorylation at both S168 and S250 positions seems to be very important for the half-life of the protein, since ablation of phosphorylation at either position leads to increased stability upon elicitation. Hence, the phosphorylation-dependent regulation of protein substrate stability appears to be a common mechanism through which MAPKs regulate plant defense responses.
Localization studies revealed that WRKY46 is localized in the nucleus. This result is consistent with the fact that it acts as a transcription factor, possibly regulating the transcription of defense-related genes. Accordingly, WRKY46 overexpression boosts the defense responsive NHL10 promoter activity (in the absence of pathogens or PAMPs). Since there are WRKY DNA-binding elements in the NHL10 promoter (Zheng et al., 2004), which have been shown to be important for regulation of PAMP-induced expression (Pecher et al., 2014), it is likely that WRKY46 acts directly as a positive transcriptional activator via such cis-elements. The compromised activation of the NHL10 promoter by WRKY46 overexpression in the mpk3 or mpk6 mutants suggests the importance of these two MAPKs, particularly MPK3, in controlling WRKY46 activity. This may seem puzzling for the water-treated samples since no enhanced MAPKs activities are usually detected prior to PAMP elicitation. However, we cannot exclude that some handling stress did, in fact, initiate a transient MAPK activation in the protoplast system and this may trigger a long-lasting effect on the subsequently expressed WRKY46 proteins. This would mean that effect of WRKY46 on the NHL10 promoter is indirect or at least acts only in synergy with the general stress-induced MAPK activities. In support of the latter, the WRKY46-induction of the NHL10 promoter in the flg22-treated protoplasts (Figure 5B, right) – where MAPKs as well as other signaling pathways are activated – is also compromised by mpk3 or mpk6 mutations. In summary, the mode of the WRKY46-MAPKs regulation of NHL10 promoter is currently unknown but it appears to be unrelated to WRKY46 protein stability or its phospho-status (Note: the phospho-mimic WRKY46 did not show enhanced transcriptional activity). Future experiments could reveal if the MPK3/MPK6 effect may be due to altered affinity of WRKY46 to the promoter or direct impact on its transcriptional activity. Alternatively, a more likely scenario would be that there may be other MPK3/MPK6 targets that influence WRKY46 activity. Furthermore, as already discussed, there may be additional kinase(s) that phosphorylate WRKY46. Likely candidates include members of the calcium-dependent protein kinase family – especially those that have been shown to regulate WRKY46 expression (Gao et al., 2013). It will be crucial to identify these kinase(s) and understand how they, in concert with MAPKs, control the multiple functions of WRKY46 in developmental processes, abiotic and biotic stress responses.
Author Contributions
AS and LE designed and performed the experiments shown. PP cloned the WRKYs into the bacterial expression system and screened them for recombinant protein expression. WH performed the LC-MS/MS determination of the phosphorylation sites. DS, AS, and JL supervised the project and all authors contributed to writing the manuscript.
Funding
AS is supported by a German Academic Exchange (DAAD) fellowship (A/11/75070). Research in our laboratory is financed by the German Research Foundation through the Collaborative Research Centre SFB648/TP-B1 “Molecular mechanisms of information processing in plants.” The BMBF project ProNET-T3 (03ISO2211B) supported LE.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Imre Somssich for providing the Gateway-compatible pENTR clones of the WRKY transcription factors, Mubashir Ahmad (University of Ulm) for helping with in-silico studies, Nicole Bauer for excellent technical assistance and all members from our laboratory for constructive discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00061
FIGURE S1 | Nuclear localization of WRKY46: (A) Confocal laser scanning micrographs of Arabidopsis Col-0 mesophyll protoplasts transfected with the indicated WRKY46-YFP constructs. To visualize the nucleus, nuclear DNA was stained with 1 μg mL-1 of 4′,6-Diamidino-2-Phenylindole (DAPI) in the presence of 0.05 % Triton-X100 (to permeabilize the membranes). Scale Bar = 10 μm. (B) Western blot analysis with α-GFP to show intactness of WRKY46-GFP fusion proteins.
