- 1National Research Council of Italy, Institute of Biosciences and Bioresources, Research Division Portici (CNR-IBBR) Portici, Italy
- 2National Research Council of Italy, Institute for Agricultural and Forestry Systems in the Mediterranean (CNR-ISAFoM), Ercolano, Italy
- 3Department of Agriculture, University of Naples “Federico II,” Portici, Italy
- 4CREA - Council for Agricultural Research and Economics, Genomics Research Centre, Fiorenzuola d'Arda, Italy
Tomato is a major crop in the Mediterranean basin, where the cultivation in the open field is often vulnerable to drought. In order to adapt and survive to naturally occurring cycles of drought stress and recovery, plants employ a coordinated array of physiological, biochemical, and molecular responses. Transcriptomic studies on tomato responses to drought and subsequent recovery are few in number. As the search for novel traits to improve the genetic tolerance to drought increases, a better understanding of these responses is required. To address this need we designed a study in which we induced two cycles of prolonged drought stress and a single recovery by rewatering in tomato. In order to dissect the complexity of plant responses to drought, we analyzed the physiological responses (stomatal conductance, CO2 assimilation, and chlorophyll fluorescence), abscisic acid (ABA), and proline contents. In addition to the physiological and metabolite assays, we generated transcriptomes for multiple points during the stress and recovery cycles. Cluster analysis of differentially expressed genes (DEGs) between the conditions has revealed potential novel components in stress response. The observed reduction in leaf gas exchanges and efficiency of the photosystem PSII was concomitant with a general down-regulation of genes belonging to the photosynthesis, light harvesting, and photosystem I and II category induced by drought stress. Gene ontology (GO) categories such as cell proliferation and cell cycle were also significantly enriched in the down-regulated fraction of genes upon drought stress, which may contribute to explain the observed growth reduction. Several histone variants were also repressed during drought stress, indicating that chromatin associated processes are also affected by drought. As expected, ABA accumulated after prolonged water deficit, driving the observed enrichment of stress related GOs in the up-regulated gene fractions, which included transcripts putatively involved in stomatal movements. This transcriptomic study has yielded promising candidate genes that merit further functional studies to confirm their involvement in drought tolerance and recovery. Together, our results contribute to a better understanding of the coordinated responses taking place under drought stress and recovery in adult plants of tomato.
Introduction
Drought conditions historically constitute the abiotic stress with the biggest impact on crop yield and agricultural productivity (Boyer, 1982; Boyer et al., 2013). Predicted climate change threatens stable crop yields, will likely require changes in agricultural practices in response to altered rainfall patterns, and increased urban consumption. Multiple projections indicate that between 20 and 60 Mha of irrigated cropland may have to be reverted to rainfed management by the end of the century (Elliott et al., 2014). Formerly irrigated crops would become entirely dependent on rainfall and vulnerable to yield loss due to drought. This reduction is predicted to be severe in southern Spain and Italy, two major producers of tomatoes (Saadi et al., 2015). This increasing vulnerability to drought requires that we develop more resilient varieties capable of surviving drought conditions while maintaining yield (Mickelbart et al., 2015). Both traditional breeding and targeted genome editing for drought tolerance require a better understanding of drought responses mechanisms (Langridge and Reynolds, 2015), which include molecular mechanisms governing the timing of stomata closure, modulation of photosynthetic performances, accumulation of osmolytes, and growth retardation (Bosco De Oliveira et al., 2012). Stomata represent the first barrier plants employ to avoid dehydration, with the trade-off of a reduced CO2 supply to the mesophyll. As water deficit intensifies, metabolic impairments impose further limitations to photosynthesis (Chaves et al., 2009). Along with plant responses to water deficit, understanding the recovery of photosynthesis upon rehydration is of paramount importance to understand water stress effects on photosynthesis (Flexas et al., 2004).
Impact of drought on gene expression has been intensely analyzed in numerous species such as Arabidopsis (Sakuraba et al., 2015), rice (Oono et al., 2014), maize (Kakumanu et al., 2012), sorghum (Dugas et al., 2011), and poplar (Barghini et al., 2015) by high-throughput transcriptomics.
A predominant role in driving drought-induced changes in gene expression is played by the hormone abscisic acid (ABA). The mechanisms of ABA perception and signal transduction are the subject of intense research and major breakthroughs have included the identification of a family of cellular receptors (Ma et al., 2009; Park et al., 2009). An increase in ABA changes the hydraulic regulation of stomata (Chaves et al., 2009), resulting in stomata closure under adverse hydraulic conditions by controlling guard cells behavior and decreasing water permeability within the leaf vascular tissue (Pantin et al., 2013). Tomato (Solanum lycopersicum L.) is one of the major horticultural crops and an important dietary source of vitamins A and C as well as carotenoids such as lycopene (Canene-Adams et al., 2005). Tomato is also considered a plant model system for fleshy fruit development (Osorio et al., 2011) and interaction with pathogens (Andolfo and Ercolano, 2015), with several tools available, including the sequenced genome (Sato et al., 2012) and its large open source genomic repository (Suresh et al., 2014). Although tomato is cultivated worldwide, it is considered sensitive to stresses of biotic and abiotic nature (Rai et al., 2013; Kissoudis et al., 2015). Most modern tomato cultivars are sensitive to water deficit, which results in reduced seed development and germination, reduced vegetative growth, and impaired reproduction (Nuruddin et al., 2003; Bartels and Sunkar, 2005; Rai et al., 2013). Galmés et al. (2011) reported that Mediterranean drought-tolerant tomato showed higher intrinsic water use efficiency under water stress compared to a tomato accession originating from a humid habitat. Such results could be due to stress induced changes in anatomical development (Galmés et al., 2013) or the abundance and activity of aquaporins facilitating CO2 transport through the mesophyll (Kaldenhoff, 2012).
The average water footprint per Kg of tomato is 215 liters, 30% of which is supplied by irrigation (Mekonnen and Hoekstra, 2011). Therefore, it is essential to develop drought tolerant, higher yielding varieties to cope with the increasing demand for tomato (Solankey et al., 2014). Only limited mapping research has been conducted on drought tolerance compared to other abiotic stresses in tomato (Foolad, 2007). Wild relatives, such as the drought tolerant Solanum pennellii, represent a valuable source of novel traits for genetic improvement. Gene expression studies in response to water stress have been carried out in leaves using microarray approaches identifying differentially expressed transcripts of genes involved in energy, plant hormones biosynthesis, and cation transporters and a number of transcription factors and signaling proteins (Gong et al., 2010; Sadder et al., 2014). However, studies that integrate the different levels of response to drought stress in tomato are under-represented.
The goal of our research was to identify in tomato the transcriptomic changes that control physiological adjustments under drought stress and recovery. In this study, we subjected plants to two drought treatments separated by a rehydration and recovery phase. We performed RNA sequencing on each of these stages to establish transcriptomes for drought and recovery. Each transcriptomic set is accompanied by physiological measurements (chlorophyll fluorescence, CO2 assimilation, and stomatal conductance), quantification of key metabolites (proline and ABA), and biometric parameters. These measurements serve to define the conditions of drought and recovery and provide a snapshot of the physiological state in each condition. We performed genome-wide comparisons of transcriptomes using cluster analysis to identify stress-induced patterns of expression and integrated them with physiological responses.
Materials and Methods
Plant Materials, Growth Conditions, and Stress Treatments
Seeds of cultivar M82 (accession LA3475) supplied by the Tomato Genetics Resource Center (TGRC, http://tgrc.ucdavis.edu/) were germinated in soil in a semi-controlled greenhouse. Air temperature (Ta, °C), humidity (RH, %), and solar radiation (Rs, W m−2) were acquired by a data logger (Spectrum Technologies, Plainfield, IL). The average air humidity and temperature were 58% and 22°C during the day and 86% and 18°C during night-time, while the cumulated daily global radiation during the day was 8.4 (±3.6) MJ/m2 day.
When seedlings had developed two true leaves (25 days after sowing seeds), they were transplanted in pots, filled with soil (one plant per pot) and fertilized after 7 days with Nitrophoska gold (Compo Agricoltura, Cesano Maderno, Italy). Plants were well irrigated for 30 days prior to start the stress treatments. Then plants were equally divided into control and stress treatment, nine replicates per treatment, and arranged in a randomized block design.
Two cycles of water deficit were performed by water withholding until soil water content of stress pots was < 1/3 of control pots, inducing a nearly complete closure of the stomata. This corresponded to 16 and 6 days of water withholding in the first and second cycle of drought, respectively. Between these two stress cycles, plants were well irrigated allowing a full recovery of soil water content and stomatal conductance. Control plants were well watered throughout the entire experimental period.
