- 1Non-timber Forest Research and Development Center, Chinese Academy of Forestry, Zhengzhou, China
- 2The Eucommia Engineering Research Center of State Forestry Administration, Zhengzhou, China
MicroRNAs (miRNAs), a group of endogenous small non-coding RNAs, play important roles in plant growth, development, and stress response processes. Eucommia ulmoides Oliver (hardy rubber tree) is one of the few woody plants capable of producing trans-1, 4-polyisoprene (TPI), also known as Eu-rubber, which has been utilized as an industrial raw material and is extensively cultivated in China. However, the mechanism of TPI biosynthesis has not been identified in E. ulmoides. To characterize small RNAs and their targets with potential biological roles involved in the TPI biosynthesis in E. ulmoides, in the present study, eight small RNA libraries were constructed and sequenced from young and mature leaves and fruits of E. ulmoides. Further analysis identified 34 conserved miRNAs belonging to 20 families (two unclassified families), and 115 novel miRNAs seemed to be specific to E. ulmoides. Among these miRNAs, fourteen conserved miRNAs and 49 novel miRNAs were significantly differentially expressed and identified as Eu-rubber accumulation related miRNAs. Based on the E. ulmoides genomic data, 202 and 306 potential target genes were predicted for 33 conserved and 92 novel miRNAs, respectively; the predicted targets are mostly transcription factors and functional genes, which were enriched in metabolic pathways and biosynthesis of secondary metabolites. Noticeably, based on the expression patterns of miRNAs and their target genes in combination with the Eu-rubber accumulation, the negative correlation of expression of six miRNAs (Eu-miR14, Eu-miR91, miR162a, miR166a, miR172c, and miR396a) and their predicted targets serving as potential regulators in Eu-rubber accumulation. This study is the first to detect conserved and novel miRNAs and their potential targets in E. ulmoides and identify several candidate genes potentially controlling rubber accumulation, and thus provide molecular evidence for understanding the roles of miRNAs in regulating the TPI biosynthesis in E. ulmoides.
Introduction
Eucommia ulmoides Oliver is a deciduous tree and a species of the only genus in the family Eucommiaceae (euasterids I: order Garryales) (Call and Dilcher, 1997). It has been used in traditional Chinese medicine and extensively cultivated across 27 provinces in China. This species is also one of the best-known trans-1,4-polyisoprene (TPI) producers (Takeno et al., 2008; Du et al., 2015). TPI, also known as Eucommia rubber (Eu-rubber), is extracted from leaves, fruits, roots, and bark of Eucommia. Its content varies in different tissues and at different plant developmental stages, with the highest content found in fruits (Du, 2003; Yoshihisa et al., 2009). Eu-rubber, which shows less flexibility and poorer low-temperature thermoplasticity than cis-polyisoprene rubber, has attracted considerable attention as a raw industrial material for various commercial uses (Weiss, 1891; Jiang et al., 2006; Chen et al., 2008). At present, rubber tree (Hevea brasiliensis) is the most widely cultivated species for commercial production of natural rubber. However, as a typical tropical rainforest plant, this species is restricted to the tropics where it has already reached the limit of production capacity (Webster and Baulkwill, 1989; Suzuki et al., 2012; Wang Y. et al., 2013; Wang, 2014). Therefore, the shortage of natural rubber and the growing worldwide demand for the same necessitate an urgent development of Eu-rubber industry, which will meet the demand by increasing the economic yield and improving the content of Eu-rubber.
Previous studies have focused primarily on pharmacological and morphological aspects of E. ulmoides, and only limited genomic and molecular information is available. Furthermore, the biosynthetic enzymes involved in the synthesis and regulation of Eu-rubber are not well studied. In plants, the precursors of polyisoprene are derived from two distinct pathways: the mevalonate (MVA) pathway in cytoplasm and the methylerythritol phosphate (MEP) pathway in plastids (Yin et al., 2011). High-molecular-weight polyisoprene is synthesized by successive condensation of the precursors via a specific prenyltransferase (Suzuki et al., 2012). Although most MEP and MVA pathway genes have been cloned and analyzed, their function in Eu-rubber accumulation is not fully understood.
Small RNAs (sRNAs) contain endogenous small interfering RNAs and microRNAs (miRNAs) that are 21–24 nucleotides (nt) long (Jones-Rhoades et al., 2006). Small interfering RNAs are processed from perfectly double-stranded RNA, and miRNAs are derived from single-stranded RNA transcripts that form imperfectly double-stranded stem loop precursor structures (Llave et al., 2002a; Khraiwesh et al., 2010; Hao et al., 2012; Szittya and Burgyan, 2013). miRNAs are the most typical plant sRNAs that negatively regulate expression of target genes by repressing gene translation or cleaving target mRNAs. miRNAs play a vital role in various biological and metabolic processes of plant growth and development, such as signal transduction and responses to biotic or abiotic stresses (Bartel, 2004; Jones-Rhoades and Bartel, 2004; Jones-Rhoades et al., 2006; Mallory and Vaucheret, 2006; Sunkar et al., 2006; Voinnet, 2009; Wu et al., 2010). Furthermore, many plant miRNAs show tissue- or stage-specific expression patterns; for example, miR159, miR167, and miR172 are expressed between male and female flowers in asparagus, and miR156g and miR169t are stage-specific in Lycium barbarum fruits (Zeng et al., 2015; Chen et al., 2016). The expression patterns of miR156f-3p, miR157a-3p, miR5021, miR5163, miR5293, and novel_miR_27 are different in Panax notoginseng roots, stems, and leaves at different developmental stages (Wei et al., 2015). miRNAs also participate in regulating terpenoid accumulation through regulating their target genes, and in Arabidopsis thaliana and tomato, flavonoid biosynthesis is affected by the expression levels of miRNAs (Davuluri et al., 2005; Gou et al., 2011; Ng et al., 2011); miR6435, miR5021, and miR1134 are specifically expressed in glandular trichomes of Xanthium strumarium and play a role in the regulation of xanthanolide biosynthesis (Fan et al., 2015); and miR156, miR828, miR858, and miR5072 regulate anthocyanin accumulation in apple peels (Qu et al., 2016). The expression of miR159b regulates latex production in H. brasiliensis (Gébelin et al., 2013), and the negative regulation of hbr-miR172 results in a dramatic increase in latex yield of the rubber tree (Pramoolkit et al., 2014). Therefore, it is important to identify miRNAs and their target genes, which is essential to understanding miRNAs-mediated gene regulation of Eu-rubber biosynthesis.
miRNAs have been thoroughly studied in many species. Although E. ulmoides has significant medicinal and economical value, miRNAs are yet to be identified in this species. Many studies have confirmed that both conserved and species-specific miRNAs are very important in different biological processes in plants. Therefore, the aim of this study was to identify the conserved and novel miRNAs and their potential target genes in E. ulmoides and explore their interactions. To achieve this, we sequenced sRNA from leaves and fruits at different developmental stages by using the Illumina platform, analyzed gene expression profiles, and investigated the functions of their targets. Quantitative real-time PCR (qPCR) was adopted to evaluate the expression levels of miRNAs and their target genes and identify candidate genes involved in Eu-rubber accumulation. Our results provide valuable information about miRNAs involved in rubber biosynthesis in E. ulmoides.
Materials and Methods
Plant Materials
Leaves and fruits of E. ulmoides “Huazhong No. 6” were collected at Non-timber Research and Development Center of CAF (113°41′37″E, 34°46′23″N, Zhengzhou, Henan, China) on May 3 (at young stage) and July 16 (at mature stage) in 2015. The samples were labeled YL (young leaf, 0.26%, the rate of Eu-rubber accumulation), ML (mature leaf, 0.29%), YF (young fruits, 1.67%), and MF (mature fruits, 0.07%). The leaves and fruits were collected from three randomly selected individuals and pooled together in a single biological sample; two biological replicates were prepared for high-throughput sequencing. All samples were immediately frozen in liquid nitrogen and stored at −80°C for further analyses.
Total RNA Isolation, sRNA Library Construction, and Sequencing
Total RNAs were extracted from YL, ML, YF, and MF using a TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA quality was monitored on 1% agarose gels. RNA concentration was measured with a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and the integrity was assessed by an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The purified RNAs were used to construct sRNA libraries and were sequenced on an Illumina HiSeq 2500 platform at Novogene Company, Beijing, China.
