- 1Laboratory of Plant Functional Genomics, School of Applied Biosciences, Kyungpook National University, Daegu, South Korea
- 2Department of Agriculture, Abdul Wali Khan University, Mardan, Pakistan
- 3Laboratory of Plant Molecular Breeding, School of Applied Biosciences, Kyungpook National University, Daegu, South Korea
Exposure of plants to different biotic and abiotic stress condition instigates significant change in the cellular redox status; resulting in the elevation of reactive nitrogen species that play signaling role in mediating defense responses. Heavy metal associated (HMA) domain containing genes are required for spatio-temporal transportation of metal ions that bind with various enzymes and co-factors within the cell. To uncover the underlying mechanisms mediated by AtHMA genes, we identified 14 Arabidopsis HMA genes that were differentially expressed in response to nitrosative stress through RNA-seq analysis. Of those 14 genes, the expression of eight HMA genes was significantly increased, whereas that of six genes was significantly reduced. We further validated the RNA-seq results through quantitative real-time PCR analysis. Gene ontology analysis revealed the involvement of these genes in biological processes such as hemostasis and transport. The majority of these nitric oxide (NO)-responsive AtHMA gene products are carrier/transport proteins. AtHMAD1 (At1g51090) showed the highest fold change to S-nitrosocystein. We therefore, further investigated its role in oxidative and nitrosative mediated stress conditions and found that AtHMAD1 has antagonistic role in shoot and root growth. Characterization of AtHMAD1 through functional genomics showed that the knock out mutant athmad1 plants were resistant to virulent Pseudomonas syringae (DC3000) and showed early induction and high transcript accumulation of pathogenesis related gene. Furthermore, inoculation of athamd1 with avirulent strain of the same bacteria showed negative regulation of R-gene mediated resistance. These results were supported by hypersensitive cell death response and cell death induced electrolyte leakage. AtHMAD1 was also observed to negatively regulate systemic acquired resistance SAR as the KO mutant showed induction of SAR marker genes. Overall, these results imply that NO-responsive AtHMA domain containing genes may play an important role in plant development and immunity.
Introduction
Nitric oxide (NO), one of the smallest diatomic molecules is a highly reactive gaseous free radical and is the center of attention for most of the plant scientists due to its marvelous regulatory role in both plants and animals (Tuteja et al., 2004). Its high diffusivity (4.8 × 10-5 cm2 S-1 in H2O) allow it to move easily in various parts of the cell like cytoplasm and lipid layer of membranes (Arasimowicz and Floryszak-Wieczorek, 2007). A number of functions for NO in plants have been reported so far. This includes its regulatory role during salinity (Valderrama et al., 2007), drought (Zhao et al., 2004) heat (Rizhsky et al., 2004), cold (Beligni and Lamattina, 1999), and UV-B radiation (Shi et al., 2005). Due to its contribution NO was named as “Molecule of the Year” by the journal Science (Culotta and Koshland, 1992). Plants continuously interact with the external environment (biotic and/or abiotic) for multiple purposes. This interaction may range from positive such as with pollinators to negative such as with pathogens and herbivores (van Dam, 2009). Two major players of the negative interaction, biotic and abiotic stresses are the prime challenges for plants. These stresses lead to the production of increased reactive oxygen species (ROS) such as hydrogen peroxide, superoxide and hydroxyl radicals in the cell (Sudhakar et al., 2001; Xiong et al., 2002). Another type of important molecules produced during stress responses are reactive nitrogen species (RNS) (Delledonne et al., 1998) that has been shown to regulate a plethora of physiological activities and plant functions including plant defense and development (Delledonne et al., 1998; Durner et al., 1998). Most of these are formed as chemical derivatives of NO, primarily because NO itself is histologically toxic, short lived, and almost instantly reacts with other molecules. S-nitrosocysteine (CysNO) and S-nitrosoglutathione (GSNO) are considered important NO donors as they spontaneously decompose in aqueous solutions to release NO (Uehara et al., 2006; Cho et al., 2009). CysNO infiltrates into the plant cells via L-type amino acid transporters (Latson, 2011) resulting in S-nitrosylation of cellular proteins (Satoh et al., 1997; Zhu et al., 2008). The cell surface associated protein disulfide isomerase has been reported to aid in the incorporation of NO or nitroso moieties into the cell (Zai et al., 1999; Shah et al., 2007). Reduced Glutathione (GSH) is one of the most abundant low molecular weight thiols in the cell that contains glutamine, glycine, and cysteine residues. Compelling evidence about the role of GSH in gene expression, signal transduction, apoptosis, and NO metabolism has surfaced during the last decade (Pastore et al., 2001; Pompella et al., 2003). Furthermore, GSH is naturally used in combination with available NO to from GSNO, in a reversible reaction. Thus, it may play roles of both NO reservoir and NO donor (Stamler et al., 1992; Lindermayr et al., 2005). The enzyme GSNO reductase (GSNOR) is the primary enzyme that controls intracellular levels of GSNO and S-nitrosylated proteins in eukaryotes through de-nitrosylation (Feechan et al., 2005; Espunya et al., 2012).
