- 1Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 2Asian Natural Environmental Science Center, The University of Tokyo, Tokyo, Japan
- 3Institute of Plant Science and Resources, Okayama University, Kurashiki, Japan
- 4Institute for Epidemiology and Pathogen Diagnostics, Julius Kühn-Institut, Quedlinburg, Germany
In this study, we investigated the barley yellow mosaic virus (BaYMV, genus Bymovirus) factor(s) responsible for breaking eIF4E-mediated recessive resistance genes (rym4/5/6) in barley. Genome mapping analysis using chimeric infectious cDNA clones between rym5-breaking (JT10) and rym5-non-breaking (JK05) isolates indicated that genome-linked viral protein (VPg) is the determinant protein for breaking the rym5 resistance. Likewise, VPg is also responsible for overcoming the resistances of rym4 and rym6 alleles. Mutational analysis identified that amino acids Ser-118, Thr-120, and His-142 in JT10 VPg are the most critical residues for overcoming rym5 resistance in protoplasts. Moreover, the rym5-non-breaking JK05 could accumulate in the rym5 protoplasts when eIF4E derived from a susceptible barley cultivar was expressed from the viral genome. Thus, the compatibility between VPg and host eIF4E determines the ability of BaYMV to infect barley plants.
Introduction
Viral diseases cause serious economic losses by reducing both the quality and quantity of crop production (Kang et al., 2005a). The breeding of resistant crop plants is the most acceptable and applicable approach to control viral diseases because it is cost effective in terms of labor and material resources and, most importantly, it has no negative impact on the environment, with the exception of imposing strong selective pressure on virus populations (Clay and Kover, 1996; Kang et al., 2005a; Zhu et al., 2012). Of all known resistances against plant viruses, a large fraction is recessively inherited, with the majority of genes encoding for eukaryotic translation initiation factors eIF4E, eIF4G, or their isoforms (Fraile and García-Arenal, 2012; Wang and Krishnaswamy, 2012; Sanfaçon, 2015).
The eIF4E-mediated resistances in crop plants confer qualitative, genotype-specific resistance to viruses in the genus Potyvirus of the family Potyviridae (Kyle and Palloix, 1997; Ruffel et al., 2002, 2005, 2006; Nicaise et al., 2003; Gao et al., 2004; Kang et al., 2005a; Bruun-Rasmussen et al., 2007; Roudet-Tavert et al., 2007; Charron et al., 2008; Andrade et al., 2009; Naderpour et al., 2010) and occur through amino acid substitutions in eIF4E proteins encoded by the alleles (Ruffel et al., 2006; Yeam et al., 2007). The transient expression of eIF4E from susceptible cultivars renders the resistant cultivars to be susceptible to a certain virus pathotype (Ruffel et al., 2002, 2006). eIF4E in eukaryotic cells is an essential translation initiation factor that recruits the small ribosomal subunit (40S) to the mRNA cap structure, an event which is considered the first step in cap-dependent translation initiation (Malys and McCarthy, 2011). Moreover, it has been hypothesized that eIF4E also plays multiple roles in diverse processes during potyvirus infection, including translation, replication and cell-to-cell movement (Robaglia and Caranta, 2006; Truniger and Aranda, 2009). However, the precise roles of eIF4E during virus infection have yet to be demonstrated.
The viruses in the family Potyviridae contain a monopartite or bipartite (in the genus Bymovirus) positive-sense RNA genome that has a genome-linked viral protein (VPg) at the 5′ end and is polyadenylated at the 3′ end (Adams et al., 2012). The genome encodes one or two polyproteins that produce 10 mature proteins by self-encoded proteinases and P3N-PIPO (pretty interesting Potyviridae ORF) protein, a frameshift product from P3 (third protein) via a polymerase slippage (Adams et al., 2005, 2012; Chung et al., 2008; Olspert et al., 2015; Rodamilans et al., 2015). eIF4E-mediated resistance against potyviruses is overcome largely by viral VPg (Truniger and Aranda, 2009) and, to a lesser extent, P1 (first protein/protease; Nakahara et al., 2010), P3 (Hjulsager et al., 2006), CI (cytoplasmic inclusion protein; Abdul-Razzak et al., 2009; Tavert-Roudet et al., 2012), and probably HC-Pro (helper component protease; Ala-Poikela et al., 2011). The interaction between the host eIF4E and VPg is required for viral infection, and the interaction between these two proteins has been shown for several potyviruses (Léonard et al., 2000; Schaad et al., 2000; Kang et al., 2005b; Beauchemin et al., 2007; Roudet-Tavert et al., 2007; Yeam et al., 2007; Charron et al., 2008; Gallois et al., 2010; Mazier et al., 2011; Estevan et al., 2014).
Barley yellow mosaic virus (BaYMV), which belongs to the genus Bymovirus in the family Potyviridae, is one of the two causal agents [the other is barley mild mosaic virus (BaMMV)] of the economically important yellow mosaic disease of winter barley (Hordeum vulgare L.) in Europe and East Asia. In addition to their bipartite RNA genome, another feature of bymoviruses that distinguishes them from the members of other genera in the family Potyviridae is their transmission in soil by the root-inhabiting vector Polymyxa graminis Ledingham, a plasmodiophoraceous parasite (Adams et al., 2012; Tamada and Kondo, 2013). Like other plasmodiophorid-transmitted viral diseases, the planting of virus-resistant cultivars is a common available way to control yellow mosaic diseases (Ordon et al., 2009). Currently, a total of 18 resistance genes (15 recessive rym genes and three dominant Rym genes) against BaYMV (and also BaMMV) have been identified in barley, of which rym4, rym5, and rym6 (rym4/5/6) resistances are recognized as allelic genes encoding for eIF4E on a region of barley chromosome 3H (Kanyuka et al., 2005; Stein et al., 2005). The emergence of a virulent bymovirus in resistant barley varieties (mainly in rym4/5/6-carrying cultivars) potentially causes a serious threat to barley production (Kühne, 2009). The rym5 resistance gene, which was originally used in Japanese barley breeding programs (Kobayashi et al., 1987; Konishi and Kaiser, 1991), was overcome by a resistance-breaking isolate (Kashiwazaki et al., 1989). Moreover, rym6-mediated resistance also was overcome by this or other virulent isolates (Sotome et al., 2010; You and Shirako, 2013). Similarly, the rym4 and, more recently, rym5 genes, which are the major source of resistance for barley varieties in European countries, have been overcome by virulent BaYMV and BaMMV isolates (Hariri et al., 2003; Habekuss et al., 2008; Kühne, 2009). By sequence comparison between resistance-breaking and resistance-non-breaking isolates, VPg is suggested to be a determinant bymoviral protein responsible for breaking barley eIF4E-mediated resistance (Kühne et al., 2003; Kanyuka et al., 2004; Habekuss et al., 2008; Nishigawa et al., 2008). In addition, both the viral VPg and the host eIF4E are involved in host cell tropism (barley or wheat) of bymoviruses (Li and Shirako, 2015).
In this study, we utilized infectious cDNA clones derived from the resistance-breaking and resistance-non-breaking isolates of BaYMV to investigate the mechanisms underlying rym4/5/6 eIF4E-mediated resistance in barley plants. Our results identified VPg as the determinant protein for breaking of rym4/5/6 resistances. Moreover, mutational analyses combined with protoplast and whole plant inoculation assays suggest that the genetic compatibility between virally encoded VPg and host eIF4E regulates BaYMV infection at both the intracellular and intercellular (cell-to-cell and/or long-distance movement) levels.
Materials and Methods
Plants
Ryofu seeds were purchased from a local Japan Agriculture Cooperative branch. Express barley (carrying rym4) and Maris Otter seeds were from the Institute for Epidemiology and Pathogen Diagnostics, Germany. Other barley (carrying rym5: Mikamo Golden, Misato Golden; carrying rym6: Haruna Nijo, Amagi Nijo, and Miho Golden; KoA) seeds were obtained from the Institute of Plant Science and Resources at Okayama University with the support in part by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Virus Purification and Sequence Determination
BaYMV virions were purified from 2 g of infected leaves by a procedure described previously (Shirako and Brakke, 1984) and suspended in RNase-free H2O. Virus suspension was treated with Proteinase K at 37°C for 30 min, and viral RNA was extracted for cDNA synthesis. First-strand cDNA was synthesized using PrimeScript® Reverse Transcriptase (Takara Bio, Japan) and then amplified by overlapping RT-PCR using PrimeSTAR® HS DNA Polymerase (Takara Bio, Japan). All amplification products were purified using the FastGeneTM Gel/PCR Extraction Kit (Nippon Genetics, Japan) and then directly submitted to a commercial sequencing service (Eurofins Operon, Japan).
Construction of Full-Length cDNA Clones of BaYMV Tochigi Isolate JT10 RNA1 and RNA2
We have isolated a rym5-breaking BaYMV isolate (named isolate JT10) from diseased leaves of a rym5-carrying barley cultivar (Mikamo Golden) in Tochigi Prefecture, Japan in 2010 (Shirako and Li, unpublished data). Construction of infectious full-length cDNA clones of Tochigi isolate JT10 RNA1 (encoding polyprotein 1, P1) and RNA2 (encoding polyprotein 2, P2) were performed as described for Kurashiki isolate JK05 (You and Shirako, 2010). To generate a full-length cDNA clone of JT10 RNA1, named pBY-JT1, we performed RT-PCR using the TB162 primer for first-strand cDNA synthesis and TB87 and TB162 primers for the PCR procedure. The amplified 7.8-kb product was purified and digested with XbaI and BamHI and then cloned into the XbaI–BamHI sites modified from the pBY-JK1 construct (You and Shirako, 2010; Figure 1A). For construction of the cDNA clone of RNA2, named pBY-JT2, we performed RT-PCR using the TB166 primer for first-strand cDNA synthesis and TB163 and TB166 primers for PCR amplification. The purified 3.7-kb PCR product was digested with BamHI and SpeI and then cloned into the BamHI–SpeI sites of the pBY-JK2 construct (You and Shirako, 2010; Figure 1A). Both recombinant DNA procedures were carried out by standard methods using Escherichia coli strain MC1061, and the clones were verified by sequencing the plasmid inserts. All primers used for cloning are listed in Supplementary Table S1.
