- 1Department of Biotechnology, TERI University, New Delhi, India
- 2National Institute of Plant Genome Research, Jawaharlal Nehru University Campus, New Delhi, India
Stress adaptation or tolerance in plants is a complex phenomenon involving changes in physiological and metabolic processes. Plants must develop elaborate networks of defense mechanisms, and adapt to and survive for sustainable agriculture. Water-deficit or dehydration is the most critical environmental factor that plants are exposed to during their life cycle, which influences geographical distribution and productivity of many crop species. The cellular responses to dehydration are orchestrated by a series of multidirectional relays of biochemical events at organelle level. The new challenge is to dissect the underlying mechanisms controlling the perception of stress signals and their transmission to cellular machinery for activation of adaptive responses. The completeness of current descriptions of spatial distribution of proteins, the relevance of subcellular locations in diverse functional processes, and the changes of protein abundance in response to dehydration hold the key to understanding how plants cope with such stress conditions. During past decades, organellar proteomics has proved to be useful not only for deciphering reprograming of plant responses to dehydration, but also to dissect stress–responsive pathways. This review summarizes a range of organellar proteomics investigations under dehydration to gain a holistic view of plant responses to water-deficit conditions, which may facilitate future efforts to develop genetically engineered crops for better adaptation.
Introduction
Agricultural productivity and food security is subject to increasing environmental constraints, particularly to water-deficit condition due to its high magnitude of damage and global impact (Bartels and Sunkar, 2005). One-third of arable land worldwide suffers from chronic or at least transient water scarcity, which is directly proportional to the reduction of crop yield. It is estimated that the world population will be about 8.5 billion by 2030, an increase of 22% from the current population, compelling an imminent rise in food demand. Additionally, the altered precipitation patterns, onset by unpredictable changes in climate, are speculated only to worsen, which is a major threat to global food security (Boyer, 1982). Hence, understanding how plants respond to water-deficit is the key factor for developing strategies for crop improvement.
Most environmental stresses are characterized by unifying feature that, at least a part of their detrimental effect on plant performance, is caused by unavailability of water. As a convergent point of multiple abiotic cues, cellular effects of dehydration stress are not only imbalances of ionic and osmotic homeostasis, but also impaired photosynthesis and cellular energy depletion, besides oxidative damage to the cellular machinery (Verslues et al., 2006). The current challenge is to identify elements involved in perception of stress and its path of translation in the cellular machinery to induce adaptive response. The present understanding of stress signaling in plants is largely based on genomic studies, which have been postulated upon comparative modulation of gene expression in response to stress (Seki et al., 2002; Shinozaki et al., 2003; Golldack et al., 2014). However, dehydration stress response is a complex multigenic phenomenon, where transcriptome expression rarely translates in equivalence with functional proteomic signatures (Gygi et al., 1999; Bohnert et al., 2006). Increasing evidence suggest a synergistic relationship between quantitative and qualitative changes in dehydration-induced proteins and physiological adaptations of plants (Bhushan et al., 2007; Pandey et al., 2010; Chen et al., 2011). The dehydration-responsive changes in the proteome can be grouped into five categories: (1) accumulation of dehydration-responsive proteins, which are characteristics of stress defense (Bartels and Sunkar, 2005; Mittler, 2006); (2) reorganization of the proteome profile among the organelles to optimize resources and achieve cellular homeostasis (Vera-Estrella, 2005; Poschet et al., 2011); (3) association network of multi-functional proteins linked to altered spatial distribution (Wu et al., 2009; Pandey et al., 2010); (4) PTM of regulatory proteins leading to activation or deactivation of metabolic pathways or isoform variants (Lata et al., 2011); and (5) reprograming of species-specific metabolic pathways, typically involved in stress tolerance (Bhushan et al., 2006; Pandey et al., 2010; Jaiswal et al., 2012). Finally, emerging evidence suggest that the phenotypic plasticity of plants, which limits stress-induced damage, is a direct result of altered proteome dynamics at the organellar level. A clear understanding of all these functional modulations can only be achieved by systematic analysis of dehydration-induced subcellular proteomes and unwinding their elaborate interaction networks.
During the past decade, a number of stress-responsive organellar proteomics studies have been carried out on variety of plant species, and increasingly agriculturally important crop species are being investigated against model plants (Pandey et al., 2008; Komatsu et al., 2009; Bhushan et al., 2011; Jaiswal et al., 2012; Hossain and Komatsu, 2013). This is primarily due to the availability of different genotypes of a crop with variable degrees of stress tolerance. Hence, comparative analysis of contrasting genotypes is a targeted approach for selectively identifying elements conferring relative tolerance against dehydration-induced damages (Bhushan et al., 2006; Pandey et al., 2008; Komatsu et al., 2009; Jaiswal et al., 2012; Verma et al., 2014). Even though numerous studies have been carried out on the plant stress response, a deeper insight into dehydration response at the sub-proteomic level is far from complete. This review will highlight the role of each subcellular compartment in sequence of events such as perception of dehydration stress, signal relay, and finally their cooperative function to combat stress. It will also critically discuss the altered dynamics of organelle crosstalk under dehydration and attempt to build a comprehensive spatiotemporal regulation of events that drive signaling pathways in plants under water-deficit.
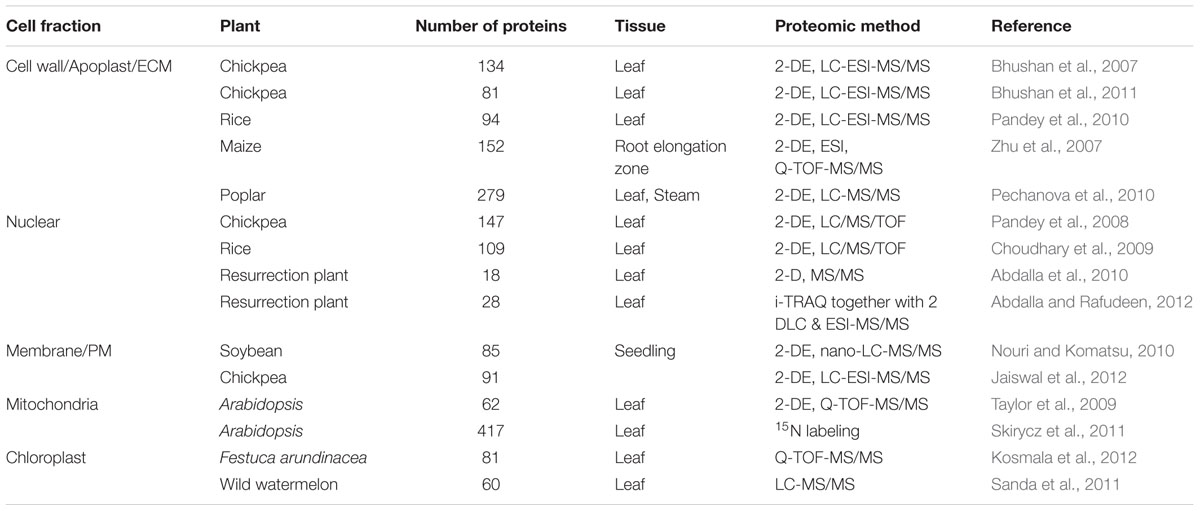
Dehydration-Responsive Cell Wall Proteome
Plant cell wall or ECM acts as a front-line defense, and is a conduit for signal transduction between the apoplast and the symplast, thereby plays a key role in cell fate decision under dehydration stress. Although proteins account for only 10% of the ECM mass, they comprise several hundreds of different molecules with diverse cellular functions (Carpita and Gibeaut, 1993). A detailed study of dehydration-responsive ECM proteome of chickpea identified 134 DRPs (Bhushan et al., 2007) and over 100 DRPs in rice (Pandey et al., 2010), presumably involved in an array of cellular functions. A similar study in maize primary root elongation zone led to the identification of water-soluble and loosely ionically bound 152 DRPs in the cell wall (Zhu et al., 2007). Pechanova et al. (2010) examined the apoplast of poplar stem and leaf tissues and reported 247 differentially regulated proteins (Table 1, Figure 1). The leaf apoplast showed abundance of proteins involved in cell wall metabolism, while the stem apoplast comprised of proteins mostly associated with cell defense. Despite the variation, the differential cell wall proteomes across the crop species showed commonalities in the essential functional protein classes viz., defense response, particularly ROS management and cell wall modification, albeit many proteins were observed to be unique to each of the crops studied. Cell wall, as emerged from these studies, is a repository of signaling molecules suggesting that the communication between the ECM and the symplast is one of the characteristic features of cellular mechanism that allows cells to respond effectively to various extracellular signals. Multivariate protein signatures such as WAKs, protein kinases, NDK, inorganic phosphatases and GTPases, among others were identified from both rice and chickpea seedlings (Bhushan et al., 2007; Pandey et al., 2010). The WAKs and protein kinases are known to relay extracellular signals through their cytoplasmic kinase domain and bind to a 2C-type protein phosphatase in the cytoplasm to form a signalosome complex (Anderson et al., 2001). NDKs identified both in rice and chickpea interact with cytosolic catalases and play a key role in relieving oxidative stress (Fukamatsu et al., 2003). Furthermore, 14-3-3 superfamily proteins were identified in the rice cell wall (Pandey et al., 2010), which have a characteristic role in stress signaling via modulating ROS accumulation (Elmayan et al., 2007). Another mechanism established for stress perception and signaling under dehydration is by induction, due to stretch-activated cytoskeletal channels. Several key proteins such as mannose lectin and aldolases were identified, which participate in compensatory mechanisms of cytoskeletal rearrangement under stress (Jewett and Sibley, 2003; Garaeva et al., 2006). Interestingly, deregulation of signaling proteins were also observed in maize and poplar tissues (Pechanova et al., 2010).