Small RNA Analysis and miRNAs Prediction
Raw sequence data were obtained via the RNA high-throughput sequencing process and screened to remove contaminated reads, sequences containing “adapters,” those without insert tags, and reads with poly-A tails. In addition, Q20, Q30, and GC content of the raw data was calculated. The sequences 18–30 nt long were used for further analysis. Clean reads of sRNA tags were mapped onto the E. ulmoides genome database (unpublished) without mismatch to analyze their expression and distribution using Bowtie (Langmead et al., 2009). The rRNA, tRNA, snRNA, and snoRNA were annotated by aligning them against the Rfam database (Rfam:http://www.sanger.ac.uk/software/Rfam) and the NCBI GenBank databases (GenBank:http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Natural antisense transcripts and trans-acting small interfering RNAs were identified by using the PlantNATsDB (http://bis.zju.edu.cn/pnatdb/) and UEA sRNA tools, respectively (Moxon et al., 2008; Chen et al., 2011). The repeat sequences were annotated by aligning them to the Repbase database by RepeatMasker (Bao et al., 2015; Smit et al., 2013-2015). The mapped sRNA tags were used to search for conserved miRNA. miRBase version 21(ftp://mirbase.org/pub/mirbase/21/) was used as a reference, and modified software miRDeep2 and srna-tools-cli were used to obtain the potential miRNA sequences and draw the secondary structures (Friedländer et al., 2011). The available software miREvo and miRDeep2 were integrated to predict novel miRNA by exploring the secondary structure, the Dicer cleavage site, and the minimum free energy of the sRNA tags unannotated in the former steps (Wen et al., 2010; Friedländer et al., 2011). To explore the occurrence of miRNA families, we used the conserved miRNAs in miFam.dat (http://www.mirbase.org/ftp.shtml), and the novel miRNA precursors were aligned to Rfam (http://www.rfam.sanger.ac.uk/search/).
Differential Expression Analysis of miRNAs
To calculate significant differences in expression patterns in miRNAs, the expressions of miRNAs were estimated using transcripts per million (TPM) according to the normalization formula: Normalized expression = Mapped read count/Total reads × 1,000,000 (Zhou et al., 2010). Differential expression analysis of the two groups of biological replicates was performed for the samples using DESeq R package (1.8.3) (Anders and Huber, 2010). The P-values were adjusted using the Benjamin & Hochberg method, and corrected P-value of 0.05 was set as a threshold for significant differential expression by default (Love et al., 2014).
Target Gene Prediction and Functional Annotation
The target genes of miRNAs were predicted with the Web-based psRNATarget program (http://plantgrn.noble.org/psRNATarget/) (Dai and Zhao, 2011). The gene ontology (GO)-biological process and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms enriched in the target gene candidates were determined using GOseq and KOBAS software, respectively (Mao et al., 2005; Young et al., 2010).
Quantitative Real-Time PCR (qPCR) Analysis
To obtain candidate genes that were potentially controlling rubber accumulation, the expression level of several miRNAs and their potential targets were selected for qPCR. Total RNA was extracted from young and mature leaves or fruits by using a TRIzol reagent (Invitrogen, USA). The extracted total RNA was reverse-transcribed into cDNA using an AMV First Strand cDNA Synthesis Kit (Sangon, Shanghai, China) to detect the expression levels of miRNAs and their target genes. For miRNA expression, stem-loop qPCR primers were designed as previously described (Varkonyi-Gasic and Hellens, 2011), and the primers for target genes were designed by using Primer 5.0 (Premier Biosoft International, Palo Alto, CA, USA). Actin α was used as an internal reference gene for normalization; the primers used for qPCR were listed in Table S1. qPCR was performed using an ABI StepOnePlus system (Applied Biosystems, Foster City, CA, USA). Each PCR was performed in a reaction volume of 20 μL containing 10 μL of SybrGreen qPCR Master Mix (2 ×), 0.4 μL of forward primer, 0.4 μL of reverse primer, 2 μL of reverse-transcribed cDNA, and 7.2 μL of ddH2O. The PCR conditions were as follows: 3 min at 95°C, 45 cycles of 95°C for 7 s, 57°C for 10 s, and 72°C for 15 s, and melt curve analysis from 60 to 95°C. All reactions were performed in triplicate for each sample, and the expression levels were calculated with the 2−ΔΔCt method (Livak and Schmittgen, 2001). All the data were subjected to an analysis of variance and the results were presented as the mean ± SD. The P-value was considered to be statistically significant if <0.05 or <0.01.
Results
Sequence Analysis of Eight sRNA Libraries
To identify the miRNAs in E. ulmoides, the libraries from young leaves, mature leaves, young fruits, and mature fruits were generated and sequenced via the Illumina HiSeq 2500 platform. By using two biological replicates of the leaf and fruit tissues collected at two different developmental stages, eight cDNA libraries (YL1, YL2, ML1, ML2, YF1, YF2, MF1, and MF2) were created from E. ulmoides, yielding approximately 98 million raw reads. After removing the low-quality sequences and adapter sequences, a total of 9,885,853 (YL1), 12,288,265 (YL2), 11,482,801 (ML1), 11,336,124 (ML2), 10,371,302 (YF1), 12,588,040 (YF2), 14,889,857 (MF1), and 12,177,627 (MF2) clean reads 18–30 nt long were generated (Table 1). To verify the quality of the libraries, we inspected the error rate and GC content of the sequencing results (Table S2). Furthermore, 60.86–72.32% of high-quality sRNA reads from each of the eight libraries were mapped to the E. ulmoides genome, suggesting no significant sequencing bias in these eight libraries. The distribution pattern of sRNA was similar among the eight libraries. The size distribution of sRNAs was uneven, with the majority (>75%) of sRNAs having 20–24 nt and those with 24 nt being the most abundant class. The second most abundant class comprised sRNAs 21 nt long, and the proportion of the sRNAs length dynamically changed in different tissues and at different developmental stages (Figure 1). Various types of non-coding RNAs, which comprised conserved miRNAs, putative novel miRNAs, rRNAs, tRNAs, snRNAs, snoRNAs, repeat-associated RNAs, natural antisense transcripts, trans-acting small interfering RNAs, and unannotated fragments, were annotated (Table S3).
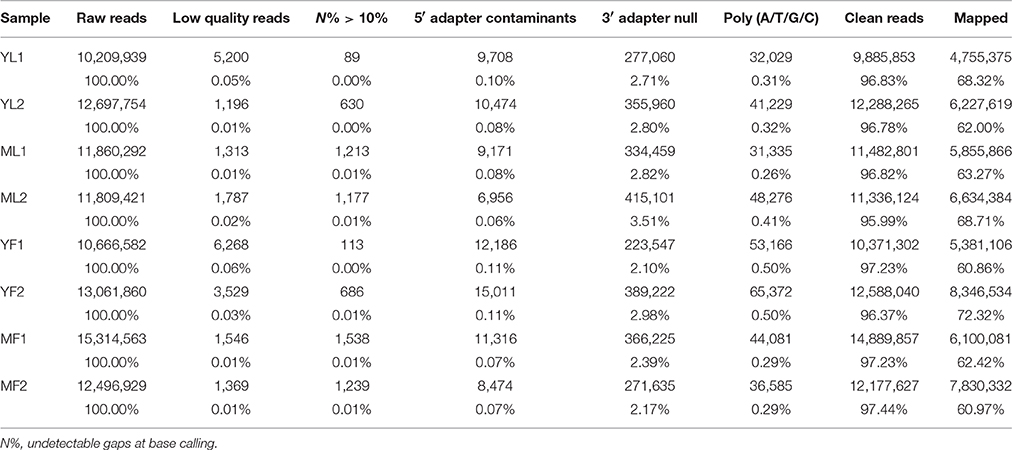
Table 1. Statistical summary of the data generated by high-throughput sequencing in Eucommia ulmoides.
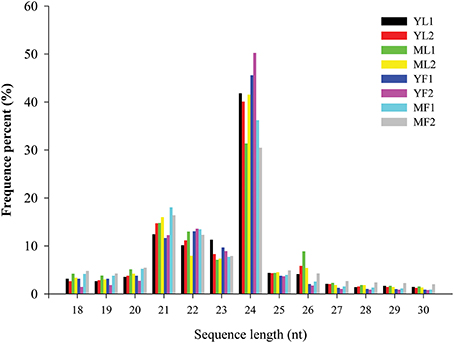
Figure 1. Length distribution and abundance of small RNAs in eight libraries of Eucommia ulmoides. X-axis, length of sRNA distribution; Y-axis, corresponding percentage of raw reads.
Identification of Conserved miRNAs
Conserved miRNA families are found in many plant species, wherein they are crucial for plant development. To identify the conserved miRNAs, the eight sRNA libraries were subjected to BLASTN search for conserved mature miRNAs and precursors in miRbase database. A total of 34 conserved miRNAs (belonging to 20 miRNA families and two unclassified families) that were orthologs of conserved miRNAs from other plant species were found in the eight libraries of E. ulmoides (Table S4). The length of the conserved miRNAs varied from 20 to 22 nt, with the majority (79.41%) having 21 nt. Interestingly, the abundance and the number of miRNA in each miRNA family were considerably different. Among these families, the most abundant was MIR159 (read counts > 10,000), whereas MIR397 and MIR6024 had the lowest abundance (read counts <10). The expression levels of the same miRNA family in each library varied significantly; for instance, the read numbers of MIR166 were 44,586 in MF1, whereas those in YF1 were only 7,381 (Figure 2A). Similarly, the expression levels of different members in each miRNA family were also different; in the miR164 family, miR164a was highly expressed in both leaf and fruit tissues, whereas the expression of only miR164c was low in fruits. In addition, the number of these miRNAs varied widely between families: 11 families had a single member, 8 families had 2 members, and 2 families possessed 3 members (Figure 2B). The conserved miRNA precursors with lengths ranging from 99 to 273 nt were identified. Nucleotide sequence analysis of these conserved miRNAs revealed a strong preference for adenosine (A) at the 10th position and for uridine (U) at the 1st position. We also analyzed the number of reads for conserved miRNAs in the eight libraries. As expected, miRNAs had a very broad range of expression, from millions of sequence reads to fewer than 10 sequence reads, and some miRNAs showed clear tissue- or stage-specific expression. The most abundant miRNA was miR159 (up to 53.55% of the total sequence reads). The expression levels of miR156a, miR159, miR396, and miR477 in mature tissues were higher than those in young tissues, whereas the expression of miR162a and miR171c was greater in young tissues. In addition, miR167a, miR172a/172c/172j, and miR395a were the most abundant in the leaf libraries, whereas miR164c, miR6024, and miR1446 were fruit-specific and miR169 was found only in mature leaves, although at a low expression level (<10).