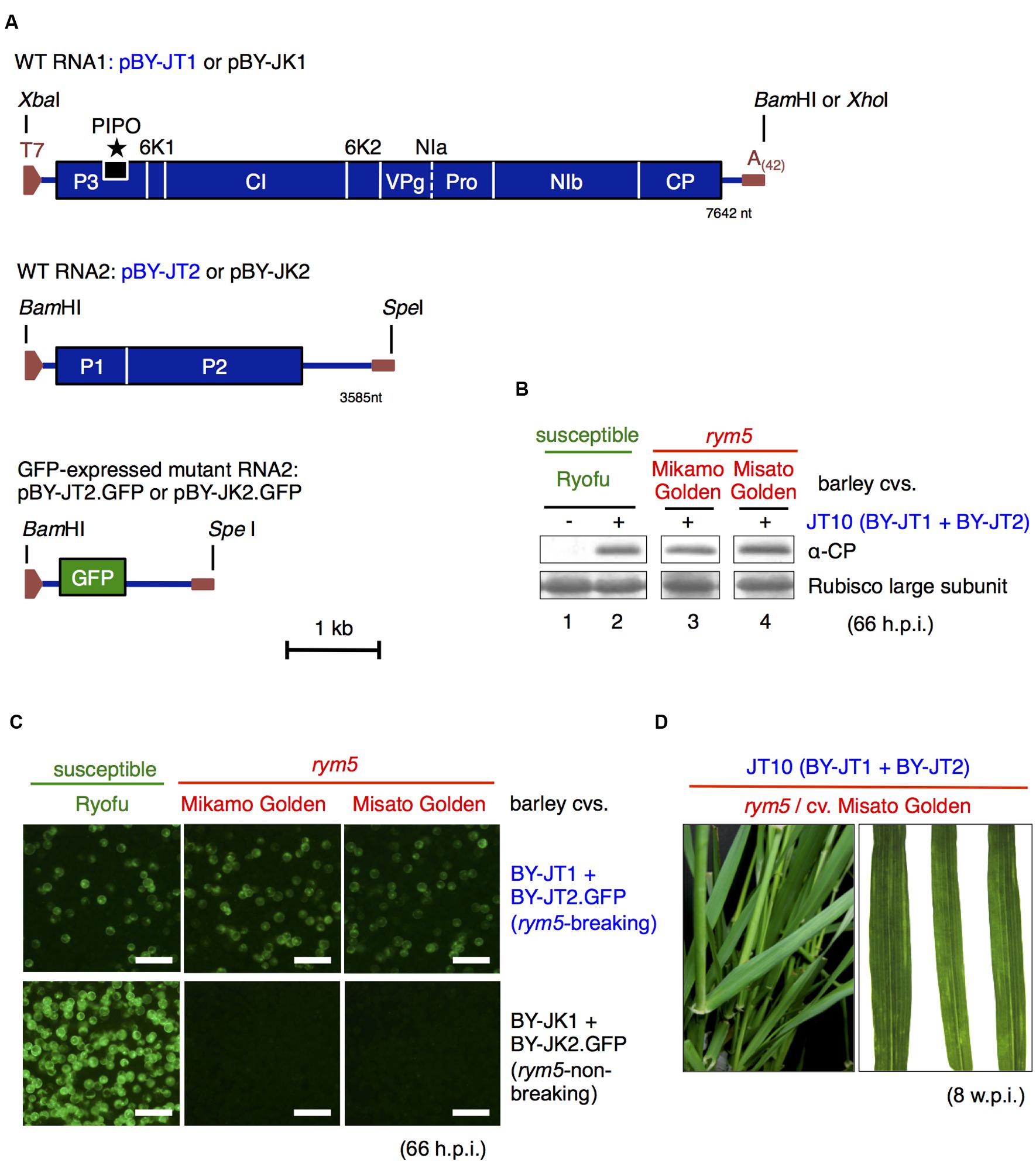
FIGURE 1. Infectivity of in vitro transcripts derived from barley yellow mosaic virus JT10 cDNA clones. (A) Schematic representation of full-length cDNA clones of JT10 (rym5-breaking) isolate (pBY-JT1 and pBY-JT2) and JK05 (rym5-non-breaking) isolate (pBY-JK1 and pBY-JK2). pBY-JT2.GFP and pBY-JK2.GFP are green fluorescent protein (GFP)-expressing RNA2 constructs generated by replacing P1 and P2 coding regions with the GFP gene. (B) Western blot analysis of coat protein (CP) accumulation in susceptible and rym5 protoplasts transfected with JT10 in vitro transcripts. The rubisco large subunits were stained by Coomassie Brilliant Blue G-250 and used as a loading control. (C) GFP fluorescence in susceptible and rym5 protoplasts transfected with in vitro transcripts of JT10 or JK05 RNA1 (pBY-JT1 or pBY-JK1) and GFP-expressing RNA2 (pBY-JT2.GFP or pBY-JK2.GFP). Bars, 200 μm. (D) Viral symptoms in upper systemic leaves of rym5 plants following inoculation with JT10 in vitro transcripts. h.p.i., hours post-inoculation; w.p.i., weeks post-inoculation.
Inoculation of Protoplasts and Leaves with in vitro Transcripts
RNA transcripts were synthesized in vitro from the linearized plasmids as described previously (Yamamiya and Shirako, 2000). Mesophyll protoplasts were prepared from 7-day-old barley seedlings according to procedures described previously (Ohsato et al., 2003; Li and Shirako, 2015). Approximately 5 × 105 cells were transfected with capped in vitro transcripts of BaYMV RNA1 and RNA2 (4 μL transcripts of each) and subsequently incubated at 15°C for 66 h in the dark.
Inoculations of barley plants with in vitro transcripts were performed as described previously (Li and Shirako, 2015). Three to seven seedlings with two to three leaf stages were inoculated in each experiment. Inoculated plants were kept in a growth chamber at 15°C with 16 h of illumination.
Construction of RNA1 Chimeras
For the mapping analysis of viral determinants for overcoming rym5 resistance, eight chimeric infectious cDNA clones were constructed for infectivity analysis: pBY-JK1.JT-XbNs [5′ untranslated region (UTR)–NsiI region from pBY-JT1], pBY-JK1.JT-MsNd (MscI–NdeI region from pBY-JT1), pBY-JK1.JT-XbKp (5′ UTR–KpnI region from pBY-JT1), pBY-JK1.JT-NsNd (NsiI–NdeI region from pBY-JT1), pBY-JK1.JT-KpNs (KpnI–NsiI region from pBY-JT1), pBY-JK1.JT-KpMs (KpnI–MscI region from pBY-JT1), pBY-JK1.JT-MsNs (MscI–NsiI region from pBY-JT1), and pBY-JK1.JT-VPg (VPg coding region from pBY-JT1; Figure 2B). These constructs were generated by replacements of the regions in Kurashiki isolate JK05 with corresponding regions of Tochigi isolate JT10. To generate pBY-JK1.JT-VPg, PCRs were performed in the presence of pBY-JK1 and pBY-JT1 as the templates. The resultant PCR products were purified, and they served as a mixed template to generate the fragment containing a VPg gene from Tochigi isolate JT10 in the background of Kurashiki isolate JK05. The final amplified fragments were purified and cloned into pBY-JK1 with AflII and SalI sites using an In-Fusion HD Cloning System CE Kit (Clontech Laboratories, USA). The primers used for this construction are listed in Supplementary Table S1. To generate the pBY-JK1.JT-MsNs construct, an MscI–NsiI fragment was released from pBY-JT1 and ligated into a similarly digested pBY-JK1. To generate the pBY-JK1.JT-KpNs construct, a KpnI–NsiI fragment was released from pBY-JT1 and ligated into a similarly digested pBY-JK1. To generate the pBY-JK1.JT-XbNs construct, an XbaI–NsiI fragment was released from pBY-JT1 and ligated into a similarly digested pBY-JK1. To generate the pBY-JK1.JT-MsNd construct, an MscI–NdeI fragment was released from pBY-JT1 and ligated into a similarly digested pBY-JK1. To generate the pBY-JK1.JT-KpMs construct, a KpnI–MscI fragment was released from pBY-JT1 and ligated into a similarly digested pBY-JK1. To generate the pBY-JK1.JT-XbKp construct, an XbaI–KpnI fragment was released from pBY-JT1 and ligated into a similarly digested pBY-JK1. To generate the pBY-JK1.JT-NsNd construct, an NsiI–NdeI fragment was released from pBY-JT1 and ligated into a similarly digested pBY-JK1.
