- 1Department of Chemistry and Geochemistry, Colorado School of Mines, Golden, CO, USA
- 2Department of Plant Biology, Carnegie Institution for Science, Stanford, CA, USA
The isolation or engineering of algal cells synthesizing high levels of medium-chain fatty acids (MCFAs) is attractive to mitigate the high clouding point of longer chain fatty acids in algal based biodiesel. To develop a more informed understanding of MCFA synthesis in photosynthetic microorganisms, we isolated several algae from Great Salt Lake and screened this collection for MCFA accumulation to identify strains naturally accumulating high levels of MCFA. A diatom, Chaetoceros sp. GSL56, accumulated particularly high levels of C14 (up to 40%), with the majority of C14 fatty acids allocated in triacylglycerols. Using whole cell transcriptome sequencing and de novo assembly, putative genes encoding fatty acid synthesis enzymes were identified. Enzymes from this Chaetoceros sp. were expressed in the cyanobacterium Synechococcus sp. PCC 7002 to validate gene function and to determine whether eukaryotic enzymes putatively lacking bacterial evolutionary control mechanisms could be used to improve MCFA production in this promising production strain. Replacement of the Synechococcus 7002 native FabH with a Chaetoceros ketoacyl-ACP synthase III increased MCFA synthesis up to fivefold. The level of increase is dependent on promoter strength and culturing conditions.
Introduction
Biologically derived diesel from water-oxidizing, photosynthetic microorganisms (PSMs) is considered an efficient and promising next-generation technology for the production of renewable fuels (Radakovits et al., 2010; Work et al., 2013). These photosynthetic organisms are capable of high photon conversion efficiencies and could be deployed so that their cultivation does not directly compete with the contemporary global food supply (Schenk et al., 2008; Mata et al., 2010). To improve commercial feasibility, research efforts have focused on areas ranging from optimizing photosynthetic yields to performing scalability assessments (Larkum et al., 2012; Ho et al., 2014; Quinn and Davis, 2015). These efforts can be combined with advances in next-generation DNA sequencing and metagenomic analysis to characterize the diversity of phototrophic life at the enzyme/molecular level, which facilitates the discovery of genetic “parts” that can be transformed into “chassis” organisms to genetically improve biotechnological phenotypes in production organisms (Kodzius and Gojobori, 2015).
While most contemporary research efforts are focused on improving biomass accumulation, relatively few efforts are targeting improved oil quality. However, lipid oil quality is a significant issue in diesel fuel utilization, as biological products typically have longer chain lengths and higher degrees of unsaturation relative to conventional petrodiesel (Durrett et al., 2008). These features lead to poor cold-flow temperature properties (longer chain lengths) and oxidative instability (higher unsaturation) (Knothe, 2005, 2008).
The genetic manipulation of fatty acid chain length in plants has succeeded in changing oil crops to synthesize more saturated, medium-chain fatty acids (MCFAs, C8-14), which are preferable for biodiesel (Voelker et al., 1992; Thelen and Ohlrogge, 2002). Expressing acyl-ACP thioesterases (acyl-ACP TEs) alone, or with ketoacyl-ACP synthase (KAS) from plants that produce MCFA, leads to MCFA accumulation in transgenic hosts (Jones et al., 1995; Leonard et al., 1998; Slabaugh et al., 1998). However, there are relatively few successful reports regarding MCFA accumulation in PSMs (Radakovits et al., 2011), and only limited information is available regarding mechanisms to increase MCFA synthesis in PSMs to produce a higher quality biofuel. Studies of TEs, the enzymes most commonly used to control fatty acid chain length, indicate differences between algal and plant TEs in terms of substrate recognition and phylogeny (Jing et al., 2011; Blatti et al., 2012; Beld et al., 2014).
In this study, we used bioprospecting, cell sorting, and fatty acid methyl ester (FAME) profiling to identify a diatom from Great Salt Lake (GSL), Utah (Chaetoceros GSL56 hereafter) that naturally accumulated high levels of C14:0 fatty acid (>16%), and initiated studies probing fatty acid synthesis (FAS) in this organism. We chose GSL as a sampling site because the growth of halophilic algae in high-salt production systems has the potential to yield strains that are productive in seawater raceways where evaporation leads to increased salt concentrations; and because unique enzymology of potential biotechnological relevance can be found in extremophiles isolated from hypersaline ecosystems (Meuser et al., 2013). Chaetoceros GSL56 accumulates among the highest levels of C14:0 in a PSM that we have observed to date. As an initial characterization of the MCFA accumulation phenotype, we probed the physiological parameters influencing C14:0 accumulation in this alga, and attained and assembled a whole-cell transcriptome to begin probing FAS enzymes. To validate gene annotations and to search for superior FAS enzymes, a targeted set of genes encoding eukaryotic FAS enzymes from this diatom were transformed into a cyanobacterium [Synechococcus sp. PCC 7002 (hereafter, Synechococcus 7002)] engineered to secrete fatty acids. We establish that a eukaryotic type III ketoacyl-ACP synthase (KASIII) enzyme can functionally replace the endogenous FAS enzyme FabH in Synechococcus 7002, and that expression of this non-native, eukaryotic enzyme improves MCFA yields under the culturing conditions used.
Materials and Methods
Strain Identification and Culturing
Chaetoceros GSL56 was isolated from Farmington Bay in Great Salt Lake (GSL), Utah, USA in 2008. Cultures were maintained and grown in f/2 medium, at 29°C in a Percival incubator (Percival, Perry, IA, USA) illuminated with white fluorescent light [∼40–120 μmol photons m-2 s-1 of photosynthetically active radiation (PAR)] using a 16/8 light/dark cycle. Cell numbers were measured using a Z2 Coulter Counter (Beckman-Coulter, Brea, CA, USA). The partial 18S rRNA gene (1162 bp) was amplified from genomic DNA and sequenced for taxonomic identification using the universal primers 360FE and 1492RE (Dawson and Pace, 2002). The 18S rRNA gene sequence was deposited in Genbank (accession no. HQ710801).
FAME Quantification
Lipids were extracted and converted into FAMEs as described previously (Radakovits et al., 2012; Work et al., 2015) with some modifications. Briefly, 1.0 ml of methanol saturated with 5% KOH (0.8 g/ml) was added to 0.5 ml fresh culture samples in 4.0 ml sample vials, sealed and incubated at 100°C for 90 min resulting in cell lysis and lipid saponification. Acid-catalyzed methylation was then carried out by adding 1.5 ml 1:16 12N HCl/MeOH to the same vial and incubating at 80°C for 6 h. FAMEs were then extracted into 1.25 ml hexane via gentle inversion. Extracts were analyzed directly by gas chromatography-flame ionization detection (GC-FID) using an Agilent 7890A gas chromatograph equipped with a DB5-ms column (Agilent Technologies, Santa Clara, CA, USA). Different concentrations of the Supelco 37 component FAME standards mix (Supelco Inc., Bellefonte, PA, USA) were analyzed by GC-FID and used for peak identification and sample quantification. Fatty acids were also extracted from representative samples spiked with C13:0 internal standards (∼80% recovery) to assess cellular fatty acid detection.
Lipid Profile Analysis by Thin Layer Chromatography and Subsequent Methyl Esterification (TLC-FAME Analysis)
Thin-layer chromatography (TLC) analysis was conducted as described previously (Vieler et al., 2007; Radakovits et al., 2011). Briefly, 10 ml of pelleted culture was resuspended in 400 μl MeOH and then sonicated for 10 min for cell lysis. Then 400 μl chloroform was added to solubilize lipids, followed by the addition of 400 μl water for phase separation. The organic layer containing lipids was transferred to new microcentrifuge tubes and dried under a N2 stream. Concentrated lipids were resuspended in 30 μl chloroform, then 4.0 μl of resuspended lipids were spotted onto HPTLC-HL normal phase, 150 mm silica gel plates (10 cm × 20 cm; Analtech, Newark, DE, USA) and developed in a TLC chamber. A series of lipid standards, which included monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG) and sulfoquinovosyldiacyl glycerol (SQDG; Lipid Products, Nutfield, UK); phos phatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylglycerol (PG; Avanti Polar Lipids, Inc., Alabaster, AL); cholesterol (Chol), palmitic acid (free fatty acid; FFA) and glyceryl trioleate (triacylglycerol; TAG; Sigma–Aldrich, St. Louis, MO, USA), were run concurrently to identify different lipid classes. The first eluent [(methyl acetate:isopropanol: chloroform:methanol:KCl (0.25%)] in a ratio of 25:25:25:10:4 (v/v/v/v/v), ran to a height of ∼5 cm from the origin. After drying, the plates were developed with a second eluent [hexane:diethylether:acetic acid in a ratio of 70:30:2 (v/v/v)] to a height of ∼8 cm from the origin. TLC plates were then sprayed with a 0.05% solution of primuline (TCI America, Portland, OR, USA) in acetone. Individual lipid bands were visualized under UV light at 365 nm. Following TLC separation, the individual lipid bands were marked and scraped from the TLC plates. The fatty esters in each lipid class were transesterified into FAMEs and analyzed by GC-FID, as described previously (Radakovits et al., 2011).
Total RNA Extraction and Transcriptome Sequencing/Assembly
Total RNA was extracted from 20 ml of Chaetoceros GSL56 cells at stationary phase that were grown in f/2 medium at 3.5% salinity, supplemented with 1.06 × 10-4 M Na2SiO3. The plant RNA reagent (Invitrogen, Grand Island, NY, USA) was used to purify RNA from cells according to the manufacturer’s instructions. Genomic DNA was removed by DNase I (RNase free, Ambion, Grand Island, NY, USA) treatment, and subsequent RNA purification was carried out using the RNeasy MinElute Cleanup Kit (Qiagen, Germantown, MD, USA). Purified RNA was first quantified using a NanoDrop ND-1000 (Thermo scientific, Grand Island, NY, USA), and then more accurately measured using the QuantiT RiboGreen RNA assay kit (Invitrogen, Grand Island, NY, USA) with fluorescence detection.
Total RNA was sequenced by the National Center for Genomic Resources (NCGR, Santa Fe, NM, USA) as part of the Marine Microbial Eukaryote Transcriptome Project (Gordon and Betty Moore Foundation; Keeling et al., 2014). The RNA library was made with an insert size of ∼200 bp, using the TruSeq RNA Library preparation kit with poly-A+ selection. RNA was sequenced from both ends (paired-end reads 2 × 50 nt) using an Illumina Hi-seq 2000 (San Diego, CA, USA). The transcriptome dataset for Chaetoceros GSL56 is currently available through NCBI1.
