- 1College of Aqua-Life Science and Technology, Shanghai Ocean University, Shanghai, China
- 2Department of Biology and Food Engineering, Bengbu University, Bengbu, China
- 3Key Laboratory of Applied Marine Biotechnology, Ningbo University, Ningbo, China
To analyze the contribution of glycerol-3-phosphate acyltransferase (GPAT) to the first acylation of glycerol-3-phosphate (G-3-P), the present study focused on a functional analysis of the GPAT gene from Lobosphaera incisa (designated as LiGPAT). A full-length cDNA of LiGPAT consisting of a 1,305-bp ORF, a 1,652-bp 5′-UTR, and a 354-bp 3′-UTR, was cloned. The ORF encoded a 434-amino acid peptide, of which 63 residues at the N-terminus defined a chloroplast transit peptide. Multiple sequence alignment and phylogeny analysis of GPAT homologs provided the convincible bioinformatics evidence that LiGPAT was localized to chloroplasts. Considering the conservation of His among the G-3-P binding sites from chloroplastidial GPATs and the substitution of His by Arg at position 195 in the LiGPAT mature protein (designated mLiGPAT), we established the heterologous expression of either mLiGPAT or its mutant (Arg195His) (sdmLiGPAT) in the GPAT-deficient yeast mutant gat1Δ. Lipid profile analyses of these transgenic yeasts not only validated the acylation function of LiGPAT but also indicated that the site-directed mutagenesis from Arg195 to His led to an increase in the phospholipid level in yeast. Semi-quantitative analysis of mLiGPAT and sdmLiGPAT, together with the structural superimposition of their G-3-P binding sites, indicated that the increased enzymatic activity was caused by the enlarged accessible surface of the phosphate group binding pocket when Arg195 was mutated to His. Thus, the potential of genetic manipulation of GPAT to increase the glycerolipid level in L. incisa and other microalgae would be of great interest.
Introduction
In plants, de novo biosynthesis of fatty acids occurs exclusively in chloroplasts. The fatty acids generated are either directly metabolized into glycolipids and PG within the chloroplast or exported across the envelope to the ER to form phospholipids and neutral lipids. To synthesize these glycerolipids in both chloroplast and ER, glycerol-3-phosphate acyltransferase (GPAT, E.C. 2.3.1.15) is first required to acylate fatty acids in the glycerol backbone of G-3-P. This enzyme localized in ER was demonstrated to be crucial for cutin, suberin, or storage oil biosynthesis in Arabidopsis thaliana (Zheng et al., 2003; Gidda et al., 2009), Ricinus communis (Cagliari et al., 2010) and Brassica napus (Chen et al., 2010). In addition, it was found that a deficiency in the chloroplastidial GPAT activity could cause a reduction (10–25%) in the PG content of Arabidopsis (Kunst et al., 1988; Xu et al., 2006). Thus, GPAT has been found to play a pivotal role in initiating all glycerolipid biosynthesis in higher plants. In comparison, functional analyses of GPAT from microalgae are rare.
To understand the features of the first step of glycerolipid biosynthesis catalyzed by GPAT in microalgae, we attempted to identify one cloned GPAT gene from an oleaginous green microalga, Lobosphaera incisa Reisigl (designated as LiGPAT). This microalga possesses a high content of photosynthetic membrane lipids as suggested by a large incised chloroplast with many parallel thylakoid membranes (Merzlyak et al., 2007; Ouyang et al., 2012, 2013b), and it has the ability to accumulate TAG to form oil bodies in cells, especially under nitrogen starvation (Khozin-Goldberg et al., 2002; Zhang et al., 2002; Tong et al., 2011; Ouyang et al., 2013b). Thus, the study of the function of the GPAT gene from L. incisa might indicate the role of GPAT in microalgae. Given that GPAT in plants can localize to the chloroplast or the ER, the subcellular localization of the encoded protein LiGPAT was analyzed by bioinformatics technique. Heterologous complementation in a GPAT deficient mutant of yeast, gat1Δ (Zheng and Zou, 2001), was used to validate the function of LiGPAT, and the yeast lipids were analyzed by lipidomic approaches using UPLC-ESI-Q-TOF-MS and multivariate data analysis. Surprisingly, we found that the conserved His in the G-3-P binding sites from chloroplastidial GPATs was substituted by Arg at position 195 in this chloroplastidial LiGPAT mature protein, and site-directed mutagenesis at this site of LiGPAT improved the phospholipid level in yeast. These findings help us to understand the characteristics of a putatively chloroplastidial GPAT in L. incisa and thus provide a strategy for genetic engineering to improve the microalgae-based production of biofuels.
Materials and Methods
Strains, Medium and Growth Conditions
Lobosphaera incisa, deposited in the Culture Collection of Algae of Charles University in Prague under ID H4301 was cultivated in BG-11 medium (Stanier et al., 1971) in 500-mL glass flasks as described previously (Ouyang et al., 2013b). During culture, the flasks were shaken several times a day by hand at regular intervals.
Cloning of cDNA and DNA Encoding LiGPAT
A pair of degenerate primers (G1 and G2) (Supplementary Table S1) for the LiGPAT gene cDNA cloning were designed based on the amino acid sequences of GPAT from Ostreococcus tauri (GenBank Accession Number 116061306) and Chlamydomonas reinhardtii (GenBank Accession Number 159473711). Total RNA isolated by TRIzol reagent (Invitrogen) from L. incisa was used to synthesize cDNA with a Reverse Transcribed Kit II (TaKaRa). The full-length cDNA of LiGPAT was amplified by a SMARTTM RACE cDNA Amplification Kit (Clontech). Two gene-specific primers (NGSP5-1 and GSP5-2) for the first 5′-RACE reaction, one gene-specific primer (GSP5-4) for the second 5′-RACE reaction, and two gene-specific primers (NGSP3-1 and GSP3-2) for the 3′-RACE reaction were designed (Supplementary Table S1). Genomic DNA extracted by the CTAB method from L. incisa (Dellaporta et al., 1983) was used to amplify both the coding region and the untranslated region of LiGPAT with four pairs of primers (Supplementary Table S1). All PCR products of the expected size were cloned into the pMD19-T cloning vector (TaKaRa). The resulting constructs were transformed into Escherichia coli DH5α and verified by sequencing. The BLAST Server1 was used to annotate the cloned sequences.
Southern Blot Analysis of LiGPAT
Genomic DNA of L. incisa was double digested with XhoI/NotI or HindIII/NotI restriction endonucleases at 37°C for 4-6 h. The digested DNA samples were fractionated on a 1.0% agarose gel and then transferred to a NC filter membrane (Millipore). A pair of primers was designed based on the conserved domain of GPAT (Supplementary Table S1). A 311-bp biotin-labeled DNA sequence was prepared to use as a probe with a North2South® Biotin Random Prime Labeling Kit (Thermo Scientific). Subsequently, the hybridization was detected by the standard Southern blot procedure (Sambrook and Russell, 2001) with a North2South Chemiluminescent Hybridization and Detection Kit (Thermo Scientific). Signals were visualized by exposure to XBT-1 film (Kodak) at room temperature for 60-120 s.
Bioinformatics Analysis
The intron and exon regions from LiGPAT were analyzed using Spidey2. Signal peptide sites of the amino acid sequence of LiGPAT were predicted by the SignalP 4.1 Server3, and the transit peptide sites were predicted by the TargetP 1.1 Server4 and the ChloroP 1.1 Server5. Conserved domains were searched in NCBI’s CDD (Marchler-Bauer et al., 2011). The PredictProtein program6 was applied to predict protein structural and functional features (Rost et al., 2004). Protein structures were performed with I-TASSER7 (Roy et al., 2010). The superimposed images of the LiGPAT tertiary structure were obtained from SuperPose 1.08 (Maiti et al., 2004) and displayed with UCSF Chimera 1.10 (Pettersen et al., 2004).
Multiple Sequence Alignment of GPAT Homologs
The available chloroplastidial GPAT amino acid sequences of Arabidopsis thaliana (GenBank Accession Number Q43307), Auxenochlorella protothecoides (GenBank Accession Number KFM22407), Chlamydomonas reinhardtii (GenBank Accession Number XP_001694977), Coccomyxa subellipsoidea C-169 (GenBank Accession Number XP_005643353), Cucurbita moschata (GenBank Accession Number BAB17755), Cyanidioschyzon merolae Strain 10D (GenBank Accession Number XP_006587606), Glycine max (GenBank Accession Number XP_006587606), Micromonas pusilla CCMP 1545 (GenBank Accession Number XP_003060587), Micromonas sp. RCC299 (GenBank Accession Number XP_002505030), Ostreococcus lucimarinus (GenBank Accession Number ABO94442), Ostreococcus tauri (GenBank Accession Number CAL52024), Phaeodactylum tricornutum (GenBank Accession Number XP_002184838), Ricinus communis (GenBank Accession Number XP_002518993), Thalassiosira pseudonana (GenBank Accession Number XP_002292905), and Volvox carteri f. nagariensis (GenBank Accession Number XP_002950506) were retrieved from GenBank. The amino acid sequences of the ER-bound GPAT isoform 4 (GenBank accession number Q9LMM0), isoform 5 (GenBank Accession Number NP_187750), isoform 6 (GenBank Accession Number NP_181346), and isoform 8 (GenBank Accession Number NP_191950) from Arabidopsis thaliana, and ER-bound GPAT from Medicago truncatula (GenBank Accession Number AES79440) and Ricinus communis (GenBank Accession Number XP_002511873) were also obtained from GenBank. Multiple sequence alignment of the chloroplastidial GPATs and the ER-bound GPATs were performed with the ClustalX program (Thompson et al., 1997). The web-based BLAST2 program (Altschul et al., 1990) at NCBI was employed to generate pairwise similarity scores of the aligned sequences. The Multiple EM for Motif Elicitation (MEME) program (Bailey et al., 2006) was used to identify conserved sequence motifs.
Phylogeny Inference
The amino acid sequences of GPAT as well as LPAAT from higher plants and microalgae were retrieved from GenBank. Three GPATs from Nitrosococcus halophilus, Bradyrhizobium japonicum, and Ralstonia pickettii DTP0602 were chosen as an arbitrary outgroup. All accession numbers are presented in the phylogeny tree. All of the conserved domain sequences annotated by searching CDD were also aligned with the ClustalX program (Thompson et al., 1997). Phylogenetic analysis was conducted using maximum likelihood (ML) methods with MEGA 6.0 (Tamura et al., 2013) by using the most appropriate model (LG + G + Γ) determined by ProtTest v3.3 (Darriba et al., 2011). Branch points were tested for significance by bootstrapping with 1,000 replications (Felsenstein, 1985; Tamura et al., 2013).