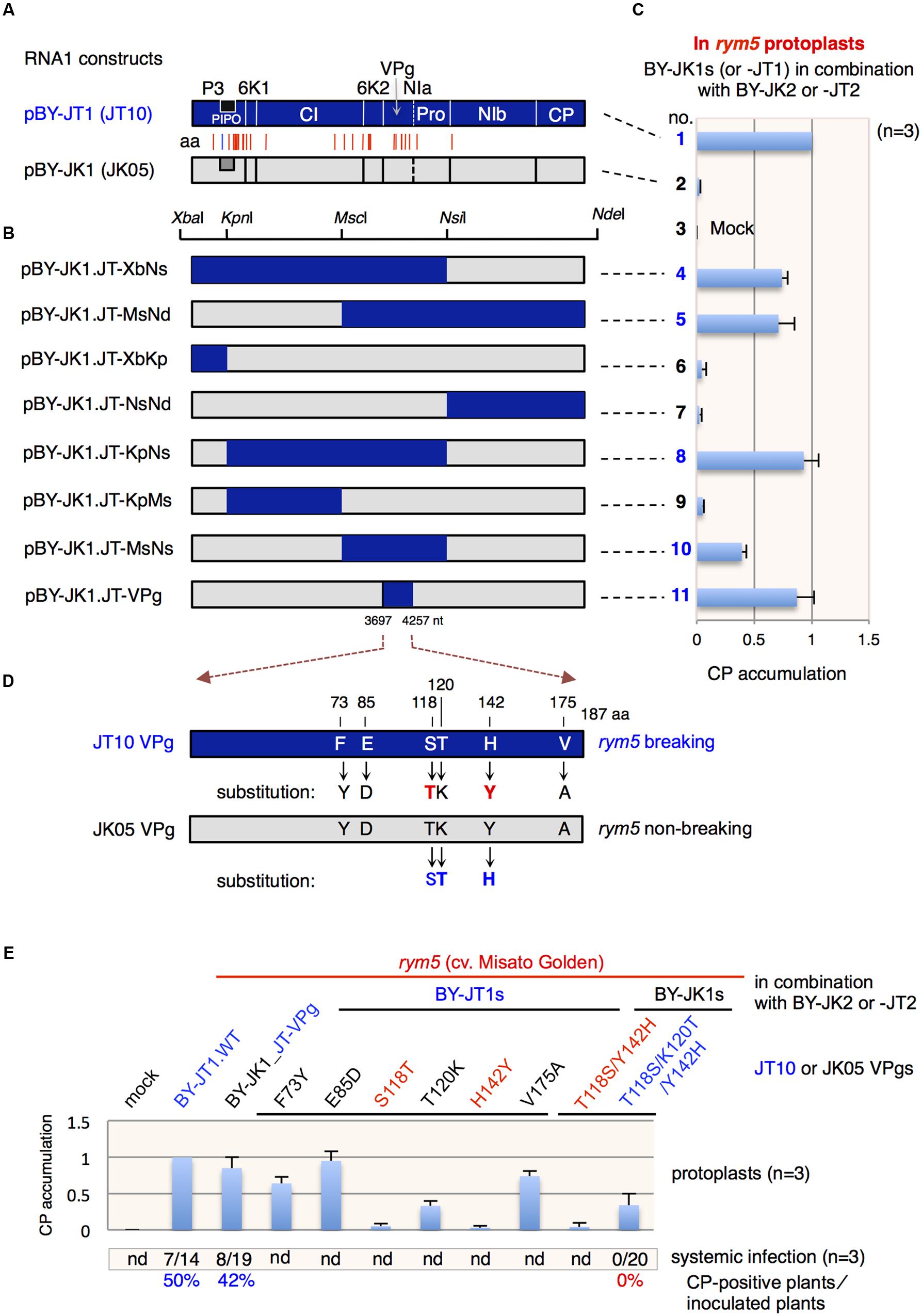
FIGURE 2. Barley yellow mosaic virus (BaYMV) VPg is responsible for breakdown of rym5 resistance in barley plants. (A) Schematic representation of RNA1 cDNA clones, pBY-JT1 (JT10) and pBY-JK1 (JK05). The positions of amino acid (aa) differences between Tochigi isolate JT10 (rym5-breaking) and Kurashiki isolate JK05 (rym5-non-breaking) are indicated with vertical red lines. A vertical blue line indicates the position of one amino acid difference in the PIPO protein. ((B) Schematic representation of chimeric cDNA clones between pBY-JT1 (JT10) and pBY-JK1 (JK05). The unique restriction sites for chimera construction are shown. (C) BaYMV coat protein (CP) accumulation relative to the CP accumulation of the wild-type JT10 virus in rym5 protoplasts transfected with chimeric RNA1 transcripts in addition to wild-type RNA2 transcripts (BY-JK2 or BY-JT2). (D) Schematic representation showing amino acid differences between JT10 VPg and JK05 VPg. The amino acid substitutions in each VPg are indicated with arrows. (E) Relative CP accumulation of JT10 or JK05 isolate having an amino acid mutation in VPg in rym5 protoplasts. The total number of CP-positive plants in the upper leaves/the total number of inoculated plants and percentage of total plants showed viral systemic infection are presented below the graph. nd, not determined. CP accumulations in transfected protoplasts or upper systemic leaves of inoculated plants were detected by western blotting (C,E). All values in bar graphs represent means with standard deviation (SD) from three independent experiments. Samples from rym5 protoplasts inoculated with JT10 were used as the reference and the relative mean values of virus mutants were calibrated against those of references.
pBY-JK1.JG-VPg, pBY-JK1.Y1A-VPg, and pBY-JK1.Y2A-VPg were constructed similarly to pBY-JK1.JT-VPg. All recombinant DNA procedures were carried out by standard methods using E. coli strain MC1061, and the clones were verified by sequencing the plasmid inserts.
Construction of RNA2 Mutants
The GFP (green fluorescent protein)-expressing RNA2 mutant pBY-JT2.GFP (Figure 1A) was derived from pBY-JT2 in the same manner as pBY-JK2.GFP replacement of the P1 and P2 coding regions between the 5′- and 3′-UTRs of RNA2 with a GFP gene (You and Shirako, 2010). The primers used for construction are listed in Supplementary Table S1.
Two eIF4E-expressing RNA2 mutants, pBY-JK2.P12/KoA_eIF4E(NIb/CP) and pBY-JK2.P12/KoA_eIF4E(P1/P2), were constructed and used in this study (Figure 4A). PCRs were performed in the presence of pBY-JK2 and the KoA eIF4E gene as separate templates (Li and Shirako, 2015). The resultant PCR products were purified, and they served as a mixed template for generation of the final expected fragments. The fragments were subsequently digested and ligated into pBY-JK2. All recombinant DNA procedures were carried out by standard methods using E. coli strain MC1061, and the clones were verified by sequencing the plasmid inserts. The primers used for each construct are listed in Supplementary Table S1.
P2 and eIF4E Antiserum Production
BaYMV P2 and barley eIF4E proteins were prepared by fusing proteins with glutathione S-transferase (GST) in E. coli cells (strain MC1061), as described by You and Shirako (2010). One rabbit was immunized for 2 mg of each purified recombinant fusion protein (GST:P2 and GST:eIF4E).
Western Blot Analysis
Western blot analysis was performed as described previously (Yamaguchi and Shirako, 2002). Polyclonal antibodies against BaYMV CP (coat protein), P2, and barley eIF4E were used as the primary antibodies. Each virus inoculation test on protoplasts or whole plants was at least triplicated, except for inoculation of BY-JK1.Y1A-VPg and Y2A-VPg mutants, which were duplicated in rym4 protoplasts, and inoculation of BY-JK1, BY-JK1.Y1A-VPg, Y2A-VPg, and JG-VPg mutants, which were only performed once or twice in rym4 and rym6 plants (Figure 3). Positive (BaYMV-infected susceptible plants) and negative (the mock plants) control samples were always included in each blot.
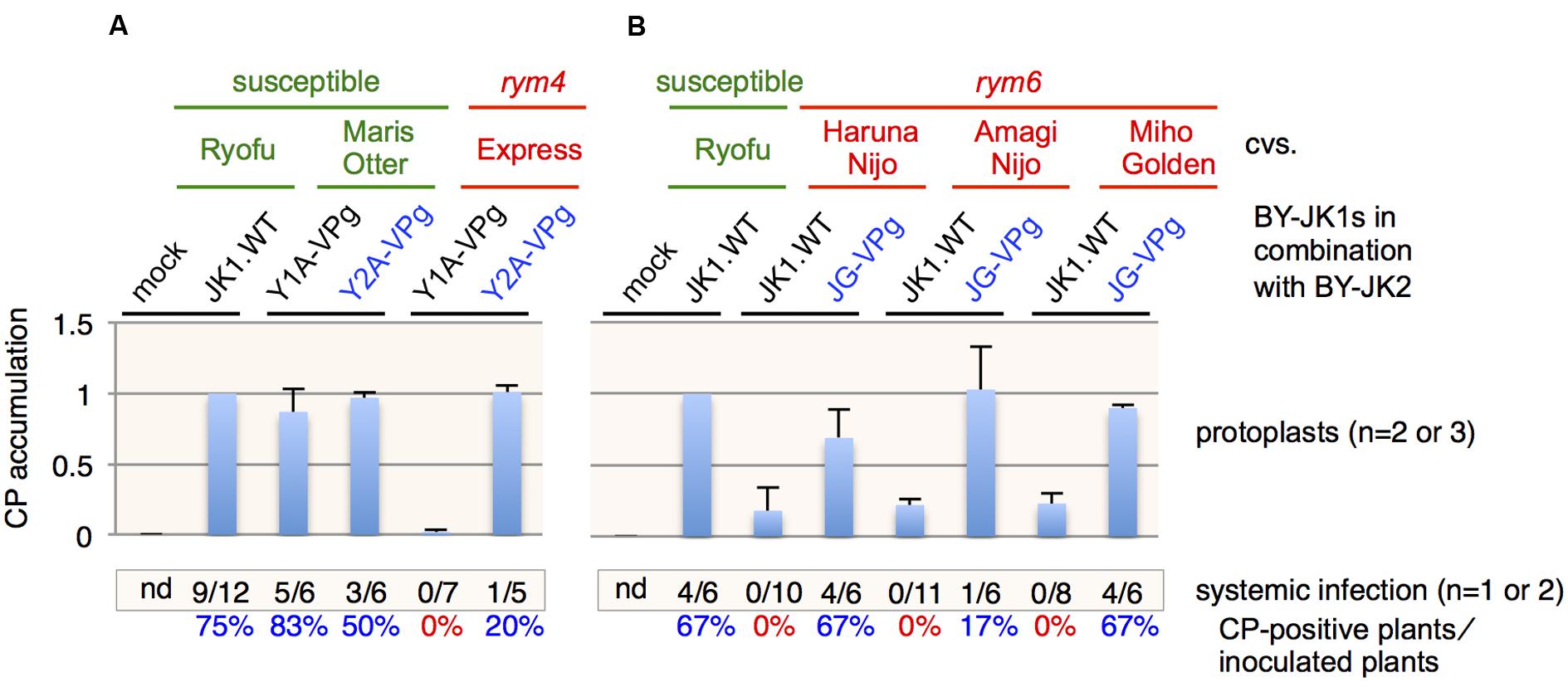
FIGURE 3. Barley yellow mosaic virus (BaYMV) VPg is responsible for breakdown of rym4 and rym6 resistances. (A,B) Relative BaYMV coat protein (CP) accumulation of the JK05 isolate with VPg replacement in rym4 (A) and rym6 (B). All values in bar graphs represent means with SD from two or three independent experiments. Samples from Ryofu protoplasts inoculated with JK05 were used as the reference.
To quantify the levels of CP accumulation in the protoplasts, the CP bands on all of the blots were scanned using ImageJ 1.42q software (Wayne Rasband National Institutes of Health, USA). The intensity of CP band of each sample was normalized with the intensity of the rubisco large subunit band of the corresponding sample, and the relative mean scores were calibrated against those of positive controls as described previously (You and Shirako, 2013).
Barley eIF4E Protein Homology Modeling
The amino acid sequence of barley KoA eIF4E was submitted to the homology-modeling server SWISS-MODEL (Arnold et al., 2006; Biasini et al., 2014)1. Construction of the three-dimensional structure of this protein was based on sequence homology to wheat eIF4E (PDB code: 2idr). The predicted structure was visualized with the software Deep View/Swiss-PdbViewer v4.1.0 (Johansson et al., 2012).
