- 1Centro Ricerca e Innovazione, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 2Dipartimento di Bioscienze, Università degli Studi di Milano, Milan, Italy
The GENOMES UNCOUPLED 1 (GUN1) gene has been reported to encode a chloroplast-localized pentatricopeptide-repeat protein, which acts to integrate multiple indicators of plastid developmental stage and altered plastid function, as part of chloroplast-to-nucleus retrograde communication. However, the molecular mechanisms underlying signal integration by GUN1 have remained elusive, up until the recent identification of a set of GUN1-interacting proteins, by co-immunoprecipitation and mass-spectrometric analyses, as well as protein–protein interaction assays. Here, we review the molecular functions of the different GUN1 partners and propose a major role for GUN1 as coordinator of chloroplast translation, protein import, and protein degradation. This regulatory role is implemented through proteins that, in most cases, are part of multimeric protein complexes and whose precise functions vary depending on their association states. Within this framework, GUN1 may act as a platform to promote specific functions by bringing the interacting enzymes into close proximity with their substrates, or may inhibit processes by sequestering particular pools of specific interactors. Furthermore, the interactions of GUN1 with enzymes of the tetrapyrrole biosynthesis (TPB) pathway support the involvement of tetrapyrroles as signaling molecules in retrograde communication.
Introduction
Upon illumination, proplastids differentiate into functional chloroplasts in developing photosynthetic tissues of cotyledons, leaves, and stems (Jarvis and López-Juez, 2013). Chloroplast biogenesis also occurs during the growth of young green tissues, as cells expand and mature chloroplasts undergo division by binary fission (Okazaki et al., 2010). This process is characterized macroscopically by rapid greening of the young chloroplast and microscopically by the concomitant formation of thylakoid membranes and the reorganization of nucleoids, i.e., DNA-containing structures without defined boundaries, which differ in number, size, and distribution within plastids at different developmental stages, and harbor the plastid gene expression (PGE) machinery (Pfalz and Pfannschmidt, 2013; Melonek et al., 2016).
At the molecular level, this rather complex biogenic transition is achieved by cytosolic synthesis of chloroplast-targeted proteins, followed by import, assembly, folding, and degradation of unfolded/misfolded proteins (Jarvis and López-Juez, 2013). Indeed, the plastid genome itself (the plastome) comprises fewer than 100 protein-coding genes, and the vast majority of the 2000–3000 proteins that make up the chloroplast proteome are encoded in the nucleus (Richly and Leister, 2004). In particular, precursor proteins carrying N-terminal transit peptides initially interact with two multiprotein complexes termed Translocon at the outer envelope membrane of chloroplasts (TOC) and Translocon at the inner envelope membrane of chloroplasts (TIC), which facilitate their active transport through the chloroplast envelope, powered by an ATP import motor, consisting of the stromal heat-shock protein 93 (Hsp93), heat-shock protein 70 (Hsp70), and heat-shock protein 90 (Hsp90; Flores-Perez and Jarvis, 2013; Inoue et al., 2013; Shi and Theg, 2013a,b). Upon translocation, proteins are exposed to different proteolytic systems of prokaryotic origin, which are responsible for protein maturation, control of protein abundance, and removal of either misfolded or damaged components. Among these, the stromal protease Clp is a multimeric complex made of chaperones and serine protease subunits, which serve general housekeeping functions. In contrast, the thylakoid-associated FtsH (Filamentous temperature sensitive H) proteases are zinc-containing metalloendopeptidases that have both chaperone and proteolytic functions, and participate in the Photosystem II repair cycle, together with the DEG serine proteases (Kato and Sakamoto, 2010; Van Wijk, 2015).
Besides translation and post-translational processes, chloroplast biogenesis also requires transcriptional coordination of thousands of nuclear genes with the expression of the comparatively few plastid genes in order to meet the needs of the developing chloroplast (Chan et al., 2016; Kleine and Leister, 2016). This is achieved through extensive exchange of information between plastids and the nucleus, for instance, via biogenic retrograde signaling—a system in which developmentally relevant stimuli in plastids induce the accumulation of specific signaling molecules that relay information to the nucleus, and in turn adjust the expression of nuclear genes to the needs of the plastids (Pogson et al., 2008; Woodson and Chory, 2008; Chan et al., 2016).
During the last 30 years, experiments with the carotenoid biosynthesis inhibitor norfluorazon (NF) and the inhibitor of plastid translation lincomycin (LIN), each of which arrests chloroplast development at the proplastid stage and represses the expression of photosynthesis-associated nuclear genes (PhANGs), have provided insights into the plastid's biogenic retrograde pathways (Oelmüller and Mohr, 1986; Oelmüller et al., 1986).
Six genome uncoupled (gun) mutants have been characterized in Arabidopsis thaliana that fail to repress transcription of the nuclear gene Lhcb1.2 after NF treatment, and are thus impaired in retrograde signaling (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003; Koussevitzky et al., 2007; Adhikari et al., 2011; Woodson et al., 2011). Five of these genes, GUN2-6, were found to be involved in tetrapyrrole biosynthesis (TPB), whereas GUN1, which encodes a nucleoid-localized pentatricopeptide repeat protein (PPR), has been shown to have a role in PGE, and to act as an integrator of multiple retrograde signals, since gun1 mutants are unique in exhibiting a gun phenotype in response to both norfluorazon and lincomycin (Gray et al., 2003; Koussevitzky et al., 2007). However, the exact molecular role of GUN1 remained enigmatic until the new insights provided by the recent identification of a set of GUN1-interacting proteins (Tadini et al., 2016; Table 1).
Based on the functions of these partners, GUN1 appears to take part in multiple processes essential for chloroplast biogenesis and maintenance of the chloroplast proteome. GUN1-mediated control of plastid ribosomal protein S1 (PRPS1) accumulation, together with co-immunoprecipitation (CoIP) of proteins involved in different steps of plastid translation, support the involvement of GUN1 in the regulation of plastid protein synthesis. Furthermore, the presence of several chaperones in the CoIP mixture suggests a role for GUN1 in the coordination of chloroplast protein import and protein degradation.
Intriguingly, several GUN1 interactors appear to accumulate to higher levels upon induction of the unfolded protein response (UPR) in Chlamydomonas reinhardtii chloroplasts, which is triggered upon conditional repression of the catalytic subunit of Clp protease (ClpP1; Ramundo et al., 2013; Ramundo and Rochaix, 2014; Rochaix and Ramundo, 2015). This finding suggests the possible involvement of GUN1 in the UPR signaling pathway.
In this review, we describe the functional roles of the different GUN1 protein partners and propose some testable hypotheses that should clarify the molecular role of GUN1 in chloroplast biogenesis and chloroplast protein homeostasis.
GUN1 is Found in Plastid Nucleoids and Interacts with the Transcriptional Machinery
GUN1 encodes a member of PPR-containing protein family, which has a small MutS-related (SMR) domain at the C-terminal end and a plastid targeting signal sequence at its N terminus. PPR motifs have been shown to mediate interactions with nucleic acids, and the SMR domain is found in proteins that act in DNA repair and recombination. However, in vivo RNA and DNA immunoprecipitation on chip (NIP-chip), as well as one-hybrid assays, have failed to detect any stable interaction of GUN1 with nucleic acids (Tadini et al., 2016), in contrast to a previous report, in which a GUN1 fragment containing both the PPR and SMR domains was shown to bind DNA in vitro (Koussevitzky et al., 2007). Nevertheless, GUN1 appears to be associated with nucleoids in the chloroplast, and more specifically with the domain of active plastid transcription, as shown by the relatively large and distinct foci of a fluorescent GUN1-YFP (Yellow Fluorescence Protein) chimera that co-localize with a Plastid Transcriptionally Active Chromosome 2-Cyan Fluorescence Protein (pTAC2-CFP) fusion in chloroplasts of mesophyll cells (Koussevitzky et al., 2007). However, although the repertoire of nucleoid-associated proteins so far identified is quite extensive, the GUN1 protein is not listed in any of the chloroplast or nucleoid/pTAC proteomes published to date (for a review see Melonek et al., 2016), most probably because it accumulates in very small amounts at specific developmental stages or under particular physiological conditions. This inference is supported by CoIP experiments with a Green Fluorescence Protein (GUN1-GFP) fusion and subsequent mass spectrometry (MS), which identified several nucleoid subunits as interactors with GUN1 (Tadini et al., 2016; Table 1).
