- State Key Laboratory of Crop Genetics and Germplasm Enhancement, College of Horticulture, Nanjing Agricultural University, Nanjing, China
To develop mutants of the ERF factor with more binding activities to the GCC box, we performed in vitro directed evolution by using DNA shuffling and screened mutants through yeast one-hybrid assay. Here, a series of mutants were obtained and used to reveal key amino acids that induce changes in the DNA binding activity of the BnaERF-B3 protein. With the BnaERF-B3-hy15 as the template, we produced 12 mutants which host individual mutation of potential key residues. We found that amino acid 156 is the key site, and the other 18 mutants host the 18 corresponding individual amino acid residues at site 156. Among the 20 individuals comprising WT (Gly156), Mu3 (Arg156), and 18 mutants with other 18 amino acid residues, Arg156 in the AP2-domain is the amino acid residue with the highest binding activity to the GCC box. The structure of the α-helix in the AP2-domain affects the binding activity. Other residues within AP2-domain modulated binding activity of ERF protein, suggesting that these positions are important for binding activity. Comparison of the mutant and wild-type transcription factors revealed the relationship of protein function and sequence modification. Our result provides a potential useful resource for understanding the trans-activation of ERF proteins.
Introduction
Plants are affected by various abiotic stresses during their life cycle. High salt, drought, and extreme temperature are major factors limiting plant growth and development, and considerably affect the efficiency of agricultural production. When plants are exposed to adverse stress, a complex signal transduction network will mediate resistance to the stress (Seki et al., 2001; Shinozaki et al., 2003; Gutterson and Reuber, 2004). Numerous transcription factors have been identified and associated with adaptive strategies of plants; these transcription factors, including those in the AP2/ERF family, bind to the DNA-binding elements at the promoter of downstream target genes and regulate gene expression (Yamasaki et al., 2013; Franco-Zorrilla et al., 2014). The AP2/ERF transcription factor family is one of the families involved in plant development, signal transduction, and stress response; this family can be further divided into four subfamilies: AP2, RAV, DREB, and ERF (Sakuma et al., 2002; Zhuang et al., 2008).
The two largest subfamilies DREB and ERF present a high similarity in their AP2-domains, but the core amino acids determine their binding activity to different cis elements (Yamaguchi-Shinozaki and Shinozaki, 1994; Fujimoto et al., 2000). Most DREB proteins bind to the DRE element, whereas the ERF subfamily factors mainly recognize the GCC box (Sakuma et al., 2002; Liu et al., 2006). The DRE element with the core sequence AGCCGAC was found to be important to the DNA-binding specificity of DREB proteins, with three positions (C4, G5, and C7) as key bases for binding affinity (Sakuma et al., 2002). In the AP2-domain of two Arabidopsis DREB proteins, Val14 and Glu19 are important to DNA-binding specificity; in particular, Val14 is critical for determining DREB-binding efficiency (Sakuma et al., 2002). Similar results were also found in the maize DREB protein (Qin et al., 2003). A GCC box has been found in the promoter region of ethylene-inducible genes, Hao and his colleagues reported that the 1st G, the 4th G, and the 6th C are essential in the GCC box for specific binding to the respective domain of ERF transcription factors (Hao et al., 1998). Some DREB and ERF factors were reported to bind to both GCC and DRE elements, such as CBF1-F (Hao et al., 2002), BnDREBIII-1 (Liu et al., 2006), and Tsi1 (Park et al., 2001).
In our previous studies, we isolated the gene BnaERF-B3-hy15 encoding an ERF transcription factor from Brassica napus L. cv. Huyou15 and obtained three BnaERF-B3 mutants by DNA shuffling (Jin et al., 2010a). The yeast one-hybrid system showed that the three modified proteins exhibit strong binding activity to the GCC box, but the original protein showed almost no binding activity. The overexpressed mutant BnaERF-B3-mu3, which shows a high binding activity to the GCC box, exhibits a higher freezing tolerance than the original BnaERF-B3 in transgenic Arabidopsis (Xiong et al., 2013). Sequence alignment results revealed that the 12 amino acid residues differ between BnaERF-B3-hy15 and the three mutants, but only Gly156 is located in the AP2-domain.
Among the 12 mutant amino acids, the amino acid that shows a high binding activity to the GCC box, is it located in the AP2-domain or in the peripheral area? Site-directed mutagenesis was repeatedly performed to identify key sites in protein sequences for functional analysis (Lu et al., 2002; Nguyen et al., 2012; Guan et al., 2014). Previous molecular analyses of DREB or ERF mutants through site-directed mutagenesis provided insights into the amino acid sequences in the AP2-domain that are decisive for protein functions (Zhao et al., 2007; Sun et al., 2008). To identify the key site (s) which determined the DNA binding activity to the GCC box, we established a series of mutants of BnaERF-B3-hy15 through site-directed mutagenesis and screened mutants through yeast one-hybrid assay. Our results revealed that Arg156 in the AP2-domain is the amino acid residue with the highest binding activity to the GCC box in the mutant and wild type BnaERF-B3. This work can be used for further research on ERF transcription factor function.
Materials and Methods
Plant Materials and Gene Cloning
The rapeseed cultivar (B. napus. L. cv. Huyou15) was used as experiment material in this study. Plants were grown in pots containing soil, vermiculite, and peat moss (1:1:1 by volume) in a growth chamber at 22°C for 20 d under a 16 h day/8 h night cycle. The BnaERF-B3-hy15 gene encoding the AP2/ERF protein was cloned from B. napus L. Huyou15 as previously described, and three mutants were obtained by Jin et al. (2010a). Two-month-old plants of A. thaliana (L.) Heynh. (ecotype columbia) were used to clone AtCBF genes.
Site-Directed Mutagenesis of the BnaERF-B3-hy15 Gene
The details of the site-directed mutagenesis of the BnaERF-B3-hy15 and CBF genes to generate mutant genes were accorded to the overlap extension PCR strategy (Xiong et al., 2004, 2006; Peng et al., 2006). The strategy for site-directed mutagenesis is shown in Figure 1. Briefly, the upstream product (Product P1) and downstream product (Product P2) were amplified by first-round PCR with two pairs of oligonucleotides, respectively. Then, the full-length mutant fragment was amplified using a second round PCR with outer pair primers. All the mutagenesis products were ligated to the pMD-19 simple-T vector (TaKaRa, Dalian, China) for DNA sequencing identification and enzyme digestion. All the primers used to clone or synthesize mutations were listed in Table S1.
