- Chongqing Engineering Research Center for Rapeseed, College of Agronomy and Biotechnology, Southwest University, Chongqing, China
Sucrose is the principal transported product of photosynthesis from source leaves to sink organs. SUTs/SUCs (sucrose transporters or sucrose carriers) and SWEETs (Sugars Will Eventually be Exported Transporters) play significant central roles in phloem loading and unloading. SUTs/SUCs and SWEETs are key players in sucrose translocation and are associated with crop yields. The SUT/SUC and SWEET genes have been characterized in several plant species, but a comprehensive analysis of these two gene families in oilseed rape has not yet been reported. In our study, 22 and 68 members of the SUT/SUCs and SWEET gene families, respectively, were identified in the oilseed rape (Brassica napus) genome through homology searches. An analysis of the chromosomal distribution, phylogenetic relationships, gene structures, motifs and the cis-acting regulatory elements in the promoters of BnSUC and BnSWEET genes were analyzed. Furthermore, we examined the expression of the 18 BnSUC and 16 BnSWEET genes in different tissues of “ZS11” and the expression of 9 BnSUC and 7 BnSWEET genes in “ZS11” under various conditions, including biotic stress (Sclerotinia sclerotiorum), abiotic stresses (drought, salt and heat), and hormone treatments (abscisic acid, auxin, cytokinin, brassinolide, gibberellin, and salicylic acid). In conclusion, our study provides the first comprehensive analysis of the oilseed rape SUC and SWEET gene families. Information regarding the phylogenetic relationships, gene structure and expression profiles of the SUC and SWEET genes in the different tissues of oilseed rape helps to identify candidates with potential roles in specific developmental processes. Our study advances our understanding of the important roles of sucrose transport in oilseed rape.
Introduction
Sucrose is the principal transported product of photosynthesis from source leaves to sink organs (Peng et al., 2014). Sucrose transporters/sucrose carriers (SUTs/SUCs) and SWEETs (Sugars Will Eventually be Exported Transporters) play significant central roles in phloem loading and unloading (Kuhn and Grof, 2010; Chen et al., 2012).
The first SUC was identified in spinach (Spinacia oleracea L.) (Riesmeier et al., 1992). Until now, cDNAs for SUCs have been isolated in almost all higher plants (Sauer and Stolz, 1994; Barker et al., 2000; Aoki et al., 2002). Previous studies have revealed that SUCs are located in the plasma membranes of the sieve element and companion cells and are expressed in various tissues from source leaves to sink tissues (Aoki et al., 2004; Sauer et al., 2004; Sivitz et al., 2005; Liesche et al., 2008; Tang et al., 2010). Phylogenetic analysis has indicated that SUCs can be divided into three types based on sequence homology and biochemical activity (Aoki et al., 2003). Briefly, type I is eudicot-specific, and functions in phloem loading (Riesmeier et al., 1994; Gottwald et al., 2000) and normal pollen function (Sivitz et al., 2008). Type II and III SUCs are found in all plant species, and monocots utilize type II SUCs for phloem loading (Slewinski et al., 2009). Type III SUCs, characterized as H+-coupled symporters (Weise et al., 2000), are localized to the vacuolar membrane (Endler et al., 2006; Reinders et al., 2008) and function in sucrose uptake into the cytoplasm (Reinders et al., 2008; Schulz et al., 2011), whereas types I and II are localized to the plasma membrane. In Arabidopsis, seven SUC-like genes and two pseudogenes have been characterized (Sauer et al., 2004). Five of these SUC-like genes (AtSUC1/2/5/8/9) belong to type I, AtSUC3 is categorized as type II, and AtSUC4 is grouped to type III. AtSUC2 is the best-characterized type I SUC, and mutational analysis has shown that SUCs are responsible for restraining plant growth and pollen germination (Gottwald et al., 2000; Sivitz et al., 2008; Srivastava et al., 2009). Antisense transformation experiments have also revealed that SUCs are responsible for the retardation of sucrose translocation, fruit size reduction, and reduced fertility in tomato (Riesmeier et al., 1994; Hackel et al., 2006).
Because SUCs load the sieve element companion cell complex with sucrose from the cell wall space, sucrose exits the symplasm with the help of sucrose transporting SWEETs (Chen et al., 2012). In marked contrast to SUCs, the newly identified class of sugar transporters called SWEETs increased our understanding of cellular sugar export. SWEET proteins consist of only seven predicted transmembrane domains (Chen et al., 2010) and have been characterized in plants, animals and humans. A phylogenetic tree of SWEET proteins from 15 plant taxa focusing on angiosperms was constructed by Eom (Kuhn and Grof, 2010). According to this tree, 17 members of the SWEET family in Arabidopsis thaliana fell into four phylogenetic clades. According to their phylogenetic relationships, SWEETs 1–3 were in clade I, SWEETs 4–8 in clade II, SWEETs 9–15 in clade III, and SWEETs 16–17 in clade IV. Clade I and II members prefer hexose transporters, and clade III members are efficient sucrose transporters. Clade IV SWEETs in Arabidopsis (AtSWEET16–17) are located on the tonoplast membrane and likely transport fructose (Chen et al., 2012; Klemens et al., 2013). AtSWEET8 and 13 feed pollen, and SWEET9 is essential for nectar secretion (Chen et al., 2010; Lin et al., 2014). SWEET11 and 12 provide sucrose to the SUTs for phloem loading. Mutations in either the AtSWEET11 or 12 genes produced no obvious phenotypes, but double mutants (atsweet11; 12) exhibited moderate defects in sucrose phloem transport and an excessive accumulation of carbohydrates in the leaves. GFP fusions indicate that AtSWEET11, 12 and 15 are expressed in the seed coat and endosperm. The triple knockout mutant exhibited a severe delay in embryo development and a wrinkled seed phenotype at maturity because of lower starch and lipid content and a smaller embryo (Lin et al., 2014). The remaining family members await characterization and could be involved in the gametophyte stage and in sugar transport in the plant.
Oilseed rape (Brassica napus L.) is a major global oil crop that is used for direct human consumption, as animal feed, and more recently, as a source of biofuel. High seed yield is one of the most important challenges in B. napus breeding, whereas the harvest index (HI) is only approximately 0.2–0.3 (Luo et al., 2015). Studies have implicated that the source and sink organs are not limiting, whereas assimilation translocation is the key limiting factor for seed yield in Brassica (Shen et al., 2005). SUCs and SWEETs may be critical genes for increasing seed yield by translocating sucrose from the source to the sink. However, information regarding SUC and SWEET in B. napus is lacking. In B. napus, the homologe of AtSUC1/5, BnSUC1C, was cloned, and its mRNA expression profiles were determined (Li et al., 2013). The results indicated that BnSUC1C is predominantly expressed in the later developmental stage of the anther. In another study, Song et al. (2015) determined the expression patterns of BnSUC family members in leaf, flower, silique, and seed development. Unfortunately, no characterized SWEET member in B. napus has been reported.
In our study, we used the BLASTP search program to query for SUC and SWEET family members in the genomes of three species, B. napus, B. rapa, and B. oleracea, using the AtSUC and AtSWEET protein sequences as the query, respectively. Their expression patterns were also determined using unpublished RNA-Seq data from our laboratory. Further analysis provided new insights into the mechanisms and regulation of assimilate allocation and a new potential for increasing crop yield.
Materials and Methods
Plant Materials and Treatments
Plants of oilseed rape (Brassica napus Zhongshuang11) were grown in field conditions in Chongqing, China. To analyse transcripts of BnSUC/BnSWEET members in different tissues, roots (R), stems (ST), senescent leaves (SL), extended leaves (EL), buds (B), flowers (F), stalk (ST), silique walls (21 DAF, SW), seeds (21 DAF, SE), and main inflorescences (MI) were collected from “ZS11”. All tissues were quickly frozen in liquid nitrogen and stored at −80°C until use.
For analysis of BnSUC/BnSWEET members under various abiotic or exogenous hormone stresses, oilseed rape seedlings (at the four-leaf stage) were grown in a greenhouse under long-day conditions (16-h light, 8-h dark) and transferred to 1/2 Hoagland solution 24 h before the induction of different stresses. A final concentration of 200 mM NaCl or 20% polyetheleneglycol-6000 (PEG-6000) was used for salt or drought stress, respectively. For heat stress, seedlings were transferred to a growth chamber at 40°C. For Sclerotinia sclerotiorum stress, methods were in accord with Li et al. (2015). For hormone treatment, 50 μM ABA (GenTel, Beijing, China), 100 μM GA3 (GenTel, Beijing, China), 10 μM BR (GenTel, Beijing, China), 75 μM NAA (GenTel, Beijing, China), 75 μM 6-BA (GenTel, Beijing, China), and 2 mM SA (GenTel, Beijing, China) were employed (Yang et al., 2009; Gao et al., 2010). Leaves were collected at 0, 3, 6, 12, 24, 48, and 72 h after salt, drought and heat treatments and 0, 0.5, 1, 3, 6, 12, and 24 h after various exogenous hormone treatments and immediately frozen in liquid nitrogen for storage at −80°C until use.