Construction of mLiGPAT and sdmLiGPAT Expression Plasmids
To construct a bacterial plasmid, the 1,131-bp of the LiGPAT mature protein coding gene (designated as mLiGPAT) with BamHI/XhoI digestion sites was amplified with a pair of primers (EcBamF and XhoR) (Supplementary Table S1). The PCR products were sticky-ended and subcloned into the BamHI/XhoI sites of pET 28a to obtain the plasmid pET-mLiG.
For heterologous expression in yeast, mLiGPAT and the site-directed mutation (Arg195His) of mLiGPAT (designated as sdmLiGPAT) expression plasmids were constructed. An approximately 1.2 kb cDNA fragment containing the BamHI/XhoI digestion sites of mLiGPAT was amplified with the primers ScBamF and XhoR (Supplementary Table S1). Site-directed mutagenesis by splicing overlap extension PCR (SOE-PCR) (Ge and Rudolph, 1997) was performed to generate sdmLiGPAT with BamHI/XhoI digestion sites using four primers ScBamF, MuScR, XhoR, and MuScF (Supplementary Table S1). The PCR products of mLiGPAT and sdmLiGPAT were sticky-ended and subcloned into the BamHI/XhoI sites of pYES2 (Invitrogen) to obtain the plasmids pY-mLiG and pY-sdmLiG, respectively. Prior to transforming the resulting plasmids into host cells, the correct orientation and in-frame fusion of all of the inserts was verified by sequencing.
Heterologous Expression of LiGPAT
To obtain soluble, recombinant mLiGPAT protein, the plasmid pET-mLiG and the vector pET 28a as a control were introduced into E. coli BL21 (DE3) pLysS (designated pmLiG/BL and pET/BL), respectively. Single colonies of pmLiG/BL or pET/BL were inoculated in LB medium. After incubation at 37°C for 12 h, the cells were collected and resuspended in LB medium with 1 mM IPTG to obtain an OD600 of 0.6. After incubation at 18°C overnight (Schein and Noteborn, 1988), soluble recombinant protein with a His-tag was expressed and detected by SDS-PAGE.
To express mLiGPAT and sdmLiGPAT in yeast for functional identification, we chose the GPAT deficient yeast mutant, gat1Δ (BY4742, Matα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, YKR067w::kanMX4) as described by Zheng and Zou (2001). This strain was purchased from EUROSCARF9. Single colonies carrying pYES2 (plasmid-only as a control) or pY-mLiG or pY-sdmLiG (designated gPY, gmLiGPAT, and gsdmLiGPAT, respectively) were inoculated into SC-uracil medium with 2% glucose. The parental strain of gat1Δ, BY4742 (Matα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0), was used as a positive control, while gat1Δ was used as a negative control. After incubation at 30°C for 30 h, cells of gat1Δ, gPY, BY4742, gmLiGPAT, and gsdmLiGPAT were collected by centrifugation and resuspended in SC-uracil medium with 2% galactose. After incubation at 30°C for 12 h to reach to the logarithmic phase, the yeast cells were transferred to 16°C and incubated for another 48 h. Cells were harvested by centrifugation at 4°C. For preparation of the yeast homogenates, the cell pellets were washed with 10 volumes of distilled H2O and then immediately frozen in liquid nitrogen and stored at -80°C until use. For total lipid extraction, the cell pellets were lyophilized and stored at -20°C until use. Each sample was collected in duplicate. Colony PCR of each sample was accomplished using the primers pYF and pYR (Supplementary Table S1) to ensure the insertion of the target gene.
Polyclonal Antibody Preparation and Purification
Soluble recombinant mLiGPAT protein was purified by Ni-affinity column chromatography (Bio-Rad) and verified by HPLC-MS. New Zealand rabbits were immunized for LiGPAT polyclonal antibody preparation. The purified mLiGPAT protein was electrophoretically transferred from SDS polyacrylamide gels to NC membranes (Millipore) for polyclonal antibody preparation (Smith and Fisher, 1984). Next, the NC blots were incubated for l h in 3% BSA in PBS before an additional incubation in the prepared LiGPAT polyclonal antibody for 16 h at 4°C. Afterward, the blots were washed several times with PBS and eluted with 0.2 M glycine-HCl (pH 2.3) for 20 min. The eluate was immediately neutralized by the addition of 1 M Tris-HCl and PBS and stored at -20°C until use.
Western Blot Analysis
Fresh cells of L. incisa were ground in liquid nitrogen, resuspended in 50-100 μL breaking buffer (25 mM Tris-HCl, pH 6.5, 50 mM NaCl, 2 mM β-mercaptoethanol) and vortexed. Frozen cell pellets of yeast were resuspended in 500 μL breaking buffer (50 mM PBS, pH 7.4, 1 mM EDTA, 5% glycerol, 1 mM PMSF) and lysed by shear force using acid-washed glass beads according to the user manual (Invitrogen). Frozen cell pellets of E. coli were resuspended in 0.1 M PBS buffer and then sonicated on ice with a probe sonicator until the suspension was partially clear.
The lysed cells were centrifuged at 20,000 × g at 4°C, and the supernatant was collected and stored at -80°C until use. The protein concentration was determined by the Bradford protein assay (Bradford, 1976).
Crude proteins from L. incisa, E. coli or yeast were electrophoretically transferred to NC membranes (Millipore) as described above, and the Western blots were performed according to the standard protocol (Sambrook and Russell, 2001). The purified LiGPAT polyclonal antibody and the secondary antibody, peroxidase-conjugated goat anti-rabbit IgG (Shanghai Youke Biotechnology Co., Ltd.), were appropriately diluted. Immunoreactive bands were visualized by the addition of DAB according to the manufacturer’s manual (Tiangen). The levels of mLiGPAT and sdmLiGPAT expressed in yeast were semi-quantified by measuring the band intensity on their corresponding blots with ImageJ software10.
Total Lipid Extraction and Fractionation
Total lipid from yeast was extracted according to Bligh and Dyer (1959) with minor modifications. Acid washed glass beads with a diameter of 0.4-0.6 mm (Omega Bio-Tek) were used to break the cell walls. The extent of lysis was observed with a microscope, keeping the degree of breakage of each sample the same as far as possible.
Phospholipids from the extracted total lipids of approximately 50 mg lyophilized yeast were separated using solid-phase extraction (Christie and Han, 2010). A 500 mg cartridge of silica gel (CNW) was first conditioned by elution with 5 mL of chloroform, and the total lipids from yeast were then applied to it. Elution with 10 mL of methanol yielded the phospholipids. This fraction was concentrated under a stream of nitrogen gas and then weighed.
UPLC-ESI-Q-TOF-MS Analysis
Reversed-phase analysis of lipids was performed on a Waters ACQUITY UPLC system using an ACQUITY UPLC BEH C8 analytical column (i.d. 2.1 × 100 mm, particle size 1.7 μm). The temperature of the sample chamber was set at 4°C, the column temperature was set at 40°C, and the injection volume was 4 μL for each analysis. A 1:4 split of the column effluent was used to achieve a flow rate of approximately 0.35 mL/min into the ESI source. To produce ions that could be readily fragmented, 0.001% lithium acetate and 0.1% formic acid were added to the mobile phase as the electrolyte. For efficient separation of the total lipids, water/tetrahydrofuran (3:1, v/v) was used as the mobile phase A and acetonitrile/methanol/tetrahydrofuran (2:1:1, v/v/v) as the mobile phase B. The initial composition of the mobile phase B was changed from 40 to 70% in 10 min and held for 7 min, then increased to 100% in 6 min and held for 1 min, and finally returned to the initial 40% in 1 min and equilibrated for 10 min. MS analysis was performed in a negative ion mode on a Waters Q-TOF Premier mass spectrometer. The mass range was from 100 to 1,200 with a scan duration of 0.3 s and an interscan delay of 0.02 s. High-purity nitrogen was used as the nebulizer and drying gas at a constant flow rate of 50 L/h, and the source temperature was set at 120°C. The capillary voltage was set at 2.6 kV, and the sampling cone voltage was set at the ramp of 35–80 V. MS/MS analysis was performed at a collision energy range of 25-35 V with argon as the collision gas. The TOF analyzer was used in a V mode and tuned for maximum resolution (>10,000 resolving power at m/z 1,000). Prior to the experiment, the instrument was calibrated with sodium formate, and the lock mass spray for precise mass determination was set with leucine enkephalin at a concentration 400 ng/μL, generating an [M-H]- ion at 554.2615 Da in ESI- mode. The lock spray frequency was set at 10 s.
Lipidomics Data Processing
The original data from the ESI- mode were acquired by the UPLC-Q-TOF-MS system and analyzed by a MassLynx 4.1 data processing system (Waters). The MarkerLynx matrices with peak numbers [based on the RT and mass-to-charge ratio (m/z)], sample names, and normalized peak intensities were exported to SIMCA-P+ 12.0 (Umetrics) and analyzed by PCA, PLS-DA, and OPLS-DA. The quality of the models PLS-DA and OPLS-DA was evaluated by two parameters, R2Y(cum) and Q2(cum). R2Y(cum) is the cumulative fraction of the sum of squares of all Y-variables that the model can explain using the latent variables, indicating the explanative ability of the model. Q2(cum) depicts the cumulative fraction of the total variation that can be predicted using the model via sevenfold cross-validation, indicating the predictability of the model. In general, R2Y(cum) and Q2(cum) values close to 1.0 indicate an excellent fit to the model, and the difference between these two values should be less than 0.3 (Wiklund, 2008). If the value of the Q2(cum) is higher than 0.9, the model is considered an excellent one (Wiklund, 2008). CV-ANOVA was systematically performed based on the PLS-DA model to rule out the non-randomness of the separation between groups. Generally, in permutation tests with 999 iterations, the intercept value of Q2 > 0.05 indicates over-fit in the original model (Kang et al., 2008; Lu et al., 2012).
Identification of Lipid Metabolites
Variables meeting two criteria, specifically, high VIP and CIJFJK excluding zero, were selected as potential lipid biomarkers, which contributed to the separation between groups (Eriksson et al., 2006; Cai et al., 2015). The lipid metabolites were identified by the RT, m/z, and the characteristic fragment ions deduced by MS/MS (Yan et al., 2010). In addition, some public databases including HMDB11, LIPID MAPS12, and METLIN13 were also used to help elucidate the putative ion structures.
