- Department of Vegetable Science, College of Horticulture, Northwest A&F University, Yangling, China
Abiotic stresses negatively affect plants growth and development by inducing protein denaturation, and autophagy degrades the damaged proteins to alleviate their toxicity, however, little is known about the involvement of autophagy in pepper (Capsicum annuum L.) tolerances to abiotic stresses. In this study, we identified autophagy-related gene (ATG) members in the whole genome of pepper by HMM method and analyzed their expression profiles in response to heat and other abiotic stresses by quantitative real-time PCR. The results showed that the CaATG contained 15 core ATG members including 29 ATG proteins with their respective conserved functional domains, involving the whole process of autophagy. Under normal environmental condition, the expression of CaATG genes showed tissue- and developmental stage-specific patterns, while under abiotic stresses of salt, drought, heat, cold and carbohydrate starvation, the accumulation of autophagosome punctate increased and the expression level of CaATG genes changed with stress type-dependent pattern, which indicates the linkage of autophagy in pepper response to abiotic stresses. After treated with heat stress, both the number of up-regulated CaATG genes and the increment of autophagosome punctate were higher in pepper thermotolerant line R9 than those in thermosensitive line B6, implying an association of autophagy with heat tolerance. In addition, CaATG6 was predicted to interact with CaHSP90 family members. Our study suggests that autophagy is connected to pepper tolerances to heat and other abiotic stresses.
Introduction
In general, plants grow in an open ecosystem with the sessile lifestyle, which makes them routinely suffer from adverse environmental conditions, mainly including high and low temperature, drought, salt, and starvation stress (Wang et al., 2003; Ahuja et al., 2010; Wang and Liu, 2010). With the aggravating of global warming in recent years, extreme high temperature climate occurs more frequently (IPCC, 2013), and it is estimated that 1°C rise in temperature could lowers the yield up to 10% (Lobell et al., 2011), thus heat stress becomes a major threat for crop production.
High temperature negatively affects plants growth and development by inducing protein misfolding, denaturation, oxidation, and aggregation (Hemantaranjan et al., 2014). Organisms develop two types of mechanisms in evolution to alleviate the injury from heat stress, i.e., effectively preventing the proteins to be damaged and efficiently removing the damaged proteins (Dokladny et al., 2015). In plants, the major executors to protect proteins under heat stress are heat shock proteins (HSPs) as molecular chaperones (Liu et al., 2012), which was verified by the studies of our research group and others in pepper (Capsicum annuum L.) (Zhu et al., 2011; Guo et al., 2014a,b), tomato (Solanum lycopersicum L.) (Bita et al., 2011), wheat (Triticum aestivum L.) (Xue et al., 2014), and cucumber (Cucumis sativus L.) (Li et al., 2014). However, the relationship between the ability to remove damaged proteins and thermotolerance of plants is still unclear and remained to be studied.
The removal of damaged proteins can be conducted by autophagy, one of the ubiquitous and highly conservative protein degradation systems in eukaryotic cells (Yoshimoto et al., 2010). During the autophagy process, the damaged proteins and organelles were wrapped into double-membrane vesicle called autophagosome and then transported into vacuoles for breakdown (Johansen and Lamark, 2011; Liu and Bassham, 2012). Recent studies divulged that autophagy participated in plant responses to environmental stresses like salt, drought, cold, oxdative, hypoxia, and osmotic stress (Slavikova et al., 2008; Liu et al., 2009; Shin et al., 2009; Kuzuoglu-Ozturk et al., 2012; Pei et al., 2014; Chen et al., 2015).
As for heat stress, although Zhou et al. (2013, 2014) found functional loss or silent expression of autophagy related genes (ATG) ATG5, ATG7, and their regulator NBR1 (neighbor of BRCA1) decreased the thermotolerance of Arabidopsis and tomato by accumulating more insoluble protein, but systematic analysis of ATGs in plant response to heat stress is limited. In our current study, 15 putative ATG members including 29 ATG proteins were identified in pepper, a temperate but thermosensitve vegetable crop, and their expression profiles in response to heat stress and other abiotic stresses were comparatively analyzed, aiming to provide a systematic base for further studies on the function and regulation of autophagy in the formation of plant thermotolerance.
Materials and Methods
Identification of CaATG Homologs
The identification of putative CaATG proteins was performed by the HMMER 3.0 software, in which the HMM profiles of pfam (http://pfam.xfam.org/) for each ATG member were generated from the published amino acid sequences of ATG members of Arabidopsis (Hanaoka et al., 2002; Kwon and Park, 2008) and rice (Xia et al., 2011), and pepper genomic protein sequences were downloaded from database PGP (V1.55) (http://peppergenome.snu.ac.kr) (Kim et al., 2014) and PGD (V2.0) (http://peppersequence.genomics.cn/page/species/index.jsp) (Qin et al., 2014). All output protein sequences were collected and then confirmed by the SMART (http://smart.embl-heidelberg.de/) and blastp program in NCBI (http://www.ncbi.nlm.nih.gov/). The candidate CaATG protein sequences from the three pepper cultivars (CM334, Zunla-1 and Chiltepin) were aligned by ClustalW2 online software (http://www.ebi.ac.uk/Tools/msa/clustalw2/) to determine the protein sequences, which were confirmed again by operating blastp. The theoretical iso-electric point (pI) and molecular weight (MW) of each CaATG protein were calculated by Compute pI/Mw tool (http://web.expasy.org/compute_pi/), and the proteins subcellular location were predicted in online software WoLF PSORT (http://wolfpsort.seq.cbrc.jp/).
Phylogenetic Analysis
The software MEGA 6 (Tamura et al., 2013) was recruited to construct the phylogenetic trees of the full-length of ATG protein sequences from pepper, rice, Arabidopsis, algae and yeast, in which the neighbor-joining method, p-distance substitution model and 1000 bootstrap replicates were selected.
Sequence Structure, Chromosomal Location, and Conserved Motifs Analysis
The exon/intron structures of CaATG genes were determined based on alignments of coding sequences with their respective genomic full-length sequences (http://peppergenome.snu.ac.kr), and the structural diagrams were generated by the online program Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.ch). MapInspect software (Ralph van Berloo, Laboratory of Plant Breeding, Wageningen University) was used to draw the chromosomal locations of CaATG genes.
Conserved motifs of the CaATG protein sequences were identified by MEME program (http://meme.nbcr.net/meme/), in which normal motif discovery mode and 20 for maximum number of motifs were selected, and the parameter of site distribution was set as zero or one occurrence per sequence.
RNA-Seq Analysis of CaATG Genes Tissue-Specific Expression
Based on the databases of genomic RNA-seq (Kim et al., 2014), the tissue and stage-specific expression profiles of 26 CaATG genes (the data of CaATG5, ATG12, and VPS34 are absent) were analyzed in 17 tissues of pepper cultivar “CM334” and seven tissues of “ECW30R,” including root, stem, leaf, pericarp and placenta at different days post-anthesis (PC-6DPA, -16DPA and -25DPA, PL-6DPA, PL-13DPA, -16DPA, PL-20DPA, and-25DPA), pericarp and placenta at mature green (PC-MG, PL-MG) and at breaker (PC-B, PL-B), pericarp and placenta at 5 and 10 days post-breaker (PC-B5 and PC-B10, PL-B5 and PL-B10).
