- Instituto de Conservación y Mejora de la Agrodiversidad Valenciana, Universitat Politècnica de València, Valencia, Spain
Eggplant (Solanum melongena) is related to a large number of wild species that are a source of variation for breeding programmes, in particular for traits related to adaptation to climate change. However, wild species remain largely unexploited for eggplant breeding. Detailed phenotypic characterization of wild species and their hybrids with eggplant may allow identifying promising wild species and information on the genetic control and heterosis of relevant traits. We characterizated six eggplant accessions, 21 accessions of 12 wild species (the only primary genepool species S. insanum and 11 secondary genepool species) and 45 interspecific hybrids of eggplant with wild species (18 with S. insanum and 27 with secondary genepool species) using 27 conventional morphological descriptors and 20 fruit morphometric descriptors obtained with the phenomics tool Tomato Analyzer. Significant differences were observed among cultivated, wild and interspecific hybrid groups for 18 conventional and 18 Tomato Analyzer descriptors, with hybrids generally having intermediate values. Wild species were generally more variable than cultivated accessions and interspecific hybrids displayed intermediate ranges of variation and coefficient of variation (CV) values, except for fruit shape traits in which the latter were the most variable. The multivariate principal components analysis (PCA) reveals a clear separation of wild species and cultivated accessions. Interspecific hybrids with S. insanum plotted closer to cultivated eggplant, while hybrids with secondary genepool species generally clustered together with wild species. Many differences were observed among wild species for traits of agronomic interest, which allowed identifying species of greatest potential interest for eggplant breeding. Heterosis values were positive for most vigor-related traits, while for fruit size values were close to zero for hybrids with S. incanum and highly negative for hybrids with secondary genepool species. Our results allowed the identification of potentially interesting wild species and interspecific hybrids for introgression breeding in eggplant. This is an important step for broadening the genetic base of eggplant and for breeding for adaptation to climate change in this crop.
Introduction
Eggplant (Solanum melongena L.) is an important vegetable in tropical and subtropical regions across the world, where it is a source of dietary fiber, micronutrients and bioactive compounds (Mennella et al., 2010; Niño-Medina et al., 2014; San José et al., 2014). At present eggplant is the sixth most important vegetable after tomato, watermelon, onion, cabbage, and cucumber and the most important Solanum crop native to the Old World (FAO, 2016). At the global level, it has been one of the crops with the greatest increase in production in the last years, with total production rising by 59% in a decade, from 31.0·106 t in 2004 to 49.3·106 t in 2013 (FAO, 2016).
The narrow genetic base of eggplant, probably a consequence of a genetic bottleneck during its domestication in Southeast Asia (Meyer et al., 2012), is a limitation to obtain major breeding advances. This limited genetic diversity contrasts with the large morphological and genetic variation present in the eggplant wild relatives (Meyer et al., 2012; Vorontsova et al., 2013; Vorontsova and Knapp, in press). Phylogenetically, eggplant is a member of the so-called “spiny solanums” group (Solanum subgenus Leptostemonum), which contains many wild species from the Old World, most of them from Africa (Vorontsova et al., 2013; Vorontsova and Knapp, in press). These wild species could represent a source of variation for developing a new generation of eggplant cultivars with dramatically improved yield and quality, as well as for addressing the challenges posed by adaptation to the climate change. In this respect, resistance and tolerance to several major diseases and pests is found among wild eggplant relatives (Daunay and Hazra, 2012; Rotino et al., 2014) and they can also be found in a wide range of environmental conditions, including desertic and semi-desertic areas, environments with extreme temperatures (Knapp et al., 2013; Vorontsova and Knapp, in press). Some eggplant wild relatives are known to possess high levels of chlorogenic acid and other bioactive compounds of interest for human health (Mennella et al., 2010; Meyer et al., 2015). However, with a few exceptions (Rotino et al., 2014; Liu et al., 2015), eggplant breeders have largely neglected the potential of wild species for eggplant breeding, and contrarily to other crops like tomato (Díez and Nuez, 2008), wild relatives have not made a relevant contribution to the development of new eggplant cultivars.
Eggplant can be crossed with a large number of wild relatives (Daunay and Hazra, 2012; Rotino et al., 2014; Plazas et al., 2016). The closest wild relative of eggplant is S. insanum (Knapp et al., 2013; Vorontsova et al., 2013), which is naturally distributed in Southeast Asia, Madagascar and Mauritius (Knapp et al., 2013; Vorontsova and Knapp, in press), where it is frequently found as a weed (Mutegi et al., 2015). Solanum insanum is considered as the wild ancestor of eggplant and is the only species in the primary genepool of cultivated eggplant (Syfert et al., 2016). Hybrids of eggplant with S. insanum are easily obtained; fruits from interspecific hybridization have many seeds, which have high germination rates, and the hybrid plants are fully fertile (Davidar et al., 2015; Plazas et al., 2016). Interspecific hybrids have also been obtained with many wild species from the secondary genepool (Daunay and Hazra, 2012; Rotino et al., 2014; Plazas et al., 2016), which includes some 50 African and Southeast Asian species (Vorontsova et al., 2013; Syfert et al., 2016). The degree of success of interspecific sexual hybridization between eggplant and secondary genepool species, as well as the hybrid fertility is variable depending on the species involved and the direction of the cross (Plazas et al., 2016).
The characterization of wild species and interspecific hybrids for traits of interest for breeders is a fundamental step for the efficient utilization of crop wild relatives in breeding. Combined data on the cultivated and wild species and their interspecific hybrids, not only allows identifying sources of variation and materials of potential interest, but also provides information on the inheritance of some traits present in the wild species, as has been demonstrated in crosses between S. incanum and eggplant (Prohens et al., 2013). Also, characterization of these materials for vigor traits may allow identification of materials potentially useful as rootstocks. In this respect, highly vigorous eggplant of wild relatives and interspecific hybrids are increasingly used for eggplant grafting, as they induce precocity and higher yield and many of them are tolerant to biotic and abiotic stresses (Gisbert et al., 2011; Daunay and Hazra, 2012). In the case of eggplant wild relatives there are a number of studies on their taxonomic and phylogenetic relationships (Vorontsova et al., 2013; Vorontsova and Knapp, in press), of resistance or tolerance to diseases and pests (Bubici and Cirulli, 2008; Daunay and Hazra, 2012; Naegele et al., 2014). However, to our knowledge there are no comprehensive studies on the morphological and agronomic traits of interest in a set of wild species of the primary and secondary genepools of eggplant and their interspecific hybrids with cultivated eggplant.
Several characterization studies in eggplant with standardized morphological and agronomic descriptors developed by the European Eggplant Genetic Resources Network (EGGNET; van der Weerden and Barendse, 2007) and the International Board for Plant Genetic Resources (IBPGR, 1990) have revealed that are suited for providing a useful morphological and agronomic characterization for eggplant breeders (Prohens et al., 2005; Muñoz-Falcón et al., 2009; Boyaci et al., 2015). EGGNET and IBPGR descriptors have been successfully used for evaluating segregating generations of interspecific crosses between eggplant and related species (Prohens et al., 2012, 2013). In addition to conventional morphological descriptors fruit phenomics data provide eggplant breeders with relevant information for evaluating the variation of the fruit morphology. In this respect, the phenomics tool Tomato Analyzer (Rodríguez et al., 2010) has revealed as useful for the detailed morphometric analysis of fruit size and shape of eggplant and related materials (Prohens et al., 2012; Hurtado et al., 2013).
Here we characterize cultivated eggplant, wild relatives from the primary and secondary genepools and interspecific hybrids between cultivated eggplant and wild relatives using conventional and Tomato Analyzer descriptors. Apart from providing a characterization of the three types of materials studied and their differences, we aim to evaluate the interest for breeding of different wild relatives using characterization data of the wild relatives and of their interspecific hybrids with eggplant. The information obtained may also provide clues on the interest of wild species and hybrids as potential rootstocks for eggplant.