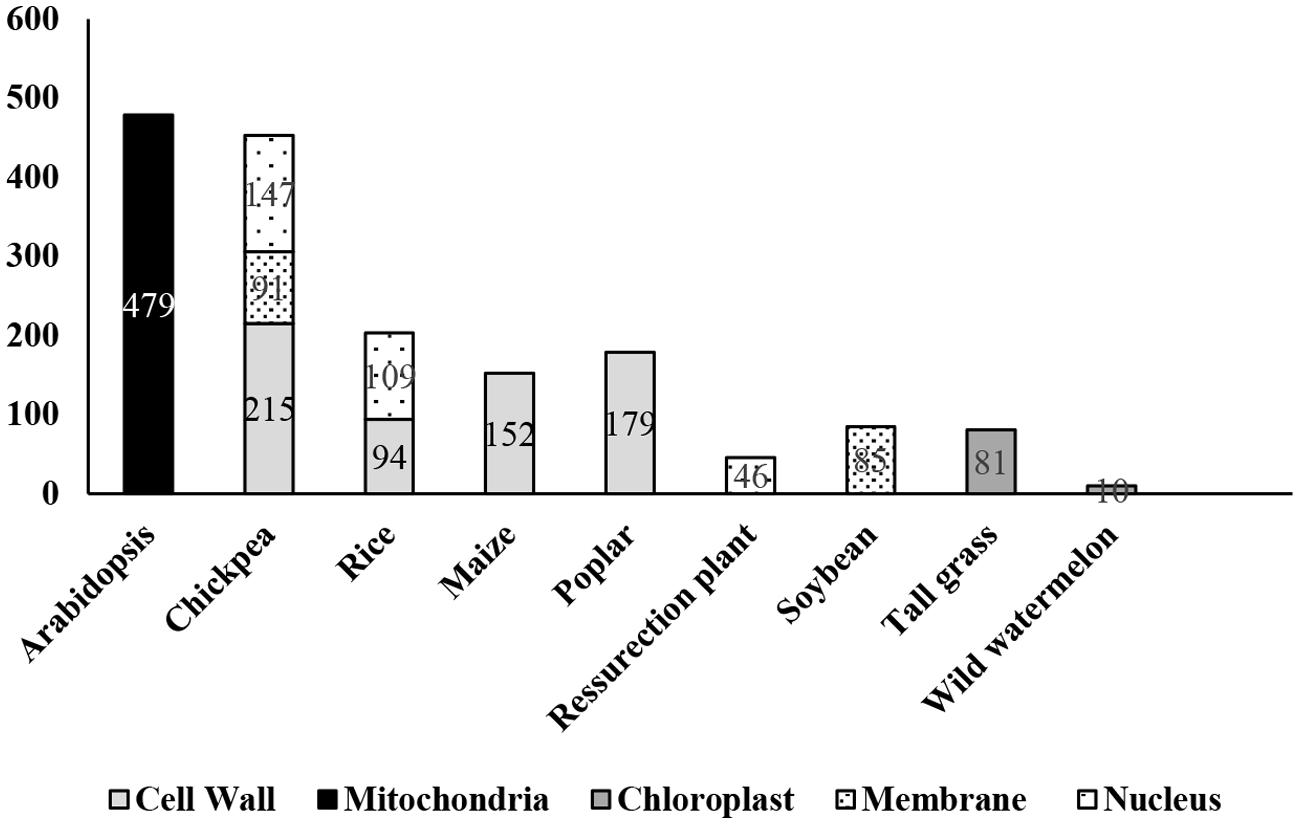
FIGURE 1. Number of dehydration-responsive proteins identified in different plant species. Numerical within the bar represents proteins reported from different proteomics studies.
The apoplast is known to be a source of ROS production (Zhu et al., 2007; Moschou et al., 2008). Dehydration-induced ROS can act as signaling molecules for stress response, but above a certain threshold they lead to oxidative injury to many cellular components. Most of the dehydration tolerance mechanisms rely primarily on protection of the cellular structure wherein an important method is the control of the level of ROS or the limitation of damage caused by ROS. It is well established that dehydration increases ROS levels, particularly O2- and H2O2 (Apel and Hirt, 2004; Papadakis and Roubelakis-Angelakis, 2005). Accumulation of apoplastic ROS can be viewed by in situ imaging with fluorescent indicator (Zhu et al., 2007). A critical screening of the cell wall proteome displayed several of the DRPs linked to antioxidative/detoxifying reactions, for example, APX, SOD, malate dehydrogenase, GPX, MDAR, DHAR, germin like protein, and oxalate oxidase, most of which showed an induced expression (Bhushan et al., 2007; Zhu et al., 2007; Pandey et al., 2010; Pechanova et al., 2010). Molecular chaperones and other proteins were also reported to be involved in the protection of cellular machinery. A novel phytoferritin, classically known as an iron storage protein, was identified in the cell wall of chickpea, which was postulated to have a significant role in ROS neutralization (Bhushan et al., 2007).
It is well established that under osmotic stress, cell wall serves as a source of sugars to maintain osmotic balance and undergoes lignification to avoid further water loss. Multiple stress-responsive proteins such as Ado-met, methyl transferases, AdoHcyase, adenosine kinase were identified (Bhushan et al., 2007; Zhu et al., 2007; Pandey et al., 2010; Pechanova et al., 2010), which are part of lignification pathway. Additionally, proteins such as cellulose synthase, beta galactosidases, xyloglucan hydrolase and hexosaminidase, and polygalacturonase that are known to utilize cell wall polysaccharides as an alternate carbon source under sugar depletion were also identified (Bhushan et al., 2007; Zhu et al., 2007; Pandey et al., 2010; Pechanova et al., 2010). Further, a better understanding of dehydration tolerance was built by a comparative cell wall proteomics study of tolerant and susceptible varieties of chickpea (Bhushan et al., 2011). The dehydration-responsive proteomes revealed that early perception, advanced signaling, notably cell wall restructuring, enhanced osmotic adjustment and better management of ROS are the keys to enhanced adaptation in plants.
Dehydration-Responsive Nuclear Proteome
Nucleus senses and physiologically responds to stress via multimodal signaling pathways, which are combinations of multiple input cues attributed by various organelles. The consequence is signal-specific response often resulting in cascade of downstream signals leading to activation of sub-responses such as hormonal modulations, systemic actions, and secondary regulations (Narula et al., 2013). In recent years, there have been several reports on the changes in nuclear proteome in varied cellular events (Bae et al., 2003; Lee et al., 2006; Salzano et al., 2006; Henrich et al., 2007; Buhr et al., 2008; Pandey et al., 2008; Repetto et al., 2008, 2012; Choudhary et al., 2009; Abdalla et al., 2010; Cooper et al., 2011; Varma and Mishra, 2011; Abdalla and Rafudeen, 2012). There have been at least four reports on dehydration-responsive nuclear proteome from three crops, one each from chickpea (Pandey et al., 2008) and rice (Choudhary et al., 2009), and two from the resurrection plant Xerophyta viscosa, (Abdalla et al., 2010; Abdalla and Rafudeen, 2012). The study in chickpea (Pandey et al., 2008) identified 147 DRNPs putatively involved in diverse functions, predominantly gene regulation and transcription, cell defense, protein degradation and chromatin remodeling, while a similar study in rice (Choudhary et al., 2009) led to the identification of over 100 DRNPs, categorized in comparable classes (Table 1, Figure 1). The DRNPs have also been elucidated in resurrection plant, capable of surviving at 5% RWC for prolonged periods. The study focussed on understanding the late protection mechanisms by comparative analysis between plants at 35% RWC vs. fully hydrated plants.
