- 1College of Horticulture, Northwest A&F University, Yangling, China
- 2Institute of Vegetables, Hangzhou Academy of Agricultural Sciences, Hangzhou, China
SQUAMOSA promoter binding protein (SBP)-box genes encode plant-specific transcription factors that are extensively involved in many physiological and biochemical processes, including growth, development, and signal transduction. However, pepper (Capsicum annuum L.) SBP-box family genes have not been well characterized. We investigated SBP-box family genes in the pepper genome and characterized these genes across both compatible and incompatible strain of Phytophthora capsici, and also under different hormone treatments. The results indicated that total 15 members were identified and distributed on seven chromosomes of pepper. Phylogenetic analysis showed that SBP-box genes of pepper can be classified into six groups. In addition, duplication analysis within pepper genome, as well as between pepper and Arabidopsis genomes demonstrated that there are four pairs of homology of SBP-box genes in the pepper genome and 10 pairs between pepper and Arabidopsis genomes. Tissue-specific expression analysis of the CaSBP genes demonstrated their diverse spatiotemporal expression patterns. The expression profiles were similarly analyzed following exposure to P. capsici inoculation and hormone treatments. It was shown that nine of the CaSBP genes (CaSBP01, 02, 03, 04, 05, 06, 11, 12, and 13) exhibited a dramatic up-regulation after compatible HX-9 strain (P. capsici) inoculation, while CaSBP09 and CaSBP15 were down-regulated. In case of PC strain (P. capsici) infection six of the CaSBP genes (CaSBP02, 05, 06, 11, 12, and 13) were arose while CaSBP14 was down regulated. Furthermore, Salicylic acid, Methyl jasmonate and their biosynthesis inhibitors treatment indicated that some of the CaSBP genes are potentially involved in these hormone regulation pathways. This genome-wide identification, as well as characterization of evolutionary relationships and expression profiles of the pepper CaSBP genes, will help to improve pepper stress tolerance in the future.
Introduction
Transcription factors (TFs) are DNA-binding proteins that regulate gene expression at the level of mRNA transcription. They are capable of activating or repressing the transcription of multiple target genes (Yang et al., 2008). In plants, TFs play essential roles in the regulation of many developmental processes (Li et al., 2013). SQUAMOSA promoter binding protein (SBP)-box genes encode a TFs that contain a highly conserved DNA-binding domain termed the SBP domain (Klein et al., 1996; Cardon et al., 1999). This domain comprises approximately 76 amino acid residues that are involved in both DNA binding and nuclear localization, including two zinc-binding sites (Yamasaki et al., 2004). The AmSBP1 and AmSBP2 genes of Antirrhinum majus were the first SBP-box genes to be discovered based on their ability to interact with the promoter sequence of the floral meristem identity gene SQUAMOSA (Klein et al., 1996). Additional SBP-box genes were later identified, isolated, and characterized in many plants, including Arabidopsis thaliana (Cardon et al., 1999), silver birch (Lannenpaa et al., 2004), Salvia miltiorrhiza (Zhang et al., 2014), rice (Xie et al., 2006), maize (Chuck et al., 2010), tomato (Salinas et al., 2012), grape (Hou et al., 2013b), and Gossypium hirsutum (Zhang et al., 2015).
SQUAMOSA promoter binding protein genes have been found to play a role in the gene regulatory network of the flower formation pathway, and many studies have revealed that these genes are closely related to flower development (Klein et al., 1996; Cardon et al., 1997; Shikata et al., 2009). Moreover, recent studies showed that SBP-box genes are involved in signal transduction and responses to abiotic and biotic stress in many species. For instance, AtSPL14 has been found to be involved in determining sensitivity to the programmed cell death-inducing fungal toxin fumonisin B1 (Stone et al., 2005). AtSPL2 (At5g43270), which is modified in transgenic Arabidopsis overexpressing the JASMONATE CARBOXYL METHYLTRANSFERASE gene (AtJMT) response to jasmonic acid mediated resistance pathway (Jung et al., 2007). VpSBP5 likely participates in regulating resistance to Erysiphe necator by activating the SA-induced systemic acquired resistance pathway and MeJA-induced wound signaling pathway in grapes (Hou et al., 2013b). However, little is currently known about the SBP-box genes in pepper, especially regarding resistance to Phytophthora blight.
Pepper (Capsicum annuum L.) is one of the most important vegetable crops worldwide. The Phytophthora blight in pepper is caused by the oomycete Phytophthora capsici, which mainly attacks the roots and is one of the most destructive diseases worldwide (Hausbeck and Lamour, 2004; Zhang et al., 2013), as it also infects tomato, eggplant, cucumber, watermelon, pumpkin, squash, cocoa, and other plants (Biles et al., 1995; Oelke and Bosland, 2003). The pathogen can affect the plant at any stage of development causing damping-off, seedling blight, and wilting, followed by plant death. Infected plants have rapidly expanding water-soaked lesions (Kousik et al., 2012). Analysis of C. annuum SBP-box (CaSBP) genes in response to P. capsici and hormones is therefore important for identification of candidate genes in pepper.
In the current study, we report the genome-wide identification and characterization of SBP-box genes in the pepper genome, including sequence alignment, phylogenetic analysis, intron-exon structure, chromosomal location, and synteny. Moreover, we investigated the expression patterns of CaSBP genes in various pepper tissues/organs, as well as the transcriptional responses of CaSBP genes in the roots of different P. capsici. Five CaSBP genes were selected based on their expression patterns after inoculation with P. capsici, and their expression profiles were assessed following treatment with different plant hormones and corresponding biosynthetic inhibitors. Our findings lay the foundation for future research into the functions of disease-related genes from the SBP-box gene family in pepper.
Materials and Methods
Identification and Annotation of SBP-Box Genes in Pepper
A hidden Markov model (HMM) profile of the SBP domain (Accession no. PF03110) was downloaded from the Pfam database1 This domain was used to query the CM334 (C. annuum) Genome Database and Zunla-1 (C. annuum) Genome Database2 (V1.55) with the BLASTP program. All hits with an E-value < 1.5e-7 were identified. All non-redundant protein sequences were searched for the SBP domain using NCBI’s conserved domain database3 Candidate CaSBP genes were aligned with DNAMAN software (Version 5.0), and genes with differing sequences between the two cultivars were identified (Guo et al., 2015). Primers (Supplementary Table S1) were designed to amplify the sequences with Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA), and CM334 and Zunla-1 sequences for the same gene were then aligned to confirm the correct sequences. In order to compute the theoretical isoelectric point (pI) and protein molecular weight (MW), the deduced amino acid sequences were analyzed using DNAStar Lasergene software (Version 7.1). Names of putative CaSBP genes were assigned based on chromosomal order.
Sequence Alignments, Phylogenetic Analysis, and Intron/Exon Structure Determination
Multiple amino acid sequence alignment was performed using DNAMAN software (Version 5.0). The sequence logo was obtained using the online platform Weblogo4 for conserved sequences. Phylogenetic trees were constructed using MEGA 6.0 with the maximum likelihood method and 1000 bootstrap replicates. Intron/exon structures were determined by aligning coding sequences to their corresponding genomic sequences. A diagram of intron/exon structures was obtained using the method described by Guo et al. (2015), which depicts both exon positions and gene lengths.
Chromosomal Location and Duplication Analysis
Chromosomal location information was derived from the pepper genome5, and genes were mapped to chromosomes using MapDraw (Liu and Meng, 2003) and their physical chromosome positions. Identification of duplicate genes within the pepper genome and between pepper and Arabidopsis was performed using the following criteria described by Gu et al. (2002): (1) the FASTA-alignable region between the two proteins had to be greater than 80% of the longer protein, and (2) the identity (I) between the two proteins had to be ≥30% if the alignable region was longer than 150 aa and ≥0.01n + 4.8 L-0.32(1+exp(-L/1000) (Rost, 1999) if otherwise, where n = 6 and L is the alignable length between the two proteins (Rost, 1999; Gu et al., 2002).
Plant Materials and Seedling Treatment
In this study, we used the pepper cultivar AA3 (provided by the pepper research group, College of Horticulture, Northwest A&F University, Yangling, China), which is susceptible to a compatible HX-9 strain and resistant to an incompatible PC strain of P. capsici. Plants were grown in a growth chamber at 22/18°C day/night temperature and 16/8 h day/night photoperiod. Various vegetative and reproductive tissues, including roots, stems, leaves, flowers, green fruits, and mature fruits were collected and stored at -80°C for tissue-specific experiments.
Pepper plants at the 8–10 true leaves stage were inoculated with compatible and incompatible strains of P. capsici using the root-drenching method, as described by Wang et al. (2013a), while control plants were inoculated with sterile distilled water. Root samples were taken at 0, 6, 12, 24, and 48 h and stored at -80°C. Seedlings were treated with 100 μM SA synthesis inhibitor (paclobutrazol, PBZ; Liu et al., 2006) or 50 μM MeJA synthesis inhibitor (salicylhydroxamic acid, SHAM; Dong et al., 2009). After 24 h of treatment, plants were treated with the corresponding inducer, 5 mM SA or 50 μM MeJA, using the method described by Yin et al. (2014). A mixture of 0.5% Tween and 0.1% alcohol was used as a control for PBZ and SHAM treatment, while PBZ and SHAM treatment alone (no inducer) was also used as an induction control. Leaves were harvested at 0, 3, 6, 9, 12, 24, and 48 h and were quickly frozen with liquid nitrogen and stored at -80°C.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated using the method described by Guo et al. (2012), and cDNA was synthesized according to the manufacturer’s instructions of PrimeScript Kit (Takara, Dalian, China). The cDNA was then diluted to 50 ng/μL with ddH2O. For quantitative real-time PCR (qRT-PCR), primer pairs (Supplementary Table S2) for CaSBP genes were designed by Primer Premier 5.0, and their specificities was assessed using NCBI Primer BLAST6 The ubiquitin binding-protein gene (UBI-3) from pepper was used as reference (Schmittgen and Livak, 2008). qRT-PCR was performed as described by Guo et al. (2015) on the iQ5.0 Bio-Rad iCycler thermocycler (Bio-Rad, Hercules, CA, USA) using SYBR Green Supermix (Takara, Dalian, China). qRT-PCR cycling conditions were as follows: pre-denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. The fluorescent signal was measured at the end of each cycle, and melting curve analysis was performed by heating the PCR product from 56 to 95°C in order to verify the specificities of the primers. Three independent biological replicates were carried out. The relative expression levels of pepper SBP genes were calculated using the -ΔΔCT method (Schmittgen and Livak, 2008).