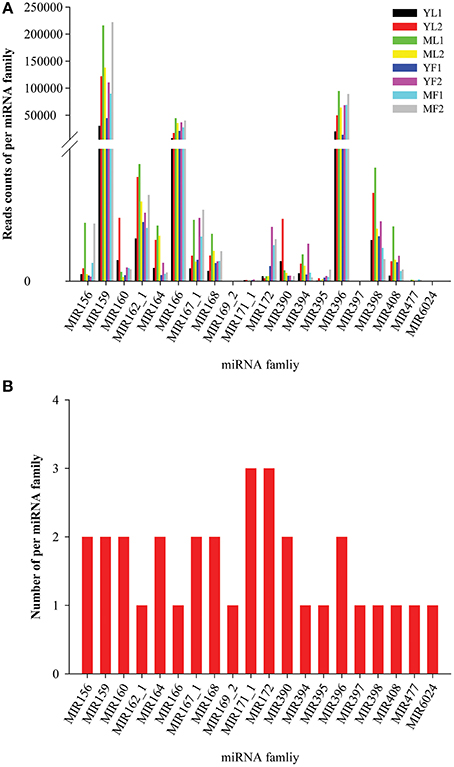
Figure 2. Abundance (A) and distribution of the member numbers (B) in each conserved miRNA family in Eucommia ulmoides.
Identification of Novel miRNAs
To search for potentially novel miRNAs in E. ulmoides, miREvo and miRDeep 2 software were used to explore the stem-loop hairpin secondary structure and the dicer cleavage site and to measure the minimum free energy of the novel miRNA sequences. A total of 115 novel hairpin miRNAs candidates between 18 and 24 nt long were identified; these miRNA candidates are likely to be new miRNAs or new members of conserved miRNA families in E. ulmoides (Table S5). The length of the precursors in novel miRNAs ranged from 51 to 296 nt, with an average of 133 nt; this was consistent with the typical length distribution of mature miRNAs. The nucleotide bias analysis showed that novel miRNAs had a similar tendency in all libraries. The majority of these novel miRNAs had a uridine residue at the first position of the 5′ terminal and they were 21 nt long, except for those in YL, which had a length of 22 nt (Figure S1). The average negative minimal folding free energy of these miRNA precursors ranged from −171 to −19.90 kcal mol−1 with a mean of −58.10 kcal mol−1. These values were much lower than the minimal folding free energy of tRNA (−27.50 kcal mol−1) and rRNA (−33 kcal mol−1) (Bonnet et al., 2004).
The abundance of miRNAs was significantly different among the identified novel miRNAs. The expression levels of most of the novel miRNAs were low, generating fewer than 500 reads—21 (<100 normalized reads) miRNAs were found at low copies, only 13 (>1000 normalized reads) miRNAs were highly expressed in both leaf and fruit tissues—thereby indicating lower expression levels than those observed for conserved miRNAs. The complementary miRNA-star sequences (miRNA*) for each novel candidate miRNA were also detected. Of 115 novel miRNAs, 70 complementary sequences were found in the eight libraries. Most miRNA* showed weak expression, and their expression levels were much lower than the expression of their corresponding miRNAs. This is consistent with the idea that miRNA* strands are degraded rapidly during the biogenesis of mature miRNAs (Rajagopalan et al., 2006; Li et al., 2013). Intriguingly, Eu-miR8* reads were extremely enriched in mature fruits, suggesting that the retention of star strands can occur in a tissue- and stage-specific manner. Eu-miR37* were found to accumulate at the levels comparable to those of their respective miRNAs, thus indicating a functional importance of this miRNA*.
Differential Expression of miRNAs in E. ulmoides
We performed a differential expression analysis between the leaf and fruit libraries to detect the miRNA expression. All tested samples were sequenced with two biological replicates and the value of the Pearson correlation coefficient r between the two biological replicates was 0.796–0.902. Those miRNAs from each of the two biological replicates with overlapping differential expression were used for subsequent analyses. miRNAs with P-values lower than 0.05 were considered significantly altered. The comparison between YF and MF, YL and ML, YL and YF, and ML and MF identified 14 conserved miRNAs and 49 novel miRNAs that were differentially expressed (Table 2). The members of differentially expressed miRNAs were different in each group and they were involved in stage- or tissue-specific expression. Among these differentially expressed miRNAs, the pairwise analysis of the miRNA abundance between libraries identified 46 (19 increased, 27 decreased), 6 (1 increased, 5 decreased), 19 (9 increased, 10 decreased), and 24 (12 increased, 12 decreased) significantly differentially expressed miRNAs in YF vs. MF, YL vs. ML, YL vs. YF, and ML vs. MF groups, respectively. There were no miRNAs common for the four groups, and 26 miRNAs were present only in the YF vs. MF group. Differentially expressed Eu-miR27, Eu-miR36, and Eu-miR126 were found only in the YL vs. YF group and miR172a, miR164a, Eu-miR4, Eu-miR33, and Eu-miR103 only in the ML vs. MF group. These results suggest that miRNAs play a specific role in the biosynthesis of rubber in E. ulmoides. In addition, the expression pattern of miRNAs also revealed a dynamically regulated expression in leaves and fruits at different developmental stages. Sixteen miRNAs were dynamically distributed in these four groups. For instance, the expression levels of Eu-miR8 were up-regulated in YF vs. YF but suppressed in YF vs. MF and ML vs. MF. These results suggest that the expression of miRNAs was significantly altered at different developmental stages, and therefore, it might play important roles in biosynthesis of Eu-rubber in specific tissues at specific growth and developmental stages. In addition, among these differentially expressed miRNAs, the expression patterns of 36 miRNAs (4 conserved and 32 novel) were inversely correlated with the accumulation rate of Eu-rubber, suggesting these miRNAs were involved in regulating the biosynthesis of Eu-rubber.
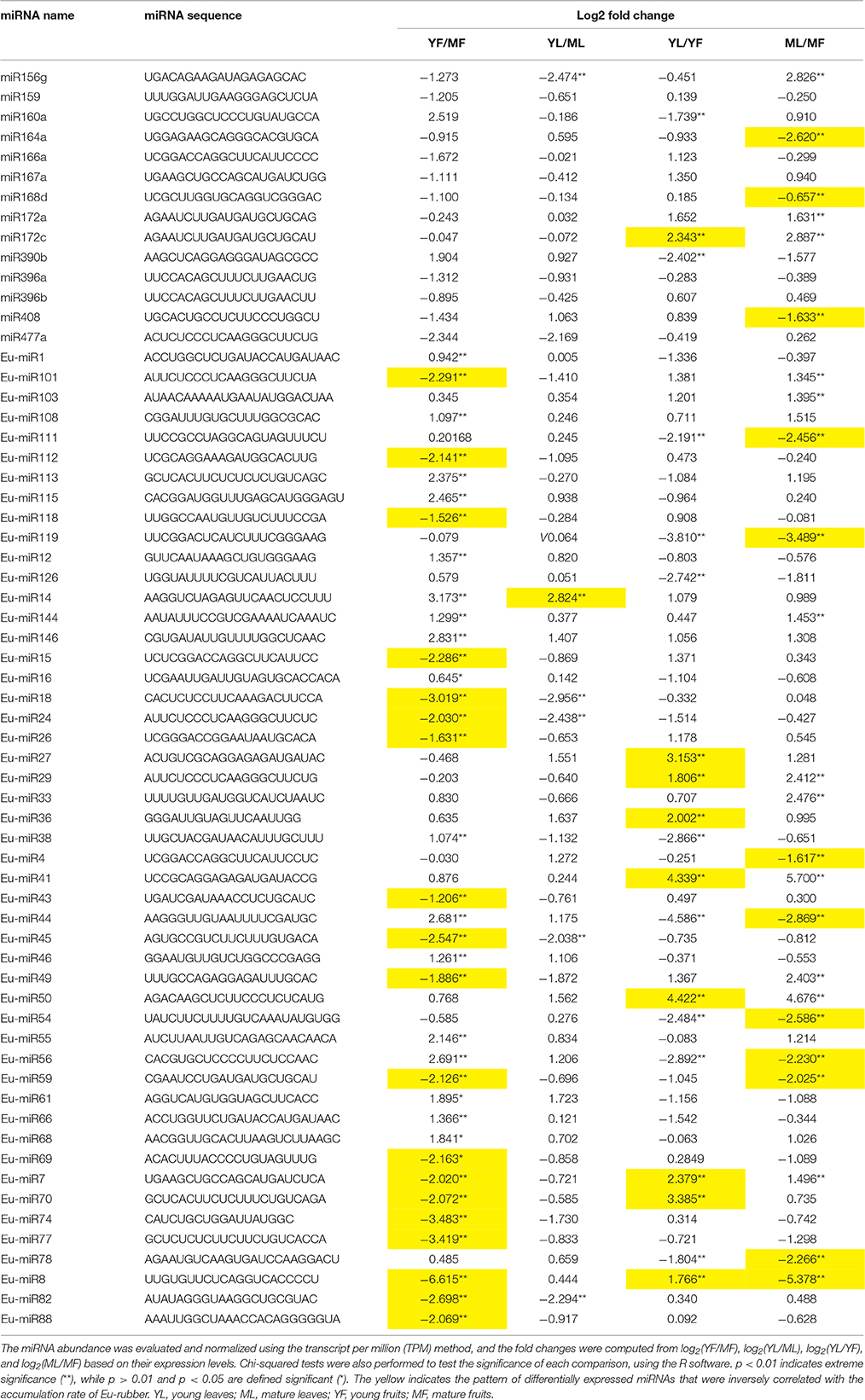
Table 2. Differential expression of miRNA genes in YF vs. MF, YL vs. ML, YL vs. YF and ML vs. MF of Eucommia ulmoides.
