- 1Institute of Crop Science, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 2Department of Biotechnology, College of Life Science, Zhejiang Sci-Tech University, Hangzhou, China
Rice is more sensitive to salinity, particularly at its early vegetative and later productive stages. Wild plants growing in harsh environments such as wild barley from Qinghai-Tibet Plateau adapt to the adverse environment with allelic variations at the loci responsible for stressful environment, which could be used for rice genetic improvement. In this study, we overexpressed HsCBL8 encoding a calcium-sensor calcineurin B-like (CBL) protein in rice. The gene was isolated from XZ166, a wild-barley (Hordeum spontanum) line originated from Qinghai-Tibet Plateau. We found that XZ166 responded to high NaCl concentration (200 mM) with more HsCBL8 transcripts than CM72, a cultivated barley line known for salinity tolerance. XZ166 is significantly different from CM72 with nucleotide sequences at HsCBL8. The overexpression of HsCBL8 in rice resulted in significant improvement of water protection in vivo and plasma membrane, more proline accumulation, and a reduction of overall Na+ uptake but little change in K+ concentration in the plant tissues. Notably, HsCBL8 did not act on some genes downstream of the rice CBL family genes, suggesting an interesting interaction between HsCBL8 and unknown factors to be further investigated.
Introduction
High salinity is one of the most prevalent abiotic stresses that pose severe reduction in plants' growth and productivity of crops such as rice (Oryza sativa) grown in coastal and irrigated lands (Martínez-Atienza et al., 2007). Approximately 6% (800 million hectares) of world's total land area has been reported as salt affected (Romeza and Flowers, 2008), mainly contributed by accumulation of salts over time in arid and semiarid regions, salts from oceans brought in by wind and rain, and weathering of the rocks (Rengasamy, 2002). Sodium chloride (NaCl) is usually considered as the major soluble salt in saline soils encountered by plants (Munns and Tester, 2008). The responses of rice to salt stress could involve the regulation of membrane integrity, ionic compartmentation, osmotic adjustment and accumulation of macromolecules (Hu et al., 2012). To cope with deleterious effects of salt stress, efforts have been made to map QTLs that respond to salt stress in rice and barley (Gao and Lin, 2013; Long et al., 2013; Ahmadi-Ochtapeh et al., 2015; Das and Rao, 2015), clone genes based on mapping and transfer elite genes from wild barley into rice (Ellis et al., 2000).
Wild barley (H. spontanum), native to Qinghai-Tibet Plateau, has more genetic diversity compared with other wild and cultivated barley species, and the plateau is also considered as the center of the origin for cultivated barley in the oriental region (Dai et al., 2012). Wild barley species thrive best to survive under harsh environments, such as drought (Ahmed et al., 2013) and salt stress (Wu et al., 2013), and could provide elite gene pool for crop improvement (Ellis et al., 2000). Some genes from cultivated barley response to stresses, for example, HvCBL4 (calcium-sensor calcineurin B-like, CBL), a close homolog of the AtCBL4, positively responded to salinity in Arabidopsis (Rivandi et al., 2011). Moreover, the overexpression of HvCBF4 (C-repeat/dehydration-responsive element binding factors, CBF/DREBs) in transgenic rice plants induced tolerance against drought, salinity and low-temperature stresses with normal plant growth (Oh et al., 2007). Maintaining low level of Na+ and narrow Na+/K+ ratio in the cytosol is critical for cellular metabolism and salt tolerance in glycophytes (Qiu et al., 2002). One such example is of tolerant rice subspecies that can exclude Na+ from the shoot and maintain a low Na+/K+ ratio (Mekawy et al., 2015). To date, only HbCIPK2 from a wild barley species (H. brevisubulatum) was reported as a positive regulator of salt and osmotic stress responses, and as a factor to control Na+/K+ homeostasis in Arabidopsis (Li et al., 2012). However, little is known about the functional attributes of the gene(s) in response to abiotic stresses in H. spontanum from Qinghai-Tibet Plateau.
Calcium ion (Ca2+), the most dynamic second messenger in all eukaryotic organisms, interacts with many binding proteins such as calmodulin (CaM), calmodulin like proteins (CAML), and CBLs. Among these proteins, CBLs, which are small Ca2+ binding proteins, mainly interact with CBL-interacting protein kinase (CIPK) as an upstream factor for downstream signaling and in response to several abiotic factors, including salt, cold, drought, proton, reactive oxygen species (ROS), abscisic acid (ABA) and gibberellins (GA) (Thoday-Kennedy et al., 2015). CBLs are most similar to the regulatory B subunit of calcineurin (CNB), a protein phosphatase in animals (Liu and Zhu, 1998). Therefore, the Ca2+ binding capacity of the CBL protein is dependent on the number and structure of the elongation factor-hand domain (EF-hand), that could affect the stability of CBL-CIPK complex (Weinl and Kudla, 2009).
The first CBL protein that reacts to salt stress is AtCBL4 (Liu and Zhu, 1998). The active complex of CBL4-CIPK24 complex is activated downstream the Na+/H+ antiporter (NHX) to induce Na+ efflux from roots under high salt conditons (Qiu et al., 2002). The interaction between AtCBL10 and CIPK24 under salt stress also activates the Na+ transporter to transfer Na+ to the vacuole of shoots or leaves (Quan et al., 2007). AtCBL1, AtCBL5, and AtCBL9 are also involved in the response signaling to salt, drought, osmotic and ROS stress (Thoday-Kennedy et al., 2015). AtCBL2 and AtCBL3 could regulate the activity of proton pump V-ATPase on the vacuole membrane to maintain the ion homeostasis in the cell. By contrast, double-cbl2/cbl3-mutant plants were found more resistant to high Na+ and low K+ conditions (Tang et al., 2012). Similar stress responses of CBLs have also been reported in rice OsCBL8 (Xiang et al., 2007), Ammopiptanthus mongolicus AmCBL1 (Chen et al., 2011), soybean GmCBL1 (Li et al., 2012), rape BnCBL1 (Chen et al., 2012), and tobacco NsylCBL10 (Dong et al., 2015). Ten CBL proteins have also been discovered in Arabidopsis, rice and populous (Albrecht et al., 2001; Kolukisaoglu et al., 2004; Zhang et al., 2008). Eight CBLs have also been detected in maize, sorghum and grape (Weinl and Kudla, 2009). Seven CBLs have been found in canola (Zhang et al., 2014).
HsCBL8 exhibits a significantly distant phylogenetic relationship between with salt-responsive AtCBL4; this relationship indicates that HsCBL8 can differ from other known plant CBLs (Liu and Zhu, 1998). This finding also shows that HsCBL8 may function in a mechanism different from that of AtCBL4. Some CBL proteins, such as AtCBL1, AtCBL4, AtCBL5, AtCBL8, and AtCBL9 modified through N-myristoylation or S-acylation (Batistic et al., 2012), can acquire functional characteristics in membrane-localized dependent signaling in plant cells. By contrast, CBLs, including AtCBL2, AtCBL3, AtCBL6, and AtCBL7, are characterized by N-myristoylation motifs (MGCXXS/T). In this study, HsCBL8 was cloned from a wild barely (H. spontanum) line XZ166 native to Qinghai-Tibet Plateau and highly tolerant to severe salt stress. This study aimed to investigate the growth performance of transgenic rice plants expressing HsCBL8 in response to salt stress and to explore the related mechanism.
Materials and Methods
Plant Materials and Their Growth Conditions
The seeds of the Qinghai-Tibetan annual wild barley line XZ166 (H. spontanum) and cultivated barley CM72 (H. vulgare L. ssp. Vulgare), Arabidopsis thaliana ecotype Columbia-0 (Col-0), and rice Zhonghua11 (ZH11, Oryza sativa) were obtained from the Provincial Key Laboratory of Gene Resources, Zhejiang University, Hangzhou, China. The seeds of XZ166 and CM72 were surface sterilized in 5% NaClO for 15 min, rinsed with tap water, and germinated on moistened fine sand in plastic pots (15 cm × 20 cm × 15 cm) for 2 days at 25°C in the dark. The plastic pots with germinated seeds were then transferred to a greenhouse under a 14 h light (420 μmol m−2 s−1) period at 25°C and 10 h dark period at 23°C daily. We adopted 70% relative humidity and watered the plants with nutrient solution (pH 6.0, mg L−1) that contained (NH4)2SO4 (48.2), MgSO4 (154.88), K2SO4 (15.9), KNO3 (18.5), KH2PO4 (24.8), Ca(NO3)2 (86.17), Fe-citrate (7), MnCl2·4H2O (0.9), ZnSO4·7H2O (0.11), CuSO4·5H2O (0.04), HBO3 (2.9), and H2MoO4 (0.01). The solution was renewed at 6-day intervals, and its value was adjusted to pH 6.0 ± 0.1 using 1 M HCl daily. CaCl2 was added with NaCl to maintain a molar ratio of Na+:Ca2+ = 10:1.
The Arabidopsis ecotype Col-0 and the T2 plants (HsCBL8Promoter-GUS) were grown for 3 weeks in the greenhouse (16 h light, 110 μmol m−2 s−1), subsequently transferred to half strength MS liquid media, and allowed to grow for 1 week on the medium. After the plants became adapted to the MS media, different treatments, including 100 μM ABA, 100 mM NaCl, and 20% PEG6000 with 5% sucrose were applied at 4°C for 12 h. Moreover, plants in different developmental stages were used for β-glucuronidase GUS histochemical staining.
The seed dormancy of ZH11 and transgenic plants (35S-HsCBL8) was terminated with treatment of 0.5% H2O2 for 12 h. The seeds were then sterilized in 5% NaClO for 20 min, rinsed with tap water, and germinated on moistened filter papers in an incubator in a 14 h light period (110 μmol m−2 s−1) and 10 h dark period at 25°C. To calculate the germination rate, 20 rice seeds per biological repeat (four repeats independently) were treated with 125 mM NaCl in 1/4 nutrient solution. Moreover, the solution was renewed daily. Data on germination were collected from the “specific day” until 70 to 80% was reached per treatment for each genotype. Then the germination rate was calculated.
