- 1South Subtropical Crop Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, China
- 2Key Laboratory of Tropical Fruit Biology, Ministry of Agriculture, Zhanjiang, China
- 3The Key Laboratory of Gene Engineering of the Ministry of Education, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
Exogenous ethylene, or ethephon, has been widely used to induce pineapple flowering, but the molecular mechanism behind ethephon induction is still unclear. In this study, we cloned four genes encoding ethylene receptors (designated AcERS1a, AcERS1b, AcETR2a, and AcETR2b). The 5′ flanking sequences of these four genes were also cloned by self-formed adaptor PCR and SiteFinding-PCR, and a group of putative cis-acting elements was identified. Phylogenetic tree analysis indicated that AcERS1a, AcERS1b, AcETR2a, and AcETR2b belonged to the plant ERS1s and ETR2/EIN4-like groups. Quantitative real-time PCR showed that AcETR2a and AcETR2b (subfamily 2) were more sensitive to ethylene treatment compared with AcERS1a and AcERS1b (subfamily 1). The relative expression of AcERS1b, AcETR2a, and AcETR2b was significantly increased during the earlier period of pineapple inflorescence formation, especially at 1–9 days after ethylene treatment (DAET), whereas AcERS1a expression changed less than these three genes. In situ hybridization results showed that bract primordia (BP) and flower primordia (FP) appeared at 9 and 21 DAET, respectively, and flowers were formed at 37 DAET. AcERS1a, AcERS1b, AcETR2a, and AcETR2b were mainly expressed in the shoot apex at 1–4 DAET; thereafter, with the appearance of BP and FP, higher expression of these genes was found in these new structures. Finally, at 37 DAET, the expression of these genes was mainly focused in the flower but was also low in other structures. These findings indicate that these four ethylene receptor genes, especially AcERS1b, AcETR2a, and AcETR2b, play important roles during pineapple flowering induced by exogenous ethephon.
Introduction
Pineapple (Ananas comosus L.) is one of the most popular tropical fruits. In China and many other pineapple-planting regions, one of the most serious problems of pineapple cultivation is natural flowering, which results in unscheduled fruiting (Bartholomew et al., 2003). In China, natural flowering is often observed in February after the pineapples have experienced low temperatures in January or the previous December. This natural flowering results in most of the fruits being harvested from May to June; furthermore, the natural flowering lasts for several weeks because it is not synchronized (Zhu et al., 2012), resulting in increased cultivation costs and decreased profit for pineapple growers.
Once reproductive development is initiated, pineapple inflorescence and fruit development continue without interruption until fruit maturation (Bartholomew et al., 2003). Therefore, to produce fruit in every month of the year, ethylene or ethephon (which degrades to produce ethylene) has been widely used to induce pineapple flowering (Bartholomew et al., 2003). Although there has been much research concerning the role of ethylene in flower development (Lin et al., 2009), the molecular mechanism behind ethylene induction of flower development is still unclear.
The gaseous plant hormone ethylene is an important regulator of plant growth and development, including seed germination, flower development, fruit ripening, leaf senescence, and response to environmental stress (Abeles et al., 1992; Ma et al., 2014; Wen, 2015). The transition from vegetative to reproductive growth is a key step in a plant’s life cycle. Flowering time is controlled by various endogenous and exogenous signals, including photoperiod, temperature, and plant age (Srikanth and Schmid, 2011; Song et al., 2013; van Doorn and Kamdee, 2014). Ethylene plays a role in the regulation of flowering timing, but its effects appear complicated (Ma et al., 2014). For example, in Arabidopsis, Ogawara et al. (2003) reported that ethylene promoted floral transition; however, the ctr1 mutant and the 1-aminocyclopropane-1-carboxylic acid (ACC)-treated wild-type showed delayed flowering, indicating that ethylene inhibited Arabidopsis flowering (Achard et al., 2007). In rice, converse effects of ethylene have also been observed. Overexpression of OsETR2 reduces ethylene sensitivity and delays floral transition, whereas the suppression of OsETR2 by RNAi enhances ethylene sensitivity and accelerates rice flowering, indicating that ethylene promotes rice flowering (Wuriyanghan et al., 2009). By contrast, transgenic lines overexpressing OsCTR2 and the osctr2 loss-of-function mutant show delayed flowering, suggesting that ethylene represses the floral transition in rice (Wang et al., 2013b). On the other hand, in pineapple (Ananas comosus), it has been proposed that flowering is triggered by a small burst of ethylene production in the meristem in response to environmental cues. AcACS2 was induced in the meristem during induction of the flowering, and silencing AcACS2 in transgenic pineapple plants results in a significant flowering delay. Therefore, AcACS2 may be the main contributor to the ethylene promotion of pineapple flowering (Trusov and Botella, 2006). These contradictory observations may be due to different growth conditions, genetic backgrounds, or mechanisms employed by these signaling components (Ma et al., 2014).
A linear signaling pathway has been established on the basis of genetic analyses of ethylene-responsive Arabidopsis mutants (Ma et al., 2014). In this pathway, the first step is the binding of ethylene to the ethylene receptors, which play a crucial role by negatively regulating ethylene responses (Chang and Stadler, 2001). In Arabidopsis thaliana, five ethylene receptor genes have been identified, including ETR1, ETR2, ERS1, ERS2, and EIN4 (Hua et al., 1998; Schaller and Kieber, 2002; Chen et al., 2005; O’Malley et al., 2005; Kendrick and Chang, 2008). Based on phylogenetic analysis, the ethylene receptors can be divided into two subfamilies that share structural features: subfamily 1 has three hydrophobic regions plus a conserved histidine kinase domain and includes ETR1 and ERS1; whereas subfamily 2 has four hydrophobic regions and a more diverged kinase domain and includes ETR2, ERS2, and EIN4 (Chang and Stadler, 2001; Schaller and Kieber, 2002; Chen et al., 2005).
In the absence of the ethylene, receptors actively suppress ethylene responses; when ethylene binds, the suppression is removed and the response occurs (Wen, 2015). All five Arabidopsis receptors are involved in ethylene signaling and have overlapping roles in regulating ethylene responses (Hua et al., 1995, 1998; Hall and Bleecker, 2003). However, recent studies showed that the five receptor isoforms are not entirely redundant in their roles (Hall and Bleecker, 2003; Binder et al., 2004, 2006; Seifert et al., 2004; O’Malley et al., 2005; Xie et al., 2006; Kevany et al., 2007; Qu et al., 2007; Plett et al., 2009a,b; Liu et al., 2010; Wang et al., 2013a; Wilson et al., 2014). For instance, only specific ethylene receptors mediate fruit ripening in tomato, but other receptors had little or no effect on fruit ripening (Tieman et al., 2000; Kevany et al., 2007). In addition, the etr1 ers1 mutant phenotype can be rescued by the expression of subfamily 1 receptors but not subfamily 2 receptors, which indicated the unique function for subfamily 1 receptors in Arabidopsis (Wang et al., 2013a).
Ethylene receptors are involved in many biological processes, such as ethylene-induced nutational bending of the apical hook, recovery of growth after ethylene treatment, nutational bending, control of flowering time and fruit maturation and response to salt stress (Merchante et al., 2013; Shakeel et al., 2013). Ethylene receptor genes can be induced by various abiotic stresses, including wounding, salt and drought treatments (Zhang et al., 1999; Zhang J. et al., 2001; Zhang J.S. et al., 2001; Xie et al., 2002; Tao et al., 2015), and ethylene receptors also regulate plant defense responses (He et al., 2005; Cao et al., 2007; Wilson et al., 2014; Tao et al., 2015). For example, under salt stress, loss of ETR1 or EIN4 promotes Arabidopsis seed germination, loss of ETR2 delays germination, and loss of either ERS1 or ERS2 has no measurable effect on germination (Wilson et al., 2014). In addition to stress responses, ethylene receptors are highly expressed in reproductive organs and may be involved in reproductive development (Hua et al., 1998; Tieman et al., 2000; Yamasaki et al., 2000; Zhang J. et al., 2001; Zhang J.S. et al., 2001; Klee, 2002, 2004; Xie et al., 2002; Rieu et al., 2003; Zhou et al., 2006; Wuriyanghan et al., 2009; Thongkum et al., 2015). For instance, reduction of the expression of the tomato ethylene receptors LeETR4 or LeETR6 increased ethylene production and promoted ripening (Tieman et al., 2000; Kevany et al., 2007), which means that climacteric fruit ripening is also controlled at the receptor level (Lin et al., 2009).
Here, we reported the isolation and characterization of four genes encoding ethylene receptors in pineapple and their promoters. Using two different treatment concentrations of ethephon, we studied the genes’ temporal and spatial expression during inflorescence induction by quantitative real-time PCR (qRT-PCR) and in situ hybridization.
Materials and Methods
Plant Materials and Treatments
Pineapples (Ananas comosus L. cv. Perola) were planted in the South Subtropical Crop Research Institute, at Zhanjiang, Guangdong province, China (20°39′ N latitude and 110°15′ E longitude). Homogenous 15-month-old plants were selected for induced flowering. In September 2012, after 5 pm in the evening, 50 ml of 200 or 1200 mg l-1 ethephon (5 or 30 ml of 40% ethephon diluted to 10 l with water, respectively) was poured over each group of plants; the control plant group was treated with 50 ml of water. Each treatment group and the control group contained at least 300 plantings. The two different concentrations of ethephon were selected because our previous study (Liu et al., 2009) showed that the application of 200 and 800 mg l-1 ethephon induced 100% pineapple (‘Zhenzhu’) flowering, but the higher ethephon concentration resulted in smaller fruits.
After ethephon treatment, the shoot apex or infiorescence of the pineapple was collected at 0, 1, 2, 5, 9, 14, 21, 28, and 37 days and either immediately frozen in liquid nitrogen followed by storage at -80°C until RNA extraction or fixed for subsequent in situ hybridization.
RT-PCR with Degenerate Primers and Cloning of PCR Products
After 2 days of ethylene treatment, the shoot apex was collected and was used to extract total RNA using the method of Fu et al. (2004). cDNA was synthesized from 2 μg of RNA using PrimeScript II first Strand cDNA Synthesis Kit (Takara Bio Inc., Dalian, China) and Oligo dT as a primer, following the manufacturer’s instructions.
This cDNA was used as a template to isolate DNA fragments encoding ethylene receptors by PCR. We used five pairs of degenerate primers (Supplementary Table S1) designed from the alignment of sequences from other plants in the program iCODEHOP v1.1 (Boyce et al., 2009). The conditions of the PCR were as follows: 94°C for 3 min, 40 cycles at 94°C for 30 s, 54°C for 1 min, and 72°C for 2 min and a final 7 min extension at 72°C using a DNA thermal cycler. The purified products were cloned into the pMD-18 T vector (Takara Bio Inc., Dalian, China) and sequenced by the Invitrogen Company (Guangzhou, China) using an ABI 3770 DNA sequencer.
Cloning of AcERS1a, AcERS1b, AcETR2a, and AcETR2b Full-Length cDNAs by RACE
From the initial partial cDNA fragments, five full-length ethylene receptor cDNA clones were produced by 5′ and 3′ RACE. The RACE reactions were performed using the SMARTerTM RACE cDNA Amplification Kit (Clontech) according to the manufacturer’s instructions.