Results
Construction of an Infectious cDNA Clone from a rym5-Breaking BaYMV Isolate
The full-length nucleotide sequences of RNA1 and RNA2 of BaYMV JT10 (rym5-breaking isolate) were determined and deposited in the GenBank database with accession numbers AB920780 for RNA1 (7642 nt) and AB920781 for RNA2 (3585 nt). The full-length cDNA clones of JT10 were constructed and named pBY-JT1 and pBY-JT2 (Figure 1A) for RNA1 and RNA2, respectively. The replicative ability of JT10 RNA transcripts (BY-JT1 RNA1 and BY-JT2 RNA2) from the two clones was examined in protoplasts isolated from rym5-carrying barley cultivars (Mikamo Golden and Misato Golden). The protoplasts isolated from barley cultivar Ryofu were used as a susceptible control. The protoplasts were transfected with JT10 transcripts and then incubated at 15°C for 66 h in the dark. After incubation, the protoplasts were harvested and subjected to western blot analyses using a CP antiserum (You and Shirako, 2010). CP accumulations were detected in all the tested protoplasts (Figure 1B), indicating virus multiplication. To monitor visually the virus infection in protoplasts, the P1–P2 polyprotein coding region in pBY-JT2 was replaced with a GFP gene-coding sequence (pBY-JT2.GFP; Figure 1A). GFP fluorescence was observed in both susceptible and rym5 protoplasts transfected with BY-JT1 RNA1 + BY-JT2.GFP RNA2, but not in rym5 protoplasts transfected with BY-JK1 RNA1 + BY-JK2.GFP RNA2 (Figure 1C), confirming the infectious nature of the transcripts.
Next, the systemic infectivity of JT10 RNA1 and RNA2 was examined at the whole plant level through manual inoculation. Yellow mosaic leaf symptoms were observed in susceptible (cultivar Ryofu) and rym5 (cultivars Mikamo Golden and Misato Golden) plants at 5–7 weeks post-inoculation (w.p.i.; Figure 1D and data not shown). RT-PCR/sequencing analysis of virus progenies confirmed that the JT10 transcripts were systemically infectious to the rym5 cultivars (data not shown). For comparison, the cultivar Ryofu plants inoculated with JK05 RNA transcripts (BY-JK1 RNA1 and BY-JK2 RNA2 of a rym5-non-breaking BaYMV isolate; You and Shirako, 2013) developed yellow mosaic symptoms 3–5 weeks faster than those inoculated with the JT10 virus (data not shown).
VPg is the Determinant Viral Protein for Breaking rym5 Resistance
Previous research showed that BaYMV RNA1, which encodes a set of proteins including the RNA polymerase, could autonomously replicate in barley protoplasts while RNA2 needs RNA1 for replication (You and Shirako, 2010; Li and Shirako, 2015). Thus, we speculated that BaYMV RNA1 encoded the most important factor(s) responsible for breaking eIF4E-mediated (rym5) resistance. Additionally, as replacing the coding region in RNA2 with the GFP gene (Figure 1A) does not affect JT10 ability to multiply in rym5 protoplasts (Figure 1C), the viral factor(s) responsible for breaking rym5 resistance thus likely reside in BaYMV RNA1. Sequence comparison showed that within RNA1, the JT10 isolate has 29 amino acid differences with the rym5-non-breaking JK05 isolate (You and Shirako, 2013). Notably, the amino acid substitutions were present in almost all of the coding proteins except for CP (Figure 2A).
To identify the viral factor(s) involved in breaking rym5 resistance, partial DNA fragments derived from pBY-JT1 were used to replace the corresponding sequences in a cDNA clone derived from JK05 RNA1 that was generated previously (You and Shirako, 2010; here renamed as pBY-JK1 and pBY-JK2 for RNA1- and RNA2-derived clones, respectively). Seven chimeras were constructed by utilizing the available unique restriction enzyme sites (Figure 2B). Leaf protoplasts isolated from cultivar Misato Golden (rym5) plants were transfected with the mixtures of in vitro transcripts synthesized using pBY-JK1-based chimeras and pBY-JK2. Western blotting indicated that four chimeric viruses (BY-JK1.JT-XbNs, BY-JK1.JT-MsNd, BY-JK1.JT-KpNs, and BY-JK1.JT-MsNs) were able to accumulate in rym5 protoplasts, although the level of the BY-JK1.JT-MsNs was lower than that of other mutants, probably due to the inhibiting effect of the chimeric sequence on virus accumulation (Figure 2C). Those infectious transcripts similarly contained the central region of the JT10 genome, which encodes for 6K2 (6 kDa protein 2), VPg and NIa (nuclear inclusion a proteinase; Figure 2B). When only the VPg portion of the JT10 genome was used to replace the corresponding region in pBY-JK1, the chimeric transcripts (BY-JK1.JT-VPg) were infectious in rym5 protoplasts (Figure 2C) and also in whole plants, in which a mild mosaic symptom appeared and CP accumulations were detected in upper systemic leaves at 4 w.p.i. (Figure 2E; data not shown). These observations confirm that VPg is responsible for breaking eIF4E-mediated resistance in barley plants.
Ser-118, Thr-120, and His-142 of VPg are Critical for Overcoming rym5 Resistance
Sequence alignment showed that six amino acid residues at positions 73, 85, 118, 120, 142, and 175 in JT10 VPg differ from the corresponding residues in JK05 VPg (Figure 2D; Table 1). To identify precisely which key amino acid(s) of VPg are important for breaking rym5 resistance, each of those six amino acids in the pBY-JT1 background was substituted into the same residues as encoded in the JK05 VPg, and then the infectivity of each virus mutant (with the presence of JT1 RNA2) was examined in rym5 protoplasts and whole plants (cultivar Misato Golden). Western blotting showed that substitution of Ser-118 and His-142 into Thr (S118T) and Tyr (H142Y), respectively, abolished JT10 ability to infect rym5 protoplast, while substitution of Thr-120 into Lys (T120K) largely reduced JT10 accumulation in rym5 protoplasts (Figure 2E). In contrast, mutation of Phe-73, Glu-85, and Val-175 (F73Y, E85D, and V175A, respectively) had no or partial effects on JT10 accumulation in rym5 protoplasts (Figure 2E). These results indicate that Ser-118 and His-142 as well as Thr-120, to a lesser degree, are critical for overcoming rym5 resistance, while Phe-73, Glu-85, and Val-175 are less important for breaking rym5 resistance at the intracellular level. Our preliminary results showed that the JT10 mutants with the substitution of Phe-73 or Glu-85 did not cause any systemic symptom in rym5 plants (data not shown). This may suggest that these residues are critical for viral systemic infection throughout whole plants.
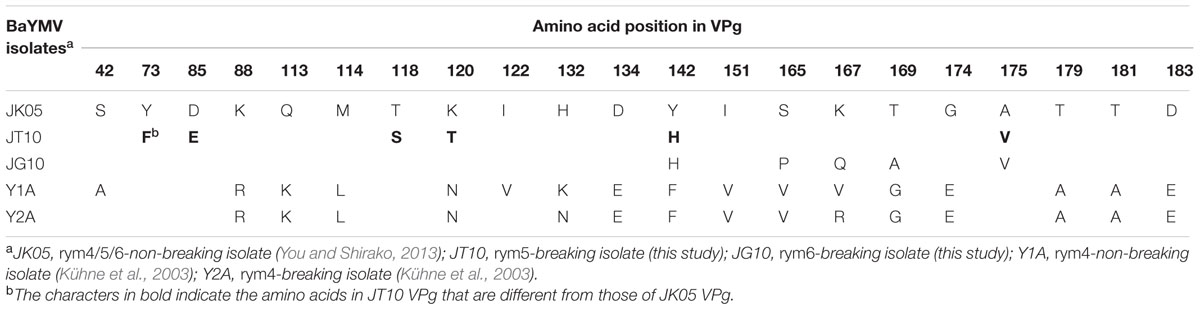
TABLE 1. Amino acid differences of VPg proteins from barley yellow mosaic virus (BaYMV) JK05, JT10, JG10, Y1A, and Y2A isolates.
In a reciprocal experiment, JK05 was able to infect rym5 protoplasts only when Thr-118, Lys-120, and Tyr-142 were simultaneously substituted (triple mutation, T118S/K120T/Y142H) but not when only Thr-118 and Tyr-142 were simultaneously substituted (double mutation, T118S/Y142H; Figure 2E). The accumulation level of JK05 with the triple mutations was low in rym5 protoplasts (Figure 2E), which is consistent with the previous observation (using JT10). Moreover, JK05 with the triple mutations was unable to infect the whole plants (Figure 2E), in line with the notion that the two other amino acid residues at the position of 73 and 85 in VPg might be essential for viral systemic infection, although they are partially important for breaking the resistance at the intracellular level.
VPg is Also the Determinant Protein Responsible for Breaking rym4 and rym6 Resistances
Since rym4/5/6 are allelic with similar eIF4E genes (Kanyuka et al., 2005; Stein et al., 2005), it is possible that VPg is also the determinant protein for breaking rym4 and rym6 resistances. In Europe, rym4 resistance has been used widely for barley breeding in recent decades (Kühne, 2009). BaYMV isolates from German Y2A but not Y1A were able to infect rym4 plants in the field (Kühne et al., 2003). When VPg genes derived from Y2A and Y1A isolates were used to replace the VPg gene in pBY-JK1, JK05 Y2A-VPg but not JK05 Y1A-VPg could infect rym4 protoplasts and plants (cultivar Express), whereas both chimeric viruses could infect susceptible barley plants (cultivar Maris Otter; Figure 3A). The BaYMV JK05 isolate could accumulate to low levels in rym6 protoplasts (cultivars Haruna Nijo, Amagi Nijo, or Miho Golden) but could not systemically infect rym6 plants (You and Shirako, 2013; Figure 3B). The BaYMV JG10 isolate is a rym6-breaking isolate from rym6 plants (cultivar Haruna Nijo) in Gumma, Japan in 2010 (see Table 1 for its VPg amino acid sequence; Shirako and Li, unpublished data). JK05 JG10-VPg accumulated to high levels in rym6 protoplasts and systemically infected all three varieties carrying rym6 (Figure 3B). Taken together, BaYMV VPg is also the determinant protein for breaking rym4 and rym6 resistances.