Prediction and Annotation of Potential Target Genes
To elucidate the functions of conserved and novel miRNAs of E. ulmoides, we predicted putative targets by using web-based psRNATarget program with default settings. A total of 202 and 306 target mRNAs were found for 33 conserved and 92 novel miRNAs, respectively (Table S6). Seventy-two (14.17%) target gene sequences exhibited no functional annotation owing to the low expression. The annotated 436 predicted target genes belonged to a large number of transcription factors and functional gene families with different biological functions. A single miRNA were predicted to regulate several target genes, which usually belong to a large gene family. In our study, the number of target genes for each differential miRNAs ranged from 1 to 16, and the highest number of target genes was predicted for miR397, whereas 22 miRNAs (3 conserved miRNAs and 19 novel miRNAs) had only one target gene. In addition, the majority of miRNAs from the same family and even some miRNAs from different families could regulate the same target genes; for example, miR160 and miR159 could regulate the expression of auxin response factors.
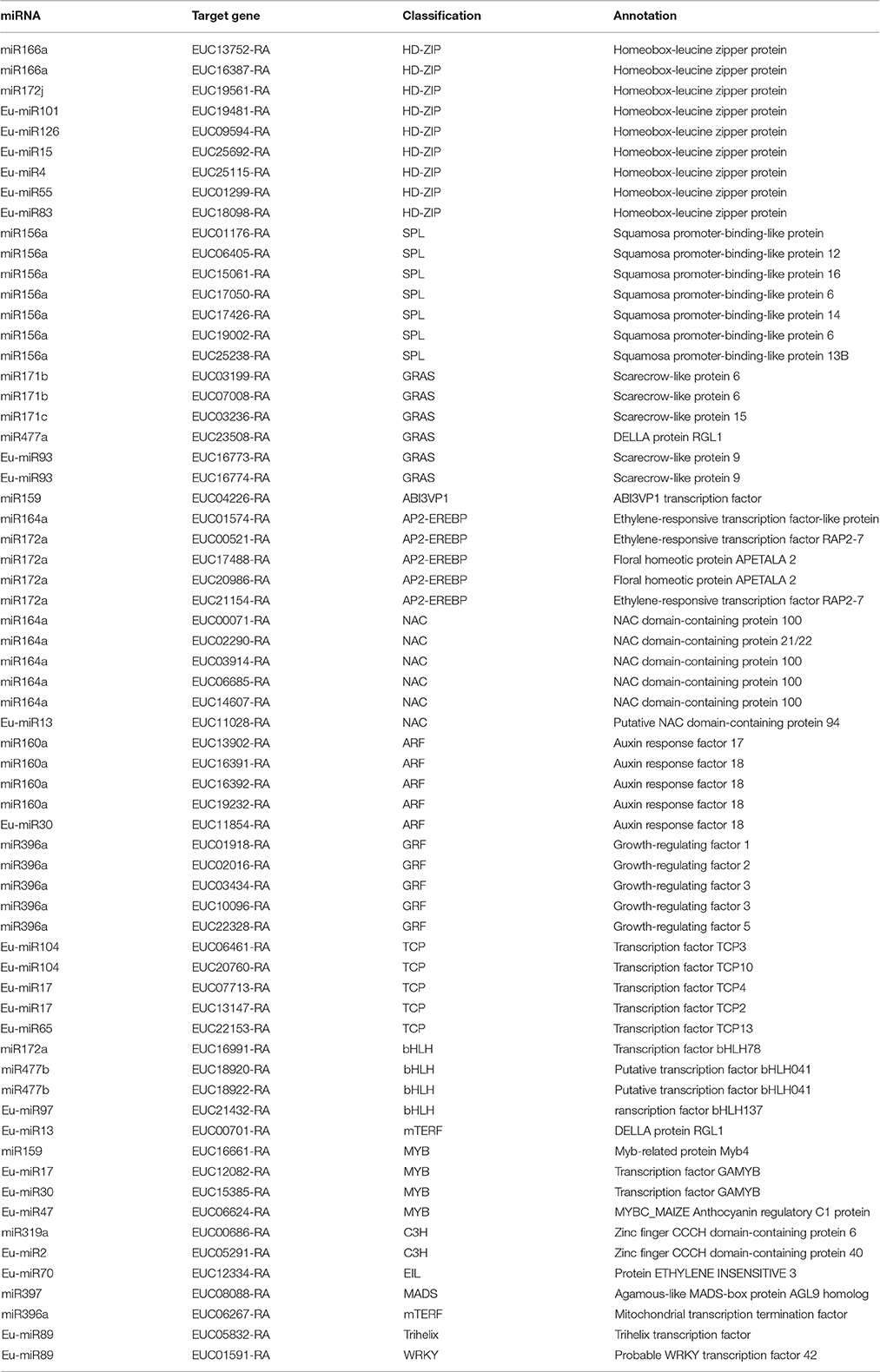
The target genes of conserved miRNAs are involved in various biological processes including transcription factor, transcriptional regulation, signal transduction, and stress responses. A total of 65 putative transcription factors that are distributed in 17 families, which are involved in the regulation of gene expression and signal transduction, were identified (Table 3). For example, homeobox-leucine zipper protein (HD-ZIP) transcription factors, which are the largest group of transcription factors in the present study, were found to serve as the main target genes of miR166a; squamosa promoter-binding protein-like (SPL) transcription factors were predicted to act as target genes of miR156; NAC and MYB transcription factors were shown to be regulated by miR164a and miR159, respectively; auxin response factors (ARFs) played an important role in adaptive responses in plant growth and development and were predicted to be modulated by miR160; and miR172a, miR172j, and miR477b were the target for basic helix-loop-helix (bHLH) transcription factors. Many functional genes involved in disease resistance were predicted to be the target genes of the differentially expressed miRNAs; for instance, G-type lectin S-receptor-like serine/threonine-protein kinase was predicted to be a target of miR172a/172c/172j; miR397 was predicted to be a target of laccase protein; and the predicted target genes of miR167a/164c/167d and miR390a were involved in leucine-rich repeat receptor-like serine/threonine protein kinase. Although the novel miRNAs were sequenced at relatively lower levels than the conserved miRNAs, 92 out of 115 novel miRNAs were found to have candidate targets. Similarly, the target genes of the novel miRNAs also consisted of transcription factors and functional genes. For example, teosinte branched1/cycloidea/proliferating cell factor1 transcription factors were found to serve as the target genes of Eu-miR17, whereas Eu-miR118 and Eu-miR121 are the targets of disease resistance proteins.
To gain insights into possible roles of differentially expressed miRNAs, putative target genes of these differentially expressed miRNAs were identified based on GO analysis and KEGG pathway analysis. GO annotation enrichment showed that the target genes could be summarized into three main categories in each group: biological processes, cellular components, and molecular functions (Figure 3). The majority of target genes involved in major biological processes were enriched in cellular process and metabolic process. In the cellular component group, the target genes were associated with cell, cell part, organelle, and membrane. Based on the molecular function category, the target genes were associated with binding and catalytic activity. The differential expression of target genes involved in various biological processes indicated that these target genes are responsible for biosynthesis of Eu-rubber from different tissues and at different developmental stages. Overall, a total of 24, 7, 23, and 20 different pathways enriched with target genes of differentially expressed miRNAs were found in the YF vs. MF, YL vs. ML, YL vs. YF, and ML vs. MF groups, respectively. The term metabolic pathways was the most dominant, followed by the biosynthesis of secondary metabolites (Table S7).