Heat and Other Abiotic Stresses Treatments
The pepper thermotolerant line R9 (sweet pepper, introduced from the World-Asia Vegetable Research and Development Center, PP0042-51) and thermosensitive line B6 (hot pepper, selected by the pepper research group, College of Horticulture, Northwest A&F University, Yangling, China) were used in this study. The pepper seedlings were grown under normal conditions (26°C/20°C day/night, photoperiod of 16 h light/8 h dark cycle, 200 μmol·m−2·s−1 illumination intensity and 70% relative humidity) in a controlled climate chamber.
Pepper seedlings at 5–6 true leaves stage were used for abiotic stresses treatments. The seedlings of control treatment were grown under normal conditions, while for heat stress (HS) treatments, seedlings of R9 and B6 were transferred into climate chamber with temperature of 40°C, and the leaves were collected at 0, 1, 3, 6, and 12 h after HS. The seedlings of R9 were kept at 4°C for 3 h for cold treatment, kept in the dark for 2 days for carbohydrate starvation (Pei et al., 2014), incubated in Hoagland's solution containing 200 mM NaCl for 3 h for salt treatment (Xiao et al., 2014), and dried for 3 h between folds of tissue pepper for drought treatment (Xia et al., 2011). The roots and stems of pepper seedlings were collected for NaCl treatment, and the roots, stems and leaves for drought treatment, while the leaves for other treatments. After collection, the samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. Each treatment was conducted with three biological replicates and each replicate contained five pepper seedlings.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from frozen samples using Plant RNA Kit (OMEGA) according to the manufacturer's instructions, and then cDNA was generated using the Primescript™first strand cDNA Synthesis Kit (Takara). Quantitative real-time PCR (qRT-PCR) for relative quantification of CaATGs expression was performed according to the method of Guo et al. (2014b), and the ubiquitin-conjugating protein gene UBI-3 (AY486137) was used as the reference gene (Wan et al., 2011). The CaATGs gene-specific primer pairs for qRT-PCR were listed in Table S1. The relative expression levels of pepper CaATG genes were calculated following the 2−ΔΔCT method (Livak and Schmittgen, 2001), in which the relative expression levels of the control seedlings were set to one and two-fold change was looked as the standard to judge the up- or down-regulation. The significance test for difference in expression levels of each CaATG gene between control and stress treatment was performed by student's t-test method at α = 0.01 level.
Generation of the Heatmap for Gene Expression Level
The heatmaps for CaATG genes expression level were created by HemI (The Cuckoo Workgroup, Wuhan, China). For the tissue-specific expression, the RNA-seq data extracted from the published databases of Kim et al. (2014) were normalized using logarithm with the base of 2, and those genes having no transcription were indicated as white color. For the expression under abiotic or heat stress, the values of 2−ΔΔCT were used directly for the up-regulated genes, while the values of −(2−ΔΔCT)−1 were employed for the down-regulated genes to make the heatmap be observed more intuitively.
Autophagosome Staining and Microscopic Observation
The combination of E-64d for autophagosomes accumulation and LysoTracker for staining have been widely used to detecting autophagic activity in a variety of organisms including plants (Moriyasu et al., 2003; Kuzuoglu-Ozturk et al., 2012; Phadwal et al., 2012; Chikte et al., 2014). After infiltrated with 100 μM E-64d (Sigma), pepper leaves and root segments were stained with 2 μM LysoTracker Red DND-99 (Invitrogen) according to the method of Liu et al. (2005). The stained autophagosome punctates were observed under fluorescent microscope (BX51; Olympus) using a 50-W mercury lamp, and punctate areas were scanned by the tool of particles analysis in ImageJ 1.46 (National Institutes of Health, Bethesda, Maryland). The Statistical Analysis System software (version 8.2, SAS Institute, Cary, NC, USA) for one-way analysis of variance (ANOVA) and the shortest significant ranges (SSR) method were used to compare the differences in punctate areas among the leaves of B6 and R9 with and without heat stress at the level α = 0.05.
Prediction of Protein-Protein Interaction Network
The interolog from Arabidopsis was used for predicting protein-protein interaction network among CaATG members and HSPs by searching in STRING database (Search Tool for the Retrieval of Interacting Genes/Proteins) (http://string-db.org). In order to improve the reliability of prediction results, the parameters with a high confidence (0.700) was set to filter the edge information of all the ATG members as well as HSPs interacting with ATGs. The output was finally imported into Cytoscape_v3.2.1 (National Institute of General Medical Sciences, MD, USA) to generate the protein-protein interaction network map.
Results
Genome-Wide Identification of ATG Genes in Pepper
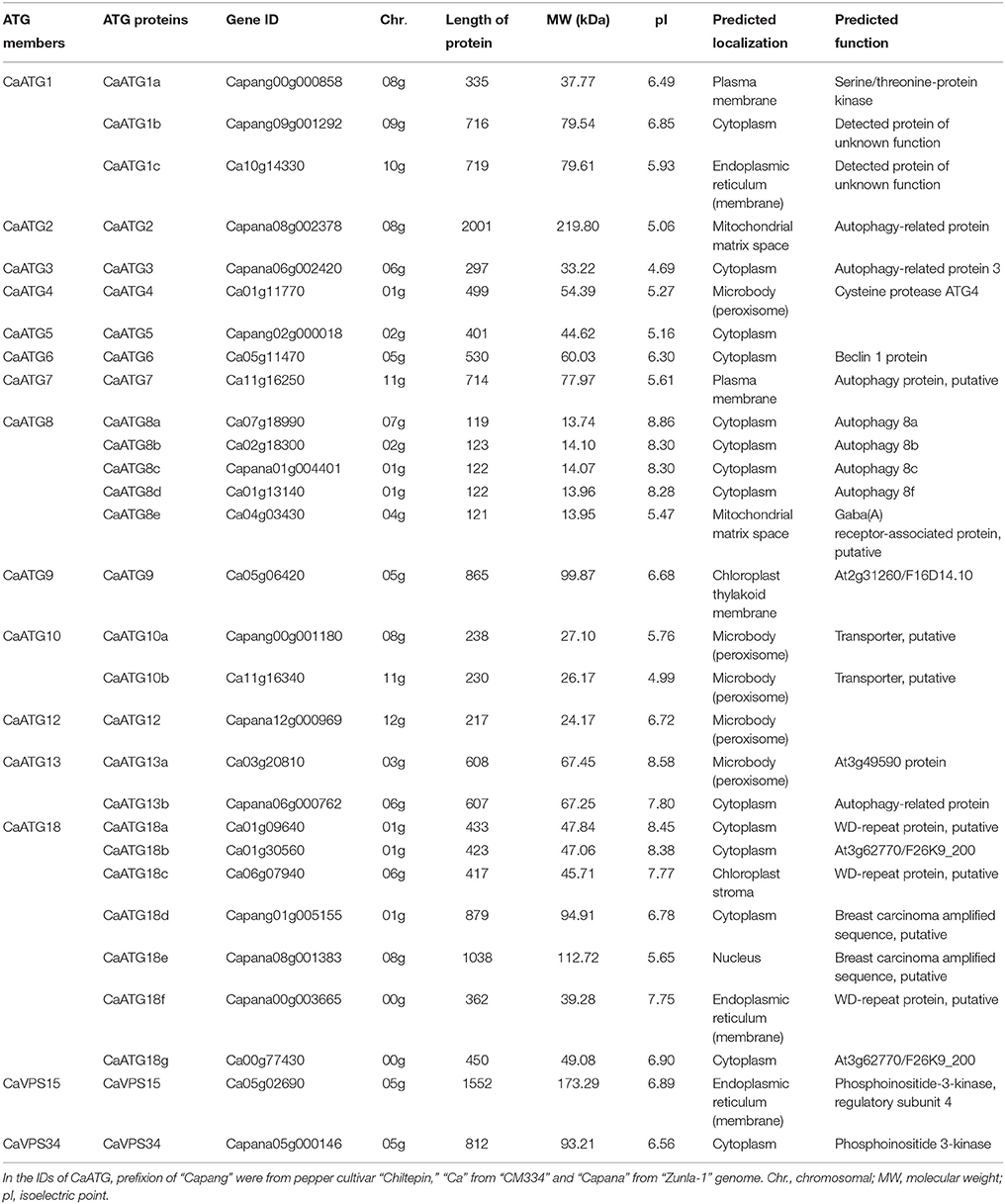
Since no published data is available about autophagy related genes in pepper so far, the sequences of ATG members from Arabidopsis (Hanaoka et al., 2002; Kwon and Park, 2008) and rice (Xia et al., 2011) were used to search the pepper genome database (http://peppergenome.snu.ac.kr). The searching results showed that 15 core ATG members containing 29 proteins with ATG domains were identified (Table 1; Table S2), involving all processes of autophagy.