Results
Genome-Wide Identification and Annotation of SBP-Box Genes in Pepper
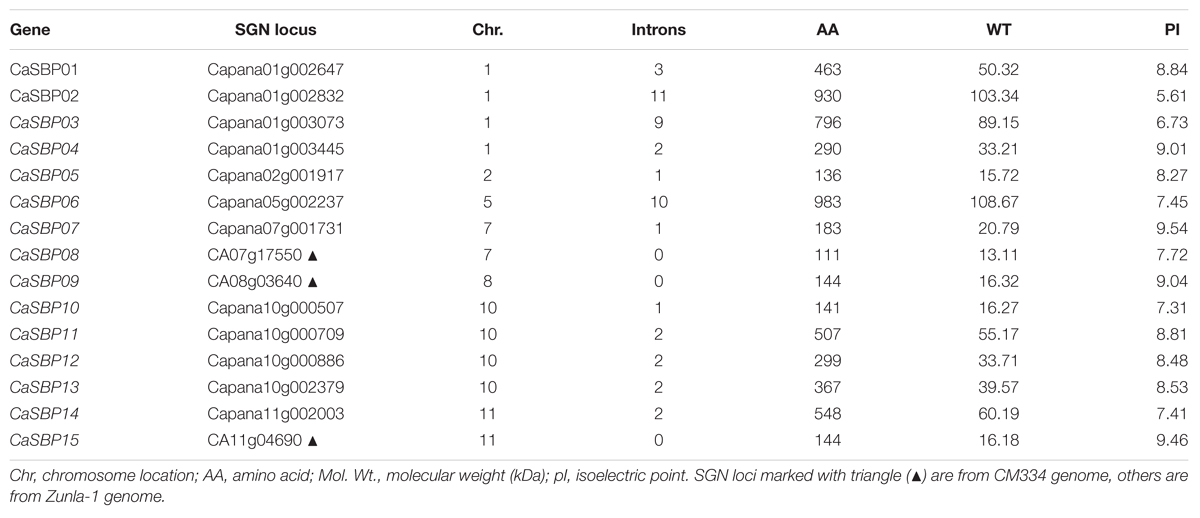
The identification of SBP-box gene family members in pepper was performed in three steps. In the first step, the HMM profile of the SBP domain was used as a BLAST query against the pepper genome. A total of 15 and 16 candidate SBP-box genes were obtained from pepper cultivars CM334 and Zunla-1, respectively. In the second step, CM334 and Zunla-1 genes were compared, and sequences were re-amplified to verify the corresponding genes. One candidate gene (Gene ID: Capana03g002994) found in Zunla-1 was discarded due to poor identification in comparison with the corresponding sequence in CM334. In the final step, each predicted SBP-box protein sequence was confirmed to have a conserved SBP domain using an NCBI search. As a result, 15 candidate SBP-box genes were confirmed and named based on their chromosomal order in pepper (Table 1). The CaSBP coding sequences ranged from 336 bp (CaSBP08) to 3024 bp (CaSBP06), while deduced proteins ranged from 111 to 983 amino acids in length and from 13.11 to 108.67 kDa in MW. The predicted isoelectric points (pI) of the CaSBPs varied from 5.61 to 9.54.
Sequence Alignments, Phylogenetic Analysis, and Intron/Exon Structure Determination
Multiple sequence alignment of full-length protein sequences was performed to analyze the domain structures of CaSBPs in detail. The SBP domain is the only conserved domain shared by all CaSBPs (Figure 1A) and was highly similar across proteins, with high or complete conservation at certain positions (Figure 1B). All CaSBPs exhibit two zinc finger-like structures (C3H, C2HC) and a highly conserved bipartite nuclear localization signal (NLS), with the exception of CaSBP08, which lacks the C2HC and NLS. In addition, CaSBP09 and CaSBP15 are also lacking C3H, as the second zinc finger-like structure partially overlaps the NLS, as previously reported (Birkenbihl et al., 2005).
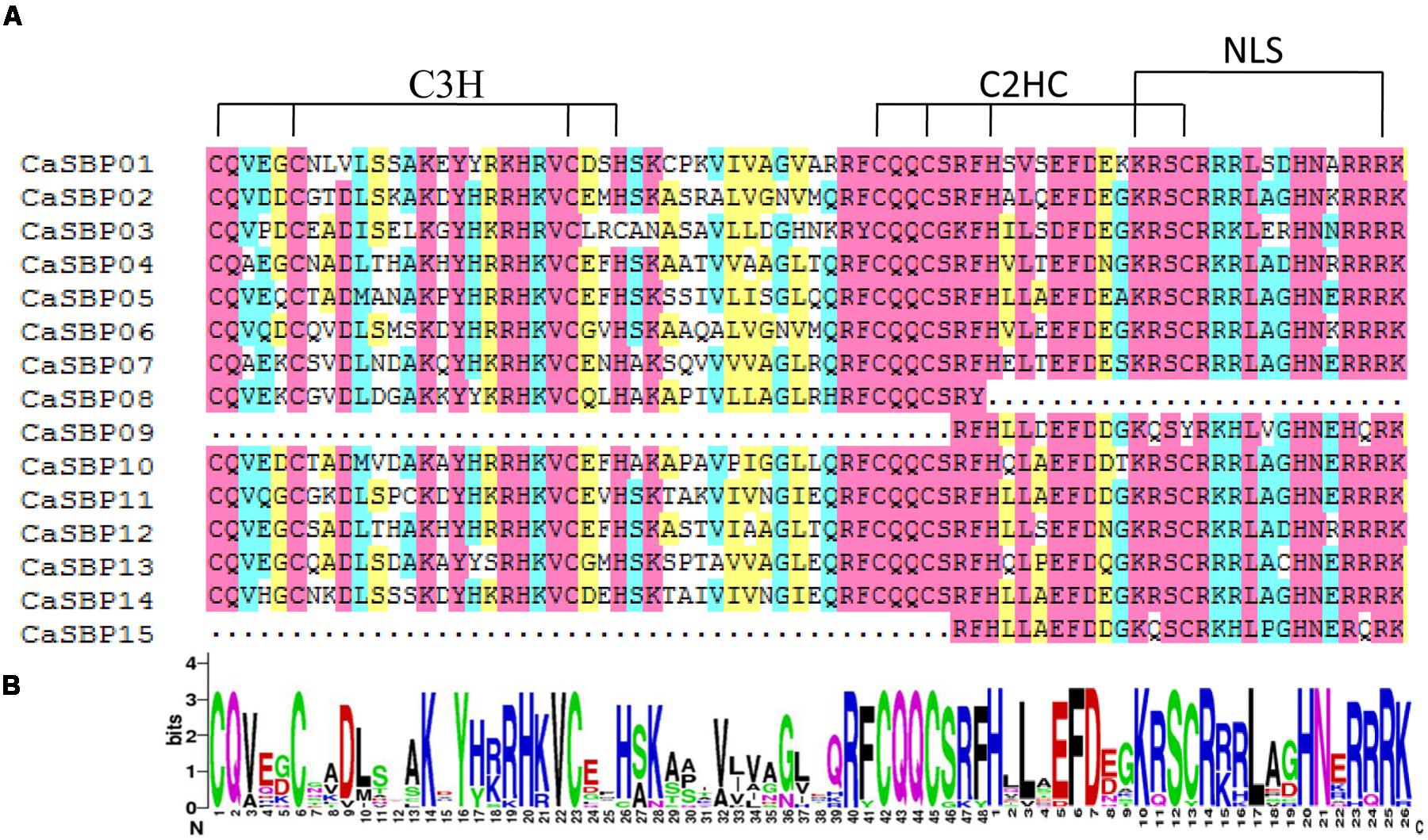
FIGURE 1. SBP domain alignment in CaSBPs. (A) Multiple alignment of the SQUAMOSA promoter binding protein (SBP) domains of pepper SBP-box proteins obtained with DNAMAN software. The two conserved zinc-finger structures (C3H, C2HC) and NLS are indicated. (B) Sequence logo of the SBP domain of CaSBPs. The overall height of each stack represents the degree of conservation at each position, while the height of the letters within each stack indicates the relative frequency of the corresponding amino acid.
To investigate the evolutionary relationship between CaSBP genes and SBP-box genes from Arabidopsis, tomato (Solanum lycopersicum), and rice (Oryza sativa), we constructed a phylogenetic tree using the maximum likelihood algorithm (Figure 2), with 17 Arabidopsis genes, 17 tomato genes, and 19 rice genes (Supplementary Table S3). Only the protein sequences of the highly conserved SBP domains were used for phylogenetic analysis, as alignment of the full-length protein sequences revealed that only the SBP domains were conserved (Hou et al., 2013a). According to the unrooted phylogenetic tree, CaSBP proteins clustered with those of the other species into six distinct groups (I–VI; Figure 2), with each group containing at least one protein from each species. The plant SBP-box gene family is evolutionarily diversified. An unrooted phylogenetic tree was also constructed using only the SBP domains from CaSBPs (Figure 3A).