Footnotes
- ^ http://david.abcc.ncifcrf.gov/
- ^ http://string-db.org/
- ^ http://www.cytoscape.org/
- ^ www.graphpad.com
References
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gómez-Gómez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Bethke, G., Pecher, P., Eschen-Lippold, L., Tsuda, K., Katagiri, F., Glazebrook, J., et al. (2012). Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25, 471–480. doi: 10.1094/MPMI-11-11-0281
Bethke, G., Unthan, T., Uhrig, J. F., Pöschl, Y., Gust, A. A., Scheel, D., et al. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 8067–8072. doi: 10.1073/pnas.0810206106
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Boudsocq, M., Willmann, M. R., Mccormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Cai, M., Qiu, D., Yuan, T., Ding, X., Li, H., Duan, L., et al. (2008). Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 31, 86–96. doi: 10.1111/j.1365-3040.2007.01739.x
Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B., Yu, J. Q., et al. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6, 287–300. doi: 10.1093/mp/sst026
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., and Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., and Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. doi: 10.1111/tpj.12958
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., and Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Ding, Z. J., Yan, J. Y., Xu, X. Y., Yu, D. Q., Li, G. X., Zhang, S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79, 13–27. doi: 10.1111/tpj.12538
dit Frey, N., Garcia, A. V., Bigeard, J., Zaag, R., Bueso, E., Garmier, M., et al. (2014). Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 15:R87. doi: 10.1186/gb-2014-15-6-r87
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Dyson, M. R., Shadbolt, S. P., Vincent, K. J., Perera, R. L., and Mccafferty, J. (2004). Production of soluble mammalian proteins in Escherichia coli: identification of protein features that correlate with successful expression. BMC Biotechnol. 4:32. doi: 10.1186/1472-6750-4-32
Eschen-Lippold, L., Bauer, N., Lohr, J., Palm-Forster, M. A., and Lee, J. (2014). Rapid mutagenesis-based analysis of phosphorylation sites in mitogen-activated protein kinase substrates. Methods Mol. Biol. 1171, 183–192. doi: 10.1007/978-1-4939-0922-3_15
Eschen-Lippold, L., Bethke, G., Palm-Forster, M. A., Pecher, P., Bauer, N., Glazebrook, J., et al. (2012). MPK11-a fourth elicitor-responsive mitogen-activated protein kinase in Arabidopsis thaliana. Plant Signal. Behav. 7, 1203–1205. doi: 10.4161/psb.21323
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Feilner, T., Hultschig, C., Lee, J., Meyer, S., Immink, R. G. H., Koenig, A., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4, 1558–1568. doi: 10.1074/mcp.M500007-MCP200
Feys, B. J., Wiermer, M., Bhat, R. A., Moisan, L. J., Medina-Escobar, N., Neu, C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613. doi: 10.1105/tpc.105.033910
Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., et al. (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815. doi: 10.1093/nar/gks1094
Gao, X., Chen, X., Lin, W., Chen, S., Lu, D., Niu, Y., et al. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9:e1003127. doi: 10.1371/journal.ppat.1003127
Gómez-Gómez, L., and Boller, T. (2002). Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256. doi: 10.1016/S1360-1385(02)02261-6
Guan, Y., Lu, J., Xu, J., Mcclure, B., and Zhang, S. (2014). Two mitogen-activated protein kinases, MPK3 and MPK6, are required for funicular guidance of pollen tubes in Arabidopsis. Plant Physiol. doi: 10.1104/pp.113.231274 [Epub ahead of print].
Hu, Y., Dong, Q., and Yu, D. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185–186, 288–297. doi: 10.1016/j.plantsci.2011.12.003
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Johnson, C. S., Kolevski, B., and Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kim, C. Y., and Zhang, S. (2004). Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J. 38, 142–151. doi: 10.1111/j.1365-313X.2004.02033.x
Lagace, M., and Matton, D. P. (2004). Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219, 185–189. doi: 10.1007/s00425-004-1253-2
Lassowskat, I., Böttcher, C., Eschen-Lippold, L., Scheel, D., and Lee, J. (2014). Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front. Plant Sci. 5:554. doi: 10.3389/fpls.2014.00554
Lee, J., Eschen-Lippold, L., Lassowskat, I., Boettcher, C., and Scheel, D. (2015). Cellular reprogramming through mitogen-activated protein kinases. Front. Plant Sci. 6:940. doi: 10.3389/fpls.2015.00940
Lee, J., Rudd, J. J., Macioszek, V. K., and Scheel, D. (2004). Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J. Biol. Chem. 279, 22440–22448. doi: 10.1074/jbc.M401099200
Li, G., Meng, X., Wang, R., Mao, G., Han, L., Liu, Y., et al. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8:e1002767. doi: 10.1371/journal.pgen.1002767
Liu, Y. D., and Zhang, S. Q. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. doi: 10.1105/tpc.104.026609