During the experiment, the soil water content (θ, m3/m3) was determined from dielectric measurements performed by a Time Domain Reflectometer (TDR100 Campbell Scientific Inc. Logan, UT) and applying the Topp's equation. The 14.2 cm trifilar probes were placed in 3 pots per treatment.
Leaf samples for molecular and biochemical analyses were collected at different time point of the experiment.
Gas Exchange and Modulated Chl a Fluorescence Emission
Net photosynthetic CO2 assimilation rate (A, μmol m−2 s−1) and stomatal conductance to water vapor (gs, mol m−2 s−1) were measured on a fully expanded, well-exposed top leaf on 5–6 plants per treatment between 10:00 a.m. and 1:00 p.m. Measurements were carried out using a portable open-system gas-exchange and modulated fluorometer analyser Li-6400XT (Li-Cor Biosciences, Lincoln, NE, USA), with CO2 inside leaf chamber set to 400 μmol CO2 mol−1 air. An artificial light source LED with emission peaks centered at 635 nm in the red and at 465 nm in the blue provided a PPFD equal to 2000 μmol (photons) m−2 s−1 (90% red, 10% blue). The instrument was also used to measure the steady-state (F′) and, upon a 0.8 s saturating light pulse emission, the maximum (F) Chl a fluorescence emission of leaves under actinic light. The software of the instrument (Li-Cor, 2011) calculated the gas-exchange parameters on the basis of von Caemmerer and Farquhar (1981) model, and the effective quantum yield of PSII photochemistry in light-adapted leaves, ΦPSII = (– F′)/ according to Genty et al. (1989).
Transient Chl a Fluorescence Emission
For fluorescence assays a continuous excitation Handy PEA fluorometer (Hansatech, Instruments Ltd., King's Lynn, Norfolk, England) was used. The excitation red light pulse for fluorescence induction (FI) was emitted by a (red) 650 nm light diode source, and applied for 1 s at the maximal available (sub-saturating) photosynthetic photon flux density (PPFD) of 3500 μmol (photon)/m2 s. Leaves were dark adapted for 30 min by means of the equipped white leaf-clips, prior to the assessment of the basal fluorescence emission (Fo), and the peak fluorescence emission (Fp) reached during the fast polyphasic raise induced by the sub-saturating excitation light pulse. As Fp is a viable approximation of the maximum fluorescence emission (Fm) (Strasser et al., 2000; Giorio et al., 2011), the dark-adapted maximum quantum yield of PSII photochemistry was calculated by the instrument software as Fv/Fm = (Fm − Fo)/Fm according to Kitajima and Butler (1975).
Biomass Determinations
Plant leaf area (PLA, m2) was measured, at the end of the experiment, on excised leaves using a scanning planimeter (Li-3100, LiCor, Lincoln, NE, U.S.A.). The instrument is equipped with a fluorescent source and a solid state scanning camera to measure the area of leaves as they move through it. For dry weight measurements, leaves and stems were oven-dried at 70°C for 10 days until a stable weight was reached. Both leaf area and total dry weight were measured on three plants.
Isolation of RNA, cDNA Synthesis, and qRT-PCR
Total RNA was extracted from leaf tissues using 1 mL of TRIZOL Reagent (Life Technologies, Carlsbad, CA, USA) per 100 mg of pulverized tissue. The homogenized samples were incubated for 5 min at room temperature and 200 μL of chloroform was added per milliliter of TRIZOL reagent. After centrifugation at 12,000 g for 15 min at 4°C, the upper aqueous phase was removed and 500 μL of 100% isopropanol was added. The samples were incubated at room temperature for 10 min and centrifuged at 12,000 g for 10 min at 4°C. The supernatant was removed and the RNA pellet was washed with 1 mL of 75% ethanol and centrifuged at 7500 g for 5 min at 4°C. The pellet was air dried and dissolved in RNase-free water. RNA quantity was measured spectrophotometrically by NanoDrop ND-1000 Spectrophotometer (NanoDropTechnologies), and integrity was verified on a denaturing MOPS/formaldehyde gel. One microgram of DNase-treated total RNA was reverse transcribed using SuperScript II Reverse Transcriptase and oligo (dT20) (Life Technologies, Carlsbad, CA, USA) by incubation at 42°C for 50 min. The reaction was stopped by heat inactivation at 70°C. The complementary DNA was diluted 1:20 and 4.5 μL was used for each qRT-PCR reaction, performed with 6.25 μL of 1X Platinum SYBR Green qPCR SuperMix (Life Technologies, Carlsbad, CA, USA) and 1.75 μL of primer mix (4.28 μM). Primers used are listed in Supplementary Table S1. Preparation of reactions was automated using the Liquid Handler Robot Tecan Freedom Evo and ABI 7900 HT (Applied Biosystems, Foster City, CA, USA) and performed with ABI 7900 HT (Applied Biosystems, Foster City, CA, USA). Cycling conditions were 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Three biological replicates with three technical repetitions were tested. Quantification of gene expression was carried out using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Elongation Factor EF1-α was used as an endogenous reference gene (Nicot et al., 2005; Corrado et al., 2007) for the normalization of the expression levels of the target genes. RNA extracted from plants grown in control condition served as calibrator sample for relative quantification of gene expression.
Proline and ABA Content Measurements
Leaf samples were collected by excising the leaf at the petiole from three biological replicates. Two technical replicates were performed for each sample. Proline content was determined according to the method of Claussen (2005). Two hundred and fifty milligrams of finely ground leaf tissue were suspended in 1.5 mL of 3% sulfosalicylic acid and filtered through a layer of glass-fiber filter (Macherey-Nagel, Ø 55 mm, Germany). One milliliter of Glacial acetic acid and 1 mL ninhydrin reagent (2.5 g ninhydrin/100 mL of a 6:3:1 solution of glacial acetic acid, distilled water and 85% ortho-phosphoric acid, respectively) were added to 1 mL of the clear filtrate. The mixture was incubated for 1 h in a boiling water bath. The reaction was terminated at room temperature for 5 min. Readings were taken immediately at a wavelength of 546 nm. The proline concentration was determined by comparison with a standard curve.
For ABA measurements, 150 mg of fine powder were extracted in distilled, autoclaved water with constant shaking at 4°C overnight in the dark. The supernatant was collected after centrifugation (10,000 × g for 10 min) and diluted 50-fold with TBS buffer (50 mM TRIS, 1 mM MgCl2, 150 mM NaCl, pH 7.8). Subsequently, ABA was analyzed by indirect enzyme-linked assay (ELISA) using the Phytodetek ABA test kit (Agdia, Elkhart, IN, USA) following the manufacturer's instructions. Color absorbance following reaction with substrate was read at 405 nm using a plate autoreader (1420 Multilabel Counter Victor3TM, PerkinElmer).
Statistical Analyses
The statistical significance of soil water content, gas-exchange and fluorescence parameters, ABA and proline contents between water treatments was evaluated through Student's t-test.
RNA Library Preparation and Library Sequencing
RNA pools of three biological replicates were used for all RNA-Seq experiments. The total RNA was DNase treated and purified using the RNeasy Plant Mini kit (Qiagen) following manufacturer's protocol (Qiagen, Valencia, CA, USA). RNA samples were analyzed quantitatively and qualitatively by NanoDrop ND-1000 Spectrophotometer (NanoDropTechnologies) and by Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
cDNA libraries were prepared with 1 μg of starting total RNA and using the Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA), according to TruSeq protocol. Library size and integrity were determined using the Agilent Bioanalyzer 2100 (Santa Clara, CA). Each library was diluted to 2 nM and denatured. Eight pM of each library was loaded onto cBot (Illumina, San Diego CA) for cluster generation with cBot Paired End Cluster Generation Kit (Illumina, San Diego, CA) and sequenced using the Illumina HiSeq 1500 with 100 bp paired-end reads in triplicate obtaining ~14 million reads for replicate. The sequencing service was provided Genomix4life Ltd (http://www.genomix4life.com) at laboratory of Molecular Medicine and Genomics (University of Salerno, Italy).