The role of NO in biotic stress has also been studied. Sensing avirulent microbial pathogen activates the production reactive oxygen intermediates (ROIs) and reactive nitrogen intermediates (RNIs) (Burniston and Wilson, 2008) leading to the activation of multiple regulatory pathways. Similar to abiotic stress, plants have a specialized defense system to respond rapidly to pathogen ingress. In most cases virulent strains of bacterial pathogens like Pst can be recognized by trans-membrane pattern recognition receptors via pathogen-associated molecular patterns (PAMPs) such as flagellin (Zipfel and Felix, 2005). This activates PAMPs triggered immunity (Jones and Dangl, 2006). On the other hand, avirulent pathogens have effector proteins coded by avr genes that are recognized by the R-Proteins of the host plant activating R-gene mediated resistance (Jones and Dangl, 2006). Once the pathogen is successfully recognized, the production of reactive species, superoxides and hydrogen peroxide (H2O2) starts at the site of infection (Apostol et al., 1989; Grant and Loake, 2000). Such an interaction between avr and R gene products results in the effector-triggered immunity (ETI) (Jones and Dangl, 2006) is often associated with localized plant cell death, also known as the hypersensitive response (HR) and generation of systemic signals to activate systemic acquired resistance (SAR) (Ryals et al., 1996). Though the mechanistic control of R-avr interactions is still being investigated, the requirement of salicylic acid (SA) has been shown in many plant-pathogen interactions (MauchMani and Slusarenko, 1996). The phyto-hormones SA, jasmonic acid (JA), and ethylene play crucial roles in plant immunity by transmitting signals and inducing expression of specific genes (Feechan et al., 2005; López et al., 2008). An important feature of the plant defense response following pathogenic attack is the hypersensitive cell death response, an intentional suicide of cells at the site of infection to prevent the spread of disease (Jabs et al., 1996; Greenberg and Yao, 2004). Yun et al. (2011) reported that AtGSNOR1 controls the level of nitrosothiol (SNO) in plants during the HR. They further suggested that in atgsnor1-3, SNO accumulation and SA production are inversely proportional; thus, the higher the level of SNO accumulation, the lower the production of SA and its β-glucoside derivative (SAG).
Metals are imperative in cellular and subcellular functions such as hydrolysis, oxidation of various molecules, and electron transfer. According to one estimate, one-third to half of all proteins require metallic ions, either as co-factors or as important structural components (Huffman and O’Halloran, 2001; Dupont et al., 2010).
Metallochaperones are specialized proteins responsible for the safe, site-specific transportation of metallic ions inside the cell (Huffman and O’Halloran, 2001; Haas et al., 2009; Robinson and Winge, 2010). For instance, heavy metal-associated isoprenylated plant proteins (HIPPs) that are metallochaperones contain a metal binding domain and play an important role in the transport of metal ions (Freund et al., 2008). The concept of metallochaperones was initially derived from the study of the CopZ protein and antioxidant protein 1 (ATX1) in bacteria and yeast, respectively. These small proteins (69 and 73 residues, respectively) have the Cys-XX-Cys metal binding motif within the first loop of a ferredoxin-like structural fold (βαββαβ) that represents the canonical heavy metal-associated domain (Freund et al., 2008; Robinson and Winge, 2010). These Cu metallochaperones have the ability to reduce the energy limit for the transfer of Cu to target proteins (Wernimont et al., 2000; Rubino and Franz, 2012).