A comparison of the studies mentioned above revealed intriguing facts on regulatory mechanisms controlling the dehydration response (Figure 2). The proteins involved in signaling and gene regulation were prominent, which comprised of transcription factors, nuclear trafficking proteins, protein kinases and phosphatases. The altered expression of well-characterized dehydration-responsive transcription factors such as WRKY, bZIP, and AP2 domain containing proteins was noticeable in rice and chickpea, but not in resurrection plant, probably because the study recorded late response. Signaling partners like protein kinase, serine–threonine kinase, histidine kinase, receptor like protein kinase, and tyrosine phosphatase were identified in rice and chickpea, while casein kinase was reported in resurrection plant. Proteins involved in nucleocytoplasmic transport such as RAN, RANbp, RAN GTPase WIP1, GTPase binding, and dynamin like proteins were found to be consistently up-regulated. These proteins play crucial role in demarcating identity of the two compartments thus ensuring directionality of transport, which is the key to signaling network (Melchior and Gerace, 1998; Vetter et al., 1999). The ubiquitous presence of chromatin assembly/remodeling proteins, for example, histone deacetylase, histone 2A, histone 2B, histone 3, histone 4 was evident. The conspicuous presence of plurifunctional protein 14-3-3 in both monocots and dicot was intriguing, due to its acknowledged role in in ABA signaling, nucleocytoplasmic trafficking and chromatin remodeling, besides its role in development (Brunet et al., 2002). The other prominent functional proteins present were enzymes involved in scavenging of ROS, synthesis of osmolytes and chaperones whose activities enable plants to curtail damage and sustain physiological activity to survive stress conditions.
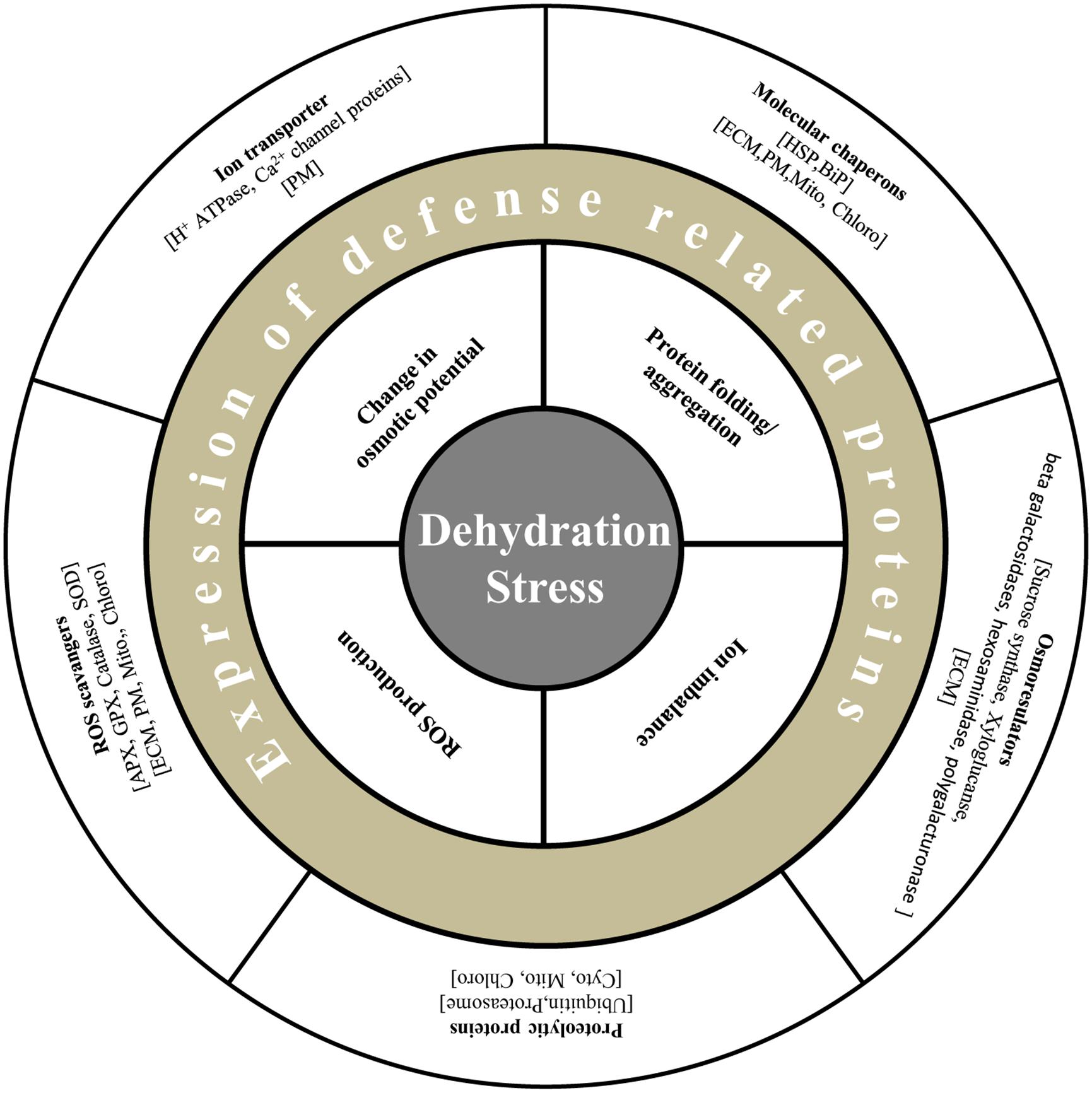
FIGURE 2. Diagrammatic representation of defense related proteins under dehydration stress in plants. Plants are able to alleviate dehydration stress by maintenance of turgor by osmoregulators such as sucrose synthase, xyloglucan, and exchange of ions by transporters such as H+ antiporters and ATPases. Generation of ROS is restricted by enzymes viz., APX, GPX, and SOD. Molecular chaperons like HSP and PIP maintain protein structure, and proteolytic enzymes degrade misfolded proteins.
In a classical proteomics study, Subba et al. (2013) examined and compared the dehydration-responsive nuclear proteome of tolerant and susceptible cultivars of chickpea. The basic stress-responsive features were found to be similar in both the varieties; however, cultivar-specific variations in terms of protein isoforms were observed for several of the common proteins. The tolerant variety was found to maintain better water status and displayed less oxidative damage. These findings highlight a coordinated response of nuclear proteome under dehydration involving both regulatory and functional protein network. Despite a large part of the identified proteins were found to be species-specific, conservation of primary elements is suggestive of a possible analogous mechanism for better adaptation.
Membrane Proteome Under Water-Deficit
Cell membranes are composed of dynamic lipid–protein matrices constituting the interface between cellular compartments. The lipid component presents a discriminating barrier to solute movement, while the membrane-associated proteins perform distinctive role in metabolism and maintenance of cellular homeostasis during stress adaptation (Jaiswal et al., 2012). Imposition of any form of osmotic stress elicits physical and biochemical signals perceived by membrane proteins at the frontier, which results in torrent of secondary subcellular signal relays. The importance of membrane proteins under abiotic stress has been well recognized and multiple studies have been carried out despite the limitation of their low abundance, hydrophobicity and complex electrophoretic properties (Michelet and Boutry, 1995; Garcia-Gomez et al., 2000; Beffagna et al., 2005; Liu et al., 2005). There have been, however, two noticeable membrane-associated proteomics studies in crop species under osmotic stress (Table 1, Figure 1). Nouri and Komatsu (2010) developed an osmotic stress responsive proteome and identified 12 and 86 differentially expressed proteins via gel-based and gel-free methods, respectively. Interestingly, proteins belonging to the classes involved in cell structure, metabolism, and protein-folding and protein synthesis were down-regulated under osmotic stress. The proteins involved in transport, signaling or defense were found to be down-regulated. Among the up-regulated proteins, most prominent were the transporters such as H+-ATPases, which build membrane potential for energy production, maintain turgor and intracellular pH (Figure 2). Another important up-regulated protein was calnexin. Curiously, the transcript abundance of calnexin did not change when compared to unstressed condition, while the immunoblot analysis confirmed the protein abundance in the plasma membrane. This phenomenon is suggestive of possible migration of classical ER-associated signaling protein, calnexin, to plasma membrane under osmotic stress.