Results
Cloning and Characterization of the LiGPAT Gene
Based on the amino acid sequences of GPAT proteins available from Ostreococcus tauri and Chlamydononas reinhardtii, a pair of degenerate primers (Supplementary Table S1) was designed, with which a 321-bp cDNA fragment was amplified from L. incisa. BLAST analysis revealed that this sequence was a GPAT homolog, and so it was designated LiGPAT. Subsequently, a 3,278-bp full-length cDNA of this gene, consisting of a 1,305-bp ORF, a 1,619-bp 5′-UTR, and a 354-bp 3′-UTR was obtained by the RACE technique. The nucleotide sequence of LiGPAT was identical to the unique annotated GPAT gene from the transcriptome of L. incisa (Ouyang et al., 2013a). A comparison of the cDNA sequence with its corresponding DNA sequence (Supplementary Figure S1A) revealed that the gene contained seven introns. The introns, 279, 371, 404, 145, 307, 211, and 745 bp beginning from the 5′-end, contained splice sites that all conformed to the GT-AG rule. Both the cDNA and the DNA sequences of LiGPAT were deposited in GenBank under the accession numbers KM670441 and KM670442, respectively. Southern blot analysis of the genomic DNA digested by either NotI/XhoI or NotI/HindIII using a 311-bp specific probe suggested that LiGPAT was a single copy gene in L. incisa (Supplementary Figure S1B). The same result for nucleus-encoded chloroplastidial GPAT genes was observed in a number of angiosperm families (Ishizaki et al., 1988; Kunst et al., 1988; Weber et al., 1991; Nishida et al., 1993; Bhella and Mackenzie, 1994).
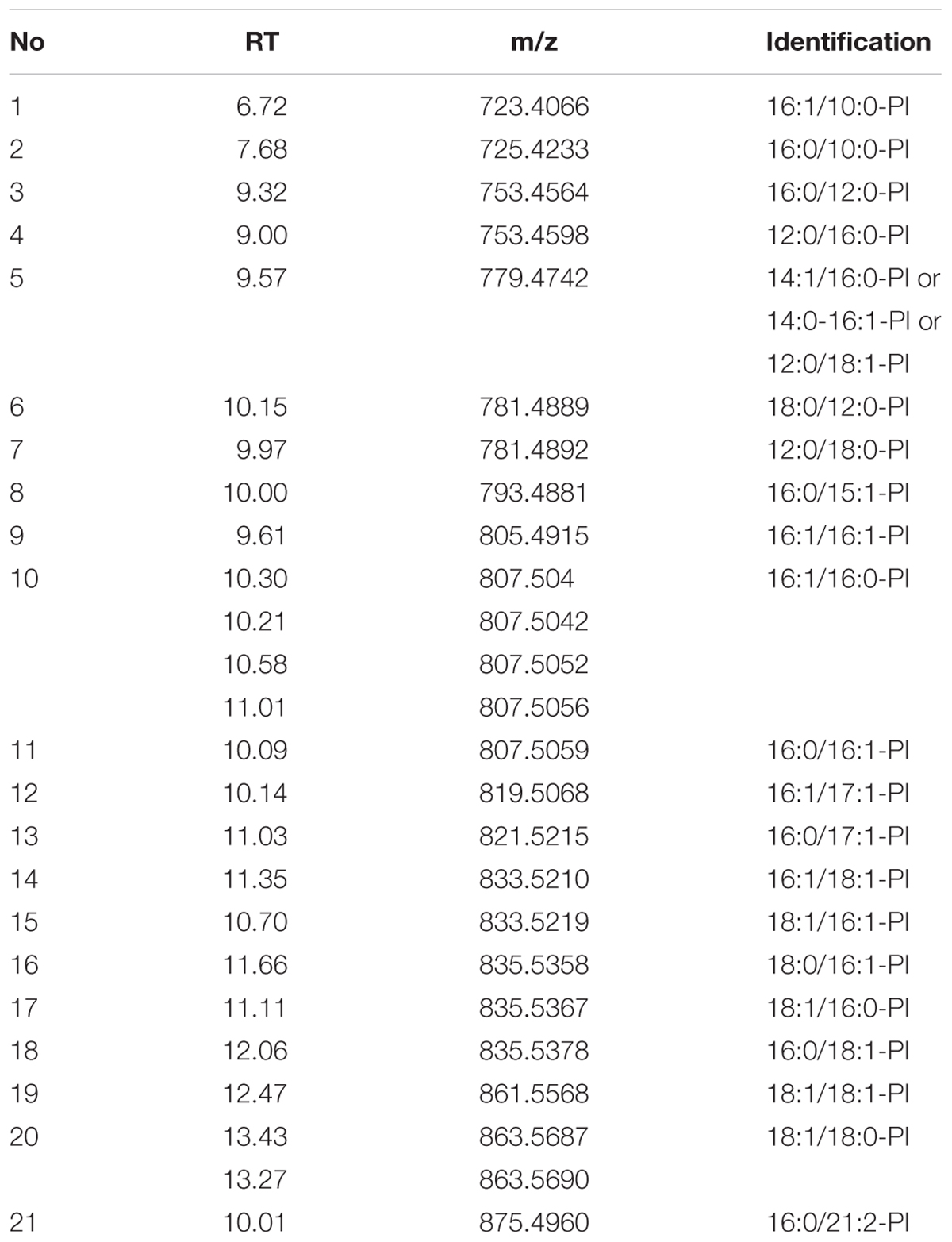
The ORF of LiGPAT encoded a 434-amino acid peptide, in which the domains GPAT_N (GenBank Accession Number cl20739) and LPLAT_GPAT (GenBank Accession Number cd07985) were annotated by searching CDD. A structural motif similar to a prokaryotic membrane lipoprotein lipid attachment site (Leu8- Cys18) was identified by the PredictProtein program (Figure 1A). This motif was also identified in a chloroplastidial form of the acetyl-CoA carboxylase of pea (Shorrosh et al., 1996) and a chloroplastidial NEF1 of Arabidopsis thaliana (Ariizumi et al., 2004). Neither the transmembrane domain nor the signal peptide in LiGPAT was predicted, whereas the N-terminal sequence of 63 residues was identified as a cTP (Figure 1A) as predicted by both the TargetP 1.1 Server and the ChloroP 1.1 Server. Characteristics of this cTP, including a high content (14.28%) of hydroxylated residues (Ser and Thr), a high content (19.05%) of hydrophobic residues (Ala and Val), the absence of the acidic residues (Asp and Glu), and very few Pro and Gly among the first 10 residues, were consistent with previously described cTP sequences (von Heijne et al., 1989). These results suggested that the LiGPAT might be a chloroplastidial GPAT.
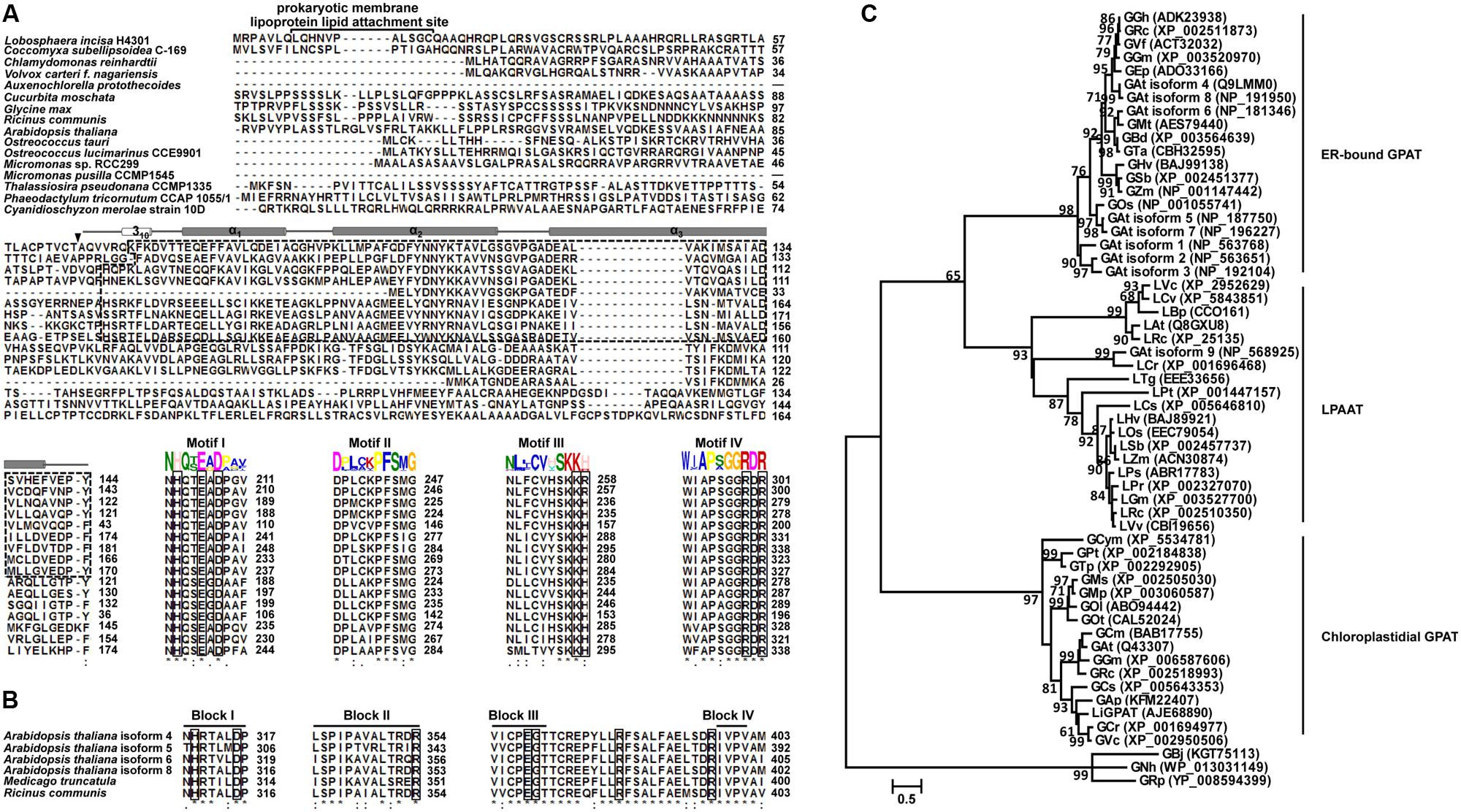
FIGURE 1. Multiple sequence alignment and phylogeny analysis of GPAT homologs. Identical amino acids are indicated by “∗”, and the important catalytic sites are boxed. (A) ClustalW alignment of the N-terminal sequences and four acyltransferase domain sequences from chloroplastidial GPATs. The identified GPAT_N domain is indicated by a dashed line box. The secondary structure assigned for the GPAT_N domain is represented by a cylinder (α-helix). A filled down arrow (▼) indicates the cleavage site of LiGPAT. The putative prokaryotic membrane lipoprotein lipid attachment site is indicated above the aligned sequences. Motifs I-IV were predicted by the MEME program. (B) ClustalW alignment of four acyltransferase domain sequences from ER-bound GPATs. Blocks I-IV were suggested by Lewin et al. (1999). (C) Phylogeny inference based on GPAT and LPAAT amino acid sequences. The phylogeny tree was inferred by the Maximum Likelihood (ML) method based on LG + G + Γ. The tree with the highest log likelihood (-6,958.7325) is shown. Bootstrap analysis was based on 1,000 re-samplings, and only support values higher than 60% are shown in the phylogeny. All accession numbers are presented in the phylogeny tree.