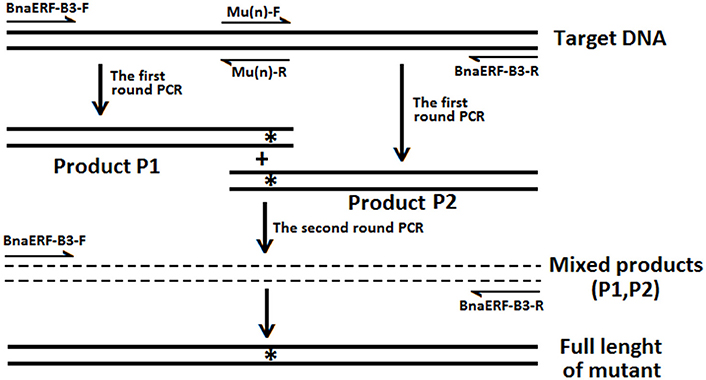
Figure 1. The strategies and primers of site-directed mutagenesis of BnaERF-B3. “n” Represent different mutants. “*” Represent the mutation sites.
Construction of Yeast Expression Vectors
The amplification mutagenesis products were inserted between the Bam HI and Sac I sites of the yeast expression vector pPC87, which was reconstructed from pPC86. The vector of pPC87 hosts the Trp synthesize gene and contains a galactose-inducible (GAL4) protein activating the domain under the control of the yeast alcohol dehydrogenase (ADH1) promoter. A 75-bp fragment containing the GCC element (5′-ACCCTC GAGCGGATAACAATTTCACACAGGGGCGGCTCTTAGGCGGCTCTTATAAGAGCCGCCGGAT CCGGGCCC-3′, GenBank No. AF394909) was inserted into the vector G222, which could synthesize Ura; the vector also contained the reporter gene LacZ under the control of the iso-1-cytochrome C (CYC1) minimal promoter. This recombinant plasmid was used as a bait plasmid in the yeast one-hybrid system (Zhang Y. et al., 2009; Jin et al., 2010a,b).
Yeast One-Hybrid and β-Galactosidase Activity Assay
The bait plasmid was transformed into the yeast strain EGY48. Subsequently, the pPC87 recombinant plasmids were transferred into yeast cells, which could grow on the medium without Ura. The transformants were cultured on SD media without Ura and Trp at 30°C for 3 d. A colony-lift filter assay was used for qualitative analysis of trans-activation activity. After the fusion protein interactions with the GCC element, the lacZ gene was induced and positive cells (blue) were detected through enzymatic staining with X-gal as the substrate. The relative β-galactosidase activity of the transformant colonies were examined with the o-nitrophenyl-β-D-galactopyranoside (ONPG) (Clontech Laboratories, Inc.).
Results
Directed Evolution and Screening the Potential Key Amino Acids Involved in DNA Binding Activity
In our previous study, we cloned a member of the ERF family gene, named BnaERF-B3-hy15; the DNA shuffling strategy was used to shuffle the BnaERF-B3-hy15 gene, and three mutants (Mu1, Mu2, and Mu3) of this gene were obtained from variant mutants. The four proteins (BnaERF-B3, Mu1, Mu2, and Mu3) were investigated using yeast one-hybrid assays, and the binding activity to the GCC box relatively varied (Jin et al., 2010a).
The 3D structure of the proteins showed that the AP2-domain consisted of three β-sheets and one α-helix, which functions in DNA binding. Nevertheless, whether the mutant site plays a key role in the binding activity remains unclear. Here, further sequencing analysis of these four genes showed that a total of 26 nucleic acid sites were altered (Figure 2, Table 1), which led to 12 amino acid site substitutions. A subsequent comparison of the amino acid sequences of four proteins showed that these proteins were highly homologous, only one different amino acid (Gly156) was found in the AP2-domain and α-helix, the remaining mutants were located in the periphery of the domain, and one amino acid (site192) differed between Mu2 (Val192) and Mu3 (Ile192) (Figure S1).
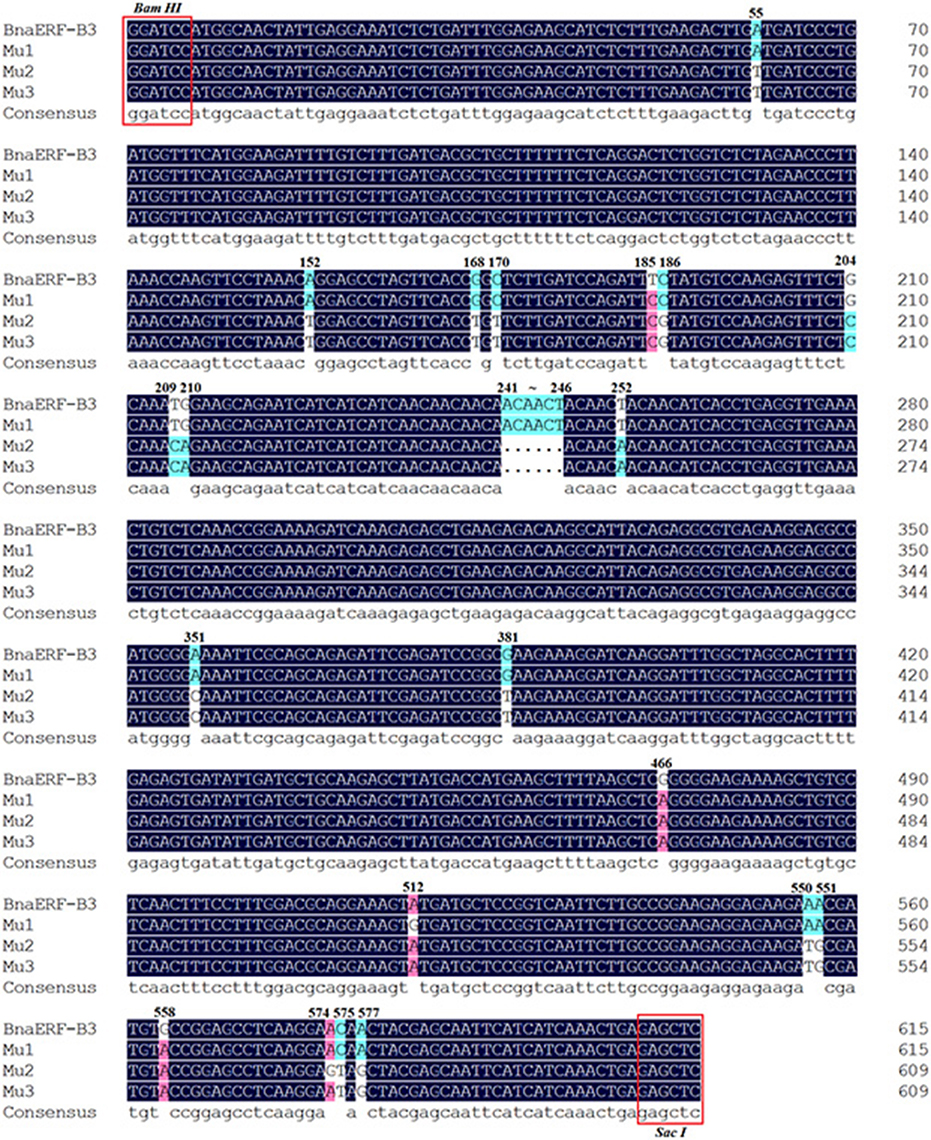
Figure 2. The mutated nucleic acid sites among the three mutants and wild-type proteins. The different color codes represent the sequence homology.