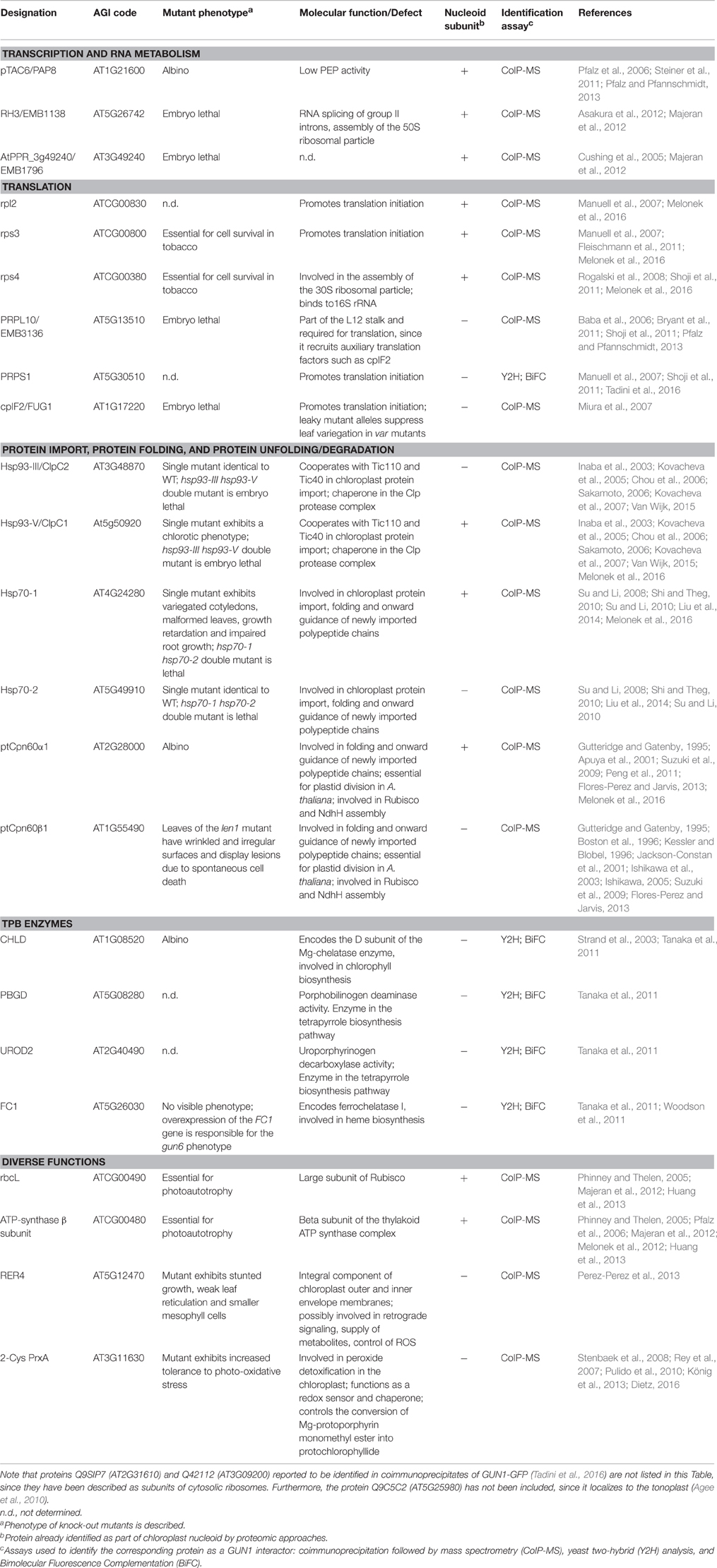
pTAC6 is among the GUN1 interactors, and it has been reported to interact directly with the plastid-encoded RNA polymerase (PEP), building together with pTAC2 and other polymerase-associated proteins (PAPs) the soluble RNA polymerase (sRNPase) complex (Pfalz et al., 2006), a central component of nucleoids (Steiner et al., 2011; Figure 1). Intriguingly, pTAC6 (also known as PAP8) contains no known domain and exhibits no homologies that could provide hints as to its function in PGE (Steiner et al., 2011). However, functional genomics analyses have indicated that homozygous pap knockout lines develop white cotyledons, fail to accumulate chlorophyll even under low light intensities, and do not produce primary leaves unless they are cultivated on MS medium supplemented with sucrose (for a review, see Pfalz and Pfannschmidt, 2013). Furthermore, analyses of PGE in pap mutants revealed strong repression of the accumulation of PEP-dependent transcripts, whereas levels of nucleus-encoded RNA polymerase (NEP)-dependent transcripts were not depleted, while some were enhanced, indicating that pTAC6/PAP8 and the other PAP proteins are essential for the activity of PEP (see Table 1).
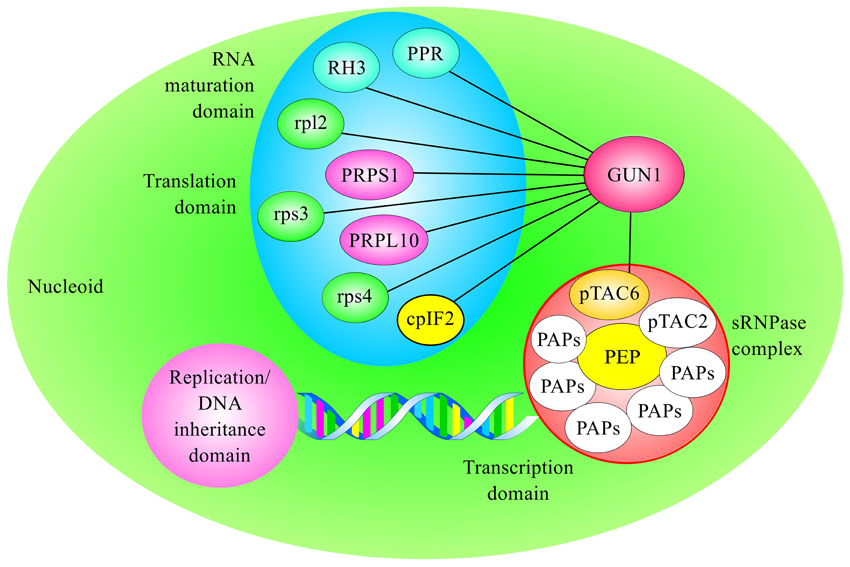
Figure 1. Schematic overview of GUN1 protein interactors involved in gene transcription, ribosome biogenesis and plastid translation. The scheme takes into account the partition of nucleoids into functional subdomains proposed by Pfalz and Pfannschmidt (2013). PPR refers to AtPPR_3g49240, also known as EMB1796, as reported in Table 1.
GUN1 Controls Plastid Translation and Ribosome Biogenesis
GUN1 also interacts with several ribosomal subunits, such as the plastid-encoded ribosomal proteins L2, S3, and S4 (rpl2, rps3, and rps4) and the nucleus-encoded plastid ribosomal protein L10 (PRPL10; Figure 1). Furthermore, yeast two-hybrid and Bimolecular Fluorescence Complementation (BiFC) assays revealed a physical interaction between GUN1 and PRPS1 (Tadini et al., 2016). Ribosomal proteins have been reproducibly detected in nucleoid and pTAC proteomes (Melonek et al., 2016), further supporting the existence of a translational subdomain within the nucleoids, as proposed by Pfalz and Pfannschmidt (2013). The homologs of PRPL10, rpl2, PRPS1, rps3, and rps4 are essential components of the protein biosynthetic machinery in Escherichia coli (Baba et al., 2006; Shoji et al., 2011) and the indispensability of rps3 and rps4 has been also proven in tobacco plastids (Rogalski et al., 2008; Fleischmann et al., 2011). Furthermore, PRPL10 is annotated as EMBryo defective 3136 (EMB3136) in the SeedGenes Project database (http://www.seedgenes.org/), and in its absence Arabidopsis embryo development arrests at the globular stage (Bryant et al., 2011). Mutants devoid of PRPS1 have not been described. However, given the conservation of PRPS1 function in prokaryotes and chloroplasts, it can be confidently assumed that complete lack of PRPS1 is lethal in Arabidopsis.
