- College of Horticulture, Northwest A&F University, Yangling, China
The NAP (NAC-like, activated by AP3/P1) transcription factor belongs to a subfamily of the NAC transcription factor family, and is believed to have an important role in regulating plant growth and development. However, there is very little information about this subfamily in Rosaceous plants. We identified seven NAP genes in the peach genome. PpNAP2 was categorized in the NAP I group, and contained a conserved transcription activation region. The other PpNAP genes belonged to the NAP II group. The expression patterns of the PpNAP genes differed in various organs and developmental stages. PpNAP1 and PpNAP2 were highly expressed in mature and senescing flowers, but not in leaves, fruits, and flower buds. PpNAP3 and PpNAP5 were only expressed in leaves. The PpNAP4 expression level was high in mature and senescing fruits, while PpNAP6 and PpNAP7 expression was up-regulated in mature and senescent leaves and flowers. During the fruit development period, the PpNAP4 and PpNAP6 expression levels rapidly increased during the S1 and S4 stages, which suggests these genes are involved in the first exponential growth phase and fruit ripening. During the fruit ripening and softening period, the PpNAP1, PpNAP4, and PpNAP6 expression levels were high during the early storage period, which was accompanied by a rapid increase in ethylene production. PpNAP1, PpNAP4, and PpNAP6 expression slowly increased during the middle or late storage periods, and peaked at the end of the storage period. Additionally, abscisic acid (ABA)-treated fruits were softer and produced more ethylene than the controls. Furthermore, the PpNAP1, PpNAP4, and PpNAP6 expression levels were higher in ABA-treated fruits. These results suggest that PpNAP1, PpNAP4, and PpNAP6 are responsive to ABA and may regulate peach fruit ripening.
Introduction
The development and maturation of plant tissues involve complex processes regulated by genetic, hormonal, and environmental factors (Wang, 2008). The NAP transcription factor is a member of a subfamily of the plant-specific NAC (NAM, ATAF1, 2.CUC2) transcription factor family, which is important in many vital biological processes during plant growth and development (Sablowski and Meyerowitz, 1998; Fernandez et al., 2006; Fan et al., 2015). Sablowski and Meyerowitz (1998) determined that AtNAP is associated with cell expansion in specific Arabidopsis thaliana flower organs, while Guo and Gan (2006) reported that AtNAP is important for leaf senescence. This was further supported by a study that revealed AtNAP regulates leaf senescence processes by directly binding to the promoter of SAG113 to form an ABA-AtNAP-SAG113 PP2C regulatory chain that controls stomatal movement and water loss in senescing leaves (Zhang and Gan, 2012). Other studies have demonstrated that NAP affects leaf senescence in bamboo (Chen et al., 2011), crocus (Kalivas et al., 2010), Festuca arundinacea (Guo et al., 2010), Asarina procumbens (Fan and Zhao, 2014), and rice (Ooka et al., 2003). Kou et al. (2012) reported that AtNAP expression increased during silique senescence in A. thaliana. Fernandez et al. (2006) observed that VvNAP may be important for grapevine flower and fruit development. In Citrus sinensis (L.) Osbeck, CitNAC expression was detected only in the fruit peel and pulp during the fruit ripening or senescence stages (Liu et al., 2009). Additionally, recent studies showed that the NAP subfamily is also important for regulating plant senescence and response to abiotic stresses (Meng et al., 2009; Zhang and Gan, 2012; Huang et al., 2013). The NAP transcription factor has been identified in various plant species, including rice (Ooka et al., 2003), bamboo (Chen et al., 2011), wheat (Cristobal et al., 2006), cotton (Meng et al., 2009), grape (Fernandez et al., 2006), maize (Fan et al., 2014), and soybean (Meng et al., 2007). However, the effect of NAP on the development of Rosaceae plants has not been studied.
Peach (Prunus persica) is an economically important crop, whose typical climacteric fruit undergoes a program of enhanced ethylene production and an associated increase in respiration rate at the onset of ripening (Barry and Giovannoni, 2007). Therefore, peach fruit softening and senescence rapidly occur after harvest, which makes storage and transport difficult. This limits peach production. A more thorough characterization of the physiological basis of peach fruit growth and ripening will enable the development of effective strategies to regulate these processes. Furthermore, peach, as a stone fruit, exhibits a typical double sigmoid growth pattern during fruit development, with distinct growth stages (S1–S4). The S1 stage corresponds to the first exponential growth phase, and is characterized by a rapid increase in cell division and elongation. In the S2 stage, which proceeds more slowly than S1, most of the dry matter is involved in pit hardening and seed and embryo growth. The S3 stage represents the second exponential growth phase, during which the fruit rapidly increases in size. Fruit ripening occurs in the final stage (S4) (Li et al., 1989; Tonutti et al., 1997; Soto et al., 2013).
In this study, we identified seven members of the peach NAP subfamily and analyzed their expression during leaf, flower, and fruit development and senescence. We revealed that members of this subfamily may function in the development and maturation of flowers and fruits, and regulate fruit softening.
Materials and Methods
Plant Materials
Peach tree (P. persica cv. ‘Qinguang 8’) samples were collected from the Experimental Station of the College of Horticulture at the Northwest A & F University in Yangling, Shaanxi, China. Samples included flowers, leaves, and fruits. Flower samples consisted of flower buds, blooming flowers, and flowers 2 days after full bloom. Young leaves were those that had just unfolded, and were collected from new shoots, while mature and senescing leaves were collected from the middle sections of new shoots. Young, mature, and senescing fruits were collected 42, 107, and 131 days after full bloom (DAFB), respectively. For fruit development analyses, young fruits were hand-picked 25 DAFB, and samples were collected every 15 days until the fruits reached commercial maturity (i.e., fruits with light green or partially red peels and slightly hard flesh). At least 20 fruits at each developmental stage were used to determine fruit weight, diameter, and gene expression.
For storage analyses, fruits with no visible defects were randomly hand picked at commercial maturity and divided into two groups. One group was soaked with 100 mM abscisic acid (ABA) for 10 min at 25 ± 1°C. The other group was soaked with water and served as the control group. Each group consisted of 120 fruits, which were kept in individual plastic bags at 25 ± 1°C. During the storage period, fruit samples were collected every 2 days, until the flesh fully softened. All samples were frozen with liquid nitrogen and stored at -80°C.
RNA Extraction and Reverse Transcription
Total RNA was extracted using cetyltrimethylammonium bromide (Chang et al., 1993), and reverse transcription was completed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara).
Identification of Peach NAP Subfamily Members
Arabidopsis thaliana, Vitis vinifera, and Solanum lycopersicum NAP gene sequences were used to search the peach genome database1 with the NCBI BLASTp tool to identify peach genes that were highly homologous to NAP subfamily genes.
Multiple Sequence Alignment, Phylogenetic Analysis, and Exon/Intron Structure Determination
The NCBI BLAST tool2 was used to assess sequence similarities. The open reading frames of PpNAP genes were analyzed using the NCBI Open Reading Frame Finder tool3 Multiple sequence alignment analyses were conducted using the DNAMAN program, and graphical annotations of consensus sequences were completed using the Weblogo online tool4 A phylogenetic tree was generated using the NJ method (with 1,000 repeats) of the MEGA 6.06 software. Genetic structure investigations were conducted using the Gene Structure Display Server online tool5 Signal peptides were analyzed with the SignalP program6 (version 3.0; Bendtsen et al., 2004). Protein molecular weights and pIs were calculated using the ExPASy Compute pI/Mw tool7
Molecular Cloning of Peach NAP Subfamily Members
To clone the PpNAP genes, Primer Premier 6.0 was used to design gene-specific primer pairs according to the peach genome sequence (Table 1). Using cDNA templates, PCR was completed with the Phanta Super-Fidelity DNA Polymerase (Vazyme) according to the manufacturer’s recommended procedure. The PCR products were isolated and purified with the MiniBEST Agarose Gel DNA Extraction Kit Ver. 4.0 (Takara). Purified products were inserted into the pMD-19T vector (Takara). Positive clones were confirmed by blue/white plaque assays. Primers for cloning and quantitative reverse transcription (qRT)-PCR were synthesized by Sangon Biotech (Shanghai) Co., Ltd, which also completed all DNA sequencing reactions.