Identification of Sucrose Transporters in B. napus, B. rapa, and B. oleracea
Brassica rapa and Brassica oleracea sucrose transporters SUC and SWEET were identified by performing a BLASTP analysis (Altschul et al., 1997) with the BRAD database (http://brassicadb.org/brad/index.php) and the B. napus genome (http://www.genoscope.cns.fr/brassicanapus/) at a cut-off value of < E−20 using the A. thaliana sucrose transporter SUC and SWEET amino acid sequences as the query sequences, respectively.
Phylogenetic Analyses of the SUC and SWEET Families in B. napus
To gain insights into the evolutionary relationships of oilseed rape SUC and SWEET proteins, we performed multiple alignments of the SUC and SWEET proteins of certain species (A. thaliana, B. rapa, B. oleracea, and B. napus). Multiple sequence alignments of the deduced amino acid sequences of the SUC and SWEET proteins were performed using the default parameters of ClustalW (Eom et al., 2015). Dendrograms were generated by the MEGA 6 program (Tamura et al., 2013) using the neighbor-joining (NJ) method and bootstrap analysis (1000 replications).
Protein Properties and Sequence Analyses
The molecular weight (MW) and isoelectric points (pI) of the presumed sucrose transporter proteins were predicted by the online ExPASy proteomics server database (http://expasy.org/). The Gene Structure Display Server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/index.php) was used to generate the exon/intron organization. Motifs were identified using the MEME program (http://meme-suite.org/). The maximum number of motifs was 25 and 29 in the BnSUC and BnSWEET proteins, respectively, and the optimum width of the motifs was set from 6 to 50. Furthermore, all identified motifs were annotated according to InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/).
Promoter Cis-Element Analysis of BnSUCs and BnSWEETs
The promoter sequences (1.5 kb upstream of the translation start site) of the BnSUC and BnSWEET genes were obtained from the B. napus genome (http://www.genoscope.cns.fr/brassicanapus/). PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to analyse the BnSUC and BnSWEET gene promoters and identify their cis-elements (Rombauts et al., 1999; Lescot et al., 2002).
RNA Isolation and Real-Time Quantitative RT-PCR
Total RNA was extracted from all samples with the RNeasy extraction Kit (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 1 μg of total RNA using M-MLV transcriptase (TaKaRa Biotechnology, Dalian, China) according to the manufacturer's instructions after the contaminated genomic DNA was removed by DNase I treatment. Real-time PCR was used to determine the expression levels of BnSUCs and BnSWEETs in different tissues and in response to various stresses. Quantitative real-time PCR was performed according to methods described in Wei et al. (2016). To obtain precise and reproducible results, each sample was replicated three times. As many studies have suggested only SWEET9 through SWEET15 function as sucrose transporters, we only performed expression analysis of BnSWEET9–BnSWEET15 in various tissues and stresses. Because of their high similarities, 18 BnSUC and 16 BnSWEET gene primers were designed for quantitative RT-PCR analysis (Table S1).
Results
Identification and Phylogenetic Analysis of Oilseed Rape SUCs and SWEETs
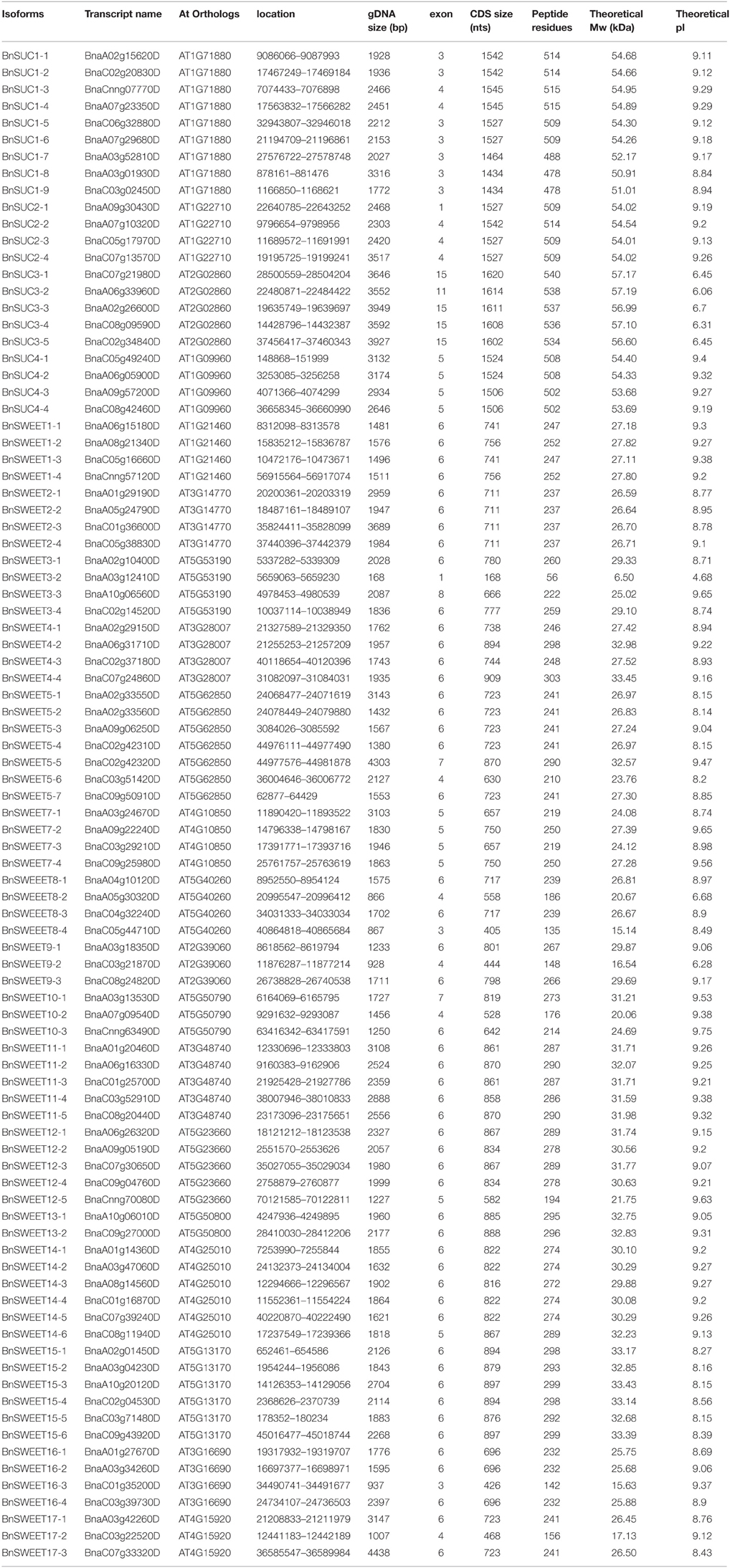
To identify all putative SUC and SWEET protein sequences in B. napus, B. rapa, and B. oleracea, BLASTP analysis was conducted. Through this approach, 22, 9, and 8 SUC genes were identified. The 22 predicted BnSUCs proteins ranged from 478 (BnaSUC1- 8) to 540 (BnaSUC3-1) amino acid (aa) residues in length, with an average length of 512 aa. The relative molecular mass varied from 50.91 kDa (BnSUC1-8) to 57.19 kDa (BnSUC3-2). The pI values ranged from 6.06 (BnSUC3-2) to 9.40 (BnSUC4-1), with 17 members exhibiting pI values >7 (Table S1). Sixty-eight, 26 and 16 SWEET proteins were identified in the B. napus, B. rapa, and B. oleracea genomes, respectively. The 68 predicted BnSWEET proteins ranged from 56 (BnSWEET3-2) to 303 (BnSWEET4-4) aa in length, with an average length of 245.5 aa. The relative molecular mass varied from 6.5 kDa (BnSWEET3-2) to 33.45 kDa (BnSWEET4-4). The pIs ranged from 4.68 (BnSWEET3-2) to 9.75 (BnSWEET10-3), with 92.6% (63) members exhibiting pI values >7, and the other three having pI values < 7 (Table 1).
To study the evolutionary relationships among oilseed rape SUC and SWEET proteins and known SUCs and SWEETs from A. thaliana, B. rapa and B. oleracea, an unrooted neighbor-joining phylogenetic tree was created using the amino acid sequences of the SUC and SWEET family proteins from oilseed rape, A. thaliana, B. rapa, and B. oleracea. Based on our multiple sequence alignment and phylogenetic analysis, SUCs cluster into three types, consistent with previous studies using angiosperms SUCs (Figure 1). Briefly, 26 SUC proteins, including 13 BnSUCs, seven AtSUCs, four BrSUCs, and two BoSUCs, were most likely type I SUCs. AtSUC3, five BnSUC3s, two BrSUC3s, and two BoSUC3s belonged to type II. Type III consisted of four BnSUC4s, two BrSUC4s, two BoSUC4s, and AtSUC4. Although the SWEET gene family is large, 126 members from A. thaliana, B. napus, B. rapa, and B. oleracea were clustered into four clades (I, II, III, and IV), containing 21, 36, 56, and 13 members, respectively (Figure 2).