Transcriptome assembly was conducted by NCGR using NCGR’s internal pipelines. Reads less than 25 bp after quality trimming were discarded, with the remaining reads assembled into contigs using ABySS (Simpson et al., 2009). All assembled contigs were subjected to gap closing using GapCloser v 1.10 (Li et al., 2008). To identify overlaps between contigs, the OLC (overlap layout consensus) assembler miraEST was used (Chevreux et al., 2004). BWA was used to align sequence reads back to assembled contigs (Li and Durbin, 2009). A final subset of contigs was created by filtering the contigs dataset by a minimal length of 150 bp. ESTScan with a Bacillariophyta scoring matrix was used to predict coding sequences (CDS) from the final subset of contigs (Iseli et al., 1999; Lottaz et al., 2003).
Transcriptome Annotation and Molecular Phylogeny
All CDS sequences were aligned against the non-redundant (nr) protein databases at the National Center for Biotechnology Information (NCBI) using the BLASTx algorithm with an E-value cutoff of 10-6. The resulting top 10 blast hits were exported into Blast2Go software v 3.0.10 for functional annotation and statistical analysis (Conesa and Götz, 2008).
Transcripts putatively involved in fatty acid metabolism, including FAS and TAG synthesis, were reexamined. Representative sequences of each gene were downloaded from NCBI and used to search contig datasets to avoid assembly error. All identified transcripts were aligned against transcriptome reads in the NCBI Sequence Read Archive (SRA) database to check assembly integrity and coverage. To identify sub-classes of FAS genes, such as β-ketoacyl-ACP synthase I, II, and III (KASI, II, and III), alignment of translated amino acids sequences was conducted using MUSCLE 3.8.31 and phylogenetic trees were generated using the Phylogeny.fr program (Dereeper et al., 2008).
KAS Complementation in Synechococcus 7002
Only one full length KASIII was annotated in the Chaetoceros GSL56 transcriptome (KASIII hereafter), which was amplified from Chaetoceros GSL56 cDNA using the primers listed in Supplementary Table S2. The amplified DNA fragment containing NdeI/HindIII restriction sites from the primers was inserted into a modified pNSI-cpcBA-YFP plasmid (Davies et al., 2014), containing a gentamicin resistance cassette (aacC1; Kovach et al., 1995) so that KASIII was positioned immediately after the cpcBA promoter (Xu et al., 2011) to form the new plasmid pNSI-cpcBA-gslKASIII-GentR. For homologous recombination, the Synechococcus 7002 fabH gene and flanking sequence (0.9 kb) was amplified from Synechococcus 7002 genomic DNA, and the resulting 2.0 kb DNA fragment inserted into plasmid pNSI-cpcBA-YFP-GentR by cloning in between the M13 forward and the M13 reverse sequences, generating plasmid pfabH. A 2.6 kb DNA fragment that included the cpcBA promoter, KASIII gene and the aacC1 marker was then amplified from pNSI-cpc-gslKASIII-GentR and inserted into pfabH to replace part (the first 500 bp) of the coding region of fabH, which is the target of gene disruption. The resulting plasmid, pfabH-cpcBA-gslKASIII-GentR, was used to transform Synechococcus 7002 for KASIII substitution assays. In order to minimize an imbalance between KASIII and other FAS enzymes in transgenic strains, another plasmid without the cpcBA promoter was created (plasmid pfabH-gslKASIII-GentR), in which KASIII expression was driven by the endogenous Synechococcus 7002 fabH promoter. This plasmid was constructed using designed primers (Supplementary Table S2), NEBuilder HiFi DNA assembly master mix (New England Biolabs Inc., MA) and Gibson cloning (Gibson et al., 2009) to avoid the use of restriction sites. A DNA fragment that contained the KASIII gene and the aacC1 marker gene was inserted into plasmid pfabH, replacing the first 500 bp of coding region of fabH at the start codon. DNA cloning was conducted in Escherichia coli DH5α cells. The genetic replacement of fabH with KASIII was conducted in a Synechococcus 7002 wild type strain and a lauric acid secreting (SA01) strain (Xu et al., 2013; Work et al., 2015).
To assess in vivo KASIII function, fully segregated transgenic strains were tested. Wild type and transgenic strains were first cultivated in 30 ml A+ medium in 125 ml flasks with the required antibiotic(s) (50 μg/ml gentamicin for SK01 and SK02, 50 μg/ml spectinomycin for SA01, 50 μg/ml gentamicin + 50 μg/ml spectinomycin for SAK01 and SAK02) at 37°C in an illuminated Percival incubator providing continuous illumination of ∼200 μmol PAR m-2 s-1 and aerated with 1% CO2/air. After 3–4 days of cultivation, all cultures were diluted in 50 ml fresh media (with antibiotics) in 250 ml flasks, and normalized to the same optical density (OD730 = 0.5). For fatty acid production, experimental cultures were cultivated at two different conditions: (A) room temperature, atmospheric CO2, and constant illumination at ∼80 μmol photons m-2 s-1 of PAR; and (B) 30°C, 1% CO2 and constant illumination at ∼80 μmol photons m-2 s-1 of PAR inside a growth chamber (Multitron, AJ125BC).
Results
Chaetoceros GSL56 Fatty Acid Profiles and Physiology
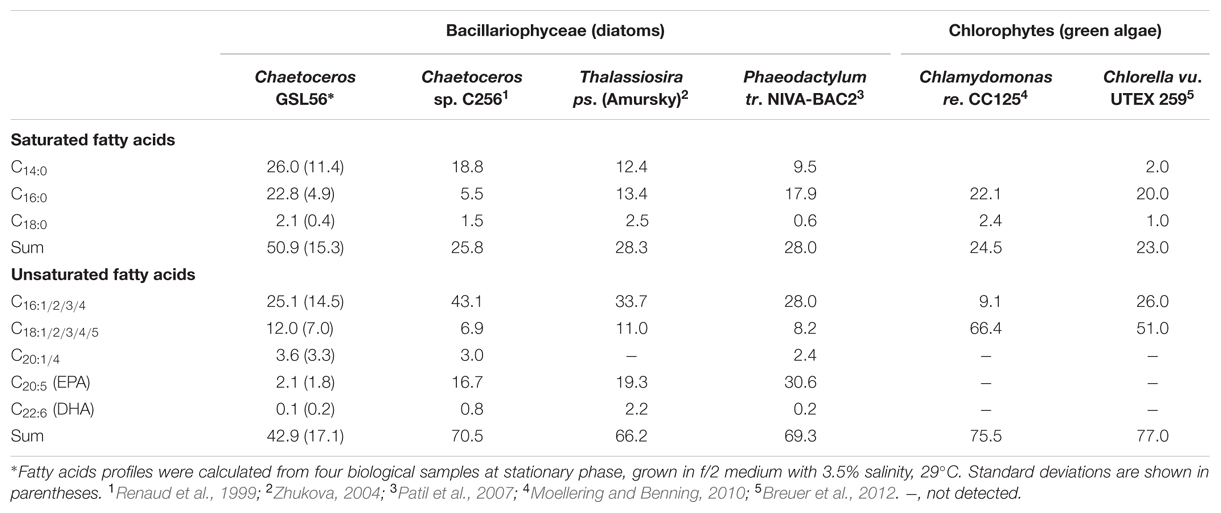
To identify organisms and enzymes capable of facilitating MCFA production in algae and cyanobacteria, we surveyed an algal collection isolated from GSL for organisms with natively high levels of MCFA (Figure 1A). Relative to the 42 halophilic algae that we isolated from this site, the ratio of C14:0 to other fatty acids was the highest in Chaetoceros GSL56 as determined by FAME analysis (Figure 1; Table 1). C14:0 fatty acid reached ∼20–35% of the total fatty acid content in this alga, depending on growth conditions and phases. Marine diatoms (e.g., Chaetoceros, Thalassiosira, and Phaeodactylum) tend to accumulate more C14:0 fatty acid (by percentage) relative to representative green algae [e.g., Chlamydomonas and Chlorella (Table 1)]. Chaetoceros GSL56 grew faster and reached higher cell numbers in medium containing 3.5% salinity, compared to lower and higher NaCl concentrations, 1.75 and 6% salinity, respectively (Supplementary Figure S1). Growth under different light intensities indicated that cells grew to a slightly higher density at lower light intensities (20 μmol PAR m-2 s-1 and 60 μmol PAR m-2 s-1) relative to a higher light intensity (120 μmol PAR m-2 s-1), where the final cell concentrations were decreased relative to cultures at lower light. Therefore, cells were cultivated in medium containing 3.5% salinity at 60 μmol PAR m-2 s-1, unless otherwise noted.