RNA Sequencing Analysis
The cleaning of the raw sequences from the RNA sequencing data was made using the Trim Galore package (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). In the first step, low-quality bases were trimmed from the 3′ end of the reads. In the second step, Cutadapt (Martin, 2011) removed adapter sequences; the default settings for paired-end was used. The quality check of the remaining sequences was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The (1) cleaned pairs, and (2) the high quality single reads obtained after the cleaning step, were used as input for the mapping to the tomato genome (version 2.40), independently. Bowtie version 2.1.0 (Langmead and Salzberg, 2012) and Tophat version 2.0.8 (Kim et al., 2013) were used for mapping. Paired and single reads, uniquely mapped, were counted, independently, per gene available from the iTAG annotation, version 2.3, using the HTSeq-count (http://www-huber.embl.de/users/anders/HTSeq/) version 0.5.4p1, in “union” default mode setting.
In order to define the set of expressed genes, raw read counts were normalized to RPKM (Reads per Kilobase per Million) and genes above the 1 RPKM cut-off were kept for the subsequent analyses. Differential expressed genes (DEGs) were found performing the negative binomial test implemented in the DESeq package (Anders and Huber, 2010) version 1.10.1, at a false discovery rate threshold (FDR) 0.01.
The k-means (MacQueen, 1967) cluster analyses were then performed on the log2 of the gene expression level (size factor normalized implemented in DESeq Package (Anders and Huber, 2010) for DEGs detected in all the stages, using 20 cluster, a number defined by the Elbow method (Thorndike, 1953), i.e., minimizing the within group variance at different cut-offs. GO enrichments were estimated via the goseq Bioconductor package (Young et al., 2010) (FDR ≤ 0.05) on each detected cluster possessing similar expression profiles in all the stages. As goseq requires gene length data, median transcript length per gene was obtained by parsing with a custom R script cDNA fasta files, as obtained from Ensembl Plants repository. GO annotations for tomato genes were obtained via BLAST2GO (Conesa and Götz, 2008) against NR databases and default settings. Unless otherwise stated, further graphical outputs were obtained with R custom scripts.
Results
To gain a comprehensive understanding of mechanisms activated by dehydration and rehydration events, we subjected tomato plants to cycles of drought stress and rewatering. Soil water status was monitored throughout the experiment as a measure of the progression of drought stress, and recovery under rewatering. Figure 1A shows values of pots containing plants of the M82 genotype. When water was withheld, pots subjected to drought stress underwent a continuous decline in soil water content (θ), which was significantly lower compared to control pots starting 2 days from the beginning of water withholding (Figure 1A). Drought continued until θ was ~22% of the control pots. This was the maximum stress point (Dr1) for the 1st cycle of drought. Reinstatement of irrigation allowed an immediate full recovery of θ to control values, which were maintained until the end of rewatering (RW). A rapid decline in θ (Figure 1A) was observed when a second cycle of drought stress was imposed and values similar to those of Dr1 were measured 6 days after the beginning of drought treatment (identified as Dr2). A graphic representation of the progression of the experiment is depicted in Figure 1B.
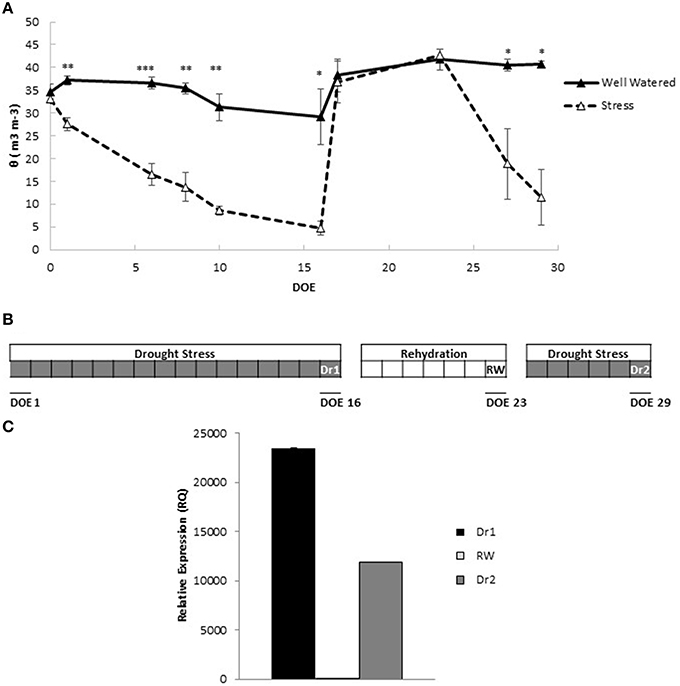
Figure 1. Experimental outline. (A) Volumetric soil water content (θ) throughout the progression of the experiment. Values represent average measurements ± SD of three replicates. Asterisks denote significant differences according to Student's t-test between well watered and stressed pots. *, **, and *** indicate significantly different values in drought stress compared to well-watered pots at p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively. (B) Schematic representation of the experimental design highlighting the points Dr1 (16 d of irrigation withholding), RW (7 days of irrigation), and Dr2 (6 days of irrigation withhold). (C) Gene expression of Solyc03g116390.2.1 in leaves after two cycles of drought stress (Dr1 and Dr2) and 1 week of rewatering (RW). RNA samples extracted from leaves of well watered plants were used as controls. Gene expression analyses were conducted by qRT-PCR. DOE, days of experiment.
For a preliminary evaluation of the efficacy of the drought stress and rehydration cycles at the cellular level, gene expression of Solyc03g116390.2.1, encoding a late embryogenesis abundant protein previously shown to be inducible by drought stress (Gong et al., 2010), was measured at Dr1, RW and Dr2. As shown in Figure 1C, a fold increase in gene expression of at least 10.000 times was observed at Dr1 and Dr2 compared to unstressed control plants. At RW, expression levels in rehydrated plants were comparable to controls, indicating that a full recovery had occurred (Figure 1C).
Physiological Effects of Drought Stress
To assess the impact of drought stress and rehydration on the physiology of tomato, leaf gas exchange and photosystem PSII efficiency were measured (Figure 2). Net CO2 assimilation rate (Figure 2A) and stomatal conductance to water vapor (Figure 2B) decreased significantly in stressed plants after 10 days of withholding water. At Dr1 CO2 assimilation (A) decreased to a minimum of 2.2 μmol m−2 s−1 in the stressed plants, 10% of the CO2 assimilation rate measured in controls (Figure 2A). Similar patterns were observed for stomatal conductance (gs), which in the stressed plants at Dr1 was as low as 0.030, compared to 0.710 mol m−2 s−1 found in the fully watered plants (Figure 2B). A moderate recovery of both A and gs was observed 1 day after rewatering when soil water content was fully restored (Figure 1A). Both parameters rose to values comparable to those of the controls at RW. Under the second treatment CO2 assimilation and stomatal conductance decreased more rapidly as compared to the previous stress cycle, reaching minimum average values of 4.0 μmol m−2 s−1 for A and 0.070 mol m−2 s−1 for gs at Dr2 (Figures 2A,B).
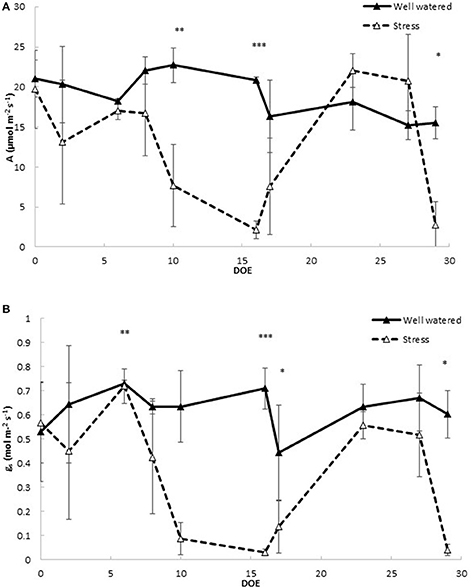
Figure 2. Leaf gas exchange parameters in well watered and drought stressed plants throughout the experiment. (A) Photosynthetic CO2 assimilation (A); (B) stomatal conductance to water vapor (gs). Values represent average measurements ± SD, n ≥ 5. *, **, and *** indicate significantly different values in drought stressed compared to well-watered plants at p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively. DOE, days of experiment.
The effective quantum yield of PSII (ϕPSII) decreased in response to the first drought treatment from an average of 0.13 in the control plants to 0.10 in the stressed plants (Figure 3A). Upon rewatering these values recovered to levels similar to controls. These values experienced even steeper declines during the 6 days of the second drought treatment (Figure 3A). The maximal quantum yield of the Photosystem II (PSII), measured in dark-adapted leaves, (Fv/Fm) remained stable throughout the experiment in the range of 0.81–0.83 in the control treatments while the values at Dr1 were lower, about 0.77 for Fv/Fm. Stressed plants partially recovered after 1 day of rewatering and completely recovered by RW (Figure 3B). Water stress reduced plant leaf area by 31% and dry matter accumulation by an average of 40% (Figures 4A,B).