Alignment and Phylogeny Analysis of GPAT Homologs
To ascertain the features of chloroplastidial GPAT amino acid sequences, a pairwise sequence alignment and a complete multiple sequence alignment were carried out separately. The results of the pairwise alignment showed a higher similarity among chloroplastidial GPAT proteins from higher plant species (67-74%) than from microalgal species (33-79%). LiGPAT was more conserved with GPATs from other Trebouxiophyceae species (Coccomyxa subellipsoidea and Auxenochlorella protothecoides) and Chlorophyceae species (Chlamydomonas reinhardtii and Volvox carteri) (56-60%) than with Mamiellophyceae species (Ostreococcus lucimarinus, Ostreococcus tauri, Micromonas pusilla, and Micromonas sp.) (42-43%), Stramenopiles species (Thalassiosira pseudonana and Phaeodactylum tricornutum) (36-43%), and Rhodophyta species (Cyanidioschyzon merolae) (33%).
The complete multiple sequence alignment identified 39 fully conserved residues that corresponded to only 8.8% of the average 430 residues. The GPAT_N domain identified in LiGPAT was also found in GPATs from Coccomyxa subellipsoidea, Chlamydomonas reinhardtii, Volvox carteri, Auxenochlorella protothecoides, Cucurbita moschata, Glycine max, Ricinus communis, and Arabidopsis thaliana (Figure 1A). The length of the domain GPAT_N was similar (74-78 amino acid residues) except for that from Auxenochlorella protothecoides, due to the incomplete sequence, but its sequence was different from others (Figure 1A). In contrast, four motifs predicted by using the MEME program were relatively conserved (Figure 1A). The H(X)4D motif in Motif I was a conserved consensus sequence among many glycerolipid acyltransferases. The residues Lys, His, Arg, and Arg in Motif III and IV in chloroplastidial GPATs (Figure 1A) were considered to form a positive pocket to bind the phosphate group of G-3-P (Lewin et al., 1999; Turnbull et al., 2001). It is worth noting that these four G-3-P binding sites were well conserved except for the His residue, which was replaced by Arg in L. incisa and Coccomyxa subellipsoidea (Figure 1A).
The ER-bound GPATs were aligned, and four acyltransferase domains were identified (Figure 1B). Interestingly, these acyltransferase domains were significantly different from those of chloroplastidial GPATs, except the His and Asp residues from the H(X)4D motif in Block I (Figure 1B). The Gly residue in Block III and the Pro residue in Block IV, both of which were suggested to be catalytically important sites (Lewin et al., 1999), were completely conserved (Figure 1B). In addition, the residues Arg in Block II and Glu and Ser in Block III were invariant among the cytoplasmic GPATs for G-3-P binding (Figure 1B).
Although the previously defined conserved domains of GPAT were similar to those of LPAAT (Heath and Rock, 1998; Lewin et al., 1999; Slabas et al., 2002), the phylogenetic tree showed an apparently different phylogenetic support between lineages of GPAT and LPAAT except that the GPAT isoform 9 from Arabidopsis thaliana was in the LPAAT clade (Figure 1C). The ER-bound GPATs and the mitochondrial GPATs formed a cluster apart from the chloroplastidial one comprising both higher plants and microalgae (Figure 1C), and this result was in agreement with a previous report (Cagliari et al., 2010). This separation could be explained by the differences in the G-3-P binding and catalytically important sites between the cytoplasmic and the chloroplastidial GPATs as mentioned above. The phylogeny also suggested a sister group relationship between the subclade, consisting of diatoms and red algae, and the one comprising higher plants and green microalgae (Figure 1C), which was consistent with the sequence similarity among these species.
These multiple-sequence alignments and phylogeny indicated that LiGPAT possessed the sequence features that conformed to those of chloroplastidial GPATs, providing further evidence that LiGPAT was localized in L. incisa chloroplasts.
Functional Identification of LiGPAT in a gat1Δ Mutant of Yeast
Multiple sequence alignment (Figure 1A) showed that the Arg195 in mLiGPAT was different from His, which was considered one of the G-3-P binding sites in most chloroplastidial GPATs. When this residue His was mutated to Ser, the biological activity of squash chloroplastidial GPAT decreased (Slabas et al., 2002). Accordingly, it was inferred that the catalytic ability of this LiGPAT might be different from (probably lower than) the one with His at position 195. Thus, to identify the function of LiGPAT, heterologous expression of mLiGPAT as well as its mutant (Arg195His) sdmLiGPAT generated by site-directed mutagenesis was performed in the GPAT-deficient yeast strain gat1Δ.
To identify the function of GPAT in yeast, the activity of this enzyme was determined in vitro, for example, by routinely using 14C-labeled G-3-P as described by Zheng and Zou (2001). Because of the inconvenience in other ordinary laboratories without any protection from irradiation, a metabolomics approach by using UPLC-ESI-Q-TOF-MS and multivariate data analysis (Fiehn et al., 2000; Wiklund et al., 2005; Van Assche et al., 2015) was employed in this study.
A PCA model with two-components was constructed, which showed that gsdmLiGPAT (site-directed mutated) clustered with the parental yeast strain BY4742 (this group was designated as the wild-type) but was clearly separated from the gat1Δ (GPAT-deficient) and plasmid-only control yeast gPY (this group was designated as deficient) (Figure 2A). In this PCA model, which could not be well validated, gmLiGPAT (mLiGPAT-transformed) did not significantly separate from the deficient group. In comparison, both the OPLS-DA [R2Y(cum) = 0.992 and Q2(cum) = 0.964] and the PLS-DA [R2Y(cum) = 0.961 and Q2(cum) = 0.922] models with high R2Y(cum) and Q2(cum) values could provide reliable support for the separation of gmLiGPAT and the wild-type group from the deficient group (Figures 2B,C). Validation of the PLS-DA model with the number of permutations equaling 999 generated intercepts of R2 = 0.54 and Q2 = -0.112 (Figure 2D), giving an additional proof of the statistically valid and well fit model because the intercept of Q2-point regression line was below zero. These statistical analyses indicated that lipid compositions of gmLiGPAT and gsdmLiGPAT indeed significantly differed from those of gat1Δ and gPY but were similar to those of the parental strain BY4742. It was concluded that the deficiency of GPAT in the mutant gat1Δ was corrected by the introduction of LiGPAT, thus confirming the acylation function of the GPAT protein from L. incisa.
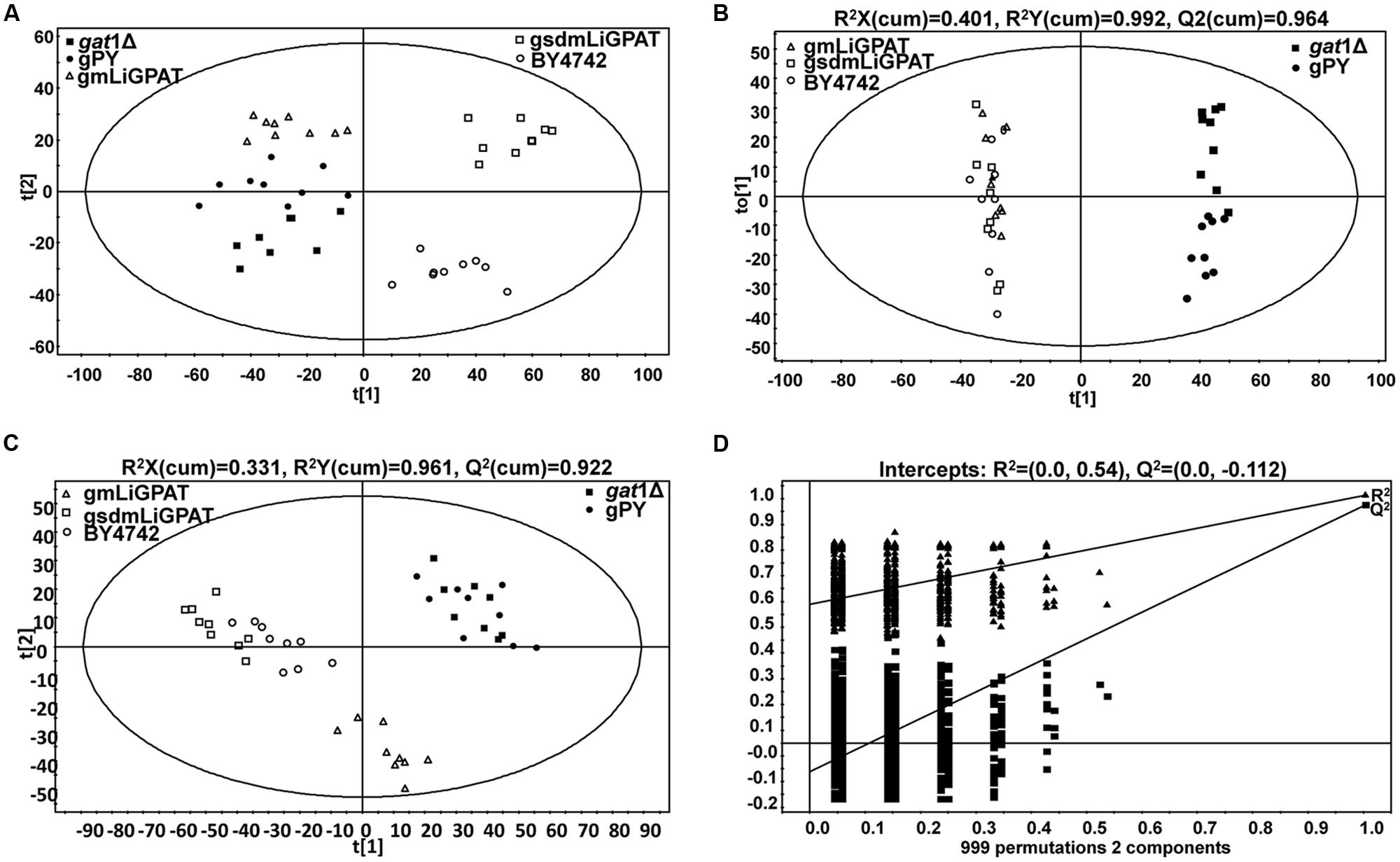
FIGURE 2. Multivariate data analysis of lipidomics data from transgenic (gPY, gmLiGPAT, and gsdmLiGPAT), GPAT-deficient (gat1Δ), and parental (BY4742) yeasts in negative ion scan mode. (A) PCA score plot of data from gat1Δ, gPY, gmLiGPAT, gsdmLiGPAT, and BY4742 for the first two components. (B) OPLS-DA score plot of data from the wild-type group (gmLiGPAT, gsdmLiGPAT, and BY4742) versus the deficient group (gPY and gat1Δ). (C) PLS-DA score plot of data from the wild-type group versus the deficient group. (D) Validation plot of PLS-DA analysis with the number of permutations equaling 999. R2 (filled triangle) is the explained variance, and Q2 (filled square) is an estimate of the predictive ability of the 7 models.