Molecular Cloning of HsCBL8
Seedlings of the XZ166 with three leaves were treated with 200 mM NaCl for 48 h, and their roots were selected for RNA extraction. Total RNA was extracted by a kit method (OMEGA, Norcross, GA RNA kit). PrimeScriptTM First Strand cDNA Synthesis Kit (Takara, Dalian China) was used to synthesize the cDNA. The genomic DNA of the XZ166 leaves was extracted using the CTAB method. The degenerate primers (Table S1) were designed based on cDNA sequences available on the National Center for Biotechnology Information (NCBI) database with accession IDs AL713904, FJ901265.1, EU085040.1, AB378095.1, XM_002524988.1, DQ907707.1, DQ201200.1, FJ901264.1, and FJ901259.1. The 25 μL PCR reaction mix used for amplification included L-Taq polymerase (Takara, Dalian China) (1.5 units), genomic DNA (25 ng), per primer 10 μM, dNTP (20 mM). PCR amplification was performed as follows: 95°C for 3 min, 10 cycles of 94°C for 30 s, and 60°C for 30 s at −0.5°C per cycle; 72°C for 60 s; and 25 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 60 s, and 72°C for 5 min. The PCR products were separated by 1.0% agarose gel electrophoresis. We obtained four wild barley CBL sequence candidates; one of which was identified as cultivated barley EST FLbaf27i23 under the Barley Database (http://www.shigen.nig.ac.jp/barley/). The coding sequences (CDSs) of its cDNA and genomic DNA were amplified using primers designed from FLbaf27i23, which contained additional Xba I and BamH I restriction sites (underlined, Table S1), respectively. The HsCBL8 promoter was cloned using inverse PCR (IPCR, http://dps.plants.ox.ac.uk/langdalelab/protocols) combined with nested PCR. Genomic DNA (2.5 μg) was cut by Xho I and Cla I independently; the DNAs ligated by T4 DNA ligase (Takara, Daliang China) were employed for amplification. The primers used are shown in Table S1. The prepared 25 μL PCR reaction mix included LTaq polymerase (Takara, Dalian China) (1.5 units), genomic DNA (50 ng), per primer (10 μM), and dNTPs (20 mM). PCR amplification was performed as follows: 95°C for 3 min, 10 cycles of 94°C for 30 s, 60°C for 30 s, −0.5°C per cycle, and 72°C for 3 min; 25 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 3 min; and 72°C for 5 min. PCR products were separated by 1.0% agarose gel electrophoresis. Then, the HsCBL8 promoter region was amplified using primers (Table S1) that contained additional Hind III and Xba I restriction sites (underlined).
Analysis of HsCBL8 CDS and Promoter Sequences
Nucleotide and protein sequences (Table S2) were analyzed by BioEdit 7.2.5 software (Abbott, Carlsbad, Canada) and NCBI (http://blast.ncbi.nlm.nih.gov/). Amino acid sequence alignment was analyzed by Clustal X, and phylogenetic trees were established by the neighborhood-joining bootstrap method; using the default parameters in the software MEGA 6.06 (http://www.megasoftware.net/index.php). The cis elements of the HsCBL8 promoter sequence (the upstream 1000 bp from the initiation codon) were predicted by PlantCARE online (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Vector Construct and Plant Transformation
The transgenic Arabidopsis plants harboring HsCBL8Promoter-GUS were generated according to the methods we previously described (Wang et al., 2014). In brief, a 2160 bp HsCBL8 promoter and GUS was inserted into pCAMBIA1300 binary vector. After verification by restrictive digestion and DNA sequencing, the construct was applied to transform wild type plants by floral dipping using Agrobacteria tumefaciens strain GV3103. Transgenic plants (T0) survived on solid Murashige and Skoog (MS) medium containing 30 mg ml−1 hygromycin were further verified by PCR genotyping. T1 and T2 seedlings were selected by PCR genotyping with the primers listed in Table S1.
pCAMBIA1300 was modified by inserting the CaMV 35S promoter-GUS (35S-GUS) region cut from pBI121 using Hind III and EcoR I and named as pCAMBIA1300-35S-GUS. The genomic DNA of the HsCBL8 coding region was inserted into the Xba I and BamH I sites of the pCAMBIA1300-35S-GUS plasmid to form expression vector pCAMBIA1300-35S-HsCBL8 (Figure 1A). This construct was transferred into Agrobacterium tumefaciens EHA105 and used for rice ZH11 transformation. Callus induction and subculture were performed on the medium supplemented with 2 mg/L 2,4-D, 3% sucrose, and 0.55% agar, named NB medium by Ozawa (2012). After a number of subcultures with an interval of 4 weeks, calli (between 1 and 3 mm in diameter) were used for transformation by co-culture with EHA105 harboring pCAMBIA1300-35S-HsCBL8. Then, the co-cultured calli were transferred onto pre-selection medium (NB supplemented with 30 mg/L hygromycin B), and incubated in the dark for 7 days. Subsequently, the differentiated calli were placed on the selection medium (NB with 50 mg/L hygromycin B) for another 14–17 days. The resistant calli were then transferred to pre-regeneration medium (NB supplemented with 2 mg/L 6-BA, 1 mg/L NAA, 5 mg/L ABA and 50 mg/L hygromycin B) and cultured in the dark for 7 days, followed by plant regeneration on medium NB supplemented with 3 mg/L 6-BA, 0.5 mg/L NAA and 50 mg/L hygromycin B, for 14–21 days under a 16 h photoperiod until shoots appeared. Regenerated plants were subsequently transferred into greenhouse and self-pollinated to produce progenies. The transgenic rice was first selected by hygromycin and PCR analysis using primers listed in Table S1, and then confirmed by Southern blot. The 35S region of the pCAMBIA1300-35S-GUS plasmid was replaced by the HsCBL8 promoter (2170 bp before the initiation codon; HsCBL8promoter) to form the vector pCAMBIA1300-ProHsCBL8-GUS using Hind III and Xba I (Figure 1B). This construct was transferred into A. tumefaciens GV3101 and used for Arabidopsis (Col-0) transformation by the floral dip method with slight modification. The Arabidopsis plants were grown in a greenhouse for about 4 weeks with a cycle of 16 h light and 8 h dark periods before transformation. GV3101 cells, cultured in Luria–Bertani (LB) medium for 18–20 h at 28°C (optical density at 600 nm [OD600]: 1.6–2.0), were collected by centrifugation and re-suspended in infiltration medium (half-strength MS medium, 5% sucrose, 0.044 μM benzylamino purine, and 0.2% Silwet L-77) to an OD600 of 0.8–1.0. Plant inflorescences were dipped in the infiltration solution for 2 min, and transferred to a dark chamber in a greenhouse for 2 days, and then cultured under normal conditions. Transgenic plants (T0) survived on solid Murashige and Skoog (MS) medium containing 30 mg ml−1 hygromycin, were further verified by PCR genotyping. T1 and T2 seedlings were selected by PCR genotyping with the primers listed in Table S1.
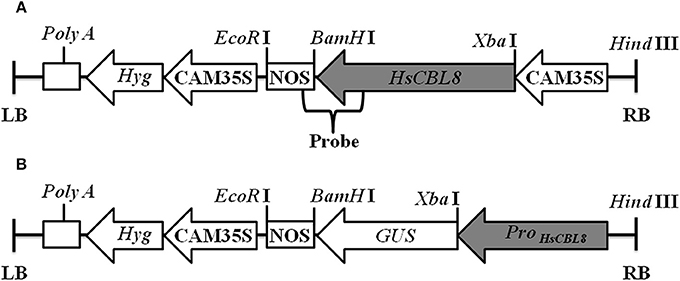
Figure 1. Linear maps of the molecular constructs 35S-HsCBL8 (A) and HsCBL8Promoter-GUS (B) in the pCAMBIA1300 vector used for transformation. LB, left border; RB, right border; Hyg, Hygromycin B phosphotransferase gene; CAM35S, Cauliflower mosaic virus 35S promoter; NOS, Nos terminator; GUS, β-Glucuronidase gene; ProHsCBL8, HsCBL8 promoter.
PCR Analysis
Both genomic and cDNA were identified. The specific primers (Table S1) from HsCBL8 genomic DNA were used to separately amplify 679 bp fragments. The specific primers (Table S1) from the HsCBL8 cDNA were used to amplify 596 bp fragments, and OsGAPDH was used as reference gene (425 bp). The prepared 20 μL PCR reaction mix contained rTaq polymerase (Takara, Dalian China) (1.0 units), genomic DNA (20 ng), per primer (10 μM), and dNTP (10 mM). The following PCR program was used: 95°C for 4 min; 25 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 60 s; and 72°C for 5 min. The PCR products were separated by 1.0% agarose gel electrophoresis.
Southern Blot Analysis
Approximately 10 μg DNA per sample was separately and completely digested by Hind III and EcoR I, separately, and the restriction fragments were separated with 0.8% (w/v) agarose gel. Then, DNA was transferred onto a Hybond N+ nylon membrane (Boehringer, Mannheim, Germany) for 12 h, and the membrane was hybridized using a specific probe, including HsCBL8 and an NOS sequence (Figure 1A). The probe (679 bp) was then amplified in accordance with primers (Table S1) using plasmid pCAMBIA1300-35S-HsCBL8. The probe was labeled with the enzyme horseradish peroxidase (Roche DIG High Prime DNA Labeling and Detection Starter kit II) for 10–12 h at 65°C. After hybridization, the membrane was washed twice with 2 × standard saline citrate (SSC) plus 0.1% sodium dodecyl sulfate (SDS) at 65°C for 10 min and washed twice with 0.5 × SSC plus 0.1% SDS for 10 min at room temperature. The membrane was covered with the detection reagent mixture for 30 min in accordance with the supplier's instructions. Finally, the membrane was wrapped in Saran wrap with 1 ml CSPD ready-to-use for 5 min and subsequently detected by a Fuji Photo Film Releases LAS-3000 Imaging System (Tokyo, Japan).
