- 1Institute of Crop Science, Nutritional Crop Physiology, University of Hohenheim, Stuttgart, Germany
- 2Key Laboratory of Plant-Soil Interaction, Ministry of Education, Center for Resources, Environment and Food Security, College Resources and Environmental Sciences, China Agricultural University, Beijing, China
The quality of edible seeds for human and animal nutrition is crucially dependent on high zinc (Zn) and iron (Fe) seed concentrations. The micronutrient bioavailability is strongly reduced by seed phytate that forms complexes with seed cations. Superior genotypes with increased seed Zn concentrations had been identified, but low micronutrient seed levels often prevail when the plants are grown in Zn-deficient soils, which are globally widespread and correlate with human Zn-deficiency. Here, seed Zn concentrations of Arabidopsis accessions grown in Zn-deficient and Zn-amended conditions were measured together with seed Fe and manganese (Mn), in a panel of 108 accessions. By applying genome-wide association, de novo candidate genes potentially involved in the seed micronutrient accumulation were identified. However, a candidate inositol 1,3,4-trisphosphate 5/6-kinase 3 gene (ITPK3), located close to a significant nucleotide polymorphism associated with relative Zn seed concentrations, was dispensable for seed micronutrients accumulation in Col-0. Loss of this gene in itpk3-1 did neither affect phytate seed levels, nor seed Zn, Fe, and Mn. It is concluded that large natural variance of micronutrient seed levels is identified in the population and several accessions maintain high seed Zn despite growth in Zn-deficient conditions.
Introduction
Zinc is an essential micronutrient for plant growth, and at the same time an important dietary source of minerals for humans (Marschner, 2011). However, plant Zn deficiency is a widespread problem due to the limited soil availability of Zn, often as a result of high CaCO3 and high pH, in many agricultural areas (Alloway, 2004; Cakmak, 2008). In addition, evidence accumulated that elevated CO2 decreased Zn concentration in plants, including grains or seeds that are consumed as food by animals and humans (McGrath and Lobell, 2013; Loladze, 2014; Myers et al., 2014). Therefore, Zn deficiency is not only a current prevalent phenomenon in plants and humans, especially those that rely mostly on plant-based diets, but its relevance will increase in the near future (Alloway, 2004; Wessells and Brown, 2012). Successful routes to Zn biofortification of edible grains involve agronomic and genetic means, but the overall potential to genetically increase grain Zn appears limited (in contrast to potentially high leaf Zn in hyperaccumulators), especially when soil Zn is low (White and Broadley, 2011).
The accumulation of minerals in edible seeds depends on a series of complex processes: the ion availability in soils, uptake efficiency by roots, translocation to the shoots, uptake and storage in the seeds (Grusak and DellaPenna, 1999; Olsen and Palmgren, 2014). The multitude of the involved processes, which are also mineral-specific, make it very hard to identify the underlying genetics that is responsible for the final mineral accumulation in seeds and use it for improving the mineral composition. While the transport proteins involved in loading or efflux of a given nutrient, such as Zn and Fe, are mostly quite substrate-specific, the uptake mechanisms and storage pathways are partially overlapping. For example, Zn and Fe may be both chelated internally and sequestered differently by the same organic molecule, such as the principle iron chelator nicotianamine (Haydon et al., 2012). Furthermore, Zn and Fe bioavailability in seeds is strongly reduced by phytate, a hexa-phosphorylated inositol that serves as a storage form of phosphorus in seeds. The strongly negatively charged phytate chelates cations, including metal micronutrients (Raboy, 2009). However, detailed analysis of barley grains revealed that Zn and Fe clearly have different speciation and Fe was co-fractionated with phytate, while most Zn was co-eluted with a sulfur containing fraction, meaning that Zn binds to peptides, rather than phytate (Persson et al., 2009). In the soil, solubilization of micronutrients in the rhizosphere involves the secretion of organic acids, with common beneficial effects on the availability of many micronutrients in the soil, such as Fe, Mn, and Zn.
In Arabidopsis, several studies using natural variation and/or recombinant inbred lines (RILs) concluded that the correlation between ion concentrations in different tissues was highly dependent on the growth conditions and target organs (Ghandilyan et al., 2009; Buescher et al., 2010; Baxter et al., 2012). Several quantitative trait locus (QTL) analyses already suggested candidate regions controlling the seed mineral nutrients and phytate content (Vreugdenhil et al., 2004; Waters and Grusak, 2008; Ghandilyan et al., 2009). Although many loci were identified as candidate regions (and candidate genes were identified) in the linkage mapping, biological proof via mutant lines are often lacking. Surprisingly, in the mentioned studies only a very limited overlap of chromosomal regions was identified, which is likely due to the use of different RIL populations, but also different growing conditions (Vreugdenhil et al., 2004; Waters and Grusak, 2008; Ghandilyan et al., 2009). Interestingly, no overlap between leaf and seed Zn concentrations was found and very little overlap in QTLs for ion concentrations when plants were grown in soil or hydroponics. This was obvious even in the same RIL populations under controlled greenhouse conditions, potentially indicating that rhizosphere-related processes and internal plant remobilization have a significant impact on Zn storage (Ghandilyan et al., 2009, 2012). The low resolution in QTL mapping further imposes difficulties to identify causal individual genes that explain a significant part of the trait variance within the population.
Genome-wide association is a powerful and efficient tool to identify the underlying genetics of a trait based on the variance in a panel of geographical-diverse genotypes of a species. In contrast to RILs, which are based on the genetics of two parents, these panels may identify larger natural variation associated with the trait. Natural genetic variation and GWA have been extensively used in Arabidopsis to dissect the genetics of various traits, including flowering, nutrition, and yield (Atwell et al., 2010; Li et al., 2010; Chao et al., 2012; Sanchez-Bermejo et al., 2015). In the recent few years, with GWA, many studies successfully identified crucial genes responsible for leaf nutrient contents, including cadmium, arsenic, sulfur, and selenium (Chao et al., 2012, 2014a,b). Though the size population of GWA must be sufficiently large, a panel with only 96 accessions was evidently sufficient to identify causal genes for the variance in several traits in practice (Atwell et al., 2010).
Since low seed zinc is primarily caused by plant growth in Zn-deficient soil, we were interested in how seed Zn concentrations of Arabidopsis accessions differ in their response to Zn deficiency. We further quantified seed Fe and Mn in addition to Zn, of 108 accessions, grown in a Zn-supplemented and Zn-scarce soil–sand mixture. Moreover, we were intrigued by the genetics underlying the natural variation in seed Zn, Fe, and Mn contents. By applying GWA, we identified candidate genes, potentially involved in the seed micronutrient accumulation. A candidate inositol 1,3,4-trisphosphate 5/6-kinase gene (ITPK3), located close to a significant nucleotide polymorphism associated with relative Zn seed concentrations, was however, dispensable for seed phytate, Zn, Fe, and Mn accumulation in the accession Col-0.
Materials and Methods
Plant Material, Soil–Sand Preparation, and Growth Conditions
The 108 Arabidopsis thaliana accessions used in this study are listed in the Supplementary Table S1. Seeds for all accessions were obtained from Dr. Karl Schmid (Stuttgart, Germany). All accessions and mutants have been previously described (Stetter et al., 2015). The At4g08170 mutant (SALK_120653C, NASC code N653925) was bought from the European Arabidopsis Stock Centre (Nottingham, UK).