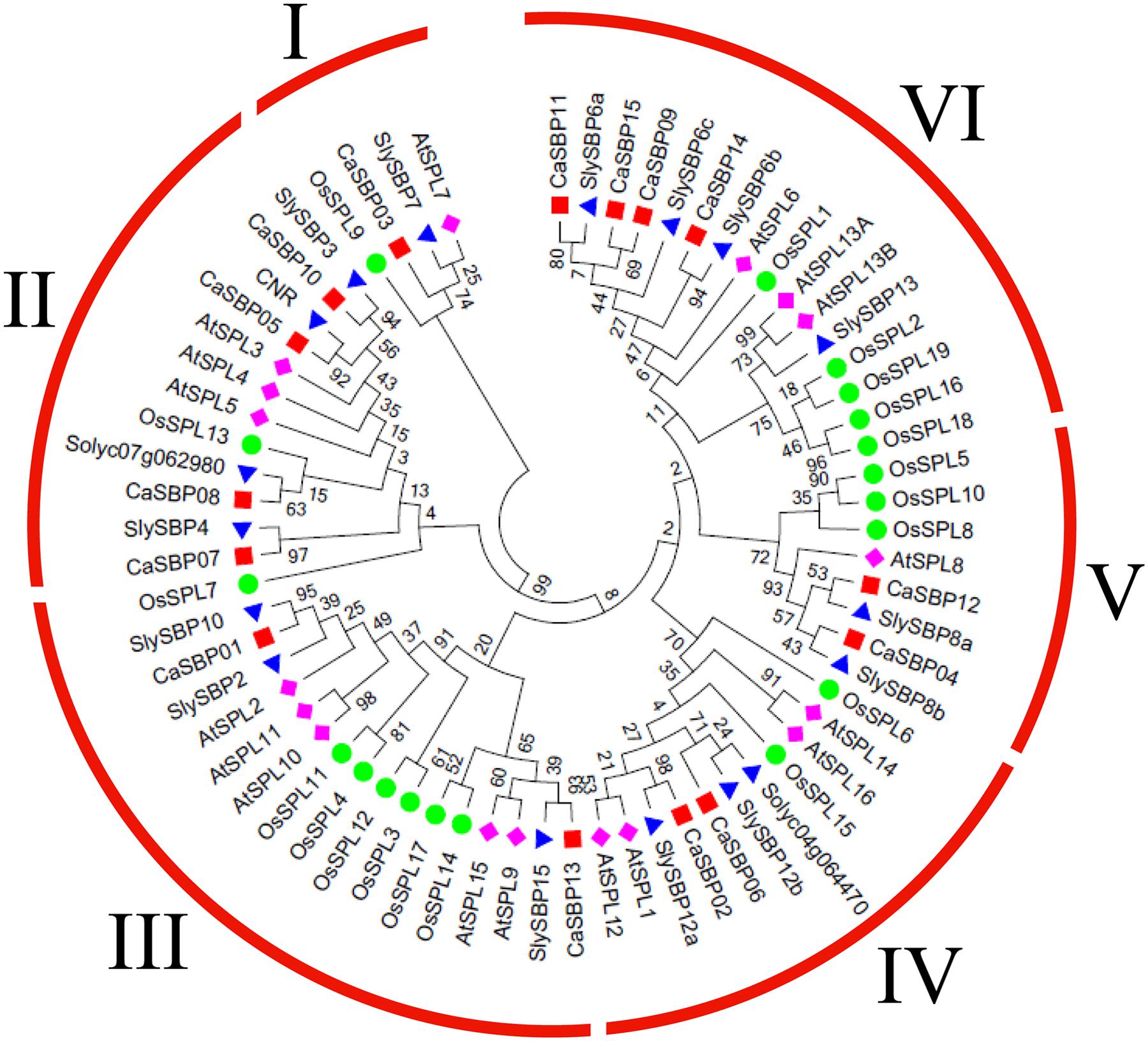
FIGURE 2. Phylogenetic analysis of pepper and other plant SBPs. A phylogenetic tree was constructed with SBP domain protein sequences from pepper, tomato, Arabidopsis, and rice. The SBP domain sequences, accession numbers/locus IDs, and data sources of all genes used for phylogenetic tree construction are listed in Supplementary Table S3.
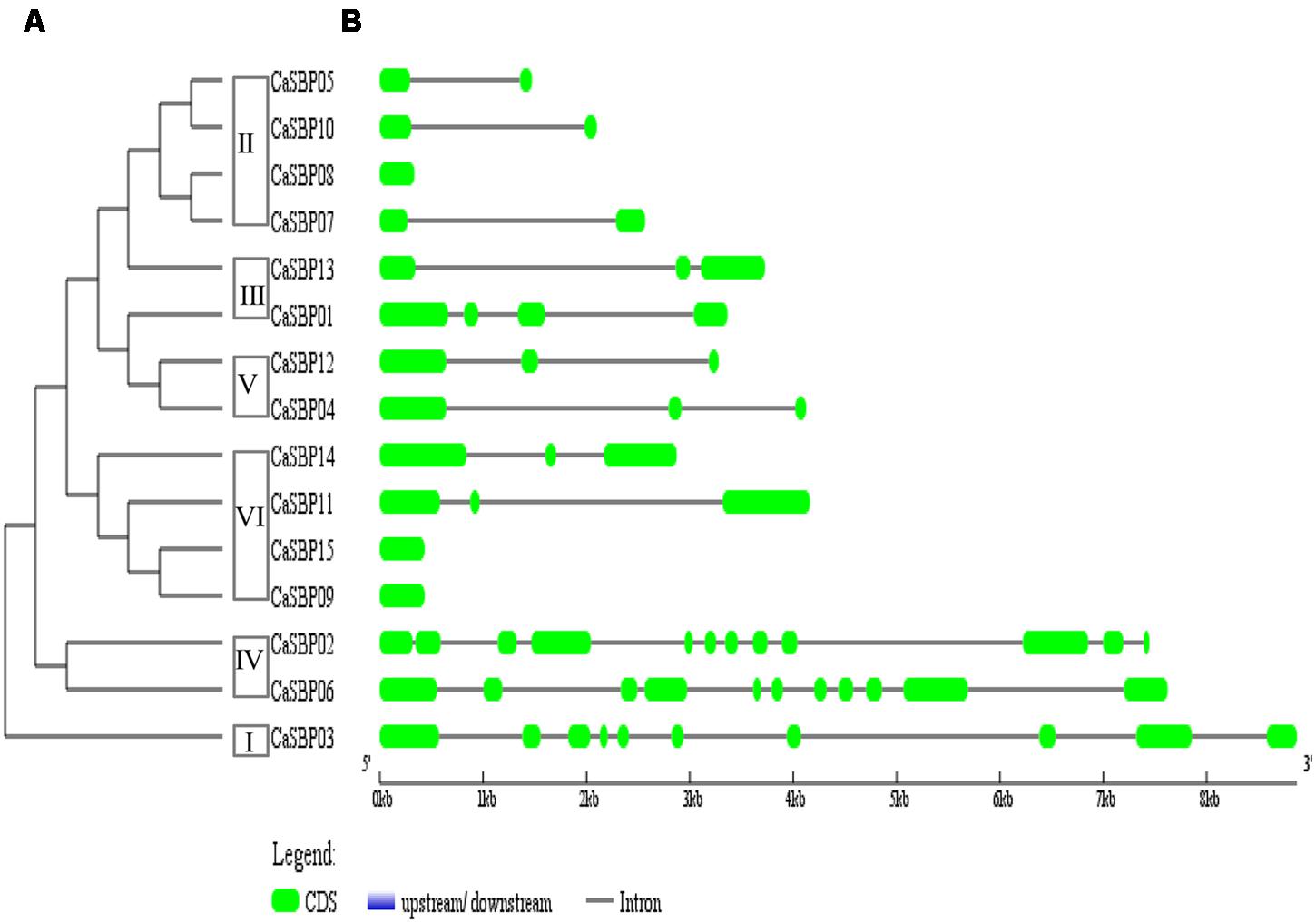
FIGURE 3. Phylogenetic analysis and intron/exon structures of CaSBP genes. (A) A phylogenetic tree was constructed with pepper SBP domain protein sequences. (B) Exons and introns are indicated by green boxes and black horizontal lines, respectively.
Intron/exon structures of all 15 CaSBP genes were generated based on genome sequences and corresponding coding sequences (Figure 3B). Intron/exon structure diagrams revealed high variation in the number of introns, from zero (CaSBP08, CaSBP09, and CaSBP15) to 11 (CaSBP02). Based on the CaSBP tree (Figure 3A), class I proteins contain nine introns, class II contains 0–1, class III contains 2–3, class IV contains 10–11, class V contains 2, and class VI contains 0–2 introns.
Chromosomal Location and Duplication Analysis
We found that CaSBP genes were located on seven of the twelve pepper chromosomes (Figure 4): chromosomes 1, 2, 5, 7, 8, 10, and 11 (Table 1). Chromosomes 1 and 10 contained the most CaSBP genes, with four genes each (CaSBP01–CaSBP04 and CaSBP10–CaSBP13, respectively), followed by chromosomes 7 and 11, with two genes each (CaSBP07–CaSBP08 and CaSBP14–CaSBP15, respectively).