Taking into consideration the function of these ribosomal proteins, it can be argued that their interaction with GUN1 has a dual purpose. On the one hand, GUN1 modulates protein synthesis by controlling the abundance of PRPS1, which, together with rps3 and rps2, has been reported to form the domain responsible for the interaction of the 30S ribosomal subunit with mRNA, promoting translation initiation (Manuell et al., 2007; Tadini et al., 2016). This role is supported further by the stable interaction of GUN1 with the chloroplast translation initiation factor 2 (cpIF2; Tadini et al., 2016), also known as FUG1, and reported to be essential for chloroplast biogenesis (Miura et al., 2007).
On the other hand, GUN1 seems to be involved in the process of ribosome biogenesis too, since nucleoid-associated ribosomes are thought to be in various stages of assembly, with several rRNA maturation steps occurring in a co-transcriptional and assembly-assisted manner, as in prokaryotic systems (Bohne, 2014). For instance, the DEAD-box-containing, ATP-dependent RNA helicase 3 (RH3), which has been functionally linked to the chloroplast nucleoid (Majeran et al., 2012), is among the proteins that interact with GUN1 (Tadini et al., 2016; see also Figure 1 and Table 1). RH3 is directly involved in the splicing of group II introns in the rpl2, trnA, trnI, and rps12 transcripts and could be coimmunoprecipitated with immature and mature 23S rRNA (Asakura et al., 2012). Furthermore, the PPR protein At3g49240 also known as AtPPR_3g49240, according to the PPR protein database (http://www.plantenergy.uwa.edu.au/applications/ppr/ppr.php), is also part of GUN1's interactors, and its maize ortholog, GRMZM2G074599_P01, has been identified in the chloroplast nucleoid (Majeran et al., 2012). The gene is annotated as embryo defective 1796 (EMB1796) in the SeedGenes database, since the complete lack of AtPPR_3g49240 leads to the arrest of embryonic development at the globular stage (Cushing et al., 2005), further supporting the essential role of GUN1 interactors in chloroplast biogenesis.
GUN1 and the Import of Chloroplast Proteins
Almost a quarter of the GUN1 interactors identified by CoIP-MS are chaperones (see Table 1), a relatively high proportion when compared with the extensive repertoire of protein functions found within the nucleoid (Melonek et al., 2016). The stromal Hsp93 and Hsp70 chaperones mediate different steps in protein import into the chloroplast stroma, whereas the 60 KD chaperonin Cpn60 is thought to be involved in the folding of newly imported mature proteins and to function downstream of Hsp93 and Hsp70 (Kessler and Blobel, 1996; Jackson-Constan et al., 2001; Flores-Perez and Jarvis, 2013). Furthermore, the two genes most highly co-regulated with GUN1 encode the proteins TIC110 and TOC159 (Tadini et al., 2016), which are part of the outer and inner chloroplast translocons, respectively, suggesting a role of GUN1 in chloroplast protein import (Figure 2).
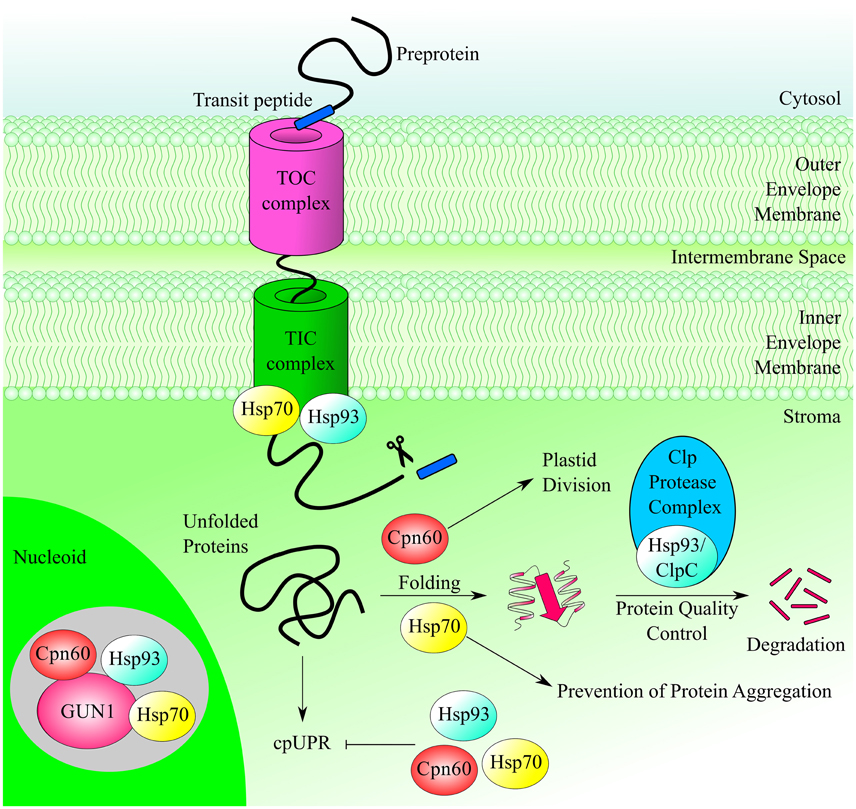
Figure 2. GUN1 interacts with different plastid chaperones. The chaperones Hsp93, Hsp70, and Cpn60 participate in different processes within the chloroplast, such as protein import, protein folding/unfolding, prevention of protein aggregation, and regulation of plastid division, and they might play a key role in the chloroplast Unfolded Protein Response (cpUPR). CoIP-MS analysis has shown that they are also part of GUN1-containing protein complexes.
The Hsp93 Chaperones
In Arabidopsis, there are two nearly identical isoforms of Hsp93, termed Hsp93-V and Hsp93-III (or ClpC1 and ClpC2, respectively) and both interact with GUN1. The two proteins are highly homologous, but Hsp93-V is expressed at much higher levels than Hsp93-III (Kovacheva et al., 2005, 2007), and only Hsp93-V has been reported as a component of the nucleoid proteome (Phinney and Thelen, 2005; Majeran et al., 2012; Melonek et al., 2012; Huang et al., 2013). Furthermore, both hsp93 single mutants are viable whereas hsp93-III hsp93-V double mutant is embryo-lethal, indicating that the two proteins have redundant functions in Arabidopsis chloroplasts (Constan et al., 2004; Sjögren et al., 2004; Kovacheva et al., 2007).
The current model for chloroplast protein import assumes that the preprotein transit peptide interacts with the TOC, and is subsequently transported through the TIC in an energy-dependent process (Shi and Theg, 2013b). In particular, the Tic110–Tic40 interaction is proposed to trigger the release of the transit peptide from Tic110 and enable the association of the preprotein with Hsp93 (Inaba et al., 2003). Tic40 then stimulates ATP hydrolysis by Hsp93, which harnesses the energy released to draw the preprotein into the stroma (Chou et al., 2006).
The Hsp70 Chaperones
Recent work has also demonstrated the involvement of Hsp70 in protein translocation into chloroplasts, as part of the translocon energy-dependent engine together with Hsp93 and Hsp90 (Inoue et al., 2013; Liu et al., 2014). Like Hsp93, Hsp70 proteins occur in two isoforms, Hsp70-1 and Hsp70-2, in the chloroplasts of Arabidopsis (Su and Li, 2008) and only Hsp70-1 was found in the proteomes of pTAC and crude nucleoids (for a review see Melonek et al., 2016). However, both Hsp70 proteins have been identified as GUN1 interactors (Tadini et al., 2016). Protein import assays using chloroplasts isolated from the Arabidopsis Hsp70 knockout mutants hsp70-1 and hsp70-2 showed that stromal Hsp70s are important for the import of both photosynthetic and non-photosynthetic precursor proteins, especially in early developmental stages (Su and Li, 2010). Furthermore, no hsp70-1 hsp70-2 double mutant has ever been isolated. Thus, the two Hsp70s are likely to have redundant functions that are essential for plant development and chloroplast biogenesis.
