Corrigendum (2): Overexpression of Rice Glutaredoxin OsGrx_C7 and OsGrx_C2.1 Reduces Intracellular Arsenic Accumulation and Increases Tolerance in Arabidopsis thaliana
Corrigendum: Overexpression of Rice Glutaredoxin OsGrx_C7 and OsGrx_C2.1 Reduces Intracellular Arsenic Accumulation and Increases Tolerance in Arabidopsis thaliana
- 1Genetics and Molecular Biology Division, Council of Scientific and Industrial Research-National Botanical Research Institute, Lucknow, India
- 2Department of Biotechnology, Kumaun University, Nainital, India
- 3Environmental Biotechnology Division, Council of Scientific and Industrial Research-National Botanical Research Institute, Lucknow, India
- 4Stockbridge School of Agriculture, University of Massachusetts, Amherst, Massachusetts
Glutaredoxins (Grxs) are a family of small multifunctional proteins involved in various cellular functions, including redox regulation and protection under oxidative stress. Despite the high number of Grx genes in plant genomes (48 Grxs in rice), the biological functions and physiological roles of most of them remain unknown. Here, the functional characterization of the two arsenic-responsive rice Grx family proteins, OsGrx_C7 and OsGrx_C2.1 are reported. Over-expression of OsGrx_C7 and OsGrx_C2.1 in transgenic Arabidopsis thaliana conferred arsenic (As) tolerance as reflected by germination, root growth assay, and whole plant growth. Also, the transgenic expression of OsGrxs displayed significantly reduced As accumulation in A. thaliana seeds and shoot tissues compared to WT plants during both AsIII and AsV stress. Thus, OsGrx_C7 and OsGrx_C2.1 seem to be an important determinant of As-stress response in plants. OsGrx_C7 and OsGrx_C2.1 transgenic showed to maintain intracellular GSH pool and involved in lowering AsIII accumulation either by extrusion or reducing uptake by altering the transcript of A. thaliana AtNIPs. Overall, OsGrx_C7 and OsGrx_C2.1 may represent a Grx family protein involved in As stress response and may allow a better understanding of the As induced stress pathways and the design of strategies for the improvement of stress tolerance as well as decreased As content in crops.
Introduction
Glutaredoxins (Grxs) are ubiquitous low molecular weight, cysteine-rich proteins that take part in diverse cellular processes including maintenance and regulation of cellular redox state, iron homeostasis and redox-dependent signaling pathways (Holmgren, 1989; Holmgren and Aslund, 1995; Lillig et al., 2008). Various Grx genes had been identified in both eukaryote and prokaryote. Based on the predicted amino acid sequences and arrangement of cysteine residues in active-site motifs, all Grxs are divided into three major classes CPYC-, CGFS-, and CC-type. The CC-type class has been identified in land plants only whereas others are found in all organisms from prokaryotes to eukaryotes (Rouhier et al., 2004, 2006; Garg et al., 2010).
Glutaredoxins are GSH-dependent redox enzymes that catalyze the reduction of disulfides through reduced GSH in a coupled system with NADPH and GR (Fernandes and Holmgren, 2004). Besides the traditional roles of thiol-disulfide oxidoreductases in oxidative stress responses, many other functions of Grxs were also reported, including roles in iron homeostasis, iron-sulfur cluster biosynthesis, and stress-related redox sensor (Michelet et al., 2008; Dalle-Donne et al., 2009; Rouhier et al., 2010; Zaffagnini et al., 2012). Protein-S-glutathionylation is also a reversible post-translational modification of thiol groups with GSH, which provides protection to the cysteine residues of proteins from irreversible oxidation and may also alter the activity of many proteins either positively or negatively (Gallogly and Mieyal, 2007). Protein glutathionylation in plants is favored by the condition of enhanced reactive oxygen species (ROS) production, where glutathionylation is a mechanism for redox regulation and signaling. Grxs are efficient catalysts for the de-glutathionylation reaction, although the fundamental mechanism of glutathionylation in vivo remains obscure (Dalle-Donne et al., 2009; Gao et al., 2010). Due to the involvement of Grxs in many cellular processes, these enzymes have been implicated in a variety of significant physiological and pathological processes (Lillig et al., 2008). In contrast, to the plenty of reports about Grxs gene structure and biochemical activity, the functions of plant Grxs are still elusive. It was known that abiotic stresses including metals and metalloids (As) exposure increase ROS production. The different ROS, including superoxide (), hydrogen peroxide (H2O2), singlet oxygen (O2), and hydroxyl radicals (OH∙), may lead to unspecific oxidation of proteins and membrane lipids and also causes DNA injury (Pastori and Foyer, 2002; Apel and Hirt, 2004). To cope with As stress-induced oxidative stress plants have evolved ROS-scavenging enzymes, such as superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX) and complex antioxidant systems, such as glutaredoxin and thioredoxin (Apel and Hirt, 2004).
High As concentrations in groundwater is a serious problem in several countries, including Bangladesh, China, India, Japan, and some parts of the United States. Crops including rice grown on soils irrigated with As-contaminated water accumulate high amount of As in their edible parts. Thus, the As contaminated rice becomes a primary source for As contamination in human and causes a serious health risk and many diseases including cancer, skin lesions (Anawar et al., 2002; Meharg and Rahman, 2003; Das et al., 2004). Also, the rice stem and husk used as fodder for animals can make a route of As entry in human. So, there is an urgent need to identify and develop an efficient strategy to minimize As accumulation in plant parts (seeds and shoot). Several studies conducted on As tolerance and metabolism in plants, such as screening of low As accumulating rice varieties, studies on transporters and genes involved in As uptake and partitioning in rice plants. Earlier reports on As uptake and translocation in plants confirmed that various genes were involved in uptake of AsV and AsIII, reduction of AsV to AsIII, transportation, chelation, vacuolar sequestration and extrusion of AsIII. Arsenate enters into the cell via OsPHT1;1 (Kamiya et al., 2013), OsPHT1;8 (Wu et al., 2011) in rice and AtPHT1;1, AtPHT1;4, AtPHT1;5, AtPHT1;7, AtPHT1;8, AtPHT1;9 in A. thaliana (Shin et al., 2004; Catarecha et al., 2007; Remy et al., 2012; LeBlanc et al., 2013; Fontenot et al., 2015). Whereas, the AsIII can enter and transported via aquaporins such as AtNIP1;1, AtNIP1;2, AtNIP5;1 (Kamiya and Fujiwara, 2009), AtNIP3;1 (Xu et al., 2008), AtNIP6;1 (Bienert et al., 2008), AtNIP7;1 (Isayenkov and Maathuis, 2008) in A. thaliana and OsNIP1;1, OsNIP2;2 (OsLsi6), OsNIP3;1 (Ma et al., 2008), OsNIP3;2 (Bienert et al., 2008), OsNIP3;3 (Katsuhara et al., 2014) in rice. OsLsi1 facilitates the influx of AsIII and organic arsenical (DMA, MMA) whereas, OsLsi2 involved in efflux and xylem loading (Ma et al., 2008). In earlier studies, As tolerance mechanism in plants mainly focuses on the uptake of As (AsIII and AsV) and formation of AsIII complex with phytochelatins, metallothionein, and glutathione to transport in cell vacuoles (Tripathi et al., 2007). However, only a few studies conducted on AsIII tolerance mediated by plant Grxs were reported. Pteris vittata Grx (PvGrx 5-6) when overexpressed in A. thaliana increases As tolerance and decreases As accumulation in transgenic plants by regulating intracellular AsIII levels during AsV stress. However, the exact mechanism was not identified (Sundaram et al., 2008, 2009; Sundaram and Rathinasabapathi, 2010). In an earlier study, we also reported that two glutaredoxins were up-regulated during both AsV and AsIII stress and might have a role in As tolerance (Chakrabarty et al., 2009; Rai et al., 2011; Dave et al., 2013; Dubey et al., 2015). Also, the upregulation of OsGrxs was found in both As sensitive and tolerant varieties, but the expression of OsGrxs was higher in sensitive varieties along with various other genes (Dave et al., 2013). Recently, we showed that the rice glutaredoxin (OsGrx) genes play a role in detoxification of arsenicals by GSH recycling and regulating intracellular AsIII levels by indirectly or directly modulating the aquaglyceroporins (AsIII channels) that conduct AsIII entry or efflux from yeast cells (Verma et al., 2016).
Here, we reported the increased As stress tolerance in OsGrxs expressing transgenic A. thaliana lines compared to wild-type (WT) lines, thus providing the evidence for the involvement of rice glutaredoxin (OsGrxs) genes in As tolerance in plants. Furthermore, during As stress, expression of the certain A. thaliana aquaporin genes (NIPs) was up-regulated in OsGrxs expressing transgenic plants compared with WT controls. Thus, these findings suggest a specific protective role of a redox protein against As stress and provide a genetic engineering strategy to improve safer crop production.
Materials and Methods
Plant Material
Rice (Oryza sativa, accessions IC-115730, and IC-3470072) seeds were germinated and let to grow in hydroponic conditions. After 1-week, seedlings were exposed to Na2HAsO4 (10 and 25 μM) and NaAsO2 (10 and 25 μM) in the hydroponic medium for another week (Dave et al., 2013).
Arabidopsis thaliana (ecotype Columbia) were grown in plastic containers, using soilrite and irrigated by Hogland’s nutrient solution. The plants were grown under 16 h light photoperiod of 150 mmol s-1 m-2 supplied by cool white fluorescent lights at 23°C constant temperature. Upon reaching 5–10 cm length, the primary inflorescences were clipped once to favor the growth of multiple secondary bolts for plants to be used for Agrobacterium-mediated transformation (Clough and Bent, 1998).
Expression Analysis of Two As Responsive OsGrxs During As Stress
The expression of OsGrxs in Na2HAsO4 (10 and 25 μM) and NaAsO2 (10 and 25 μM) stress were examined. RNA was extracted from rice roots using Qiagen RNeasy (Qiagen, USA) and cDNA was prepared using RevertAid First Strand cDNA synthesis kit (Thermo Scientific, USA). This cDNA was used as a template for real-time PCR described in the latter sections. Primers used in this study were listed in Supplementary Table S1.
Construction of Expression Vector and Agrobacterium-Mediated Transformation
The open reading frame of the OsGrx_C7 (312 bp) and OsGrx_C2.1 (402 bp) cDNA clone was amplified using the gene specific primers. The PCR product cloned in the pTZ vector to obtain pTZ-OsGrx_C7 and pTZ-OsGrx_C2.1 and verified by sequencing. Both OsGrxs were cloned in XbaI – SacI linearized plant expression vector pBI121 to derive pBI121-OsGrx_C7 and pBI121-OsGrx_C2.1 (Supplementary Figure S1A). The pBI121-OsGrx_C7, pBI121-OsGrx_C2.1 and pBI121 were transferred into Agrobacterium tumefaciens GV3101 strain via freeze-thaw method (Jyothishwaran et al., 2007). Plants were inoculated with Agrobacterium tumefaciens strain GV3101, carrying pBI121-OsGrx_C7, pBI121-OsGrx_C2.1 or pBI121 by the floral dip method (Clough and Bent, 1998). The aerial floral parts of the plants were dipped for 1 min with gentle shaking in 100 mL of 5% (w/v) sucrose with Agrobacterium tumefaciens cells at 0.8 A600 and 0.02% (v/v) of the surfactant Silwet L-77 (Union Carbide, Danbury, CT, USA). After inoculation, plants were covered with a transparent plastic to maintain humidity for 24 h and then kept at growth chamber till seed set. Putative transformants (T1) were selected based on their resistance to kanamycin (50 mg L-1) in the ½ × MS medium with 2% sucrose and 0.8% (w/v) agar. Further, the presence of the transgene was verified using a PCR procedure on isolated genomic DNA as a template with OsGrxs gene-specific primers. The putative transgenic plants were grown in controlled environments in growth chamber and seeds were collected from individual plants. From the T2 seeds, several plants were selfed and their progeny analyzed for homozygosity. Homozygous T3 lines identified from these analyzes were used for evaluations of transgene expression and functional validation.