To obtain the 5′ cDNA regions of the ethylene receptor genes, the primer sets AcETR1SP1 and UPM, AcERS1SP1 and UPM, AcETR2SP1 and UPM, AcEIN4SP1 and UPM, and AcERS2SP1 and UPM were used for the first-round PCR of the 5′ RACE for AcETR1, AcERS1, AcETR2, AcEIN4, and AcERS2, respectively. The primer sets AcETR1SP2 and NUP, AcERS1SP2 and NUP, AcETR2SP2 and NUP, AcEIN4SP2 and NUP, and AcERS2SP2 and NUP were then used for nested PCR. To obtain the 3′ cDNA regions of the ethylene receptor genes, the primer sets AcETR1SP3 and UPM, AcERS1SP3 and UPM, AcETR2SP3 and UPM, AcEIN4SP3 and UPM, and AcERS2SP3 and UPM were used for the first-round PCR of the 3′ RACE. The primer sets AcETR1SP2 and NUP, AcERS1SP2 and NUP, AcETR2SP2 and NUP, AcEIN4SP2 and NUP, and AcERS2SP2 and NUP were then used for nested PCR. The conditions for the first-round 5′ RACE and 3′ RACE PCR experiments were as described by Li et al. (2012), and primers listed in Supplementary Table S1.
The sequencing and BLAST results showed that the cDNAs from the AcERS2 primer set and the AcEIN4 primer set were the same; they are typical undifferentiated ETR2/EIN4-type receptors found throughout monocots and more similar to ETR2 than EIN4 (Gallie, 2015), therefore both were designated AcETR2b. In addition, the cDNA clone from the AcETR1 primer set belonged to ERS1, not ETR1, so it was designated AcERS1b; the cDNA clone from AcERS1 was designated AcERS1a.
According to the 5′ RACE and 3′ RACE results, the specific primer sets AcERS1aSP5 and AcERS1aSP6, AcERS1bSP5 and AcERS1bSP6, AcETR2aSP5 and AcETR2aSP6, and AcETR2bSP5 and AcETR2bSP6 (Supplementary Table S1) were used to obtain the full-length cDNAs of AcERS1a, AcERS1b, AcETR2a, and AcETR2b, respectively. The full-length cDNAs were amplified via PCR using the 5′ RACE-Ready cDNA as the pineapple template, the conditions of the PCR were as described by Li et al. (2012). The PCR products were purified and cloned into the pMD-18T vector and sequenced.
Isolation of the AcERS1a, AcERS1b, AcETR2a, and AcETR2b Genes from Pineapple Genomic DNA
Genomic DNA from pineapple leaves was extracted using the Plant DNAout kit (Tiandz, Beijing, China) following the manufacturer’s instructions. The primer sets are listed in Supplementary Table S1. The specific primers sets AcERS1aSP5 and AcERS1aSP6, AcERS1bSP5 and AcERS1bSP6, AcETR2aSP5 and AcETR2aSP6, and AcETR2bSP5 and AcETR2bSP6 (Supplementary Table S1), were used to amplify the AcERS1a, AcERS1b, AcETR2a, and AcETR2b genes, respectively, from pineapple genomic DNA.
PCR amplification was carried out under the following conditions: 3 min at 94°C, followed by 30 cycles of 10 s at 98°C and 8 min at 68°C. The purified products were cloned into the pMD-18T vector and sequenced.
Promoters Isolation
Self-formed adaptor PCR (SEFA PCR; Wang et al., 2007) was used to isolate the promoter of AcERS1a, while SiteFinding-PCR (Tan et al., 2005) was used to isolate the promoters of AcERS1b, AcETR2a, and AcETR2b. Genomic DNA from pineapple leaves was extracted as above. Based on the genomic DNA sequences of AcERS1a, AcERS1b, AcETR2a, and AcETR2b, primers for use in SEFA PCR and SiteFinding-PCR were designed (listed in Supplementary Table S1). The PCR reactions were performed as described by Tan et al. (2005) and Wang et al. (2007) with slight modifications. In the SEFA PCR, only three primers were used in the PCR (i.e., AcERS1a-GSP1, AcERS1a-GSP2, and AcERS1a-GSP3); for the SiteFinding-PCR, long Taq DNA polymerase was purchased from Takara (Dalian, China), and GSP3 was not used in the PCR. The purified products were cloned into the pMD-18T vector and sequenced.
Bioinformatics Analysis
The nucleotide sequences, deduced amino acid sequences, and open reading frames (ORFs) were analyzed online1. The genomic DNA of the five Arabidopsis thaliana ethylene receptor genes was obtained from TAIR2. The deduced amino acid sequences were compared using BLAST against the NCBI’s Conserved Domain Database (CDD)3 to find the conserved domains. MW and pI predictions for the deduced AcERS1a, AcERS1b, AcETR2a, and AcETR2b oligopeptides were carried out with the Compute pI/MW tool4. Sequence alignment was performed using ClustalW5. The ClustalW-produced alignment file was formatted using the BOXSHADE program6. Phylogenetic analysis and construction of a neighbor-joining tree were performed using the MEGA 4.0 software using the bootstrap method with 1,000 bootstrap iterations. Putative cis-acting elements upstream of the start codons of AcERS1a, AcERS1b, AcETR2a, and AcETR2b were identified by searching the PLACE databases7 (Higo et al., 1999).
Quantitative Real-Time PCR
AcERS1a, AcERS1b, AcETR2a, and AcETR2b transcript levels were determined by quantitative real-time PCR. Total RNA was extracted using the method of Fu et al. (2004). First-strand cDNA was prepared from 1 μg of total RNA with oligo dT primers using the PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio Inc., Dalian, China) according to the manufacturer’s protocol and stored at -20°C. Real-time PCR was performed using the Thermo Scientific DyNAmo ColorFlash SYBR Green qPCR Kit (Thermo) on a Stratagene Mx3005P quantitative PCR machine. The following program was used: 95°C for 7 min, followed by 40 cycles of 95°C for 5 s, 56°C for 15 s, and 72°C for 25 s. As an internal control, levels of actin were quantified in parallel with the target genes (Li et al., 2012). Normalization and fold changes were calculated using the ΔΔCt method, three independent biological replicates were used in the analysis. Primer sets are listed in Supplementary Table S2.
In Situ Hybridization
Samples were fixed in formaldehyde and acetic acid (50% ethanol, 5% acetic acid, and 3.7% formaldehyde) for 24 h at 4°C, and the hybridization and immunological detection steps were performed as described by Dai et al. (2007). Nine-micrometer thick sample sections were cut with a microtome; all of the sections in this study were cut longitudinally. The AcERS1a, AcERS1b, AcETR2a, and AcETR2b probes were amplified using gene-specific primers (Supplementary Table S2). The PCR fragments were inserted into the NcoI and SalI sites of pGE-T (Tiangen, China) and transcribed in vitro from either the T7 or SP6 promoter for sense or antisense strand synthesis, respectively, using the Digoxigenin RNA Labeling kit (Roche).
Subcellular Localization
To analyze the subcellular localization, the ORFs of AcERS1a, AcERS1b, AcETR2a, and AcETR2b without termination codon were obtained by PCR amplification using specific primers (Supplementary Table S3), and subsequently cloned into the pGFP2 vector at the XhoI/KpnI sites using In-Fusion HD Cloning Kits (Clontech) according to the manufacturer’s instructions, which resulted in the 35S::gene-GFP vectors under the control of the CaMV 35S promoter. The fusion constructs and the control GFP vector were transformed into tobacco BY2 protoplasts by PEG method as described by Shan et al. (2012). GFP fluorescence was observed with a fluorescence microscope (Zeiss Axioskop 2 Plus).
Statistical Analysis
The data were analyzed using analysis of variance (ANOVA) followed by Duncan’s multiple range test or an independent-samples t-test at a 5% significance level.
Results
Effect of Different Concentrations of Ethephon on Pineapple Flowering and Fruiting
Flowering was observed approximately 2.5 months after ethephon treatment; however, at that time, none of the control plants were flowering. The control plants flowered naturally during the following February. A previous study showed that higher ethephon concentrations inhibited the growth of ‘Zhenzhu’ pineapple fruits and that smaller fruits were harvested (Liu et al., 2009). However, for ‘Perola’ pineapples, 200 or 1200 mg l-1 ethephon had no effect on the flowering rate (both were 100%), flowering time (the time to the first flower appearing after ethylene treatment), or fruit qualities, such as fruit weight and number of fruitlets (Table 1). The different reactions to higher ethephon concentrations may be due to the different sensitivities to ethephon treatment between ‘Perola’ and ‘Zhenzhu’ pineapples. Therefore, we studied only the ethylene receptor genes’ in vivo transcript profiles for the 200 mg l-1 ethephon treatment.
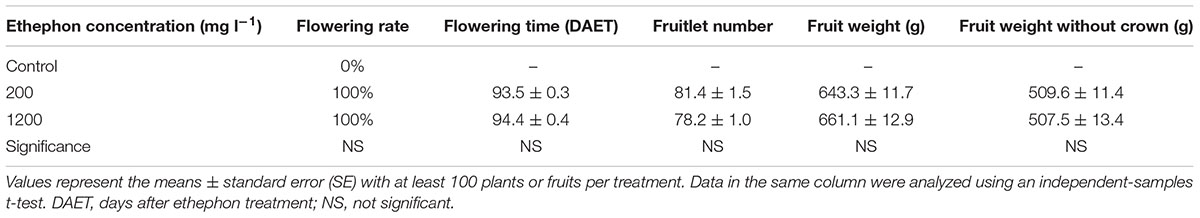
TABLE 1. Effect of different concentrations of ethephon on pineapple flowering time and fruit weight.
Isolation and Characterization of the cDNA Clones of AcERS1a, AcERS1b, AcETR2a, and AcETR2b from Pineapple
Using different primer sets (Supplementary Table S1), five different cDNA fragments were obtained from RT-PCR, and using RACE extension, five full-length cDNA clones of pineapple ethylene receptor genes were obtained. However, the sequencing and BLAST results showed that the cDNAs from the AcERS2 primer set and the AcEIN4 primer set were the same and did not contain the conserved histidine in the H-motif; therefore, they belong to the undifferentiated ETR2/EIN4 group and are more similar to ETR2 than EIN4 (Gallie, 2015). Both were designated AcETR2b (the cDNA clone from the AcETR2 primer set was designated AcETR2a). On the other hand, the BLAST results showed that the cDNA clone from the AcETR1 primer set belonged to ERS1 and not ETR1, so it was designated AcERS1b. The cDNA clone from AcERS1 was designated AcERS1a.
The corresponding full-length AcERS1a, AcERS1b, AcETR2a, and AcETR2b cDNA clones were 2304, 2411, 2558, and 2635 bp long and contained ORFs of 1938, 1905, 2268, and 2277 bp, respectively. The AcERS1a, AcERS1b, AcETR2a, and AcETR2b genes encode a putative protein of 645, 634, 755, and 758 amino acids with a predicted MW of 72.11, 71.28, 83.19, and 84.53 kDa and a pI of 6.39, 6.77, 8.71, and 6.67, respectively. The sequences of AcERS1a, AcERS1b, AcETR2a, and AcETR2b have been submitted to the GenBank database (accession numbers KM062066, KM062063, KM062064, and KM062065, respectively).
The lengths of the genomic DNA sequences of AcERS1a, AcERS1b, AcETR2a, and AcETR2b are shown in Figure 1. Comparisons between the cDNA sequences and the genomic sequences revealed that the genomic DNA lengths of AcERS1a, AcERS1b, and AtERS1 were different, but their genomic DNA structures were similar, all had five exons and four introns in their genomic DNA (Figure 1A). Similarly, although the genomic DNA lengths of AcETR2a and AcETR2b were different, both had two exons and one intron (Figure 1B). The corresponding 5′ untranslated regions (UTRs) of AcERS1a, AcERS1b, AcETR2a, and AcETR2b were 1900, 1101, 32, and 122 bp, respectively. When the UTRs were compared with the genes’ cDNAs, there were 1734 and 882 bp-long introns located in the genes’ 5′ UTRs located 40 and 41 bp from the start codons of the AcERS1a and AcERS1b genes, respectively.
