- 1Plant Genomic Network Research Team, RIKEN Center for Sustainable Resource Sciences, Yokohama, Japan
- 2Kihara Institute for Biological Research, Yokohama City University, Yokohama, Japan
- 3CREST, Japan Science and Technology Agency, Saitama, Japan
Drought stress has a negative impact on crop yield. Thus, understanding the molecular mechanisms responsible for plant drought stress tolerance is essential for improving this beneficial trait in crops. In the current study, a transcriptional analysis was conducted of gene regulatory networks in roots of soil-grown Arabidopsis plants in response to a drought stress treatment. A microarray analysis of drought-stressed roots and shoots was performed at 0, 1, 3, 5, 7, and 9 days. Results indicated that the expression of many drought stress-responsive genes and abscisic acid biosynthesis-related genes was differentially regulated in roots and shoots from days 3 to 9. The expression of cellular and metabolic process-related genes was up-regulated at an earlier time-point in roots than in shoots. In this regard, the expression of genes involved in oxidative signaling, chromatin structure, and cell wall modification also increased significantly in roots compared to shoots. Moreover, the increased expression of genes involved in the transport of amino acids and other solutes; including malate, iron, and sulfur, was observed in roots during the early time points following the initiation of the drought stress. These data suggest that plants may utilize these signaling channels and metabolic adjustments as adaptive responses in the early stages of a drought stress. Collectively, the results of the present study increases our understanding of the differences pertaining to the molecular mechanisms occurring in roots vs. shoots in response to a drought stress. Furthermore, these findings also aid in the selection of novel genes and promoters that can be used to potentially produce crop plants with increased drought tolerance.
Introduction
Adverse environmental factors, such as drought stress, severely limit agricultural production and reduce the yield and quality of crop plants. Water scarcity is predicted to increase as an outcome of climate change, and thus poses a serious challenge to agricultural production worldwide. Understanding the molecular response of plants to a drought stress and utilizing this knowledge for developing different molecular approaches to ameliorate the harmful effects of water deficit has always been an important objective for molecular breeders (Xiong et al., 2002; Umezawa et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006; Seki et al., 2007; Shinozaki and Yamaguchi-Shinozaki, 2007; Hirayama and Shinozaki, 2010).
Plants sense changes in the environment and modify cellular physiology in a complex, integrated manner by upregulating the expression of various combinations of regulatory and functional genes. Despite a comprehensive knowledge of mechanisms governing cellular responses, our understanding of the early events in the perception of stress signals is relatively poor (Urao et al., 1999; Wohlbach et al., 2008). Drought stress triggers significant molecular and physiological changes in plants, such as adjustments of metabolism and osmotic potential, and reducing leaf turgor pressure, which lead to a reduction or cessation of growth (Tardieu et al., 2014). Although water deficiency inhibits plant growth at whole plant level, roots can grow under low water potentials that completely inhibit stem and leaf growth (Spollen and Sharp, 1991; Spollen et al., 1993; Chazen and Neumann, 1994; Wu and Cosgrove, 2000; Sharp et al., 2004). Since increasing root surface area facilitates water absorption, it is plausible that differences between roots and shoots may have evolved in response to water scarcity as an adaptation strategy to dry conditions (Wu and Cosgrove, 2000; Sharp et al., 2004). Cellular and molecular responses underlying adaptation to environmental stresses have been extensively studied and are governed by changes in gene expression (Matsui et al., 2008; Liu et al., 2014). Changes in the expression of a large number of genes belonging to diverse functional groups, such as transcription factors, protein kinases, and phosphatases, all contribute to the signal transduction that occurs in plants in response to and adaptation to a drought stress (Kreps et al., 2002; Seki et al., 2002, 2007; Xiong et al., 2002; Shinozaki et al., 2003; Matsui et al., 2008; Hirayama and Shinozaki, 2010).
Drought stress response has been extensively studied in Arabidopsis and the subject has been comprehensively reviewed (Iuchi et al., 2001; Xiong et al., 2002; Seki et al., 2007; Shinozaki and Yamaguchi-Shinozaki, 2007; Matsui et al., 2008; Harb et al., 2010; Osakabe et al., 2014). Stress-responsive genes are comprised of enzymes regulating osmotic pressure, aquaporins, detoxifying enzymes, late embryogenesis abundant proteins, reactive oxygen species scavengers and chaperones that protect the integrity of cell membranes and ensure ion transport/balances. Additionally, functionally diverse transcription factors and protein kinases, that regulate gene expression and signal transduction, are also an integral component of the drought stress response (Wei et al., 2009). The molecular response of plants to drought stress has been categorized into abscisic acid (ABA) dependent and ABA independent pathways (Yamaguchi-Shinozaki and Shinozaki, 2006). ABA biosynthesis, transport and accumulation all increase in response to a water deficit. The increased ABA content in leaves triggers stomata closure, ultimately decreasing the rate of gas exchange, respiration, and photosynthetic activity (Yamaguchi-Shinozaki and Shinozaki, 2006). An increase in the endogenous ABA content also induces the expression of a number of stress-related genes in plants (Yamaguchi-Shinozaki and Shinozaki, 2006). Briefly, the ABA signaling pathway affects plant adaptation to stress by regulating the internal water status in plants (Osakabe et al., 2014). The ABA independent pathway is mainly regulated by dehydration-responsive element/C-repeat (DRE/CRT) and DRE-/CRT-binding protein 2 (DREB2) transcription factors (Yamaguchi-Shinozaki and Shinozaki, 2006).
As previously mentioned, the root system is the first to perceive drought stress signals. Therefore, root development is significantly affected by water availability in the soil. Most studies in Arabidopsis, however, have explored transcriptomic changes in whole plants by only investigating shoots on soil-grown plants or air-dried roots. Therefore, at the present time, the drought response of roots in soil-grown plants remains largely unknown. To fill this gap, changes in the expression of genes in roots and shoots of soil-grown plants in response to a progressive drought stress were characterized and compared by sampling plants at 0, 1, 3, 5, 7, and 9 days of a drought stress. This provided the opportunity to dissect the molecular response of shoots vs. roots to a drought stress. The objective of the study was to obtain information that could be used to develop new strategies for developing drought tolerant plants.
Materials and Methods
Plant Material and Growth Conditions
Seeds of Arabidopsis thaliana (Col-0 ecotype) were grown on MS medium at 22°C under 16-h-light/8-h-dark (40–80 μmol photons m−2 s−1) for 9 days. Plantlets were then transferred to ceramics granular soil (size 2.5L, Sakatanotane, Japan) and grown for 8 days at 22°C (16 h light/8 h dark cycle, 60 μmol m−2 s−1 photon flux density). The drought treatment was then commenced by removing excess water from the trays and ceasing any subsequent watering. Roots and shoots were harvested separately at 0, 1, 3, 5, 7, and 9 days after the onset of drought treatment. Plants were removed from soil and roots and shoots of 12 plants were harvested from 3 pots for each biological replication. All samples were collected at noon. After harvesting, samples were immediately placed in liquid nitrogen and stored at −80°C until RNA extraction.
Microarray Analysis
RNA was extracted from all biological replicates with the mirVana™ miRNA Isolation Kit (Ambion, USA) according to the manufacturer's instructions. The microarray analyses were carried out as previously described (Nguyen et al., 2015). Briefly, fluorescent-labeled (Cy3) cRNAs were prepared from 400 ng total RNA from each sample using a Quick Amp labeling kit (Agilent Technologies) and subsequently hybridized to an Agilent Arabidopsis custom microarray (GPL19830). Three biological replicates were processed for each treatment, with the exception of roots 7 and 9 days as well as shoot 1 and 3 days, where four biological replications were processed, giving a total of 40 hybridizations. Arrays were scanned with a microarray scanner (G2505B, Agilent Technologies) and the R 2.12.1 software program (R Core Team). RMA normalization was performed for the obtained signals of the microarray probes using limma package (Ritchie et al., 2015). A student's t-test (p-value) was performed as a parametric test and the Benjamini and Hochberg False Discovery Rate (FDR; q-value) procedure was used to control the certainty level (Benjamini and Hochberg, 1995). Genes with at least a 2-fold change in expression and having a q < 0.1 were considered to be differentially expressed. The microarray data has been deposited to GenBank with accession number GSE76827.
MapMan and Gene Ontology (GO) Analysis
The average log2 value of all biological replicates was calculated separately for roots and shoots for individual annotations at each time point. Gene ontology analyses were carried out using the PANTHER (protein annotation through evolutionary relationship) classification system database maintained at http://pantherdb.org/ (Mi et al., 2013). The GO analyses were performed for molecular function, protein classification and pathway. To further validate the results, the normalized log2 values were then used to compare the transcriptomic changes using MapMan 3.6.0RC1(Thimm et al., 2004). PageMan analysis was also performed using MapMan 3.6.0RC1 which included a Wilcoxon test with BH correction (Thimm et al., 2004).
Real Time PCR
Real time PCR analysis was performed for RD29A (AT5G52310), NCED2 (AT4G18350), NCED3 (AT3G14440), and GolS4 (AT1G60470) genes with standard curve method in order to confirm that plants were experiencing water stress and to confirm the results obtained by microarray analysis. cDNA for each sample was synthesized from 200ng RNA using QuantiTect Rev. Transcription Kit according to the manufacturer's protocol (QIAGEN, USA). For NCED2 the forward and reverse primers were 5′-CGCCGGTTTGGTT TACTTTA-3′ and 5′-GCGTGAAGCTCC TTCGTAAC-3′ respectively. Forward and reverse primers used for NCED3 were 5′-ACTCATGCTATT CTACGCCAGAG-3′ and 5′-ACCAACGGTTT TTAAATCTCCAT-3′, respectively. For RD29A the forward and reverse primers were 5′-TGGATCTGAAGAA CGAATCTGATATC-3′ and 5′-GGTCTT CCCTTCGCCAGAA-3′, respectively. For GolS4 the forward and reverse primers were 5′- TTGCCATGGCTTA TTACGTTC -3′ and 5′-AAACAGTCCATCACGGCATAG-3′, respectively. Actin 2, used as an internal control, was amplified using the forward and reverse primers 5′-TGAAGTGTGATGTGGATATCAGG-3′ and 5′-GATTTCTTTGCTCATACGGTCAG-3′, respectively.
Results
General Transcriptional Changes in Roots and Shoots during Early Drought Stress
The transcriptional changes in roots and shoots of soil grown plant subjected to progressive drought were analyzed. The water retention capacity of ceramics granular soil is poor due to its large pore size, thus the drought stress increased rapidly relative to normal soil, and all plants had died by day 10 of the drought stress (data not shown). The pots started to dry around day 5 but no morphological symptoms of drought stress were observed by that time (Figure S1). By day 9 the plants appeared wilted (Figures 1A–E), however, plants could recover if they were watered (data not shown). In roots and shoots, no genes were observed to be significantly up or down-regulated in either roots or shoots on day 1 of the drought treatment. At day 3 of the drought treatment, 497 genes were significantly up-regulated in roots (Figure 1 and Table S1), while 292 genes were significantly down-regulated (Figure S2 and Table S2). At 5, 7, and 9 days of the drought treatment, the number of up-regulated genes in roots was 824, 1,854, and 3,007 respectively (Figure 1). The number of down-regulated genes in roots at days 5, 7, and 9 of the drought treatment were 899, 2327, and 3742, respectively (Figure S2 and Table S2). In total, 3539 genes were up-regulated and 4154 genes were down-regulated in roots. Similar to roots, no genes were observed to be significantly up- or down-regulated in shoots on day 1 of the drought treatment. On day 3 of the drought treatment, 122 genes were significantly up-regulated in shoots (Figure 1 and Table S3), while 91 genes were significantly down-regulated (Figure S2 and Table S4). On days 5, 7, and 9 of the drought treatment, the number of up-regulated genes in shoots was 961, 2549, and 4126, respectively (Figure 1). On the other hand, the number of down-regulated genes was 528, 2442, and 4848, respectively (Figure S2 and Table S2). In total, 4763 genes were up-regulated and 5213 genes were down-regulated in shoots. The expression of 1906 genes was up-regulated in both roots and shoots (Table S5), while the expression of 2218 genes was down-regulated (Table S6) in both roots and shoots (Figure S2). To determine the reliability of results of the microarray analysis, the expression of four genes (NCED2, NCED3, RD29A, and GolS4) that were up-regulated by drought stress was examined by real time PCR. The results of the real time PCR analysis confirmed the results obtained using the microarray (Figure 2). Genes that were up-regulated in roots at least 4-fold on day 3 of the drought stress, as compared to 0 day, are listed in Table 1. These genes belong to diverse functional groups, including oxygenases, cytochrome P450 family proteins, Multidrug And Toxin Extrusion (MATE) efflux transporters, and RD29A, RD29B (AT5G52300), etc., and may play an important role in early drought response.