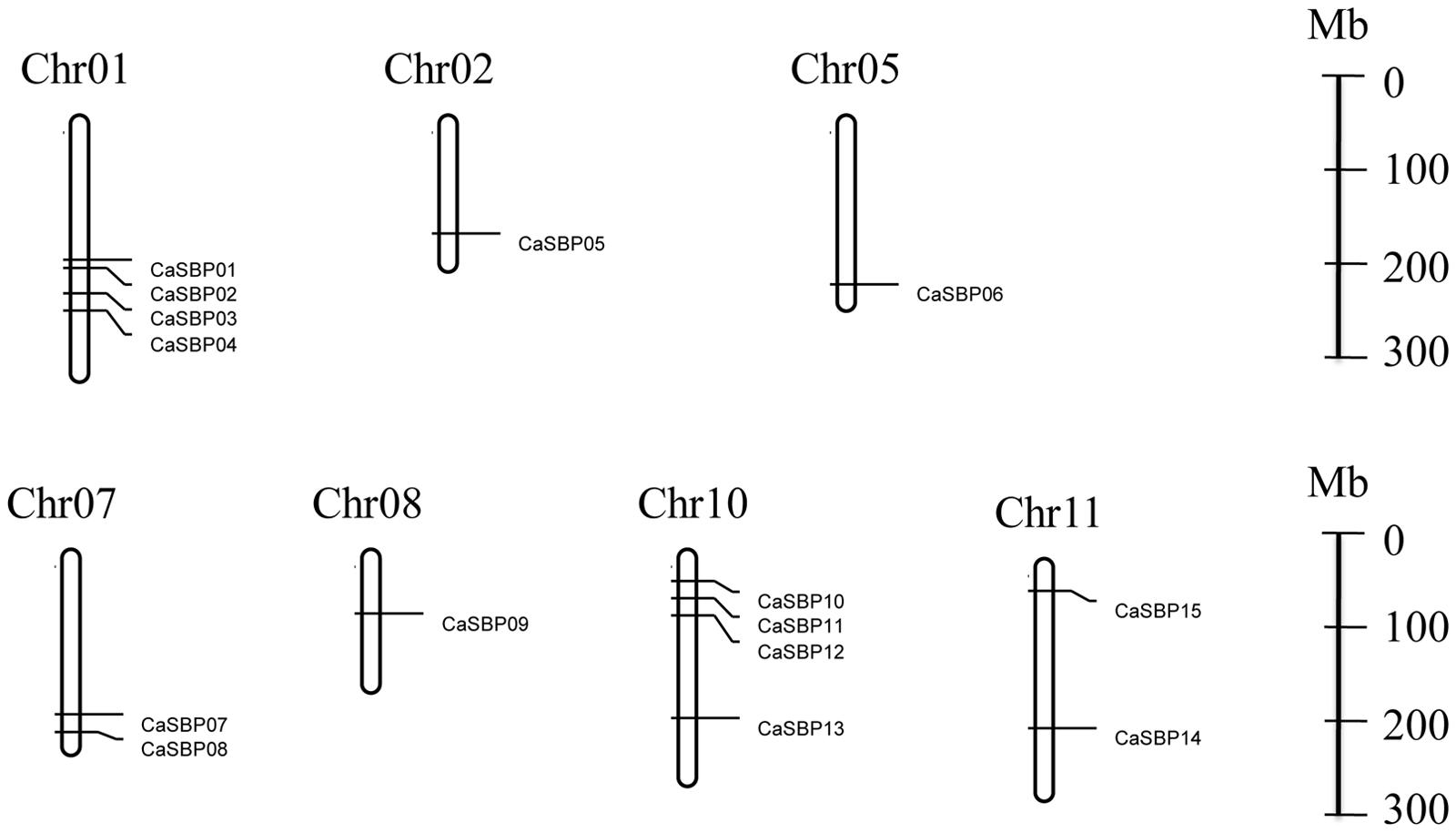
FIGURE 4. Chromosome mapping of SBP genes in pepper. Chromosome numbers are indicated at the top of each bar. Scale is represented in megabases (Mb).
Duplication analysis, using the criteria described by Gu et al. (2002), confirmed that four pairs of pepper SBP-box genes (CaSBP02/06, CaSBP04/12, CaSBP05/10, and CaSBP09/15) were the result of interchromosomal segmental duplications (Figure 5). Because Arabidopsis is a popular model plant and the functions of several Arabidopsis SBP-box genes have been well characterized, we also used the same criteria to identify SBP-box gene orthologs between the pepper and Arabidopsis genomes to further study the origin, evolutionary history, and putative function of the pepper SBP-box genes. Based on this analysis, we identified ten pairs of CaSBP–AtSPL orthologs (CaSBP01–AtSPL2, CaSBP02–AtSPL1/12, CaSBP03–AtSPL7, CaSBP04/12–AtSPL8, CaSBP05/10–AtSPL3, and CaSBP06–AtSPL1/12) (Figure 6), indicating that many of pepper SBP-box genes and their Arabidopsis counterparts appear to be derived from a common ancestor. According to these results, we were able to infer the functions of several pepper SBP-box genes based on their Arabidopsis homologs, facilitating research into the roles of SBP-box genes in pepper.
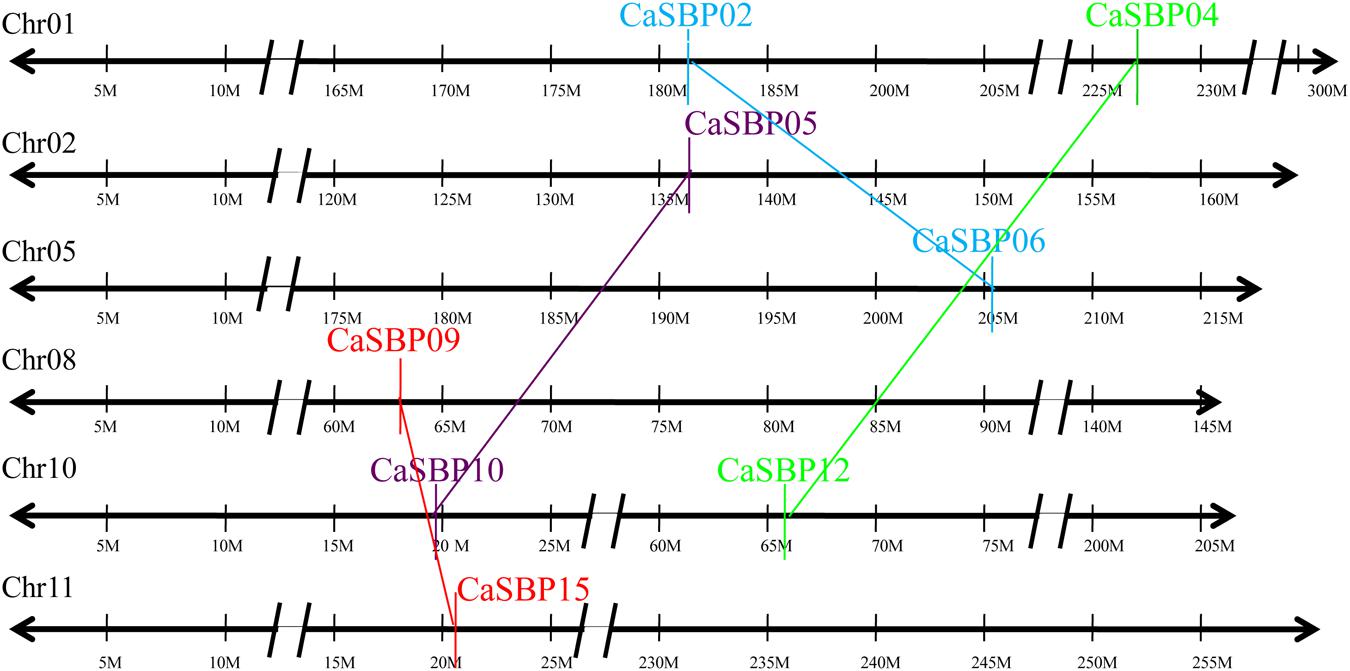
FIGURE 5. Duplication analysis of pepper SBP-box genes. The positions of duplicated CaSBP genes are depicted on pepper chromosomes 1, 2, 5, 8, 10, and 11. Colored lines connecting two chromosomal regions indicate duplicated regions between pepper chromosomes.
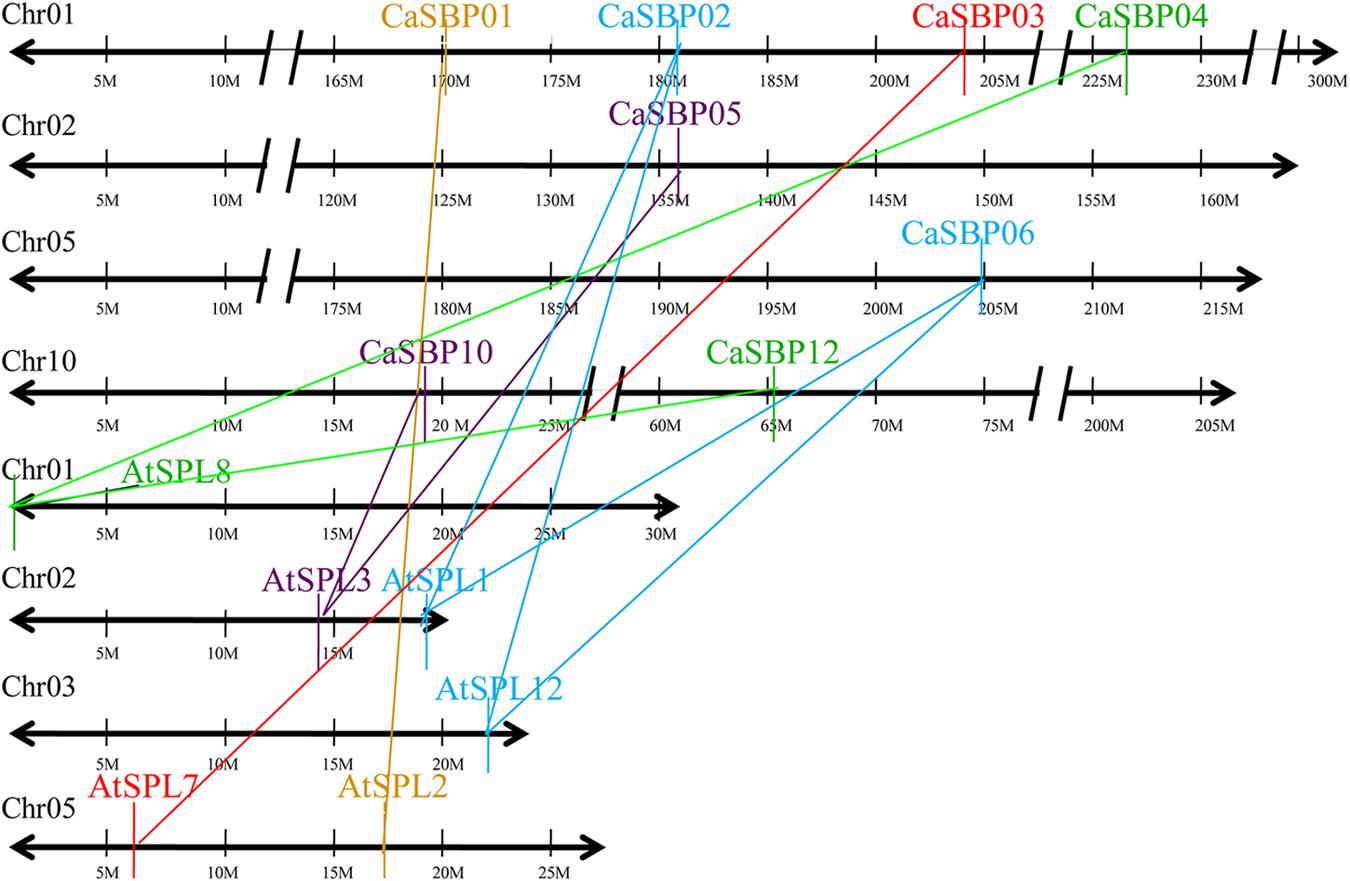
FIGURE 6. Duplication analysis of SBP-box genes between pepper and Arabidopsis genomes. The positions of related CaSBP and AtSPL genes are depicted on pepper chromosomes 1, 2, 5, and 10 and Arabidopsis chromosomes 1, 2, 3, and 5. Colored lines connecting two chromosomal regions indicate duplicated regions between pepper and Arabidopsis chromosomes.