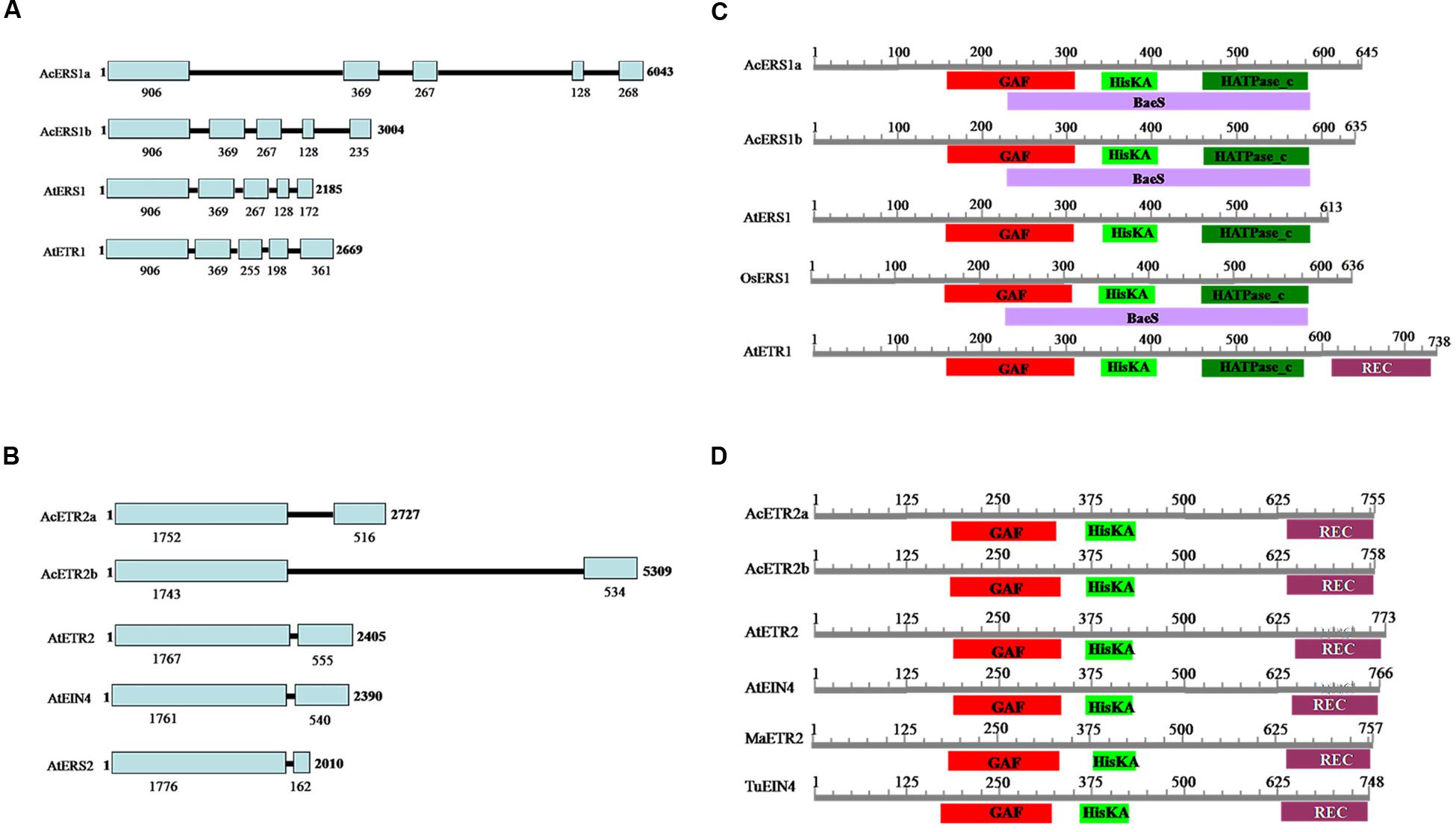
FIGURE 1. Pineapple (Ananas comosus L.) ethylene receptors. (A) Comparison of the genomic structures of AcERS1a (KR610528) and AcERS1b (KR610529) with AtETR1 (TAIR: AT1G66340) and AtERS1 (TAIR: AT2G40940). (B) Comparison of the genomic structures of AcETR2a (KR610530) and AcETR2b (KR610531) with AtETR2 (TAIR: AT3G23150), AtEIN4 (TAIR: AT3G04580) and AtERS2 (TAIR: AT1G04310). (C) Domain structures of AcERS1a, AcERS1b, and other ERS1s and ETR1 proteins. (D) Domain structures of AcETR2a, AcETR2b, and other ETR2 and EIN4 proteins. The sequences used in the domain structures were obtained from the GenBank databases: AcERS1a (AIT52526), AcERS1b (AIT52523), AcETR2a (AIT52524), and AcETR2b (AIT52525) from Ananas comosus (Ac); AtERS1 (AAC49090), AtETR1 (AAA70047), AtETR2 (AAC62208), and AtEIN4 (AAD02485) from Arabidopsis thaliana (At); MaETR2 (XP_009386224) from Musa acuminata AAA Group (Ma); OsERS1 (AAB72193), from Oryza sativa (Os); and TuEIN4 (EMS56298) from Triticum urartu (Tu).
Homology searches were performed with the BLAST and BLASTX algorithms to confirm sequence identity. The results indicated that the deduced amino acid sequence of AcERS1a and AcERS1b shared a high sequence identity with the ETR1 and ERS1 proteins of diverse plant species, and AcETR2a and AcETR2b shared high sequence identity with the genes encoding ETR2s or EIN4s of diverse plant species.
A search of the CDD (Marchler-Bauer et al., 2015) for conserved protein domains indicated that both the predicted amino acid sequences of AcERS1a and AcERS1b contained a GAF domain, a HisKA (histidine kinase A) domain, a HATPase_c (histidine kinase-like ATPase) domain, and a BaeS (signal transduction histidine kinase) domain. These four domains are also found in OsERS1 (AAB72193), but the BaeS domain is not found in AtERS1 and AtETR1 (Figure 1C, Supplementary Figure S1).
Similarly, a search of the CDD for conserved protein domains indicated that both AcETR2a and AcETR2b contained a GAF domain, a HisKA domain and a REC (signal receiver domain) domain, which was similar to MaETR2, AtETR2, and AtEIN4 (Figure 1D; Supplementary Figure S1).
To evaluate the molecular evolutionary relationships of ethylene receptors in other species, we retrieved 46 different ethylene receptor proteins from several plant species through BLASTP searches, and a phylogenetic tree was constructed using the neighbor-joining method. As shown in Figure 2, phylogenetic analysis based on multiple alignment showed that there were two distinct subfamilies: AcERS1a and AcERS1b fell into a cluster with AtERS1 and AtETR1 (subfamily 1), whereas AcETR2a and AcETR2b fell into the other cluster with AtETR2, AtEIN4, and AtERS2 (subfamily 2).
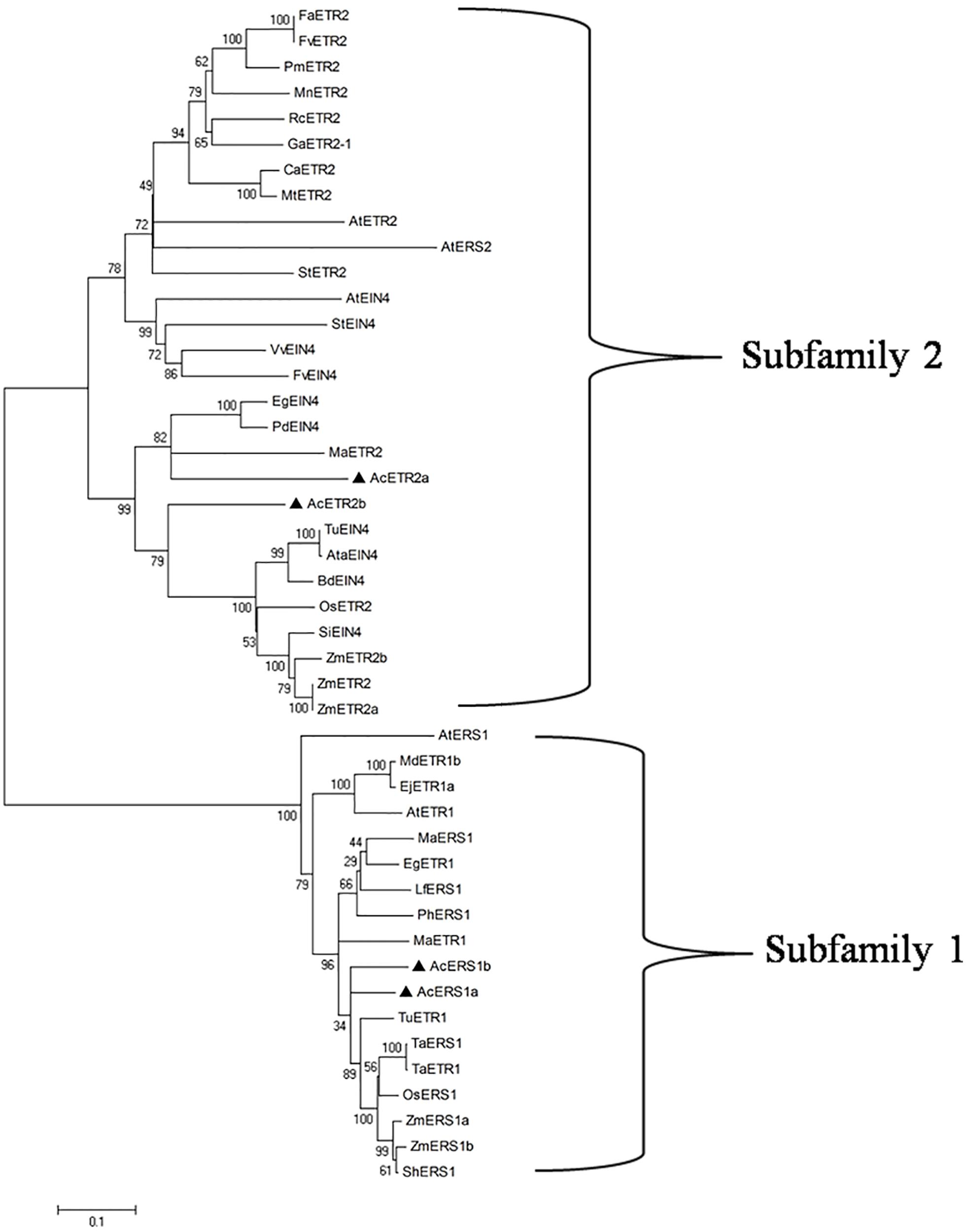
FIGURE 2. Phylogenetic analysis of ethylene receptors. The phylogenetic tree of the ethylene receptor proteins. Using the ClustalW program, the tree was constructed using the neighbor-joining method after sequence alignment. The scale bar corresponds to 0.1 amino acid substitutions per residue. The sequences used in this alignment were obtained from the GenBank databases: AcERS1a (AIT52526), AcERS1b (AIT52523), AcETR2a (AIT52524), and AcETR2b (AIT52525) from Ananas comosus (Ac); AtERS1 (AAC49090), AtETR1 (AAA70047), AtETR2 (AAC62208), AtEIN4 (AAD02485), and AtERS2 (AAC62209) from Arabidopsis thaliana (At); AtaEIN4 (EMT13419) from Aegilops tauschii (Ata); BdEIN4 (XP_003580931) from Brachypodium distachyon (Bd); CaETR2 (XP_004495309) from Cicer arietinum (Ca); EgETR1 (XP_010914501) and EgEIN4 (XP_010935766) from Elaeis guineensis (Eg); EjETR1a (AGE15296) from Eriobotrya japonica (Ej); FaETR2 (CAC48386) from Fragaria × ananassa (Fa); FvETR2 (XP_004288459) and FvEIN4 (XP_004302073) from Fragaria vesca subsp. Vesca (Fv); GaETR2-1 (AGG55710) from Gossypium arboretum (Ga); LfERS1 (ABD66593) from Lilium formosanum × Lilium longiflorum (Lf); MaERS1 (BAF44104) and MaETR2 (XP_009386224) from Musa acuminata AAA Group (Ma); MdETR1b (AAW69924) from Malus domestica (Md); MnETR2 (EXB98165) from Morus notabilis (Mn); MtETR2 (KEH42695) from Medicago truncatula (Mt); OsERS1 (AAB72193) and OsETR2 (AAN15203) from Oryza sativa (Os); PdEIN4 (XP_008787409) from Phoenix dactylifera (Pd); PhERS1 (AAD04949) from Phalaenopsis hybrid cultivar (Ph); PmETR2 (XP_008224393) from Prunus mume (Pm); RcETR2 (XP_002529316) from Ricinus communis (Rc); ShERS1 (ADJ66722) from Saccharum hybrid cultivar ROC20 (Sh); SiEIN4 (XP_004975078) from Setaria italica (Si); StETR2 (XP_006350680) and StEIN4 (XP_006345886) from Solanum tuberosum (St); TaERS1 (ADJ67795) and TaETR1 (AHL44971) from Triticum aestivum (Ta); TuETR1 (EMS66524) and TuEIN4 (EMS56298) from Triticum urartu (Tu); VvEIN4 (XP_002270757) from Vitis vinifera (Vv); and ZmERS1a (AFW67901), ZmERS1b (DAA50871), ZmETR2 (NP_001104852), ZmETR2a (AY359580), and ZmETR2b (AY359581) from Zea mays (Zm).
