- Key Laboratory of Biology and Genetic Resources of Tropical Crops, Ministry of Agriculture, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
In plants MADS-box transcription factors (TFs) play important roles in growth and development. However, no plant MADS-box gene has been identified to have a function related to secondary metabolites regulation. Here, a MADS-box TF gene, designated as HbMADS4, was isolated from Hevea brasiliensis by the yeast one-hybrid experiment to screen the latex cDNA library using the promoter of the gene encoding H. brasiliensis small rubber particle protein (HbSRPP) as bait. HbMADS4 was 984-bp containing 633-bp open reading frame encoding a deduced protein of 230 amino acid residues with a typical conserved MADS-box motif at the N terminus. HbMADS4 was preferentially expressed in the latex, but little expression was detected in the leaves, flowers, and roots. Its expression was inducible by methyl jasmonate and ethylene. Furthermore, transient over-expression and over-expression of HbMADS4 in transgenic tobacco plants significantly suppressed the activity of the HbSRP promoter. Altogether, it is proposed that HbMADS4 is a negative regulator of HbSRPP which participates in the biosynthesis of natural rubber.
Introduction
The MADS-box transcription factors (TFs) family is found throughout eukaryotes (Theissen et al., 2000). The MADS-box TF proteins contain a typical conserved MADS-box motif and dimerization domain at the N-terminal. The MADS-box TFs family contains much more number of members in plants than in other eukaryotes (Grimplet et al., 2016). In plants, MADS-box TFs are best known as regulators of reproductive development, such as flowering induction, flower development, or fruit development (Theissen et al., 2000; Parenicová et al., 2003; Adamczyk et al., 2007; Arora et al., 2007; Itkin et al., 2009; Smaczniak et al., 2012a,b; Puig et al., 2013). MADS-box TFs have also been found extensively in vegetative tissues, embryo, root, trichome, etc, suggesting their diverse functions in plant growth and development processes (Alvarez-Buylla et al., 2000; Ku et al., 2008). MADS-box TFs are also involved in biotic or abiotic stress response regulation (Arora et al., 2007; Khong et al., 2015). However, no plant MADS-box gene has been identified to have a function related to secondary metabolites regulation.
Hevea brasiliensis Müll. Arg (rubber tree) is the only plant widely cultivated to produce natural rubber (NR) (van Beilen and Poirier, 2007). NR (a cis 1, 4-polyisoprene) biosynthesis occurs on rubber particles suspended in rubber tree laticifers (d’Auzac et al., 1989). Latex (cytoplasmic content of laticifers) contants 30–50% cis-polyisoprene which is synthetized through the mevalonate pathway (Chow et al., 2007, 2012; Sando et al., 2008). The NR biosynthesis is a typical isoprenoid secondary metabolism which is similar to the isoprenoid biosynthesis of other plants (Chow et al., 2007). NR is generated by three main steps including initiation, elongation and termination (Puskas et al., 2006). The prenyltransferase, rubber elongation factor and small rubber particle protein play important roles during elongation, giving rise to the long chains of cis-polyisoprene (Dennis and Light, 1989; Oh et al., 1999; Asawatreratanakul et al., 2003; Berthelot et al., 2012). H. brasiliensis small rubber particle protein (HbSRPP) is a major latex protein and obviously participates in NR biosynthesis (Oh et al., 1999; Berthelot et al., 2012). In H. brasiliensis, the general metabolic pathway leading to rubber biosynthesis is now clear (Chow et al., 2007; Sando et al., 2008), but the molecular regulation of NR rubber in H. brasiliensis is limited (Wang et al., 2013). Therefore, the identification and functional study of regulation of NR biosynthesis-related gene may elucidate the molecular mechanisms of NR biosynthesis in rubber tree. Here, we report identification and functional analysis of the rubber tree HbMADS4 that encodes a MADS-box TF. HbMADS4 activated the HbSRPP promoter in yeast. Gel shift assay and co-transfection results revealed HbMADS4 and HbSRPP promoter interaction and supression of HbSRPP expression, indicating that HbMADS4 maybe a negative transcription regulator of HbSRPP involved in NR biosynthesis.
Materials and Methods
Plant Material
Hevea brasiliensis cultivar CATAS7-33-97 was planted in the experimental plantation of the Chinese Academy of Tropical Agriculture Sciences (Danzhou Hainan, PR, China). The epicormic shoots were treated by methyl jasmonate (MeJA) and Ethylene (ET) as described previously (Hao and Wu, 2000). Latex, roots, leaves, and flowers were collected and then immediately frozen into liquid nitrogen.
Yeast One-Hybrid
Latex RNA was isolated as described previously (Tang et al., 2007). Latex cDNAs synthesis was performed according to the instructions of cDNA Synthesis Kit (Fermentas). The promoter of HbSRPP was amplified by PCR with the primers (Guo et al., 2014). The bait vector was constructed by inserting the promoter of HbSRPP into pAbAi vector (Clontech) at the site of Hind III/Xho I. Latex cDNAs, the Sma I-linearized pGADT7-Rec prey vector and the bait vector were introduced into the yeast strain Y1HGold (Clontech). The transformed yeast cells were grown on SD/-Leu selective medium containing 450 ng/ml Aureobasidin A (AbA) at 30 for 3 d. The positive colonies were further analyzed by colony PCR and sequence analysis.
To confirm the binding specificity of HbMADS4 with the HbSRPP, HbMADS4 was amplified by PCR from latex cDNA. HbMADS4 was ligated into the pGADT7-Rec vector, generating pGADT7-HbMADS4. pGADT7-HbMADS4 and pHbSRPP-AbAi were co-transformed into the Y1HGold yeast strain. pGADT7-Rec53+p53-AbAi, pAbAi-HbSRPP, pGADT7-HbMADS4 and pGADT7-HbMADS4 + pAbAi were used as control. Transformed yeast cells were cultured on SD/-Leu selective medium containing 450 ng/ml AbA for 3 d at 30°C.
Phylogenetic Analysis
A total of 38 MADS-box IF sequences from other plants were downloaded from NCBI database. The phylogenetic tree was constructed by employing MEGA version 4.0 using the neighbor-joining method with 1000 bootstrap replicates.
Quantitative PCR (qPCR)
Total RNA was isolated as described previously (Tang et al., 2007). First strand cDNA was synthesized using cDNA synthesis kit according to the manufacturer’s instruction (Fermentas). Quantitative PCR (qPCR) was performed according to Wang’s method (Wang et al., 2013). The primer pairs used for the HbMADS4 were 5′-CACAGCTGTATGTACTTACCTATC-3′) and MR (5′-CACAGCTGT ATGTACTTACCTATC-3′). HbACT7 was used as internal control as recommended previously (Li et al., 2014). The details of experimental manipulations and data analysis were as described previously (Wang et al., 2013).
Subcellular Localization
The HbMADS4 ORF was amplified by PCR using the following primer pairs F1 (ACGCC ATGGTATGACAAGGCAGAAAATCCAGA) and R1 (ATTGAGATCTATCTGGGAAGGGTAA CCCCAAT), which contained Nco I/Bgl II site (underlined), respectively. The PCR products were ligated into the pCAMBIA1302 vector at the site of Nco I /Bgl II, generating CaMV35S::HbMADS4-GFP. CaMV35S::HbMADS4-GFP and pCAMBIA1302 were transformed into onion epidermal cells vis Agrobacterium-mediated transformation. The introduced cells were cultured on MS medium in darkness at 25°C for 24 h and then visualized by confocal microscopy.