Expression Profiles of CaSBP Genes in Pepper Tissues
In order to provide additional information on the functions of SBP-box genes in pepper, we investigated their expression profiles in various organs and at different stages of fruit development in cultivar AA3 via qRT-PCR with transcript-specific primers (Supplementary Table S2). Generally, the expression patterns of CaSBP genes can be classified into two types (Figure 7). The minority of CaSBP genes, specifically CaSBP01, CaSBP08, CaSBP09, and CaSBP10, exhibited low-level, constitutive expression in all pepper tissues/organs examined. The remaining CaSBP genes were only expressed in certain tissues or organs. CaSBP02 was the most highly expressed SBP-box gene in the examined tissues. In general, the expression of CaSBP genes was highest in the leaf, followed by the stem, root, green fruit, mature fruit, and flowers.
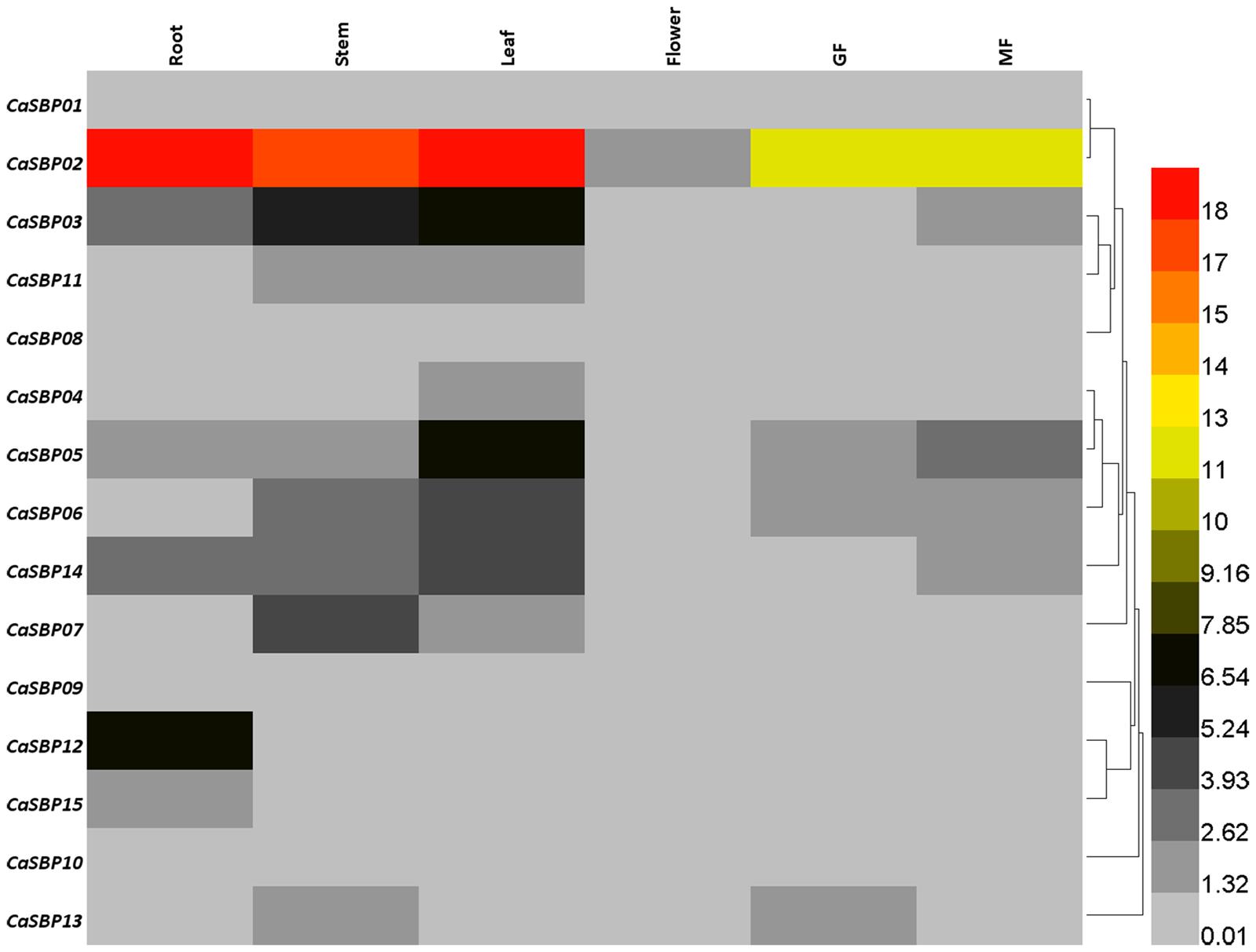
FIGURE 7. Tissue-specific expression analysis of pepper SBP-box genes. Analyzed tissues included root, stem, leaf, flower, green fruit (GF), and mature fruit (MF).
Expression Analysis of CaSBP Genes under P. capsici and Hormone Treatments
To investigate the effect of P. capsici infection on the expression of CaSBP genes, roots from the AA3 cultivar were inoculated with compatible and incompatible P. capsici strains, and changes in gene expression were analyzed using qRT-PCR (Figure 8). The results indicate that after inoculation with either the compatible or incompatible strain, four CaSBP genes (CaSBP02, CaSBP05, CaSBP06, and CaSBP13) were up-regulated 0–24 h post-inoculation and subsequently down-regulated, while CaSBP04 was up-regulated 0–12 h and then down-regulated. Similarly, CaSBP14 was up-regulated 0–6 h post-inoculation and subsequently down-regulated. Following inoculation with just the incompatible strain, four genes (CaSBP01, CaSBP03, CaSBP05, and CaSBP08) exhibited down-regulation 0–12 h post-inoculation, followed by up-regulation to 24 h and subsequent down-regulation again. CaSBP10 and CaSBP11 exhibited the same pattern but following inoculation with the compatible strain only. Following compatible strain inoculation, four genes (CaSBP01, CaSBP02, CaSBP03, and CaSBP12) were up-regulated 0–24 h and subsequently down-regulated, while two genes (CaSBP07 and CaSBP09) were up-regulated 0–6 h after inoculation with the incompatible strain and then down-regulated. Moreover, CaSBP09 exhibited consistent down-regulation following inoculation with the compatible strain, and CaSBP12 exhibited up-regulation 0–48 h after inoculation with the incompatible strain. Generally, the expression patterns of CaSBPs after inoculation with P. capsici can be divided into five categories. The first and second categories contain one gene each, CaSBP04 and CaSBP10, whose expression peaked at 12 and 48 h, respectively, after inoculation with either the compatible or incompatible strain. The third category contains seven genes (CaSBP01–CaSBP03, CaSBP05, CaSBP06, CaSBP11, and CaSBP13) whose expressions peaked 24 h after inoculation with either the compatible or incompatible strain. The fourth category contains two genes, CaSBP08 and CaSBP12, whose expressions peaked earlier following inoculation with the compatible strain than following inoculation with the incompatible strain. The fifth category contains four genes (CaSBP07, CaSBP09, CaSBP14, and CaSBP15), whose expressions were down-regulated 12 h after inoculation with either the compatible or incompatible strain.
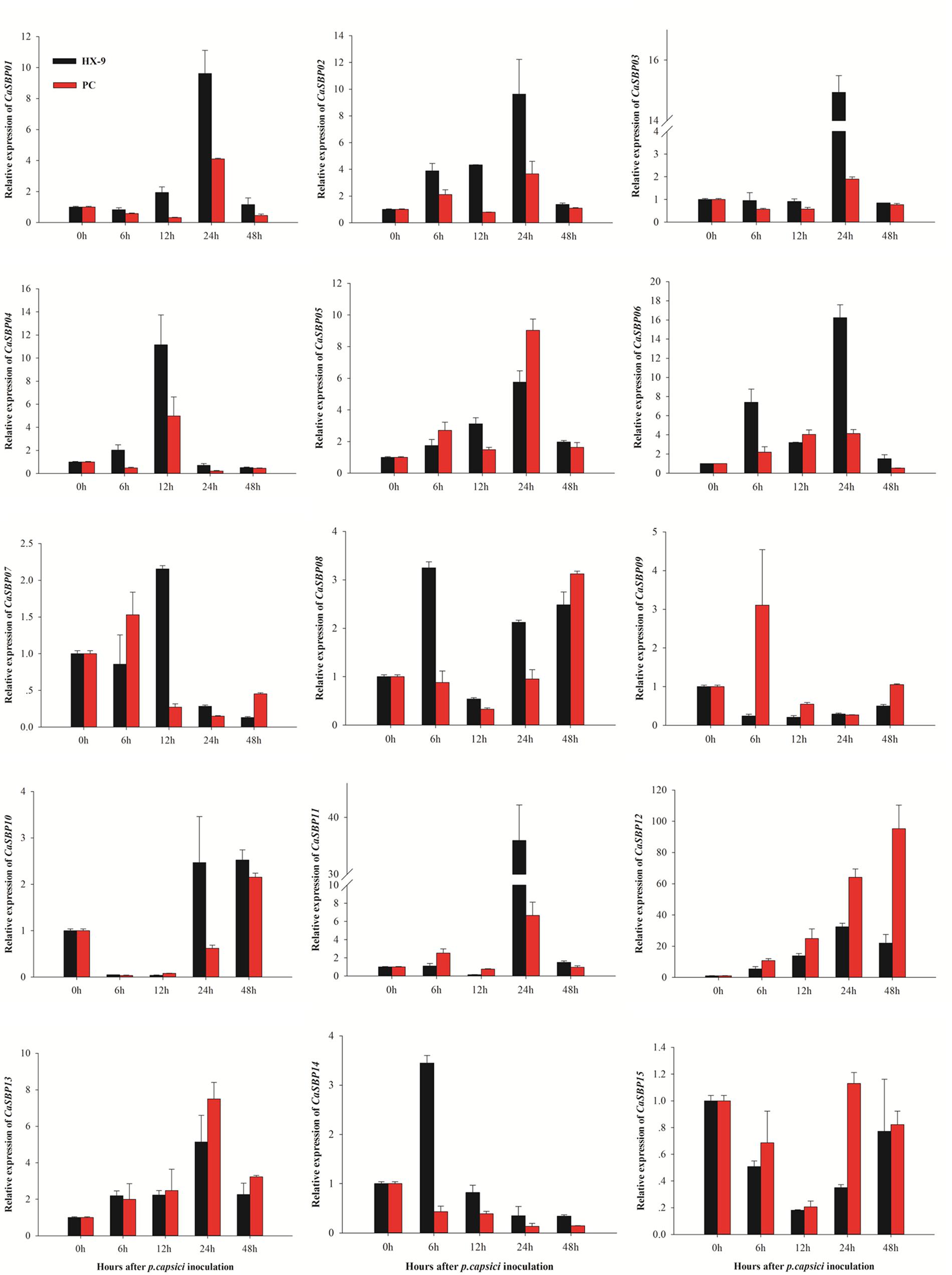
FIGURE 8. Expression profiles of CaSBPs in response to inoculation with compatible or incompatible Phytophthora capsici strains. Mean values and SDs for three replicates are shown.
