- Repository of Tomato Genomics Resources, Department of Plant Sciences, University of Hyderabad, Hyderabad, India
Type II pyridoxal phosphate-dependent decarboxylase (PLP_deC) enzymes play important metabolic roles during nitrogen metabolism. Recent evolutionary profiling of these genes revealed a sharp expansion of histidine decarboxylase genes in the members of Solanaceae family. In spite of the high sequence homology shared by PLP_deC orthologs, these enzymes display remarkable differences in their substrate specificities. Currently, limited information is available on the gene repertoires and substrate specificities of PLP_deCs which renders their precise annotation challenging and offers technical challenges in the immediate identification and biochemical characterization of their full gene complements in plants. Herein, we explored their evolutionary trails in a comprehensive manner by taking advantage of high-throughput data accessibility and computational approaches. We discussed the premise that has enabled an improved reconstruction of their evolutionary lineage and evaluated the factors offering constraints in their rapid functional characterization, till date. We envisage that the synthesized information herein would act as a catalyst for the rapid exploration of their biochemical specificity and physiological roles in more plant species.
Introduction
Pyridoxal 5′-phosphate (PLP), one of the active prosthetic group of vitamin B6; is a coenzyme with unequaled catalytic versatility. This is involved in a plethora of biochemical reactions like transamination (transfer of amino groups), decarboxylation (removal of a carboxyl group at the β- or γ-carbon), deamination (removal of an amine group), interconversion of L and D-amino acids, and racemization. PLP-dependent enzymes are mainly involved in the regulation of biosynthesis of amino acids, amino acid-derived metabolites, amino sugars and other amine-containing compounds (Facchini et al., 2000; Eliot and Kirsch, 2004). It has been found that the enzymatic versatility of these enzymes is achieved by the covalent binding of PLP group to an amino group of an active lysine in their substrates (John, 1995). Depending on their protein structures, all PLP-dependent enzymes have been classified in at least five structural groups (Percudani and Peracchi, 2003; Milano et al., 2013). Among those, Type I group, the most common structure, is present in aminotransferases, decarboxylases, and enzymes that catalyze α-, β- or γ-eliminations. Type II encode the enzymes involved in β-elimination reactions. Type III is mainly alanine racemase-specific whereas type IV enzymes generally include D-alanine aminotransferase. Type V fold represents the most distinct group among five types and includes mostly glycogen and starch phosphorylases (Percudani and Peracchi, 2003).
Among the PLP-dependent enzymes, type II PLP decarboxylases (henceforth mentioned as ‘PLP_deC’) form an important group of ‘Carboxy-Lyases.’ This group is comprised of L-glutamate decarboxylase (GAD), aromatic L-amino acid decarboxylases (commonly mentioned as AADs/AAADs/AADCs in the published literature), and serine decarboxylase (SDC; group II), prokaryotic forms of ornithine, lysine and arginine decarboxylases (group III), and the eukaryotic forms of ornithine and lysine decarboxylases (group IV; Sandmeier et al., 1994). In this mini-review, we first briefly summarized the available information on their functional roles in plants. Then we highlighted the long prevailed challenges associated with the annotation of PLP_deCs. Unavailability of high quality and comprehensive sequence datasets remained one of the main reasons for their incorrect annotation in the past, therefore we explored their full complement in 52 plant species and analyzed their evolutionary lineage. Taken together, the summary presented here would help us in improving their annotation in large number of plant species; an important factor which has long impeded their rapid functional and biochemical characterization in plants.
An Overview of the Roles of PLP_deCs in Plant Development
Available expression data indicate that PLP_deCs exhibit developmental, tissue-specific, and inducible transcripts accumulation during plant development (De Luca et al., 1988; Aerts et al., 1992; Pasquali et al., 1992; Facchini et al., 1996; Maldonado-Mendoza et al., 1996; Lopez-Meyer and Nessler, 1997; Liu et al., 2012). In addition, several PLP_deCs have been characterized for their roles in plant development (De Luca et al., 1988; Facchini and De Luca, 1995; Facchini et al., 2000; Torrens-Spence et al., 2014a). For example, GADs, which catalyze the conversion of L-glutamate to γ-aminobutyric acid (GABA); a non-protein amino acid, are involved in a range of cellular processes, including pollen-tube development in Arabidopsis and Picea wilsonii (Palanivelu et al., 2003; Ling et al., 2013), vascular development in pine (Molina-Rueda et al., 2015), stem elongation (Baum et al., 1996), cytosolic pH regulation, balancing the carbon/nitrogen, defense and protection against biotic and abiotic stresses (Bouche et al., 2004). The main mechanism that contributes to GABA production involves decarboxylation of glutamate via GADs in plants (Akihiro et al., 2008; Takayama and Ezura, 2015). Of these GADs, two GADs, including SlGAD2 and SlGAD3 have been identified as the major contributors of GABA conversion in tomato fruits. It has been established that differential activities of these enzymes during fruit development is the main reason of the higher glutamate content in the ripened tomato fruits and their peculiar ‘umami’ taste (Rolin et al., 2000; Carrari and Fernie, 2006; Akihiro et al., 2008; Saito et al., 2008; Osorio et al., 2011). The role of these GADs in determining fruit quality through GABA production may be conserved as increased GAD expression has also been reported during ripening in other fruits such as Chinese berry (Myrica rubra; Feng et al., 2012). Further, Ca2+/calmodulin (CaM) has been identified as one of the main signaling mediators which are responsible for the conversion of glutamate into GABA. A C-terminal calmodulin-binding domain (CaMBD) in GADs of the majority of plants has been suggested to be both required and responsible for Ca2+/CaM-dependent activation of the oligomerized GAD complexes in plants (Zik et al., 2006; Akama and Takaiwa, 2007). However, evidence also suggests that its presence is not universally essential for such activity as several GADs, lacking a typical CaMBD, have been found to function independent of Ca2+/CaM in rice and apple (Akama et al., 2001; Fait et al., 2008; Trobacher et al., 2013).