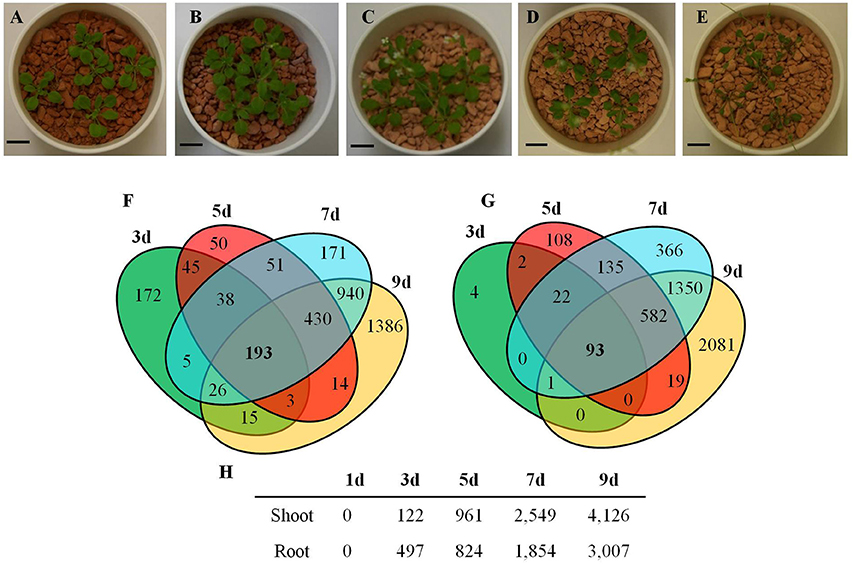
Figure 1. Arabidopsis genes up-regulated by a progressive drought stress. (A–E) Plants grown in ceramics soil under drought conditions, (A) Day 0, (B) Day 3, (C) Day 5, (D) Day 7, and (E) Day 9 of the drought stress treatment. (F) Venn diagram of the number of genes up-regulated by drought stress in roots. (G) Venn diagram of the number of genes up-regulated by drought stress in shoots. (H) Number of genes up-regulated in roots and shoots during a progressive drought stress. (A–E) Scale bar = 1cm. (F–H) Genes with at least a 2-fold increase in expression and having a FDR < 0.1 were considered to be up-regulated.
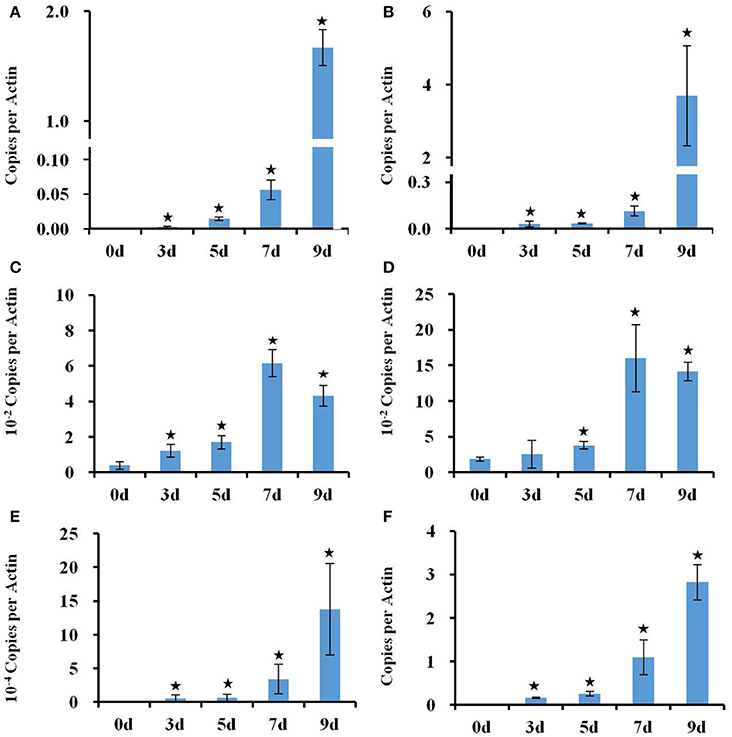
Figure 2. Real time PCR analysis of the expression of RD29A, NCED3, NCED2, and GolS4 genes in response to a progressive drought stress. (A,B) Expression of RD29A gene in roots (A) and shoots (B), (C,D) Expression of NCED3 in roots (C) and shoots (D). Expression of NCED2 (E) and GolS4 (F) in roots. Actin 2 expression was used to normalize the expression of all genes. Column bars with an asterisk above are significantly different from Day 0 based on a t-test (p < 0.05).
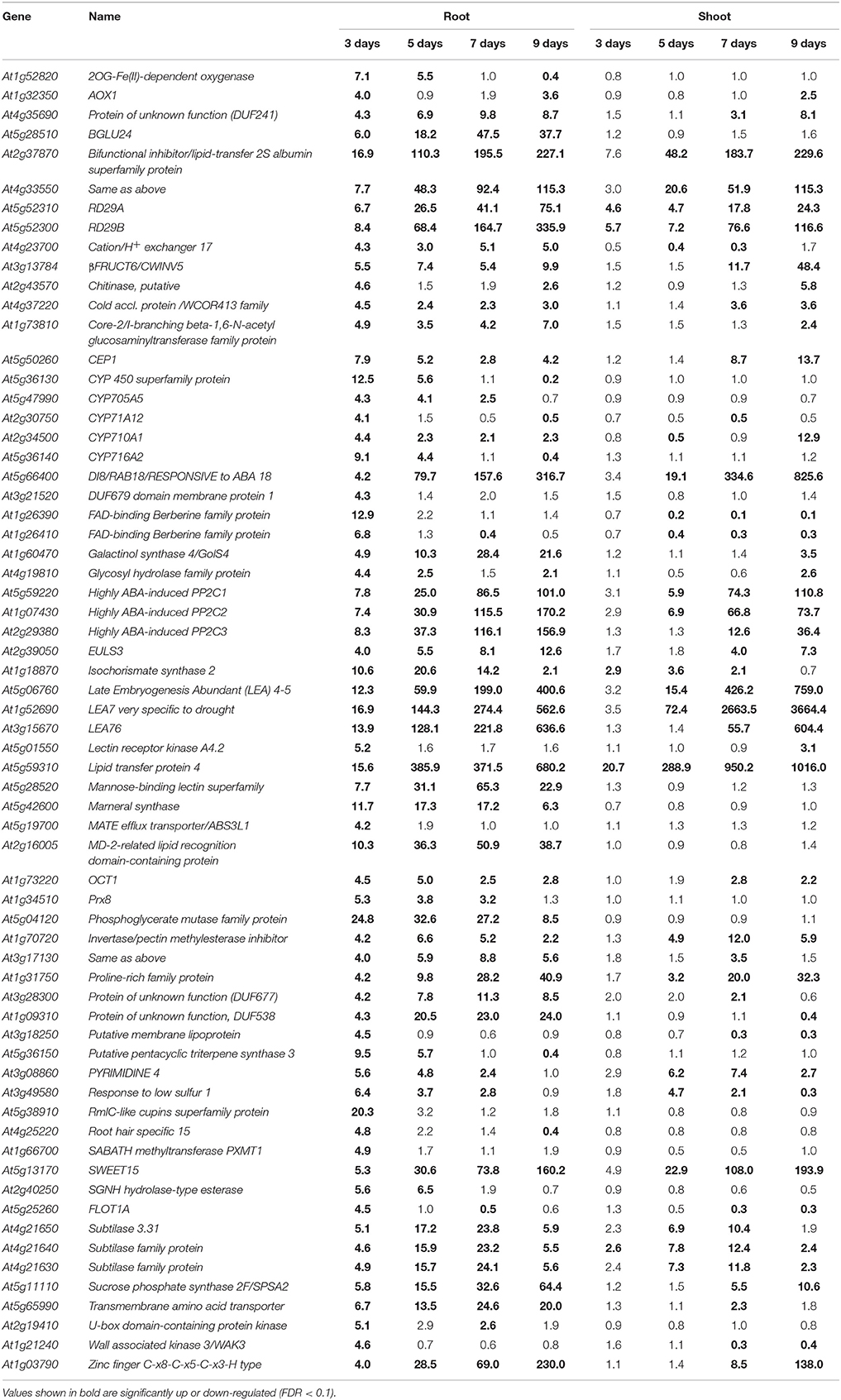
Table 1. Arabidopsis genes highly up-regulated in roots during the early stages of a progressive drought stress.
In general, the drought-inducible genes were up-regulated in roots at a very early stage of the drought stress treatment (days 3–5), while in shoot tissue this response was slightly delayed (days 5–7). For example, the expression of protein phosphatase 2C1 (PP2C1; AT5G59220), PP2C2 (AT1G07430), and PP2C3 (AT2G29380) was significantly up-regulated in roots on days 3–9 of the drought stress. The expression of PP2C1 and PP2C2 was up-regulated in shoots on days 5–9, while the expression of PP2C3 was up-regulated on days 7–9 of the drought stress (Table S1). The expression of DREB2A (AT5G05410) was up-regulated in roots from day 5 to 9, and in shoots from day 7 to 9. DREB2B (AT3G11020) was up-regulated in roots from day 7 to 9, and only at day 9 in shoots (Table S1).
Our data revealed that 1633 genes were specifically up-regulated in roots (Figure S2) and was examined in relation to previously published tiling microarray results (Matsui et al., 2008). In comparison to the genes up-regulated in response to 2 or 10 h of drought stress identified in the tiling array, (Matsui et al., 2008), the current analysis identified 1353 new genes (83% of total genes specifically up-regulated in roots) that were specifically up-regulated in roots in response to drought stress (Table S1). The newly identified genes were members of diverse gene families such as major facilitator super family (MFS) transporters [AT1G08900, AT1G30560, AT1G33440, AT1G72140, AT1G80530, AT2G26690, AT2G34355, AT3G20460, AT3G45680, AT3G47960, AT4G19450, STP8 (AT5G26250), AT5G27350, and AT5G62680], MATE efflux transporters (AT1G71140, AT5G17700, AT5G19700, and AT5G38030), microRNA genes [MIR156b (AT4G30972), MIR161 (AT1G48267), MIR162b (AT5G23065), MIR164 (AT5G01747), MIR167c (AT3G04765), MIR168b (AT5G45307), MIR396a (AT2G10606), MIR402 (AT1G77235), MIR777a (AT1G70645), and MIR848a (AT5G13887)], various transcription factors (MYB, NAC domain, WRKY, etc.), ABA biosynthesis-related genes (NCED5, NCED9), pectin biosynthesis/modification-related genes, pre-tRNA genes, and various S-adenosyl-L-methionine (SAM) dependent transferases (Figure 3 and Table S1). In comparison to the tiling array conducted by Matsui et al. (2008), 1,724 additional genes were identified in the current study that were specifically down-regulated in roots (Table S2). Moreover, our data also revealed the differential regulation of several genes in roots vs. shoots (Tables S1, S3) that have been already reported to be involved in drought stress response (Huang et al., 2008; Matsui et al., 2008).