To investigate the expression patterns of CaSBPs in response to treatment with various signal molecules, five representative genes (CaSBP04, CaSBP10–12, and CaSBP15), one from each of the five categories above, were treated with SA inhibitor (PBZ) or MeJA inhibitor (SHAM), and changes in gene expression were analyzed using qRT-PCR (Figure 9). Results showed that the expression of all five genes was rapidly down-regulated 0–6 h after treatment with SA inhibitor (PBZ) or MeJA inhibitor (SHAM), reaching the lowest level at 6 h. After 24 h of treatment, the corresponding inducer (SA or MeJA) was applied. Subsequently, the expression levels of the five genes after SA treatment peaked at 12 h, with the exception of CaSBP11, which peaked at 48 h. Following MeJA treatment, expression levels of the five genes peaked earlier than 12 h.
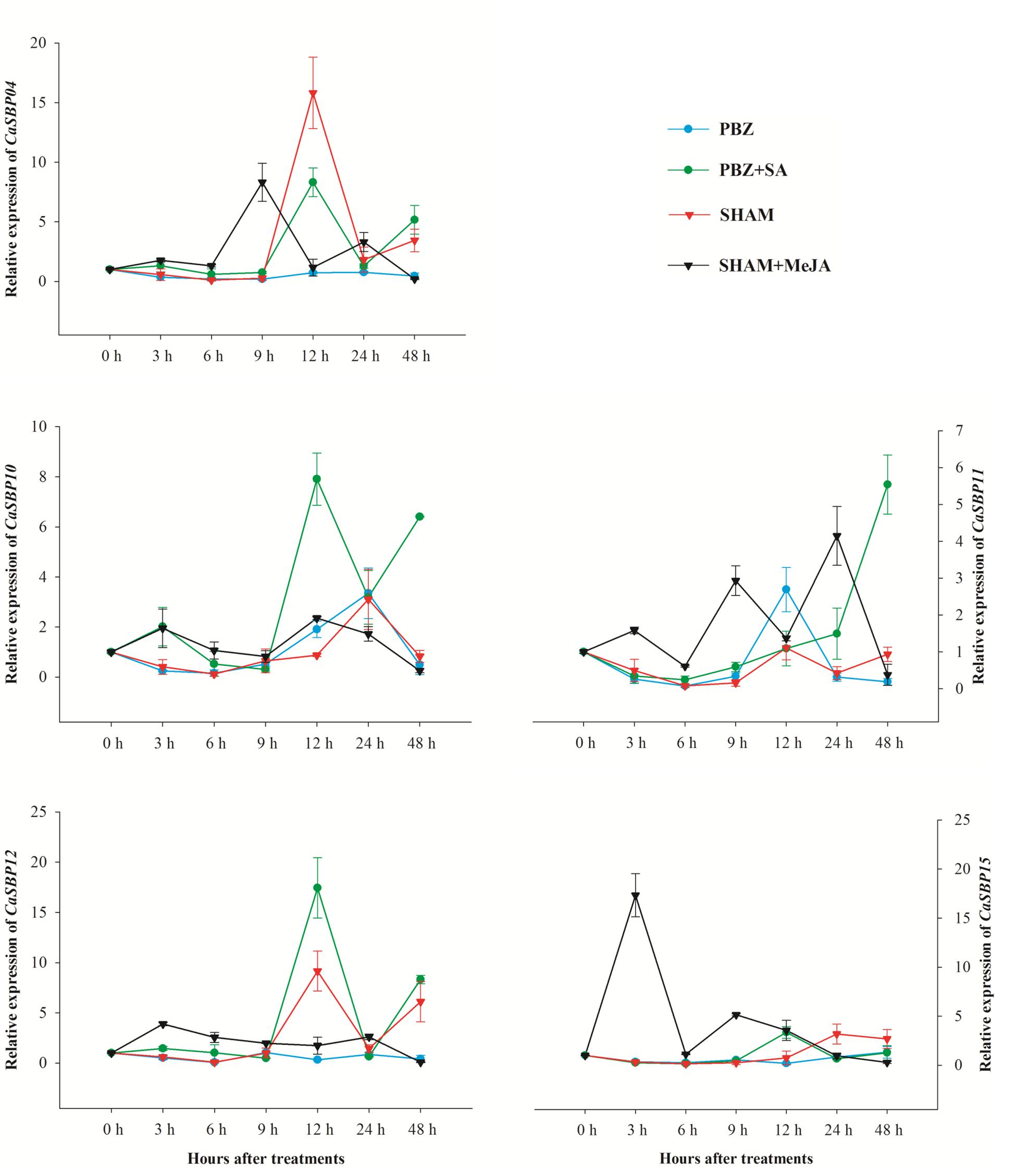
FIGURE 9. Expression profiles of CaSBPs in response to treatment with SA or MeJA hormones and the corresponding inhibitors PBZ or SHAM. Mean values and SDs for three replicates are shown.
Discussion
Most evidence suggests that SBP-box genes play central roles in plant development, signal transduction, and defense processes (Schwarz et al., 2008; Shikata et al., 2009; Hou et al., 2013b). Benefitting from the availability of genome sequences, the functions of SBP-box genes have been characterized in many plants, including Arabidopsis, S. miltiorrhiza (Zhang et al., 2014), rice (Yang et al., 2008), tomato (Yang et al., 2008), Populus trichocarpa (Li and Lu, 2014), grape (Hou et al., 2013a), apple (Li et al., 2013), G. hirsutum (Zhang et al., 2015), Prunus mume (Xu et al., 2015), castor bean (Zhang and Ling, 2014), and citrus (Shalom et al., 2015). However, the functions of pepper SBP-box TFs are still unknown. In this study, through genome-wide identification and molecular cloning, we discovered the first set of CaSBP genes (Table 1). In total, we identified 15 CaSBPs in pepper, a number similar to that found in S. miltiorrhiza (Zhang et al., 2014), P. mume (Xu et al., 2015), castor bean (Zhang and Ling, 2014), and citrus (Shalom et al., 2015).
Phylogenetic tree analysis showed that SBPs from representative plants are clustered into six groups, with CaSBP genes distributed across all six groups (Figure 2). In addition, each group contains at least one gene from Arabidopsis, tomato, and rice. CaSBP genes are more closely related to genes from tomato or Arabidopsis than to rice SBP-box genes, reflecting the fact that Arabidopsis, tomato, and pepper are eudicots and diverged more recently from a common ancestor (Li et al., 2013). These results indicate that although plant SBP-box genes may be derived from a common ancestor, many have undergone distinct patterns of differentiation with the divergence of different lineages. Gene structure analyses showed that within the same phylogenetic group, most CaSBP genes shared similar intron/exon structures, indicating that the evolution of SBP domains may be closely related to the diversification of gene structures, as described previously in tomato (Wan et al., 2013), rice (Xie et al., 2006), apple (Li et al., 2013), and grape (Hou et al., 2013a). CaSBP genes are distributed across seven of the twelve pepper chromosomes, with no CaSBP genes on chromosomes 3, 4, 6, 9, or 12. Similarly, only chromosomes 6, 8, 9, and 11 lack SBP genes in tomato, suggesting that SBP genes may have been widely distributed across the genome of the Solanaceae common ancestor.
Gene duplication events include tandem, segmental, and whole-genome duplications, and they have played crucial roles in the evolution of various organisms (Xu et al., 2012). In the SBP-box gene family, there are two pairs of Arabidopsis (AtSPL1/12 and AtSPL4/5), six pairs of rice genes (OsSPL1/6, OsSPL3/12, OsSPL4/11, OsSPL5/10, OsSPL14/17, and OsSPL16/18), eight pairs of apple genes (MdSBP1B/9, MdSBP4A-B/20, MdSBP8/27A-B, MdSBP10/21, MdSBP10/22, MdSBP11/21, MdSBP12/23, and MdSBP13/15), and six pairs of grape genes (VvSBP2/15, VvSBP3/12, VvSBP5/7, VvSBP9/11, VvSBP9/18, and VvSBP11/18) located within segmental duplications (Xie et al., 2006; Li et al., 2013; Hou et al., 2013a). Similarly, we used the criteria described by Gu et al. (2002) and confirmed that four pairs of pepper SBP-box genes (CaSBP02/06, CaSBP04/12, CaSBP05/10, and CaSBP09/15) are located in putative segmental duplications. Therefore, it is clear that segmental duplications have played an important role in the expansion of the plan SBP-box gene family.