The Cpn60 Chaperonins
After preproteins delivered to the stroma have been processed, they may require accessory factors to enable them to fold into their functional conformation, or to reach their final intra-organellar destination. The stromal molecular chaperones Hsp70, Cpn60, and Cpn10 are all believed to mediate the folding or onward guidance of newly imported polypeptide chains (Boston et al., 1996; Jackson-Constan et al., 2001). In particular, immunoprecipitation experiments have revealed that Cpn60 operates in close proximity with Tic110 (Kessler and Blobel, 1996), while import experiments have shown a transient association of mature, newly imported proteins with the Cpn60-Tic110 complex, suggesting that Tic110 can recruit Cpn60 in an ATP-dependent manner for the folding of proteins upon their arrival in the stroma. It has also been suggested that stromal Hsp70 and Cpn60 act sequentially to facilitate the maturation of imported proteins, particularly those destined for the thylakoid membranes (Madueno et al., 1993; Tsugeki and Nishimura, 1993; Peng et al., 2011). The Arabidopsis genome encodes two members of the Cpn60α family, denoted ptCpn60α1 and ptCpn60α2, and four members of Cpn60β, known as ptCpn60β1–β4 (Suzuki et al., 2009). Two of them, ptCpn60α1 and ptCpn60β2, have been linked to the nucleoid proteome (Melonek et al., 2016), and ptCpn60α1 and ptCpn60β1 are among the GUN1 interactors identified via the CoIP-MS strategy (see Table 1). The complete loss of ptCpn60α1, in the mutant termed schlepperless (slp), causes retardation of embryo development before the heart stage and an albino seedling phenotype, indicating that ptCpn60α1 is essential for chloroplast biogenesis (Apuya et al., 2001). Conversely, plants devoid of ptCpn60β1, also known as lesion initiation 1 (len1), have leaves with wrinkled and irregular surfaces and undergo localized, spontaneous cell death in the absence of pathogen attack, i.e., lesion formation, under short-day conditions (Ishikawa et al., 2003).
Other Functions of Plastid Chaperones
Besides their roles in plastid protein import, all GUN1-interacting chaperones are present in the stroma at significant amounts relative to their association with the chloroplast import apparatus and perform various other functions together with different protein complexes (Figure 2). For instance Hsp93, also termed ClpC, acts as a regulatory chaperone in the Clp protease complex, the most abundant stromal protease with general household functions (Sakamoto, 2006; Van Wijk, 2015). Clp substrates are selected through various signals intrinsic to amino acid sequences and the ATP-dependent ClpC chaperone activity helps to progressively unfold selected substrates that are delivered to the ClpPR core for degradation into small peptides (~8–10 amino acids long; Olinares et al., 2011).
Similarly, Cpn60 forms a large oligomeric protein complex (>600 KDa) that promotes the assembly of Rubisco (Gutteridge and Gatenby, 1995). In particular, it has been observed that the large subunit of Rubisco (RbcL) is specifically associated with Cpn60 before assembly into the holoenzyme and that the Cpn60-RbcL complex is an obligatory intermediate. Furthermore, Cpn60 proteins have been shown to be essential for plastid division in A. thaliana (Suzuki et al., 2009). Thus, mesophyll cells in ptcpn60α1-1 (a missense mutant) and ptcpn60β1-1 (a protein null) plants, contain fewer and larger chloroplasts, indicating that normal levels of plastid Cpn60 are required for the correct folding of the stromal plastid division proteins and/or regulation of FtsZ (Filamentous temperature-sensitive Z) polymer dynamics (Suzuki et al., 2009).
The same holds true for the Hsp70 proteins, which are also involved in modulation of protein activity, regulation of protein degradation and prevention of irreversible protein aggregation when they are free in the stroma (Su and Li, 2008). Potentially GUN1 can be involved in a multitude of activities, besides plastid protein import, thus further investigations are needed to clarify the functional significance of GUN1–chaperone interactions.
GUN1 and the Chloroplast Unfolded Protein Response (cpUPR)
Chaperones, together with enzymes that process and degrade proteins, are also necessary to maintain protein folding homeostasis in the various compartments of eukaryotic cells. Distinct signal transduction pathways, known as unfolded protein responses (UPRs), have evolved to couple the unfolded/misfolded protein load to the expression of specific chaperones and enzymes that promote folding and the disposal of misfolded proteins in each compartment.
The unfolded protein response was first discovered in the endoplasmic reticulum (ER) in yeast, where inhibition of protein folding leads to the transcriptional up-regulation of several chaperones (Cox et al., 1993), and subsequently in mitochondria, where accumulation of unfolded proteins in the mitochondrial matrix stimulates the expression of nuclear gene transcripts coding for mitochondrial chaperones (Aldridge et al., 2007; Lin and Haynes, 2016). Compared to yeast and metazoans, studies of plant UPRs are less advanced, and molecular details are known mainly for the ER-dependent UPR, which shows certain similarities with the process in multicellular eukaryotes, as well as plant-specific features (Ruberti et al., 2015). Recently, the possible existence of a chloroplast UPR (cpUPR) has been investigated in the green alga Chlamydomonas reinhardtii. Taking advantage of a repressible chloroplast gene expression system (Rochaix et al., 2014), Ramundo et al. (2014) induced the selective gradual depletion of the essential stromal Clp protease, in order to follow the early and late events caused by the decrease in its abundance. Temporal profiles of gene expression and protein accumulation revealed a marked increase in levels of chaperones, including Hsp70B, upon Clp depletion. Similar data have also been reported for Arabidopsis, where up-regulation of chloroplast chaperones and protein-sorting components occurred upon constitutive repression of Clp (Rudella et al., 2006; Zybailov et al., 2009). In particular, characterization of total leaf proteomes of WT and clpr2-1 highlighted differential expression of 768 proteins. The largest functional category quantified (with 205 proteins) comprised proteins involved in translation, folding and degradation. Strikingly, all the chaperones interacting with GUN1, including Hsp93, Hsp70, Cpn60, as well as the DEAD box RNA helicase RH3, are among those up-regulated (by between 1.6- and 8.5-fold) in clpr2-1 leaves, whereas no significant change in the chloroplast ribosomal protein population was observed (Zybailov et al., 2009).
Taken together, these findings suggest that disruption of protein homeostasis in organelles can be sensed and transduced to the nucleus to induce the expression of a specific set of factors responsible for promoting folding and monitoring protein quality control (Ramundo and Rochaix, 2014; Rochaix and Ramundo, 2015). After entering the higher plant chloroplast, these factors are able to interact with the nucleoid-associated GUN1 protein (Figure 2), which might therefore play a role in the cpUPR process.
GUN1 and Chloroplast Metabolism
The large subunit of ribulose bisphosphate carboxylase (RbcL) and the β subunit of the ATP synthase are also among the interactors of GUN1 identified by CoIP-MS analysis (Tadini et al., 2016). Because of their relatively high abundance in the chloroplast proteome, it is tempting to assume that these proteins are simply contaminants. However, RbcL and subunits of the ATP synthase have been repeatedly identified in the pTAC/nucleoid proteomes, even though different procedures were employed for isolation of crude nucleoid fractions and highly purified pTAC complexes (for a review see Melonek et al., 2016), thus suggesting these proteins might have a dual localization to the chloroplast stroma and nucleoids. The nucleoid association of RbcL and ATP synthase, i.e., of proteins that are not directly involved in core nucleoid functions, might also indicate that nucleoids also monitor photosynthesis and energy metabolism and respond appropriately to any perturbations (Figure 3).