Quantitative Reverse Transcriptase-PCR (qRT-PCR)
Transcript quantification was done by qRT-PCR using 7500 fast real-time PCR system (Applied Biosystems, USA). Total RNA, isolated using Qiagen RNeasy (Qiagen, USA) was used for cDNA synthesis. cDNA synthesis was done by using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). This cDNA was used as a template for quantifying the transcript level of OsGrxs using the gene-specific primers using 2× Master SYBR® Green (Applied Biosystem, USA). Primers for an AtTub6 were included as an internal control. The relative shift in the threshold cycle (ddCt) value was determined by subtracting the average dCt of Grx – average dCt of tubulin (n = 3). Moreover, expression was calculated by the 2ˆ-ddct method.
Glutaredoxin Assay
Soluble protein was extracted from leaves using method previously described by Fouad and Rathinasabapathi (2006). Grx activity was determined with a coupled enzyme reaction described by Holmgren (1989). In this assay, NADPH-dependent reduction of 2-hydroxyethyl disulfide (HED) in the presence of glutathione reductase (GR) was monitored at 340 nm using a UV-visible spectrophotometer (Perkin Elmer, USA). The assay mixture contained 100 mg mL-1 of bovine serum albumin, 1 mM GSH, 6 μg mL-1 yeast GR, 0.4 mM NADPH, 0.1 M Tris–HCl, 2 mM ethylene diamine tetraacetic acid (EDTA), pH 8.0, and 0.7 mM HED in a total volume of 800 μL. Non-enzymatic NADPH-dependent reduction of HED was measured for the background. Total protein was estimated by Bradford method. The enzyme activity was expressed as nmol mg-1 protein min-1.
Evaluation of As Tolerance of OsGrxs Transformed A. thaliana
Arsenic resistance during germination was evaluated by plating the transgenic seeds in ½ × MS medium containing 2% (w/v) sucrose and 0.8% (w/v) agar with or without 25 μm sodium arsenite and 250 μm sodium arsenate. The seeds were surface sterilized using 2% (v/v) sodium hypochlorite and washing thoroughly with sterilized water. Approximately 100 seeds were plated and then kept at 4°C for 48 h for stratification, and then transferred at 23°C for 3 days under light for calculating the germination rate under control and the As treatment conditions.
Root growth was evaluated by germinated seed on ½ × MS agar medium. Three-day old seedlings were transferred onto vertical plates containing ½ × MS agar medium, containing varying concentrations of sodium arsenate (50, 100, 250, and 500 μM) or sodium arsenite (5, 10, 25, and 50 μM). By marking and measuring the root tip at the beginning and end of a 10 days period, root length and fresh weight were calculated.
To evaluate the whole plant As tolerance, OsGrxs expressing transgenic lines and WT were grown in individual containers under identical conditions. A. thaliana plants were irrigated with or without sodium arsenate (250 μM) and sodium arsenite (25 μM) in Hoagland nutrient solution (Hoagland and Arnon, 1950). The As-containing medium was added to the potting medium without touching the aerial portion of the plants. After 6 weeks, growth difference was observed by measuring the aboveground biomass dry weight of completely dried samples.
Measurement of As Accumulation in Transgenic A. thaliana
Arsenic accumulation for short-term was assayed by growing A. thaliana plant for 7 days in hydroponic medium with or without 25 μM AsIII and 250 μM AsV. Samples were washed with 10 mM EDTA and distilled water, dried, mineralized in HNO3 and As was analyzed using ICP-MS. Simulated irrigation experiment assayed long-term exposure effect on the plant’s aerial parts. Fourteen-days old plants were transferred in soilrite and grown under controlled environments, watered with nutrients solution containing 25 μM AsIII and 250 μM AsV every week till plant completes its life cycle. Subsequently, plants were thoroughly rinsed with 10 mM EDTA and distilled water, dried, and digested in HNO3 and amount of As in the samples was determined using ICP-MS (Perkin Elmer, USA) (Supplementary Table S2).
Expression Analysis of A. thaliana Aquaporin Genes
Seeds of A. thaliana WT (Columbia-0) and transgenic lines were sterilized and germinated as described in previous sections. After 14 days, plants were transferred to hydroponics culture medium with 10 μM, 25 μM AsIII and 100 μM, 250 μM AsV under controlled environments (16 h light: 8 h dark cycle, 22°C, 150 mMm-1s-1 light intensity and 70% relative humidity). After 7 days, plants were harvested, root and shoot tissues used for RNA isolation and extraction of total protein.
Total RNA was extracted from A. thaliana plants using RNeasy Plant Mini Kit according to the manufacturer’s instructions (Qiagen). The qRT-PCR were performed using 2 × SYBR Green Master Mix and ABI7500 fast (Applied Biosystem, USA) in a total volume of 20 μl. The reaction contains 1 μl of each primer (5 μM), 7 μl of nuclease-free water, and 10 μl of 2× Master SYBR® Green (Applied Biosystem, USA) and cDNA as template. Reaction condition at 95°C for 10 min followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, 72°C for 15 s. The reaction without template designated as a negative control in the same PCR run for each primer pair. For each of the two independent RNA extractions, measurements of gene expression were obtained in triplicate.
Determination of Total GSH and GSSG in Transgenic A. thaliana Lines under As Stress
Arabidopsis thaliana plants were grown as described in above section. Plants were washed and re-suspended in 100 mM potassium phosphate buffer (pH 7.8), with 5% 5-sulfosalicylic acid, centrifuged for 30 min at 5000 × g at 4°C and the supernatant was used to determine total free GSH concentration.
The reaction mixture contains the cell extract (10 μL), 0.50 mM DTNB, and 0.25 mM NADPH in 100 mM potassium phosphate buffer with 5 mM EDTA, pH 7.8. The reaction was started by adding 0.2 U of GR and kinetics of DTNB conversion to TNB were followed spectrophotometrically at 412 nm. GSH concentrations were calculated from standard curves obtained with various GSH and GSSG concentrations based on the rate of TNB formation and expressed as nanomoles of GSH/mg of protein. GSH: GSSG ratios were calculated using the equation GSH/GSSG = 2(total GSH-GSSG)/GSSG (Rahman et al., 2007).
Data Analysis
Quantitative data were analyzed by analysis of variance using Graph Pad Prism program. All values reported in this work are means of at least five individual replications unless mentioned otherwise. Experiments were repeated three times. Results were expressed as means followed by corresponding standard errors.
Results
Two Rice Glutaredoxin (OsGrx) Genes Induced during As Stress
Global gene expression analysis of rice during As stress showed up-regulation of two genes, LOC_Os01g27140 (OsGrx_C7), and LOC_Os01g40500 (OsGrx_C2.1) designated as members of Grx protein family in rice (Chakrabarty et al., 2009). The expression of OsGrxs was analyzed in As tolerant (IC-340072) and sensitive (IC-115730) rice cultivars (Dave et al., 2013) under AsIII stress. To further extend this analysis, the qRT-PCR analysis done with a different set of As treated samples and results suggested that the expression of OsGrx_C7 and OsGrx_C2.1 get induced after exposure to both AsIII and AsV. Both OsGrxs show high mRNA accumulation under AsV stress compared to AsIII. However, sensitive cultivar (IC-115730) showed higher expression level as compared to tolerant cultivar. Also, OsGrxC_7 have higher expression level than OsGrxC_2.1 under As stress (Figure 1). Thus, our result indicated that both OsGrxs might involve directly or indirectly in As tolerance and metabolism.
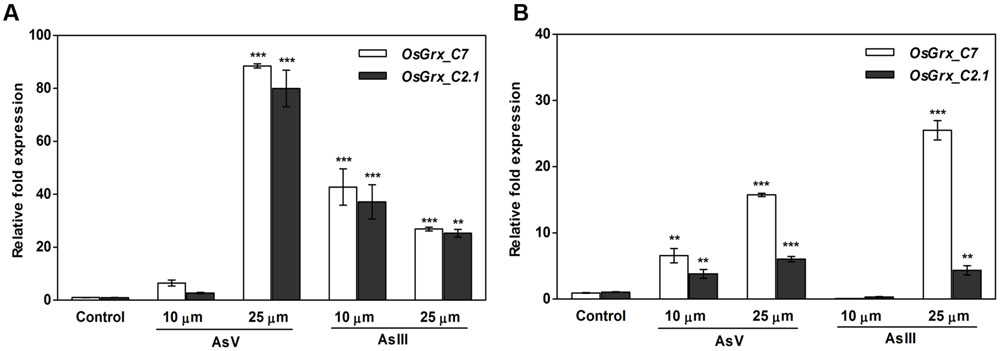
FIGURE 1. Quantitative Reverse Transcriptase-PCR (qRT-PCR) analysis of As-responsive glutaredoxin gene revealed that both OsGrxs upregulated under As stress. For qRT-PCR analysis, the rice (Oryza sativa) seedlings of (A) sensitive (IC 115730) and (B) tolerant (IC 340072), treated with AsV and AsIII were used for RNA isolation. Expression analysis was performed using OsGrx_C7 and OsGrx_C2.1 gene specific primers and rice actin as an internal control. Asterisks indicate significant differences (∗∗∗P < 0.001, ∗∗P < 0.01). Error bar mean ± SE.
Overexpression of OsGrxs in Transgenic A. thaliana
Transgenic A. thaliana lines were obtained, that ectopically express OsGrx_C7 and OsGrx_C2.1 under the transcriptional control of the CaMV 35S promoter. Expression of the transgene was analyzed in several independent lines. The transgenic lines showed varying expression levels of OsGrx_C7 and OsGrx_C2.1. Three of each OsGrxs expressing lines with similar expression pattern were selected for further analysis (Supplementary Figure S1B). The growth and development of transgenic lines and WT were comparable in pots or on half-strength MS plates.
OsGrxs Expressing Transgenic A. thaliana Lines Display Tolerance to AsIII and AsV
Germination of WT seeds were significantly inhibited on As containing medium, but OsGrxs expressing lines were least affected (Supplementary Figure S2A). In root growth bioassay there was no phenotypic change observed between WT and transgenic lines grown on ½ × MS medium without As. Transgenic lines were further grown in ½ × MS medium supplemented with 5 μM, 10 μM, 25 μM AsIII and 50 μM, 100 μM, 250 μM AsV. Although, WT as well as transgenic lines, showed root growth inhibition in the presence of the AsV and AsIII in a concentration-dependent manner (Figures 2A,B and 3A,B). OsGrxs overexpressing lines showed less root growth inhibition compared to WT, which was more significant at 25 μM AsIII and 250 μM AsV concentrations. The tolerance was also visible in aerial tissues, as transgenic lines grew better and attained significantly higher fresh weight as compared to WT (Figure 4). Root hair growth and elongation severely inhibited in WT plants as compared to transgenic lines under AsIII toxicity (Figure 5; Supplementary Figure S2B).