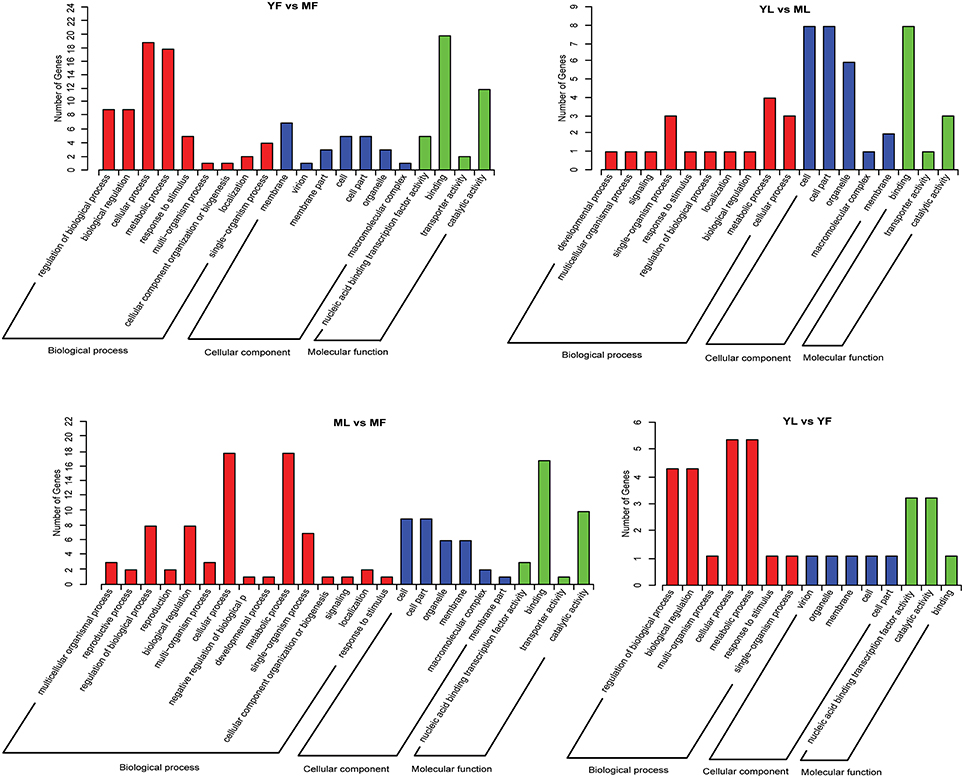
Figure 3. Gene ontology (GO) enrichment analysis of target genes for the differentially expressed miRNAs in Eucommia ulmoides.
To further evaluate the relationship between miRNAs and their target genes and confirm candidate genes involved in rubber accumulation, we performed qPCR to analyze the miRNAs and their targets, which may participate in rubber accumulation of E. ulmoides according to our sequencing data. The qPCR results for all tested miRNAs were consistent with the sequencing data and suggested a negative correlation between miRNAs expression and their putative target genes (Figure 4). Five miRNAs (Eu-miR14, Eu-miR91, miR162a, miR166a, and miR172c) were significantly upregulated in young leaves, whereas their target genes were downregulated. Moreover, miR166a and miR396a were negatively correlated with their putative targets in the YF vs. MF group. Eu-rubber is a secondary metabolite whose biosynthesis might be controlled by these target genes. The putative target gene of Eu-miR91 was predicted to be 1-deoxy-D-xylulose-5-phosphate synthase (DXS), the first rate-limiting enzyme in the MEP pathway, which was associated with the biosynthesis of terpenoids. The predicted target genes of miR166a, miR172c, and miR396a encoded transcription factors HD-ZIP, AP2-EREBP, and GRF. Moreover, Eu-miR14 and miR162a were predicted to target the genes of the protein bonzai 3 (BON 3) and the endoribonuclease dicer homolog 1 (DCL 1), respectively.
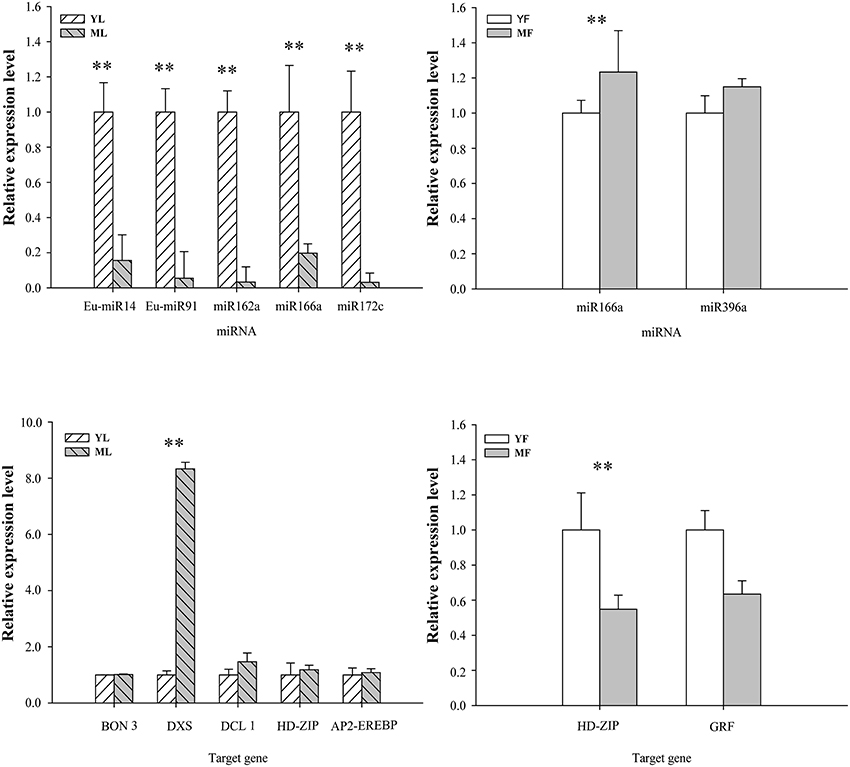
Figure 4. qPCR analysis expression levels of miRNAs and their targets in Eucommia ulmoides. All data were subjected to analysis of variance (ANOVA) and the data represent the mean ± S.D. The results were considered statistically significant at (*p <0.05) or (**p <0.01).
Discussion
Eu-rubber is an important industrial gum found in the leaves, fruits, roots, and bark of E. ulmoides. Previous studies mainly centered on pharmacological applications, genetic diversity, and transcriptional levels of this species (Wang et al., 2006; Chen et al., 2012; Suzuki et al., 2012; Huang et al., 2013; Feng et al., 2015). In the present study, we identified miRNAs and associated target genes involved in Eu-rubber biosynthesis and thus provide valuable information about molecular regulatory mechanisms of Eu-rubber biosynthesis. Thus, eight sRNA libraries from young and mature leaves and fruits of E. ulmoides were constructed and sequenced. These libraries generated more than 95 million clean reads; of those, 60% were perfectly matched to the E. ulmoides genome, indicating the overall good quality of the sequence data. The 24 nt class exhibited the highest abundance, which was consistent with the distribution patterns of sRNAs reported in Nicotiana benthamiana (Baksa et al., 2015), Panax notoginseng (Wei et al., 2015), Medicago sativa (Fan et al., 2014), Citrus trifoliata (Song et al., 2010), Taxus chinensis (Qiu et al., 2009), and Asparagus officinalis (Chen et al., 2016). A total of 34 conserved miRNAs and 115 novel miRNAs were identified. Most of the identified miRNAs are highly conserved in most land plants, which suggests that they are deeply conserved in the plant kingdom and have a possible important role in plant growth and development (Cuperus et al., 2011).
Analyzing the temporal and spatial expression patterns of miRNAs would provide useful information about their molecular functions. The miRNAs previously identified in opium poppy, Ricinus communis, Jatropha curcas, Medicago sativa, and Panax notoginseng exhibited tissue- and stage-specific expression patterns, and therefore, play an important role in development and stress adaptation (Unver et al., 2010; Sun, 2012; Wang C. M. et al., 2012; Xu et al., 2013; Fan et al., 2014; Wei et al., 2015). The analysis of the conserved miRNAs showed that MIR159, MIR166, and MIR396 were highly expressed in both leaf and fruit tissues, which was consistent with the expression patterns in tobacco (Baksa et al., 2015). In this study, MIR159 (miR159/319) was the most abundant miRNA, accounting for more than 50% of the total sequence reads, thereby suggesting that MIR159 may have specialized functions in Eu-rubber biosynthesis. MIR159 targets four MYB proteins, which contain a conserved MYB DNA binding domain and are one of the larger group of plant protein family that play a regulatory role in developmental processes and defense responses (Wu and Poethig, 2006; Fornara and Coupland, 2009). In Arabidopsis, MYB transcription factors are involved in plant leaf development (Palatnik et al., 2003; Millar and Gubler, 2005), whereas in Hevea brasiliensis, a MYB transcription factor induces programmed cell death in rubber tree bark, and its expression is significantly decreased in the latex of trees affected by tapping panel dryness (Gébelin et al., 2013). MIR396 was predicted to target 17 growth-regulating factor genes encoding respective putative transcription factors, which play a regulatory role in plant growth and development as well as in defense responses (Wu and Poethig, 2006). In E. ulmoides, miR396 is expressed in high levels in mature tissues; this pattern of miR396 accumulation is in accord with that observed in Arabidopsis and H. brasiliensis, where it is exclusively expressed in mature leaves (Wang et al., 2011; Lertpanyasampatha et al., 2012). In addition, different conserved miRNAs, even those in the same family, exhibited different expression levels in various tested tissues, suggesting their specific potential roles in growth and development of E. ulmoides. Most of the newly identified miRNAs showed lower abundance levels than the conserved miRNAs, which suggested that these novel species-specific miRNAs are either young miRNAs that arose recently through evolution or they are more volatile than the other miRNAs (Jones-Rhoades et al., 2006; Rajagopalan et al., 2006; Song et al., 2010; Cuperus et al., 2011; Mao et al., 2012; Chen et al., 2016).