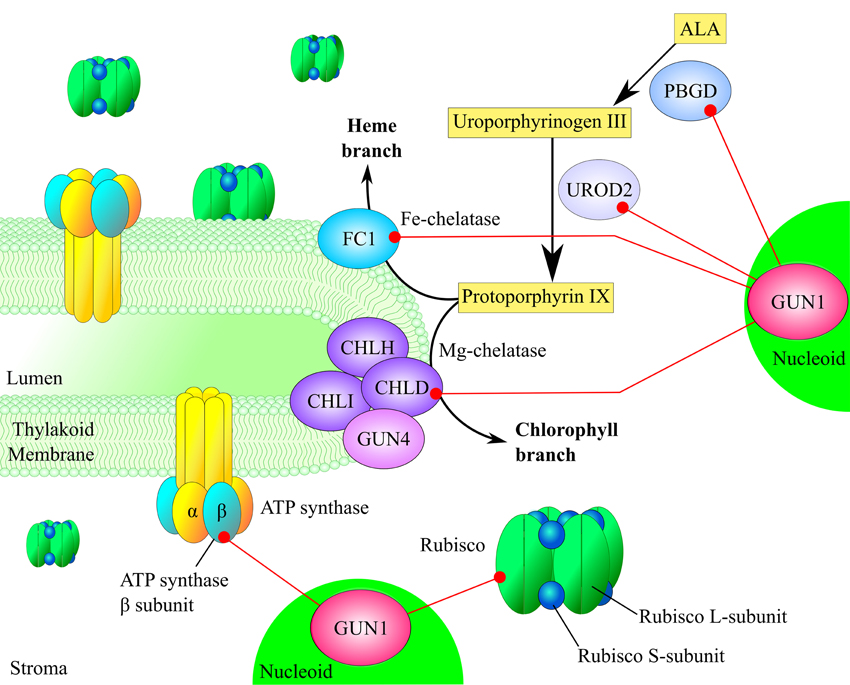
Figure 3. GUN1 is involved in photosynthesis and tetrapyrrole biosynthesis. The large subunit of Rubisco and the β-subunit of the thylakoid ATP synthase have been coimmunoprecipitated with GUN1, supporting a role for GUN1 in coordinating nucleoid activities with chloroplast metabolism. GUN1 also interacts with four enzymes of the tetrapyrrole biosynthesis pathway, i.e., the D subunit of Mg chelatase (CHLD), porphobilinogen deaminase (PBGD), uroporphyrinogen III decarboxylase (UROD2), and ferrochelatase I (FC1), as shown by yeast two-hybrid and Bimolecular Fluorescence Complementation. Note that the proteins RER4 and 2-Cys PrxA have not been included in this scheme for reasons of clarity.
Unlike RbcL and the ATP synthase β subunit, RETICULATA-RELATED 4 (RER4), an integral component of the chloroplast envelope membranes with three transmembrane α-helices, has never been identified in the pTAC/nucleoid proteome, although it appears to be an interactor of GUN1 (Table 1). The mutant rer4-1 exhibits leaf reticulation, having green veins that stand out against paler intervein tissue, with fewer and smaller mesophyll cells than those of the wild type leaves (Perez-Perez et al., 2013). The molecular function of RER4 remains to be established. However, some hints as to its role in the chloroplast can be derived from features of the rer4-1 mutant phenotype. A possible involvement of RER4 in retrograde signaling is suggested by the altered growth and development of mesophyll cells. Alternatively, the absence of RER4 might deplete the supply of essential metabolites during early stages of leaf development, which could explain the aberrant mesophyll structure. Furthermore, RER4 has been suggested to be involved in the control of reactive oxygen species (ROS), since the reticulated pigmentation of the rer4-1 mutant grown under long-day conditions can be rescued by a short-day photoperiod, which markedly dampens ROS accumulation.
The 2-Cys peroxiredoxin A (2-Cys Prx A; see also Table 1), another interactor with GUN1, appears also to have a role in ROS scavenging (Rey et al., 2007; Pulido et al., 2010; Dietz, 2016) and, like RER4, it has never been reported to be part of the pTAC/nucleoid proteome (Pfalz et al., 2006; Majeran et al., 2012; Huang et al., 2013). 2-Cys Prx A and the highly homologous 2-Cys Prx B function as redox sensors and chaperones, thanks to the flexibility of their protein structure (König et al., 2013), and they have been shown to control the conversion of Mg-protoporphyrin monomethyl ester into protochlorophyllide (Stenbaek et al., 2008).
The involvement of GUN1 in TPB is further supported by its interaction with four TPB enzymes, namely subunit D of Mg chelatase (CHLD), porphobilinogen deaminase (PBGD), uroporphyrinogen III decarboxylase (UROD2), and ferrochelatase I (FC1), as demonstrated by both yeast two-hybrid and BiFC assays (Tadini et al., 2016; Figure 3). Interestingly, mutants defective in three of these GUN1 interactors—CHLD, PBGD, and FC1—have themselves been described as gun mutants (Strand et al., 2003; Huang and Li, 2009; Woodson et al., 2011), but have never been identified in crude nucleoid preparations, unlike subunit I of Mg chelatase (CHLI; Melonek et al., 2012; Huang et al., 2013).
GUN1 and Plastid Protein Homeostasis: Some Testable Hypotheses
The recent identification of the GUN1 protein's partners in chloroplasts of Arabidopsis by means of CoIP-MS studies as well as in yeast two-hybrid and BiFC assays (Tadini et al., 2016) strongly suggests a major role for GUN1 in plastid protein homeostasis (Figure 4). This regulatory role involves proteins that are, in most cases, members of multimeric protein complexes and whose functions are often context-dependent. Furthermore, most GUN1 interactors appear to participate in four major processes:
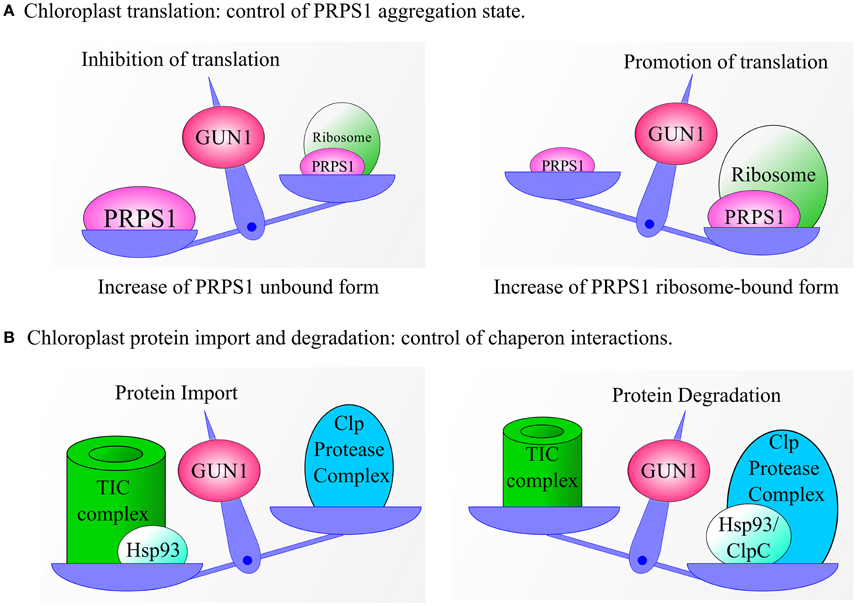
Figure 4. Models explaining GUN1-dependent regulation of chloroplast translation, protein import and protein degradation. (A) GUN1 controls the abundance of PRPS1 and its aggregation state. Increased levels of free PRPS1 prevent loading of mRNAs onto the ribosome and inhibit polysome formation, thus reducing overall rates of protein synthesis in the plastid. Conversely, when PRPS1 binds to ribosomes, polysome formation, and protein translation are stimulated. (B) Under certain conditions, the interaction between GUN1 and the Hsp93/ClpC protein might serve to bring the chaperone into close proximity with the TIC complex, thus favoring plastid protein import and reducing protein degradation. Alternatively, GUN1 could favor the interaction of Hsp93/ClpC with the Clp protease, thus promoting protein degradation at the expense of protein import. Note that a similar pattern of behavior can also be proposed for the other GUN1-interacting chaperones.