Materials and Methods
Plant Material
The plant material included six accessions of cultivated eggplant (S. melongena), 21 accessions of a total of 12 wild species, and 45 interspecific hybrids between the eggplant accessions and seven of the wild species (Table 1). The eggplant accessions include materials from both the Occidental (Ivory Coast) and Oriental (Sri Lanka) cultivated genepools (Vilanova et al., 2012; Cericola et al., 2013). Among the wild relatives, three accessions belong to the primary genepool (GP1) S. insanum, and 18 accessions to secondary genepool (GP2) species, namely S. anguivi (n = 2), S. campylacanthum (n = 3), S. dasyphyllum (n = 1), S. incanum (n = 1), S. lichtensteinii (n = 2), S. lidii (n = 2), S. linnaeanum (n = 2), S. pyracanthos (n = 1), S. tomentosum (n = 1), S. vespertilio (n = 2), and S. violaceum (n = 1). All the accessions are deposited at the germplasm bank of the Universitat Politècnica de València (València, Spain). The 45 interspecific hybrids were obtained after reciprocal crossings between cultivated eggplant and wild relatives (Plazas et al., 2016) resulting in 18 hybrids between eggplant and primary genepool species and 27 hybrids between eggplant and secondary genepool species (Table 1). Five plants per accession or interspecific hybrid were grown under open field conditions during the summer season of 2015 at the Universitat Politècnica de València (Valencia, Spain; GPS coordinates of the plot: 39° 28′ 55″ N, 0° 22′ 11″ W; altitude 7 m a.s.l.). Plants were spaced 1.2 m between rows and 1.0 m within the row and distributed according to a completely randomized design. Drip irrigation was applied and 80 g plant−1 of a 10N–2.2P–24.9K plus micronutrients fertilizer (Hakaphos Naranja; Compo Agricultura, Barcelona, Spain) was applied during the whole cultivation period through the irrigation system. Plants were trained with bamboo canes and pruned when needed. Weeds were removed manually and no phytosanitary treatments were needed.
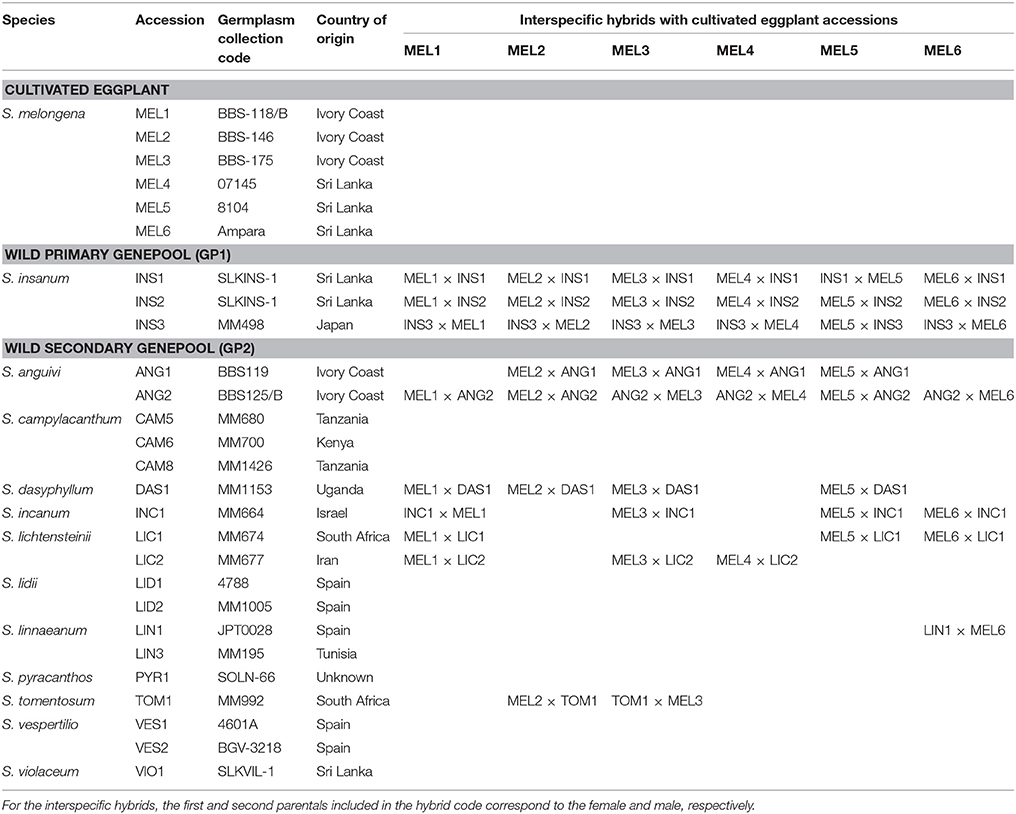
Table 1. Accessions of cultivated eggplant (Solanum melongena) and wild relatives of the primary and secondary genepools, and interspecific hybrids between cultivated eggplant and wild relatives used for the morphological and phenomics characterization.
Characterization
All plants were characterized using 27 conventional morphological descriptors based on EGGNET (van der Weerden and Barendse, 2007) and IBPGR (IBPGR, 1990) descriptors (Table 2). These morphological descriptors describe different traits of the whole plant (4), leaf (7), inflorescence and flower (7) and fruit (9) and in general display limited GxE interaction (IBPGR, 1990). Except for descriptors concerning the whole plant (e.g., plant growth habit), for which one measurement was taken per plant (i.e., one measurement per replicate), five measurements were taken from each individual plant in order to obtain individual plant averages for the conventional morphological descriptors (i.e., five measurements per replicate). Using a similar approach, five fruits per plant (replicate), collected at the commercially ripe stage (i.e., physiologically immature) for cultivated eggplant and at a similar physiological stage (when they had attained full size but was not physiologically mature) in the case of wild species and interspecific hybrids, were cut opened longitudinally and scanned using an HP Scanjet G4010 photo scanner (Hewlett Packard, Palo Alto, CA, USA) at a resolution of 300 dpi. Scanned images were subjected to fruit morphometric analysis with the fruit shape phenomics tool Tomato Analyzer version 4 software (Rodríguez et al., 2010). A total of 20 fruit morphometric descriptors were recorded using this tool (Table 2).
Data Analyses
For each trait, the mean, range and coefficient of variation (CV, %) were calculated using average accession or hybrid values of cultivated eggplant (n = 6), wild relatives (n = 21) and interspecific hybrids (n = 45). Means of each accession or hybrid were subjected to analyses of variance (ANOVA) to detect differences among the three groups considered. Significance of differences among group means was evaluated using the Student-Newman-Keuls multiple range test at P = 0.05. Heterosis over mid parent (H; %) for the traits of greater agronomic importance was studied in the interspecific hybrids using formula H = 100 × ((F1 − MP)/MP), where F1 = hybrid mean, and MP = mean of the parents. Values of H above 100% indicate that the hybrid is superior to the highest parent, and therefore present positive heterosis over the highest parent. Principal components analyses (PCA) were performed using pairwise Euclidean distances among accession or hybrid means for standardized characterization data. All the statistical analyses were performed using the Statgraphics Centurion XVI software (StatPoint Technologies, Warrenton, VA, USA).