To better understand the functions of miRNAs, it is necessary to predict and annotate their target genes. The prediction in this study showed that these target genes have a wide range of functions, with the majority of the targets being transcription factors related to plant growth and development, including HD-ZIP, SPL, NAC, MYB, ARFs, bHLH, and WRKY transcription factors. Previous research indicated that miRNA families in different plants are highly conserved, and perform analogous regulatory functions (Zhao et al., 2010). miR166 was predicted to regulate HD-ZIP transcription factors, which are unique to plant kingdom; they are involved in organ morphogenesis and regulation of meristem growth in Arabidopsis thaliana (Green et al., 2005; Wang H. et al., 2013). miR164 regulates NAC-domain gene necessary for normal embryonic, vegetative, and floral development in Arabidopsis (Mallory et al., 2004). bHLH3 transcription factor, a target gene of miR828, regulates the accumulation of anthocyanins in apple after debagging (Qu et al., 2016). miR156 is highly conserved in plants and predicted to target the SPL transcription factor, which plays an important part in the spatiotemporal regulation of sesquiterpene biosynthesis in A. thaliana and Pogostemon cablin (Yu et al., 2015). miR171 targets mRNAs coding for the scarecrow-like proteins, which is a transcription factor that has been implicated in gibberellins synthesis in Arabidopsis (Llave et al., 2002b). WRKY transcription factors have been reported to play roles in regulating the biosynthesis of terpenoids in various plants including Panax quinquefolius and P. notoginseng (Eulgem et al., 2000; Ma et al., 2009; Wang J. et al., 2012; Yang et al., 2012; Wei et al., 2015). In our study, miR166a, miR156, miR164a, miR159, miR160, miR172, and Eu-miRn89 were predicted to target HD-ZIP, SPL, NAC, MYB, ARFs, bHLH, and WRKY transcription factors, respectively. The conservation of putative target genes suggests that these transcription factors also share the same function in the growth and development of E. ulmoides, especially in the synthesis of secondary metabolites. Furthermore, several miRNA targets encode proteins involved in responses to environmental stresses, such as heat shock protein (HSP). HSP, which is induced by heat shock stress conditions and is critical in cellular homeostasis under adverse environmental conditions, is the target of miR397, Eu-miR1, and Eu-miR166.
The qPCR analysis was used to identify candidate miRNAs and their predicted target genes, which were associated with regulation genes involved in the accumulation of rubber in E. ulmoides. The expression patterns of five (Eu-miR14, Eu-miR91, miR162a, miR166a, and miR172c) and three (miR166a, miR396a, and miR396b) miRNAs, which were consistent with the accumulation of Eu-rubber in YL vs. ML and YF vs. MF, respectively, were selected. miRNAs usually negatively regulate the expression of mRNA, and the selected miRNAs expression patterns exhibited inverse correlation with their corresponding targets, suggesting that miRNAs regulate the accumulation of Eu-rubber by post-transcriptionally regulating target genes to enhance rubber content in E. ulmoides. In plants, terpenoid is synthesized from isopentenyl diphosphate and its allylic isomer dimethylallyl diphosphate via two different pathways, the MVA pathway in the cytosol and the MEP pathway in cellular plastids, where the DXS gene catalyzes the first rate-limiting enzyme for isopentenyl diphosphate synthesis in the MEP pathway (Pulido et al., 2012). And previous reports have suggested that the MEP pathway is also involved in rubber biosynthesis in E. ulmoides and Hevea brasiliensis (Ko et al., 2003; Chow et al., 2007; Bamba et al., 2010). Several miRNAs were computationally predicted to directly target known genes in the terpenoid backbone biosynthesis pathway, including DXS (Wei et al., 2015). Therefore, in the present work, DXS was predicted to be a target of Eu-miR91, the expression levels of Eu-miR91 in young leaves was higher than that in mature leaves, and the expression of the target gene DXS was a perfect inverse to that of Eu-miR91, which suggest that reduced expression of Eu-miR91 could promote the accumulation of rubber by DXS gene. The negative correlation between the expression of miR166a, miR172c, and miR396a and their target genes encoding transcription factors were also revealed. These miRNAs may regulate Eu-rubber accumulation via suppression of their target gene expression. Previous studies have showed that Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper class I (HDZip I) gene, can regulatethe cell growth during leaf development (Hur et al., 2015). Our results indicated that the HD-ZIP expression enhanced rubber content by promoting leaf and fruit growth. The negative correlation between the expression of miR166a and its target genes encoding HD-ZIP transcription factor was revealed, therefore implying that miR166a participate in the process of rubber biosynthesis in E. ulmoides via regulating the HD-ZIP gene. AP2-EREBP is the majority of stress responsive genes involved in ethylene responses, and has been characterized that positively regulates rubber biosynthesis in Hevea brasiliensis (Li et al., 2016). In our results, the expression of the target gene AP2-EREBP was a perfect inverse to that of miR172c—the expression of miR172c was down regulated in mature leaves, whereas the expression of AP2-EREBP was up regulated, suggesting that miR172c promotes the accumulation of rubber via AP2-EREBP in E. ulmoides. miR396 is conserved among dicot and monocot plants, and it targets the GRF genes encoding putative transcription factors involved in growth and development. A previous study showed that miR396 is abundant in mature tissues (Wang et al., 2011; Lertpanyasampatha et al., 2012), which was corroborated by the lower expression of miR396a in young fruits compared to that in in mature fruits observed in the present study. Moreover, the expression of miR396a was negatively correlated to GRF transcription factor, implying that miR396 might be negatively correlated with the accumulation of Eu-rubber. Our study is the first to associate these three transcription factors, HD-ZIP, AP2-EREBP, and GRF, with the accumulation of rubber, and thus, these transcription factors are candidate genes that regulate Eu-rubber accumulation in E. ulmoides.
Eu-rubber accumulation is a complex process involving many precursors and a series of regulatory genes. Thus, it is simplistic to assume a simple correlation between miRNA expression and their targets when identifying the mechanism of Eu-rubber accumulation, and further characterization of these miRNAs is required.
Conclusions
In conclusion, the present study is the first genome-wide investigation of miRNAs and their targets in E. ulmoides. Eight sRNA libraries were generated and sequenced from E. ulmoides leaves and fruits at both young and mature stages using high-throughput sequencing technology. We identified 34 conserved miRNAs and 115 novel miRNAs and characterized their expression patterns, target genes, and function. Furthermore, six miRNAs and their targets that were involved in the accumulation of Eu-rubber were revealed. Although the foundation of the complex miRNA-mediated regulatory networks remains to be resolved, this miRNA dataset will help to elucidate the gene regulatory networks in E. ulmoides and other species. Therefore, this study provides a valuable source for studying complex gene regulatory functions of miRNAs, especially in the rubber accumulation of E. ulmoides.
Author Contributions
LW performed the experiments, data analysis and drafted the manuscript. HD and TW helped to conceive the study and participated in the design and coordination. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AP2-EREBP, ethylene-responsive transcription factor; ARFs, auxin response factors; bHLH, basic helix-loop-helix; BON 3, protein bonzai 3; DCL 1, endoribonuclease dicer homolog 1; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; Eu-rubber, Eucommia rubber; GO, gene ontology; GRF, Growth-regulating factor; HD-ZIP, homeobox-leucine zipper protein; HSP, heat shock protein; KEGG, Kyoto Encyclopedia of Genes and Genomes; MEP, methylerythritol phosphate; miRNA, microRNA; miRNA*, miRNA-star sequences; MVA, mevalonate; SPL, squamosa promoter-binding protein-like; sRNA, small RNA; TPI, trans-1, 4-polyisoprene.
Acknowledgments
This work was supported by Twelve-Five science and technology support program (No. 2012BAD21B0502) and the Forestry Industry Research Special Funds for Public Welfare Projects (201004029).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01632/full#supplementary-material
Table S1. Primers used in qPCR analysis of miRNAs and target genes.
Table S2. Quality evaluation of sequencing data.
Table S3. Number of reads for each classified small RNA identified in Eucommia ulmoides.
Table S4. Conserved miRNAs identified from Eucommia ulmoides. Expression profiles of conserved miRNAs were estimated by transcript per million (TPM). Heat map colors represent absolute normalized levels of miRNA expression ranging from less than 10 RPM to more than 1000 RPM, as indicated in the color key.