Chloroplast Protein Synthesis
A wealth of evidence accumulated during the last two decades supports the primacy of plastid protein synthesis in the control of chloroplast gene expression (Choquet and Wollman, 2002; Manuell et al., 2007; Tiller and Bock, 2014; Sun and Zerges, 2015). In this context, GUN1 has been suggested to regulate translation in plastids by modulating the abundance and binding affinity of PRPS1 (Tadini et al., 2016). In particular, PRPS1 is the only ribosomal protein that shuttles between ribosome-bound and ribosome-free forms (Merendino et al., 2003; Delvillani et al., 2011), the latter being more abundant in plants that lack GUN1. Based on observations in E. coli, where the unbound form is thought to inhibit translation by competing with ribosomes for mRNAs (Delvillani et al., 2011), it can be argued that the GUN1-dependent equilibrium between the two PRPS1 states has an important role in controlling polysome assembly and protein synthesis in chloroplasts (Figure 4A). However, further investigations are needed to clarify this issue. For instance, lines characterized by the ectopic expression of PRPS1 or carrying PRPS1 constructs under the control of inducible promoters, coupled with assays aimed to measure the translation rate in plastids, should allow us to verify the role of PRPS1 in modulating protein synthesis. Furthermore, GUN1 controls the abundance of PRPS1 at the post-transcriptional level. This suggests the involvement of an as yet unidentified plastid protease in this aspect of GUN1 function. In addition, the significance of the interaction of GUN1 with other ribosomal proteins, factors involved in ribosome biogenesis and regulators of plastid protein synthesis remains to be elucidated.
Chloroplast Protein Import and Degradation
Based on the observations reported above, it appears that GUN1 may well control the interactions of a sub-set of chaperones, promoting plastid protein import when their association with the TIC complex is favored, and stimulating protein degradation, folding/unfolding when they interact with proteases or other protein complexes in the stroma or in the thylakoid membranes (Figure 4B). Such a regulatory mechanism would enable GUN1 to coordinate protein translocation across the chloroplast envelope with protein degradation in the stroma, as well as with plastid division, thus modulating the protein content of the chloroplast in accordance with physiological requirements.
Relatively simple biochemical analyses can be used to verify the importance of GUN1 in influencing the interactions of the stromal chaperones, such as protein complex fractionation via sucrose-gradient ultracentrifugation and/or Blue-Native PAGE coupled with two-dimensional (2D) SDS-PAGE, and immunoblot analyses. Furthermore, the interactions of GUN1 with chaperones should be shown to occur at the plastid envelope and protein import efficiency should be tested in chloroplasts isolated from gun1 and WT seedlings in order to implicate GUN1 in regulating plastid protein import.
Retrograde Signal Induction
GUN1 may well be a master regulator of plastid-to-nucleus communication in A. thaliana, as it appears to integrate signals derived from perturbations in PGE, TPB, and redox state, in order to modulate nuclear gene expression. Indeed, components of all three pathways have been shown to interact with GUN1, suggesting that signal integration might take place through physical interaction.
Due to the limited abundance of GUN1, as indicated by the fact that the protein has yet to be detected in plastid proteome studies, it is tempting to disregard the idea that its physical interaction with PGE-, TPB-, and redox-related proteins could lead to protein sequestration and directly to differences in protein translation, TPB, and redox balance (Koussevitzky et al., 2007; Pogson et al., 2008; Woodson and Chory, 2008; Kleine and Leister, 2016). Nevertheless, a direct association with GUN1 could control protein abundance through post-transcriptional mechanisms, as in the case of PRPS1 and CHLD (Tadini et al., 2016). Thus, control of CHLD and possibly of FC1 levels could alter the tetrapyrrole flux and influence the abundance of the tetrapyrrole intermediate Mg-protoporphyrin IX (Mg-ProtoIX), or the tetrapyrrole product Fe-protoporphyrin IX (heme), which have been reported to act as negative and positive retrograde signals, respectively (for a review, see Chan et al., 2016). Alternatively, the interaction of GUN1 with the near-identical paralogs ClpC1 and ClpC2 could contribute to the coordination of plastid protein content with tetrapyrrole biosynthesis. Indeed, the activity of the stromal Clp protease has been shown to modulate tetrapyrrole flux by controlling (i) the accumulation of chlorophyll a oxygenase, which converts chlorophyll a into chlorophyll b (Nakagawara et al., 2007), and (ii) the level of glutamyl-tRNA reductase (GluTR), thus regulating the rate-limiting reaction in tetrapyrrole synthesis—the conversion of glutamate-1-semialdehyde into 5-aminolevulinic acid (Apitz et al., 2016).
Therefore, accurate determination of tetrapyrrole intermediates should be performed in gun1 mutant and WT backgrounds. The analyses should be restricted to young seedlings or even to different developmental stages of the chloroplast, in line with the roles of tetrapyrrole and GUN1-mediated signaling in chloroplast development.
Concluding Remarks
In the past decade, substantial progress has been made in elucidating retrograde signaling, with the identification of multiple retrograde pathways and more than 40 components involved at different levels in chloroplast-to-nucleus communication. Nevertheless, the molecular function of GUN1 has remained unclear until the recent identification of the GUN1 protein's partners. Based on the functional roles of GUN1 interactors and the embryo lethal or albino phenotypes of most of the corresponding knock-out mutants, we have learned that GUN1 plays a role in chloroplast biogenesis, possibly by controlling protein turnover and protein import, and through the coordination of plastid and nuclear gene expression. Furthermore, GUN1 could have a role in the cpUPR process. Nonetheless, the involvement of GUN1 in plastid biogenesis and protein homeostasis is only just beginning to be understood. For instance, other approaches will be needed to validate the GUN1's protein partners identified by CoIP-MS. The use of a GUN1-GFP protein chimera, expressed under the control of a strong constitutive promoter such as the Cauliflower Mosaic Virus 35S (35S-CaMV), is indeed prone to the identification of false interactors. CoIP-MS studies using a GUN1 specific antibody appears to be the ideal strategy to identify protein partners. Alternatively, the use of GUN1 chimeras under the control of GUN1 native promoter is also practicable. Moreover, we do not know whether all these activities take place within one GUN1-containing nucleoid or if there are different nucleoids/locations for each GUN1-dependent function. The developmental stages of the chloroplast itself may even show distinct patterns of compartmentalization of the different functions. In addition, GUN1's interactions with its diverse partners might have quite different functional consequences: (i) promote specific functions, by bringing enzymes into close proximity with their own substrates and, ultimately, controlling the enzyme abundance, (ii) inhibit processes by sequestering sub-pools of specific proteins and, also in this case, controlling their abundance.
We are confident that future work, based on the exciting breakthroughs discussed in this Review, will shed new light on the molecular functions of GUN1 and its involvement in chloroplast biogenesis and protein homeostasis.
Author Contributions
MC, LT, CP, RF, and PP participated to the organization of the manuscript. MC and PP designed and conceived the pictures. PP wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by ERA-NET Cofund FACCE SURPLUS (BarPLUS grant id. 93). We thank Paul Hardy for critical reading of the manuscript. The authors wish to acknowledge the reviewers for the detailed and helpful comments to the manuscript.