Expression of eIF4E gene Derived from a Susceptible Barley Cultivar Enables JK05 Multiplication in rym5 Protoplasts
Considering that the compatibility between VPg and host eIF4E plays an important role in determining BaYMV ability to infect barley plants, we anticipated that the expression of eIF4E gene derived from a susceptible barley cultivar could facilitate the accumulation of non-breaking isolates in rym5 protoplasts. To this end, the eIF4E gene derived from susceptible cultivar KoA (KoA eIF4E) was inserted into JK05 RNA2 (pBY-JK2) downstream of the P2 gene with the addition of either the NIb/CP or the P1/P2 protease cleavage site to release eIF4E from the polyprotein or to express the P2 fusion product (P1 proteinase most probably only acts in cis; Figures 4A,B). Protoplasts from susceptible (cultivar Ryofu) and rym5 (cultivar Misato Golden) plants were transfected with JK05 RNA1 and eIF4E-expressing RNA2 transcripts. As expected, CP accumulations were detected in rym5 protoplasts after transfection with either JK05 RNA1 + JK05 RNA2 KoA_eIF4E(NIb/CP) or + JK05 RNA2 KoA_eIF4E(P1/P2) transcripts (Figure 4C). The CP accumulations of those KoA_eIF4E-expressing viruses in rym5 protoplasts were not as abundant as those in susceptible protoplasts but were rather comparable to JT10 accumulations in rym5 protoplasts (Figure 4C). These results demonstrated that expression of KoA eIF4E from viral genome, to some extent, could assist incompatible JK05 (rym5-non-breaking isolate) in accumulating in rym5 protoplasts. Note that although both KoA eIF4E-carrying transcripts were infectious in rym5 protoplasts, as expected, free eIF4E (∼24 kDa) was efficiently released by the NIb/CP but not the P1/P2 cleavage sites (Figure 4D). Thus, eIF4E also could be functional in the form of a P2-eIF4E fusion protein (∼94 kDa). Nevertheless, expression of KoA eIF4E from the viral genome did not enable JK05 infection in whole plants (Figure 4C), indicating that some other resistance factor(s) should be involved in rym5 plants.
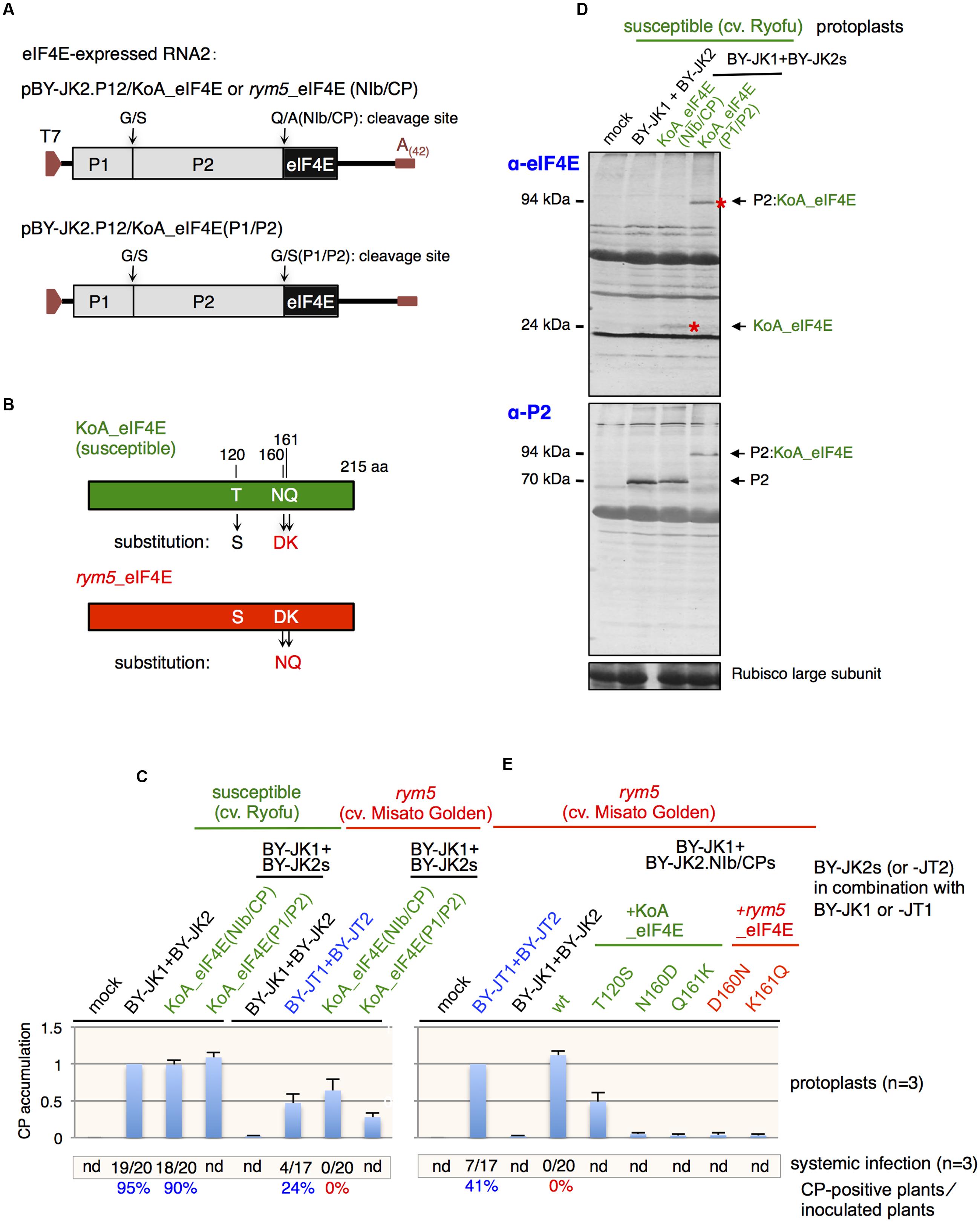
FIGURE 4. Expression of eIF4E from viral genome. (A) Schematic representation of eIF4E-expressing JK05 RNA2 constructs. (B) Schematic representation showing major amino acid differences between KoA (susceptible) and rym5 (resistance) eIF4E alleles. The amino acid substitutions in each eIF4E are indicated with arrows. (C,E) Barley yellow mosaic virus (BaYMV) coat protein (CP) accumulation in susceptible and rym5 protoplasts transfected with eIF4E-expressing JK05 RNA2 (BY-JK2) transcripts in addition to JK05 RNA1 (BY-JK1) transcripts. All values in bar graphs represent means with SD from three independent experiments. Samples from Ryofu or rym5 protoplasts inoculated with JK05 (C) or JT10 (E) were used as the reference. (D) Western blot analysis of barley eIF4E and BaYMV P2 accumulation in protoplasts from cultivar Ryofu (susceptible) transfected with eIF4E-expressing JK05 RNA2 (BY-JK2) transcripts plus JK05 RNA1 (BY-JK1) transcripts. The red asterisks mark the bands of eIF4E and P2:eIF4E fusions on the blots.
Sequence alignment showed that the three amino acid residues at positions 120 (Thr), 160 (Asn), and 161 (Gln) in eIF4E from rym5-carrying cultivars are different from those corresponding residues in eIF4E from susceptible cultivars (Figure 4E; Table 2). Substitution of both Asn-160 and Gln-161 into Asp and Lys (N160D and Q161K), respectively, abolished KoA eIF4E activity to facilitate JK05 infection in rym5 (cultivar Misato Golden) protoplasts, while substitution of Thr-120 into Ser (T120S) only reduced virus accumulation (Figure 4E). Nevertheless, substitutions of either Asp-160 to Asn (D160N) or Lys-161 to Gln (K161Q) in rym5 eIF4E were not sufficient to enable JK05 accumulation in rym5 protoplasts (Figure 4E). Thus, the simultaneous presence of both Asn-160 and Gln-161 in rym5 eIF4E might be required for facilitating BaYMV infection.
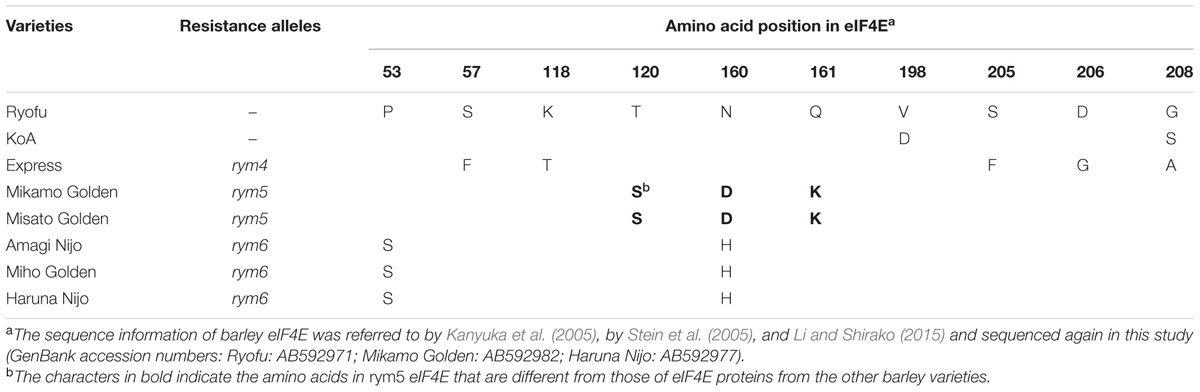
TABLE 2. Amino acid differences of eIF4E proteins from several barley varieties carrying different resistance alleles.
Discussion
In the present study, viral factor(s) responsible for breaking rym5 resistance were identified by genomic mapping analysis using infectious cDNA clones derived from BaYMV JK05 (rym5-non-breaking) and JT10 (rym5-breaking) isolates (Figures 1 and 2). Along with the presence of JK05 RNA2, the JK05 RNA1 chimera (BY-JK1.JT-VPg) with the JK05 VPg gene replaced with that of JT10 isolate accumulated in rym5 protoplasts and was infectious to rym5 plants at the whole plant level (Figure 2). Likewise, the RNA1 VPg chimeras with the JK05 VPg gene replaced with those of rym4 and rym6 resistance-breaking isolates (Y2A and JG) each individually disrupted the corresponding resistance at the whole plant level (Figure 3; and summarized in Table 3). Thus, BaYMV VPg is the determinant protein responsible for breaking eIF4E-mediated rym4/5/6 resistance in barley plants. Furthermore, we tried to identify precisely the key amino acid(s) of VPg required for breaking these eIF4E-mediated resistances. Substitutions of at least three amino acids at the positions of 118, 120, and 142 in VPg of rym5-non-breaking isolate JK05 are required for overcoming rym5 resistance in protoplasts, but they were not sufficient for the establishment of viral systemic infection in rym5 plants (Figure 2). These results indicate that mutations of numerous amino acid residues in VPg are required for complete breakdown of eIF4E-mediated resistance in barley plants. VPg has some typical features of intrinsically disordered proteins and is known to be linked to the genome of some positive-sense RNA plant viruses (e.g., the members of Potyviridae and Secoviridae families and Sobemovirus, Polerovirus, and Enamovirus genera) and vertebrate ones (e.g., the members of Picornaviridae and Caliciviridae families; Jiang and Laliberté, 2011). In potyviruses, VPg is a hub protein that interacts with several proteins both of viral and host origin, likely including eIF4E (see below), and appears to be involved in diverse viral processes, such as translation, replication, cell-to-cell and/or long-distance movement (Jiang and Laliberté, 2011; Revers and García, 2015).