Results
Differences between Eggplant, Wild Relatives and Interspecific Hybrids
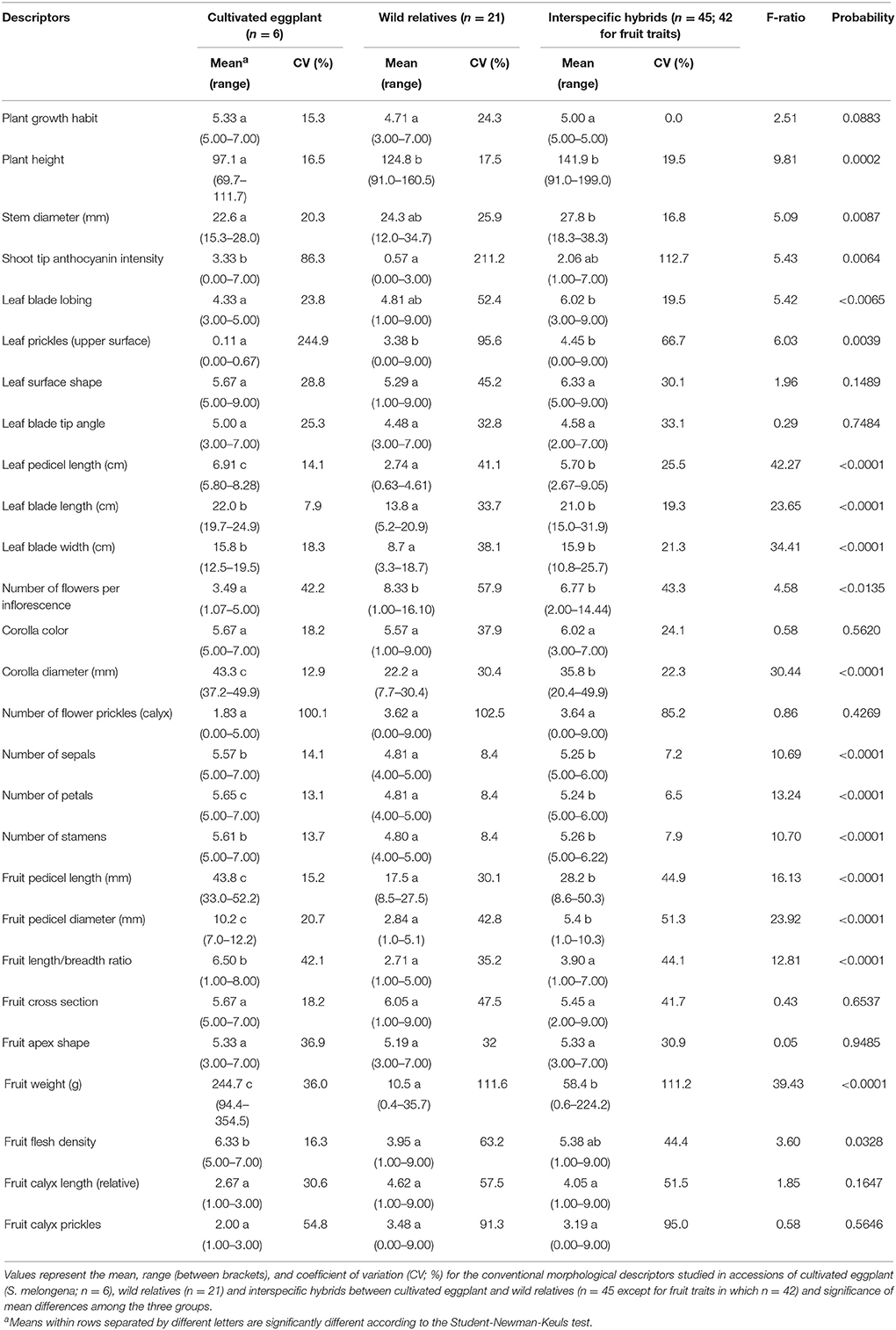
Significant differences (P < 0.05) were found among average values for the groups constituted by cultivated eggplant, wild relatives and interspecific hybrids for 18 out of the 27 conventional descriptors (Table 3). Generally, wild species and interspecific hybrids had larger plant size, greater leaf prickliness, more flowers per inflorescence, and less elongated fruits than the cultivated species. The cultivated species and interspecific hybrids had more anthocyanin pigmentation, larger leaf size, and greater number of flower parts than the wild species. Flower, fruit pedicel and fruit size had the greater average values in the cultivated species, while the smaller ones were for the wild species, with the interspecific hybrids having intermediate values. The three groups overlap for all conventional descriptors except for Leaf Pedicel Length, Corolla Diameter, Fruit Pedicel Length, Fruit Pedicel Diameter, and Fruit Weight, in which all the accessions of the cultivated species presented higher values than any of the wild species.
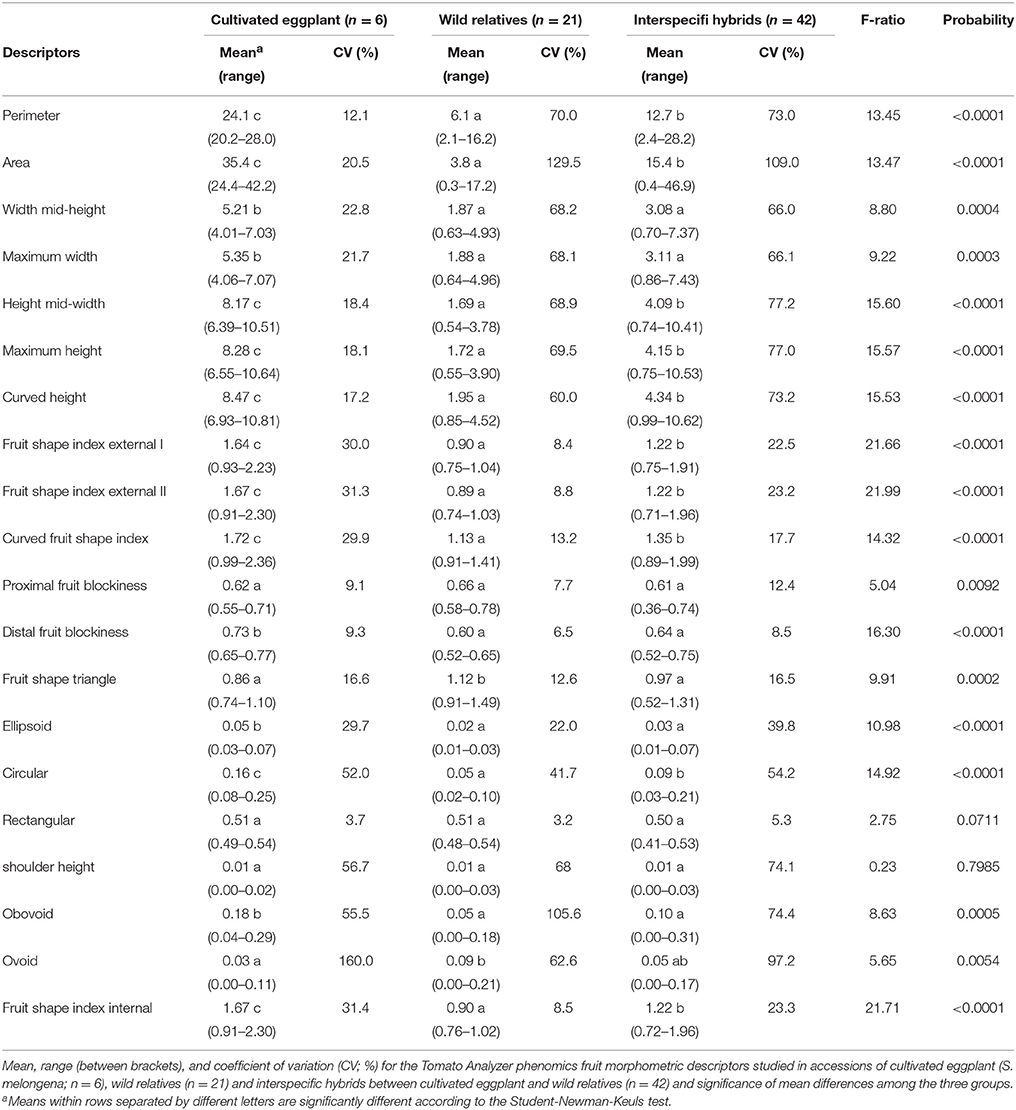
All Tomato Analyzer descriptors evaluated, except two (Rectangular and Shoulder Height) displayed significant (P < 0.05) differences among average values for the three groups (Table 4). For the eight Tomato Analyzer descriptors related to fruit size the cultivated eggplant presented significantly higher values than wild species, while for Ovoid it had lower values; interspecific hybrids presented intermediate values, in most cases being significantly different from both cultivated eggplant and wild species (Table 4). Cultivated eggplant had greater Distal Fruit Blockiness and Ellipsoid values than either wild species or interspecific hybrids, while wild species had higher values for Triangular than either cultivated species or interspecific hybrids. Similarly to conventional descriptors, the three groups overlap in the ranges of variation for all Tomato Analyzer descriptors except for Perimeter, Area, Height Mid-width, Maximum Height, Curved Height and Circular, in which there is no overlap between the range of variation of cultivated and wild species, with the values of the former being larger than those of the latter (Table 4).
Variation in Eggplant, Wild Relatives, and Interspecific Hybrids
Variation for the conventional and Tomato Analyzer descriptors was found in the materials studied (Tables 3, 4; Figure 1). For most traits, more variation both in terms of range and CV was found in the wild species, compared to the cultivated eggplant accessions. For all conventional descriptors there was more variation in the wild species than in the cultivated eggplant, except for Shoot Tip Anthocyanin Intensity, the number of flower parts. Conversely, in the case of Tomato Analyzer descriptors, the range of variation was greater in wild species than in the cultivated eggplant for only six out of the 20 descriptors evaluated (Perimeter, Width Mid-height, Maximum Width, Rectangular, and Ovoid), while for the CV the wild species had a greater value than cultivated eggplant for nine of the descriptors, of which seven are related to fruit size (Table 4).