AADs represent the second important category of PLP_deC enzymes which catalyze the decarboxylation of aromatic L-amino acids. These enzymes are mainly involved in the biosynthesis of secondary metabolites in plants (De Luca et al., 1988; Tieman et al., 2006; Lehmann and Pollmann, 2009). The best investigated enzymes in this category are Dopa decarboxylase (DDC), L-tryptophan decarboxylase (TDC), L-tyrosine decarboxylase (TYDC), and histidine decarboxylase (HDC) (Facchini et al., 2000; Torrens-Spence et al., 2014a). Whereas TDC catalyzes decarboxylation of tryptophan to tryptamine and other mono-terpenoid indole alkaloids such as serotonin (5-hydroxytryptamine), TYDC mediates conversion of L-tyrosine to tyramine (Lopez-Meyer and Nessler, 1997; Facchini et al., 2000; Asano et al., 2012). Due to their significance in the production of secondary metabolites, these genes have also been used in the genetic manipulation studies aiming at improving the contents of pharmaceutically important bio-molecules in transgenic plants and/or cell lines, which is summarized in Table 1. Similarly, HDC, which catalyzes the conversion of histidine to histamine, has been found to participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde in tomatoes (Picton et al., 1993; Tieman et al., 2006). Their recent characterization in tomato and pepper further identified four HDC ripening-preferential homologs, including HDC9, HDC10, HDC11, and HDC12; which may be involved in the similar biochemical conversions to regulate the overall fruit quality (Kumar et al., 2015).
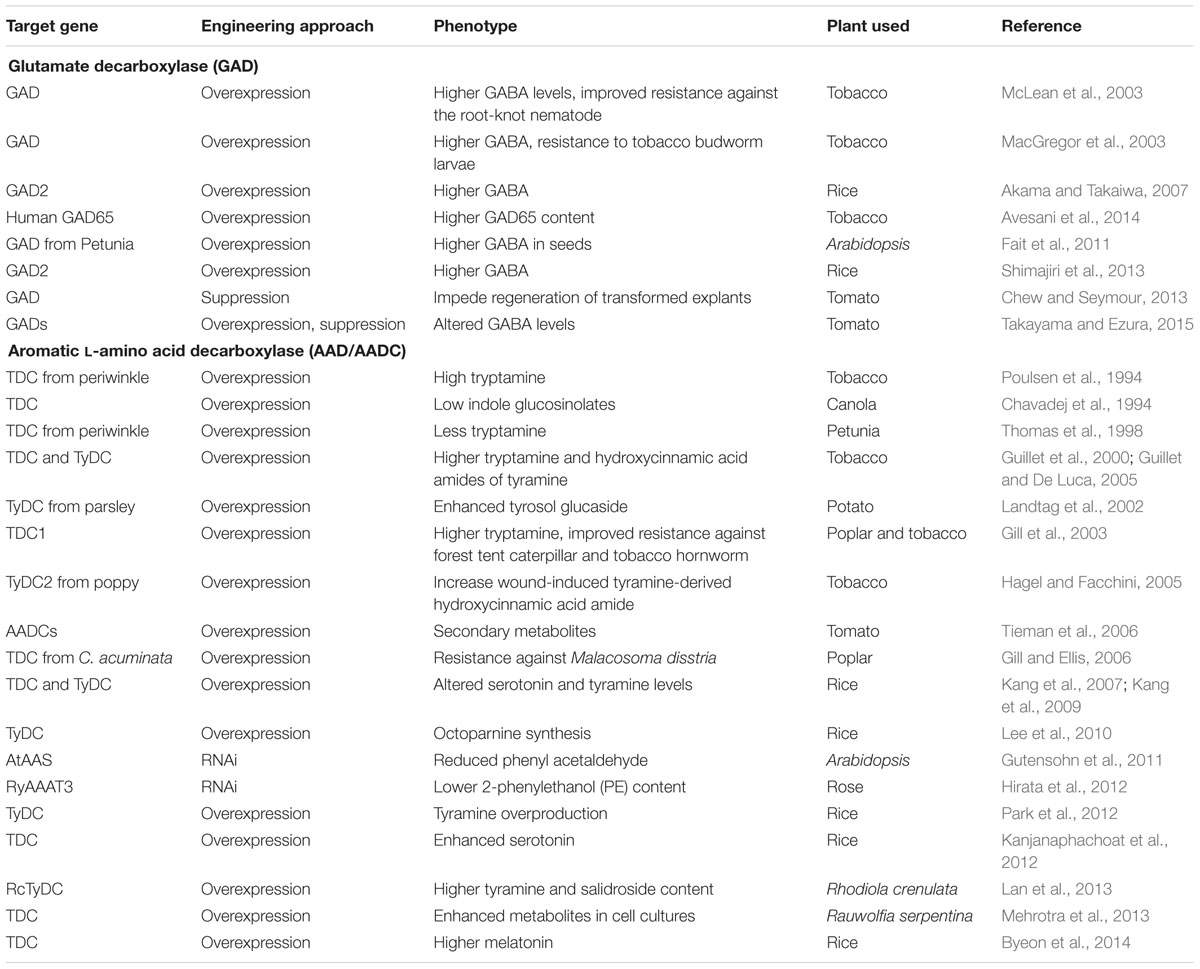
TABLE 1. List of the selected examples where PLP_deCs were undertaken in the genetic manipulation studies.
PLP_deC members of the third group encode SDC. These enzymes catalyze the conversion of serine to ethanolamine (EA) in plants. EA may act as a precursor of phosphatidylethanolamine (PE) and phosphatidylcholine (PC); the major phospholipids in eukaryotic membranes (Gibellini and Smith, 2010). Similar to the other PLP_deCs, these enzymes also determine the levels of secondary metabolites such as choline in plants (Mudd and Datko, 1989; Rontein et al., 2001).
