- Department of Biology, University of Mississippi, Oxford, MS, USA
An advantage of analyzing abscission in genetically tractable model plants is the ability to make use of classic genetic tools such as suppression analysis. We have investigated the regulation of organ abscission by carrying out suppression analysis in Arabidopsis flowers. Plants carrying mutations in the NEVERSHED (NEV) gene, which encodes an ADP-ribosylation factor GTPase-activating protein, retain their outer floral organs after fertilization. Mutant alleles of CAST AWAY (CST), which encodes a receptor-like cytoplasmic kinase, were found to restore organ abscission in nev flowers in an allele-specific manner. To further explore the basis of the interactions between CST and NEV, we tested whether the site of a nev mutation is predictive of its ability to be suppressed. Our results suggest instead that the strength of a nev allele influences whether organ abscission can be rescued by a specific allele of CST.
Introduction
Plants have the astonishing ability to release their floral organs, leaves, fruit, and seeds at programmed points in their life cycle or in response to signals from their environment. Within Arabidopsis flowers, the series of events leading to organ abscission is genetically tractable. Analysis using this model system has revealed the influence of organ boundary genes in establishing the placement of abscission zones (Wang et al., 2006; González-Carranza et al., 2007; Gómez-Mena and Sablowski, 2008; McKim et al., 2008; Gubert et al., 2014), the critical roles played by hormones such as jasmonic acid (Kim et al., 2013) and managers of membrane traffic (Liljegren et al., 2009; Liu et al., 2013), and a signaling module that regulates the cell separation phase of organ abscission (Fang and Fernandez, 2002; Cho et al., 2008; Stenvik et al., 2008; Shi et al., 2011; Gubert and Liljegren, 2014; Patharkar and Walker, 2015; Santiago et al., 2016; Taylor et al., 2016). Central components in this module include a secreted peptide, INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and redundant leucine-rich repeat receptor-like kinases, HAESA (HAE) and HAESA-like2 (HSL2), that activate a MAP kinase cascade leading to organ abscission.
We have used suppression analysis as a genetic tool to identify additional genes that control the abscission process in Arabidopsis flowers. Starting with the nevershed (nev) mutant which blocks organ shedding due to defects in membrane traffic (Liljegren et al., 2009), we screened for second-site mutations that would restore organ abscission in the presence of the original mutation. The nev-3 allele chosen for this screen (Figure 1A) changes an invariant arginine in the encoded protein known to be essential for ADP-ribosylation factor GTPase-activating activity (Luo et al., 2007). Multiple alleles of genes encoding three receptor-like kinases—EVERSHED (EVR), SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1), and CAST AWAY (CST)—were found to rescue abscission in nev flowers (Leslie et al., 2010; Lewis et al., 2010; Burr et al., 2011). Mutations in these receptor-like kinases are also able to reverse nev-mediated alterations in the structure of the Golgi apparatus and associated trans-Golgi network. Additional analyses suggest that activation of organ abscission is modulated by inhibitory interactions between CST and EVR with HAE/HSL2 (Burr et al., 2011; Gubert and Liljegren, 2014). Recent studies have demonstrated that SERK1 and two related receptor-like kinases act as co-receptors of HAE/HSL2 (Meng et al., 2016; Santiago et al., 2016). We have proposed that CST and EVR may prevent the signaling that leads to organ abscission by sequestering HAE/HSL2 at the cell surface and promoting their internalization prior to activation by IDA (Burr et al., 2011). As NEV is thought to function in the cycling of HAE/HSL2 to the plasma membrane, disruption of CST or EVR activity may restore organ abscission in nev flowers by shifting the balance of stabilized HAE/HSL2 receptors at the cell surface from an excessive pool of internalized, inactive receptors in endosomal compartments (Burr et al., 2011; Bryan et al., 2012; Liljegren, 2012).