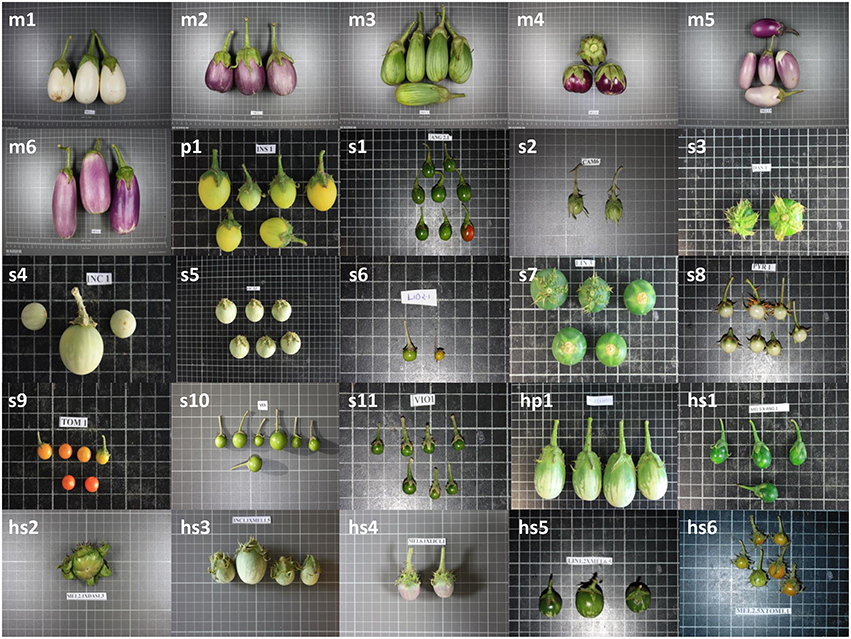
Figure 1. Fruit samples of the materials used. This include: Cultivated eggplant (S. melongena) accessions MEL1 (m1) to MEL6 (m6); wild species of primary genepool S. insanum (p1); wild species of secondary genepool S. anguivi (s1), S. campylacanthum (s2), S. dasyphyllum (s3), S. incanum (s4), S. lichtensteinii (s5), S. lidii (s6), S. linnaeanum (s7), S. pyracanthos (s8), S. tomentosum (s9), S. vespertilio (s10), and S. violaceum (s11); interspecific hybrids between eggplant and primary genepool species S. insanum (hp1); and, interspecific hybrids between eggplant and secondary genepool species S. anguivi (hs1), S. dasyphyllum (hs2), S. incanum (hs3), S. lichtensteinii (hs4), S. linnaeanum (hs5), and S. tomentosum (hs6). Fruits are not depicted at the same scale; the size of the grid cells is 1 × 1cm.
For interspecific hybrids a large range of variation was observed for many conventional descriptors, with variation parameters generally larger than those of the cultivated species and smaller than those of the wild species. In this respect, the range of variation was larger than that of the cultivated eggplant for all but nine conventional descriptors, while compared to wild species it was larger for 11 descriptors (Table 3). The coefficient of variation for conventional descriptors was also larger than in the cultivated species for all traits except nine (Plant Growth Habit, Stem Diameter, Leaf Blade Lobing, Leaf Prickles, Number of Flower Prickles, Number of Sepals, Number of Petals, Number of Stamens, and Fruit Apex Shape) and larger than that of the wild species for eight descriptors (Plant Height, Leaf Blade Tip Angle, Fruit Pedicel Length, Fruit Pedicel Diameter, Fruit Length/Breadth Ratio, Fruit Cross Section, Fruit Apex Shape, and Fruit Calyx Prickles; Table 3).
Regarding the variation for Tomato Analyzer traits, the range of variation in the interspecific hybrids was greater than those of cultivated eggplant and wild species for all traits except five in the case of cultivated eggplant, which correspond to fruit shape indexes and Circular, and only one (Ovoid) in the case of wild species (Table 4). Also, larger values were obtained in the CV for Tomato Analyzer descriptors in the interspecific hybrids compared to the cultivated species for all traits but seven. When compared to wild species the interspecific hybrids also presented higher CV for all traits, except four (Table 4).
Multivariate Analysis
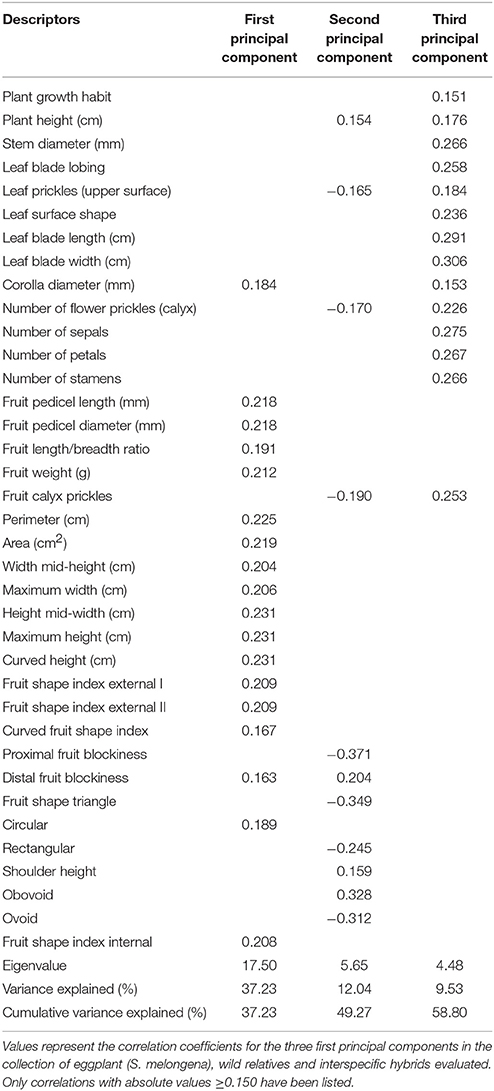
The three first components of the principal components analysis made with all conventional and Tomato Analyzer descriptors accounted for 58.8% accounted of the total variation among accession means, with the first, second and third component accounting, respectively for 37.2, 12.0, and 9.5% of the total variation (Table 5). The first principal component was positively correlated to Corolla diameter, fruit size and to elongated fruit shape (Table 5). The second principal component was positively correlated to Plant Height and to obovoid fruit shape. The third principal component was positively correlated to Plant Growth Habit (i.e., prostrate habit), to multiple plant, leaf and corolla size traits, to a higher number of flower parts (sepals, petals and stamens) and to an increased prickliness in leaves, and flower and fruit calyces (Table 5).
The projection of eggplant, wild species and interspecific hybrids in the PCA plot reveals that although considerable diversity exists in both eggplant (black squares) and wild species (white symbols), the interspecific hybrids (gray symbols) present a more scattered distribution in the PCA plot (Figures 2, 3). Interspecific hybrids with the primary genepool species S. insanum plot closer to the cultivated eggplant and are intermingled with it the PCA graphs. On the contrary, interspecific hybrids with secondary genepool species plot closer to the wild species and are also intermingled with them (Figures 2, 3). The first component separates the group formed by eggplant and the interspecific hybrids with the primary genepool species S. insanum, which present positive values for this component, from the group formed by all the wild species and interspecific hybrids with secondary genepool species. Among the interspecific hybrids with secondary genepool species, those with S. incanum and S. lichtensteinii are the closest to eggplant in this first component (Figures 2, 3). When considering the second component all eggplant accessions but one have positive values, while interspecific hybrids with S. insanum are equally distributed in the positive and negative values of this second component (Figure 2). Primary genepool wild species S. insanum and all secondary genepool species, except S. campylacanthum, S. pyracanthos, S. tomentosum and one accession of each of S. anguivi and S. lidii have negative values for this second component. When considering interspecific hybrids with secondary genepool species, although they are intermingled with the wild species for this second component most of the hybrids present positive values for this second component, with the exceptions being the hybrids with S. lichtensteinii (four out of five), S. linnaeanum and one of each of the interspecific hybrids with each of the species S. anguivi and S. incanum (this latter with a value very close to 0). Amazingly, the highest values for this second component correspond to interspecific hybrids with S. anguivi (Figure 2). For the third component both eggplant and the interspecific hybrids with S. insanum are scattered and display positive or negative values (Figure 3). Most wild species accessions have negative values for this third component, except the accessions of S. dasyphyllum, S. linnaeanum, S. pyracanthos, and S. violaceum, as well as one accession of S. incanum (with values close to 0). The lowest values for this component are those of S. lidii, S. vespertilio and S. tomentosum (Figure 3). On the other hand all interspecific hybrids with secondary genepool species, with the exception of two interspecific hybrids with S. anguivi, present positive values for this third component. In this case, the highest values for the third component correspond to interspecific hybrids with S. dasyphyllum, S. lichtensteinii, and S. incanum (Figure 3).