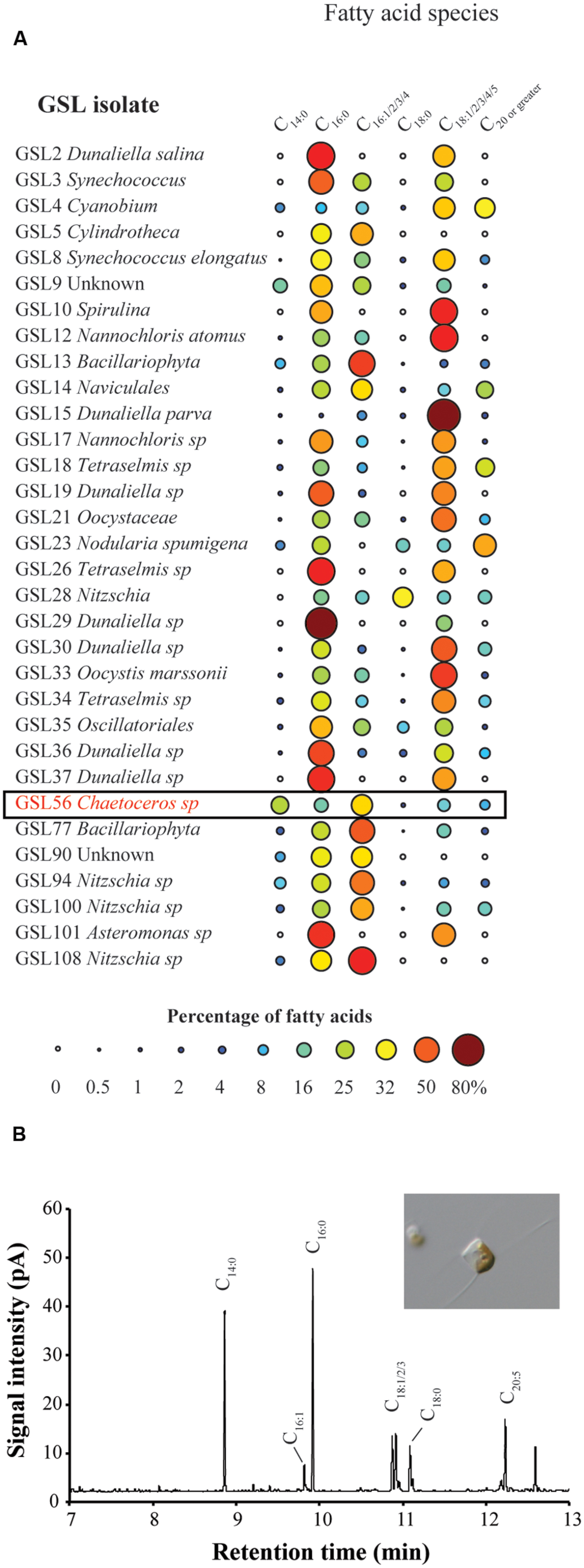
FIGURE 1. Fatty acid screen of GSL culture collection (A) and representative GC-FID fatty acid methyl ester (FAME) profile derived from Chaetoceros GSL56 fatty acids (B). The size of each circle in (A) is proportional to the percentage of total fatty acid represented by the corresponding fatty acid species. Representative microscopic image from Chaetoceros GSL56 grown in f/2 medium is inset in (B).
To further understand the biosynthesis and physiology of C14:0 fatty acid in Chaetoceros GSL56, specifically whether C14:0 accumulates predominately in membranes or storage products, lipids classes were extracted from cultures at stationary phase and subjected to TLC-FAME analysis (Figure 2). TLC analysis showed that the dominant lipid classes in Chaetoceros GSL56 include two phospholipids (PC and PG), SQDG, DGDG, MGDG, free fatty acids (FFAs), and triacylglyceride (TAG) (Figure 2A). The FFAs may represent artifacts of sample preparation. Sequential FAME analysis revealed that ∼66% of C14:0 fatty acid was stored in TAG and the second largest portion was in SQDG (∼10%; Figure 2B). Compared to total fatty acid distributions, about 50% of total fatty acids were found in TAG and 8% was in SQDG, demonstrating enrichment in these two lipids classes (Figure 2C).
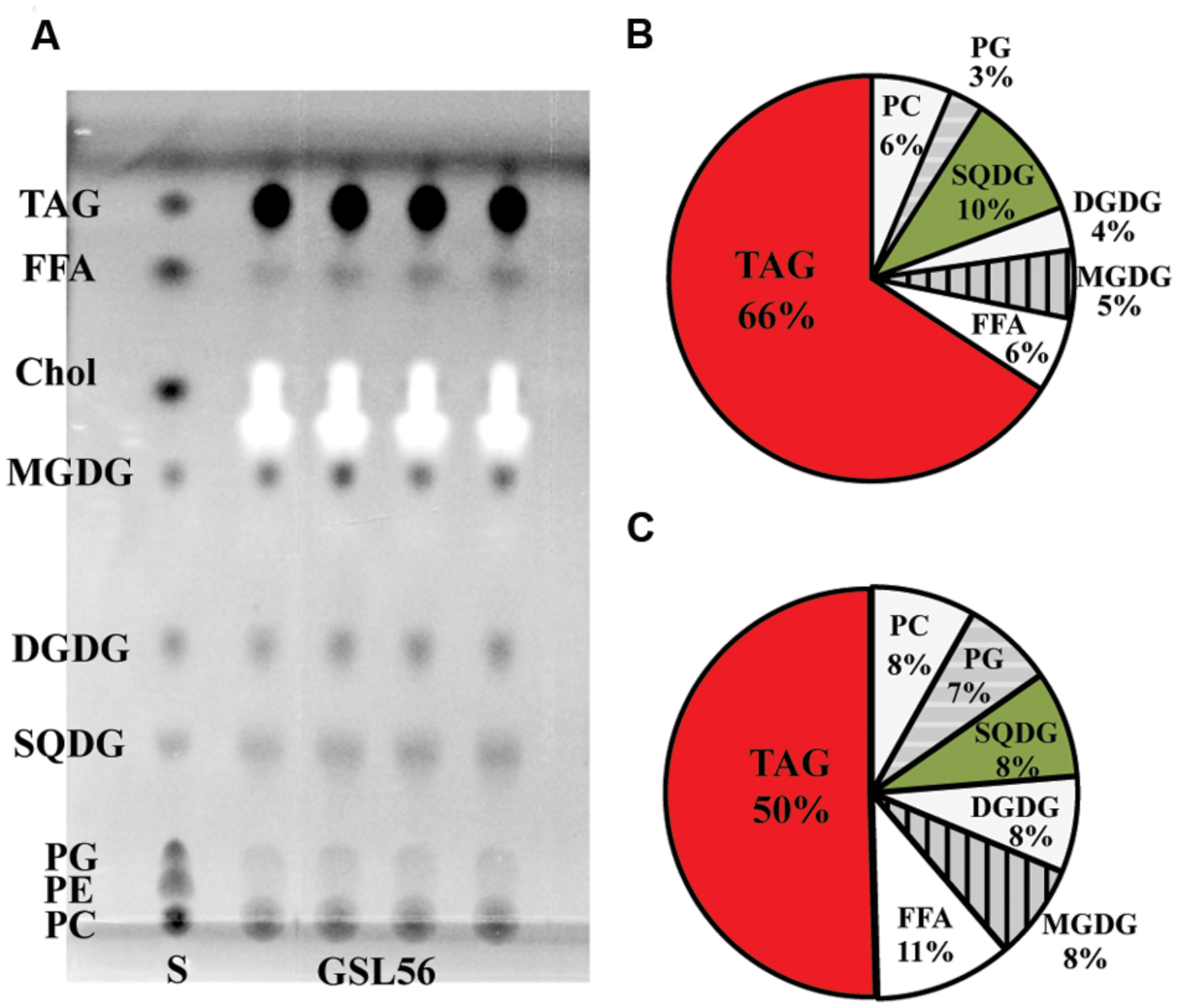
FIGURE 2. Chaetoceros GSL56 lipid classes separated by thin-layer chromatograpy (A), C14:0 distribution within the lipid classes (B), and the relative distribution of all lipids classes (C). Four lipid samples extracted from GSL56 cells were run concurrently with lipid standards (S) on TLC plates. Each data set represents two biological replicates. Abbreviations: Chol, cholesterol; DGDG, digalactosyldiacylglycerol; FFA, free fatty acid; MGDG, mono-galactosyldiacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol; TAG, triacylglycerol.
Transcriptome Analysis and Functional Annotation
To investigate the fatty acid biosynthetic pathway underpinning high C14:0 fatty acid content in Chaetoceros GSL56, we obtained a whole-cell transcriptome and assembled the data into expressed genes. Whole transcriptome sequencing was carried out using a cDNA library obtained from cells at stationary phase. A total of 36,745,292 raw reads were generated from sequencing the cDNA library using Illumina HiSeq 2000 (Table 2). The paired raw reads are available in the NCBI Sequence Read Archive (SRA) database with the accession number SRX551298. After trimming and cleaning sequences for adapters and primers, preprocessed sequences were assembled into 16,977 contiguous sequences (contigs) with sizes ranging from 150 to 18,638 bp. A subset of 14,914 contigs was predicted to contain protein CDS by ESTscan (150–17,385 nt in length, average = 1,720 nt, 14,088 unique), and 11,176 (∼80%) of these unique sequences had homologs when aligned against the nr database in NCBI using BlastX (E-value ≤ 10-6). The top five species contributing to predicted transcript annotation were Thalassiosira pseudonana (diatom, 16%), Phaeodactylum tricornutum (diatom, 15%), Ectocarpus siliculosus (brown algae, 6%), Phytophthora infestans (oomycete, 4%) and Aureococcus anophagefferens (heterokont, 4%) (Supplementary Figure S2).
The most represented annotations belonging to each GO category are listed in Supplementary Figure S2. Transcripts predicted to encode proteins in the molecular function category are more abundant than in the other two categories, cellular component and biological process. In the molecular function category, the GO term “protein binding” is the most represented, and in cellular component and biological process, the GO terms “integral component of membrane” and “oxidation-reduction process” are the most represented, respectively.
Analysis of Fatty Acid Biosynthesis Pathway
We expected that the predominant control of MCFA chain lengths occurs in the fatty acid biosynthetic pathway since C14:0 is present in all lipids classes. Genes putatively involved in the de novo FAS pathway were first identified (Figure 3; Supplementary Table S3). Examination of gene isozymes with putative functions in fatty acid initiation and elongation indicates an underrepresentation of the genes in the upstream portion of this pathway, when compared to Chlamydomonas reinhardtii and two diatoms – T. pseudonana and P. tricornutum (Miller et al., 2010; Boyle et al., 2012; Dyhrman et al., 2012). In the transcript assembly, we could only identify a single gene encoding an acyl-carrier protein (ACP), which acts as a tether shuttling growing fatty acids to multiple components of the fatty acid synthase complex. The predicted polypeptide is 72% identical to an ACP from the diatom P. tricornutum (GenBank: EEC50984.1; Figure 4B). In both the C. reinhardtii and T. pseudonana transcriptomes, two ACPs with relatively high expression levels are found. Two putative monomeric acetyl-CoA carboxylase (ACCase) enzymes were identified in the Chaetoceros GSL56 transcriptome (transcript ID: 5820 and 12896) that have 82–85 and 72–76% similarities with representatives in P. tricornutum and T. pseudonana, respectively. Heteromeric ACCase enzymes that have dissociated enzymatic subunits were not identified, whereas they were identified in C. reinhardtii. The abundances of these two ACCase transcripts in Chaetoceros GSL56 was low as indicated by calculated reads per kilobase per million mapped reads (RPKM) values (13 and 0.4), in contrast to ACCase transcript abundance in C. reinhardtii (RPKM 246.5 and 250; Miller et al., 2010). It should be noted that a direct comparison of RPKM values between species may be compromised due to different genome sizes and different isozymes. Thus, we compared the RPKM of ACCase to other genes involved in the fatty acid biosynthetic pathway within the same species. Whereas the ACCase ranks high in the fatty acid biosynthesis pathway in C. reinhardtii (Miller et al., 2010; Lv et al., 2013), it is among the least abundant transcripts in Chaetoceros GSL56. In contrast to ACP and ACCase, the genes involved in fatty acid elongation reactions are more abundant in Chaetoceros GSL56 relative to C. reinhardtii. For example, 11 unique isozymes were identified for the 3-ketoacyl reductase (KAR) enzymes, with the highest RPKM value equal to 245, whereas in C. reinhardtii only one copy of this gene was found. Transcripts encoding hydroxyacyl-ACP dehydratase (HAR) and enoyl-ACP reductase (EAR) were also identified at high abundance (RPKM: 267 and 131, respectively).