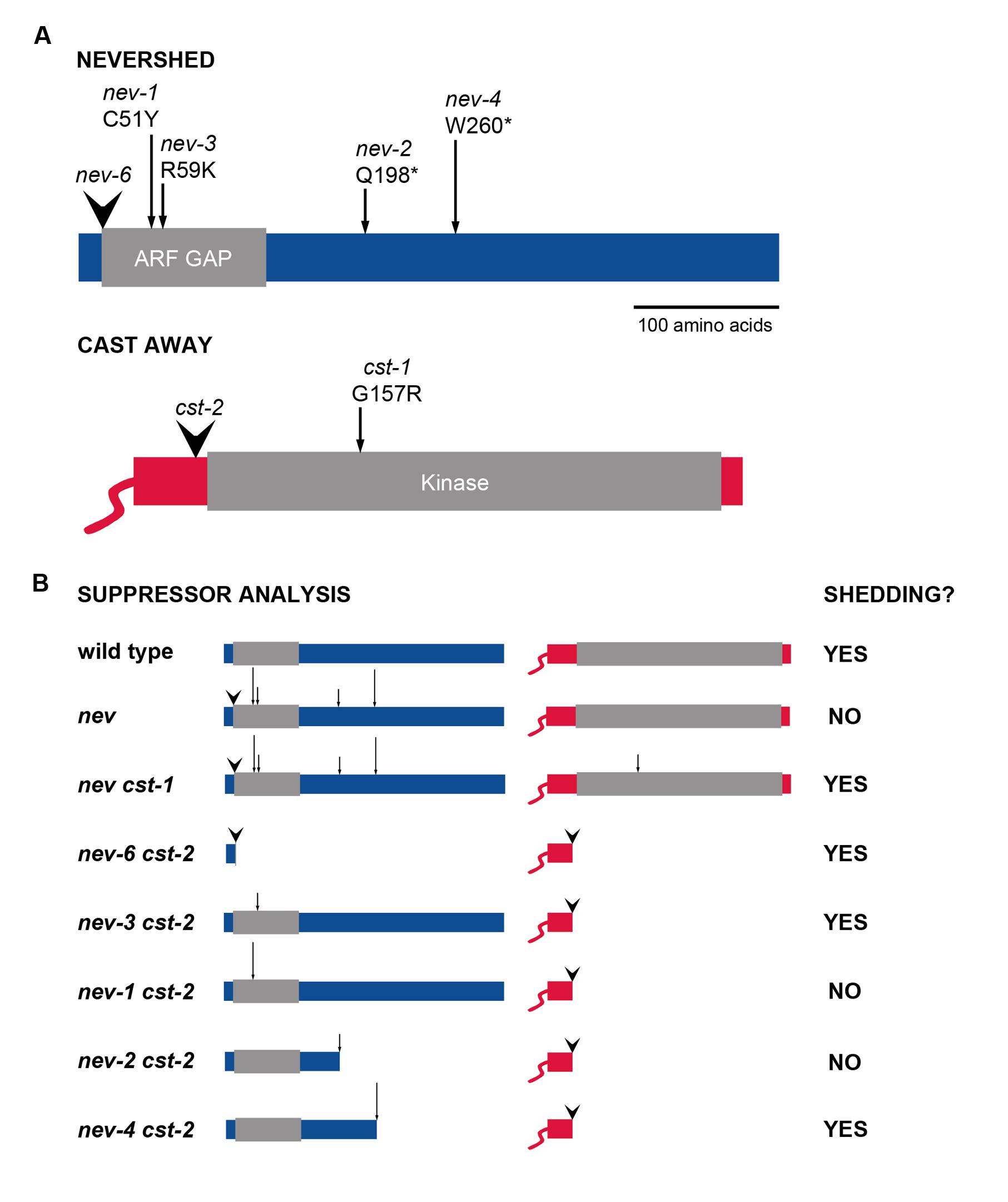
FIGURE 1. Alleles of NEV and CST used in suppressor analysis of organ abscission. (A) The sites of the mutations analyzed are indicated within the encoded NEV and CST proteins (Liljegren et al., 2009; Burr et al., 2011). T-DNA insertions are marked by arrowheads and point mutations by arrows. (B) Diagram of the nev cst allele combinations tested for rescue of organ shedding (Burr et al., 2011; this study).