Role of PLP_deCs in Stresses
Growing evidences suggest that transcript levels of PLP_deC genes are also influenced by both abiotic (Menke et al., 1999; Lee et al., 2010; Akcay et al., 2012; Liu et al., 2012; Al-Quraan et al., 2013; Hyun et al., 2014; Hu et al., 2015; Kumar et al., 2015) and biotic stresses (Kawalleck et al., 1993; Facchini et al., 1996; Lopez-Meyer and Nessler, 1997; Li et al., 2013; Yogendra et al., 2014). Further, plant hormones such as IAA (Aerts et al., 1992; Goddijn et al., 1992), ABA, salicylic acid, and ethylene (Turano and Fang, 1998; Wang et al., 2000; Kumar et al., 2015) and metal ions Ni2+, Mn2+, Cu2+, Fe3+, and Mg2+ also modulate their expression (Fujimori and Ohta, 2003; Yang et al., 2013). The elevated GABA levels have been implicated in improving plant survival under abiotic stresses such as salinity and hypoxic conditions in tomato (Yin et al., 2010; Mae et al., 2012). It also improves plant resistance to the northern root-knot nematode in tobacco (McLean et al., 2003), and to fungal pathogens in rice (Forlani et al., 2014). Similarly, TDC mediated enhanced alkaloids production is known to confer resistance in transgenic poplar and tobacco plants against their specific herbivores (Gill et al., 2003). Enhanced amino acid metabolism through transcriptional activation has been proposed to be the underlying molecular mechanisms for such improved tolerance. In this context, transcription factor OsMYB55 has been found to impart its function by regulating OsGAD3 activity via directly binding to its promoter and activating GABA production under hyperthermia in OsMYB55-overexpression transgenic plants (El-Kereamy et al., 2012). Altogether, one of the main roles of PLP_deC enzymes appears to be in stress alleviation via controlling the production of secondary metabolites in plants.
Challenges Associated with the Identification and Annotation of PLP_deCs in Plants
Plant PLP_deCs share a common evolutionary lineage, however, significant sequence divergence has resulted in an intricate evolutionary relationships between the orthologous enzymes and their functional divergence. Since only a limited number of PLP_deC enzymes have been characterized, till date, elucidation of the complete range of their physiological roles in more plant species remains a monumental task. Their functions have been predicted on the basis of their sequence homology to the already characterized closest PLP_deCs, however, this approach is not always infallible (Thornton et al., 2000). For example, AtSDC was initially characterized as a HDC member (Rontein et al., 2003). A similar survey of the SDC-like protein homologs in GenBank revealed an incorrect annotation of their several homologs as HDC-like/AADs (Torrens-Spence et al., 2013). More recently, we also erroneously annotated tomato AtSDC homolog as SlHDC1 (Kumar et al., 2015). It is noteworthy to mention that upon phylogenetic analysis, SDC-like members were closely placed with HDC proteins in the same major clade and the discrepancies observed in their annotation might have occurred due to the higher sequence similarity between the members of two classes.
Until recently, both SDC and SDC-like enzymes were considered to be functionally conserved. However, new biochemical evidence suggests that these enzymes are functionally diverged in plants as the two SDC-like enzymes in chickpea and Medicago truncatula have been found to have unusual aldehyde synthase activity (Torrens-Spence et al., 2014b). Despite acting on the aromatic amino acids, these proteins demonstrated limited homology to the other characterized plant AADs and their preferred substrates were discovered to be the bulky hydrophobic amino acids (Facchini et al., 2000; Lehmann and Pollmann, 2009; Torrens-Spence et al., 2014b). Similar to SDCs, the sequence and the phylogenetic ambiguity between TYDC and TDC members especially that of rice in the previous studies makes annotation of plant AADs challenging (Kumar et al., 2015). In brief, the major challenges associated with the annotation of plant PLP_deCs are, first; presence of multiple gene models predicted in a genome under the same gene model name; second; lack of a high quality genome sequence for a few published draft genomes, third; the diverse substrate preferences of PLP_deC enzymes, fourth; availability of the limited information on their biochemical activities and preferred substrates, and fifth; a high sequence similarity between PLP_deCs especially between HDC and SDC-like proteins and TDCs and TYDCs.
Similarly, as a consequence of the high sequence similarity between TDC and TYDC or HDC and SDC members, elucidation of their preferred substrate specificities is always difficult. The situation gets more complicated by the fact that the activities of TYDCs and TDCs can change by changing a single active site residue; even without altering their substrate selectivity. Substitution of a tyrosine residue to phenylalanine in an active site catalytic loop of plant AADs was found to alter their decarboxylase activity to aldehyde synthase chemistry (Torrens-Spence et al., 2013). Catalytic promiscuity (the ability of a single enzyme to catalyze different chemical reactions) or loose substrate specificity of PLP_deC enzymes augments this situation further, implying that an organism may have more PLP-dependent activities than the actual number of genes, encoding these enzymes. It can also complicate the present scenario regarding their annotation (Percudani and Peracchi, 2003). One of the solutions to this problem lies in computation of newer and better optimized bioinformatics pipelines for the identification of putative active site residues; especially by training them on the available information of the already characterized plant PLP_deCs (De Luca et al., 1988; Facchini and De Luca, 1995; Facchini et al., 2000; Torrens-Spence et al., 2014a). Notably, a similar approach has successfully resulted in the identification of a glycine as the key residue in TDC sequences, whereas a serine occupied the same position in the same conserved motif in the TYDC sequences in Papaver somniferum and Catharanthus roseus (Torrens-Spence et al., 2014a). Besides the identification of such key residues, four additional residues which did not have any obvious role in governing their indolic or phenolic substrate specificity were also identified. This finding implicated that the activity of plant PLP_deCs is governed by a small number of residues.