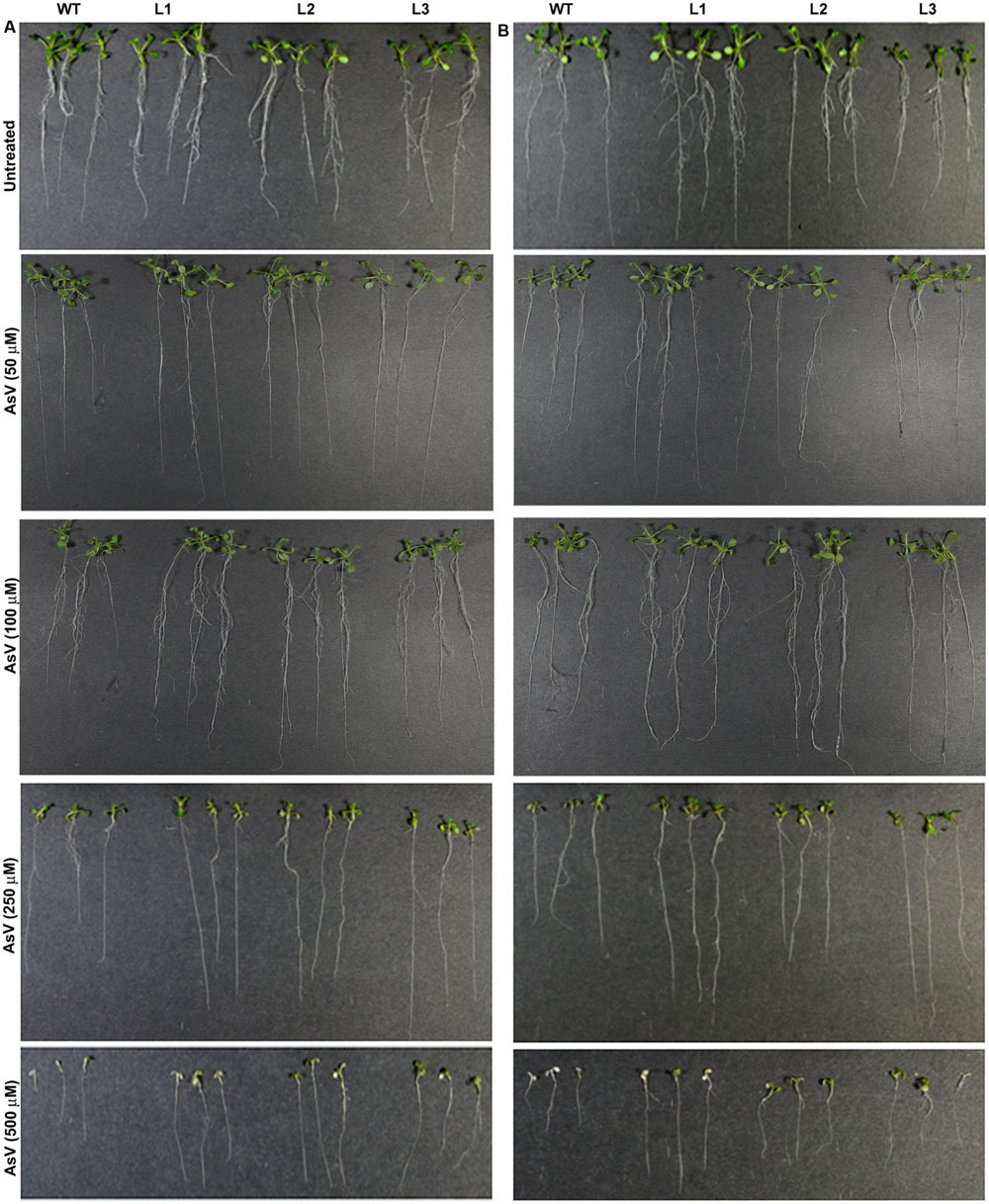
FIGURE 2. Enhanced arsenate (AsV) tolerance in A. thaliana plants transformed with (A) OsGrx_C7 and (B) OsGrx_C2.1. Phenotypic changes in WT and transgenic A. thaliana plants carrying OsGrx_C7 and OsGrx_C2.1 observed after grown vertically on plates of ½ × MS medium without As and with 50 μM AsV, 100 μM AsV, 250 μM AsV, and 500 μM AsV, for 10 days. (n = 5 plants per treatment per line and repeated five times).
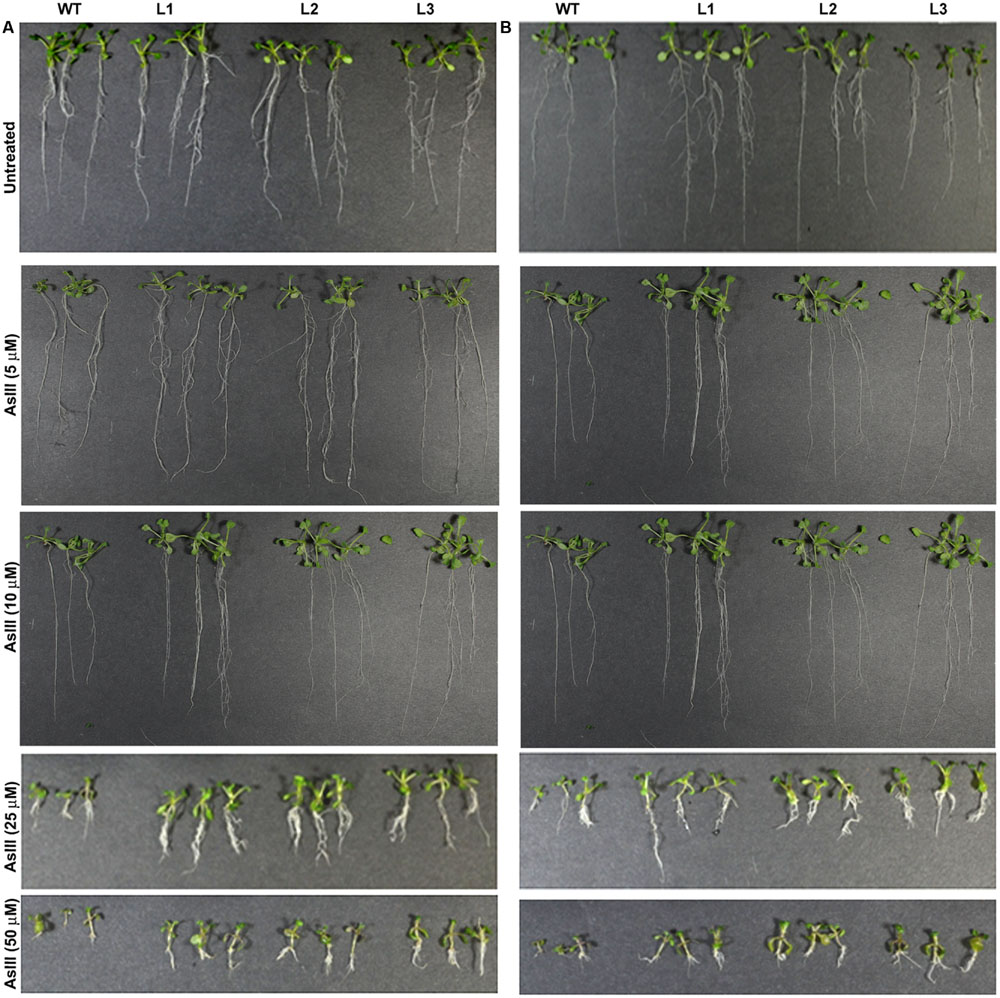
FIGURE 3. Enhanced arsenite (AsIII) tolerance in A. thaliana plants transformed with (A) OsGrx_C7 and (B) OsGrx_C2.1. Phenotypic changes in WT and transgenic A. thaliana plants carrying OsGrx_C7 and OsGrx_C2.1 observed after grown vertically on plates of ½ × MS medium without As and with 5 μM AsIII, 10 μM AsIII, 25 μM AsIII, and 50 μM AsIII for 10 days. (n = 5 plants per treatment per line and repeated five times).
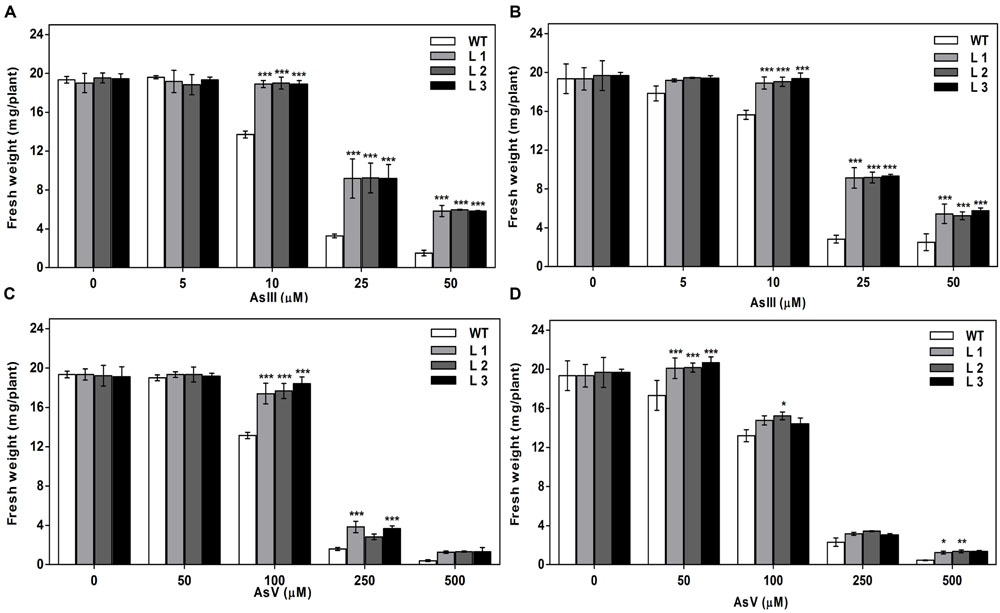
FIGURE 4. Expression of OsGrxs affects the biomass accumulation during arsenic (AsIII and AsV) stress. (A) Fresh weight of WT and OsGrx_C7 transformed plants and (B) Fresh weight of WT and OsGrx_C2.1 transformed plants during AsIII stress, (C) Fresh weight of WT and OsGrx_C7 transformed plants and (D) Fresh weight of WT and OsGrx_C2.1 transformed plants during AsV stress. The A. thaliana plants carrying OsGrx_C7 and OsGrx_C2.1 and WT plants were grown on plates of ½ × MS medium containing AsIII (5, 10, 25, and 50 μM) and AsV (50, 100, 250, and 500 μM) for 10 days. (n = 5 plants per treatment per line and repeated five times). Asterisks indicate significant differences: (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
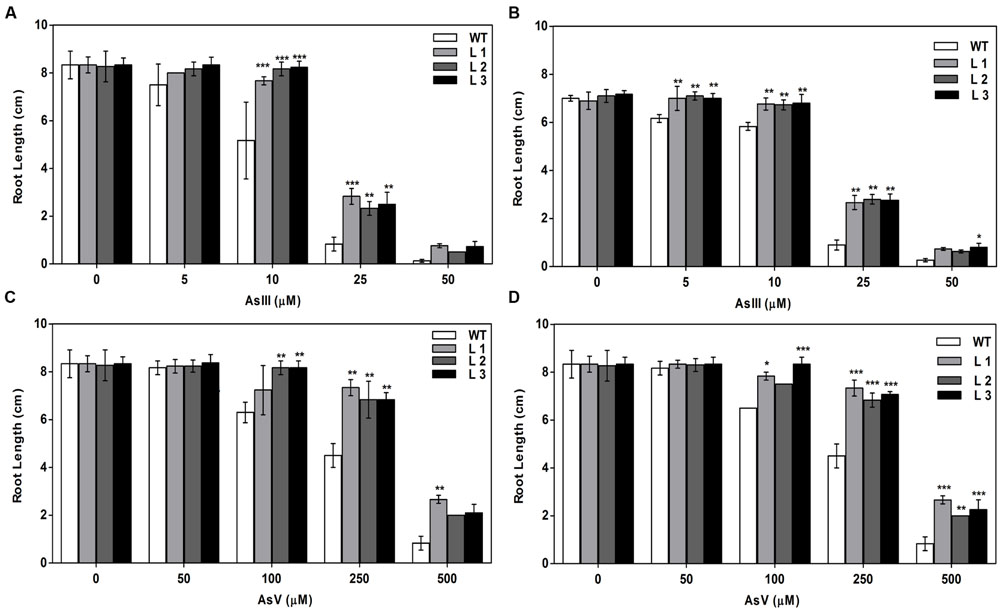
FIGURE 5. Expression of both OsGrxs affects plant growth (root length) during As (AsIII and AsV) stress. (A) Root length of WT and OsGrx_C7 transformed plants, and (B) Root length of WT and OsGrx_C2.1 transformed plants during AsIII stress, (C) Root length of WT and OsGrx_C7 transformed plants and (D) Root length of WT and OsGrx_C2.1 transformed plants during AsV stress. The A. thaliana plants carrying OsGrx_C7 and OsGrx_C2.1 and WT plants were grown on plates of ½ × MS medium containing AsIII (5, 10, 25, and 50 μM) and AsV (50, 100, 250, and 500 μM) for 10 days. (n = 5 plants per treatment per line and repeated five times). Asterisks indicate significant differences: (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
The As response of the transgenic lines was examined in 6 weeks old plants. For this, 12-day-old seedlings were transferred to pots and allowed to grow for two more weeks. Then, the plants were irrigated with or without sodium arsenate (250 μM) and sodium arsenite (25 μM) in Hoagland nutrient solution for 6 weeks. The phenotypic effects of As treatments after 6 weeks are shown in Figure 6. Both AsIII and AsV resulted in retardation of seedling development in WT seedlings. The transgenic seedlings showed less severe symptoms and continued to grow. Fresh biomass, dry weight and seeds from these plants grown on As for 6 weeks revealed higher biomass and seeds yield in transgenic lines compared to WT plants grown on AsIII and AsV. These results indicate that transgenic plants displayed improved tolerance to As stress.