Copper also plays a vital role in plant defense to pathogens. A good example is the interaction between rice and Xanthomonas oryzae pv. oryzae (Xoo), which causes bacterial blight disease. Xoo is sensitive to Cu; however, it has the ability to secrete an effector molecule that in turn activates transcription of the rice susceptibility gene XA13. This further induces Cu removal from xylem vessels of the infected plant, by promoting its binding with copper transporters COPT1 and COPT5. This, in turn, facilitates the spread of X. oryzae pv oryzae through plant vessels. This has been proven through the reverse genetics approach and a recessive allele of XA13 has been found to confer increased resistance to X. oryzae pv oryzae infection (Freund et al., 2008). HMA domain containing genes are also reportedly involved in the interaction between plants and fungi. An example is OsHIPP05 (Os04g0401000) that shows susceptibility to virulent Magnaporthe oryzae, while showing enhanced resistance to the avirulent race of the same pathogen (Fukuoka et al., 2009; Nakao et al., 2011). Another study showed that overexpression of rice HIPP05 makes the non-host plant Arabidopsis susceptible to M. oryzae (Nakao et al., 2011).
Metallic ions containing proteins can also play a role in transferring NO bioactivity. The nitrosylation of metallo-co-factors is considered mostly NO dependent rather than GSNO dependent (Foster et al., 2009; Yun et al., 2016) indicating the importance of NO in post-translational modifications (Foster et al., 2009; Yun et al., 2016). Furthermore, the key player of the SA signaling pathway in plants, the transcriptional co-activator NPR1, has been reported to bind with transition metals via Cys residues at its C-terminus (Wu et al., 2012). Therefore, the interaction of NO with bound transition metals might regulate NPR1 during redox changes (Tada, 2009).
Although various roles of heavy metal associated (HMA) domain containing proteins have been described, however, little information is available about their roles in the protection of plants against pathogens and environmental stresses. In the present study, we adopted a combined in silico and biological approach to examine structure and putative function of the Arabidopsis thaliana HMA domain containing gene (AT1G51090-AtHMAD1) that showed differential expression in response to CysNO in a separate study involving transcriptomic analysis (Hussain et al., 2016). We also sought to determine its possible role in the regulation of oxidative and nitrosative stress and plant defense against virulent and avirulent pathogens. Collectively, these findings will lay a foundation for future studies on HMA domain containing proteins and will uncover their role in plant growth and defense.
Materials and Methods
In silico Data Analysis of HMA Domain Containing Genes
All the A. thaliana HMA (AtHMA) domain-containing genes that showed differential expression toward CysNO-mediated transcriptome analysis (Hussain et al., 2016) were identified and heat map showing the signal intensities of differentially expressed genes (up and down-regulated) was generated using R (version 3.3.1). Sequences for all AtHMA genes were retrieved from the Arabidopsis Information Resource (TAIR) database1. All sequences were aligned by the neighbor-joining method using ClustalW in BioEdit v7.2.5 (Hall, 1999). Promoter analysis of 1.5-kb region upstream of the transcription initiation site was also conducted using PlantCare (Lescot et al., 2002) to identify cis-regulatory elements. Cis-elements were then mapped using the Regulatory Sequence Analysis Tool (Medina-Rivera et al., 2015). Gene interaction analysis was performed using the protein–protein interaction network STRING (version 10.02). We also performed Gene Ontology (GO) analysis to determine putative involvement of NO-responsive AtHMA proteins in biological processes, molecular function and their protein class through the GO consortium database3 using A. thaliana as reference genome (Mi et al., 2013). The results so obtained at P-value < 0.05 were selected and compiled into pie charts.