References
Adhikari, N. D., Froehlich, J. E., Strand, D. D., Buck, S. M., Kramer, D. M., and Larkin, R. M. (2011). GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23, 1449–1467. doi: 10.1105/tpc.110.082503
Agee, A. E., Surpin, M., Sohn, E. J., Girke, T., Rosado, A., Kram, B. W., et al. (2010). MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 152, 120–132. doi: 10.1104/pp.109.145078
Aldridge, J. E., Horibe, T., and Hoogenraad, N. J. (2007). Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE 2:e874. doi: 10.1371/journal.pone.0000874
Apitz, J., Nishimura, K., Schmied, J., Wolf, A., Hedtke, B., Van Wijk, K. J., et al. (2016). Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol. 170, 2040–2051. doi: 10.1104/pp.15.01945
Apuya, N. R., Yadegari, R., Fischer, R. L., Harada, J. J., Zimmerman, J. L., and Goldberg, R. B. (2001). The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60alpha gene. Plant Physiol. 126, 717–730. doi: 10.1104/pp.126.2.717
Asakura, Y., Galarneau, E., Watkins, K. P., Barkan, A., and Van Wijk, K. J. (2012). Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 159, 961–974. doi: 10.1104/pp.112.197525
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008. doi: 10.1038/msb4100050
Bohne, A. V. (2014). The nucleoid as a site of rRNA processing and ribosome assembly. Front. Plant Sci. 5:257. doi: 10.3389/fpls.2014.00257
Boston, R. S., Viitanen, P. V., and Vierling, E. (1996). Molecular chaperones and protein folding in plants. Plant Mol. Biol. 32, 191–222. doi: 10.1007/BF00039383
Bryant, N., Lloyd, J., Sweeney, C., Myouga, F., and Meinke, D. (2011). Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 155, 1678–1689. doi: 10.1104/pp.110.168120
Chan, K. X., Phua, S. Y., Crisp, P., McQuinn, R., and Pogson, B. J. (2016). Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. doi: 10.1146/annurev-arplant-043015-111854
Choquet, Y., and Wollman, F. A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529, 39–42. doi: 10.1016/S0014-5793(02)03260-X
Chou, M. L., Chu, C. C., Chen, L. J., Akita, M., and Li, H. M. (2006). Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175, 893–900. doi: 10.1083/jcb.200609172
Constan, D., Froehlich, J. E., Rangarajan, S., and Keegstra, K. (2004). A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136, 3605–3615. doi: 10.1104/pp.104.052928
Cox, J. S., Shamu, C. E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206. doi: 10.1016/0092-8674(93)90648-A
Cushing, D. A., Forsthoefel, N. R., Gestaut, D. R., and Vernon, D. M. (2005). Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221, 424–436. doi: 10.1007/s00425-004-1452-x
Delvillani, F., Papiani, G., Dehò, G., and Briani, F. (2011). S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acids Res. 39, 7702–7715. doi: 10.1093/nar/gkr417
Dietz, K. J. (2016). Thiol-based peroxidases and ascorbate peroxidases: why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol. Cells 39, 20–25. doi: 10.14348/molcells.2016.2324
Fleischmann, T. T., Scharff, L. B., Alkatib, S., Hasdorf, S., Schöttler, M. A., and Bock, R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23, 3137–3155. doi: 10.1105/tpc.111.088906
Flores-Pérez, U., and Jarvis, P. (2013). Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 1833, 332–340. doi: 10.1016/j.bbamcr.2012.03.019
Gray, J. C., Sullivan, J. A., Wang, J. H., Jerome, C. A., and Maclean, D. (2003). Coordination of plastid and nuclear gene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 135–144. discussion 144–145. doi: 10.1098/rstb.2002.1180
Gutteridge, S., and Gatenby, A. A. (1995). Rubisco synthesis, assembly, mechanism, and regulation. Plant Cell 7, 809–819. doi: 10.1105/tpc.7.7.809
Huang, M., Friso, G., Nishimura, K., Qu, X., Olinares, P. D., Majeran, W., et al. (2013). Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J. Proteome Res. 12, 491–504. doi: 10.1021/pr300952g
Huang, Y. S., and Li, H. M. (2009). Arabidopsis CHLI2 can substitute for CHLI1. Plant Physiol. 150, 636–645. doi: 10.1104/pp.109.135368
Inaba, T., Li, M., Alvarez-Huerta, M., Kessler, F., and Schnell, D. J. (2003). atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278, 38617–38627. doi: 10.1074/jbc.M306367200
Inoue, H., Li, M., and Schnell, D. J. (2013). An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 110, 3173–3178. doi: 10.1073/pnas.1219229110
Ishikawa, A. (2005). Tetrapyrrole metabolism is involved in lesion formation, cell death, in the Arabidopsis lesion initiation 1 mutant. Biosci. Biotechnol. Biochem. 69, 1929–1934. doi: 10.1271/bbb.69.1929
Ishikawa, A., Tanaka, H., Nakai, M., and Asahi, T. (2003). Deletion of a chaperonin 60 beta gene leads to cell death in the Arabidopsis lesion initiation 1 mutant. Plant Cell Physiol. 44, 255–261. doi: 10.1093/pcp/pcg031
Jackson-Constan, D., Akita, M., and Keegstra, K. (2001). Molecular chaperones involved in chloroplast protein import. Biochim. Biophys. Acta 1541, 102–113. doi: 10.1016/S0167-4889(01)00148-3
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Kato, Y., and Sakamoto, W. (2010). New insights into the types and function of proteases in plastids. Int. Rev. Cell Mol. Biol. 280, 185–218. doi: 10.1016/S1937-6448(10)80004-8
Kessler, F., and Blobel, G. (1996). Interaction of the protein import and folding machineries of the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 93, 7684–7689. doi: 10.1073/pnas.93.15.7684
Kleine, T., and Leister, D. (2016). Retrograde signaling: organelles go networking. Biochim. Biophys. Acta. 1857, 1313–1325. doi: 10.1016/j.bbabio.2016.03.017
König, J., Galliardt, H., Jütte, P., Schäper, S., Dittmann, L., and Dietz, K. J. (2013). The conformational bases for the two functionalities of 2-cysteine peroxiredoxins as peroxidase and chaperone. J. Exp. Bot. 64, 3483–3497. doi: 10.1093/jxb/ert184
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. doi: 10.1126/science.1140516
Kovacheva, S., Bédard, J., Patel, R., Dudley, P., Twell, D., Ríos, G., et al. (2005). In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41, 412–428. doi: 10.1111/j.1365-313X.2004.02307.x
Kovacheva, S., Bédard, J., Wardle, A., Patel, R., and Jarvis, P. (2007). Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50, 364–379. doi: 10.1111/j.1365-313X.2007.03060.x
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. doi: 10.1126/science.1079978
Lin, Y. F., and Haynes, C. M. (2016). Metabolism and the UPR(mt). Mol. Cell 61, 677–682. doi: 10.1016/j.molcel.2016.02.004
Liu, L., McNeilage, R. T., Shi, L. X., and Theg, S. M. (2014). ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26, 1246–1255. doi: 10.1105/tpc.113.121822
Madueno, F., Napier, J. A., and Gray, J. C. (1993). Newly imported rieske iron-sulfur protein associates with Both Cpn60 and Hsp70 in the chloroplast stroma. Plant Cell 5, 1865–1876. doi: 10.1105/tpc.5.12.1865
Majeran, W., Friso, G., Asakura, Y., Qu, X., Huang, M., Ponnala, L., et al. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 158, 156–189. doi: 10.1104/pp.111.188474
Manuell, A. L., Quispe, J., and Mayfield, S. P. (2007). Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol. 5:e209. doi: 10.1371/journal.pbio.0050209
Melonek, J., Matros, A., Trösch, M., Mock, H. P., and Krupinska, K. (2012). The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell 24, 3060–3073. doi: 10.1105/tpc.112.099721
Melonek, J., Oetke, S., and Krupinska, K. (2016). Multifunctionality of plastid nucleoids as revealed by proteome analyses. Biochim. Biophys. Acta. 1864, 1016–1038. doi: 10.1016/j.bbapap.2016.03.009
Merendino, L., Falciatore, A., and Rochaix, J. D. (2003). Expression and RNA binding properties of the chloroplast ribosomal protein S1 from Chlamydomonas reinhardtii. Plant Mol. Biol. 53, 371–382. doi: 10.1023/B:PLAN.0000006941.56233.42
Miura, E., Kato, Y., Matsushima, R., Albrecht, V., Laalami, S., and Sakamoto, W. (2007). The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19, 1313–1328. doi: 10.1105/tpc.106.049270
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2053–2058. doi: 10.1073/pnas.98.4.2053
Nakagawara, E., Sakuraba, Y., Yamasato, A., Tanaka, R., and Tanaka, A. (2007). Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J. 49, 800–809. doi: 10.1111/j.1365-313X.2006.02996.x