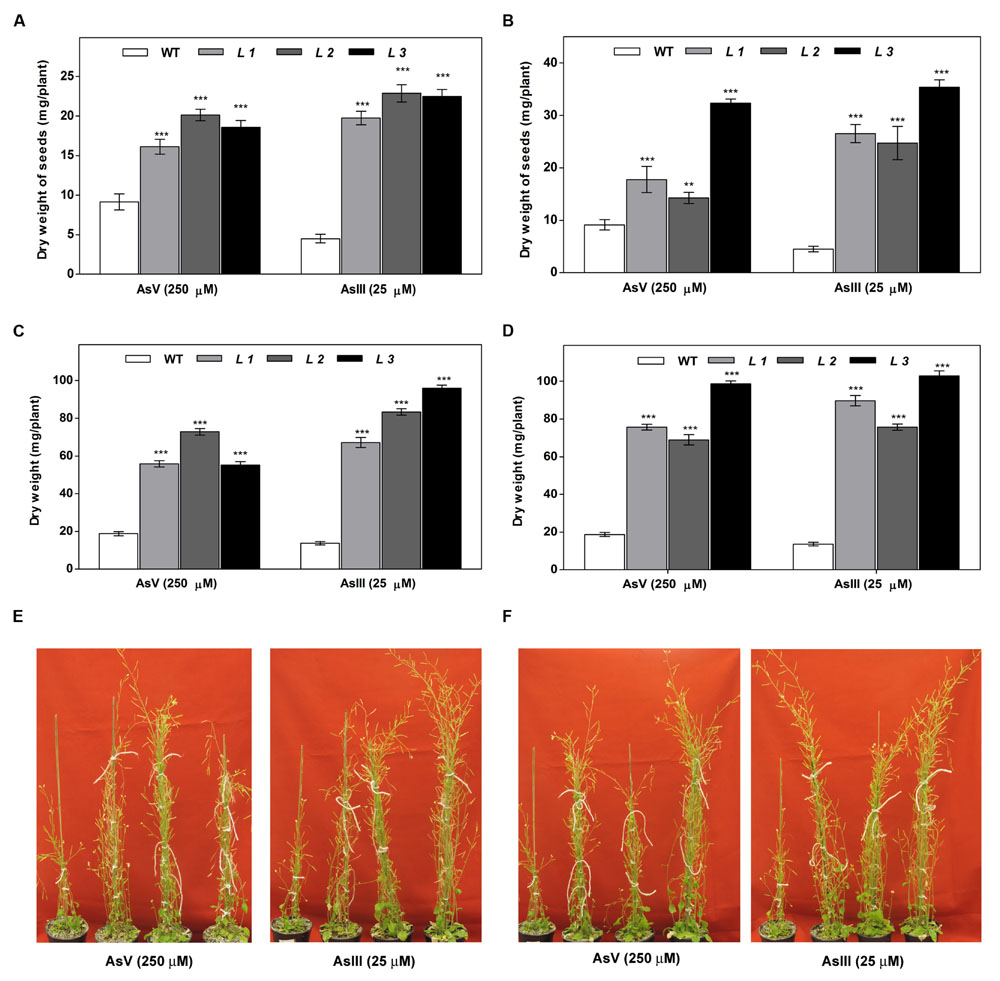
FIGURE 6. Higher biomass accumulation in above-ground tissues of transgenic A. thaliana plants after long-term cultivation in soil under As (AsIII and AsV) stress. (A) The dry weight of seeds of WT and OsGrx_C7 transformed plants, (B) Dry weight of seeds of WT and OsGrx_C2.1 transformed plants, (C) Dry weight of WT and OsGrx_C7 transformed plants, (D) Dry weight of WT and OsGrx_C2.1 transformed plants. The A. thaliana plants carrying (E) OsGrx_C7, (F) OsGrx_C2.1 and WT plants grown in soil spiked with 250 μM As(V) and 25 μM As(III) for 45 days (n = 8 plants per treatment per line and repeated three times). Asterisks indicate significant differences: (∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
Overexpression of OsGrxs reduces As Accumulation in A. thaliana
The OsGrxs expressing transgenic plants grown in hydroponics medium containing AsIII and AsV. The whole plant was digested using concentrated HNO3 for ICP-MS analysis. The results of ICP-MS analysis displayed significantly less As accumulation in the transgenic plants (Figures 7A,B). Arsenic accumulation in seeds and shoot tissues were also measured after long-term exposure of AsIII and AsV in soilrite irrigated with As-contaminated water (described in above section). ICP-MS analysis showed ca. 38% reduced As accumulation in seeds under AsV (Figure 7C) and AsIII stress (Figure 7D) compared to WT. Whereas, in shoot tissues (Shoot + rosette leaves) ca. 40% less As accumulated after exposure to AsV (Figure 7E) and AsIII (Figure 7F) compared to WT. A decrease in As content in seeds and shoot tissues of OsGrxs expressing transgenic lines confirms the role of OsGrxs in lower As uptake and higher AsIII extrusion.
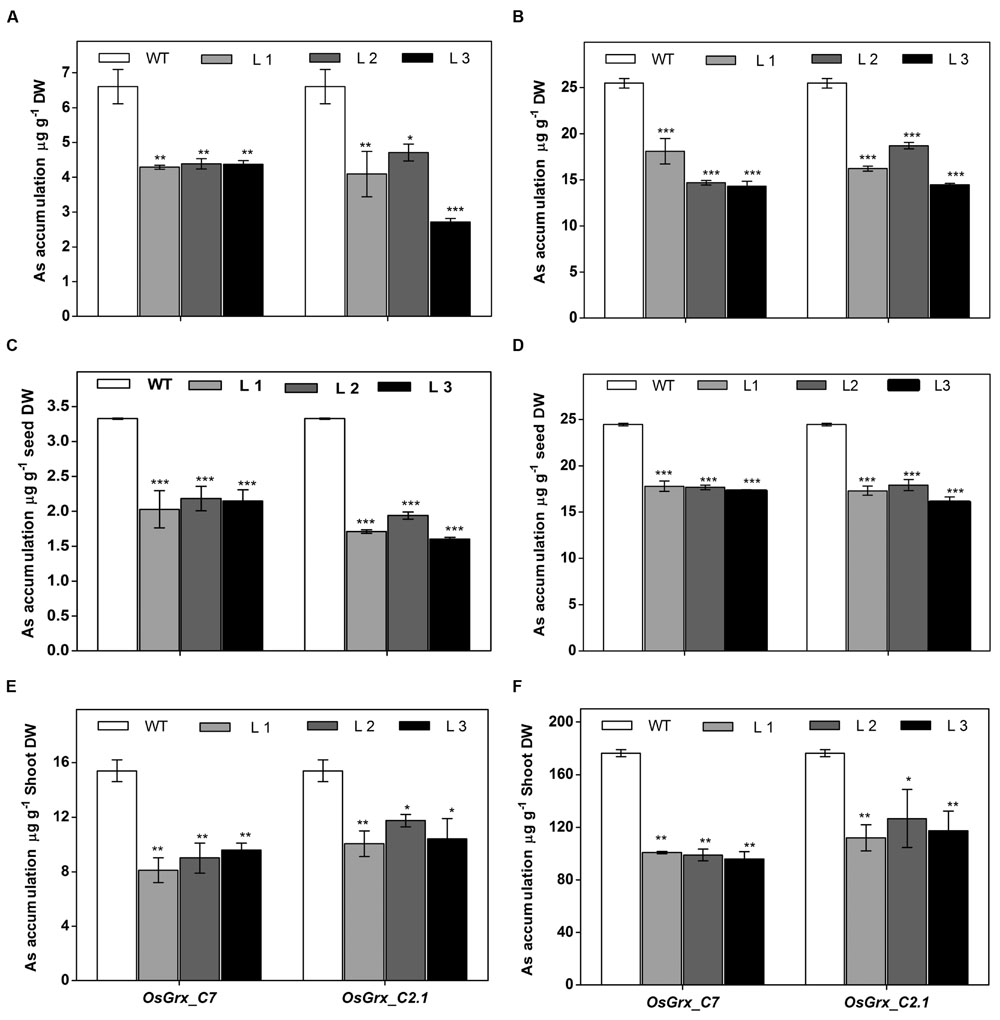
FIGURE 7. Heterologous expression of OsGrxs displayed significantly reduced arsenic accumulation in A. thaliana. Arsenic accumulation in plants grown in hydroponics with (A) 25 μM AsIII, (B) 250 μM AsIII for 7 days and whole plant was digested and subjected to ICPMS analysis (n = 10 plants per treatment per line and repeated three times). Arsenic accumulation in seeds when plants are grown in soil spiked with (C) 25 μM AsIII, (D) 250 μM AsV for 45 days. Arsenic accumulation in rosette leaves + stalks when plants are grown in soil spiked with (E) 25 μM AsIII and (F) 250 μM AsV for 45 days (n = 8 plants per treatment per line and repeated three times). Asterisks indicate significant differences: (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
Altered Expression of As Transporters in OsGrxs Expressing A. thaliana Transgenic Lines
To elucidate the rationale behind tolerance of transgenic lines to AsIII and AsV, the expression of aquaporin’s subfamily noduline-26 like intrinsic proteins (NIP), involved in AsIII transport in plants, were also examined. OsGrxs expressing lines displayed enhanced expression of AtNIP1;1, AtNIP2;1, AtNIP3;1, and AtNIP7;1 during AsV stress in both shoot (Figures 8 and 9) and root (Figures 10 and 11). However, AtNIP6;1 was slightly upregulated only in shoots under AsV stress while, AtNIP5;1 showed downregulation in roots of OsGrx_C2.1 expressing transgenic A. thaliana. It has been already reported that plant NIPs are involved in translocation of As(OH)3 against a concentration gradient (Bienert et al., 2008). AsV, being a phosphate analog, is taken up in plants via phosphate transporter and get reduced to AsIII, which might create concentration gradient for AsIII and transported by NIPs. Increased expression of NIPs in transgenic root indicated that they might involve in the extrusion of As(OH)3 in medium (Figure 10).