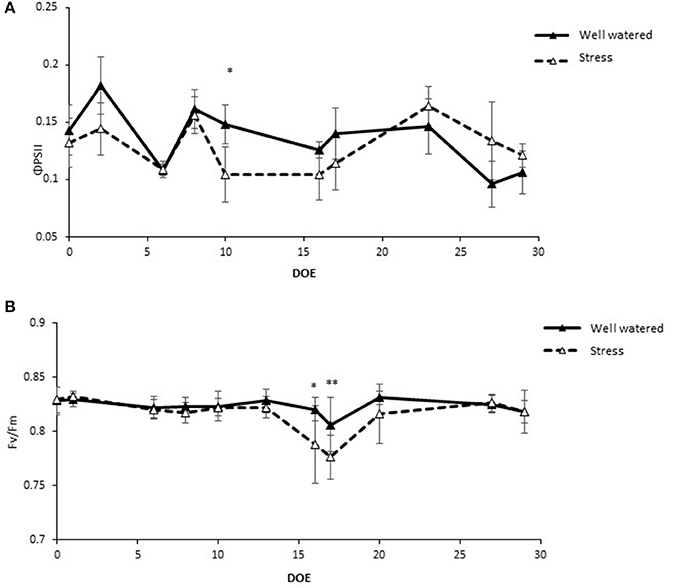
Figure 3. Chlorophyll fluorescence parameters in well watered and drought stressed plants throughout the experiment. (A) Quantum yield of PSII (ΦPSII); (B) Maximum quantum yield of PSII (Fv/Fm). Values represent average measurements ± SD, n ≥ 5. * and ** indicate significantly different values in drought stress compared to well-watered plants at p ≤ 0.05 and p ≤ 0.01, respectively. DOE, days of experiment.
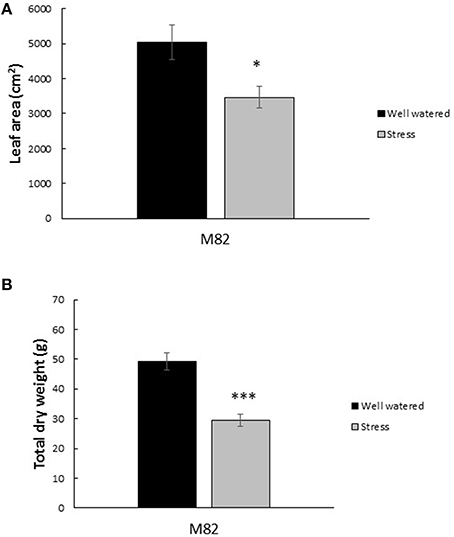
Figure 4. Impact of drought stress cycles on biometric parameters. Average Leaf Area (A) and total dry weight (B) in well watered and drought stress treatments (n = 3). For dry weight measurements, roots, stems, leaves, and fruits were included. Measurements were taken at the end of the experiment. * and *** indicate significantly different values in drought stress compared to well-watered plants at p ≤ 0.05 and p ≤ 0.001, respectively.
Proline and ABA Content in Drought Stressed Tomato
Well known metabolic alterations induced by drought stress include leaf accumulation of the osmolyte proline (Claussen, 2005) and the hormone ABA (Sharp and LeNoble, 2002). We therefore measured proline and ABA content at several time points in our experiment by a spectrophotometer and ELISA assay, respectively (Figure 5). After 13 days of drought stress, there was a slight accumulation of proline in the stressed plants. Under more severe stress conditions (Dr1) the amount of proline in the stressed plants was about 10 fold higher than the control M82. Proline amount in the stressed plants decreased in response to rewatering. However, values were still higher than controls for both genotypes at RW. The accumulation of proline reached the highest observed values of 4.5 at Dr2 (Figure 5A).
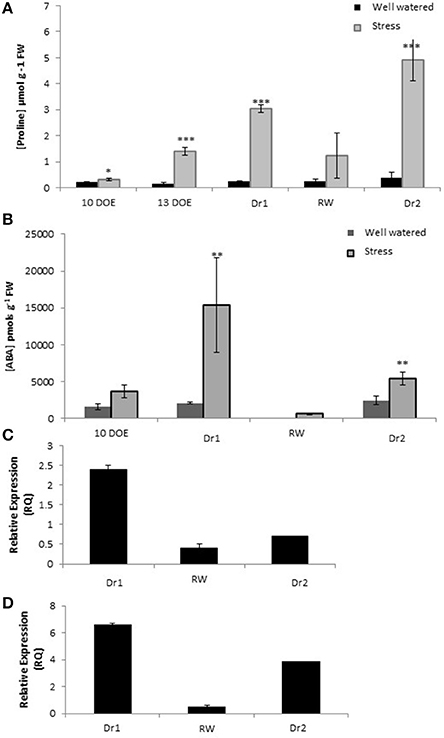
Figure 5. Quantification of Proline (A) and ABA content (B) in leaves; Gene expression of rate limiting enzymes P5CS (C) and NCED (D). DOE, days of experiment. *, ** and *** indicate significantly different values in drought stressed compared to well-watered plants at p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively.
Ten days of drought stress was not sufficient to elicit accumulation of leaf ABA as compared to the controls (Figure 5A). As the stress became more severe leaf ABA content was as high as 15,367 picomols/g fresh weight at Dr1. After rewatering, ABA levels decreased to values lower than those measured in the control plants at day 10 of experiment. At the end of second drought cycle (6 days of stress, Dr2), ABA content was 10 fold higher than that measured at RW, and about twice the 10-day stress values of first drought period (Figure 5B).
To investigate the correlation between proline and ABA accumulation and transcription of related biosynthetic genes we measured the gene expression of two rate-limiting steps in their biosynthetic processes. Expression of Solyc08g043170.2.1 and Solyc07g056570.1.1 encoding a Pyrroline-5-carboxylate synthetase (P5CS) and a 9-cis-epoxycaratenoid dioxygenase (NCED) respectively was evaluated by qPCR. In Dr1, P5CS was induced, while in RW and Dr2 no significant up-regulation was observed (Figure 5C). NCED was induced at comparable levels at Dr1 and Dr2, while at RW expression levels were similar to the controls (Figure 5D).
Transcriptomic Perturbations in Response to Drought Stress and Rehydration
To identify genes whose expression were altered by drought stress and rewatering in leaves, which could result in the observed physiological alterations, we carried out transcriptome sequencing. We used the Illumina platform on RNA samples extracted from leaves of M82 well-watered plants (WW) as well as in Dr1, RW, and Dr2 (Supplementary Table S2). RNA sequencing derived data were then subjected to gene expression analyses followed by clustering of genes showing similar trends of expression by comparison of the four conditions, and by gene ontology (GO) enrichment analysis. By comparing the different treatments, we identified 966 genes that showed differential expression in at least one of the comparisons, which were therefore considered as Differentially Expressed Genes (DEGs) (Supplementary Table S3).
The analysis highlighted that a large number of DEGs were down-regulated during drought stress. Comparative analysis of drought stressed (Dr1 and Dr2) vs. watered plants (WW and RW) revealed 119 DEGs common to all 4 comparisons (Supplementary Table S4). These included several histone encoding genes (e.g., Solyc10g008910), cell wall modifying enzymes (e.g., Solyc04g082140) as well as heat shock proteins (e.g., Solyc11g020330) (Supplementary Table S4).
In order to classify transcripts based on their behavior in WW, Dr1, RW, and Dr2, a cluster analysis was performed on the DEGs using normalized expression values of the DEGs in each of the experimental conditions. Twenty clusters were identified which grouped transcripts with similar expression trends (Supplementary Table S5). RNA sequencing results and the cluster analysis were validated using qRT-PCR on genes selected from different clusters (Supplementary Table S6, Figure 6). Figures 6A,B show normalized expression values from RNA sequencing and qRT-PCR experiments, respectively. As shown in Figure 6C, a good correlation was observed between the two sets of results. Seven clusters of DEGs were selected for further investigation based on their similar expression patterns (Supplementary Table S5, Figures 7A,B). Among them, clusters 1, 2, 14, 17, and 18 included genes with a higher expression level in WW and RW, while the remaining two clusters 7 and 20 were composed of transcripts with higher expression in Dr1 and Dr2 stressed plants (Figure 7A). Interestingly, clusters containing genes repressed during drought contained several histone variants and chlorophyll binding proteins. Several heat shock proteins and a heat shock factor also appeared to be induced by drought (Table 1). GO enrichment analyses were performed on clusters 1, 2, 14, 17, and 18 and clusters 7 and 20 independently (Figure 7B, Supplementary Tables S7, S8). These analyses showed that genes related to photosynthetic light harvesting (such as Chlorophyll a/b binding protein, Solyc08g067320) and to modification of cell wall (i.e., Pectinesterase, Solyc09g075350) were down-regulated in Dr1 and Dr2. Several genes encoding sucrose and starch metabolic processes were also down-regulated (Supplementary Table S6).