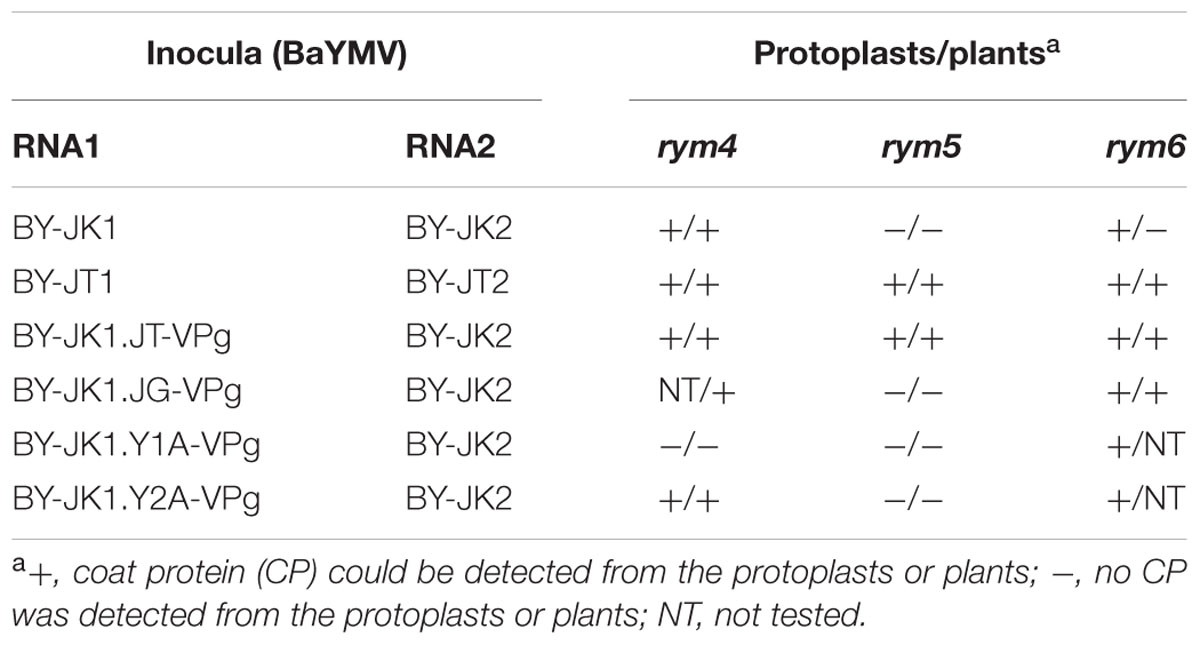
TABLE 3. Summary of accumulation and systemic infectivity of barley yellow mosaic virus (BaYMV) VPg mutants in rym4/5/6 protoplasts or plants.
The role of barley eIF4E during BaYMV infection was examined by the expression of host eIF4E from the viral genome RNA. When an eIF4E derived from a susceptible cultivar was expressed from JK05 RNA2, CP accumulation was detected by western blot analysis (Figure 4), demonstrating that the presence of compatible eIF4E enables the multiplication of a rym5-non-breaking BaYMV isolate in rym5 cells. In the case of potyviruses, the host eIF4E has been shown to affect virus infection, but there is no direct evidence that shows eIF4E function in virus multiplication at the intracellular level (Schaad et al., 2000; Gao et al., 2004). Our study provided the experimental evidence showing host eIF4E functions in plant virus multiplication at the intracellular level, although it remains unclear how eIF4E participates in this process. The current models for the eIF4E roles in potyvirus infection at the intracellular level proposes that eIF4E acts in concert with VPg and other viral (such as P1 and HC-Pro) and host [such as eIF4G and poly(A)-binding protein] factors to facilitate viral genome replication, translation and/or safeguarding virus translation/replication (Wang and Krishnaswamy, 2012; Mäkinen and Hafrén, 2014). In addition, eIF4E is also proposed to be involved in potyviral intracellular trafficking, cell-to-cell (or long-distance) movement via interaction with VPg, CI protein, and eIF4G (Wang and Krishnaswamy, 2012; Mäkinen and Hafrén, 2014). Interestingly, our results provide the notion that barley eIF4E also plays a role(s) in BaYMV cell-to-cell movement and/or systemic virus accumulation because the substitutions of multiplication-related amino acids in VPg from the non-breaking isolate were insufficient for breaking barley eIF4E-mediated resistance at the whole plant level (Figure 2). As progression of BaYMV infection is rather slow in barley plants, it is difficult to observe cell-to-cell movement processes, such as those observed for pea seed-borne mosaic virus (PSbMV, a potyvirus) in pea plants (Gao et al., 2004).
Our study indicates that the compatibility between VPg and eIF4E proteins is required for virus multiplication (and virus cell-to-cell movement), suggesting that between barley eIF4E and BaYMV VPg there might be a direct interaction or an indirect interaction with an interposed viral or host factor(s). However, a direct or indirect interaction between these two proteins was not detected by a yeast two-hybrid (Y2H) system or by co-immunoprecipitation assays (Li and Shirako, unpublished data). The inability to detect a VPg–eIF4E interaction might be due to a likely occurred misfolding of VPg to interact with eIF4E in Y2H-system, the low temperature requirement for BaYMV replication (You and Shirako, 2012), the instability of host eIF4E (Monzingo et al., 2007) or a possible occurrence of a weak interaction below detection limit. The failure to demonstrate the interaction between pea eIF4E and VPg encoded by PSbMV, which is also a low temperature-adapted virus (Congdon et al., 2016; Jones, 2016), was reported previously (Gao et al., 2004). Nonetheless, we speculate that interaction between BaYMV VPg and eIF4E does exist in host barley. The amino acid substitutions in barley eIF4E may disrupt its interaction with VPg and then result in resistance to BaYMV infection. Compared to an eIF4E from a susceptible cultivar, five substitutions at amino acid positions 57, 118, 205, 206, and 208 were found in rym4 eIF4E; three substitutions at positions 120, 160, and 161 were found in rym5 eIF4E; and two substitutions at positions 53 and 161 were found in rym6 eIF4E (Kanyuka et al., 2005; Hofinger et al., 2011; Table 2; Li and Shirako, unpublished data). According to the three-dimensional model of barley eIF4E (Kanyuka et al., 2005; Stein et al., 2005; Figure 5), all amino acid substitutions are located at or near the cap structure binding domain. As predicted in wheat eIF4E (Monzingo et al., 2007), amino acids at positions 160 and 161 are close to two key amino acids, 158 and 163, which are predicted to be located at a cap structure binding region, suggesting that amino acids at position 160 and 161 also may be involved in the binding of the RNA cap structure. Substitutions in either of these two positions may result in resistance to viral infection, as occurred in rym5 and rym6 (Table 2). In addition, the amino acid at position 118 was important for stabilizing the structure of the protein, and the substitution at this position, as occurred in rym4 eIF4E, also may affect the function of eIF4E (Table 2).
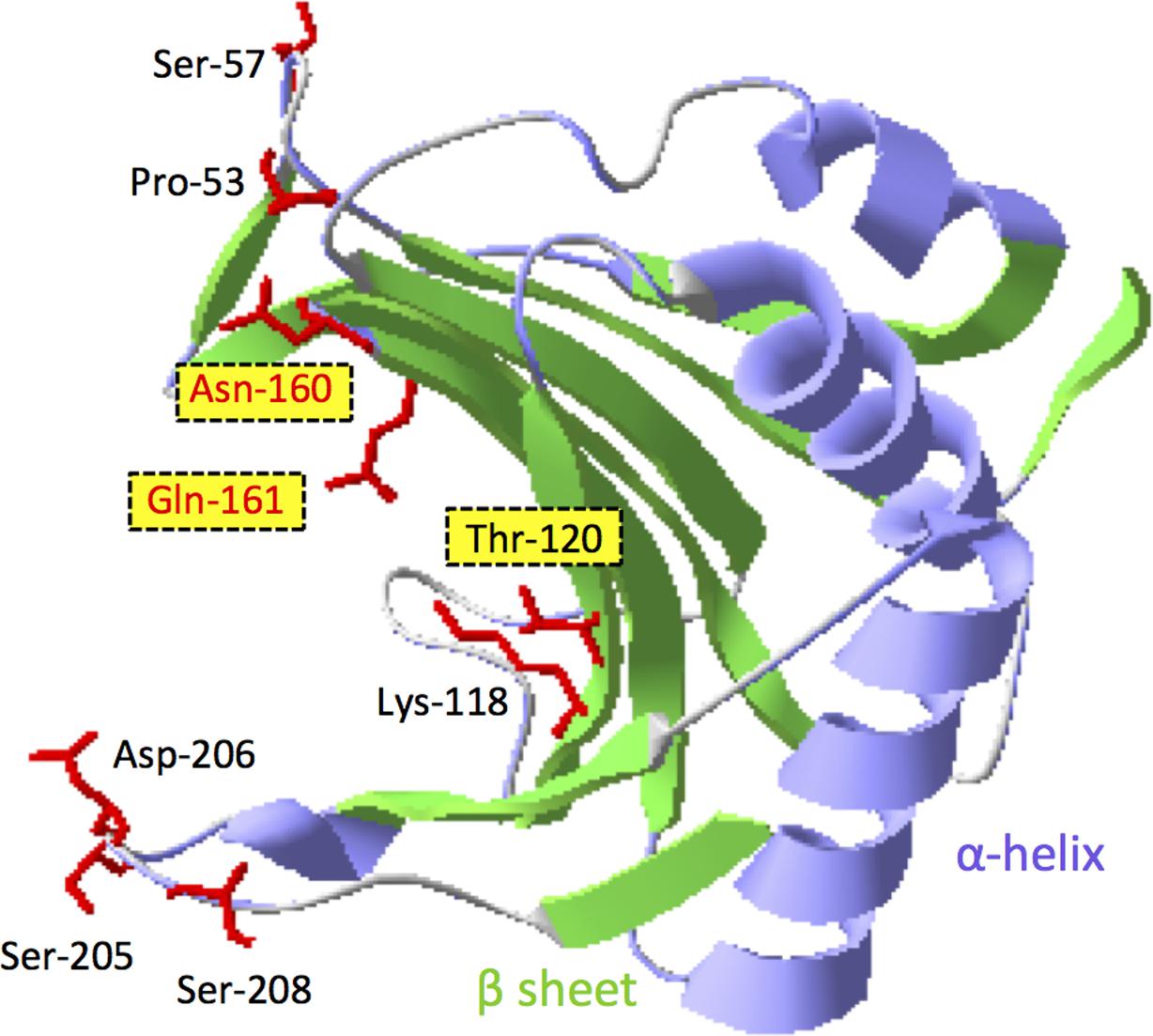
FIGURE 5. Three-dimensional (3D) model of barley KoA eIF4E protein. The putative 3D structure of KoA eIF4E was predicted by the homology-modeling server SWISS-MODEL. The N-terminal 39 amino acids of KoA eIF4E are invisible in this model. The putative cap-binding “pocket” is located at the left side of the eIF4E molecule. The amino acid residues of KoA eIF4E highlighted with the red side chains indicate the positions of amino acid differences among the eIF4E proteins encoded by rym4, rym5, and rym6 plants (see Table 2). The residues highlighted with yellow boxes were mutated in this study. The 3D image was generated using Swiss-PdbViewer.