Among the 15 ATG members in pepper, there were seven proteins in ATG18 (CaATG18a-18g), five in ATG8 (CaATG8a-8e), three in ATG1 (CaATG1a-1c), two in ATG10 (CaATG10a/10b), and ATG13 (CaATG13a/13b) respectively, and one in each of the other ATG members (ATG2, ATG3, ATG4, ATG5, ATG6, ATG7, ATG9, ATG12, VPS15, and VPS34) (Table 1). The CaATG proteins varied from 119 to 2001 amino acid (aa) in length, from 13.744 to 219.801 kDa in molecular weight and from 4.69 to 8.86 in predicted isoelectric points (Table 1).
The 29 CaATG genes were unevenly distributed on the 12 chromosomes of pepper, in which 6 CaATG genes were located on Chr01, and 5 genes on Chr08, 4 on Chr05, 3 on Chr06, 2 on Chr02, 07, and 11 respectively, and only 1 on each of the remaining 5 chromosomes respectively (Figure S1).
Phylogenetic Relationship, Conserved Domains, and Sequence Structure of CaATG Genes
Phylogenetic trees of the 15 ATG members were constructed respectively based on the ATG protein sequences from five species including pepper, Arabidopsis, rice, algae and yeast (Figure 1). Compared to those from algae and yeast, the ATG proteins from three higher plant species were basically clustered together. Twenty-six out of twenty-nine pepper ATG proteins (89.6%) except CaATG18a/18b/18d showed the nearest phylogenetic relationship with those of Arabidopsis, agreeing with their botanical classifications.
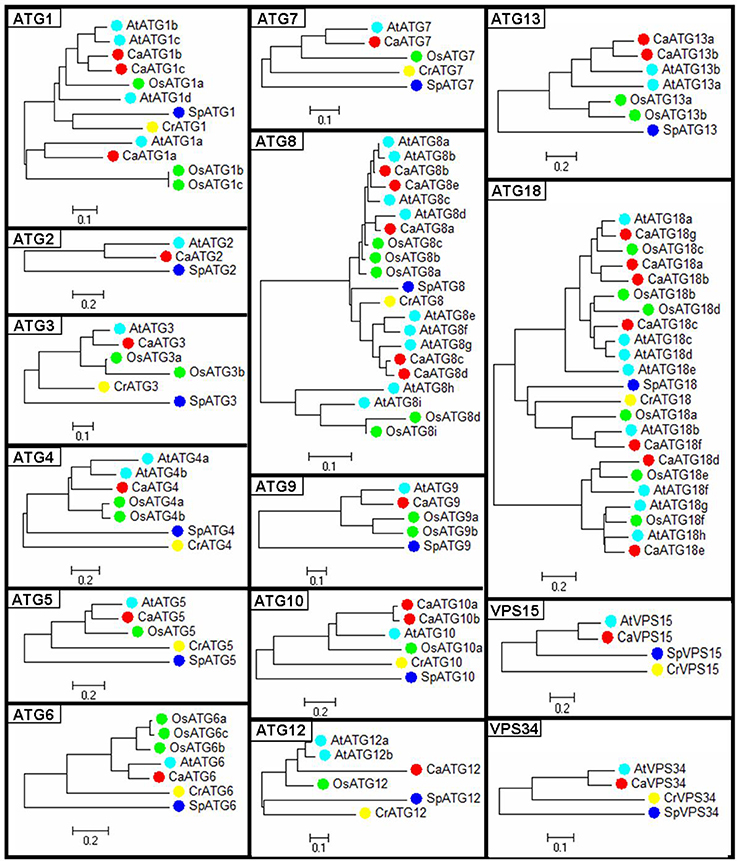
Figure 1. Phylogenetic tree of ATG proteins from pepper. Ca, Capsicum annuum L.; At, Arabidopsis thaliana; Os, Oryza sativa; Cr, Chlamydomonas reinhardtii; Sp, Saccharomyces cerevisiae.
The classification of 29 CaATG proteins was coincident with the motifs distribution identified by MEME web server (http://meme-suite.org/tools/meme) (Figures 2A,B, Table S3). For instance, the conserved domains of motif1-motif2 and motif13-motif4 were observed in all of the five CaATG8 proteins and all of the eight CaATG18 proteins, respectively. Similar structure of motif15-motif14-motif8 at N-terminal was also presented in CaATG1a, 1b, and 1c. However, diversity in motifs and their distribution were found. For example, CaATG8e did not contain motif18 like the other 4 ATG8 proteins, and CaATG1a lacked the C-terminal structure of motif19-motif20 as shown in ATG1b and c (Figures 2A,B).