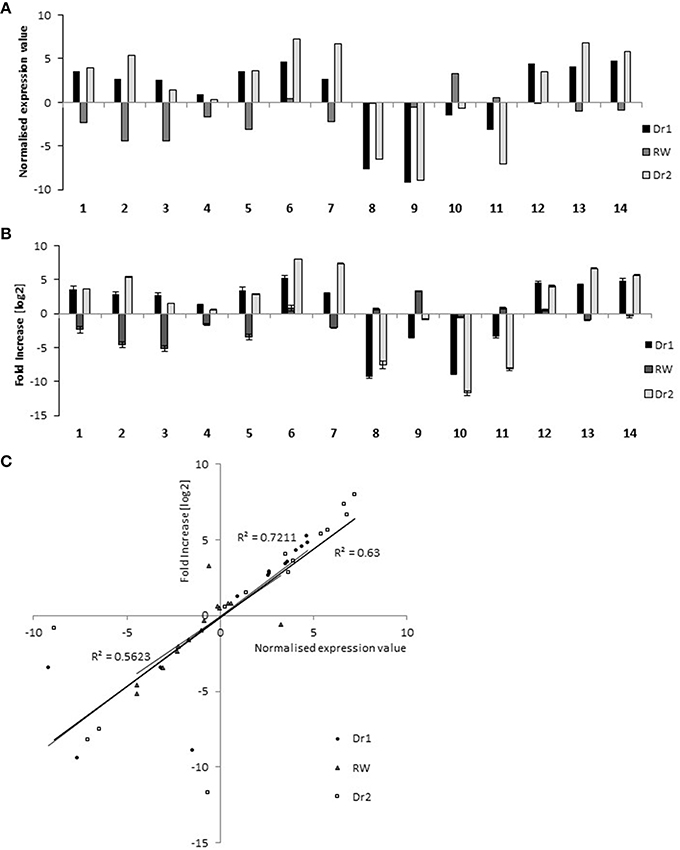
Figure 6. qRT-PCR validation of RNA sequencing data on 14 selected genes (Supplementary Table S6). (A) Expression value detected by RNA-seq method. (B) Expression analysis conducted by qRT-PCR. Data have been plotted on a log2 scale. (C) Correlation between RNA-Sequencing and qRT-PCR data. The normalized expression value obtained with RNA sequencing (x axis) were compared to the log2 of fold increase by qRT-PCR (y axis). RNA from well-watered control plants was used as calibrator sample.
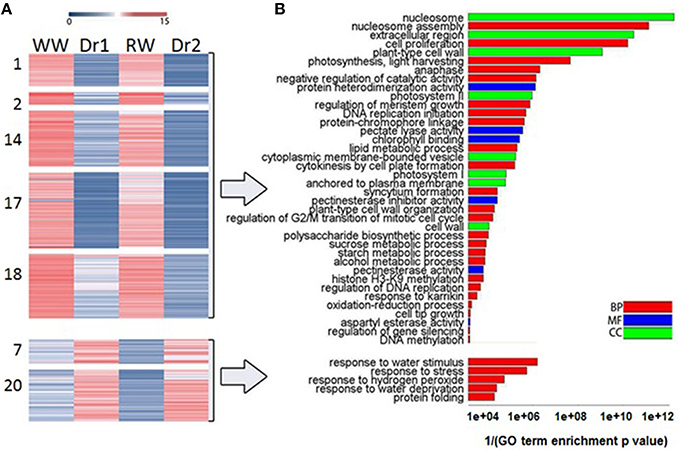
Figure 7. (A) Heatmap of selected clusters of Differentially Expressed Genes showing their expression behavior. Red and blue indicate higher and lower expression values, respectively. (B) Barplot showing GO Enrichment Analyses (goseq R package, FDR ≤ 0.05) of clusters 1, 2, 14, 17, 18 and 7, 20 independently, plotting GO terms (y axis) and the reciprocal of enrichment p value (x axis). Colors indicate GO ontology: red for Biological Process (BP), blue for Molecular Function (MF), and green for Cellular Component (CC).
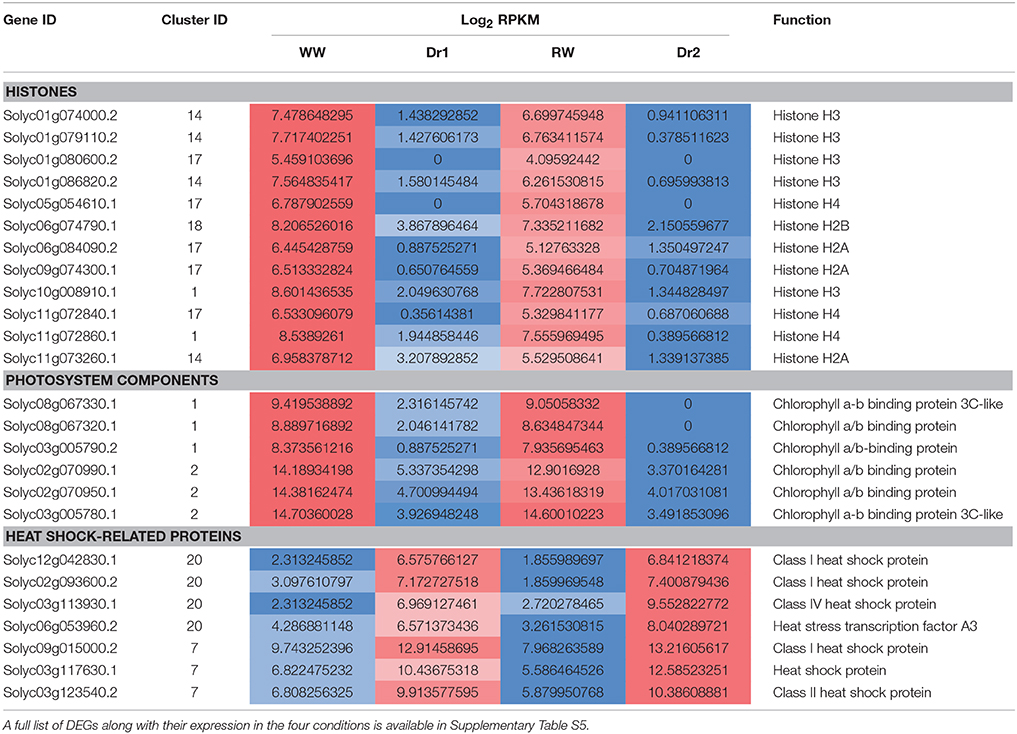
Table 1. Expression values (Log2 RPKM) in WW, Dr1, RW, Dr2 of DEGs belonging to selected functional categories (down, histones, and chlorophyll binding proteins; up, heat shock proteins).
GO categories enriched in clusters 7 and 20, instead, were more specifically related to stress, including classes such as response to water stimulus (members included dehydrin, Solyc01g109920.2) and water deprivation (including genes such as 2 NAC domain encoding IPR003441- Solyc12g013620.1/Solyc07g063410.2) (Figure 7, Supplementary Table S8). Transcripts coding for proteins involved in protein folding (Peptidyl-prolyl cis-trans isomerase Solyc09g092690.2 and heat shock protein Solyc03g117630.1) were also induced by water stress.