Jaiswal et al. (2014) investigated the dehydration-responsive membrane-associated proteome of chickpea by 2-DE coupled with mass spectrometry. Proteomic analysis revealed 184 proteins significantly altered their intensities over dehydration treatments. The DRPs were categorized into 23 classes that were mainly related to generation of precursor metabolites and energy, protein metabolic process, transport and photosynthesis, among others. The dominant DRPs included various units of ATP synthase and light harvesting antenna complexes associated with PSI and PSII, besides oxygen evolving complex. Most of the DRPs associated with these classes were down-regulated, which might cause reduced ROS production, thus preventing cellular damage. The proteins majorly up-regulated were molecular chaperones such as HSPs and BiP. One of the interesting proteins identified was a SUN superfamily nuclear envelope protein. The SUN superfamily proteins are known to ‘bridge’ across the inner and outer nuclear membranes and physically connect the nucleus to every major component of cytoskeleton and serve as both mechanical adaptors and nuclear envelope receptors (Tzur et al., 2006). Functional characterization of CaSUN1 (Jaiswal et al., 2014) confirmed its participation in osmotic stress response, suggesting that SUN superfamily protein might be the signaling connect between cytoplasmic and nucleoplasmic activities for inducing stress response.
Reprograming of Mitochondrial and Plastid Proteome Under Dehydration
Cellular homeostasis is often disrupted by changes in the extracellular environment that uncouple biochemical pathways and result in undesirable accumulation of ROS. Under osmotic stress, reduction in CO2 fixation rate leads to decline in regeneration of NADP+ by Calvin cycle. Overreduction of electron transport chain in both plastids and mitochondria forms singlet oxygen (O2∗) which impair photosynthesis, creating a metabolic imbalance and generating oxidative distress (Mittler, 2006). Mitochondria and plastid coordinate to limit ROS accumulation and uphold energy balance to ensure cell survival under stress. A comprehensive study by Taylor et al. (2009) combined data from proteomic screening and GFP-targeting analysis to create a list of proteins from these organelles. Data curation yielded 279 non-redundant proteins, of which 5% belonged to peroxisome, 22% to mitochondria and 73% to chloroplast. Mitochondria isolated from stressed pea seedlings maintained their electron transport chain activity, while there was an apparent change in the activity of uncoupling proteins. There was ubiquitous activation of ROS detoxification pathways such as ascorbate/glutathione cycle and SOD-mediated detoxification of O2∗. Protection of existing matrix enzymes by synthesis of soluble protein-folding molecular chaperones like HSP22, HSP70, and HSP90 was also observed (Figure 2). However, repression of proteins associated with carbon assimilation in chloroplast could be an adjustment of the altered energy levels in plant under osmotic stress.
Chloroplast proteome was investigated following dehydration and subsequent watering in tall grass (Festuca arundinacea), between a high and low drought tolerant variety (Kosmala et al., 2012). Under water-deficit conditions, the rate of CO2 fixation is reduced, while higher rate of light reaction acts as a source of ROS leading to decrease in photosynthetic efficiency (Mittler, 2006). This phenomenon of photoinhibition results in degradation of proteins such as D1 in PSII. An ATP-dependent zinc FtH metalloprotease was identified, which might be involved in removal of damaged D1 protein from PS II. Another novel class of proteins identified was lipocalins, which had earlier been reported to be associated with chilling stress. These proteins are known to protect the thylakoid membrane from oxidative stress. Further, proteins like fibrillins, which maintain structural integrity of thylakoid membrane, were also detected. Altogether, these proteins were shown to be involved either directly in photosynthetic reactions or in protection of photosynthetic apparatus under stress. Sanda et al. (2011) investigated the effect of water-deficit on photosynthetic electron transport chain and identified 60 proteins that changed in abundance (Table 1, Figure 1). Most of the proteins identified were either chaperones or proteins related to electron transport chain. Interestingly, the integrity of PSI and PSII could largely be maintained even though carbon fixation rate was subdued.
A significant work by Skirycz et al. (2011), using 15N-labeling, established the interdependence of mitochondria and plastid function in Arabidopsis subjected to water-deficit condition. This study complemented the transcriptomic data (Skirycz et al., 2011) and provided novel insights into the underlying mechanism of stress responses. The proteome data clearly entailed the cooperative mechanism between chloroplast and mitochondria, which maintains the physiological balance under stress. The dehydration-responsive down-regulated proteins were found to belong mainly to primary metabolism, particularly photosynthesis, photorespiration, glycolysis, TCA cycle, and mitochondrial electron transport chain. On the contrary, the enzymes involved in redox homeostasis such as thioredoxin, APX, and ribosomal proteins were overrepresented.
Protein Phosphorylation Under Dehydration Stress
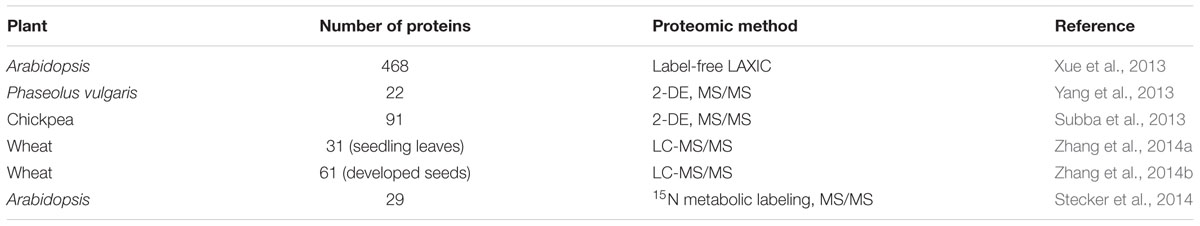
Protein phosphorylation is the central post-translational process which co-ordinates the synchronization of stress signals by regulating the protein pool dynamics between the cytosol and rest of the organelles (Pawson and Scott, 2005; Bonhomme et al., 2012). In recent years, the development of sensitive mass spectrometric techniques in conjunction with strategies to enrich the phosphorylated peptides/proteins led to the identification of a large repertoire of phosphoproteins from several plant species (Table 2, Figure 1). Most efforts in plant phosphoproteomics analyses have focused on the identification of protein phosphorylation in developmental stages (Chitteti and Peng, 2007), cellular compartments (Nuhse, 2004; Ito et al., 2009; Jones et al., 2009), chromatin structure (Tan et al., 2007) and only recently has the focus extended to stress response (Subba et al., 2013; Xue et al., 2013; Stecker et al., 2014; Zhang et al., 2014a,b).
Xue et al. (2013) introduced a novel mass spectrometry-based label-free quantitation method that facilitated systematic profiling of plant phosphoproteome changes with high efficiency and accuracy. In Arabidopsis, 468 up-regulated phosphopeptides representing 497 phosphosites showed significant changes under osmotic stress induced by mannitol and ABA treatment. Several known and novel components in the osmotic stress response pathway were identified. Phosphoproteomic analysis of polyethylene glycol-induced osmotic stress in root tips of common bean (Phaseolus vulgaris) was carried out by Yang et al. (2013), which led to the identification of 22 DRPs, of which 10 were found to be phosphorylated. Subba et al. (2013) identified 91 putative phosphoproteins in chickpea under dehydration, presumably involved in a variety of functions including cell defense and rescue, photosynthesis and photorespiration, molecular chaperones, and ion transport. Multiple sites of phosphorylation were predicted on several key elements, which include both the regulatory as well as the functional proteins. A novel protein DREPP (developmentally regulated plasma membrane polypeptide) was found to be differentially regulated under dehydration stress. Two significant studies on wheat developing seeds and leaves were conducted recently by Zhang et al. (2014a,b). The comparative phosphoproteome analysis revealed 63 unique phosphopeptides, corresponding to 61 phosphoproteins in the developing seeds, while 31 proteins showed significant changes in phosphorylation level in the leaves. Functional analysis indicated that some of these proteins might be involved in signal transduction, embryo and endosperm development of grains, and dehydration response and defense under water-deficit conditions. Stecker et al. (2014) identified protein phosphorylation events under osmotic stress using 15 N-metabolic labeling and untargeted mass spectrometry. The results indicated that regulatory proteins such as members of the MAPK family are specifically phosphorylated in response to osmotic stress and highlighted the utility of targeted phosphoproteomic analysis in understanding protein regulation networks.