Quantitative Reverse Transcription PCR Assays
The qRT-PCR was conducted using the iQ5 real-time PCR system (Bio-Rad). The gene-specific primers (Table 1) were designed using the Beacon Designer 8.0 software (Premier Biosoft International). Each primer pair (Tm 60°C) was designed to amplify an approximately 200-bp fragment. For each sample, 1 μL cDNA, 1 μL each primer, 2 μL double-distilled water, and 5 μL 2x SYBR Premix ExTaq II (Takara) were used in a total volume of 10 μL. The two-step RT-PCR was completed using the manufacturer’s recommended program, but the annealing temperature was changed to 60°C. Samples were heated at 95°C for 10 s, cooled to 65°C for 15 s, and finally heated to 95°C at a rate of 0.1°C s–1 for melting curve analyses. The specific transcript accumulation was analyzed using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Peach 18S ribosomal RNA was used to normalize data. The amplification, melt curve and melt park of 18s ribosomal gene in all samples can be seen in Supplementary Figure S1. Each sample was analyzed in triplicate.
Flesh Firmness and Ethylene Production
Flesh firmness of five randomly selected fruits was measured using the GY-4 firmness meter equipped with a 8-mm diameter probe. A small epicarp segment was peeled from two places of each fruit to enable probe attachment. Three biological replicates were measured. Ethylene production was determined as described by Liguori et al. (2004) using the Trace GC Ultra gas chromatograph (Thermo Fisher Scientific). The oven, injector, and detector temperatures were 90, 110, and 140°C, respectively.
Search for Cis-Acting Elements in the Promoters of Peach NAP Genes
Upstream regions (2000 bp upstream of the transcription start site) of selected peach NAP genes were used to search the PlantCARE database for putative cis-acting elements (Lescot et al., 2002).
Statistical Analyses
Gene expression levels were subjected to analysis of variance using SAS. Values are provided as the mean ± standard error (n = 3). The overall least significant difference (p < 0.05) was calculated and used to separate means.
Results
Identification of Peach NAP Subfamily Members
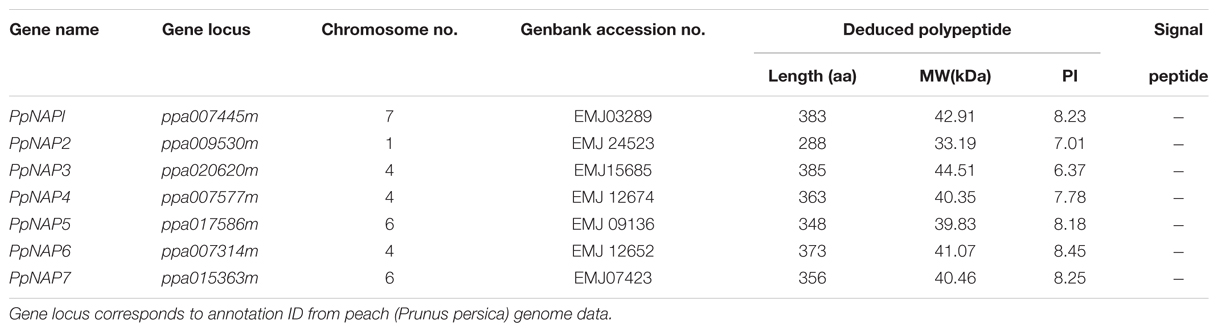
Seven NAP genes were detected in the peach genome with query IDs of ppa007445m, ppa009530m, ppa020620m, ppa007577m, ppa017586m, ppa007314m, and ppa015363m, which corresponded to PpNAP1, PpNAP2, PpNAP3, PpNAP4, PpNAP5, PpNAP6, and PpNAP7, respectively. These peach NAP genes contain a conserved NAC domain structure at the N-terminus, and the domain can be divided into A, B, C, D, and E subdomains. The conserved amino acid sequences in the A, B C, D, and E subdomains were LPPGFRFHPTDEELIVHYL, IIAEVDIYKFDPWELP, EWYFFSPRDRKYPNGARP NRAAVSGYWKATGTDK, VGVKKALVFYKGRPPKGYKT-DWIMHEYRL, and SMRLDDWVLCRIYKK, respectively (Figure 1). Furthermore, according to Fan et al. (2015), the NAP subfamily could be divided into two groups (NAP I and NAP II). Because of the presence of the relatively conserved transcription activation region, PpNAP2 was included in the NAP I group, while the other PpNAP genes were included in the NAP II group (Figure 1). The PpNAP genes were highly homologous to NAP genes from other species. Similar to other NAP genes, PpNAP1–6 consisted of three exons and two introns, while PpNAP7 contained two exons and one intron (Figure 2). The deduced polypeptide sequences ranged from 288 to 385 amino acids, with predicted molecular weights between 33.19 and 44.51 kDa. The predicted pIs of PpNAP genes were from 6.37 to 8.45. None of the identified peach NAP genes contained signal peptide sequences according to SignalP analysis (Table 1).
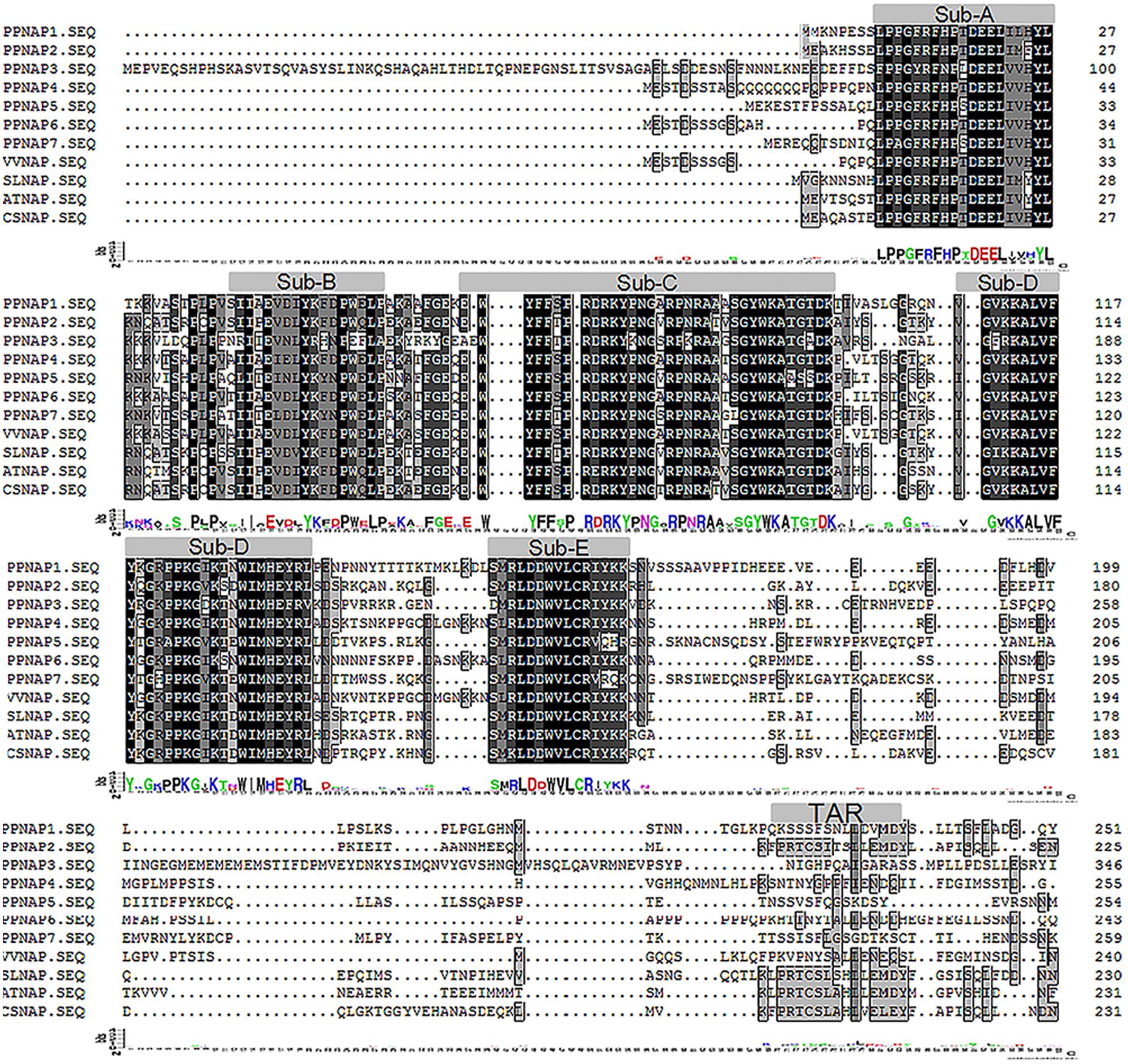
FIGURE 1. Multiple sequence alignments of PpNAP proteins and NAP proteins from other plants. The accession numbers of the proteins homologous to AtNAP are provided in Supplementary Table S2.