GUS Analysis
Histochemical staining of GUS activity was performed as follows. Plant tissues were incubated at 37°C overnight in GUS staining buffer (2 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid in 50 mM sodium Pi buffer, pH 7.2) containing 0.1% Triton X-100, 2 mM K4Fe(CN)6, 2 mM K3Fe(CN)6, and 10 mM EDTA. The stained seedlings were sequentially transferred to 50, 70, and 100% (v/v) ethanol to remove chlorophyll. The stained materials were examined with a dissecting microscope (Leica MZ95) with a digital camera. GUS activity was also determined by fluorometric assay (Gallagher, 1992).
Transgenic Rice Treated with High NaCL
The young rice seedlings (30 days) were treated with 125 mM NaCl in half-strength nutrient solution. Fresh weights were calculated at 0, 8, 16, and 24 h; plant phenotypes were photographed at 2, 4, and 6 days, and all leaves at 0, 24, and 72 h were used for analysis of proline, malondialdehyde (MDA), and Na+ and K+ contents, and rate of electrolyte leakage (REL). Plants without NaCl treatment were used as control. Meanwhile, proline content was determined as follows: 0.2 g fresh leaves were chopped and immersed in 5 ml of 3% sulphosalicylic acid at 100°C to extrac free proline. Proline content was then measured by UV Spectrophotometer (UV1000, LabTech) at 520 nm. MDA content was determined by the thiobarbituric acid (TBA) reaction. Leaves (0.2 g) were homogenized with 10 ml of 10% trichloroacetic acid (TCA) and centrifuged at 4000 rpm for 10 min. Part of the supernatant (2 ml) was mixed with 2 ml 0.6% TBA and incubated in 100°C water for 15 min, then cooled immediately on ice and centrifuged at 4000 rpm for 10 min. OD450, OD532, and OD600 were determined by UV spectrophotometry (UV1000, LabTech). MDA concentration (MDAC) were calculated through the formula; MDAC (μmol/L) = [6.45 (OD532 − OD600) − 0.56 OD450] × V × m, where V is the volume of the total extraction liquid and m is the sample weight. The leaves and roots were collected and dehydrated in a Muffle furnace at 500°C for 6 h. These ash samples were then extracted in 50% HNO3 overnight. The Na+ and K+ contents were measured by atomic absorption spectrophotometry (Z-5000 Polarized Zeeman, Hitachi, Japan). Membrane permeability can be reflected by the REL. Fresh leaves were collected and washed three times with deionized water to remove surface-adhered electrolytes. Each sample was divided equally and placed into two sealed vials containing 10 ml of deionized water. One vial was incubated at 25°C on a rotary shaker for 3 h, and then the electrical conductivity (EC1) was measured with a conductivity meter (162A, Thermo Orion, USA). Another vial was autoclaved at 100°C for 20 min, and the electrical conductivity (EC2) was determined by a conductivity meter. The REL can then be defined as REL = (EC1/EC2).
In these analyses, four biological replicates were performed independently, and four plants were used in per replicate.
Real-Time PCR Analysis
XZ166 and CM72 plants (with three leaves, about 10 days) were treated with 200 mM NaCl, and the roots and shoots of three plants were separately collected at 0, 1, 6, 12, 24, and 48 h. Similarly, ZH11 and transgenic (35S-HsCBL8) seedlings (30 days) were treated with 125 mM NaCl, and shoot of three plants were separately collected at 0, 3, and 6 h. Three biological replicates were carried out independently. The primers adopted are listed in Table S1. HvGapdh and actin mRNAs were separately employed as internal controls for barley and rice. Real-time PCR was performed using the CFX96 system (Bio-Rad, America) with iTaq Universal SYBR® Green Supermix (Bio-Rad). The PCR program was implemented as follows: 95°C for 3 min and 40 cycles of 95°C for 15 s, 58°C for 15 s, and 72°C for 15 s.
Statistical Analysis
Statistical analysis for real-time PCR was conducted using the software CFX Manager (version 2.1), and the relative expression level of per gene was calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001). Moreover, student's t-test was applied for each parameter studied. Differences were considered significant at P <0.05 and highly significant at P <0.01.
Results
Cloning of HsCBL8
The first 494 bp cDNA sequence amplified with degenerate PCR primers (Table S1) was found similar to the full-length cDNA sequence of the gene FLbaf27i23 in barley (H. vulgare subsp. vulgare) (http://www.shigen.nig.ac.jp/barley/). BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis of the coding sequence (CDS) predicted 915 nucleotides, which code HvCBL8 with 305 amino acids (Figure 2A). A fragment of 873 bp at the CDS was cloned from wild barley XZ166 using primers based on FLbaf27i23, which encodes the HsCBL8 protein of 291 amino acids (Figure 2A, NCBI, AEM44693.1). We performed IPCR of the 2388 bp fragment (NCBI ID, HQ696007.1) upstream the initial codon. Moreover, 31 cis elements of the upstream 1000 bp from initiation codon were predicted by PlantCARE. Ten of these elements were light response elements, five were MeJA response elements, four were ABA response elements, four were endosperm expression elements, three were drought response elements, two were GA-responsive and circadian control elements, and one was a salicylic acid (SA) response element (Table 1). The predicted cis elements of the HsCBL8 promoter sequence indicated that HsCBL8 could be induced by different stresses (Table 1).
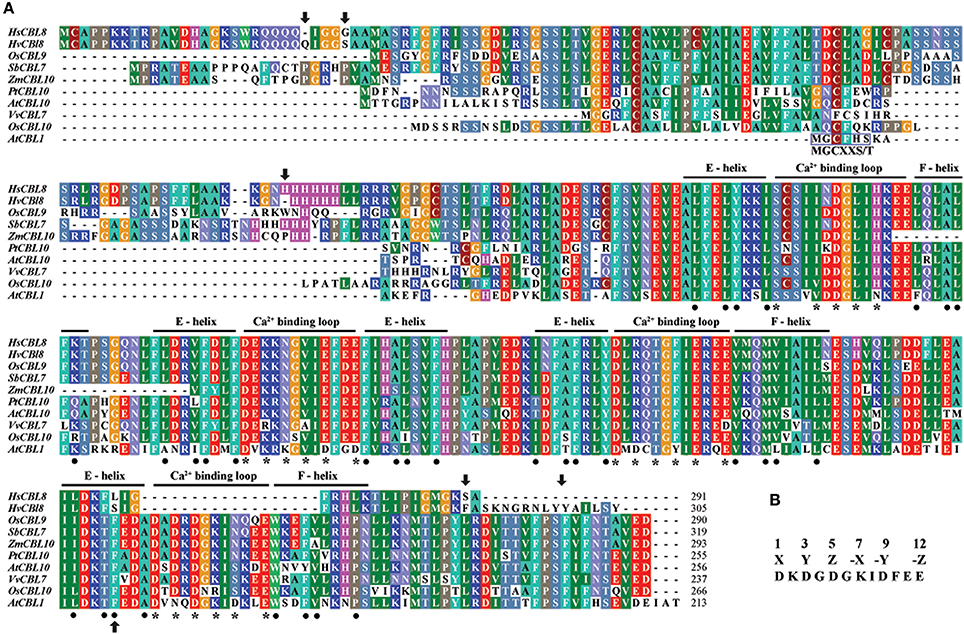
Figure 2. Analysis of the CBL amino-acid sequences. (A) Alignment of the amino-acid sequences belonging to the sub-cluster (II-I-I-II) of some species, including HsCBL8, HvCBL8, OsCBL9, SbCBL7, ZmCBL10, PtCBL10, AtCBL10, VvCBL7, and OsCBL10. AtCBL1 was used as the control. The four EF–hand calcium-binding motifs are clearly indicated. Asterisks and dots denote the amino-acid residues important for calcium binding and the EF–hand structure, respectively. Arrows show the amino acids altered between HsCBL8 and HvCBL8. Conserved myristoylation motifs (MGCXXS/T) in the N-termini of AtCBL1 was marked by a blue box. (B) EF–hand consensus sequence in accordance with Kolukisaoglu et al. (2004).

Table 1. Prediction of cis-elements responding to environment stimuli in HsCBL8 promoter (the upstream 1000 bp from the initiation codon) using software PlantCARE.
Phylogenetic Analysis of HsCBL8
To study the phylogenetic relationship of HsCBL8, the amino acid sequence of HsCBL8 was compared with 71 CBL genes from plant species including monocot (rice, sorghum, maize and barley), dicot (Arabidopsis, grape and populous), and protozoan (Naegleria fowleri). The corresponding IDs are presented in Table S2. Accordingly, members of the family CBL family could be clustered into two groups (Figure 3). Group I only comprised four members. Three belonged to simple plants and N. fowleri, and one represented Arabidopsis (AtCBL5). By contrast, Group II was divided into two sub-clusters: II-I and II–II. Moreover, II-I was grouped into II-I-I and II-I-II, whereas II-I-I was sub-clustered into II-I-I-I and II-I-I-II. HsCBL8 was found closely related to rice OsCBL9 (63.70%) and thus placed in the group II-I-I-II, which included 12 CBLs (Figure 3). Results revealed that most of CBLs belonging to monocots were slightly deviant phylogenetically from dicot species, whereas the CBLs of lower plants were found in close relation to higher plant species in each group (Figure 3). Moreover, HsCBL8 was significantly different to HvCBL8 in terms of a 14-amino-acids deletion at its nitrite end, and five other changed sites, including three changed, one deletion, and one more amino acid (Figure 2A). Normally, the CBL protein contains four EF–hand domains for Ca2+ ion binding (Figure 2B), but the last domain was lost in HsCBL8 and HvCBL8 (Figure 2A). Obviously, a conserved myristoylation motif (MGCXXS/T) in the N-terminus of AtCBL1 was not discovered in the group II-I-I-II (Figure 2), including HsCBL8 (Figure 2A).