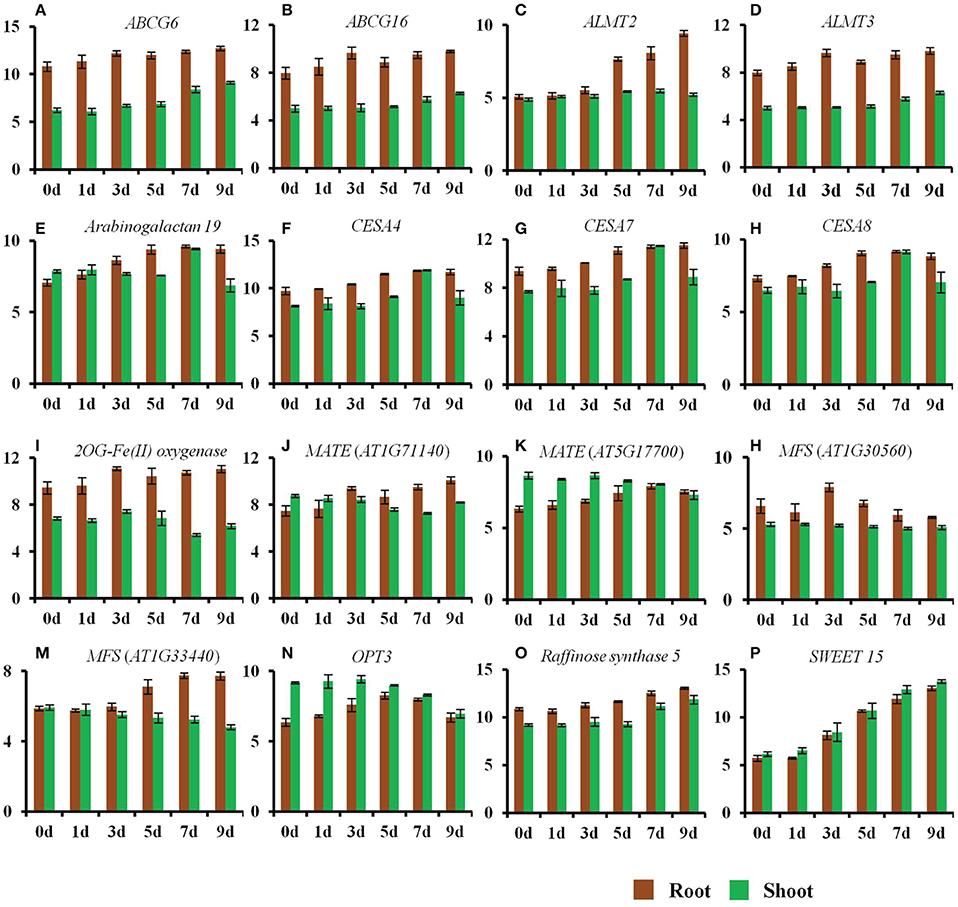
Figure 3. Changes in the expression of genes by a progressive drought stress. The normalized log2 values were used to plot the expression of genes regulated by a progressive drought stress. (A) ABCG6, (B) ABCG16, (C) ALMT2, (D) ALMT3, (E) Arabinogalactan protein 19, (F) CESA4, (G) CESA7, (H) CESA8, (I) 2OG-Fe(II) oxygenase (AT4G10500), (J) MATE transporter (AT1G71140), (K) MATE transporter (AT5G17700), (L) MFS transporter (AT1G30560), (M) MFS transporter (AT1G33440), (N) OPT3, (O) Raffinose synthase 5, (P) SWEET 15. Error bars represent standard deviation.
Gene Ontology (GO) and MapMan Analysis
GO enrichment analysis revealed that the majority of the up-regulated genes in roots and shoots on day 3 of the drought treatment belonged to catalytic activity (GO:0003824; Figure 4), however, in roots a significant number of up-regulated genes were also identified as related to transport (GO:0005215) and structural molecular activity (GO:0005198). As the drought stress progressed, genes belonging to molecular binding (GO:0005488), transport (GO:0005215), and transcription factors (GO:0001071) were also up-regulated in shoots (Figure 4). At day 7 and 9 of the drought stress, the response of roots and shoots seemed very similar (Figure 4). The number of up-regulated genes in roots on day 3 of the drought stress was higher than the number of up-regulated genes in shoot tissue, while an opposite trend was observed on days 5–9 of the drought stress (Figure 1). GO enrichment analysis indicated that different transporters/transport related genes were significantly up-regulated in roots compared to shoots on day 3 of the drought stress (Figure 4). The number of up-regulated genes involved in structural molecule activity (GO:0005198) was also higher in roots compared to shoots (Figure 4). GO enrichment analysis for protein classification revealed that a number of calcium binding proteins (PC00060) were also up-regulated in roots on day 3 of drought stress (Figure S3). The most striking differences observed between roots and shoots were at day 3 of drought stress for pathway analysis. In shoots, genes involved in general transcription regulation (P00023) and transcription regulation by bZIP transcription factor (P00055) were recognized by GO analysis, while in roots genes belonging to 28 pathways were recognized (Figure S4). Majority of genes up regulated or down regulated both in roots and shoots were classified as engaged in metabolic or cellular process according to GO analysis (Figure S5).
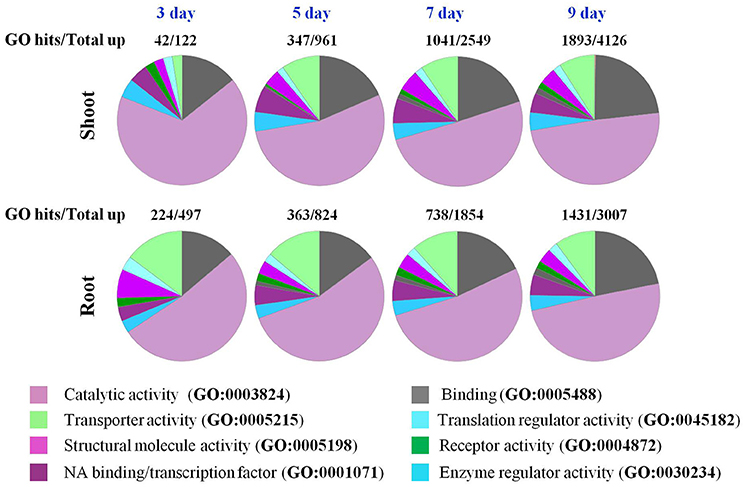
Figure 4. GO enrichment analysis of genes in roots and shoots of Arabidopsis plants that respond to a progressive drought stress. Go functional classification was performed using the panther classification system maintained at http://pantherdb.org/.
The MapMan and GO analyses are comparable to each other, which serves as a justification for comparative analysis (Klie and Nikoloski, 2012). MapMan and PageMan analysis were done to validate the GO enrichment analysis and to categorize the genes in more detail. MapMan analysis indicated that a greater number of genes categorized as cell wall biosynthesis related genes, lipid metabolism related genes, and genes involved in secondary metabolism were also up-regulated in roots compared to shoots on day 3 of the drought stress (Figures S6, S7). MapMan analysis revealed that genes involved in photosynthesis/light reactions were significantly down-regulated in shoots starting at day 5 of the drought stress (Figures S6, S7). In shoots, genes involved in minor CHO metabolism and cell wall synthesis were up-regulated at days 7 and 9 of the drought stress, while almost all the genes involved in photosynthesis/light reactions were significantly down-regulated (Figure S7).
PageMan analysis of roots revealed that bins related to major carbohydrate (CHO) metabolism, cell wall synthesis, and DNA and chromatin structure were significantly up-regulated, while bins relating to amino acid metabolism, and nucleic acid metabolism were down-regulated (Figure S8). Bins related to mitochondrial electron transport (shoots), amino acid metabolism (roots and shoots), nucleotide metabolism (roots and shoots) were significantly down-regulated at an early stage of drought stress (from day 3), while bins related to development and RNA synthesis/transcription (shoot) became significantly up-regulated as the drought treatment progressed (Figure S8).
Changes in the Expression of Cell Wall and Suberin Synthesis Genes
Roots rapidly sense changes in water potential and significantly alter roots architecture in an attempt to acquire more water in order to maintain a non-detrimental water potential. This is evident by the changes in the expression of genes belonging to cell wall, suberin, and lignin biosynthesis. Expression of ABC transporters involved in lignin transport also increased in roots (Table S1). The expression of cellulose synthase (CES) genes, CESA4 (AT5G44030), CESA7 (AT5G17420), and CESA8 (AT4G18780) was significantly up-regulated in roots on days 5–9 of the drought stress (Figure 3). These genes have been reported to contribute to secondary cell wall synthesis (Carpita, 2011). In contrast, the expression of genes involved in primary cell wall synthesis (CESA1; AT4G32410, CESA3; AT5G05170, CESA6; AT5G64740) was not altered in roots. Moreover, the expression of ABCG6 (AT5G13580) and ABCG16 (AT3G55090), was significantly up-regulated in roots at a very early stage of the drought stress (Figure 3). These genes belong to a set of five Arabidopsis ABCG transporters that are required for synthesis of an effective suberin barrier in roots and seed coats (ABCG2; AT2G37360, ABCG6, and ABCG20; AT3G53510) and for synthesis of an intact pollen wall (ABCG1; AT2G39350 and ABCG16) (Yadav et al., 2014). The expression of arabinogalactan protein 19 (AT1G68725) was also up-regulated in roots from day 3 to 9 of the drought stress (Figure 3). This gene contributes to plant growth, as mutants for this gene show reduced height, altered leaf shape and size, and lighter color (Yang et al., 2007).
Regulation of Osmoprotectant Biosynthesis-Related Genes
The expression of genes involved in the biosynthesis of osmoprotectants changed significantly during the early stages of the drought stress, particularly in roots. Raffinose and galactinol are involved in tolerance to drought, high salinity, and cold stresses. Galactinol synthase (GolS) catalyzes the first step in the biosynthesis of raffinose (Taji et al., 2002). Seven GolS (GolS1-7) members have been reported in Arabidopsis. The expression of GolS1 (AT2G47180) and GolS2 (AT1G56600) is up-regulated by drought stress. Plants over expressing GolS2 exhibit increased levels of endogenous galactinol and raffinose, and are tolerant to drought stress (Taji et al., 2002). The data in the current study indicate that the expression of GolS1 was specifically up-regulated (Table S3), while that of GolS3 (AT1G09350) was specifically down-regulated in shoots on day 7 and 9 of the drought stress (Table S4). The expression of GolS2 was up-regulated in roots from days 5 to 9 of the drought stress, while in shoots it was up-regulated from day 3 to 9 (Table S1). The expression of GolS4 was significantly up-regulated in roots from day 3 to 9, while in shoots it was up-regulated only on day 9 of the drought stress (Table 1). The expression of raffinose synthase 5 (RS5; AT5G40390) was up-regulated in roots and shoots on days 7 and 9 of the drought stress (Figure 3). Changes in the expression of genes involved in proline synthesis were also observed. The expression of P5CS1 (AT2G39800) was up-regulated in roots and shoots from day 5 to 9 (Table S1), while the expression of P5CS2 (AT3G55610) was specifically up-regulated in shoots on day 7 and 9 (Table S3).