Even though the above-mentioned studies were carried out in distinct species and different tissues using diverse methods of dehydration and varied methods of analysis, three major clusters of phosphorylated proteins were identified ubiquitously. Signaling proteins were the most prominent viz., stress induced MAPK pathway, ABA responsive SnRKs, calcium dependent protein kinases, casein kinase and protein phosphatase 2C, besides others. The second cluster consisted primarily of ribosomal proteins and proteins involved in degradation process, indicating that translation and turnover of proteins is tightly regulated by PTMs (Figure 2). Other major class consisted of chaperones, LEA proteins and dehydrin, among others involved in protecting the cellular components. These data are particularly important, at least in part, owing to the fact that the currently available stress-responsive phosphoproteins in plant are under-represented. Subcellular phosphoproteomics studies, under stress, are future imperative for understanding in detail the crosstalk between organelles orchestrated by PTM events, which cannot be elucidated by genomic investigations.
Dehydration-Responsive Proteome Network
Exposure of plants to water-deficit conditions leads to wide range of changes in protein expression levels. Protein composition of a cell is representative of multiple inflections such as protein sorting, translocation, post-translational modification, and protein degradation, all of which are influenced by stress conditions. More importantly, the distribution of proteins in various organelles under stress and their crosstalk to regulate stress responses is the key to understanding cellular mechanisms, which may not be accurately predicted based on genome expression profiling. In this review, the proteome changes in major organelles under osmotic stress have been re-visited, which can be summarized under three adaptive mechanisms: stress perception and signaling, defense response and metabolic regulation and detailed as follows:
Stress Perception And Signaling
Perception of osmotic stress, as it emerged from the analysis, is a complex phenomenon that comprises multitude of signaling pathways. The primary inducer of all stress-sensing mechanisms is any kind of change in intracellular osmotic balance. Alteration in cell turgor pressure leads to activation of proteins in the cell membrane and extracellular matrix viz., WAKs and receptor kinases, which may interact with signaling proteins specifically 14-3-3, MAP kinases and protein kinases in the cytosol. The interactions relay the signal to activate several families of transcription factors in the nucleus including WRKY, AP2, DREB, EREBP, RF2B, and leucine zipper. These cues also activate Ca2+ channels and ROS secondary signaling pathways, which further exemplify the stress signal via CDPKs to ensue modulation of stress-responsive components such as annexins, calnexins, and calmodulins. Annexin and calnexin are known to translocate to the cell membrane under dehydration, which enhances association with other molecules in the membrane, both resulting in the activation of the downstream signaling cascade (Lee et al., 2006; Jia et al., 2009). Both the proteins are also essential part of ABA-mediated signaling pathway. The reactive oxygen moieties induce the ABA-mediated pathways by activation of signaling proteins like SnRKs, protein phosphatase 2C and intermediates like 14-3-3 proteins in the cell membrane and cytosol, FCA receptor in the nuclear membrane and transcription factor such as tubby like protein (Figure 3). Proteins involved in nucleocytoplasmic transport such as RAN, WIP1, dyamin and RANbP ensure the directionality of signal relays and facilitate the overall signaling network.
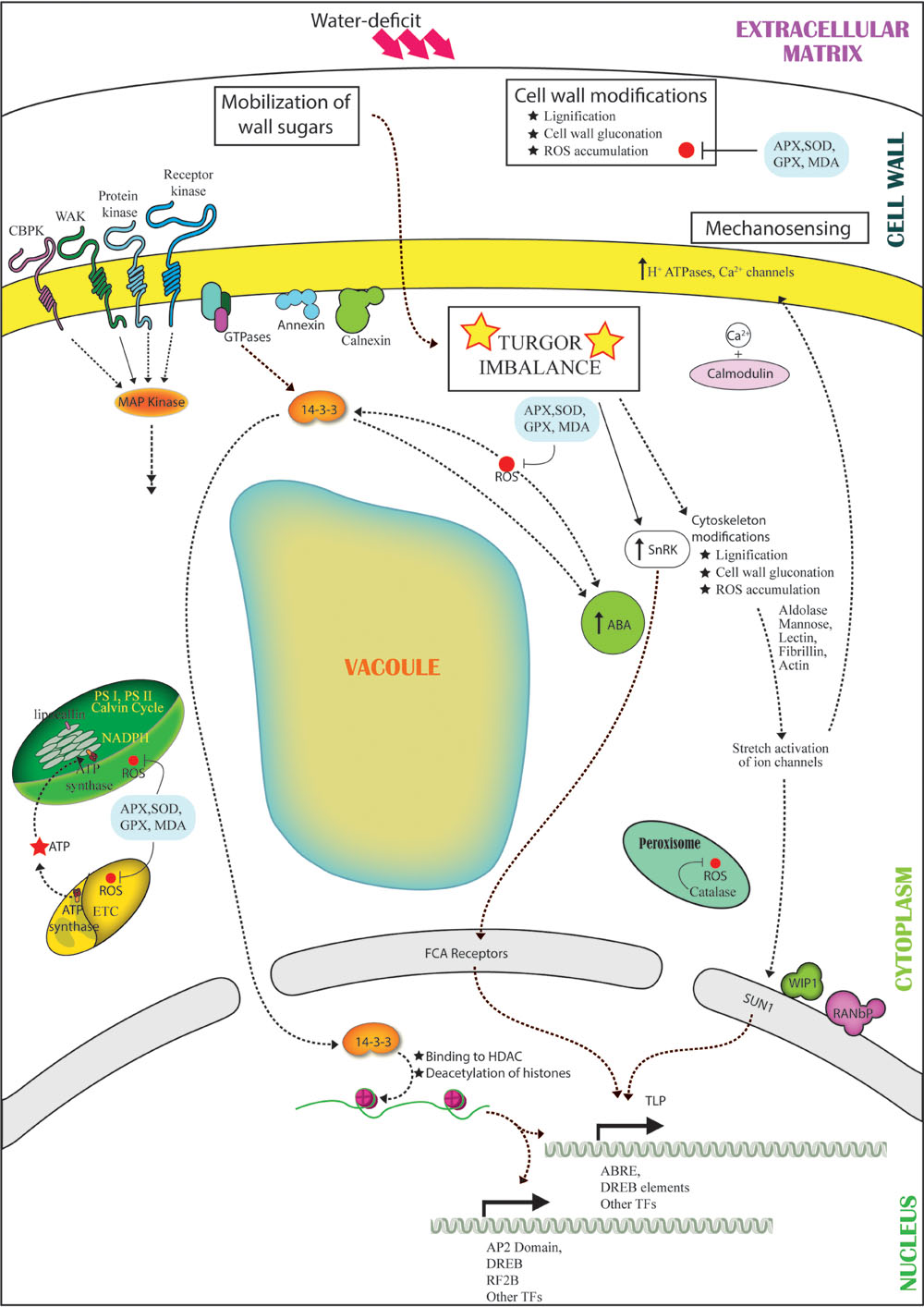
FIGURE 3. Representation of the cross talk among different pathways under water-deficit conditions. Dehydration stress leads to turgor imbalance and production of ROS. The turgor imbalance is mechanosensed, leading to modification in cytoskeleton proteins (lectins, actin, fibrillin) and sugars (mannose, aldolase) in turn leading to stretch-activation of ion channels (H+ ATPases, Ca2+) and activation of Ca2+/calmodulin signaling pathway. Wall sugars are mobilized and activate SnRK. Cell wall undergoes modifications like lignification and contraction. The stress is sensed by receptors (CBPK, WAKs, protein kinases, receptor kinases), which interact with GTPase. GTPase activates ABA and 14-3-3. Level of 14-3-3 increases with ROS production and parallely interacts and activates ABA and SnRK. 14-3-3 translocates to the nucleus and interacts with ACDH leading to choromatin remodeling and activation of transcription factors (AP2, DREB, RF2B, TLP). ABA interacts with the FCA receptors, and nuclear envelope protein SUN1 detects the ion channel activation owing to mechanosensing further leading to activation of TFs.