Evolution of PLP_deCs in Plants
The evolutionary profiling of PLP_deCs using the available sequencing data of plant genomes can further help in the correct identification and functional elucidation of more orthologs in additional plant species. It has been proposed that the number of PLP_deC genes in an organism depends on its adaptation to the specific nutrient sources. With their roles in diverse aspects of plant development and in both abiotic and biotic stress responses, identification of more such genes in additional species and exploration of the coding sequences, especially corresponding to the key residues determining the specific activities of the encoded enzymes or the C-terminal domains such as CaMBD in GADs, among PLP_deC orthologs would help bridge the crucial knowledge gaps existing in current understanding of their precise functions and underlying molecular mechanisms in plants. An investigation of their full complement in 52 plant species, representing the major clades of the species tree of plants, revealed a clear expansion of these genes in land plants over their aquatic ancestors (Figure 1). It is conceivable that the sharp expansion in the PLP_deC complements in early land plants was necessitated by the requirement of additional nutrient sources for their successful acclimatization in the new environment. It was observed that green algae such as Chlamydomonas reinhardtii and Volvox carteri had only three PLP_deCs, without any HDC member. These genes underwent a slight expansion in microalga Coccomyxa subellipsoideaC169 which resulted in the origin of HDC gene in this member of chlorophyta. Identification of more PLP_deCs in the non-vascular plant such as Physcomitrella patens (15) and their further expansion in the earliest vascular plants, such as Selaginella moellendorffii (21), suggested that these genes might have contributed to fulfill the additional N requirement. The PLP_deC complement remained similar in gymnosperms (Picea abies) and the most basal angiosperm (Amorella trichopoda). However, it again showed a noticeable expansion in monocots as 89 such genes were identified in wheat. It is believable that such high number of these genes in wheat may be due to the presence of three genomes in the hexaploid wheat. Besides wheat, a significant expansion of PLP_deC genes also occurred in its tetraploid relative switchgrass (Panicum virgatum). However, lack of such expansion in the other tetraploid Setaria complicated the evolutionary trends associated with these genes in monocots. Furthermore, over 60% of the total PLP_deCs in wheat and Setaria fell in AAD category, suggesting that this class was preferentially retained in this species during evolution.
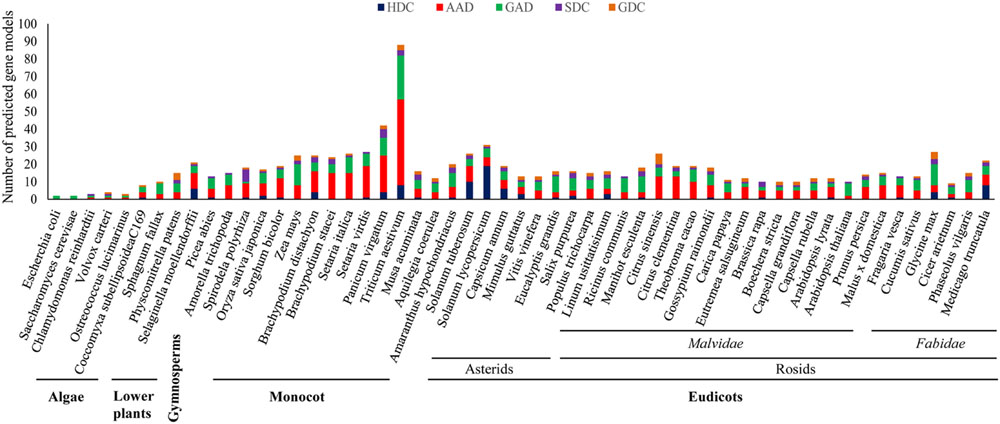
FIGURE 1. Identification of PLP_deC genes in 52 plant species, including members of algae, non-vascular lower plants, gymnosperm, and angiosperms, representing all the major clades of the species trees of plants, revealed a clear expansion of these genes from algae to land plants during evolution. HDC, histidine decarboxylase; AAD, aromatic L-amino acid decarboxylase; GAD, glutamate decarboxylase; SDC, serine decarboxylase; GDC, glycine decarboxylase.
Altogether, an expansion in the PLP_deC complements evidently favored the evolution of land plants. Generally, GAD and AAD over HDC and SDC members seem to have been preferred for such expansion, except in a few species with more HDC gene copies such as tomato and potato during evolution in plants. Comparatively, more AADs and GADs were identified in monocots than dicots, however, a few dicot species such as Eucalyptis grandis in asterids clade and Ricinus communis, Manihot esculenta, and Glycine max in rosids clade also had more GAD members. Closer examination of the C-terminal of the identified GAD proteins revealed that majority of them contained the CaMBD domain. For example, all tomato GADs possessed this domain suggesting that the oligomerized GAD complexes in tomato might be activated only in Ca2+/CaM-dependent manner (Zik et al., 2006; Akama and Takaiwa, 2007). However, lack of a typical CaMBD in a few GAD members in both monocots and dicots further suggested that such GAD enzymes might have evolved to function normally even in the absence of a conserved CaMBD domain. Finally, the evolution of HDC genes in land plants is intriguing as this class was found to be the most diverse in term of the strength of their members, which varied from none (in many plant species) to 19 in tomato. Although each class was found to be expanded at least in few monocot and/or dicot species, it remains unclear how these genes would have benefitted these plants and thus warrant further studies.
Conclusion and Future Prospects
We discussed the evolutionary trends associated with PLP_deC genes in plants. A clear expansion of the members of the different PLP_deC subclasses was found to accompany the evolution of land plants from their aquatic ancesters. Expansion of a certain subclass of PLP_deC genes such as HDC in tomato or other AADs in wheat raised important questions on their relevance and unknown functions in these species. The analysis of their evolutionary profiles presented herein would help to annotate PLP_deC orthologs in more plant species. A combined approach, including the biochemical characterization method, improved computational tools, especially trained on the already characterized PLP_deCs, and information of the 3-D structures of the representatives of each subclass is required to elucidate their precise functions in plants.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was financially supported by grants received from the Department of Science and Technology (DST). Author is grateful to DST for INSPIRE-Faculty Award (grant number IFA-LSPA-15) and thanks Prof. R. P. Sharma, Department of Plant Sciences, University of Hyderabad, for his support in carrying out the research work. Author is grateful to Dr. Hriyadesh Prakash, University of Hyderabad, for critical reading of the manuscript.
