- 1Institute of Biotechnology, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 2Center of Analysis and Measurement, Zhejiang University, Hangzhou, China
Calmodulin-binding transcription activators (CAMTAs) play important roles in various plant biological processes including disease resistance and abiotic stress tolerance. Oilseed rape (Brassica napus L.) is one of the most important oil-producing crops worldwide. To date, compositon of CAMTAs in genomes of Brassica species and role of CAMTAs in resistance to the devastating necrotrophic fungal pathogen Sclerotinia sclerotiorum are still unknown. In this study, 18 CAMTA genes were identified in oilseed rape genome through bioinformatics analyses, which were inherited from the nine copies each in its progenitors Brassica rapa and Brassica oleracea and represented the highest number of CAMTAs in a given plant species identified so far. Gene structure, protein domain organization and phylogentic analyses showed that the oilseed rape CAMTAs were structurally similar and clustered into three major groups as other plant CAMTAs, but had expanded subgroups CAMTA3 and CAMTA4 genes uniquely in rosids species occurring before formation of oilseed rape. A large number of stress response-related cis-elements existed in the 1.5 kb promoter regions of the BnCAMTA genes. BnCAMTA genes were expressed differentially in various organs and in response to treatments with plant hormones and the toxin oxalic acid (OA) secreted by S. sclerotiorum as well as the pathogen inoculation. Remarkably, the expression of BnCAMTA3A1 and BnCAMTA3C1 was drastically induced in early phase of S. sclerotiorum infection, indicating their potential role in the interactions between oilseed rape and S. sclerotiorum. Furthermore, inoculation analyses using Arabidopsis camta mutants demonstrated that Atcamta3 mutant plants exhibited significantly smaller disease lesions than wild-type and other Atcamta mutant plants. In addition, compared with wild-type plants, Atcamta3 plants accumulated obviously more hydrogen peroxide in response to the PAMP chitin and exhibited much higher expression of the CGCG-box-containing genes BAK1 and JIN1, which are essential to the PAMP triggered immunity (PTI) and/or plant resistance to pathogens including S. sclerotiorum. Our results revealed that CAMTA3 negatively regulated PTI probably by directly targeting BAK1 and it also negatively regulated plant defense through suppressing JA signaling pathway probably via directly targeting JIN1.
Introduction
Calcium is a ubiquitous second messenger used by plants to regulate a variety of biological processes in response to a wide range of environmental and developmental stimuli (Galon et al., 2010; Reddy et al., 2011). In response to these stimuli, Ca2+ signals are decoded and transmitted by several types of Ca2+ sensor proteins including calmodulins (CaMs), calcineurin B-like proteins (CBLs), and calcium-dependent protein kinases (CDPKs/CPKs; Kudla et al., 2010; Du et al., 2011). CaM can bind to certain transcription factors such as calmodulin-binding transcription activators (CAMTAs).
CAMTAs, also referred to as signal-responsive (SR) proteins, are thought to exist in all multicellular organisms (Bouché et al., 2002; Rahman et al., 2016). Taking advantage of the rapid developing of plant genome sequencing, CAMTA family has been identified at genome-wide level in over 40 plant species (Bouché et al., 2002; Choi et al., 2005; Koo et al., 2009; Yang et al., 2012; Shangguan et al., 2014; Wang et al., 2015; Yang et al., 2015; Yue et al., 2015; Rahman et al., 2016). Nevertheless, composition of CAMTAs in many economically important crop species such as Brassica species is still unknown.
CAMTAs contain multiple functional domains including a CG-1 DNA-binding domain, an ankyrin (ANK) repeat domain, an IQ (Isoleucine glutamine) domain, and a CaM binding (CaMB) domain that are located in turn from the N terminus to the C terminus (Bouché et al., 2002; Choi et al., 2005; Finkler et al., 2007; Rahman et al., 2016). Most of CAMTAs also possess a TIG (Transcription-associated immuno globulin-like) domain (Rahman et al., 2016). CAMTAs specifically recognize and bind to (A/C/G)CGCG(T/C/G) or (A/C)CGTGT cis-elements in the promoter region of target genes, thereby regulate their expression (Yang and Poovaiah, 2002; Choi et al., 2005; Du et al., 2009). The biological functions of CAMTAs are being revealed but mainly in Arabidopsis, rice and tomato. The functions of CAMTAs were dependent on their interaction with Ca2+/CaM (Choi et al., 2005; Du et al., 2009). Arabidopsis CAMTA3 negatively regulates accumulation of salicylic acid and host plant resistance to both bacterial (Du et al., 2009) and fungal pathogens (Galon et al., 2008; Nie et al., 2012) as well as nonhost resistance to bacterial pathogen Xanthomonas oryzae pv. oryzae, probably via tuning CBP60G, EDS1, and NDR1-mediated defense signaling and reactive oxygen species (ROS) accumulation (Rahman et al., 2016). AtCAMTA3 signaling is modulated by ubiquitination process during regulation of plant immunity (Zhang et al., 2014). Similarly, a rice CAMTA OsCBT-1 negatively regulates rice resistance to blast fungal pathogen and leaf blight bacterial pathogen (Koo et al., 2009). Besides, AtCAMTA3 also plays important roles in plant defense against insect herbivore, glucose metabolism and ethylene-induced senescence in Arabidopsis (Laluk et al., 2012; Qiu et al., 2012). Arabidopsis CAMTA1, CAMTA2, and CAMTA3 contribute to low temperature and freezing tolerance by activation of CBF (C-repeat/DRE binding factor) transcription factors (Doherty et al., 2009; Kim et al., 2013). Tomato CAMTAs are differentially expressed during fruit development and ripening processes and in responsive to biotic and abiotic stimuli (Yang et al., 2012, 2013). Silencing of SlSR1 and SlSR3L enhances resistance to bacterial and fungal pathogens while silencing of SlSR1L leads to decreased drought stress tolerance (Li et al., 2014). Collectively, these reports clearly demonstrate that CAMTAs, especially CAMTA3, are important regulators of plant resistance to biotrophic pathogens. Nevertheless, their role in plant resistance to necrotrophic pathogens remains poorly understood.
Oilseed rape (Brassica napus L.) is one of the most important oil crops worldwide. Despite relatively extensive studies of CAMTAs in several model plant species, little is known about this gene family in oilseed rape and other Brassica species. Only one CAMTA sequence has been identified in oilseed rape to date (Bouché et al., 2002). In this study, taking advantage of completion of the oilseed rape genome sequence (Chalhoub et al., 2014), we systemically identified the CAMTA gene family in B. napus genome and performed comprehensive sequence analyses as well as functional analyses in disease resistance. Our results demonstrated that oilseed rape genome contained the highest number of CAMTAs in a given plant species identified so far. BnCAMTA3A1 and BnCAMTA3C1 were likely to be the functional homologs of AtCAMTA3 functioning in disease resistance. Furthermore, using Arabidopsis camta mutants, we revealed that CAMTA3 negatively regulated chitin-triggerred immunity and plant defense to the devastating necrotrophic pathogen Sclerotinia sclerotiorum, probably via directly targeting BAK1 and JIN1.
Materials and Methods
Identification of CAMTA Proteins in Brassica Species
To identify CAMTA protein sequences in oilseed rape, the six Arabidopsis CAMTAs were used as query to search by BLASTP program against B. napus genome databases deposited in NCBI (http://www.ncbi.nlm.nih.gov/) and the GNEOSCOPE (http://www.genoscope.cns.fr/spip/). All retrieved non-redundant sequences were collected, and subjected to conserved domain analysis using the Pfam (http://pfam.sanger.ac.uk/) and NCBI-CDD (http://www.ncbi.nlm.nih.gov/cdd) databases. These sequences were compared with Arabidopsis and tomato CAMTA proteins using ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default settings and were viewed by GeneDoc. Those containing a CG-1 domain, an ANK repeat domain and a CaMB domain were recognized as CAMTA proteins. CAMTAs in oilseed rape were named in accordance with their phylogenetic relationship to six Arabidopsis CAMTAs. Identification of CAMTAs in B. rapa and B. oleracea, two progenitor species of B. napus, was performed similarly.
Gene Structure, Protein Domain, and Phylogenetic Analyses of BnCAMTA Genes
The gene structure was analyzed online by the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/index.php; Guo et al., 2007). A schematic diagram of protein domain structures with functional motifs was constructed using Domain Illustrator software (http://dog.biocuckoo.org/; Ren et al., 2009). The sequence logos of CaMB domain were generated using the Geneious software (v6.1.6) package (http://www.geneious.com/). Multiple sequence alignments of the full-length CAMTA proteins from representative plant species were conducted using ClustalW. The phylogenetic tree was constructed using MEGA 5.0 (Tamura et al., 2011) with maximum likelihood (ML) method and a bootstrap test was performed with 1000 replicates.
