- 1School of Life Sciences, Chongqing University, Chongqing, China
- 2College of Agronomy and Biotechnology, Southwest University, Chongqing, China
Target of rapamycin (TOR), a master sensor for growth factors and nutrition availability in eukaryotic species, is a specific target protein of rapamycin. Rapamycin inhibits TOR kinase activity viaFK506 binding protein 12 kDa (FKBP12) in all examined heterotrophic eukaryotic organisms. In Arabidopsis, several independent studies have shown that AtFKBP12 is non-functional under aerobic condition, but one study suggests that AtFKBP12 is functional during anaerobic growth. However, the functions of AtFKBP12 have never been examined in parallel under aerobic and anaerobic growth conditions so far. To this end, we cloned the FKBP12 gene of humans, yeast, and Arabidopsis, respectively. Transgenic plants were generated, and pharmacological examinations were performed in parallel with Arabidopsis under aerobic and anaerobic conditions. ScFKBP12 conferred plants with the strongest sensitivity to rapamycin, followed by HsFKBP12, whereas AtFKBP12 failed to generate rapamycin sensitivity under aerobic condition. Upon submergence, yeast and human FKBP12 can significantly block cotyledon greening while Arabidopsis FKBP12 only retards plant growth in the presence of rapamycin, suggesting that hypoxia stress could partially restore the functions of AtFKBP12 to bridge the interaction between rapamycin and TOR. To further determine if communication between TOR and auxin signaling exists in plants, yeast FKBP12 was introduced into DR5::GUS homozygous plants. The transgenic plants DR5/BP12 were then treated with rapamycin or KU63794 (a new inhibitor of TOR). GUS staining showed that the auxin content of root tips decreased compared to the control. DR5/BP12 plants lost sensitivity to auxin after treatment with rapamycin. Auxin-defective phenotypes, including short primary roots, fewer lateral roots, and loss of gravitropism, occurred in DR5/BP12 plants when seedlings were treated with rapamycin+KU63794. This indicated that the combination of rapamycin and KU63794 can significantly inhibit TOR and auxin signaling in DR5/BP12 plants. These studies demonstrate that TOR is essential for auxin signaling transduction in Arabidopsis.
Introduction
Rapamycin (also known as sirolimus), a macrolide antibiotic, is produced by the soil bacterium, Streptomyces hydroscopicus. All tested heterotrophic eukaryotes, including yeast, nematodes (Caenorhabditiselegans), fruit flies (Drosophila), and mammals, are sensitive to rapamycin (Xiong and Sheen, 2015). Rapamycin feeding mimics nutrition restriction and energy depletion and thus significantly extends the lifespans of mammals and plants (Fontana et al., 2010; Ren et al., 2012). Rapamycin is therefore a research focus of human preventive medicine. Interestingly, most plants are insensitive to rapamycin under aerobic growth condition (Xu et al., 1998; Menand et al., 2002; Ren et al., 2012; Montané and Menand, 2013). This insensitivity might involve self-immune mechanisms. From an evolutionary perspective, billions of years separate the beginnings of yeast and the development of human cells, yet human cell exposure to rapamycin produces the same nutrition starvation phenomenon as found in yeast. This indicates a highly conserved target for rapamycin in eukaryotes. TOR1 and TOR2 genes, targets of rapamycin, have been identified in budding yeast and this has allowed advanced TOR studies (Cafferkey et al., 1993; Kunz et al., 1993; Sabatini et al., 1994; Chen et al., 1995; Loewith et al., 2002). Since its initial discovery, the TOR gene has been isolated from all examined eukaryotic organisms. Most eukaryotic organisms contain only one TOR gene, whereas two and three TOR genes exist in yeast and Leishmania major, respectively (Madeira Da Silva and Beverley, 2010). Disruption of the TOR gene is lethal in eukaryotes, indicating that TOR is required for life in eukaryotic cells (Wullschleger et al., 2006). Disruption of the TOR signal is one of the major causes of nutrition-related diseases in animals and humans, including diabetes, cancer, and cardiovascular disease (Zagouri et al., 2012; Cornu et al., 2013). TOR function is highly conserved from yeast to humans, and it controls key biological processes such as ribosome biogenesis, protein synthesis, three carboxylic acid cycles, and stress responses (Fontana et al., 2010; Cornu et al., 2013).
The 12-KDa FK506-binding protein 12 (FKBP12) is the receptor protein of rapamycin and it mediates the interaction between TOR and rapamycin (Brown et al., 1994). In yeast and mammals, rapamycin first forms a heterogeneous complex with FKBP12 and then specifically targets and binds to the FRB domain of TOR to form a rapamycin-FKBP12-TOR complex that in turn inhibits the kinase activity of TOR (Chiu et al., 1994; Sabatini et al., 1994; Choi et al., 1996). In this rapamycin-FKBP12-TOR system, FKBP12 plays a crucial role by being directly involved in rapamycin recognition and binding. Mutations in the FKBP12 gene result in rapamycin insensitivity in yeast (Koltin et al., 1991). TOR deletion is lethal, and all tested fungi and animals are sensitive to rapamycin (Heitman et al., 1991; Loewith et al., 2002; Wullschleger et al., 2006). Based on the rapamycin-FKBP12 negative regulation system of TOR, TOR and its signaling pathway in yeast and animals have been extensively studied. The structure and function of FKBP12 proteins are highly conserved and human FKBP12 can functionally complement that of yeast (Koltin et al., 1991). Although the amino acid sequences of plant FKBP12s are relatively similar to those of yeast and mammals, wild-type (WT) Arabidopsis is insensitive to rapamycin and does not express any detectable phenotypes even at high concentrations (20 μg/mL rapamycin) under aerobic condition (Xu et al., 1998; Menand et al., 2002; Mahfouz et al., 2006; Sormani et al., 2007; Ren et al., 2012; Montané and Menand, 2013). Various genetic, biochemical, yeast two hybrid (Y2H), and pharmacological analyses have demonstrated that plant FKBP12 does not have the ability to form rapamycin/FKBP12/TOR complexes (Xu et al., 1998; Sormani et al., 2007; Moreau et al., 2012; Ren et al., 2012; Montané and Menand, 2013). However, overexpression of AtFKBP12 can dramatically enhance rapamycin sensitivity during anaerobic growth (Xiong and Sheen, 2012). This observation invalidates the common belief that TOR signaling in Arabidopsis is always insensitive to rapamycin, but the AtFKBP12 transgenic plants have never been carefully examined in parallel under aerobic and anaerobic growth condition so far.
Growth factors such as insulin and IGF in mammals are the key signals determining cell growth, proliferation, differentiation, and fate. TOR has a close relationship with growth factors (Wang et al., 2006; Vander Haar et al., 2007; Feng and Levine, 2010). In plants, auxin is the major phytohormone and growth factor controlling the cell cycle, division, elongation, differentiation, growth, and development (Teale et al., 2006). Auxin and growth factors can activate the TOR signaling pathway to modulate specific mRNA transcription, translation, translation re-initiation, and selective protein synthesis by phosphorylating key downstream regulators such as S6 ribosomal protein kinase (S6K), S6 ribosomal protein, and eukaryotic initiation factor 4E (eIF4E; Dinkova et al., 2000; Beltrán-Peña et al., 2002; Bögre et al., 2013; Schepetilnikov et al., 2013; Villa-Hernández et al., 2013). Our recent observations indicate that many auxin signaling associated genes are differentially expressed in Arabidopsis seedlings treated with AZD8055 (one of asTORis; Dong et al., 2015). TOR therefore appears to play a crucial role in mediating auxin signals. However, one study reported that auxin is decoupled from TOR activation in Arabidopsis (Xiong et al., 2013). To clarify the issues concerning FKBP12 functions and the relationship between auxin and TOR signaling in plants, we cloned FKBP12 from human (HsFKBP12), yeast (ScFKBP12) and Arabidopsis (AtFKBP12) and generated overexpression transgenic lines of Arabidopsis. The functions of HsFKBP12, ScFKBP12 and AtFKBP12 were tested in parallel for their sensitivity to rapamycin under aerobic and anaerobic conditions. Our results demonstrate that plants can acquire rapamycin sensitivity by overexpression of HsFKBP12 or ScFKBP12, and ScFKBP12 showed more pronounced effects than HsFKBP12 under both aerobic and anaerobic growth conditions. No rapamycin response was observed from transgenic plants containing P35S::AtFKBP12 during aerobic growth, but the rapamycin sensitivity can be partially rescued in response to submergence. Double transgenic plants named after DR5/BP12s, which contained P35S::ScFKBP12 and DR5:: GUS, were generated. When DR5/BP12 plants were simultaneously treated with rapamycin and KU63794 (KU), normal growth and development of the roots was inhibited and the root geotropism was also altered. This was a classic phenotype of defective auxin signaling. GUS staining showed that auxin distribution was reduced in DR5/BP12 plants in the presence of the TOR inhibitor. TOR significantly influenced the concentration and response of auxin in the plant and altered the downstream signal response processes.