Isolation and Identification of the Promoters
Using SEFA PCR (Wang et al., 2007), we isolated a 5114-bp fragment upstream of the start codon of AcERS1a, and using SiteFinding-PCR (Tan et al., 2005), we isolated 3502-bp, 1592-bp and 1149-bp fragments upstream of the start codon of AcERS1b, AcETR2a, and AcETR2b (promoter sequences listed in the Supplementary Figure S2).
Using the PLACE web tools, several putative cis-regulatory elements were deciphered from the promoter sequences of AcERS1a, AcERS1b, AcETR2a, and AcETR2b. The types of cis-regulatory elements between AcERS1a and AcERS1b were similar, and their differences were mostly in the number of repeats (Supplementary Table S4). TATA box sequence elements, which are required for critical and precise transcription initiation, were found in large numbers in all promoters. Several other types of regulatory elements were found in the four promoters, including cis-acting elements involved in the response to ethylene, cytokinin and auxin (Supplementary Table S4). In general, more cis-regulatory elements and repeats were found in the promoter regions of AcERS1a and AcERS1b than of AcETR2a and AcETR2b; the detailed comparisons of the putative cis elements in these promoters are listed in Supplementary Table S4.
Expression of AcERS1a, AcERS1b, AcETR2a, and AcETR2b during Pineapple Flowering Formation
For the control, the relative expression levels of AcERS1a, AcERS1b, AcETR2a, and AcETR2b were similar; none significantly changed during the whole period. However, for the ethylene treatment groups, the expression pattern of AcERS1a was different from the other three genes.
For the 200 mg l-1 ethylene treatment, the relative expression levels of AcERS1a were up-regulated compared with the control at 1 DAET (less than 1.5-fold), 5 DAET (approximately 2-fold) and 37 DAET (approximately 1.5-fold; Figure 3). The 1200 mg l-1 ethylene treatment response was similar to the 200 mg l-1 ethylene treatment. Compared with the control, the relative expression levels of AcERS1a were up-regulated at 1 and 2 DAET (less than 1.5-fold) when treated with 1200 mg l-1 ethylene (Figure 3).
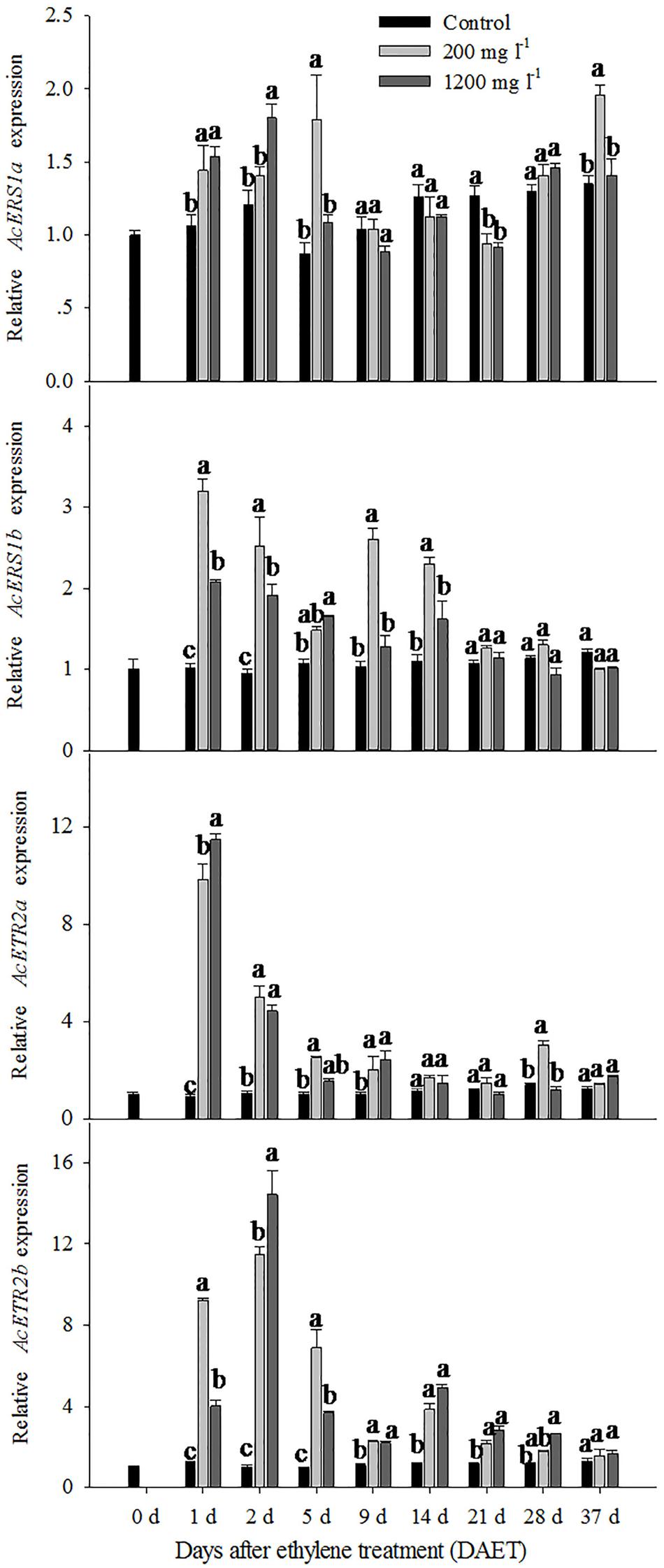
FIGURE 3. The expression of AcERS1a, AcERS1b, AcETR2a, and AcETR2b during pineapple (Ananas comosus) flower transition. The expression of AcERS1a, AcERS1b, AcETR2a, and AcETR2b was evaluated using quantitative RT-PCR (qRT-PCR). Expression levels were normalized using actin as the endogenous control gene. The values represent the means ± SE from three independent experiments. For each gene, values within every time point followed by the same letter are not significantly different (P > 0.05) between treatments according to Duncan’s multiple range test.
The expression pattern of AcERS1b was different than AcERS1a. For the 200 and 1200 mg l-1 ethylene treatment, the relative expression level of AcERS1b was significantly (p < 0.05) up-regulated at 1 DAET and remained up-regulated until 14 DAET. Then, the relative expression level decreased sharply and showed similar expression to the control after 21 DAET. During this period, the relative change in expression level of 1200 mg l-1 was lower than the treatment with 200 mg l-1, except at 5 DAET (Figure 3).
Compared with AcERS1a and AcERS1b (subfamily 1), AcETR2a and AcETR2b (subfamily 2) were more sensitive to ethylene treatment (Figure 3). For the 200 mg l-1 and 1200 mg l-1 ethylene treatments, the relative expression levels of AcETR2a were similar; both were sharply up-regulated and reached their peaks at 1 DAET (10.9-fold and 12.7-fold, respectively). Thereafter, the expression levels decreased but remained significantly up-regulated (p < 0.05) at 2–9 DAET compared with the control (Figure 3).
Both the 200 and 1200 mg l-1 ethylene treatments significantly up-regulated (p < 0.05) the relative expression levels of AcETR2b from 1 to 21 DAET compared with the control. The expression of the 1200 mg l-1 ethylene treatment peaked at 2 DAET, but the expression of the 200 mg l-1 treatment peaked at 1 day (Figure 3).
In Situ Hybridization of AcERS1a, AcERS1b, AcETR2a, and AcETR2b
To study the spatial expression of AcERS1a, AcERS1b, AcETR2a, and AcETR2b during pineapple inflorescence induction, we performed in situ hybridization using the Digoxigenin RNA Labeling kit. Based on the results of Table 1, we only studied the in vivo transcripts profiles of the 200 mg l-1 ethephon treatment group.
The histological results showed there was no new structure development until 5 DAET. At least two BP were observed (Figures 4d, 5c, 6c, and 7c) at 9 DAET, and more BP developed at 14 DAET. From 21 to 28 DAET, FP developed, each located between two bracts (Figures 4f,g, 5e,f, 6e,f, and 7e,f). Finally, at 37 DAET, flowers were formed at the inflorescence (Figures 4h, 5g, 6g, and 7g).
The spatial expression patterns of AcERS1b, AcETR2a, and AcETR2b were similar, but AcERS1a was lower, especially in the later period. For the earliest stages after ethephon treatment (from 0–5 DAET), AcERS1a, AcERS1b, AcETR2a, and AcETR2b were mainly expressed at the shoot apex. Following the appearance of BP and FP, higher expression of these genes was found in these new structures. When the flower formed at 37 DAET, these genes were mostly expressed in the flower, with minor expression in other locations (Figures 4–7).