Prediction of cis-Acting Elements in the BnCAMTA Genes
To investigate cis-elements in the promoter sequences of the BnCAMTA genes, 1.5 kb sequences upstream of the initiation codon (ATG) were collected and subjected to stress response-related cis-acting element online prediction analysis with Signal Scan search program in the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html) and the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Plant Material and Hormone Treatments
Oilseed rape plants were grown in growth room at 22–23°C with a 16/8 h day/night photoperiod. Arabidopsis plants of Col-0 and six CAMTA mutants (Atcamta1-6) were grown in a growth chamber at 20–21°C under a 15/9 h day/night photoperiod. For BnCAMTA gene expression analyses, leaves of 4-week-old plants were sprayed with hormones SA (1 mM) and JA (200 μM) as well as a chemical OA (1 mM), the toxin secreted by the pathogen S. sclerotiorum, or 0.01% ethonal (the solvent for the above chemicals) as a control, and collected at 0, 4, 12, and 24 h after treatment. In addition, various organs of oilseed rape plants including root, stem, cotyledone, and true leaves were also sampled for gene expression analysis. All samples were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
S. sclerotiorum Inoculation and Plant Resistance Analyses
Leaves of 4 week-old B. napus and Arabidopsis plants were inoculated with mycelial plugs of 3 mm diameter of S. sclerotiorum as described (Saand et al., 2015a). The inoculated leaves were collected at 0, 6, and 12 h post inoculation for RNA extraction and gene expression analyses. The necrosis symptoms of the inoculated leaves were investigated and the size of lesions was measured. The inoculation analysis was performed three times, each in at least 6 plants for each treatment and gene backgrounds. For the statistical analysis of the lesion size data, ANOVA (analysis of variance) analysis was performed with SPSS software (Version 19.0, IBM, USA). Significant difference between the mean values of three independent experiments was determined with Duncan's multiple range test (DMRT; p < 0.05).
Detection of Chitin-Triggered Hydrogen Peroxide
The hydrogen peroxide (H2O2) elicited by chitin (100 μg mL−1, Sigma, USA) in leaf discs of Atcamta3 mutant and wild type Col-0 plants were measured using a Microplate Luminometer (TITERTEK BERTHOLD, Germany) following previously described protocol (Saand et al., 2015a). For each experiment, 10 leaves were collected for each genotype. All experiments were conducted three times independently. The quantitative measurement data were statistically analyzed using SPSS software and represent means ± standard error.
RNA Isolation and Gene Expression Analyses
Total RNA was extracted with Trizol reagent (TAKARA, Japan) following the manufacturer's instructions. RNA was treated with DNase I (TAKARA, Japan) and reverse-transcribed into cDNA using the PrimeScript RT reagent kit (TAKARA, Japan). The obtained cDNAs were used for gene expression analyses with semiquantitative reverse transcription PCR (RT-PCR) and quantitative real time PCR (qRT-PCR). Semiquantitative RT-PCR was performed following the program: 94°C for 5 min, followed by 32 or 28 (for internal control gene) cycles of denaturation for 50 s at 94°C, annealing for 50 s at 55°C, extension for 20 s at 72°C, and a final extension for 10 min at 72°C. The obtained products were analyzed by electrophoresis on a 1.5% agarose gel and detected under ultraviolet light. The qRT-PCR was conducted in StepOne Real-Time PCR System (Applied Biosystems, USA) using SYBER Premix Ex Taq reagents (TaKaRa, Japan) following the program: 95°C for 30 s, 95°C for 5 s, and 60°C for 45 s for 40 cycles. To normalize the sample variance, B. napus β -Tubulin and Arabidopsis ACTIN8 genes served as internal controls. Relative gene expression values were calculated using the 2−ΔΔCt method. The primers used for gene expression analyses are listed at Table S1. For the statistical analysis of the gene expression data, ANOVA analysis was performed with SPSS software (Version 19.0, IBM, USA). Significant difference between mean values was determined with DMRT (p < 0.05).
Results
Identification of CAMTA Genes in B. napus and Its Two Progenitor Species
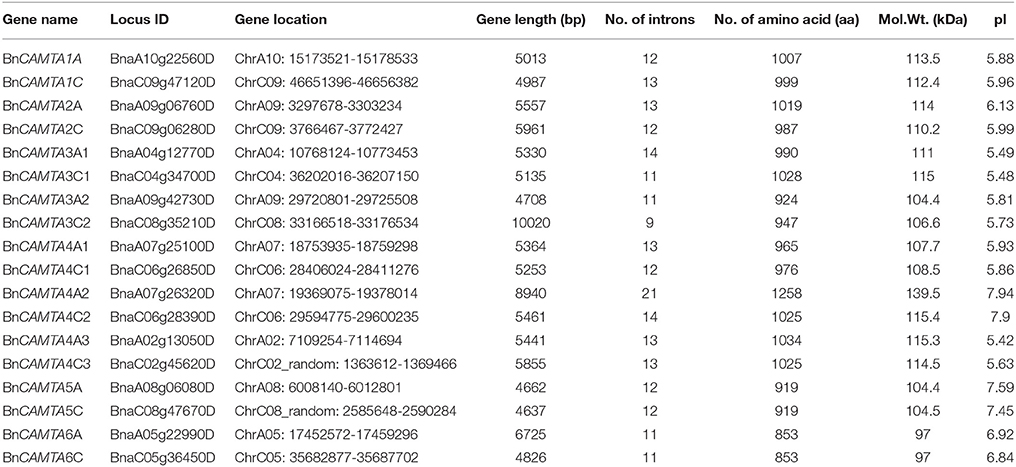
To identify CAMTA genes in B. napus, the six Arabidopsis CAMTAs were used as query to BLASTP search in the complete genome of B. napus. Based on domain composition analyses for the retrieved candidate sequences, a total of 18 CAMTA sequences were identified in B. napus genome, representing the highest number of CAMTAs in a given plant species identified so far. They were named in accordance with their phylogenetic relationship with the six Arabidopsis CAMTAs and the location in subgenomes (A or C). The comprehensive information of BnCAMTA genes, including locus ID, gene location, length and intron number, predicted protein size, molecular weight, and isoelectric point (pI), is listed in Table 1. The length of the BnCAMTA gene sequences was 4.6–6.0 kb with three exceptions BnCAMTA3C2 (10.0 kb), BnCAMTA4A2 (8.9 kb), and BnCAMTA6A (6.7 kb), which contained significantly longer genomic sequence due to possessing an extraordinarily large intron (Table 1; Figure 1). The size of predicted BnCAMTA proteins was 919–1034 amino acids (aa) except BnCAMTAs 6A and 6C with 853 aa and BnCAMTA4A2 with 1258 aa (Table 1). BnCAMTA4A2 was larger due to carrying an extra N terminal sequence (Figure 1A). BnCAMTA proteins varied obviously in their pI value. The majority of them (12 out of 18) owned a pI of lower than 6.2, two of them (BnCAMTAs 6A and 6C) had a pI of near 7.0, while the remaining four (BnCAMTAs 4C2, 4A2, 5A, and 5C) possessed a pI of higher than 7.4 (Table 1), implying that while most of the BnCAMTA proteins are acidic, some of them are neutral or basic. Collectively, these results indicated that B. napus genome possesses much more CAMTA genes than other plant species, and their physico-chemical characteristics were generally conserved but with obvious exceptions.
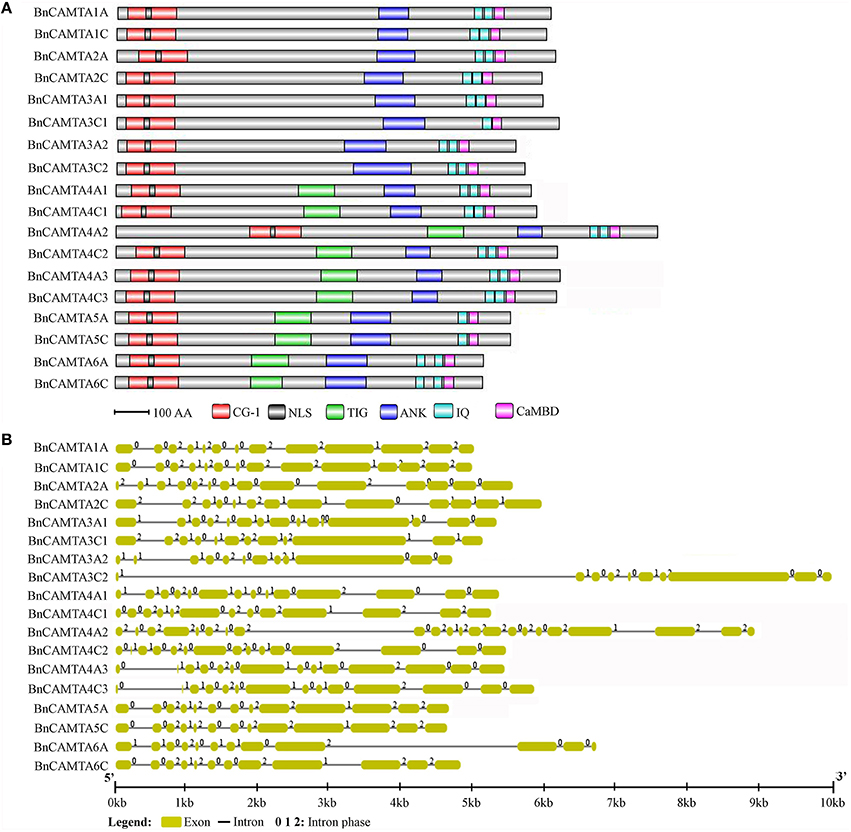
Figure 1. Protein domain composition and gene structure of BnCAMTAs. (A) Schematic representation of domains of BnCAMTA proteins. Analyses of conserved domains were performed using the Pfam database (http://pfam.janelia.org/). NLS motifs were searched by Motif scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan). CaMBDs were analyzed using the Calmodulin Target Database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/). The domain structures of BnCAMTAs were drawn to scale using Domain Graph software (http://dog.biocuckoo.org/). Abbreviations: CG-1, CG-1 DNA-binding domain; NLS, nuclear localization signal motif; TIG, transcription-associated immuno globulin-like domain; ANK, ankyrin repeat domain; IQ, isoleucine glutamine motif; CaMBD, calmodulin-binding domain. (B) Exon-intron structure of BnCAMTA genes. The exons and introns are indicated by blue boxes gray lines, respectively. The BnCAMTA gene structures were drawn to scale using the Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/).