Materials and Methods
Plant Materials and Growth Conditions
In the present study, wild-type (WT) Arabidopsis Columbia (Columbia-0) ecotype and transgenic lines were grown in growth chambers with 16 h light/8 h dark photoperiod at 22°C. Transgenic plants were generated using the floral dipping method (Zhang et al., 2006). In all cases, A. thaliana seeds were surface-sterilized for 5 min in 70% ethanol, and then washed in 10% sodium hypochlorite containing 0.3% Tween-20. Finally, seeds were washed five times with sterilized water. Before sowing on plates, the seeds were incubated for 2 days at 4°C in the dark.
RNA Extraction and Generation of Overexpression Constructs
Total RNA was extracted from Arabidopsis Columbia (Columbia-0) and human cells using TRIzol (Invitrogen), following the manufacturer's protocol. cDNA was synthesized by using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Transgen). The full-length coding sequence of HsFKBP12 and AtFKBP12 was amplified by TransStarTaq DNA Polymerase (Transgen) using the corresponding primers (Supplementary Table 1). PCR primers for amplification of the full-length genes from Arabidopsis were designed based on its cDNA sequence (http://www.Arabidopsis.org, http://www.ncbi.nlm.nih.gov). A NotI site at the 5′ end of the forward primer and a Sbf I site at the 3′ end of reverse primer were introduced. The remaining steps of plasmid construction were performed as described elsewhere (Ren et al., 2012).
Quantitative Real-Time PCR and Semi-RT PCR
Samples were collected and frozen in liquid nitrogen for total RNA extraction using the RNAprep Pure Plant Kit (TianGen Biotech). Approximately 1 μg of total RNA was then used for reverse transcription. qRT-PCR was performed using the TransStart Top Green qPCRSuperMix (TransgenBiotech) kit, following the manufacturer's protocol and using a two-step method for PCR reaction. The PCR parameters included 94°C pre-denaturation for 30 s, followed by 40 cycles of the following two steps: 5 s at 94°C and 30 s at 60°C. Plant actin2 was used as constitutive reference. The primers used in the present study were listed in Supplementary Table 1.
EasyTaq DNA Polymerase (TransGen Biotech) was used for semi-RT PCR, following the manufacturer's protocol. The PCR parameters included 94°C pre-denaturation for 5 min, followed by 24 cycles for the following three steps: 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C. This was then followed by incubation for 10 min at 72°C for final extension.
Generation of Promoter-GUS Constructs
The genomic DNA of Arabidopsis was extracted from Arabidopsis Columbia (Columbia-0) using one step Plant DNA Extraction Reagent (BIOTEKE), following the manufacturer's protocol. Based on our previous PRPS6B:: GUS construct (Ren et al., 2012), the PRPS6B was replaced by the promoter of AtFKBP12 (1.1 kb) at the AsisI and NotI restriction sites, respectively. Then PAtFKBP12:: GUS construct was transferred into the destination vector, pEarleyGate303, via LR recombination reactions. The primers of PAtFKBP12 are listed in the Supplementary Table 1.
Western Blotting
Total protein was extracted using a cell lysis buffer for Western and IP (Biyuntian), which contained 20 mMTris (pH7.5), 150 mMNaCl, 1%Triton X-100, sodium pyrophosphate, β-glycerophosphate, EDTA, Na3VO4, and leupeptin. Approximately 1 mM of phenylmethylsulphonyl fluoride (PMSF), 1 × Protease Inhibitor Cocktail (Sigma), and 1 × Phosphatase Inhibitor Cocktail 3 (Sigma) were added to the cell lysis buffer for Western and IP prior to the extraction process. The supernatant was collected after centrifugation at 12,000 rpm and quantified by using the RC/DC protein assay kit (BioRad). Approximately 40 μg of total protein was loaded onto a 15% SDS-PAGE gel. After electrophoresis and electroblotting, PVDF membranes (GE) were blocked with milk blocking buffer (Cwbio) for 12 h (overnight) at 4°C and then incubated in blocking buffer containing anti-HA or anti-Myc (dilution: 1:1000; Earthox, E022010-01, E022050-01) for 2 h at room temperature. The PVDF membrane was washed three times (10 min each) with PBST (Cwbio). The secondary antibody [HRP, goat anti-mouse IgG (H+L) Earthox, E030110-01] was diluted 1:10,000 with PBST. Incubation and elution of the secondary antibody were performed as previously described. Proteins were detected using the cECL Western Blot Kit (Cwbio).
Rapamycin Sensitivity Test
Our previous study have shown that low concentrations of rapamycin severely inhibit the growth of BP12-2 (Ren et al., 2012). In current work, a rapamycin concentration gradient from 0 to 5 μM was added to 0.5 MS medium, and DMSO was used as control. Four kinds of plant seeds, including the WT, BP12-2, P35S:: AtFKBP12, and P35S:: HsFKBP12were surface-sterilized and sowed on the medium. After 10 days, differences between various treatments and materials were assessed, and fresh weight and root length were measured. Under liquid growth condition, 3 ml 0.5 MS liquid medium with DMSO or rapamycin was added into 6-well plate. The liquid medium was refreshed every 2 days.
Fresh Weight and Root Growth Measurements
Plants were grown on 0.5 MS medium with different TOR inhibitors. After 10 days growth, all Petri dishes were photographed, and the fresh weight of plants was determined by weighing. Root length measurements were repeated four times per treatment by using the ImageJ software, and measurements for fresh weight and lateral root density were repeated four times. The measurement for lateral root initiation index was as described elsewhere (Dubrovsky et al., 2009).
β-GUS Staining
GUS staining was performed as previously described (Menand et al., 2002) at 6 h for PAtFKBP12:: GUS, 30 min for 35S:: GUS, and 1 h for DR5/BP12-OE11.
Results
Overexpression of ScFKBP12 and HsFKBP12, but Not AtFKBP12, Rescues Arabidopsis Rapamycin Sensitivity under Aerobic Growth Condition
Most plants are resistant to rapamycin in solid medium (Xu et al., 1998; Menand et al., 2002; Ren et al., 2012; Montané and Menand, 2013). Two hypotheses are proposed for rapamycin insensitivity in plants. One hypothesis is that rapamycin resistance in plants is due to the low expression of FKBP12. Another hypothesis is the currently accepted opinion that plant FKBP12s cannot bind to rapamycin. To study the first hypothesis, the AtFKBP12 promoter was amplified from Arabidopsis genomic DNA, and the PAtFKBP12::GUS plasmid was constructed and transformed into WT Arabidopsis. A total of 12 independent transgenic plants containing PAtFKBP12:: GUS were obtained and confirmed by DNA-PCR using GUS specific primers (Supplementary Table 1). GUS staining showed that the GUS signal of PAtFKBP12:: GUS plants was much lower than that of P35S:: GUS transgenic plants, which were generated in a previous study (Figure 1A; Ren et al., 2011). Total RNA from different tissues of Arabidopsis, including roots, stems, leaves, flowers, and siliques was extracted to determine tissue-specific expression of the AtFKBP12 gene. qRT-PCR analysis showed that AtFKBP12was constitutively expressed in Arabidopsis (Figure 1B). The expression level of AtFKBP12 was higher in the leaves and flowers than in the roots, stems, and siliques.
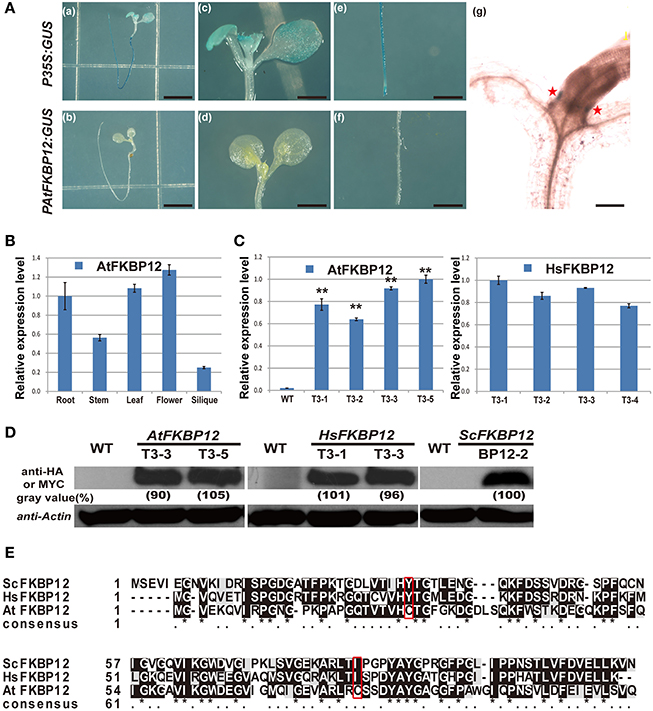
Figure 1. The expression pattern of AtFKBP12 in Arabidopsis and the identification of transgenic lines of AtFKBP12, HsFKBP12 and PAtFKBP12::GUS in Arabidopsis. (A) The GUS staining of PAtFKBP12::GUS and P35S::GUS transgenic lines. 5 DAG (days after germination) seedlings were stained. (a,b) bar = 0.5 cm; (c–f) bar = 2 mm; (g) bar = 0.2 mm. (B) qRT-PCR to detect expression pattern of AtFKBP12 in different Arabidopsis tissues. (C) qRT-PCR to detect expression level of AtFKBP12 or HsFKBP12 in transgenic lines. Asterisks denote Student's t-test significance compared with WT (*P < 0.05;**P < 0.01). (D) The protein expression level in three FKBP12 transgenic lines. AtFKBP12 and HsFKBP12 used anti-HA antibody; BP12-2 used anti-MYC antibody. (E) The multiple protein sequence alignment of ScFKBP12, HsFKBP12, and AtFKBP12. Significant mutations were marked with red box.