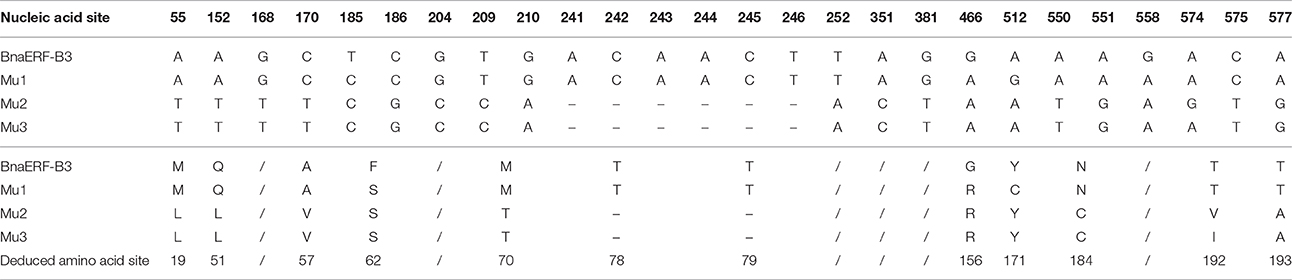
Table 1. Mutated nucleic acid sites and deduced amino acid sites among the three mutants and wild-type proteins.
Characterization of the Role of Each of 12 Amino Acid Site Substitutions in the Binding Activity of the GCC Box through Site-Directed Mutagenesis
To identify the amino acid substitution site that may be essential for the binding activity of the mutants, a series of site-directed mutants of BnaERF-B3 were generated according to the sequence alignment results. The strategy for site-directed mutagenesis is shown in Figure 1. All the 11 mutants (Mu4 to Mu14) were confirmed through sequencing, then cloned into the expression vector pPC87, and transformed to yeast, which hosts another vector with the GCC box sequence. Results of the yeast one-hybrid screening showed that the clones of Mu10, where Gly156 was substituted with Arg, appeared dark blue on X-gal-containing plates, but the 10 other mutants (Mu4 to Mu9 and Mu11 to Mu14) did not appear obviously blue (Figure 3). The β-galactosidase activity assay was used to calculate relative activity. As shown in Figure 4, Mu10 showed a 13-fold increase over BnaERF-B3. The differences in the trans-activation activity of BnaERF-B3 and the mutants showed that the mutation of Gly156 located in the AP2-domain significantly increased the binding activity. However, the other mutations outside the AP2-domain may have minimal or no influence on the binding activity. Our results indicated that the amino acid residues at site 156 may have an important role in binding to the GCC box.
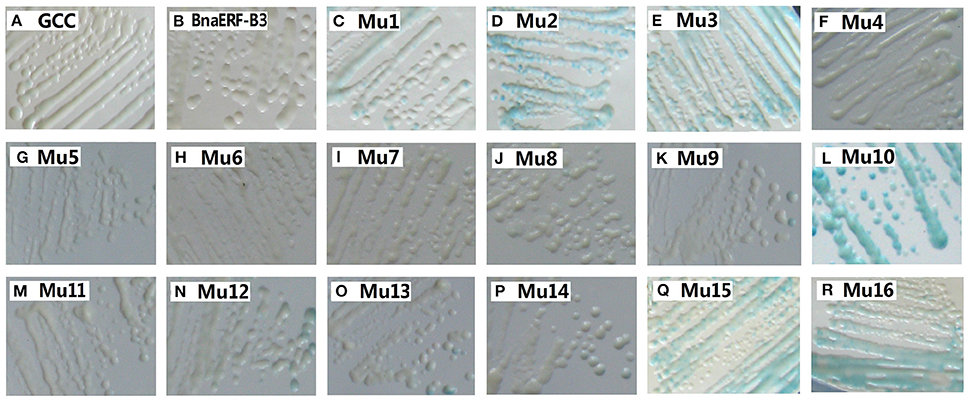
Figure 3. Yeast-one hybrid analysis of activities of BnaERF-B3 and mutants. The yeast cells were examined with X-gal after growing on SD/-Ura /-Trp- medium for 2 d at 30°C. GCC: yeast reporter cells carry GCC box (as a control); Mu1–Mu3: the mutants by Jin et al. (2010a). The information of the substitution site and amino acid of all Mu4 to Mu16 were as follows: Mu4: Met19Leu; Mu5: Gln51Leu; Mu6: Ala57Val; Mu7: Phe62Ser; Mu8: Met70Thr; Mu9: Thr78–, Thr79–; Mu10: Gly156Arg; Mu11: Tyr171Cys; Mu12: Asn184Cys; Mu13: Thr192Val, Thr193Ala; Mu14: Thr192Ile, Thr193Ala; Mu15: Gly156Arg, Thr192Val; Mu16: Gly156Arg, Thr192Ile.