Contrasting behaviors are shown by the pair of cst mutant alleles we identified with regard to their ability to rescue abscission in nev flowers (Burr et al., 2011). The cst-1 allele introduces a missense mutation (G157R) near the ATP-binding site within the CST kinase domain (Figure 1A), abolishing the kinase activity of the mutant protein. Organ shedding in nev-3. nev-2, and nev-6 flowers is recessively rescued by two copies of the cst-1 allele (Figure 1B; Burr et al., 2011). The cst-2 allele contains a T-DNA insertion immediately upstream of the kinase domain, and is predicted to encode a truncated protein (Figure 1A). One copy of cst-2 dominantly restores organ abscission in nev-3 and nev-6 flowers, but nev-2 flowers retain their organs even if both copies of cst-2 are present (Figure 1B; Burr et al., 2011).
As these results were partially consistent with the allele-specific mechanism of conformational suppression, in which a suppressor mutation restores a physical interaction between two proteins, we designed a study to determine whether the location of a nev mutation would be predictive of its ability to be rescued by the cst alleles. Specifically, we tested whether alleles that independently affect either the ARF GAP domain or the C-terminal region of NEV would mimic the distinct interactions of nev-3 and nev-2 with cst-2.
Materials and Methods
Plants
The mutant alleles used in this study and methods for genotyping cst-1 and cst-2 have been described previously (Liljegren et al., 2009; Burr et al., 2011). nev-1 and nev-4 were genotyped as described in Supplementary Table S1. The nev-1. nev-4, and cst-1 mutants were isolated from the Ler ecotype; the cst-2 mutant was isolated from the Col ecotype. Since the nev cst-2 double mutants would be analyzed in a mixed Ler/Col background, a cst-1 stock backcrossed once into the Col ecotype was used to generate the nev cst-1 double mutants. Plants were grown at 21°C with 50% humidity and a 16-h photoperiod.
Imaging
Digital images were taken with a PowerShot SX160 IS (Canon, Melville, NY, USA) or Alpha Innotech gel documentation system (ProteinSimple, San Jose, CA, USA). Image brightness and contrast were adjusted with Photoshop CS6 (Adobe, Mountain View, CA, USA).
RT/PCR
Wild-type and mutant inflorescences with flowers through stage 15 (Smyth et al., 1990) were ground in liquid nitrogen, and RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s instructions. Specific regions of wild-type and mutant cDNAs were synthesized using gene-specific primers (described in Supplementary Table S2) and SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. A subset of the RNA samples were pre-treated with DNase using the Ambion DNA-free Kit (Thermo Fisher Scientific, Waltham, MA, USA) prior to cDNA synthesis. To confirm the presence of the cst-2 transcript, a second round of amplification was performed with a nested primer. In addition, replicates were carried out with and without reverse transcriptase.
Results
Allele-Specific Suppression of nev-Mediated Abscission Defects
Previously, we discovered that while one copy of the cst-2 allele is sufficient to rescue organ shedding in nev-3 (R59K) flowers, abscission in nev-2 (Q198*) flowers cannot be restored by either one or two copies of cst-2 (Figure 1B; Burr et al., 2011). Whereas the nev-3 mutation affects an arginine residue essential for the enzymatic activity of the ARF GAP domain, the protein encoded by nev-2 is predicted to be truncated downstream of the ARF GAP domain (Figure 1A; Luo et al., 2007; Liljegren et al., 2009). Both copies of the cst-1 allele are required to suppress the abscission defects of nev-3 and nev-2 flowers (Figure 1B; Burr et al., 2011). Based on these results, we hypothesized that if CST and NEV function in a complex, the ARF GAP domain of NEV might facilitate this interaction (Burr et al., 2011).