Table S5. Predicted novel miRNAs and their pre-miRNA sequences in Eucommia ulmoides. Expression profiles of conserved miRNAs were estimated by transcript per million (TPM). Heat map colors represent absolute normalized levels of miRNA expression ranging from less than 10 RPM to more than 1000 RPM, as indicated in the color key.
Table S6. Target prediction of miRNAs in Eucommia ulmoides.
Table S7. KEGG pathway analysis of genes for differentially expressed miRNAs in Eucommia ulmoides.
Figure S1. First nucleotide bias of novel miRNA in Eucommia ulmoides young leaves (A), mature leaves (B), young fruits (C), and mature fruits (D).
Availability of Supporting Data
The small RNA sequencing raw data were deposited in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under the accession number SRP074871.
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Baksa, I., Nagy, T., Barta, E., Havelda, Z., Várallyay, É., Silhavy, D., et al. (2015). Identification of Nicotiana benthamiana microRNAs and their targets using high throughput sequencing and degradome analysis. BMC Genomics 16:1025. doi: 10.1186/s12864-015-2209-6
Bamba, T., Murayoshi, M., Gyoksen, K., Nakazawa, Y., Okumoto, H., Katto, H., et al. (2010). Contribution of mevalonate and methylerythritol phosphate pathways to polyisoprenoid biosynthesis in the rubber-producing plant Eucommia ulmoides Olive. Z. Naturforsch. 65c, 363–372.
Bao, W., Kojima, K. K., and Kohany, O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 6:11. doi: 10.1186/s13100-015-0041-9
Bartel, D. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bonnet, E., Wuyts, J., Rouzé, P., and Van de Peer, Y. (2004). Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20, 2911–2917. doi: 10.1093/bioinformatics/bth374
Call, V., and Dilcher, D. (1997). The fossil record of Eucommia (Eucommiaceae) in North America. Am. J. Bot. 84, 798–814. doi: 10.2307/2445816
Chen, D., Yuan, C., Zhang, J., Zhang, Z., Bai, L., Meng, Y., et al. (2011). PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res. 40, D1187–D1193. doi: 10.1093/nar/gkr823
Chen, J., Zheng, Y., Qin, L., Wang, Y., Chen, L., He, Y., et al. (2016). Identification of miRNAs and their targets through high-throughput sequencing and degradome analysis in male and female Asparagus officinalis. BMC Plant Biol. 16:80 doi: 10.1186/s12870-016-0770-z
Chen, R., Harada, Y., Bamba, T., Nakazawa, O., and Gyokusen, K. (2012). Overexpression of an isopentenyl diphosphate isomerase gene to enhance trans-polyisoprene production in Eucommia ulmoides Oliver. BMC Biotechnol. 12:78. doi: 10.1186/1472-6750-12-78
Chen, R., Namimatsu, S., Nakadozono, Y., Bamba, T., Nakazawa, Y., and Gyokusen, K. (2008). Efficient regeneration of Eucommia ulmoides Oliver plant from hypocotyls explant. Biol. Plant. 52, 713–717. doi: 10.1007/s10535-008-0137-x
Chow, K. S., Wan, K. L., Isa, M. N. M., Bahari, A., Tan, S. H., Harikrishna, K., et al. (2007). Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J. Exp. Bot. 58, 2429–2440. doi: 10.1093/jxb/erm093
Cuperus, J. T., Fahlgren, N., and Carrington, J. C. (2011). Evolution and functional diversification of MIRA genes. Plant Cell 23, 431–442. doi: 10.1105/tpc.110.082784
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Davuluri, G. R., van Tuinen, A., Fraser, P. D., Manfredonia, A., Newman, R., Burgess, D., et al. (2005). Fruit-specific RNAi-mediated suppression of DET1 enhance scarotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 23, 890–895. doi: 10.1038/nbt1108
Du, H. Y. (2003). Guutta-Percha-Content Character, its Variance and Selection of Superior Clone Associated with Eucommia Ulmoides Olov. dissertation, Central-South Forestry University, Changsha.
Du, H. Y., Hu, W. Z., and Yu, R. (2015). The Report on Development of China's Eucommia Rubber Resources and Industry (2014-2015). Beijing: Social Sciences Academic Press.
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fan, R., Li, Y., Li, C., and Zhang, Y. (2015), Differential microRNA analysis of glandular trichomes young leaves in Xanthium strumarium L. reveals their putative roles in regulating terpenoid biosynthesis. PLoS ONE 10:e0139002. doi: 10.1371/journal.pone.0139002
Fan, W., Zhang, S., Du, H., Sun, X., Shi, Y., and Wang, C. (2014). Genome-wide identification of different dormant Medicago sativa L. microRNAs in response to fall dormancy. PLoS ONE 9:e114612. doi: 10.1371/journal.pone.0114612
Feng, H., Zhou, H., and Oouyang, D. (2015). Chemical constituents and pharmacology of Eucommia ulmoides Oliv. Chin. J. Clin. Pharmacol. Ther. 20, 713–720.
Fornara, F., and Coupland, G. (2009). Plant phase transitions make a SPLash. Cell 138, 625–627. doi: 10.1016/j.cell.2009.08.011
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2011). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Gébelin, V., Leclercq, J., Kuswanhadi, Argout, X., Chaidamsari, T., Hu, S., et al. (2013). The small RNA profile in latex from Hevea brasiliensistrees is affected by tapping panel dryness. Tree Physiol. 33, 1084–1098. doi: 10.1093/treephys/tpt076
Gou, J.-Y., Felippes, F. F., Liu, C.-J., Weigel, D., and Wang, J.-W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Green, K. A., Prigge, M. J., Katzman, R. B., and Clark, S. E. (2005). CORONA, a member of the Class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17, 691–704. doi: 10.1105/tpc.104.026179
Hao, D., Yang, L., Xiao, P., and Liu, M. (2012). Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis. Physiol. Plant 146, 388–403. doi: 10.1111/j.1399-3054.2012.01668.x
Huang, H. Y., Du, H. Y., Wuyun, T. N., and Liu, P. F. (2013). Development of SSR molecular markers based on transcriptome sequencing of Eucommia ulmoides. Sci. Silvae Sinica 49, 176–181. doi: 10.11707/j.1001-7488.20130523
Hur, Y. S., Um, J. H., Kim, S. H., Kim, K., Park, H. J., Lim, J. S., et al. (2015). Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomainleucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 205, 316–328. doi: 10.1111/nph.12998
Jiang, J. H., Kai, G. Y., Cao, X. Y., Chen, F. M., He, D. N., and Liu, Q. (2006). Molecular cloning of a HMG-CoA reductase gene from Eucommia ulmoides Oliver. Biosci. Rep. 26, 171–181. doi: 10.1007/s10540-006-9010-3
Jones-Rhoades, M. W., and Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. doi: 10.1016/j.molcel.2004.05.027
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53, doi: 10.1146/annurev.arplant.57.032905.105218
Khraiwesh, B., Arif, M. A., Seumel, G. I., Ossowski, S., Weigel, D., Reski, R., et al. (2010). Transcriptional control of gene expression by microRNAs. Cell 140, 111–122. doi: 10.1016/j.cell.2009.12.023
Ko, J. H., Chow, K. S., and Han, K. H. (2003). Transcriptome analysis reveals novel features of the molecular events occurring in the laticifers of Hevea brasiliensis (para rubber tree). Plant Mol. Biol. 53, 479–492. doi: 10.1023/B:PLAN.0000019119.66643.5d
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lertpanyasampatha, M., Gao, L., Kongsawadworakul, P., Viboonjun, U., Chrestin, H., Liu, R., et al. (2012). Genome-wide analysis of microRNAs in rubber tree (Hevea brasiliensis L.) using high-throughput sequencing. Planta 236, 437–445. doi: 10.1007/s00425-012-1622-1
Li, D., Wang, L., Liu, X., Cui, D., Chen, T., Zhang, H., et al. (2013). Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS ONE 8:e55107. doi: 10.1371/journal.pone.0055107
Li, H. L., Guo, D., Zhu, J. H., Wang, Y., Chen, X. T., and Peng, S. Q. (2016). Comparative transcriptome analysis of latex reveals molecular mechanisms underlying increased rubber yield in Hevea brasiliensis self-rooting juvenile clones. Front. Plant Sci. 7:1204. doi: 10.3389/fpls.2016.01204
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Llave, C., Kasschau, K. D., Rector, M. A., and Carrington, J. C. (2002a). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. doi: 10.1105/tpc.003210
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002b). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1101/002832
Ma, D., Pu, G., Lei, C., Ma, L., Wang, H., Guo, Y., et al. (2009). Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. doi: 10.1093/pcp/pcp149
Mallory, A. C., and Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38, S31–S36. doi: 10.1038/ng1791