To better understand the composition of CAMTAs in the tetraploid B. napus, CAMTAs in its two progenitor species B. rapa and B. oleracea was also identified using similar approaches. The results showed that the two Brassica species exhibited similar CAMTA composition, both containing 9 CAMTAs (Table S1). Comparison analysis indicated that B. napus genome possessed exactly the total copies of CAMTAs in its two progenitor species.
Chromosomal Location of BnCAMTA Gene Family
The 18 BnCAMTA genes were mapped on 14 oilseed rape chromosomes (Figure S1). Among them, eight were scattered each on one chromosome (A02, C02_random, A04, C04, A05, C05, A08, and A010), while the remaining 10 were distributed in five chromosomes (C06, A07, C08, A09, and C09) with each two genes in one chromosome. BnCAMTAs 4C1 and 4C2 as well as BnCMTAs 4A1 and 4A2 were located nearly each other on Chromosomes C06 and A07, respectively, while BnCMTAs 2A and 3A2 as well as BnCAMTAs 1C and 2C were distributed distantly on the two ends of Chromosomes A09 and C09, respectively (Figure S1). In addition, BnCAMTAs 3C2 and 5C lay in Chromosome C08 although the precise position of BnCAMTA5C in this chromosome remained unclear (tentatively called ChrC08_random). This result suggested that gene duplication and recombination occurred, most obviously for BnCAMTA4s, and contributed to CAMTA gene expansion in B. napus.
Conserved Domain and Gene Structural Analyses of BnCAMTAs
The CAMTA proteins consist of multiple predicted functional domains, evolutionally conserved in amino acid sequence and organization order. The domain structure analyses revealed that all the 18 BnCAMTA proteins contained a CG-1 DNA-binding domain in the N-terminal portion, an ankyrin repeat (ANK) domain in the middle, one or two IQ motifs and a calmodulin binding (CaMB) domain in the C-terminal region (Figure 1A). In addition, 10 BnCAMTAs belonging to subgroups 4, 5, and 6 contained a TIG domain, located between the N-terminal CG-1 domain and the ANK domain (Figure 1A). All BnCAMTA proteins were predicted to contain a nuclear localization signal (NLS) in the N-terminus of the protein, consistent with their role as transcription factors that function in the nucleus (Figure 1A). This result indicated that the domain composition of CAMTAs in B. napus is similar to those in other plant species (Rahman et al., 2016).
Further, the exon-intron structure of the BnCAMTA genes was analyzed. The result demonstrated that the exon-intron configuration of most BnCAMTA genes was highly conserved with 11-14 introns, as observed for that of CAMTA genes in other plant species (Rahman et al., 2016). The exceptions were BnCAMTA3C2 and BnCAMTA4A2 genes. Both contained an intron with an unusual large size. Additionally, BnCAMTA3C2 had only nine introns while BnCAMTA4A2 possessed 21 introns (Figure 1B). Whether they exhibit distinct function from the others remains further study.
Conservation of CaMB Domain of BnCAMTAs
CaMB domain is indispensable to CAMTAs. To understand the conservation of this domain in BnCAMTAs, the corresponding sequence regions were aligned and compared with that in well-studied Arabidopsis and tomato CAMTAs. The alignment revealed a conserved motif for functional residues as W X V X(2) L X K X(2) [LI] R W R X K X(3) [LF] [RKIV] X (Figure 2). Except for minor variation in some positions such as the 11th and 21st positions, the motif for BnCAMTAs generally fitted the one reported for Arabidopsis and tomato CAMTAs (W X V X(2) L X K X(2) [LF] R W R X [KR] X(3) [FL] R X). In this motif for BnCAMTAs, the 11th hydrophobic residue was dominated by L except two sequences (BnCAMTAs 6A and 6C) as I. Similarly, the 21st position was dominated by R, but it was K in BnCAMTAs 4A1, 4C1, 4A3, and 4C3 and I and V in BnCAMTAs 6A and 6C, respectively (Figure 2). Collectively, these data demonstrated that the motif of CaMB domain was highly conserved in CAMTA proteins of oilseed rape and other plant species.
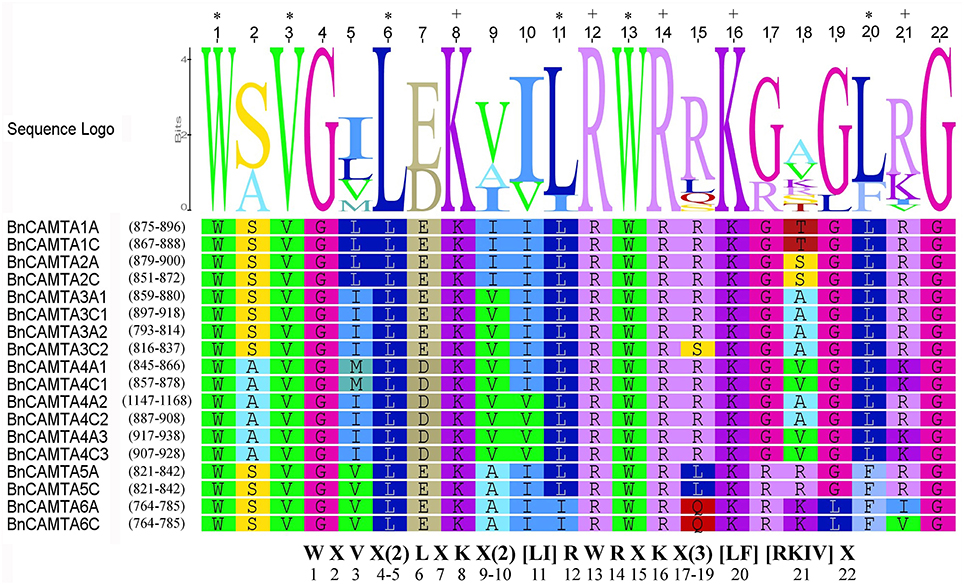
Figure 2. Sequence logo of the CaMB domain of 18 BnCAMTA proteins. The “*” and “+” indicate conserved hydrophobic and positively charged residues in the CaMB domain. The conserved motif for these functional amino acids is shown at the bottom. The sequence logo of BnCAMTAs was drawn using Geneous R6 software (v6.1.6).
Phylogenetic Relationship of BnCAMTA Genes
To gain insight into the phylogenetic relationship of BnCAMTA genes, a phylogenetic tree based on maximum-likelihood (ML) methods was constructed for 18 B. napus CAMTAs along with those from B. rapa, B. oleracea, Arabidopsis and tomato (Table 1, Table S2, and Figure 3). The phylogenetic analysis indicated that 18 BnCAMTAs clustered into three groups (I–III) with CAMTAs from other plant species with strong bootstrap support. All four memebers of BnCAMTA subgroups 5 and 6 constituted group I, together with CAMTA subgroups 5 and 6 from other plant species. All six members of BnCAMTA subgroup 4 gathered into group II along with CAMTA4s from other plant species, while all 8 non-TIG BnCAMTAs (all members of BnCAMTA subgroups 1, 2, and 3) formed group III, together with CAMTA subgroups 1, 2, and 3 from the other species (Figure 3). This phylogenetic tree revealed that all copies of CAMTAs in the two progenitors B. rapa and B. oleracea were well-inherited in B. napus. The similar clustering pattern was also obtained when the phylogenetic tree was reconstructed for the CAMTA proteins only from Arabidopsis and oilseed rape (Figure S2). It is noteworthy that different members of BnCAMTA subgroups 3 and 4 exhibited distinguishable phylogenetic distance to Arabidopsis CAMTA3 and CAMTA4. BnCAMTAs 3A1 and 3C1 were phylogenetically closer to AtCAMTA3 than BnCAMTAs 3A2 and 3C2. Similarly, BnCAMTAs 4A3 and 4C3 were phylogenetically closer to AtCAMTA4 than the other four BnCAMTA4s (Figure S2). These results indicated that CAMTA3 and CAMTA4 genes had been expanded in the three Brassica species compared with Arabidopsis although they belong to the same family (Brassicaceae). It is intriguing to probe whether members of the same subgroups function similarly or differentially considering that the pivotal role of AtCAMTA3 in plant defense has been unveiled.