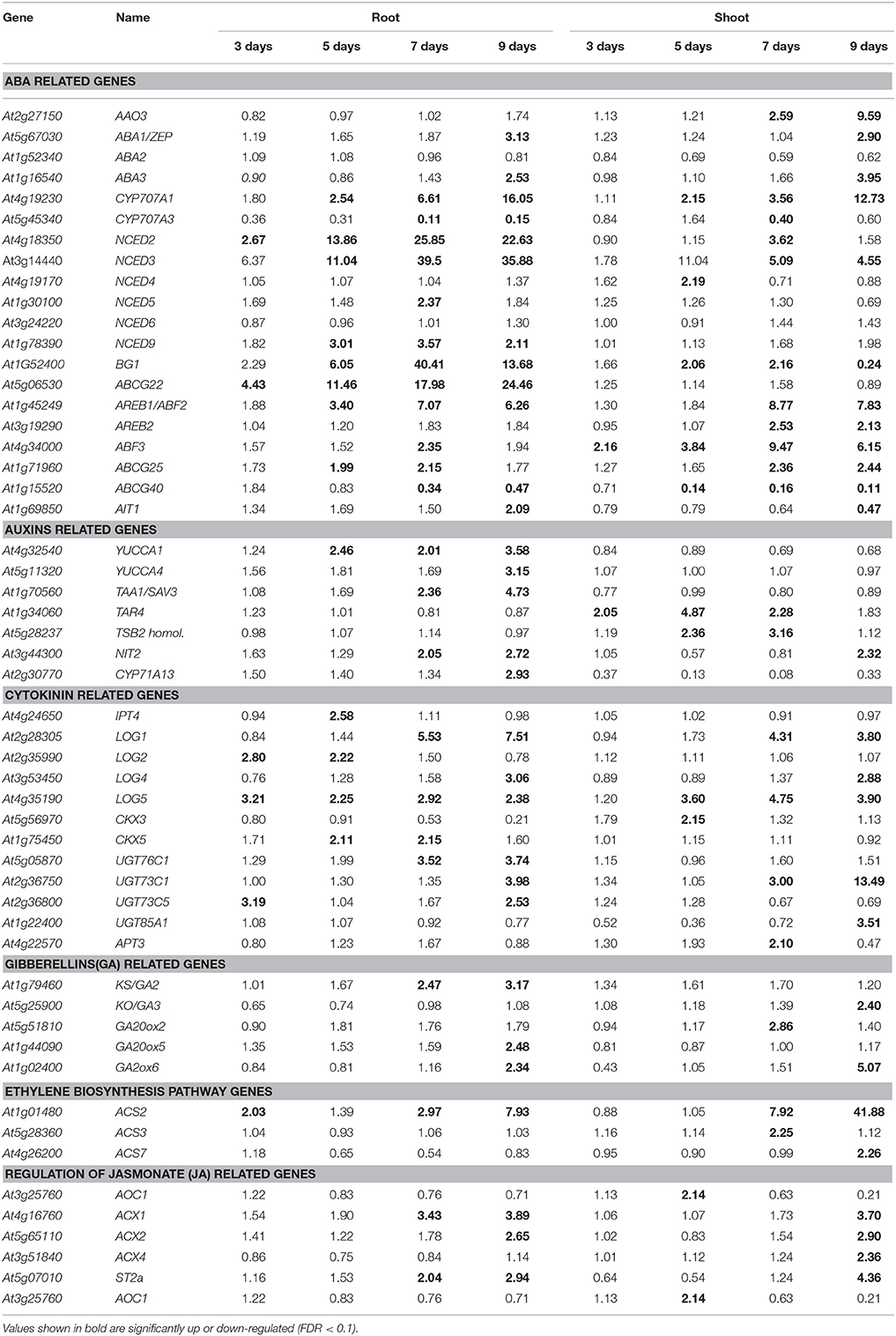
Transcriptional Changes in ABA and Other Hormone-Related Genes
A number of hormone-related genes were significantly up-regulated in roots and shoots (Table 2). Among genes in the ABA biosynthesis pathway, NCED2 was up-regulated on day 3, while CYP707A1 (AT4G19230), NCED3, and NCED9 (AT1G78390) were up-regulated in roots from day 5 to 9 of the drought stress (Table 2). Thus, it is reasonable to conclude that ABA biosynthesis was up-regulated around day 5 of the drought stress. The up-regulation of NCED2 and NCED3 occurred earlier in roots than in shoot tissue. In contrast, the expression of AAO3 was specifically up-regulated in shoots. The expression of transcription factors involved in ABA response also changed differentially in roots and shoots. The expression of AREB1/ABF2 (AT1G45249), AREB2/ABF4 (AT3G19290), and ABF3 (AT4G34000) was reported to be up-regulated in vegetative tissues in response to drought, high salinity, and ABA (Fujita et al., 2005). In the present study, the expression of AREB1/ABF2 was up-regulated in roots from day 5 to 9 of the drought stress, and from day 7 to 9 in shoots. The expression of AREB2/ABF4 was specifically up-regulated in shoots on day 7 and 9, while the expression of ABF3 was up-regulated in roots on day 7. On the other hand, it was up-regulated from day 3 to 9 of the drought stress in shoots (Table 2 and Table S3).
Similarly, the expression of transporters involved in ABA transport and ABA-induced stomatal closure also responded differently in roots and shoots. An increase in the expression of ABCG25 (AT1G71960) was observed in roots on day 7, while the increase in shoots occurred on day 7–9 of the drought stress. The expression of ABCG22 (AT5G06530) was specifically up-regulated in roots. The expression of ABCG40 (AT1G15520) was significantly down-regulated in both roots and shoots in response to the drought stress. Down-regulation of ABCG40 was observed in roots on days 7 and 9, and from day 5 to 9 in shoots (Table 2). The expression of the ABA transporter, AIT1 (AT1G69850), was significantly up-regulated in roots on day 9 of the drought stress, while it was significantly down-regulated in shoots (Table 2).
The expression of auxin biosynthesis-related genes also displayed differential patterns of expression in roots vs. shoots (Table 2). The expression of YUCCA1 (AT4G32540) and tryptophan aminotransferase of Arabidopsis 1 (TAA1/SAV3; AT1G70560) was specifically up-regulated in roots. The expression of NITrilase 2 (NIT2; AT3G44300) was up-regulated in roots on days 7 and 9 and only on day 9 in shoots (Table 2). The expression of tryptophan aminotransferase related 4 (TAR4; AT1G34060) and the Tryptophan Synthase Beta subunit (TSB2; AT5G28237) homolog was specifically up-regulated in shoots. It appears that auxins are up-regulated at later stages in roots in response to a drought response relative to ABA biosynthesis. Among the cytokinin biosynthesis-related genes, the expression of LOG2 (AT2G35990), CKX5 (AT1G75450), UGT76C1 (AT5G05870), and UGT73C5 (AT2G36800) was up-regulated specifically in roots, while the up-regulation of LOG5 (AT4G35190) was delayed in shoots compared to roots (Table 2). Among the gibberellin (GK) related genes, the expression of GA2 (AT1G79460) was up-regulated on days 7 and 9 of the drought stress, while the expression of GA20ox5 (AT1G44090) and GA20ox6 (AT1G02400) was specifically up-regulated on day 9. Among the ethylene biosynthesis-related genes, only the expression of ACS2 (AT1G01480) was up-regulated in roots from day 3 to 9 of the drought stress, while the expression of other ethylene biosynthesis-related genes in roots did not change in response to drought stress (Table 2). Among the jasmonate (JA) related genes, the expression of ACX1 (AT4G16760) and ST2a (AT5G07010) was up-regulated on days 7 and 9, while the expression of ACX2 (AT5G65110) was up-regulated only on day 9 in roots. The majority of genes involved in brassinosteroid synthesis were down-regulated.
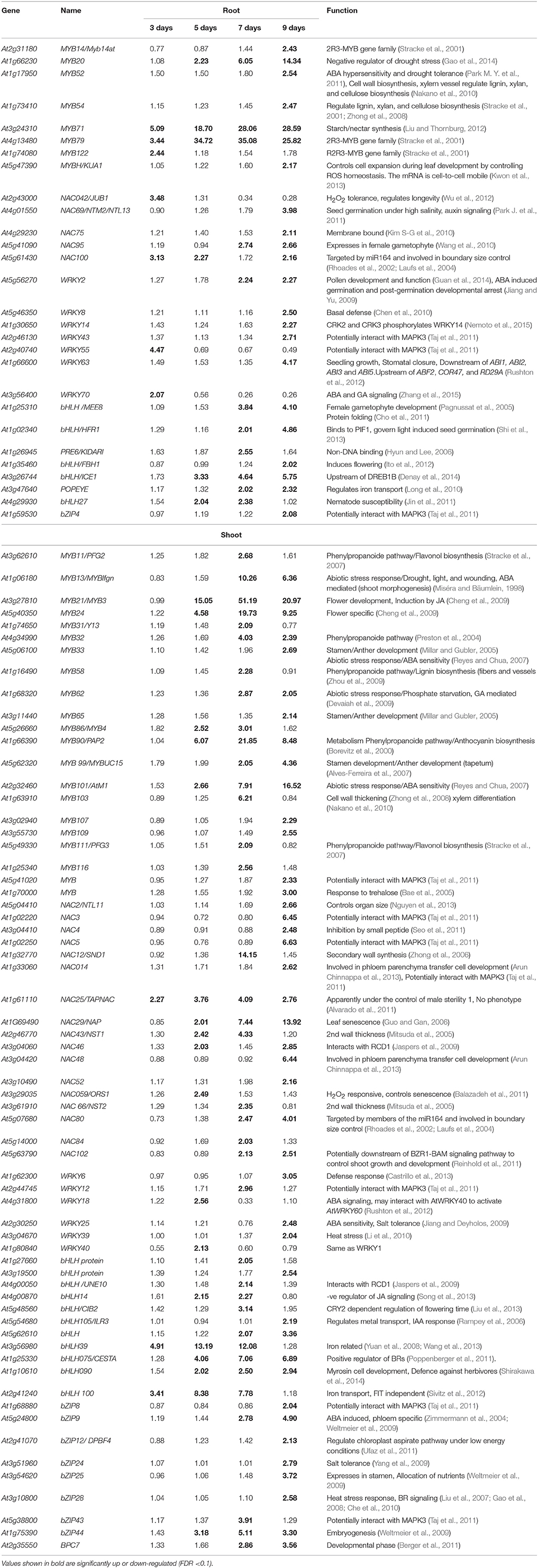
Changes in the Expression of Transcription Factors
The role of various transcription factors (TFs), such as DREB, AREB, MYC, and NAC, in regulating drought response has been previously reviewed (Yamaguchi-Shinozaki and Shinozaki, 2005, 2006; Nakashima et al., 2014). Therefore, changes in the expression of these transcription factors will not be discussed in detail. The present study focuses on TFs that were differentially up-regulated either in roots vs. shoots. The expression of eight MYB family members was specifically up-regulated in roots (Table 3). Among these, the expression of MYB79 (AT4G13480) and MYB71 (AT3G24310) was up-regulated on days 3–9, while MYB20 (AT1G66230) was up-regulated on days 7 and 9 of the drought stress. The expression of MYB122 (AT1G74080), was up-regulated only at the 3rd day of drought stress, while expression of MYB14 (AT2G31180), MYB52 (AT1G17950), MYB54 (AT1G73410), and MYBH (AT5G47390) was up-regulated only at the 9th day of drought stress. The expression of NAC95 (AT5G41090), WRKY2 (AT5G56270), and MEE8 (AT1G25310) was specifically up-regulated in roots on days 7 and 9. The expression of ICE1 (AT3G26744) was up-regulated in roots from day 5 to 9 day of the drought stress (Table 3).