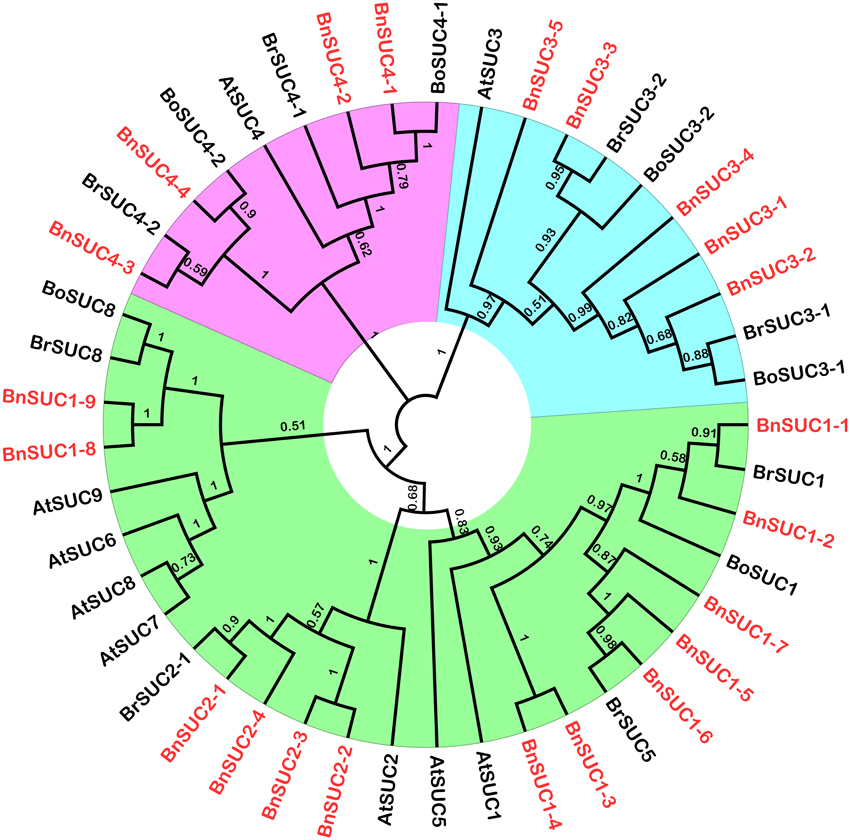
Figure 1. The phylogenetic analysis of SUC proteins from A. thaliana, B. oleracea, B. rape and B. napus. Forty-five SUCs were used to construct the NJ tree with 1000 bootstraps based on the protein sequences. The SUC proteins were grouped into three distinct types.
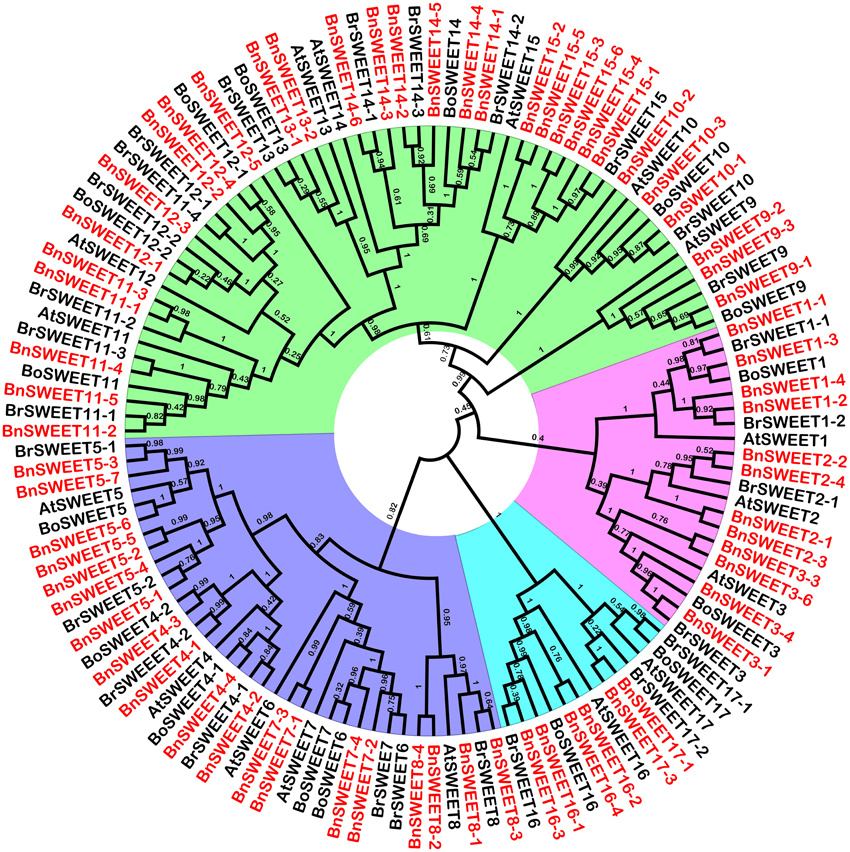
Figure 2. The phylogenetic analysis of SWEET proteins from A. thaliana, B. oleracea, B. rape and B. napus. One hundred and twenty-six. SWEETs proteins were used to construct the NJ tree with 1000 bootstraps based on the protein sequences. The SWEET proteins were clustered into four clades.
The Chromosomal Location, Gene Structure, and Conserved Motifs of Oilseed Rape SUCs and SWEETs
The locations of the BnSUC and BnSWEET genes are shown in Figure 3. Approximately 50% of the two gene families are located on the A or C genomes, and the precise chromosomal positions of the BnSUC and BnSWEET genes are listed in Table S1. The 22 BnSUCs are located on 14 chromosomes in B. napus. Chromosome A07 contains 3 SUC genes, whereas chromosomes A02, A03, A06, C02, C07, and C08 each contain two SUC genes. Chromosomes A09, C03, C05, C06, A09_random, C05_random and Cnn_random each contain one SUC gene (Figure 3). As shown in Figure 3, 68 BnSWEET genes are located on A09_random, C09_random, Cnn_random, and on 18 of the 19 chromosomes (the exception is chromosome C06). Chromosome A03 contains the largest number (8) of BnSWEET genes, followed by chromosome C03, which contains six members. Chromosome A02 and C02 each contain five members. Five chromosomes each contain four genes, and another five chromosomes (A09, A10, C05, C08, and Cnn_random) each contain three members. Chromosome A05 and A08 each contain two members, and the other five chromosomes (A04, A07, C04, C03_random, and C09_random) each contain only one member.
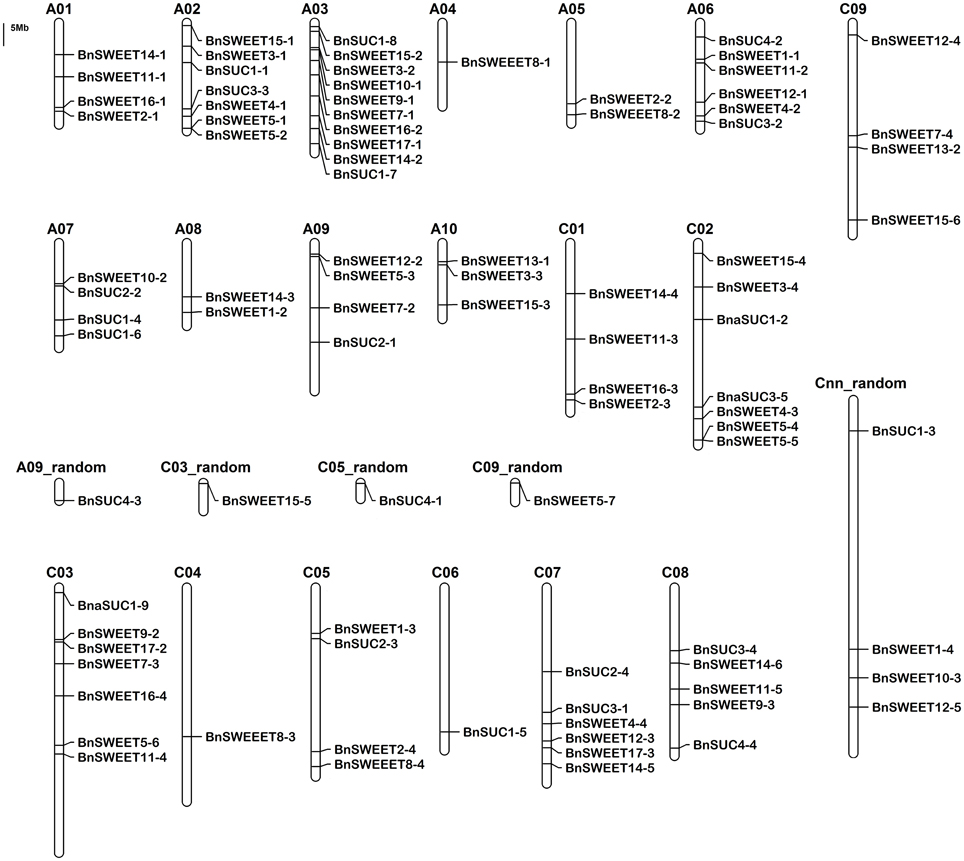
Figure 3. The distribution of the BnSUC and BnSWEET genes in the B. napus genome. The chromosomal position of each BnSUC and BnSWEET gene was mapped according to the B. napus genome. The chromosome number is indicated at the top of each chromosome. The scale is in megabases (Mb).