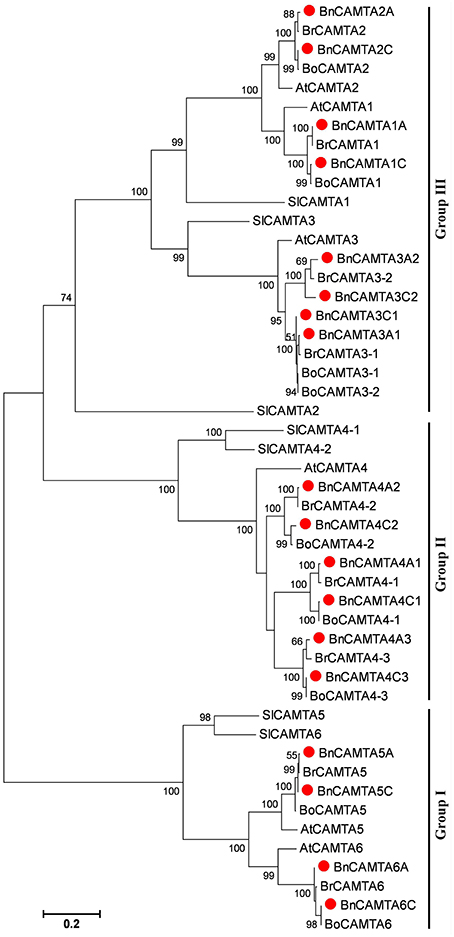
Figure 3. Phylogenetic tree of oilseed rape CAMTAs along with homologs from its progenitors B. rapa and B. oleracea as well as Arabidopsis and tomato. Bootstrap values are displayed on the branches. Oilseed rape CAMTAs are marked with a solid red circle before the protein names. The tree was generated using the MEGA5 program by maximum likelihood (ML) methods.
Prediction of cis-Acting Elements in Promoters of BnCAMTA Genes
Nine well-defined and stress response-related cis-acting elements (DRE/CRT, ABRE, AuxRE, SARE, G- box, W-box, CG-box, P1BS, and SURE) were scanned in 1.5 kb sequences upstream of the ATG of BnCAMTA genes to obtain preliminary clues on how the BnCAMTA genes respond expressionally to stress stimuli. The results showed that there were various stress/stimulus response-related cis-acting elements in the promoter of BnCAMTA genes (Table 2). Analyses in both PLACE and PlantCARE databases predicted that BnCAMTA genes widely contained ABA responsive element (ABRE) and G-box element, some carried W-box element while a few possessed auxin responsive element (AuxRE) in their promoters (Table 2). Moreover, search in PLACE database predicted that some BnCAMTA genes owned additional cis-emelmets such as dehydration and cold responsive element (DRE/CRT), auxin responsive element (AuxRE), SA-responsive element (SARE), phosphate starvation-responsive element (P1BS), and sulfur-responsive element (SURE) in their promoters (Table 2). In addition, four BnCAMTA genes possessed 1–3 copies of CAMTA-recognizable CG-box elements according to the prediction result in the PLACE database, suggesting that CAMTAs might regulate their own gene transcription. Interestingly, every BnCAMTA gene contained at least one type of stress response-related cis-element, but the type of cis-element(s) in BnCAMTA genes was distinguishable (Table 2). Collectively, the stress-responsive cis-element analyses indicated that the BnCAMTAs are likely to be involved in plant response to various stresses and hormone signals.
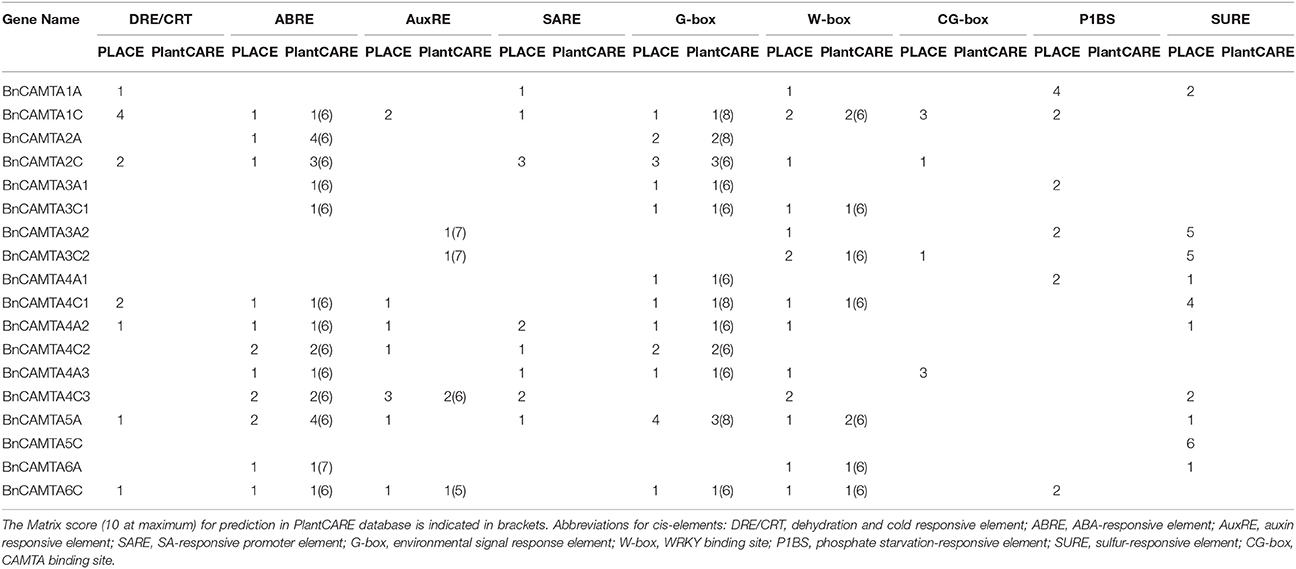
Table 2. Predicted stress response-related cis-elements in the 1.5 kb sequence upstream of ATG of the BnCAMTA genes.
Constitutive Expression of BnCAMTA Genes in Various Tissues of B. napus
To obtain a clue for the possible functions of the BnCAMTA genes, their expression profiles in different tissues or organs, including cotyledons of 1-week-old seedlings as well as roots, stems, and leaves of 4-week-old plants, were analyzed by semiquantitative RT-PCR. The results showed that different BnCAMTA genes exhibited distinct expression patterns. Seven out of 18 BnCAMTA genes (BnCAMTAs 2A, 4C1, 4C3, 5A, 5C, 6A, and 6C) were expressed highly in all investigated organs. Five genes (BnCAMTAs 1A, 2C, 3A1, 3C1, and 4A1) were expressed highly in stem, cotyledon and leaves but only weakly or even not in root. BnCAMTA4C2 gene was expressed highly in stem but weakly in all other organs, while the remaining five BnCAMTA genes (BnCAMTAs 1C, 3A2, 3C2, 4A2, and 4A3) were only very weakly expressed in all types of organs (Figure 4), since their transcripts were detected only in the second round of RT-PCR using products of the first round PCR as template (Figure S3). Collectively, these expression data provided evidence to support that BnCAMTA genes play distinct roles in plant development.

Figure 4. Constitutive expression patterns of BnCAMTA genes in various tissues. Expression patterns of the BnCAMTA genes in root (R), stem (S), cotyledon (C) and true leaf (L) were analyzed by semiquantitative RT-PCR. The oilseed rape β -Tubulin gene served as a loading control gene. The profile of electrophoresis on a 1.5% agarose gel of the products obtained from 32 cycles (28 cycles for control) of PCR was shown.
Expression of BnCAMTA Genes in Response to Hormone and Chemical Treatments
To obtain a clue on functions of BnCAMTA genes, expressional response of these genes to multifunctional hormones SA and JA as well as oxalic acid (OA), the toxin secreted by the pathogen S. sclerotiorum was detected by RT-qPCR. From treatment perspective, SA strongly induced expression of BnCAMTAs 3A2, 3C2, and 4C1 by over 4 folds in at least one time point, and moderately induced expression of BnCAMTAs 4A1, all 5s and 6s by around 2 folds at the early time point (4 hpi) as well as that of BnCAMTAs 1A and 4C2 at the late time point (24 hpi), but repressed expression of the remaining BnCAMTA genes. JA strongly induced expression of BnCAMTAs 3C2, 4A2 and all two 5s by over 4 folds at 12 hpi, and moderately induced expression of BnCAMTAs 1A, 1C, 2C, 3A1, 3A2, 4C2, and 6s at 12 and/or 24 hpi, but suppressed expression of the remaining BnCAMTA genes. OA generally induced expression of BnCAMTA genes at 24 hpi however, repressed expression of three subgroup 4 BnCAMTA genes (4A2, 4A3, and 4C3; Figure 5). From gene perspective, expression of most of the BnCAMTA genes was up-regulated by these three stimuli, although the level of alteration varied in response to different stimulus. However, expression of BnCAMTA genes 4A3 and 4C3 was significantly down-regulated by all stimuli. In addition, expression of BnCAMTA genes 1C, 2A, 2C, 3A1, 3C1, and 4A2 was reduced by SA, while expression of BnCAMTA2A and BnCAMTA4A2 was repressed by JA and OA, respectively (Figure 5). The results indicated that the BnCAMTA genes widely but differentially respond at expression level to the three defense and stress-related signaling molecules SA, JA and OA.