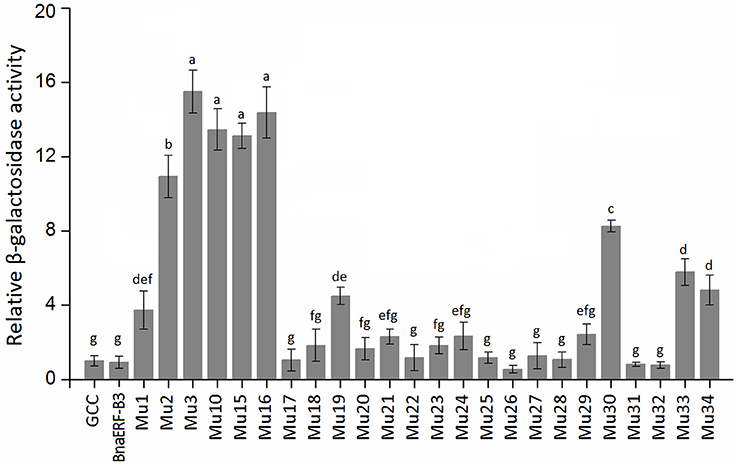
Figure 4. The relative β-galactosidase activities of BnaERF-B3 and mutants. Three independent clones were chose to culture in liquid SD/-Ura/-Trp medium for testing the β-galactosidase relative activity with o-nitrophenyl-β-D-galactopyranoside (ONPG) as the substrate. The data are the means ± SD of three replicates. Different letters on the vertical bars indicate significant difference at 0.05 levels.
Only one amino acid differed between Mu2 and Mu3, but the binding activity of these mutants varied. We further designed two sets of site-directed mutagenesis (Thr192Val and Thr192Ile) based on Mu10, which hosts Gly156Arg. Yeast one-hybrid assay and β-galactosidase activity were also used to test the activity of Mu15 (Gly156Arg + Thr192Val) and Mu16 (Gly156Arg + Thr192Ile; Figures 3, 4). Although Mu16 showed a slightly higher increase in activity than Mu15, both mutants exhibited higher activity than BnaERF-B3. This result implied that the mutation of T192 may minimally affect the activity, whereas the mutation of Arg156 played a dominant role.
Characterization of the Role of Each Altered Amino Acid in the 156th Site in Terms of the Binding Activity to the GCC Box through Site-Directed Mutagenesis
The mutation at site 156 from Gly to Arg greatly increased the activity, but we cannot determine whether the activity will be constant even if the 156th amino acid was mutated to the 18 other kinds of amino acid. To investigate the other amino acid residues that affect the binding activity, we replaced the 156th amino acid with the other 18 remaining amino acids into BraERF-B3 and the corresponding mutants were named Mu17 to Mu34.
Yeast one-hybrid analysis results showed that the transformant cells of various mutants had different activity, in which some clones appeared colorless, weakly stained blue, or stained deep blue (Figure 5). The results of quantitative analysis of β-galactosidase activity showed that Mu30 (Gly156Gln) had higher binding ability, followed by Mu33 (Gly156Lys), Mu34 (Gly156His), and Mu19 (Gly156Leu) (Figure 4). However, the activity of all the mutants was lower than that of Mu10 (Gly156Arg). These results illustrated that the space conformation of different amino acids caused conformational changes in the active domain, and the transcription factor showed different levels of activity. Therefore, the 156th amino acid is essential to maintain protein activity. Arg, Gln, Lys, His, and Leu are candidate amino acids for promoting BraEFR-B3 binding to the GCC box; among which, Arg exhibits the highest potential.
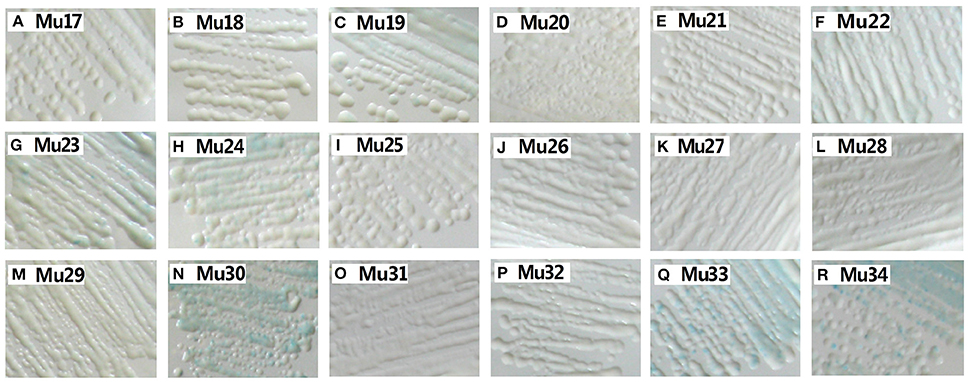
Figure 5. Yeast-one hybrid analysis of trans-activation activity of mutants of Mu17 to Mu34. The yeast cells were examined with X-gal after growing on SD/-Ura /-Trp- medium for 2 d at 30°C. The information of the substitution site and amino acid of all Mu17 to Mu34 were as follows: Mu17: Gly156Ala; Mu18: Gly156Val; Mu19: Gly156Leu; Mu20: Gly156Ile; Mu21: Gly156Pro; Mu22: Gly156Phe; Mu23: Gly156Tyr; Mu24: Gly156Trp; Mu25: Gly156Ser; Mu26: Gly156Thr; Mu27: Gly156Cys; Mu28: Gly156Met; Mu29: Gly156Asn; Mu30: Gly156Gln; Mu31: Gly156Asp; Mu32: Gly156Glu; Mu33: Gly156Lys; Mu34: Gly156His.
Mutation in the 156th Amino Acid Changes the Structure
Numerous studies have proved that the structure of amino acids considerably affects the protein structure and conformation. Studies on ERF proteins also showed that sequence diversity in the ERF domain can indirectly affect the binding activity to the GCC box (Ohta et al., 2001; Jin et al., 2010a). Among the 12 amino acid site substitutions, only Gly156Arg is located in the AP2-domain. The mutant hosting Gly156Arg (Mu10) showed the highest binding activity to the GCC box, followed by Gly156Gln (Mu30), Gly156Lys (Mu33), Gly156His (Mu34), and Gly156Leu (Mu19). However, the mutants Gly156Thr (Mu26), Gly156Asp (Mu31), and Gly156Glu (Mu32) showed no detectable binding activity to the GCC box. The 3D structures of the mutant and wild-type BnaERF-B3 were derived using the Swiss–Model server, with AtERF1 as the template (Figure 6). Comparison of the mutated amino acid residue sequences and the structure of BnaERF-B3 indicated that the 3D structures have no visible differences in the three β-sheets. However, an obvious change in the α-helix structure occurred when the amino acid residues were substituted. In the mutants which had high binding activity, more N atom (blue ball) were be found in models. The different amino acids in site 156th represented different conformation, which may lead to activity change.