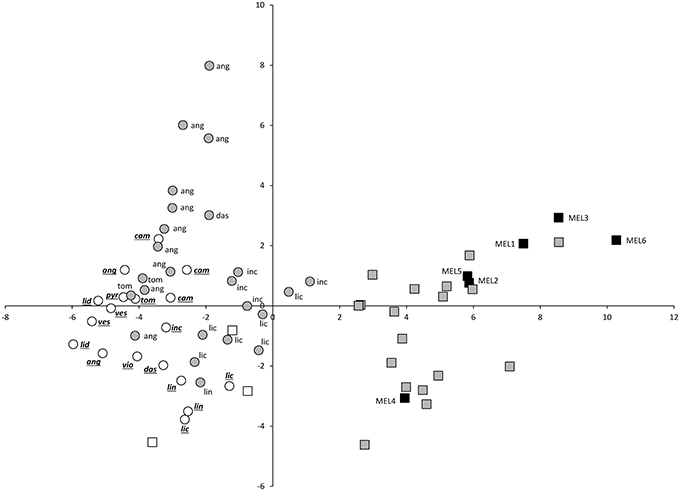
Figure 2. First (X-axis) and second (Y-axis) principal components (37.2 and 12.0% of the total variation explained, respectively) scatterplot of cultivated eggplant, wild relatives and interspecific hybrids based on 27 conventional and 20 Tomato Analyzer morphological descriptors. Cultivated eggplant (S. melongena) is represented by black squares and the “MEL” accession code, primary genepool species S. insanum by white squares, interspecific hybrids between eggplant and S. insanum by gray squares, secondary genepool species by white circles (with species codes in underlined italics), and interspecific hybrids between eggplant and secondary genepool species by gray circles (with wild species codes in normal font). For secondary genepool species and their hybrids with eggplant, the following codes are used: ang (S. anguivi), cam (S. campylacanthum), das (S. dasyphyllum), inc (S. incanum), lic (S. lichtensteinii), lid (S. lidii), lin (S. linnaeanum), pyr (S. pyracanthos), tom (S. tomentosum), ves (S. vespertilio), vio (S. violaceum).
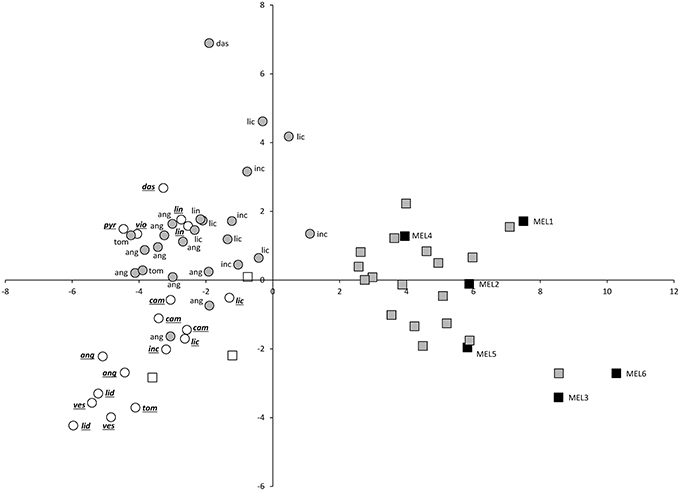
Figure 3. First (X-axis) and thrid (Y-axis) principal components (37.2 and 9.5% of the total variation explained, respectively) scatterplot of cultivated eggplant, wild relatives and interspecific hybrids based on 27 conventional and 20 Tomato Analyzer morphological descriptors. Cultivated eggplant (S. melongena) is represented by black squares and the “MEL” accession code, primary genepool species S. insanum by white squares, interspecific hybrids between eggplant and S. insanum by gray squares, secondary genepool species by white circles (with species codes in underlined italics), and interspecific hybrids between eggplant and secondary genepool species by gray circles (with wild species codes in normal font). For secondary genepool species and their hybrids with eggplant, the following codes are used: ang (S. anguivi), cam (S. campylacanthum), das (S. dasyphyllum), inc (S. incanum), lic (S. lichtensteinii), lid (S. lidii), lin (S. linnaeanum), pyr (S. pyracanthos), tom (S. tomentosum), ves (S. vespertilio), vio (S. violaceum).
Traits of Agronomic Interest in Wild Species
The 12 wild species evaluated presented considerable differences for traits of agronomic interest (Table 6). For example, important differences were found for vegetative traits. For example, the tallest plants were those of S. anguivi, which also presented thick stems (Table 6). Important differences were also found for Leaf Blade Lobing. The greatest leaf prickliness was observed S. dasyphyllum, S. pyracanthos, and S. violaceum, while S. anguivi and S. tomentosusm did not present prickles in the leaves. The largest leaf blades were those of S. dasyphyllum and S. campylacanthum, while the smallest were those of S. tomentosum, with a Leaf Blade Length of 5.2 cm (Table 6). When considering flower and fruit traits, the two species with a larger number of flowers per inflorescence were S. lidii and S. vespertilio, with more than 13 flowers/inflorescence, while the smaller number was S. insanum (Table 6). Important differences were also observed for Corolla Color. All wild species had five petals (and sepals and stamens), except S. lidii and S. vespertilio, which had only four. The largest fruits were those of S. incanum and S. lichtensteinii, with average values above 25 g, more than 10-fold heavier than those of S. anguivi, S. lidii, S. pyracanthos, S. tomentosum, S. vespertilio, and S. violaceum. The highest calyx prickliness was observed in S. linnaeanum, S. pyracanthos, and S. violaceum, while S. anguivi, S. lidii, and S. vespertilio did not present calyx prickles (Table 6). The most elongated fruit were those of S. incanum, while the most flattened ones were those of S. dasyphyllum and S. lidii (Table 6).
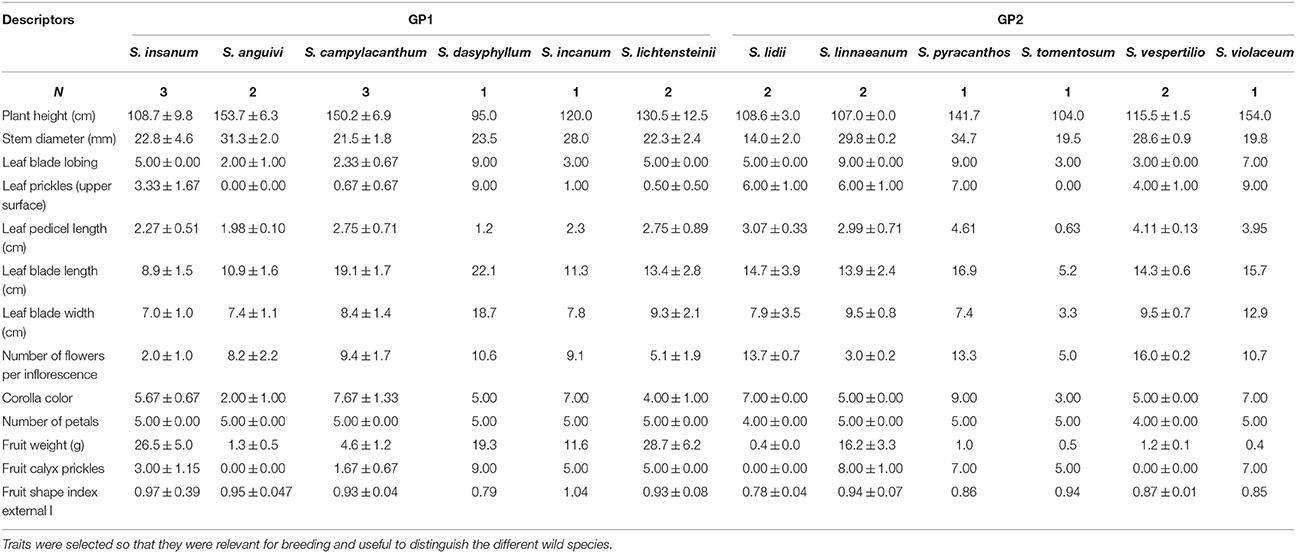
Table 6. Average (±SE) values based on accession means for selected traits in the 12 wild species (one from the primary genepool GP1, S. insanum; and 11 from the secondary genepool GP2) evaluated.