Soil–sand mixtures of a Zn-scarce soil from a C-horizon of a loess soil (0.7 mg kg-1 Zn, Supplementary Table S6) was mixed at 1:1 ratio with quartz sand (0.6–1.2 mm diameter), which was washed with HCl (rinsed with tap water, pH < 1 adjusted with HCl, incubated for over 1 day, rinsed with deionized water to pH > 5) to wash out trace nutrients, biological contaminations and dust. The soil–sand mix was fertilized with 1.1 g kg-1 NH4NO3, 0.9 g kg-1 K2SO4, 2.1 g kg-1 MgSO4 and 1.6 g kg-1 Ca(H2PO4)2. 200 g of soil–sand per plant was placed in the pot before watering with 7–8 ml micronutrients, according to a modified Hoagland’s solution (1 mM NH4NO3, 1 mM KH2PO4, 0.5 mM MgSO4, 1 mM CaCl2, 0.1 mM Na2EDTA-Fe, 2 μM ZnSO4, 9 μM MnSO4, 0.32 μM CuSO4, 46 μM H3BO3, 0.016 μM Na2MoO4). In addition, 3 mg kg-1 Zn was added into soil for the control treatment (+Zn). As a control potting soil, Einheitserde EET, Einheitserde- und Humuswerke, Sinntal-Jossa, Germany, was used.
Seeds were stratified at 4°C for 3 days to promote germination. All plants were cultivated in controlled greenhouse (GWA) or growth chambers (mutant experiment). The growth conditions were set as: long days (16 h light/8 h dark), 23°C light/20°C dark, 120–140 μmol m-2 s-1 and 65% humidity.
Identification of T-DNA Insertions
The At4g08170 mutant (SALK_120653C, NASC code N653925) carried a T-DNA insertion in the intron in wild-type line Col-0. Homozygous mutant was confirmed by two PCRs on genomic DNA, with using T-DNA annealing primer LBb1.3 (ATTTTGCCGATTTCGGAAC), and gene-specific primers (LP: ACCAATTGAACAAACACAGGC, RP: AGATGGTGGTAAATTGCACAAG). Briefly, DNA of wild-type and the T-DNA mutant seedlings was extracted with the DNeasy Plant Mini Kit (Qiagen, Germany). One PCR with LP and RP primes produced a 1200 bp product for wild-type, but no product for the homozygous mutant. The other PCR with LBb1.3 (SALK) and RP primers produced a 750 bp product for the homozygous mutant, but not for wild-type. In addition, qRT-PCR was conducted on extracted and reverse transcribed mRNA to check for the transcripts in wild-type and mutant seeds (qRT-PCR protocol described below).
Measurement of Minerals and Phytate Content
Six randomized replicates per accession were analyzed and three replicates for the mutant experiment. Seeds were harvested after 3–4 months growth and then stored in closed microfuge tubes. Over 10 μg seeds were digested with 2.5 ml 69% HNO3 and 2 ml 30% HCl for 1 h. The samples were placed in a microwave at 170°C for 25 min, followed by 200°C for 40 min. The extracts were measured with ICP-MS (Inductively Coupled Plasma – Mass Spectrometry) to determine Zn, Fe, and Mn concentration. The Zn deficiency response was calculated as: [(+Zn) – (-Zn)]/+Zn * 100. The relative Zn was calculated as: Zn/(Zn + Fe + Mn) * 100.
The Phytic Acid (Total Phosphorus) Assay Kit (Megazyme, Ireland) was modified to measure seed total phosphorus (sum of free orthophosphate and P in phytic acid) and phytate content. In brief, approximate 0.1 g seeds were digested in 2 ml 0.66M HCl in microfuge tubes and shaked overnight, before centrifugation at 13,000 rpm for 10 min. One milliliter of the supernatant was transferred to a fresh 2 ml microfuge tube and neutralized by the addition of 1 ml 0.75 M NaOH. The enzymatic dephosphorylation reaction was conducted as recommended in the protocol and measured with absorbance at 655 nm.
Genome-Wide Association (GWA)
Genome-wide association was conducted using the online web application GWAPP (Seren et al., 2012), which is a user-friendly and powerful tool to carry out the association mapping. For the Zn deficiency response, we transformed the phenotypes using a logarithmic transformation, which yields extreme value to be less extreme. The AMM (Segura et al., 2012) approach was used to correct for the population structure for all association mapping.
Gene Annotation Analysis
The gene annotation analysis was applied using MapMan (Usadel et al., 2009). The gene list was generated from GWA of different phenotypes (described above). Briefly, 12 phenotypes were used for association mapping, SNPs with –log10(p-value) ≥ 5, filtering for minor allele frequency (MAF) ≥ 0.1. All genes located within ±20 kb of the SNPs were selected as the gene list. TAIR 10 was used as the reference database (The Arabidopsis Information Resource1).
Gene Ontology Enrichment Analysis
The gene ontology enrichment analysis was applied using the online web application agriGO (Du et al., 2010), (Supplementary Table S3). Singular Enrichment Analysis (SEA) was conducted for cellular components, molecular function and biological process. TAIR 10 was used as the reference database (The Arabidopsis Information Resource1). False discovery rate (FDR) < 5% was chosen, yellow color indicates significance.
Quantitative RT-PCR Analysis
Harvested seeds were stored at room temperature in closed microfuge tubes. Total RNA of seeds was extracted using the innuPREP Plant RNA Kit (Analytik Jena, Germany) after homogenized with 30 μg seeds (Retsch, Germany). Around 1 μg total RNA was used to synthesis a cDNA library using the QuantiTect Reverse Transcription Kit (Qiagen, Germany). Primers were ordered from Invitrogen (USA). The primers used were AT5G16760 (ITPK1, 5′-TAGGGATGCCAAAGATGCTAATA-3′ and 5′-GTCCCAGAAGAACTCAGTCAACA-3′); AT4G33770 (ITPK2, 5′-CAAGGTATTTGTGGTGGGTGAT-3′ and 5′-GAGGGTCCAAGTCTGCGTTAT-3′); AT4G08170 (ITPK3, 5′-ATCGTCGCCGTGTTCGTTAGT-3′ and 5′-AAACGGACCCTGCTCTGAAAGT-3′); AT2G43980 (ITPK4, 5′-GCCATCTCGGGTAGAGGACTTT-3′ and 5′-AGCAGTTCAGTTCAATGGACAAGA-3′). The primers of ITPK1, ITPK2, and ITPK4 were used previously (Tang et al., 2013). For the PCR procedure, a 15 μl reaction was carried out, containing 6 μl 20X diluted cDNA, 7.5 μl SYBR Green Supermix (KAPA Biosystems, USA), 0.3 μl forward primers, 0.3 μl reverse primers, and 0.9 μl RNase-free H2O. The reaction was conducted in 384-well plates in RT-PCR systems (Bio-Rad, USA). The standard protocol was set as: 3 min at 95°C, followed by 44 cycles of 3 s at 95°C and 15 s at 60°C, and then 5 s at 65°C for the melt curve. Two reference genes SAND (FW: CAGACAAGGCGATGGCGATA, RV: GCTTTCTCTCAAGGGTTTCTGGGT) and PDF2 (FW: TAACGTGGCCAAAATGATGC, RV: GTTCTCCACAACˆCGCTTGGT) were used. Reactions were performed in three technical replicates and 3–4 biological replicates. Relative transcript levels were calculated with the 2-ΔΔCt method by the Bio-Rad software (Livak and Schmittgen, 2001). All kits and homogenizer were used according to the manufacturer’s instructions.