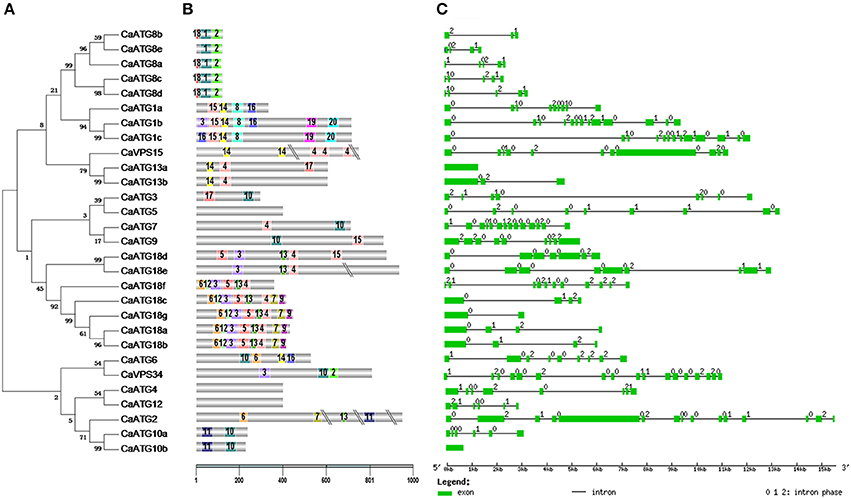
Figure 2. Conserved domains and sequence structure of CaATG genes. (A) Phylogenetic analysis of CaATG proteins. Unrooted phylogenetic tree built on the basis of full-length of ATG protein sequences; (B) Motifs of CaATG proteins identified by MEME tools. Different motifs are indicated by different borders and colors; (C) Exon/intron organizations of CaATG genes. Green boxes indicate the exon regions and lines indicate introns. The length of the boxes and lines are scaled based on the length of genes. Numbers 0, 1, and 2 represent introns in phases 0, 1, and 2 respectively.
The structures in exon/intron distribution CaATG genes were further analyzed based on the alignments of their coding region sequences with respective genomic full-length sequences. Except CaATG10b and CaATG13a, the other 27 CaATG genes contained multiple introns with irregular intron phases for 0, 1, and 2 (Figure 2C).
By subcellular localization prediction software WoLF PSORT, it was predicted that 14 of the 29 CaATG proteins were located in the cytoplasm, five in microbody (peroxisome), three in endoplasmic reticulum, two in plasma membrane (CaATG1a and CaATG7) and mitochondrial matrix space (CaATG2 and CaATG8e), and one in chloroplast thylakoid membrane (CaATG9), chloroplast stroma (CaATG18c), and nucleus (CaATG18e), respectively (Table 1).
RNA-Seq Analysis of CaATG Genes Tissue- and Developmental Stage-Specific Expression
To investigate the spatial and temporal expression patterns of ATG genes in pepper, we retrieved the data of 26 CaATG genes from the genomic RNA-seq databases of different tissues (root, stem and leaf) and fruit developmental stages (pericarp and placenta at various stages) of pepper cultivar “CM334” and “ECW30R.” As a whole, the expression of 26 CaATG genes showed tissue- and organ-specific pattern (Figure 3, Table S4), in which CaATG2, 10a, 10b, and 18a showed a relatively low expression level, while CaATG8a and CaATG8c remained at higher expression levels in each tested tissue. Similar to the results in Arabidopsis (Sláviková et al., 2005), except CaATG1a, 8a, 8c, 10a, 13b, 18b, most of the CaATG genes were abundantly transcribed in root and stem, meaning the higher activity of autophagy in plant root and stem under normal growth condition. In addition, some CaATG genes, such as CaATG3, 4, 6, 13a, and 18g, showed higher expression levels in reproductive organs, while CaVPS15 only highly expressed in PC-MG, PC-B10 of “CM334” and PL-20DPA of “ECW30R.” Interestingly, with the increment in days after anthesis or breaker, a majority of the CaATG genes displayed up-regulated expression in pepper pericarp and placenta, suggesting the linkage of autophagy to the development and maturity of pepper fruit.
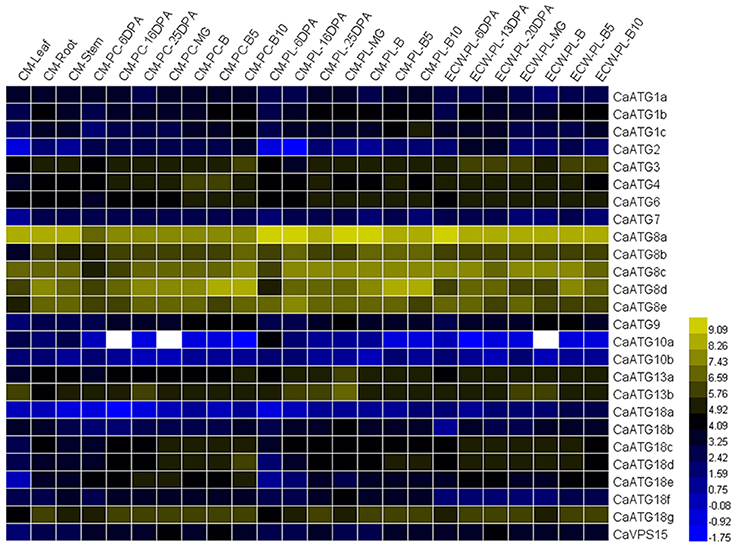
Figure 3. RNA-seq analysis of CaATG genes tissue- and developmental stage-specific expression. Raw data were retrieved from RNA-seq data of “CM334” (CM) and “ECW30R” (ECW). PC, pericarp; PL, placenta; DPA, days post anthesis; MG, mature green; B, breaker. The RNA-seq data were normalized using logarithm with the base of 2, and those of genes no transcription were indicated as white color.
Autophagy Responses to Abiotic Stresses in Pepper
The combination of LysoTracker Red as a probe and E-64d as an accumulator was used to detect the formation of autophagosome in pepper. When E-64d was absent, the punctate dots of autophagosome could not be accumulated (Figures 4A,B), while the punctates became visualized with the treatment of E64-d under both of normal (Figures 4C,D) and abiotic stresses conditions (Figures 4E-N). Compared to the less number under normal conditions (Figures 4C,D), punctate dots dramatically increased with abiotic stresses of salt (Figures 4E,F), drought (Figures 4G,H), heat (Figures 4I,J), cold (Figures 4K,L), and carbohydrate starvation (Figures 4M,N) in pepper leaves or roots.
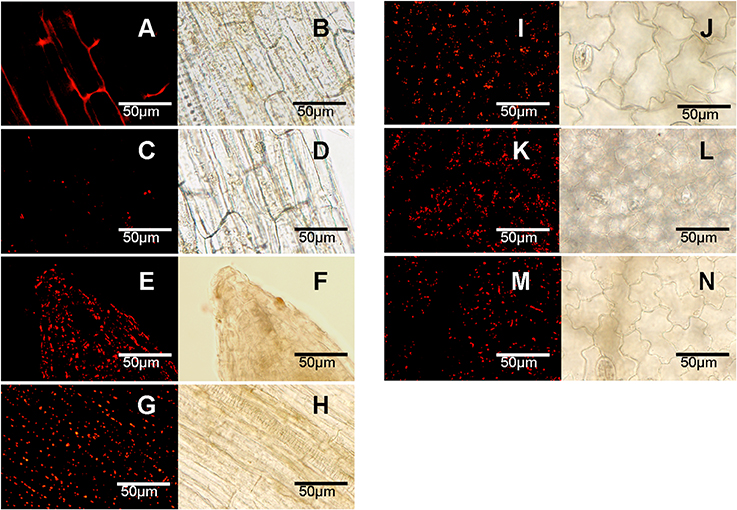
Figure 4. Accumulation of autophagosomes under abiotic stresses (labeled with LysoTracker Red). (A–D) Normal conditions; (E,F) salt stress of NaCl 3h; (G,H) drought stress of dried 3 h between folds of tissue pepper; (I,J) heat stress of 40°C 3 h; (K,L) cold stress of 4°C 3 h; (M,N) carbohydrate starvation stress of dark 2 days. (A–H) root; (I–N) leaf. (A,B) no E164d treatment; (C–N) infiltrated with E164d; (A,C,E,G,I,K,M), stained punctates under fluorescence; (B,D,F,H,J,L,N), under corresponding white light. The experiments were repeated three times with similar results. Scale bar = 50 μm.