Figure 6. The molecular modeling of the conserved AP2 domain among the BnaERF-B3 and mutants. The different residues are indicated as ball and stick representation. Red ball represents O atom, blue ball represents N atom, and gray ball represents C atom.
Identification of Key Sites in AP2-Domain for Binding Activity to the GCC Box
Previous studies have showed that some residues in AP2-domain of ERFs were indispensable for binding with GCC box (Jofuku et al., 1994; Allen et al., 1998; Cao et al., 2001; Hao et al., 2002; Liu et al., 2006). Sequence alignment revealed that these residues were not found on the BnaERF-B3. The mutant Mu3 of BnaERF-B3 showed highest binding activity to GCC box, it is reasonable to deduce that Arg156 is responsible for the binding activity of GCC box. To further validate the residues in the AP2-domain regions that might be responsible for the activity of ERFs to activate transcription, a series of site-directed mutants (Mu35–Mu42) were further constructed according to the different sites of sequence alignment of Mu3 and other ERFs, which reported by other researchers (Jofuku et al., 1994; Allen et al., 1998; Cao et al., 2001; Hao et al., 2002; Liu et al., 2006). All the mutants were transformed into yeast cells and the interaction with GCC box were quantified with X-gal assay.
Figure 7 showed that the mutants still showed weak or strong binding activity to GCC box, except Mu39. The synthetic mutant Mu43, which contained all the mutation sites of Mu35 to Mu42, exhibited a higher binding activity. On the basis of Gly156 was replaced by Arg, the other amino acid sites have little effect on the binding with GCC box. β-galactosidase activity assay also showed a consistent result (Figure 8). Mu36 (Trp116Ser) had the highest binding activity, followed by Mu43, Mu35 (Pro115Asn). In tobacco, the substitution of Pro9 or Trp10 with Asn or Ser in AP2-domain led to increase of the binding activity of NtERF2-F (Hao et al., 2002). Together, these results strongly suggested that the binding specificity with cis element can be altered by amino acid exchange mutation, and also demonstrated that Arg156 is an important determinant of the Bna-ERF-B3 binding specificity.
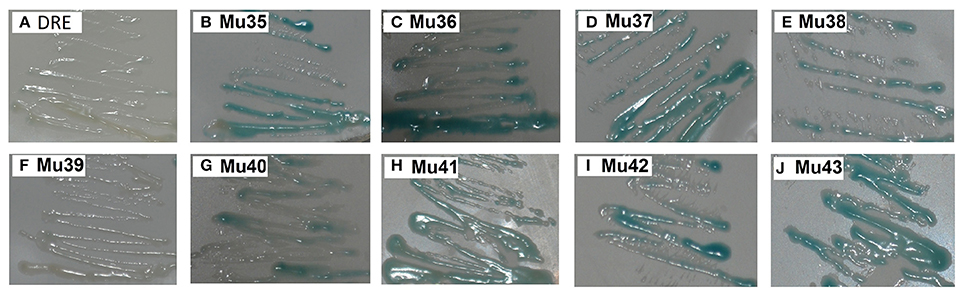
Figure 7. Yeast one-hybrid analysis of activities of mutants of and Mu35 to Mu43. The yeast cells were examined with X-gal after growing on SD/-Ura /-Trp- medium for 2 d at 30°C. DRE: yeast reporter cells carry DRE element (as a control). The information of the substitution site and amino acid of all Mu35 to Mu42 were as follows: Mu35: Pro115Asn; Mu36: Trp116Ser; Mu37: Ala120Val; Mu38: Asp125Glu; Mu39: Gly136Glu; Mu40: Gly136Ser; Mu41: Ser140Thr; Mu42: Ala144Val. Mu43 contained all the mutation sites of Mu35 to Mu42.
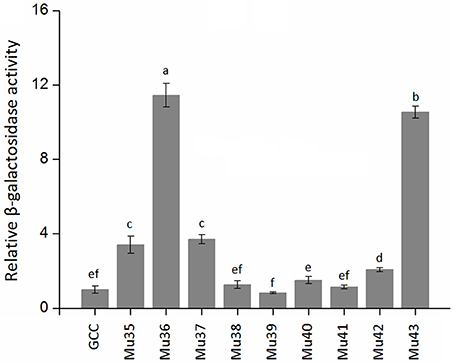
Figure 8. The relative β-galactosidase activities of Mu35 to Mu43. The data are the means ± SD of three replicates. Different letters on the vertical bars indicate significant difference at 0.05 levels.
Identification of Gly in AP2-Domain of DREBs for Binding Activity to the DRE cis Element
As the members of AP2/ERF family, ERFs and DREBs have high similarity on amino acid sequences. Cao et al. (2001) and Hao et al. (2002) have reported that Val14 and Glu19 residues were crucial in the regulation of the binding activity of DREB1A to the DRE cis element, while Ala14 was an important determinant of the ERF binding specificity. Three DREB proteins (CBF1, CBF2, and CBF3) in Arabidopsis have been identified to bind with DRE element (Liu et al., 1998). Amino acid sequence analysis of BnaERF-B3 and these three DREB proteins indicated that the AP2-domains were also highly homologous. We found that in the AP2-domains of CBFs of Arabidopsis, the corresponding amino acids of Gly156 in BnaERF-B3 were all Arg. To explore whether Gly can affect the binding activity of DREB proteins, three mutants MuCBF1 (Arg95Gly), MuCBF2 (Arg98Gly), and MuCBF3 (Arg98Gly) of Arabidopsis were obtained. Yeast one-hybrid (Figure 9A) and β-galactosidase activity (Figure 9B) showed that these three mutants also had the strong binding ability with DRE element. This result showed Gly did not affect the binding affinity of DREBs to DRE cis element.