Previous studies have determined that gene divergence and duplication events are the major reasons for evolutionary momentum (Vision et al., 2000; Bowers et al., 2003). In our studies, eight pairs of segmental duplications were identified for the following BnSUCs: BnSUC1-1/BnSUC1-2, BnSUC1-3/BnSUC1-4, BnSUC1-5/BnSUC1-6, BnSUC1-8/BnSUC1-9, BnSUC2-1/BnSUC2-4, BnSUC2-2/BnSUC2-3, BnSUC4-1/BnSUC4-2, and BnSUC4-3/BnSUC4-4 (Figure 4). Twenty-three sister gene pairs (including two tandem duplicates: BnSWEET5-1/BnSWEET5/2 and BnSWEET5-4/BnSWEET5/5) were identified for the BnSWEET gene family and are shown in Figure 5.
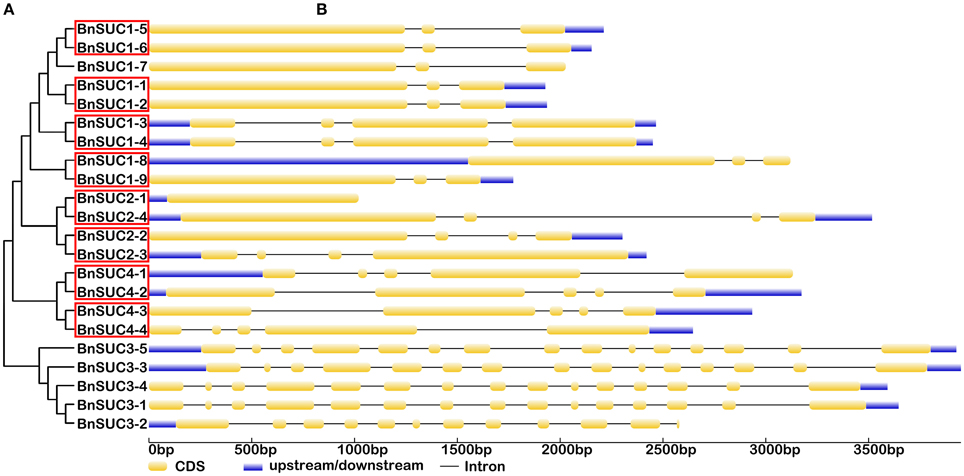
Figure 4. The exon-intron structure of the BnSUC genes according to their phylogenetic relationships. An unrooted phylogenetic tree was constructed with 1000 bootstraps based on the full-length sequences of BnSUC (A). Exon-intron structure analyses of the BnSUC genes were performed using the online tool GSDS (B). The lengths of the exons and introns of each BnSUC gene are proportional. Eight segmental duplicates are highlighted by the red box.
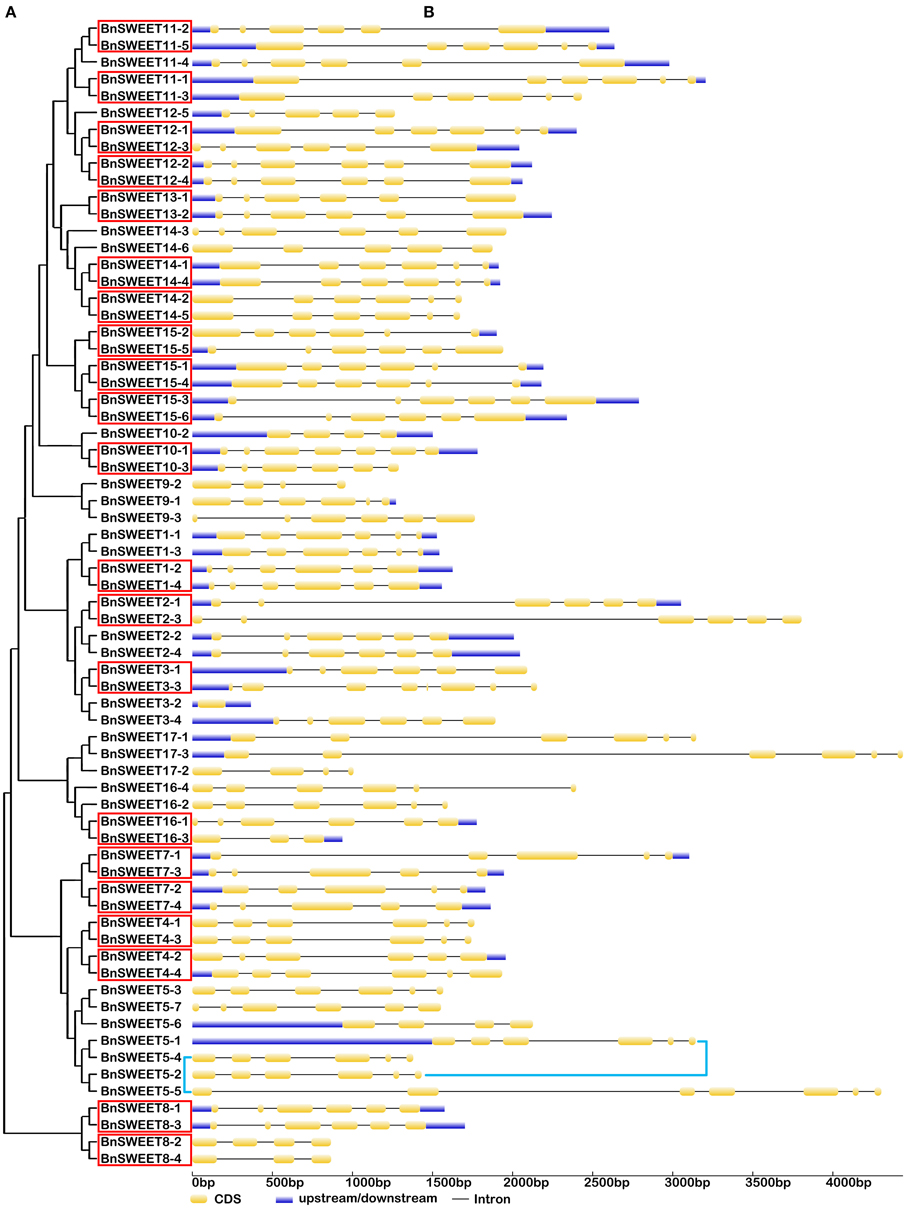
Figure 5. The exon-intron structure of the BnSWEET genes according to their phylogenetic relationships. An unrooted phylogenetic tree was constructed with 1000 bootstraps based on the full-length sequences of BnSWEET (A). Exon-intron structure analyses of the BnSWEET genes were performed using the online tool GSDS (B). The lengths of the exons and introns of each BnSWEET gene are proportional. Twenty-one segmental duplicates are highlighted by red boxes. Two tandem duplicates are highlighted by blue lines.
Gene structural diversity and conserved motif divergence play important roles in the evolution of the SWEET/SUC gene family (Hu et al., 2010; Xu et al., 2012). To further examine the structural features of oilseed rape SUC and SWEET genes, we compared their exon/intron organizations. As shown in Figure 4, the most closely related BnSUC genes within the same type shared similar gene structures in terms of either intron numbers or exon lengths. For example, type I genes had one to four exons. Seven of the 13 type I genes had three exons, and the rest had five exons, except BnSUC2-1, which had only one exon. By contrast, all type III genes had five exons, and type II members had 15 exons, except BnSUC3-2, which had 11 exons. We analyzed the gene structures of BnSWEET members using the same approach (Figure 5). Most members (75%) had six exons, and 1, 2, 6, 5, 2, and 1 had 8, 7, 5, 4, 3, and 1 exons, respectively. Overall, 83.3, 57.9, 83.3, and 71.4% of the members in clades I, II, III, and IV contained six exons, respectively.
Twenty-five and 29 putative protein motifs were predicted using the MEME program for oilseed rape BnSUC (Figure 6) and BnSWEET (Figure 7) proteins, respectively. For BnSUC proteins, motifs 1-13 were observed in all 22 BnSUC proteins. Motifs 15, 17, 22, and 23 were only detected in the type I subgroup. Motifs 14, 19, and 25 were detected only in type II. Motif 16 was only detected in the type III subgroup. According to InterProScan annotation, motifs 1-8 were SUC1-RELATED sucrose transport proteins. Motifs 1-5 were observed in almost all of the BnSWEET proteins and accounted for 92.6, 89.7, 91.2, 92.6, and 85.3% of the motifs, respectively. Motifs 1-3 were annotated as SWEET sugar transporters. Notably, BnSWEET3-2, BnSWEET8-2, BnSWEET8-4, BnSWEET9-2, BnSWEET10-2, BnSWEET16-3, and BnSWEET17-2 were the most diverse, consistent with their gene structures. Generally, proteins with similar motif compositions were clustered in the same class, indicating similar functions among members of the same class.