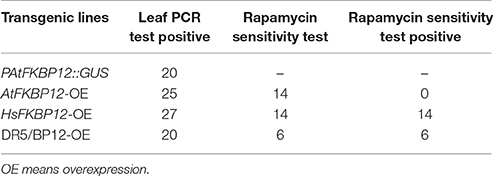
To further examine whether rapamycin insensitivity in Arabidopsis in solid medium results from the low expression of AtFKBP12, transgenic lines containing P35S::AtFKBP12 were prepared. A total of 25 independent lines were identified by PCR analysis of leaf tissue. A total of 14 independent lines (OE1–14) were selected for expression profiling of AtFKBP12 by semi-qPCR. The AtFKBP12transcript was detected in all 14 independent lines, whereas the expression levels of AtFKBP12varied among the different lines. Four lines with the highest AtFKBP12 expression levels were selected for qRT-PCR analysis (Figure 1C). For the rapamycin sensitivity assay, all transgenic lines were grown on solid medium supplemented with 5 μM rapamycin, and the results demonstrated that all lines, including the highest overexpression lines (OE1, OE3, OE5, and OE14) and the lowest overexpression lines (OE2 and OE6) were insensitive to rapamycin under aerobic condition (Table 1). These findings indicate that the expression level of AtFKBP12 was not related to rapamycin insensitivity in plants. This observation was consistent with most previous studies (Xu et al., 1998; Menand et al., 2002; Mahfouz et al., 2006; Sormani et al., 2007; Ren et al., 2012; Montané and Menand, 2013).
To verify this result, we cloned HsFKBP12 from human cells and generated 25 independent P35S::HsFKBP12 transgenic lines. OE1–OE14 were selected for semi-qPCR and qRT-PCR analyses and treated with 5 μM rapamycin similar to that of AtFKBP12 lines (Table 1, Figure 1C). In contrast to the P35S::AtFKBP12 lines, all 14 P35S::HsFKBP12 lines (OE1-OE14) responded to rapamycin (Table 1). In previous studies, we have extensively characterized the rapamycin sensitivity of BP12-2, which has a single copy of P35S::ScFKBP12 and is hypersensitive to rapamycin (Ren et al., 2012). In the present study, AtFKBP12-OE3, AtFKBP12-OE5, HsFKBP12-OE1, HsFKBP12-OE3 showed higher expression levels among all tested transgenic lines of AtFKBP12 and HsFKBP12, respectively. Western blot results confirmed that the expression of AtFKBP12 and HsFKBP12 proteins were similar and significantly higher (Figure 1D and Supplementary Figure 1A). We therefore selected them for subsequent rapamycin sensitivity testing using procedures described earlier. Figure 2 shows that all tested lines had normally growth on 1/2MS solid medium with DMSO, indicating that overexpression of AtFKBP12, ScFKBP12, and HsFKBP12 had no obvious effect on plant growth. However, in the presence of rapamycin, BP12-2 plants showed hypersensitivity, followed by HsFKBP12-OE1 and OE3, whereas the AtFKBP12 transgenic lines and WT plants did not respond to rapamycin treatment (Figure 2A). The administration of a low concentration of rapamycin (0.1 μM) resulted in a delay in the growth and development of BP12-2, and its fresh weight decreased by nearly 50% compared to the WT (Figure 2E). Administration of 0.1 μM rapamycin did not result in strong detectable inhibitory effects on HsFKBP12-OE1 and OE3. However, with increased rapamycin concentration, the HsFKBP12 transgenic lines also exhibited strong inhibitory effects (Figure 2A). When the concentration of rapamycin reached to 5μM, its inhibitory effect on HsFKBP12-OEs was pronounced (Figures 2A–C). Root and root hair length significantly decreased, leaf size was reduced, and plant fresh weight decreased by >50% (Figures 2D,E). At high concentration of rapamycin, the growth suppression of BP12-2 was still stronger than in HsFKBP12-OE lines. In contrast, the P35S::AtFKBP12 transgenic lines did not show clear inhibitory effects and had phenotypes similar to those of the WT control (Figure 2).
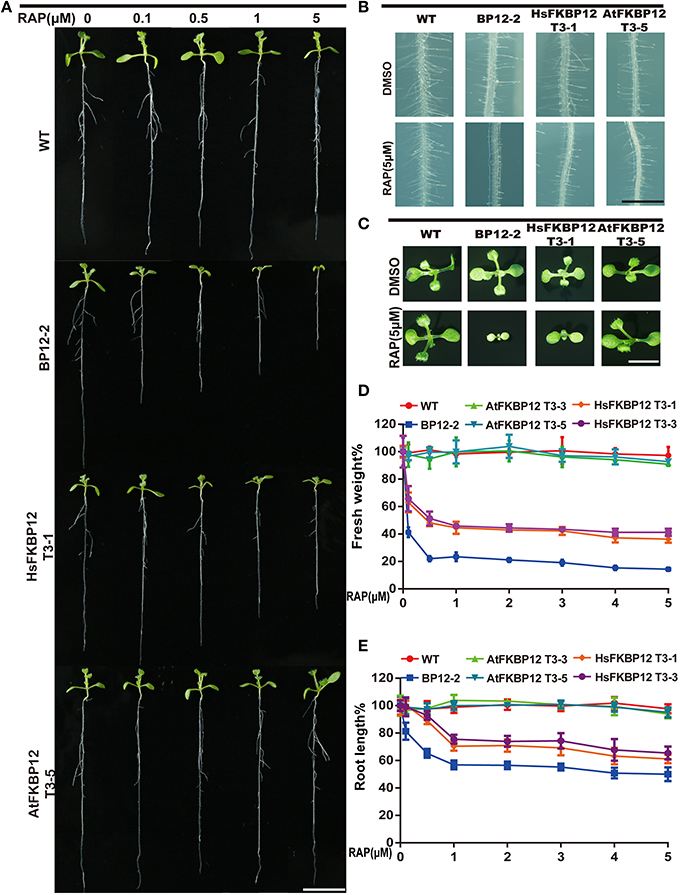
Figure 2. Rapamycin sensitivity tests of the transgenic plants with ScFKBP12, HsFKBP12, and AtFKBP12. (A) Dose-dependent effect of rapamycin on the growth and development of WT and three transgenic plants. The concentration of rapamycin ranged from 0 to 5 μM (10 DAG). Bar = 1 cm. (B,C) The root hair and leaf growth of WT and three transgenic plants on 0.5 MS medium supplied with DMSO or rapamycin (5 μM). (B) Bar = 1 mm; (C) Bar = 0.5 cm. (D,E) Quantitative analysis and comparison of the root length and the fresh weight (%) of WT and three transgenic plants after treatment with rapamycin. Error bars indicate ±SD for quadruplication.
Submergence Can Partially Restore Rapamycin Sensitivity of AtFKBP12 in Arabidopsis
Unlike above observations, previous study showed that AtFKBP12 is able to bridge the interaction between rapamycin and TOR in liquid culture (Xiong and Sheen, 2012). To further examine this observation and the functions of ScFKBP12, HsFKBP12 and AtFKBP12 in parallel under anaerobic growth condition, the sterile seeds of WT, BP12-2, HsFKBP12-OE1 and AtFKBP12-OE5 have been germinated in 0.5xMS liquid medium containing 10 μM rapamycin and grown for 8 days, respectively. The Supplementary Figure 2 showed that the seedling growth of BP12-2, HsFKBP12-OE1, and AtFKBP12-OE5 were indistinguishable from WT plants in terms of its response to submergence in 0.5xMS liquid medium with DMSO. However, the cotyledon greening, primary root elongation and fresh weight of BP12-2, followed byHsFKBP12-OE1,have been significantly arrested in the presence of rapamycin (Supplementary Figure 2). These results indicate that the combination of rapamycin and hypoxia could generate additive inhibition effects on TOR activity which are in agreement with recent observations in mammals (Bedogni et al., 2005; Agarwal et al., 2016; Damerill et al., 2016). Importantly, submergence can partially inhibit seedling growth of AtFKBP12-OE5 when compared with WT in the presence of 10 μM rapamycin (Supplementary Figure 2). This observation likely suggests that anaerobic stress caused by submergence can partially rescue the functions of AtFKBP12 to interact with TOR and rapamycin.