Recombinant HbMADS4 Protein and Purification of HbMADS4
The coding regions of HbMADS4 was amplified with the following primer pairs: 5′- ACGTG GATCCATGACAAGGCAGAAAATCCAGA 3′ (with the BamHI site underlined) and 5′-AT TGCTCGAGTCAATCTGGGAAGGGTAACCCC-3′ (with the XhoI site underlined). PCR products were ligateded into pET28a (+) vector (Novagen, Madison, WI, USA) at the site of BamHI/XhoI sites. The constructed plasmid was introduced into Escherichia coli strain Rosetta (DE3). Protein biosynthesis was induced with 1.0 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 37°C. After inducting for 4 h, bacteria were harvested and resuspended in the extraction buffer containing 200 mM Tris/HCl pH 8.0, 5 mM 2-mercaptoethanol, 0.1% Triton X-100 and lysozyme (100 μg ml-1), and incubated at 30°C for 30 min. The extract was centrifuged at 12000 g for 30 min at 4°C. The supernatant was precipitated and purified on a HiTrap affinity column (GE Healthcare).
Electrophoretic Mobility Shift Assay (EMSA)
Electrophoretic Mobility Shift Assay was performed using the Electrophoretic Mobility Shift Assay kit (Invitrogen, Carlsbad, CA, USA). The DNA-protein binding reaction was performed by incubating purified HbMADS4 with double-stranded HbSRPP promoter nucleotides at room temperature for 30 min, and then analyzed by polyacrylamide gel electrophoresis. The gel was stained with SYBR Green EMSA stain for visualizing DNA, the same gel was stained with SYPRO Ruby EMSA stain for monitoring protein.
Dual-Luciferase (Dual-LUC) Assay
The assay was performed according to Hellens’s method (Hellens et al., 2005). In brief, the HbSRPP promoter was cloned into pGreen-II vector, generating pGreen-HbSRPP. HbMADS4 was inserted pGreen-II62SK, generating pGreenII62SK-HbMADS4. Two constructs were introduced into Agrobacterium tumefaciens (strain GV3101). The introduced Agrobacterium cells harboring pGreenII-HbSRPP were mixed with the Agrobacterium strains harboring pGreenII62SK-HbMADS4, in a volume ratio of 1:2. The Agrobacterium mixtures were infiltrated into the abaxial side of tobacco leaves. After culturing for 3 days, total protein was extracted from the infected area. The fluorescent values of LUC and REN were detected using the Dual-Luciferase Reporter Assay System (Promega) following the manufacture’s manual. The value of LUC was normalized to that of REN. Three biological repeats were measured.
Plant Transformation and GUS Assay
The promoter of HbSRPP was inserted into the pCAMBIA1381 vector at the site of BamH I /Hind III, generating pHbSRPP::GUS (PH). The construct was introduced into the A. tumefaciens strain GV3103. Tobacco transformation was performed using a leaf disc vis Agrobacterium-mediated method (Horsch et al., 1985). The T0 transgenic plants were selected hygromycin resistance and GUS histochemical staining. The T1 seedings (HP) were grown on MS medium containing hygromycin. For the effector construct, the HbMADS4 cDNA was inserted into the pBI121 vector at a site of Xba I/Sac I, under the control of the CaMV 35S promoter. The effector was transformed into HP by Agrobacterium -mediated method. Co-transformed plants (CTPs) were selected by hygromycin and kanamycin resistance, further tested by RT-PCR. HbMADS4 was amplified with a specific primer pairs P1 (5′-CTGCAACTGGGAAGCTCTTTGAGT-3′) and P2 (5′-GCAGCTCAAGAGAAG GTTGATCT-3′). The NtACT was used as an internal control parallel in the reactions. NtACT was amplified with a specific primers AF (5′-TGTCAGCAACTGGGACGATATGG-3′) and AR (5′- GA GTCATCTTCTCTCTGTTGGC-3′). CTPs were separately assayed for GUS activity by fluorometry as described previously (Jefferson, 1987). Three biological repeats were measured. Protein content was determined using the Bradford protein assay (Bradford, 1976). Data were subjected to ANOVA.
Results
Cloning of HbMADS4
In order to screen novel TFs which regulate HbSRPP, we performed yeast one-hybrid assay with the rubber tree latex cDNA library using the HbSRPP promoter as bait. 48 positive colonies were obtained and sequenced. BLAST analysis showed one cDNA (designated as HbMADS4, GenBank accession No. KX100586) encoding a MADS-box protein. The binding specificity of the MADS-box protein with the HbSRPP promoter was determined by the one-to-one interaction analysis. As shown in Figure 1, only the yeast clones harboring HbMADS4 and pAbAi-HbSRPP or positive control could grow on the SD/-Leu selective medium containing 450 ng/ml AbA, indicating that HbMADS4 bound to the HbSRPP promoter and activated transcription in yeast.
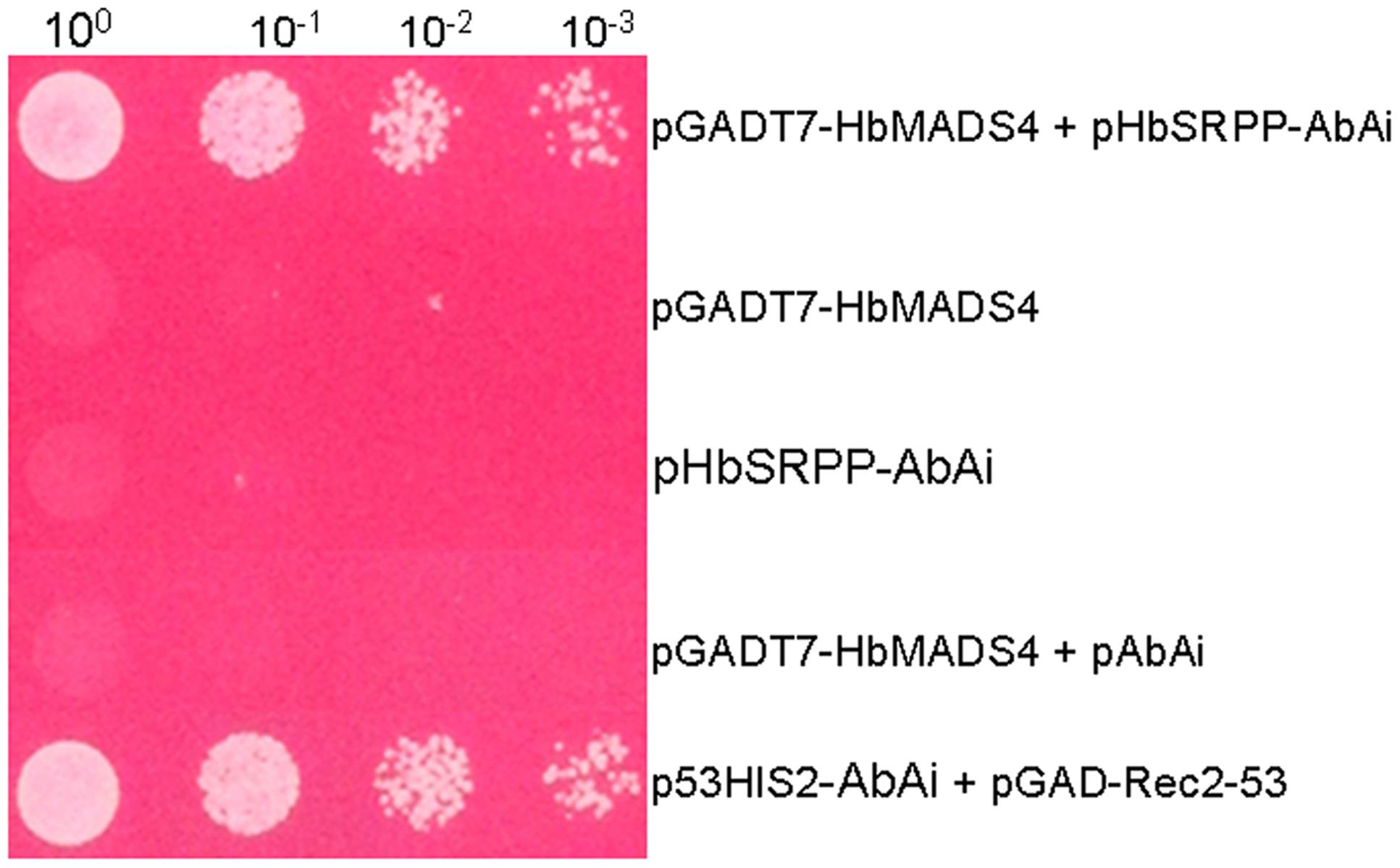
FIGURE 1. Activation of HbSRPP promoter in yeast by HbMADS4. Yeast cells carrying pGADT7- HbMADS4+pHbSRPP-AbAi, pGADT7-Rec53+p53-AbAi, pHbSRPP-AbAi, pGADT7-HbMADS4, and pGADT7-HbMADS4+pAbAi were grown in SD/-Leu selective medium containing 450 ng/ml AbA for 3 days at 30°C.