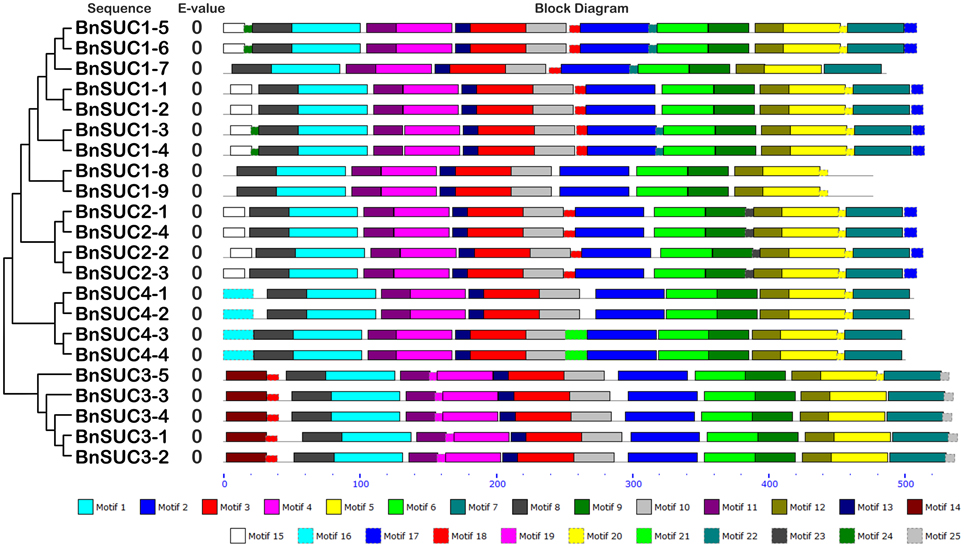
Figure 6. The conserved motifs of the BnSUC proteins according to their phylogenetic relationships. The conserved motifs of the BnSUC proteins were identified by MEME. Gray lines represent the non-conserved sequences, and each motif is indicated by a colored box numbered at the bottom. The lengths of motifs in each protein are proportional.
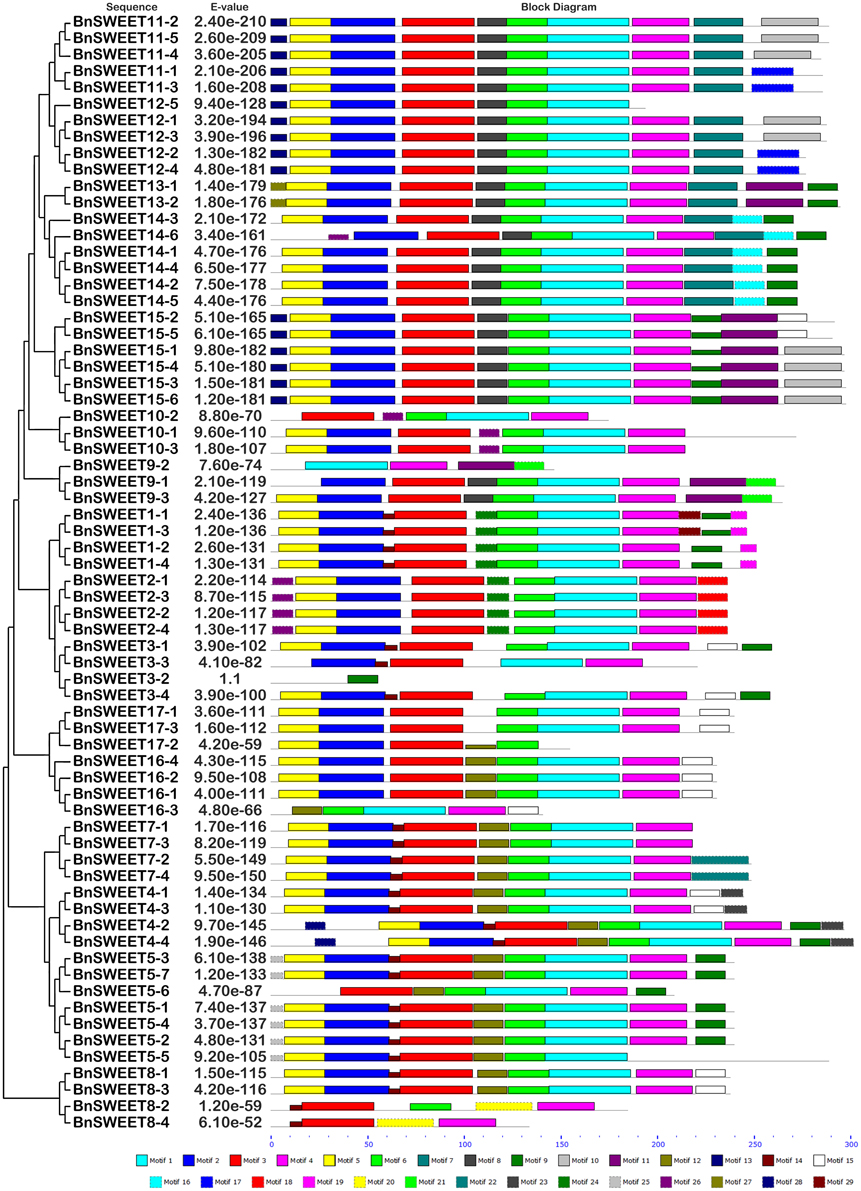
Figure 7. The conserved motifs of the BnSWEET proteins according to their phylogenetic relationships. The conserved motifs in the BnSWEET proteins were identified by MEME. Gray lines represent the non-conserved sequences, and each motif is indicated by a colored box numbered at the bottom. The lengths of motifs in each protein are proportional.
The Cis-Acting Regulatory Elements in the Promoter of BnSUCs and BnSWEETs
Cis-regulatory elements, which act as binding sites for TFs, are critical for determining the expression patterns of genes (Liu et al., 2013). To further understand the transcriptional regulation and potential functions of BnSUCs and BnSWEETs, the 1500 bp regions upstream of the transcriptional start codons were analyzed using the PLACE database to identify cis-regulatory elements. Ninety-eight types of cis-regulatory elements were detected in the promoters of BnSUC genes, including the TATA-box, CAAT-box, and light responsive elements. In addition, some types of elements are potentially responsive to stresses, including the heat shock element (HSE), the low temperature-responsive element (LTR), the defense and stress-responsive element (TC-rich repeats), the MYB binding site involved in drought-inducibility (MBS), and elements that act in response to hormones, such as the gibberellin-responsive element (GARE-motif), the SA response (TCA-element), the MeJA response (CGTCA motif), and the ABA response (ABRE). All 22 BnSUC genes contained 5-19 cis-elements related to stress or hormone responses. Nineteen and 17 BnSUC genes contained the GARE motif (gibberellin-responsive element) and the CGTCA motif (MeJA response) in their promoter regions, respectively, suggesting that BnSUC genes may play key roles in responses to gibberellin and MeJA. In addition, 18, 16, and 15 BnSUC genes contained TC-rich repeats (defense and stress-responsiveness), MBSs (drought stress), and HSEs (heat stress), respectively. Among the 22 BnSUC genes, BnSUC1-2 may be involved in responses to low temperature stress, as implied by the presence of 10 LTR elements (Table S2).
As listed in Table S3, the top seven elements involved in stress and hormone responses were detected in the promoters of BnSWEETs. There were 90 instances of the heat shock element (HSE), 103 instances of the defense and stress-responsive element (TC-rich repeats), 80 instances of the MYB binding site involved in drought-inducibility (MBS), 42 instances of the gibberellin-responsive element (GARE-motif), 61 instances of the SA response (TCA-element), 73 instances of the MeJA response (CGTCA motif), and 58 instances of the ABA response element (ABRE).
Expression Profiles of Selected BnSUC and BnSWEET Genes in Different Tissues
To investigate the functions of BnSUC and BnSWEET genes, their gene expression profiles in different tissues were determined by qRT-PCR. Tissues were sampled from roots, stems, senescent leaves, extended leaves, buds, flowers, stalk, silique walls, seeds, and main inflorescences of “ZS11” to explore the expression patterns of selected BnSUCs and BnSWEETs. As illustrated in Figure 8, the selected BnSUC genes including 11 type I, 5 type II, and 2 type III members were mainly expressed in extended leaves and flowers, except 2 type II genes (BnSUC3-1, which was mainly expressed in roots and stalks, and BnSUC3-4, which was mainly expressed in main inflorescences and stems). Therefore, BnSUC3 might be involved in different functions with other members.