Generation of Double Transgenic DR5/BP12 Plants Containing Both P35S::ScFKBP12 and DR5:: GUS
Differing observations on the relationship between TOR and auxin signaling were reported by different groups (Dinkova et al., 2000; Beltrán-Peña et al., 2002; Schepetilnikov et al., 2013; Xiong et al., 2013; Dong et al., 2015). The above results indicated that under aerobic and anaerobic growth conditions, the rapamycin sensitivity of Arabidopsiscanbe rescued by ScFKBP12 in the most significant manner. ScFKBP12 was therefore selected for subsequent analysis of TOR signaling in Arabidopsis. To further investigate the relationship between TOR and auxin, we introduced the P35S:: ScFKBP12 construct (with a BASTA selection marker) into DR5:: GUS homozygous Arabidopsis lines, which have been used as an auxin signal reporter (Xiang et al., 2011). A total of 20 independent double transgenic lines containing both DR5:: GUS and P35S:: ScFKBP12 were generated by screening T0 transgenic seeds sown on 0.5 MS medium plate containing BASTA (10mg/L). DNA-PCR analysis indicated that these lines contained both DR5::GUS and P35S::ScFKBP12 constructs. We named these double transgenic plants as DR5/BP12-OE lines (Table 1). The T2 generation seeds of these 20 DR5/BP12-OE lines were collected for the BASTA segregation assay. The results showed that 6 of 20 lines displayed a 3:1 Mendelian segregation ratio for BASTA resistance and sensitivity, indicating that these 6 lines likely contained a single copy of P35S:: ScFKBP12. T3 generation seeds of these 6 lines were then selected for rapamycin sensitivity testing, which showed that all 6 lines were similarly sensitive to rapamycin treatment (Table 1). Semi-qPCR and qRT-PCR analyses indicated that DR5/BP12-OE5 and DR5/BP12-OE11 exhibited higher levels of ScFKBP12 expression than the other lines (Figures 3F,G). They were therefore selected for subsequent experiments.
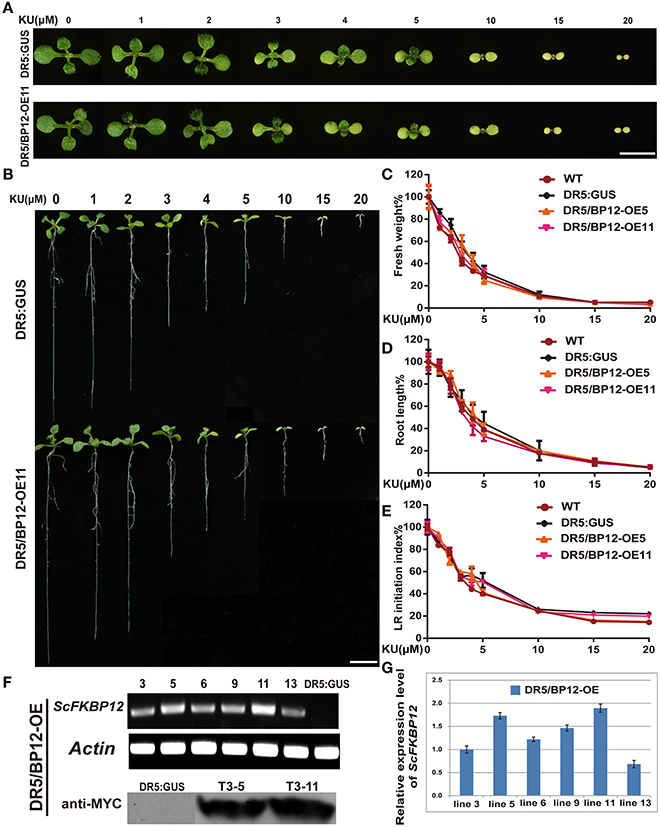
Figure 3. KU sensitivity tests of WT, DR5::GUS and DR5/BP12-OE transgenic lines and the identification of transgenic lines of DR5/BP12-OE transgenic lines. (A) The leaf formation of DR5::GUS and DR5/BP12-OE11 after treatment with different concentration of KU (10 DAG). Bar = 0.5 cm. (B) Dose-dependent inhibitory effect of KU on WT, DR5:: GUS and DR5/BP12-OE plants growth. The KU concentration ranged from 0 to 20 μM (10 DAG). Bar = 1 cm. (C–E) The quantitative analysis and comparison of fresh weight (%), root length (%) and lateral root initiation index (%) (LR initiation index) of DR5::GUS and DR5/BP12-OE plants after treating with KU at different concentration. Error bars indicate ±SD for quadruplication. (F,G) Identification of transgenic lines of DR5/BP12-OE lines by semi-qPCR, western blot (F, note: Since some results of the western blot located between DR:GUS and T3-5 are not associated with this manuscript, part of the membrane has been cut out), and qRT-PCR (G).
The Combination of Low-Concentration Rapamycin and KU Significantly Inhibits Seedling Growth and Development
When DR5/BP12-OE5 and DR5/BP12-OE11 were treated with concentrations of rapamycin ranging from 5 to 20 μM, the plants showed no obvious growth differences related to increasing of rapamycin concentration. So 5 μM seemed to be the saturation concentration of rapamycin (Supplementary Figure 3). This result indicated that rapamycin alone did not inhibit TOR function, even at higher concentrations. To proceed further, studies using second-generation TOR inhibitors to rapamycin-BP12-2 systems should be conducted. AsTORis have been developed and extensively utilized as second-generation TOR inhibitors both in basic scientific research studies and clinical trials (Benjamin et al., 2011). AsTORis directly interact with the kinase domain of TOR by competing with ATP, thereby inhibiting the kinase activity of TOR. More than 30 asTORis have been developed to date. Different asTORis have different structures and different IC50 (half maximal inhibitory concentration). KU63794 (KU), Torin1 and AZD8055 represent mild, moderate and strong inhibitors of TOR, respectively. They have frequently been used in TOR studies on mammalian systems and in plants (Montané and Menand, 2013; Schepetilnikov et al., 2013; Xiong et al., 2013; Dong et al., 2015; Li et al., 2015). However, Torin1 and AZD8055 show some off-target effects on PI3Ks (Thoreen et al., 2009; Chresta et al., 2010; Liu et al., 2010). In contrast, KU displayed the highest specificity toward TOR with no detectable off-target effects (García-Martínez et al., 2009). KU was therefore selected as an ATP competitive inhibitor of TOR for this study.
The inhibitory effect of KU was initially tested using concentration-gradient treatments (Figure 3). KU inhibited plant growth in a dose-dependent manner. When the KU concentration was <2 μM, root length of DR5/BP12-OE5 and DR5/BP12-OE11 was reduced compared to the DMSO control but no obvious differences in leaf development were observed. With higher (10–20 μM) KU concentrations, strong inhibitory effects were observed. Root development in DR5::GUS, DR5/BP12-OE5, and DR5/BP12-OE11 was inhibited. Leaf formation was impaired, and the true leaves were smaller and chlorotic (Figures 3A,B). The fresh weight of the whole plant decreased significantly. The 20 μM KU concentration completely inhibited plant growth. Phenotype and related data analysis (Figures 3A–E) indicated that the concentration range of 3–5 μM was critical to plant growth, and possibly the IC50 of KU.
We then tested various combinations of rapamycin and KU. Rapamycin combined with KU in the medium produced inhibition in the development of DR5/BP12-OE5 and DR5/BP12-OE11 in a dose-dependent manner (Supplementary Figure 4). When the concentration of KU remained at 1 μM, the WT lines showed no inhibitory effect at any rapamycin concentration, whereas DR5/BP12-OE5 and DR5/BP12-OE11showed severe inhibitory effects on root length (Supplementary Figures 4A,D), leaf development (Supplementary Figure 4B), and fresh weight (Supplementary Figure 4D). In contrast, the combination of 5 μM KU with gradually increased rapamycin concentrations resulted in a more pronounced inhibitory effect that was phenotypically expressed in the DR5/BP12-OEtreated lines (Figure 4A). Comparison of the combination of 1 μM KU plus rapamycin with 5 μM KU plus rapamycin showed that the latter combination had greater growth inhibition (Figure 4). With 0.5 μM rapamycin and 5 μM KU, growth and development of DR5/BP12-OE5andDR5/BP12-OE11 was completely terminated, and nearly no root growth or true leaf formation were observed.
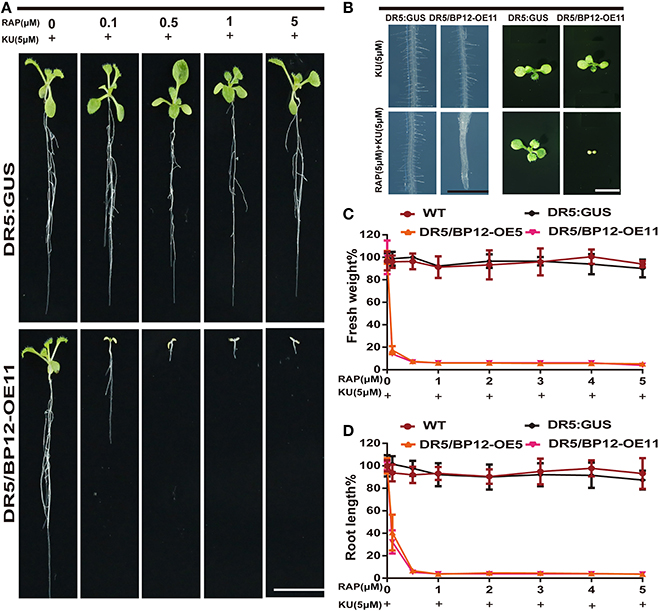
Figure 4. The inhibition of Arabidopsis by combined rapamycin with KU. (A) The growth of whole plant of DR5:: GUS and DR5/BP12-OE11 after treated with rapamycin and KU (10 DAG). Rapamycin concentration ranged from 0 to 5 μM, whereas KU was used at a final concentration of 5 μM. Bar = 1 cm. (B) The inhibitory effect of KU or rapamycin plus KU on root hair development and leaf formation of DR5:: GUS and DR5/BP12-OE11. Bar = 1 mm at the left and 0.5 cm in the right. (C,D) The quantitative analysis and comparison of root length (%) and fresh weight (%) of WT, DR5::GUS and DR5/BP12-OE plants after treatment with rapamycin or/and KU. Error bars indicate ±SD for quadruplication.