Molecular Characterization of HbMADS4
HbMADS4 was 984-bp containing 633-bp open reading frame encoding a deduced protein of 230 amino acid residues protein with a predicted molecular mass of 23.71 kD. The deduced HbMADS4 protein contained a MADS-box domain which is typical for the MADS-box TFs (Figure 2). However, the entire HbMADS4 protein sequence has limited identity with other MADS-box protein reported in rubber tree (Figure 2). HbMADS4 should be a new member of the rubber tree MADS-box family. HbMADS4 amino acid sequences and MADS-box from other different species were compared and a phylogenetic tree showed that HbMADS4 is members of the StMADS class subfamily (Figure 3).

FIGURE 2. Alignment of HbMADS4 and other rubber tree MADS-box transcription factors (TFs). The amino acid sequence of HbMADS4 was aligned with HbMADS1 (GenBank accession no. GU142913), HbMADS2 (GU142914), and HbMADS3 (GU142915). Black and gray shadings indicate conserved amino acid residues.
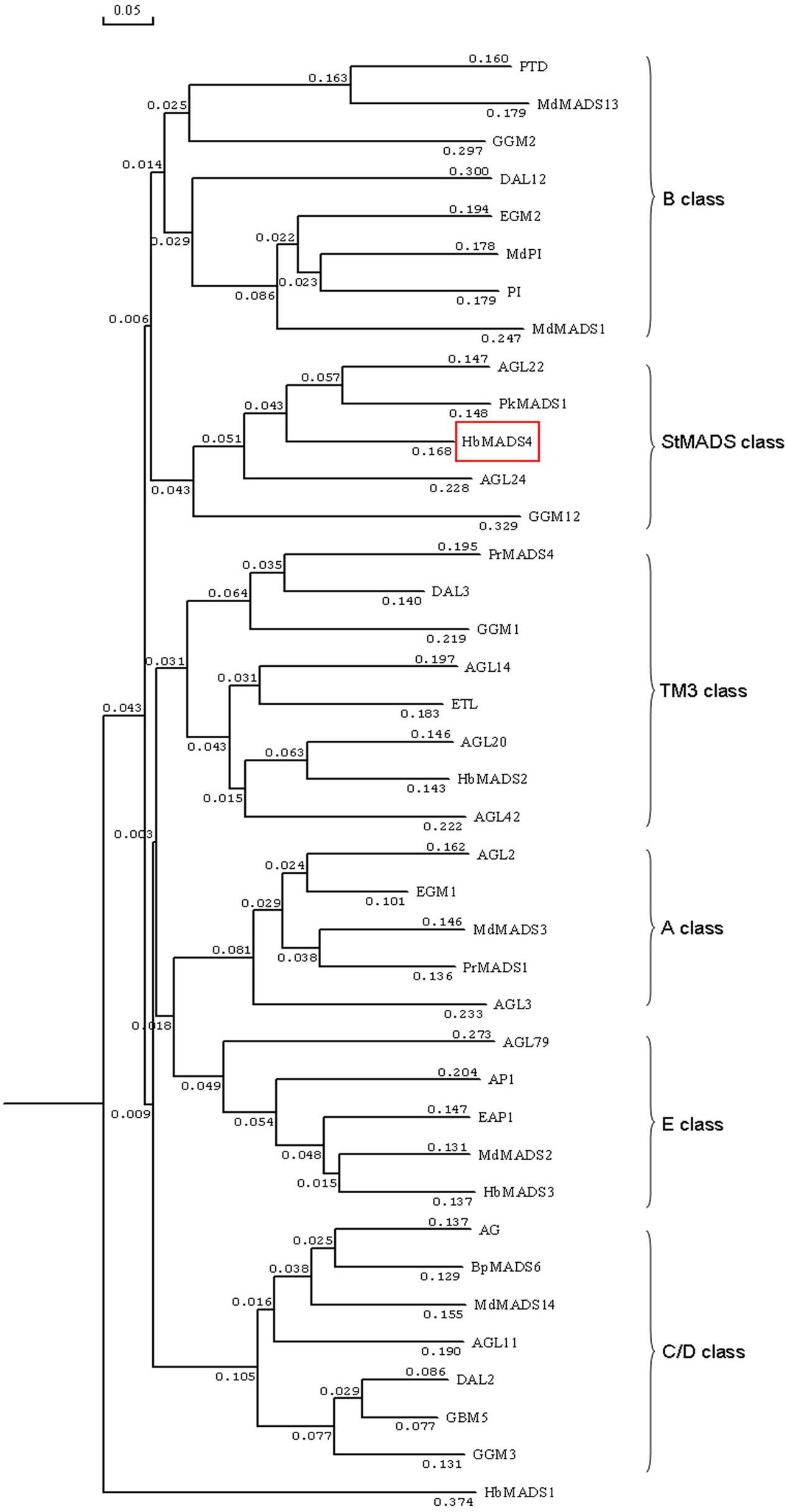
FIGURE 3. Phylogenetic analysis of HbMADS4 with other MADS-box TFs by MEGA version 2.1 from CLUSTAL W alignments. The neighbor-joining method was used to construct the tree. The MADS-box TFs used in the evolutionary analysis are retrieved from Genbank including AG (Arabidopsis thaliana, P17839), AGL2 (A. thaliana, P29382), AGL3 (A. thaliana, 30678072), AGL11 (A. thaliana, GI12229648), AGL14 (A. thaliana, Q38838), AGL20 (A. thaliana, O64645), AGL22 (A. thaliana, Q9FVC1), AGL24 (A. thaliana,CAB79364), AGL42(A. thaliana AAN52777), AGL79 (A. thaliana, AAN52802), AP1 (A. thaliana, S27109), BpMADS6 (Betula pendula, CAA67968), DAL2 (Picea abies, S51934), DAL3 (P. abies, S51936), DAL12 (P. abies, AAF18375), EAP1 (Eucalyptus globules, AAG24909), EGM1(E. grandis, AAC78282), EGM2(E. grandis, AF029976), ETL(E. globulus, AAD16052), GBM5 (Ginkgo biloba, AAM76208), GGM1 (Gnetum gnemon, CAB44447), GGM2 (G. gnemon, CAB44448), GGM3 (G. gnemon, CAB44449), GGM12 (G. gnemon, CAB44458), HbMADS1 (Hevea brasiliensis,GU142913), HbMADS2 (H. brasiliensis, GU142914), HbMADS3 (H. brasiliensis,GU142915), HbMADS4 (H. brasiliensis, KX100586), MdMADS1 (Malus domestica, AAC25922), MdMADS2 (M. domestica, AAC83170), MdMADS3 (M. domestica, AAD51422), MdMADS13 (M. domestica, CAC80856), MdMADS14 (M. domestica, CAC80857), MdPI (M. domestica CAC28021), PkMADS1 (Paulownia kawakamii, AAF22455), PI (A. thaliana, NP_197524), PTD (Populus trichocarpa, AAC13695), PrMADS1 (Pinus radiata, T09569), and PrMADS4 (P. radiate, AAB80807).