The expression profiles of 29 CaATG genes responding to above abiotic stresses were further investigated thoroughly in sweet pepper line R9. The fold-change of CaATG genes expression levels under various stresses was shown in Figure 5 and Table S5. Out of 29 CaATGs, 16 CaATG genes (CaATG1a, 1c, 5, 6, 7, 8a, 8b, 8d, 13b, 18c, 18d, 18e, 18f, 18g, VPS15, and VPS34) were unaffected in some cases and up-regulated in others, and the expression of CaATG3 did not show significant response to any stress treatment, while the rest of 12 genes showed stress-dependent changes patterns in their expression levels.
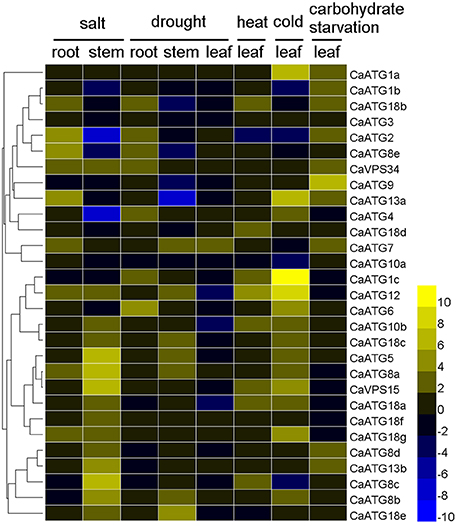
Figure 5. Expression profiles of CaATG genes in response to abiotic stresses. Pepper seedlings of R9 at 5–6 true leaves stage were used for salt (incubated in Hoagland's solution containing 200 mM NaCl for 3 h), drought (dried for 3 h between folds of tissue pepper), heat (kept at 40°C for 3 h), cold (kept at 4°C for 3 h), and carbohydrate starvation (kept in the dark for 2 days) treatments. The relative expression levels of pepper CaATG genes were calculated following the 2−ΔΔCT method and the relative expression levels of the control seedlings were set to 1. In the heatmap, the values of 2−ΔΔCT were used directly for the up-regulated genes, while the values of −(2−ΔΔCT)−1 were employed for the down-regulated genes. Expression data were obtained from three biological replicates.
Under salt stress treatment, in pepper root, nine CaATG genes (CaATG2, 7, 8a, 8e, 12, 13a, 18b, 18g, and VPS34) were up-regulated, and no one showed down-regulation. In contrast, in stem, 15 genes (especially CaATG5, 8a, 8c, and 15) were induced while four (especially CaATG2 and 4) were suppressed. For drought stress, the gene expression fold-change of CaATGs were lower in leaf than those in root and stem, and seven genes (CaATG1c, 2, 4, 6, 8e, 18b, and VPS34) in root, seven (CaATG5, 7, 8a, 8b, 12, 18c, and 18e) in stem and one (CaATG7) in leaf were up-regulated, while four (CaATG8e, 9, 13a, and 18b) in stem, three (CaATG10b, 12, and 18a) in leaf, and no one in root were down-regulated, which was similar to that under salt stress. After heat stress, eight genes were significantly up-regulated but CaATG2 was down-regulated. For cold stress, 14 CaATG genes particularly CaATG1a, 1c, 12, and 13a were up-regulated, and four (CaATG1b, 2, 8c, and 10a) showed slightly down-regulation. As for carbohydrate starvation stress, 10 genes (CaATG1a, 1b, 2, 7, 18d, 9, 13a, 13b, 18b, and CaVPS34) especially CaATG9 were up-regulated.
Heat-Induced Autophagosome Punctates Accumulation and ATG Genes Expression in Two Pepper Lines with Different Thermal Tolerance
As shown in Figure 6A, pepper line R9 showed higher ability in tolerance and restoration against heat stress than line B6. After 40°C heat treatment, the punctate dots were markedly induced in both lines (Figure 6B), and the area of fluorescent signals in R9 was significantly larger than that in B6 (Figure 6C), although the punctate accumulation did not show significant difference between R9 and B6 under normal temperature condition.
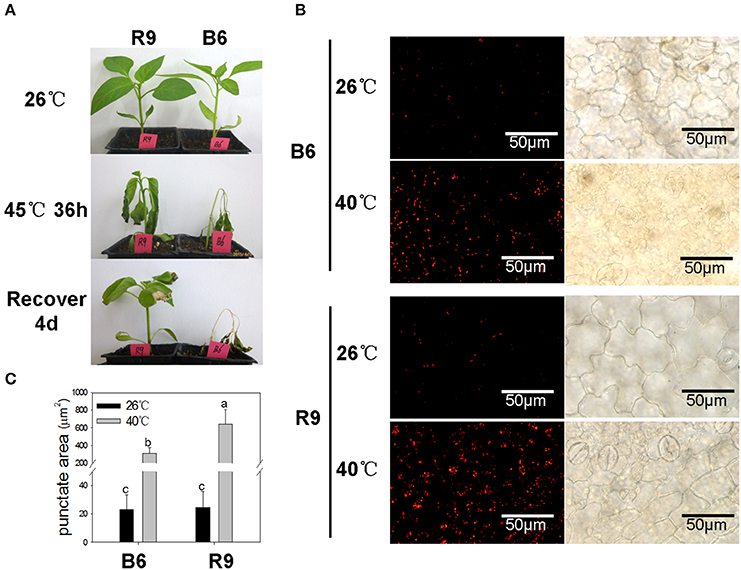
Figure 6. Heat-induced autophagosome punctates accumulation in pepper. R9, thermotolerant line; B6, thermosensitive line. (A) Phenotype observation of R9 and B6 seedlings under heat stress. When grown to 4–5 true leaves under normal temperature condition (26°C/20°C, day/night), the pepper seedlings were treated with 45°C for 36 h, and then transferred into normal conditions to recover for 4 days. (B) Accumulation of autophagosomes punctuates in pepper leaves under heat stress of 40°C 3 h. The experiments were repeated three times with similar results. Scale bar = 50 μm. (C) The punctate area of Lysotracker Red stained autolysosome-like structures in B6 and R9 leaves under normal conditions and heat stress. The different lower-case letters showed the difference of punctate areas between R9 and B6 under normal and heat stress conditions using the shortest significant ranges (SSR) method at the level α = 0.05.