Plant Material and Growth Conditions
Seeds of the Arabidopsis (Col-0) athmad1 (AT1G51090) loss of function mutant were obtained from Nottingham Arabidopsis Stock Centre (NASC)4 and grown either on 1/2 Murashige and Skoog (MS) medium or soil at 23 ± 2°C, under long day conditions (16 h light and 8 h dark). All plants were genotyped at the rosette leaf stage (4 weeks old plants unless stated otherwise) in order to identify homozygous lines. Arabidopsis wild type (WT) and all other lines used in the study were of the Col-0 genetic background. Perturbations in the cellular levels of ROS and RNS levels affect metal-containing enzymes such as peroxidases and catalases (Clarke et al., 2000; Fourcroy et al., 2004). Since our study included experiments to determine the role of HMA containing genes in oxidative and nitrosative stress in plants, we used the Arabidopsis gsnor1-3 mutant as a comparative control.
Redox Stress Assay
Arabidopsis athmad1 plants were tested for various growth parameters under oxidative and nitrosative stress conditions, as described by Sharma et al. (2016). Arabidopsis WT and athmad1 seeds were surface sterilized and germinated on 1/2 strength MS medium in at least three replicates each, containing 2 mM H2O2, 1 μM methyl viologen (Medina-Rivera et al., 2015), 0.75 mM S-nitrosoglutathione (GSNO), and 0.75 mM S-nitrosocysteine (CysNO). Data on hypocotyl emergence, cotyledon development frequency (CDF), and root and shoot length were recorded 1 week after treatment.
Pathogen Inoculation
Virulent Pseudomonas syringae pv. tomato (Jaffrey et al., 2001) strain DC3000 and avirulent Pst DC3000 expressing the avrB effector were grown, maintained, and inoculated, as described previously (Yun et al., 2011). Briefly, both the bacterial strains were cultured on LB (Luria-Bertani)-agar media with appropriate antibiotics for selection (100 μg/mL rifampicin for virulent Pst DC3000 and kanamycin [50 μg/mL] and rifampicin for Pst DC3000 [avrB]), following which a single colony was transferred to LB broth and incubated at 28°C overnight. Both bacterial strains were then harvested by centrifugation at 13,000 rpm for 1 min in 10 mM MgCl2, and syringe-infiltrated into the abaxial side of leaves at a concentration of 5 × 105 CFU (unless stated otherwise). Control plants were only infiltrated with 10 mM MgCl2. Plants were observed regularly for disease symptoms and leaf samples were collected for further analysis.
Pathogenicity Assessment and Electrolyte Leakage Assay
Leaf samples (1 cm each) from plants inoculated with Pst DC3000 were collected, crushed in 1 mL of sterile 10 mM MgCl2 2 and 4 days post inoculation (DPI). The homogenate was diluted 10 times and spread on LB-agar plates containing appropriate antibiotics. Plates were incubated at 28°C for 48 h and colonies were counted. Ion or electrolyte leakage was measured as described by Dellagi et al. (1998) with some modifications. Conductivity was measured 0, 1, 2, 4, 6, 8, 10, 12, and 24 h post infiltration (hpi), using a portable conductivity meter (HURIBA Twin Cond B- 173, Japan).
Quantitative Real-Time PCR (qRT-PCR) Analysis
RNA was extracted from the inoculated plants using Trizol® (Invitrogen, USA) as described by Imran et al. (2016). For gene expression analysis, cDNA was synthesized using the DiaStarTM RT kit (SolGent, Korea) according to the manufacturer’s instructions. PCR was performed in the EcoTM real-time PCR machine (Illumina, USA) using 2X Quantispeed SYBR Kit (PhileKorea) along with 100 ng of template DNA and 10 nM of each primer in a final volume of 20 μL. A “no template control” was used as a negative control that contained distilled water instead of template DNA. Two-step PCR reactions were performed for 40 cycles using primers given in Supplementary Table S1.