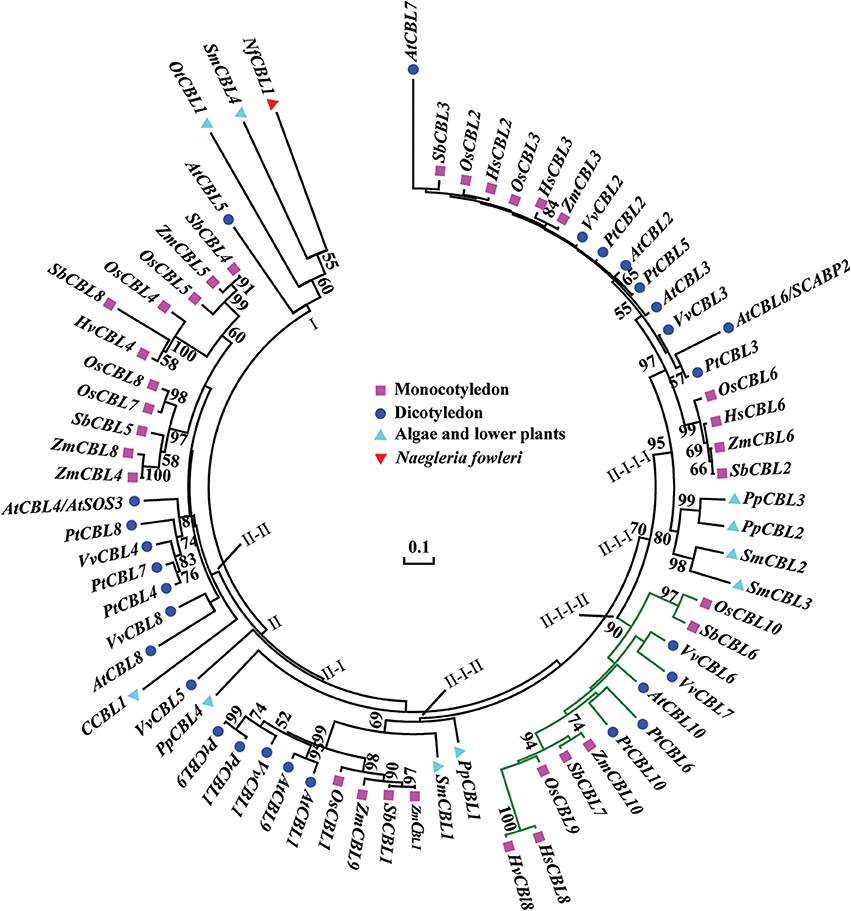
Figure 3. Phylogenetic relationships of CBL proteins from algae, lower plants, higher plants, and protozoan Naegleria fowleri (Nf). The NCBI accession numbers of the CBL proteins in this paper are presented in Table S2. The bar shows the evolutionary distance, and the number at the nodes represents the reliability percentage (%; only >50% is shown) of the bootstrap values based on 1000 replications. Codification: Hs, Hordeum spontaneum; Hv, H. vulbare; Os, Oryza sativa; Zm, Zea mays; Sb, Sorghum bicolor; At, Arabidopsis thaliana; Pt, Populus trichocarpa; Vv, Vitis vinifera; Ot, Ostreococcus tauri; C, Chlorella sp.; Pp, Physcomitrella patens; Sm, Selaginella moellendorfii.
Expression Analysis of HsCBL8 in Barley under Salt Stress
To evaluate whether HsCBL8 in XZ166 and HvCBL8 in CM72 could respond to salt, we conducted real time reverse-transcription PCR analysis to test their expression patterns in the control and under salt stress. Results revealed that the expression of both the genes in their respective plant species, were significantly induced in their shoots and roots, respectively, in response to 200 mM Nacl stress (Figure 4). Overall, HsCBL8 was expressed to significantly higher levels in both shoots and roots than that in HvCBL8. The expression profile of both the genes in the shoot generally followed a sigmoid curve in general. The expression level was enhanced in the first hour of NaCl application, reaching its peak after 6 h and then gradually declining with prolonged salt stress from 12 to 48 h, respectively (Figure 4A). In the roots, the expression of the CBL8s in both XZ166 and CM72 was greatly suppressed (Figure 4B) during first hour of NaCl treatment. However, both the genes were highly expressed at 3 and 6 h of stress treatment. Afterward, a gradual decline was observed in the expression pattern after up to 48 h of stress treatment. However, significant differences in transcriptional levels were observed between HvCBL8 and HsCBL8 at 3, 24, and 48 h in the roots (Figure 4B), revealing the variation in the degree of sensitivity of these genes to salt stress.
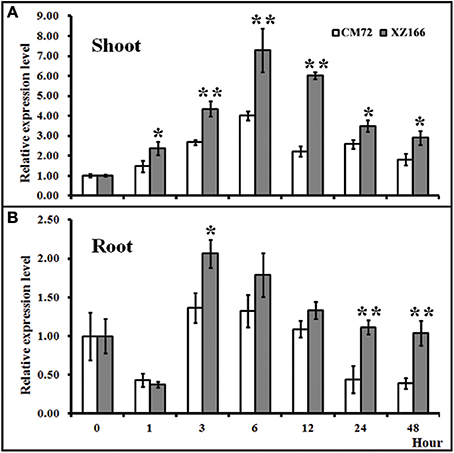
Figure 4. Relative expression levels of CBL8 between XZ166 and CM72 plants treated with 200 mM NaCl. (A) Shoot, and (B) root. The expression level for 0 h treatment was set to 1. Values include the means of three independent replicates. *Indicates a statistically significant difference at p <0.05 and **signifies a highly significant difference compared with those of the CM72.
HsCBL8 Transgenic Rice and Its Response to Salt Stress
To study the role of HsCBL8 in rice, an Agrobacterium strain EHA105 harboring the plasmid pCAMBIA1300-35S-HsCBL8, was transformed into rice ZH11 using embryo calli induced from mature seeds. A Southern blot analysis using two restriction enzymes, Hind III and EcoR I, revealed the presence of two copies (T1-1 and T1-3) of the T1 transgenic line of rice (Figure 5). Of these transgenic plants, 20 T2 seedlings of T1-1 and 20 T2 seedlings T1-3 were verified by PCR and resulted in the detection of 9 homozygous transgene (35S-HsCBL8) plants with 679 bp band length. Furthermore, T3 seeds from T2 homozygous lines namely T2-1-3 (L1) and T2-3-9 (L2) (Figure S1), were selected for further analysis to evaluate the functional attributes of the gene for its tissue-specific expression.
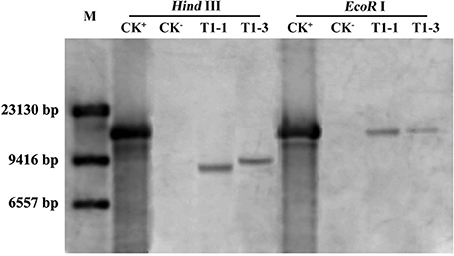
Figure 5. Southern blot analysis of T1 transgenic rice plants. M, DNA marker (λ-DNA/Hind III); CK+, pCAMBIA1300-35S-HsCBL8 (12.9 kbp); CK−, ZH11; T1-1 and T1-3 selected transgenic lines with one copy of HsCBL8 integrated into the rice genome.
To evaluate the possible role of HsCBL8 in salt tolerance, the seedling growth and morphology of the transgenic rice lines (L1 and L2) were compared with that of the control (ZH11) plants (Figure 6). The comparison was conducted under normal and 125 mM NaCl stress conditions. We found insignificant differences in seed germination (Figure 6A) and seedling fresh weight (Figure 6C) between the transgenic and ZH11 plants under the normal growth conditions as evident from the seedling growth (Figure 6B). By contrast, fairly obvious and significant variations in seed germination rate as well as seedling fresh weight were observed between the transgenic lines and non-transgenic ZH11 under 125 mM NaCl stress (Figures 6A,C). The overall germination rate of both transgenic and ZH11 seeds were reduced under salt stress relative to that of the untreated seeds. However, the lines L1 and L2 thrived best at around 75% germination which was 20% higher than that of ZH11 (Figure 6A). This finding was evident from the stunted growth of the radicles and plumules of the ZH11 seedlings under salt stress (Figure 6B). Moreover, the rate of water loss from the transgenic lines was significantly lower than that of ZH11 when the application of salt stress was prolonged from 0 to 24 h. Obvious differences between the fresh weights of seedlings of transgenic lines and ZH11 were observed at 16 (p <0.05) and 24 h (p <0.01) of salt stress (Figure 6C). Furthermore, we tested salt tolerance in the plants at the well-grown vegetative stage. We then planted the seedlings of L1, L2 and ZH11 for 30 days in normal conditions hydroponically. After 30 days of growth, we applied 125 mM NaCl stress, initially for 2 days, 4 days, and finally 6 days (Figure 6D). The affects of NaCl treatment were significantly prominent on the leaves of the ZH11 plants, especially on the older leaves. The degree of severity of the effect was increased with the exposure time of the ZH11 plants to the treatment from 2 to 6 days compared with that in the transgenic lines. This effect was manifested as withering of older leaves, increased water loss, and curling of young leaves in the ZH11 plants compared with those in the L1 and L2 plants. The transgenic plants showed better performance as exhibited by the strong vitality of the top new leaves, even after 6 days of NaCl treatment (Figure 6D). These results indicate the obvious role of HsCBL8 in salt tolerance.