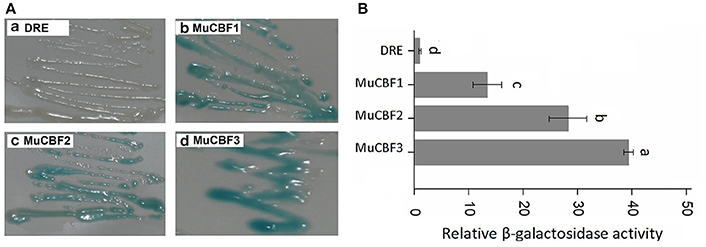
Figure 9. Yeast one-hybrid analysis of MuCBF1, MuCBF2, and MuCBF3. (A) The binding activities of MuCBF1, MuCBF2, and MuCBF3. (B) The relative β-galactosidase activities of MuCBF1, MuCBF2, and MuCBF3. DRE: yeast reporter cells carry DRE element (as a control). The data are the means ± SD of three replicates. Different letters on the vertical bars indicate significant difference at 0.05 levels.
Discussion
Amino acid sequences can determine the higher-order of protein structure when the specific conformation of the protein exhibits biological function. A single amino acid mutation may cause functional disorder in a protein. Sickle cell anemia is a disease caused by the replacement of a Glu acid residue with Val at the sixth position of the β-chain of hemoglobin (Mehanna, 2001). A study on the recombinant encephalomyocarditis (EMC) virus showed that a point mutation at position 776 (Ala776Thr) of the EMC virus polyprotein resulted in a loss of viral diabetogenicity (Jun et al., 1997). The modification of peptides and proteins has attracted wide spread attention and has become an important research direction; several enzymes and proteins have been successfully modified to increase their efficiency, investigate gene functions, and realize molecular breeding (Lassner and Bedbrook, 2001; Eijsink et al., 2005; Xiong et al., 2007a, 2011; Reinstädler et al., 2010). Common strategies include random-priming in vitro recombination, gene shuffling, site-directed mutagenesis, and semi-rational design (Stemmer, 1994; Xiong et al., 2007b,c, 2010; Tian et al., 2013, 2015). Site-directed mutagenesis is widely used in research on proteins to reveal the relationship between structure and function. Several studies have been performed to explore key amino acid residues. For example, when residue Ala363 was changed to Val, the cytochrome CYP2B2 mutant protein exhibited an altered metabolite profile; by contrast, substitution of Ala478 with Gly yielded a higher catalytic efficiency for lidocaine oxidation than that of CYB2B2 (Domanski and Halpert, 2001). A mutation at Glu160 in the active site of trichosanthin caused a significant loss of potency, whereas substitution of Gln156 with Ala only slightly decreased the activity (Wong et al., 1994).
Transcription factors can directly regulate downstream genes by specifically binding cis-acting elements in the promoter of target genes at their DNA-binding site. Various studies have confirmed that transcription factors play pivotal roles in many processes during plant growth, such as integration of various signal transduction pathways and responses to abiotic/biotic stresses (Kasuga et al., 1999; Yamaguchi-Shinozaki and Shinozaki, 2006; Nakashima et al., 2012). ERF transcription factors were shown to be excellent candidates for improving multiple stress resistance by regulating the expression of various stress-inducible genes (Gutterson and Reuber, 2004; Xu et al., 2011; Mizoi et al., 2012; Rong et al., 2014). The special DNA-binding domains of ERFs have been well characterized, which mainly recognize the cis-acting element AGCCGCC, known as the GCC box (Ohme-Takagi and Shinshi, 1995; Hao et al., 1998; Zhang G. et al., 2009).
Our previous work found that an ERF member named BnaERF-B3 could not activate the expression of the reporter gene in yeast, whereas the mutants of BnaERF-B3 (Mu1, Mu2, and Mu3) showed binding activity with GCC box (Jin et al., 2010a). Overexpressing BnaERF-B3-mu3 exhibited more freezing tolerance than the original BnaERF-B3 in transgenic Arabidopsis (Xiong et al., 2013). To identify the key amino acids that are crucial to the binding activity of BnaERF-B3, site-directed mutagenesis was used to generate a series of mutants.
We first produced 11 BnaERF-B3 mutants containing mutant amino acid residues. The relative activity of Mu10 was higher than that of BnaERF-B3. The results showed that the mutation in the amino acid Arg156 was a potential key site involved in binding activity. The mutations with other amino acids outside the AP2-domain were not essential for binding activity. The key amino acid Arg156 was located in the α-helix. Although ERF transcription factor members present high similarity in the conserved AP2-domain, the differences of amino acid residues in their domains lead to ERFs with different levels of binding activity to the GCC box. The conserved Ala37 in the α-helix was confirmed to be essential for binding to the GCC box (Liu et al., 2006). Thr178 was participated in forming α-helix structures (Allen et al., 1998). Other studies also strongly suggest that the α-helix was important for AP2-domain function in ERF proteins and may participate in protein–protein interactions (Jofuku et al., 1994; Büttner and Singh, 1997).
Besides Arg156, when Gly156 was substituted with other four amino acids (Gln, Lys, His, and Leu), the proteins also showed high binding activity. However, proteins hosting other three amino acids (Thr, Asp, and Glu) showed no detectable binding activity. As compared with Gly, the other aliphatic amino acids (Leu, Val, and Ile) seemed improve the affinity of ERF factor with GCC box, especially Leu. Here, we also found that the introduction of basic amino acid residues (Arg, Gln, Lys, and His) caused the increasing of binding activity, while replacements with acidic amino acid residues (Asp and Glu) appeared to have less impact. This result showed that acidic amino acid residues might interfere with the DNA binding while other types of amino acids seem to be compatible or advantageous. Previous studies also found that in some experiments acidic amino acid-rich amino-terminus did not have nucleic acid binding activity (Frédéric et al., 2011). The residues in AP2-domain of ERF genes were indispensable for binding with GCC box. Multiple sequence analysis results showed that several amino acids, such as Asp43 and Asn57, are highly conserved in the AP2-domain of ERF proteins (Hao et al., 1998; Nakano et al., 2006). Gly174 was necessary for AP2 genetic functions; the replacement of Gly residues with either Ser or Glu may perturb its function (Jofuku et al., 1994). Two mutations of NtERF2-F, ERFpn and ERFws, exhibited a higher binding activity with GCC box (Hao et al., 2002). A series of mutations was further performed to explore the optimal amino acid in AP2-domain for BnaERF-B3 binding to the GCC box. Analysis of the result revealed that the amino acid residues of AP2-domain are sufficient to affect the binding activity of ERF to GCC box. Especially Pro115Asn and Trp116Ser can obviously increase the binding activity. The change in the protein activity may be due to the different amino acids that cause a conformational change in the protein.