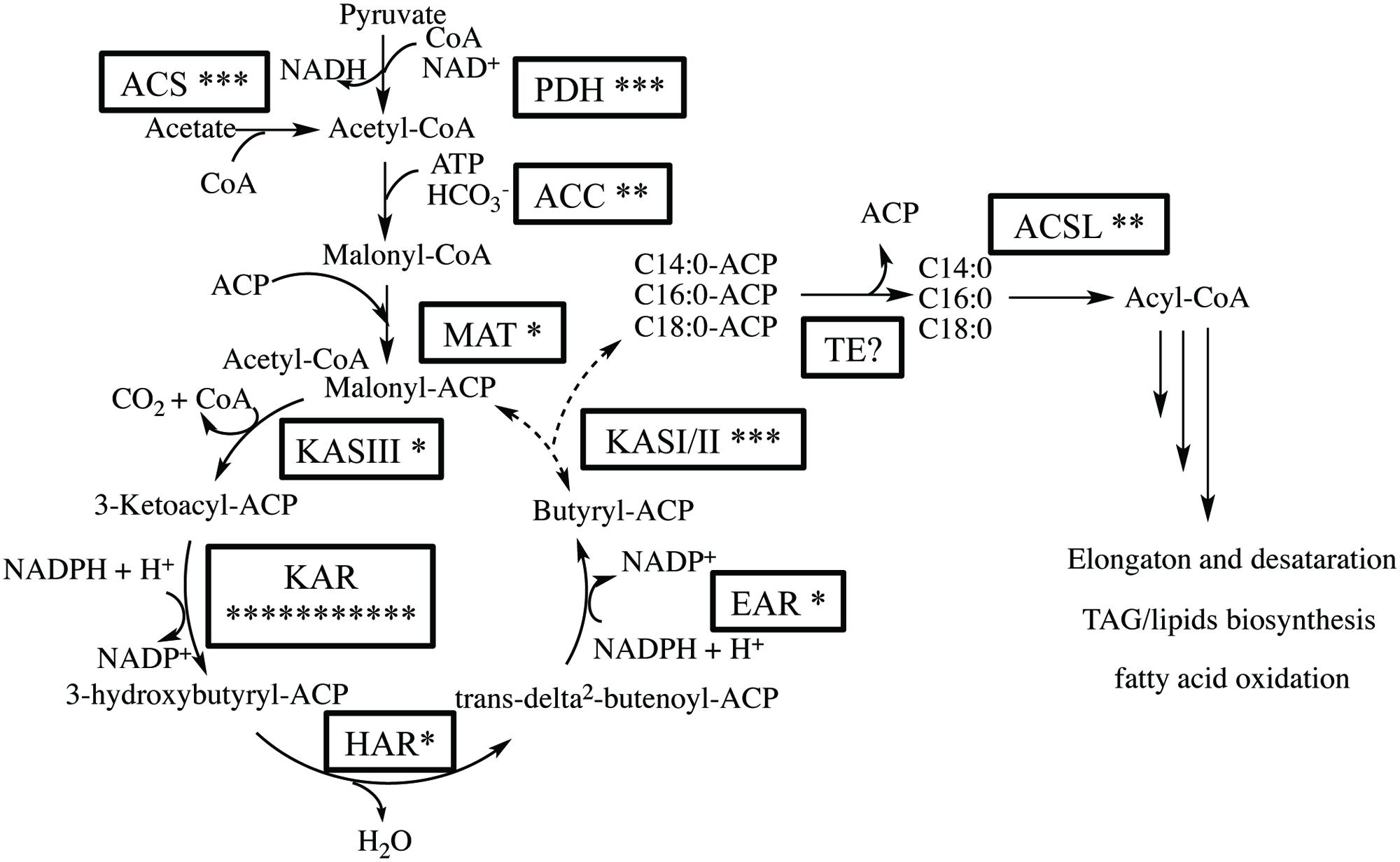
FIGURE 3. Putative fatty acid synthesis (FAS) pathway in Chaetoceros GSL56 based on functional annotation. The asterisks attached to the enzyme name indicate the putative number of isozymes encoded by genes present in the transcriptome. Enzyme abbreviations: ACC, acetyl-CoA carboxylase; ACS, acetyl-CoA synthetase; ACSL, long chain acyl-CoA synthase; EAR, enoyl-ACP reductase; TE, acyl-ACP thioesterase; HAR, hydroxyacyl-ACP dehydratase; KAR, β-ketoacyl-ACP reductase; KAS, β-ketoacyl-ACP synthase; MAT, malonyl-CoA-ACP transacylase; PDH, pyruvate dehydrogenase E1.
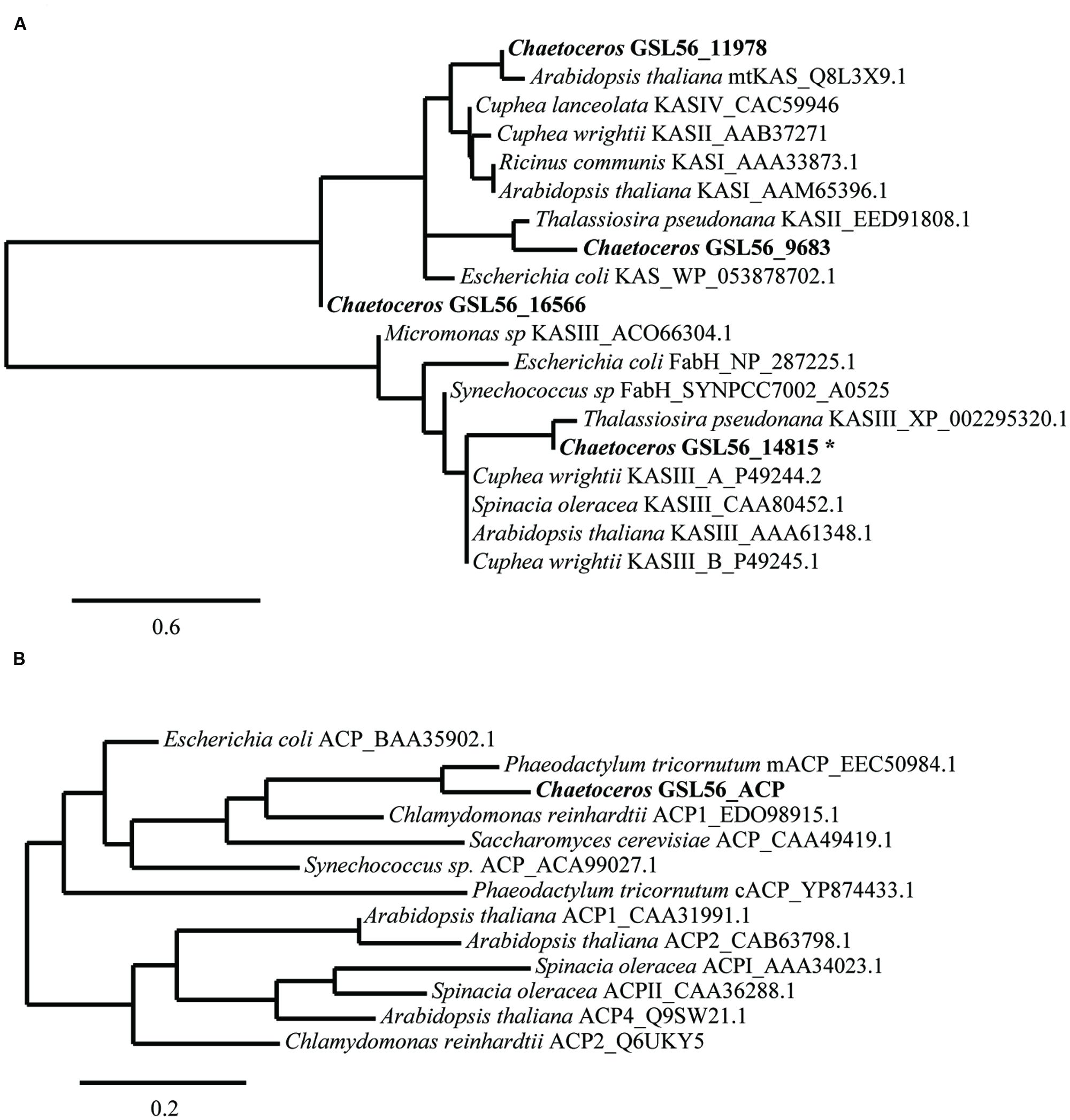
FIGURE 4. Sequence similarities between Chaetoceros GSL56 β-ketoacyl-ACP synthase (KAS) (A) and acyl carrier proteins (ACP) (B) relative to indicated plant, bacteria, and algae representatives. Scale bars represent the branch lengths: the number of amino acid substitutions per site. The asterisk indicates the sequence that was used in KASIII substitution assay.