Analysis of HbMADS4 Expressions
The HbMADS4 transcript levels were detected in different rubber tree tissues by qPCR. qPCR detected a signification level of HbMADS4 in the latex, and the level was low in the leaves, flowers, and roots (Figure 4A). Signaling pathways, especially those of MeJA and ET are actively implicated in the regulation of latex regeneration (Zhu and Zhang, 2009; Duan et al., 2010; Tang et al., 2013). To examine the expression patterns of the HbMADS4 in response to MeJA and ET, the rubber tree shoots were treated by MeJA or ET, respectively. As shown in Figure 4B, JA markedly up-regulated HbMADS4 expression. The expression of HbMADS4 increased and reached the highest level at 9 h, and then decreased at 24 h. The expression of HbMADS4 was also induced by ET. The expression of HbMADS4 began to increase at 6 h and reached the highest level at 24 h, and then remarkably decreased at 48 h. (Figure 4B).
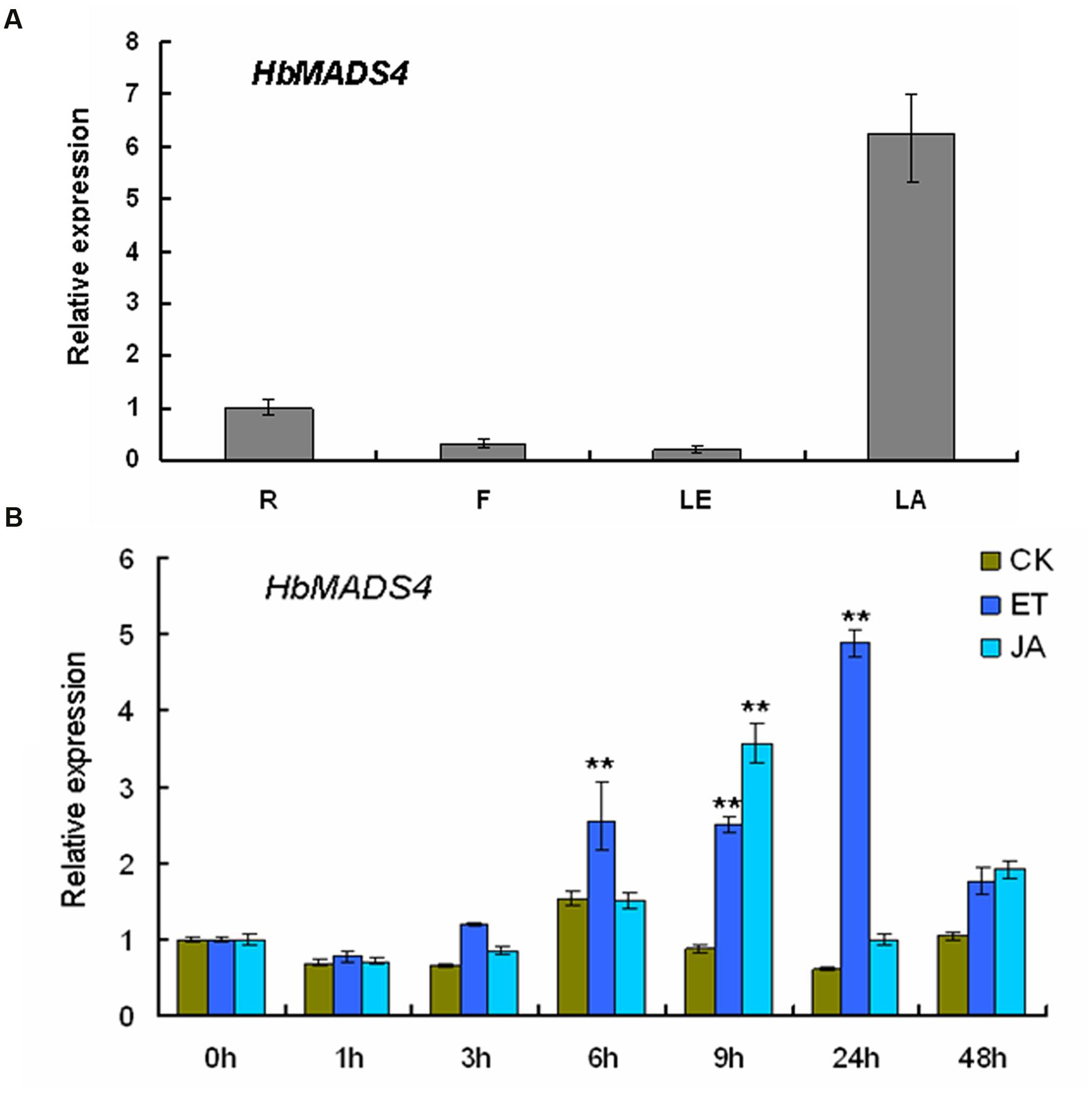
FIGURE 4. Transcription patterns of HbMADS4. (A) Differential expression of HbMADS4 in different rubber tree. R, roots; B, bark; LE, leaves; LA, latex. (B) Expression patterns of HbMADS4 respond to JA and ET treatment. The y axis is the scale of the relative transcript abundance level. The x axis is the time course of JA and ET treatment. An average of three independent biological replicates of each time was performed. Data are presented as mean ± SE (n = 3). The significant difference was assessed by ANOVA (two asterisks corresponding to P < 0.01).
Subcellular Localization of HbMADS4
To study the subcelluar location of the HbMADS4 protein, the green fluorescent protein (GFP) was used as a reporter. GFP was fuse in frame to 5′ end of the ORF of HbMADS4 in the pCAMBIA1302, resulting construct GFP-HbMADS4. GFP-HbMADS4 or pCAMBIA1302 was introduced into onion epidermal cells. As shown in Figure 5, GFP-HbMADS4 fusion protein was located in the nuclei of onion epidermal cell. In contrast, in the control GFP fluorescence was observed throughout the onion epidermal cell. These results indicate that HbMADS4 is a nuclear-localized protein.