Oelmüller, R., Levitan, I., Bergfeld, R., Rajasekhar, V. K., and Mohr, H. (1986). Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168, 482–492. doi: 10.1007/BF00392267
Oelmüller, R., and Mohr, H. (1986). Photooxidative destruction of chloroplasts and its consequences for expression of nuclear genes. Planta 167, 106–113. doi: 10.1007/BF00446376
Okazaki, K., Kabeya, Y., and Miyagishima, S. Y. (2010). The evolution of the regulatory mechanism of chloroplast division. Plant Signal. Behav. 5, 164–167. doi: 10.4161/psb.5.2.10461
Olinares, P. D., Kim, J., and van Wijk, K. J. (2011). The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta 1807, 999–1011. doi: 10.1016/j.bbabio.2010.12.003
Peng, L., Fukao, Y., Myouga, F., Motohashi, R., Shinozaki, K., and Shikanai, T. (2011). A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol. 9:e1001040. doi: 10.1371/journal.pbio.1001040
Pérez-Pérez, J. M., Esteve-Bruna, D., González-Bayón, R., Kangasjarvi, S., Caldana, C., Hannah, M. A., et al. (2013). Functional redundancy and divergence within the Arabidopsis RETICULATA-RELATED gene family. Plant Physiol. 162, 589–603. doi: 10.1104/pp.113.217323
Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K. J., and Oelmüller, R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18, 176–197. doi: 10.1105/tpc.105.036392
Pfalz, J., and Pfannschmidt, T. (2013). Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18, 186–194. doi: 10.1016/j.tplants.2012.11.003
Phinney, B. S., and Thelen, J. J. (2005). Proteomic characterization of a triton-insoluble fraction from chloroplasts defines a novel group of proteins associated with macromolecular structures. J. Proteome Res. 4, 497–506. doi: 10.1021/pr049791k
Pogson, B. J., Woo, N. S., Förster, B., and Small, I. D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. doi: 10.1016/j.tplants.2008.08.008
Pulido, P., Spínola, M. C., Kirchsteiger, K., Guinea, M., Pascual, M. B., Sahrawy, M., et al. (2010). Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J. Exp. Bot. 61, 4043–4054. doi: 10.1093/jxb/erq218
Ramundo, S., Casero, D., Mühlhaus, T., Hemme, D., Sommer, F., Crèvecoeur, M., et al. (2014). Conditional depletion of the chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell 26, 2201–2222. doi: 10.1105/tpc.114.124842
Ramundo, S., Rahire, M., Schaad, O., and Rochaix, J. D. (2013). Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in chlamydomonas. Plant Cell 25, 167–186. doi: 10.1105/tpc.112.103051
Ramundo, S., and Rochaix, J. D. (2014). Chloroplast unfolded protein response, a new plastid stress signaling pathway? Plant Signal. Behav. 9:e972874. doi: 10.4161/15592316.2014.972874
Rey, P., Becuwe, N., Barrault, M. B., Rumeau, D., Havaux, M., Biteau, B., et al. (2007). The Arabidopsis thaliana sulfiredoxin is a plastidic cysteine-sulfinic acid reductase involved in the photooxidative stress response. Plant J. 49, 505–514. doi: 10.1111/j.1365-313X.2006.02969.x
Richly, E., and Leister, D. (2004). An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329, 11–16. doi: 10.1016/j.gene.2004.01.008
Rochaix, J. D., and Ramundo, S. (2015). Conditional repression of essential chloroplast genes: evidence for new plastid signaling pathways. Biochim. Biophys. Acta 1847, 986–992. doi: 10.1016/j.bbabio.2014.11.011
Rochaix, J. D., Surzycki, R., and Ramundo, S. (2014). Tools for regulated gene expression in the chloroplast of Chlamydomonas. Methods Mol. Biol. 1132, 413–424. doi: 10.1007/978-1-62703-995-6_28
Rogalski, M., Schöttler, M. A., Thiele, W., Schulze, W. X., and Bock, R. (2008). Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20, 2221–2237. doi: 10.1105/tpc.108.060392
Ruberti, C., Kim, S. J., Stefano, G., and Brandizzi, F. (2015). Unfolded protein response in plants: one master, many questions. Curr. Opin. Plant Biol. 27, 59–66. doi: 10.1016/j.pbi.2015.05.016
Rudella, A., Friso, G., Alonso, J. M., Ecker, J. R., and Van Wijk, K. J. (2006). Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18, 1704–1721. doi: 10.1105/tpc.106.042861
Sakamoto, W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57, 599–621. doi: 10.1146/annurev.arplant.57.032905.105401
Shi, L. X., and Theg, S. M. (2010). A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22, 205–220. doi: 10.1105/tpc.109.071464
Shi, L. X., and Theg, S. M. (2013a). The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833, 314–331. doi: 10.1016/j.bbamcr.2012.10.002
Shi, L. X., and Theg, S. M. (2013b). Energetic cost of protein import across the envelope membranes of chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 110, 930–935. doi: 10.1073/pnas.1115886110
Shoji, S., Dambacher, C. M., Shajani, Z., Williamson, J. R., and Schultz, P. G. (2011). Systematic chromosomal deletion of bacterial ribosomal protein genes. J. Mol. Biol. 413, 751–761. doi: 10.1016/j.jmb.2011.09.004
Sjögren, L. L., Macdonald, T. M., Sutinen, S., and Clarke, A. K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136, 4114–4126. doi: 10.1104/pp.104.053835
Steiner, S., Schröter, Y., Pfalz, J., and Pfannschmidt, T. (2011). Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157, 1043–1055. doi: 10.1104/pp.111.184515
Stenbaek, A., Hansson, A., Wulff, R. P., Hansson, M., Dietz, K. J., and Jensen, P. E. (2008). NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 582, 2773–2778. doi: 10.1016/j.febslet.2008.07.006
Strand, A., Asami, T., Alonso, J., Ecker, J. R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. doi: 10.1038/nature01204
Su, P. H., and Li, H. M. (2008). Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 146, 1231–1241. doi: 10.1104/pp.107.114496
Su, P. H., and Li, H. M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531. doi: 10.1105/tpc.109.071415
Sun, Y., and Zerges, W. (2015). Translational regulation in chloroplasts for development and homeostasis. Biochim. Biophys. Acta 1847, 809–820. doi: 10.1016/j.bbabio.2015.05.008
Susek, R. E., Ausubel, F. M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. doi: 10.1016/0092-8674(93)90459-4
Suzuki, K., Nakanishi, H., Bower, J., Yoder, D. W., Osteryoung, K. W., and Miyagishima, S. Y. (2009). Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol. 9:38. doi: 10.1186/1471-2229-9-38
Tadini, L., Pesaresi, P., Kleine, T., Rossi, F., Guljamow, A., Sommer, F., et al. (2016). GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 170, 1817–1830. doi: 10.1104/pp.15.02033
Tanaka, R., Kobayashi, K., and Masuda, T. (2011). Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book 9:e0145. doi: 10.1199/tab.0145
Tiller, N., and Bock, R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120. doi: 10.1093/mp/ssu022
Tsugeki, R., and Nishimura, M. (1993). Interaction of homologues of Hsp70 and Cpn60 with ferredoxin-NADP+ reductase upon its import into chloroplasts. FEBS Lett. 320, 198–202. doi: 10.1016/0014-5793(93)80585-I
Van Wijk, K. J. (2015). Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu. Rev. Plant Biol. 66, 75–111. doi: 10.1146/annurev-arplant-043014-115547
Woodson, J. D., and Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. doi: 10.1038/nrg2348
Woodson, J. D., Perez-Ruiz, J. M., and Chory, J. (2011). Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21, 897–903. doi: 10.1016/j.cub.2011.04.004
Zybailov, B., Friso, G., Kim, J., Rudella, A., Rodríguez, V. R., Asakura, Y., et al. (2009). Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteomics 8, 1789–1810. doi: 10.1074/mcp.M900104-MCP200
Keywords: nucleoid, GUN1, protein homeostasis, retrograde signaling, biogenic control
Citation: Colombo M, Tadini L, Peracchio C, Ferrari R and Pesaresi P (2016) GUN1, a Jack-Of-All-Trades in Chloroplast Protein Homeostasis and Signaling. Front. Plant Sci. 7:1427. doi: 10.3389/fpls.2016.01427
Received: 21 July 2016; Accepted: 07 September 2016;
Published: 22 September 2016.
Edited by:
Fiammetta Alagna, Research Council for Agriculture and Agricultural Economy, ItalyReviewed by:
Lynn Richardson, Michigan State University, USASilvia Ramundo, University of California, San Francisco & Carnegie Institution for Science & Howard Hughes Medical Institute, USA
Copyright © 2016 Colombo, Tadini, Peracchio, Ferrari and Pesaresi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paolo Pesaresi, paolo.pesaresi@unimi.it
 Monica Colombo1
Monica Colombo1 Luca Tadini
Luca Tadini Paolo Pesaresi
Paolo Pesaresi