To investigate whether other nev alleles that alter critical residues in the ARF GAP domain show similar interactions with the cst alleles, we analyzed nev-1 cst-1 and nev-1 cst-2 double mutants (Figure 2). nev-1 is a missense allele (C51Y) that alters the third essential cysteine within the Cys-x2-Cys-x(16,17)-Cys-x2-Cys zinc finger motif of the ARF GAP domain (Figure 1A; Liljegren et al., 2009). While cst-1 is able to recessively suppress the shedding defects of nev-1 flowers, the floral organs of the nev-1 cst-2 double mutant remain firmly attached (Figures 2A–D). These results indicate that despite their close proximity within the ARF GAP domain and indistinguishable single mutant phenotypes, the nev-1 and nev-3 alleles do not behave equivalently when interacting with cst-2 (Figures 1A,B).

FIGURE 2. Rescue of organ shedding in nev cst flowers is allele-dependent. The outer organs of wild-type (WT) flowers are shed by floral stage 17 (A), and stay attached in nev-1 (B) and nev-4 (E) flowers. Abscission is restored recessively in nev-1 cst-1 (C) and nev-4 cst-1 (F) flowers. Although it behaves dominantly in nev-3 and nev-6 flowers (Burr et al., 2011), the cst-2 allele is unable to suppress the abscission defects of nev-1 flowers (D), and rescues organ shedding recessively in nev-4 flowers (G,H). Scale bars = 1 cm.
We also tested whether another nev allele that introduces a stop codon downstream of the ARF-GAP domain exhibits similar interactions with the cst alleles. Like nev-2. nev-4 is a nonsense allele (W260*) predicted to encode an abbreviated protein with an intact ARF-GAP domain (Figure 1A; Liljegren et al., 2009). As with all nev alleles tested, cst-1 recessively rescues organ abscission in nev-4 flowers (Figures 2E,F). However, unlike nev-2 cst-2 flowers (Figure 1B), the shedding defects of nev-4 flowers can also be suppressed with two copies of cst-2 (Figures 2G,H). Therefore, despite the shared features of the nev-2 and nev-4 alleles, they interact with cst-2 in distinct modes (Figures 1A,B).
Analysis of nev and cst Transcripts
To examine whether there are qualitative differences in expression of the nev and cst mutant transcripts compared to wild-type, RT/PCR experiments were carried out on total RNA isolated from the inflorescences of wild-type and mutant plants. Oligos located in exon 11 of NEV and exon 6 of CST were used to synthesize the first strand of the cDNAs, and specific regions of the transcripts were subsequently amplified (see Supplementary Table S2). Substantial differences were not observed for the transcript levels of either missense (nev-3. nev-1. cst-1) or nonsense (nev-4. nev-2) alleles of NEV and CST compared to wild-type (Supplementary Figures S1A and S2A). In contrast, comparable levels of correctly spliced transcripts were not apparent in either of the insertional alleles (nev-6. cst-2) analyzed (Supplementary Figures S1A and S2A).
To test for the presence of altered transcripts in nev-6 and cst-2 flowers, oligos located upstream of the T-DNA insertion sites were used to synthesize cDNA fragments from DNase-treated RNA samples (Supplementary Table S2; Supplementary Figures S1B and S2B). Similar levels of a NEV cDNA product including part of exon 1 were observed in all nev mutants and wild-type plants examined (Supplementary Figure S1B). Since the nev-6 T-DNA insertion is located in intron 1, these results indicate that an altered mutant transcript is produced in nev-6 flowers that may encode an abbreviated protein. Reduced levels of nested products including parts of exon 1 and 2 of CST were observed in cst-2 flowers compared to cst-1 and wild-type (Supplementary Table S2; Supplementary Figure S2B). These results are consistent with the production of a truncated cst-2 protein.
Discussion
Here we report further evidence that the cst-1 and cst-2 alleles differ in their ability to restore organ shedding in nev flowers. While cst-1 recessively suppresses each of the five nev alleles tested, cst-2 suppresses nev-3 and nev-6 dominantly, nev-4 recessively, and fails to rescue nev-1 and nev-2 (Burr et al., 2011; this study).
These results highlight the complexity of interpreting the mechanisms of allelic suppression. Although the ultimate goal for many geneticists carrying out suppression analysis is to find instances of conformational suppression, whereby allele-specific rescue reflects a restored physical interaction between two mutant proteins, this scenario is actually rare in practice (Manson, 2000). Indeed, our selection of the nev-3 missense allele (Figure 1A) as the genetic background for this screen was driven by an interest in identifying a mutant version of an unknown protein that might interact with and restore the ARF GAP enzymatic activity of the nev mutant protein.