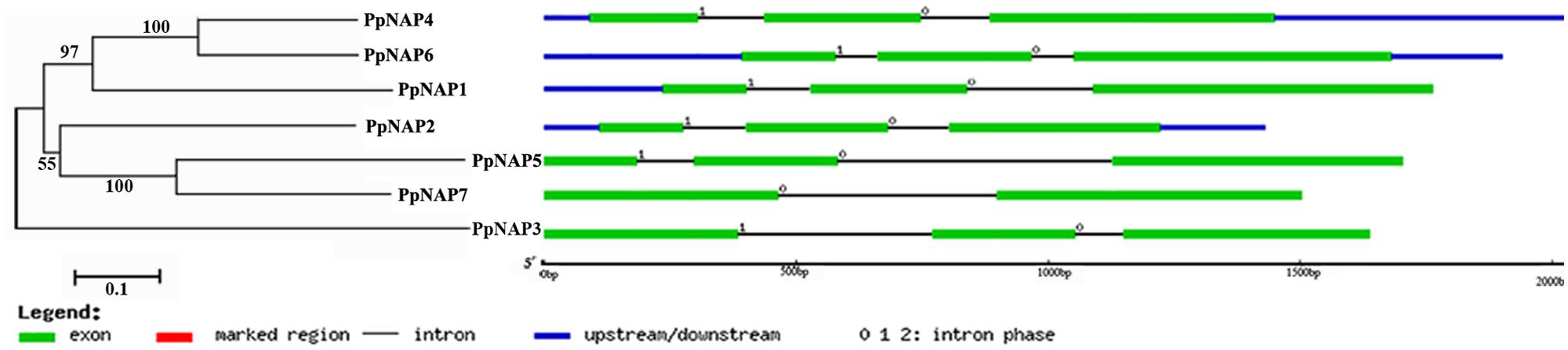
FIGURE 2. The structures of PpNAP genes. The numbers 0, 1, and 2 represent introns in phases 0, 1, and 2, respectively.
Phylogenetic Analysis of the Peach NAP Subfamily Members
To evaluate the evolutionary relationships among NAP subfamily members, cluster analyses were completed using the amino acid sequences encoded by the identified PpNAP genes and by NAC genes from potato, tomato, pepper, orange, grape, rice, A. thaliana, and bamboo using the MEGA 6.06 software. Phylogenetic analyses revealed that all PpNAPs are clustered in the NAP subfamily (Figure 3). PpNAP2 was similar to citrus, A. thaliana, and western balsam poplar NAPs, while PpNAP4 and PpNAP6 were similar to NAPs from grape and wheat. In contrast, PpNAP3, PpNAP5, and PpNAP7 were not particularly similar to NAPs of other plants. Additionally, the deduced amino acid sequences were more highly conserved among PpNAP4, PpNAP5, PpNAP6, and PpNAP7, while the similarities among PpNAP1, PpNAP2, and PpNAP3 were less than 28%.
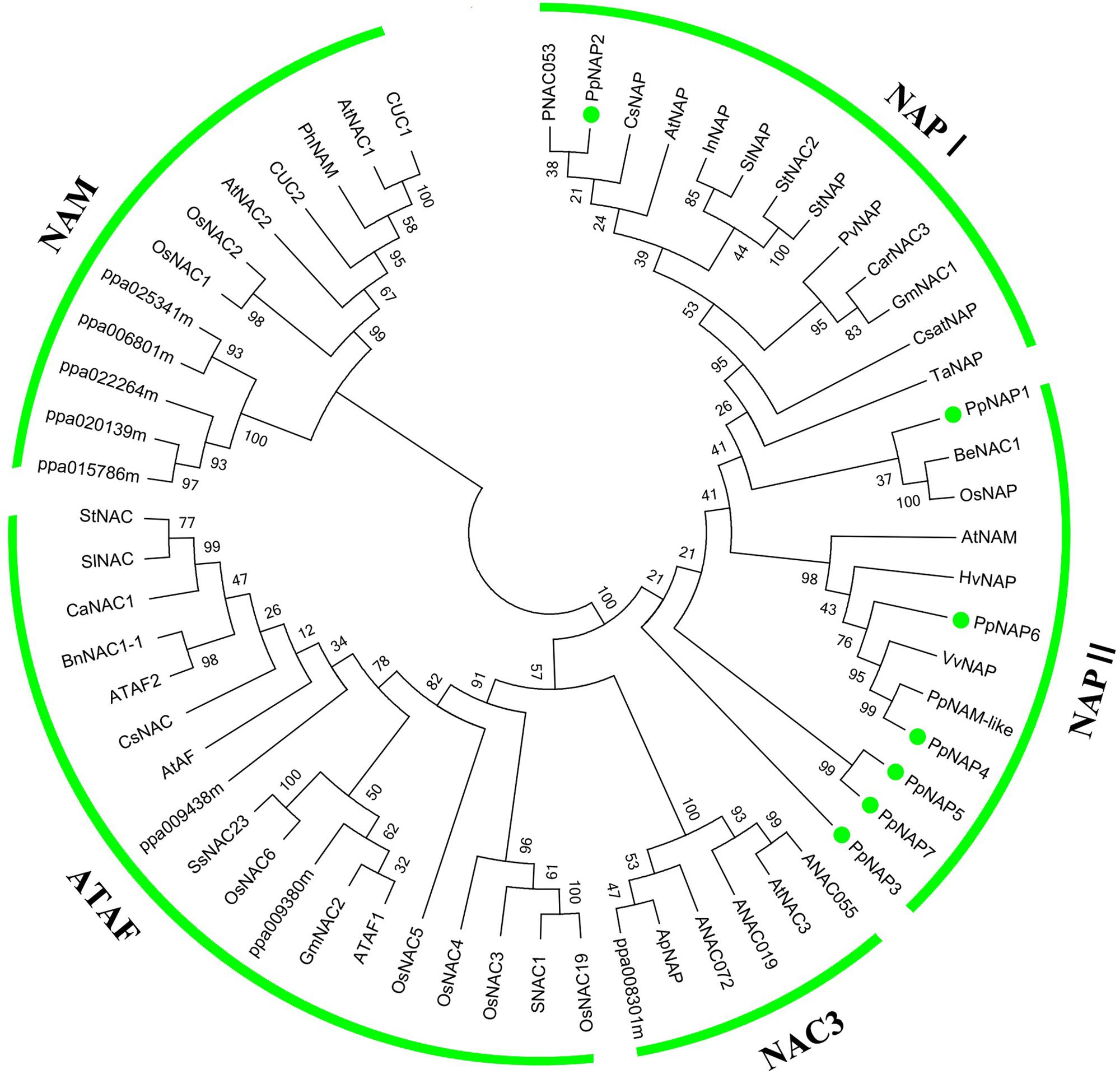
FIGURE 3. Phylogenetic tree of PpNAP with NAC transcription factors from different plants. The unrooted phylogenetic tree was constructed using the NJ method of the MEGA 6.06 program. GenBank accession numbers are provided in Supplementary Table S3.