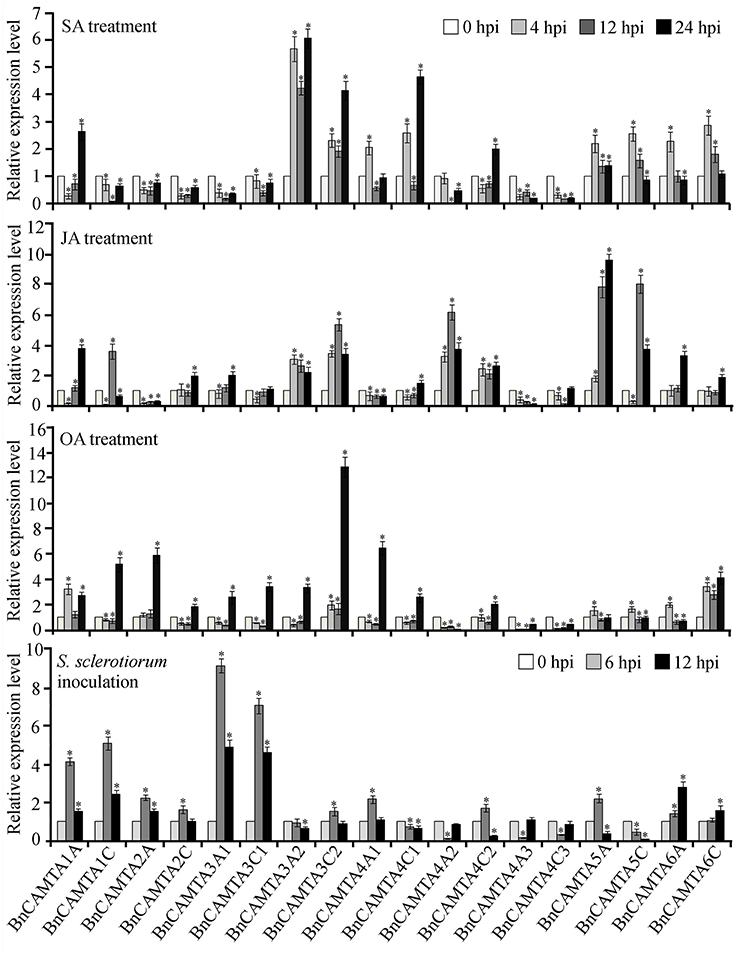
Figure 5. Expression profiles of BnCAMTA genes in response to SA, JA and OA treatments and S. sclerotiorum inoculation. SA (salicylic acid, 1 mM), JA (jasmonic acid, 200 μM), and OA (oxalic acod, 1 mM)-sprayed and S. sclerotiorum-inoculated leaves of 4-week-old oilseed rape plants were sampled at the indicated time points for qRT-PCR analyses with β -Tubulin gene serving as a loading control gene. Data represent the mean ± SE of three independent experiments. Significant difference between mean values is indicated as a “*” (p < 0.05, DMRT).
Expression of BnCAMTA Genes during the Early Phase of S. sclerotiorum Infection
To probe the potential roles of BnCAMTAs in resistance to S. sclerotiorum, expression of the 18 BnCAMTA genes in oilseed rape leaves after S. sclerotiorum inoculation was inspected by qRT-PCR. The result showed that expression of six BnCAMTA genes (1A, 1C, 2A, 3A1, 3C1, and 6A) was significantly increased by over 2 folds after pathogen inoculation, peaking at 6 hpi except BnCAMTA6A, which reached a maximum at 12 hpi (Figure 5). Among them, BnCAMTA3A1 and BnCAMTA3C1 exhibited the most drastic change in expression. Their transcripts were increased by 9.1 and 7.0 folds, respectively, at 6 hpi. In addition, expression of six other BnCAMTA genes (2C, 3C2, 4A1, 4C2, 5A, and 6C) was also up-regulated in at least one time points but only at a change fold of less than 2.0. On the contrary, expression of four subgroup 4 BnCAMTA genes (4A2, 4A3, 4C1, and 4C3) and BnCAMTA5C was strongly decreased at the early time point of pathogen inoculation (6 hpi; Figure 5). These results confirmed that BnCAMTA genes are differentially transcriptionally responsive to S. sclerotiorum infection at the early phase.
Exogenous Supply of SA, JA, and OA Altered Resistance against S. sclerotiorum in Oilseed Rape Plants
The observation that some BnCAMTA genes are strongly responsive to SA, JA, and OA treatments and S. sclerotiorum inoculation prompted us to investigate the effect of these chemicals on resistance to S. sclerotiorum in oilseed rape plants. Leaves of oilseed rape plants were treated with these chemicals and inoculated with S. sclerotiorum at 4 h after treatments. As shown in Figure 6, SA and JA treatments obviously enhanced oilseed rape resistance to S. sclerotiorum, but OA treatment reduced plant resistance, as manifested by that S. sclerotiorum caused necrotic lesions were significantly smaller (1.4 and 1.6 cm at diameter) in SA- and JA-treated leaves, but larger (2.6 cm at diameter) in OA-treated leaves, than those (2.2 cm at diameter) in mock-inoculated control leaves at 36 hpi (Figure 6). This result indicated that SA and JA are associated with oilseed rape resistance to S. sclerotiorum.

Figure 6. Effect of exogenous treatment with SA, JA, and OA on resistance against S. sclerotiorum in oilseed rape plants. (A) The necrotic disease symptoms caused by S. sclerotiorum inoculation in leaves pretreated with 0.01% ethanol (mock), SA (1 mM), JA (200 μM), and OA (1 mM), respectively, at 4 h prior to pathogen inoculation. The photographs were taken at 36 hpi. (B) Statistical analysis of the lesion diameter. Data represent the mean ± SE of three independent experiments. Significant difference between mean values is indicated as small letters (p < 0.05, DMRT).
Arabidopsis CAMTA3 Negatively Regulated Resistance to S. sclerotiorum
To further explore the role of CAMTAs in plant resistance, we preformed inoculation analyses in six Atcamta mutants to examine their response to the devastating necrotrophic fungal pathogen S. sclerotiorum. Results of the inoculation analyses showed that the S. sclerotiorum caused necrotic lesions in the camta3 plants were significantly smaller (0.86 cm at diameter) than those in wild-type and the other mutant plants (over 1.17 cm at diameter) at 24 hpi (Figure 7), demonstrating that the camta3 mutant plants were more resistant to S. sclerotiorum in comparison with wild-type and the other camta mutant plants. This result revealed that CAMTA3 plays a negative role in plant resistance to S. sclerotiorum.
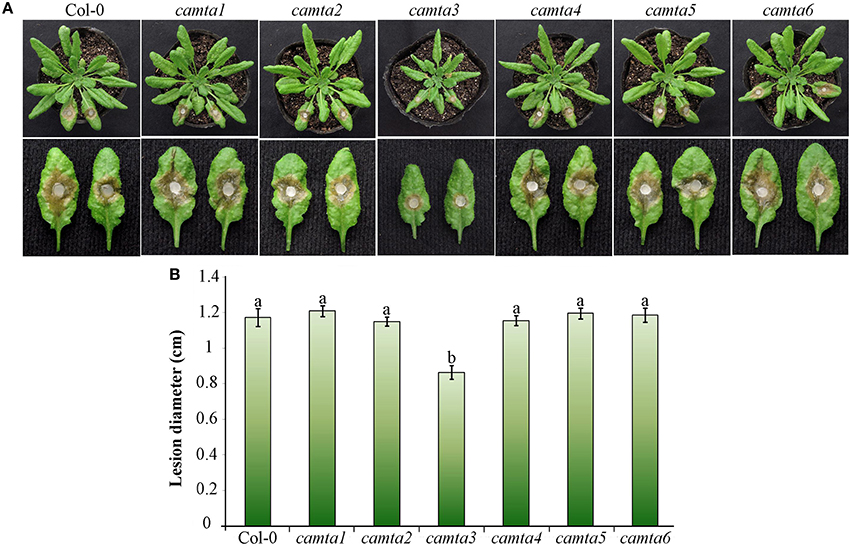
Figure 7. Atcamta3 mutant plants exhibited enhanced resistant to S. sclerotiorum. (A) The necrotic disease symptoms of the Atcamta mutants and Col-0 wild-type plants (upper panel) and detached leaves (lower panel) after inoculated with S. sclerotiorum. Photographs were taken at 30 hpi. (B) Statistical analysis of the lesion diameter. Data represent the mean ± SE of three independent experiments. Significant difference between mean values is indicated as small letters (p < 0.05, DMRT).