The infectivity assays carried out using both protoplasts and whole plants were useful for elucidating resistance mechanisms. It becomes clear whether the step in the viral multiplication or movement was blocked during the resistance responses. In many examples of potyvirus infection, compatibility between VPg and eIF4E has been shown. Our present data reveal that both barley eIF4E and BaYMV VPg function in virus multiplication and/or, presumably, virus cell-to-cell movement. The knowledge regarding amino acid residues in eIF4E and VPg that are critical for virus infection can be utilized in the crop breeding programs as well as in the surveillance of emerging resistance-breaking virus variants. Lastly, this work provides a basis for further understanding of other recessive resistance mechanisms in plants.
Author Contributions
HL and YS conceived the work plan, conducted the experiments and data analysis, and drafted the first draft of the manuscript. TK and YS contributed materials and reagents. HL and HK drew tables and figures. First draft of the manuscript was edited and approved by all authors.
Funding
This work was supported by a research aid from the University of Tokyo (grant number 3001090101). HL was supported by a doctoral scholarship program from the Japanese Government (grant number 111522).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ida Bagus Andika for helpful discussions and critical reading of the manuscript, the two reviewers for their valuable suggestions and comments, the Tochigi Prefectural Agricultural Experiment Station (Tochigi Branch) for providing BaYMV-infected leaves of cultivar Mikamo Golden, the Sapporo Breweries Ltd., Gumma, Japan for providing BaYMV-infected leaves of cultivar Haruna Nijo, and the Barley and Wild Plant Resource Center of the Institute of Plant Science and Resources, Okayama University and the Institute for Epidemiology and Pathogen Diagnostics, Germany for providing the barley varieties.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01449
Footnotes
References
Abdul-Razzak, A., Guiraud, T., Peypelut, M., Walter, J., Houvenaghel, M.-C., Candresse, T., et al. (2009). Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E-mediated resistance against Lettuce mosaic potyvirus. Mol. Plant Pathol. 10, 109–113. doi: 10.1111/j.1364-3703.2008.00513.x
Adams, M., Zerbini, F., French, R., Rabenstein, F., Stenger, D., and Valkonen, J. (2012). “Family Potyviridae,” in Virus Taxonomy, 9th Report of the International Committee for Taxonomy of Viruses, eds A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (San Diego, CA: Elsevier Academic Press), 1069–1089.
Adams, M. J., Antoniw, J. F., and Beaudoin, F. (2005). Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 6, 471–487. doi: 10.1111/j.1364-3703.2005.00296.x
Ala-Poikela, M., Goytia, E., Haikonen, T., Rajamäki, M.-L., and Valkonen, J. P. T. (2011). Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF (iso) 4E and eIF4E and contains a 4E binding motif. J. Virol. 85, 6784–6794. doi: 10.1128/JVI.00485-11
Andrade, M., Abe, Y., Nakahara, K. S., and Uyeda, I. (2009). The cyv-2 resistance to Clover yellow vein virus in pea is controlled by the eukaryotic initiation factor 4E. J. Gen. Plant Pathol. 75, 241–249. doi: 10.1007/s10327-009-0163-3
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Beauchemin, C., Boutet, N., and Laliberté, J.-F. (2007). Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81, 775–782. doi: 10.1128/JVI.01277-06
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Bruun-Rasmussen, M., Møller, I. S., Tulinius, G., Hansen, J. K. R., Lund, O. S., and Johansen, I. E. (2007). The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum. Mol. Plant Microbe Interact. 20, 1075–1082. doi: 10.1094/MPMI-20-9-1075
Charron, C., Nicolaï, M., Gallois, J. L., Robaglia, C., Moury, B., Palloix, A., et al. (2008). Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. doi: 10.1111/j.1365-313X.2008.03407.x
Chung, B. Y. W., Miller, W. A., Atkins, J. F., and Firth, A. E. (2008). An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U.S.A. 105, 5897–5902. doi: 10.1073/pnas.0800468105
Clay, K., and Kover, P. X. (1996). The red queen hypothesis and plant/pathogen interactions. Annu. Rev. Phytopathol. 34, 29–50. doi: 10.1146/annurev.phyto.34.1.29
Congdon, B. S., Coutts, B. A., Renton, M., and Jones, R. A. C. (2016). Pea seed-borne mosaic virus: stability and wind-mediated contact transmission in field pea. Plant Dis. 10, 953–958. doi: 10.1094/PDIS-11-15-1249-RE
Estevan, J., Maréna, A., Callot, C., Lacombe, S., Moretti, A., Caranta, C., et al. (2014). Specific requirement for translation initiation factor 4E or its isoform drives plant host susceptibility to Tobacco etch virus. BMC Plant Biol. 14:67. doi: 10.1186/1471-2229-14-67
Fraile, A., and García-Arenal, F. (2012). “Virus-plant co-evolution,” in eLS (Chichester: John Wiley & Sons Ltd). doi: 10.1002/9780470015902.a0023723
Gallois, J.-L., Charron, C., Sánchez, F., Pagny, G., Houvenaghel, M.-C., Moretti, A., et al. (2010). Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso) 4E and eIF(iso) 4G. J. Gen. Virol. 91, 288–293. doi: 10.1099/vir.0.015321-0
Gao, Z., Johansen, E., Eyers, S., Thomas, C. L., Noel Ellis, T. H., and Maule, A. J. (2004). The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40, 376–385. doi: 10.1111/j.1365-313X.2004.02215.x
Habekuss, A., Kühne, T., Krämer, I., Rabenstein, F., Ehrig, F., Ruge-Wehling, B., et al. (2008). Identification of Barley mild mosaic virus isolates in Germany breaking rym5 resistance. J. Phytopathol. 156, 36–41.
Hariri, D., Meyer, M., and Prud’homme, H. (2003). Characterization of a new barley mild mosaic virus pathotype in France. Eur. J. Plant Pathol. 109, 921–928. doi: 10.1023/B:EJPP.0000003663.32298.f4
Hjulsager, C. K., Olsen, B. S., Jensen, D. M., Cordea, M. I., Krath, B. N., Johansen, I. E., et al. (2006). Multiple determinants in the coding region of Pea seed-borne mosaic virus P3 are involved in virulence against sbm-2 resistance. Virology 355, 52–61. doi: 10.1016/j.virol.2006.07.016
Hofinger, B. J., Russell, J. R., Bass, C. G., Baldwin, T., dos Reis, M., Hedley, P. E., et al. (2011). An exceptionally high nucleotide and haplotype diversity and a signature of positive selection for the eIF4E resistance gene in barley are revealed by allele mining and phylogenetic analyses of natural populations. Mol. Ecol. 20, 3653–3668. doi: 10.1111/j.1365-294X.2011.05201.x
Jiang, J., and Laliberté, J. F. (2011). The genome-linked protein VPg of plant viruses—a protein with many partners. Curr. Opin. Virol. 1, 347–354. doi: 10.1016/j.coviro.2011.09.010
Johansson, M. U., Zoete, V., Michielin, O., and Guex, N. (2012). Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics 13:173. doi: 10.1186/1471-2105-13-173
Jones, R. A. C. (2016). Future scenarios for plant virus pathogens as climate change progresses. Adv. Virus Res. 95, 87–147. doi: 10.1016/bs.aivir.2016.02.004
Kang, B.-C., Yeam, I., and Jahn, M. M. (2005a). Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. doi: 10.1146/annurev.phyto.43.011205.141140
Kang, B.-C., Yeam, I., Frantz, J. D., Murphy, J. F., and Jahn, M. M. (2005b). The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42, 392–405. doi: 10.1111/j.1365-313X.2005.02381.x
Kanyuka, K., Druka, A., Caldwell, D. G., Tymon, A., McCallum, N., Waugh, R., et al. (2005). Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol. Plant Pathol. 6, 449–458. doi: 10.1111/j.1364-3703.2005.00294.x
Kanyuka, K., McGrann, G., Alhudaib, K., Hariri, D., and Adams, M. J. (2004). Biological and sequence analysis of a novel European isolate of Barley mild mosaic virus that overcomes the barley rym5 resistance gene. Arch. Virol. 149, 1469–1480. doi: 10.1007/s00705-004-0318-7
Kashiwazaki, S., Ogawa, K., Usugi, T., Omura, T., and Tsuchizaki, T. (1989). Characterization of several strains of barley yellow mosaic virus. Ann. Phytopathol. Soc. Japan 55, 16–25. doi: 10.3186/jjphytopath.55.16
Kobayashi, S., Yoshida, H., and Sotome, K. (1987). Breeding for resistance to yellow mosaic disease in malting barley. Barley Genet. Newsl. 5, 667–672.