Various TFs were also specifically up-regulated in shoots. The expression of NAC25/TAPNAC (AT1G61110) was significantly up-regulated from day 3 to 9, while the expression of bHLH100 (AT2G41240) was significantly elevated from day 3 to 7 of the drought stress (Table 3). The expression of MYB21 (AT3G27810), MYB24 (AT5G40350), MYB90 (AT1G66390), MYB101 (AT2G32460), NAC29 (AT1G69490), bHLH075/CESTA (AT1G25330), bHLH090 (AT1G10610), and bZIP44 (AT1G75390) significantly increased specifically in shoots from day 5 to 9 of the drought stress.
Changes in the Expression of Solute Transport-Related Genes
The expression of genes related to the transport of amino acids and other solutes including, malate, iron (Fe), and sulfur (S) changed significantly in both roots and shoots in response to the drought stress treatment. The expression of the malate transporters ALMT2 (AT1G08440), ALMT3 (AT1G18420), and ALMT10 (AT4G00910) was up-regulated in roots during the early stages of the drought stress (Figure 3 and Table S1). The expression of the sucrose family transporter gene SWEET15 (AT5G13170) was also significantly up-regulated in both roots and shoots (Figure 3). The expression of a MATE family member, ZRZ (ZRIZI; AT1G58340), which is involved in communicating a leaf-borne signal that determines the rate of organ initiation (Burko et al., 2011), was also up-regulated in roots.
Genes related to the transport of Fe, S, and other solutes were also differentially regulated in roots and shoots. Among these genes, those related to Fe transport were of particular interest. The expression of genes principally responsible for Fe uptake from the soil, i.e., iron regulated transporter 1 (IRT1; AT4G19690) and ferric reduction oxidase 2 (FRO2; AT1G01580) was significantly down-regulated in roots from day 5 to 9, indicating that plants were not uptaking Fe from soil during that time (Table S1). FRO2 reduces ferric to ferrous to increase its solubility and facilitates Fe uptake by IRT1 in plants (Jeong and Connolly, 2009). The expression of Fe transporter IRT3 (AT1G60960) was also down-regulated in roots on days 7 and 9, and on day 9 in shoots. On the other hand, genes regulating Fe distribution within a plant body were significantly up-regulated during the early stages of the drought stress. The expression of nicotianamine (NA) synthase 2 (NAS2: AT5G56080), which encodes a metal chelator NA, was up-regulated on day 3 and subsequently down-regulated on days 7 and 9. The expression of oligopeptide transporter 3 (OPT3; AT4G16370), involved in Fe distribution within a plant body (Stacey et al., 2008), was very significantly up-regulated in roots from day 3 to 7 of the drought stress. The expression of OPT3 in shoots was down-regulated on day 9. The expression of IRT2 (AT4G19680) was up-regulated in roots on day 3 and then down-regulated on days 7 and 9, while in shoots it was down-regulated on day 9 of the drought stress. The expression of a gene coding a Fe-S cluster biosynthesis family protein (AT2G36260) was significantly down-regulated in roots from day 5 to 9, while the expression of another gene coding an Fe-S cluster biosynthesis protein (AT2G16710) increased in both roots and shoots (Table S1). The expression of a mitochondrial Fe reductase, FRO8 (AT5G50160) increased in roots from day 5 to 9. The expression of the metal transporters YSL2 (AT5G24380) and VIT1 (AT2G01770) increased in roots on day 7 of the drought stress (Table S1), while the expression of FRO4 (AT5G23980) decreased in roots and increased in shoots (Tables S2, S3). Many bHLH transcription factors reported to be involved in Fe homeostasis were differentially regulated in roots and shoots. The expression of the transcription factors regulating Fe uptake/translocation POPEYE (AT3G47640) and BRUTUS (BTS; AT3G18290) was up-regulated in roots on days 7 and 9 (Table 3 and Table S1), while the expression of bHLH115 (AT1G51070) was down-regulated in roots on days 7 and 9 (Table S2). The expression of bHLH38 (AT3G56970) increased in roots and shoots from day 5 to 7, while that of bHLH39 (AT3G56980) increased specifically in shoots from day 3 to 7 of the drought stress. The expression of bHLH101 (AT5G04150) increased in roots on day 3, and from day 5 to 9 in shoots. Collectively, these results suggest that Fe distribution within a plant body can significantly change during the course of a drought stress.
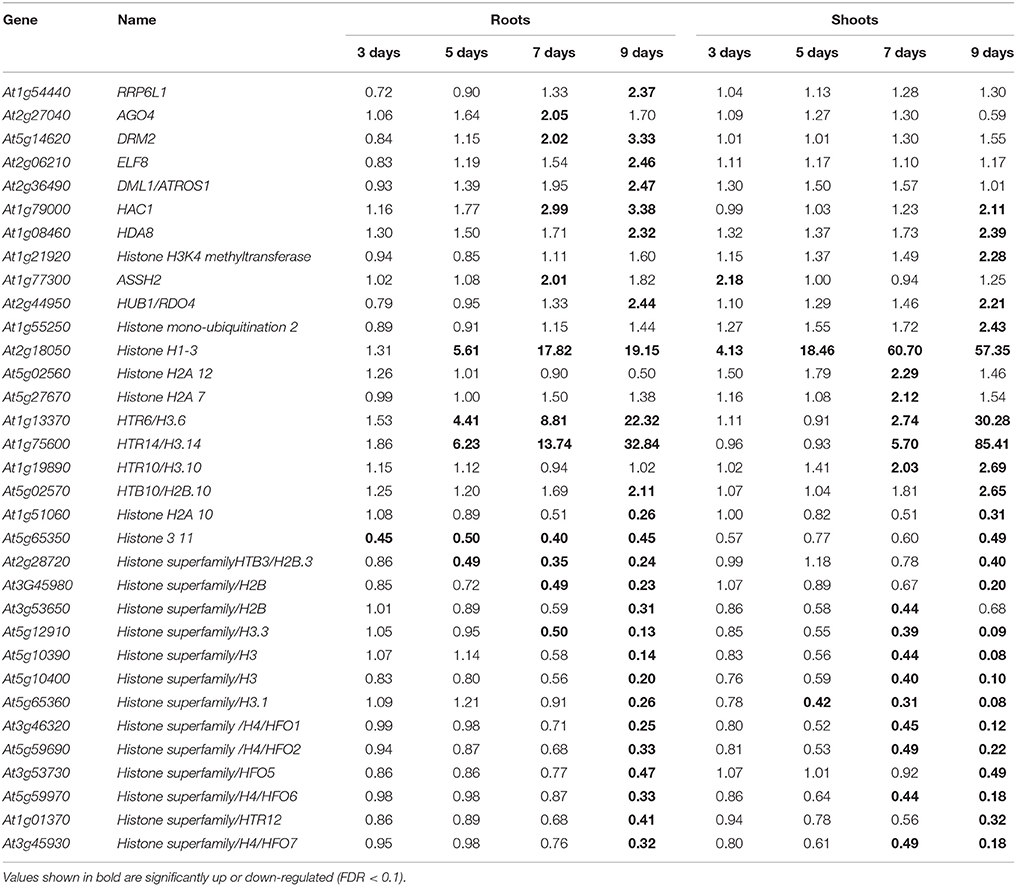
Changes in the Expression of Genes Related to Transcription Regulation and Chromatin Synthesis/Modification
In Arabidopsis, genes controlling epigenetic changes that occur in response to abiotic stresses have been reported (Kim et al., 2015). In the present study, we focused on the differential expression of genes related to chromatin structure or chromatin modification in both roots and shoots. The expression of AtRRP6L1 (AT1G54440), which controls DNA methylation; Early Flowering 8 (ELF8; AT2G06210), which is putatively involved in regulating gene expression; and Demeter Like 1 (DML1; AT2G36490), a repressor of transcriptional silencing; was significantly up-regulated in roots on day 9 of the drought stress. The expression of AGO4 (AT2G27040), which is involved in siRNA-mediated gene silencing, was up-regulated in roots on day 7; and DRM2 (AT5G14620; methyl transferase) was up-regulated on days 7 and 9 of the drought stress (Table 4). The expression of Histone DeAcetylase 8 (HDA8; AT1G08460) was up-regulated in both roots and shoots on day 9. Changes in the expression of various histone protein-related genes were also observed. The expression of histone H1-3 (AT2G18050) was significantly up-regulated in roots from day 5 to 9, and from day 3 to 9 in shoots. On the other hand, the up-regulation of HTR6/H3.6 (AT1G13370) and HTR14/H3.14 (AT1G75600) occurred earlier in roots than in shoots. The expression of HTR10/H3.10 (AT1G19890) was specifically up-regulated in shoots (Table 4). Many histone-related genes were also significantly down-regulated in both roots and shoots (Table 4), indicating that chromatin structure changes significantly in plants under drought stress conditions.
Discussion
Transcriptomic changes in Arabidopsis in response to drought stress have been previously reported (Kreps et al., 2002; Seki et al., 2002, 2007; Shinozaki et al., 2003; Huang et al., 2008; Matsui et al., 2008). Global changes of gene expression from both roots and shoots of drought-stressed Arabidopsis plants under soil conditions, however, has not been investigated. Thus, our data provide new information pertaining to the differential regulation of genes in shoots vs. roots in response to drought stress. It should be noted that plants started flowering during drought stress (Figure S1), thus numerous genes and transcription factors related to flowering were also up-regulated in shoots. This could be a potential interference in understanding the drought responsive genes, particularly in shoot tissue.
Root to Shoot Signaling During a Drought Stress
The differential regulation of ABA biosynthesis- and transport-related genes highlights the importance of root to shoot signaling in response to drought stress. NCEDs are considered to be limiting factors in ABA synthesis and signaling, and the suppression of NCED3 results in severe sensitivity to drought (Iuchi et al., 2001). The expression of NCED3 has been reported to be up-regulated in both roots and shoots in response to drought stress (Behnam et al., 2013). In the current study, the expression of NCED5 and NCED9 was specifically up-regulated in roots. Additionally, the induction of NCED2 in roots occurred earlier in the response to drought stress than it did in shoots (Table 1). The Arabidopsis genes involved in ABA transport have also been characterized. ABCG25 is a drought- and ABA-inducible plasma membrane protein that exports ABA from the vascular system (Kuromori et al., 2010). Our data indicated that expression of ABCG25 was up-regulated in both roots and shoots by the drought stress. ABCG40 is a plasma membrane ABA influx transporter, which is highly expressed in guard cells (Kang et al., 2010). ABCG40 knockout mutants (atabcg40) exhibit defects in stomatal closure in response to osmotic stress and application of ABA (Kang et al., 2010). In the current data, the expression of ABCG40 was significantly down-regulated in both roots and shoots in response to drought stress (Table 1). The down-regulation of ABCG40 in roots was observed on days 7 and 9 of the drought stress, whereas it was down-regulated from day 5 to 9 in shoots (Table 2).
AIT1, a member of the nitrate transporter gene family, also transports ABA (Kanno et al., 2012) and its expression was differentially regulated in roots and shoots in the current study (Table 1). Our data indicates that AIT1, ABCG25, and ABCG40 are differentially regulated in response to drought stress, thus it will be important to determine if additional ABA transporters are involved in ABA transport in response to drought stress. Arabidopsis ABA-deficient mutants are more sensitive to drought stress than abcg25 and abcg40 mutants, suggesting that additional transporters with redundant functions may also be involved in ABA transport (Osakabe et al., 2014). Passive ABA transport may also contribute to signaling (Seo and Koshiba, 2011). The specific up-regulation of NCED5 and NCED9 in roots, as well as the earlier induction of NCED2 in roots than in shoots, indicate that ABA signaling may originate in roots. It has been suggested that the root to shoot transport of ABA is not required since ABA produced in leaves effectively triggers ABA signaling and stomatal closure (Christmann et al., 2007). The differential up-regulation of genes involved in ABA synthesis and transport in roots vs. shoots suggest that ABA may be transported from roots to shoots. The specific up-regulation of ABCG22 in roots further supports this idea. Although the substrate of ABCG22 has not been determined, ABCG22 has been reported to be involved in the regulation of stomata, and knock down mutants of ABCG22 (atabcg22) exhibit lower leaf temperature and are drought sensitive (Kuromori et al., 2011). While ABCG22 has been reported to be expressed in aerial organs (Kuromori et al., 2011), the specific up-regulation of ABCG22 in roots in response to a drought stress in the current study suggests that it may also be involved in root to shoot signaling to control stomatal closure.