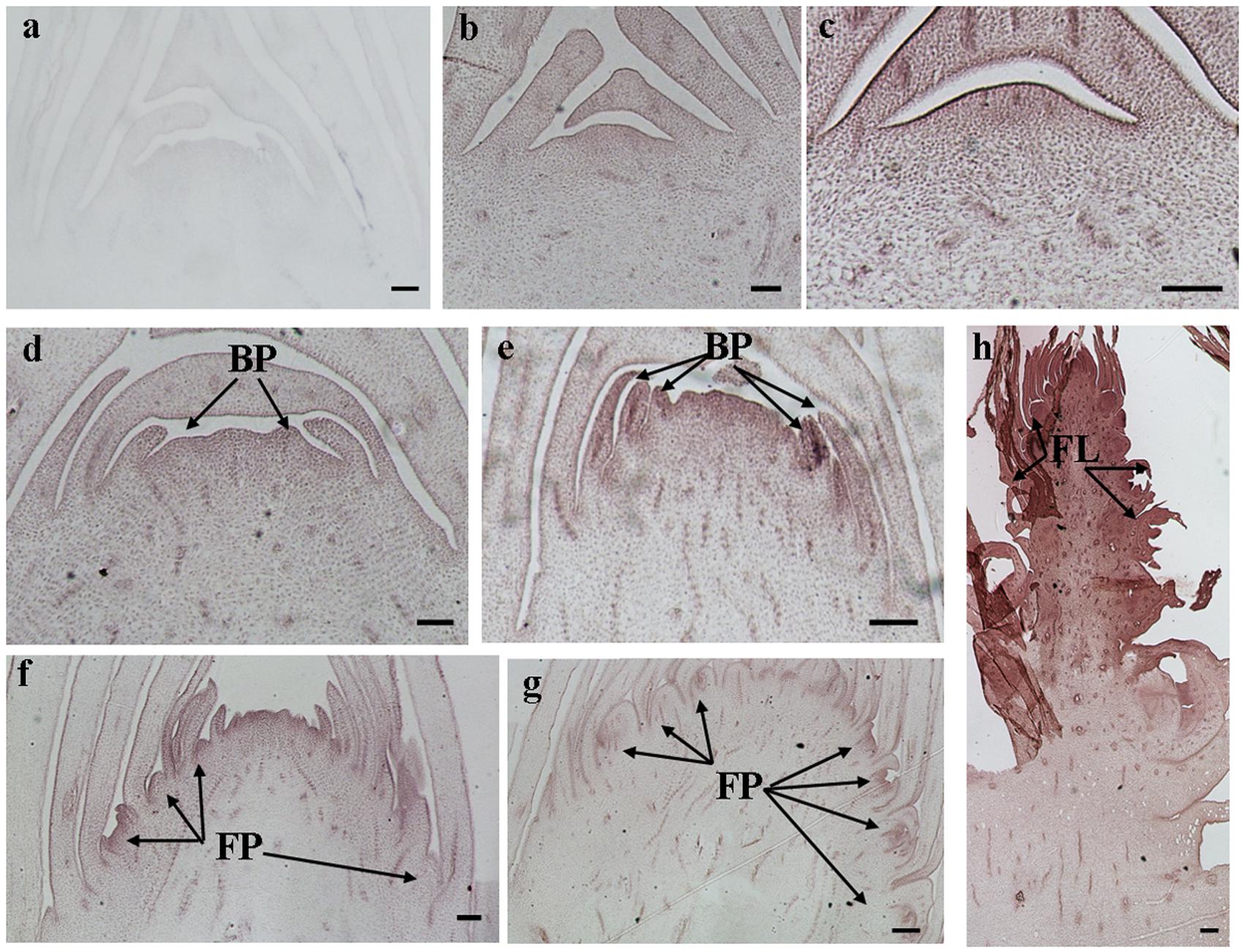
FIGURE 4. In situ detection of AcERS1a transcripts. (a–h) Longitudinal sections of shoot apices and inflorescences sampled at 9 and 1, 5, 9, 14, 21, 28, and 37 days after ethephon treatment, respectively. Sections were hybridized with AcERS1a antisense (b–h) or sense (a) probes. BP, bract primordia; FP, flower primordia; FL, flower. Bars = 100 μm.
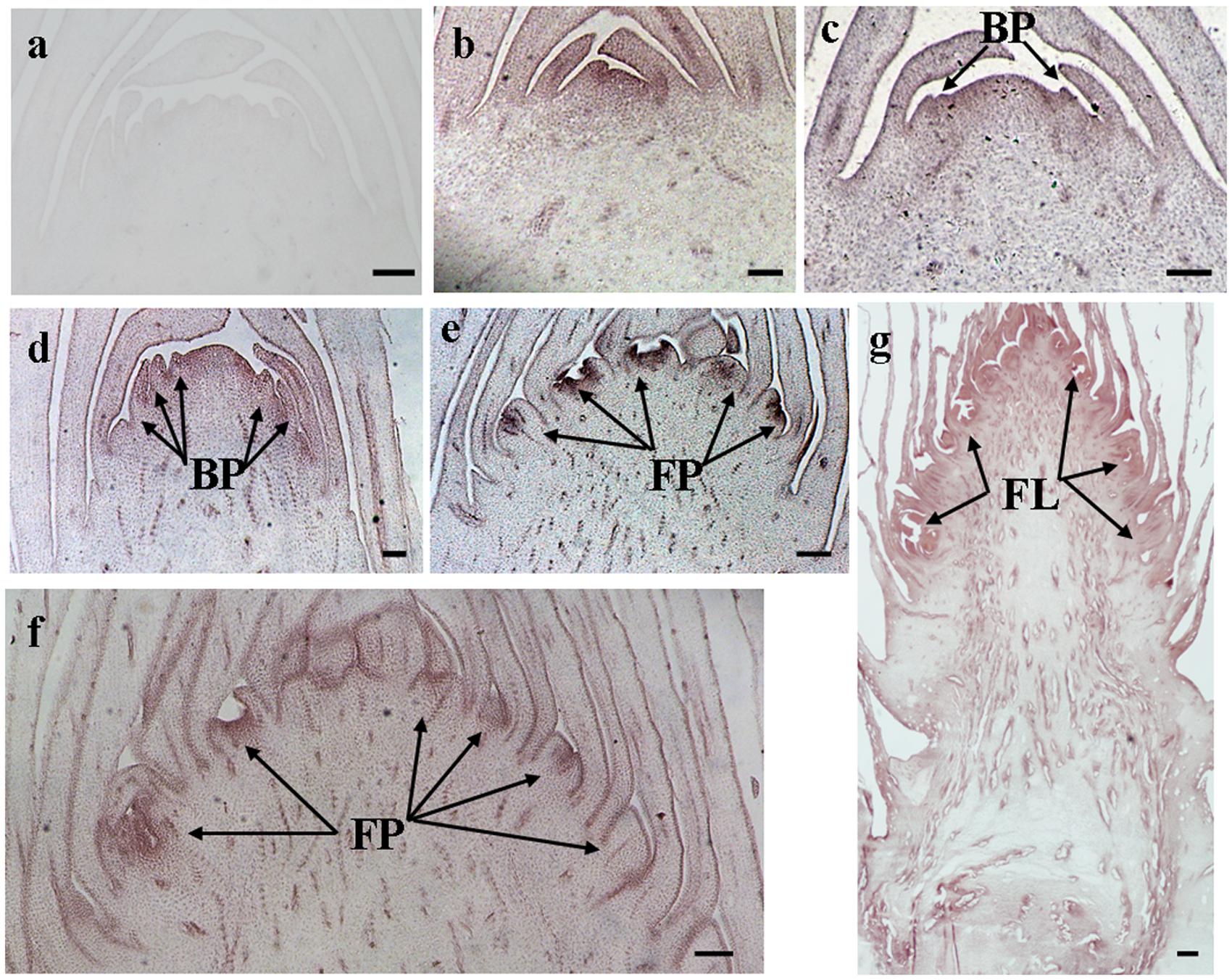
FIGURE 5. In situ detection of AcERS1b transcripts. (a–g) Longitudinal sections of shoot apices and inflorescences sampled at 14 and 1, 9, 14, 21, 28, and 37 days after ethephon treatment, respectively. Sections were hybridized with AcERS1b antisense (b–g) or sense (a) probes. BP, bract primordia; FP, flower primordia; FL, flower. Bars = 100 μm.
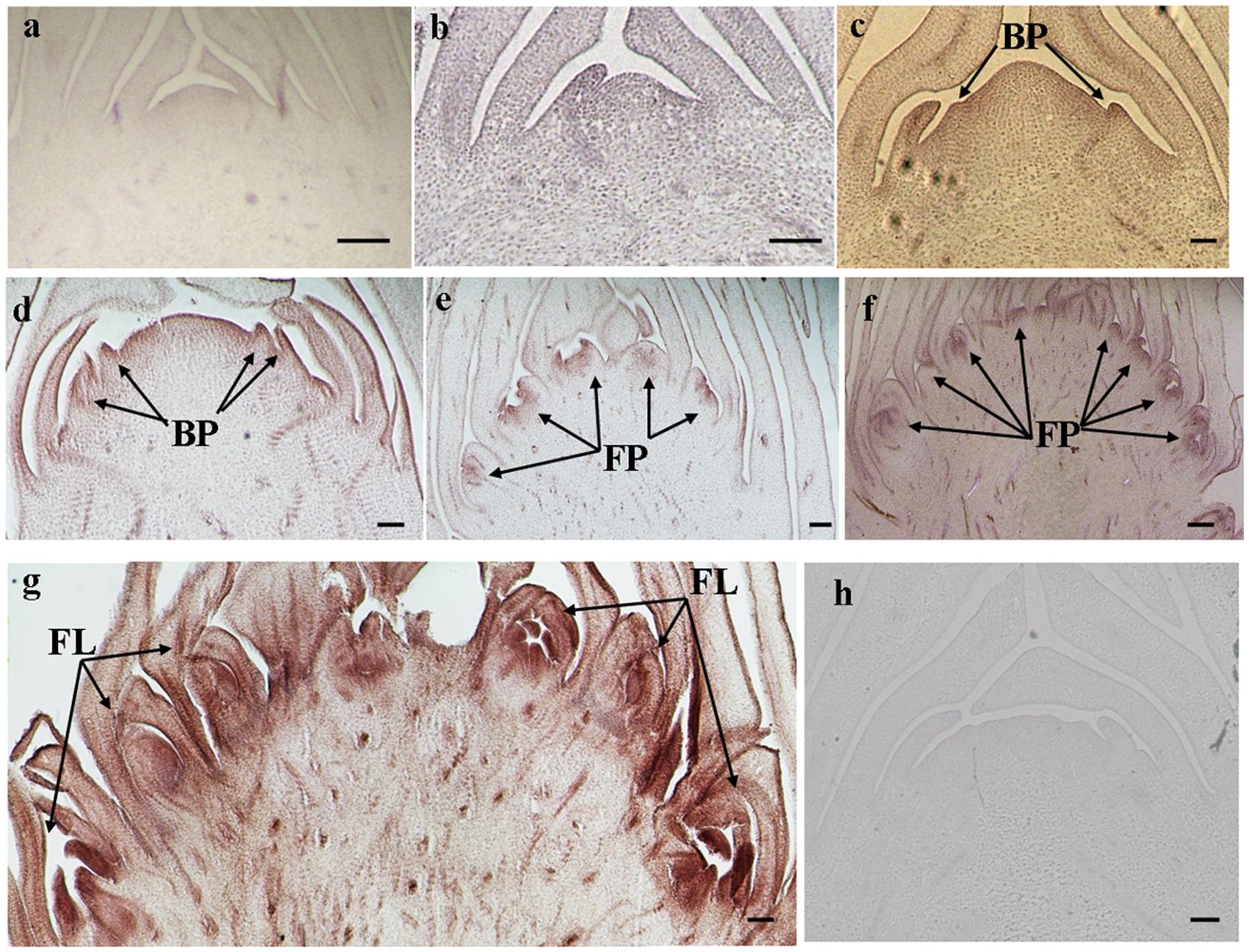
FIGURE 6. In situ detection of AcETR2a transcripts. (a–h) Longitudinal sections of shoot apices and inflorescences sampled at 0 and 5, 9, 14, 21, 28, 37, and 9 days after ethephon treatment, respectively. Sections were hybridized with AcETR2a antisense (a–g) or sense (h) probes. BP, bract primordia; FP, flower primordia; FL, flower. Bars = 100 μm.