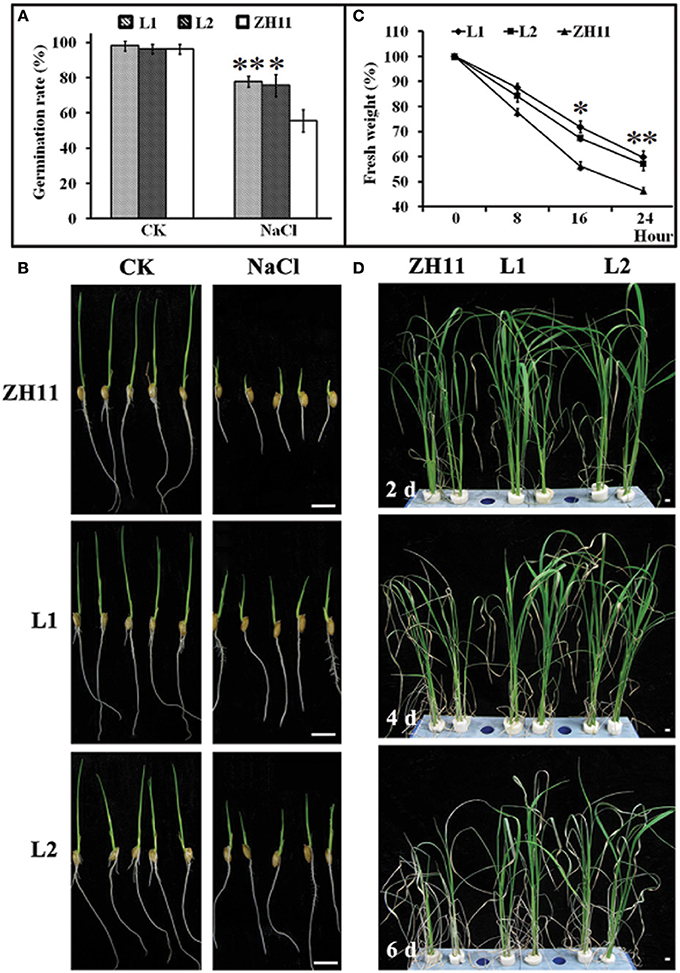
Figure 6. Phenotypic changes of transgenic rice lines induced by salt stress. (A) Germination rate of seeds treated with 125 mM NaCl for 5 days, and morphological characteristics of rice young seedlings (B). (C) Water loss, 2 week pre-cultured plants were treated with 125 mM NaCl. (D) Photos of rice shoots with treatment of 125 mM NaCl, the seedlings were pre-cultured for 30 days in normal conditions. Values include the means ± standard deviation (SD) (n = 3). *(p <0.05) and **(p <0.01) indicate that these values were statistically significantly and highly significantly different respectively, compared with that of the ZH11. Bar = 1 cm. CK, normal condition; L1 and L2, transgenic lines.
Role of HsCBL8 in the Salt Stress Response in Transgenic Rice
Generally, the accumulation of compatible osmolytes such as proline, are associated to the stress tolerance of the corresponding plant (Ahmed et al., 2013; Mekawy et al., 2015). Meanwhile, quantification of MDA concentrations and REL is considered as an efficient indicator of the structural integrity of the plant membranes in response to abiotic factors, such as salt stress. To evaluate the functional attributes of overly expressed HsCBL8 for rice tolerance against salt stress, we analyzed the proline and MDA concentrations, and rate of REL. Accordingly, the HsCBL8 overexpression in the transgenic rice resulted in the accumulation of 35 and 56% additional proline content in the L1 and L2 lines, respectively, after 24 h of salt stress compared with that in ZH11 (Figure 7A). Moreover, plant exposure to plants to salt stress for 72 h resulted in an overall increase in proline content in both the transgenic and ZH11 plants compared with those in 24 h. Even so, 33% and 25% accumulation of additional proline content was observed in L1 and L2 respectively, relative to that in ZH11. This result indicates the possible role of HsCBL8 in salt tolerance. On the contrary, HsCBL8 overexpression significantly reduced the MDA content and REL in the transgenic lines compared with the non-transgenic ZH11 plants under salt stress. This reduction in MDA content was in the order of 36% (in both transgenic lines) at 24 h, but 31 and 29% in L1 and L2, respectively, at 72 h of salt stress compared with the contents in ZH11 (Figure 7B). Similarly, 33 and 46% reduction in REL was observed in L1 and L2 respectively, at 24 h of salt treatment, whereas, reduction of 23% and 24% reduction in REL was noted in L1 and L2, respectively, at 72 h of salt stress compared with that in the ZH11 plants (Figure 7C).
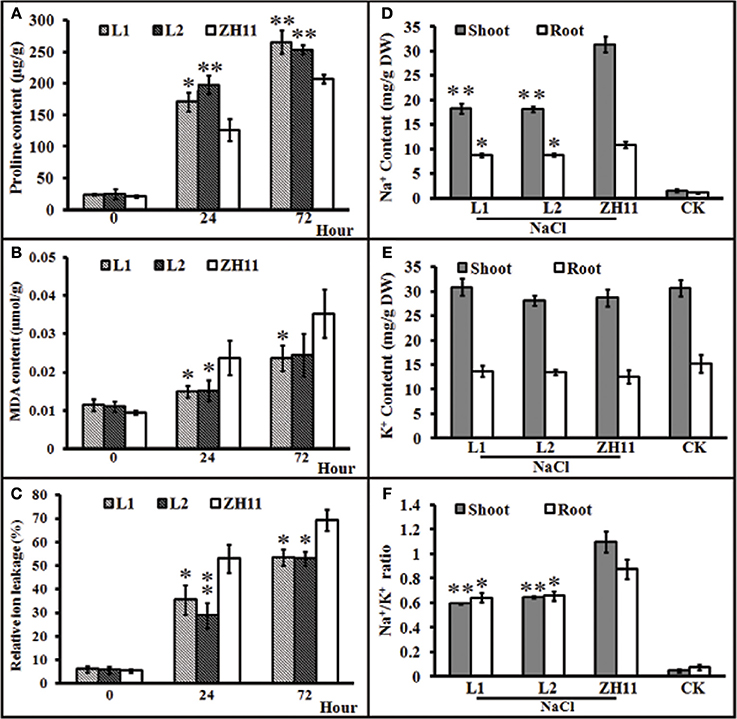
Figure 7. Characteristics of transgenic rice lines and ZH11 treated with salt stresses. (A–C), physiological indexes reflecting salt tolerance: (A) proline content, (B) MDA content, and (C) relative ion leakage. Plants pre-cultured for 2 weeks were treated with 125 mM NaCl. (D–F), Na+ and K+ contents (mg/g DW) of shoots and roots of transgenic and ZH11 plants. (D) Na+ and (E) K+ levels and (F) Na+/K+ ratio. Na+ and K+ contents were measured after 6 days of 125 mM NaCl treatment. The treated plants were pre-cultured for 30 days. Values include the means ± SD (n = 4). *(p <0.05) and **(p <0.01) indicate that these values were statistically significantly and highly significantly different, respectively, compared with that of ZH11 or CK. CK and ZH11 growing in normal conditions; L1 and L2, transgenic rice lines.
Na+ and K+ Contents in the HsCBL8 Transgenic Rice
Generally, the ability of a crop variety to inhibit the uptake of potentially toxic ions such as Na+ and preferential uptake of K+ to keep the balance between ions is regarded as a desirable trait for salt tolerance. To investigate whether HsCBL8 could induce the salt tolerance trait in transgenic rice, we analyzed the status of Na+ and K+ ions and their comparative ratio in roots and shoots under control, as well as salt stress conditions. Plants of both transgenic as well as ZH11 pre-grown for 30 days under normal condition were exposed to 125 mM NaCl stress for 6 days. After 6 days, analysis of the ions revealed insignificant differences in Na+ and K+ concentration among unstressed transgenic lines and ZH11. By contrast, under salt stress, Na+ accumulation was generally noted at higher content in the shoots than in the roots in either of the transgenic lines and ZH11 plants. However, the Na+ contents in ZH11 plants were significantly higher in the shoots (42%) as well as the roots (18.5%) than in both of the transgenic lines (Figure 7D). On the other hand, salt stress induced a slight change in root and shoot K+ contents between the transgenic lines and ZH11 plants applied with or without NaCl treatment (Figure 7E). Consequently, the Na+/K+ ratio in both shoot and root was lower in transgenic lines than that in ZH11 plants (Figure 7F), indicating that the overexpression of HsCBL8 in rice could regulate and induce salt tolerance.
Expression Levels of Some Stress Responding Genes in the Transgenic Rice Harboring HsCBL8
To investigate whether HsCBL8 could regulate the salt-responsive genes in rice, we performed the gene expression profiling of the 35S-HsCBL8 transgenic lines (L1 and L2) in comparison with the non-transgenic plants under normal conditions and salt stress. The genes selected were either specifically salt responsive (OsSOS2/OsCIPK24, OsNHX1, and OsSOS1) and/or responsive to multiple stresses (abiotic or biotic) (OsCIPK15, OsRD29A, and OsDREB2A). Under normal growth conditions (unstressed, 0 h), we observed extremely weak expression, as well as insignificant variations in the transcriptional levels of all the selected genes in both ZH11 and transgenic plants. This result indicates that HsCBL8 exerted an almost negligible interaction with these genes (Figures 8A–F) in the unstressed environment. At the onset of stress application (125 mM NaCl) initially for 3 h, the overall relative expression level of the genes was enhanced relative to that in the unstressed plants. However, the difference between the transgenic lines and ZH11 was insignificant. Moreover, duration of salt stress exposure was prolonged to 6 h, all the genes were found substantially up-regulated compared with the unstressed plants (0 h) or those subjected to salt stress for 3 h. However, a a comparison between the transgenic lines and ZH11 at 6 h of salt stress revealed that the overexpression of HsCBL8 in both the transgenic rice lines up-regulated some of the genes, such as OsSOS2/OsCIPK24, OsCIPK15, and OsRD29A, to a higher level than that in ZH11 (Figures 8A,B,E). Even so, the overall variation was insignificant, similar to the one observed in plants exposed to 3 h of salt stress or the untreated plants. These results indicate that the overexpression of HsCBL8 in rice could not affect the regulation of salt-responsive genes significantly, showing the negligible interaction of HsCBL8 with all the selected genes expressed in response to salt stress.