To understand which lipid mainly contributed to the separation of the wild-type group from the deficient group, 35 potential lipid biomarkers (VIP ranged from 38.34 to 2.59 with an average of 7.80) (Table 1) were selected according to both VIP values and the corresponding 95% confidence interval based on a jack-knife procedure (Eriksson et al., 2006). A total of 29 of these selected biomarkers were subsequently identified to be (PI, lyso-PI, PG, PS, PC, and PE (Table 1). Among these metabolites, PI accounted for 55.17% (16–29) and possessed relatively high VIP (ranging from 2.83 to 38.34 with an average of 9.613) (Table 1), indicating that this phospholipid was the main contributor to the separation. The relative abundance of most PI species from the wild-type group was higher than from the deficient group (Figure 3). This result was roughly consistent with the previous report (Redón et al., 2011) that the main increase of PI was observed when the yeast strain was cultured under low temperature. Therefore, the total relative abundance of PI species in the wild-type was compared with that in the deficient group. All of the PI species, 21 in total, were subsequently identified (Table 2), and the relative abundance was compared. The results showed that the total relative abundance of PI from gmLiGPAT was higher, although not significantly higher (P > 0.05), than from gat1Δ and gPY, whereas the total relative abundance of PI from gsdmLiGPAT was significantly higher (P < 0.01) than from gmLiGPAT, gat1Δ, and gPY, but there was no significant difference (P > 0.05) from BY4742 (Figure 4C). It was thus predicted that the site-directed mutagenesis of LiGPAT Arg195His might enhance the catalytic activity of this protein and result in an increase in the phospholipid level in yeast. The subsequent measurement of phospholipid content showed that the phospholipid level from gsdmLiGPAT was higher than from gmLiGPAT or BY4742 (Figure 4D), thus supporting the prediction.
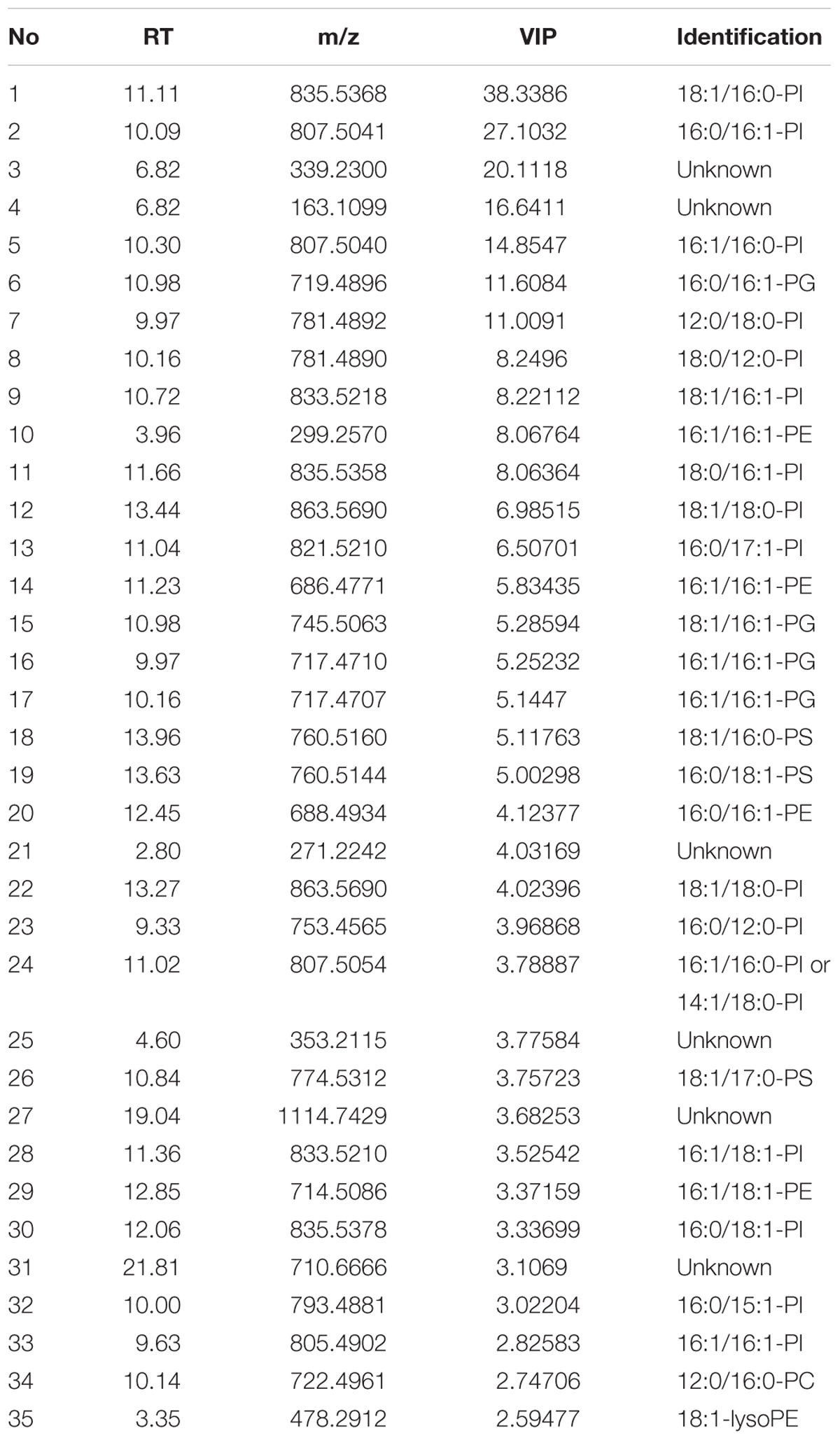
TABLE 1. Identification of the top 35 metabolites contributing to differences between the wild-type group (gmLiGPAT, gsdmLiGPAT, and BY4742) and the deficient group (gat1Δ and gPY).
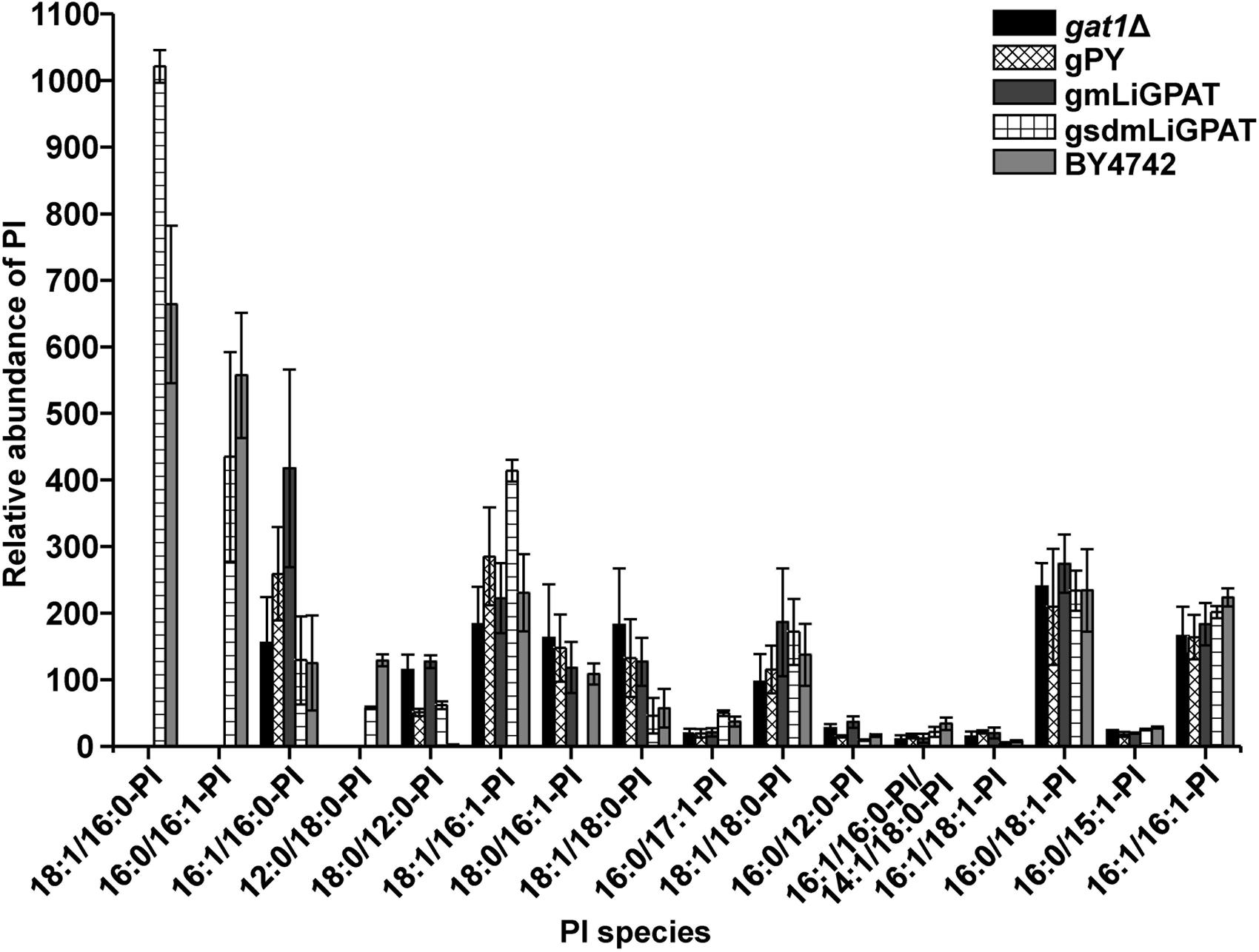
FIGURE 3. Relative abundance of PI species in the top 35 VIP from gat1Δ, gPY, gmLiGPAT, gsdmLiGPAT, and BY4742.