Discussion
In the present study, we have provided a detailed picture of physiological, metabolic and molecular adjustments employed by adult plants of tomato when exposed to events of prolonged water stress. The progression of stress was assessed by detailed monitoring of soil water content and stomatal conductance (gs). Leaf water status was strongly impaired by drought stress, as indicated by the very low values of gs observed at Dr1 and Dr2 (Figure 2B). Lutfor Rahman et al. (1999) found in four tomato genotypes that after ca. Ten days of interrupted irrigation, soil water content and gs decreased to values quite similar to our data, and a leaf water potential ranging from −10 to −14 MPa, indicating severe water stress. On this basis, a condition of severe stress can be hypothesized when plants approached Dr1 or Dr2. As expected (Hsiao et al., 1976), both leaf area and dry weight of the entire plant were significantly reduced in response to water deficit (Figure 4). The reduction in photosynthetic rate also likely contributed to plant growth reduction (Figure 2; Chaves et al., 2002). We found that the CO2 concentration inside the leaf (data not shown) was not affected during the first week of drought treatment, when soil water deficit (Figure 1) had no clear effect on both A and gs (Figure 2). During subsequent days of stress, both stomatal conductance (Figure 2B) and intercellular [CO2] (data not shown) showed a decrease. This indicates the action of stomatal limitation to A, while concomitant non-stomatal limitations, especially during progression to Dr1 should not be ruled out (Tezara et al., 1999; Cifre et al., 2005). The efficiency of the electron transport in the photosystems also decreased as stress progressed (Figure 3). The low Fv/Fm ratio at Dr1 likely indicates that photoinactivation of PSII may have occurred (Baker, 2008). Therefore, we conclude that photochemical limitations contributed to the decline in CO2 assimilation along with the altered metabolic content under severe drought (Cifre et al., 2005; Chaves et al., 2009).
These inferences are consistent with the effects observed in response to rewatering. Upon rehydration, a prompt recovery of CO2 assimilation in response to stomata reopening would indicate no significant impairment of the photosynthetic machinery (Cornic, 2000).
Chaves et al. (2009) reviewed that a recovery in CO2 assimilation of about 50% within a day from rewatering indicates severe water stress and that a few more days are required to reestablish the photosynthetic machinery. Recently, a complete recovery of the photosynthetic rate 24 h after rewatering was observed when mild drought conditions were applied (Nilsen et al., 2014). The moderate recovery observed here in both A and gs after 1 day of soil rewatering is therefore an additional indication of severe drought stress conditions.
One of the goals of this study was it identify potential factors controlling growth under drought stress conditions. At the molecular level, several transcription factors were up-regulated during drought stress, which could also help to explain the observed drought-induced growth reduction, in addition to the induction of stress responses (Supplementary Table S5). We found that two isoforms of subunit A of Nuclear factor Y (NF-Y), encoding orthologs of Arabidopsis NF-YA7 and NF-YA10 were also induced by drought stress. NF-Y is a heterotrimetric transcription factor whose subunit A is responsible for binding to DNA promoter sequences containing the CCAAT-box. Over-expression of NF-YAs, including NF-YA7 and NF-YA10, causes a dwarf phenotype and an increase in stress tolerance. Expression of NF-YA transcripts is stress-inducible and is inhibited in Arabidopsis in control conditions by miR169, a microRNA present in several isoforms (Li et al., 2008; Leyva-González et al., 2012). The changes in gene expression we observed in NF-Y isoforms likely play a role in growth retardation under the imposed drought conditions.
Our results also yielded other GO categories that contribute to growth and development. GO categories enriched in gene clusters down-regulated by drought stress included the cellular compartment “plant-type cell wall,” “cell wall” and the molecular function “pectate lyase activity” and “pectinesterase activity,” suggesting that cell wall modifying activities are repressed after prolonged drought stress. These categories included genes encoding cell wall modifying enzymes such as two laccase-22 (Solyc02g065170.2; Solyc07g052240.2), several pectinesterases and four expansins (Solyc05g007830.2, Solyc06g005560.2, Solyc06g076220.2, Solyc07g054170.2).
We observed that the tomato heat stress transcription factor HsfA3 was also up-regulated in drought stress conditions. In Arabidopsis HsfA3 is regulated by the dehydration-responsive element binding protein 2A (DREB2A). Plants overexpressing DREB2A are drought and heat tolerant and show increased levels of HsfA3 (Yoshida et al., 2008). HsfA3 is a powerful driver of heat shock protein expression and probably accounts for the observed up-regulation of several members of the Heat shock protein family, in several species including tomato (Table 1; Supplementary Table S5; Schramm et al., 2008; Li et al., 2013). Over expression of HSFA4a and HSFA9, two targets of HsfA3, has been reported to increase desiccation tolerance in Helianthus (Personat et al., 2014). HsfA3 may play a role in stress tolerance beyond heat tolerance and merits further study based on our results.
We found the presence of high levels of ABA (Figure 5) and of several targets of the ABA signal transduction pathway among the drought up-regulated genes. Included in these were RD29B (Solyc03g025810.2) and several LEA proteins indicating that the pathway is active in tomato after prolonged drought stress. The concomitant up-regulation of inhibitors of the ABA signaling cascade such as putative orthologs of Arabidopsis PP2CA (Solyc03g096670.2), MFT (SELFPRUINING 2G, Solyc02g079290.2), and AFP3 (Solyc05g012210.2; Garcia et al., 2008) was also observed. The increased expression of these genes indicates that a negative feedback loop is also in place. This is analogous to reports in Arabidopsis plants exposed to moderate drought stress, where the up-regulation of effectors of the ABA response also led to expression of negative regulatory components (Clauw et al., 2015).
Drought stress caused induction of three NAC domain-containing transcription factors (Supplementary Table S5), two of which encode JA2 (Solyc12g013620) and JA2like (JA2L, Solyc07g063140). These have been recently shown to have antagonistic roles in stomatal movements in tomato during pathogen attack (Du et al., 2014). JA2 is induced by ABA and promotes stomatal closure through induction of expression of the ABA biosynthetic gene NCED1. In contrast, JA2L is induced by the bacterial virulence factor coronatine and is proposed to have a role in stomatal reopening by regulating the expression of genes involved in Salicylic acid metabolism (Du et al., 2014). The presence of both JA2 and JA2L in our list of drought-induced genes indicates that these two TFs might also be involved in abiotic stress-triggered stomatal movements and represents a further indication that antagonistic pathways concur to the final balance of physiological adjustments.
The GO Enrichment analysis of the categories over-represented in clusters showing interesting patterns allowed for the identification of specific functions regulated by drought stress (Figure 7, Supplementary Tables S7, S8). Several GO categories related to photosynthesis, such as “photosystem I,” “photosystem II,” “chlorophyll binding,” and “photosynthesis, light harvesting” were enriched in clusters containing genes with higher expression in well-watered rather than drought stressed samples. This possibly indicates a reduced synthesis of components of the photosynthetic machinery under stress conditions (Clusters 1, 2, 14, 17, 18; Table 1). These differences in expression correlated with the low photosynthetic assimilation rate observed in drought stressed plants (Figure 2). A similar down-regulation of photosynthetic genes was observed in a progressive drought stress treatment on Arabidopsis plants. Moderate drought stress reduced the photosynthesis rate while expression of photosynthetic genes was not affected (Harb et al., 2010). This suggests that the mode of drought stress application and the severity of the stress influence the impact on the photosynthetic machinery. Concomitant with the downregulation of components of the photosystem under drought, we observed an upregulation of a FtsH homolog (Solyc03g112590.2) with a predicted chloroplast target peptide. FtsHs are ATP-dependent zinc metalloproteases, which, in chloroplasts, have been suggested to be involved in the turnover of the oxidized D1 protein of the PSII reaction center during recovery from photoinhibition (Lindahl et al., 2000; Bailey et al., 2002). Recently, Zhang et al. (2014) observed a progressive and constant upregulation of FtsH in Medicago truncatula subjected to drought stress and suggested a role in the repair of PSII damages resulting from drought-induced oxidative stress (Zhang et al., 2014).
Reduction of leaf growth occurs as a result of reduced cell division and/or cell expansion. Analysis of the GO enrichment categories suggests that both cell division and expansion are affected in tomato during drought stress. Categories such as “Cell Proliferation,” “Anaphase,” “Regulation of DNA replication” were enriched in well-watered rather than drought stressed samples, indicating that cell division is very likely repressed during drought.
We also observed that histone s gene families were the most down-regulated group of functional categories (Table 1). Histone expression is regulated during the progression of the cell cycle and tightly connected to DNA replication (Meshi et al., 2000; Rattray and Müller, 2012). The repression of the expression of several H3 and H4 isoforms (Table 1, Supplementary Tables S5, S6), concomitant with repression of genes such as a DNA topoisomerase, two DNA polymerase, single-stranded DNA-binding replication protein A large subunit, single-stranded DNA binding protein p30 could be an indication of a reduction in cell division. This is in all probability one of the factors leading to growth reduction in stress conditions. The observed repression of histone expression under drought is consistent with a recent report in rice showing that expression of several histone isoforms was reduced upon salt and drought treatment (Hu and Lai, 2015).