PpNAP Gene Expression in Various Organs at Different Developmental Stages
To investigate the potential functions of PpNAP genes during peach development, transcription level changes in different organs were analyzed using qRT-PCR. The PpNAP expression patterns were different among various organs and developmental stages (Figure 4). The PpNAP expression levels in leaves were lower than those in flowers and fruits. The expression of PpNAP6 and PpNAP7 rapidly increased in maturing and senescing leaves. The PpNAP1, PpNAP4, and PpNAP5 genes were more highly expressed in young and senescent leaves than in mature leaves. In contrast, PpNAP3 transcript levels were high in mature leaves, while PpNAP2 expression remained relatively stable and at low levels (Figure 4A).
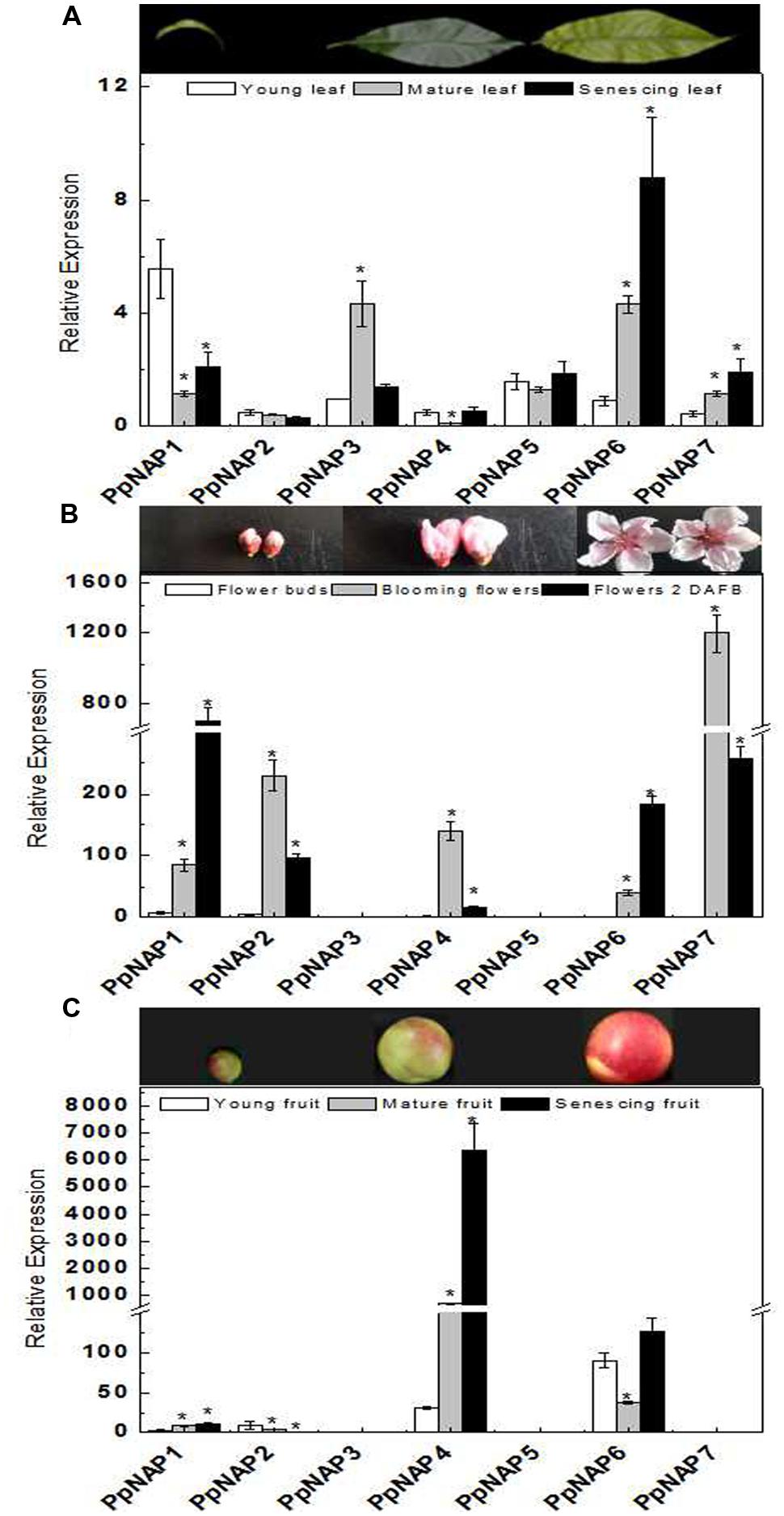
FIGURE 4. Quantitative reverse transcription PCR analysis of selected peach NAP genes in leaves (A), flowers (B), and fruits (C) with different developmental stage. The value for each sample is the mean of three replicates. Vertical bars indicate standard error. The primer sequences are provided in Supplementary Table S1. Bars correspond to the mean ± standard error (n = 3). Asterisks indicate significant differences in young leaves (A), flower buds (B), and young fruits (C) at p < 0.05 according to the Student’s t-test.
The expression levels of PpNAP1 and PpNAP6 were rapidly up-regulated in blooming and 2 DAFB flowers. PpNAP2, PpNAP4, and PpNAP7 were highly expressed in blooming flowers, but expressed at very low levels in flower buds. Similarly, PpNAP3 and PpNAP5 expression were almost undetectable in flowers at all developmental stages (Figure 4B).
The PpNAP4 and PpNAP6 expression levels were higher than those of the other PpNAP genes in fruits. The higher expression levels were most obvious for PpNAP4 in mature and senescent fruits and PpNAP6 in young and senescing fruits. PpNAP1 and PpNAP2 were expressed at low levels, while PpNAP3, PpNAP5, and PpNAP7 expression was barely detectable (Figure 4C).
PpNAP Gene Expression Profiles During Fruit Development
To confirm the accuracy of the predicted cDNA sequences and further explore the biological functions of PpNAP1, PpNAP2, PpNAP4, and PpNAP6 in fruit, we designed specific primer pairs using the peach genome sequence for cloning and expression analyses in mature ‘Qinguang 8’ fruits. The cDNA sequences of PpNAP2, PpNAP4, and PpNAP6 were consistent with the corresponding genome sequences, while that of PpNAP1 was 48 nucleotides longer than expected (see Supplementary Material). The expression of Pp-ACO1 is strictly related to the transition between the pre-climacteric and climacteric stage. We have analyzed the expression of Pp-ACO1 during developmental stage, and the result showed the obvious enhance of Pp-ACO1 expression at S4 stage (Supplementary Figure S2).