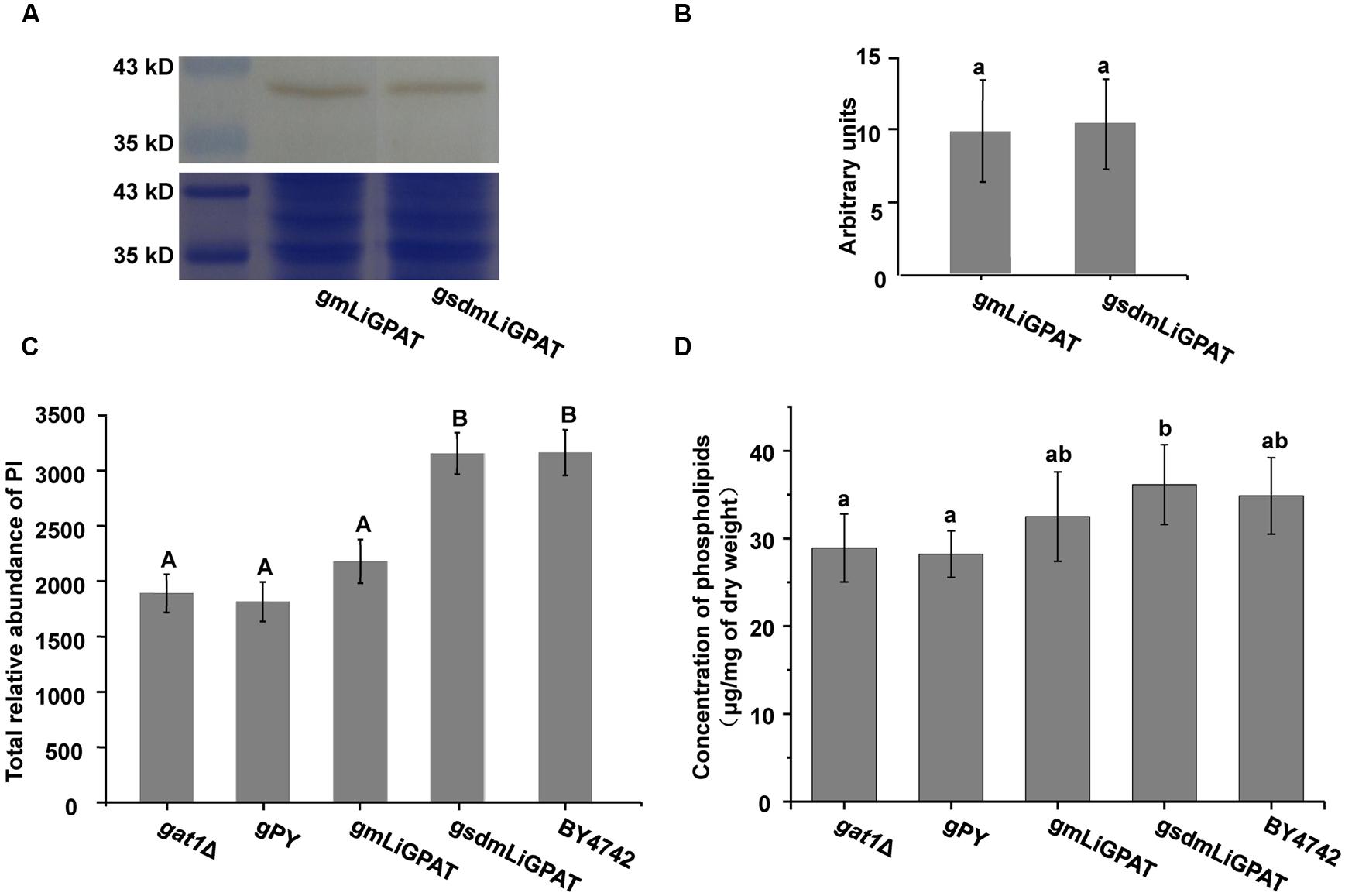
FIGURE 4. The effect of site-directed mutagenesis of LiGPAT from Arg195 to His in yeast. (A) Western blot analysis of gmLiGPAT and gsdmLiGPAT with the LiGPAT antibody. (B) The density of the blots of gmLiGPAT and gsdmLiGPAT were measured with ImageJ software and expressed in arbitrary optical density units. Values are the mean ± SD, n = 3. The average levels of gmLiGPAT and gsdmLiGPAT showed no difference (P > 0.05). (C) Comparison of the total relative abundance of all PI species among gat1Δ, gPY, gmLiGPAT, gsdmLiGPAT, and BY4742. Values with the same letter showed no significant difference (P > 0.05); the others showed significant differences (P < 0.01). (D) Comparison of the concentration of phospholipids among gat1Δ, gPY, gmLiGPAT, gsdmLiGPAT, and BY4742. Values with the same letter showed no significant difference (P > 0.05); the others showed significant differences (P < 0.05).
Discussion
Plastid-Localized GPAT from L. incisa
Glycerol-3-phosphate acyltransferases targeting the chloroplast, cytoplasm, and mitochondrion have been recognized in plants. The chloroplastidial GPAT localized in the stroma is a soluble protein, and it can utilize acyl-(acyl-carrier protein) as the acyl donor (Joyard and Douce, 1977). In contrast, the cytoplasmic form targeted to the ER is hydrophobic, and it is able to utilize acyl-CoA as the acyl donor (Frentzen et al., 1990). Genes of both chloroplastidial and ER-bound GPAT from several higher plants have been cloned, a total of 10 from Arabidopsis (Zheng et al., 2003; Xu et al., 2006; Gidda et al., 2009; Cagliari et al., 2010; Chen et al., 2010; Yang et al., 2012), 9 from Ricinus communis (Cagliari et al., 2010), and at least 2 from Glycine max (Eskandari et al., 2013), for instance. Examination of the algal genomes indicated that the microalgae Chlamydomonas reinhardtii, Ostreococcus tauri, Cyanidioschyzon merolae strain 10D, Phaeodactylum tricornutum CCAP 1055/1, and Thalassiosira pseudonana CCMP1335 were missing the recognizable extraplastidial GPAT homologs (Lykidis and Ivanova, 2008). It was suggested that the GPATs in these microalgae might have dual localization in both chloroplast and ER (Lykidis and Ivanova, 2008).
The present study provides the convincible bioinformatics evidence that one GPAT from the green microalga L. incisa is localized to chloroplasts (Figures 1A,C). In the latest NCBI database, there were deposited putative green microalgal GPATs, which were similar to the ER-bound GPAT9 from Arabidopsis. Therefore, it was inferred that the GPATs from L. incisa and the above-mentioned microalgae might be only localized to chloroplasts. Obviously, this idea would be more convincing and significant with analysis of accurate subcellular localization of the chloroplastidial GPATs and function of the cytoplasmic ones from these green microalgae.
Site-Directed Mutagenesis of LiGPAT Resulted in an Increase of the Phospholipid Level in Yeast
The phospholipid level of the yeast transformed with the site-directed mutated LiGPAT (Arg195His) was higher than that of yeast transformed with the original LiGPAT (Figure 4D), indicating that the catalytic ability of LiGPAT was improved by site-directed mutagenesis. To explore whether this improvement resulted from an increased level of protein expression or an increased enzymatic activity, semi-quantitative analysis of LiGPAT introduced into yeast was performed using western blots with purified LiGPAT polyclonal antibody. The reliability of this antibody was supported by western blot analysis of the total proteins extracted from L. incisa and transformed E. coli pmLiG/BL (Supplementary Figure S2). Comparison of the band intensity on the blots indicated that the expression levels of mLiGPAT and sdmLiGPAT were not significantly difference (P > 0.05) (Figures 4A,B), suggesting that the site-directed mutagenesis from Arg195 to His had no effect on protein expression level but could enhance the enzymatic activity of LiGPAT. This prompted us to investigate the relationship between protein structure and enzymatic activity of LiGPAT because the mutated residue Arg is situated in the G-3-P binding pocket (Figure 1A).
The crystal structure of squash chloroplastidial GPAT protein (PDB entry 1K30) was the only structure solved with high-resolution that elucidated the structure-function relationship of GPAT (Heath and Rock, 1998; Turnbull et al., 2001; Tamada et al., 2004). Accordingly, 3D models of mLiGPAT and sdmLiGPAT (Figure 5) were developed by using the I-TASSER server, which was an integrated platform for automated protein structure and function prediction based on the sequence-to-structure-to-function paradigm (Roy et al., 2010). The secondary structural elements of the mLiGPAT and sdmLiGPAT proteins were organized into two domains (Figure 5A), which were found well conserved in Cucurbita moschata, Chlamydomonas reinhardtii, Arabidopsis thaliana, and Glycine max (Turnbull et al., 2001; Misra and Panda, 2013). Domain I is the GPAT_N, and it forms a four-helix bundle (consisting of the 310 helix linking residues 7–10 and helices α1–3) with a simple square, right-handed up–down-up–down topology (Figures 1A and 5A). A loop region called the “interlinking loop” linked the small Domain I and the large Domain II. Domain II comprises the alternating α/β secondary structural elements (Figure 5A) and positively charged residues, which constitutes a positively charged pocket for binding the phosphate group of G-3-P. These residues were well conserved in most plants, except that the His residue at position 195 in mLiGPAT was substituted by Arg (Figure 1A). However, this replacement did not change the charge property and secondary structure of the pocket from mLiGPAT compared to that from its site-directed mutant sdmLiGPAT (Figure 5A). Structural superimposition of the binding sites for the phosphate group of G-3-P from mLiGPAT and sdmLiGPAT was illustrated by atom (Figure 5B) and surface (Figure 5C). The modeling indicated that the side-chain conformation of residues at positions 195 and 238 were different between these two proteins, suggesting a smaller accessible surface of the phosphate group binding pocket from mLiGPAT than from sdmLiGPAT. Therefore, changes of the side-chain conformation might be responsible for the difference in the enzymatic activity of mLiGPAT and sdmLiGPAT when they function in yeast.