Instead, our results suggest that the cst-1 and cst-2 alleles restore organ abscission in nev flowers through distinct suppression mechanisms. We have found that the kinase-dead CST protein encoded by cst-1 recessively suppresses the abscission defects of all nev alleles tested, including nev-6, which is predicted to produce an abbreviated protein missing the ARF GAP domain (Figure 1A; Supplementary Figure S1B). Suppression of a deletion (or null) allele of the original gene by an extragenic suppressor is considered strong evidence of bypass suppression (Prelich, 1999). Bypass suppression occurs when a second site mutation creates an alternate opportunity to cover the function disabled by the first mutation (Manson, 2000; Michels, 2002). Another hallmark of bypass suppression is that it is not allele-specific (Manson, 2000), which fits with the observed behavior of the nev cst-1 double mutants (Figure 1B). Considering that interactions between CST and HAE were detected in subdomains of the plasma membrane via biomolecular fluorescence complementation assays (Burr et al., 2011), the cst-1 allele may consistently rescue organ shedding in nev flowers due to the failure of the kinase-deficient cst-1 protein to promote the internalization of the HAE/HSL2 receptors from the cell surface. The recessive nature of the cst-1 suppression suggests that the reduced amount of the functional CST kinase in nev flowers heterozygous for cst-1 is sufficient to remove enough of the HAE/HSL2 receptors from this plasma membrane pool to prevent activation of the MAP kinase module leading to organ abscission. Redelivery of HAE/HSL2 to the cell surface after internalization is predicted to be disrupted in each of the nev mutant alleles tested (Figure 1A).
We have found that cst-2, which may produce reduced levels of an abbreviated, membrane-associated protein without a kinase domain (Figure 1A; Supplementary Figure S2B), rescues organ abscission in nev-3. nev-4, and nev-6 flowers but not in nev-1 or nev-2 flowers (Figure 1B). This allele-specific outcome may result from a dominant-negative mutation enacting a gradient of suppression (Manson, 2000; Burr et al., 2011). Under this scenario, the predicted strength of the nev alleles tested would range from nev-6 and nev-3 (relatively weak; rescued by one copy of cst-2) to nev-4 (intermediate, rescued by two copies of cst-2) to nev-1 and nev-2 (strong, not rescued by cst-2). Relative differences in the activities of nev mutant proteins may impact the ratio of HAE/HSL2 receptors trapped in the endosomal compartments and thereby influence the ease of cst-2 mediated suppression (Figure 1B). While a truncated nev-6 mutant protein without an ARF GAP domain would not be expected to retain more function than the nev-2 mutant protein (Figure 1), intronic T-DNA insertions can be spliced out in a fraction of the transcripts produced, leading to synthesis of functional protein (Chehab et al., 2011; Rodriguez et al., 2014). Although we did not detect notable levels of correctly spliced NEV transcripts in nev-6 flowers (Supplementary Figure S1A), it is likely that even a small amount of functional protein is sufficient to promote abscission. Indeed, it has been previously observed that the petals of nev-7 flowers detach more readily than those of nev-3 flowers (Liu et al., 2013). Like nev-6, the nev-7 allele contains a T-DNA insertion in the first intron (Liljegren et al., 2009).
It is striking that the nev-1 and nev-2 alleles can be recessively rescued by cst-1 but not by cst-2. These results, in addition to the dominant suppression of nev-3 and nev-6 by cst-2, suggest that the truncated cst-2 protein may exhibit an altered set of interactions with receptor-like kinase complexes than the cst-1 protein. Future analysis of the expression, localization, and ability of the cst-2 and cst-1 mutant proteins to form heteromeric complexes with EVR and HAE may reveal additional clues to the unique mechanisms underlying their restoration of the signaling leading to organ abscission in nev flowers.