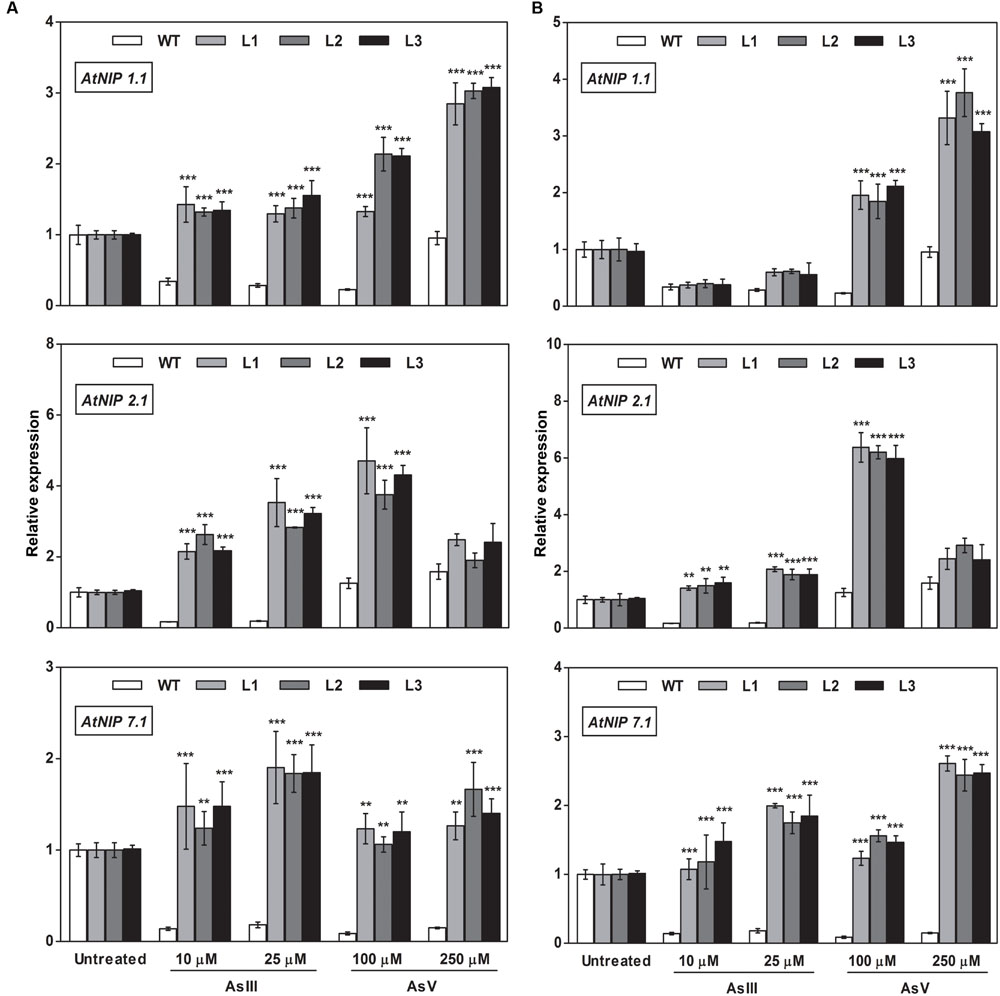
FIGURE 8. Heterologous expression of OsGrxs increases the mRNA level of A. thaliana aquaporins (AtNIP1.1; AtNIP2.1; AtNIP7.1) in the shoot. Transgenic A. thaliana plants carrying (A) OsGrx_C7 and (B) OsGrx_C2.1 were grown on ½ × MS medium and transferred to hydroponic medium supplemented with 10 μM AsIII, 25 μM AsIII and 100 μM AsV, 250 μM AsV, grown for 1 week. Samples were harvested and washed thoroughly with RNase-free water to remove As attached on the plant surface. Total RNA of A. thaliana shoot was extracted for qRT-PCR analysis. Asterisks indicate significant differences: (∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
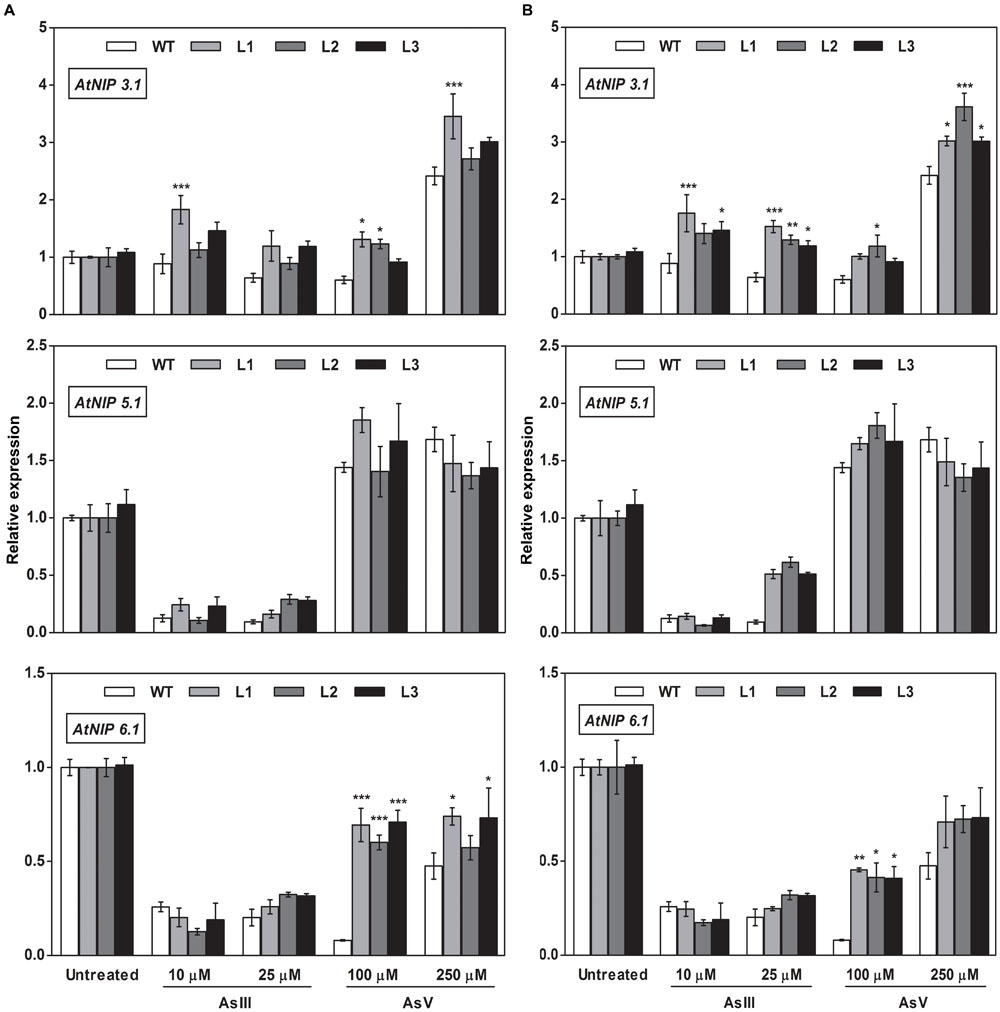
FIGURE 9. Overexpression of OsGrxs in A. thaliana alters the mRNA level of aquaporins (AtNIP3.1; AtNIP5.1; AtNIP 6.1) in shoot tissues. Transgenic A. thaliana plants carrying (A) OsGrx_C7 and (B) OsGrx_C2.1 were grown on ½ × MS medium and transferred to hydroponic medium supplemented with 10 μM AsIII, 25 μM AsIII and 100 μM AsV, 250 μM AsV, grown for 1 week. Samples were harvested and washed thoroughly with RNase-free water to remove As attached on the plant surface. Total RNA of A. thaliana shoot was extracted for qRT-PCR analysis. Relative expressions of AtNIPs (AtNIP3.1; AtNIP5.1; AtNIP6.1) were represented. Asterisks indicate significant differences: (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
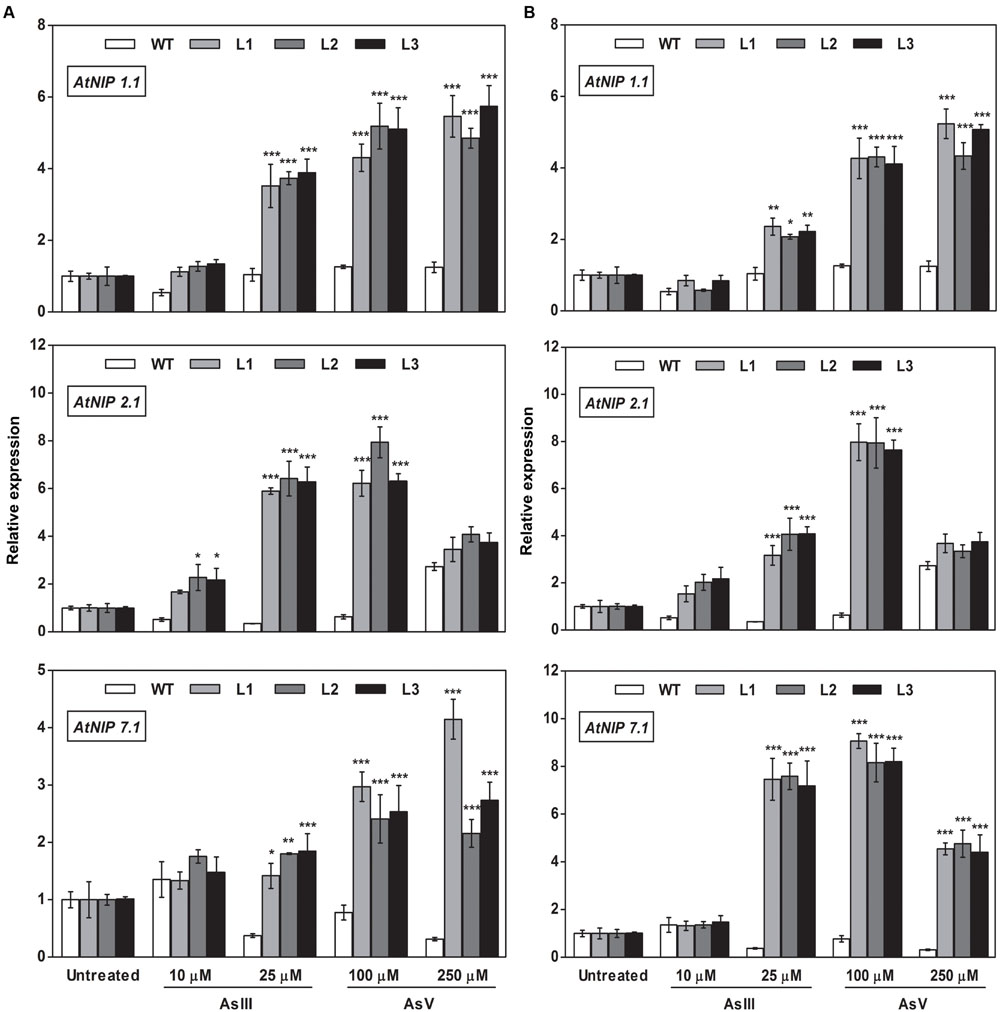
FIGURE 10. Overexpression of OsGrxs in A. thaliana increases the mRNA level of aquaporins (AtNIP1.1; AtNIP2.1; AtNIP7.1) in the root. Transgenic A. thaliana plants carrying (A) OsGrx_C7 and (B) OsGrx_C2.1 were grown on ½ × MS medium and transferred to hydroponic medium supplemented with 10 μM AsIII, 25 μM AsIII and 100 μM AsV, 250 μM AsV, grown for 1 week. Samples were harvested and washed thoroughly with RNase-free water to remove As attached on the plant surface. Total RNA extracted from roots of A. thaliana for qRT-PCR analysis. Relative expressions of AtNIPs (AtNIP1.1; AtNIP2.1; AtNIP7.1) were represented. Asterisks indicate significant differences: (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
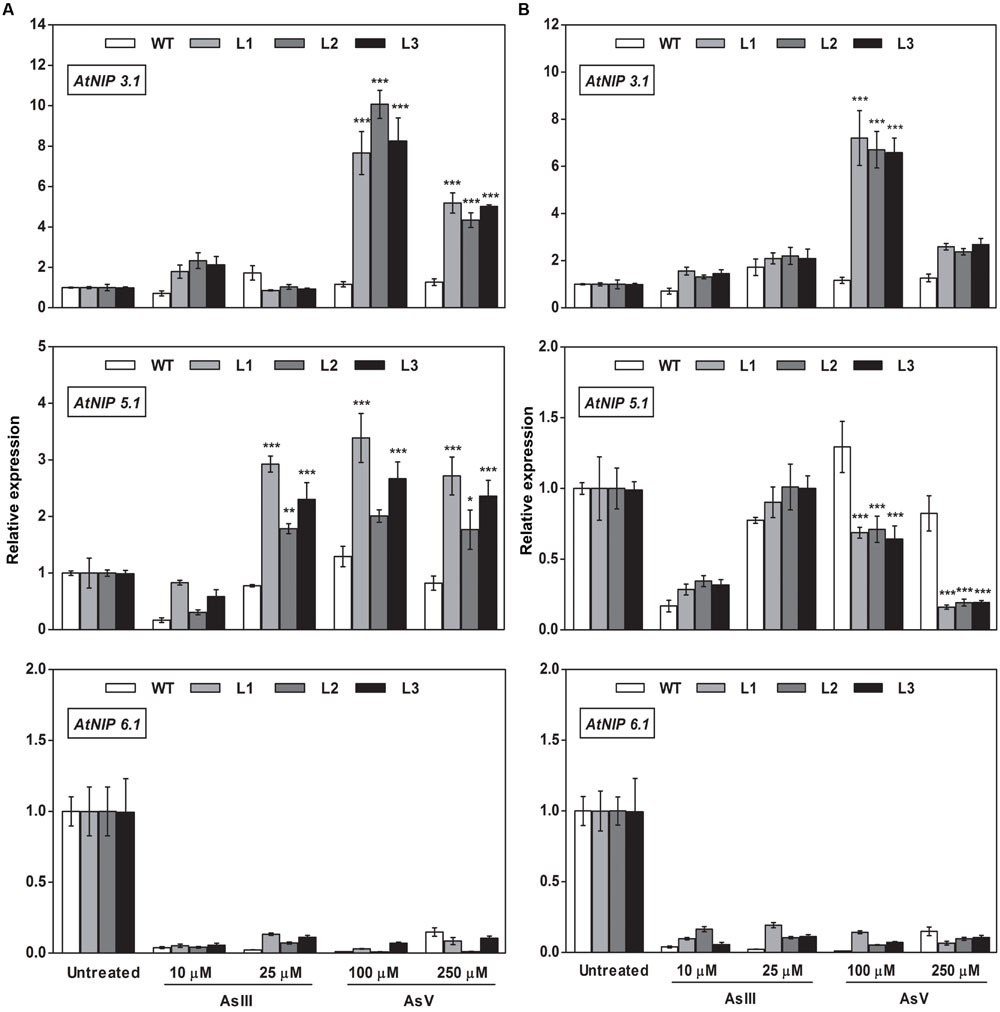
FIGURE 11. Overexpression of OsGrxs alters the mRNA level of A. thaliana aquaporins (AtNIP3.1; AtNIP5.1; AtNIP 6.1) in the root. Transgenic A. thaliana plants carrying (A) OsGrx_C7 and (B) OsGrx_C2.1 were grown on ½ × MS medium and transferred to hydroponic medium supplemented with 10 μM AsIII, 25 μM AsIII and 100 μM AsV, 250 μM AsV, grown for 1 week. Samples were harvested and washed thoroughly with RNase-free water to remove As attached on the plant surface. Total RNA was extracted from roots of A. thaliana for qRT-PCR analysis. Relative expressions of AtNIPs (AtNIP3.1; AtNIP5.1; AtNIP6.1) were represented. Asterisks indicate significant differences: (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
During AsIII stress AtNIPs have different expression pattern, OsGrx_C7 and OsGrx_C2.1 expressing lines displayed enhanced expression of AtNIP1;1, AtNIP2;1 AtNIP7;1 in both root and shoot. During AsIII exposure, NIPs were upregulated to transport and extrude AsIII in the medium. The concentration gradient might be created by depletion of AsIII in medium (uptake and sequestered in plant vacuole) or by extracellular GSH binds to AsIII. Also, transgenic lines displayed unaltered expression of AtNIP3.1, AtNIP6.1 in OsGrx_C7 and OsGrx_C2.1 expressing lines during AsIII stress. AtNIP5.1 showed higher expression in OsGrx_C7 transgenic root. However, in untreated condition expression levels of mRNA transcripts for these genes are least affected in transgenic lines. The altered transcript level of aquaporin in the presence of OsGrxs under AsV and AsIII stress confirmed that both OsGrxs alter the expression of aquaporin for As tolerance and AsIII extrusion.
The Oxidoreductase Activity of OsGrxs Increases after As Treatment
To test whether, As tolerance in transgenic A. thaliana was correlated to OsGrxs activity, glutaredoxin activity was measured in, soluble fractions of leaf protein from plants exposed to AsIII (25 μM) and AsV (250 μM). OsGrx_C7 expressing transgenic lines had c.a 27%, and OsGrx_C2.1 expressing transgenic lines had c.a. 30% Grx-specific activity compared with WT in the control condition. On exposure of As, glutaredoxin activity in OsGrx_C7 expressing transgenic lines was increased up to c.a 40% and c.a 35% compared to WT in AsIII and AsV stress, respectively. Moreover, OsGrx_C2.1 expressing transgenic lines showed increased glutaredoxin activity up to c.a 35% and c.a 30% compared to WT in AsIII and AsV stress respectively (Figures 12A,B).