Substrate specificity of some key enzymes in Chaetoceros GSL56 is likely to lead to C14:0 fatty acid accumulation. In plants, two enzymes, ketoacyl-ACP synthase (KASI/II, KASIII) and acyl-ACP TEs (FatA/B) typically show different enzymatic activities toward different fatty acid chain lengths, which contribute to fatty acid chain length determination (Jones et al., 1995; Leonard et al., 1998; Abbadi et al., 2000; Dehesh et al., 2001). Therefore, we initially attempted to identify genes encoding these two enzyme classes in Chaetoceros GSL56. To maximize gene identification, protein queries that consisted of homologous genes from diatoms and several other algae were conducted. Putative genes were then aligned with well-characterized genes for further annotation (Figure 4). There are four distinct transcripts annotated as KAS. Based on sequence alignments with KAS genes from plants and bacteria (Figure 4A), one transcript (14815) likely encodes a plant type KASIII enzyme, and it is not related to the other KAS enzymes. Transcript (9683) is predicted to be a KASII enzyme that is similar to KASII from P. tricornutum. Transcript 11978 has homology with plant type KASI/II/IV, while the fourth transcript (16566) has only tenuous KAS similarity. Unambiguous TE identification is complicated since there are often no clear homologs to the conical plant-type enzymes found in diatoms. As expected, a homolog of this type of an acyl-ACP TE was not identified in Chaetoceros GSL56 (data not shown); however, genes encoding putative thioester hydrolyzing enzymes were found (transcripts: 2647, 3750, 4370, 9448, and 9539).
KASIII in Chaetoceros GSL56 Substitutes for FabH in Synechococcus sp. PCC7002
As shown above, Chaetoceros GSL56 effectively accumulates high levels of MCFA. However, the data also demonstrate that this strain has poor growth and biomass productivity metrics (Supplementary Figure S1). We therefore examined whether enzymatic “parts” from Chaetoceros GSL56 could be used in a biofuel “chassis” organism (Synechococcus 7002) to improve a biofuel phenotype, in this case MCFA synthesis and secretion. First, each of the genes potentially encoding thioester hydrolyzing enzymes were cloned and transformed into Synechococcus 7002 concomitantly with fadD disruption to enable fatty acid secretion, an approach that was successfully used previously for genuine thioesterase enzymes (Work et al., 2015); however, none of the encoded enzymes showed acyl-ACP TE activity in Synechococcus 7002 (data not shown). We then explored whether expression of KASIII (functional homolog of endogenous FabH) could influence FAS in this cyanobacterium. A recent report concluded that FabH is the rate-limiting step in FAS in Synechococcus 7002 (Kuo and Khosla, 2014). Plasmids were designed for concurrently knocking out the native Synechococcus 7002 fabH gene while inserting KASIII from Chaetoceros GSL56 in its place. Enzyme expression was driven either at native levels by the endogenous fabH promoter (SK01) or at very high levels by the cpcBA promoter (SK02; Xu et al., 2011). Fully segregated transgenic strains were obtained through repeating streaking of single colonies on A+ plates containing the required antibiotics (Figure 5). Controls using a plasmid that had yellow fluorescent protein instead of KASIII from Chaetoceros GSL56 were also generated, but these mutants were not able to reach homoplasmy, indicating that FabH (or a functional replacement) is essential for Synechococcus 7002 viability. Relative to the wildtype Synechococcus 7002, the two transgenic strains expressing KASIII (SK01 and SK02) showed no major differences in either fatty acid profiles or total fatty acid production when transformed into the wildtype background (data not shown). We then transformed a Synechococcus 7002 strain SA01 (Work et al., 2015) that is able to secrete lauric acid (C12:0) due to the expression of the medium chain acyl-ACP TE from Umbellularia californica (UcFatB; Table 3). As the SA01 strain is already able to secrete FFAs, we probed whether enhanced MCFA synthesis would occur when the rate-limiting endogenous FabH was replaced with the Chaetoceros GSL56 KASIII. KASIII substitution/overexpression in other systems has been shown to influence fatty acid profiles/yields, putatively by changing the rate of fatty acid initiation relative to downstream enzyme activities, and/or by catalyzing elongation reactions for short chain (but not long chain) acyl-ACP substrates (Abbadi et al., 2000; Dehesh et al., 2001; González-Mellado et al., 2010).
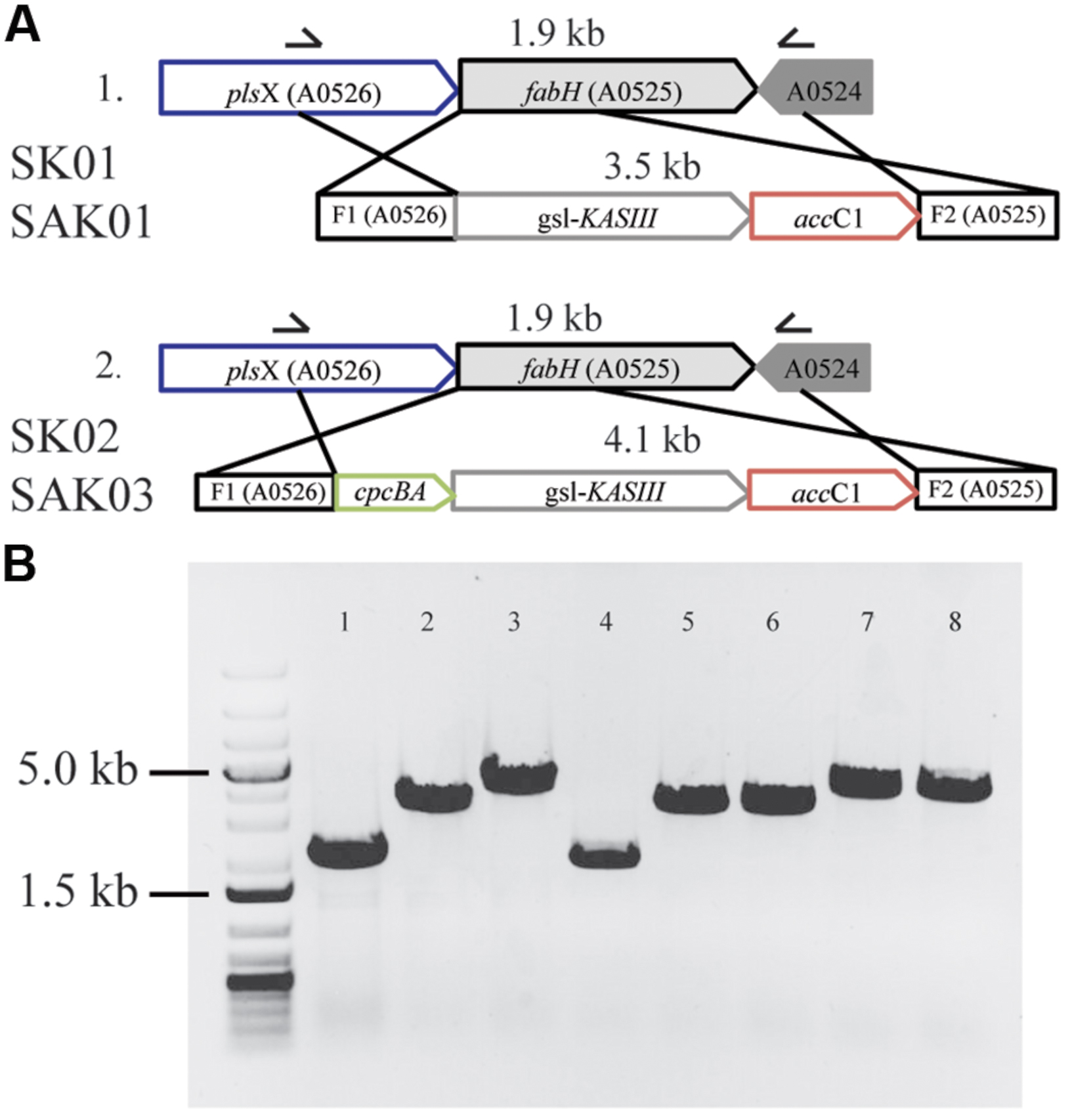
FIGURE 5. Substitution of KASIII from Chaetoceros GSL56 replacing FabH in Synechococcus 7002. (A) Schematic of homologous recombination for KASIII gene complementation. (B) Whole cell PCR with gene specific primers demonstrating the integration of recombinant genes in Synechococcus 7002 transformants. From left to right: molecular size marker: 1 kb plus DNA ladder; 1: Synechococcus 7002 wildtype strain; 2: SK01; 3: SK02; 4: SA01; 5–6: two different lines of SAK01; 7–8: two different lines of SAK03.
Enhanced Lauric Acid Production in Transgenic Strains
When FabH was replaced by the Chaetoceros GSL56 KASIII enzyme in the Synechococcus 7002 SA01 background, both new transgenic strains (SAK01 and SAK03) showed enhanced levels of C12:0 fatty acid production/secretion relative to SA01 (Figure 6; Table 4). An increase was observed under both of the two different culturing conditions tested. For cells grown at room temperature without CO2 augmentation (growth condition #1), SA01, SAK01, and SAK03 all grew similarly (Figure 6A). Despite no noticeable growth differences between SA01 and the KASIII expressing mutants, C12:0 fatty acid accumulated at much higher levels per ml of culture and in the relative percentage of all fatty acids. The enhancement ranged from 1.1 to 5-fold depending on growth phases and the promoters used. In SA01, the highest levels of C12:0 accumulation occurred at day 11 (11.5 mg/L, 20% of total fatty acids) and started to slightly decrease thereafter (Figures 6B,C). However, in the KASIII expressing strains, the C12:0 fatty acid levels continue to show increases even at day 20. The relative percentages of C12:0 fatty acid also continued to increase during the experimental period. We also tested SA01, SAK01, and SAK03 at higher temperature (30°C) and supplemented with 1% CO2 (growth condition #2). All strains grew faster than in growth condition #1. Under these conditions, the transgenic strains (SAK01/03) showed defective growth relative to SA01 (Figure 6D); and unexpectedly, all strains showed decreased C12:0 fatty acid productivity and a lower percentage of C12:0 than in growth condition #1 (Figures 6E,F). The highest amount of C12:0 (54 mg/L, 30% of all fatty acids) was produced from SAK03 with KASIII expression driven by the cpcBA promoter. In sum, expression of KASIII in Synechococcus 7002 and co-expression of a medium chain specific thioesterase enhances MCFA synthesis (C12:0, Figure 7) in Synechococcus 7002 under the growth conditions tested.