Maldonado-Bonilla, L. D., Eschen-Lippold, L., Gago-Zachert, S., Tabassum, N., Bauer, N., Scheel, D., et al. (2014). The Arabidopsis tandem zinc finger 9 protein binds RNA and mediates pathogen-associated molecular pattern-triggered immune responses. Plant Cell Physiol. 55, 412–425. doi: 10.1093/pcp/pct175
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
Meng, X., and Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Moreau, M., Degrave, A., Vedel, R., Bitton, F., Patrit, O., Renou, J. P., et al. (2012). EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora. Mol. Plant Microbe Interact. 25, 421–430. doi: 10.1094/MPMI-05-11-0111
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Palm-Forster, M. A., Eschen-Lippold, L., and Lee, J. (2012). A mutagenesis-based screen to rapidly identify phosphorylation sites in mitogen-activated protein kinase substrates. Anal. Biochem. 427, 127–129. doi: 10.1016/j.ab.2012.05.015
Pecher, P., Eschen-Lippold, L., Herklotz, S., Kuhle, K., Naumann, K., Bethke, G., et al. (2014). The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ’VQ-motif’-containing proteins to regulate immune responses. New Phytol. 203, 592–606. doi: 10.1111/nph.12817
Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. doi: 10.1016/S0092-8674(00)00213-0
Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z., Gerstein, M., Snyder, M., et al. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23, 80–92. doi: 10.1101/gad.1740009
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Ranf, S., Eschen-Lippold, L., Pecher, P., Lee, J., and Scheel, D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68, 100–113. doi: 10.1111/j.1365-313X.2011.04671.x
Reddy, P. S., Kavi Kishor, P. B., Seiler, C., Kuhlmann, M., Eschen-Lippold, L., Lee, J., et al. (2014). Unraveling regulation of the small heat shock proteins by the heat shock factor HvHsfB2c in barley: its implications in drought stress response and seed development. PLoS ONE 9:e89125. doi: 10.1371/journal.pone.0089125
Robatzek, S., and Somssich, I. E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28, 123–133. doi: 10.1046/j.1365-313X.2001.01131.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sethi, V., Raghuram, B., Sinha, A. K., and Chattopadhyay, S. (2014). A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26, 3343–3357. doi: 10.1105/tpc.114.128702
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sinha, A. K., Jaggi, M., Raghuram, B., and Tuteja, N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6, 196–203. doi: 10.4161/psb.6.2.14701
van Verk, M. C., Bol, J. F., and Linthorst, H. J. (2011). WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11:89. doi: 10.1186/1471-2229-11-89
Wang, F., Yang, Y., Wang, Z., Zhou, J., Fan, B., and Chen, Z. (2015). A critical role of lyst-interacting protein5, a positive regulator of multivesicular body biogenesis, in plant responses to heat and salt stresses. Plant Physiol. 169, 497–511. doi: 10.1104/pp.15.00518
Wang, H., Liu, Y., Bruffett, K., Lee, J., Hause, G., Walker, J. C., et al. (2008). Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20, 602–613. doi: 10.1105/tpc.108.058032
Wang, H., Ngwenyama, N., Liu, Y., Walker, J. C., and Zhang, S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73. doi: 10.1105/tpc.106.048298
Weyhe, M., Eschen-Lippold, L., Pecher, P., Scheel, D., and Lee, J. (2014). Ménage à trois: the complex relationships between mitogen-activated protein kinases, WRKY transcription factors and VQ-motif-containing proteins. Plant Signal. Behav. 9:e29519. doi: 10.4161/psb.29519
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zheng, M. S., Takahashi, H., Miyazaki, A., Hamamoto, H., Shah, J., Yamaguchi, I., et al. (2004). Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to Cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement. Planta 218, 740–750. doi: 10.1007/s00425-003-1169-2
Keywords: mitogen-activated protein kinase, WRKY transcription factors, phosphorylation, protein stability, defense, pathogen-associated molecular patterns (PAMPs)
Citation: Sheikh AH, Eschen-Lippold L, Pecher P, Hoehenwarter W, Sinha AK, Scheel D and Lee J (2016) Regulation of WRKY46 Transcription Factor Function by Mitogen-Activated Protein Kinases in Arabidopsis thaliana. Front. Plant Sci. 7:61. doi: 10.3389/fpls.2016.00061
Received: 07 October 2015; Accepted: 14 January 2016;
Published: 04 February 2016.
Edited by:
Olivier Lamotte, Centre National de la Recherche Scientifique, FranceReviewed by:
Hirofumi Yoshioka, Nagoya University, JapanAndrea Pitzschke, University of Salzburg, Austria
Copyright © 2016 Sheikh, Eschen-Lippold, Pecher, Hoehenwarter, Sinha, Scheel and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justin Lee, jlee@ipb-halle.de
†Present address: Arsheed H. Sheikh, School of Life Sciences, Gibbet Hill Campus, The University of Warwick, Coventry, CV4 7AL, UK; Pascal Pecher, Department of Cell and Developmental Biology, John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK
‡These authors have contributed equally to this work.
 Arsheed H. Sheikh
Arsheed H. Sheikh Lennart Eschen-Lippold
Lennart Eschen-Lippold Pascal Pecher
Pascal Pecher Wolfgang Hoehenwarter
Wolfgang Hoehenwarter Alok K. Sinha
Alok K. Sinha Dierk Scheel
Dierk Scheel Justin Lee
Justin Lee