TOR Inhibitors Affect Arabidopsis Auxin Signals
Auxin related plant effects could be distinguished from DR5/BP12-OE11 treated with rapamycin plus KU (Figures 5A,B). Severe growth and development defects, as well as geotropism in DR5/BP12-OE5 and DR5/BP12-OE11 were seen when the combined concentration of rapamycin and KU reached 5 μM. These findings suggested that severe inhibition of TOR would also affect the auxin signal pathway of plants. To investigate inhibitory effects of TOR on auxin, we examined the GUS signals of DR5/BP12-OE11 treated with rapamycin or/and KU (5 μM), with DMSO used as control (Figure 5C). The content of auxin in the root tips of inhibitor-treated lines significantly decreased compared to the control, indicating that the auxin levels were affected by TOR inhibitors. Next, four auxin biosynthesis-related genes, ATT1, TAR2, YUCCA1, and YUCCA2, were selected for mRNA expression analysis following TOR inhibitor treatment. ATT1 and TAR2 are genes involved with the first step of auxin biosynthesis. They catalyze the formation of indole-3-pyruvic acid (IPA) from tryptophan. YUCCA1 and YUCCA2 catalyze the formation of IAA from IPA (Mashiguchi et al., 2011). Compared to the DMSO control lines, ATT1, TAR2, YUCCA1, and YUCCA2 were all significantly down regulated in TOR inhibitor-treated lines (Figure 5D and Supplementary Figure 4E). The TOR inhibitors therefore reduced the auxin levels in Arabidopsis. The observed down-regulation of auxin levels was also consistent with the disruption in root development in TOR inhibitor-treated DR5/BP12-OE5 and DR5/BP12-OE11. We added exogenous auxin to assess the response of rapamycin-treated DR5/BP12-OE5 and DR5/BP12-OE11. Figure 6 shows that the low concentration of exogenous auxin effectively promoted lateral root and root hair development in the absence of a TOR inhibitor. Under rapamycin treatment, no changes were observed in DR5:: GUS, whereas the DR5/BP12-OE5 and DR5/BP12-OE11 lines showed severe impairment of lateral root formation and root hair elongation (Figures 6B–D). The addition of different doses of exogenous auxin resulted in partial restoration of root hair development in low-concentration inhibitor-treated plants (Figures 6B,D). With 5 μM of rapamycin, lateral root development in the DR5/BP12-OE5 and DR5/BP12-OE11 lines was completely inhibited regardless of supplementation with higher amounts of exogenous auxin. These results indicate that TOR is involved in the auxin response.
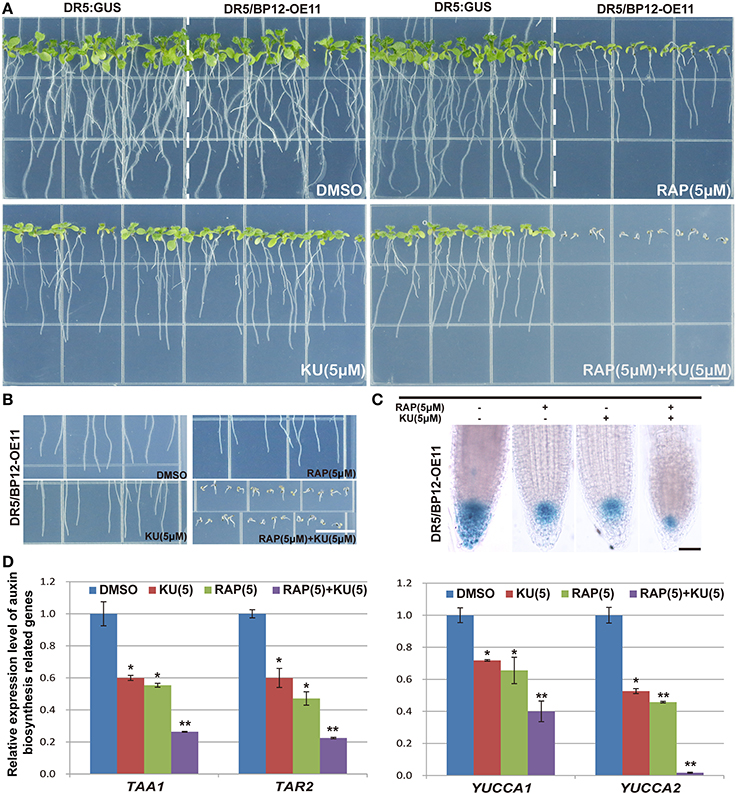
Figure 5. The inhibition of DR5:: GUS and DR5/BP12-OE11 by rapamycin or/and KU affect auxin levels. (A) The phenotype of DR5::GUS and DR5/BP12-OE11 treated with rapamycin or/and KU. DMSO was used as control (12 DAG). Bar = 1 cm. (B) The changed gravitropism of plant root. Plants grew for 12 din 0.5 MS medium with different TOR inhibitors or DMSO. Bar = 1 cm. (C) Rapamycin or/and KU affect auxin distribution in root tip, DR5:: GUS reporter was used as a marker of auxin distribution. Plants grew for 7 d in 0.5MS medium with different TOR inhibitors or DMSO. Bar = 0.1 mm. (D) Detection expression level of auxin synthesis-related genes by qRT-PCR. DR5/BP12-OE11 grew 12 days in 0.5 MS medium with different TOR inhibitors [RAP (5 μM), KU (5 μM), RAP (5 μM) +KU (5 μM); DMSO was used as control]. Each value represents the mean± SD of 3 independent experiments. Asterisks denote Student's t-test significance compared with control (*P < 0.05;**P < 0.01).
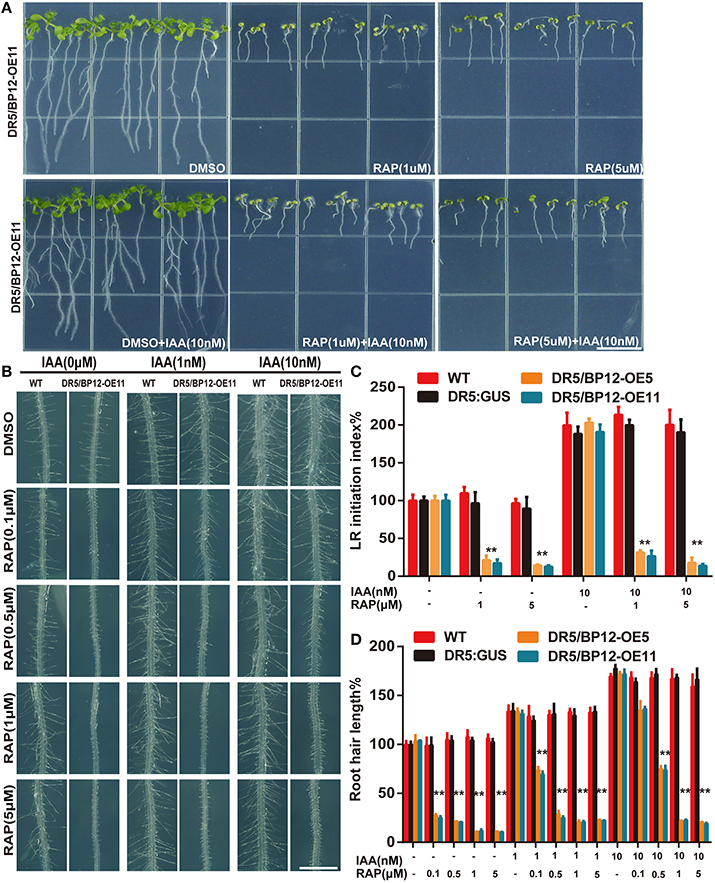
Figure 6. TOR was required for exogenous IAA response in Arabidopsis. (A) The phenotype of DR5/BP12-OE11 treated with IAA and rapamycin (8 DAG). Bar = 1 cm. (B) The root hair elongation of DR5:: GUS and DR5/BP12-OE11 with exogenous IAA and rapamycin. Bar = 1 mm. Error bars indicate ±SD for quadruplication. (C,D) The quantitative analysis and comparison of LR initiation index (%) and root hair length (%) of WT, DR5::GUS and DR5/BP12-OE plants after treatment with IAA and rapamycin. Error bars indicate ±SD for quadruplication. Asterisks denote Student's t-test significance compared with DR5:: GUS (**P < 0.01).