Heterosis in Interspecific Hybrids
Interspecific hybrids between eggplant and its wild relatives generally displayed positive heterosis for plant size traits, with average heterosis values of up to 90.5% for Plant height and 46.2% for Stem diameter in the hybrids of eggplant with S. dasyphyllum (Table 7). The only negative value observed for these traits was for Stem Diameter in the interspecific hybrid with S. linnaeanum. Most interspecific hybrids presented higher prickliness than their parent species, and in consequence, very high average values for heterosis for Leaf Prickles are observed, with values between 91.0% for S. dasyphyllum and 800.0% for S. tomentosum. Leaf size traits were also, in general, heterotic in the interspecific hybrids, with the exception of Leaf Pedicel Length in S. dasyphyllum and S. linnaeanum. The same phenomenon was observed for the Number of Flowers per Inflorescence, with values of up to 87.7% in the hybrids with S. tomentosum (Table 7). The pigmentation of the corolla (Corolla Color) also presented average positive heterosis values in the hybrids of eggplant with five out of the seven wild species, the exception being interspecific hybrids with S. anguivi and S. tomentosum. The number of flower parts, represented by the Number of Petals, displayed low absolute values for heterosis in all cases (Table 7).
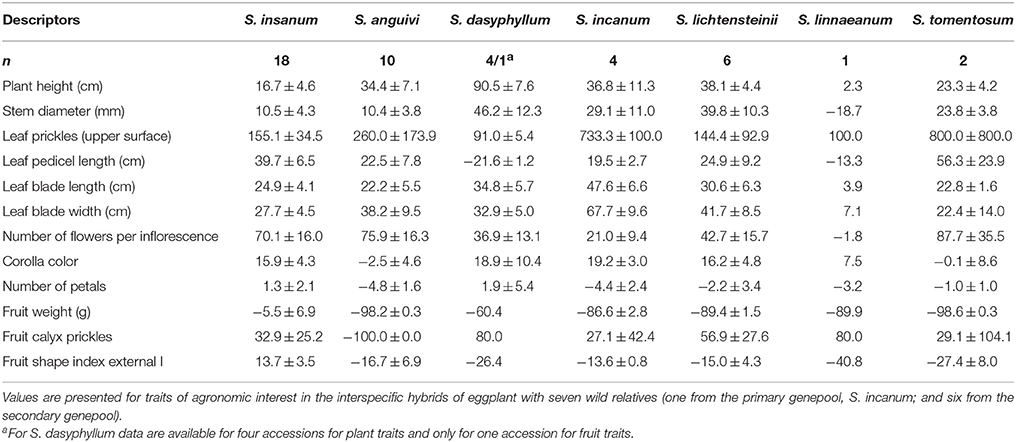
Table 7. Heterosis over mid parent values (%; ±SE) based on accession and interspecific hybrid means.
Regarding Fruit Weight, considerable differences were observed between the hybrids with the primary genepool species (S. insanum) on one hand, and the hybrids with secondary genepool species on the other. In this respect, while the hybrids with S. insanum displayed small negative average heterosis (−5.5%), not significantly different from 0, in the case of secondary genepool species, the heterosis for Fruit Weight is highly negative, with values between −60.4% for hybrids with S. dasyphyllum to −98.6% in hybrids with S. tomentosum (Table 7). As occurred for Leaf Prickles, positive heterosis values, although of smaller magnitude, were observed for Fruit Calyx Prickles, with the exception of the hybrids with S. anguivi, which did not present prickles in the calyx, and in consequence had a heterosis value of −100%. Finally, for fruit shape, the hybrids with primary genepool species S. insanum presented positive heterosis, while those with secondary genepool species had negative heterosis values (Table 7).
Discussion
Crop wild relatives are widely recognized as an invaluable genetic resource for breeding, in particular for broadening the genetic base of crops with narrow genetic diversity, and as sources of variation for traits of interest in breeding crops, including adapting them to the challenges posed by climate change (Dempewolf et al., 2014). Modern varieties of many important crops carry introgressions from wild species resulting from breeding programmes performed in the last 100 years (Hajjar and Hodgkin, 2007). One of the most outstanding examples is tomato, where modern commercial hybrids carry different combinations of 15 different introgressions from different wild species (Díez and Nuez, 2008; Sabatini et al., 2013). However, in the case of eggplant, despite being one of the most important vegetables and being intercrossable with many wild relatives, there are few reports on the use of the variation available in the wild species for eggplant breeding (Daunay and Hazra, 2012; Rotino et al., 2014; Liu et al., 2015) and no modern commercial varieties of eggplant carrying introgressions from wild species are known to us.
In our study we have evaluated six accessions of cultivated eggplant, 21 accessions of 12 wild species, and 45 interspecific hybrids of cultivated eggplant with seven wild species. This represents the largest study up to now on morphological and agronomic traits for breeding of this type of materials. As expected, many differences were found within and among cultivated eggplant, wild relatives and the interspecific hybrids for the conventional descriptors used, confirming the utility of the EGGNET (van der Weerden and Barendse, 2007) and IBPGR (1990) conventional morphological descriptors and Tomato Analyzer traits (Rodríguez et al., 2010) used for evaluating eggplant wild relatives and interspecific hybrids (Prohens et al., 2013).
Also, many differences were found for the traits studied among cultivated eggplant, wild species and interspecific hybrids. Although many of the wild species of eggplant thrive in arid and semi-arid conditions (Knapp et al., 2013; Vorontsova and Knapp, in press), when grown under the favorable conditions of cultivated environments, the wild species and their interspecific hybrids generally display a high vigor, expressed as average values for plant height and stem diameter above those of cultivated eggplant. This is of interest for developing new rootstocks, which generally require having high vigor (Gisbert et al., 2011), and opens the way to exploiting several to the wild species evaluated and interspecific hybrids as potential new rootstocks for eggplant. Another important trait of agronomic interest for which there were considerable differences among groups was prickliness, which was much greater in wild species and interspecific hybrids, confirming that alleles from the cultivated eggplant are recessive (Doganlar et al., 2002; Gramazio et al., 2014; Portis et al., 2015). The number of flowers per inflorescence was also much greater in wild species and interspecific hybrids. This trait is very important in eggplant breeding, as a reduced value of this trait results in increased fruit size uniformity (Sękara and Bieniasz, 2008). Also, fruit size and shape, which are of great relevance for breeding (Daunay and Hazra, 2012; Portis et al., 2015), also differed considerably among the three groups, with the interspecific hybrids presenting intermediate values, although on most cases they were closer to those of the wild species, indicating dominance of the genes of the latter (Doganlar et al., 2002).
The much higher variation observed in wild species and interspecific hybrids for vegetative, flower and inflorescence traits compared to cultivated eggplant was expected, as we were comparing a single species with an admixture of different wild species or hybrids, which present a much higher genetic diversity (Meyer et al., 2012; Särkinen et al., 2013; Vorontsova et al., 2013). However, for traits related to the fruit size and shape much higher variation was observed in the cultivated eggplant than in the wild species, confirming the general observation that the morphological variation in the organ for which a crop is domesticated (in this case the fruit) increases during domestication (Meyer and Purugganan, 2014). Amazingly, in the case of interspecific hybrids a larger variation was found for most fruit size and shape traits than in the cultivated eggplant. Although most interspecific hybrids were more similar to the wild species, in some cases they were intermediate, revealing that different genic control mechanisms must exist for fruit size and shape among the wild relatives of eggplant. In this respect, the multivariate analysis clearly shows that interspecific hybrids with the primary genepool species S. insanum are morphologically closer to the cultivated eggplant, while the hybrids with secondary genepool species present a general morphology closer to that of the wild species. These results may support the hypothesis that S. insanum is the wild ancestor of cultivated eggplant (Knapp et al., 2013), as domestication should be easier when genes for domestication traits from the wild species display intermediate dominance rather than full dominance.