Statistical Analysis
Data analysis, graphs and statistics were done by using Microsoft Excel and R2. The statistics of mineral and phytate concentrations, and relative transcript levels was performed by one-way ANOVA. Broad-sense heritability was calculated as genotypic variance divided by total variance (Visscher et al., 2008). The total variance was partitioned into genetic variance and residuals.
Results
Natural Variation of Seed Mineral Nutrients in Response to Zinc Deficiency
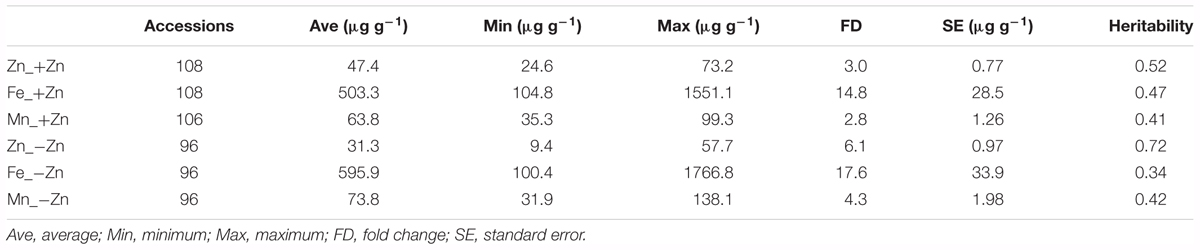
Of a larger panel of Arabidopsis thaliana accessions, 108 accessions produced sufficient seeds under +Zn and 96 under –Zn to reliably quantify the seed mineral nutrients (Table 1). Seed Zn, Fe, and Mn concentrations were measured from plants grown in a Zn-sufficient (+Zn) and a Zn-deficient (-Zn) soil. In general, the seed Fe concentration was 10- to 20-fold higher than that of Zn, and around eightfold higher than Mn (Table 1). The variation in Zn and Mn concentration was also smaller than those for Fe, concerning either the fold change (FC) or standard error (SE). The SE of the Fe concentration was large, 28.5 μg g-1 and 33.9 μg g-1 in +Zn and -Zn, respectively, much larger than that of Zn and Mn. The heritability ranged from 0.34 to 0.72 for different elements. As expected, -Zn decreased the Zn concentration from 47.4 μg g-1 to 31.3 μg g-1, but generally increased the Fe and Mn concentrations.
A distinct distribution pattern was observed of seed Zn concentration in +Zn and -Zn. The majority of genotypes had around 40 μg g-1 and 35 μg g-1 for +Zn and -Zn, respectively (Figure 1A). However, for Fe and Mn, the distribution patterns across +Zn and -Zn were overlapping (Figures 1B,C). In addition, the comparison between +Zn and -Zn conditions confirmed that all Arabidopsis accessions had lower Zn concentrations in -Zn, but Fe and Mn concentrations were more variable between -Zn and +Zn conditions (Figures 1D–F).
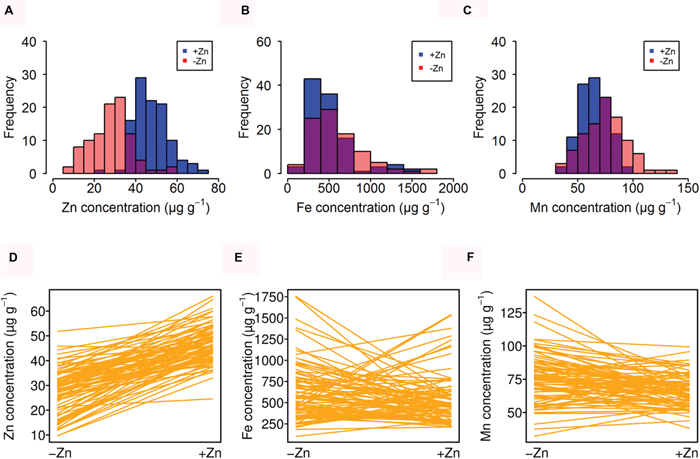
FIGURE 1. Natural variation of seed mineral nutrients in different Zn soils. (A–C) Distribution of average Zn, Fe, and Mn concentration for all accessions grown in -Zn (red) and +Zn (blue) soils. The overlap is shown in different color. (D–F) Reaction norms of Zn, Fe, and Mn concentration for all accessions. Every line indicates one accession.
To assess how seed Zn, Fe, and Mn were reduced by Zn deficiency, we quantified the Zn response, Fe response, and Mn response. This response was calculated as a relative Zn, Fe or Mn concentration according to [(+Zn) – (-Zn)]/+Zn * 100. Overall, the Fe response and the Mn response were positively correlated, and most accessions increased Fe and Mn seed storage when encountering Zn deficiency (Figures 1D–F and 2).
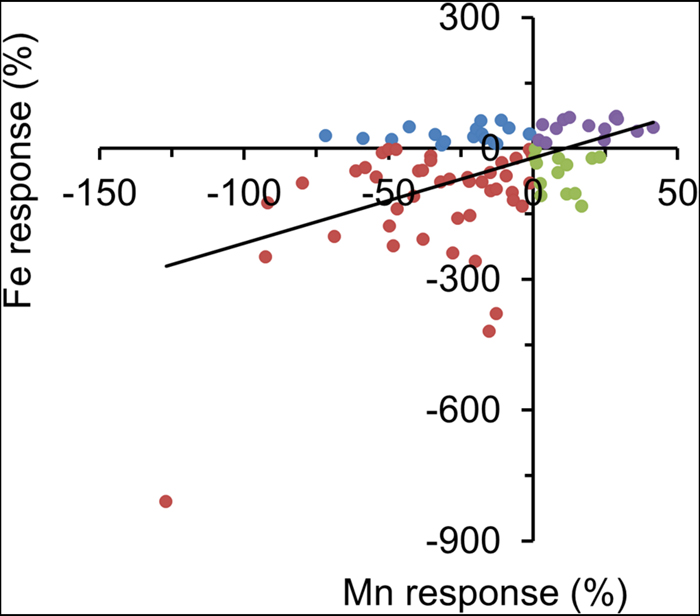
FIGURE 2. Fe and Mn concentration responses to Zn deficiency for all accessions. Different color dots indicate different -Zn response behavior in accessions. For example, red dots indicate these accessions increased seed Fe and Mn concentration in -Zn soils.