The qRT-PCR results revealed that PpNAP4 expression in the mesocarp rapidly increased during the S1 fruit development stage (25–55 DAFB; Figures 5A,D), increased slowly during S2 and S3 (55–102 DAFB; Figures 5A,D), and significantly increased during S4, where it was maintained at a high level (102–121 DAFB; Figures 5A,D). In contrast, PpNAP6 expression rapidly increased during S1, but decreased in S2, remained stable during S3, and increased during S4 (Figures 5A,E). The expression levels of PpNAP1 and PpNAP2 were low during fruit development, with elevated expression levels only during S3 (85–100 DAFB) and S1 (25–55 DAFB), respectively (Figures 5A–C).
PpNAP Gene Expression Profiles During Fruit Ripening and Softening
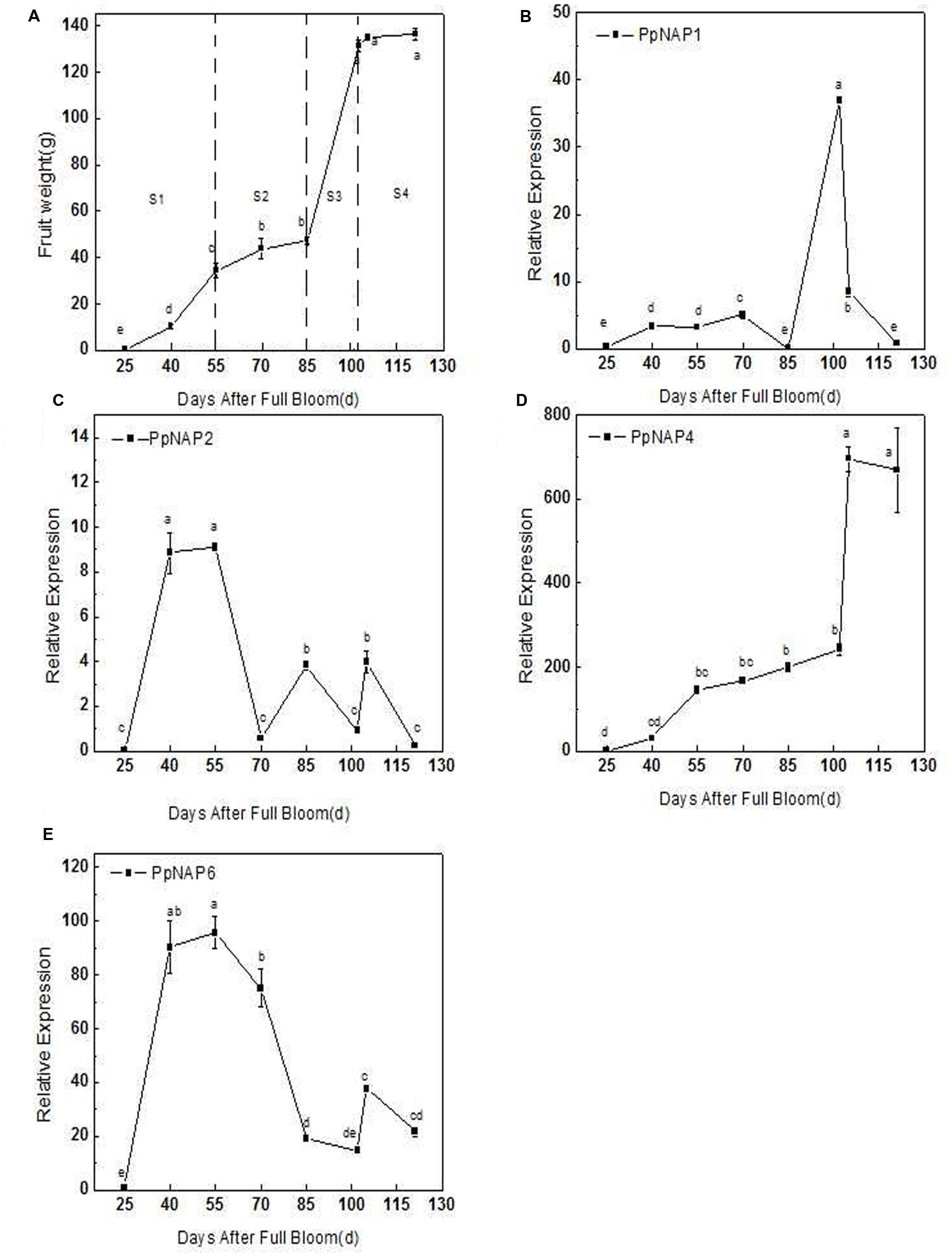
FIGURE 5. Average fruit weight after full bloom (A), quantitative reverse transcription PCR analysis of selected peach NAP genes during fruit development (B–E). Bars correspond to the mean ± standard error (n = 3). The overall least significant difference (p < 0.05) was calculated and used to separate means.
The firmness, ethylene production, and PpNAP expression of commercially mature ‘Qinguang 8’ fruits were measured during fruit storage. In the first 2 days after harvest (DAH), fruit firmness decreased slowly, while from 2 to 8 DAH, fruit firmness declined rapidly (Figure 6A). Ethylene production doubled from 0 to 2 DAH, increased slowly from 2 to 6 DAH, and then decreased considerably (Figure 6B).
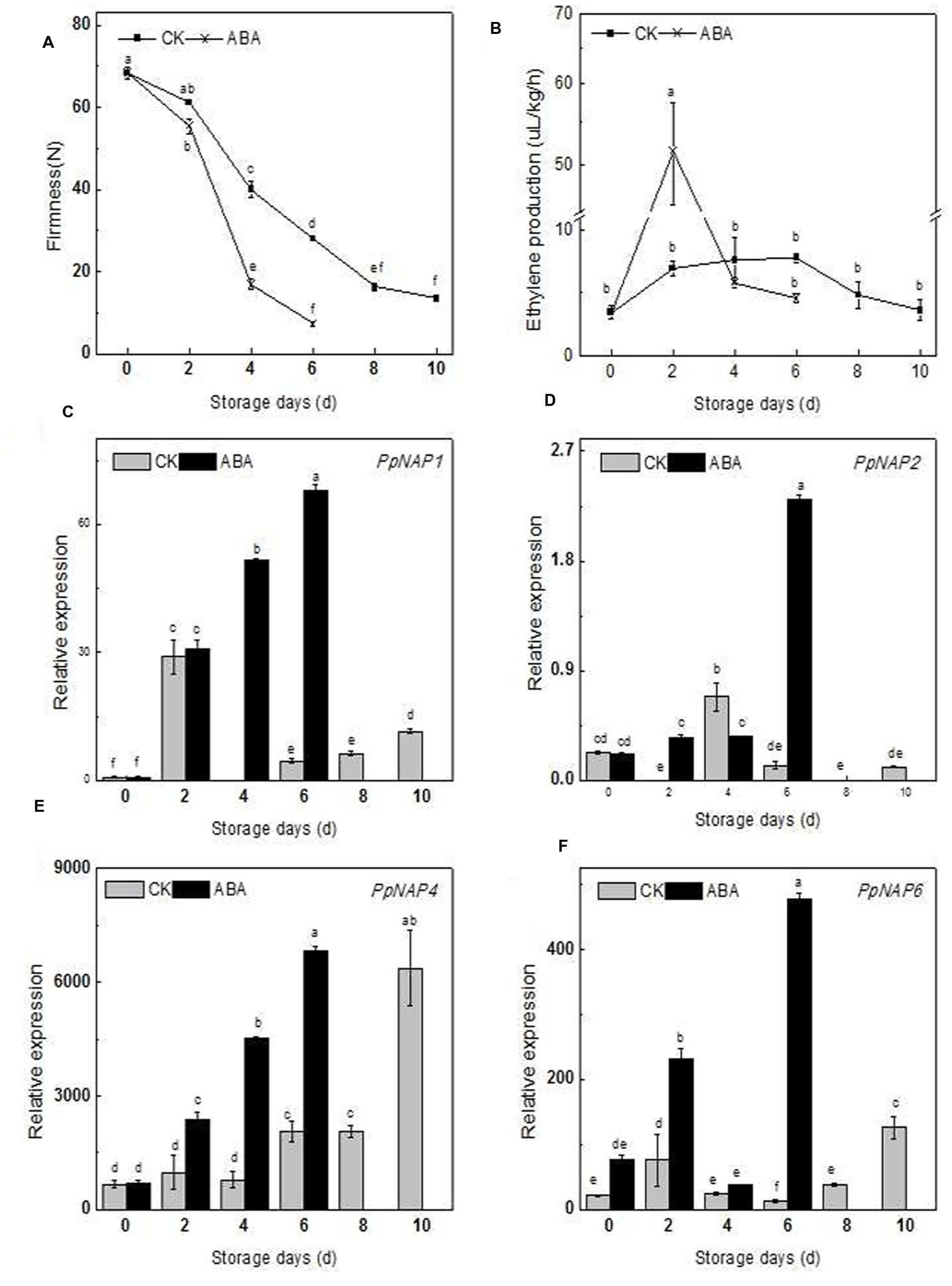
FIGURE 6. Firmness (A), ethylene production (B) and expression levels of selected peach NAP genes in control and ABA-treated fruits during the storage period (C–F). Bars correspond to the mean ± standard error (n = 3). The overall least significant difference (p < 0.05) was calculated and used to separate means.