The time-course expression profiles of CaATG genes at 0, 1, 3, 6, and 12 h during heat stress were analyzed (Figure 7, Table S6). In leaves of pepper thermotolerant line R9, the expression of 26 out of 29 CaATG genes were up-regulated, while only CaATG8d and VPS34 were weakly down-regulated, and no obvious change was observed in CaATG18f. Among the 26 up-regulated genes, 24 genes reached respective expression peaks at 1 h of heat stress, and CaATG8c, 18b at 3 and 6 h, respectively. The expression of almost all genes went down to their original levels at 12 h after heat stress treatment. For the thermosensitive line B6, 17 in 29 ATG genes were up-regulated especially CaATG1a and CaATG10a, and five genes were down-regulated. Among the 17 up-regulated genes, 16 genes showed the similar dynamic change patterns with those in R9, and only CaATG8d was up-regulated in B6 but down-regulated in R9. It is worth noting that compared with those in B6, 11 genes were specifically up-regulated in R9, among them five genes did not display obvious changes and six genes especially CaATG6 were down-regulated in B6.
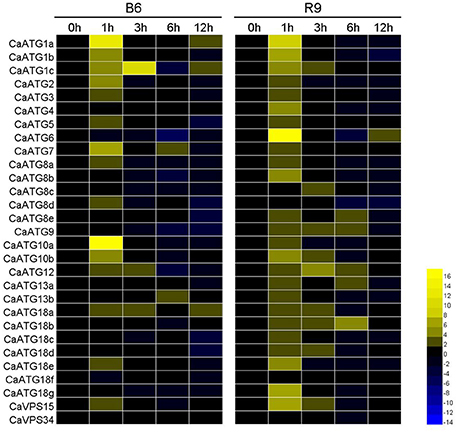
Figure 7. Expression profiles of CaATG genes during heat stress in pepper leaf. R9, thermotolerant line; B6, thermosensitive line. The relative expression levels of pepper CaATG genes were calculated following the 2−ΔΔCT method and the relative expression levels of the control seedlings at 0 h were set to 1. In the heatmap, the values of 2−ΔΔCT were used directly for the up-regulated genes, while the values of −(2−ΔΔCT)−1 were employed for the down-regulated genes. Expression data were derived from three biological replicates.
Prediction of Protein-Protein Interaction Network
It is well known that the proteostasis controlled by the cooperation of autophagy and heat shock response is essential for heat tolerance of organisms (Dokladny et al., 2015), therefore the protein-protein interaction network of CaATGs and the interaction between CaATGs and CaHSPs were predicated based on the interolog from Arabidopsis (Figure 8). Among the CaATGs except CaATG18e (having no interaction partner), CaATG18c (only interacting with CaATG2) and CaATG18d (only interacting with CaATG18a/18b/18g), the other CaATGs possessed complex interaction networks, especially CaATG7 (having 17 interaction partners) and CaATG12 (15 interaction partners). Interestingly, CaATG6 could interact with almost all of other CaATG members except CaATG13 and CaATG8, furthermore, CaATG6 was predicted as a bridge to connect ATGs and HSPs, and all of the HSPs belonged to HSP90 family, including CA10g01090 (At5g56010.1, HSP81-3), CA07g20630 (At5g56000.1, Hsp81.4), CA06g05770 (At5g52640.1, HSP90.1), CA00g30210 (At2g04030.1, Hsp88.1), CA07g04920 (At3g07770.1, Hsp89.1), and CA04g21920 (At4g24190.1, Hsp90.7).
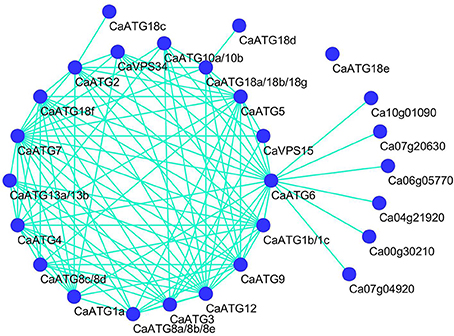
Figure 8. Prediction protein-protein interation network among CaATG members and HSPs. CaATG10a and 10b were both homologous with Arabidopsis AtATG10 (At3g07525.2), CaATG13a and 13b with AtATG13 (At3g49590.3), CaATG18a, 18b, and 18g with AtATG18a (At3g62770.1), CaATG1b and 1c with AtATG1b (At2g37840.1), CaATG8a, 8b, and 8e with AtATG8c (At1g62040.2), CaATG8c and 8d with AtATG8f (At4g16520.1), other CaATG proteins have single homolog each.
Discussion
During recent years the autophagy, a ubiquitous process in eukaryotic cells for the degradation of damaged proteins and organelles, has been extensively investigated in the field of organisms fighting against disease or unfavorable environments (Klionsky, 2007). As a complex biological process, autophagy requires the pipelined collaboration of a set of ATG proteins with different functions (Chung, 2011).
In our study, a total of 15 core ATG members including 29 proteins with respective typical functional domains were identified based on the comprehensive genome sequencing data of pepper (Kim et al., 2014; Qin et al., 2014) (Table 1; Table S2), which is consistent with the 30 ATG proteins in Arabidopsis (Kwon and Park, 2008), 33 in rice (Xia et al., 2011), and 31 in maize (Li et al., 2015). In addition, same to the above three plant species, ATG14 and 27 are also missing in pepper, however, another absent ATG member ATG16 in pepper can be identified in the other three species, although the low sequence homology with its yeast counterpart, which suggests that the functional homologs of ATG16 maybe exist in pepper but characterize with higher diversity for identifying with regular BLAST research. Furthermore, it is noteworthy that unlike having two ATG4 proteins in Arabidopsis (Hanaoka et al., 2002), rice (Xia et al., 2011), maize (Chung et al., 2009), and wheat (Pei et al., 2014), only one ATG4 was identified in pepper, which is consistent with the result in tobacco (Zhou et al., 2015), suggesting a solanaceae crops-specificity in the number of ATG4.
Among the 15 ATG members in pepper, CaATG1, 8, 10, 13, and 18 are presented as gene families (Table 1), which are also observed in Arabidopsis (Kwon and Park, 2008), maize (Li et al., 2015), and rice (Xia et al., 2011). Although owning their respective conserved ATG domains (Table S2), same predicted subcellular localizations (except CaATG10a and 10b) (Table 1), in some degree, different proteins in the same CaATG members displayed differences in expression profiles during growth, development (Figure 3, Table S4) and responses to abiotic stresses (Figure 5, Table S5), suggesting multiple functions of homologous ATG genes (Sláviková et al., 2005), which may be related to the diversity in motifs and their distribution in ATGs proteins (Figures 2A,B, Table S3). For ATG1, the protein kinase domain (motif15-motif14-motif8) at N-terninal are conserved in the three CaATG1 proteins (Table S2), but the C-terninal regulatory domain (including motif19-motif20), functioning in forming a complex with ATG13 (Cheong et al., 2008), is missed in CaATG1a (Figures 2A,B), and this truncation is also observed in Arabidopsis (Suttangkakul et al., 2011) and maize (Li et al., 2015). Additionally, CaATG18 possesses a conservative N-terninal WD domain (motif13-motif4) in all 7 ATG18 proteins (Table S2), however, both CaATG18d and CaATG18e had a longer C-terninal extension (Figures 2A,B). These results suggest the possible functional divergence of different proteins in the same member, which need further research to verify.