References
Aerts, R. J., Alarco, A. M., and De Luca, V. (1992). Auxins induce tryptophan decarboxylase activity in radicles of catharanthus seedlings. Plant Physiol. 100, 1014–1019. doi: 10.1104/pp.100.2.1014
Akama, K., Akihiro, T., Kitagawa, M., and Takaiwa, F. (2001). Rice (Oryza sativa) contains a novel isoform of glutamate decarboxylase that lacks an authentic calmodulin-binding domain at the C-terminus. Biochim. Biophys. Acta 1522, 143–150. doi: 10.1016/S0167-4781(01)00324-4
Akama, K., and Takaiwa, F. (2007). C-terminal extension of rice glutamate decarboxylase (OsGAD2) functions as an autoinhibitory domain and overexpression of a truncated mutant results in the accumulation of extremely high levels of GABA in plant cells. J. Exp. Bot. 58, 2699–2707. doi: 10.1093/jxb/erm120
Akcay, N., Bor, M., Karabudak, T., Ozdemir, F., and Turkan, I. (2012). Contribution of Gamma amino butyric acid (GABA) to salt stress responses of Nicotiana sylvestris CMSII mutant and wild type plants. J. Plant Physiol. 169, 452–458. doi: 10.1016/j.jplph.2011.11.006
Akihiro, T., Koike, S., Tani, R., Tominaga, T., Watanabe, S., Iijima, Y., et al. (2008). Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol. 49, 1378–1389. doi: 10.1093/pcp/pcn113
Al-Quraan, N. A., Sartawe, F. A., and Qaryouti, M. M. (2013). Characterization of gamma-aminobutyric acid metabolism and oxidative damage in wheat (Triticum aestivum L.) seedlings under salt and osmotic stress. J. Plant Physiol. 170, 1003–1009. doi: 10.1016/j.jplph.2013.02.010
Asano, T., Kobayashi, K., Kashihara, E., Sudo, H., Sasaki, R., Iijima, Y., et al. (2012). Suppression of camptothecin biosynthetic genes results in metabolic modification of secondary products in hairy roots of Ophiorrhiza pumila. Phytochemistry 91, 128–139. doi: 10.1016/j.phytochem.2012.04.019
Avesani, L., Merlin, M., Gecchele, E., Capaldi, S., Brozzetti, A., Falorni, A., et al. (2014). Comparative analysis of different biofactories for the production of a major diabetes autoantigen. Transgenic Res. 23, 281–291. doi: 10.1007/s11248-013-9749-9
Baum, G., Lev-Yadun, S., Fridmann, Y., Arazi, T., Katsnelson, H., Zik, M., et al. (1996). Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J. 15, 2988–2996.
Bouche, N., Fait, A., Zik, M., and Fromm, H. (2004). The root-specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in Arabidopsis. Plant Mol. Biol. 55, 315–325. doi: 10.1007/s11103-004-0650-z
Byeon, Y., Park, S., Lee, H. Y., Kim, Y. S., and Back, K. (2014). Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J. Pineal Res. 56, 275–282. doi: 10.1111/jpi.12120
Carrari, F., and Fernie, A. R. (2006). Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 57, 1883–1897. doi: 10.1093/jxb/erj020
Chavadej, S., Brisson, N., Mcneil, J. N., and De Luca, V. (1994). Redirection of tryptophan leads to production of low indole glucosinolate canola. Proc. Natl. Acad. Sci. U.S.A. 91, 2166–2170. doi: 10.1073/pnas.91.6.2166
Chew, B. L., and Seymour, G. B. (2013). The effects of glutamate decarboxylase (GAD) RNAi knockout in tissue cultured transgenic tomato (Solanum lycopersicum). Plant Omics 6, 13–17.
De Luca, V., Fernandez, J. A., Campbell, D., and Kurz, W. G. (1988). Developmental regulation of enzymes of indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 86, 447–450. doi: 10.1104/pp.86.2.447
Eliot, A. C., and Kirsch, J. F. (2004). Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383–415. doi: 10.1146/annurev.biochem.73.011303.074021
El-Kereamy, A., Bi, Y. M., Ranathunge, K., Beatty, P. H., Good, A. G., and Rothstein, S. J. (2012). The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 7:e52030. doi: 10.1371/journal.pone.0052030
Facchini, P. J., and De Luca, V. (1995). Expression in Escherichia coli and partial characterization of two tyrosine/dopa decarboxylases from opium poppy. Phytochemistry 38, 1119–1126. doi: 10.1016/0031-9422(94)00814-A
Facchini, P. J., Huber-Allanach, K. L., and Tari, L. W. (2000). Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54, 121–138. doi: 10.1016/S0031-9422(00)00050-9
Facchini, P. J., Johnson, A. G., Poupart, J., and De Luca, V. (1996). Uncoupled defense gene expression and antimicrobial alkaloid accumulation in elicited opium poppy cell cultures. Plant Physiol. 111, 687–697. doi: 10.1104/pp.111.3.687
Fait, A., Fromm, H., Walter, D., Galili, G., and Fernie, A. R. (2008). Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci. 13, 14–19. doi: 10.5363/tits.13.2_14
Fait, A., Nesi, A. N., Angelovici, R., Lehmann, M., Pham, P. A., Song, L., et al. (2011). Targeted enhancement of glutamate-to-gamma-aminobutyrate conversion in Arabidopsis seeds affects carbon-nitrogen balance and storage reserves in a development-dependent manner. Plant Physiol. 157, 1026–1042. doi: 10.1104/pp.111.179986