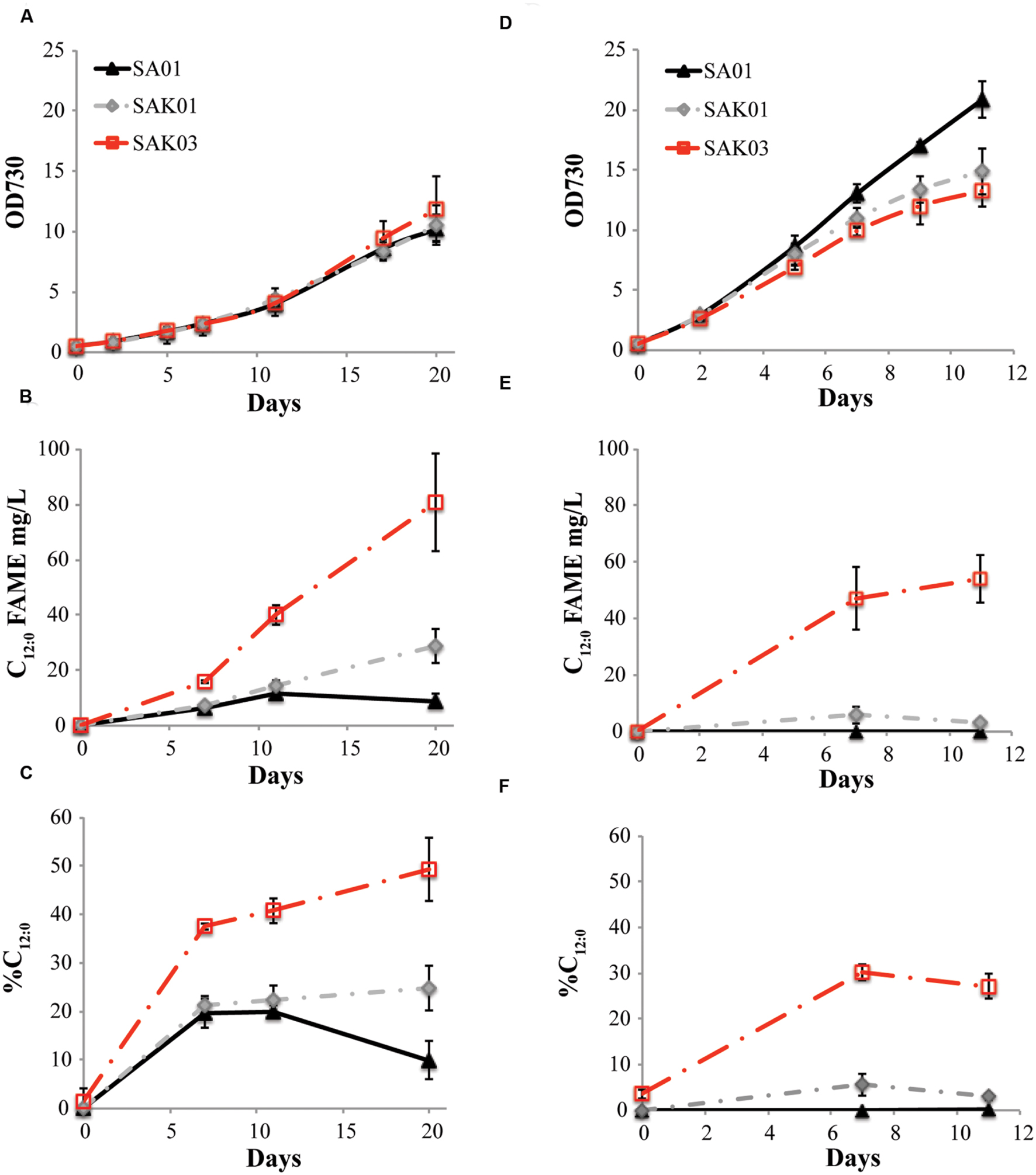
FIGURE 6. Comparison of C12:0 fatty acid production using two different growth conditions. (A,D) optical density of cultures measured at 730 nm, (B,E) C12:0 fatty acid production, and (C,F) percentage of C12:0 fatty acid relative to total fatty acids. Growth condition #1 (A–C): room temperature, atmospheric CO2; growth condition #2 (D–F), 30°C, 1% CO2. Data represent three independent biological replicates.
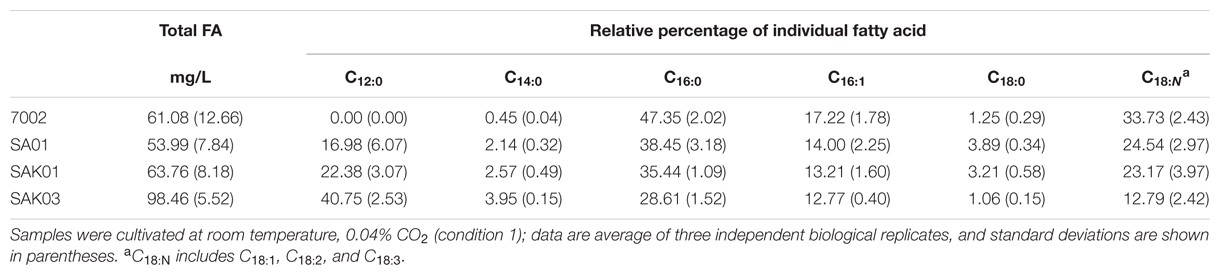
TABLE 4. Fatty acid profiles of Synechococcus 7002 wildtype strain and transgenic strains from KASIII substitution assays sampled at day 11.
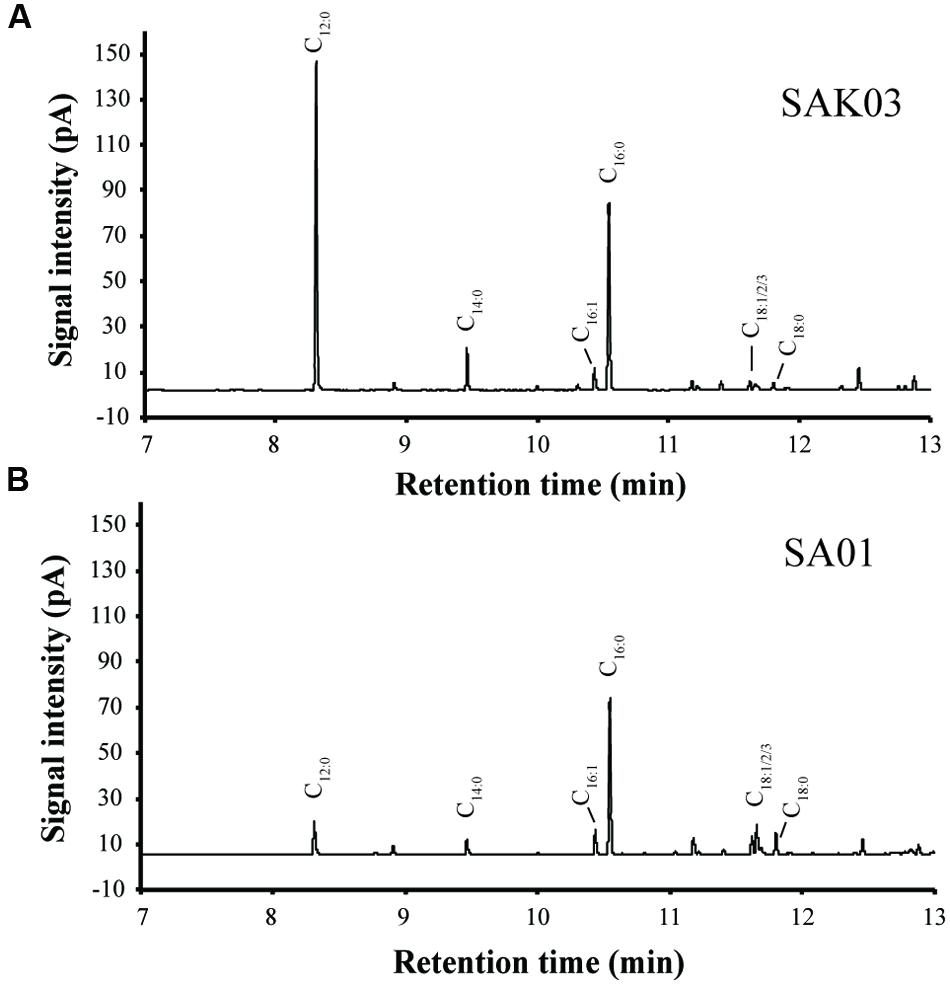
FIGURE 7. Representative GC-FID chromatograms of FAMEs from the Synechococcus 7002 mutants SAK03 (A) and SA01 (B). FAMEs were extracted from batch cultures grown at room temperature and harvested on day 20.
Discussion
Medium chain fatty acids are desirable in both the fuel and chemical industries (Battey et al., 1989). However, large quantities of MCFAs are only available in some specific oilseeds, such as coconut and Cuphea. Expression of MCFA specific enzymes can lead to the accumulation of shorter chain fatty acids in crop plants, providing a way to increase MCFA productivity (Voelker et al., 1992). In this study, we explored FAS in a halophilic alga (Chaetoceros GSL56) that is enriched for MCFA, and used whole cell transcriptome sequencing to identify genes encoding putative FAS enzymes. Significantly, we identified a gene encoding a KASIII enzyme that when expressed in Synechococcus 7002 is able to functionally replace the native cyanobacterial FabH enzyme, and when co-expressed in a C12:0 MCFA secreting strain results in increased levels of MCFA synthesis.
Chaetoceros GSL56 primarily synthesizes C14:0, C16:0, C18:0, C18:N, and C20:5 fatty acids that are differentially incorporated into all lipid classes (Table 1; Figures 1 and 2). Depending on growth conditions, saturated C14:0 fatty acids range from ∼15–40% of all fatty acids in Chaetoceros GSL56 which is amongst the highest native C14:0 levels that we have observed in an alga (Figure 1A). Additional marine algae have also been documented to contain MCFA, primarily within the Bacillariophyta and Haptophyta phyla (Chen et al., 2007; Guihéneuf et al., 2010). Analysis of individual lipid classes in Chaetoceros GSL56 revealed that MCFAs are preferentially incorporated into TAG (Figure 2; Supplementary Table S1), which is consistent with the MCFA profiles in plant seeds and the other diatom species (Battey and Ohlrogge, 1989; Alonso et al., 2000; Chen et al., 2007). In contrast to neutral oil droplets, the phospholipids and galactolipids, which are used as membrane constituents, are enriched in long chain fatty acids (C16-20).