During storage, the PpNAP1, PpNAP4, and PpNAP6 expression levels exhibited similar trends. Expression increased during the early storage period and was highest at 2 DAH, which coincided with the first peak of ethylene release. The expression levels subsequently declined to varying degrees. This was followed by an increasing trend from 4 or 6 DAH to 10 DAH (Figures 6C,E,F). PpNAP4 and PpNAP6 expression levels were highest at the end of the storage period (Figures 6E,F). In contrast, PpNAP2 expression was maintained at a low level throughout the storage period, with highest expression levels at 4 DAH (Figure 6D).
Effects of ABA Treatment on PpNAP Gene Expression, Ethylene Release, and Fruit Firmness
The firmness of the ABA-treated fruits was lower than that of the control fruits in the first 2 DAH, after which the firmness of the treated and control fruits decreased significantly, with treated fruits softening faster. The maximum storage periods for treated and control fruits were 6 and 10 days, respectively (Figure 6A).
After ABA treatment, the release rate of endogenous ethylene sharply increased and peaked at 2 DAH, with higher peak rates for treated fruits than for controls (Figure 6B). The PpNAP1, PpNAP4, and PpNAP6 expression levels increased following ABA treatment for the duration of the storage period (Figures 6E,F). PpNAP2 expression increased following ABA treatment at 2 and 6 DAH (Figure 6D).
Sequence Analysis of NAP Promoters for Fruit-Specific Expression
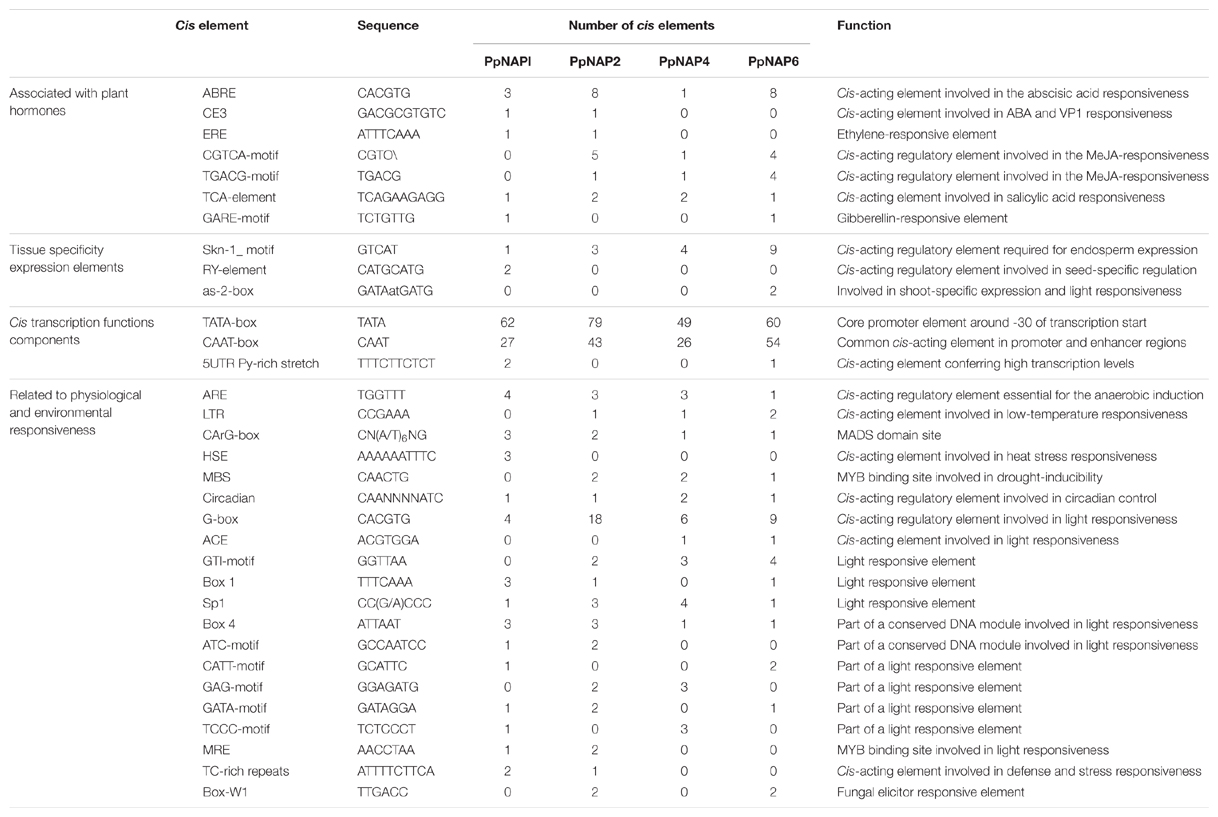
PlantCARE database were used to identify cis-acting elements in the promoter regions of four NAP genes specifically expressed in fruit. The detected cis-acting elements were categorized in the following four classes: (1) involved in the perception of plant hormones, such as ABA, ethylene, methyl jasmonate, salicylic acid, and gibberellic acid; (2) related to expression elements specific to particular tissues, such as endosperm, seed, and shoot; (3) involved in transcription activation and enhancement, such as the TATA-box, CAAT-box, and 5′ untranslated region pyrimidine-rich stretch; and (4) associated with responses to environmental and physiological stimuli, such as drought, low temperature, heat stress, anaerobic conditions, light, fungal elicitors, and other stresses (Table 2).
Among the identified cis-acting elements associated with hormone-related responses, the ABA-responsive element was present (one to eight copies) in all studied promoters, while the coupling element 3 was detected only in the PpNAP1 and PpNAP2 promoters. The CGTCA and TGACG cis-acting element motifs responsive to methyl jasmonate were detected (one to four or five copies) in all promoters except for that of PpNAP1, while the MADS-domain site CArG-box was present in all promoters (one to three copies).