The regulatory functions of auxin in plants include apical dominance, tropism, root germination, stem extension, organ differentiation, and aging (Teale et al., 2006; Zhao, 2010). In the presence of auxin, AUX/IAA repressor proteins are rapidly degraded by the ubiquitination pathway and then the transcription of primary auxin response genes (SAUR, GH3, and AUX/IAA family) are activated within a few minutes (Dharmasiri and Estelle, 2004; Wang and Estelle, 2014). In the present study, we utilized auxin response genes as qRT-PCR markers for auxin responses in DR5/BP12-OE11, which was exposed to TOR inhibitors. Based on previous results of expression profiling analysis (Dong et al., 2015), six genes were selected for gene expression analysis at the mRNA level. Figure 7 shows that all auxin response genes were down-regulated by rapamycin or KU treatment. This differential expression was most obvious when the DR5/BP12-OE11 line was treated with rapamycin plus KU. To further study the effects of TOR inhibitors on auxin signaling, two auxin biosynthesis-related genes and two primary auxin response genes were chosen as markers to study the impact of TOR inhibitors on auxin signals at different time points (Supplementary Figure 5). The results show that the four selected genes were down-regulated at 10 min after treatment. This result indicates that TOR inhibitors could rapidly affect the expression of primary auxin response genes by down-regulation the expression of auxin biosynthesis-related and responsive genes to block auxin signaling. However, YUCCA genes and SAURs were up-regulated with longer treatment times. This result may be due to the feedback regulation of YUCCA genes in the lack of auxin (Suzuki et al., 2015). The up-regulation of YUCCA genes might indicate that auxin levels were down-regulated by TOR inhibitors in Arabidopsis. In contrast to rapamycin or KU alone, rapamycin plus KU could quickly inhibit auxin signaling and disrupt that feedback system. This might explain why rapamycin plus KU strongly inhibit the growth and development of Arabidopsis. The treatment with TOR inhibitors caused a significant change in the transcriptional expression levels of these genes. Therefore, TOR is crucial in auxin homeostasis and significantly affected the auxin signal pathway in plants.
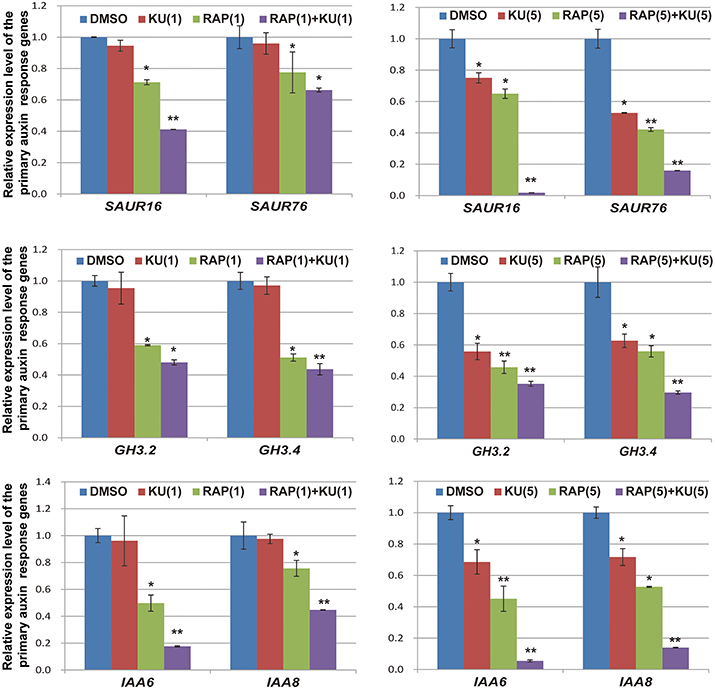
Figure 7. The expression level of primaryauxin response gene family AUX/IAAs, SAURs, and GH3s were affected by TOR specific inhibitors. DR5/BP12-OE11 grew in 0.5 MS medium containing TOR inhibitors [RAP (5 μM), KU (5 μM), RAP (5 μM) + KU (5 μM); DMSO was used as control)] for 12 days. Each value represents the mean± SD of 3 independent experiments. Asterisks denote Student's t-test significance compared with control (*P < 0.05; **P < 0.01).
Discussion
Hypoxia Stress Could Enhance the Interactions between TOR, FKBP12 and Rapamycin in Arabidopsis
Several independent studies have shown that plant FKBP12 is non-functional on solid medium (Xu et al., 1998; Menand et al., 2002; Mahfouz et al., 2006; Sormani et al., 2007; Leiber et al., 2010; Ren et al., 2012), but all these groups did not examine the functions of plant FKBP12s under anaerobic growth condition. Only one group has shown that FKBP12 is functional in during anaerobic growth (Xiong and Sheen, 2012), but they did not set any solid medium control in their experiments. In this study, we re-examined the functions of AtFKBP12 along with HsFKBP12 and ScFKBP12 in Arabidopsis under both anaerobic and aerobic growth conditions. Upon submergence, overexpression of FKBP12s can significantly block the growth of Arabidopsis in the presence of rapamycin. More importantly, hypoxia caused by submergence can partially rescue the functions of AtFKBP12 to interact with TOR and rapamycin. Since Arabidopsis is land plant but not aquatic plant, the biochemical and physiological processes of the submerged seeds and seedlings should be quite different from seeds or seedling growing under aerobic condition. Recent study also revealed that submergence can lead to hypoxia and anaerobic respiration in plant cells (Chen et al., 2015). It is possible that the protein structure of AtFKBP12 can be altered to contribute to the interaction between rapamycin, FKBP12 and TOR under hypoxia stress. However, the underlying mechanism is still remains to be defined. It should be noted that the large amount of literatures already showed that rapamycin treatment can generate the effects similar to hypoxic stress and hypoxia inducible factor-1α (Hif1α), a key mediator of cellular adaptation to hypoxia, can be efficiently inhibited by rapamycin in mammals (Bedogni et al., 2005; Agarwal et al., 2016; Damerill et al., 2016). Based on these observations, it is not surprising that TOR signaling might also act as a key target of hypoxia in Arabidopsis as observed in mammals (Hudson et al., 2002; Arsham et al., 2003). Altogether, the combination of rapamycin and hypoxia can generate additive inhibition effects on plant growth in the presence of exogenous or endogenous FKBP12s.
Our results also demonstrated that the rapamycin treatment, ScFKBP12 transgenic lines showed the strongest drug sensitivity. This might be explained in terms of amino acid sequences (Figure 1E). Amino acid sequence alignment showed relatively high similarity among ScFKBP12, HsFKBP12, and AtFKBP12. The glycine residues and the drug-binding pocket are all conserved (Clardy, 1995). However, two amino acids of AtFKBP12, tyrosine at position 25 and isoleucine at position 79, were changed into cysteine (labeled with a red box in Figure 1E), which affected the protein structure and its biological activity (Xu et al., 1998). The existence of two cysteines might also lead to the formation of a disulfide bond, which would be a critical modification of protein structure and corresponding enzymatic activity. Comparison the amino acid sequence between ScFKBP12 andHsFKBP12, twokey residuesGln54and Glu55in HsFKBP12interact to rapamycin were changed into Gly60 and Gln61 in ScFKBP12 (Supplementary Figure 1B; Choi et al., 1996; Sormani et al., 2007). Additionally, many different amino acids were also observed in N-terminal of the proteins. These amino acid differences likely lead to the different affinity rapamycin binding between HsFKBP12 and ScFKBP12 in Arabidopsis, but the detailed molecular mechanism remains to be further defined.
The Combination of Rapamycin and KU Significantly Inhibits Arabidopsis Growth
Rapamycin can inhibit TOR activity through rapamycin-FKBP12-FRB/TOR complex formation and display a plateau effect (Sormani et al., 2007; Ren et al., 2012). The second generation TOR inhibitors (asTORis) inhibit TOR by targeting the catalytic site of the kinase domain and competing with ATP in a dose-dependent manner. KU is able to inhibit TORC1 and TORC2 with an IC50 of 10 nM in vitro but it inhibits cell growth at 3 μM in MEF cells (García-Martínez et al., 2009) and the IC50 of rapamycin is 0.1nM in HEK293 cells (Edwards and Wandless, 2007). The IC50 values of KU and rapamycin in Arabidopsisare ~5 and ~1 μM, respectively. This suggests that the influx/efflux transporters and pharmacological dynamics of TOR inhibitor drugs are quite different in animal and plant cells. Rapamycin and KU precipitates occurred in MS medium solution and plates when the drugs were used at high concentrations (>5 μM). The solubility of KU and rapamycin is poor even in DMSO. This would act to reduce experimental accuracy. An important strategy would be to combine these two different kinds of TOR inhibitors with different inhibitory mechanisms to suppress TOR function. The applications and studies of Torin1, AZD8055, and KU in plants have shown that KU is the most moderate inhibitor, but one with very high TOR specificity. Therefore, we tried the combination of rapamycin with KU to evaluate efficiency of TOR inhibition. We found that TOR is strongly inhibited by a combination of rapamycin and KU at low concentrations (1 μM + 1 μM). A single inhibitor could not produce this level of inhibitory effect at a 10-folds higher concentration. The inhibitor-combination application effectively lowered the dosage of inhibitor and reduced the experiment costs. Plant growth was completely terminated once the concentration of rapamycin and KU was 5 μM. However, the relationship between rapamycin and KU during the actual process of inhibition remains elusive and requires further investigation.