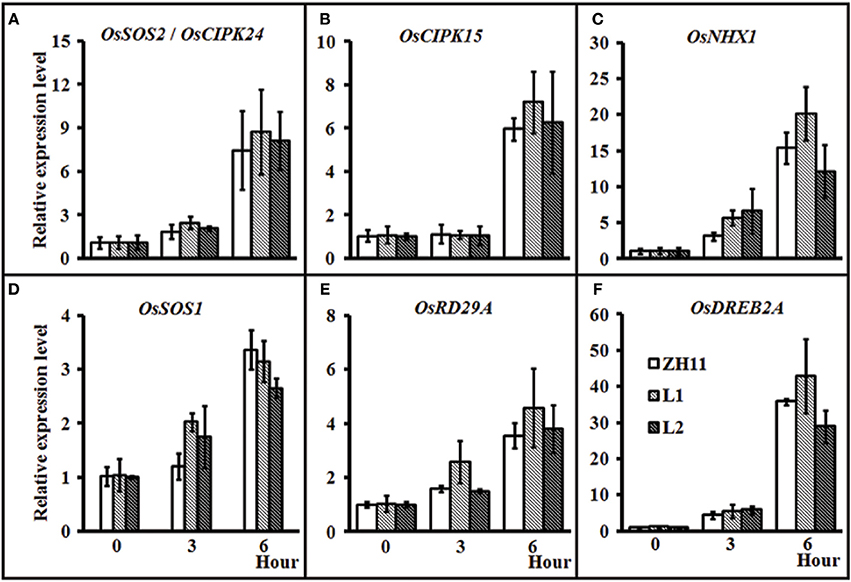
Figure 8. Relative expression of some stress-responsive genes in rice ZH11 and two transgenic lines with treatment of 125 mM NaCl for 0, 3, and 6 h. The plants were pre-cultured for 3 weeks in hydroponic solution. The expression level of each gene for WT (0 h) was set to 1. Values include the means ± SD (n = 3). Actin1 (NCBI ID: NC_008396) was used as reference gene. NCBI ID: OsSOS2 (A), DQ248963; OsCIPK15 (B), AB264037; OsNHX1 (C), AB021878; OsSOS1 (D), AY785147; OsDREB2A (E), JQ341059, and OsRD29A (F) (Rao et al., 2011).
GUS Activity Driven by the HsCBL8 Promoter in Response to Abiotic Stress in Transgenic Arabidopsis
To have more detailed perspective of expression pattern of HsCBL8 in response to various abiotic stresses, the promoter fragment of HsCBL8 was fused with the GUS reporter gene and transferred to Arabidopsis to develop HsCBL8Pro-GUS transgenic lines (Figure 1B). The expression of GUS activity was examined in 17 independent transgenic lines. Except for three lines (with zero GUS activity), the other 14 lines exhibited similar tissue-specific expression pattern. However, the intensity of the colors varied among lines, probably because of the position effect. The GUS expression pattern of a representative line (TGUS-7) is presented herein (Figures 9A–J). Under normal conditions, GUS signals were extremely weak in the cotyledon apex at the second day after germination (Figure 9A), whereas, the hypocotyl stele was stained light blue at the third day (Figure 9B), showing the activity of GUS promoted in hypocotyl and cotyledons (Figure 9C) and further strengthened in the vascular bundles of elder rosette leaves (Figures 9D,J). However, no activity was observed in the newly formed root tissues, including lateral root tips (Figures 9D,J), instead the blue stain was observed within the root stele tissue (Figure 9J) and the junction tissue of tap and lateral roots (Figure 9J). Furthermore, GUS activity at the reproductive phase was detected in sepals, filaments, and tissues between the style and stigma, and tissues between the silique and silique petiole but no GUS signal was observed in the petals and seeds (Figures 9E–H).
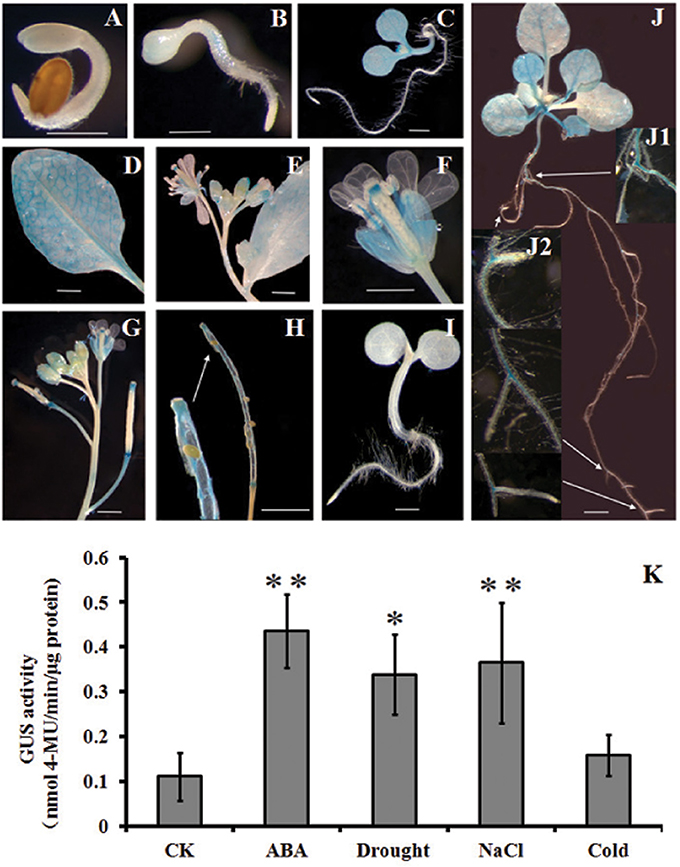
Figure 9. Histochemical analysis of GUS activity in transgenic Arabidopsis plants. (A–J) GUS staining at different developmental stages; (K), GUS activity responding to ABA, drought, NaCl, and cold treatments. (A) 2-day-old seedlings, (B) 3-day-old seedling, (C) 6-day-old seedling, (D) rosette leaf, (E,F) flowers, (G) young siliques, (H) mature siliques, (I) Col-0, and (J) 2-week-old seedlings. (K) 4-week-old seedlings supplemented with 100 μM ABA, 20% PEG6000, and 100 mM NaCl. For cold treatment, the seedlings were transferred to a 4°C growth chamber in the dark. GUS activity was assayed after 12 h treatment. The values are expressed as means ± SD (n = 4). *Indicates statistically significant difference at (p <0.05) and **indicates a highly statistically significant difference at (p <0.01) compared with CK. Bar = 5 mm. The line with arrow shows the magnified tissue portion.
Some cis elements in the HsCBL8 promoter sequence predicted by PlantCARE, such as ABRE and MBS (Table 1), normally respond to ABA signal and drought stress, respectively. To evaluate the response of the HsCBL8 gene to ABA, drought, NaCl and cold stress, GUS activity was measured by fluorometric assay (Figure 9K). Four-week-old Arabidopsis seedlings were supplemented with 100 μM ABA, 20% PEG6000, and 100 mM NaCl to apply ABA, drought and salt stress, respectively. For cold treatment, the seedlings were transferred to a growth chamber under dark conditions at 4°C. GUS activity was assayed after 12 h treatment. Accordingly, compared with untreated plants, GUS activity was observed to significantly (p <0.01) higher levels in the plants treated with ABA (294%), NaCl (230%), and PEG6000 (p <0.05; 205%), respectively, whereas, an increase of only 43% was obtained with cold treatment, indicating the possible response of HsCBL8 to these stresses.
Discussion
Hordeum spontanum, derived from Qinghai-Tibet Plateau (origin center of the cultivated barley) is known as a highly salt-tolerant species (Dai et al., 2012; Wu et al., 2013). Being a member of same crop family, studies on the elite genes of this species in rice could help us better understand of the salt-tolerance mechanisms in wild barley. To obtain insight about the salt stress responses in rice related to elite gene(s) from wild barley, we selected the HsCBL8 gene and thoroughly analyzed its functional relations with other genes in response to the salt stress regime. Initially, we conducted the phylogenetic analysis of HsCBL8 and revealed that the encoded protein belongs to the group of CBL proteins only modified with N-myristoylation or S-acylation (Batistic et al., 2012), (Figure 2A). Thus, these proteins are involved in close phylogenetic relationships with the members of group II-I-I-II (Figure 2) besides 10 other CBL proteins. This group was clustered into a clade defined as a transmembrane (TM) helix (Kleist et al., 2014) in accordance with AtCBL10, which harbors a unique transmembrane-spanning region (Kim et al., 2007). Moreover, ZmCBL10, OsCBL9, SbCBL7, and HsCBL8 also harbor long N-terminal extensions (Figure 2A), indicating that the long N-terminal amino-acid sequence of HsCBL8 may be involved in sub-cellular localization. However, we could not find the transmembrane domain in the HsCBL8 protein sequence similar to that in AtCBL10. Therefore, further investigation must to be conducted to confirm the assumption.
In general, each CBL protein harbors four EF–hand motifs (Figures 2A,B), but not all of the CBL proteins contain four canonical EF–hand motifs. For instance, five CBL proteins in Arabidopsis (CBL2, CBL3, CBL4, CBL5, and CBL8) do not possess any canonical EF hand, whereas, CBL6, CBL7 and CBL10 contain one and CBL1 and CBL9 contain two canonical EF hands (Batistic et al., 2012). Herein, both HsCBL8 and HvCBL8 lack the fourth EF hands (Figure 2A). Moreover, the structural and numeral differences in the EF hands demonstrate variations in Ca2+ binding affinity and interactions between the CBL and CIPK proteins. For example, crystal structure analyses of AtCBL2 and AtCBL4 revealed that AtCBL2 bound only to CIPK14 with four Ca2+ ions in all four EF hands (Akaboshi et al., 2008), whereas, AtCBL4 bound only to CIPK24 with two Ca2+ ions in two EF hands (Sánchez-Barrena et al., 2007). Therefore, the model representing the cooperative binding of the EF hand and Ca2+ ion in HsCBL8 could not be deduced from the current results. This finding suggests that some known downstream factors of CBLs were not regulated by HsCBL8 (Figure 9). Instead HsCBL8 might interact with other factors to regulate downstream genes responding to salt stress in transgenic rice.