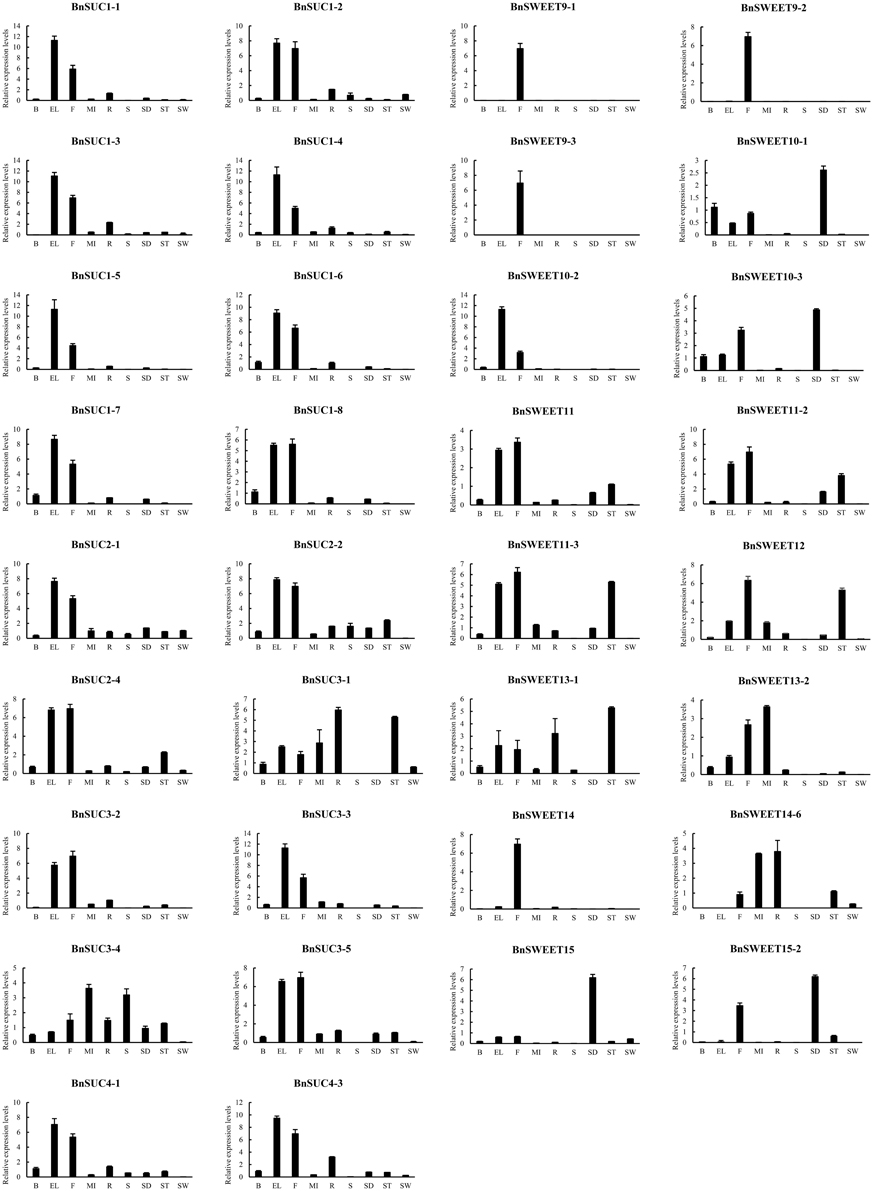
Figure 8. The tissue-specific expression patterns of selected BnSUC and BnSWEET genes in different tissues. The expression patterns of the selected BnSUC and BnSWEET genes in the nine indicated organs were analyzed by qRT-PCR. R: roots; S: stems; EL: extended leaves; B: buds; F: flowers; ST: stalk; SW: silique walls; SE: seeds; MI: main inflorescences.
The expression patterns of selected BnSWEETs in different tissues varied. Three BnSWEET9 genes and all BnSWEET14 genes were mostly expressed in flowers, indicating that BnSWEET9 and BnSWEET14 function in flower development. All BnSWEET15 members were highly expressed in developing seeds. The rest of the selected BnSWEET genes were expressed in a variety of tissues, particularly in extended leaves, developing seeds, stalks and main inflorescences. The three BnSWEET10 genes had different expression patterns, that is, BnSWEET10-1, and BnSWEET10-3 were mainly expressed in developing seeds, buds, flowers, and extended leaves, whereas BnSWEET10-2 was highly expressed in extended leaves and flowers but was not expressed in developing seeds and other tissues. Neither BnSWEET11 nor BnSWEET12 expression was detected in stems and silique walls, whereas BnSWEET13-1, and BnSWEET13-2 transcripts were not detected in developing seeds and silique walls. The high expression of BnSWEETs in specific tissues may indicate specific roles in the corresponding tissues.
Response of Selected BnSUC and BnSWEET Genes to Various Abiotic, Biotic, and Exogenous Hormone Stresses
To obtain insight on the roles of BnSUC and BnSWEET genes responding to various stresses, oilseed rape seedlings of ZS11 were subjected to salt, drought, heat, and S. sclerotiorum stresses. As shown in Figure 9, only BnSUC1-2, BnSWEET10-3 and BnSWEET12 were upregulated under salt stress, and the remaining genes were downregulated. BnSUC1-2, BnSWEET10-3, BnSWEET12, and BnSWEET14 were upregulated after 3 h under drought stress and then immediately decreased, except BnSWEET12. For heat stress, 11 of 16 genes were upregulated after 3 h and decreased immediately, except BnSUC1-2 and BnSUC2-4. For S. sclerotiorum infections, BnSUC1-1, BnSUC1-5, BnSUC2-2, BnSUC2-4, BnSWEET9-2, and BnSWEET11 were upregulated after 48 h.
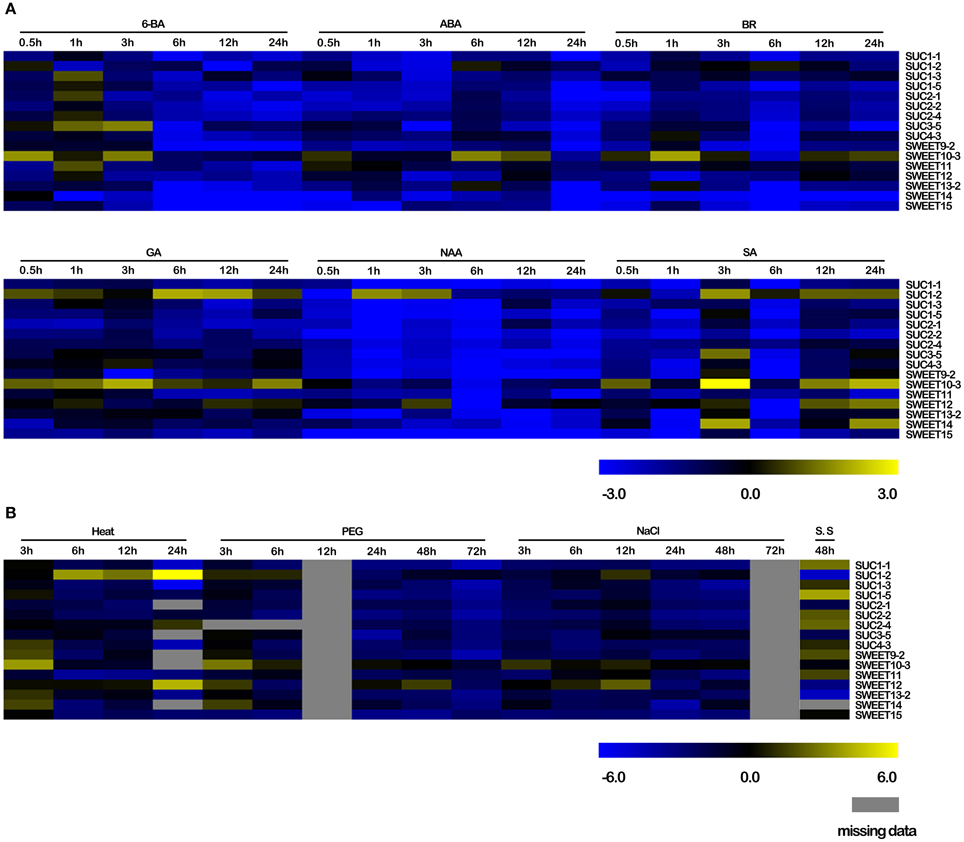
Figure 9. Responses of selected BnSUC and BnSWEET genes to various abiotic, biotic and exogenous hormone stresses. (A) 6-BA, 6-benzylaminopurine, a cytokinin, ABA, abscisic acid, BR, brassinolide, GA, gibberellin, NAA, auxin, SA, salicylic acid, (B) Heat, heat stress, NaCl, salt stress, PEG, drought stress, S.S, Sclerotinia sclerotiorum stress.
Plant hormones such as SA, auxin, NAA, BR, GA, and ABA play important roles in the regulation of developmental processes (Yang et al., 2012; Curaba et al., 2014). In our study, hormone treatments resulted in a wide variety of changes in the transcript levels of sucrose transporter genes in oilseed rape as determined by qRT-PCR. Briefly, all members were upregulated under GA stress, particularly BnSUC1-2, BnSUC3-5, BnSUC4-3, BnSWEET10-3, BnSWEET12, and BnSWEET13-2. These genes were also induced by ABA, BR, and 6-BA stresses, whereas only BnSUC1-2 and BnSWEET10-3 were induced under SA stress, and only BnSUC1-2 and BnSWEET12 were upregulated by NAA stress at some time points (Figure 9). These results indicate that hormones are involved in the regulation of sucrose transporters in oilseed rape.
Discussion
Sucrose transporters have important functions in plant growth and development, particularly in vascular tissues. Recently, preliminary analyses of the SUC and SWEET gene families have been conducted in many plant species (Aoki et al., 2004; Hackel et al., 2006; Baker et al., 2012; Chen et al., 2012). The results of these studies shed light on the functions of the SUC and SWEET genes. However, the SUC and SWEET gene families have not been studied in B. napus, an important oilseed crop. In our study, we analyzed the phylogenetics, intron-exon organization, chromosomal locations, conserved motifs, and expression patterns of the BnSUC and BnSWEET genes in various tissues and in stress responses.