Discussion
Identified NAP Subfamily Members and Sequence Analyses
The NAP is a transcription factor with crucial roles in many biological processes during plant growth and development (Fan et al., 2015). In this study, we identified seven NAP genes in the peach genome that were homologous to NAP genes from three other plant species. However, Fan et al. (2015) reported that there are four NAP genes in peach, corresponding to the PpNAP1, PpNAP2, PpNAP4, and PpNAP6 genes identified in our study. We detected three more NAP genes, namely PpNAP3, PpNAP5, and PpNAP7. Multiple sequence alignments revealed that the seven peach NAP proteins contained the five typical NAC subdomains, and were very similar to other NAP proteins (Figure 1). Phylogenetic analyses indicated that all seven PpNAP genes clustered in the NAP subfamily, with only PpNAP2 belonging to the NAP I group, while the others belonged to the NAP II group. This suggests the function of PpNAP2 may differ from that of the other members.
Tissue-Specific Expression of PpNAP Genes
The expression levels of the identified peach NAP genes were measured in leaves, flowers, and fruits, as well as during maturation and senescence. The results indicated the genes had different expression patterns, which suggests they may have different roles in various physiological pathways. PpNAP1 and PpNAP2 had relatively high expression levels in blooming flowers and flowers 2 DAFB, but low levels in leaves, fruits, and flower buds (Figures 4A–C). Therefore, these genes may be involved in regulating flower maturation and aging. PpNAP3 and PpNAP5 expression was observed in leaves, but was almost undetectable in flowers and fruits (Figures 4A–C), which indicates they may be associated with leaf development. The expression of PpNAP4 was rapidly up-regulated and maintained at high levels during fruit maturation and senescence (Figure 4C), suggesting that this gene may play a key role in regulating peach fruit ripening and softening. PpNAP6 and PpNAP7 expression levels were up-regulated in mature and senescent leaves and flowers (Figures 4A,B). Therefore, they may be associated with the maturation and senescence of leaves and flowers. These results suggest that the expression of PpNAP genes depends on tissue type, which is supported by the results of related studies in other plants. For example, the expression of VvNAP was observed only in grapevine flowers and fruits, and not in vegetative organs such as leaves, shoots, or roots (Fernandez et al., 2006). The expression patterns of AtNAP differed among stamens, fertilized flowers, and developing siliques in A. thaliana (Sablowski and Meyerowitz, 1998). In Mikania micrantha, the MmNAP gene was observed to be specifically expressed in stems, petioles, shoots, and leaves, but not in roots (Li et al., 2012).
Possible Roles of NAP Subfamily Members in Fruit Development and Softening
During fruit development, the PpNAP4 and PpNAP6 expression levels increased rapidly in stages S1 and S4. However, in the S2 stage, PpNAP4 expression slowly increased while PpNAP6 expression levels decreased (Figure 5). During the S3 stage, PpNAP4 and PpNAP6 expression levels stabilized. Because of the association of the NAP gene with cell division and expansion of stamens and petals (Sablowski and Meyerowitz, 1998), our results suggest that PpNAP4 and PpNAP6 are likely involved in the first exponential growth phase and fruit ripening.
During the fruit ripening and softening process, the expression of PpNAP1, PpNAP4, and PpNAP6 increased considerably in the first 2 DAH, which was accompanied by an increase in ethylene production. Furthermore, the expression of PpNAP1, PpNAP4, and PpNAP6 tended to increase during the middle or late storage periods, and was highest at the end of the storage period. These results are consistent with those for AtNAP (Kou et al., 2012), VvNAP (Fernandez et al., 2006), and CitNAC (Liu et al., 2009). Therefore, the functions of PpNAP1, PpNAP4, and PpNAP6 are probably similar to those of AtNAP, VvNAP, and CitNAC, and involve activities related to peach fruit ripening and senescence.
The accumulation of ABA plays a key role in the regulation of peach fruit ripening and senescence (Zhang et al., 2009b), and stimulates ethylene biosynthesis and ripening in tomato fruits (Zhang et al., 2009a). Peach fruits treated with ABA during the S4 fruit development stage exhibited accelerated ripening and up-regulated expression of the ethylene biosynthesis genes ACS1 and ACO1 (Soto et al., 2013). Compared with control fruits, ABA-treated fruits softened faster and released more ethylene, ultimately resulting in a shorter maximum storage period. These results are similar to those observed for tomato (Zhang et al., 2009a), and suggest that ABA may stimulate ethylene biosynthesis. Additionally, the expression levels of PpNAP1, PpNAP4, and PpNAP6 increased in ABA-treated fruit (Figures 6A,B), which was similar to the response of AtNAP in ABA-treated siliques (Kou et al., 2012). In rice and A. thaliana leaves, NAP gene expression was also induced by exogenous ABA (Chen et al., 2014; Yang et al., 2014). Therefore, the ABA-responsive PpNAP1, PpNAP4, and PpNAP6 genes may regulate peach fruit ripening and softening. However, the specific regulatory mechanism requires further characterization.
Analysis of Promoter Sequences of Selected Peach NAP Genes
Because of their involvement in regulating transcription, gene promoters contain important cis-acting elements (Zhu and Li, 1997). To characterize the possible regulatory mechanisms of NAP genes during fruit development, maturation, and softening, we analyzed the promoters of four fruit-specific NAP genes. Several motifs associated with responses to phytohormones and environmental factors were detected. These motifs included the ABA-responsive element and coupling element 3, the CGTCA and TGACG motifs associated with responses to methyl jasmonate, and the TCA-element related to responses to salicylic acid (Table 2). Exogenous ABA can up-regulate NAP expression in A. thaliana and rice (Liang et al., 2014; Yang et al., 2014). Zhou et al. (2013) reported that OsNAP can regulate leaf senescence by affecting jasmonic acid signaling pathways, and that overexpressing OsNAP increases the production of endogenous jasmonic acid in rice. Other studies have demonstrated that hormones, including ABA, jasmonic acid, and salicylic acid, have important regulatory roles during fruit ripening and softening (Creelman and Mullet, 1995; Zhang et al., 2003, 2009a). Therefore, it can be inferred that PpNAP genes regulate peach fruit development and softening by influencing specific hormone signal transduction pathways. Moreover, genes containing the MADS-box motif have key roles in flower and fruit development and maturation (Adamczyk and Fernandez, 2009; Smaczniak et al., 2012). We also observed that one to three copies of the MADS-domain site CArG-box were present in the promoters of four NAP genes, indicating that PpNAP and MADS-box genes may interact to regulate peach fruit development and ripening. However, the specific regulatory mechanisms of PpNAP genes that affect peach fruits require further study.
Author Contributions
CZ, MH, HL, and DZ: Design and interpretation of all experiments. FL, JL, and MQ: Performed all plant physiological and biochemical experiments. CZ, FL and LC: Wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Minghui Lu for English language modifying. This research was supported by grants from National Science Foundation of China (Grant No. 31572079) and the Natural Science Foundation of Shaanxi Province, China (Grant No. 2015JM3103).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00147
FIGURE S1 | The amplification (A), melt curve (B) and melt park (C) of 18s ribosomal gene in all samples.
FIGURE S2 | Quantitative reverse transcription PCR analysis of PpACO1genes in fruits with different developmental stage. Arrow indicates the time of harvest(121 DAFB).
Footnotes
- ^ www.rosaceae.org/species/prunus_persica/genome_v1.0
- ^ http://www.ncbi.nlm.nih.gov/BLAST/
- ^ http://www.ncbi.nlm.nih.gov/gorf/gorf.html.
- ^ http://weblogo.berkeley.edu/logo.cgi.
- ^ http://gsds.cbi.pku.edu.cn.
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ web.expasy.org/compute_pi/.