Cellular mechanosensing of osmotic imbalance may also be a potential mode of perception of stress acuity. The stretch activation of cytoskeleton associated proteins due to changes in the osmotic pressure and ion imbalance created by dehydration may lead to physical signal relays to modulate regulatory nuclear proteins. Further, identification of LINC (linker of nucleoskeleton and cytoskeleton) complex members such as SUN-domain protein, WIP1 and RANbP in the nucleus reinforce the theory of dehydration-induced mechanotransmission (Figure 3).
Defense Response
The mechanisms of dehydration avoidance vs. dehydration tolerance form the basis for understanding and interpreting dehydration-responsive events. The primary response of plants under water-deficit is to avoid further loss of water. The proton pumps associated with the plasma membrane facilitate stomatal closure under dehydration by mediating the efflux of K+ and Ca2+, which regulates the activity of plasma membrane H+-ATPase to maintain homeostasis of intracellular ions (Komatsu et al., 2009). Furthermore, lignification of cell wall is a preventive measure, which aids in preserving the intracellular moisture content and ion balance. Multiple stress-responsive proteins such as Ado-met, methyl transferase, AdoHcyase, adenosine kinase are known to be a part of lignification pathway. An alternate mechanism to maintain turgor under dehydration is the breakdown of cell wall polysaccharides by glycosyl hydrolases such as beta galactosidase, xyloglucan hydrolase and hexosaminidase, identified in the ECM, to direct sugars to cytoplasm (Bhushan et al., 2007; Pandey et al., 2010). Additionally, up-regulated proteins such as Δ1-pyrroline-5-carboxylate synthase, involved in biosynthesis of osmoprotectants and sugar metabolizing enzymes like sucrose synthase were also evident (Figures 2 and 3).
The production of ROS under dehydration can act as signaling molecules for stress response, although overproduction of ROS results in cellular damage. The survival of plant largely depends on the ROS accumulation under stress against the detoxification process mediated by ROS-scavenging proteins (Figures 2 and 3). As evident from proteome data, the electron transport chain in the chloroplast is repressed to avoid oxidative damage to the cell. Reduced expression of oxygen evolving complex proteins, PSI and PSII associated proteins might also enhance cell defense. Furthermore, accumulation of ROS-scavenger proteins such as SOD, ascorbate peroxidase, catalase and glutathione peroxidase under dehydration was noticed in all subcellular compartments. Proteins like lipocalin and fibrillins in the chloroplast were observed to be overexpressed under dehydration and are known to protect thylakoid membranes and PSII, respectively, against photoinhibition induced oxidative stress (Levesque-Tremblay et al., 2009; Singh et al., 2011).
Disturbance in the intracellular pH due to change in solute concentration under dehydration has an adverse effect on protein folding. Accumulation of molecular chaperons such as HSPs and Bip helps in protecting the existing proteins via folding and refolding of the misfolded proteins. Accumulation of RNA-binding proteins, dehydrins and LEA proteins could be additional support for protection of cellular translational apparatus. Furthermore, misfolded proteins, unassembled subunits of multimeric proteins and/or mutated proteins are degraded via intracellular proteolysis, particularly in the cytosol and the nucleus as revealed by phosphoproteome analysis (Figure 2).
Metabolic Regulations
When subjected to water-deficit conditions, plants dynamically alter growth rates and redistribute resources for better survival. Growth reduction increases the survival rate under severe stress but under moderate stress, it can be counterproductive. The proteome data establishes that primary metabolism pathways such as plastidial ATPase, Calvin cycle, and photorespiration are down-regulated, but mitochondrial ATP synthesis is up-regulated, indicating the importance of mitochondria in assisting plastid functions during water-deficit conditions (Skirycz et al., 2011). Under stress, plants try to achieve a balance by preserving the integrity of PSI and PSII, while compromising on overall carbon fixation rates (Sanda et al., 2011). The role of mitochondria in supporting plastid function by recycling reducing equivalents and redirecting its ATP pool is a classic example of optimization of resources under stress. This altered metabolic dynamics among the organelles under stress, emphasizes the importance of proteome-based approaches for systematic understanding of biological processes.
Concluding Remarks
Screening of organellar proteomes under water-deficit conditions revealed new and interesting insights into modulations of stress response in plants (Figure 3). The number of organellar proteomics studies available to date is limited and much work is required to enrich the database. Studies on cell membrane are important to fill the missing links in the signaling pathway. Besides, studies on fractionated cytosolic proteomes are rare and should be taken up to build the remaining chain of events. Moreover, cross-species comparisons for all the compartments and tissue-specific studies are required to postulate a comprehensive mechanism of dehydration progression and the plant responses. We, however, hope that this summary will not only be beneficial in understanding molecular basis of acquisition of dehydration tolerance at the organellar level, but will also highlight the importance of studying changes in compartment-specific protein abundance.
Author Contributions
NC conceived the study. DG, YR, and SG collated the data. DG, SC, and NC discussed the study and wrote the article.
Funding
This work was supported by grants from the Council of Scientific and Industrial Research [CSIR; Grant No. 38(1385/14/EMR-II)], Govt. of India and the National Institute of Plant Genome Research, New Delhi, India.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Mr. Jasbeer Singh for illustrations and graphical representation in the manuscript.
Abbreviations
2-DE, two-dimensional electrophoresis; ABA, abscisic acid; APX, ascorbate peroxidise; ATP, adenosine triphosphate; DRNPs, dehydration-responsive nuclear proteome; DRPs, dehydration-responsive proteins; ECM, extra cellular matrix; GFP, green florescent protein; GPX, glutathione peroxidise; GTPase, guanosine triphosphatase; H2O2, hydrogen peroxide; HSP, heat shock protein; iTRAQ, isobaric tag for relative and absolute quantification; LEA, late embryogenesis abundance; MAPK, mitogen activated protein kinase; MDA, malondialdehyde; NADP, nicotinamide adenine dinucleotide phosphate; NDK, nucleoside diphosphate kinase; PSI & PSII, photosystem I & II; PTMs, post-translational modifications; RAN, ras-related nuclear protein; ROS, reactive oxygen species; RWC, relative water content; SOD, superoxide dismutase; TCA, tricarboxylic acid; WAKs, wall associated kinases.