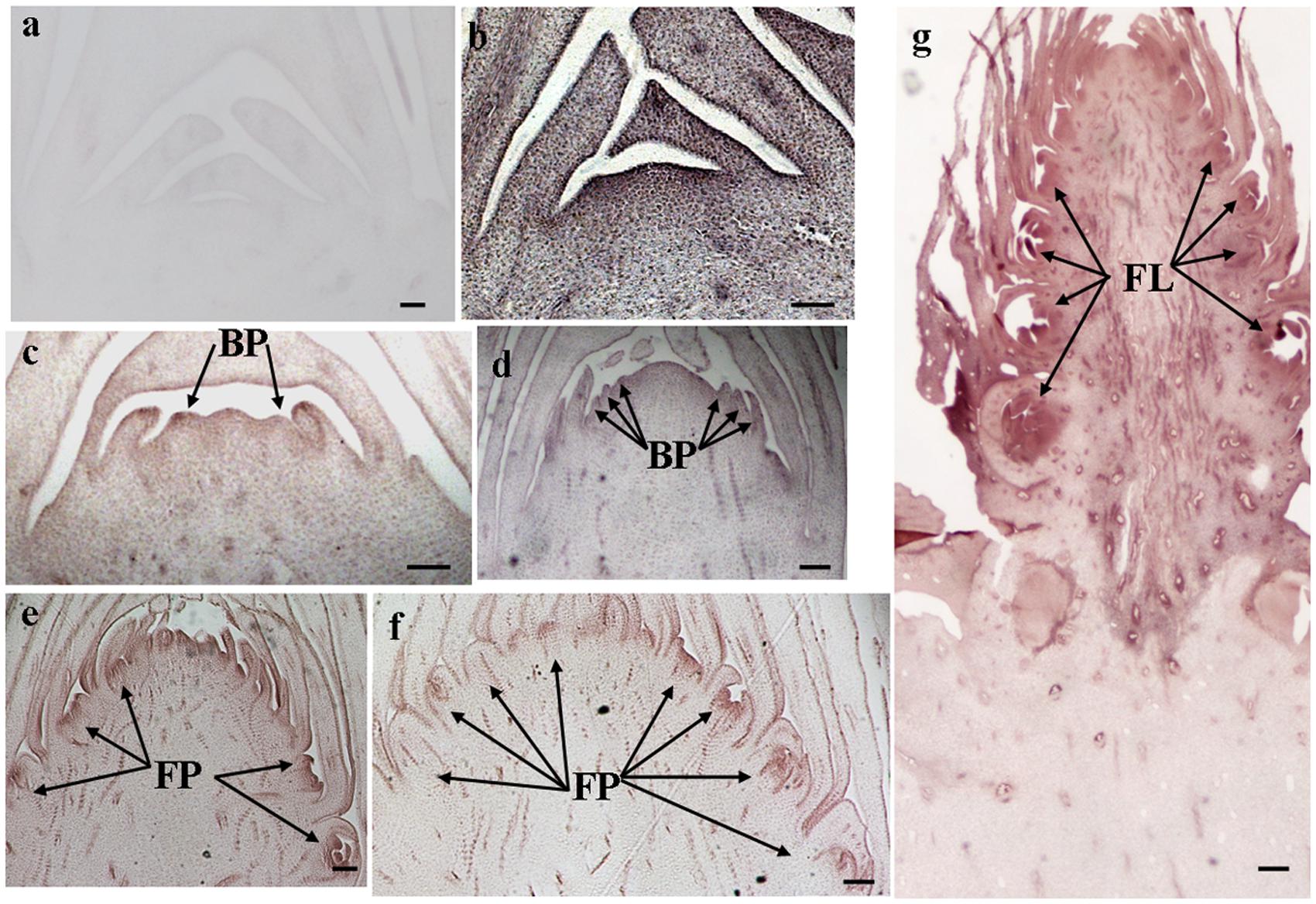
FIGURE 7. In situ detection of AcETR2b transcripts. (a–g) Longitudinal sections of shoot apices and inflorescences sampled at 0 and 1, 9, 14, 21, 28, and 37 days after ethephon treatment, respectively. Sections were hybridized with AcETR2b antisense (b–g) or sense (a) probes. BP, bract primordia; FP, flower primordia; FL, flower. Bars = 100 μm.
Subcellular Localization of AcERS1a, AcERS1b, AcETR2a, and AcETR2b
To examine the subcellular localization of AcERS1a, AcERS1b, AcETR2a, and AcETR2b in vivo, the ORFs without termination codons were fused in-frame with the GFP gene. By transient analysis using tobacco BY-2 protoplasts, the fluorescence of the AcERS1a, AcERS1b, AcETR2a, and AcETR2b proteins were localized in the cytoplasm (Figure 8), which is in agreement with the subcellular location of five Arabidopsis ethylene receptors in tobacco epidermal leaf cells (Grefen et al., 2008).
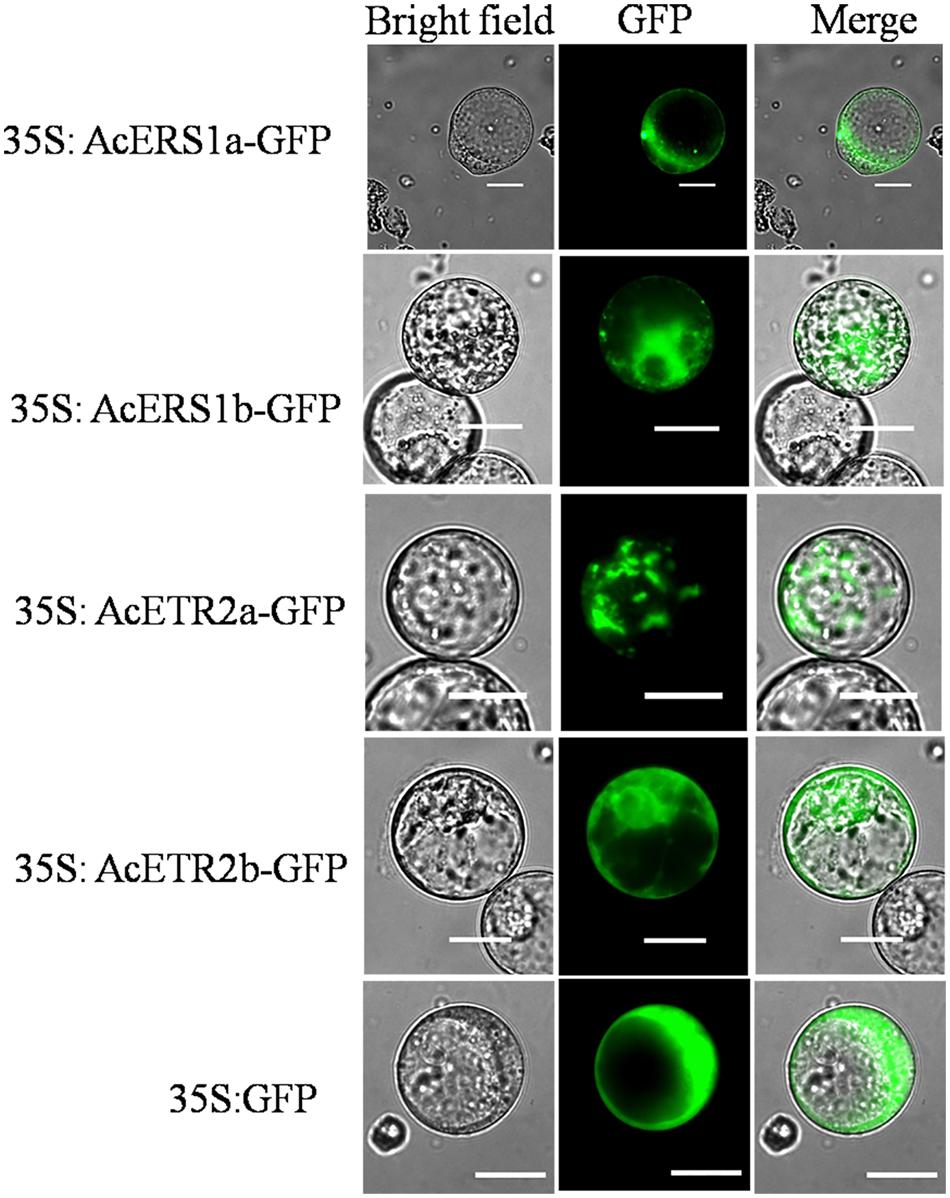
FIGURE 8. Subcellular localization of AcERS1a, AcERS1b, AcETR2a, and AcETR2b in tobacco BY-2 protoplasts. Protoplasts were transiently transformed with AcERS1a, AcERS1b, AcETR2a, and AcETR2b–GFP constructs or GFP vector using a modified PEG method. GFP fluorescence was observed with a fluorescence microscope. Bars = 25 μm.
Discussion
Unlike many other plants, pineapples can be forced to flower by the exogenous application of the gaseous hormone ethylene (or ethephon). This method has been widely used in pineapple production (Bartholomew et al., 2003), but the molecular mechanism of ethylene induction remains unclear.
Recent study of the evolution of ethylene receptors showed that in monocots, there are only two types of ethylene receptors: subfamily 1 receptors, which lack a receiver domain (S1-R or ERS1-like), and subfamily 2 receptors, which contain a receiver domain (S2+R or EIN4/ETR2-like), and ERS2 evolved specifically in the Brassicaceae (Gallie, 2015). In this study, we first designed five degenerate primers sets based on the alignments of the ethylene receptor sequences from other plants (Supplementary Table S1), and we attempted to cover all ethylene receptor genes in pineapple. Although five full-length cDNA clones were obtained by RT-PCR amplification and RACE extension, the sequence analysis and BLAST results showed that the cDNA clone from the AcETR1 primer set (Supplementary Table S1) belonged to ERS1, not ETR1. The cDNA clones from the AcERS2 primer set and the AcEIN4 primer set (Supplementary Table S1) were the same; both were typical undifferentiated ETR2/EIN4-type receptors found throughout monocots (Figure 2), (Gallie, 2015) and were more similar to ETR2 than EIN4. Therefore, these cDNA clones were designated AcETR2b (Supplementary Figure S2). We found there were only two ERS1s (AcERS1a and AcERS1b) and two ETR2s (AcETR2a and AcETR2b) but not other types of ethylene receptors in pineapple, which agreed with the results of Gallie (2015).
When the genomic DNAs of AcERS1a and AcERS1b were compared with their respective cDNAs, there were two introns located in the 5′ UTRs of the AcERS1a and AcERS1b genes. This result was similar to the result for the AtERS1 gene, where a 236-bp long intron was located in the 5′ UTR, 29 bp from the start codon, and for the CcETR1 gene, where a 978-bp long intron was located in the 5′ UTR (Bustamante-Porras et al., 2007).
The qRT-PCR results showed that the relative expression levels of AcERS1b, AcETR2a, and AcETR2b were significantly up-regulated (p < 0.05) after ethylene treatment (both at 200 and 1200 mg l-1) from 1–9 DAET and even at 14–21 DAET (Figure 3). From the histological results, BP growth developed at 9 DAET, and FP growth initiated at 21 DAET. By contrast, AcERS1a, AcERS1b, AcETR2a, and AcETR2b transcripts were initially detected mainly at the shoot apex in the early stages (0–9 DAET) and then in the BP and FP (14–28 DAET). When the flower formed at 37 DAF, these genes were mostly expressed in the flowers (Figures 4–7). The combined results of the qRT-PCRs and in situ hybridizations show that these four ethylene receptor genes play an important role during ethephon induction of pineapple flowering. However, to verify these genes’ function directly, transgenic pineapple or Arabidopsis plants and pineapple ethylene receptors knockout lines should be developed to study their flowering behavior, especially in response to ethylene, which should be addressed in a further study.
Ethylene receptors are negative regulators of the ethylene signal transduction pathway, that is, they inhibit the pathway when not bound to ethylene (Shakeel et al., 2013). Ethylene induces pineapple flowering, which means that the expression of ethylene receptors should be down-regulated in response to ethylene treatment. In our study, however, after treatment with ethephon, the relative expression of the pineapple ethylene receptors was not down-regulated but initially significantly up-regulated (Figure 3) and then decreases to the same level as the control. Ethylene treatment changed the expression of ethylene receptors differently for different plants. For example, the relative expressions of OsERS2 (Yau et al., 2004), LeETR4 and LeETR6 (Kevany et al., 2007), AdETR1 (Yin et al., 2008) and DenERS1 (Thongkum et al., 2015) decreased after ethylene treatment, whereas the relative expressions of AtERS1, AtERS2, and AtETR2 increased (Hua et al., 1998). The transcription of AcETR1, AcEIN4 (Hua et al., 1998), PhETR1, and PhETR2 (Dervinis et al., 2000) was not changed by ethylene treatment. RNA levels are not always positively correlated with protein levels. In tomato, for example, the expression of NR, LeETR4 and LeETR6 is low and constitutive throughout immature fruit development, and there is a significant increase in the transcription of NR, LeETR4 and LeETR6 at the onset of ripening. However, in contrast to mRNA expression, the receptor proteins levels were highest in immature fruits and significantly decreased to a lower level at the onset of ripening and thereafter because of protein degradation through the 26S proteasome-dependent pathway (Kevany et al., 2007). Therefore, whether the pineapple ethylene receptor protein is also degraded is the subject of the next study.