Histological Staining
Pseudomonas syringae pv. tomato expressing the avrB effector proteins activate R gene-mediated (RPM1) defense in A. thaliana, thereby triggering hypersensitive resistance (Leister et al., 1996). The HR of athmad1 plants to Pst DC3000 (avrB) infiltration was quantified via trypan blue staining as described by Koch and Slusarenko (1990). Cell death was quantified (arbitrary units) in terms of the intensity of trypan blue staining using Adobe Photoshop CS65, as described earlier (Yun et al., 2011).
SNO Measurement
As mentioned previously, published evidence suggests an inter-relationship between NO and metal biology in plants under basal, as well as induced conditions. Arabidopsis atgsnor1-3 accumulates comparatively higher levels of SNO (Feechan et al., 2005). This prompted us to investigate the S-nitrosothiol levels in athmad1 before and after inoculation with Pst DC3000 (avrB). For this purpose, Arabidopsis WT, atgsnor1-3, and athmad1 plants were syringe- infiltrated with 5 × 105 CFU mL-1 Pst DC3000 (avrB). Leaf samples were collected and ground in KPI buffer (pH 5.5) after 0, 6, 12, and 24 h post infiltration. SNO measurements were performed in an NO analyzer (NOA-280i, Sievers, USA).
Foliar SA Application
For foliar application of SA, we used the method described by Gao et al. (2011) with minor modifications. Four-week-old soil grown WT, atgsnor1-3, and athmad1 plants were uniformly sprayed with 1 mM SA. Control plants were sprayed with distilled water. Leaf samples were collected at 0, 12, and 24 h after spraying for further analysis.
Statistical Analysis
All the data was statistically analyzed using analysis of variance followed by standard error through Student Statistics 9 software. Student t-test was performed using Microsoft Excel program.
Results
Validation of RNA-Seq Data through qRT PCR and GO-Enrichment Analysis
The overall gene expression profile of AtHMA genes that showed differential response to CysNO through RNA-seq analysis (Figure 1A, Suplementary Table 2) were further validated through qRT PCR analysis (Figure 1B). All of the six randomly selected up-regulated HMA genes showed significantly high transcript accumulation, whereas two down-regulated genes showed significantly reduced transcript accumulation compared to buffer (Figure 1B). A correlation coefficient of 0.67 shows high congruence of RNA-Seq and qRT-PCR experiments. All these genes were then analyzed for GO terms of Biolgoical processes, Protein class and Molecular function usigng GO Consortium6. The results indicated that 93.30% of genes were involved in cellular processes, 2% in metabolic processes, 33% in biological regulation (Hemostasis), and 93.30 % are involved in localizaition [transport (Figure 1C)].