To verify this hypothesis, we investigated the growth performance of transgenic rice lines with HsCBL8 expression in response to salt stress. We observed that the constitutive expression of HsCBL8 significantly improved the tolerance against salt stress in rice. Germination rate was the first index that showed the obvious role of HsCBL8 against salt stress in the transgenic rice treated with 125 mM NaCl (Figure 6A). These findings followed the similar trends reported for other CBLs in various plant species, such as the transgenic Arabidopsis harboring 35S-AtCBL5 (Cheong et al., 2010) and the poplar containing 35S-PeCBL10 (Li D. D. et al., 2013). Moreover, scholars reported that OsCBL2 may be involved in a GA-signaling pathway that leads to the vacuolation of the aleurone cell (Hwang et al., 2005). Meanwhile, the attributes of AtCBL1 in terms of positive response to gibberellins (Li Z.-Y. et al., 2013) and negative response to ABA (Pandey et al., 2008) during seed germination and seedling development could provide evidence supporting the similar characteristics of HsCBL8 under the salt stress condition. The overexpression of HsCBL8 in our study also decreased Na+ uptake partly in rice (Figures 7D–F). Moreover, AtCBL10, which shares the same cluster with HsCBL8 (Figure 3), responded similarly to salt stress by modulating the activities of Na+ transporters (Kim et al., 2007; Quan et al., 2007). AtCBL10's homologs PeCBL10 (Li D. D. et al., 2013) and NsylCBL10 (Dong et al., 2015) in roots, whereas, PtCBL10A and PtCBL10B (Tang et al., 2014) in shoots also showed salt tolerance by regulating Na+ balance, indicating that HsCBL8 might regulate some Na+ transporters or channels involved in the efflux of excess Na+ under salt stress. However, K+ uptake was slightly affected in the transgenic rice with HsCBL8 (Figure 7E). Recent evidence showed that AtCBL10 may directly compete with CIPK23 for binding to AKT1 and negatively modulate AKT1 activity (Ren et al., 2013) but AtCBL1/9-CIPK23-AKT1 (K+ transporter) might enhance K+ uptake under low-K+ conditions (Xu et al., 2006; Li et al., 2014). These findings suggested that HsCBL8 may lessen the antagonism of high Na+ to K+ uptake. Nevertheless, the underlying mechanism remains unknown. Unexpectedly, the transcriptional levels of some known downstream factors of CBLs in transgenic rice were not regulated by HsCBL8 (Figure 8), including the Na+ transporters OsNHX1 and OsSOS1. Thus, HsCBL8 might interact with other known or unknown factors for improving salt tolerance, membrane protection (Figures 7B,C), promoting proline accumulation (Figure 7A), and so on.
To confirm the functional attributes of HsCBL8 in plant growth and development, we conducted GUS staining. Staining under normal conditions resulted in the appearance of blue stains in cotyledon, hypocotyl stele, leaves, root stele, and at the junction of the lateral root, sepal, filament, tissues between style and stigma, and tissue between silique and silique petiole (Figures 9A–J). Thus, the localization of HsCBL8 was found similar to that of other CBLs. For instance, the presence of AtCBL3 is in roots, leaves, siliques and seeds (Tang et al., 2012); OsCBL2 is in the aleurone layer induced by GA (Hwang et al., 2005); whereas, AtCBL1, AtCBL2, AtCBL3, and AtCBL9 were involved in seed germination and seedling and pollen development (Pandey et al., 2008; Li D. D. et al., 2013; Mähs et al., 2013; Zhou et al., 2015). Moreover, the overexpression of soybean GmCBL1 promoted hypocotyl elongation under normal light conditions in Arabidopsis (Li Z.-Y. et al., 2013). HsCBL8 may also involve light and GA response cis elements in its promoter sequence (Table 1), but few is known about this aspect and further study is needed.
Some genes from CBL family were considered stress responsive either directly or indirectly, by interacting with other genes (Kim, 2013). Therefore, the expression levels of the genes specifically responsive to salt stress, including OsSOS2/OsCIPK24, OsNHX1, and OsSOS1 (Fukuda et al., 1999), were analyzed. Besides the genes expressed under salt-specific stress, microbe-associated molecular pattern-induced defense genes (OsCIPK15, Kurusu et al., 2010, ABA- and osmotic-stress-responsive genes (Rao et al., 2011), and drought-, high salt- and cold-stress-responsive (OsDREB2A, Dubouzet et al., 2003 genes were also analyzed. As evident in the results (Figure 4), the application of salt stress induced HsCBL8 expression in the wild barley. Notably, the expression of HsCBL8 declined significantly at the first hour in root and increased significantly afterwards. Possibly, it takes time for plants adapting themselves to adverse environments, a process described as “shook” in some literatures (Munns and Tester, 2008; Shavrukov, 2013). The promoter analysis of HsCBL8 revealed the presence of some stress response cis domains (Table 1), including ABA- and drought-response sequences, which was confirmed from the GUS activity analysis, as the promoter was activated by ABA, drought, and salt stress (Figure 9K), indicating the involvement of HsCBL8 in upstream signaling pathway responding to these abiotic stresses.
ABA is widely considered as a stress-response phytohormone. A study on plants with CBL mutation revealed that AtCBL9 was more sensitive to exogenous ABA and could regulate ABA bio-synthesis as the young seedlings of cbl9 mutant accumulated additional ABA (Pandey et al., 2004). However, the plants overexpressing AtCBL5 did not alter their response to ABA (Cheong et al., 2010), indicating that AtCBL5 may regulate downstream genes independent from ABA. Moreover, we reported that AtCIPK15 could coordinate with several CBL proteins responding to ABA, including AtCBL1, 2, 3, 5, 8, and 9 (Batistic et al., 2012; Kim, 2013). These findings could suggest that HsCBL8 might enhance tolerance against salt stress in rice by acting antagonistically to ABA biosynthesis or signaling. These observations were further confirmed when the overexpression of HsCBL8 improved seed germination ratios in rice (Figure 6A). Furthermore, HsCBL8 might regulate GA signaling against ABA.
In conclusion, HsCBL8 derived from Qinghai-Tibetan wild barley was overexpressed in transgenic rice. This gene exhibited a significantly increased tolerance to high salinity through the systemic regulation of seed germination, proline accumulation, plasma membrane protection, and decreased overall Na+ uptake. No defect was observed in the germination, seedling, and reproductive stages of the transgenic plants under salt stress. This finding indicated that H. spontanum from Qinghai-Tibet plateau can provide a significant gene pool for cereal crop improvement (Ellis et al., 2000; Dai et al., 2012). Our study successfully demonstrated the involvement of HsCBL8 in regulating various physiological processes that induce salt stress tolerance in rice. Our study could be used as a basis for further research on the potential functional attributes of HsCBL8 in response to other abiotic stress factors and its interaction with other unknown factors.
Author Contributions
WG, LJ, and GZ conceived and designed the experiment and contributed reagents/materials/analysis tools. WG and TC conducted the experiments. WG, TC and NH analyzed the data. WG, TC, LJ and NH wrote the manuscript. WG, TC, NH, GZ, and LJ reviewed, edited and approved the manuscript.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China [LY13C130001]; the Science Foundation of Zhejiang Sci-Tech University [14042008-Y]; the Zhejiang Provincial Top Key Discipline of Biology [2012A03-C], and Jiangsu Collaborative Innovation Center for Modern Crop Production.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms. Mei Li and Mr. Xianyin Zhang for their technical assistance, and EnPaper company for language improvement.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01678/full#supplementary-material
Table S1. Primers used in this program.
Table S2. Protein sequences of CBLs used in phylogenetic analysis.
Figure S1. Screening of homozygous T2 transgenic plants by PCR using: (A) 679 bp sequence of HsCBL8, and (B) RT-PCR analysis of the HsCBL8 T3 transgenic lines using 575 bp sequence of HsCBL8, and OsGAPDH (BCBI ID: NM_001067432) as reference gene. Six T3 seeds (B) come from T2 line T2-1-3 and T2-3-9 in (A), respectively. M, DNA marker. CK+, pCAMBIA1300-35S-HsCBL8; CK−, ZH11.
References
Ahmadi-Ochtapeh, H., Soltanloo, H., Ramezanpour, S. S., Naghavi, M. R., Nikkhah, H. R., and Yoosefi Rad, S. (2015). QTL mapping for salt tolerance in barley at seedling growth stage. Biol. Plantarum 59, 283–290. doi: 10.1007/s10535-015-0496-z
Ahmed, I. M., Cao, F., Zhang, M., Chen, X., Zhang, G., and Wu, F. (2013). Difference in yield and physiological features in response to drought and salinity combined stress during anthesis in tibetan wild and cultivated barleys. PLoS ONE 8:e77869. doi: 10.1371/journal.pone.0077869
Akaboshi, M., Hashimoto, H., Ishida, H., Saijo, S., Koizumi, N., Sato, M., et al. (2008). The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J. Mol. Biol. 377, 246–257. doi: 10.1016/j.jmb.2008.01.006
Albrecht, V., Ritz, O., Linder, S., Harter, K., and Kudla, J. (2001). The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20, 1051–1063. doi: 10.1093/emboj/20.5.1051
Batistic, O., Rehers, M., Akerman, A., Schlucking, K., Steinhorst, L., Yalovsky, S., et al. (2012). S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 22, 1155–1168. doi: 10.1038/cr.2012.71
Chen, J. H., Sun, Y., Sun, F., Xia, X. L., and Yin, W. L. (2011). Tobacco plants ectopically expressing the Ammopiptanthus mongolicus AmCBL1 gene display enhanced tolerance to multiple abiotic stresses. Plant Growth Regul. 63, 259–269. doi: 10.1007/s10725-010-9523-4
Chen, L., Ren, F., Zhou, L., Wang, Q.-Q., Zhong, H., and Li, X.-B. (2012). The Brassica napus calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 63, 6211–6222. doi: 10.1093/jxb/ers273
Cheong, Y. H., Sung, S. J., Kim, B.-G., Pandey, G. K., Cho, J.-S., Kim, K.-N., et al. (2010). Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol. Cells 29, 159–165. doi: 10.1007/s10059-010-0025-z
Dai, F., Nevo, E., Wu, D., Comadran, J., Zhou, M., Qiu, L., et al. (2012). Tibet is one of the centers of domestication of cultivated barley. Proc. Natl. Acad. Sci. U.S.A. 109, 16969–16973. doi: 10.1073/pnas.1215265109
Das, G., and Rao, G. J. N. (2015). Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 6:698. doi: 10.3389/fpls.2015.00698
Dong, L., Wang, Q., Manik, S. M. N., Song, Y., Shi, S., Su, Y., et al. (2015). Nicotiana sylvestris calcineurin B-like protein NsylCBL10 enhances salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 34, 2053–2063. doi: 10.1007/s00299-015-1851-4
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Ellis, R. P., Forster, B., Robinson, D., Handley, L., Gordon, D., Russell, J., et al. (2000). Wild barley: a source of genes for crop improvement in the 21st century? J. Exp. Bot. 51, 9–17. doi: 10.1093/jexbot/51.342.9
Fukuda, A., Nakamura, A., and Tanaka, Y. (1999). Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim. Biophys. Acta 1446, 149–155. doi: 10.1016/S0167-4781(99)00065-2
Gallagher, S. R. (1992). “Quantitation of GUS activity by fluorometry,” in GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression, ed S. R. Gallagher (New York, NY: Academic Press Inc.), 47–59.