Arabidopsis CAMTA3 Negatively Regulated Chitin-Elicited Accumulation of Hydrogen Peroxide
To provide some insights into the mechanisms of AtCAMTA3 to regulate plant resistance, we inspected effect of AtCAMTA3 on accumulation of hydrogen peroxide induced by the PAMP chitin, which exists in the cell wall of the fungal pathogen S. sclerotiorum. In response to 100 μg mL−1 chitin, atcamta3 mutant plants accumulated much higher level of hydrogen peroxide, peaking at over 1200 RLU, than wild-type plants, culminating at about 600 RLU under current measuring system (Figure 8). This result demonstrated that AtCAMTA3 negatively regulates chitin-triggered PTI as manifested by its negative regulation on chitin-triggered accumulation of hydrogen peroxide.
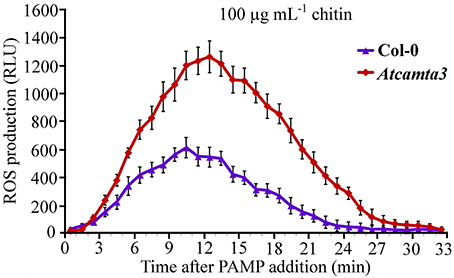
Figure 8. Atcamta3 mutant plants accumulated higher level of chitin-elicited H2O2. Dynamics of H2O2 accumulation in response to chitin elicitation in leaves of Col-0 and Atcamta3 mutant plants was shown. Chitin-triggered H2O2 bursts were measured using luminol-based assay in leaf discs of Col-0 and Atcamta3 mutant plants. Data are shown as relative luminal units (RLU) and represent the mean ± SE of three independent experiments.
Arabidopsis CAMTA3 Negatively Regulated the Expression of a Set of CGCG-Box Containing Defense Signaling Genes
To further elucidate the mechanisms of AtCAMTA3 in regulating resistance to S. sclerotiorum, we examined the expression of four putative or confirmed AtCAMTA3 targeted genes (EDS1, NDR1, BAK1, and JIN1) and three defense signaling pathway marker genes (PR1, PDF1.2, and VSP1) in wild-type and Atcamta3 mutant plants before and after inoculating with S. sclerotiorum. These four genes were selected for this study because they are known to play important roles in plant resistance and PTI to S. sclerotiorum and/or other pathogens (Guo and Stotz, 2007; Du et al., 2009; Perchepied et al., 2010; Nie et al., 2012; Zhang et al., 2013; Macho and Zipfel, 2014). Meanwhile, EDS1 and NDR1 are targets of AtCAMTA3 (Du et al., 2009; Nie et al., 2012). Here, we found that the two PTI and/or S. sclerotiorum resistance regulatory genes BAK1 and JIN1 also contained a CGCG cis-element in the region of –173 to –168 (ACGCGT) and –262 to –257 (CCGCGT), respectively, of their promoters (Figure 9A), they are therefore the potential targets of CAMTA3. Semiquantitative RT-PCR analysis revealed that the expression of EDS1, NDR1, BAK1, and JIN1 in Atcamta3 plants was obviously increased compared with the wild-type plants (Figure 9B), demonstrating that AtCAMTA3 negatively regulates chitin-triggered immunity and resistance to S. sclerotiorum probably via negatively and directly regulating the expression of EDS1, NDR1, BAK1, and JIN1. Moreover, expression of PR1, PDF1.2, and VSP1, marker genes of SA, ethylene and JA defense signaling pathways was obviously higher in Atcamta3 plants than in wild type plants (Figure 9B), indicating that AtCAMTA3 negatively regulates resistance to S. sclerotiorum probably through modulating SA, ethylene and JA defense signaling pathways. In addition, in Atcamta3 plants, transcripts of EDS1, NDR1, BAK1, and JIN1 were still increased in response to S. sclerotiorum inoculation at 6 hpi (Figure 9B), suggesting that factor(s) other than AtCAMTA3 might respond to S. sclerotiorum inoculation to promote the expression of these defense signaling genes in Atcamta3 plants.
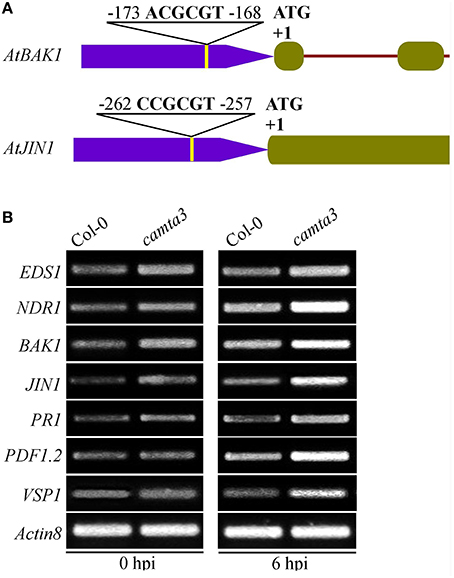
Figure 9. Expression patterns of AtCAMTA3 target genes EDS1 and NDR1 and putative target genes BAK1 and JIN1 in Atcamta3 mutant plants. (A) Schematic representation of CGCG elements in promoter regions of the BAK1 and JIN1 genes. (B) Expression patterns of four confirmed or putative AtCAMTA3-targeted genes and three defense signaling pathway marker genes (PR1, PDF1.2, and VSP1) in Col-0 wild-type and Atcamta3 mutant plants at 0 and 6 h post S. sclerotiorum inoculation. Gene expression was examined by semiquantitative RT-PCR with the Arabidopsis ACTIN8 gene serving as a loading control gene. The profile of electrophoresis on a 1.5% agarose gel of the products obtained from 32 cycles (28 cycles for control) of PCR was shown.
Discussion
Composition and Functions of CAMTA Gene Family in Oilseed Rape
In this study, we found that oilseed rape possesses a total of 18 CAMTAs. Oilseed rape is thus the plant species containing the highest number of CAMTAs among over 40 plant species whose CAMTA family has been identified to date (Bouché et al., 2002; Choi et al., 2005; Koo et al., 2009; Yang et al., 2012; Shangguan et al., 2014; Wang et al., 2015; Yang et al., 2015; Yue et al., 2015; Rahman et al., 2016). The number of CAMTA genes in oilseed rape is 3 folds as many as that in Arabidopsis, which is consistent with the ratio of total number of transcription factors in oilseed rape to that in Arabidopsis (Chalhoub et al., 2014). An important reason that oilseed rape carries so high number of CAMTAs is that oilseed rape is a tetraploid of the two progenitors B. rapa and B. oleracea, and thus contains copies of genes from both progenitors. As a matter of fact, we found that B. rapa and B. oleracea each own 9 CAMTAs, while the oilseed rape subgenomes A and C, which correspond to genomes of B. rapa and B. oleracea, respectively, each contain 9 CAMTAs with identical composition of subgroups as observed as B. rapa and B. oleracea (Table 1, Table S1, and Figure 3). Another reason that oilseed rape carries much higher number of CAMTAs than Arabidopsis is that oilseed rape genome has undergone CAMTA gene expansion compared with Arabidopsis genome. The subgenomes A and C of oilseed rape and genomes of B. rapa and B. oleracea, each contain 9 CAMTAs with 2 CAMTA3s and 3 CAMTA4s (Table 1, Table S1, and Figure 3), demonstrating that genomes of all three Brassica species have expanded CAMTA3 and CAMTA4 genes and this expansion in oilseed rape occurred before formation of oilseed rape. Expansion of CAMTA3 and CAMTA4 genes in Brassica species is unique in all 11 species belonging to Brassicaceae, Caricaece, Malvaceae, Rutaceae, and Myrtaceae of Rosids whose CAMTA family has been identified to date (Figures 1, 2 in Rahman et al., 2016). The reason and significance of this expansion are unclear. While the function of CAMTA4 remains unknown, CAMTA3 in Arabidopsis has been well-recognized to play important roles in host and nonhost resistance against various pathogens (Galon et al., 2008; Du et al., 2009; Nie et al., 2012; Rahman et al., 2016). Therefore, whether both members of CAMTA3 in B. rapa and B. oleracea and all four in oilseed rape function in disease resistance is worthy of experimental clarification. In view of our comprehensive expression analyses, different members of subgroups BnCAMTA3 and BnCAMTA4 exhibit distinct expression profiles both constitutively in various tissues and in response to hormone treatments and pathogen inoculation. BnCAMTA3A1 and BnCAMTA3C1 are highly expressed in stem, cotyledon and true leaf while BnCAMTA3A2 and BnCAMTA3C2 are nearly not expressed in all these tissues (Figure 4). Meanwhile, BnCAMTA3A1 and BnCAMTA3C1 are not obviously responsive to SA and JA treatments but strongly responsive to S. sclerotiorum inoculation, while conversely, BnCAMTA3A2 and BnCAMTA3C2 are highly responsive to SA and JA treatments but not significantly responsive to S. sclerotiorum inoculation (Figure 5). Moreover, the gene structure of BnCAMTA3C2 is distinct to the other members of BnCAMTA3 subgroup (Table 1; Figure 1B). Similarly, distict expression patterns both constitutively in various tissues and in response to hormone treatments and pathogen inoculation are also observed for different members of BnCAMTA4 genes (Figures 4, 5). The gene structure of BnCAMTA4A2 and the pI value of BnCAMTA4A2 and BnCAMTA4C2 are distinguishable from the other members of BnCAMTA4 subgroup (Table 1; Figure 1B). Therefore, different members of subgroups BnCAMTA3 and BnCAMTA4 are most likely to play different roles in development, abiotic stress tolerance, and disease resistance. This seems to be also the case for functions of different subgroups of the CAMTA gene family in oilseed rape considering their distinct expression profiles both constitutively in various tissues and in response to diverse abiotic and biotic stimuli.