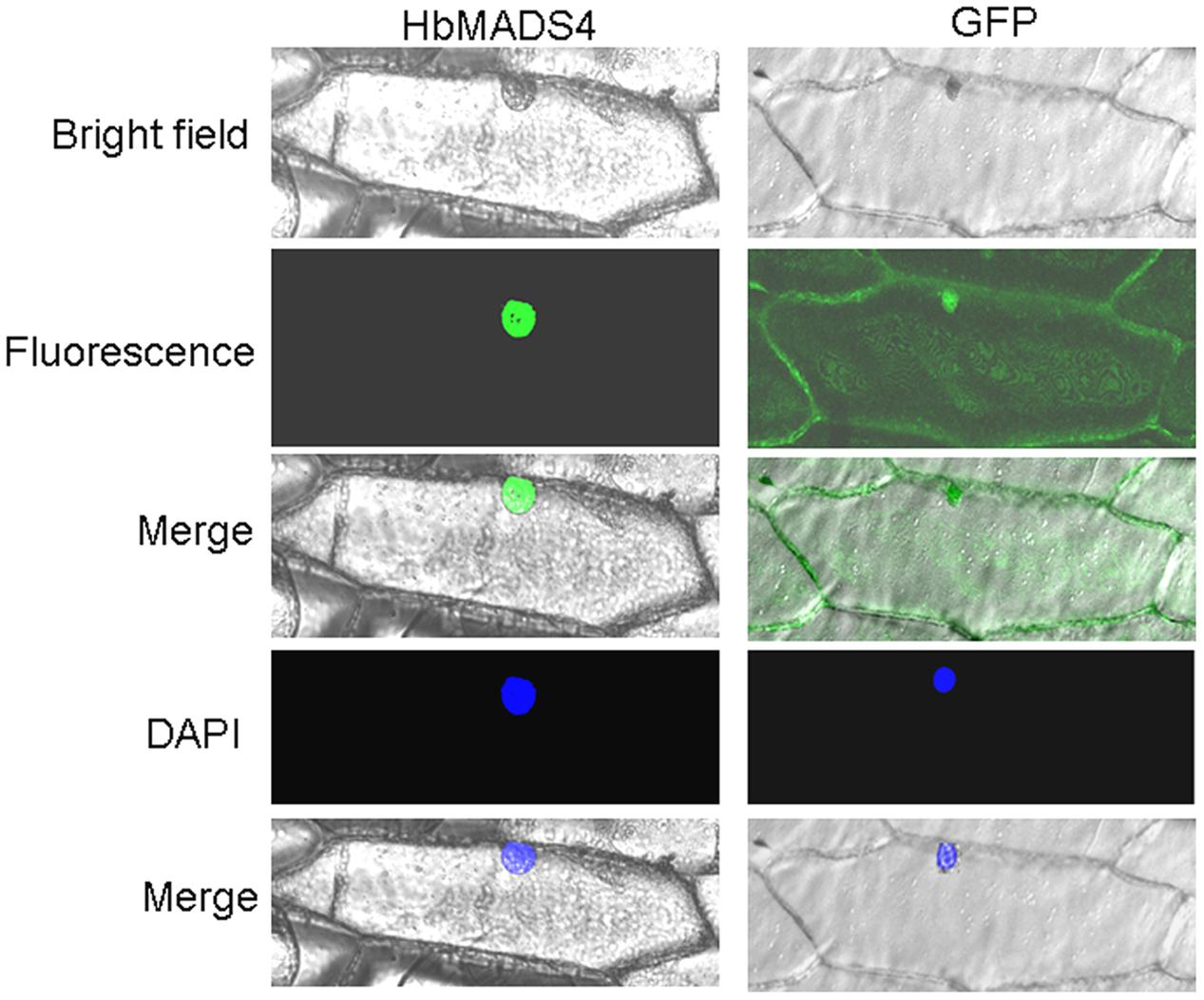
FIGURE 5. Nuclear localization of HbMADS4. The right panel, the corresponding fluorescence, bright field, merged fluorescence image, and DAPI image of GFP control; the left panel, the corresponding fluorescence, bright field, merged fluorescence image, and DAPI image of HbMADS4-GFP.
HbMADS4 Can Interact with the HbSRPP Promoter In Vitro
To investigate whether HbMADS4 binds to the HbSRPP promoter in vitro, we performed EMSA using recombinant HbMADS4 protein (Figure 6A) and 1050-bp HbSRPP promoter nucleotide probes. As shown in Figure 6B,C, HbMADS4 was able to bind with HbSRPP promoter and cause mobility shifts. Moreover the quantity of shifted protein-DNA complexes increased gradually, and the quantity of free DNA levels decreased accordingly (Figure 6B, lanes 2–6). The results showed that in vitro HbMADS4 was able to recognize and interact with the HbSRPP promoter.
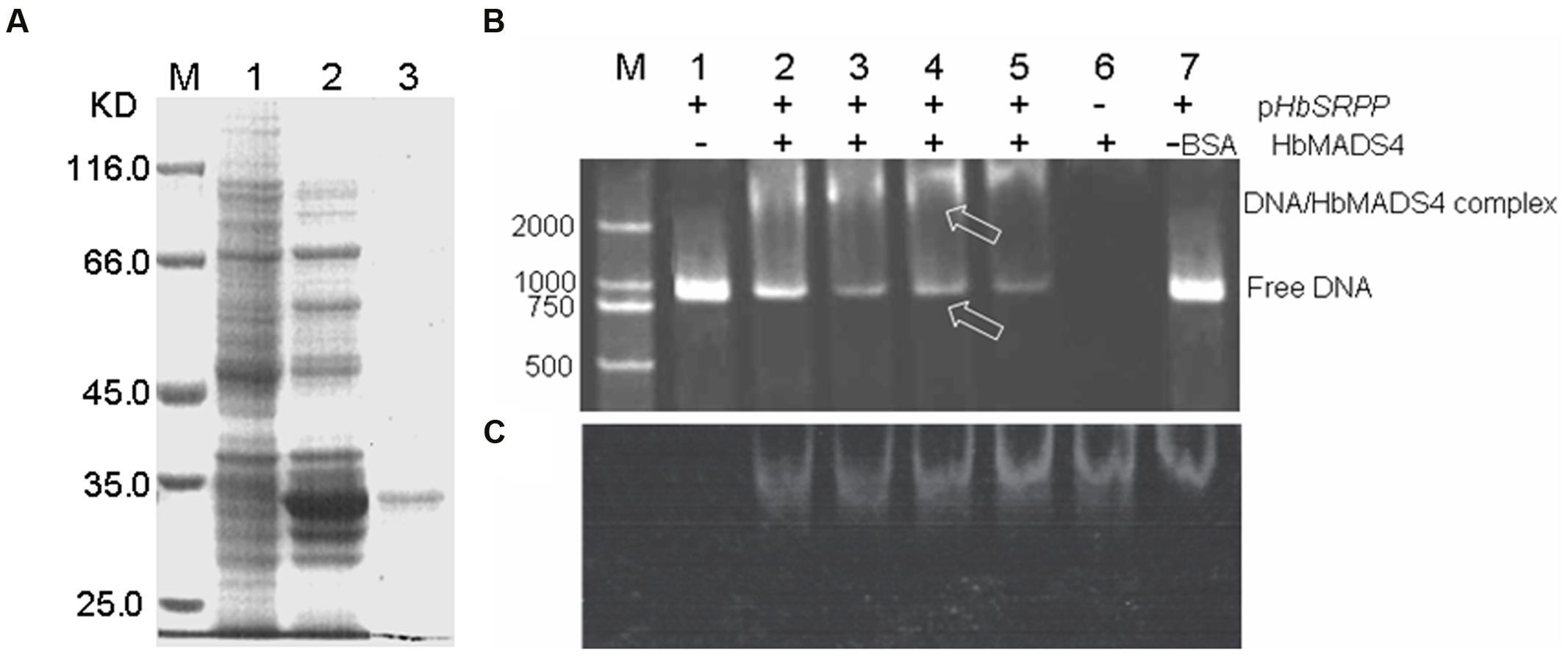
FIGURE 6. HbMADS4 binding to the promoter of HbSRPP as analyzed by EMSA. (A) Over-expression of HbMADS4 in Escherichia. coli. Expression vector (pET- HbMADS4) was constructed in which the fusion protein was driven by the T7 promoter, made IPTG-inducible, and transformed into E. coli BL21 (DE3). Expression was induced by the addition of 0.2 mM IPTG, and total cell proteins were analyzed after 5 h by SDSPAGE. M molecular markers; 1. E. coli cells harboring pET-HbMADS4 not induced; 2, E. coli cells harboring pET-HbMADS4 after 5 h of induction; 3. the purified HbMADS4 fusion protein by E. coli cells harboring pET-HbMADS4 after 5 h of induction. (B) The HbSRPP promoter with HbMADS4 protein stained with SYBR green EMA for visualizing DNA. (C) The same gel as in (B) stained with SYPRO Ruby EMSA for visualizing protein. M DNA maker (DL2000). Lane 1. the promoter of HbSRPP DNA only; Lanes. 2–5 the promoter of HbSRPP DNA with increasing amounts of HbMADS4 protein (50, 100, 150, and 200 ng); Lane 6. 200 ng HbMADS4 protein only; Lane 7. 200 ng of bovine serum albumin (BSA) and the promoter of HbSRPP DNA, respectively. The arrows indicated the HbMADS4-DNA complex or free DNA.