TOR Was Required for Plant Growth in Respond to Auxin
Auxin plays an important role in the growth and development of plants. Auxin activates TOR and promotes the translation reinitiation of mRNA (Schepetilnikov et al., 2013). The TOR inhibitor Torin1 can interfere with the auxin redistribution (DR5-GFP) in root tips and root gravitropic responses indicating a close connection between TOR and auxin (Dinkova et al., 2000; Schepetilnikov et al., 2013). Some key genes associated with auxin signaling transduction are significantly mis-expressed when TOR is suppressed by AZD8055 (Dong et al., 2015). Although the approaches, TOR inhibitors (rapamycin/KU vs. Torin1) and plant materials (AtFKBP12, HsFKBP12 and ScFKBP12 transgenic plants vs. TOR RNAi plants) we used are quite different between ours and (Schepetilnikov et al., 2013), two independent studies really reach to similar conclusion in which TOR plays a crucial role in the auxin signaling transduction (Schepetilnikov et al., 2013). However, Xiong et al. (2013) demonstrated that auxin is decoupled from TOR signaling activation, and thus auxin signaling (DR5-GFP) doesn't alter when TOR is suppressed (Xiong et al., 2013). These conflicting observations likely caused by different experimental conditions. Xiong et al. (2013), transiently treated the seedlings only for few hours, whereas we and (Schepetilnikov et al., 2013), grow seedlings for several days (Schepetilnikov et al., 2013; Xiong et al., 2013). We observed that the strong growth and gravity defects occurred in root when DR5/BP12-OE5 and DR5/BP12-OE11 was treated with rapamycin plus KU. The auxin distribution significantly decreased in root tip when TOR was suppressed. These results were highly consistent with the observation of Schepetilnikov et al., but not with Xiong et al. (Schepetilnikov et al., 2013; Xiong et al., 2013). Although Xiong et al., emphasized that TOR was decoupled from auxin signaling in Arabidopsis, many auxin signaling associated genes were also differentially expressed in tor seedlings (Xiong et al., 2013). Importantly, some overlapping differentially expressed genes were found in TOR suppression plants by independent groups (Ren et al., 2012; Xiong et al., 2013; Dong et al., 2015), indicating that TOR acts as a key player in auxin signaling transduction in Arabidopsis. However, the underlying molecular mechanism involving TOR and auxin signals remains unclear. With establishment of the DR5/BP12 system in this study, a highly potent, inducible, and selective TOR suppression system is now available for advanced studies on the relationship between TOR and auxin in plants.
Author Contributions
MR, KD, and LY designed the experiments. KD, XZ, KZ, and WW performed the experiments. MR, PD, and JZ analyzed the data. MR, KD, and LY wrote the manuscript text.
Funding
National Basic Research Program of China (No. 2013CB127100); Fundamental Research Funds for the Central Universities (No. CDJZR14295501); Chongqing Frontier and Applied Basic Research (No. CSTC2014JCYJA80012); the Open Project Funding of State Key Laboratory of Cotton Biology (No. CB2014A08 and CB2015A14); Promote Scientific Research and Cooperation and High level Personnel Training Project in America and Oceania supported by Ministry of Education (No. 0903005109094/003) and National Natural Science Foundation of China (No. 31200903).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Grants: National Basic Research Program of China (No. 2013CB127100), National Natural Science Foundation of China (No. 31200903), Chongqing Frontier and Applied Basic Research (No. CSTC2014JCYJA80012), and Promote Scientific Research and Cooperation and High level Personnel Training Project in America and Oceania supported by Ministry of Education (No. 0903005109094/003). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00291
Supplementary Figure 1. The protein expression level in three FKBP12 transgenic lines (A) and the multiple protein sequence alignment of ScFKBP12, HsFKBP12, and AtFKBP12 (B). Key residues were marked with red box.
Supplementary Figure 2. Rapamycin sensitivity tests of the transgenic plants with ScFKBP12, HsFKBP12, and AtFKBP12 inanaerobic growth conditions (8 DAG). (A) The effect of rapamycin on the growth and development of WT and three transgenic plants during anaerobic growth condition (8 DAG). Bar = 0.5 cm. (B) The leaf growth of WT and three transgenic plants on 0.5 MS liquid medium supplied with DMSO or rapamycin (5 μM). (B) Bar = 0.5 cm. (C) Quantitative analysis and comparison of the fresh weight (%) of WT and three transgenic plants after treatment with rapamycin. Error bars indicate ±SD for quadruplication. Asterisks denote Student's t-test significance compared with control (*P < 0.05;**P < 0.01).
Supplementary Figure 3. Rapamycin cannot completely inhibit DR5/BP12-OE lines growth even with very high concentration (20 μM) (10 DAG).
Supplementary Figure 4. The inhibition of Arabidopsis by combined rapamycin with KU. (A) The growth of whole plant of DR5:: GUS and DR5/BP12-OE11 after treated with rapamycin and KU (10 DAG). Rapamycin concentration ranged from 0 to 5 μM, whereas KU was used in a final concentration of 1 μM. Bar = 1 cm. (B) The inhibition effect of rapamycin or KU or rapamycin plus KU on root hair development and leaf formation of DR5::GUS and DR5/BP12-OE11. Bar = 1 mm at the left and 0.5 cm in the right. (C,D) The quantitative analysis and comparison of root length and fresh weight (%) of DR5::GUS and DR5/BP12-OE plants after treatment with rapamycin or/and KU. (E) Detection expression level of auxin synthesis-related genes by qRT-PCR. DR5/BP12-OE11 grew 12 days in 0.5 MS medium with different TOR inhibitors [RAP (1 μM), KU (1 μM), RAP (1 μM) +KU (1 μM); DMSO was used as control]. Each value represents the mean± SD of 3 independent experiments. Asterisks denote Student's t-test significance compared with control (*P < 0.05;**P < 0.01).
Supplementary Figure 5. The expression level of auxin biosynthesis-related genes and primaryauxin response genes were affected by TOR specific inhibitors in short time treatment. DR5/BP12-OE11grew in 0.5 MS medium for 10 days. Seedlings were transferred into 0.5 MS medium containing TOR inhibitors [RAP (5 μM), KU (5 μM), RAP (5 μM)+KU (5 μM); DMSO was used as control]for different time points (10 min, 30 min, 1 h, 2 h, 3 h, 6 h,12 h, 24 h), then root was collected for RNA extraction. Each value represents the mean ± SD of 3 independent experiments.
Supplementary Table 1. Primers were used in this study.
References
Agarwal, S., Loder, S., Brownley, C., Cholok, D., Mangiavini, L., Li, J., et al. (2016). Inhibition of Hif1alpha prevents both trauma-induced and genetic heterotopic ossification. Proc. Natl. Acad. Sci. U.S.A. 113, E338–E347. doi: 10.1073/pnas.1515397113
Arsham, A. M., Howell, J. J., and Simon, M. C. (2003). A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 278, 29655–29660. doi: 10.1074/jbc.M212770200
Bedogni, B., Welford, S. M., Cassarino, D. S., Nickoloff, B. J., Giaccia, A. J., and Powell, M. B. (2005). The hypoxic microenvironment of the skin contributes to Akt-mediated melanocyte transformation. Cancer Cell 8, 443–454. doi: 10.1016/j.ccr.2005.11.005
Beltrán-Peña, E., Aguilar, R., Ortiz-Lopez, A., Dinkova, T. D., and De Jimenez, E. S. (2002). Auxin stimulates S6 ribosomal protein phosphorylation in maize thereby affecting protein synthesis regulation. Physiol. Plant. 115, 291–297. doi: 10.1034/j.1399-3054.2002.1150216.x
Benjamin, D., Colombi, M., Moroni, C., and Hall, M. N. (2011). Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 10, 868–880. doi: 10.1038/nrd3531
Bögre, L., Henriques, R., and Magyar, Z. (2013). TOR tour to auxin. EMBO J. 32, 1069–1071. doi: 10.1038/emboj.2013.69
Brown, E. J., Albers, M. W., Shin, T. B., Ichikawa, K., Keith, C. T., Lane, W. S., et al. (1994). A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758. doi: 10.1038/369756a0
Cafferkey, R., Young, P. R., Mclaughlin, M. M., Bergsma, D. J., Koltin, Y., Sathe, G. M., et al. (1993). Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13, 6012–6023. doi: 10.1128/MCB.13.10.6012
Chen, J., Zheng, X. F., Brown, E. J., and Schreiber, S. L. (1995). Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. U.S.A. 92, 4947–4951. doi: 10.1073/pnas.92.11.4947
Chen, L., Liao, B., Qi, H., Xie, L. J., Huang, L., Tan, W. J., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246. doi: 10.1080/15548627.2015.1112483
Chiu, M. I., Katz, H., and Berlin, V. (1994). RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. U.S.A. 91, 12574–12578. doi: 10.1073/pnas.91.26.12574
Choi, J., Chen, J., Schreiber, S. L., and Clardy, J. (1996). Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273, 239–242. doi: 10.1126/science.273.5272.239