Role and Mechanism of AtCAMTA3 in PTI and Resistance to the Necrotrophic Pathogen S. sclerotiorum
Role of CAMTAs in disease resistance against a wide range of biotrophic pathogens in various plants has been reported. These pathosystems include Arabidopsis against bacterial pathogens Pst DC3000 (Du et al., 2009) and Xoo (Rahman et al., 2016) as well as the fungal pathogen Golovinomyces cichoracearum (Nie et al., 2012), and rice against the bacterial pathogen Xoo and the fungal pathogen Magnaporthe grisea (Koo et al., 2009). However, Function of CAMTAs in plant disease resistance against necrotrophic pathogens has only been reported for one pathogen Botrytis cinerea (Galon et al., 2008; Li et al., 2014). In this study, using camta mutants, we demonstrate that AtCAMTA3 negatively regulates the resistance to the typical necrotrophic pathogen S. sclerotiorum, which is one of the most devastating fungal pathogens and causes the most imprtant disease, the white mold disease, in one of the most important oil-producing crops oilseed rape (Bolton et al., 2006). Additionally, oilseed rape CAMTA genes 1A, 1C, 3A1, and 3C1 are strongly responsive to S. sclerotiorum inoculation but differentially respond to the treatment with SA and JA, which play important roles in resistance to S. sclerotiorum (Guo and Stotz, 2007; Perchepied et al., 2010). Therefore, these four CAMTA genes may also play a role in resistance to S. sclerotiorum in oilseed rape. Taken together, these studies reveal that CAMTA genes, especially CAMTA3, contribute greatly to resistance against both biotrophic and necrotrophic pathogens in various plant species.
In addition, in this study, we provide some new intriguing points for the mechanisms of CAMTA3 to regulate PTI and S. sclerotiorum resistance. First, BAK1 might be the target of CAMTA3. BAK1 is a pivotal receptor kinase in PTI triggered by diverse PAMPs such as bacterial PAMP fig22 and fungal PAMP chitin (Macho and Zipfel, 2014). More importantly, it is also required for PTI triggered by SCFE1, a putative PAMP purified from S. sclerotiorum (Zhang et al., 2013). Interestingly, we found in this study that the AtBAK1 gene contains a CGCG cis-element in the region of –173 to –168 (ACGCGT) of its promoter (Figure 9A). Furthermore, expression of AtBAK1 is greatly enhanced in Atcamta3 mutant plants compared with wild-type plants (Figure 9B). Moreover, Atcamta3 mutant plants accumulate much higher level of chitin-elicited hydrogen peroxide than wild-type plants (Figure 8). Collectively, our results indicate that AtCAMTA3 negatively regulates the resistance to S. sclerotiorum probably via suppressing AtBAK1-meadited PTI. Second, CAMTA3 may target JIN1/MYC2 to directly modulate JA signaling thereby regulating plant defense against pathogens including S. sclerotiorum. JA signaling pathway is one of the most important plant defense pathways. This pathway is essential to the reisitance to S. sclerotiorum (Guo and Stotz, 2007; Perchepied et al., 2010). As a key component of JA signaling pathway, JIN1 is indispensible for the resistance to S. sclerotiorum (Guo and Stotz, 2007). We found that the JIN1 gene contains a CGCG cis-element in the region of −262 to −257 (CCGCGT) of its promoter (Figure 9A). Further, expression of AtJIN1 is greatly enhanced in Atcamta3 mutant plants than in wild-type plants (Figure 9B). Together, our results suggest that AtCAMTA3 may modulate the JA signaling pathway via direct targeting JIN1 and thereby regulates the resistance to pathogens including S. sclerotiorum. In these scenarios, it will be very intriguing to confirm whether CAMTA3 can indeed direct bind and regulate expression of BAK1 and JIN1 by other approaches such as ChIP and EMSA assays. Finally, we oberved that expression of CAMTA3-targeted EDS1 and NDR1 genes is obviously increased in Atcamta3 mutant plants than in wild-type plants (Figure 9B) as reported previously (Du et al., 2009; Nie et al., 2012; Rahman et al., 2016). These genes act upstream of SA signaling, which play a role in resistance to S. sclerotiorum (Guo and Stotz, 2007). Thus, EDS1 and NDR1 genes may also contribute to this resistance. The confirmation of function of these genes in this resistance will clarify the significance of CAMTA3-targeting of these two genes in resistance to S. sclerotiorum.
Based on our findings and the previously published reports (Benn et al., 2014; Rahman et al., 2016), we propose a schematic model for CAMTA3-mediated signaling in plants in response to pathogens and PAMPs (Figure 10). In this model, stimuli including pathogens such as S. sclerotiorum and Xoo as well as PAMPs such as chitin and flg22, may activate nucleotidyl cyclase (NC) to generate cyclic nucleotides including cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP), which activate Ca2+ channels such as cyclic nucleotide gated channels (CNGCs), leading to cytosolic Ca2+ influx (Qi et al., 2010; Ma and Berkowitz, 2011; Saand et al., 2015a,b). The cytosolic Ca2+ elevations are transduced by various Ca2+ sensor proteins including CaM, which activates CAMTA3. The activated CAMTA3 directly binds to the CGCG cis-elements in the promoter of defense-related target genes including EDS1, NDR1, CBP60g, EIN3, and JIN1 and regulate their expression, which modulates the accumulation and signaling of SA, ET, and JA, and thereby alters disease resistance. Simultaneously, increased cytosolic Ca2+ would activate calcium-dependent protein kinases (CDPKs), which subsequently phosphorylate and activate RBOHD/F, resulting in ROS accumulation and thereby affecting hypersensitive response (HR) and plant disease resistance. Intriguingly, CAMTA3 may target BAK1 to modulate the recognition complex, a beginning step for plant response to pathogens and PAMPs (Figure 10).
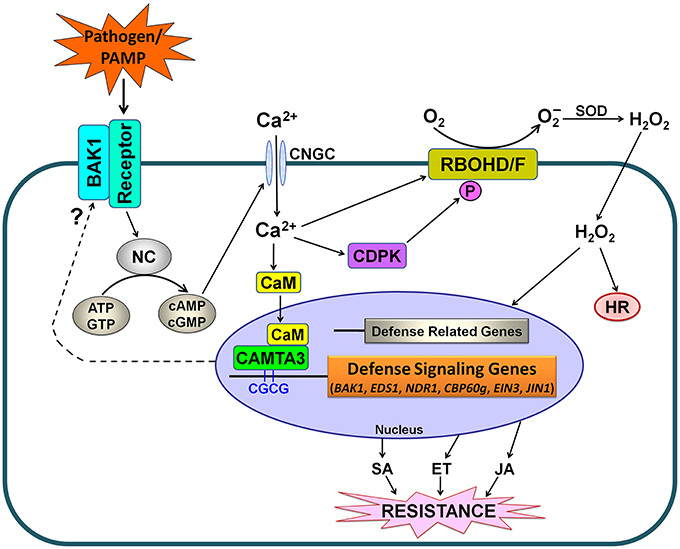
Figure 10. Simplified schematic work model for CAMTA3-mediated defense signaling in plants. PAMPs from pathogens or exogenously supply are recognized by plant receptor proteins. Activation of the recognition complex generally requires BAK1. This recognition may lead to the activation of NC and consequently a rise in the concentration of cyclic nucleotides (cAMP and cGMP), resulting in cytosolic Ca2+ influx through opening of Ca2+ channels such as CNGCs. The cytosolic Ca2+ transients modulate CDPK-promoted and RBOHD/F-mediated ROS accumulation as well as CAMTA3-mediated denfense signaling. The activated CAMTA3 directly binds to the CGCG cis-elements in the promoter of defense-related target genes including EDS1, NDR1, CBP60g, EIN3, and JIN1 and regulate their expression, which modulates the accumulation and signaling of SA, ET and JA, and thereby alters disease resistance. Additionally, CAMTA3 may target BAK1 to mudulate the recognition complex, reflecting its global control of plant defense through regulating expression of the target genes at multiple nodes of the defense network. Abbreviations: BAK1, BRI1-associated receptor kinase; CaM, calmodulin; CAMTA, calmodulin binding transcriptional activator; CDPK, calcium-dependent protein kinases; CNGC, cyclic nucleotide gated channel; ATP, adenosine triphosphate; GTP, guanosine triphosphate; cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; NC, nucleotidyl cyclase; HR, hypersensitive response; SOD, superoxide dismutase; SA, salicylic acid; ET, ethylene; JA, jasmonic acid.