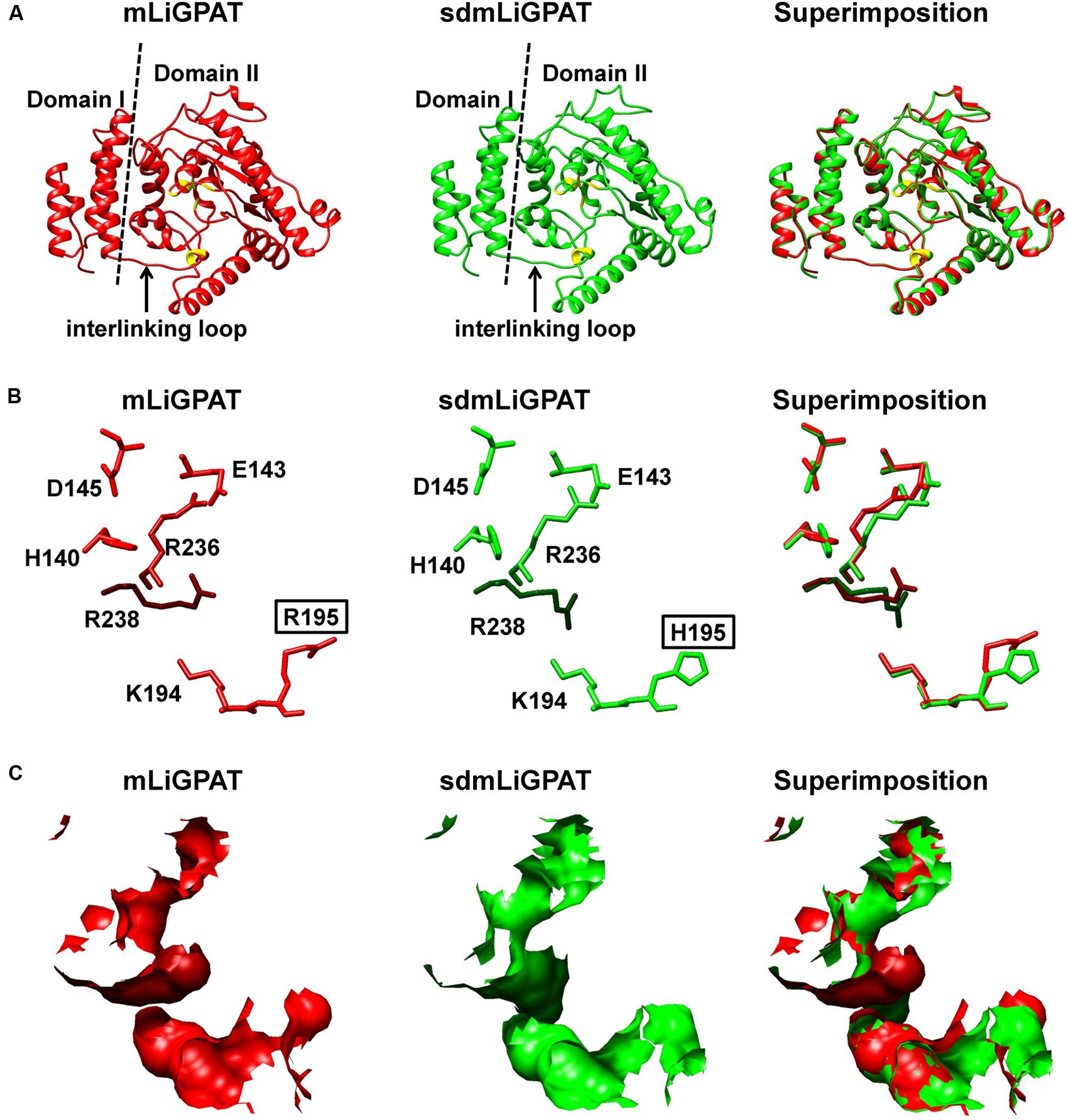
FIGURE 5. Superimposition of 3D structure models of the LiGPAT mature protein (red) and its mutant (Arg195His) (green). (A) Schematic ribbon representation showing the arrangements of α-helices, β-sheets, and loops. Domain I (left) and Domain II (right) are separated by the dashed line. The “interlinking loop” region is indicated by arrow. Residues composing the positively charged G-3-P binding pocket are in yellow. Atom (B) and surface (C) representation displaying the superimposition of the positively charged G-3-P binding pocket.
In brief, acylation by GPAT is considered to be the rate-limiting step in the glycerolipid synthesis pathway and to regulate fatty acid flux through the pathway (Coleman and Lee, 2004; Wendel et al., 2009). In Arabidopsis, RNAi of the chloroplastidial GPAT in the ats1-1 mutant background led to small leaves (Xu et al., 2006). Thus, it was inferred that the low growth rate of L. incisa (Ouyang et al., 2013b) might be partially associated with the relatively low enzymatic activity of LiGPAT. Recently, a modification of the GPAT-coding gene together with four other genes has been documented to improve the TAG content in Chlorella minutissima UTEX 2219 (Hsieh et al., 2012). Hence, genetic manipulation of the G-3-P binding sites of GPAT could be taken as a breakthrough to increase the growth rate and glycerolipid content of L. incisa and other microalgae.
Author Contributions
Z-GZ and L-LO designed the study and wrote the paper. L-LO carried out the experiments and she and Z-GZ were involved in data analysis. HL assisted with heterologous expression of LiGPAT in yeast. X-JY and J-LX helped design the lipidomic experiments and interpret the data. Z-GZ gave the final approval of the version to be published. All authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professors Chengwu Zhang from Jinan University for providing Lobosphaera incisa H4301. This work was supported by the National Natural Science Foundation of China (31402274, 31172389), China Postdoctoral Science Foundation (2014M551381), and the Special Project of Marine Renewable Energy from the State Oceanic Administration (SHME2011SW02).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00286
Abbreviations
AA, arachidonic acid; CDD, Conserved Domain Database; CIJFJK, jack-knifed confidence interval; cTP, chloroplast transit peptide; CV-ANOVA, cross-validated analysis of variance; DAB, diaminobenzidine; ER, endoplasmic reticulum; G-3-P, Glycerol-3-phosphate; GPAT, Glycerol-3-phosphate acyltransferase; LPAAT, lysophosphatidic acid acyltransferase; m/z, mass-to-charge ratio; NC, nitrocellulose; OPLS-DA, orthogonal projection to latent structures with discriminant analysis; PC, phosphatidylcholine; PCA, principal components analysis; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PLS-DA, projection to latent structures with discriminant analysis; PS, phosphatidylserine; RT, retention time; SDS-PAGE, SDS polyacrylamide gel electrophoresis; TAG, triacylglycerol; TAP, Tris acetate phosphate; UPLC-ESI-Q-TOF-MS, ultra performance liquid chromatography-electron spraying ionization-qadrupole-time-of-flight-mass spectrometry; VIP, variable importance in the projection.
Footnotes
- ^http://blast.ncbi.nlm.nih.gov/
- ^http://www.ncbi.nlm.nih.gov/spidey/
- ^http://www.cbs.dtu.dk/services/SignalP/
- ^http://www.cbs.dtu.dk/services/TargetP/
- ^http://www.cbs.dtu.dk/services/ChloroP/
- ^http://www.predictprotein.org
- ^http://zhanglab.ccmb.med.umich.edu/I-TASSER/
- ^http://wishart.biology.ualberta.ca/SuperPose/
- ^http://web.uni-frankfurt.de/fb15/mikro/euroscarf/
- ^rsb.info.nih.gov/ij/
- ^http://www.hmdb.ca
- ^http://www.lipidmaps.org
- ^http://metlin.scripps.edu
References
Altschul, S. F., Gish, W., Miller, W., Myer, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1006/jmbi.1990.9999
Ariizumi, T., Hatakeyama, K., Hinata, K., Inatsugi, R., Nishida, I., Sato, S., et al. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39, 170–181. doi: 10.1111/j.1365-313x.2004.02118.x
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Bhella, R. S., and Mackenzie, S. L. (1994). Nucleotide sequence of a cDNA from Carthamus tinctorius encoding a glycerol-3-phosphate acyltransferase. Plant Physiol. 106, 1713–1714. doi: 10.1104/pp.106.4.1713
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein using the principal of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Cagliari, A., Margis-Pinheiro, M., Loss, G., Mastroberti, A. A., de Araujo Mariath, J. E., and Margis, R. (2010). Identification and expression analysis of castor bean (Ricinus communis) genes encoding enzymes from the triacylglycerol biosynthesis pathway. Plant Sci. 179, 499–509. doi: 10.1016/j.plantsci.2010.07.015
Cai, D., Li, D., Zhao, S., Dou, X., Wang, F., Huang, G., et al. (2015). A correlation between diet and longevity characterization by means of element profiles in healthy people over 80 years from a chinese longevous region. Biol. Trace Elem. Res. 165, 18–29. doi: 10.1007/s12011-015-0233-7
Chen, X., Truksa, M., Snyder, C. L., El-Mezawy, A., Shah, S., and Weselake, R. J. (2010). Three homologous genes encoding sn-glycerol-3-phosphate acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus. Plant Physiol. 155, 851–865. doi: 10.1104/pp.110.169482
Christie, W. W., and Han, X. (2010). Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. Bridgwater: The Oily Press.
Coleman, R. A., and Lee, D. P. (2004). Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43, 134–176. doi: 10.1016/S0163-7827(03)00051-1
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Dellaporta, S. L., Wood, J., and Hick, J. B. (1983). A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. doi: 10.1007/bf02712670
Eriksson, L., Johansson, E., Kettaneh-Wold, N., Trygg, J., Wikström, C., and Wold, S. (2006). Multi- and Megavariate Data Analysis. Basic principles and applications, Part I. Umeå: Umetrics AB.
Eskandari, M., Cober, E. R., and Rajcan, I. (2013). Using the candidate gene approach for detecting genes underlying seed oil concentration and yield in soybean. Theor. Appl. Genet. 126, 1839–1850. doi: 10.1007/s00122-013-2096-7
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Fiehn, O., Kopka, J., Dörmann, P., Altmann, T., Trethewey, R. N., and Willmitzer, L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18, 1157–1161. doi: 10.1038/81137
Frentzen, M., Neuburger, M., Joyard, J., and Douse, R. (1990). Intraorganelle localization and substrate specificities of the mitochondrial acyl-CoA:sn-glycerol-3-phosphate O-acyl-transferase and acy-CoA:1-acyl-sn-glycerol-3-phosphate O-acyltransferase from potato tubers and pea leaves. Eur. J. Biochem. 187, 395–402. doi: 10.1111/j.1432-1033.1990.tb15317.x
Ge, L., and Rudolph, P. (1997). Simultaneous introduction of multiple mutations using overlap extension PCR. Biotechniques 22, 28–30.
Gidda, S. K., Shockey, J. M., Rothstein, S. J., Dyer, J. M., and Mullen, R. T. (2009). Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 47, 867–879. doi: 10.1016/j.plaphy.2009.05.008
Heath, R. J., and Rock, C. O. (1998). A conserved histidine is essential for glycerolipid acyltransferase catalysis. J. Bacteriol. 180, 1425–1430.