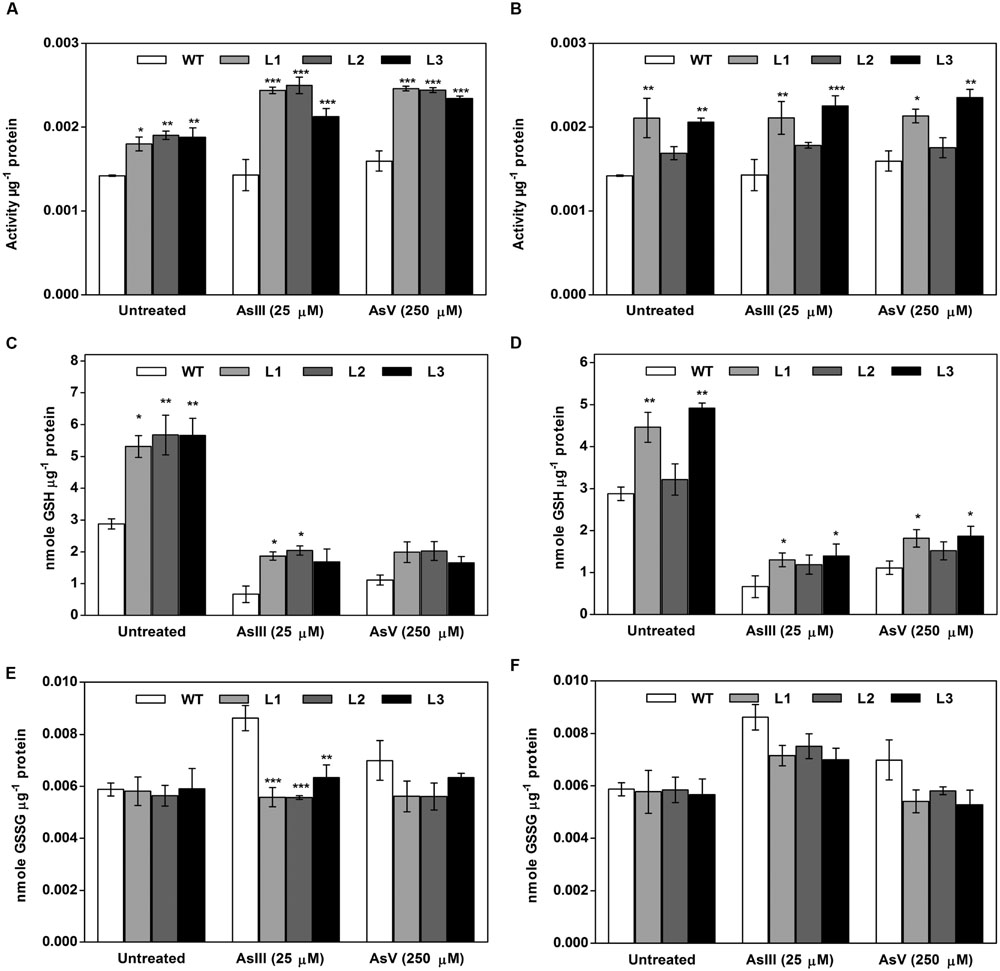
FIGURE 12. Both OsGrxs displayed enhanced Grx activity and intracellular GSH content during As stress in transgenic A. thaliana compared to WT. (A) Enhanced Grx activity in OsGrx_C7 transformed plants, (B) enhanced Grx activity in OsGrx_C2.1 transformed plants. (C) Intracellular GSH content in OsGrx_C7 transformed plants, (D) intracellular GSH in OsGrx_C2.1 transformed plants, (E) intracellular GSSG content in OsGrx_C7 transformed plants, (F) intracellular GSSG in OsGrx_C2.1 transformed plants. Transgenic A. thaliana plants were grown on ½ × MS medium and transferred to hydroponic medium supplemented with 25 μM AsIII and 250 μM AsV, grown for 1 week. Samples were harvested and washed thoroughly with water to remove As attached on the plant surface. The plants were homogenized in potassium phosphate buffer pH 7.8 (100 mM, ice-cold) with 5% 5-sulfosalicylic acid and used to determine intracellular glutathione (oxidized and reduced). The reaction started by adding 0.2 U of GR and the kinetics of DTNB conversion to TNB were followed spectrophotometrically at 412 nm. GSH concentrations calculated from standard curves obtained with various GSH and GSSG concentrations. Asterisks indicate significant differences: (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Error bars, mean ± SE.
Heterologous Expression of OsGrxs Maintains the Cellular Glutathione Level under As Stress
In planta expression of OsGrxs displayed high GSH content compared to WT plants. Under AsIII stress, intracellular GSH concentration was increased up to 65% in OsGrx_C7 expressing plants and up to 45% in OsGrx_C2.1 expressing plants compared to WT. While on AsV exposure, intracellular GSH concentration was increased c.a 35% and c.a 30% in plants expressing OsGrx_C7 and OsGrx_C2.1 respectively compared to WT (Figures 12C,D). Expression of OsGrxs displayed decreases in GSSG content compared to WT plants (Figures 12E,F).
Discussion
Members of Grx protein family in rice, A. thaliana as well other higher plants reported to involve in reversible glutathionylation and iron metabolism (Lillig et al., 2008). The involvement of OsGrxs in As transport is still unexplored. Here, we described two rice glutaredoxins (OsGrx_C7 and OsGrx_C2.1) had a significant role in As tolerance. Results showed that transgenic expression of both OsGrxs in A. thaliana was sufficient to increase plant tolerance to As. Earlier studies reported that expression of OsGrx_C7 and OsGrx_C2.1 was induced during As stress in rice in addition to defense and stress responsive genes, transporters, heat-shock proteins, metallothioneins, sulfate-metabolizing proteins (Norton et al., 2008; Chakrabarty et al., 2009; Gautam et al., 2012; Nguyen et al., 2014). In this study, similar results were observed through RT-PCR analysis, upon AsIII and AsV exposure (Figure 1) supporting the possible involvement of OsGrxs in As metabolism and transport. These results suggested that on As exposure both OsGrxs show upregulation to cope with redox imbalance created by arsenic, as heavy metals such as arsenate, arsenite, chromium, cadmium, leads to increasing oxidative stress in cells which create redox imbalance and cellular damage (Stohs and Bagchi, 1995). The expression of both OsGrxs was higher in sensitive varieties due to higher As uptake and translocation compared to tolerant variety. These results also supported by previous studies of our group where sensitive rice variety showed higher upregulation of glutaredoxins and genes involved in thiol metabolism due to greater As influx (13-fold) than tolerant genotype at 25-μM AsIII exposure in hydroponics (Dave et al., 2013). In contrast to sensitive variety, tolerant genotype displayed relatively low expression of glutaredoxins, genes involved in thiol metabolism, and less phytochelatin accumulation that was enough to detoxify the low levels of As. However, phytochelatin accumulation was also greater in the sensitive varieties, but it was not adequate to detoxify the high As content, and therefore it leads to strong up-regulation of the antioxidant gene including As responsive glutaredoxins (Dave et al., 2013).