The Evolution of the Oilseed Rape SUC and SWEET Genes
Polyploidization is considered an outstanding power of eukaryotic evolution and the main contributor to evolutionary events (Dun et al., 2014). After the Brassica genus of plants formed from A. thaliana, it triplicated its genome, with an expansion in gene numbers (Lysak et al., 2005, 2007). Generally, three syntenic copies of each gene in A. thaliana should be detected in a diploid Brassica species, such as B. rapa (Wang et al., 2011; Cheng et al., 2013; Dun et al., 2014), and B. oleracea (Cheng et al., 2014; Liu et al., 2014). Thus, the allotetraploid B. napus, which was generated from the spontaneous hybridization of B. rapa and B. oleracea approximately 7500–12,500 years ago (Chalhoub et al., 2014), should contain six copies of each Arabidopsis gene. However, the B. rapa (Mun et al., 2009; Wang et al., 2011), and B. napus (Chalhoub et al., 2014; Cheng et al., 2014) genomes only contain 1.5–2 and 2–6 copies of each gene in Arabidopsis, respectively, because of genome shrinkage and gene loss. In our study, a total of 22 SUC and 68 SWEET genes were identified in the genome of the oilseed rape.
Our molecular characterizations revealed great variations. The molecular masses and pIs varied markedly among subfamilies, whereas the SUC and SWEET proteins clustered in the same subfamily and shared closer molecular masses and pIs. Consistent with previous studies (Reinders et al., 2012; Chong et al., 2014), the members of the SUC and SWEET gene families in the genomes of A. thaliana, B. napus, B. rapa, and B. oleracea were classified into three and four groups, respectively. Variation in exon–intron structure plays a significant role in the evolution of gene families (Rogozin et al., 2005; Xu et al., 2012). Our studies of the SUC and SWEET gene families provide an explanation for this diversification in gene structure. Type II BnSUC3s contain more exons than type I and type III BnSUCs, indicating their diverse functions. These data, along with detected motif data, suggest similar origins, and evolutionary patterns for the SUC and SWEET genes of different species.
Expression Patterns of SUC and SWEET Genes in Various Tissues and Stress Responses
Comprehensive gene expression analyses of sucrose transporter family genes have revealed that SUCs have distinct expression patterns in various tissues of Arabidopsis, maize, wheat and grape (Meyer et al., 2004; Slewinski et al., 2009; Afoufa-Bastien et al., 2010; Mukherjee et al., 2015). Consistent with these previous results, BnSUCs also exhibited differential expression patterns in various tissues. Notably, BnSUC2 in the type I subclass was highly expressed in different tissues, particularly flowers and extended leaves, suggesting specific roles required in these tissue types. According to previous studies, AtSUC3, which was clustered into type II with the BnSUC3s, functions as a sugar signal-transducer. However, SUC1/2 were only observed in eudicot species, and these genes are necessary for phloem loading (Srivastava et al., 2008; Gould et al., 2012) and normal pollen function (Sivitz et al., 2008). Type III SUCs act as sucrose carriers and are localized to the vacuolar membrane (Endler et al., 2006; Schneider et al., 2012).
All clade III members of BnSWEETs were highly expressed in the extended leaves, flowers and developing seeds. Similar patterns have been observed in other plant species (Xuan et al., 2013; Lin et al., 2014). Some BnSWEETs exhibited high expression levels in specific tissues, implying specific roles required in these tissue types. For example, BnSWEET9-1/9-2/9-3, and BnSWEET14 were highly specifically expressed in flowers, and BnSWEET15 was abundantly expressed in developing seeds.
Several studies have examined regulators of sucrose transporters. Meyer et al. (2004) observed that AtSUC3 expression is strongly induced upon wounding of Arabidopsis tissues. Chincinska et al. (2008) subsequently determined that StSUT4 expression in wild-type plants was induced by GA and ET. Mukherjee et al. (2015) demonstrated that ABA negatively regulates sucrose import into the endosperm by repressing TaSUT1 in wheat using physiological, molecular and biochemical approaches. In our study, expression changes in 16 selected genes were detected under heat, drought, salt, S. sclerotiorum, and six hormone stresses. Notably, sucrose transporter genes in oilseed rape were positively regulated by cytokinin (6-BA) and GA, consistent with previous reports (Chincinska et al., 2008). By contrast, these selected genes were mostly downregulated under ABA, NAA, SA, NaCl, and PEG stresses. These results indicate that sucrose transporter genes (BnSUCs and BnSWEETs) are regulated by plant hormones and abiotic and biotic stresses. These results may facilitate improvements in crop yield.
Author Contributions
JL conceived and designed the experiments. HJ, KL, and BY performed the experiments. HJ, KL, BY, TW, LZ, AZ, JW, LL, and CQ analyzed the data. HJ and KL wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program) 2015CB150201, the National Natural Science Foundation of China (U1302266, 31271756) and the “111” Project (B12006).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01464
References
Afoufa-Bastien, D., Medici, A., Jeauffre, J., Coutos-Thevenot, P., Lemoine, R., Atanassova, R., et al. (2010). The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 10:245. doi: 10.1186/1471-2229-10-245
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Aoki, N., Hirose, T., Scofield, G. N., Whitfeld, P. R., and Furbank, R. T. (2003). The sucrose transporter gene family in rice. Plant Cell Physiol. 44, 223–232. doi: 10.1093/pcp/pcg030
Aoki, N., Scofield, G. N., Wang, X. D., Patrick, J. W., Offler, C. E., and Furbank, R. T. (2004). Expression and localisation analysis of the wheat sucrose transporter TaSUT1 in vegetative tissues. Planta 219, 176–184. doi: 10.1007/s00425-004-1232-7
Aoki, N., Whitfield, P., Hoeren, F., Scofield, G., Newell, K., Patrick, J., et al. (2002). Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol. Biol. 50, 453–462. doi: 10.1023/A:1019846832163
Baker, R. F., Leach, K. A., and Braun, D. M. (2012). SWEET as sugar: new sucrose effluxers in plants. Mol. Plant 5, 766–768. doi: 10.1093/mp/sss054
Barker, L., Kuhn, C., Weise, A., Schulz, A., Gebhardt, C., Hirner, B., et al. (2000). SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12, 1153–1164. doi: 10.1105/tpc.12.7.1153
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Chen, L. Q., Qu, X. Q., Hou, B. H., Sosso, D., Osorio, S., Fernie, A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
Cheng, F., Mandakova, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Cheng, F., Wu, J., and Wang, X. (2014). Genome triplication drove the diversification of Brassica plants. Hortic. Res. 1:14024. doi: 10.1038/hortres.2014.24
Chincinska, I. A., Liesche, J., Krugel, U., Michalska, J., Geigenberger, P., Grimm, B., et al. (2008). Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiol. 146, 515–528. doi: 10.1104/pp.107.112334
Chong, J., Piron, M. C., Meyer, S., Merdinoglu, D., Bertsch, C., and Mestre, P. (2014). The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 65, 6589–6601. doi: 10.1093/jxb/eru375
Curaba, J., Singh, M. B., and Bhalla, P. L. (2014). miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 65, 1425–1438. doi: 10.1093/jxb/eru002
Dun, X., Shen, W., Hu, K., Zhou, Z., Xia, S., Wen, J., et al. (2014). Neofunctionalization of duplicated Tic40 genes caused a gain-of-function variation related to male fertility in Brassica oleracea lineages. Plant Physiol. 166, 1403–1419. doi: 10.1104/pp.114.246470
Endler, A., Meyer, S., Schelbert, S., Schneider, T., Weschke, W., Peters, S. W., et al. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141, 196–207. doi: 10.1104/pp.106.079533
Eom, J. S., Chen, L. Q., Sosso, D., Julius, B. T., Lin, I. W., Qu, X. Q., et al. (2015). SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 25, 53–62. doi: 10.1016/j.pbi.2015.04.005
Gao, Y., Li, T., Liu, Y., Ren, C., Zhao, Y., and Wang, M. (2010). Isolation and characterization of gene encoding G protein α subunit protein responsive to plant hormones and abiotic stresses in Brassica napus. Mol. Biol. Rep. 37, 3957–3965. doi: 10.1007/s11033-010-0054-x
Gottwald, J. R., Krysan, P. J., Young, J. C., Evert, R. F., and Sussman, M. R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. U.S.A. 97, 13979–13984. doi: 10.1073/pnas.250473797
Gould, N., Thorpe, M. R., Pritchard, J., Christeller, J. T., Williams, L. E., Roeb, G., et al. (2012). AtSUC2 has a role for sucrose retrieval along the phloem pathway: evidence from carbon-11 tracer studies. Plant Sci. 188, 97–101. doi: 10.1016/j.plantsci.2011.12.018
Hackel, A., Schauer, N., Carrari, F., Fernie, A. R., Grimm, B., and Kuhn, C. (2006). Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 45, 180–192. doi: 10.1111/j.1365-313X.2005.02572.x
Hu, R., Qi, G., Kong, Y., Kong, D., Gao, Q., and Zhou, G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10:145. doi: 10.1186/1471-2229-10-145
Klemens, P. A., Patzke, K., Deitmer, J., Spinner, L., Le Hir, R., Bellini, C., et al. (2013). Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 163, 1338–1352. doi: 10.1104/pp.113.224972
Kuhn, C., and Grof, C. P. (2010). Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 13, 288–298. doi: 10.1016/j.pbi.2010.02.001
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, F., Yan, L., Lai, J., Ma, C., Gautam, M., and Fu, T. (2013). Molecular cloning and mRNA expression profile of sucrose transporter gene BnSUT1C from Brassica napus L. Indian J. Exp. Biol. 51, 1130–1136.