Hsieh, H. J., Su, C. H., and Chien, L. J. (2012). Accumulation of lipid production in Chlorella minutissima by triacylglycerol biosynthesis-related genes cloned from Saccharomyces cerevisiae and Yarrowia lipolytica. J. Microbiol. 50, 526–534. doi: 10.1007/s12275-012-2041-5
Ishizaki, O., Nishida, I., Agata, K., Eguchi, G., and Murata, N. (1988). Cloning and nucleotide sequence of cDNA for the plastid glycerol-3-phosphate acyltransferase from squash. FEBS Lett. 238, 424–430. doi: 10.1016/0014-5793(88)80525-8
Joyard, D., and Douce, R. (1977). Site of synthesis of phosphatidic acid and diacylglycerol in spinach chloroplasts. Biochim. Biophys. Acta 486, 273–285. doi: 10.1016/0005-2760(77)90023-6
Kang, J., Choi, M.-Y., Kang, S., Kwon, H. N., Wen, H., Lee, C. H., et al. (2008). Application of a 1H nuclear magnetic resonance (NMR) metabolomics approach combined with orthogonal projections to latent structure-discriminant analysis as an efficient tool for discriminating between Korean and Chinese herbal medicines. J. Agric. Food Chem. 56, 11589–11595. doi: 10.1021/jf802088a
Khozin-Goldberg, I., Bigogno, C., Shrestha, P., and Cohen, Z. (2002). Nitrogen starvation induces the accumulation of arachidonic acid in the freshwater green alga Parietochloris incisa (Trebouxiophyceae). J. Phycol. 38, 991–994. doi: 10.1046/j.1529-8817.2002.01160.x
Kunst, L., Browse, J., and Somerville, C. (1988). Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 85, 4143–4147. doi: 10.1073/pnas.85.12.4143
Lewin, T. M., Wang, P., and Coleman, R. A. (1999). Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38, 5764–5771. doi: 10.1021/bi982805d
Lu, N., Wei, D., Chen, F., and Yang, S.-T. (2012). Lipidomic profiling and discovery of lipid biomarkers in snow alga Chlamydomonas nivalis under salt stress. Eur. J. Lipid Sci. Technol. 114, 253–265. doi: 10.1002/ejlt.201100248
Lykidis, A., and Ivanova, N. (2008). “Genomic prospecting for microbial biodiesel production,” in Bioenergy, eds J. D. Wall, C. S. Harwood, and A. Demain (Washington: ASM Press), 407–418.
Maiti, R., Van Domselaar, G. H., Zhang, H., and Wishart, D. S. (2004). SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 32, W590–W594. doi: 10.1093/nar/gkh477
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Merzlyak, M. N., Chivkunova, O. B., Gorelova, O. A., Reshetnikova, I. V., Solovchenko, A. E., Khozin-Goldberg, I., et al. (2007). Effect of nitrogen starvation on optical properties, pigments, and arachidonic acid content of the unicellular green alga Parietochloris incisa (Trebouxiophyceae, Chlorophyta). J. Phycol. 43, 833–843. doi: 10.1111/j.1529-8817.2007.00375.x
Misra, N., and Panda, P. K. (2013). In search of actionable targets for agrigenomics and microalgal biofuel production: sequence-structural diversity studies on algal and higher plants with a focus on GPAT protein. OMICS 17, 173–186. doi: 10.1089/omi.2012.0094
Nishida, I., Tasaka, Y., Shiraisi, H., and Murata, N. (1993). The gene and the RNA for the precursor to the plastid-located glycerol-3-phosphate acyltransferase of Arabidopsis thaliana. Plant Mol. Biol. 21, 267–277. doi: 10.1007/bf00019943
Ouyang, L.-L., Chen, S.-H., Li, Y., and Zhou, Z.-G. (2013a). Transcriptome analysis reveals unique C4-like photosynthesis and oil body formation in an arachidonic acid-rich microalga Myrmecia incisa Reisigl H4301. BMC Genomics 14:396. doi: 10.1186/1471-2164-14-396
Ouyang, L.-L., Du, D. H., Yu, S. Y., Li, C. Y., Zhang, C. W., Gao, H. J., et al. (2012). Expressed sequence tags analysis revealing the taxonomic position and fatty acid biosynthesis in an oleaginous green microalga, Myrmecia incisa Reisigl (Trebouxiophyceae, Chlorophyta). Chin. Sci. Bull. 57, 3342–3352. doi: 10.1007/s11434-012-5159-2
Ouyang, L.-L., Li, H., Liu, F., Tong, M., Yu, S. Y., and Zhou, Z.-G. (2013b). “Accumulation of arachidonic acid in a green microalga, Myrmecia incisa H4301, enhanced by nitrogen starvation and its molecular regulation mechanism,” in Arachidonic acid: Dietary Sources and General Functions, eds G. G. Dumancas, B. S. Murdianti, and E. A. Lucas (New York, NY: Nova Science Publishers), 1–20.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Redón, M., Guillamón, J. M., Mas, A., and Rozès, N. (2011). Effect of growth temperature on yeast lipid composition and alcoholic fermentation at low temperature. Eur. Food Res. Technol. 232, 517–527. doi: 10.1007/s00217-010-1415-3
Rost, B., Yachdav, G., and Liu, J. (2004). The predict protein server. Nucleic Acids Res. 32, W321–W326. doi: 10.1093/nar/gkh377
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Schein, C. H., and Noteborn, M. H. M. (1988). Formation of soluble recombinant proteins in Escherichia coli is favored by lower growth temperature. Nat. Biotechnol. 6, 291–294. doi: 10.1038/nbt0388-291
Shorrosh, B. S., Savage, L. J., Soll, J., and Ohlrogge, J. B. (1996). The pea chloroplast membrane-associated protein, IEP96, is a subunit of acetyl-CoA carboxylase. Plant J. 10, 261–268. doi: 10.1046/j.1365-313x.1996.10020261.x
Slabas, A. R., Kroon, J. T. M., Scheirer, T. P., Gilroy, J. S., Hayman, M., Rice, D. W., et al. (2002). Squash glycerol-3-phosphate (1)-acyltransferase: alteration of substrate selectivity and identification of arginine and lysine residues important in catalytic activity. J. Biol. Chem. 277, 43918–43923. doi: 10.1074/jbc.m206429200
Smith, D. E., and Fisher, P. A. (1984). Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J. Cell Biol. 99, 20–28. doi: 10.1083/jcb.99.1.20
Stanier, R. Y., Kunisawa, M. M., and Cohen-Bazir, G. (1971). Purification and properties of unicellular blue-green algae (order Chlorococcales). Bacteriol. Rev. 35, 171–201.
Tamada, T., Feese, M. D., Ferri, S. R., Kato, Y., Yajima, R., and Toguri, T. (2004). Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea. Acta Crystallogr. D Biol. Crystallogr. 60, 13–21. doi: 10.1107/s0907444903020778
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tong, M., Yu, S. Y., Ouyang, L.-L., and Zhou, Z.-G. (2011). Comparison of increased arachidonic acid content in Myrmecia incisa cultured during the course of nitrogen or phosphorus starvation. J. Fish. China 35, 763–773. doi: 10.3724/SP.J.1231.2011.17114
Turnbull, A. P., Rafferty, J. B., Sedelnikova, S. E., Slabas, A. R., Schierer, T. P., Kroon, J. T. M., et al. (2001). Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase. Structure 9, 347–353. doi: 10.1016/S0969-2126(01)00595-0
Van Assche, R., Temmerman, L., Dias, D. A., Boughton, B., Boonen, K., Braeckman, B. P., et al. (2015). Metabolic profiling of a transgenic Caenorhabditis elegans Alzheimer model. Metabolomics 11, 477–486. doi: 10.1007/s11306-014-0711-5
von Heijne, G., Steppuhn, J., and Herrmann, R. G. (1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180, 535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x
Weber, S., Wolter, F. P., Buck, F., Frentzen, M., and Heinz, E. (1991). Purification and cDNA sequencing of an oleate-selective acyl-ACP:sn-glycerol-3-phosphate acyltransferase from pea chloroplasts. Plant Mol. Biol. 17, 1067–1076. doi: 10.1007/bf00037145
Wendel, A. A., Lewin, T. M., and Coleman, R. A. (2009). Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochim. Biophys. Acta 1791, 501–506. doi: 10.1016/j.bbalip.2008.10.010
Wiklund, S., Karlsson, M., Antti, H., Johnels, D., Sjöström, M., Wingsle, G., et al. (2005). A new metabonomic strategy for analysing the growth process of the poplar tree. Plant Biotechnol. J. 3, 353–362. doi: 10.1111/j.1467-7652.2005.00129.x
Xu, C., Yu, B., Cornish, A. J., Froehlich, J. E., and Benning, C. (2006). Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3-phosphate acyltransferase. Plant J. 47, 296–309. doi: 10.1111/j.1365-313X.2006.02790.x
Yan, X. J., Li, H. Y., Xu, J. L., and Zhou, C. X. (2010). Analysis of phospholipids in microalga Nitzschia closterium by UPLC-Q-TOF-MS. Chin. J. Oceanol. Limnol. 28, 106–112. doi: 10.1007/s00343-010-9263-3
Yang, W., Simpson, J. P., Li-Beisson, Y., Beisson, F., Pollard, M., and Ohlrogge, J. B. (2012). A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiol. 160, 638–652. doi: 10.1104/pp.112.201996
Zhang, C.-W., Cohen, Z., Khozin-Goldberg, I., and Richmond, A. (2002). Characterization of growth and arachidonic acid production of Parietochloris incisa comb. nov (Trebouxiophyceae, Chlorophyta). J. Appl. Phycol. 14, 453–460. doi: 10.1023/A:1022375110556
Zheng, Z., Xia, Q., Dauk, M., Shen, W., Selvaraj, G., and Zou, J. (2003). Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15, 1872–1887. doi: 10.1105/tpc.012427
Keywords: Lobosphaera incisa H4301, glycerol-3-phosphate acyltransferase (GPAT), plastid, site-directed mutagenesis, UPLC-Q-TOF-MS, glycerolipid
Citation: Ouyang L-L, Li H, Yan X-J, Xu J-L and Zhou Z-G (2016) Site-Directed Mutagenesis from Arg195 to His of a Microalgal Putatively Chloroplastidial Glycerol-3-Phosphate Acyltransferase Causes an Increase in Phospholipid Levels in Yeast. Front. Plant Sci. 7:286. doi: 10.3389/fpls.2016.00286
Received: 05 July 2015; Accepted: 22 February 2016;
Published: 10 March 2016.
Edited by:
Flavia Vischi Winck, University of São Paulo, BrazilReviewed by:
Yves Waché, AgroSup Dijon/University of Burgundy, FranceVladimir I. Titorenko, Concordia University, Canada
Copyright © 2016 Ouyang, Li, Yan, Xu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Gang Zhou, zgzhou@shou.edu.cn
 Long-Ling Ouyang
Long-Ling Ouyang Hui Li2
Hui Li2