Correlations between Mineral Nutrients
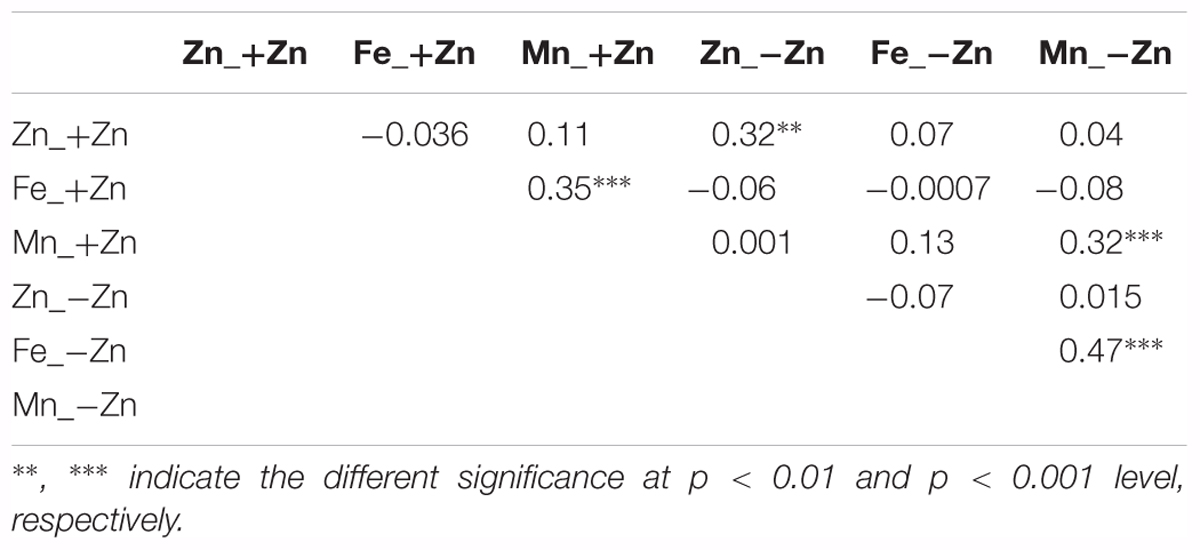
Seed Zn concentration was neither correlated with Fe nor Mn. In +Zn, plant growth and seed micronutrients were apparently not limited by soil Zn availability (Table 2). In contrast, positive correlations were found for Fe and Mn in both +Zn and -Zn. Interestingly, the Zn concentration in +Zn was positively correlated with that of -Zn, as well as with Mn, but not with Fe. This indicated that genetic factors controlled seed Zn and Mn accumulation more tightly, while the Fe accumulation was more sensitive to the environment. The Zn deficiency response of seed Zn, Fe and Mn all extremely negatively correlated with the mineral nutrients concentration in -Zn, but much less correlated with that of +Zn (Figure 3).
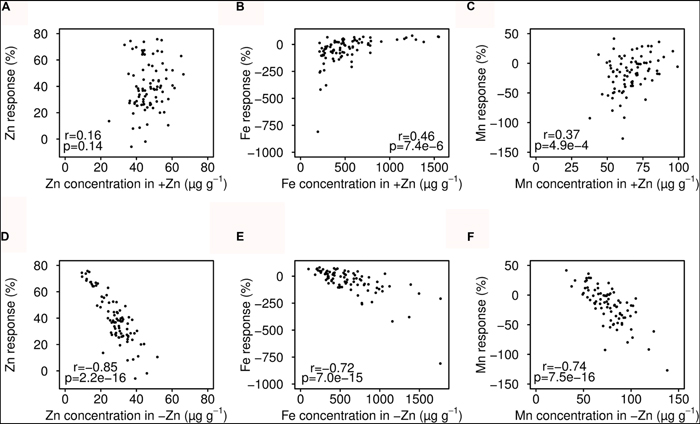
FIGURE 3. Relationship between seed mineral nutrient concentrations and Zn deficiency response. The Zn deficiency response was calculated as: [(+Zn) – (-Zn)]/+Zn * 100. (A–C) Correlation of mineral nutrients in +Zn and response. (D–F) Correlation of mineral nutrients in -Zn and response.
Natural Variation of Relative Seed Zinc Concentration
In order to quantify whether synergetic or antagonistic uptake between Zn and other mineral nutrients occurred, especially for other heavy metal-like micronutrients Fe and Mn, we additionally calculated the relative seed Zn concentration, which has no direct mechanistic meaning, but takes into account relative speciation differences. It was calculated as Zn/(Zn + Fe + Mn) * 100, which is also a proxy of the relative differences between Zn and Fe, because of the much higher concentrations of Fe. The distribution and norm reaction pattern of relative Zn was different from that of the seed Zn concentration (Figures 1A,C and 4A,B, Supplementary Figure S3). The relative Zn in +Zn did not correlate with that in -Zn, but in accordance with the Zn response, the relative Zn response was also highly determined by the Zn-deficient condition (Figure 4C–E).
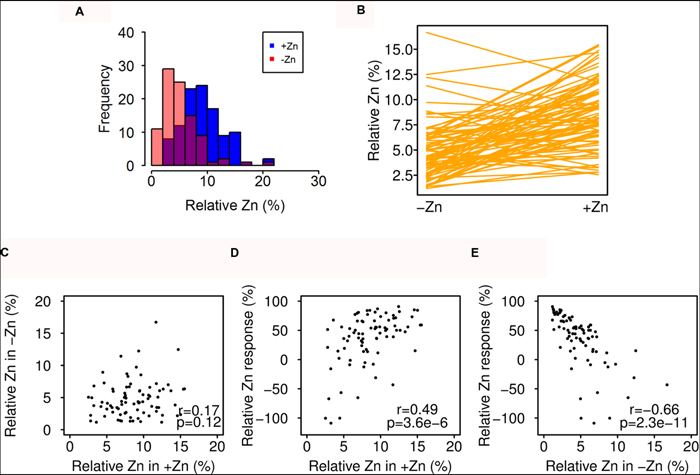
FIGURE 4. Natural variation in relative Zn concentration response to Zn deficiency. (A) Distribution of relative Zn in -Zn (red) and +Zn (blue) soils. The overlap is shown in different color. (B) Reaction norms of relative Zn for all accessions. (C–E) Correlation of relative Zn in +Zn, -Zn, and Zn-deficiency response. The relative Zn was calculated as: Zn/(Zn + Fe + Mn) * 100.
Genome-Wide Association Mapping
To uncover the underlying genetics of seed nutrients accumulation and the response to -Zn, we carried out GWA using GWAPP, a user-friendly web application (Seren et al., 2012). GWAPP has already included about 206,000 SNPs and 1386 individuals for analysis. As several accessions in this study were not included in GWAPP, the GWAS was performed on 96, 108 or the overlapping 82 accessions (Supplementary Table S2). This population might be large enough to identify causal genetic factors involved in the phenotypic diversity of this population, as previously reported (Atwell et al., 2010; Stetter et al., 2015). The AMM model was used to eliminate the noise of population structure, mainly due to isolation by distance (Platt et al., 2010; Segura et al., 2012; Seren et al., 2012). A stringent P-value cutoff with 5% FDR was instituted for quantifying the significance. Only SNPs with minor allele frequency (MAF) ≥ 0.1 were presented in the Manhattan plot, to reduce potential false positive SNPs.