Autophagy is widely involved in plants growth, development, and stress responses (Yoshimoto et al., 2004; Bassham et al., 2006), which is also supported by our results. According to the RNA-seq data published by Kim et al. (2014), the 26 CaATG genes expressed in a tissue- and developmental stage-specific patterns, in which some genes, like CaATG8a and 8c, showed high expression levels in all the tested tissues and all the developmental processes of pepper fruit (Figure 3, Table S4). Under abiotic stresses of salt, drought, heat, cold, and starvation, the accumulation of autophagosome punctates increased markedly (Figure 4), showing the possibility of autophagy participation the pepper response to abiotic stresses. However, the expression levels of some CaATG genes were differently regulated by various stresses (Figure 5, Table S5). For instance, CaATG1b was induced by starvation but inhibited by salt and cold, and CaATG2 was up-regulated by salt, drought and starvation but down-regulated by heat and cold, which were also observed in CaATG8c, 10b, 12, and 18b. Furthermore, this differential regulation existed even in different tissues under the same stress. The expression of CaATG8e was enhanced in root but inhibited in stem by both of salt and drought, while in case of drought stress, the transcription of CaATG12 and 18b were induced in stem and root but inhibited in leaf and stem, respectively. These results indicate that autophagy may be regulated by distinct pathways during different abiotic stresses or in different tissues (Lv et al., 2014).
Compared to other abiotic stresses like drought, salt, starvation and cold, the roles of autophagy in heat stress are seldom reported in plant. The major negative consequence of high temperature on organism cell is protein misfolding and denaturation (Hemantaranjan et al., 2014), therefore, it can be hypothesized that autophagy plays an important role in plant heat responses. Zhou et al. (2013, 2014) found that heat stress can induce ATG genes expression and autophagosome punctates accumulation in both of Arabidopsis and tomato, and similar results were also observed in two pepper lines B6 and R9 in our study (Figures 6, 7). However, under heat stress condition, the number of up-regulated CaATG genes and the increment of punctate area in thermotolerant line R9 was obviously higher than those in thermosensitive line B6, which demonstrating an association of autophagy with heat tolerance in pepper.
In our previous and present research work, we have also found that under heat stress the expression of CaHSP70-1 (Guo et al., 2014a), CaHsf family members (Guo et al., 2014b) were more abundantly transcribed in R9 than those in B6. Therefore, we speculate that pepper heat tolerance may be regulated by the cooperation between autophagy and heat shock response (HSR), the two aspects maintaining proteostasis, which is supported by the predicted interaction between autophagy (CaATG6) and HSR (CaHSP90 family) (Figure 8). Xu et al. (2011) reported that the functional interaction of HSP90 and ATG6 (Beclin 1) plays a role in controlling TLR (Toll-like receptor)-mediated autophagy in response to microbial infections, however, Dokladny et al. (2013) thought HSR can negatively regulate autophagy by HSP70 in mammals. Because HSP70 and HSP90 routinely work together in conducting their chaperone activities (Clerico et al., 2015), it is possible that ATG6, HSP70 and HSP90 jointly regulate autophagy in response to the changing environmental conditions. Additionally, due to the obvious up-regulation in R9 but down-regulation in B6 (Figure 7), CaATG6 is speculated playing a key role in the regulation of heat tolerance in pepper.
While the accumulation of dysfunctional proteins induced by adverse environmental condition cause damage to plants growth and development, efficient operation of protein degradation system including autophagy is necessary for survival. Combining the data presented in this study, we suggest an association between autophagy and pepper tolerance to heat and other abiotic stresses, which provides proof and reference for understanding plant tolerant mechanisms to environment stresses. However, future extensive researches are required to investigate the signal pathway to activate the autophagy process under unfavorable conditions.
Author Contributions
YZ, MG, and ML contributed to the experimental design. YZ, MG, HW, J. Lu, J. Liu, and CZ performed the research. YZ performed the bioinformatics analysis and drafted the manuscript. ML and ZG revised the paper and contributed reagents/materials/analysis tools. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31572114), the Shaanxi Agriculture Science and Technology Projects (Grant No. 2014K01114101), the Basic Fund for Scientific Research of Northwest A&F University (Grant No. 2452015141), the Tang Zhongying Fund for Breeding of Northwest A&F University, and the Opening Fund of Key Laboratory for Crop Biotechnology of Xinjiang Uygur Autonomous Region (Grant No. XJYS030212014103).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00131
Table S1. The CaATGs gene-specific primers for RT-qPCR.
Table S2. ATG domains of CaATG proteins in pepper.
Table S3. Motif sequences in CaATG proteins indentified by MEME tools.
Table S4. Expression data of CaATGs against RNA-seq data of each tissue.
Table S5. Expression data of CaATG genes in response to abiotic stresses.
Table S6. Expression data of CaATG genes during heat stress in peppe leaf.
Figure S1. Chromosomal localization of the 29 CaATG genes.
Abbreviations
HSPs, heat shock proteins; Hsf, heat shock transcription factor; ATG, AuTophaGy; GFP, green fluorescent protein; qRT-PCR, quantitative real-time PCR; HSR, heat shock response.
References
Ahuja, I., de Vos, R. C. H., Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Bassham, D. C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L. J., et al. (2006). Autophagy in development and stress responses of plants. Autophagy 2, 2–11. doi: 10.4161/auto.2092
Bita, C. E., Zenoni, S., Vriezen, W. H., Mariani, C., Pezzotti, M., and Gerats, T. (2011). Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomato plants. BMC Genomics. 12:384. doi: 10.1186/1471-2164-12-384
Chen, L., Liao, B., Qi, H., Xie, L. J., Huang, L., Tan, W., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246. doi: 10.1080/15548627.2015.1112483
Cheong, H., Nair, U., Geng, J., and Klionsky, D. J. (2008). The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 19, 668–681. doi: 10.1091/mbc.E07-08-0826
Chikte, S., Panchal, N., and Warnes, G. (2014). Use of lysotracker dyes: a flow cytometric study of autophagy. Cytometry A. 85, 169–178. doi: 10.1002/cyto.a.22312
Chung, T. (2011). See how I eat my greens¡ÂłAutophagy in plant cells. J. Plant Biol. 54, 339–350. doi: 10.1007/s12374-011-9176-5
Chung, T., Suttangkakul, A., and Vierstra, R. D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149, 220–234. doi: 10.1104/pp.108.126714
Clerico, E. M., Tilitsky, J. M., Meng, W., and Gierasch, L. (2015). How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588. doi: 10.1016/j.jmb.2015.02.004
Dokladny, K., Myers, O. B., and Moseley, P. L. (2015). Heat shock response and autophagy-cooperation and control. Autophagy 11, 200–213. doi: 10.1080/15548627.2015.1009776
Dokladny, K., Zuhl, M. N., Mandell, M., Bhattacharya, D., Schneider, S., Deretic, V., et al. (2013). Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J. Biol. Chem. 288, 14959–14972. doi: 10.1074/jbc.M113.462408
Guo, M., Yin, Y. X., Ji, J. J., Ma, B. P., Lu, M. H., and Gong, Z. H. (2014a). Cloning and expression analysis of heat–shock transcription factor gene CaHsfA2 from pepper (Capsicum annuum L.). Genet. Mol. Res. 13, 1865–1875. doi: 10.4238/2014.March.17.14
Guo, M., Zhai, Y. F., Lu, J. P., Chai, L., Chai, W. G., Gong, Z. H., et al. (2014b). Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int. J. Mol. Sci. 15, 19741–19759. doi: 10.3390/ijms151119741
Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., et al. (2002). Leaf senescence and starvation–induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. doi: 10.1104/pp.011024
Hemantaranjan, A., Bhanu, A. N., Singh, M. N., Yadav, D. K., Patel, P. K., Singh, R., et al. (2014). Heat stress responses and thermotolerance. Adv. Plants Agric. Res. 1:12. doi: 10.15406/apar.2014.01.00012
IPCC (2013). “Climate change: the physical science basis,” in Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge, UK; New York, NY: Cambridge University Press), 3–32.