Chresta, C. M., Davies, B. R., Hickson, I., Harding, T., Cosulich, S., Critchlow, S. E., et al. (2010). AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70, 288–298. doi: 10.1158/0008-5472.CAN-09-1751
Clardy, J. (1995). The chemistry of signal transduction. Proc. Natl. Acad. Sci. U.S.A. 92, 56–61. doi: 10.1073/pnas.92.1.56
Cornu, M., Albert, V., and Hall, M. N. (2013). mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 23, 53–62. doi: 10.1016/j.gde.2012.12.005
Damerill, I., Biggar, K. K., Shehab, M. A., Li, S. S.-C., Jansson, T., and Gupta, M. B. (2016). Hypoxia Increases IGFBP-1 Phosphorylation Mediated by mTOR Inhibition. Mol. Endocrinol. 30, 201–216. doi: 10.1210/me.2015-1194
Dharmasiri, N., and Estelle, M. (2004). Auxin signaling and regulated protein degradation. Trends Plant Sci. 9, 302–308. doi: 10.1016/j.tplants.2004.04.003
Dinkova, T. D., Aguilar, R., and Sanchez De Jimenez, E. (2000). Expression of maize eukaryotic initiation factor (eIF) iso4E is regulated at the translational level. Biochem. J. 351(Pt 3), 825–831. doi: 10.1042/bj3510825
Dong, P., Xiong, F., Que, Y., Wang, K., Yu, L., Li, Z., et al. (2015). Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 6:677. doi: 10.3389/fpls.2015.00677
Dubrovsky, J. G., Soukup, A., Napsucialy-Mendivil, S., Jeknic, Z., and Ivanchenko, M. G. (2009). The lateral root initiation index: an integrative measure of primordium formation. Ann. Bot. 103, 807–817. doi: 10.1093/aob/mcn267
Edwards, S. R., and Wandless, T. J. (2007). The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain. J. Biol. Chem. 282, 13395–13401. doi: 10.1074/jbc.M700498200
Feng, Z., and Levine, A. J. (2010). The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 20, 427–434. doi: 10.1016/j.tcb.2010.03.004
Fontana, L., Partridge, L., and Longo, V. D. (2010). Extending healthy life span–from yeast to humans. Science 328, 321–326. doi: 10.1126/science.1172539
García-Martínez, J. M., Moran, J., Clarke, R. G., Gray, A., Cosulich, S. C., Chresta, C. M., et al. (2009). Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem. J. 421, 29–42. doi: 10.1042/BJ20090489
Heitman, J., Movva, N. R., and Hall, M. N. (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909. doi: 10.1126/science.1715094
Hudson, C. C., Liu, M., Chiang, G. G., Otterness, D. M., Loomis, D. C., Kaper, F., et al. (2002). Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 22, 7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002
Koltin, Y., Faucette, L., Bergsma, D. J., Levy, M. A., Cafferkey, R., Koser, P. L., et al. (1991). Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 11, 1718–1723. doi: 10.1128/MCB.11.3.1718
Kunz, J., Henriquez, R., Schneider, U., Deuter-Reinhard, M., Movva, N. R., and Hall, M. N. (1993). Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585–596. doi: 10.1016/0092-8674(93)90144-F
Leiber, R. M., John, F., Verhertbruggen, Y., Diet, A., Knox, J. P., and Ringli, C. (2010). The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22, 1898–1908. doi: 10.1105/tpc.109.073007
Li, L., Song, Y., Wang, K., Dong, P., Zhang, X., Li, F., et al. (2015). TOR-inhibitor insensitive-1 (TRIN1) regulates cotyledons greening in Arabidopsis. Front. Plant Sci. 6:861. doi: 10.3389/fpls.2015.00861
Liu, Q., Chang, J. W., Wang, J., Kang, S. A., Thoreen, C. C., Markhard, A., et al. (2010). Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benz o[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J. Med. Chem. 53, 7146–7155. doi: 10.1021/jm101144f
Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., et al. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468. doi: 10.1016/S1097-2765(02)00636-6
Madeira Da Silva, L., and Beverley, S. M. (2010). Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc. Natl. Acad. Sci. U.S.A. 107, 11965–11970. doi: 10.1073/pnas.1004599107
Mahfouz, M. M., Kim, S., Delauney, A. J., and Verma, D. P. (2006). Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18, 477–490. doi: 10.1105/tpc.105.035931
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. doi: 10.1073/pnas.1108434108
Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., et al. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U.S.A. 99, 6422–6427. doi: 10.1073/pnas.092141899
Montané, M. H., and Menand, B. (2013). ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 64, 4361–4374. doi: 10.1093/jxb/ert242
Moreau, M., Azzopardi, M., Clement, G., Dobrenel, T., Marchive, C., Renne, C., et al. (2012). Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481. doi: 10.1105/tpc.111.091306
Ren, M., Qiu, S., Venglat, P., Xiang, D., Feng, L., Selvaraj, G., et al. (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155, 1367–1382. doi: 10.1104/pp.110.169045
Ren, M., Venglat, P., Qiu, S., Feng, L., Cao, Y., Wang, E., et al. (2012). Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24, 4850–4874. doi: 10.1105/tpc.112.107144
Sabatini, D. M., Erdjument-Bromage, H., Lui, M., Tempst, P., and Snyder, S. H. (1994). RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43. doi: 10.1016/0092-8674(94)90570-3
Schepetilnikov, M., Dimitrova, M., Mancera-Martinez, E., Geldreich, A., Keller, M., and Ryabova, L. A. (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32, 1087–1102. doi: 10.1038/emboj.2013.61
Sormani, R., Yao, L., Menand, B., Ennar, N., Lecampion, C., Meyer, C., et al. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7:26. doi: 10.1186/1471-2229-7-26
Suzuki, M., Yamazaki, C., Mitsui, M., Kakei, Y., Mitani, Y., Nakamura, A., et al. (2015). Transcriptional feedback regulation of YUCCA genes in response to auxin levels in Arabidopsis. Plant Cell Rep. 34, 1343–1352. doi: 10.1007/s00299-015-1791-z
Teale, W. D., Paponov, I. A., and Palme, K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859. doi: 10.1038/nrm2020
Thoreen, C. C., Kang, S. A., Chang, J. W., Liu, Q., Zhang, J., Gao, Y., et al. (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032. doi: 10.1074/jbc.M900301200
Vander Haar, E., Lee, S. I., Bandhakavi, S., Griffin, T. J., and Kim, D. H. (2007). Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9, 316–323. doi: 10.1038/ncb1547
Villa-Hernández, J. M., Dinkova, T. D., Aguilar-Caballero, R., Rivera-Cabrera, F., Sanchez De Jimenez, E., and Perez-Flores, L. J. (2013). Regulation of ribosome biogenesis in maize embryonic axes during germination. Biochimie 95, 1871–1879. doi: 10.1016/j.biochi.2013.06.011
Wang, L., Rhodes, C. J., and Lawrence, J. C. Jr. (2006). Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J. Biol. Chem. 281, 24293–24303. doi: 10.1074/jbc.M603566200
Wang, R., and Estelle, M. (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21, 51–58. doi: 10.1016/j.pbi.2014.06.006
Wullschleger, S., Loewith, R., and Hall, M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. doi: 10.1016/j.cell.2006.01.016
Xiang, D., Yang, H., Venglat, P., Cao, Y., Wen, R., Ren, M., et al. (2011). POPCORN functions in the auxin pathway to regulate embryonic body plan and meristem organization in Arabidopsis. Plant Cell 23, 4348–4367. doi: 10.1105/tpc.111.091777
Xiong, Y., Mccormack, M., Li, L., Hall, Q., Xiang, C., and Sheen, J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. doi: 10.1038/nature12030
Xiong, Y., and Sheen, J. (2012). Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287, 2836–2842. doi: 10.1074/jbc.M111.300749
Xiong, Y., and Sheen, J. (2015). Novel links in the plant TOR kinase signaling network. Curr. Opin. Plant Biol. 28, 83–91. doi: 10.1016/j.pbi.2015.09.006
Xu, Q., Liang, S., Kudla, J., and Luan, S. (1998). Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J. 15, 511–519. doi: 10.1046/j.1365-313X.1998.00232.x
Zagouri, F., Sergentanis, T. N., Chrysikos, D., Filipits, M., and Bartsch, R. (2012). mTOR inhibitors in breast cancer: a systematic review. Gynecol. Oncol. 127, 662–672. doi: 10.1016/j.ygyno.2012.08.040
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Keywords: target of rapamycin, FKBP12, rapamycin, KU63794, auxin
Citation: Deng K, Yu L, Zheng X, Zhang K, Wang W, Dong P, Zhang J and Ren M (2016) Target of Rapamycin Is a Key Player for Auxin Signaling Transduction in Arabidopsis. Front. Plant Sci. 7:291. doi: 10.3389/fpls.2016.00291
Received: 22 September 2015; Accepted: 23 February 2016;
Published: 11 March 2016.
Edited by:
Martin Huelskamp, University of Cologne, GermanyReviewed by:
Rossana Henriques, Centre for Research in Agricultural Genomics, SpainZoltan Magyar, Hungarian Academy of Sciences-Biological Research Centre, Hungary
Copyright © 2016 Deng, Yu, Zheng, Zhang, Wang, Dong, Zhang and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maozhi Ren, renmaozhi@cqu.edu.cn
†These authors have contributed equally to this work.
 Kexuan Deng1†
Kexuan Deng1† Maozhi Ren
Maozhi Ren