Twelve phenotypes, representing seed micronutrient concentrations and relative values, were used for GWA, however, only a single significant SNP in the GWA of relative -Zn was found, 4G_5149241 (Supplementary Table S2; Supplementary Figure S4; Figure 5A). This SNP consists of G/T nucleotides and the accessions containing the allele G had higher relative Zn in -Zn, compared to those accessions containing the allele T (Figures 5B,C). Interestingly, most accessions (12 of 14) that contain allele “G” locate to coastal regions (Figure 5D). This SNP locates 14 kb downstream of AT4G08170 (ITPK3), a gene encoding a inositol 1,3,4-trisphosphate 5/6-kinase 3 protein. The amino acid sequence of ITPK3 is not changed by the SNP. ITPK3 belongs to a small gene family with four members that are involved in phosphorylation of inositol 1,3,4-trisphosphate, in the pathway to generate the end-product phytate (inositol hexakisphosphate), a chelator of micronutrients, preferably found in seeds (Raboy, 2009).
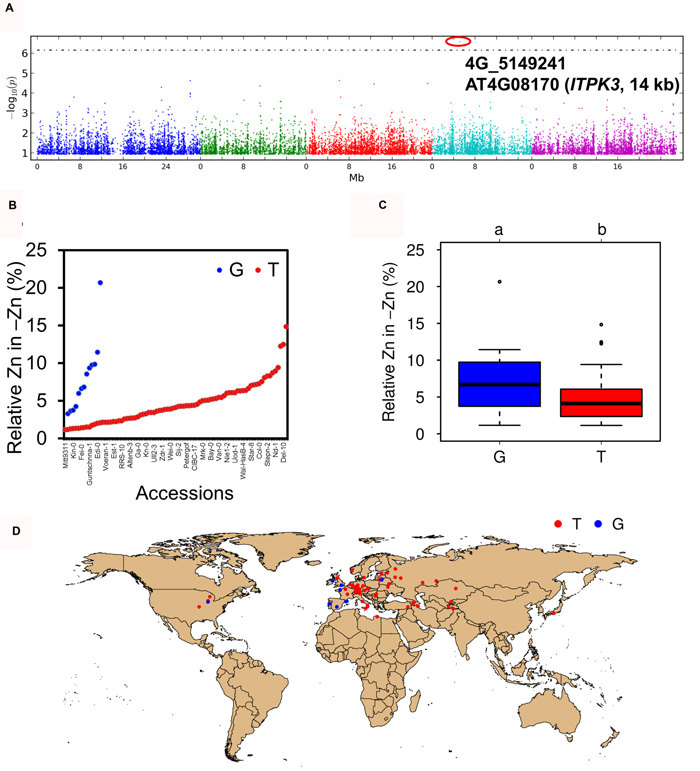
FIGURE 5. Genome-wide association of relative Zn concentration. (A) Manhattan plot of relative Zn concentration for -Zn. The value of -Zn response was used after logarithmic transformation. SNPs with minor allele count (MAC) ≥ 15 were presented. The 5% FDR threshold was denoted by a dashed line. (B,C) The diversity and relative -Zn for identified SNP (4G_5149241), which is associated with AT4G08170 (ITPK3). “a” and “b” above the figures indicate the significant difference at p < 0.05 level. (D) Geographical distribution of “T” (red) and “G” (blue) accessions. Every dot indicates one accession.
Validation of the ITPK
Because of the interaction of seed micronutrients and phytate, we considered the ITPK3, identified from the GWA, as the best candidate gene and isolated a homozygous loss-of-function allele, to investigate its impact on seed micronutrients. A T-DNA mutant line in the Col-0 background (itpk3-1) was isolated from the SALK collection, so the itpk3-1 is in a “T” background genotype at 4G_5149241 (as Col-0 has a “T” in the significant SNP). PCR analysis verified the T-DNA insertion in the largest intron within gene (Figure 6A). RT-PCR confirmed that the ITPK3 expression was lost in the itpk3-1 mutant seeds, both in +Zn and -Zn (Figure 6B). The ITPK gene family includes four members (Tang et al., 2013), of which only ITPK1 is also significantly expressed in seeds. Intriguingly, the ITPK1 transcript level was also reduced in the itpk3-1 mutant, especially in -Zn (Figure 6B). As the inositol 1,3,4-trisphosphate kinase carries out an intermediate step in phytate synthesis, we analyzed the free orthophosphate and phytate concentrations as well. However, neither total P (the sum of free orthophosphate and phytate), nor phytate, were decreased in both +Zn and -Zn in the itpk3-1 mutant (Figure 6C). Potentially as a result of unchanged phytate, seed Zn, Fe, Mn, and RZn concentrations were all not significantly different in the itpk3-1 mutant, despite a minor trend for lower Fe in the mutant (Figure 6D).
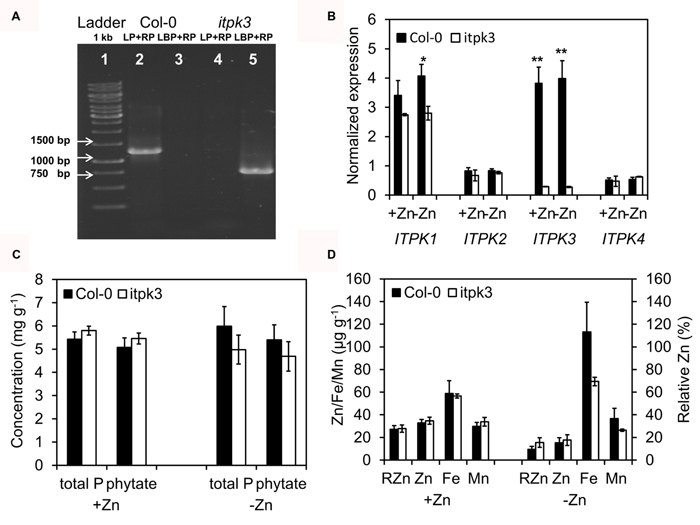
FIGURE 6. Loss of IPTK3 does not affect seed phytate and micronutrients. (A) Homozygous confirmation of itpk3 mutant by PCR. The band size of second and fifth column are around 1200 and 750 bp, respectively. (B) RT-PCR analysis of ITPK1-4 transcripts level in Col-0 and itpk3. * and ** indicate the significant difference at p < 0.05 and p < 0.01 level. (C) Total P and phytate concentration in Col-0 and itpk3-1. (D) Zn, Fe, Mn, RZn in Col-0 and itpk3.
Identification of De Novo Candidate Genes and Gene Ontology Enrichment Analysis
As the ITPK3 was apparently not responsible for natural variance in the seed Zn, Fe, or Mn accumulation, we next tried to identify de novo candidate genes and processes by over-representation analysis of trait-associated genes. The genes were annotated with MapMan (Usadel et al., 2009) and broad distributed SNPs that were highly associated with the phenotypes were selected, by the criterion of MAF ≥ 0.1 and –log (p_value) ≥ 5. With this, 69 SNPs were identified and 487 annotated genes (471 unique genes) in their vicinity (±20 kb around the SNP; Supplementary Tables S2 and S3). Genes related to transport, cell wall, lipid metabolism, or inositol phosphate synthesis were over-represented in this analysis (Supplementary Table S4). Among other genes, YSL6 (AT3G27020, YELLOW STRIPE LIKE 6), a gene encoding a metal-chelate transporter, was identified in the -Zn condition (Supplementary Table S3).