Suppression of the HbSRPP Promoter by HbMADS4
Given the fact that in vitro HbMADS4 was able to interact with the HbSRPP promoter, we asked if HbMADS4 can regulate the HbSRPP promoter in plant. A dual-luciferase assay system was employed for this purpose (Figure 7A). The level of the luciferase activity controlled by HbMADS4 and HbSRPP promoters was elevated (Figure 7B), the expression of HbMADS4 resulted more than 6 folds decrease of the luciferase activity. The result shows that transient over-expression of HbMADS4 suppressed the HbSRPP promoter.
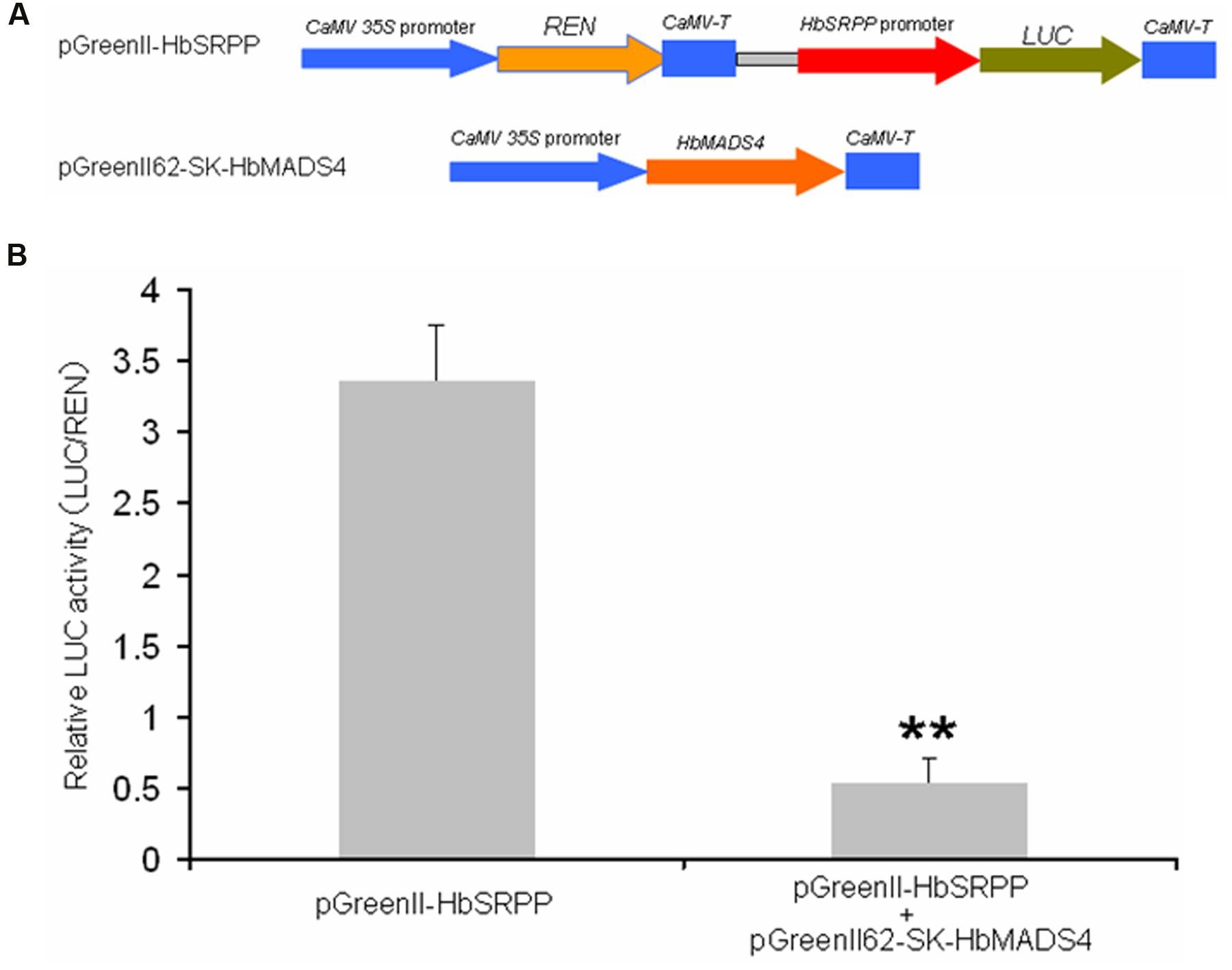
FIGURE 7. Repression of HbSRPP promoter in transient expression system by HbMADS4. (A) Schematic diagrams of the transient expression vectors used in the transient expression analysis. (B) Relative LUC activity from transient expression analysis of HbSRPP promoter co-infiltrated with a plasmid containing gene for HbMADS4 fused to the 35S promoter.
It is very difficult to obtain transgenics in rubber tree. To further test whether HbMADS4 can regulate the HbSRPP promoter, tobacco plants were co-transformed with the vector carrying the HbMADS4 cDNA fused to the CaM 35S promoter, and a PH construct (Figure 8A). Plants transformed with PH and CTPs with PH and CaMV35S::HbMADS4 were selected by hygromycin and kanamycin resistance, CTPs were further detected by RT-PCR (Figure 8B) and GUS histochemical staining (Figure 8C). Over-expression of HbMADS4 suppressed the HbSRPP promoter in the CTPs compared with in the PH. As shown in Figure 8D, in CTPs (i.e., CTP1, 5, 8, 9, 18, 19, 20, and 21), expression of HbMADS4 resulted in a more than 4 folds decrease of HbSRPP promoter activity. The results show that the over-expression of HbMADS4 strongly suppressed the HbSRPP promoter.