Gao, J.-P., and Lin, H.-X. (2013). “QTL analysis and map-based cloning of salt tolerance gene in rice,” in Rice Protocols, ed Y. Yang (Totowa, NJ: Humana Press), 69–82.
Hu, S., Tao, H., Qian, Q., and Guo, L. (2012). Genetics and molecular breeding for salt-tolerance in rice. Rice Genom. Genet. 3, 39–49. doi: 10.5376/rgg.2012.03.0007
Hwang, Y. S., Bethke, P. C., Cheong, Y. H., Chang, H. S., Zhu, T., and Jones, R. L. (2005). A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiol. 138, 1347–1358. doi: 10.1104/pp.105.062703
Kim, B. G., Waadt, R., Cheong, Y. H., Pandey, G. K., Dominguez-Solis, J. R., Schültke, S., et al. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52, 473–484. doi: 10.1111/j.1365-313X.2007.03249.x
Kim, K.-N. (2013). Stress responses mediated by the CBL calcium sensors in plants. Plant Biotechnol. Rep. 7, 1–8. doi: 10.1007/s11816-012-0228-1
Kleist, T. J., Spencley, A. L., and Luan, S. (2014). Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 5:187. doi: 10.3389/fpls.2014.00187
Kolukisaoglu, U., Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Kurusu, T., Hamada, J., Nokajima, H., Kitagawa, Y., Kiyoduka, M., Takahashi, A., et al. (2010). Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol. 153, 678–692. doi: 10.1104/pp.109.151852
Li, D. D., Xia, X. L., Yin, W. L., and Zhang, H. C. (2013). Two poplar calcineurin B-like proteins confer enhanced tolerance to abiotic stresses in transgenic Arabidopsis thaliana. Biol. Plant. 57, 70–78. doi: 10.1007/s10535-012-0251-7
Li, J., Long, Y., Qi, G.-N., Li, J., Xu, Z.-J., Wu, W.-H., et al. (2014). The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26, 3387–3402. doi: 10.1105/tpc.114.123455
Li, R., Zhang, J., Wu, G., Wang, H., Chen, Y., and Wei, J. (2012). HbCIPK2, a novel CBL-interacting protein kinase from halophyte Hordeum brevisubulatum, confers salt and osmotic stress tolerance. Plant Cell Environ. 35, 1582–1600. doi: 10.1111/j.1365-3040.2012.02511.x
Li, Z.-Y., Xu, Z.-S., Chen, Y., He, G.-Y., Yang, G.-X., Chen, M., et al. (2013). A novel role for Arabidopsis CBL1 in affecting plant responses to glucose and gibberellin during germination and seedling development. PLoS ONE 8:e56412. doi: 10.1371/journal.pone.0056412
Li, Z. Y., Xu, Z. S., He, G. Y., Yang, G. X., Chen, M., Li, L. C., et al. (2012). Overexpression of soybean GmCBL1 enhances abiotic stress tolerance and promotes hypocotyl elongation in Arabidopsis. Biochem. Biophys. Res. Commun. 427, 731–736. doi: 10.1016/j.bbrc.2012.09.128
Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, N. V., Dolstra, O., Malosetti, M., Kilian, B., Graner, A., Visser, R. G. F., et al. (2013). Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 126, 2335–2351. doi: 10.1007/s00122-013-2139-0
Mähs, A., Steinhorst, L., Han, J. P., Shen, L. K., Wang, Y., and Kudla, J. (2013). The calcineurin B-like Ca2+ sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis. Mol. Plant 6, 1149–1162. doi: 10.1093/mp/sst095
Martínez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J.-K., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012. doi: 10.1104/pp.106.092635
Mekawy, A. M., Assaha, D. V., Yahagi, H., Tada, Y., Ueda, A., and Saneoka, H. (2015). Growth, physiological adaptation, and gene expression analysis of two Egyptian rice cultivars under salt stress. Plant Physiol. Biochem. 87, 17–25. doi: 10.1016/j.plaphy.2014.12.007
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Oh, S. J., Kwon, C. W., Choi, D. W., Song, S. I., and Kim, J. K. (2007). Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol. J. 5, 646–656. doi: 10.1111/j.1467-7652.2007.00272.x
Ozawa, K. (2012). “A high-efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.),” in Transgenic Plants: Methods and Protocols, eds J. M. Dunwell and A. C. Wetten (Totowa, NJ: Humana Press), 51–57.
Pandey, G. K., Cheong, Y. H., Kim, K.-N., Grant, J. J., Li, L., Hung, W., et al. (2004). The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16, 1912–1924. doi: 10.1105/tpc.021311
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B.-G., and Luan, S. (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1, 238–248. doi: 10.1093/mp/ssn003
Qiu, Q.-S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19, 1415–1431. doi: 10.1105/tpc.106.042291
Rao, X.-L., Zhang, X.-H., Li, R.-J., Shi, H.-T., and Lu, Y.-T. (2011). A calcium sensor-interacting protein kinase negatively regulates salt stress tolerance in rice (Oryza sativa). Funct. Plant Biol. 38, 441–450. doi: 10.1071/FP10205
Ren, X. L., Qi, G., Feng, H., Zhao, S., Zhao, S., Wang, Y., et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74, 258–266. doi: 10.1111/tpj.12123
Rengasamy, P. (2002). Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: an overview. Aust. J. Exp. Agric. 42, 351–361. doi: 10.1071/EA01111
Rivandi, J., Miyazaki, J., Hrmova, M., Pallotta, M., Tester, M., and Collins, N. C. (2011). A SOS3 homologue maps to HvNax4, a barley locus controlling an environmentally sensitive Na+ exclusion trait. J. Exp. Bot. 62, 1201–1216. doi: 10.1093/jxb/erq346
Romeza, J., and Flowers, T. (2008). Ecology. Crops for a salinized world. Science 322, 1478–1480. doi: 10.1126/science.1168572
Sánchez-Barrena, M. J., Fujii, H., Angulo, I., Martinez-Ripoll, M., Zhu, J. K., and Albert, A. (2007). The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 26, 427–435. doi: 10.1016/j.molcel.2007.04.013
Shavrukov, Y. (2013). Salt stress or salt shock: which genes are we studying? J. Exp. Bot. 64, 119–127. doi: 10.1093/jxb/ers316
Tang, R.-J., Liu, H., Yang, Y., Yang, L., Gao, X.-S., Garcia, V. J., et al. (2012). Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res. 22, 1650–1665. doi: 10.1038/cr.2012.161
Tang, R. J., Yang, Y., Yang, L., Liu, H., Wang, C. T., Yu, M. M., et al. (2014). Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant Cell Environ. 37, 573–588. doi: 10.1111/pce.12178
Thoday-Kennedy, E., Jacobs, A., and Roy, S. (2015). The role of the CBL–CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 76, 3–12. doi: 10.1007/s10725-015-0034-1
Wang, Z., Chen, M., Chen, T., Xuan, L., Li, Z., Du, X., et al. (2014). TRANSPARENT TESTA2 regulates embryonic fatty acid biosynthesis by targeting FUSCA3 during the early developmental stage of Arabidopsis seeds. Plant J. 77, 757–769. doi: 10.1111/tpj.12426
Weinl, S., and Kudla, J. (2009). The CBL–CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 184, 517–528. doi: 10.1111/j.1469-8137.2009.02938.x
Wu, D., Shen, Q., Cai, S., Chen, Z.-H., Dai, F., and Zhang, G. (2013). Ionomic responses and correlations between elements and metabolites under salt stress in wild and cultivated barley. Plant Cell Physiol. 54, 1976–1988. doi: 10.1093/pcp/pct134
Xiang, Y., Huang, Y., and Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144, 1416–1428. doi: 10.1104/pp.107.101295
Xu, J., Li, H.-D., Chen, L.-Q., Wang, Y., Liu, L.-L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Zhang, H., Yang, B., Liu, W.-Z., Li, H., Wang, L., Wang, B., et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 14:8. doi: 10.1186/1471-2229-14-8
Zhang, H., Yin, W., and Xia, X. (2008). Calcineurin B-Like family in Populus: comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul. 56, 129–140. doi: 10.1007/s10725-008-9293-4
Keywords: salt tolerance, HsCBL8, rice, Arabidopsis, Hordeum spontanum
Citation: Guo W, Chen T, Hussain N, Zhang G and Jiang L (2016) Characterization of Salinity Tolerance of Transgenic Rice Lines Harboring HsCBL8 of Wild Barley (Hordeum spontanum) Line from Qinghai-Tibet Plateau. Front. Plant Sci. 7:1678. doi: 10.3389/fpls.2016.01678
Received: 12 August 2016; Accepted: 25 October 2016;
Published: 10 November 2016.
Edited by:
Vasileios Fotopoulos, Cyprus University of Technology, CyprusReviewed by:
Meixue Zhou, University of Tasmania, AustraliaLiezhao Liu, Southwest University, China
Copyright © 2016 Guo, Chen, Hussain, Zhang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lixi Jiang, jianglx@zju.edu.cn
†These authors have contributed equally to this work.
 Wanli Guo
Wanli Guo Tianlong Chen
Tianlong Chen Nazim Hussain
Nazim Hussain Guoping Zhang
Guoping Zhang Lixi Jiang
Lixi Jiang