Pineapple natural flowering can occur under different environmental conditions, including low temperatures, short photoperiods, geotropic stimulation and other stress conditions (Bartholomew et al., 2003; Van de Poel et al., 2009). These environmental cues trigger ACC synthase gene expression, resulting in increased ACC production and ethylene biosynthesis (Botella et al., 2000; Trusov and Botella, 2006). In pineapple, ethylene acts as the signal for switching from the vegetative to the flowering stage (Burg and Burg, 1966; Min and Bartholomew, 1997); thus, these environmental cues induce pineapple flowering. Ethylene receptor genes can be induced by various abiotic stresses (Zhang et al., 1999; Zhang J. et al., 2001; Zhang J.S. et al., 2001; Xie et al., 2002; Tao et al., 2015). In this study, a higher concentration of ethephon might be perceived as one type of stress by pineapple seedlings, which resulted in the expression of ethylene receptors being sharply increased after ethephon treatment (Figure 3). On the other hand, our previous study showed that after ethephon application, the liberation of endogenous ethylene in both the shoot apex and D-leaf increased sharply until the 8th day when it reached the peak (Liu et al., 2011); thus, at a higher concentration of endogenous ethylene, more ethylene receptors were bound to ethylene, resulting in a gradual decrease of the transcription of the receptors (Figure 3). The reduction in ethylene receptor levels can increase the sensitization of plants to ethylene; subsequently, a lower concentration of ethylene can stimulate the ethylene response (Tieman et al., 2000; Cancel and Larsen, 2002; Wang et al., 2013a) and induce pineapple flowering. In this study, we found that the new BP emerged at 9 DAET (Figures 4d, 5c, 6c, and 7c), immediately following (or at the same time as) the ethylene production peak.
Cis-elements play important roles in the regulation of gene expression by controlling the efficiency of promoters. In this study, a range of putative cis-regulatory elements that are responsive to ethylene, auxin, cytokinin, gibberellin, light, and abiotic stress was found (Supplementary Table S4). During the period of ethylene-induced pineapple flowering, ERELEE4, LECPLEACS2, and GCCCORE motifs were found in the promoters of these pineapple ethylene receptors (Supplementary Table S4), which suggests a potential regulation of these ethylene receptor genes by ethylene. In addition, among the cis-elements related to plant hormones, there were 10 different types of cis-elements involved in the auxin response (Supplementary Table S4). Because the shoot apex is one of the main sites of auxin synthesis (Normanly et al., 1993; Ljung et al., 2001), auxin may be involved in the expression of AcERS1a, AcERS1b, AcETR2a, and AcETR2b. On the other hand, there were multiple ARR1AT motifs in all four promoters (44, 36, 16, and 21 ARR1AT motifs in the promoters of AcERS1a, AcERS1b, AcETR2a, and AcETR2b, respectively; Supplementary Table S4). ARR1AT binds to Arabidopsis response regulator 1 (ARR1), which is involved in cytokinin signaling (Ross et al., 2004) and may be a link between cytokinin signaling and meristem regulation (Bao et al., 2009). By contrast, pineapple flowering originates from the shoot apex meristem (Figures 4–7), indicating that cytokinin may be involved in the regulation of the four ethylene receptors’ expression and further flower formation in pineapple.
In Arabidopsis, the five ethylene receptors (Grefen et al., 2008) and REVERSION-TO-ETHYLENE SENSITIVITY 1 (RTE1) are localized at the ER and the Golgi (Dong et al., 2008); our result of subcellular localization showed that AcERS1a, AcERS1b, AcETR2a, and AcETR2b were mainly localized to the cytoplasm (Figure 8). In addition, other important components of the ethylene signaling pathway, such as EIN2 (ETHYLENE INSENSITIVE 2) and CTR1 (constitutive triple response 1), are localized to the endoplasmic reticulum (ER; Gao et al., 2003; Bisson et al., 2009). At the membrane, CTR1, EIN2, and RTE1 regulate the ethylene receptors via physical interactions (Ma et al., 2014). Therefore, further studies should focus on these genes’ interactions with the ethylene receptors in pineapple.
Conclusion
Our results showed that ethylene receptor genes (i.e., AcERS1a, AcERS1b, AcETR2a, and AcETR2b) may play an important role during inflorescence development in pineapple because the relative expression of these genes was significantly increased by ethephon treatment (Figure 3), and the expression was mainly focused in the newly formed BP, FP, and flower structures (Figures 4–7).
Author Contributions
Y-HL and G-MS conceived and designed the experiments. Y-HL conducted experiments and performed the RT-PCR, quantitative real-time PCR, subcellular localization and in situ hybridization. Q-SW, S-HL, and H-NZ participated in the preparation of the plant material and the field experiments. XH participated in writing the manuscript and subcellular localization. ZZ performed part of the data analysis. All of authors in this study read and approved the manuscript.
Funding
This work was financially supported by the Chinese Special Fund for Public Welfare Industry (201203021), Natural Science Foundation of Hainan (20163108) and the Chinese Special Fund of Basic Scientific Research Project for State Level and Public Welfare-Scientific Research Institutes (sscri 201014).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Daniel R. Gallie (University of California, Riverside) for help of the analysis of pineapple ethylene receptors and Dr. Yu Zhao (Huazhong Agricultural University) for help of in situ hybridization. We thank Dr. Torsten Möhlmann (Technische Universität Kaiserslautern) for kindly providing pGFP2 plasmids.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00710
FIGURE S1 | Ethylene receptors of pineapple.
FIGURE S2 | Sequences of pineapple promoters.
Table S1 | Oligonucleotide primers used in the experiments for isolate DNA fragments, rapid amplification of cDNA ends, full-length cDNA cloning and promoters isolation.
Table S2 | Primers for quantitative real-time PCR and in situ hybridization.
Table S3 | Primers for subcellular localization.
Table S4 | Putative cis elements identified in AcERS1a, AcERS1b, AcETR2a and AcETR2b by PLACE. Numbers 1, 2, 3… represents the repeats of cis-elements while 0 indicate the absence of that particular cis-element.
Abbreviations
BP, bract primordial; DAET, days after ethylene treatment; EIN4, ETHYLENE INSENSITIVE 4; ERS1, ETHYLENE RESPONSE SENSOR 1; ERS2, ETHYLENE RESPONSE SENSOR 1; ETR1, ETHYLENE RESPONSE 1; ETR2, ETHYLENE RESPONSE 2; FP, flower primordia; GFP, green fluorescent protein; MW, molecular weight; pI, isoelectric point; PLACE, plant cis-acting regulatory DNA elements; qRT-PCR, quantitative RT-PCR; RACE, rapid amplification of cDNA ends; TAIR, The Arabidopsis information resource; UTRs, untranslated regions.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov
- ^ http://www.arabidopsis.org/
- ^ http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
- ^ http://www.expasy.org/tools/protparam.html
- ^ http://www.ebi.ac.uk/clustalw/
- ^ http://www.ch.embnet.org/software/BOX_form.html
- ^ http://www.dna.affrc.go.jp/PLACE/signalscan.html
References
Abeles, F. B., Morgan, P. W., and Saltveit, M. E. Jr. (1992). Ethylene in Plant Biology, 2nd Edn. New York, NY: Academic Press.
Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., et al. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U.S.A. 104, 6484–6489. doi: 10.1073/pnas.0610717104
Bao, Y., Dharmawardhana, P., Arias, R., Allen, M. B., Ma, C., and Strauss, S. H. (2009). WUS and STM-based reporter genes for studying meristem development in poplar. Plant Cell Rep. 28, 947–962. doi: 10.1007/s00299-009-0685-3
Bartholomew, D. P., Paull, R. E., and Rohrbach, K. G. (2003). The Pineapple: Botany, Production and Uses. Wallingford, CT: CABI Publishing.
Binder, B. M., O’Malley, R. C., Wang, W., Moore, J. M., Parks, B. M., Spalding, E. P., et al. (2004). Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol. 136, 2913–2920.
Binder, B. M., O’Malley, R. C., Wang, W., Zutz, T. C., and Bleecker, A. B. (2006). Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol. 142, 1690–1700. doi: 10.1104/pp.106.087858
Bisson, M. M. A., Bleckmann, A., Allekotte, S., and Groth, G. (2009). EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem. J. 424, 1–6. doi: 10.1042/BJ20091102
Botella, M., Del Amor, F., Amorós, A., Serrano, M., Martínez, V., and Cerdá, A. (2000). Polyamine, ethylene and other physico-chemical parameters in tomato (Lycopersicon esculentum) fruits as affected by salinity. Physiol. Plant. 109, 428–434. doi: 10.1034/j.1399-3054.2000.100409.x
Boyce, R., Chilana, P., and Rose, T. M. (2009). iCODEHOP: a new interactive program for designing COnsensus-DEgenerate Hybrid Oligonucleotide Primers from multiply aligned protein sequences. Nucleic Acids Res. 37, W222–W228. doi: 10.1093/nar/gkp379
Burg, S. P., and Burg, E. A. (1966). Auxin-induced ethylene formation: its relation to flowering in pineapple. Science 152, 1269. doi: 10.1126/science.152.3726.1269
Bustamante-Porras, J., Campa, C., Poncet, V., Noirot, M., Leroy, T., Hamon, S., et al. (2007). Molecular characterization of an ethylene receptor gene (CcETR1) in coffee trees, its relationship with fruit development and caffeine content. Mol. Genet. Genomics 277, 701–712. doi: 10.1007/s00438-007-0219-z
Cancel, J. D., and Larsen, P. B. (2002). Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol. 129, 1557–1567. doi: 10.1104/pp.003780
Cao, W. H., Liu, J., He, X. J., Mu, R. L., Zhou, H. L., Chen, S. Y., et al. (2007). Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143, 707–719. doi: 10.1104/pp.106.094292
Chang, C., and Stadler, R. (2001). Ethylene hormone receptor action in Arabidopsis. Bioessays 23, 619–627. doi: 10.1002/bies.1087
Chen, Y. F., Etheridge, N., and Schaller, G. E. (2005). Ethylene signal transduction. Ann. Bot. 95, 901–915. doi: 10.1093/aob/mci100
Dai, M., Hu, Y., Zhao, Y., Liu, H., and Zhou, D. X. (2007). A WUSCHELLIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144, 380–390. doi: 10.1104/pp.107.095737
Dervinis, C., Clark, D. G., Barrett, J. E., and Nell, T. A. (2000). Effect of pollination and exogenous ethylene on accumulation of ETR1 homologue transcripts during flower petal abscission in geranium (Pelargonium × hortorum LH Bailey). Plant Mol. Biol. 42, 847–856. doi: 10.1023/A:1006409827860
Dong, C. H., Rivarola, M., Resnick, J. S., Maggin, B. D., and Chang, C. (2008). Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 53, 275–286. doi: 10.1111/j.1365-313X.2007.03339.x
Fu, X., Deng, S., Su, G., Zeng, Q., and Shi, S. (2004). Isolating high-quality RNA from mangroves without liquid nitrogen. Plant Mol. Biol. Rep. 22, 197–197. doi: 10.1007/BF02772728
Gallie, D. R. (2015). Appearance and elaboration of the ethylene receptor family during land plant evolution. Plant Mol. Biol. 87, 521–539. doi: 10.1007/s11103-015-0296-z
Gao, Z. Y., Chen, Y. F., Randlett, M. D., Zhao, X. C., Findell, J. L., Kieber, J. J., et al. (2003). Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 278, 34725–34732. doi: 10.1074/jbc.M305548200
Grefen, C., Stadele, K., Ruzicka, K., Obrdlik, P., Harter, K., and Horak, J. (2008). Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 1, 308–320. doi: 10.1093/mp/ssm015
Hall, A. E., and Bleecker, A. B. (2003). Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell 15, 2032–2041. doi: 10.1105/tpc.013060
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hua, J., Chang, C., Sun, Q., and Meyerowitz, E. M. (1995). Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269, 1712–1714. doi: 10.1126/science.7569898
Hua, J., Sakai, H., Nourizadeh, S., Chen, Q. G., Bleecker, A. B., Ecker, J. R., et al. (1998). EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10, 1321–1332. doi: 10.1105/tpc.10.8.1321
Kendrick, M. D., and Chang, C. (2008). Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485. doi: 10.1016/j.pbi.2008.06.011
Kevany, B. M., Tieman, D. M., Taylor, M. G., Cin, V. D., and Klee, H. J. (2007). Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51, 458–467. doi: 10.1111/j.1365-313X.2007.03170.x
Klee, H. J. (2002). Control of ethylene-mediated processes in tomato at the level of receptors. J. Exp. Bot. 53, 2057–2063. doi: 10.1093/jxb/erf062
Klee, H. J. (2004). Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol. 135, 660–667. doi: 10.1104/pp.104.040998
Li, Y. H., Zou, M. H., Feng, B. H., Huang, X., Zhang, Z., and Sun, G. M. (2012). Molecular cloning and characterization of the genes encoding an auxin efflux carrier and the auxin influx carriers associated with the adventitious root formation in mango (Mangifera indica L.) cotyledon segments. Plant Physiol. Biochem. 55, 33–42. doi: 10.1016/j.plaphy.2012.03.012
Lin, Z., Zhong, S., and Grierson, D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336. doi: 10.1093/jxb/erp204
Liu, Q., Xu, C., and Wen, C. K. (2010). Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Boil. 10:60. doi: 10.1186/1471-2229-10-60
Liu, S. H., Zang, X. P., and Sun, G. M. (2011). Changes in endogenous hormone concentrations during inflorescence induction and development in pineapple (Ananas comosus cv. Smooth Cayenne) by ethephon. Afr. J. Biotech. 10, 10892–10899.