Johansen, T., and Lamark, T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. doi: 10.4161/auto.7.3.14487
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper providesinsights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Klionsky, D. J. (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937. doi: 10.1038/nrm2245
Kuzuoglu-Ozturk, D., Yalcinkaya, O. C., Akpinar, B. A., Mitou, G., Korkmaz, G., Gozuacik, D., et al. (2012). Autophagy–related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta 236, 1081–1092. doi: 10.1007/s00425-012-1657-3
Kwon, S., and Park, O. K. (2008). Autophagy in plants. J. Plant Biol. 51, 313–320. doi: 10.1007/BF03036132
Li, F. Q., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, H., Liu, S., Yi, C., Wang, F., Zhou, J., Xia, X., et al. (2014). Hydrogen peroxide mediates abscisic acid–induced HSP70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ. 37, 2768–2780. doi: 10.1111/pce.12360
Liu, G. T., Wang, J. F., Cramer, G., Dai, Z. W., Duan, W., Xu, H. G., et al. (2012). Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol. 12:174. doi: 10.1186/1471-2229-12-174
Liu, Y., and Bassham, D. C. (2012). Autophagy: pathways for self–eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, Y., Schiff, M., Czymmek, K., Talloczy, Z., Levine, B., and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. doi: 10.1016/j.cell.2005.03.007
Liu, Y. M., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 254, 402–408. doi: 10.1006/meth.2001.1262
Lobell, D. B., Schlenker, W., and Costa-Roberts, J. (2011). Climate trends and global crop production since 1980. Science 333, 616–620. doi: 10.1126/science.1204531
Lv, X., Pu, X., Qin, G., Zhu, T., and Lin, H. (2014). The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 19, 905–921. doi: 10.1007/s10495-014-0981-4
Moriyasu, Y., Hattori, M., Jauh, G. Y., and Rogers, J. C. (2003). Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process. Plant Cell Physiol. 44, 795–802. doi: 10.1093/pcp/pcg100
Pei, D., Zhang, W., Sun, H., Wei, X. J., Yue, J. Y., and Wang, H. Z. (2014). Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 33, 1697–1710. doi: 10.1007/s00299-014-1648-x
Phadwal, K., Alegre-Abarrategui, J., Watson, A. S., Pike, L., Anbalagan, S., Hammond, E. M., et al. (2012). A novel method for autophagy detection in primary cells Impaired levels of macroautophagy in immunosenescent T cells. Autophagy 8, 677–689. doi: 10.4161/auto.18935
Qin, C., Yu, C. S., Shen, Y. O., Fang, X. D., Chen, L., Min, J. M., et al. (2014). Whole–genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Shin, J. H., Yoshimoto, K., Ohsumi, Y., Jeon, J. S., and An, G. (2009). OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol.Cells 27, 67–74. doi: 10.1007/s10059-009-0006-2
Slavikova, S., Shy, G., Yao, Y., Glozman, R., Levanony, H., Pietrokovski, S., Elazar, Z., et al. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56, 2839–2849. doi: 10.1093/jxb/eri276
Slavikova, S., Ufaz, S., Avin-Wittenberg, T., Levanony, H., and Galili, G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59, 4029–4043. doi: 10.1093/jxb/ern244
Suttangkakul, A., Li, F., Chung, T., and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779. doi: 10.1105/tpc.111.090993
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Wan, H. J., Yuan, W., Ruan, M. Y., Ye, Q. J., Wang, R. Q., Li, Z. M., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 416, 24–30. doi: 10.1016/j.bbrc.2011.10.105
Wang, W. X., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, Y., and Liu, Y. (2010). Progress in plant autophagy. Chin. J. Cell Biol. 32, 677–689. Available online at: http://www.cjcb.org/arts.asp?id=1326
Xia, K. F., Liu, T., Ouyang, J., Wang, R., Fan, T., and Zhang, M. Y. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18, 363–377. doi: 10.1093/dnares/dsr024
Xiao, H. J., Yin, Y. X., Chai, W. G., and Gong, Z. H. (2014). Silencing of the CaCP gene delays salt- and osmotic-induced leaf senescence in Capsicum annuum L. Int. J. Mol. Sci. 15, 8316–8334. doi: 10.3390/ijms15058316
Xu, C. F., Liu, J., Hsu, L. C., Luo, Y. P., Xiang, R., and Chuang, T. H. (2011). Functional interaction of heat shock protein 90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J. 25, 2700–2710. doi: 10.1096/fj.10-167676
Xue, G. P., Sadat, S., Drenth, J., and McIntyre, C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. doi: 10.1093/jxb/ert399
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshimoto, K., Takano, Y., and Sakai, Y. (2010). Autophagy in plants and phytopathogens. FEBS Lett. 584, 1350–1358. doi: 10.1016/j.febslet.2010.01.007
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B., Yu, J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9:e1003196. doi: 10.1371/journal.pgen.1003196
Zhou, J., Wang, J., Yu, J. Q., and Chen, Z. (2014). Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci. 5:174. doi: 10.3389/fpls.2014.00174
Zhou, X. M., Zhao, P., Wang, W., Zou, J., Cheng, T. H., Peng, X. B., et al. (2015). A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 22, 245–257. doi: 10.1093/dnares/dsv012
Keywords: Capsicum annuum L., autophagy-related genes, genome-wide identification, heat stress, expression profiles
Citation: Zhai Y, Guo M, Wang H, Lu J, Liu J, Zhang C, Gong Z and Lu M (2016) Autophagy, a Conserved Mechanism for Protein Degradation, Responds to Heat, and Other Abiotic Stresses in Capsicum annuum L. Front. Plant Sci. 7:131. doi: 10.3389/fpls.2016.00131
Received: 01 November 2015; Accepted: 25 January 2016;
Published: 09 February 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Mingyong Zhang, South China Botanical Garden, ChinaMohamed Hamed Arisha, Zagazig University, Egypt
Copyright © 2016 Zhai, Guo, Wang, Lu, Liu, Zhang, Gong and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minghui Lu, xnjacklu@nwsuaf.edu.cn
 Yufei Zhai
Yufei Zhai Meng Guo
Meng Guo Hu Wang
Hu Wang Jinping Lu
Jinping Lu Jinhong Liu
Jinhong Liu Chong Zhang
Chong Zhang Zhenhui Gong
Zhenhui Gong Minghui Lu
Minghui Lu