With the growing accessibility of approaches to identify the transcriptomes of abscission zone cells in model as well as crop plants using laser capture microdissection (Cai and Lashbrook, 2006, 2008; Agustí et al., 2009) and RNA sequencing (Niederhuth et al., 2013; Kim et al., 2016; Sundaresan et al., 2016), the agronomic value of using model plants to study abscission is under debate (Patterson et al., 2015). The recent discovery that drought-triggered leaf abscission is dependent on the activities of IDA, HAE/HSL2, and NEV (Patharkar and Walker, 2016) significantly enhances the usefulness of Arabidopsis as a model system. Furthermore, until analysis of gene function is feasible in crop plants, parallel approaches to investigate the functions of abscission zone-enriched genes in model plants with reverse genetic approaches will be crucial.
Forward genetic screens, when carefully designed, are also expected to provide novel insights regarding the regulation of organ abscission. In addition to our discovery of a set of receptor-like kinases that modulate organ abscission via proposed interactions with HAE and HSL2, the homeodomain transcription factor BREVIPEDICELLUS (BP) was found to act downstream of the IDA-HAE/HSL2 signaling module through suppression analysis of ida flowers (Shi et al., 2011). While bp mutants display enlarged abscission zones (Wang et al., 2006), it is noteworthy that the cst. evr, and serk1 mutants do not present phenotypes on their own, yet alleles of each are able to rescue organ shedding in the context of nev flowers (Leslie et al., 2010; Lewis et al., 2010; Burr et al., 2011). Suppression analysis of a weak hae hsl2 mutant has revealed that mutations in either of two mannosyltransferases that normally mediate degradation of the mutant hsl2 protein in the ER may restore abscission by allowing this partially functioning receptor to escape to the cell surface (Baer et al., 2016). Understanding the threshold levels at which organs are released in sensitized mutants like nev. ida, and hae hsl2 may inform the future design of nuanced solutions to control abscission in crop plants.
Author Contributions
SL designed the experiments. All authors performed the experiments and contributed in preparing the figures.
Funding
This research was supported by National Science Foundation grants to SL (IOS-1239311; IOS-1453733) and to the Mississippi EPSCoR program (EPS-0903787).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Christian Burr and Iris Chen for initiating this study; Brad Jones, Sara Patterson, and Timothy Tranbarger for helpful conversations; and Adam Harris, Greta Parker, Victoria McClearn, Jill Thiede, Hayden Malone, and Charles McCrory for technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01588
References
Agustí, J., Merelo, P., Cercós, M., Tadeo, F. R., and Talón, M. (2009). Comparative transcriptional survey between laser-microdissected cells from laminar abscission zone and petiolar cortical tissue during ethylene-promoted abscission in citrus leaves. BMC Plant Biol. 9:127. doi: 10.1186/1471-2229-9-127
Baer, J., Taylor, I., and Walker, J. C. (2016). Disrupting ER-associated protein degradation suppresses the abscission defect of a weak hae hsl2 mutant in Arabidopsis. J. Exp. Bot. 67:18. doi: 10.1093/jxb/erw313
Bryan, A., Racolta, A., Tax, F., and Liljegren, S.J. (2012). “The social network: receptor kinases and cell fate determination,” in Receptor-Like Kinases in Plants: from Development to Defense, Vol. 13, eds B. Kemmerling and F. Tax (Berlin: Springer), 41–65.