Whether the identified chromosomal regions were specifically enriched in +Zn, -Zn, and the Zn deficiency response conditions, was further analyzed (Supplementary Table S3). There was no specific enrichment found for +Zn and -Zn, however, genes were significantly enriched in carbohydrate binding for the Zn deficiency response (Supplementary Figure S1).
Discussion
Hidden hunger represents a major thread to humanity and describes the phenomenon of malnutrition and micronutrient deficiencies (e.g., of Zn and Fe) even in the presence of sufficient calories in the diet. Billions of humans are at least at some time periods affected by insufficient nutrients in their grains/seeds-based diet, as a result of the limited soil mineral availability (Cakmak, 2008). A further decrease in seed micronutrients is observed with increased CO2, aggravating the global problem further in the future, but also suggesting that genetic improvements for individual micronutrients may be possible (Loladze, 2014; Myers et al., 2014). Zn deficiency, both in plants and humans, is a widespread problem resulting from low fertility soils in many regions, including China, India, and Tukey (Alloway, 2004; Cakmak, 2008). Therefore, using a diverse Arabidopsis population, a soil experiment was conducted to simulate the natural situation of Zn deficiency. Plants mobilize Zn from soils with high pH and high CaCO3 by rhizosphere processes that are probably not relevant and neglected, when plants are growing in optimal soils or hydroponics solutions.
Substantial natural variation of the seed Zn, Fe, and Mn concentration was encountered in different Arabidopsis accessions, since several biological processes are involved to affect the nutrients accumulation in seeds (Grusak and DellaPenna, 1999; Olsen and Palmgren, 2014). Minerals have to be mobilized in the soils, are transported to the roots, are translocated to the shoots and are redistributed into different tissues, including seeds. While Zn and Mn concentrations were similar as in recent studies (Table 1; Figure 1), the Fe concentration, surprisingly, was much higher and not in agreement with earlier studies in different growth conditions or different tissues (Ghandilyan et al., 2009; Buescher et al., 2010; Baxter et al., 2012). We therefore re-analyzed seed Fe concentrations in 10 randomly chosen accessions (including Col-0) grown in controlled conditions in a growth chamber and well supplied potting soil. We then compared their Fe concentrations using the same extraction and analytics procedures. Seed Fe was much lower when grown in high nutrient potting soil and highly similar to previous studies. The Fe concentration ranged from 140 to 1083 μg g-1 in this study, but only from 133 to 300 μg g-1 when grown in potting soil (Supplementary Figure S2), with the accession Edi-0 having extremely high seed Fe concentrations, both when grown in the sand–soil mix (1083 μg g-1) or potting soil (400 μg g-1). The abnormal high Fe may thus be caused by the soil or other environmental factors in the greenhouse (light humidity, temperature). This indicated that Fe was highly sensitive to the chosen Zn-deficient soil and, as expected, that the seed uptake of Fe probably strongly depends on the soil conditions. Indeed, a lack of correlation between +Zn and -Zn with the seed Fe concentration also implied that Fe was subject to adaptation to the environment.
The growth of the plants under Zn-deficient soil conditions decreased the seed Zn concentration for most accessions, validating that the -Zn condition imposed a deficiency condition to the plants. It was expected that -Zn increases the Fe and Mn accumulation, as such correlations are often found, as the IRT3 and other Zn transporters are overexpressed due to -Zn (Assuncao et al., 2010). However, this was observed only in a subset of the accessions, which increased the Fe and Mn accumulation in seeds under -Zn (Figures 1 and 2). By contrast, Figures 1E,F also show that many accessions even decreased the Fe and Mn under -Zn. As a consequence, many accessions had positive values in the Fe or Mn response (Figure 2). Furthermore, significant correlations were found only between certain conditions and Zn, Fe, and Mn (Table 2). These results demonstrated that seed nutrients accumulation is more complex than the per se the root uptake. Ghandilyan et al. (2009) and Baxter et al. (2012) found that the concentrations of Arabidopsis leaf minerals are a poor proxy for seed minerals. Interestingly, few accessions maintained relatively high Zn concentrations in -Zn soils. For example, the Zn concentration of Ts-1 was 37.16 μg g-1 and 39.33 μg g-1 in +Zn and in -Zn, respectively. This implied that there are genetic backgrounds in which high Zn is maintained in seeds even under -Zn growth. How these genotypes manage higher seed Zn is an interesting target for future research, to genetically improve the seed Zn content in the adverse Zn condition.
Certain correlations between different mineral concentrations are reliably encountered, irrespective of the growth conditions and tissues (Ghandilyan et al., 2009; Baxter et al., 2012). In this study, we found positive correlation between Fe and Mn in +Zn and -Zn, especially for the response (Table 2; Figure 2). This suggests that Fe and Mn share common seed uptake mechanisms to face the adverse soil -Zn condition. However, the positive correlation between phenotypes doesn’t mean that the same SNPs/QTLs are found in GWA or QTL analysis (Vreugdenhil et al., 2004; Ghandilyan et al., 2009). As frequently found with polygenic phenotypes that are controlled by many small-effect genetic factors, only few SNPs/genes were identified in common, although high correlation was found between the phenotypes (Supplementary Table S3).
Small population size and polygenic phenotypes clearly limit the power of GWA in this study to identify significant SNPs (Ogura and Busch, 2015). As a result, only one significant SNP was identified. However, non-significant SNPs are also often promising candidates, as reported before (Filiault and Maloof, 2012; van Rooijen et al., 2015). Therefore, we arbitrary selected the top-associated and broadly distributed SNPs (-logP ≥ 5 and MAF ≥ 0.1) as potential candidates. Indeed, some valuable genes were identified within 20 kb on either side of the SNPs. Then, 20 kb was proven to be an effective distance to identify promising genes (Chao et al., 2012; Filiault and Maloof, 2012). For instance, the metal-chelate transporter gene YSL6 (YELLOW STRIPE LIKE 6) was identified as a candidate in the -Zn condition (Supplementary Table S3). YSL6 was proven to be involved in iron release from chloroplasts in Arabidopsis (Conte et al., 2013; Divol et al., 2013). Notably, YSL2, YSL3, YSL7, YSL8 were all identified in candidate regions responsible for the seed accumulation of Zn, Fe, or Cu in previous QTL studies in Arabidopsis (Waters and Grusak, 2008).