References
Adamczyk, B. J., and Fernandez, D. E. (2009). MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 149, 1713–1723. doi: 10.1104/pp.109.135806
Barry, C. S., and Giovannoni, J. J. (2007). Ethylene and fruit ripening. J. Plant Growth Regul. 26, 143–159. doi: 10.1007/s00344-007-9002-y
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1007/BF02670468
Chen, X., Wang, Y., Lv, B., Li, J., Luo, L., Lu, S., et al. (2014). The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 55, 604–619. doi: 10.1093/pcp/pct204
Chen, Y., Qiu, K., Kuai, B., and Ding, Y. (2011). Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Physiol. Plant. 142, 361–371. doi: 10.1111/j.1399-3054.2011.01472.x
Creelman, R. A., and Mullet, J. E. (1995). Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. U.S.A. 92, 4114–4119. doi: 10.1073/pnas.92.10.4114
Cristobal, U., Assaf, D., Tzion, F., Ann, B., and Jorge, D. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. doi: 10.1126/science.1133649
Fan, J. P., and Zhao, R. (2014). Cloning and expression pattern analysis of the ApNAP gene in Asarina procumbens. Plant Sci. J. 32, 251–258.
Fan, K., Shen, H., Bibi, N., Li, F., Yuan, S., Wang, M., et al. (2015). Molecular evolution and species-specific expansion of the NAP members in plants. J. Integr. Plant Biol. 57, 673–687. doi: 10.1111/jipb.12344
Fan, K., Wang, M., Miao, Y., Ni, M., Bibi, N., Yuan, S., et al. (2014). Molecular evolution and expansion analysis of the NAC transcription factor in Zea mays. PLoS ONE 9:e111837. doi: 10.1371/journal.pone.0111837
Fernandez, L., Ageorges, A., and Torregrosa, L. (2006). A putative NAP homolog specifically expressed during grapevine flower and berry development. Vitis 45, 51–52.
Guo, Y., and Gan, S. S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Guo, Y., Wei, Q., and Kuai, B. (2010). Cloning and characterization of the AtNAP orthologous in Festuca arundinacea. J. Fudan Univ. 49, 544–551.
Huang, G. Q., Li, W., Zhou, W., Zhang, J. M., Li, D. D., Gong, S. Y., et al. (2013). Seven cotton genes encoding putative nac domain proteins are preferentially expressed in roots and in responses to abiotic stress during root development. Plant Growth Regul. 71, 101–112. doi: 10.1007/s10725-013-9811-x
Kalivas, A., Pasentsis, K., Argiriou, A., and Tsaftaris, A. S. (2010). Isolation, characterization, and expression analysis of an NAP-like cDNA from Crocus (Crocus sativus L.). Plant Mol. Biol. Rep. 28, 654–663. doi: 10.1007/s11105-010-0197-x
Kou, X., Watkins, C. B., and Gan, S. S. (2012). Arabidopsis AtNAP regulates fruit senescence. J. Exp. Bot. 63, 6139–6147. doi: 10.1093/jxb/ers266
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van, de Peer Y, et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, D. M., Wang, J. H., Peng, S. L., Zhu, G. F., and Lu, F. B. (2012). Molecular cloning and characterization of two novel NAC genes from Mikania micrantha (Asteraceae). Genet. Mol. Res. 11, 4383–4401. doi: 10.4238/2012.September.19.3
Li, S. H., Huguet, J. G., Schoch, P. G., and Orlando, P. (1989). Response of peach tree growth and cropping to soil water deficit at various phenological stages of fruit development. J. Hortic. Sci. 64, 541–552.
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Liguori, G., Weksler, A., Zutahi, Y., Lurie, S., and Kosto, I. (2004). Effect of 1-methylcyclopropene on ripening of melting flesh peaches and nectarines. Postharvest Biol. Technol. 31, 263–268. doi: 10.1016/j.postharvbio.2003.09.007
Liu, Y. Z., Baig, M. N. R., Fan, R., Ye, J. L., Cao, Y. C., and Deng, X. X. (2009). Identification and expression pattern of a novel NAM, ATAF, and CUC-like gene from Citrus sinensis Osbeck. Plant Mol. Biol. Rep. 27, 292–297. doi: 10.1007/s11105-008-0082-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Meng, C., Cai, C., Zhang, T., and Guo, W. (2009). Characterization of six novel nac genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci. 176, 352–359. doi: 10.1016/j.plantsci.2008.12.003
Meng, Q., Zhang, C., Gai, J., and Yu, D. (2007). Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.). J. Plant Physiol. 164, 1002–1012. doi: 10.1016/j.jplph.2006.05.019
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Sablowski, R. W., and Meyerowitz, E. M. (1998). A homolog of no apical meristem is an immediate target of the floral homeotic genes apetala3/pistillata. Cell 92, 93–103. doi: 10.1016/S0092-8674(00)80902-2
Smaczniak, C., Immink, R. G., Angenent, G. C., and Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Soto, A., Ruiz, K. B., Ravaglia, D., Costa, G., and Torrigiani, P. (2013). Aba may promote or delay peach fruit ripening through modulation of ripening– and hormone-related gene expression depending on the developmental stage. Plant Physiol. Biochem. 64, 11–24. doi: 10.1016/j.plaphy.2012.12.011
Tonutti, P., Bonghi, C., Ruperti, B., and Ramina, A. (1997). “The modulation of ethylene biosynthesis and ACC oxidase gene expression during peach fruit development and fruitlet abscission,” in Biology and Biotechnology of the Plant Hormone Ethylene, eds A. K. Kanellis, C. Chang, H. Kende, and D. Grierson (Dordrecht: Springer Netherlands), 149–153.
Yang, J., Worley, E., and Udvardi, M. (2014). A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26, 4862–4867. doi: 10.1105/tpc.114.133769
Zhang, K., and Gan, S. S. (2012). An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 158, 961–969. doi: 10.1104/pp.111.190876
Zhang, M., Leng, P., Zhang, G., and Li, X. (2009a). Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J. Plant Physiol. 166, 1241–1252. doi: 10.1016/j.jplph.2009.01.013
Zhang, M., Yuan, B., and Leng, P. (2009b). The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 60, 1579–1588. doi: 10.1093/jxb/erp026
Zhang, Y., Chen, K., Zhang, S., and Ferguson, I. (2003). The role of salicylic acid in postharvest ripening of kiwifruit. Postharvest Biol. Technol. 28, 67–74. doi: 10.1016/S0925-5214(02)00172-2
Zhou, Y., Huang, W., Liu, L., Chen, T., Fei, Z., and Lin, Y. (2013). Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 13:132. doi: 10.1186/1471-2229-13-132
Keywords: Prunus persica, NAP subfamily, fruit, development, ripening
Citation: Li F, Li J, Qian M, Han M, Cao L, Liu H, Zhang D and Zhao C (2016) Identification of Peach NAP Transcription Factor Genes and Characterization of their Expression in Vegetative and Reproductive Organs during Development and Senescence. Front. Plant Sci. 7:147. doi: 10.3389/fpls.2016.00147
Received: 09 September 2015; Accepted: 28 January 2016;
Published: 16 February 2016.
Edited by:
Mario Pezzotti, University of Verona, ItalyReviewed by:
Claudio Bonghi, University of Padova, ItalyAthanasios Tsaftaris, Centre for Research and Technology Hellas (CE.R.T.H), Greece
Copyright © 2016 Li, Li, Qian, Han, Cao, Liu, Zhang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caiping Zhao, zhcc@nwsuaf.edu.cn.
 Fang Li
Fang Li Jinjin Li
Jinjin Li Ming Qian
Ming Qian Mingyu Han
Mingyu Han Caiping Zhao
Caiping Zhao