Liu, S. H., Zang, X. P., Sun, G. M., and Ye, C. H. (2009). Integrated effects of ethrel on pineapple’s (Zhenzhu cv.) flower induction. Guangdong Agr. Sci. 8, 78–80.
Ljung, K., Östin, A., Lioussanne, L., and Sandberg, G. (2001). Developmental regulation of indole-3-acetic acid turnover in Scots pine seedlings. Plant Physiol. 125, 464–475. doi: 10.1104/pp.125.1.464
Ma, B., Chen, H., Chen, S. Y., and Zhang, J. S. (2014). “Roles of ethylene in plant growth and responses to stresses,” in Phytohormones: A Window to Metabolism, Signaling and Biotechnological Applications, eds L.-S. P. Tran and S. Pal (New York, NY: Springer Science+Business Media), 81–118.
Marchler-Bauer, A., Derbyshire, M. K., and Gonzales, N. R. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Merchante, C., Alonso, J. M., and Stepanova, A. N. (2013). Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560. doi: 10.1016/j.pbi.2013.08.001
Min, X. J., and Bartholomew, D. P. (1997). Temperature affects ethylene metabolism and fruit initiation and size of pineapple. Acta Hortic. 425, 329–338. doi: 10.17660/ActaHortic.1997.425.36
Normanly, J., Cohen, J. D., and Fink, G. R. (1993). Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc. Natl. Acad. Sci. U.S.A. 90, 10355–10359. doi: 10.1073/pnas.90.21.10355
Ogawara, T., Higashi, K., Kamada, H., and Ezura, H. (2003). Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J. Plant Physiol. 160, 1335–1340. doi: 10.1078/0176-1617-01129
O’Malley, R. C., Rodriguez, F. I., Esch, J. J., Binder, B. M., O’Donnell, P., Klee, H. J., et al. (2005). Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 41, 651–659. doi: 10.1111/j.1365-313X.2004.02331.x
Plett, J. M., Cvetkovska, M., Makenson, P., Xing, T., and Regan, S. (2009a). Arabidopsis ethylene receptors have different roles in Fumonisin B 1-induced cell death. Physiol. Mol. Plant Pathol. 74, 18–26. doi: 10.1016/j.pmpp.2009.08.004
Plett, J. M., Mathur, J., and Regan, S. (2009b). Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana. J. Exp. Bot. 60, 3923–3933. doi: 10.1093/jxb/erp228
Qu, X., Hall, B. P., Gao, Z., and Schaller, G. E. (2007). A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 7:3. doi: 10.1186/1471-2229-7-3
Rieu, I., Wolters-Arts, M., Derksen, J., Mariani, C., and Weterings, K. (2003). Ethylene regulates the timing of anther dehiscence in tobacco. Planta 217, 131–137.
Ross, E. J., Stone, J. M., Elowsky, C. G., Arredondo-Peter, R., Klucas, R. V., and Sarath, G. (2004). Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 55, 1721–1731. doi: 10.1093/jxb/erh211
Schaller, G. E., and Kieber, J. J. (2002). Ethylene. Arabidopsis Book 1: e0071. doi: 10.1199/tab.0071
Seifert, G. J., Barber, C., Wells, B., and Roberts, K. (2004). Growth regulators and the control of nucleotide sugar flux. Plant Cell 16, 723–730. doi: 10.1105/tpc.019661
Shakeel, S. N., Wang, X., Binder, B. M., and Schaller, G. E. (2013). Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. AoB Plants 5:lt010. doi: 10.1093/aobpla/plt010
Shan, W., Kuang, J. F., Chen, L., Xie, H., Peng, H. H., Xiao, Y. Y., et al. (2012). Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 63, 5171–5187. doi: 10.1093/jxb/ers178
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod- and temperature sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Tan, G., Gao, Y., Shi, M., Zhang, X., He, S., Chen, Z., et al. (2005). SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res. 33, e122. doi: 10.1093/nar/gni124
Tao, J. J., Chen, H. W., Ma, B., Zhang, W. K., Chen, S. Y., and Zhang, J. S. (2015). The role of ethylene in plants under salinity stress. Front. Plant Sci. 6:1059. doi: 10.3389/fpls.2015.01059
Thongkum, M., Burns, P., Bhunchoth, A., Warin, N., Chatchawankanphanich, O., and van Doorn, W. G. (2015). Ethylene and pollination decrease transcript abundance of an ethylene receptor gene in Dendrobium petals. J. Plant Physiol. 176, 96–100. doi: 10.1016/j.jplph.2014.12.008
Tieman, D. M., Taylor, M. G., Ciardi, J. A., and Klee, H. J. (2000). The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc. Natl. Acad. Sci. U.S.A. 97, 5663–5668. doi: 10.1073/pnas.090550597
Trusov, Y., and Botella, J. R. (2006). Silencing of the ACC synthase gene ACACS2 causes delayed flowering in pineapple [Ananas comosus (L.) Merr.]. J. Exp. Bot. 57, 3953–3960. doi: 10.1093/jxb/erl167
Van de Poel, B., Ceusters, J., and De Proft, M. P. (2009). Determination of pineapple (Ananas comosus, MD-2 hybrid cultivar) plant maturity, the efficiency of flowering induction agents and the use of activated carbon. Sci. Hortic. 120, 58–63. doi: 10.1016/j.scienta.2008.09.014
van Doorn, W. G., and Kamdee, C. (2014). Flower opening and closure: an update. J. Exp. Bot. 65, 5749–5757. doi: 10.1093/jxb/eru327
Wang, F., Cui, X., Sun, Y., and Dong, C. H. (2013a). Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep. 32, 1099–1109. doi: 10.1007/s00299-013-1421-6
Wang, Q., Zhang, W., Yin, Z., and Wen, C. K. (2013b). Rice CONSTITUTIVE TRIPLE-RESPONSE2 is involved in the ethylene-receptor signalling and regulation of various aspects of rice growth and development. J. Exp. Bot. 64, 4863–4875. doi: 10.1093/jxb/ert272
Wang, S., He, J., Cui, Z., and Li, S. (2007). Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl. Environ. Microbial. 73, 5048–5051. doi: 10.1128/AEM.02973-06
Wilson, R. L., Kim, H., Bakshi, A., and Binder, B. M. (2014). The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol. 165, 1353–1366. doi: 10.1104/pp.114.241695
Wuriyanghan, H., Zhang, B., Cao, W. H., Ma, B., Lei, G., Liu, Y. F., et al. (2009). The ethylene receptor ETR2 delays floral transition and affects starch accumulations in rice. Plant Cell 21, 1473–1494. doi: 10.1105/tpc.108.065391
Xie, C., Zhang, Z. G., Zhang, J. S., He, X. J., Cao, W. H., He, S. J., et al. (2002). Spatial expression and characterization of a putative ethylene receptor protein NTHK1 in tobacco. Plant Cell Physiol. 43, 810–815. doi: 10.1093/pcp/pcf095
Xie, F., Liu, Q., and Wen, C. K. (2006). Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol. 142, 492–508. doi: 10.1104/pp.106.082628
Yamasaki, S., Fujii, N., and Takahashi, H. (2000). The ethylene-regulated expression of CS-ETR2 and CS-ERS genes in cucumber plants and their possible involvement with sex expression in flowers. Plant Cell Physiol. 41, 608–616. doi: 10.1093/pcp/41.5.608
Yau, C. P., Wang, L., Yu, M., Zee, S. Y., and Yip, W. K. (2004). Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. J. Exp. Bot. 55, 547–556. doi: 10.1093/jxb/erh055
Yin, X. R., Chen, K. S., Allan, A. C., Wu, R. M., Zhang, B., Lallu, N., et al. (2008). Ethylene-induced modulation of genes associated with the ethylene signalling pathway in ripening kiwifruit. J. Exp. Bot. 59, 2097–2108. doi: 10.1093/jxb/ern067
Zhang, J., Xie, C., Wu, X., Du, B., and Chen, S. (2001). Tobacco two-component gene NTHK2. Chinese Sci. Bull. 46, 574–577. doi: 10.1007/BF02900413
Zhang, J. S., Xie, C., Liu, F., Liu, F. H., and Chen, S. Y. (1999). A novel tobacco gene coding for a product similar to bacterial two-component regulators. Chinese Sci. Bull. 44, 1025–1029. doi: 10.1007/BF02886023
Zhang, J. S., Xie, C., Shen, Y. G., and Chen, S. Y. (2001). A two-component gene (NTHK1) encoding a putative ethylene-receptor homolog is both developmentally and stress-regulated in tobacco. Theor. Appl. Genet. 102, 815–824. doi: 10.1007/s001220000469
Keywords: Ananas comosus L., ethylene, ethylene receptors, flowering, inflorescence development
Citation: Li Y-H, Wu Q-S, Huang X, Liu S-H, Zhang H-N, Zhang Z and Sun G-M (2016) Molecular Cloning and Characterization of Four Genes Encoding Ethylene Receptors Associated with Pineapple (Ananas comosus L.) Flowering. Front. Plant Sci. 7:710. doi: 10.3389/fpls.2016.00710
Received: 16 January 2016; Accepted: 09 May 2016;
Published: 24 May 2016.
Edited by:
Jaime Prohens, Universitat Politècnica de València, SpainReviewed by:
Panagiotis Kalaitzis, Mediterranean Agronomic Institute of Chania, GreeceJimmy R. Botella, University of Queensland, Australia
Copyright © 2016 Li, Wu, Huang, Liu, Zhang, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun-He Li, liyunhe16@163.com; Guang-Ming Sun, gm-sun@163.com
 Yun-He Li
Yun-He Li Qing-Song Wu1
Qing-Song Wu1