The study of individual wild species suggests that S. anguivi, S. campylacanthum, S. pyracanthos, and S. violaceum may be of interest for increasing the vigor of cultivated eggplant or for being used as rootstocks. Also, wild eggplant species use to have undesirable traits (e.g., prickliness, small fruit size, etc.) that have to be removed during the breeding (Rotino et al., 2014). In this case, the most desirable wild species are those that are most similar to the crop for these traits. For example, the lack of prickles or very low prickliness of S. anguivi, S. campylacanthum, and S. tomentosum is a very favorable trait for breeders (Daunay and Hazra, 2012). Regarding fruit weight, the wild species with greater fruit weight should be the most interesting for breeders in order to recover fruit size in few backcross generations. In this case, S. insanum, S. dasyphyllum, and S. lichtensteinii should be the most interesting candidates if a rapid recovery of fruit size is desired. In any case, Prohens et al. (2013) showed that fruit size recovers quickly even in first backcrosses with the wild species S. incanum, which has an intermediate fruit size among wild species.
Although differences were observed among interspecific hybrids from different wild species, hybrids were in general vigorous, displaying heterosis for vigor traits. This phenomenon had already been described in interspecific hybrids with S. incanum (Gisbert et al., 2011; Prohens et al., 2013), and our results suggest that this is a common phenomenon in the hybrids between eggplant and wild relatives. Amazingly, most interspecific hybrids were highly heterotic for prickliness, with heterosis values over 100%. Prickles even appeared in interspecific hybrids with wild species that were not prickly, like S. tomentosum. In previous works, heterosis for prickliness had already been described in interspecific crosses in eggplant (Prohens et al., 2012; Devi et al., 2015; Plazas et al., 2016). Several studies with segregating populations of S. linnaeanum and S. insanum show that differences in prickliness between cultivated eggplant and wild relatives is under the control of a few QTL (Doganlar et al., 2002; Gramazio et al., 2014) and therefore prickliness should be easily removed in backcross generations. Although for fruit size traits negative heterosis was generally observed in the interspecific hybrids, indicating a greater similarity to the wild species, interspecific hybrids with primary genepool species S. insanum presented values close to zero, similarly to intraspecific hybrids of eggplant (Rodríguez-Burruezo et al., 2008), indicating intermediate dominance and values intermediate between both parental species. However, hybrids with wild species from the secondary genepool displayed highly negative heterosis, in some cases close to 100% like in interspecific hybrids with S. anguivi and S. tomentosum, suggesting that in these materials it may be more difficult to recover fruit size in the backcross generations.
In conclusion, the characterization with conventional descriptors and the Tomato Analyzer phenomics tool has allowed a detailed characterization of eggplant, close wild relatives and their interspecific hybrids. The high variation among wild species identified sources of variation and most promising species for traits of interest for eggplant breeding. The fact that interspecific hybrids with primary genepool species S. insanum are intermediate or close to eggplant for many traits, may facilitate the use of this species in introgression breeding and supports previous evidence that this species is the ancestor of cultivated eggplant. Also, the high vigor of most interspecific hybrids may be directly exploited by using them as rootstocks. The information obtained here on phenotypic characteristics and heterosis of wild species and interspecific hybrids is of interest for eggplant breeding. Given the adaptation of many wild species to stressful conditions, their utilization in eggplant breeding may result in the development of a new generation of cultivars adapted to climate change challenges.
Author Contributions
JP, SV, PG, and MP conceived and designed the research; PK and MP performed the phenotypic and phenomics characterization; PK, JP, and PG analyzed the data; JP, SV, PG, and MP wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was undertaken as part of the initiative “Adapting Agriculture to Climate Change: Collecting, Protecting and Preparing Crop Wild Relatives” which is supported by the Government of Norway. The project is managed by the Global Crop Diversity Trust with the Millennium Seed Bank of the Royal Botanic Gardens, Kew and implemented in partnership with national and international gene banks and plant breeding institutes around the world. For further information see the project website: http://www.cwrdiversity.org/. This work has also been funded in part by European Union's Horizon 2020 research and innovation programme under grant agreement No 677379 (G2P-SOL) and from Spanish Ministerio de Economía y Competitividad and FEDER (grant AGL2015-64755-R). Prashant Kaushik is grateful to ICAR for a pre-doctoral grant. Pietro Gramazio is grateful to Universitat Politècnica de València for a pre-doctoral (Programa FPI de la UPV-Subprograma 1/2013 call) contract.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Boyaci, H. F., Topcu, V., Tepe, A., Yildirim, I. K., Oten, M., and Aktas, A. (2015). Morphological and molecular characterization and relationships of Turkish local eggplant heirlooms. Not. Bot. Horti Agrobot. Cluj Napoca 43, 100–107. doi: 10.15835/nbha4319773
Bubici, G., and Cirulli, M. (2008). Screening and selection of eggplant and wild related species for resistance to Leveillula taurica. Euphytica 164, 339–345. doi: 10.1007/s10681-008-9663-z
Cericola, F., Portis, E., Toppino, L., Barchi, L., Acciarri, N., Ciriaci, T., et al. (2013). The population structure and diversity of eggplant from Asia and the Mediterranean basin. PLOS ONE 8:e73702. doi: 10.1371/jounal.pone.0073702
Daunay, M. C., and Hazra, P. (2012). “Eggplant,” in Handbook of Vegetables, eds K. V. Peter and P. Hazra (Houston, TX: Studium Press), 257–322.
Davidar, P., Snow, A. A., Rajkumar, M., Pasquet, R., Daunay, M. C., and Mutegi, E. (2015). The potential for crop to wild hybridization in eggplant (Solanum melongena, Solanaceae) in southern India. Am. J. Bot. 102, 129–139. doi: 10.3732/ajb.1400404
Devi, C. P., Munshi, A. D., Behera, T. K., Choudhary, H., Vinod, Gurung, B., et al. (2015). Cross compatibility in interspecific hybridization of eggplant, Solanum melongena, with its wild relatives. Sci. Hort. 193, 353–358. doi: 10.1016/j.scienta.2015.07.024
Dempewolf, H., Eastwood, R. H., Guarino, L., Khoury, C. K., Müller, J. V., and Toll, J. (2014). Adapting agriculture to climate change: a global initiative to collect, conserve, and use crop wild relatives. Agroecol. Sustain. Food 38, 369–377. doi: 10.1080/21683565.2013.870629
Díez, M. J., and Nuez, F. (2008). “Tomato,” in Handbook of Plant Breeding: Vegetables II, eds J. Prohens and F. Nuez (New York, NY: Springer), 249–323.
Doganlar, S., Frary, A., Daunay, M. C., Lester, R. N., and Tanksley, S. (2002). Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161, 1713–1726. doi: 10.1007/s10681-014-1234-x
FAO (2016). FAOSTAT Production Databases. Available online at: http://faostat3.fao.org/home/E (Accessed 11 February, 2016).