In the present study, we chose to utilize a heterologous transgenic expression method to evaluate the function of OsGrxs in plants and demonstrated that overexpression of any one of two As-induced OsGrxs genes can confer As resistance in plants (Figures 2 and 3). The differences in relative root growth were significant and reproducible (Figure 4). Furthermore, lines expressing these two genes showed lower As content in whole plants (hydroponics), shoots (leaves + shoot) and seeds of transgenic A. thaliana (Figure 7). Sundaram et al. (2009) also reported a similar observation, where a Grx from Pteris vittata (PvGRX5) expressed in A. thaliana, resulted in more As tolerance compared with control lines based on germination, root growth, and whole plant growth. PvGRX5 expressing lines also contained significantly lower total As compared to control lines following treatment with AsV. However, they have failed to identify how PvGRX5 affects AsV uptake, AsIII translocation and efflux.
Based on our earlier work on yeast (Verma et al., 2016), we raised the question whether the As uptake and efflux regulated by OsGrxs affected the transcript level of aquaporins and other As-related transporters. In the present study, we demonstrated that A. thaliana NIPs including AtNIP1;1, AtNIP2;1 and AtNIP7;1 were upregulated during AsV stress (Figures 8 and 10). These NIPs are known to involve in AsIII uptake and translocation. Thus, confirmed that both OsGrxs were involved in AsV tolerance by regulating the transcript level of aquaporins. Both OsGrxs are also involved in AsV reduction (Verma et al., 2016) and might create AsIII gradient to extrude via aquaporins down to concentration gradient. In previous reports, it was demonstrated that aquaporins transported AsIII in the form of As(OH)3 that are not transported by active transporters such as ABCC (plants) and Ycf1, Acr3 (Yeast) (Wysocki et al., 1997). In our study, we also found that OsGrx_C7 and OsGrx_C2.1 expressing lines displayed enhanced expression of AtNIP1;1, AtNIP2;1 and AtNIP7;1 during AsIII stresses (Figures 8 and 10). It has been reported that the expression of several A. thaliana NIPs, AtNIP5;1, AtNIP6;1, and AtNIP7;1, enhances the ability of yeast cells to extrude AsIII (Bienert et al., 2008; Isayenkov and Maathuis, 2008). Earlier it was also known that rice NIP aquaporin OsNIP2;1 (Lsi1) is also able to efflux AsIII out of rice root cells into the external medium (Zhao et al., 2010). Whereas, suppressing the expression of OsNIP2;1 in rice and AtNIP1;1, and AtNIP7;1 in A. thaliana resulted in the decreased uptake of AsIII (Isayenkov and Maathuis, 2008; Ma et al., 2008; Kamiya et al., 2009). These studies suggested that NIPs facilitate the bidirectional movement of AsIII in plants. Recently Xu et al. (2015) reported that AtNIP3;1 is involved in AsIII uptake and root-to-shoot translocation in A. thaliana, probably as a passive and bidirectional AsIII transporter. Here, the expression of AtNIP3;1 was not as high as compared with other NIPs during AsIII stress (Figures 9 and 11). We also observed tissue-specific as well as Grx specific modulation of few NIPs. Thus, our results suggest that overexpression of rice Grxs modulates the expression of different NIPs in a tissue-specific manner and some of the NIPs might involve in the extrusion of AsIII from the roots of A. thaliana upon exposure to AsIII and AsV stresses. Moreover, we have searched the different data sets provided by various authors, in the data set of Chakrabarty et al. (2009), we observed the co-expression of the glutaredoxins [Os01g27140 (OsGrx_C7) and Os02g40500 (OsGrx_C2.1)] with the aquaporins in Rice genome array (Supplementary Figure S3). These results support our data that overexpression of OsGrxs alters the expression pattern of certain aquaporins (NIPs) for AsIII extrusion.
The As detoxification involves transport of As(GS)3 complex from the cytosol to the extracellular medium and or vacuolar lumen by active transporters. GSH might also be exported into the extracellular medium under As stress (Thorsen et al., 2012), lead to GSH depletion in the cell cytosol (Meharg and Hartley-Whitaker, 2002; Tripathi et al., 2007, 2012). Measurement of total GSH and GSSG during As stress gave the evidence of the involvement of both OsGrxs in maintaining of cellular GSH content (Figure 12). Thus, both OsGrxs maintain cellular redox status either by increasing GSH production or by GSH recycling. Grxs are also known to involve in defense activities by controlling the expression of antioxidant genes. Grxs able to catalyzed reversible glutathionylation of protein during adverse condition to protect cellular proteins damage by ROS (Klatt and Lamas, 2000). Grxs act by interacting with target proteins, altering their redox states and function. Grxs also control the expression of several other genes by regulating the DNA-binding activities of redox-sensitive transcription factors such as AP-1, NF-kβ, and OxyR (Hirota et al., 1997; Luikenhuis et al., 1998; Zheng et al., 1998; Harper et al., 2001). Here, we propose that both OsGrxs maintain the redox status of the cells and modulates the expression of AtNIPs under As stress, which might be accounted for the extrusion of intracellular AsIII and minimize the translocation of toxic As to edible parts of plants (Figure 7). Many others researchers had altered As tolerance in transgenic plants via over-expressing specific genes encoding enzymes involved in the synthesis of phytochelatins in heavy metal detoxification (Dhankher et al., 2002; Gasic and Korban, 2007). However, the characterization of OsGrxs represents the first successful attempt toward a genetic dissection of AsIII tolerance mechanism in the rice and is likely independent of phytochelatins and possibly involves aquaporins. Thus, our results strongly suggest that OsGrxs could be used to engineer transgenic crops with an ability to accumulate low As levels in seeds and shoot parts.
It is the first report on identification and characterization of a member of Grx family protein, in rice involved in As transport. These OsGrxs participate in reducing AsIII uptake and AsIII extrusion by altering the aquaporins (AtNIPs) transcripts in A. thaliana under As stress. OsGrxs also play a crucial role in maintaining the intracellular GSH pool and redox status of the cells under As stress, indicating that the thiol/disulfide couple is ideally suited to redox modulation, serving as redox sensor and a switch to alter protein structures and activities leading to modulation of several gene functions. Although the mechanistic details are not entirely clear, it is likely that there are several independent and interacting genes are involved. Further analysis will help to decipher the exact mechanism involved in OsGrxs mediated As metabolism in plants. However, the observations of this study are of central importance for the development of rice genotypes with less As accumulation in their edible parts through upregulation of OsGrxs to meet the perspective of As safe rice.
Author Contributions
DC, SM, and PV conceived and designed the experiments. All experiments performed by PV and SV. DC, SM, and PV analyzed the data. DC, RT, VP, and OD revised the paper. DC, PV, RT, OD, and SV wrote the paper. All authors have read and approve of the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors acknowledge the DST, Govt. of India, for funding as project DST File no. SR/SO/PS/81/2010 and CSIR for BSC 0107. PV and SV thankfully acknowledge Council of Scientific and Industrial Research (CSIR), India for senior research fellowship. Support by the Director CSIR-NBRI is gratefully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00740
References
Anawar, H. M., Akai, J., Mostofa, K. M., Safiullah, S., and Tareq, S. M. (2002). Arsenic poisoning in groundwater: health risk and geochemical sources in Bangladesh. Environ. Int. 27, 597–604. doi: 10.1016/S0160-4120(01)00116-7
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bienert, G. P., Thorsen, M., Schussler, M. D., Nilsson, H. R., Wagner, A., Tamas, M. J., et al. (2008). A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biol. 6:26. doi: 10.1186/1741-7007-6-26
Catarecha, P., Segura, M. D., Franco-Zorrilla, J. M., Garcia-Ponce, B., Lanza, M., Solano, R., et al. (2007). A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19, 1123–1133. doi: 10.1105/tpc.106.041871
Chakrabarty, D., Trivedi, P. K., Misra, P., Tiwari, M., Shri, M., Shukla, D., et al. (2009). Comparative transcriptome analysis of arsenate and arsenite stresses in rice seedlings. Chemosphere 74, 688–702. doi: 10.1016/j.chemosphere.2008.09.082
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dalle-Donne, I., Rossi, R., Colombo, G., Giustarini, D., and Milzani, A. (2009). Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96. doi: 10.1016/j.tibs.2008.11.002
Das, H. K., Mitra, A. K., Sengupta, P. K., Hossain, A., Islam, F., and Rabbani, G. H. (2004). Arsenic concentrations in rice, vegetables, and fish in Bangladesh: a preliminary study. Environ. Int. 30, 383–387. doi: 10.1016/j.envint.2003.09.005
Dave, R., Singh, P. K., Tripathi, P., Shri, M., Dixit, G., Dwivedi, S., et al. (2013). Arsenite tolerance is related to proportional thiolic metabolite synthesis in rice (Oryza sativa L.). Arch. Environ. Contam. Toxicol. 64, 235–242. doi: 10.1007/s00244-012-9818-8
Dhankher, O. P., Li, Y. J., Rosen, B. P., Shi, J., Salt, D., Senecoff, J. F., et al. (2002). Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat. Biotechnol. 20, 1140–1145. doi: 10.1038/nbt747
Dubey, A. K., Kumar, N., Sahu, N., Verma, P. K., Chakrabarty, D., Behera, S. K., et al. (2015). Response of two rice cultivars differing in their sensitivity towards arsenic, differs in their expression of glutaredoxin and glutathione S transferase genes and antioxidant usage. Ecotoxicol. Environ. Saf. 124, 393–405. doi: 10.1016/j.ecoenv.2015.10.017
Fernandes, A. P., and Holmgren, A. (2004). Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox. Signal. 6, 63–74. doi: 10.1089/152308604771978354
Fontenot, E. B., DiTusa, S. F., Kato, N., Olivier, D. M., Dale, R., Lin, W. Y., et al. (2015). Increased phosphate transport of Arabidopsis thaliana Pht1;1 by site-directed mutagenesis of tyrosine 312 may be attributed to the disruption of homomeric interactions. Plant Cell Environ. 38, 2012–2022. doi: 10.1111/pce.12522
Fouad, W. M., and Rathinasabapathi, B. (2006). Expression of bacterial L-aspartate-alpha-decarboxylase in tobacco increases beta-alanine and pantothenate levels and improves thermotolerance. Plant Mol. Biol. 60, 495–505. doi: 10.1007/s11103-005-4844-9
Gallogly, M. M., and Mieyal, J. J. (2007). Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 7, 381–391. doi: 10.1016/j.coph.2007.06.003
Gao, X.-H., Zaffagnini, M., Bedhomme, M., Michelet, L., Cassier-Chauvat, C., Decottignies, P., et al. (2010). Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii: kinetics and specificity in deglutathionylation reactions. FEBS Lett. 584, 2242–2248. doi: 10.1016/j.febslet.2010.04.034
Garg, R., Jhanwar, S., Tyagi, A. K., and Jain, M. (2010). Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 17, 353–367. doi: 10.1093/dnares/dsq023
Gasic, K., and Korban, S. S. (2007). Transgenic Indian mustard (Brassica juncea) plants expressing an Arabidopsis phytochelatin synthase (AtPCS1) exhibit enhanced As and Cd tolerance. Plant Mol. Biol. 64, 361–369. doi: 10.1007/s11103-007-9158-7
Gautam, N., Verma, P. K., Verma, S., Tripathi, R. D., Trivedi, P. K., Adhikari, B., et al. (2012). Genome-wide identification of rice class I metallothionein gene: tissue expression patterns and induction in response to heavy metal stress. Funct. Integr. Genomics 12, 635–647. doi: 10.1007/s10142-012-0297-9
Harper, R., Wu, K., Chang, M. M., Yoneda, K., Pan, R., Reddy, S. P., et al. (2001). Activation of nuclear factor-kappa b transcriptional activity in airway epithelial cells by thioredoxin but not by N-acetyl-cysteine and glutathione. Am. J. Respir. Cell Mol. Biol. 25, 178–185. doi: 10.1165/ajrcmb.25.2.4471
Hirota, K., Matsui, M., Iwata, S., Nishiyama, A., Mori, K., and Yodoi, J. (1997). AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl. Acad. Sci. U.S.A. 94, 3633–3638. doi: 10.1073/pnas.94.8.3633
Hoagland, D. R., and Arnon, D. I. (1950). The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 347, 32.