Feng, C., Chen, M., Xu, C. J., Bai, L., Yin, X. R., Li, X., et al. (2012). Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genomics 13:19. doi: 10.1186/1471-2164-13-19
Forlani, G., Bertazzini, M., and Giberti, S. (2014). Differential accumulation of gamma-aminobutyric acid in elicited cells of two rice cultivars showing contrasting sensitivity to the blast pathogen. Plant Biol. (Stuttg) 16, 1127–1132. doi: 10.1111/plb.12165
Fujimori, K., and Ohta, D. (2003). Heavy metal induction of Arabidopsis serine decarboxylase gene expression. Biosci. Biotechnol. Biochem. 67, 896–898. doi: 10.1271/bbb.67.896
Gibellini, F., and Smith, T. K. (2010). The Kennedy pathway–De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428. doi: 10.1002/iub.337
Gill, R. I., Ellis, B. E., and Isman, M. B. (2003). Tryptamine-induced resistance in tryptophan decarboxylase transgenic poplar and tobacco plants against their specific herbivores. J. Chem. Ecol. 29, 779–793. doi: 10.1023/A:1022983529555
Gill, R. I. S., and Ellis, B. E. (2006). Over-expression of tryptophan decarboxylase gene in poplar and its possible role in resistance against Malacosoma disstria. New For. 31, 195–209. doi: 10.1007/s11056-005-2719-1
Goddijn, O. J., De Kam, R. J., Zanetti, A., Schilperoort, R. A., and Hoge, J. H. (1992). Auxin rapidly down-regulates transcription of the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol. Biol. 18, 1113–1120. doi: 10.1007/BF00047714
Guillet, G., and De Luca, V. (2005). Wound-inducible biosynthesis of phytoalexin hydroxycinnamic acid amides of tyramine in tryptophan and tyrosine decarboxylase transgenic tobacco lines. Plant Physiol. 137, 692–699. doi: 10.1104/pp.104.050294
Guillet, G., Poupart, J., Basurco, J., and De Luca, V. (2000). Expression of tryptophan decarboxylase and tyrosine decarboxylase genes in tobacco results in altered biochemical and physiological phenotypes. Plant Physiol. 122, 933–943. doi: 10.1104/pp.122.3.933
Gutensohn, M., Klempien, A., Kaminaga, Y., Nagegowda, D. A., Negre-Zakharov, F., Huh, J. H., et al. (2011). Role of aromatic aldehyde synthase in wounding/herbivory response and flower scent production in different Arabidopsis ecotypes. Plant J. 66, 591–602. doi: 10.1111/j.1365-313X.2011.04515.x
Hagel, J. M., and Facchini, P. J. (2005). Elevated tyrosine decarboxylase and tyramine hydroxycinnamoyltransferase levels increase wound-induced tyramine-derived hydroxycinnamic acid amide accumulation in transgenic tobacco leaves. Planta 221, 904–914. doi: 10.1007/s00425-005-1484-x
Hirata, H., Ohnishi, T., Ishida, H., Tomida, K., Sakai, M., Hara, M., et al. (2012). Functional characterization of aromatic amino acid aminotransferase involved in 2-phenylethanol biosynthesis in isolated rose petal protoplasts. J. Plant Physiol. 169, 444–451. doi: 10.1016/j.jplph.2011.12.005
Hu, X., Xu, Z., Xu, W., Li, J., Zhao, N., and Zhou, Y. (2015). Application of gamma-aminobutyric acid demonstrates a protective role of polyamine and GABA metabolism in muskmelon seedlings under Ca(NO3)2 stress. Plant Physiol. Biochem. 92, 1–10. doi: 10.1016/j.plaphy.2015.04.006
Hyun, T. K., Eom, S. H., Han, X., and Kim, J. S. (2014). Evolution and expression analysis of the soybean glutamate decarboxylase gene family. J. Biosci. 39, 899–907. doi: 10.1007/s12038-014-9484-2
John, R. A. (1995). Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta 1248, 81–96. doi: 10.1016/0167-4838(95)00025-P
Kang, K., Kim, Y. S., Park, S., and Back, K. (2009). Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 150, 1380–1393. doi: 10.1104/pp.109.138552
Kang, S., Kang, K., Lee, K., and Back, K. (2007). Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 227, 263–272. doi: 10.1007/s00425-007-0614-z
Kanjanaphachoat, P., Wei, B. Y., Lo, S. F., Wang, I. W., Wang, C. S., Yu, S. M., et al. (2012). Serotonin accumulation in transgenic rice by over-expressing tryptophan decarboxylase results in a dark brown phenotype and stunted growth. Plant Mol. Biol. 78, 525–543. doi: 10.1007/s11103-012-9882-5
Kawalleck, P., Keller, H., Hahlbrock, K., Scheel, D., and Somssich, I. E. (1993). A pathogen-responsive gene of parsley encodes tyrosine decarboxylase. J. Biol. Chem. 268, 2189–2194.
Kumar, R., Jiwani, G., Paree, A., Sravankumar, T., Khurana, A., and Sharma, A. K. (2015). Evolutionary profiling of group IIPLP-dependent decarboxylase gene family suggests expansion and functional diversification of histidine decarboxylases in tomato. Plant Genome 9, 1–15.