Konishi, T., and Kaiser, R. (1991). Genetic difference in barley yellow mosaic virus resistance between Mokusekko 3 and Misato Golden. Jpn. J. Breed. 41, 499–505. doi: 10.1270/jsbbs1951.41.499
Kühne, T. (2009). Soil-borne viruses affecting cereals: known for long but still a threat. Virus Res. 141, 174–183. doi: 10.1016/j.virusres.2008.05.019
Kühne, T., Shi, N., Proeseler, G., Adams, M. J., and Kanyuka, K. (2003). The ability of a bymovirus to overcome the rym4-mediated resistance in barley correlates with a codon change in the VPg coding region on RNA1. J. Gen. Virol. 84, 2853–2859.
Kyle, M., and Palloix, A. (1997). Proposed revision of nomenclature for potyvirus resistance genes in Capsicum. Euphytica 97, 183–188. doi: 10.1023/A:1003009721989
Léonard, S., Plante, D., Wittmann, S., Daigneault, N., Fortin, M. G., and Laliberté, J.-F. (2000). Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74, 7730–7737. doi: 10.1128/JVI.74.17.7730-7737.2000
Li, H., and Shirako, Y. (2015). Association of VPg and eIF4E in the host tropism at the cellular level of Barley yellow mosaic virus and Wheat yellow mosaic virus in the genus Bymovirus. Virology 476, 159–167. doi: 10.1016/j.virol.2014.12.010
Mäkinen, K., and Hafrén, A. (2014). Intracellular coordination of potyviral RNA functions in infection. Front. Plant Sci. 5:110. doi: 10.3389/fpls.2014.00110
Malys, N., and McCarthy, J. E. (2011). Translation initiation: variations in the mechanism can be anticipated. Cell. Mol. Life Sci. 68, 991–1003. doi: 10.1007/s00018-010-0588-z
Mazier, M., Flamain, F., Nicolaï, M., Sarnette, V., and Caranta, C. (2011). Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS ONE 6:e29595. doi: 10.1371/journal.pone.0029595
Monzingo, A. F., Dhaliwal, S., Dutt-Chaudhuri, A., Lyon, A., Sadow, J. H., Hoffman, D. W., et al. (2007). The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol. 143, 1504–1518. doi: 10.1104/pp.106.093146
Naderpour, M., Lund, O. S., Larsen, R., and Johansen, E. (2010). Potyviral resistance derived from cultivars of Phaseolus vulgaris carrying bc-3 is associated with the homozygotic presence of a mutated eIF4E allele. Mol. Plant Pathol. 11, 255–263. doi: 10.1111/j.1364-3703.2009.00602.x
Nakahara, K. S., Shimada, R., Choi, S.-H., Yamamoto, H., Shao, J., and Uyeda, I. (2010). Involvement of the P1 cistron in overcoming eIF4E-mediated recessive resistance against Clover yellow vein virus in pea. Mol. Plant Microbe Interact. 23, 1460–1469. doi: 10.1094/MPMI-11-09-0277
Nicaise, V., German-Retana, S., Sanjuán, R., Dubrana, M.-P., Mazier, M., Maisonneuve, B., et al. (2003). The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 132, 1272–1282. doi: 10.1104/pp.102.017855
Nishigawa, H., Hagiwara, T., Yumoto, M., Sotome, T., Kato, T., and Natsuaki, T. (2008). Molecular phylogenetic analysis of Barley yellow mosaic virus. Arch. Virol. 153, 1783–1786. doi: 10.1007/s00705-008-0163-1
Ohsato, S., Miyanishi, M., and Shirako, Y. (2003). The optimal temperature for RNA replication in cells infected by Soil-borne wheat mosaic virus is 17°C. J. Gen. Virol. 84, 995–1000. doi: 10.1099/vir.0.19021-0
Olspert, A., Chung, B. Y. W., Atkins, J. F., Carr, J. P., and Firth, A. E. (2015). Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 16, 995–1004. doi: 10.15252/embr.201540509
Ordon, F., Habekuss, A., Kastirr, U., Rabenstein, F., and Kühne, T. (2009). Virus resistance in cereals: sources of resistance, genetics and breeding. J. Phytopathol. 157, 535–545. doi: 10.1111/j.1439-0434.2009.01540.x
Revers, F., and García, J. A. (2015). Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. doi: 10.1016/bs.aivir.2014.11.006
Robaglia, C., and Caranta, C. (2006). Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. doi: 10.1016/j.tplants.2005.11.004
Rodamilans, B., Valli, A., Mingot, A., San León, D., Baulcombe, D., López-Moya, J. J., et al. (2015). RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J. Virol. 89, 6965–6967. doi: 10.1128/JVI.00337-15
Roudet-Tavert, G., Michon, T., Walter, J., Delaunay, T., Redondo, E., and Le Gall, O. (2007). Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HC-Pro. J. Gen. Virol. 88, 1029–1033. doi: 10.1099/vir.0.82501-0
Ruffel, S., Dussault, M.-H., Palloix, A., Moury, B., Bendahmane, A., Robaglia, C., et al. (2002). A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32, 1067–1075. doi: 10.1046/j.1365-313X.2002.01499.x
Ruffel, S., Gallois, J. L., Lesage, M. L., and Caranta, C. (2005). The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 274, 346–353. doi: 10.1007/s00438-005-0003-x
Ruffel, S., Gallois, J. L., Moury, B., Robaglia, C., Palloix, A., and Caranta, C. (2006). Simultaneous mutations in translation initiation factors eIF4E and eIF (iso) 4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87, 2089–2098. doi: 10.1099/vir.0.81817-0
Sanfaçon, H. (2015). Plant translation factors and virus resistance. Viruses 7, 3392–3419. doi: 10.3390/v7072778
Schaad, M. C., Anderberg, R. J., and Carrington, J. C. (2000). Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273, 300–306. doi: 10.1006/viro.2000.0416
Shirako, Y., and Brakke, M. K. (1984). Two purified RNAs of soil-borne wheat mosaic virus are needed for infection. J. Gen. Virol. 65, 119–127. doi: 10.1099/0022-1317-65-1-119
Sotome, T., Kawada, N., Kato, T., Sekiwa, T., Nishigawa, H., Natsuaki, T., et al. (2010). The current and new strains of barley yellow mosaic virus (BaYMV) in Tochigi prefecture. Jpn. J. Crop Sci. 79, 29–36. doi: 10.1626/jcs.79.29
Stein, N., Perovic, D., Kumlehn, J., Pellio, B., Stracke, S., Streng, S., et al. (2005). The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 42, 912–922. doi: 10.1111/j.1365-313X.2005.02424.x
Tamada, T., and Kondo, H. (2013). Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors. J. Gen. Plant Pathol. 79, 307–320. doi: 10.1007/s10327-013-0457-3
Tavert-Roudet, G., Abdul-Razzak, A., Doublet, B., Walter, J., Delaunay, T., German-Retana, S., et al. (2012). The C terminus of lettuce mosaic potyvirus cylindrical inclusion helicase interacts with the viral VPg and with lettuce translation eukaryotic initiation factor 4E. J. Gen. Virol. 93, 184–193. doi: 10.1099/vir.0.035881-0
Truniger, V., and Aranda, M. A. (2009). Recessive resistance to plant viruses. Adv. Virus Res. 75, 119–159. doi: 10.1016/S0065-3527(09)07504-6
Wang, A., and Krishnaswamy, S. (2012). Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. doi: 10.1111/j.1364-3703.2012.00791.x
Yamaguchi, Y., and Shirako, Y. (2002). Engineering of a Sagiyama alphavirus RNA-based transient expression vector. Microbiol. Immunol. 46, 119–129. doi: 10.1111/j.1348-0421.2002.tb02668.x
Yamamiya, A., and Shirako, Y. (2000). Construction of full-length cDNA clones to Soil-borne wheat mosaic virus RNA1 and RNA2, from which infectious RNAs are transcribed in vitro: virion formation and systemic infection without expression of the N-terminal and C-terminal extensions to the capsid protein. Virology 277, 66–75. doi: 10.1006/viro.2000.0587
Yeam, I., Cavatorta, J. R., Ripoll, D. R., Kang, B.-C., and Jahn, M. M. (2007). Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19, 2913–2928. doi: 10.1105/tpc.107.050997
You, Y., and Shirako, Y. (2010). Bymovirus reverse genetics: requirements for RNA2-encoded proteins in systemic infection. Mol. Plant Pathol. 11, 383–394. doi: 10.1111/j.1364-3703.2010.00613.x
You, Y., and Shirako, Y. (2012). Influence of amino acid at position 132 in VPg on replication and systemic infection of Barley yellow mosaic virus. Virus Res. 166, 121–124. doi: 10.1016/j.virusres.2012.03.001
You, Y., and Shirako, Y. (2013). Evaluation of host resistance to Barley yellow mosaic virus infection at the cellular and whole-plant levels. Plant Pathol. 62, 226–232. doi: 10.1111/j.1365-3059.2012.02616.x
Keywords: barley yellow mosaic virus, eIF4E, rym4/5/6, resistance-breaking, VPg, virus replication, virus movement, mesophyll protoplast
Citation: Li H, Kondo H, Kühne T and Shirako Y (2016) Barley Yellow Mosaic Virus VPg Is the Determinant Protein for Breaking eIF4E-Mediated Recessive Resistance in Barley Plants. Front. Plant Sci. 7:1449. doi: 10.3389/fpls.2016.01449
Received: 09 August 2016; Accepted: 12 September 2016;
Published: 30 September 2016.
Edited by:
Mario Tavazza, Italian National Agency for New Technologies, Energy and Sustainable Economic Development, ItalyReviewed by:
Helene Sanfacon, Agriculture and Agriculture-Food Canada, CanadaJuan Jose Lopez-Moya, Centre for Research in Agricultural Genomics, Spanish National Research Council – Catalan Institute for Food and Agricultural Research and Technology – Autonomous University of Barcelona – University of Barcelona, Spain
Copyright © 2016 Li, Kondo, Kühne and Shirako. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huangai Li, huangaili@yahoo.com Yukio Shirako, ushirako@mail.ecc.u-tokyo.ac.jp
†Present address: Huangai Li, School of Life Sciences, Tsinghua University, Beijing, China
 Huangai Li
Huangai Li Hideki Kondo
Hideki Kondo Thomas Kühne4
Thomas Kühne4 Yukio Shirako
Yukio Shirako