Holmgren, A., and Aslund, F. (1995). Glutaredoxin. Methods Enzymol. 252, 283–292. doi: 10.1016/0076-6879(95)52031-7
Isayenkov, S. V., and Maathuis, F. J. (2008). The Arabidopsis thaliana aquaglyceroporin AtNIP7;1 is a pathway for arsenite uptake. FEBS Lett. 582, 1625–1628. doi: 10.1016/j.febslet.2008.04.022
Jyothishwaran, G., Kotresha, D., Selvaraj, T., Srideshikan, S. M., Rajvanshi, P. K., and Jayabaskaran, C. (2007). A modified freeze-thaw method for efficient transformation of Agrobacterium tumefaciens. Curr. Sci. 93, 770–772.
Kamiya, T., and Fujiwara, T. (2009). Arabidopsis NIP1;1 transports antimonite and determines antimonite sensitivity. Plant Cell Physiol. 50, 1977–1981. doi: 10.1093/pcp/pcp130
Kamiya, T., Islam, M. R., Duan, G. L., Uraguchi, S., and Fujiwara, T. (2013). Phosphate deficiency signaling pathway is a target of arsenate and phosphate transporter OsPT1 is involved in as accumulation in shoots of rice. Soil Sci. Plant Nutr. 59, 580–590. doi: 10.1080/00380768.2013.804390
Kamiya, T., Tanaka, M., Mitani, N., Ma, J. F., Maeshima, M., and Fujiwara, T. (2009). NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J. Biol. Chem. 284, 2114–2120. doi: 10.1074/jbc.M806881200
Katsuhara, M., Sasano, S., Horie, T., Matsumoto, T., Rhee, J., and Shibasaka, M. (2014). Functional and molecular characteristics of rice and barley NIP aquaporins transporting water, hydrogen peroxide and arsenite. Plant Biotechnol. 31, 213–219. doi: 10.5511/plantbiotechnology.14.0421a
Klatt, P., and Lamas, S. (2000). Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur. J. Biochem. 267, 4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x
LeBlanc, M. S., McKinney, E. C., Meagher, R. B., and Smith, A. P. (2013). Hijacking membrane transporters for arsenic phytoextraction. J. Biotechnol. 163, 1–9. doi: 10.1016/j.jbiotec.2012.10.013
Lillig, C. H., Berndt, C., and Holmgren, A. (2008). Glutaredoxin systems. Biochim. Biophys. Acta 1780, 1304–1317. doi: 10.1016/j.bbagen.2008.06.003
Luikenhuis, S., Perrone, G., Dawes, I. W., and Grant, C. M. (1998). The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell 9, 1081–1091. doi: 10.1091/mbc.9.5.1081
Ma, J. F., Yamaji, N., Mitani, N., Xu, X. Y., Su, Y. H., McGrath, S. P., et al. (2008). Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. U.S.A. 105, 9931–9935. doi: 10.1073/pnas.0802361105
Meharg, A. A., and Hartley-Whitaker, J. (2002). Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 154, 29–43. doi: 10.1046/j.1469-8137.2002.00363.x
Meharg, A. A., and Rahman, M. M. (2003). Arsenic contamination of Bangladesh paddy field soils: implications for rice contribution to arsenic consumption. Environ. Sci. Technol. 37, 229–234. doi: 10.1021/es0259842
Michelet, L., Zaffagnini, M., Vanacker, H., Le Marechal, P., Marchand, C., Schroda, M., et al. (2008). In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J. Biol. Chem. 283, 21571–21578. doi: 10.1074/jbc.M802331200
Nguyen, Q.-T. T., Huang, T.-L., and Huang, H.-J. (2014). Identification of genes related to arsenic detoxification in rice roots using microarray analysis. Int. J. Biosci. Biochem. Bioinforma. 4, 22–27. doi: 10.7763/IJBBB.2014.V4.304
Norton, G. J., Lou-Hing, D. E., Meharg, A. A., and Price, A. H. (2008). Rice-arsenate interactions in hydroponics: whole genome transcriptional analysis. J. Exp. Bot. 59, 2267–2276. doi: 10.1093/jxb/ern097
Pastori, G. M., and Foyer, C. H. (2002). Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 129, 460–468. doi: 10.1104/pp.011021
Rahman, I., Kode, A., and Biswas, S. K. (2007). Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165. doi: 10.1038/nprot.2006.378
Rai, A., Tripathi, P., Dwivedi, S., Dubey, S., Shri, M., Kumar, S., et al. (2011). Arsenic tolerances in rice (Oryza sativa) have a predominant role in transcriptional regulation of a set of genes including sulphur assimilation pathway and antioxidant system. Chemosphere 82, 986–995. doi: 10.1016/j.chemosphere.2010.10.070
Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Sa-Correia, I., and Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Rouhier, N., Couturier, J., and Jacquot, J. P. (2006). Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 57, 1685–1696. doi: 10.1093/jxb/erl001
Rouhier, N., Couturier, J., Johnson, M. K., and Jacquot, J.-P. (2010). Glutaredoxins: roles in iron homeostasis. Trends Biochem. Sci. 35, 43–52. doi: 10.1016/j.tibs.2009.08.005
Rouhier, N., Gelhaye, E., and Jacquot, J. P. (2004). Plant glutaredoxins: still mysterious reducing systems. Cell Mol. Life. Sci. 61, 1266–1277. doi: 10.1007/s00018-004-3410-y
Shin, H., Shin, H. S., Dewbre, G. R., and Harrison, M. J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Stohs, S. J., and Bagchi, D. (1995). Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18, 321–336. doi: 10.1016/0891-5849(94)00159-H
Sundaram, S., and Rathinasabapathi, B. (2010). Transgenic expression of fern Pteris vittata glutaredoxin PvGrx5 in Arabidopsis thaliana increases plant tolerance to high temperature stress and reduces oxidative damage to proteins. Planta 231, 361–369. doi: 10.1007/s00425-009-1055-7
Sundaram, S., Rathinasabapathi, B., Ma, L. Q., and Rosen, B. P. (2008). An arsenate-activated glutaredoxin from the arsenic hyperaccumulator fern Pteris vittata L. regulates intracellular arsenite. J. Biol. Chem. 283, 6095–6101. doi: 10.1074/jbc.M704149200
Sundaram, S., Wu, S., Ma, L. Q., and Rathinasabapathi, B. (2009). Expression of a Pteris vittata glutaredoxin PvGRX5 in transgenic Arabidopsis thaliana increases plant arsenic tolerance and decreases arsenic accumulation in the leaves. Plant Cell Environ. 32, 851–858. doi: 10.1111/j.1365-3040.2009.01963.x
Thorsen, M., Jacobson, T., Vooijs, R., Navarrete, C., Bliek, T., Schat, H., et al. (2012). Glutathione serves an extracellular defence function to decrease arsenite accumulation and toxicity in yeast. Mol. Microbiol. 84, 1177–1188. doi: 10.1111/j.1365-2958.2012.08085.x
Tripathi, R. D., Srivastava, S., Mishra, S., Singh, N., Tuli, R., Gupta, D. K., et al. (2007). Arsenic hazards: strategies for tolerance and remediation by plants. Trends Biotechnol. 25, 158–165. doi: 10.1016/j.tibtech.2007.02.003
Tripathi, R. D., Tripathi, P., Dwivedi, S., Dubey, S., Chatterjee, S., Chakrabarty, D., et al. (2012). Arsenomics: omics of arsenic metabolism in plants. Front. Physiol. 3:275. doi: 10.3389/fphys.2012.00275
Verma, P. K., Verma, S., Meher, A. K., Pande, V., Mallick, S., Bansiwal, A. K., et al. (2016). Overexpression of rice glutaredoxins (OsGrxs) significantly reduces arsenite accumulation by maintaining glutathione pool and modulating aquaporins in yeast. Plant Physiol. Biochem. 106, 208–217. doi: 10.1016/j.plaphy.2016.04.052
Wu, Z. C., Ren, H. Y., McGrath, S. P., Wu, P., and Zhao, F. J. (2011). Investigating the Contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 157, 498–508. doi: 10.1104/pp.111.178921
Wysocki, R., Bobrowicz, P., and Ulaszewski, S. (1997). The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J. Biol. Chem. 272, 30061–30066. doi: 10.1074/jbc.272.48.30061
Xu, W., Dai, W., Yan, H., Li, S., Shen, H., Chen, Y., et al. (2015). Arabidopsis NIP3;1 plays an important role in arsenic uptake and Root-to-Shoot translocation under arsenite stress conditions. Mol. Plant 8, 722–733. doi: 10.1016/j.molp.2015.01.005
Xu, X. Y., McGrath, S. P., Meharg, A. A., and Zhao, F. J. (2008). Growing rice aerobically markedly decreases arsenic accumulation. Environ. Sci. Technol. 42, 5574–5579. doi: 10.1021/es800324u
Zaffagnini, M., Bedhomme, M., Marchand, C. H., Couturier, J. R., Gao, X. H., Rouhier, N., et al. (2012). Glutaredoxin s12: unique properties for redox signaling. Antioxid. Redox. Signal. 16, 17–32. doi: 10.1089/ars.2011.3933
Zhao, F.-J., McGrath, S. P., and Meharg, A. A. (2010). Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annu. Rev. Plant Biol. 61, 535–559. doi: 10.1146/annurev-arplant-042809-112152
Keywords: arsenic, GSH, OsGrxs, glutaredoxin, Oryza sativa, aquaporin
Citation: Verma PK, Verma S, Pande V, Mallick S, Deo Tripathi R, Dhankher OP and Chakrabarty D (2016) Overexpression of Rice Glutaredoxin OsGrx_C7 and OsGrx_C2.1 Reduces Intracellular Arsenic Accumulation and Increases Tolerance in Arabidopsis thaliana. Front. Plant Sci. 7:740. doi: 10.3389/fpls.2016.00740
Received: 03 February 2016; Accepted: 16 May 2016;
Published: 01 June 2016.
Edited by:
Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Gaurav Sablok, University of Technology Sydney, AustraliaLi-Song Chen, Fujian Agriculture and Forestry University, China
Copyright © 2016 Verma, Verma, Pande, Mallick, Deo Tripathi, Dhankher and Chakrabarty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Debasis Chakrabarty, chakrabartyd@nbri.res.in
 Pankaj K. Verma
Pankaj K. Verma Shikha Verma
Shikha Verma Veena Pande2
Veena Pande2 Shekhar Mallick
Shekhar Mallick Rudra Deo Tripathi
Rudra Deo Tripathi Om P. Dhankher
Om P. Dhankher Debasis Chakrabarty
Debasis Chakrabarty