Li, J., Zhao, Z., Hayward, A., Cheng, H., and Fu, D. (2015). Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica 205, 483–489. doi: 10.1007/s10681-015-1417-0
Liesche, J., Schulz, A., Krugel, U., Grimm, B., and Kuhn, C. (2008). Dimerization and endocytosis of the sucrose transporter StSUT1 in mature sieve elements. Plant Signal. Behav. 3, 1136–1137. doi: 10.4161/psb.3.12.7096
Lin, I. W., Sosso, D., Chen, L. Q., Gase, K., Kim, S. G., Kessler, D., et al. (2014). Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508, 546–549. doi: 10.1038/nature13082
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Liu, Y., Wang, L., Xing, X., Sun, L., Pan, J., Kong, X., et al. (2013). ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 54, 944–959. doi: 10.1093/pcp/pct047
Luo, X., Ma, C. Z., Yue, Y., Hu, K. N., Li, Y. Y., Duan, Z. Q., et al. (2015). Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genomics 16:379. doi: 10.1186/s12864-015-1607-0
Lysak, M. A., Cheung, K., Kitschke, M., and Bures, P. (2007). Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 145, 402–410. doi: 10.1104/pp.107.104380
Lysak, M. A., Koch, M. A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. doi: 10.1101/gr.3531105
Meyer, S., Lauterbach, C., Niedermeier, M., Barth, I., Sjolund, R. D., and Sauer, N. (2004). Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol. 134, 684–693. doi: 10.1104/pp.103.033399
Mukherjee, S., Liu, A., Deol, K. K., Kulichikhin, K., Stasolla, C., Brule-Babel, A., et al. (2015). Transcriptional coordination and abscisic acid mediated regulation of sucrose transport and sucrose-to-starch metabolism related genes during grain filling in wheat (Triticum aestivum L.). Plant Sci. 240, 143–160. doi: 10.1016/j.plantsci.2015.09.010
Mun, J. H., Kwon, S. J., Yang, T. J., Seol, Y. J., Jin, M., Kim, J. A., et al. (2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10:R111. doi: 10.1186/gb-2009-10-10-r111
Peng, D., Gu, X., Xue, L. J., Leebens-Mack, J. H., and Tsai, C. J. (2014). Bayesian phylogeny of sucrose transporters: ancient origins, differential expansion and convergent evolution in monocots and dicots. Front. Plant Sci. 5:615. doi: 10.3389/fpls.2014.00615
Reinders, A., Sivitz, A. B., Starker, C. G., Gantt, J. S., and Ward, J. M. (2008). Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol. Biol. 68, 289–299. doi: 10.1007/s11103-008-9370-0
Reinders, A., Sivitz, A. B., and Ward, J. M. (2012). Evolution of plant sucrose uptake transporters. Front. Plant Sci. 3:22. doi: 10.3389/fpls.2012.00022
Riesmeier, J. W., Willmitzer, L., and Frommer, W. B. (1992). Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713.
Riesmeier, J. W., Willmitzer, L., and Frommer, W. B. (1994). Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 13, 1–7.
Rogozin, I. B., Sverdlov, A. V., Babenko, V. N., and Koonin, E. V. (2005). Analysis of evolution of exon-intron structure of eukaryotic genes. Brief. Bioinformatics. 6, 118–134. doi: 10.1093/bib/6.2.118
Rombauts, S., Dehais, P., Van Montagu, M., and Rouze, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296.
Sauer, N., Ludwig, A., Knoblauch, A., Rothe, P., Gahrtz, M., and Klebl, F. (2004). AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J. 40, 120–130. doi: 10.1111/j.1365-313X.2004.02196.x
Sauer, N., and Stolz, J. (1994). SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 6, 67–77.
Schneider, S., Hulpke, S., Schulz, A., Yaron, I., Holl, J., Imlau, A., et al. (2012). Vacuoles release sucrose via tonoplast-localised SUC4-type transporters. Plant Biol. (Stuttg). 14, 325–336. doi: 10.1111/j.1438-8677.2011.00506.x
Schulz, A., Beyhl, D., Marten, I., Wormit, A., Neuhaus, E., Poschet, G., et al. (2011). Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 68, 129–136. doi: 10.1111/j.1365-313X.2011.04672.x
Shen, J. X., Fu, T. D., Yang, G. S., Ma, C. Z., and Tu, J. X. (2005). Genetic analysis of rapeseed self-incompatibility lines reveals significant heterosis of different patterns for yield and oil content traits. Plant Breed. 124, 111–116. doi: 10.1111/j.1439-0523.2004.01069.x
Sivitz, A. B., Reinders, A., and Ward, J. M. (2005). Analysis of the transport activity of barley sucrose transporter HvSUT1. Plant Cell Physiol. 46, 1666–1673. doi: 10.1093/pcp/pci182
Sivitz, A. B., Reinders, A., and Ward, J. M. (2008). Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 147, 92–100. doi: 10.1104/pp.108.118992
Slewinski, T. L., Meeley, R., and Braun, D. M. (2009). Sucrose transporter1 functions in phloem loading in maize leaves. J. Exp. Bot. 60, 881–892. doi: 10.1093/jxb/ern335
Song, J., Jiang, L., and Jameson, P. E. (2015). Expression patterns of Brassica napus genes implicate IPT, CKX, sucrose transporter, cell wall invertase, and amino acid permease gene family members in leaf, flower, silique, and seed development. J. Exp. Bot. 66, 5067–5082. doi: 10.1093/jxb/erv133
Srivastava, A. C., Dasgupta, K., Ajieren, E., Costilla, G., McGarry, R. C., and Ayre, B. G. (2009). Arabidopsis plants harbouring a mutation in AtSUC2, encoding the predominant sucrose/proton symporter necessary for efficient phloem transport, are able to complete their life cycle and produce viable seed. Ann. Bot. 104, 1121–1128. doi: 10.1093/aob/mcp215
Srivastava, A. C., Ganesan, S., Ismail, I. O., and Ayre, B. G. (2008). Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148, 200–211. doi: 10.1104/pp.108.124776
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, C., Huang, D., Yang, J., Liu, S., Sakr, S., Li, H., et al. (2010). The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of Hevea brasiliensis (para rubber tree). Plant Cell Environ. 33, 1708–1720. doi: 10.1111/j.1365-3040.2010.02175.x
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wei, L., Jian, H., Lu, K., Filardo, F., Yin, N., Liu, L., et al. (2016). Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 14, 1368–1380. doi: 10.1111/pbi.12501
Weise, A., Barker, L., Kuhn, C., Lalonde, S., Buschmann, H., Frommer, W. B., et al. (2000). A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12, 1345–1355. doi: 10.1105/tpc.12.8.1345
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Xuan, Y. H., Hu, Y. B., Chen, L. Q., Sosso, D., Ducat, D. C., Hou, B. H., et al. (2013). Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. U.S.A. 110, E3685–E3694. doi: 10.1073/pnas.1311244110
Yang, B., Jiang, Y., Rahman, M. H., Deyholos, M. K., and Kav, N. N. (2009). Identification and expression analysis of wrky transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 9:68. doi: 10.1186/1471-2229-9-68
Yang, X., Zhang, X., Yuan, D., Jin, F., Zhang, Y., and Xu, J. (2012). Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 12:110. doi: 10.1186/1471-2229-12-110
Keywords: Brassica napus, sucrose transporters, SUC, SWEET, expression analysis, stresses response
Citation: Jian H, Lu K, Yang B, Wang T, Zhang L, Zhang A, Wang J, Liu L, Qu C and Li J (2016) Genome-Wide Analysis and Expression Profiling of the SUC and SWEET Gene Families of Sucrose Transporters in Oilseed Rape (Brassica napus L.). Front. Plant Sci. 7:1464. doi: 10.3389/fpls.2016.01464
Received: 16 May 2016; Accepted: 14 September 2016;
Published: 28 September 2016.
Edited by:
Chengdao Li, Murdoch University, AustraliaReviewed by:
Xiaoming Wu, Oil Crops Research Institute Chinese Academy of Agricultural Sciences, ChinaQing-Yong Yang, Huazhong Agricultural University, China
Copyright © 2016 Jian, Lu, Yang, Wang, Zhang, Zhang, Wang, Liu, Qu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiana Li, ljn1950@swu.edu.cn
†These authors have contributed equally to this work.
 Hongju Jian
Hongju Jian Kun Lu†
Kun Lu† Liezhao Liu
Liezhao Liu Jiana Li
Jiana Li