Burr, C. A., Leslie, M. E., Orlowski, S. K., Chen, I., Wright, C. E., Daniels, M. J., et al. (2011). CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol. 156, 1837–1850. doi: 10.1104/pp.111.175224
Cai, S., and Lashbrook, C. C. (2006). Laser capture microdissection of plant cells from tape-tranferred paraffin sections promote recovery of structurally intact RNA for global gene profiling. Plant J. 48, 628–637. doi: 10.1111/j.1365-313X.2006.02886.x
Cai, S., and Lashbrook, C. C. (2008). Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 146, 1305–1321. doi: 10.1104/pp.107.110908
Chehab, E. W., Kim, S., Savchenko, T., Kliebenstein, D., Dehesh, K., and Braam, J. (2011). Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol. 156, 770–778. doi: 10.1104/pp.111.174169
Cho, S. K., Larue, C. T., Chevalier, D., Wang, H., Jinn, T. L., Zhang, S., et al. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 15629–15634. doi: 10.1073/pnas.0805539105
Fang, S.-C., and Fernandez, D. E. (2002). Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol. 130, 78–89. doi: 10.1104/pp.004721
Gómez-Mena, C., and Sablowski, R. (2008). ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 20, 2059–2072. doi: 10.1105/tpc.108.059188
González-Carranza, Z. H., Rompa, U., Peters, J. L., Bhatt, A. M., Wagstaff, C., Stead, A. D., et al. (2007). HAWAIIAN SKIRT: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol. 144, 1370–1382. doi: 10.1104/pp.106.092288
Gubert, C. M., Christy, M. E., Ward, D. L., Groner, W. D., and Liljegren, S. J. (2014). ASYMMETRIC LEAVES1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 14:195. doi: 10.1186/s12870-014-0195-5
Gubert, C. M., and Liljegren, S. J. (2014). HAESA and HAESA-LIKE2 activate organ abscission downstream of NEVERSHED and EVERSHED in Arabidopsis flowers. Plant Signal. Behav. 9:e29115. doi: 10.4161/psb.29115
Kim, J., Dotson, B., Rey, C., Lindsey, J., Bleecker, A. B., Binder, B. M., et al. (2013). New clothes for the jasmonic acid receptor COI1: delayed abscission, meristem arrest and apical dominance. PLoS ONE 8:e60505. doi: 10.1371/journal.pone.0060505
Kim, J., Yang, J., Yang, R., Sicher, R. C., Chang, C., and Tucker, M. L. (2016). Transcriptome analysis of soybean leaf abscission identifies transcriptional regulators of organ polarity and cell fate. Front. Plant Sci. 7:125. doi: 10.3389/fpls.2016.00125
Leslie, M. E., Lewis, M. W., Youn, J.-Y., Daniels, M. J., and Liljegren, S. J. (2010). The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development 137, 467–476. doi: 10.1242/dev.041335
Lewis, M. W., Leslie, M. E., Fulcher, E. H., Darnielle, L., Healy, P., Youn, J. Y., et al. (2010). The SERK1 receptor-like kinase regulates organ separation in Arabidopsis flowers. Plant J. 5, 817–828. doi: 10.1111/j.1365-313X.2010.04194.x
Liljegren, S. J. (2012). Organ abscission: exit strategies require signals and moving traffic. Curr. Opin. Plant Biol. 15, 670–676. doi: 10.1016/j.pbi.2012.09.012
Liljegren, S. J., Leslie, M. E., Darnielle, L., Lewis, M. W., Taylor, S. M., Luo, R., et al. (2009). Regulation of membrane trafficking and organ separation by the NEVERSHED ARF GAP protein. Development 136, 1909–1918. doi: 10.1242/dev.033605
Liu, B., Butenko, M. A., Shi, C. L., Bolivar, J. L., Winge, P., Stenvik, G. E., et al. (2013). NEVERSHED and INFLORESCENCE DEFICIENT IN ABSCISSION are differentially required for cell expansion and cell separation during floral organ abscission in Arabidopsis thaliana. J. Exp. Bot. 64, 5345–57. doi: 10.1093/jxb/ert232
Luo, R., Ahvazi, B., Amariei, D., Shroder, D., Burrola, B., Losert, W., et al. (2007). Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem. J. 402, 439–447. doi: 10.1042/BJ20061217
Manson, M. D. (2000). Allele-specific suppression as a tool to study protein-protein interactions in bacteria. Methods 20, 18–34. doi: 10.1006/meth.1999.0902