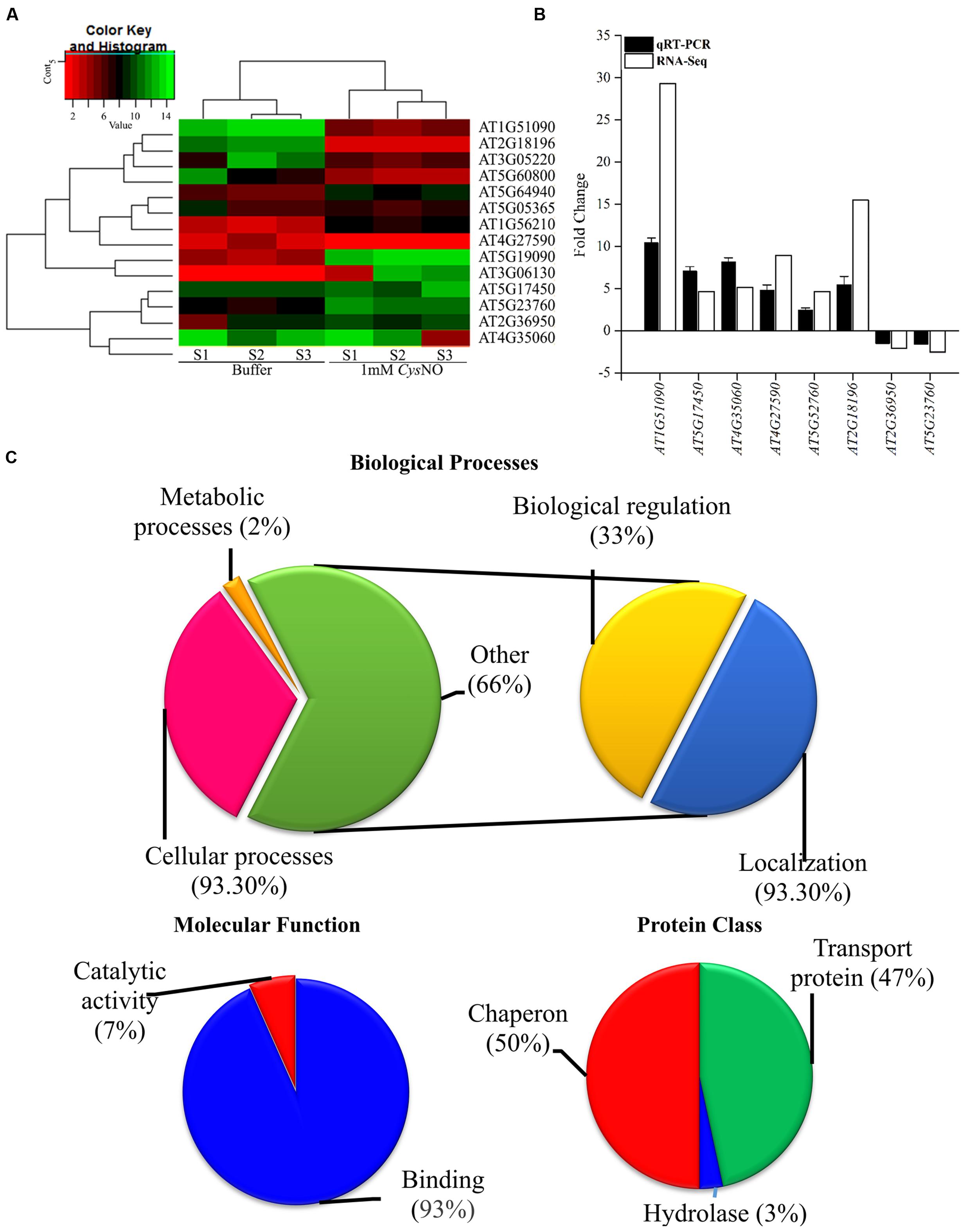
FIGURE 1. NO-responsive heavy metal associated domain genes. (A) Heat map of the AtHMA genes showing the signal intensities of differentially expressed genes in response to 1 mM CysNO through RNA-seq analysis. (B) Validation of RNA-seq data through qRT PCR analysis of randomly selected DEGs. (C) GO-enrichment analysis showing role of AtHMA genes in different biological processes and their classification into different protein classes.
Conserved Domain and Promoter Analysis for Cis-Regulatory Elements
Analysis for the presence of conserved motifs showed the presence of a highly conserved Cys-XX-Cys motif in 11 of the 14 gene products (Supplementary Figure S1E; blue box). Furthermore, the genes having Cys-XX-Cys also contained three highly conserved lysine (K) residues, and other important conserved residues (Supplementary Figure S1E).
Cis-regulatory elements are non-coding DNA sequences present in the promoters of various genes to which various regulatory proteins bind in order to control gene expression. These elements reportedly play a major role in transcriptional control of genes (Wittkopp and Kalay, 2012) involved in defense related pathways (Caarls et al., 2015) and plant responses toward pathogens (Rushton and Somssich, 1998). Therefore, we analyzed promoter sequences of all the AtHMA genes for the presence of cis-regulatory elements. We found 19 cis-regulatory elements in the promoters at variable distances form the transcriptional initiation site. These elements included ABRE (TACGTG) involved in regulation of osmotic and drought stress responsiveness genes (Kim et al., 2011); the CAAT-box (CAAT/CAAAT), responsible for tissue specific activity (Shirsat et al., 1989); AT-rich sequences (TAAAATACT) and EIRE (TTCGACC) involved in maximal elicitor-mediated activation (Fukuda, 1997); the TATA-box, a core promoter element for enhanced transcription (Mukumoto et al., 1993); ERE (ATTTCAAA) that are ethylene-responsive elements (Ohme-Takagi and Shinshi, 1995); and the MYB binding site (MBS; CAACTG) involved in drought-stress regulation (Singh and Laxmi, 2015), (Figure 2). This suggests the importance of AtHMA domain containing proteins in biotic and abiotic stress tolerance.