The amino acid residues of AP2-domains are also conserved in DREB subgroup as well as ERF subgroup. To assess the functional significance of the corresponding residue of ERF subgroup in binding to DRE element, three mutants MuCBF1 (Arg95Gly), MuCBF2 (Arg98Gly), and MuCBF3 (Arg98Gly) were obtained. The result revealed that the substitution of Arg with Gly did not affect the binding activity of the DREB transcription factors to the DRE cis element. The findings in this study can be used for further research on AP2/ERF protein functions and plant molecular breeding in the future.
Author Contributions
AX initiated and designed the research. JZ, ML, BW, YL, and AX performed the experiments. JZ, ML, and AX analyzed the data. AX contributed reagents/materials/analysis tools. JZ and ML wrote the paper. JZ, ML, and AX revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was supported by the National Natural Science Foundation of China (31570691), Natural Science Foundation of Jiangsu Province (BK20130027) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01603/full#supplementary-material
Figure S1. Alignment of amino acid sequences in AP2-domain of BnaERF-B3 and the mutants. The numbers above the amino acids corresponding to the position of the difference amino acid between BnaERF-B3 and the mutants. The α-helix and β-sheet regions are marked with black bar and arrows, respectively.
Table S1. Primers used for cloning or synthesizing mutations. F, Forward primer; R, Reverse primer. – represents the site was lacking. Due to the mutation site of Mu13, Mu14, Mu15, and Mu16 was near to the termination, so we only designed the reverse primer.
References
Allen, M. D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. doi: 10.1093/emboj/17.18.5484
Büttner, M., and Singh, K. B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 5961–5966. doi: 10.1073/pnas.94.11.5961
Cao, Z., Li, J., Chen, F., Li, Y., Zhou, H., and Liu, Q. (2001). Effect of two conserved amino acid residues on DREB1A function. Biochemistry (Mosc) 66, 623–627. doi: 10.1023/A:1010251129429
Domanski, T. L., and Halpert, J. R. (2001). Analysis of mammalian cytochrome P450 structure and function by site-directed mutagenesis. Curr. Drug Metab. 2, 117–137. doi: 10.2174/1389200013338612
Eijsink, V. G., Gåseidnes, S., Borchert, T. V., and van den Burg, B. (2005). Directed evolution of enzyme stability. Biomol. Eng. 22, 21–30. doi: 10.1016/j.bioeng.2004.12.003
Franco-Zorrilla, J. M., López-Vidriero, I., Carrasco, J. L., Godoy, M., Vera, P., and Solano, R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111, 2367–2372. doi: 10.1073/pnas.1316278111
Frédéric, G., Nicolas, L., Thierry, C., Pascal, G., Céline, J., Fabienne, F. O., et al. (2011). Proline- and acidic amino acid-rich basic leucine zipper proteins modulate peroxisome proliferator-activated receptor (ppar) activity. Proc. Natl. Acad. Sci. U.S.A. 108, 4794–4799. doi: 10.1073/pnas.1002862108
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Guan, Y., Meng, X., Khanna, R., LaMontagne, E., Liu, Y., and Zhang, S. (2014). Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 10:e1004384. doi: 10.1371/journal.pgen.1004384
Gutterson, N., and Reuber, T. L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7, 465–471. doi: 10.1016/j.pbi.2004.04.007
Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273, 26857–26861. doi: 10.1074/jbc.273.41.26857
Hao, D., Yamasaki, K., Sarai, A., and Ohme-Takagi, M. (2002). Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 41, 4202–4208. doi: 10.1021/bi015979v
Jin, X. F., Zhu, B., Peng, R. H., Jiang, H. H., Chen, J. M., Zhuang, J., et al. (2010a). Optimizing the binding activity of the AP2/ERF transcription factor with the GCC box element from Brassica napus by directed evolution. BMB Rep. 43, 567–572. doi: 10.5483/BMBRep.2010.43.8.567
Jin, X. F., Xiong, A. S., Peng, R. H., Liu, J. G., Gao, F., Chen, J. M., et al. (2010b). OsAREB1, an ABRE-binding protein responding to ABA and glucose, has multiple functions in Arabidopsis. BMB Rep. 43, 34–39. doi: 10.5483/BMBRep.2010.43.1.034
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Jun, H. S., Kang, Y., Notkins, A. L., and Yoon, J. W. (1997). Gain or loss of diabetogenicity resulting from a single point mutation in recombinant encephalomyocarditis virus. J. Virol. 71, 9782–9785.
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Lassner, M., and Bedbrook, J. (2001). Directed molecular evolution in plant improvement. Curr. Opin. Plant Biol. 4, 152–156. doi: 10.1016/S1369-5266(00)00152-7
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, dreb1 and dreb2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1409. doi: 10.1105/tpc.10.8.1391
Liu, Y., Zhao, T. J., Liu, J. M., Liu, W. Q., Liu, Q., Yan, Y. B., et al. (2006). The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett. 580, 1303–1308. doi: 10.1016/j.febslet.2006.01.048
Lu, Y., Zen, K. C., Muthukrishnan, S., and Kramer, K. J. (2002). Site-directed mutagenesis and functional analysis of active site acidic amino acid residues d142, d144 and e146 in manduca sexta (tobacco hornworm) chitinase. Insect Biochem. Mol. Biol. 32, 1369–1382. doi: 10.1016/S0965-1748(02)00057-7
Mehanna, A. (2001). Sickle cell anemia and antisickling agents then and now. Curr. Med. Chem. 8, 79–88. doi: 10.2174/0929867013373778
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nguyen, X. C., Kim, S. H., Lee, K., Kim, K. E., Liu, X. M., Han, H. J., et al. (2012). Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep. 31, 737–745. doi: 10.1007/s00299-011-1192-x
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/tpc.13.8.1959
Park, J. M., Park, C. J., Lee, S. B., Ham, B. K., Shin, R., and Paek, K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2–type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Peng, R. H., Xiong, A. S., and Yao, Q. H. (2006). A direct and efficient PAGE-mediated overlap extension PCR method for gene multiple-site mutagenesis. Appl. Microbiol. Biotechnol. 73, 234–240. doi: 10.1007/s00253-006-0583-3
Qin, F., Li, J., Zhang, G., Zhao, J., Chen, S., and Liu, Q. (2003). Isolation and structural analysis of DRE-binding transcription factor from maize (Zea mays L.). Acta Bot Sin 45, 331–339.