Although Chaetoceros GSL56 has a promising MCFA phenotype, this alga has poor growth metrics relative to biotechnologically relevant strains. We therefore identified enzymes in the MCFA biosynthetic pathway that could be used as “parts” in more biotechnologically promising strains. Synechococcus 7002 is a cyanobacterium with among the fastest growth rates of any PSM, and has become the recent focus of several biotechnology efforts (McNeely et al., 2010; Mendez-Perez et al., 2011; Davies et al., 2014; Work et al., 2015). We therefore transferred selected components of the Chaetoceros GSL56 FAS machinery into Synechococcus 7002 to (i) verify functional annotations and (ii) determine whether enzymes from Chaetoceros GSL56 could be expressed in a cyanobacterium to increase MCFA synthesis. Based on precedence in plant studies, (Voelker et al., 1992; Dehesh, 2001), we targeted substrate specific enzymes, such as acyl-ACP TEs and β-ketoacyl ACP synthase (KAS), to influence MCFA synthesis (Voelker et al., 1992; Leonard et al., 1998).
Initially, we probed whether five genes encoding putative thioester hydrolysis enzymes contained acyl-ACP TE activity that could be used to produce MCFAs in Synechococcus 7002. Expression of each of these genes individually while concurrently knocking out the gene encoding the fatty acid recycling enzyme FadD (Kaczmarzyk and Fulda, 2010; Liu et al., 2011; Ruffing, 2014; Work et al., 2015), did not result in the production of any MCFA in Synechococcus 7002. This may be because the enzymes tested were not genuine acyl-ACP TEs, or because they did not fold into functional enzymes in Synechococcus 7002. We then targeted the KAS enzyme family. The β-ketoacyl ACP synthase III (KASIII) functionally replaced the FabH enzyme in Synechococcus 7002, confirming the annotation of KASIII as a genuine β-ketoacyl ACP synthase III. Furthermore, expression of KASIII in a Synechococcus 7002 strain that coexpresses a plant-sourced medium chain specific thioesterase (UcfatB) produced up to 40% of total fatty acid as lauric acid (SAK03), which is approximately four times more than corresponding strains expressing the native FabH enzyme. Enhanced lauric acid production in transgenic strains may be due to, (i) an increased acyl-ACP pool size since fabH is the rate-limiting enzyme of fatty acid synthase in Synechococcus 7002 (Kuo and Khosla, 2014); (ii) modified overall FAS rate that better matches thioesterase activity (Dehesh et al., 2001); and/or, (iii) concentrated short to medium chain acyl-ACPs pools caused by KASIII (Abbadi et al., 2000). The exact mechanism of KASIII enhancement, and testing whether KASIII participates in MCFA synthesis in Chaetoceros GSL56, is the subject of further investigation.
The euryhaline cyanobacterium Synechococcus 7002, is a particularly promising host for FFA production, because it shows a unique high tolerance to FFAs (Ruffing, 2014). However, the total metabolic flux to FAS represents a small portion (∼5–10%) of cell dry weight, indicating evolutionary limitations to fatty acid productivities in this host (Work et al., 2015). Replacing FabH, which is the kinetically rate-limiting enzyme in FAS, with KASIII did not improve fatty acid production when transformed into the wild-type host (data not shown), suggesting that downstream elements that traffic fatty acids to membrane lipids may now be rate limiting. However, the expression of acyl-ACP thioesterase allows cleavage of MCFA from acyl-ACP, and has been reported to increase malonyl-ACP turnover rates and reduce the inhibition of acyl-ACP in other systems (Magnuson et al., 1993). Transgenic strains expressing KASIII and thioesterase (SAK01 and SAK03) accumulated higher amounts of C12:0 and C14:0 fatty acids, relative to their parental strain SA01, and these increases are consistent with UcfatB hydrolytic activity (Voelker and Davies, 1994). Levels of MCFA are influenced by promoter strength, indicating a correlation between KASIII expression levels and MCFA production.
It is still undetermined how Chaetoceros GSL56 regulates C14 fatty acid production as we were unable to identify an acyl-ACP TE with C14:0 specificity in this alga, which is consistent with other studies in red algae and diatoms (Beld et al., 2014). Future studies of the role of KASIII on medium chain FAS, as well as the substrate specificities of other enzymes, such as acyltransferase, will be important in understanding fatty acid chain length regulation in this and other diatoms.
In future research, we also intend to explore whether, the expression of other eukaryotic FAS enzymes in Synechococcus 7002, which typically has less than 10% of its biomass in fatty acyl lipids, results in further yield improvements. It is possible that these eukaryotic enzymes, which did not evolve under the regulatory mechanisms used by cyanobacteria, remain active outside of the metabolic context that evolved to limit FAS in cyanobacteria such as Synechococcus 7002.
Author contributions
HG designed and transformed Synechococcus 7002 strains, performed batch experiments and the majority of biochemical assays, and compiled this manuscript. RJ participated in GSL strain screening, generated data in Figure 1 and revised this manuscript. FD participated in designing transformation vectors and revised this manuscript. LS and PS developed methods for extracting and quantitating internal standards. MP provided the conception of this work, oversaw and edited this manuscript. All authors revised the manuscript for intellectual content.
Funding
Funding support was provided by the Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences and Biosciences Division (award DE-SC0008547). Analytical support was provided by the National Science Foundation through the Major Research Instrumentation Program (CHE-1229156).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor Bonnie Baxter at Westminster College in Salt Lake City, UT, and Dr. Susan Brown at the University of Hawaii for isolating and providing Chaetoceros GSL56. We would like to thank the National Center for Genome Resources, the Gordon and Betty Moore Foundation’s Marine Microbiology Initiative, and the International Marine Microbial Eukaryote Research Community for the sequenced Chaetoceros GSL56 transcriptome. We’d like to thank Patrick Eduafo and Dongze Li for their assistance in TLC-FAME analysis, Shelby Hollmaier for screening fatty acid profiles of the GSL culture collection, and thank Dr. Randor Radakovits for preparing Chaetoceros GSL56 for whole cell transcriptome sequencing.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00690
Footnotes
References
Abbadi, A., Brummel, M., and Spener, F. (2000). Knockout of the regulatory site of 3-ketoacyl-ACP synthase III enhances short- and medium-chain acyl-ACP synthesis. Plant J. 24, 1–9. doi: 10.1046/j.1365-313x.2000.00841.x
Alonso, D. L., Belarbi, E.-H., Fernández-Sevilla, J. M., Rodríguez-Ruiz, J., and Grima, E. M. (2000). Acyl lipid composition variation related to culture age and nitrogen concentration in continuous culture of the microalga Phaeodactylum tricornutum. Phytochemistry 54, 461–471. doi: 10.1016/S0031-9422(00)00084-4
Battey, J. F., and Ohlrogge, J. B. (1989). A comparison of the metabolic fate of fatty acids of different chain lengths in developing oilseeds. Plant Physiol. 90, 835–840. doi: 10.1104/pp.90.3.835
Battey, J. F., Schmid, K. M., and Ohlrogge, J. B. (1989). Genetic engineering for plant oils: potential and limitations. Trends Biotechnol. 7, 122–126. doi: 10.1016/0167-7799(89)90088-7
Beld, J., Blatti, J., Behnke, C., Mendez, M., and Burkart, M. (2014). Evolution of acyl-ACP thioesterases and β-ketoacyl-ACP synthases revealed by protein–protein interactions. J. Appl. Phycol. 26, 1619–1629. doi: 10.1007/s10811-013-0203-4
Blatti, J. L., Beld, J., Behnke, C. A., Mendez, M., Mayfield, S. P., and Burkart, M. D. (2012). Manipulating fatty acid biosynthesis in microalgae for biofuel through protein-protein interactions. PLoS ONE 7:e42949. doi: 10.1371/journal.pone.0042949
Boyle, N. R., Page, M. D., Liu, B., Blaby, I. K., Casero, D., Kropat, J., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825. doi: 10.1074/jbc.M111.334052
Breuer, G., Lamers, P. P., Martens, D. E., Draaisma, R. B., and Wijffels, R. H. (2012). The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 124, 217–226. doi: 10.1016/j.biortech.2012.08.003
Chen, G.-Q., Jiang, Y., and Chen, F. (2007). Fatty acid and lipid class composition of the eicosapentaenoic acid-producing microalga, Nitzschia laevis. Food Chem. 104, 1580–1585. doi: 10.1016/j.foodchem.2007.03.008
Chevreux, B., Pfisterer, T., Drescher, B., Driesel, A. J., Müller, W. E. G., Wetter, T., et al. (2004). Using the miraEST assembler for reliable and automated mrna transcript assembly and snp detection in sequenced ESTs. Genome Res. 14, 1147–1159. doi: 10.1101/gr.1917404
Conesa, A., and Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008:619832. doi: 10.1155/2008/619832
Davies, F. K., Work, V. H., Beliaev, A. S., and Posewitz, M. C. (2014). Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2:21. doi: 10.3389/fbioe.2014.00021
Dawson, S. C., and Pace, N. R. (2002). Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. U.S.A. 99, 8324–8329. doi: 10.1073/pnas.062169599
Dehesh, K. (2001). How can we genetically engineer oilseed crops to produce high levels of medium-chain fatty acids? Eur. J. Lipid Sci. Technol. 103, 688–697. doi: 10.1002/1438-9312(200110)103:10<688::AID-EJLT688>3.0.CO;2-9
Dehesh, K., Tai, H., Edwards, P., Byrne, J., and Jaworski, J. G. (2001). Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 125, 1103–1114. doi: 10.1104/pp.125.2.1103
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Durrett, T. P., Benning, C., and Ohlrogge, J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x
Dyhrman, S. T., Jenkins, B. D., Rynearson, T. A., Saito, M. A., Mercier, M. L., Alexander, H., et al. (2012). The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE 7:e33768. doi: 10.1371/journal.pone.0033768
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
González-Mellado, D., Wettstein-Knowles, P., Garcés, R., and Martínez-Force, E. (2010). The role of β-ketoacyl-acyl carrier protein synthase III in the condensation steps of fatty acid biosynthesis in sunflower. Planta 231, 1277–1289. doi: 10.1007/s00425-010-1131-z
Guihéneuf, F., Fouqueray, M., Mimouni, V., Ulmann, L., Jacquette, B., and Tremblin, G. (2010). Effect of UV stress on the fatty acid and lipid class composition in two marine microalgae Pavlova lutheri (Pavlovophyceae) and Odontella aurita (Bacillariophyceae). J. Appl. Phycol. 22, 629–638. doi: 10.1007/s10811-010-9503-0
Ho, S.-H., Ye, X., Hasunuma, T., Chang, J.-S., and Kondo, A. (2014). Perspectives on engineering strategies for improving biofuel production from microalgae — a critical review. Biotechnol. Adv. 32, 1448–1459. doi: 10.1016/j.biotechadv.2014.09.002
Iseli, C., Jongeneel, C. V., and Bucher, P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 99, 138–148.