Comparative genomic analysis is a relatively rapid and effective way to transfer genomic knowledge acquired in one taxon to another, whose genome structure, function, and/or evolution are less known (Lyons et al., 2008). Thus, putative functions of pepper SBP-box genes can be inferred via comparison with orthologs in well-studied model plants such as Arabidopsis. In this study, duplication analysis between pepper and Arabidopsis indicated that ten pairs of SBP-box genes (CaSBP01/AtSPL02, CaSBP02-06/AtSPL1-12, CaSBP03/AtSPL7, CaSBP04-12/AtSPL8, and CaSBP05-10/AtSPL3) are located in syntenic genomic regions and represent putative orthologs (Figure 6). To date, the majority of Arabidopsis SBP-box genes, including AtSPL2 (Shikata et al., 2009), AtSPL3 (Yamaguchi et al., 2009), AtSPL4 (Jung et al., 2011), AtSPL5 (Jung et al., 2011), AtSPL6 (Padmanabhan et al., 2013), AtSPL7 (Yamasaki et al., 2009), AtSPL8 (Zhang et al., 2007; Xing et al., 2010), AtSPL9 (Cui et al., 2014), AtSPL10 (Shikata et al., 2009), AtSPL11 (Shikata et al., 2009), AtSPL13 (Martin et al., 2010), AtSPL14 (Stone et al., 2005), and AtSPL15 (Schwarz et al., 2008) have been functionally characterized. Therefore, the functions of several CaSBP gene homologs, such as CaSBP01–CaSBP05, CaSBP10, and CaSBP12, can be predicted based on their Arabidopsis counterparts. Further experiments are necessary to confirm these functions.
In order to further reveal the possible roles of CaSBP genes in pepper growth and development, the expression profile of each CaSBP gene was investigated in six different tissues. Results indicate that CaSBP genes exhibit different expression patterns (Figure 7). While a few CaSBP genes (CaSBP01, CaSBP08–CaSBP10) demonstrated low-level, constitutive expression in all tissues or organs examined, the majority were limited to certain tissues/organs, with CaSBP02 exhibiting the highest expression across all tissues. The transcription levels of CaSBP03, CaSBP05, and CaSBP06 were also higher than other CaSBP genes in root, stem, and leaf, consistent with the results of previous sequencing in hot peppers (Kim et al., 2014). In addition, the expression of CaSBP genes in flowers and fruits was lower than that in roots, stems, and leaves, similar to results from grapes (Hou et al., 2013a), which may indicate that CaSBP genes play a role in the transition from vegetative to reproductive growth. Unlike MdSBP genes in apple (Li et al., 2013), however, CaSBP expression patterns were not correlated with gene location, gene length, gene structure, or gene sequence.
Most CaSBP genes were up-regulated after inoculation with compatible and incompatible P. capsici. Specifically, CaSBP02, CaSBP05, CaSBP06, CaSBP11, CaSBP12, and CaSBP13 exhibited significantly higher expression under P. capsici stress conditions in pepper roots (Figure 8). In addition, the transcript levels of CaSBP05, CaSBP12, and CaSBP13 were up-regulated more rapidly and more intensely following inoculation with the strain than with the compatible strain. Recent studies have indicated that a novel peroxidase (CanPOD) and oxysterol-binding protein (CanOBP) genes, which are involved in the defense response to P. capsici infection, exhibit expression patterns similar to these CaSBPs (Liu, 2009; Wang et al., 2013b). Moreover, similar expression patterns are also found in some defense-related genes – such as the disease-associated protein gene (CABPR1), β-1,3-glucanase gene (CABGLU), and peroxidase gene (CAPO1) – in pepper roots after inoculation with compatible and incompatible P. capsici (Wang, 2013). However, according to Kim and Hwang (2000), the expression of CABPR1 is higher in the compatible interaction than in the incompatible interaction. While differences in expression changes between CaSBP and CABPR1 genes may be due to differences in inoculation of the P. capsici strains or to differences in the compatibility systems, it suggests that these genes are related to the pepper’s resistance to P. capsici. Phylogenetic tree analysis showed that CaSBP02 and CaSBP06 exhibited a close relationship with AtSPL14, which has been found to be involved in programmed cell death and plays a role in sensitivity to fumonisin B1 (Stone et al., 2005). Moreover, the ortholog of AtSPL14 and VpSBP5 is likely to participate in regulating resistance to E. necator (Hou et al., 2013a). It also has been reported that AtSPL genes are co-expressed with two TFs, TGA1, and WRKY65, which are induced by pathogens and regulate the expression of several stress-responsive genes, such as pathogenesis-related 1 protein (PR-1) and GLUTATHIONE S-TRANSFERASE 6 (GST6; Wang et al., 2009). Based on the above results, we speculate that these SBP genes may be involved in disease resistance, but this will need to be verified.
The signal transduction pathway mediated by salicylic acid (SA) and methyl jasmonate (MeJA) is linked to the plant defense response (Thomma et al., 2001; An et al., 2008; Choi and Hwang, 2011). SA typically mediates basal defense to biotrophic pathogens (Thomma et al., 2001), while MeJA generally controls defensive reactions to necrotrophs (Glazebrook, 2005). Therefore, we investigated the responses of five representative CaSBPs (CaSBP04, CaSBP10, CaSBP11, CaSBP12, and CaSBP15) to plant hormone signals by examining their transcript levels in pepper leaves upon treatment with SA or MeJA and their corresponding biosynthesis inhibitors. The expression levels of most genes peaked at 12 h following SA treatment, the exception being CaSBP11, which peaked at 48 h. Following MeJA treatment, the maximum expression of all five genes occurred earlier than after SA treatment. It has been reported that SA and MeJA can induce the expression of defense-related gene PR-1 in tobacco (Xu et al., 1994; Vidal et al., 1997). Moreover, SA induces the recruitment of trans-activating TGA factors to the promoter of a defense gene in Arabidopsis (Johnson et al., 2003). The Arabidopsis SBP-box gene AtSPL2 and the grape SBP-box gene VpSBP5 also exhibit responsiveness to biotic stress signaling hormones (Jung et al., 2007; Hou et al., 2013b). Therefore, we speculate that these genes may be involved in the response to various plant stress hormones, particularly the MeJA-induced necrotroph pathway.
Conclusion
In this study, we identified SBP-box genes in pepper and analyzed them via sequence alignment, phylogenetic analysis, intron/exon structure, chromosomal location, and duplication analysis. We also assessed the expression profiles of pepper SBP genes across different tissues (root, stem, leaf, flower, and fruit) and under infection with both compatible and incompatible P. capsici strains and hormone treatment. Most CaSBP genes are expressed at low levels under normal circumstances and are induced by P. capsici and hormones, indicating that these genes may be involved in the resistance pathways mediated by P. capsici, SA, and MeJA. Candidate pepper SBP-box genes from this analysis should be further functionally characterized for deeper understanding of the precise regulatory checkpoints that operate during stress responses.
Author Contributions
H-XZ, W-GC, and Z-HG conceived and designed the experiments. H-XZ, J-HJ, Y-MH, D-WL, B-YL, and AK performed the experiments. H-XZ analyzed the data. W-GC and Z-HG contributed reagents/materials/analysis tools. H-XZ wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31272163, 31401879), the Shaanxi Provincial Science and Technology Coordinating Innovative Engineering Project (2012KTCL02-09) and the Hangzhou Municipal Science and Technology Commission Project (20140932H01).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00504
Footnotes
- ^http://www.sanger.ac.uk.
- ^http://peppergenome.snu.ac.kr/blast.php
- ^http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
- ^http://weblogo.berkeley.edu
- ^http://peppergenome.snu.ac.kr/
- ^http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome.
References
An, S. H., Sohn, K. H., Choi, H. W., Hwang, I. S., Lee, S. C., and Hwang, B. K. (2008). Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228, 61–78. doi: 10.1007/s00425-008-0719-z
Biles, C. L., Brunton, B. D., Wall, M. M., and Rivas, M. (1995). Phytophthora capsici zoospore infection of pepper fruit in various physical environments. Proc. Okla. Acad. Sci. 75, 1–5.
Birkenbihl, R. P., Jach, G., Saedler, H., and Huijser, P. (2005). Functional dissection of the plant-specific SBP-Domain: overlap of the DNA-binding and nuclearnlocalization domains. J. Mol. Biol. 352, 585–596. doi: 10.1016/j.jmb.2005.07.013
Cardon, G., Höhmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. doi: 10.1016/S0378-1119(99)00308-X
Cardon, G. H., Höhmann, S., Nettesheim, K., Saedler, H., and Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3 a novel gene involved in the floral transitionl. Plant J. 12, 367–377. doi: 10.1046/j.1365-313X.1997.12020367.x
Choi, D. S., and Hwang, B. K. (2011). Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23, 823–842. doi: 10.1105/tpc.110.082081
Chuck, G., Whipple, C., Jackson, D., and Hake, S. (2010). The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137, 1243–1250. doi: 10.1242/dev.048348
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Dong, T. X., Cai, K. Z., and Zeng, R. S. (2009). Effects of methyl jasmonate(MeJA)on photosynthetic traits of rice seedlings under drought stress. Ecol. Environ. 18, 1872–1876.