FIGURE 2. Promoter analysis for the presence of cis-regulatory elements. A total of 14 HMA domain containing genes were analyzed for the presence of cis-regulatory elements, 1.5 kb upstream of the transcription initiation site. The resultant cis-regulatory elements were mapped using Regulatory Sequence Analysis Tool (Shirsat et al., 1989) (http://floresta.eead.csic.es/rsat/) for better presentation.
Metallo-Protein Interaction
As AtHMA proteins are mostly involved in heavy metal transport (de Abreu-Neto et al., 2013); therefore, to determine the possible ligand-binding interactions of AtHMA proteins, we performed an in silico experiment to analyze the protein sequence of AtHMAD1 for its ability to bind various metal ligands using RaptorX (Kallberg et al., 2012) which is a web portal for protein structure and function prediction using protein sequences. Our results showed that the first domain (1–80 amino acids) of AtHMAD1 can bind to different metal ligands at different rates (Supplementary Figure S1D), as represented by pocket multiplicity (PM; the frequency at which a particular pocket was found in a set of ligand- protein structures). The highest PM was observed for Cu at sequence residues N15, C16, S17, and C19 with a PM rate above 40 (Kallberg et al., 2012). This suggests that AtHMAD1 might be involved in Cu (or other metal ligands) binding, which is a characteristic feature of metallochaperones. The strong in silico binding interaction suggests the possible role of AtHMAD1 in metal transportation in and out of the cells.
AtHMAD1 Positively Regulates Shoot Growth and Negatively Regulates Root Growth under Oxidative and Nitrosative Stress Conditions
A number of growth parameters like hypocotyl emergence, CDF, shoot and root length in WT and loss-of-function mutant athmad1 plants were studied to determine the possible role of AtHMAD1 in plant growth and development. CDF represents the number of developed, green seedlings with the first cotyledonary leaves (under nitrosative stress, seeds of several Arabidopsis genotypes may germinate but ultimately die, e.g., atgsnor1-3 and other NO-related genotypes; Yun et al., 2011). Under oxidative (2 mM H2O2, 1 μM methyl viologen,) and nitrosative stress (0.75 mM GSNO, and 0.75 mM CysNO), athmad1 plants showed a significant reduction in hypocotyl emergence (Supplementary Figure S3), CDF (Figure 3B), and shoot length (Figures 3A,C) compared to WT. In contrast, athmad1 plants showed an increase in root length under nitrosative stress; however, compared to WT, a significant reduction in root length was observed in athmad1 plants under conditions of oxidative stress induced by H2O2 and MV (Figures 3A,D).