Conclusion
In the present study, we have identified and characterized 18 CAMTA genes in oilseed rape genome. They were inherited from the nine copies each in its progenitors B. rapa and B. oleracea and represented the highest number of CAMTAs in a given plant species identified to date. The oilseed rape CAMTAs clustered into three major groups and had expanded subgroups CAMTA3 and CAMTA4 uniquely in rosids species, which occurred before formation of oilseed rape. Comprehensive expression analyses indicated that BnCAMTA genes are likely to play distinct roles in development, abiotic stress tolerance and disease resistance. Among the four BnCAMTA3 genes, BnCAMTA3A1 and BnCAMTA3C1 are most probably the functional homologs of AtCAMTA3 and contribute to plant defense. Furthermore, functional analyses employing Arabidopsis camta mutants revealed that CAMTA3 negatively regulates PAMP triggered immunity (PTI) probably by directly targeting BAK1 and it also negatively regulates plant defense against pathogens such as S. sclerotiorum through suppressing JA signaling pathway probably via directly targeting JIN1. Our findings provide some insights into the composition of CAMTAs and their roles and functional mechanisms in plant defense.
Author Contributions
HR and XZ conducted the bioinformatics and phylogenetic analyses. HR and YX carried out the gene expression and functional analysis, designed and analyzed all statistical data. XC conceived of the study, and participated in its design and coordination. XC and HR prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Liquan Du, College of Life and Environmental Sciences, Hangzhou Normal University, China, for providing seeds of six Arabidopsis CAMTA knockout lines (Atcamta1-6). This work was financially supported by grants from the Genetically Modified Organisms Breeding Major Projects (no. 2014ZX0800905B), the National Natural Science Foundation of China (no. 31371892), the Zhejiang Provincial Natural Science Foundation of China (no. LZ12C14002) and the Special Fund for Agro-scientific Research in the Public Interest (no. 201103016).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00581
References
Benn, G., Wang, C. Q., Hicks, D. K., Stein, J., Guthrie, C., and Dehesh, K. (2014). A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J. 80, 82–92. doi: 10.1111/tpj.12620
Bolton, M. D., Thomma, B. P., and Nelson, B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. doi: 10.1111/j.1364-3703.2005.00316.x
Bouché, N., Scharlat, A., Snedden, W., Bouchez, D., and Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861. doi: 10.1074/jbc.M200268200
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Choi, M. S., Kim, M. C., Yoo, J. H., Moon, B. C., Koo, S. C., Park, B. O., et al. (2005). Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280, 40820–40831. doi: 10.1074/jbc.M504616200
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S., et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Du, L., Yang, T., Puthanveettil, S., and Poovaiah, B. W. (2011). “Decoding of calcium signal through calmodulin: calmodulin-binding proteins in plants,” in Coding and Decoding of Calcium Signals in Plants, ed S. Luan (Berlin; Heidelberg: Springer), 177–233.
Finkler, A., Ashery-Padan, R., and Fromm, H. (2007). CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett. 581, 3893–3898. doi: 10.1016/j.febslet.2007.07.051
Galon, Y., Finkler, A., and Fromm, H. (2010). Calcium-regulated transcription in plants. Mol. Plant 3, 653–669. doi: 10.1093/mp/ssq019
Galon, Y., Nave, R., Boyce, J. M., Nachmias, D., Knight, M. R., and Fromm, H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582, 943–948. doi: 10.1016/j.febslet.2008.02.037
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: a gene structure display server. Yichuan 29, 1023–1026. doi: 10.1360/yc-007-1023
Guo, X., and Stotz, H. U. (2007). Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol. Plant Microbe Interact. 20, 1384–1395. doi: 10.1094/mpmi-20-11-1384
Kim, Y., Park, S., Gilmour, S. J., and Thomashow, M. F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75, 364–376. doi: 10.1111/tpj.12205
Koo, S. C., Choi, M. S., Chun, H. J., Shin, D. B., Park, B. S., Kim, Y. H., et al. (2009). The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Mol. Cells 27, 563–570. doi: 10.1007/s10059-009-0081-4
Kudla, J., Batistic, O., and Hashimoto, K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell 22, 541–563. doi: 10.1105/tpc.109.072686
Laluk, K., Prasad, K. V., Savchenko, T., Celesnik, H., Dehesh, K., Levy, M., et al. (2012). The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol. 53, 2008–2015. doi: 10.1093/pcp/pcs143
Li, X., Huang, L., Zhang, Y., Ouyang, Z., Hong, Y., Zhang, H., et al. (2014). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 14:286. doi: 10.1186/s12870-014-0286-3
Ma, W., and Berkowitz, G. A. (2011). Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 190, 566–572. doi: 10.1111/j.1469-8137.2010.03577.x
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Nie, H., Zhao, C., Wu, G., Wu, Y., Chen, Y., and Tang, D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158, 1847–1859. doi: 10.1104/pp.111.192310
Perchepied, L., Balagué, C., Riou, C., Claudel-Renard, C., Rivière, N., Grezes-Besset, B., et al. (2010). Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. Mol. Plant Microbe Interact. 23, 846–860. doi: 10.1094/mpmi-23-7-0846
Qi, Z., Verma, R., Gehring, C., Yamaguchi, Y., Zhao, Y., Ryan, C. A., et al. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. U.S.A. 107, 21193–21198. doi: 10.1073/pnas.1000191107
Qiu, Y., Xi, J., Du, L., Suttle, J. C., and Poovaiah, B. W. (2012). Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol. Biol. 79, 89–99. doi: 10.1007/s11103-012-9896-z
Rahman, H., Yang, J., Xu, Y. P., Munyampundu, J. P., and Cai, X. Z. (2016). Phylogeny of plant CAMTAs and role of AtCAMTAs in nonhost resistance to Xanthomonas oryzae pv. oryzae. Front. Plant Sci. 7:177. doi: 10.3389/fpls.2016.00177
Reddy, A. S., Ali, G. S., Celesnik, H., and Day, I. S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032. doi: 10.1105/tpc.111.084988
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., and Yao, X. (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273. doi: 10.1038/cr.2009.6
Saand, M. A., Xu, Y. P., Li, W., Wang, J. P., and Cai, X. Z. (2015a). Cyclic nucleotide gated channel gene family in tomato: genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 6:303. doi: 10.3389/fpls.2015.00303
Saand, M. A., Xu, Y. P., Munyampundu, J. P., Li, W., Zhang, X. R., and Cai, X. Z. (2015b). Phylogeny and evolution of plant cyclic nucleotide-gated ion channel (CNGC) gene family and functional analyses of tomato CNGCs. DNA Res. 22, 471–483. doi: 10.1093/dnares/dsv029
Shangguan, L., Wang, X., Leng, X., Liu, D., Ren, G., Tao, R., et al. (2014). Identification and bioinformatic analysis of signal responsive/calmodulin-binding transcription activators gene models in Vitis vinifera. Mol. Biol. Rep. 41, 2937–2949. doi: 10.1007/s11033-014-3150-5
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Wang, G., Zeng, H., Hu, X., Zhu, Y., Chen, Y., Shen, C., et al. (2015). Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil 386, 205–221. doi: 10.1007/s11104-014-2267-6
Yang, T., Peng, H., Whitaker, B. D., and Conway, W. S. (2012). Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 12:19. doi: 10.1186/1471-2229-12-19
Yang, T., Peng, H., Whitaker, B. D., and Jurick, W. M. (2013). Differential expression of calcium/calmodulin-regulated SlSRs in response to abiotic and biotic stresses in tomato fruit. Physiol. Plant. 148, 445–455. doi: 10.1111/ppl.12027
Yang, T., and Poovaiah, B. W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058. doi: 10.1074/jbc.M207941200
Yang, Y., Sun, T., Xu, L., Pi, E., Wang, S., Wang, H., et al. (2015). Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments. Fronti. Plant Sci. 6:459. doi: 10.3389/fpls.2015.00459
Yue, R., Lu, C., Sun, T., Peng, T., Han, X., Qi, J., et al. (2015). Identification and expression profiling analysis of calmodulin-binding transcription activator genes in maize (Zea mays L.) under abiotic and biotic stresses. Fronti. Plant Sci. 6:576. doi: 10.3389/fpls.2015.00576
Zhang, L., Du, L., Shen, C., Yang, Y., and Poovaiah, B. W. (2014). Regulation of plant immunity through ubiquitin-mediated modulation of Ca2+-calmodulin-AtSR1/CAMTA3 signaling. Plant J. 78, 269–281. doi: 10.1111/tpj.12473
Keywords: Brassica napus, CAMTA, disease resistance, PAMP triggered immunity, Sclerotinia sclerotiorum
Citation: Rahman H, Xu Y-P, Zhang X-R and Cai X-Z (2016) Brassica napus Genome Possesses Extraordinary High Number of CAMTA Genes and CAMTA3 Contributes to PAMP Triggered Immunity and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7:581. doi: 10.3389/fpls.2016.00581
Received: 04 February 2016; Accepted: 14 April 2016;
Published: 04 May 2016.
Edited by:
Naser A. Anjum, University of Aveiro, PortugalReviewed by:
Zhongyun Piao, Shenyang Agricultural University, ChinaChris Gehring, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2016 Rahman, Xu, Zhang and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Zhong Cai, xzhcai@zju.edu.cn
 Hafizur Rahman
Hafizur Rahman You-Ping Xu
You-Ping Xu Xuan-Rui Zhang
Xuan-Rui Zhang Xin-Zhong Cai
Xin-Zhong Cai