Lan, X., Chang, K., Zeng, L., Liu, X., Qiu, F., Zheng, W., et al. (2013). Engineering salidroside biosynthetic pathway in hairy root cultures of Rhodiola crenulata based on metabolic characterization of tyrosine decarboxylase. PLoS ONE 8:e75459. doi: 10.1371/journal.pone.0075459
Landtag, J., Baumert, A., Degenkolb, T., Schmidt, J., Wray, V., Scheel, D., et al. (2002). Accumulation of tyrosol glucoside in transgenic potato plants expressing a parsley tyrosine decarboxylase. Phytochemistry 60, 683–689. doi: 10.1016/S0031-9422(02)00161-9
Lee, J. H., Kim, Y. J., Jeong, D. Y., Sathiyaraj, G., Pulla, R. K., Shim, J. S., et al. (2010). Isolation and characterization of a Glutamate decarboxylase (GAD) gene and their differential expression in response to abiotic stresses from Panax ginseng C. A. Meyer. Mol. Biol. Rep. 37, 3455–3463. doi: 10.1007/s11033-009-9937-0
Lehmann, T., and Pollmann, S. (2009). Gene expression and characterization of a stress-induced tyrosine decarboxylase from Arabidopsis thaliana. FEBS Lett. 583, 1895–1900. doi: 10.1016/j.febslet.2009.05.017
Li, X., Kim, Y. B., Uddin, M. R., Lee, S., Kim, S. J., and Park, S. U. (2013). Influence of light on the free amino acid content and gamma-aminobutyric acid synthesis in Brassica juncea seedlings. J. Agric. Food Chem. 61, 8624–8631. doi: 10.1021/jf401956v
Ling, Y., Chen, T., Jing, Y., Fan, L., Wan, Y., and Lin, J. (2013). gamma-Aminobutyric acid (GABA) homeostasis regulates pollen germination and polarized growth in Picea wilsonii. Planta 238, 831–843. doi: 10.1007/s00425-013-1938-5
Liu, W., Chen, R., Chen, M., Zhang, H., Peng, M., Yang, C., et al. (2012). Tryptophan decarboxylase plays an important role in ajmalicine biosynthesis in Rauvolfia verticillata. Planta 236, 239–250. doi: 10.1007/s00425-012-1608-z
Lopez-Meyer, M., and Nessler, C. L. (1997). Tryptophan decarboxylase is encoded by two autonomously regulated genes in Camptotheca acuminata which are differentially expressed during development and stress. Plant J. 11, 1167–1175. doi: 10.1046/j.1365-313X.1997.11061167.x
MacGregor, K. B., Shelp, B. J., Peiris, S., and Bown, A. W. (2003). Overexpression of glutamate decarboxylase in transgenic tobacco plants deters feeding by phytophagous insect larvae. J. Chem. Ecol. 29, 2177–2182. doi: 10.1023/A:1025650914947
Mae, N., Makino, Y., Oshita, S., Kawagoe, Y., Tanaka, A., Aoki, K., et al. (2012). Accumulation mechanism of γ-aminobutyric acid in tomatoes (Solanum lycopersicum L.) under low O2 with and without CO2. J. Agric. Food Chem. 60, 1013–1019. doi: 10.1021/jf2046812
Maldonado-Mendoza, I. E., Lopez-Meyer, M., Galef, J. R., Burnett, R. J., and Nessler, C. L. (1996). Molecular analysis of a new member of the opium poppy tyrosine/3,4-dihydroxyphenylalanine decarboxylase gene family. Plant Physiol. 110, 43–49. doi: 10.1104/pp.110.1.43
McLean, M. D., Yevtushenko, D. P., Deschene, A., Van Cauwenberghe, O. R., Makhmoudova, A., Potter, J. W., et al. (2003). Overexpression of glutamate decarboxylase in transgenic tobacco plants confers resistance to the northern root-knot nematode. Mol. Breed. 11, 277–285. doi: 10.1023/A:1023483106582
Mehrotra, S., Srivastava, V., Rahman, L. U., and Kukreja, A. K. (2013). Overexpression of a Catharanthus tryptophan decarboxylase (tdc) gene leads to enhanced terpenoid indole alkaloid (TIA) production in transgenic hairy root lines of Rauwolfia serpentina. Plant Cell Tissue Organ Cult. 115, 377–384. doi: 10.1007/s11240-013-0369-0
Menke, F. L., Champion, A., Kijne, J. W., and Memelink, J. (1999). A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18, 4455–4463. doi: 10.1093/emboj/18.16.4455
Milano, T., Paiardini, A., Grgurina, I., and Pascarella, S. (2013). Type I pyridoxal 5′-phosphate dependent enzymatic domains embedded within multimodular nonribosomal peptide synthetase and polyketide synthase assembly lines. BMC Struct. Biol. 13:26. doi: 10.1186/1472-6807-13-26
Molina-Rueda, J. J., Pascual, M. B., Pissarra, J., and Gallardo, F. (2015). A putative role for gamma-aminobutyric acid (GABA) in vascular development in pine seedlings. Planta 241, 257–267. doi: 10.1007/s00425-014-2157-4
Mudd, S. H., and Datko, A. H. (1989). Synthesis of Ethanolamine and Its regulation in Lemna paucicostata. Plant Physiol. 91, 587–597. doi: 10.1104/pp.91.2.587
Osorio, S., Alba, R., Damasceno, C. M., Lopez-Casado, G., Lohse, M., Zanor, M. I., et al. (2011). Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425. doi: 10.1104/pp.111.175463
Palanivelu, R., Brass, L., Edlund, A. F., and Preuss, D. (2003). Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114, 47–59. doi: 10.1016/S0092-8674(03)00479-3
Park, S., Lee, K., Kim, Y. S., Chi, Y. T., Shin, J. S., and Back, K. (2012). Induced tyramine overproduction in transgenic rice plants expressing a rice tyrosine decarboxylase under the control of methanol-inducible rice tryptophan decarboxylase promoter. Bioprocess Biosyst. Eng. 35, 205–210. doi: 10.1007/s00449-011-0615-y
Pasquali, G., Goddijn, O. J., De Waal, A., Verpoorte, R., Schilperoort, R. A., Hoge, J. H., et al. (1992). Coordinated regulation of two indole alkaloid biosynthetic genes from Catharanthus roseus by auxin and elicitors. Plant Mol. Biol. 18, 1121–1131. doi: 10.1007/BF00047715
Percudani, R., and Peracchi, A. (2003). A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 4, 850–854. doi: 10.1038/sj.embor.embor914
Picton, S., Gray, J. E., Payton, S., Barton, S. L., Lowe, A., and Grierson, D. (1993). A histidine decarboxylase-like mRNA is involved in tomato fruit ripening. Plant Mol. Biol. 23, 627–631. doi: 10.1007/BF00019310
Poulsen, C., Goddijn, O. J. M., Hoge, J. H. C., and Verpoorte, R. (1994). Anthranilate synthase and chorismate mutase activities in transgenic tobacco plants overexpressing tryptophan decarboxylase from catharanthus-roseus. Transgenic Res. 3, 43–49. doi: 10.1007/BF01976026
Rolin, D., Baldet, P., Just, D., Chevalier, C., Biran, M., and Raymond, P. (2000). NMR study of low subcellular pH during the development of cherry tomato fruit. Aust. J. Plant Physiol. 27, 61–69.