References
Abdalla, K. O., Baker, B., and Rafudeen, M. S. (2010). Proteomic analysis of nuclear proteins during dehydration of the resurrection plant Xerophyta viscosa. Plant Growth Regul. 62, 279–292. doi: 10.1007/s10725-010-9497-2
Abdalla, K. O., and Rafudeen, M. S. (2012). Analysis of the nuclear proteome of the resurrection plant Xerophyta viscosa in response to dehydration stress using iTRAQ with 2DLC and tandem mass spectrometry. J. Proteomics 75, 2361–2374. doi: 10.1016/j.jprot.2012.02.006
Anderson, C. M., Wagner, T. A., Perret, M., He, Z. H., He, D., and Kohorn, B. D. (2001). WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 47, 197–206. doi: 10.1023/A:1010691701578
Apel, K., and Hirt, H. (2004). REACTIVE OXYGEN SPECIES: metabolism. Oxidative Stress and Signal Transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bae, M. S., Cho, E. J., Choi, E.-Y., and Park, O. K. (2003). Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 36, 652–663. doi: 10.1046/j.1365-313X.2003.01907.x
Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Beffagna, N., Buffoli, B., and Busi, C. (2005). Modulation of reactive oxygen species production during osmotic stress in Arabidopsis thaliana cultured cells: involvement of the plasma membrane Ca2+-ATPase and H+-ATPase. Plant Cell Physiol. 46, 1326–1339. doi: 10.1093/pcp/pci142
Bhushan, D., Jaiswal, D. K., Ray, D., Basu, D., Datta, A., Chakraborty, S., et al. (2011). Dehydration-responsive reversible and irreversible changes in the extracellular matrix: comparative proteomics of chickpea genotypes with contrasting tolerance. J. Proteome Res. 10, 2027–2046. doi: 10.1021/pr200010f
Bhushan, D., Pandey, A., Chattopadhyay, A., Choudhary, M. K., Chakraborty, S., Datta, A., et al. (2006). Extracellular matrix proteome of chickpea (Cicer arietinum L.) illustrates pathway abundance, novel protein functions and evolutionary perspect. J. Proteome Res. 5, 1711–1720. doi: 10.1021/pr060116f
Bhushan, D., Pandey, A., Choudhary, M. K., Datta, A., Chakraborty, S., and Chakraborty, N. (2007). Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell Proteomics 6, 1868–1884. doi: 10.1074/mcp.M700015-MCP200
Bohnert, H. J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms-getting genomics going. Curr. Opin. Plant Biol. 9, 180–188. doi: 10.1016/j.pbi.2006.01.003
Bonhomme, L., Valot, B., Tardieu, F., and Zivy, M. (2012). Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol. Cell Proteomics 11, 957–972. doi: 10.1074/mcp.M111.015867
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Brunet, A., Kanai, F., Stehn, J., Xu, J., Sarbassova, D., Frangioni, J. V., et al. (2002). 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156, 817–828. doi: 10.1083/jcb.200112059
Buhr, N., Carapito, C., Schaeffer, C., Kieffer, E., Van Dorsselaer, A., and Viville, S. (2008). Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells. Electrophoresis 29, 2381–2390. doi: 10.1002/elps.200700738
Carpita, N. C., and Gibeaut, D. M. (1993). Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x
Chen, J., Zhang, Y., Wang, C., Lü, W., Jin, J. B., and Hua, X. (2011). Proline induces calcium-mediated oxidative burst and salicylic acid signaling. Amino Acids 40, 1473–1484. doi: 10.1007/s00726-010-0757-2
Chitteti, B. R., and Peng, Z. (2007). Proteome and phosphoproteome dynamic change during cell dedifferentiation in Arabidopsis. Proteomics 7, 1473–1500. doi: 10.1002/pmic.200600871
Choudhary, M. K., Basu, D., Datta, A., Chakraborty, N., and Chakraborty, S. (2009). Dehydration-responsive nuclear proteome of rice (Oryza sativa L.) illustrates protein network, novel regulators of cellular adaptation, and evolutionary perspective. Mol. Cell Proteomics 8, 1579–1598. doi: 10.1074/mcp.M800601-MCP200
Cooper, B., Campbell, K. B., Feng, J., Garrett, W. M., and Frederick, R. (2011). Nuclear proteomic changes linked to soybean rust resistance. Mol. Biosyst. 7, 773–783. doi: 10.1039/c0mb00171f
Elmayan, T., Fromentin, J., Riondet, C., Alcaraz, G., Blein, J.-P., and Simon-Plas, F. (2007). Regulation of reactive oxygen species production by a 14-3-3 protein in elicited tobacco cells. Plant Cell Environ. 30, 722–732. doi: 10.1111/j.1365-3040.2007.01660.x
Fukamatsu, Y., Yabe, N., and Hasunuma, K. (2003). Arabidopsis NDK1 is a component of ROS signaling by interacting with three catalases. Plant Cell Physiol. 44, 982–989. doi: 10.1093/pcp/pcg140
Garaeva, L. D., Pozdeeva, S. A., Timofeeva, O. A., and Khokhlova, L. P. (2006). Cell-wall lectins during winter wheat cold hardening. Russ. J. Plant Physiol. 53, 746–750. doi: 10.1134/S1021443706060033
Garcia-Gomez, B. I., Campos, F., Hernandez, M., and Covarrubias, A. A. (2000). Two bean cell wall proteins more abundant during water deficit are high in proline and interact with a plasma membrane protein. Plant J. 22, 277–288. doi: 10.1046/j.1365-313x.2000.00739.x
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. (1999). Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730. doi: 10.1128/MCB.19.3.1720
Henrich, S., Cordwell, S. J., Crossett, B., Baker, M. S., and Christopherson, R. I. (2007). The nuclear proteome and DNA-binding fraction of human Raji lymphoma cells. Biochim. Biophys. Acta 1774, 413–432. doi: 10.1016/j.bbapap.2006.12.011
Hossain, Z., and Komatsu, S. (2013). Contribution of proteomic studies towards understanding plant heavy metal stress response. Front. Plant Sci. 3:310. doi: 10.3389/fpls.2012.00310
Ito, J., Taylor, N. L., Castleden, I., Weckwerth, W., Millar, A. H., and Heazlewood, J. L. (2009). A survey of the Arabidopsis thaliana mitochondrial phosphoproteome. Proteomics 9, 4229–4240. doi: 10.1002/pmic.200900064
Jaiswal, D. K., Mishra, P., Subba, P., Rathi, D., Chakraborty, S., and Chakraborty, N. (2014). Membrane-associated proteomics of chickpea identifies Sad1/UNC-84 protein (CaSUN1), a novel component of dehydration signaling. Sci. Rep. 4:4177. doi: 10.1038/srep04177
Jaiswal, D. K., Ray, D., Subba, P., Mishra, P., Gayali, S., Datta, A., et al. (2012). Proteomic analysis reveals the diversity and complexity of membrane proteins in chickpea (Cicer arietinum L.). Proteome Sci. 10:59. doi: 10.1186/1477-5956-10-59
Jewett, T. J., and Sibley, L. D. (2003). Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell 11, 885–894. doi: 10.1016/S1097-2765(03)00113-8
Jia, X. Y., He, L. H., Jing, R. L., and Li, R. Z. (2009). Calreticulin: Conserved protein and diverse functions in plants. Physiol. Plant. 136, 127–138. doi: 10.1111/j.1399-3054.2009.1223.x
Jones, A. M. E., MacLean, D., Studholme, D. J., Serna-Sanz, A., Andreasson, E., Rathjen, J. P., et al. (2009). Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J. Proteomics 72, 439–451. doi: 10.1016/j.jprot.2009.02.004
Komatsu, S., Wada, T., Abaléa, Y., Nouri, M.-Z., Nanjo, Y., Nakayama, N., et al. (2009). Analysis of plasma membrane proteome in soybean and application to flooding stress response. J. Prot. Res. 8, 4487–4499. doi: 10.1021/pr9002883
Kosmala, A., Perlikowski, D., Pawlowicz, I., and Rapacz, M. (2012). Changes in the chloroplast proteome following water deficit and subsequent watering in a high- and a low-drought-tolerant genotype of Festuca arundinacea. J. Exp. Bot. 63, 6161–6172. doi: 10.1093/jxb/ers265
Lata, C., Bhutty, S., Bahadur, R. P., Majee, M., and Prasad, M. (2011). Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. J. Exp. Bot. 62, 3387–3401. doi: 10.1093/jxb/err016
Lee, B. J., Kwon, S. J., Kim, S.-K., Kim, K.-J., Park, C.-J., Kim, Y.-J., et al. (2006). Functional study of hot pepper 26S proteasome subunit RPN7 induced by Tobacco mosaic virus from nuclear proteome analysis. Biochem. Biophys. Res. Commun. 351, 405–411. doi: 10.1016/j.bbrc.2006.10.071
Levesque-Tremblay, G., Havaux, M., and Ouellet, F. (2009). The chloroplastic lipocalin AtCHL prevents peroxidation and protects Arabidopsis against oxidative stress. Plant J. 60, 691–702. doi: 10.1111/j.1365-313X.2009.03991.x
Liu, H., Wang, M., and Chou, K.-C. (2005). Low-frequency Fourier spectrum for predicting membrane protein types. Biochem. Biophys. Res. Commun. 336, 737–739. doi: 10.1016/j.bbrc.2005.08.160
Melchior, F., and Gerace, L. (1998). Two-way trafficking with Ran. Trends Cell Biol. 8, 175–179. doi: 10.1016/S0962-8924(98)01252-5
Michelet, B., and Boutry, M. (1995). The plasma membrane H+ -ATPase (A highly regulated enzyme with multiple physiological functions). Plant Physiol. 108, 1–6.