Gisbert, C., Prohens, J., Raigón, M. D., Stommel, J. R., and Nuez, F. (2011). Eggplant relatives as sources of variation for developing new rootstocks: effects of grafting on eggplant yield and fruit apparent quality and composition. Sci. Hortic. 128, 14–22. doi: 10.1016/j.scienta.2010.12.007
Gramazio, P., Prohens, J., Plazas, M., Andújar, I., Herraiz, F. J., Castillo, E., et al. (2014). Location of chlorogenic acid biosynthesis pathway and polyphenol oxidase genes in a new interspecific anchored linkage map in eggplant. BMC Plant Biol. 14:350. doi: 10.1186/s12870-014-0350-z
Hajjar, R., and Hodgkin, T. (2007). The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156, 1–13. doi: 10.1007/s10681-007-9363-0
Hurtado, M., Vilanova, S., Plazas, M., Gramazio, P., Herraiz, F. J., Andújar, I., et al. (2013). Phenomics of fruit shape in eggplant (Solanum melongena L.) using Tomato Analyzer software. Sci. Hortic. 164, 625–632. doi: 10.1016/j.scienta.2013.10.028
Knapp, S., Vorontsova, M. S., and Prohens, J. (2013). Wild relatives of the eggplant (Solanum melongena L.: Solanaceae): new understanding of species names in a complex group. PLoS One 8:e57039. doi: 10.1371/journal.pone.0057039
Liu, J., Zheng, Z. S., Zhou, X. H., Feng, C., and Zhuang, Y. (2015). Improving the resistance of eggplant (Solanum melongena) to Verticillium wilt using wild species Solanum linnaeanum. Euphytica 201, 463–469. doi: 10.1007/s10681-014-1234-x
Mennella, G., Lo Scalzo, R., Fibiani, M., D'Alessandro, A., Francese, G., Toppino, L., et al. (2010). Chemical and bioactive quality traits during fruit ripening in eggplant (S. melongena L.) and allied species. J. Agric. Food Chem. 47, 11821–11831. doi: 10.1021/jf3037424
Meyer, R. S., Karol, K. G., Little, D. P., Nee, M. H., and Litt, A. (2012). Phylogeographic relationships among Asian eggplants and new perspectives on eggplant domestication. Mol. Phylogenet. Evol. 63, 685–701. doi: 10.1016/j.ympev.2012.02.006
Meyer, R. S., and Purugganan, M. D. (2014). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. doi: 10.1038/nrg3605
Meyer, R. S., Whitaker, B. D., Little, D. P., Wu, S. B., Kennelly, E. J., Long, C. L., et al. (2015). Parallel reductions in phenolic constituents resulting from the domestication of eggplant. Phytochemistry 115, 194–206. doi: 10.1016/j.phytochem.2015.02.006
Muñoz-Falcón, J. E., Prohens, J., Vilanova, S., and Nuez, F. (2009). Diversity in commercial varieties and landraces of black eggplants and implications for broadening the breeders' gene pool. Ann. Appl. Biol. 154, 453–465. doi: 10.1111/j.1744-7348.2009.00314.x
Mutegi, E., Snow, A. A., Rajkumar, M., Pasquet, R., Ponniah, H., Daunay, M. C., et al. (2015). Genetic diversity and population structure of wild/weedy eggplant (Solanum insanum, Solanaceae) in southern India: implications for conservation. Am. J. Bot. 102, 140–148. doi: 10.3732/ajb.1400403
Naegele, R. P., Boyle, S., Quesada-Ocampo, L. M., and Hausbeck, M. K. (2014). Genetic diversity, population structure, and resistance to Phytophthora capsici of a worldwide collection of eggplant germplasm. PLOS ONE 9:e95930. doi: 10.1371/journal.pone.095930
Niño-Medina, G., Muy-Rangel, D., Gardea-Béjar, A., González-Aguilar, G., Heredia, B., Báez-Sanudo, M., et al. (2014). Nutritional and nutraceutical components of commercial eggplant types grown in Sinaloa, Mexico. Not. Bot. Horti Agrobot. Cluj Napoca 42, 538–544. doi: 10.15835/nbha4229573
Plazas, M., Vilanova, S., Gramazio, P., Rodríguez-Burruezo, A., Fita, A., Herraiz, F. J., et al. (2016). Interspecific hybridization between eggplant and wild relatives from different genepools. J. Am. Soc. Hortic. Sci. 141, 34–44.
Portis, E., Cericola, F., Barchi, L., Toppino, L., Acciarri, N., Pulcini, L., et al. (2015). Association mapping fro fruit, plant and leaf morphology traits in eggplant. PLOS ONE 10:e0135200. doi: 10.1371/jounal.pone.0135200
Prohens, J., Blanca, J. M., and Nuez, F. (2005). Morphological and molecular variation in a collection of eggplants from a secondary center of diversity: implications for conservation and breeding. J. Am. Soc. Hort. Sci. 130, 54–63.
Prohens, J., Plazas, M., Raigón, M. D., Seguí-Simarro, J. M., Stommel, J. R., and Vilanova, S. (2012). Characterization of interspecific hybrids and first backcross generations from crosses between two cultivated eggplants (Solanum melongena and S. aethiopicum Kumba group) and implications for eggplant breeding. Euphytica 186, 517–538. doi: 10.1007/s10681-012-0652-x
Prohens, J., Whitaker, B. D., Plazas, M., Vilanova, S., Hurtado, M., Blasco, M., et al. (2013). Genetic diversity in morphological characters and phenolic acids content resulting from an interspecific cross between eggplant, Solanum melongena, and its wild ancestor (S. incanum). Ann. Appl. Biol. 162, 242–257. doi: 10.1111/aab.12017
Rodríguez, G. R., Moyseenko, J. B., Robbins, M. D., Huarachi Morejón, N., Francis, D. M., and van der Knaap, E. (2010). Tomato analyzer: a useful software application to collect accurate and detailed morphological and colorimetric data from two-dimensional objects. J. Vis. Exp. 37:e1856. doi: 10.3791/1856
Rodríguez-Burruezo, A., Prohens, J., and Nuez, F. (2008). Performance of hybrids between local varieties of eggplant (Solanum melongena) and its relation to the mean of parents and to morphological and genetic distances among parents. Eur. J. Hortic. Sci. 73, 76–83.
Rotino, G. L., Sala, T., and Toppino, L. (2014). “Eggplant,” in Alien Gene Transfer in Crop Plants, Vol. 2, eds A. Pratap and J. Kumar (New York, NY: Springer), 381–409.
Sabatini, E., Beretta, M., Sala, T., Acciarri, N., Milc, J., and Pecchioni, N. (2013). “Molecular breeding,” in Genetics, Genomics and Breeding of Tomato, eds B. E. Liedl, J. A. Labate, J. R. Stommel, A. Slade, and C. Kole (Boca Raton, FL: CRC Press), 228–303.
San José, R., Sánchez-Mata, M. C., Cámara, M., and Prohens, J. (2014). Eggplant fruit composition as affected by the cultivation environment and genetic constitution. J. Sci. Food Agric. 94, 2774–2784. doi: 10.1002/jsfa.6623
Särkinen, T., Bohs, L., Olmstead, R. G., and Knapp, S. (2013). A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol. Biol. 13:214. doi: 10.1186/1471-2148-13-214
Sękara, A., and Bieniasz, M. (2008). Pollination, fertilization and fruit formation in eggplant (Solanum melongena L.). Acta Agrobot. 61, 107–113. doi: 10.5586/aa.2008.014
Syfert, M. M., Castañeda-Álvarez, N. P., Khoury, C. K., Särkinen, T., Sosa, C. C., Achicanoy, H. A., et al. (2016). Crop wild relatives of the brinjal eggplant (Solanum melongena): poorly represented in genebanks and many species at risk of extinction. Am. J. Bot. 103, 635–651. doi: 10.3732/ajb.1500539
van der Weerden, G. M., and Barendse, G. W. M. (2007). A web-searchable database developed for the EGGNET project and applied to the Radboud University Solanaceae database. Acta Hortic. 745, 503–506. doi: 10.17660/actahortic.2007.745.37
Vilanova, S., Manzur, J. P., and Prohens, J. (2012). Development and characterization of genomic SSR markers in eggplant and their application to the study of diversity and relationships in a collection of different cultivar types and origins. Mol. Breed. 30, 647–660. doi: 10.1007/s11032-011-9650-2
Vorontsova, M. S., and Knapp, S. (in press). A revision of the spiny solanums, Solanum subgenus Leptostemonum (Solanaceae) in Africa Madagascar. Syst. Bot Monogr.
Keywords: descriptors, genepools, intespecific hybrids, introgression breeding, phenomics, Solanum melongena, Tomato Analyzer
Citation: Kaushik P, Prohens J, Vilanova S, Gramazio P and Plazas M (2016) Phenotyping of Eggplant Wild Relatives and Interspecific Hybrids with Conventional and Phenomics Descriptors Provides Insight for Their Potential Utilization in Breeding. Front. Plant Sci. 7:677. doi: 10.3389/fpls.2016.00677
Received: 30 March 2016; Accepted: 02 May 2016;
Published: 19 May 2016.
Edited by:
Rodomiro Ortiz, Swedish University of Agricultural Sciences, SwedenReviewed by:
Michael Benjamin Kantar, University of Hawaii, USAEzio Portis, DISAFA–University of Torino, Italy
Copyright © 2016 Kaushik, Prohens, Vilanova, Gramazio and Plazas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaime Prohens, jprohens@btc.upv.es
 Prashant Kaushik
Prashant Kaushik Jaime Prohens
Jaime Prohens Santiago Vilanova
Santiago Vilanova Pietro Gramazio
Pietro Gramazio Mariola Plazas
Mariola Plazas