Reinstädler, A., Müller, J., Czembor, J. H., Piffanelli, P., and Panstruga, R. (2010). Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant Biol. 10:31. doi: 10.1186/1471-2229-10-31
Rong, W., Qi, L., Wang, A., Ye, X., Du, L., Liang, H., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., et al. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. doi: 10.1105/tpc.13.1.61
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Stemmer, W. P. (1994). Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391. doi: 10.1038/370389a0
Sun, S., Yu, J. P., Chen, F., Zhao, T. J., Fang, X. H., Li, Y. Q., et al. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 283, 6261–6271. doi: 10.1074/jbc.M706800200
Tian, Y. S., Xu, J., Peng, R. H., Xiong, A. S., Xu, H., Zhao, W., et al. (2013). Mutation by DNA shuffling of 5-enolpyruvylshikimate-3-phosphate synthase from Malus domestica for improved glyphosate resistance. Plant Biotechnol. J. 11, 829–838. doi: 10.1111/pbi.12074
Tian, Y. S., Xu, J., Zhao, W., Xing, X. J., Fu, X. Y., Peng, R. H., et al. (2015). Identification of a phosphinothricin-resistant mutant of rice glutamine synthetase using DNA shuffling. Sci. Rep. 5:15495. doi: 10.1038/srep15495
Wong, K. B., Ke, Y. B., Dong, Y. C., Li, X. B., Guo, Y. W., Yeung, H. W., et al. (1994). Structure/function relationship study of Gln156, Glu160 and Glu189 in the active site of trichosanthin. Eur. J. Biochem. 221, 787–791. doi: 10.1111/j.1432-1033.1994.tb18792.x
Xiong, A. S., Jiang, H. H., Zhuang, J., Peng, R. H., Jin, X. F., Zhu, B., et al. (2013). Expression and function of a modified AP2/ERF transcription factor from Brassica napus enhances cold tolerance in transgenic Arabidopsis. Mol. Biotechnol. 53, 198–206. doi: 10.1007/s12033-012-9515-x
Xiong, A. S., Peng, R. H., Cheng, Z. M., Li, Y., Liu, J. G., Zhuang, J., et al. (2007a). Concurrent mutations in six amino acids in β-glucuronidase improve its thermostability. Protein Eng. Des. Sel. 20, 319–325. doi: 10.1093/protein/gzm023
Xiong, A. S., Peng, R. H., Liu, J. G., Zhuang, J., Qiao, Y. S., Xu, F., et al. (2007b). High efficiency and throughput system in directed evolution in vitro of reporter gene. Appl. Microbiol. Biotechnol. 74, 160–168. doi: 10.1007/s00253-006-0659-0
Xiong, A. S., Peng, R. H., Zhuang, J., Chen, J. M., Zhang, B., Zhang, J., et al. (2011). A thermostable β-glucuronidase obtained by directed evolution as a reporter gene in transgenic plants. PLoS ONE 6:e26773. doi: 10.1371/journal.pone.0026773
Xiong, A. S., Peng, R. H., Zhuang, J., Liu, J. G., Gao, F., Xu, F., et al. (2007c). A semi-rational design strategy of directed evolution combined with chemical synthesis of DNA sequences. Biol. Chem. 388, 1291–1300. doi: 10.1515/BC.2007.153
Xiong, A. S., Yao, Q. H., Peng, R. H., and Cheng, Z. M. (2010). “Directed in vitro evolution of reporter genes based on semi-rational design and high-throughput screening,” in In vitro Mutagenesis Protocols, ed J. Braman (Totowa, NJ: Springer; Humana Press), 239–256.
Xiong, A. S., Yao, Q. H., Peng, R. H., Duan, H., Li, X., Fan, H. Q., et al. (2006). PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 1, 791–797. doi: 10.1038/nprot.2006.103
Xiong, A. S., Yao, Q. H., Peng, R. H., Li, X., Fan, H. Q., Cheng, Z. M., et al. (2004). A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic Acids Res. 32:e98. doi: 10.1093/nar/gnh094
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yamasaki, K., Kigawa, T., Seki, M., Shinozaki, K., and Yokoyama, S. (2013). DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 18, 267–276. doi: 10.1016/j.tplants.2012.09.001
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, Y., Chen, C., Jin, X. F., Xiong, A. S., Peng, R. H., Hong, Y. H., et al. (2009). Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep. 42, 486–492. doi: 10.5483/BMBRep.2009.42.8.486
Zhao, T. J., Liu, Y., Yan, Y. B., Feng, F., Liu, W. Q., Zhou, H. M., et al. (2007). Identification of the amino acids crucial for the activities of drought responsive element binding factors (DREBs) of Brassica napus. FEBS Lett. 581, 3044–3050. doi: 10.1016/j.febslet.2007.05.067
Keywords: arginine, Brassica napus L., ERF factor, GCC element, site-directed mutagenesis, yeast-one hybrid
Citation: Zhuang J, Li M-Y, Wu B, Liu Y-J and Xiong A-S (2016) Arg156 in the AP2-Domain Exhibits the Highest Binding Activity among the 20 Individuals to the GCC Box in BnaERF-B3-hy15, a Mutant ERF Transcription Factor from Brassica napus. Front. Plant Sci. 7:1603. doi: 10.3389/fpls.2016.01603
Received: 19 July 2016; Accepted: 11 October 2016;
Published: 27 October 2016.
Edited by:
Agnieszka Ludwików, Adam Mickiewicz University in Poznań, PolandCopyright © 2016 Zhuang, Li, Wu, Liu and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ai-Sheng Xiong, xiongaisheng@njau.edu.cn
†These authors have contributed equally to this work.
 Jing Zhuang
Jing Zhuang Meng-Yao Li†
Meng-Yao Li† Ai-Sheng Xiong
Ai-Sheng Xiong