McKim, S. M., Stenvik, G. E., Butenko, M. A., Kristiansen, W., Cho, S. K., Hepworth, S. R., et al. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135, 1537–1546. doi: 10.1242/dev.012807
Meng, X., Zhou, J., Tang, J., Li, B., de Oliveira, M. V. V., Chai, J., et al. (2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 14, 1330–1338. doi: 10.1016/j.celrep.2016.01.023
Michels, C. V. (2002). “Suppression analysis,” in Genetic Techniques for Biological Research: A Case Study approach, ed. C. A. Michels (Chichester: John Wiley & Sons), 91–98. doi: 10.1002/0470846623.ch8
Niederhuth, C. E., Patharkar, O. R., and Walker, J. C. (2013). Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-Seq. BMC Genomics 14:37. doi: 10.1186/1471-2164-14-37
Patterson, S. E., Bolivar-Medina, J. L., Falbel, T. G., Hedtcke, J. L., Nevarez-McBride, D., Maule, A. F., et al. (2015). Are we on the right track: can our understanding of abscission in model systems promote or derail making improvements in less studied crops? Front. Plant Sci. 6:1268. doi: 10.3389/fpls.2015/01268
Patharkar, O. R., and Walker, J. C. (2015). Floral organ abscission is regulated by a positive feedback loop. Proc. Natl. Acad. Sci. U.S.A. 112, 2906–2911. doi: 10.1073/pnas.1423595112
Patharkar, O. R., and Walker, J. C. (2016). Core mechanisms regulating developmentally timed and environmentally triggered abscission. Plant Physiol. 172, 510–520. doi: 10.1104/pp.16.01004
Prelich, G. (1999). Suppression mechanisms: themes from variations. Trends Genet. 15, 261–266. doi: 10.1016/S0168-9525(99)01749-7
Rodriguez, M. C., Wawrzynska, A., and Sirko, A. (2014). Intronic T-DNA insertion in Arabidopsis NBR1 conditionally affects wild-type transcript level. Plant Signal. Behav. 9:e975659. doi: 10.4161/15592324.2014.975659
Santiago, J., Brandt, B., Wildhagen, M., Hohmann, U., Hothorn, L. A., Butenko, M. A., et al. (2016). Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5:e15075. doi: 10.7554/eLife.15075
Shi, C. L., Stenvik, G. E., Vie, A. K., Bones, A. M., Pautot, V., Proveniers, M., et al. (2011). Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23, 2553–2567. doi: 10.1105/tpc.111.084608
Smyth, D. R., Bowman, J. L., and Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. doi: 10.1105/tpc.2.8.755
Stenvik, G. E., Tandstad, N. M., Guo, Y., Shi, C. L., Kristiansen, W., Holmgren, A., et al. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805–1817. doi: 10.1105/tpc.108.059139
Sundaresan, S., Philosoph-Hadas, S., Riov, J., Mugasimangalam, R., Kuravadi, N. A., Kochanek, B., et al. (2016). De novo transcriptome sequencing and development of abscission zone-specific microarray as a new molecular tool for analysis of tomato organ abscission. Front. Plant Sci. 6:1258. doi: 10.3389/fpls.2015.01258
Taylor, I., Wang, Y., Seitz, K., Baer, J., Bennewitz, S., Mooney, B. P., et al. (2016). Analysis of phosphorylation of the receptor-like protein kinase HAESA during Arabidopsis floral abscission. PLoS ONE 11:e0147203. doi: 10.1371/journal.pone.0147203
Keywords: abscission, cell separation, shedding, NEV, CST, ARF GAP, RLCK
Citation: Groner WD, Christy ME, Kreiner CM and Liljegren SJ (2016) Allele-Specific Interactions between CAST AWAY and NEVERSHED Control Abscission in Arabidopsis Flowers. Front. Plant Sci. 7:1588. doi: 10.3389/fpls.2016.01588
Received: 04 May 2016; Accepted: 07 October 2016;
Published: 21 October 2016.
Edited by:
Timothy John Tranbarger, Institute of Research for Development, FranceReviewed by:
Xiyin Wang, North China Institute of Science and Technology, ChinaShelley Hepworth, Carleton University, Canada
Jian Huang, University of Wisconsin–Milwaukee, USA
Copyright © 2016 Groner, Christy, Kreiner and Liljegren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah J. Liljegren, liljegren@olemiss.edu
 William D. Groner
William D. Groner Catherine M. Kreiner
Catherine M. Kreiner Sarah J. Liljegren
Sarah J. Liljegren