Transcription Factors Differentially Regulate the Transcriptome in Roots and Shoots
Changes in the expression of various transcription factors were observed in roots and shoots. As plants were flowering during drought stress, the expression of various TF putatively involved in response to flowering was also up-regulated in shoots (Table 3). The role of MYB family transcription factors in controlling primary and secondary metabolism, development, cell fate and identity, and responses to different biotic and abiotic stresses has been reported (Lippold et al., 2009; Zhou et al., 2009; Dubos et al., 2010; Nakano et al., 2010; Park M. Y. et al., 2011; Katiyar et al., 2012; Liu and Thornburg, 2012; Wang and Dixon, 2012; Arun Chinnappa et al., 2013; Kwon et al., 2013; Gao et al., 2014; Kosma et al., 2014; Baldoni et al., 2015). In addition to being involved in floral development, MYB transcription factors also play a significant role in plant adaptation to drought stress, including the regulation of stomatal movement, and the induction of suberin synthesis in cuticles (Lippold et al., 2009; Park M. Y. et al., 2011; Gao et al., 2014; Baldoni et al., 2015). Synthesis of lignin and suberin in plants is controlled by VND6 and SND1, and the involvement of many MYB proteins in this process has been previously reported (Ohashi-Ito et al., 2010). MYB58 and MYB63 are known to regulate the lignin biosynthetic pathway (Zhou et al., 2009), while MYB52, MYB54, MYB85, MYB42, MYB43, MYB69, and MYB20 have been suggested to be involved in the regulation of secondary cell wall synthesis (Zhong et al., 2008). In the current study, the expression of MYB61, which influences lignin deposition (Newman et al., 2004), was up-regulated in both roots and shoots from day 5 to 7 of the drought stress. MYB41 has been recognized as a key regulator in cell wall expansion and modification under stress conditions (Lippold et al., 2009; Kosma et al., 2014). The up-regulation of MYB41 was observed in roots on day 7 and 9 and on day 9 in shoots.
Since various genes putatively involved in lignin and suberin biosynthesis, and secondary wall modifications were up-regulated in roots from day 3 to 9 of the drought stress; it seems reasonable that other MYB proteins may also be controlling the lignin/suberin biosynthesis in roots. The expression of MYB20 was specifically up-regulated in roots on days 7 and 9 of the drought stress. ABA-dependent stomatal closure is impaired in plants over expressing MYB20, resulting in an increased susceptibility to drought stress. An opposite phenotype is associated with a MYB20 knockout mutation, indicating that MYB20 may act as a negative regulator of ABA-mediated stomatal closure (Gao et al., 2014). It is plausible that specific up-regulation of MYB20 in roots may be involved in ABA sensing or signaling. The protein bHLH122 plays an important role in drought and osmotic stress tolerance in Arabidopsis and in the repression of ABA catabolism. bHLH122 can bind directly to G-box/E-box cis-elements in the CYP707A3 promoter and repress its expression. Furthermore, up-regulation of bHLH122 substantially increases cellular ABA levels (Liu et al., 2014). The expression of bHLH122 was up-regulated in roots from day 3 to 9 of the drought stress and from day 5 to 9 in shoots. Importantly, the suppression of CYP707A3 was also observed in roots and shoots (Table 1). We suggest that the differential regulation of MYB (particularly MYB71 and MYB79), bHLHs (such as ICE1, bHLH27, bHLH075, bHLH090, bHLH100), WRKY, and NAC transcription factors in both roots and shoots (Table 3), indicate that these transcription factors may differentially regulate root and shoot response to a drought stress.
Genes Related to Osmoprotectant Synthesis and Solute Transport Are Differentially Regulated in Roots and Shoots in Response to a Drought Stress
The expression of a variety of genes involved in the synthesis of proline, galactinol, and raffinose were differentially expressed in roots and shoots in response to the drought stress. Our data confirms that the expression of GolS1 and GolS2 is up-regulated by drought stress (Taji et al., 2002). In addition, our microarray analysis revealed that the expression of GolS4 was significantly up-regulated in roots from day 3 to 9 (Table 1). Differential regulation of genes involved in proline synthesis in roots and shoots was also observed in the current study. The expression of P5CS1 was up-regulated in roots and shoots from day 5 to 9 of the drought stress (Table S1), while the expression of P5CS2 was specifically up-regulated in shoots on days 7 and 9 (Table S3).
Malate and mannitol concentrations change in response to a water deficit and have been suggested to play a prominent role in osmotic adjustment in response to a water deficit (Lance and Rustin, 1984; Popp and Polania, 1989; Tarczynski et al., 1993; Martinoia and Rentsch, 1994; Tschaplinski and Tuskan, 1994; Karakas et al., 1997). Our results indicate that expression levels of various malate transporters, MATE family efflux transporters and MSF transporters were significantly up-regulated in roots (Table S1). The MATE and MSF family members transport a diverse range of substrates. These results indicate that during early drought stress, several transporters putatively involved in malate, amino acids, and ion transport are up-regulated and that the up-regulation of these transporters in roots could contribute to osmotic adjustments and stress signaling. The importance of differential regulation of S metabolism under drought stress has been well recognized (Chan et al., 2013), however, changes in Fe metabolism in response to drought stress has not been extensively discussed. Iron deficiency triggers a complex set of reactions in plants in order to increase Fe uptake from the soil, including developmental and physiological changes. Over the past decade, many transporters in Arabidopsis involved in the absorption and distribution of Fe have been identified (Conte and Walker, 2011). The transcription factor FIT1 (bHLH029) controls the expression of the Fe uptake machinery genes FRO2 and IRT1 in roots. In the current study, the expression of FIT1 was down-regulated in roots from day 5 to 9 of the drought stress and on day 9 in shoots. Similarly, the expression of FRO2, IRT1, and IRT3 was also down-regulated. In contrast, the expression of bHLH38, bHLH39, bHLH100, and bHLH101 was significantly up-regulated. bHLH038 and bHLH039 interact with FIT, while bHLH100 and bHLH101 do not regulate FIT target genes and are reported to play a crucial role in the distribution of Fe within a plant (Yuan et al., 2008; Sivitz et al., 2012; Kobayashi et al., 2014). BTS is a negative regulator of Fe deficiency response and interacts with bHLH104, ILR3, and bHLH115 (Long et al., 2010). Down-regulation of FIT-dependent response and up-regulation of NAS2, OPT3, IRT2, YSL2, and FRO8 suggest that the distribution of Fe within plant/cell changes significantly in response to a drought stress. There is increasing evidence that the genes involved in Fe deficiency response in plants are regulated by different plant hormones such as ABA, auxin, ethylene, GK and JA (Kobayashi et al., 2014). ABA improves Fe utilization by increasing root to shoot translocation of Fe under Fe deficiency (Lei et al., 2014). It would be interesting to investigate if the root to shoot translocation of Fe and ABA is synchronized under drought stress.
Changes in the availability of Fe significantly alters plants metabolism and could trigger localized signals (Bashir et al., 2011; Vigani et al., 2013b, 2016). Our data indicate that the expression of several 2OG-Fe(II) oxygenases was up-regulated in roots during the early stages of the drought stress. In plants, 2OG-Fe(II) oxygenases are involved in the synthesis of phytosiderophores (Nakanishi et al., 2000) and numerous other biosynthetic pathways. It was recently suggested that plant 2OG-Fe(II) oxygenases may play a role in Fe sensing and metabolic reprogramming in response to Fe-deficient conditions (Vigani et al., 2013a; Bashir et al., 2014). The up-regulation of different 2OG dioxygenases in roots observed in the current study suggests that these genes may also be involved in signaling under drought stress conditions.
Changes in the Expression of Genes Related to Chromatin Synthesis/Modification
Transcriptional and post-transcriptional regulation of RNA facilitates the adjustment of plants to various abiotic stresses. Small RNAs, alternative splicing, and RNA-binding proteins are known to regulate plant stress responses (Nakaminami et al., 2012). Differential changes in the expression of various genes related to these mechanisms were observed in roots and shoots (Table 4). Modifications in chromatin structure could also significantly alter gene expression in plants responding to different abiotic stresses (Chinnusamy and Zhu, 2009; Kim J-M et al., 2010; Kim et al., 2012, 2015). The differential expression of genes involved in RNA regulation, histone modification, and several other histone-related genes observed in the current study (Table 4) indicates that epigenetic responses to a drought stress may also be differentially controlled in roots and shoots. Moreover, in addition to genes involved in acetylation, methylation, and demethylation; changes in the expression of genes encoding different histone proteins could also contribute to transcriptional changes that occur in response to a drought stress.
Conclusions
Current studies indicate that a large number of genes belonging to diverse functional groups are differentially regulated in roots and shoots in response to a progressive drought stress. Thus, dissecting the root and shoot transcriptome may provide novel insights to understand the regulation of genes in response to different abiotic stresses. Transcriptional changes during early drought stress in roots were of particular interest. Genes involved in ABA synthesis, ABA and solute transport were up-regulated during early drought stress in roots. Various members of MFS transporters family, MATE efflux transporters, microRNA genes, suberin, pectin and secondary cell wall biosynthesis/modification-related genes, pre-tRNA genes, and various S-adenosyl-L-methionine (SAM) dependent transferases were also significantly up-regulated in roots. Moreover, our data also revealed the differential regulation of several genes involved in drought stress response and chromatin changes. The identification of genes that are highly responsive at the early stages of a drought stress, and that are differentially regulated in roots and shoots, significantly advances our understanding about early drought stress response in roots and shoots. These results can serve as an aid in the selection of root- and shoot-specific genes/promoters that could be utilized to potentially develop drought tolerant plants through molecular breeding.