This study has sought to establish the transcriptomic profile of severe drought treatment in tomato and subsequent recovery with physiological and metabolic measurements. This combined data provides a comprehensive picture of the plant's status under stress and opens avenues to better understand the mechanisms involved in drought tolerance.
Overall, the results presented in this work indicate that drought stress causes the repression of several genes implicated in photosynthesis/light harvesting and plant growth, which was concomitant with a reduction of CO2 assimilation, electronic transport efficiency through the photosystems and a growth reduction. In addition, we found several genes up-regulated by drought stress that encoded late effectors and early regulators of ABA signaling. The expression of these genes was in parallel with an increase of ABA and osmolyte concentration as well as closure of stomata. Therefore, gene expression and physiological responses are intimately interconnected. Analysis of these results has yielded interesting candidate genes that could play novel roles in drought tolerance and adaptation.
Author Contributions
MV, AC, MC, RA, PG, GB, and SG conceived the work and designed experiments. PI, PP, GG, CM, RN, HB, CC, PB, GB performed experiments, analyzed the results, contributed figures. PI, MV, RA, PG, GB, SG wrote the paper. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all lab members for helpful discussions and Gaetano Guarino for technical assistance. This work was supported by the Italian Ministry of University and Research, projects GenoPOM (DM17732) and GenoPOM-PRO (PON02_00395_3082360), and by the Italian Ministry of Economy and Finance (Legge n.191/2009), to the National Research Council, project “Innovazione e Sviluppo del Mezzogiorno - Conoscenze Integrate per Sostenibilità ed Innovazione del Made in Italy Agroalimentare” (CISIA).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00371
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Andolfo, G., and Ercolano, M. R. (2015). Plant innate immunity multicomponent model. Front. Plant Sci. 6:987. doi: 10.3389/fpls.2015.00987
Bailey, S., Thompson, E., Nixon, P. J., Horton, P., Mullineaux, C. W., Robinson, C., et al. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277, 2006–2011. doi: 10.1074/jbc.M105878200
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Ann. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Barghini, E., Cossu, R. M., Cavallini, A., and Giordani, T. (2015). Transcriptome analysis of response to drought in poplar interspecific hybrids. Genom. Data 3, 143–145. doi: 10.1016/j.gdata.2015.01.004
Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. CRC. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Bosco De Oliveira, A., Mendes Alencar, N. L., and Gomes-Filho, E. (2012). “Physiological and biochemical responses of semiarid plants subjected to water stress” in Water Stress, ed M. R. Rahman (Rijeka: InTech), 43–58.
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Boyer, J. S., Byrne, P., Cassman, K. G., Cooper, M., Delmer, D., Grene, T., et al. (2013). The U.S. drought of 2012 in perspective: A call to action. Global Food Security 2, 139–143. doi: 10.1016/j.gfs.2013.08.002
Canene-Adams, K., Campbell, J. K., Zaripheh, S., Jeffery, E. H., and Erdman, J. W. (2005). The tomato as a functional food. J. Nutr. 135, 1226–1230.
Chaves, M. M., Flexas, J., and Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560. doi: 10.1093/aob/mcn125
Chaves, M. M., Pereira, J. S., Maroco, J., Rodrigues, M. L., Ricardo, C. P. P., Osório, M. L., et al. (2002). How plants cope with water stress in the field? Photosynthesis and growth. Ann. Bot. 89, 907–916. doi: 10.1093/aob/mcf105
Cifre, J., Bota, J. M., Escalona, J. M., Medrano, H., and Flexas, J. (2005). Physiological tools for irrigation scheduling in grapevine (Vitis vinifera L.): an open gate to improve water-use efficiency? Agric. Ecosyst. Environ. 106, 159–170. doi: 10.1016/j.agee.2004.10.005
Claussen, W. (2005). Proline as a measure of stress in tomato plants. Plant Sci. 168, 241–248. doi: 10.1016/j.plantsci.2004.07.039
Clauw, P., Coppens, F., De Beuf, K., Dhondt, S., Van Daele, T., Maleux, K., et al. (2015). Leaf responses to mild drought stress in natural variants of Arabidopsis thaliana. Plant Physiol. 167, 800–816. doi: 10.1104/pp.114.254284
Conesa, A., and Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832. doi: 10.1155/2008/619832
Cornic, G. (2000). Drought stress inhibits photosynthesis by decreasing stomatal aperture–not by affecting ATP synthesis. Trends Plant Sci. 5, 187–188. doi: 10.1016/S1360-1385(00)01625-3
Corrado, G., Sasso, R., Pasquariello, M., Iodice, L., Carretta, A., Cascone, P., et al. (2007). Systemin regulates both systemic and volatile signaling in tomato plants. J. Chem. Ecol. 33, 669–681. doi: 10.1007/s10886-007-9254-9
Du, M., Zhai, Q., Deng, L., Li, S., Li, H., Yan, L., et al. (2014). Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26, 3167–3184. doi: 10.1105/tpc.114.128272
Dugas, D. V., Monaco, M. K., Olsen, A., Klein, R. R., Kumari, S., Ware, D., et al. (2011). Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genomics 12:514. doi: 10.1186/1471-2164-12-514
Elliott, J., Deryng, D., Müller, C., Frieler, K., Konzmann, M., Gerten, D., et al. (2014). Constraints and potentials of future irrigation water availability on agricultural production under climate change. Proc. Natl. Acad. Sci. U.S.A. 111, 3239–3244. doi: 10.1073/pnas.1222474110
Flexas, J., Bota, J., Cifre, J., Escalona, M. J., Galmés, J., Gulías, J., et al. (2004). Understanding down-regulation of photosynthesis under water stress: future prospects and searching for physiological tools for irrigation management. Ann. Appl. Biol. 144, 273–283. doi: 10.1111/j.1744-7348.2004.tb00343.x
Foolad, M. R. (2007). Genome mapping and molecular breeding of tomato. Int. J. Plant Genom. 52:64358. doi: 10.1155/2007/64358
Galmés, J., Conesa, M. À., Ochogavía, J., Perdomo, J. A., Francis, D. M., Ribas-Carbó, M., et al. (2011). Physiological and morphological adaptations in relation to water use efficiency in mediterranean accessions of Solanum lycopersicum. Plant Cell Environ. 34, 245–260. doi: 10.1111/j.1365-3040.2010.02239.x
Galmés, J., Ochogavía, J., Gago, J., Roldán, E. J., Cifre, J., and Conesa, M. À. (2013). Leaf responses to drought stress in mediterranean accessions of Solanum lycopersicum: anatomical adaptations in relation to gas exchange parameters. Plant Cell Environ. 36, 920–935. doi: 10.1111/pce.12022
Garcia, M. E., Lynch, T., Peeters, J., Snowden, C., and Finkelstein, R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67, 643–658. doi: 10.1007/s11103-008-9344-2
Genty, B., Briantais, J. M., and Baker, N. R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990, 87–92. doi: 10.1016/S0304-4165(89)80016-9
Giorio, P., Nuzzo, V., Guida, G., and Albrizio, R. (2011). Black leaf-clips increased minimum fluorescence emission in clipped leaves exposed to high solar radiation during dark adaptation. Photosynthetica 50, 467–471. doi: 10.1007/s11099-012-0042-6
Gong, P., Zhang, J., Li, H., Yang, C., Zhang, C., Zhang, X., et al. (2010). Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 61, 3563–3575. doi: 10.1093/jxb/erq167
Harb, A., Krishnan, A., Ambavaram, M. M. R., and Pereira, A. (2010). Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154, 1254–1271. doi: 10.1104/pp.110.161752
Hsiao, T. C., Acevedo, E., Fereres, E., and Henderson, D. W. (1976). Water stress, growth, and osmotic adjustment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 273, 479–500. doi: 10.1098/rstb.1976.0026
Hu, Y., and Lai, Y. (2015). Identification and expression analysis of rice histone genes. Plant Physiol. Biochem. 86, 55–65. doi: 10.1016/j.plaphy.2014.11.012
Kakumanu, A., Ambavaram, M. M. R., Klumas, C., Krishnan, A., Batlang, U., Myers, E., et al. (2012). Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-seq. Plant Physiol. 160, 846–867. doi: 10.1104/pp.112.200444
Kaldenhoff, R. (2012). Mechanisms underlying CO2 diffusion in leaves. Curr. Opin. Plant Biol. 15, 276–281. doi: 10.1016/j.pbi.2012.01.011
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Kissoudis, C., Chowdhury, R., van Heusden, S., van de Wiel, C., Finkers, R., Visser, R. G. F., et al. (2015). Combined biotic and abiotic stress resistance in tomato. Euphytica 202, 317–332. doi: 10.1007/s10681-015-1363-x
Kitajima, M., and Butler, W. L. (1975). Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim. Biophys. Acta 376, 105–115. doi: 10.1016/0005-2728(75)90209-1
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Langridge, P., and Reynolds, M. P. (2015). Genomic tools to assist breeding for drought tolerance. Curr. Opin. Biotechnol. 32, 130–135. doi: 10.1016/j.copbio.2014.11.027
Leyva-González, M. A., Ibarra-Laclette, E., Cruz-Ramírez, A., and Herrera-Estrella, L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7:e48138. doi: 10.1371/journal.pone.0048138
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Iida, K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Li, Z., Zhang, L., Wang, A., Xu, X., and Li, J. (2013). Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt hypersensitivity in germination in transgenic Arabidopsis. PLoS ONE 8:e54880. doi: 10.1371/journal.pone.0054880
Lindahl, M., Spetea, C., Hundal, T., Oppenheim, A. B., Adam, Z., and Andersson, B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12, 419–431. doi: 10.1105/tpc.12.3.419
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lutfor Rahman, S. M., Nawata, E., and Sakuratani, T. (1999). Effect of water stress on growth, yield and eco-physiological responses of four tomato (Lycopersicon esculentum Mill.) cultivars. J. Jpn. Soc. Hortic. Sci. 68, 499–504. doi: 10.2503/jjshs.68.499
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
MacQueen, J. B. (1967). “Some methods for classification and analysis of multivariate observations,” in Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability, Vol. 1 (Berkeley, CA), 281–297.