Rontein, D., Nishida, I., Tashiro, G., Yoshioka, K., Wu, W. I., Voelker, D. R., et al. (2001). Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J. Biol. Chem. 276, 35523–35529. doi: 10.1074/jbc.M106038200
Rontein, D., Rhodes, D., and Hanson, A. D. (2003). Evidence from engineering that decarboxylation of free serine is the major source of ethanolamine moieties in plants. Plant Cell Physiol. 44, 1185–1191. doi: 10.1093/pcp/pcg144
Saito, T., Matsukura, C., Sugiyama, M., Watahiki, A., Ohshima, I., Iijima, Y., et al. (2008). Screening for gamma-aminobutyric acid (GABA)-rich tomato varieties. J. Jpn. Soc. Hort. Sci. 77, 242–250. doi: 10.2503/jjshs1.77.242
Sandmeier, E., Hale, T. I., and Christen, P. (1994). Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 221, 997–1002. doi: 10.1111/j.1432-1033.1994.tb18816.x
Shimajiri, Y., Oonishi, T., Ozaki, K., Kainou, K., and Akama, K. (2013). Genetic manipulation of the gamma-aminobutyric acid (GABA) shunt in rice: overexpression of truncated glutamate decarboxylase (GAD2) and knockdown of gamma-aminobutyric acid transaminase (GABA-T) lead to sustained and high levels of GABA accumulation in rice kernels. Plant Biotechnol. J. 11, 594–604. doi: 10.1111/pbi.12050
Takayama, M., and Ezura, H. (2015). How and why does tomato accumulate a large amount of GABA in the fruit? Front. Plant Sci. 6:612. doi: 10.3389/fpls.2015.00612
Thomas, J. C., Saleh, E. F., Alammar, N., and Akroush, A. M. (1998). The indole alkaloid tryptamine impairs reproduction in Drosophila melanogaster. J. Econ. Entomol. 91, 841–846. doi: 10.1093/jee/91.4.841
Thornton, J. M., Todd, A. E., Milburn, D., Borkakoti, N., and Orengo, C. A. (2000). From structure to function: approaches and limitations. Nat. Struct. Biol. 7(Suppl.), 991–994. doi: 10.1038/80784
Tieman, D., Taylor, M., Schauer, N., Fernie, A. R., Hanson, A. D., and Klee, H. J. (2006). Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. U.S.A. 103, 8287–8292. doi: 10.1073/pnas.0602469103
Torrens-Spence, M. P., Lazear, M., Von Guggenberg, R., Ding, H., and Li, J. (2014a). Investigation of a substrate-specifying residue within Papaver somniferum and Catharanthus roseus aromatic amino acid decarboxylases. Phytochemistry 106, 37–43. doi: 10.1016/j.phytochem.2014.07.007
Torrens-Spence, M. P., Liu, P., Ding, H., Harich, K., Gillaspy, G., and Li, J. (2014b). Biochemical evaluation of the decarboxylation and decarboxylation-deamination activities of plant aromatic amino acid decarboxylases. J. Biol. Chem. 288, 2376–2387. doi: 10.1074/jbc.M112.401752
Torrens-Spence, M. P., Von Guggenberg, R., Lazear, M., Ding, H., and Li, J. (2013). Diverse functional evolution of serine decarboxylases: identification of two novel acetaldehyde synthases that uses hydrophobic amino acids as substrates. BMC Plant Biol. 14:247. doi: 10.1186/s12870-014-0247-x
Trobacher, C. P., Zarei, A., Liu, J., Clark, S. M., Bozzo, G. G., and Shelp, B. J. (2013). Calmodulin-dependent and calmodulin-independent glutamate decarboxylases in apple fruit. BMC Plant Biol. 13:144. doi: 10.1186/1471-2229-13-144
Turano, F. J., and Fang, T. K. (1998). Characterization of two glutamate decarboxylase cDNA clones from Arabidopsis. Plant Physiol. 117, 1411–1421. doi: 10.1104/pp.117.4.1411
Wang, R., Guegler, K., Labrie, S. T., and Crawford, N. M. (2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12, 1491–1509. doi: 10.2307/3871145
Yang, R., Yin, Y., Guo, Q., and Gu, Z. (2013). Purification, properties and cDNA cloning of glutamate decarboxylase in germinated faba bean (Vicia faba L.). Food Chem. 138, 1945–1951. doi: 10.1016/j.foodchem.2012.11.050
Yin, Y. G., Tominaga, T., Iijima, Y., Aoki, K., Shibata, D., Ashihara, H., et al. (2010). Metabolic alterations in organic acids and γ-aminobutyric acid in developing tomato (Solanum lycopersicum L.) fruits. Plant Cell Physiol. 51, 1300–1314. doi: 10.1093/pcp/pcq090
Yogendra, K. N., Pushpa, D., Mosa, K. A., Kushalappa, A. C., Murphy, A., and Mosquera, T. (2014). Quantitative resistance in potato leaves to late blight associated with induced hydroxycinnamic acid amides. Funct. Integr. Genomics 14, 285–298. doi: 10.1007/s10142-013-0358-8
Keywords: PLP decarboxylase, evolution, plants, phylogeny, fruits, gene expression, potato, gene complement
Citation: Kumar R (2016) Evolutionary Trails of Plant Group II Pyridoxal Phosphate-Dependent Decarboxylase Genes. Front. Plant Sci. 7:1268. doi: 10.3389/fpls.2016.01268
Received: 22 April 2016; Accepted: 10 August 2016;
Published: 23 August 2016.
Edited by:
Randeep Rakwal, University of Tsukuba, JapanReviewed by:
Pascal Braun, Technische Universität München, GermanyRamesh Katam, Florida A&M University, USA
Copyright © 2016 Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rahul Kumar, rksl@uohyd.ac.in rahulpmb@gmail.com
 Rahul Kumar
Rahul Kumar