Author Contributions
SR, KB, AM, and MS designed the study, SR, KB, and MT performed the research. AM, SR, and KB, analyzed the data, SR, KB, AM, and MS discussed the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Japan Science and Technology Agency (JST), Core Research for Evolutionary Science and Technology (CREST), grants from RIKEN, Japan [to MS]. KB is supported through the RIKEN FPR program and SR is supported by the RIKEN JRA program. We are grateful to Kei Iida (Graduate School of Medicine, Kyoto University) for supporting our microarray analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00180
References
Alvarado, V. Y., Tag, A., and Thomas, T. L. (2011). A cis regulatory element in the TAPNAC promoter directs tapetal gene expression. Plant Mol. Biol. 75, 129–139. doi: 10.1007/s11103-010-9713-5
Alves-Ferreira, M., Wellmer, F., Banhara, A., Kumar, V., Riechmann, J. L., and Meyerowitz, E. M. (2007). Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145, 747–762. doi: 10.1104/pp.107.104422
Arun Chinnappa, K. S., Nguyen, T. T. S., Hou, J., Wu, Y., and McCurdy, D. W. (2013). Phloem parenchyma transfer cells in Arabidopsis – an experimental system to identify transcriptional regulators of wall ingrowth formation. Front. Plant Sci. 4:102. doi: 10.3389/fpls.2013.00102
Bae, H., Herman, E., Bailey, B., Bae, H.-J., and Sicher, R. (2005). Exogenous trehalose alters Arabidopsis transcripts involved in cell wall modification, abiotic stress, nitrogen metabolism, and plant defense. Physiol. Plant. 125, 114–126. doi: 10.1111/j.1399-3054.2005.00537.x
Balazadeh, S., Kwasniewski, M., Caldana, C., Mehrnia, M., Zanor, M. I., Xue, G.-P., et al. (2011). ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4, 346–360. doi: 10.1093/mp/ssq080
Baldoni, E., Genga, A., and Cominelli, E. (2015). Plant MYB transcription factors: their role in drought response mechanisms. Int. J. Mol. Sci. 16, 15811–15851. doi: 10.3390/ijms160715811
Bashir, K., Hanada, K., Shimizu, M., Seki, M., Nakanishi, H., and Nishizawa, N. K. (2014). Transcriptomic analysis of rice in response to iron deficiency and excess. Rice 7, 18. doi: 10.1186/s12284-014-0018-1
Bashir, K., Ishimaru, Y., Shimo, H., Nagasaka, S., Fujimoto, M., Takanashi, H., et al. (2011). The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun. 2, 322. doi: 10.1038/ncomms1326
Behnam, B., Iuchi, S., Fujita, M., Fujita, Y., Takasaki, H., Osakabe, Y., et al. (2013). Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res. 20, 315–324. doi: 10.1093/dnares/dst012
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
Berger, N., Dubreucq, B., Roudier, F., Dubos, C., and Lepiniec, L. (2011). Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23, 4065–4078. doi: 10.1105/tpc.111.087866
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2393. doi: 10.1105/tpc.12.12.2383
Burko, Y., Geva, Y., Refael-Cohen, A., Shleizer-Burko, S., Shani, E., Berger, Y., et al. (2011). From organelle to organ: ZRIZI MATE-type transporter is an organelle transporter that enhances organ initiation. Plant Cell Physiol. 52, 518–527. doi: 10.1093/pcp/pcr007
Carpita, N. C. (2011). Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1 → 4)-β-D-glycans. Plant Physiol. 155, 171–184. doi: 10.1104/pp.110.163360
Castrillo, G., Sánchez-Bermejo, E., de Lorenzo, L., Crevillén, P., Fraile-Escanciano, A., Tc, M., et al. (2013). WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 25, 2944–2957. doi: 10.1105/tpc.113.114009
Chan, K. X., Wirtz, M., Phua, S. Y., Estavillo, G. M., and Pogson, B. J. (2013). Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci. 18, 18–29. doi: 10.1016/j.tplants.2012.07.005
Chazen, O., and Neumann, P. M. (1994). Hydraulic signals from the roots and rapid cell-wall hardening in growing maize (Zea mays L.) leaves are primary responses to polyethylene glycol-induced water deficits. Plant Physiol. 104, 1385–1392.
Che, P., Bussell, J. D., Zhou, W., Estavillo, G. M., Pogson, B. J., and Smith, S. M. (2010). Signaling from the endoplasmic reticulum activates brassinosteroid. Sci. Signal. 3, ra69 doi: 10.1126/scisignal.2001140
Chen, L., Zhang, L., and Yu, D. (2010). Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant Microbe Interact. 23, 558–565. doi: 10.1094/MPMI-23-5-0558
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., et al. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5:e1000440. doi: 10.1371/journal.pgen.1000440
Chinnusamy, V., and Zhu, J.-K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Cho, E. J., Yuen, C. Y., Kang, B.-H., Ondzighi, C. A., Staehelin, L. A., and Christopher, D. A. (2011). Protein disulfide isomerase-2 of Arabidopsis mediates protein folding and localizes to both the secretory pathway and nucleus, where it interacts with maternal effect embryo arrest factor. Mol. Cells 32, 459–475. doi: 10.1007/s10059-011-0150-3
Christmann, A., Weiler, E. W., Steudle, E., and Grill, E. (2007). A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 52, 167–174. doi: 10.1111/j.1365-313X.2007.03234.x
Conte, S. S., and Walker, E. L. (2011). Transporters contributing to iron trafficking in plants. Mol. Plant 4, 464–476. doi: 10.1093/mp/ssr015
Denay, G., Creff, A., Moussu, S., Wagnon, P., Thévenin, J., Gérentes, M.-F., et al. (2014). Endosperm breakdown in Arabidopsis requires heterodimers of the basic helix-loop-helix proteins ZHOUPI and INDUCER OF CBP EXPRESSION 1. Development 141, 1222–1227. doi: 10.1242/dev.103531
Devaiah, B. N., Madhuvanthi, R., Karthikeyan, A. S., and Raghothama, K. G. (2009). Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2, 43–58. doi: 10.1093/mp/ssn081
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Gao, H., Brandizzi, F., Benning, C., and Larkin, R. M. (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 16398–16403. doi: 10.1073/pnas.0808463105
Gao, S., Zhang, Y. L., Yang, L., Song, J. B., and Yang, Z. M. (2014). AtMYB20 is negatively involved in plant adaptive response to drought stress. Plant Soil 376, 433–443. doi: 10.1007/s11104-013-1992-6
Guan, Y., Meng, X., Khanna, R., LaMontagne, E., Liu, Y., and Zhang, S. (2014). Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 10:e1004384. doi: 10.1371/journal.pgen.1004384
Guo, Y., and Gan, S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Harb, A., Krishnan, A., Ambavaram, M. M., and Pereira, A. (2010). Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154, 1254–1271. doi: 10.1104/pp.110.161752
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Huang, D., Wu, W., Abrams, S. R., and Cutler, A. J. (2008). The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007. doi: 10.1093/jxb/ern155
Hyun, Y., and Lee, I. (2006). KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61, 283–296. doi: 10.1007/s11103-006-0010-2
Ito, S., Song, Y. H., Josephson-Day, A. R., Miller, R. J., Breton, G., Olmstead, R. G., et al. (2012). FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 3582–3587. doi: 10.1073/pnas.1118876109
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jaspers, P., Blomster, T., Brosche, M., Salojärvi, J., Ahlfors, R., Vainonen, J. P., et al. (2009). Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 60, 268–279. doi: 10.1111/j.1365-313X.2009.03951.x
Jeong, J., and Connolly, E. L. (2009). Iron uptake mechanisms in plants: functions of the FRO family of ferric reductases. Plant Sci. 176, 709–714. doi: 10.1016/j.plantsci.2009.02.011
Jiang, W., and Yu, D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9:96. doi: 10.1186/1471-2229-9-96
Jiang, Y., and Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Jin, J., Hewezi, T., and Baum, T. J. (2011). The Arabidopsis bHLH25 and bHLH27 transcription factors contribute to susceptibility to the cyst nematode Heterodera schachtii. Plant J. 65, 319–328. doi: 10.1111/j.1365-313X.2010.04424.x
Kang, J., Hwang, J.-U., Lee, M., Kim, Y.-Y., Assmann, S. M., Martinoia, E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360. doi: 10.1073/pnas.0909222107
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Karakas, B., Ozias-Akins, P., Stushnoff, C., Suefferheld, M., and Rieger, M. (1997). Salinity and drought tolerance of mannitol-accumulating transgenic tobacco. Plant Cell Environ. 20, 609–616. doi: 10.1111/j.1365-3040.1997.00132.x
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kim, J.-M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kim, J.-M., To, T. K., Ishida, J., Matsui, A., Kimura, H., and Seki, M. (2012). Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53, 847–856. doi: 10.1093/pcp/pcs053
Kim, J.-M., To, T. K., Nishioka, T., and Seki, M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33, 604–611. doi: 10.1111/j.1365-3040.2009.02076.x
Kim, S.-G., Lee, S., Seo, P. J., Kim, S.-K., Kim, J.-K., and Park, C.-M. (2010). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95, 56–65. doi: 10.1016/j.ygeno.2009.09.003
Klie, S., and Nikoloski, Z. (2012). The choice between MapMan and Gene Ontology for automated gene function prediction in plant science. Front. Genet. 3:115. doi: 10.3389/fgene.2012.00115
Kobayashi, T., Nakanishi Itai, R., and Nishizawa, N. (2014). Iron deficiency responses in rice roots. Rice 7, 27. doi: 10.1186/s12284-014-0027-0
Kosma, D. K., Murmu, J., Razeq, F. M., Santos, P., Bourgault, R., Molina, I., et al. (2014). AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J. 80, 216–229. doi: 10.1111/tpj.12624
Kreps, J. A., Wu, Y., Chang, H.-S., Zhu, T., Wang, X., and Harper, J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141. doi: 10.1104/pp.008532
Kuromori, T., Miyaji, T., Yabuuchi, H., Shimizu, H., Sugimoto, E., Kamiya, A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. U.S.A. 107, 2361–2366. doi: 10.1073/pnas.0912516107
Kuromori, T., Sugimoto, E., and Shinozaki, K. (2011). Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 67, 885–894. doi: 10.1111/j.1365-313X.2011.04641.x
Kwon, Y., Kim, J. H., Nguyen, H. N., Jikumaru, Y., Kamiya, Y., Hong, S.-W., et al. (2013). A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 64, 3911–3922. doi: 10.1093/jxb/ert223
Lance, C., and Rustin, P. (1984). The central role of malate in plant metabolism. Physiol. Végétale 22, 625–641.
Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. doi: 10.1242/dev.01320
Lei, G. J., Zhu, X. F., Wang, Z. W., Dong, F., Dong, N. Y., and Zheng, S. J. (2014). Abscisic acid alleviates iron deficiency by promoting root iron reutilization and transport from root to shoot in Arabidopsis. Plant Cell Environ. 37, 852–863. doi: 10.1111/pce.12203
Li, S., Zhou, X., Chen, L., Huang, W., and Yu, D. (2010). Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 29, 475–483. doi: 10.1007/s10059-010-0059-2
Lippold, F., Sanchez, D. H., Musialak, M., Schlereth, A., Scheible, W.-R., Hincha, D. K., et al. (2009). AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 149, 1761–1772. doi: 10.1104/pp.108.134874
Liu, G., and Thornburg, R. W. (2012). Knockdown of MYB305 disrupts nectary starch metabolism and floral nectar production. Plant J. 70, 377–388. doi: 10.1111/j.1365-313X.2011.04875.x
Liu, J.-X., Srivastava, R., Che, P., and Howell, S. H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19, 4111–4119. doi: 10.1105/tpc.106.050021
Liu, W., Tai, H., Li, S., Gao, W., Zhao, M., Xie, C., et al. (2014). bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 201, 1192–1204. doi: 10.1111/nph.12607
Liu, Y., Li, X., Li, K., Liu, H., and Lin, C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9:e1003861. doi: 10.1371/journal.pgen.1003861
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., and Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236. doi: 10.1105/tpc.110.074096
Martinoia, E., and Rentsch, D. (1994). Malate compartmentation-responses to a complex metabolism. Annu. Rev. Plant Biol. 45, 447–467. doi: 10.1146/annurev.pp.45.060194.002311
Matsui, A., Ishida, J., Morosawa, T., Mochizuki, Y., Kaminuma, E., Endo, T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. doi: 10.1093/pcp/pcn101
Mi, H., Muruganujan, A., and Thomas, P. D. (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386. doi: 10.1093/nar/gks1118
Millar, A. A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Miséra, S., and Bäumlein, H. (1998). Ectopic expression of a novel MYB gene modifies the architecture of the Arabidopsis inflorescence. Plant J. 13, 729–742. doi: 10.1046/j.1365-313X.1998.00072.x
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Nakaminami, K., Matsui, A., Shinozaki, K., and Seki, M. (2012). RNA regulation in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 149–153. doi: 10.1016/j.bbagrm.2011.07.015
Nakano, Y., Nishikubo, N., Goué, N., Ohtani, M., Yamaguchi, M., Katayama, Y., et al. (2010). MYB transcription factors orchestrating the developmental program of xylem vessels in Arabidopsis roots. Plant Biotechn. 27, 267–272. doi: 10.5511/plantbiotechnology.27.267
Nakanishi, H., Yamaguchi, H., Sasakuma, T., Nishizawa, N. K., and Mori, S. (2000). Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Mol. Biol. 44, 199–207. doi: 10.1023/A:1006491521586
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Nemoto, K., Takemori, N., Seki, M., Shinozaki, K., and Sawasaki, T. (2015). Members of the plant CRK-superfamily are capable of trans-/auto-phosphorylation of tyrosine residues. J. Biol. Chemis. 290, 16665–16677. doi: 10.1074/jbc.M114.617274
Newman, L. J., Perazza, D. E., Juda, L., and Campbell, M. M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37, 239–250. doi: 10.1046/j.1365-313X.2003.01953.x
Nguyen, A. H., Matsui, A., Tanaka, M., Mizunashi, K., Nakaminami, K., Hayashi, M., et al. (2015). Loss of Arabidopsis 5′–3′ exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol. 56, 1762–1772. doi: 10.1093/pcp/pcv096
Nguyen, H. M., Schippers, J. H., Gõni−Ramos, O., Christoph, M. P., Dortay, H., Hoorn, R. A., et al. (2013). An upstream regulator of the 26S proteasome modulates organ size in Arabidopsis thaliana. Plant J. 74, 25–36. doi: 10.1111/tpj.12097
Ohashi-Ito, K., Oda, Y., and Fukuda, H. (2010). Arabidopsis vascular-related nac-domain6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. doi: 10.1105/tpc.110.075036
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L.-S. P. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Pagnussat, G. C., Yu, H.-J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. doi: 10.1242/dev.01595
Park, J., Kim, Y.-S., Kim, S.-G., Jung, J.-H., Woo, J.-C., and Park, C.-M. (2011). Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 156, 537–549. doi: 10.1104/pp.111.177071
Park, M. Y., Kang, J.-,y., and Kim, S. Y. (2011). Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol. Cells 31, 447–454. doi: 10.1007/s10059-011-0300-7
Popp, M., and Polania, J. (1989). Compatible solutes in different organs of mangrove trees. Ann. Sci. Forest. 46, 842s–844s. doi: 10.1051/forest:198905ART0185
Poppenberger, B., Rozhon, W., Khan, M., Husar, S., Adam, G., Luschnig, C., et al. (2011). CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30, 1149–1161. doi: 10.1038/emboj.2011.35
Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., and Parish, R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995. doi: 10.1111/j.1365-313X.2004.02280.x
Rampey, R. A., Woodward, A. W., Hobbs, B. N., Tierney, M. P., Lahner, B., Salt, D. E., et al. (2006). An Arabidopsis Basic Helix-Loop-Helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174, 1841–1857. doi: 10.1534/genetics.106.061044
Reinhold, H., Soyk, S., Šimková, K., Hostettler, C., Marafino, J., Mainiero, S., et al. (2011). β-Amylase–Like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 23, 1391–1403. doi: 10.1105/tpc.110.081950
Reyes, J. L., and Chua, N.-H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Seki, M., Narusaka, M., Ishida, J., Nanjo, T., Fujita, M., Oono, Y., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. doi: 10.1046/j.1365-313X.2002.01359.x
Seki, M., Umezawa, T., Urano, K., and Shinozaki, K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10, 296–302. doi: 10.1016/j.pbi.2007.04.014
Seo, M., and Koshiba, T. (2011). Transport of ABA from the site of biosynthesis to the site of action. J. Plant Res. 124, 501–507. doi: 10.1007/s10265-011-0411-4
Seo, P. J., Hong, S.-Y., Kim, S.-G., and Park, C.-M. (2011). Competitive inhibition of transcription factors by small interfering peptides. Trends Plant Sci. 16, 541–549. doi: 10.1016/j.tplants.2011.06.001
Sharp, R. E., Poroyko, V., Hejlek, L. G., Spollen, W. G., Springer, G. K., Bohnert, H. J., et al. (2004). Root growth maintenance during water deficits: physiology to functional genomics. J. Exp. Bot. 55, 2343–2351. doi: 10.1093/jxb/erh276
Shi, H., Zhong, S., Mo, X., Liu, N., Nezames, C. D., and Deng, X. W. (2013). HFR1 Sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25, 3770–3784. doi: 10.1105/tpc.113.117424
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Shirakawa, M., Ueda, H., Nagano, A. J., Shimada, T., Kohchi, T., and Hara-Nishimura, I. (2014). FAMA is an essential component for the differentiation of two distinct cell types, myrosin cells and guard cells, in Arabidopsis. Plant Cell 26, 4039–4052. doi: 10.1105/tpc.114.129874
Sivitz, A. B., Hermand, V., Curie, C., and Vert, G. (2012). Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7:e44843. doi: 10.1371/journal.pone.0044843
Song, S., Qi, T., Fan, M., Zhang, X., Gao, H., Huang, H., et al. (2013). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS ONE 9:e1003653. doi: 10.1371/journal.pgen.1003653
Spollen, W. G., and Sharp, R. E. (1991). Spatial distribution of turgor and root growth at low water potentials. Plant Physiol. 96, 438–443. doi: 10.1104/pp.96.2.438
Spollen, W., Sharp, R., Saab, I., and Wu, Y. (1993). “Regulation of cell expansion in roots and shoots at low water potentials,” in Water Deficits: Plant Responses from Cell to Community, eds A. C. Smith and H. Griffiths (Oxford: BIOS Scientific Publishers), 37–52.
Stacey, M. G., Patel, A., McClain, W. E., Mathieu, M., Remley, M., Rogers, E. E., et al. (2008). The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 146, 589–601. doi: 10.1104/pp.107.108183
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Taj, G., Sharma, S., Gaur, V. S., and Kumar, A. (2011). Prediction of downstream interaction of transcription factors with MAPK3 in Arabidopsis thaliana using protein sequence information. Int. J. Bioinf. Res. 3, 167–177. doi: 10.9735/0975-3087.3.1.167-177
Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., et al. (2002). Important roles of drought-and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29, 417–426. doi: 10.1046/j.0960-7412.2001.01227.x
Tarczynski, M. C., Jensen, R. G., and Bohnert, H. J. (1993). Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 259, 508–508. doi: 10.1126/science.259.5094.508
Tardieu, F., Parent, B., Caldeira, C. F., and Welcker, C. (2014). Genetic and physiological controls of growth under water deficit. Plant Physiol. 164, 1628–1635. doi: 10.1104/pp.113.233353
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Tschaplinski, T., and Tuskan, G. (1994). Water-stress tolerance of black and eastern cottonwood clones and four hybrid progeny. II. Metabolites and inorganic ions that constitute osmotic adjustment. Can. J. Forest Res. 24, 681–687. doi: 10.1139/x94-091
Ufaz, S., Shukla, V., Soloveichik, Y., Golan, Y., Breuer, F., Koncz, Z., et al. (2011). Transcriptional control of aspartate kinase expression during darkness and sugar depletion in Arabidopsis: involvement of bZIP transcription factors. Planta 233, 1025–1040. doi: 10.1007/s00425-011-1360-9
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17, 113–122. doi: 10.1016/j.copbio.2006.02.002
Urao, T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, M., Hirayama, T., et al. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11, 1743–1754. doi: 10.1105/tpc.11.9.1743
Vigani, G., Bashir, K., Ishimaru, Y., Lehmann, M., Casiraghi, M., Nakanishi, H., et al. (2016). Knocking down Mitochondrial Iron Transporter (MIT) reprograms primary and secondary metabolism in rice plants. J. Exp. Bot. doi: 10.1093/jxb/erv531. [Epub ahead of print].
Vigani, G., Morandini, P., and Murgia, I. (2013a). Searching iron sensors in plants by exploring the link among 2′-OG-dependent dioxygenases, the iron deficiency response and metabolic adjustments occurring under iron deficiency. Front. Plant Sci. 4:169. doi: 10.3389/fpls.2013.00169
Vigani, G., Zocchi, G., Bashir, K., Philippar, K., and Briat, J.-F. (2013b). Signals from chloroplasts and mitochondria for iron homeostasis regulation. Trends Plant Sci. 18, 305–311. doi: 10.1016/j.tplants.2013.01.006
Wang, D., Zhang, C., Hearn, D. J., Kang, I.-H., Punwani, J. A., Skaggs, M. I., et al. (2010). Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biol. 10:110. doi: 10.1186/1471-2229-10-110
Wang, H.-Z., and Dixon, R. A. (2012). On–off switches for secondary cell wall biosynthesis. Mol. Plant 5, 297–303. doi: 10.1093/mp/ssr098
Wang, N., Cui, Y., Liu, Y., Fan, H., Du, J., Huang, Z., et al. (2013). Requirement and functional redundancy of Ib Subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6, 503–513. doi: 10.1093/mp/sss089
Wei, W., Huang, J., Hao, Y.-J., Zou, H.-F., Wang, H.-W., Zhao, J.-Y., et al. (2009). Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS ONE 4:e7209. doi: 10.1371/journal.pone.0007209
Weltmeier, F., Rahmani, F., Ehlert, A., Dietrich, K., Schütze, K., Wang, X., et al. (2009). Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 69, 107–119. doi: 10.1007/s11103-008-9410-9
Wohlbach, D. J., Quirino, B. F., and Sussman, M. R. (2008). Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20, 1101–1117. doi: 10.1105/tpc.107.055871
Wu, A., Allu, A. D., Garapati, P., Siddiqui, H., Dortay, H., Zanor, M.-I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species–responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506. doi: 10.1105/tpc.111.090894
Wu, Y., and Cosgrove, D. J. (2000). Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 51, 1543–1553. doi: 10.1093/jexbot/51.350.1543
Xiong, L., Schumaker, K. S., and Zhu, J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. doi: 10.1105/tpc.000596
Yadav, V., Molina, I., Ranathunge, K., Castillo, I. Q., Rothstein, S. J., and Reed, J. W. (2014). ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 26, 3569–3588. doi: 10.1105/tpc.114.129049
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, J., Sardar, H. S., McGovern, K. R., Zhang, Y., and Showalter, A. M. (2007). A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. Plant J. 49, 629–640. doi: 10.1111/j.1365-313X.2006.02985.x
Yang, O., Popova, O. V., Süthoff, U., Lüking, I., Dietz, K.-J., and Golldack, D. (2009). The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436, 45–55. doi: 10.1016/j.gene.2009.02.010
Yuan, Y., Wu, H., Wang, N., Li, J., Zhao, W., Du, J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397. doi: 10.1038/cr.2008.26
Zhang, L., Gu, L., Ringler, P., Smith, S., Rushton, P. J., and Shen, Q. J. (2015). Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 236, 214–222. doi: 10.1016/j.plantsci.2015.04.014
Zhong, R., Demura, T., and Ye, Z.-H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z.-H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhou, J., Lee, C., Zhong, R., and Ye, Z.-H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. doi: 10.1105/tpc.108.063321
Keywords: abiotic stresses, abscisic acid, Arabidopsis thaliana, drought, microarray, transcription factors
Citation: Rasheed S, Bashir K, Matsui A, Tanaka M and Seki M (2016) Transcriptomic Analysis of Soil-Grown Arabidopsis thaliana Roots and Shoots in Response to a Drought Stress. Front. Plant Sci. 7:180. doi: 10.3389/fpls.2016.00180
Received: 24 December 2015; Accepted: 02 February 2016;
Published: 23 February 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Biswapriya Biswavas Misra, University of Florida, USAByeong-ha Lee, Sogang University, South Korea
Copyright © 2016 Rasheed, Bashir, Matsui, Tanaka and Seki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Motoaki Seki, motoaki.seki@riken.jp
 Sultana Rasheed
Sultana Rasheed Khurram Bashir
Khurram Bashir Akihiro Matsui
Akihiro Matsui Maho Tanaka
Maho Tanaka Motoaki Seki
Motoaki Seki