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Gu, Z. L., Cavalcanti, A., Chen, F. C., Bouman, P., and Li, W. H. (2002). Extent of gene duplication in the genomes of drosophila, nematode,and yeast. Mol. Biol. Evol. 19, 256–262. doi: 10.1093/oxfordjournals.molbev.a004079
Guo, M., Lu, J. P., Zhai, Y. F., Chai, W. G., Gong, Z. H., and Lu, M. H. (2015). Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 15:151. doi: 10.1186/s12870-015-0512-7
Guo, W. L., Chen, R. G., Gong, Z. H., Yin, Y. X., Ahmed, S. S., and He, Y. M. (2012). Exogenous abscisic acid increases antioxidant enzymes and related gene expression in pepper (Capsicum annuum) leaves subjected to chilling stress. Genet. Mol. Res. 11, 4063–4080. doi: 10.4238/2012.September.10.5
Hausbeck, M. K., and Lamour, K. H. (2004). Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis. 88, 1292–1303. doi: 10.1094/PDIS.2004.88.12.1292
Hou, H., Li, J., Gao, M., Singer, S. D., Wang, H., Mao, L., et al. (2013a). Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 8:e59358. doi: 10.1371/journal.pone.0059358
Hou, H., Yan, Q., Wang, X. P., and Xu, H. (2013b). A SBP-Box gene VpSBP5 from Chinese wild vitis species responds to erysiphe necator and defense signaling molecules. Plant Mol. Biol. Rep. 31, 1261–1270. doi: 10.1007/s11105-013-0591-2
Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15, 1846–1858. doi: 10.1105/tpc.012211
Jung, C., Yeu, S. Y., Koo, Y. J., Kim, M., Choi, Y. D., and Cheong, J. J. (2007). Transcript profile of transgenic Arabidopsis constitutively producing methyl jasmonate. J. Plant Biol. 50, 12–17. doi: 10.1007/BF03030594
Jung, J. H., Seo, P. J., Kang, S. K., and Park, C. M. (2011). miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 76, 35–45. doi: 10.1007/s11103-011-9759-z
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kim, Y. J., and Hwang, B. K. (2000). Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant. 108, 51–60. doi: 10.1034/j.1399-3054.2000.108001051x
Klein, J., Saedler, H., and Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250, 7–16. doi: 10.1007/BF02191820
Kousik, C. S., Donahoo, R. S., and Hassell, R. (2012). Resistance in watermelon rootstocks to crown rot caused by Phytophthora capsici. Crop Prot. 39, 18–25. doi: 10.1016/j.cropro.2012.04.004
Lannenpaa, M., Janonen, I., Holtta-Vuori, M., Gardemeister, M., Porali, I., and Sopanen, T. (2004). A new SBP-box gene BpSPL1 in silver birch (Betula pendula). Physiol. Plant. 120, 491–500. doi: 10.1111/j.0031-9317.2004.00254.x
Li, C. L., and Lu, S. F. (2014). Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 14:131. doi: 10.1186/1471-2229-14-131
Li, J., Hou, H. M., Li, X. Q., Xiang, J., Yin, X. J., Gao, H., et al. (2013). Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.). Plant Physiol. Biochem. 70, 100–114. doi: 10.1016/j.plaphy.2013.05.021
Liu, H. T., Liu, Y. Y., Pan, Q. H., Yang, H. R., Zhan, J. C., and Huang, W. D. (2006). Novel interrelationship between salicylic acid, abscisic acid, and PIP2-specific phospholipase C in heat acclimation-induced thermotolerance in pea leaves. J. Exp. Bot. 57, 3337–3347. doi: 10.1093/jxb/erl098
Liu, K. K. (2009). Studies on the Resistance and its Mechanisms of Pepper to Phytophthora capsici. Ph.D. thesis, Northwest A & F University, Yangling District.
Liu, R. H., and Meng, J. L. (2003). MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25, 317–321.
Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H. B., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroupspapaya, poplar, and grape: coge with rosids. Plant Physiol. 148, 1772–1781. doi: 10.1104/pp.108.124867
Martin, R. C., Asahina, M., Liu, P. P., Kristof, J. R., Coppersmith, J. L., Pluskota, W. E., et al. (2010). The regulation of post-germinative transition from the cotyledon- to vegetative-leaf stages by microRNA-targeted SQUAMOSA PROMOTER-BINDING PROTEIN LIKE13 in Arabidopsis. Seed Sci. Res. 20, 89–96. doi: 10.1017/S0960258510000073
Oelke, L. M., and Bosland, P. W. (2003). Differentiation of race specific resistance to Phytophthora root rot and foliar blight in Capsicum annuum. J. Am. Soc. Hortic. Sci. 128, 213–218.
Padmanabhan, M. S., Ma, S., Burch-Smith, T. M., Czymmek, K., Huijser, P., and Dinesh-Kumar, S. P. (2013). Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 9:e1003235. doi: 10.1371/journal.ppat.1003235
Rost, B. (1999). Twilight zone of protein sequence alignments. Protein Eng. 12, 85–94. doi: 10.1093/protein/12.2.85
Salinas, M., Xing, S. P., Hohmann, S., Berndtgen, R., and Huijser, P. (2012). Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 235, 1171–1184. doi: 10.1007/s00425-011-1565-y
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schwarz, S., Grande, A. V., Bujdoso, N., Saedler, H., and Huijser, P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67, 183–195. doi: 10.1007/s11103-008-9310-z
Shalom, L., Shlizerman, L., Zur, N., Doron-Faigenboim, A., Blumwald, E., and Sadka, A. (2015). Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front. Plant Sci. 6:389. doi: 10.3389/fpls.2015.00389
Shikata, M., Koyama, T., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50, 2133–2145. doi: 10.1093/pcp/pcp148
Stone, J. M., Liang, X., Nekl, E. R., and Stiers, J. J. (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 41, 744–754. doi: 10.1111/j.1365-313X.2005.02334.x
Thomma, B. P., Penninckx, I. A., Broekaert, W. F., and Cammue, B. P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. doi: 10.1016/S0952-7915(00)00183-7
Vidal, S., Leon, I. P. D., Denecke, J., and Palva, E. T. (1997). Salicylic acid and the plant pathogen Erwinia carotovora induce defense genes via antagonistic pathway. Plant J. 11, 115–123. doi: 10.1046/j.1365-313X.1997.11010115.x
Wan, H. J., Yuan, W., Yu, K., Liu, Y. F., Li, Z. M., Ye, Q. J., et al. (2013). Genome-wide identification, structure characterization and expressionanalysis of SBP gene family in tomato. Mol. Plant Breed. 11, 299–306.
Wang, J. E. (2013). Expression Analysis and Functional Identification of CaRGA1 and CaPOD Genes Induced by Phytophthora capsici in Pepper. Ph.D. thesis, Northwest A&F University, Yangling District.
Wang, J. E., Li, D. W., Zhang, Y. L., Zhao, Q., He, Y. M., and Gong, Z. H. (2013a). Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. Eur. J. Plant Pathol. 136, 625–638. doi: 10.1007/s10658-013-0193-8
Wang, J. E., Liu, K. K., Li, D. W., Zhang, Y. L., Zhao, Q., He, Y. M., et al. (2013b). A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophthora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 14, 3158–3177. doi: 10.3390/ijms14023158
Wang, Y., Hu, Z. L., Yang, Y. X., Chen, X. Q., and Chen, G. P. (2009). Function annotation of an SBP-box gene in Arabidopsis based on analysis of co-expression networks and promoters. Int. J. Mol. Sci. 10, 116–132. doi: 10.3390/ijms10010116
Xie, K. B., Wu, C. Q., and Xiong, L. Z. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
Xing, S. P., Salinas, M., Hohmann, S., Berndtgen, R., and Huijser, P. (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935–3950. doi: 10.1105/tpc.110.079343
Xu, G. X., Guo, C. C., Shan, H. Y., and Kong, H. Z. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Xu, Y., Chang, P. F. L., Liu, D., Narasimhan, M. N., Raghothama, K. G., Hasegawa, P. M., et al. (1994). Plant defense genes are synergistically induced by ethylene and methyl Jasmonate. Plant Cell 6, 1077–1085. doi: 10.1105/tpc.6.8.1077
Xu, Z. D., Sun, L. D., Zhou, Y. Z., Yang, W. R., Cheng, T. R., Wang, J., et al. (2015). Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genomics 290, 1701–1715. doi: 10.1007/s00438-015-1029-3
Yamaguchi, A., Wu, M. F., Yang, L., Wu, G., Poethig, R. S., and Wagner, D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17, 268–278. doi: 10.1016/j.devcel.2009.06.007
Yamasaki, H., Hayashi, M., Fukazawa, M., Kobayashi, Y., and Shikanai, T. (2009). SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 347–361. doi: 10.1105/tpc.108.060137
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2004). A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 337, 49–63. doi: 10.1016/j.jmb.2004.01.015
Yang, Z. F., Wang, X. F., Gu, S. L., Hu, Z. Q., Xu, H., and Xu, C. W. (2008). Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407, 1–11. doi: 10.1016/j.gene.2007.02.034
Yin, Y. X., Guo, W. L., Zhang, Y. L., Ji, J. J., Xiao, H. J., Yan, F., et al. (2014). Cloning and characterisation of a pepper aquaporin, CaAQP, which reduces chilling stress in transgenic tobacco plants. Plant Cell Tissue Organ Cult. 118, 431–444. doi: 10.1007/s11240-014-0495-3
Zhang, L. S., Wu, B., Zhao, D. G., Li, C. L., Shao, F. G., and Lu, S. F. (2014). Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J. Integr. Plant Biol. 56, 38–50. doi: 10.1111/jipb.12111
Zhang, S. D., and Ling, L. Z. (2014). Genome-wide identification and evolutionary analysis of the SBP-box gene family in castor bean. PLoS ONE 9:e86688. doi: 10.1371/journal.pone.0086688
Zhang, X. H., Dou, L. L., Pang, C. Y., Song, M. Z., Wei, H. L., Fan, S. L., et al. (2015). Genomic organization, differential expression, and functional analysis of the SPL gene family in Gossypium hirsutum. Mol. Genet. Genomics 290, 115–126. doi: 10.1007/s00438-014-0901-x
Zhang, Y., Schwarz, S., Saedler, H., and Huijser, P. (2007). SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 63, 429–439. doi: 10.1007/s11103-006-9099-6
Keywords: Capsicum annuum L., SBP-box family genes, Phylogenetic analysis, Phytophthora capsici, hormone treatments
Citation: Zhang H-X, Jin J-H, He Y-M, Lu B-Y, Li D-W, Chai W-G, Khan A and Gong Z-H (2016) Genome-Wide Identification and Analysis of the SBP-Box Family Genes under Phytophthora capsici Stress in Pepper (Capsicum annuum L.). Front. Plant Sci. 7:504. doi: 10.3389/fpls.2016.00504
Received: 09 October 2015; Accepted: 29 March 2016;
Published: 15 April 2016.
Edited by:
Rex Brennan, The James Hutton Institute, UKReviewed by:
Javier Sanchez, University of Zurich, SwitzerlandSyed Noor Muhammad Shah, Gomal University, Pakistan
Copyright © 2016 Zhang, Jin, He, Lu, Li, Chai, Khan and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Hui Gong, zhgong@nwsuaf.edu.cn
 Huai-Xia Zhang1
Huai-Xia Zhang1 Bo-Ya Lu
Bo-Ya Lu Wei-Guo Chai
Wei-Guo Chai Zhen-Hui Gong
Zhen-Hui Gong