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Moschou, P. N., Paschalidis, K. A., Delis, I. D., Andriopoulou, A. H., Lagiotis, G. D., Yakoumakis, D. I., et al. (2008). Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. Plant Cell 20, 1708–1724. doi: 10.1105/tpc.108.059733
Narula, K., Datta, A., Chakraborty, N., and Chakraborty, S. (2013). Comparative analyses of nuclear proteome: extending its function. Front. Plant Sci. 4:100. doi: 10.3389/fpls.2013.00100
Nouri, M.-Z., and Komatsu, S. (2010). Comparative analysis of soybean plasma membrane proteins under osmotic stress using gel-based and LC MS/MS-based proteomics approaches. Proteomics 10, 1930–1945. doi: 10.1002/pmic.200900632
Nuhse, T. S. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16, 2394–2405. doi: 10.1105/tpc.104.023150
Pandey, A., Chakraborty, S., Datta, A., and Chakraborty, N. (2008). Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol. Cell Proteomics 7, 88–107. doi: 10.1074/mcp.M700314-MCP200
Pandey, A., Rajamani, U., Verma, J., Subba, P., Chakraborty, N., Datta, A., et al. (2010). Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: a proteomic approach. J. Proteome Res. 9, 3443–3464. doi: 10.1021/pr901098p
Papadakis, A. K., and Roubelakis-Angelakis, K. A. (2005). Polyamines inhibit NADPH oxidase-mediated superoxide generation and putrescine prevents programmed cell death induced by polyamine oxidase-generated hydrogen peroxide. Planta 220, 826–837. doi: 10.1007/s00425-004-1400-9
Pawson, T., and Scott, J. D. (2005). Protein phosphorylation in signaling-50 years and counting. Trends Biochem. Sci. 30, 286–290. doi: 10.1016/j.tibs.2005.04.013
Pechanova, O., Hsu, C.-Y., Adams, J. P., Pechan, T., Vandervelde, L., Drnevich, J., et al. (2010). Apoplast proteome reveals that extracellular matrix contributes to multistress response in poplar. BMC Genomics 11:674. doi: 10.1186/1471-2164-11-674
Poschet, G., Hannich, B., Raab, S., Jungkunz, I., Klemens, P. A. W., Krueger, S., et al. (2011). A novel Arabidopsis vacuolar glucose exporter is involved in cellular sugar homeostasis and affects the composition of seed storage compounds. Plant Physiol. 157, 1664–1676. doi: 10.1104/pp.111.186825
Repetto, O., Rogniaux, H., Firnhaber, C., Zuber, H., Küster, H., Larré, C., et al. (2008). Exploring the nuclear proteome of Medicago truncatula at the switch towards seed filling. Plant J. 56, 398–410. doi: 10.1111/j.1365-313X.2008.03610.x
Repetto, O., Rogniaux, H., Larré, C., Thompson, R., and Gallardo, K. (2012). The seed nuclear proteome. Front. Plant Sci. 3:289. doi: 10.3389/fpls.2012.00289
Salzano, A. M., Paron, I., Pines, A., Bachi, A., Talamo, F., Bivi, N., et al. (2006). Differential proteomic analysis of nuclear extracts from thyroid cell lines. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 833, 41–50. doi: 10.1016/j.jchromb.2005.12.025
Sanda, S., Yoshida, K., Kuwano, M., Kawamura, T., Munekage, Y. N., Akashi, K., et al. (2011). Responses of the photosynthetic electron transport system to excess light energy caused by water deficit in wild watermelon. Physiol. Plant. 142, 247–264. doi: 10.1111/j.1399-3054.2011.01473.x
Seki, M., Narusaka, M., Ishida, J., Nanjo, T., Fujita, M., Oono, Y., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. doi: 10.1046/j.1365-313X.2002.01359.x
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Singh, R. K., Anandhan, S., Singh, S., Patade, V. Y., Ahmed, Z., and Pande, V. (2011). Metallothionein-like from Cicer microphyllumis regulated by multiple abiotic stresses. Protoplasma 248, 839–847. doi: 10.1007/s00709-010-0249-y
Skirycz, A., Memmi, S., De Bodt, S., Maleux, K., Obata, T., Fernie, A. R., et al. (2011). A reciprocal 15 N-labeling proteomic analysis of expanding Arabidopsis leaves subjected to osmotic stress indicates importance of mitochondria in preserving plastid functions. J. Proteome Res. 10, 1018–1029. doi: 10.1021/pr100785n
Stecker, K. E., Minkoff, B. B., and Sussman, M. R. (2014). Phosphoproteomic analyses reveal early signaling events in the osmotic stress response. Plant Physiol. 165, 1171–1187. doi: 10.1104/pp.114.238816
Subba, P., Kumar, R., Gayali, S., Shekhar, S., Parveen, S., Pandey, A., et al. (2013). Characterisation of the nuclear proteome of a dehydration-sensitive cultivar of chickpea and comparative proteomic analysis with a tolerant cultivar. Proteomics 13, 1973–1992. doi: 10.1002/pmic.201200380
Tan, F., Li, G., Chitteti, B. R., and Peng, Z. (2007). Proteome and phosphoproteome analysis of chromatin associated proteins in rice (Oryza sativa). Proteomics 7, 4511–4527. doi: 10.1002/pmic.200700580
Taylor, N. L., Tan, Y.-F., Jacoby, R. P., and Millar, A. H. (2009). Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J. Proteomics 72, 367–378. doi: 10.1016/j.jprot.2008.11.006
Tzur, Y. B., Wilson, K. L., and Gruenbaum, Y. (2006). SUN-domain proteins: “Velcro” that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 7, 782–788. doi: 10.1038/nrm2003
Varma, P., and Mishra, R. K. (2011). Dynamics of nuclear matrix proteome during embryonic development in Drosophila melanogaster. J. Biosci. 36, 439–459. doi: 10.1007/s12038-011-9081-6
Vera-Estrella, R. (2005). Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 139, 1507–1517. doi: 10.1104/pp.105.067850
Verma, J. K., Gayali, S., Dass, S., Kumar, A., Parveen, S., Chakraborty, S., et al. (2014). OsAlba1, a dehydration-responsive nuclear protein of rice (Oryza sativa L. ssp. indica), participates in stress adaptation. Phytochemistry 100, 16–25. doi: 10.1016/j.phytochem.2014.01.015
Verslues, P. E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J., and Zhu, J.-K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539. doi: 10.1111/j.1365-313X.2005.02593.x
Vetter, I. R., Nowak, C., Nishimoto, T., Kuhlmann, J., and Wittinghofer, A. (1999). Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46. doi: 10.1038/17969
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xue, L., Wang, P., Wang, L., Renzi, E., Radivojac, P., Tang, H., et al. (2013). Quantitative measurement of phosphoproteome response to osmotic stress in Arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol. Cell Proteomics 12, 2354–2369. doi: 10.1074/mcp.O113.027284
Yang, Z.-B., Eticha, D., Führs, H., Heintz, D., Ayoub, D., Van Dorsselaer, A., et al. (2013). Proteomic and phosphoproteomic analysis of polyethylene glycol-induced osmotic stress in root tips of common bean (Phaseolus vulgaris L.). J. Exp. Bot. 64, 5569–5586. doi: 10.1093/jxb/ert328
Zhang, M., Lv, D., Ge, P., Bian, Y., Chen, G., Zhu, G., et al. (2014a). Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteomics 109, 290–308. doi: 10.1016/j.jprot.2014.07.010
Zhang, M., Ma, C.-Y., Lv, D.-W., Zhen, S.-M., Li, X.-H., and Yan, Y.-M. (2014b). Comparative phosphoproteome analysis of the developing grains in bread wheat (Triticum aestivum L.) under well-watered and water-deficit conditions. J. Proteome Res. 13, 4281–4297. doi: 10.1021/pr500400t
Zhu, J., Alvarez, S., Marsh, E. L., Lenoble, M. E., Cho, I.-J., Sivaguru, M., et al. (2007). Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol. 145, 1533–1548. doi: 10.1104/pp.107.107250
Keywords: adaptive responses, crop yield, dehydration, subcellular proteome, stress signals, spatiotemporal regulation
Citation: Gupta DB, Rai Y, Gayali S, Chakraborty S and Chakraborty N (2016) Plant Organellar Proteomics in Response to Dehydration: Turning Protein Repertoire into Insights. Front. Plant Sci. 7:460. doi: 10.3389/fpls.2016.00460
Received: 19 February 2016; Accepted: 24 March 2016;
Published: 13 April 2016.
Edited by:
Dominique Job, Centre National de la Recherche Scientifique, FranceReviewed by:
Martin Hajduch, Slovak Academy of Sciences, SlovakiaMohamed Suhail Rafudeen, University of Cape Town, South Africa
Copyright © 2016 Gupta, Rai, Gayali, Chakraborty and Chakraborty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Niranjan Chakraborty, nchakraborty@nipgr.ac.in
 Deepti B. Gupta
Deepti B. Gupta Yogita Rai
Yogita Rai Saurabh Gayali
Saurabh Gayali Subhra Chakraborty
Subhra Chakraborty Niranjan Chakraborty
Niranjan Chakraborty