Another very interesting candidate gene was ITPK3 (AT4G08170), located at 14 kb from the significant SNP 4G_5149241. It was identified for Relative Zn, which may be influenced by Zn, Mn, and Fe specification, i.e., different binding or compartmentation. ITPK family proteins are enzymes that further phosphorylate tri-phospho inositols, forming intermediate substrates for IP6 (phytic acid) production. Phytate, the salt of phytic acid, chelates cations in seeds, including Zn and Fe, and impairs their bioavailability (Raboy, 2003, 2009). Phytate, Zn and Fe are typically enriched and co-localized in the aleuron laver and the embryo, which had been taken as indication that they directly interact. However, more detailed analysis in barley grains indicated that the majority of phytate co-fractionates with Fe, but not with Zn, showing that Zn is not directly bound to phytate (Persson et al., 2009). Hence, ITPK3 was considered as a strong candidate gene that might be involved in mineral accumulation, especially of iron, and their relative values in seeds. However, the isolated null-mutant itpk3-1 had identical total phosphorus and phytate concentrations as the wild-type, Col-0, even under Zn-deficiency (Figure 6). This is in agreement with a previous study (Tang et al., 2013). Furthermore, Zn, Fe, Mn, and the RZn were also maintained in the itpk3-1 seeds, despite a minor tendency for reduced iron levels, excluding this gene as a major determinant of seed micronutrients. Thus, the identified significant SNP 4G_5149241 may just represent a false positive signal or neighboring genes may be causal. For example, this SNP locates in the third intron of uncharacterized AT4G08150 (KNAT1, KNOTTED-LIKE FROM ARABIDOPSIS THALIANA) and true knock-out alleles for all nearby genes and analysis may be required to establish whether this SNP is just a false positive (Supplementary Table S5). The mixed model used in GWA has been proven to be an appropriate method to at least partially overcome the population structure in Arabidopsis, reducing the FDR and yielding true associated SNPs (Segura et al., 2012; Seren et al., 2012). However, significantly associated SNP may still just represent “noise,” which may be overcome by redoing the linkage mapping with the candidate region (Chao et al., 2014a). Furthermore, it is still possible that ITPK3 was indeed the crucial gene responsible for variation in the seed micronutrients, but that its function was dispensable in the Col-0 background. This means that ITPK3 might have a different role in other accessions, probably because of the expression of a redundant, other ITPK gene. Alternatively, as Col-0 is an accession with “T” allele, ITPK3 might only be involved in the seed nutrient accumulation in “G” allele accessions. This study included fourteen “G” allele accessions, and intriguingly, 12 accessions were collected close to the coast (Figure 5D). It remains unclear whether this observation is meaningful, but a sodium transporter AtHKT1;1 allele was associated with coastal and saline soils in a previous study and AtHKT1;1 is crucial for salt tolerance (Baxter et al., 2010).
The gene annotation and enrichment analysis further provided some potentially valuable information on candidates. Forty-two unique genes involved in transport, cell wall, lipid metabolism, or inositol phosphate synthesis were identified (Supplementary Table S4). Furthermore, the enrichment analysis performed with the genes identified in candidate regions in +Zn, -Zn, and the Zn response (Supplementary Table S4) encountered significant enrichment in genes from the Zn response in the GO term 0030246 (carbohydrate binding). Whether this process, rather than individual genes, are involved in maintaining high seed Zn in Zn-deficiency, may be investigated also with mutants.
Overall, this GWA for seed micronutrient accumulation with a Zn-deficient soil identified substantial variation in seed micronutrients, but the gene ITPK3 was apparently not causal for the differences in the dominant accessions with the allele “T.” Carbohydrate binding may be a novel process putatively involved in the seed micronutrient accumulation and crucial for the response to Zn deficiency. Whether ITPK3 influences seed nutrients accumulation in “G” allele accessions and the association of this SNP with coastal localization in these accessions could be investigated in the future.
Author Contributions
XC and UL conceived and designed the experiments. XC performed the experiments. XC and UL analyzed the data. XC, LY, and UL wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Karl Schmidt (Stuttgart, Germany) and the Arabidopsis stock center for seeds, and Dr. Huaiyu Yang for initial lab work. We also thank the China Scholarship Council for support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01070
FIGURE S1 | Gene ontology enrichment analysis of genes from the Zn-deficiency response. Ten of 220 genes in input list and 190 of 28397 genes in reference (TAIR 10). p-value is 0.000595.
FIGURE S2 | Comparison of Fe concentration in this study (+Zn) and in garden soil.
FIGURE S3 | Histograms of Zn response (A) and relative Zn response (B).
FIGURE S4 | Q–Q plot of relative -Zn produced in GWAPP.
TABLE S1 | List of Arabidopsis accessions used in this study.
TABLE S2 | List of identified SNPs.
TABLE S3 | List of annotated genes.
TABLE S4 | List of de novo candidate genes.
TABLE S5 | List of genes locate within 20 kb on either side of significant SNP 4G_5149241.
TABLE S6 | The characteristics of soil used in this study.
Abbreviations
Fe, iron; GWA, genome-wide association; Mn, manganese; RZn, relative Zn; SNP, single nucleotide polymorphism; Zn, zinc.
Footnotes
References
Assuncao, A. G., Herrero, E., Lin, Y. F., Huettel, B., Talukdar, S., Smaczniak, C., et al. (2010). Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. U.S.A. 107, 10296–10301. doi: 10.1073/pnas.1004788107
Atwell, S., Huang, Y. S., Vilhjalmsson, B. J., Willems, G., Horton, M., Li, Y., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631. doi: 10.1038/nature08800
Baxter, I., Brazelton, J. N., Yu, D., Huang, Y. S., Lahner, B., Yakubova, E., et al. (2010). A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6:e1001193. doi: 10.1371/journal.pgen.1001193
Baxter, I., Hermans, C., Lahner, B., Yakubova, E., Tikhonova, M., Verbruggen, N., et al. (2012). Biodiversity of mineral nutrient and trace element accumulation in Arabidopsis thaliana. PLoS ONE 7:e35121. doi: 10.1371/journal.pone.0035121
Buescher, E., Achberger, T., Amusan, I., Giannini, A., Ochsenfeld, C., Rus, A., et al. (2010). Natural genetic variation in selected populations of Arabidopsis thaliana is associated with ionomic differences. PLoS ONE 5:e11081. doi: 10.1371/journal.pone.0011081
Cakmak, I. (2008). Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil 302, 1–17. doi: 10.1007/s11104-007-9466-3
Chao, D. Y., Baraniecka, P., Danku, J., Koprivova, A., Lahner, B., Luo, H., et al. (2014a). Variation in sulfur and selenium accumulation is controlled by naturally occurring isoforms of the key sulfur assimilation enzyme ADENOSINE 5’-PHOSPHOSULFATE REDUCTASE2 across the Arabidopsis species range. Plant Physiol. 166, 1593–1608. doi: 10.1104/pp.114.247825