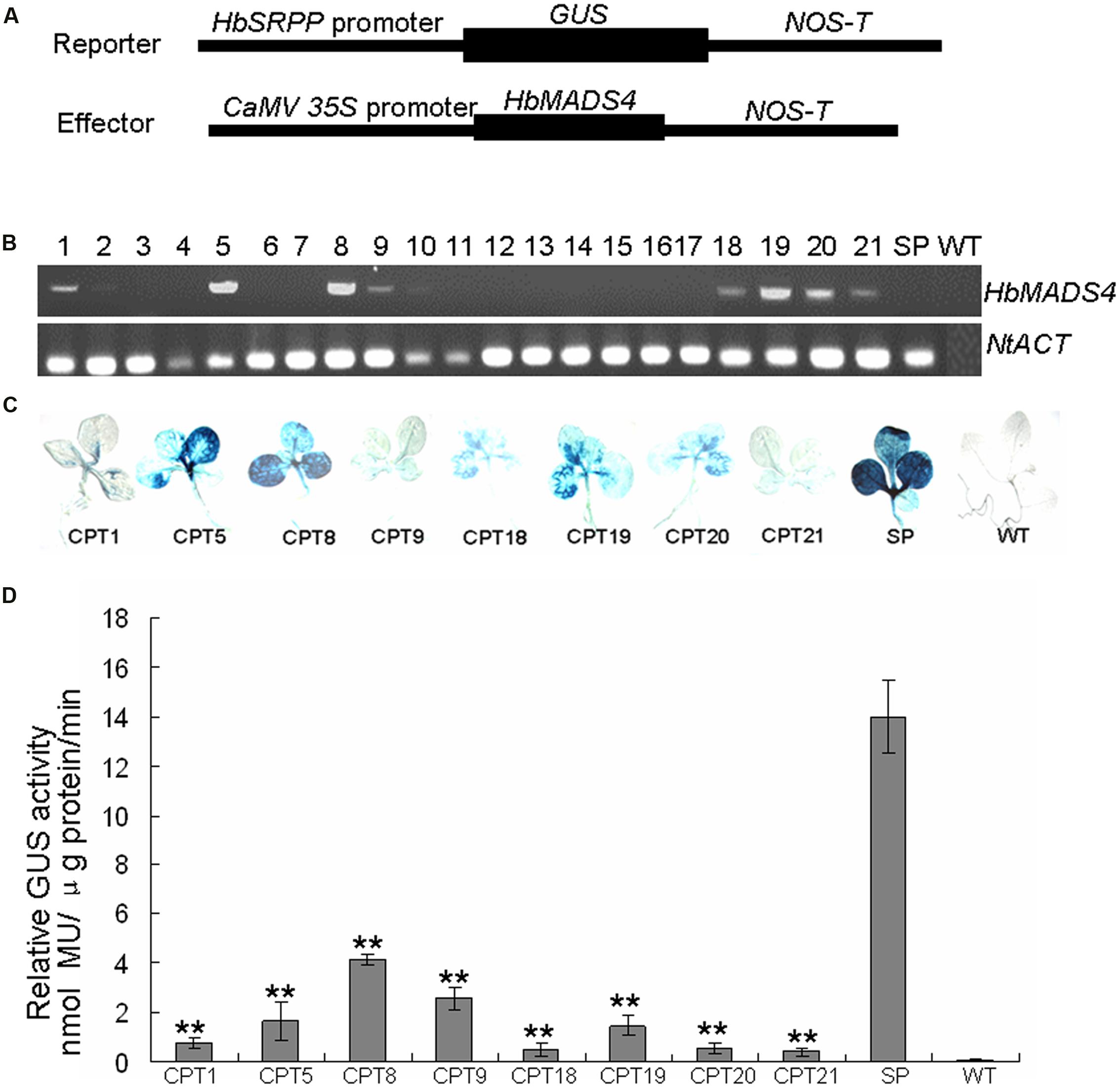
FIGURE 8. Repression of HbSRPP promoter in transgenic plants by HbMADS4. (A) Schematic diagrams of the reporter and effector constructs used in the experiment. (B) The co-transformed plants (CTPs) were examined by RT-PCR. (C) Histochemical staining analysis of the different CTPs (i.e., CPT1, 5, 8, 9, 18, 19, 20, and 24), wild-type (WT) and plant transformed with pHbSRPP::GUS (PH) were used as controls. (D) Quantitative determination of the GUS activities in the different cCTPs (i.e., CPT1, 5, 8, 9, 18, 19, 20, and 24), WT and PH were used as controls. Three replicates were included for each sample. Data are presented as mean ± SE (n = 3), and the significant difference was assessed by ANOVA (two asterisks corresponding to P < 0.01).
Discussion
MADS-box genes play important roles during plant growth and development, especially floral organ and fruit development. To date, many plant MADS-box genes have been identified in controlling reproductive development processes (Smaczniak et al., 2012a,b). Three MADS-box genes had been isolated from H. brasiliensis. The three genes were differentially expressed during somatic embryogenesis of rubber tree (Li et al., 2011). In this study, HbMADS4 was isolated from H. brasiliensis by the yeast one-hybrid experiment to screen the latex cDNA library using the HbSRPP promoteras bait. HbMADS4 was preferentially expressed in latex, but little in leaves, roots and flower. Laticifers are of importance in NR production since within them NR is formed and stored. Genes highly expressed in the laticifers maybe involved in rubber synthesis (Oh et al., 1999; Han et al., 2000). HbMADS4 was preferentially expressed in latex implies that active expression of HbMADS4 might play important role in rubber synthesis.
HbSRPP is a major rubber particles protein of 22.4 kD in the latex (Yeang et al., 1996; Oh et al., 1999). HbSRPP has been suggested to be tightly associated with small rubber particles and also involved in rubber biosynthesis (Berthelot et al., 2012). HbSRPP was susceptible to play a role in latex coagulation (Wititsuwannakul et al., 2008). HbSRPP is highly expressed in the laticifers (Han et al., 2000; Ko et al., 2003) and is regulated by MeJA, ABA, GA, cold, heat, and wounding (Guo et al., 2014). HbWRKY1, a WRKY TF, was found to negatively regulate HbSRPP expression (Wang et al., 2013). WRKY TFs specifically bind to the W-box (T)TGAC(C/T), which contains the invariant TGAC core (Ciolkowski et al., 2008). There are three W-boxes at position +60, +84, and +487 in the HbSRPP promoter region. The MADS TFs binds to the CArG-box (West et al., 1998). There are two CArG-boxes at position +502 and +702 in the HbSRPP promoter region. HbMADS4 interacts with the HbSRPP promoter in vitro and suppress the activity of the HbSRPP promoter in transgenic tobacco. The results strongly indicate that HbMADS4 is a transcriptional suppressor of HbSRPP. It will be of great interest to elucidate HbSRPP is regulated by MADS-box TFs in rubber tree.
Conclusion
A novel MADS-box TF gene, designated as HbMADS4, was isolated from H. brasiliensis HbMADS4 was preferentially expressed in the latex, but little expression was detected in the leaves, flowers, and roots. Its expression was inducible by MeJA and ethylene. HbMADS4 bound to the HbSRPP promoter. Over-expression of HbMADS4 in transgenic tobacco plants significantly suppressed the activity of the HbSRP promoter. Altogether, it is proposed that HbMADS4 is a negative regulator of HbSRPP which participating in the biosynthesis of NR.
Author Contributions
H-LL and S-QP designed the research, L-RW, H-LL, DG, YW, J-HZ, and X-TC performed the research, and HL-L and S-QP wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This research was supported by National Natural Science Foundation of China (No. 31170634, 31670611).
References
Adamczyk, B. J., Lehti-Shiu, M. D., and Fernandez, D. E. (2007). The MADS domainfactors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 50, 1007–10019. doi: 10.1111/j.1365-313X.2007.03105.x
Alvarez-Buylla, E. R., Liljegren, S. J., Pe’laz, S., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466. doi: 10.1111/j.1365-313X.2000.00891.x
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Asawatreratanakul, K., Zhang, Y. W., Wititsuwannaku, D., Wititsuwannaku, R., Takahashi, S., Rattanapittayaporn, A., et al. (2003). Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis. Eur. J. Biochem. 270, 4671–4680. doi: 10.1046/j.1432-1033.2003.03863.x
Berthelot, K., Lecomte, S., Estevez, Y., Coulary-Salin, B., Bentaleb, A., Cullin, C., et al. (2012). Rubber elongation factor (REF), a major allergen component in Hevea brasiliensis latex has amyloid properties. PLoS ONE 7:48065. doi: 10.1371/journal.pone.0048065
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chow, K. S., Mat-Isa, M. N., Bahari, A., Ghazali, A. K., Alias, H., Mohd-Zainuddin, Z., et al. (2012). Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J. Exp. Bot. 63, 1863–1871. doi: 10.1093/jxb/err363
Chow, K. S., Wan, K. L., Isa, M. N., Bahari, A., Tan, S. H., Harikrishna, K., et al. (2007). Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J. Exp. Bot. 58, 2429–2440. doi: 10.1093/jxb/erm093
Ciolkowski, I., Wanke, D., Birkenbihl, R., and Somssich, I. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
d’Auzac, J., Jacob, J. L., and Chrestin, H. (1989). “The composition of latex from Hevea brasiliensis as laticiferous cytoplasm ,” in Physiology of the Rubber Tree Latex, eds J. d’Auzac and J. L. Jacob (Boca Raton, FL: CRC Press), 35–42.