Jing, F., Cantu, D. C., Tvaruzkova, J., Chipman, J. P., Nikolau, B. J., Yandeau-Nelson, M. D., et al. (2011). Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 12:44. doi: 10.1186/1471-2091-12-44
Jones, A., Davies, H. M., and Voelker, T. A. (1995). Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell 7, 359–371. doi: 10.1105/tpc.7.3.359
Kaczmarzyk, D., and Fulda, M. (2010). Fatty acid activation in cyanobacteria mediated by Acyl-Acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 152, 1598–1610. doi: 10.1104/pp.109.148007
Keeling, P. J., Burki, F., Wilcox, H. M., Allam, B., Allen, E. E., Amaral-Zettler, L. A., et al. (2014). The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. doi: 10.1371/journal.pbio.1001889
Knothe, G. (2005). Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process. Technol. 86, 1059–1070. doi: 10.1016/j.fuproc.2004.11.002
Knothe, G. (2008). “Designer” biodiesel: optimizing fatty ester composition to improve fuel properties†. Energy Fuels 22, 1358–1364.
Kodzius, R., and Gojobori, T. (2015). Marine metagenomics as a source for bioprospecting. Mar. Genom. 24, 21–30. doi: 10.1016/j.margen.2015.07.001
Kovach, M. E., Elzer, P. H., Steven Hill, D., Robertson, G. T., Farris, M. A., Roop Ii, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kuo, J., and Khosla, C. (2014). The initiation ketosynthase (FabH) is the sole rate-limiting enzyme of the fatty acid synthase of Synechococcus sp. PCC 7002. Metab. Eng. 22, 53–59. doi: 10.1016/j.ymben.2013.12.008
Larkum, A. W. D., Ross, I. L., Kruse, O., and Hankamer, B. (2012). Selection, breeding and engineering of microalgae for bioenergy and biofuel production. Trends Biotechnol. 30, 198–205. doi: 10.1016/j.tibtech.2011.11.003
Leonard, J. M., Knapp, S. J., and Slabaugh, M. B. (1998). A Cupheaβ-ketoacyl-ACP synthase shifts the synthesis of fatty acids towards shorter chains in Arabidopsis seeds expressing Cuphea FatB thioesterases. Plant J. 13, 621–628. doi: 10.1046/j.1365-313X.1998.00066.x
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Liu, X., Sheng, J., and Curtiss, I. I. I. R. (2011). Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 6899–6904. doi: 10.1073/pnas.1103014108
Lottaz, C., Iseli, C., Jongeneel, C. V., and Bucher, P. (2003). Modeling sequencing errors by combining hidden markov models. Bioinformatics 19, ii103–ii112. doi: 10.1093/bioinformatics/btg1067
Lv, H., Qu, G., Qi, X., Lu, L., Tian, C., and Ma, Y. (2013). Transcriptome analysis of Chlamydomonas reinhardtii during the process of lipid accumulation. Genomics 101, 229–237. doi: 10.1016/j.ygeno.2013.01.004
Magnuson, K., Jackowski, S., Rock, C. O., and Cronan, J. E. (1993). Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57, 522–542.
Mata, T. M., Martins, A. A., and Caetano, N. S. (2010). Microalgae for biodiesel production and other applications: a review. Renew. Sustain. Energy Rev. 14, 217–232. doi: 10.1016/j.rser.2009.07.020
McNeely, K., Xu, Y., Bennette, N., Bryant, D. A., and Dismukes, G. C. (2010). Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 76, 5032–5038. doi: 10.1128/aem.00862-10
Mendez-Perez, D., Begemann, M. B., and Pfleger, B. F. (2011). Modular synthase-encoding gene involved in α-Olefin biosynthesis in Synechococcus sp. Strain PCC 7002. Appl. Environ. Microbiol. 77, 4264–4267. doi: 10.1128/aem.00467-11
Meuser, J., Baxter, B., Spear, J., Peters, J., Posewitz, M., and Boyd, E. (2013). Contrasting patterns of community assembly in the stratified water column of great salt lake, Utah. Microb. Ecol. 66, 268–280. doi: 10.1007/s00248-013-0180-9
Miller, R., Wu, G., Deshpande, R. R., Vieler, A., Gärtner, K., Li, X., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154, 1737–1752. doi: 10.1104/pp.110.165159
Moellering, E. R., and Benning, C. (2010). RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9, 97–106. doi: 10.1128/ec.00203-09
Patil, V., Källqvist, T., Olsen, E., Vogt, G., and Gislerød, H. (2007). Fatty acid composition of 12 microalgae for possible use in aquaculture feed. Aquac. Int. 15, 1–9. doi: 10.1007/s10499-006-9060-3
Quinn, J. C., and Davis, R. (2015). The potentials and challenges of algae based biofuels: a review of the techno-economic, life cycle, and resource assessment modeling. Bioresour. Technol. 184, 444–452. doi: 10.1016/j.biortech.2014.10.075
Radakovits, R., Eduafo, P. M., and Posewitz, M. C. (2011). Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 13, 89–95. doi: 10.1016/j.ymben.2010.10.003
Radakovits, R., Jinkerson, R. E., Darzins, A., and Posewitz, M. C. (2010). Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9, 486–501. doi: 10.1128/ec.00364-09
Radakovits, R., Jinkerson, R. E., Fuerstenberg, S. I., Tae, H., Settlage, R. E., Boore, J. L., et al. (2012). Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat. Commun. 3, 686. doi: 10.1038/ncomms1688
Renaud, S. M., Thinh, L.-V., and Parry, D. L. (1999). The gross chemical composition and fatty acid composition of 18 species of tropical Australian microalgae for possible use in mariculture. Aquaculture 170, 147–159. doi: 10.1016/S0044-8486(98)00399-8
Ruffing, A. M. (2014). Improved free fatty acid production in Cyanobacteria with Synechococcus sp. PCC 7002 as Host. Front. Bioeng. Biotechnol. 2:17. doi: 10.3389/fbioe.2014.00017
Schenk, P., Thomas-Hall, S., Stephens, E., Marx, U., Mussgnug, J., Posten, C., et al. (2008). Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenergy Res. 1, 20–43. doi: 10.1007/s12155-008-9008-8
Simpson, J. T., Wong, K., Jackman, S. D., Schein, J. E., Jones, S. J. M., and Birol, Ý (2009). ABySS: a parallel assembler for short read sequence data. Genome Res 19, 1117–1123. doi: 10.1101/gr.089532.108
Slabaugh, M. B., Leonard, J. M., and Knapp, S. J. (1998). Condensing enzymes from Cuphea wrightii associated with medium chain fatty acid biosynthesis. Plant J. 13, 611–620. doi: 10.1046/j.1365-313X.1998.00065.x
Thelen, J. J., and Ohlrogge, J. B. (2002). Metabolic engineering of fatty acid biosynthesis in plants. Metab. Eng. 4, 12–21. doi: 10.1006/mben.2001.0204
Vieler, A., Wilhelm, C., Goss, R., Süß, R., and Schiller, J. (2007). The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem. Phys. Lipids 150, 143–155. doi: 10.1016/j.chemphyslip.2007.06.224
Voelker, T., Worrell, A., Anderson, L., Bleibaum, J., Fan, C., Hawkins, D., et al. (1992). Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science 257, 72–74. doi: 10.1126/science.1621095
Voelker, T. A., and Davies, H. M. (1994). Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J. Bacteriol. 176, 7320–7327.
Work, V. H., Bentley, F. K., Scholz, M. J., D’Adamo, S., Gu, H., Vogler, B. W., et al. (2013). Biocommodities from photosynthetic microorganisms. Environ. Progress Sustain. Energy 32, 989–1001. doi: 10.1002/ep.11849
Work, V. H., Melnicki, M. R., Hill, E., Davies, F. K., Kucek, L., Beliaev, A., et al. (2015). Lauric acid production in a glycogen-less Synechococcus sp. PCC 7002 mutant. Front. Bioeng. Biotechnol. 3:48. doi: 10.3389/fbioe.2015.00048
Xu, Y., Alvey, R., Byrne, P., Graham, J., Shen, G., and Bryant, D. (2011). “Expression of genes in cyanobacteria: adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002,” in Photosynthesis Research Protocols, ed. R. Carpentier (New York, NY: Humana Press), 273–293.
Xu, Y., Tiago Guerra, L., Li, Z., Ludwig, M., Charles Dismukes, G., and Bryant, D. A. (2013). Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain PCC 7002: cell factories for soluble sugars. Metab. Eng. 16, 56–67. doi: 10.1016/j.ymben.2012.12.002
Keywords: medium chain fatty acids, Chaetoceros, type III ketoacyl-ACP synthase, Synechococcus sp. PCC 7002, lauric acid production, RNAseq
Citation: Gu H, Jinkerson RE, Davies FK, Sisson LA, Schneider PE and Posewitz MC (2016) Modulation of Medium-Chain Fatty Acid Synthesis in Synechococcus sp. PCC 7002 by Replacing FabH with a Chaetoceros Ketoacyl-ACP Synthase. Front. Plant Sci. 7:690. doi: 10.3389/fpls.2016.00690
Received: 25 January 2016; Accepted: 05 May 2016;
Published: 26 May 2016.
Edited by:
Jeffrey Leblond, Middle Tennessee State University, USAReviewed by:
Qing Liu, Commonwealth Scientific and Industrial Research Organisation, AustraliaJeremy Dahmen, Matrix Genetics, USA
Copyright © 2016 Gu, Jinkerson, Davies, Sisson, Schneider and Posewitz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew C. Posewitz, mposewit@mines.edu
 Huiya Gu
Huiya Gu Robert E. Jinkerson
Robert E. Jinkerson Fiona K. Davies
Fiona K. Davies Lyle A. Sisson1
Lyle A. Sisson1 Philip E. Schneider
Philip E. Schneider Matthew C. Posewitz
Matthew C. Posewitz