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mekonnen, M. M., and Hoekstra, A. Y. (2011). The green, blue and grey water footprint of crops and derived crop products. Hydrol. Earth Syst. Sci. 15, 1577–1600. doi: 10.5194/hess-15-1577-2011
Meshi, T., Taoka, K. I., and Iwabuchi, M. (2000). Regulation of histone gene expression during the cell cycle. Plant Mol. Biol. 43, 643–657. doi: 10.1023/A:1006421821964
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Nicot, N., Hausman, J.-F., Hoffmann, L., and Evers, D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. doi: 10.1093/jxb/eri285
Nilsen, E. T., Freeman, J., Grene, R., and Tokushisa, J. (2014). A rootstock provides water conservation for a grafted commercial tomato (Solanum lycopersicum L.) line in response to mild-drought conditions: a focus on vegetative growth and photosynthetic parameters. PLoS ONE 9:e115380. doi: 10.1371/journal.pone.0115380
Nuruddin, M., Madramootoo, C. A., and Dodds, G. T. (2003). Effects of water stress at different growth stages on greenhouse tomato yield and quality. Hortic. Sci. 38, 1389–1393.
Oono, Y., Yazawa, T., Kawahara, Y., Kanamori, H., Kobayashi, F., Sasaki, H., et al. (2014). Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS ONE 9:e96946. doi: 10.1371/journal.pone.0096946
Osorio, S., Alba, R., Damasceno, C. M. B., Lopez-Casado, G., Lohse, M., Zanor, M. I., et al. (2011). Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425. doi: 10.1104/pp.111.175463
Pantin, F., Monnet, F., Jannaud, D., Costa, J. M., Renaud, J., Muller, B., et al. (2013). The dual effect of abscisic acid on stomata New Phytol. 197, 65–72. doi: 10.1111/nph.12013
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Lumba, S., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Personat, J.-M., Tejedor-Cano, J., Prieto-Dapena, P., Almoguera, C., and Jordano, J. (2014). Co-overexpression of two Heat Shock Factors results in enhanced seed longevity and in synergistic effects on seedling tolerance to severe dehydration and oxidative stress. BMC Plant Biol. 14:56. doi: 10.1186/1471-2229-14-56
Rai, G. K., Rai, N. P., Rathaur, S., Kumar, S., and Singh, M. (2013). Expression of rd29A::AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiol. Biochem. 69, 90–100. doi: 10.1016/j.plaphy.2013.05.002
Rattray, A. M. J., and Müller, B. (2012). The control of histone gene expression. Biochem. Soc. Trans. 40, 880–885. doi: 10.1042/BST20120065
Saadi, S., Todorovic, M., Tanasijevic, L., Pereira, L. S., Pizzigalli, C., and Lionello, P. (2015). Climate change and mediterranean agriculture: impacts on winter wheat and tomato crop evapotranspiration, irrigation requirements and yield. Agric. Water Manag. 147, 103–115. doi: 10.1016/j.agwat.2014.05.008
Sadder, M., Alsadon, A., and Wahb-Allah, M. (2014). Transcriptomic analysis of tomato lines reveals putative stress-specific biomarkers. Turk. J. Agric. For. 38, 700–715. doi: 10.3906/tar-1312-17
Sakuraba, Y., Kim, Y. S., Han, S. H., Lee, B. D., and Paek, N. C. (2015). The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27, 1771–1787. doi: 10.1105/tpc.15.00222
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Schramm, F., Larkindale, J., Kiehlmann, E., Ganguli, A., Englich, G., Vierling, E., et al. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53, 264–274. doi: 10.1111/j.1365-313X.2007.03334.x
Sharp, R. E., and LeNoble, M. E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53, 33–37. doi: 10.1093/jexbot/53.366.33
Solankey, S. S., Singh, R. K., Baranwal, D. K., and Singh, D. K. (2014). Integrated genomics, physio-chemical and breeding approaches for improving heat and drought tolerance in tomato. Int. J. Veget. Sci. 125, 625–645. doi: 10.1080/19315260.2014.902414
Strasser, R., Srivastava, A., and Tsimilli-Michael, M. (2000). “The fluorescence transient as a tool to characterize and screen photosynthetic samples” in Probing Photosynthesis: Mechanism, Regulation and Adaptation, ed M. Yunus (London: CRC Press), 445–483.
Suresh, B. V., Roy, R., Sahu, K., Misra, G., and Chattopadhyay, D. (2014). Tomato genomic resources database: an integrated repository of useful tomato genomic information for basic and applied research. PLoS ONE 9:e86387. doi: 10.1371/journal.pone.0086387
Tezara, W., Mitchell, V. J., Driscoll, S. D., and Lawlor, D. W. (1999). Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Nature 401, 914–917. doi: 10.1038/44842
Thorndike, R. L. (1953). Who belongs in the family? Psychometrika 18, 267–276. doi: 10.1007/BF02289263
von Caemmerer, S., and Farquhar, G. D. (1981). Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153, 376–387. doi: 10.1007/BF00384257
Yoshida, T., Sakuma, Y., Todaka, D., Maruyama, K., Qin, F., Mizoi, J., et al. (2008). Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 368, 515–521. doi: 10.1016/j.bbrc.2008.01.134
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Keywords: ABA, gene-expression cluster analysis, photosynthesis, proline, RNA sequencing, stomatal conductance, water stress
Citation: Iovieno P, Punzo P, Guida G, Mistretta C, Van Oosten MJ, Nurcato R, Bostan H, Colantuono C, Costa A, Bagnaresi P, Chiusano ML, Albrizio R, Giorio P, Batelli G and Grillo S (2016) Transcriptomic Changes Drive Physiological Responses to Progressive Drought Stress and Rehydration in Tomato. Front. Plant Sci. 7:371. doi: 10.3389/fpls.2016.00371
Received: 16 December 2015; Accepted: 10 March 2016;
Published: 31 March 2016.
Edited by:
Olivier Lamotte, CNRS UMR Agroécologie, FranceReviewed by:
Suleyman I. Allakhverdiev, Russian Academy of Sciences, RussiaGiridara Kumar Surabhi, Regional Plant Resource Centre, India
Copyright © 2016 Iovieno, Punzo, Guida, Mistretta, Van Oosten, Nurcato, Bostan, Colantuono, Costa, Bagnaresi, Chiusano, Albrizio, Giorio, Batelli and Grillo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefania Grillo, grillo@unina.it
 Paolo Iovieno
Paolo Iovieno Paola Punzo
Paola Punzo Gianpiero Guida2
Gianpiero Guida2 Michael J. Van Oosten
Michael J. Van Oosten Roberta Nurcato
Roberta Nurcato Hamed Bostan
Hamed Bostan Chiara Colantuono
Chiara Colantuono Antonello Costa
Antonello Costa Paolo Bagnaresi
Paolo Bagnaresi Maria L. Chiusano
Maria L. Chiusano Rossella Albrizio
Rossella Albrizio Pasquale Giorio
Pasquale Giorio Giorgia Batelli
Giorgia Batelli Stefania Grillo
Stefania Grillo