Mallory, A. C., Dugas, D. V., Bartel, D. P., and Bartel, B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. doi: 10.1016/j.cub.2004.06.022
Mao, W., Li, Z., Xia, X., Li, Y., and Yu, J. (2012). A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS ONE 7:e33040. doi: 10.1371/journal.pone.0033040
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Millar, A. A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Moxon, S., Schwach, F., MacLean, D., Dalmay, T., Studholme, D. J., and Moulton, V. (2008). A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24, 2252–2253. doi: 10.1093/bioinformatics/btn428
Ng, D. W., Zhang, C., Miller, M., Palmer, G., Whiteley, M., Tholl, D., et al. (2011). cis-and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23, 1729–1240. doi: 10.1105/tpc.111.083915
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Pramoolkit, P., Lertpanyasampatha, M., Viboonjun, U., Kongsawadworakul, P., Chrestin, H., and Narangajavana, J. (2014). Involvement of ethylene-responsive microRNAs and their targets in increased latex yield in the rubber tree in response to ethylene treatment. Plant Physiol. Biochem. 84, 203–212. doi: 10.1016/j.plaphy.2014.09.016
Pulido, P., Perello, C., and Rodriguez-Concepcion, M. (2012). New insights into plant isoprenoid metabolism. Mol. Plant 5, 964–967. doi: 10.1093/mp/sss088
Qiu, D., Pan, X., Wilson, I. W., Li, F., Liu, M., Teng, W., et al. (2009). High through put sequencing technology reveals that the taxoid elicitor methyl jasmonate regulates microRNA expression in Chinese yew (Taxus chinensis). Gene 436, 37–44. doi: 10.1016/j.gene.2009.01.006
Qu, D., Yan, F., Meng, R., Jiang, X., Yang, H., Gao, Z., et al. (2016). Identification of microRNAs and their targets associated with fruit-bagging and subsequent sunlight re-exposure in the “Granny Smith” apple exocarp using high-throughput sequencing. Front. Plant Sci. 7:27. doi: 10.3389/fpls.2016.00027
Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Smit, A. F. A., Hubley, R., and Green, P. (2013-2015). RepeatMasker Open-4.0. Available online at: http://www.repeatmasker.org
Song, C., Wang, C., Zhang, C., Korir, N. K., Yum, H., Mam, Z., et al. (2010). Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11:431. doi: 10.1186/1471-2164-11-431
Sun, G. L. (2012). MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 80, 17–36. doi: 10.1007/s11103-011-9817-6
Sunkar, R., Kapoor, A., and Zhu, J. K. (2006). Posttranscriptional induction of two Cu/ Zn superoxide dismutase genes in Arabidopsisis mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. doi: 10.1105/tpc.106.041673
Suzuki, N., Uefuji, H., Nishikawa, T., Mukai, Y., Yamashita, A., Hattori, M., et al. (2012). Construction and analysis of EST libraries of the trans-polyisoprene producing plant, Eucommia ulmoides Oliver. Planta 236, 1405–1407. doi: 10.1007/s00425-012-1679-x
Szittya, G., and Burgyan, J. (2013). RNA interference-mediated intrinsic antiviral immunity in plants. Curr. Top. Microbiol. Immunol. 371, 153–181. doi: 10.1007/978-3-642-37765-5_6
Takeno, S., Bamba, T., Nakazawa, Y., Fukusaki, E., Okazawa, A., and Kobayashi, A. (2008). Quantification of trans-1,4-polyisoprene in Eucommia ulmoides by Fourier transform infrared spectroscopy and pyrolysis-gas chromatography/mass spectrometry. J. Biosci. Bioeng. 105, 355–359. doi: 10.1263/jbb.105.355
Unver, T., Parmaksız, I., and Dundar, E. (2010). Identification of conserved micro-RNAs and their target transcripts in opium poppy (Papaver somniferum L.). Plant Cell Rep. 29, 757–769. doi: 10.1007/s00299-010-0862-4
Varkonyi-Gasic, E., and Hellens, R. (2011). “Quantitative stem-loop RT-PCR for detection of microRNAs,” in RNAi and Plant Gene Function Analysis, eds H. Kodama and A. Komamine (New York, NY: Humana Press), 145–157.
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell. 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wang, A. Q., Huang, L. Q., Shao, A. J., Cui, G. H., Chen, M., and Tong, C. H. (2006). Genetic diversity of Eucommia ulmoides by RAPD analysis. J. Chinese Materia Medica 31, 1583–1586.
Wang, C. M., Liu, P., Sun, F., Li, L., Liu, P., Ye, J., et al. (2012). Isolation and Identification of miRNAs in Jatropha curcas. Int. J. Biol. Sci. 8, 418–429. doi: 10.7150/ijbs.3676
Wang, H., Li, G. B., Zhang, D. Y., Lin, J., Shen, B. L., Han, J. L., et al. (2013). Biological functions of HD-Zip transcription factors. Hereditas 35, 1179–1188. doi: 10.3724/SP.J.1005.2013.01179
Wang, J., Yang, X., Xu, H., Chi, X., Zhang, M., and Hou, X. (2012). Identification and characterization of microRNAs and their target genes in Brassica oleracea. Gene 505, 300–308. doi: 10.1016/j.gene.2012.06.002
Wang, L.-F. (2014). Physiological and molecular responses to variation of light intensity in rubber tree (Hevea brasiliensis Muell. Arg.). PLoS ONE 9:e89514. doi: 10.1371/journal.pone.0089514
Wang, L., Gu, X., Xu, D., Wang, W., Wang, H., Zeng, M., et al. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62, 761–773. doi: 10.1093/jxb/erq307
Wang, Y., Guo, D., Li, H.-L., and Peng, S.-Q. (2013). Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP. Plant Physiol. Biochem. 71, 283–289. doi: 10.1016/j.plaphy.2013.07.020
Wei, R. M., Qiu, D. Y., Wilson, I. W., Zhao, H., Liu, S. F., Miao, J. H., et al. (2015). Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing. BMC Genomics 16:835. doi: 10.1186/s12864-015-2010-6
Weiss, F. E. (1891). VIII. The Caoutchouc-containing cells of Eucommia ulmoides, Oliver. Trans. Linnean Soc. London 3, 243–254.
Wen, M., Shen, Y., Shi, S., and Tang, T. (2010). miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics 13:140. doi: 10.1186/1471-2105-13-140
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. doi: 10.1242/dev.02521
Wu, L., Zhou, H., Zhang, Q., Zhang, J., Ni, F., Liu, C., et al. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell. 38, 465–475. doi: 10.1016/j.molcel.2010.03.008
Xu, W., Cui, Q., Li, F., and Liu, A. (2013). Transcriptome-wide identification and characterization of microRNAs from castor bean (Ricinus communis L.). PLoS ONE. 8:e69995. doi: 10.1371/journal.pone.0069995
Yang, C. Q., Fang, X., Wu, X. M., Mao, Y. B., Wang, L. J., and Chen, X. Y. (2012). Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 54, 703–712. doi: 10.1111/j.1744-7909.2012.01161.x
Yin, T., Cao, X., Miao, Q., Li, C., Chen, X., Zhou, M., et al. (2011). Molecular cloning and functional analysis of an organ-specific expressing gene coding for farnesyl diphosphate synthase from Michelia chapensis Dandy. Acta Physiol. Plant. 33, 137–144. doi: 10.1007/s11738-010-0529-3
Yoshihisa, N., Takeshi, B., and Tuyoshi, T. (2009). Production of Eucommia-rubber from Eucommia ulmoides Oliv. (Hardy Rubber Tree). Plant Biotechnol. 26, 71–79. doi: 10.5511/plantbiotechnology.26.71
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). goseq: Gene Ontology Testing for RNA-seq Datasets. R Bioconductor.
Yu, Z. X., Wang, L. J., Zhao, B., Shan, C. M., Zhang, Y. H., Chen, D. F., et al. (2015). Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol. Plant 8, 98–110. doi: 10.1016/j.molp.2014.11.002
Zeng, S., Liu, Y., Pan, L., Hayward, A., and Wang, Y. (2015). Identification and characterization of miRNAs in ripening fruit of Lycium barbarum L. using high-throughput sequencing. Front. Plant Sci. 6:778. doi: 10.3389/fpls.2015.00778
Zhao, C. Z., Xia, H., Frazier, T. P., Yao, Y. Y., Bi, Y. P., Li, A. Q., et al. (2010). Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol. 10:3. doi: 10.1186/1471-2229-10-3
Keywords: Eucommia ulmoides, trans-1, 4-polyisoprene, miRNAs, high-throughput sequencing, target genes
Citation: Wang L, Du H and Wuyun T-n (2016) Genome-Wide Identification of MicroRNAs and Their Targets in the Leaves and Fruits of Eucommia ulmoides Using High-Throughput Sequencing. Front. Plant Sci. 7:1632. doi: 10.3389/fpls.2016.01632
Received: 13 July 2016; Accepted: 17 October 2016;
Published: 08 November 2016.
Edited by:
Sagadevan G. Mundree, Queensland University of Technology, AustraliaReviewed by:
Taras P. Pasternak, Albert Ludwigs University of Freiburg, GermanyIsaac Njaci, Biosciences East and Central Africa, Kenya
Copyright © 2016 Wang, Du and Wuyun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ta-na Wuyun, tanatanan@163.com
 Lin Wang
Lin Wang Hongyan Du
Hongyan Du Ta-na Wuyun
Ta-na Wuyun