Chao, D. Y., Chen, Y., Chen, J., Shi, S., Chen, Z., Wang, C., et al. (2014b). Genome-wide association mapping identifies a new arsenate reductase enzyme critical for limiting arsenic accumulation in plants. PLoS Biol. 12:e1002009. doi: 10.1371/journal.pbio.1002009
Chao, D. Y., Silva, A., Baxter, I., Huang, Y. S., Nordborg, M., Danku, J., et al. (2012). Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet. 8:e1002923. doi: 10.1371/journal.pgen.1002923
Conte, S. S., Chu, H. H., Rodriguez, D. C., Punshon, T., Vasques, K. A., Salt, D. E., et al. (2013). Arabidopsis thaliana Yellow Stripe1-Like4 and Yellow Stripe1-Like6 localize to internal cellular membranes and are involved in metal ion homeostasis. Front. Plant Sci. 4:283. doi: 10.3389/fpls.2013.00283
Divol, F., Couch, D., Conejero, G., Roschzttardtz, H., Mari, S., and Curie, C. (2013). The Arabidopsis YELLOW STRIPE LIKE4 and 6 transporters control iron release from the chloroplast. Plant Cell 25, 1040–1055. doi: 10.1105/tpc.112.107672
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. doi: 10.1093/nar/gkq310
Filiault, D. L., and Maloof, J. N. (2012). A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genet. 8:e1002589. doi: 10.1371/journal.pgen.1002589
Ghandilyan, A., Ilk, N., Hanhart, C., Mbengue, M., Barboza, L., Schat, H., et al. (2009). A strong effect of growth medium and organ type on the identification of QTLs for phytate and mineral concentrations in three Arabidopsis thaliana RIL populations. J. Exp. Bot. 60, 1409–1425. doi: 10.1093/jxb/erp084
Ghandilyan, A., Kutman, U. B., Kutman, B. Y., Cakmak, I., and Aarts, M. G. M. (2012). Genetic analysis of the effect of zinc deficiency on Arabidopsis growth and mineral concentrations. Plant Soil 361, 227–239. doi: 10.1007/s11104-012-1334-0
Grusak, M. A., and DellaPenna, D. (1999). Improving the nutrient composition of plants to enhance human nutrition and health. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 133–161. doi: 10.1146/annurev.arplant.50.1.133
Haydon, M. J., Kawachi, M., Wirtz, M., Hillmer, S., Hell, R., and Kramer, U. (2012). Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis. Plant Cell 24, 724–737. doi: 10.1105/tpc.111.095042
Li, Y., Huang, Y., Bergelson, J., Nordborg, M., and Borevitz, J. O. (2010). Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 21199–21204. doi: 10.1073/pnas.1007431107
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loladze, I. (2014). Hidden shift of the ionome of plants exposed to elevated CO2 depletes minerals at the base of human nutrition. Elife 3:e02245. doi: 10.7554/eLife.02245
McGrath, J. M., and Lobell, D. B. (2013). Reduction of transpiration and altered nutrient allocation contribute to nutrient decline of crops grown in elevated CO(2) concentrations. Plant Cell Environ. 36, 697–705. doi: 10.1111/pce.12007
Myers, S. S., Zanobetti, A., Kloog, I., Huybers, P., Leakey, A. D., Bloom, A. J., et al. (2014). Increasing CO2 threatens human nutrition. Nature 510, 139–142. doi: 10.1038/nature13179
Ogura, T., and Busch, W. (2015). From phenotypes to causal sequences: using genome wide association studies to dissect the sequence basis for variation of plant development. Curr. Opin. Plant Biol. 23, 98–108. doi: 10.1016/j.pbi.2014.11.008
Olsen, L. I., and Palmgren, M. G. (2014). Many rivers to cross: the journey of zinc from soil to seed. Front. Plant Sci. 5:30. doi: 10.3389/fpls.2014.00030
Persson, D. P., Hansen, T. H., Laursen, K. H., Schjoerring, J. K., and Husted, S. (2009). Simultaneous iron, zinc, sulfur and phosphorus speciation analysis of barley grain tissues using SEC-ICP-MS and IP-ICP-MS. Metallomics 1, 418–426. doi: 10.1039/b905688b
Platt, A., Vilhjalmsson, B. J., and Nordborg, M. (2010). Conditions under which genome-wide association studies will be positively misleading. Genetics 186, 1045–1052. doi: 10.1534/genetics.110.121665
Raboy, V. (2003). myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64, 1033–1043. doi: 10.1016/S0031-9422(03)00446-1
Raboy, V. (2009). Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 177, 281–296. doi: 10.1016/j.plantsci.2009.06.012
Sanchez-Bermejo, E., Zhu, W., Tasset, C., Eimer, H., Sureshkumar, S., Singh, R., et al. (2015). Genetic architecture of natural variation in thermal responses of Arabidopsis. Plant Physiol. 169, 647–659. doi: 10.1104/pp.15.00942
Segura, V., Vilhjalmsson, B. J., Platt, A., Korte, A., Seren, U., Long, Q., et al. (2012). An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44, 825–830. doi: 10.1038/ng.2314
Seren, U., Vilhjalmsson, B. J., Horton, M. W., Meng, D., Forai, P., Huang, Y. S., et al. (2012). GWAPP: a web application for genome-wide association mapping in Arabidopsis. Plant Cell 24, 4793–4805. doi: 10.1105/tpc.112.108068
Stetter, M. G., Schmid, K., and Ludewig, U. (2015). Uncovering genes and ploidy involved in the high diversity in root hair density, length and response to local scarce phosphate in Arabidopsis thaliana. PLoS ONE 10:e0120604. doi: 10.1371/journal.pone.0120604
Tang, Y., Tan, S., and Xue, H. (2013). Arabidopsis inositol 1,3,4-trisphosphate 5/6 kinase 2 is required for seed coat development. Acta Biochim. Biophys. Sin. 45, 549–560. doi: 10.1093/abbs/gmt039
Usadel, B., Poree, F., Nagel, A., Lohse, M., Czedik-Eysenberg, A., and Stitt, M. (2009). A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 32, 1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x
van Rooijen, R., Aarts, M. G., and Harbinson, J. (2015). Natural genetic variation for acclimation of photosynthetic light use efficiency to growth irradiance in Arabidopsis. Plant Physiol. 167, 1412–1429. doi: 10.1104/pp.114.252239
Visscher, P. M., Hill, W. G., and Wray, N. R. (2008). Heritability in the genomics era–concepts and misconceptions. Nat. Rev. Genet. 9, 255–266. doi: 10.1038/nrg2322
Vreugdenhil, D., Aarts, M. G. M., Koornneef, M., Nelissen, H., and Ernst, W. H. O. (2004). Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant Cell Environ. 27, 828–839. doi: 10.1111/j.1365-3040.2004.01189.x
Waters, B. M., and Grusak, M. A. (2008). Quantitative trait locus mapping for seed mineral concentrations in two Arabidopsis thaliana recombinant inbred populations. New Phytol. 179, 1033–1047. doi: 10.1111/j.1469-8137.2008.02544.x
Wessells, K. R., and Brown, K. H. (2012). Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS ONE 7:e50568. doi: 10.1371/journal.pone.0050568
Keywords: micronutrient, zinc deficiency, seed, genome-wide association, natural variation
Citation: Chen X, Yuan L and Ludewig U (2016) Natural Genetic Variation of Seed Micronutrients of Arabidopsis thaliana Grown in Zinc-Deficient and Zinc-Amended Soil. Front. Plant Sci. 7:1070. doi: 10.3389/fpls.2016.01070
Received: 18 March 2016; Accepted: 07 July 2016;
Published: 26 July 2016.
Edited by:
Rattan Yadav, Aberystwyth University, UKReviewed by:
Mark G. M. Aarts, Wageningen University and Research Centre, NetherlandsPu Huang, Donald Danforth Plant Science Center, USA
Copyright © 2016 Chen, Yuan and Ludewig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uwe Ludewig, u.ludewig@uni-hohenheim.de
 Xiaochao Chen
Xiaochao Chen