Dennis, M. S., and Light, D. R. (1989). Rubber elongation factor from Hevea brasiliensis. Identification, characterization and role in rubber biosynthesis. J. Biol. Chem. 264, 8608–8617.
Duan, C., Rio, M., Leclercq, J., Bonnot, F., Oliver, G., and Montoro, P. (2010). Gene expression pattern in response to wounding, methyljasmonate and ethylene in the bark of Hevea brasiliensis. Tree Physiol. 30, 1349–1359. doi: 10.1093/treephys/tpq066
Grimplet, J., Martínez-Zapater, J. M., and Carmona, M. J. (2016). Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genomics 17:80. doi: 10.1186/s12864-016-2398-7
Guo, D., Li, H. L., Tang, X., and Peng, S. Q. (2014). Molecular and functional characterization of the HbSRPP promoter in response to hormones and abiotic stresses. Transgenic Res. 23, 331–340. doi: 10.1007/s11248-013-9753-0
Han, K. H., Shin, D. H., Yang, J., Kim, I. J., Oh, S. K., and Chow, K. S. (2000). Genes expressed in the latex of Hevea brasiliensis. Tree Physiol. 20, 503–510. doi: 10.1093/treephys/20.8.503
Hao, B. Z., and Wu, J. L. (2000). Laticifer differentiation in Hevea brasiliensis: induction byexogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43. doi: 10.1006/anbo.1999.0995
Hellens, R. G., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. doi: 10.1186/1746-4811-1-13
Horsch, R., Fry, J. E., Hovmann, N., Eichholtz, D., Rogers, S., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Itkin, M., Seybold, H., Breitel, D., Rogachev, I., Meir, S., and Aharoni, A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60, 1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 242–250. doi: 10.1007/BF02667740
Khong, G. N., Pati, P. K., Richaud, F., Parizot, B., Bidzinski, P., Mai, C. D., et al. (2015). OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol. 169, 2935–2949. doi: 10.1104/pp.15.01192
Ko, J. H., Chow, K. S., and Han, K. H. (2003). Transcriptome analysis reveals novel features of the molecular events occurring in the laticifers of Hevea brasiliensis (para rubbertree). Plant Mol. Biol. 5, 479–492. doi: 10.1023/B:PLAN.0000019119.66643.5d
Ku, A. M., Huang, Y. S., Wang, Y. S., Ma, D., and Yeh, K. W. (2008). IbMADS1 (Ipomoea batatas MADS-box 1 gene) is involved in tuberous root initiation in sweet potato (Ipomoea batatas). Ann. Bot. 102, 57–67. doi: 10.1093/aob/mcn067
Li, H. H., Wang, Y., Guo, D., Tian, W. M., and Peng, S. Q. (2011). Three MADS-box genes of Hevea brasiliensis expressed during somatic embryogenesis and in the laticifer cells. Mol. Biol. Rep. 38, 4045–4052. doi: 10.1007/s11033-010-0523-2
Li, H. L., Guo, D., Yang, Z. P., Tang, X., and Peng, S. Q. (2014). Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomic 104, 14–23. doi: 10.1016/j.ygeno.2014.04.004
Oh, S. K., Kang, H., Shin, D. H., Yang, J., Chow, K. S., Yeang, H. Y., et al. (1999). Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J. Biol. Chem. 274, 17132–17138. doi: 10.1074/jbc.274.24.17132
Parenicová, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Puig, J., Meynard, D., Khong, G. N., Pauluzzi, G., Guiderdoni, E., and Gantet, P. (2013). Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr. Patterns 13, 160–170. doi: 10.1016/j.gep.2013.02.004
Puskas, J. E., Gautriaud, E., Deffieux, A., and Kennedy, J. P. (2006). Natural rubber biosynthesis-a living carbocationic polymerization. Prog. Polym. Sci. 31, 533–548. doi: 10.1016/j.progpolymsci.2006.05.002
Sando, T., Takaoka, C., Mukai, Y., Yamashita, A., Hattori, M., Ogasawara, N., et al. (2008). Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 72, 2049–2060. doi: 10.1271/bbb.80165
Smaczniak, C., Immink, R. G., Angenent, G. C., and Kaufmann, K. (2012a). Developmental and evolutionary diversity of plant MADS domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Smaczniak, C., Immink, R. G. H., Muiño, J. M., Blanvillain, R., Busscher, M., Busscher-Lange, J., et al. (2012b). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. U.S.A. 109, 1560–1565. doi: 10.1073/pnas.1112871109
Tang, C., Qi, G., Li, H., Zhang, C., and Wang, Y. (2007). A convenient and efficient protocol for isolating high-quality RNA from latex of Hevea brasiliensis (para rubber tree). J. Biochem. Biophys. Methods 70, 749–754. doi: 10.1016/j.jbbm.2007.04.002
Tang, C., Xiao, X., Li, H., Fan, Y., Yang, J., Qi, J., et al. (2013). Comparative analysis of latex transcriptome reveals putative molecular mechanisms underlying super productivity of Hevea brasiliensis. PLoS ONE 8:e75307. doi: 10.1371/journal.pone.0075307
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Münster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
van Beilen, J. B., and Poirier, Y. (2007). Establishment of new crops for the production of natural rubber. Trends Biotechnol. 25, 522–529. doi: 10.1016/j.tibtech.2007.08.009
Wang, Y., Guo, D., Li, H. L., and Peng, S. Q. (2013). Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP. Plant Physiol. Biochem. 71, 283–289. doi: 10.1016/j.plaphy.2013.07.020
West, A. G., Sharrocks, A. D., Causier, B. E., and Davies, B. (1998). DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res. 26, 5277–5287. doi: 10.1093/nar/26.23.5277
Wititsuwannakul, R., Rukseree, K., Kanokwiroon, K., and Wititsuwannakul, D. (2008). A rubber particle protein specific for Hevea latex lectin binding involved in latex coagulation. Phytochemistry 69, 1111–1118. doi: 10.1016/j.phytochem.2007.12.007
Yeang, H. Y., Cheong, K. F., Sunderasan, E., Hamzah, S., Chew, N. P., Hamid, S., et al. (1996). The 14.6kd rubber elongation factor (Hevb1) and 24 kd(Hevb3)rubber particle proteins are recognized by IgE from patients with spina bifida and latex allergy. J. Allergy Clin. Immunol. 98, 628–639. doi: 10.1016/S0091-6749(96)70097-0
Keywords: Hevea brasiliensis, small rubber particle protein, promoter, MADS-box transcription factor, negative regulator, natural rubber
Citation: Li H-L, Wei L-R, Guo D, Wang Y, Zhu J-H, Chen X- T and Peng S-Q (2016) HbMADS4, a MADS-box Transcription Factor from Hevea brasiliensis, Negatively Regulates HbSRPP. Front. Plant Sci. 7:1709. doi: 10.3389/fpls.2016.01709
Received: 12 July 2016; Accepted: 31 October 2016;
Published: 15 November 2016.
Edited by:
Diego Rubiales, Spanish National Research Council, SpainReviewed by:
Rosa Rao, University of Naples Federico II, ItalyFlavia Vischi Winck, University of São Paulo, Brazil
Copyright © 2016 Li, Wei, Guo, Wang, Zhu, Chen and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Qing Peng, shqpeng@163.com
†These authors have contributed equally to this work.
 Hui-Liang Li†
Hui-Liang Li† Shi-Qing Peng
Shi-Qing Peng