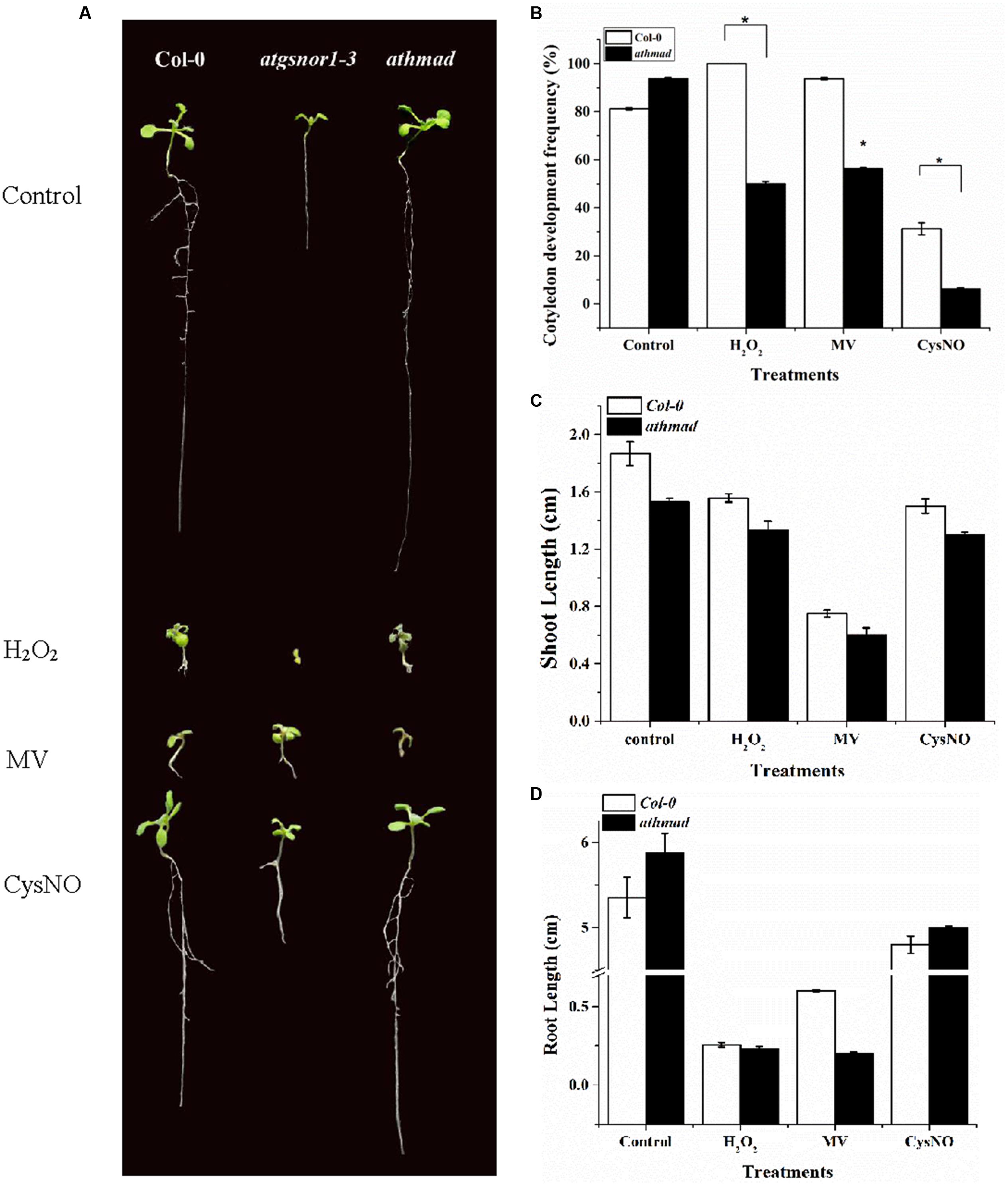
FIGURE 3. Role of AtHMAD1 in plant development. (A) Phenotypes of the indicated genotypes under control and redox stress conditions. (B) In response to oxidative and nitrosative stress athmad1 plants showed reduced CDF (C), shoot length, and (D) root length compared to untreated control plants. All data points represent means of three replicates. The experiment was repeated three times with similar results. Error bars represent the standard error. Significant interactions are indicated by an asterisk (Student’s t-test with a 95% confidence level).
AtHMAD1 Negatively Regulates Plant Basal Defense
To investigate whether AtHMAD1 is required for basal defense, we inoculated athmad1 plants, as well as the atgsnor1-3 and WT control plants with virulent Pst DC3000. The atgsnor1-3 line that is reportedly susceptible to Pst DC3000 (Feechan et al., 2005) exhibited significantly higher levels of bacterial growth compared to WT plants. The athmad1 plants showed significant resistance toward this pathogen compared to the WT plants (Figures 4A,B) suggesting that AtHMAD1 negatively regulates plant defense. This was further confirmed by rapid and high levels of transcript accumulation of the plant defense marker gene PR1 following pathogen inoculation, as evidenced by qRT-PCR analysis (Figure 4C).
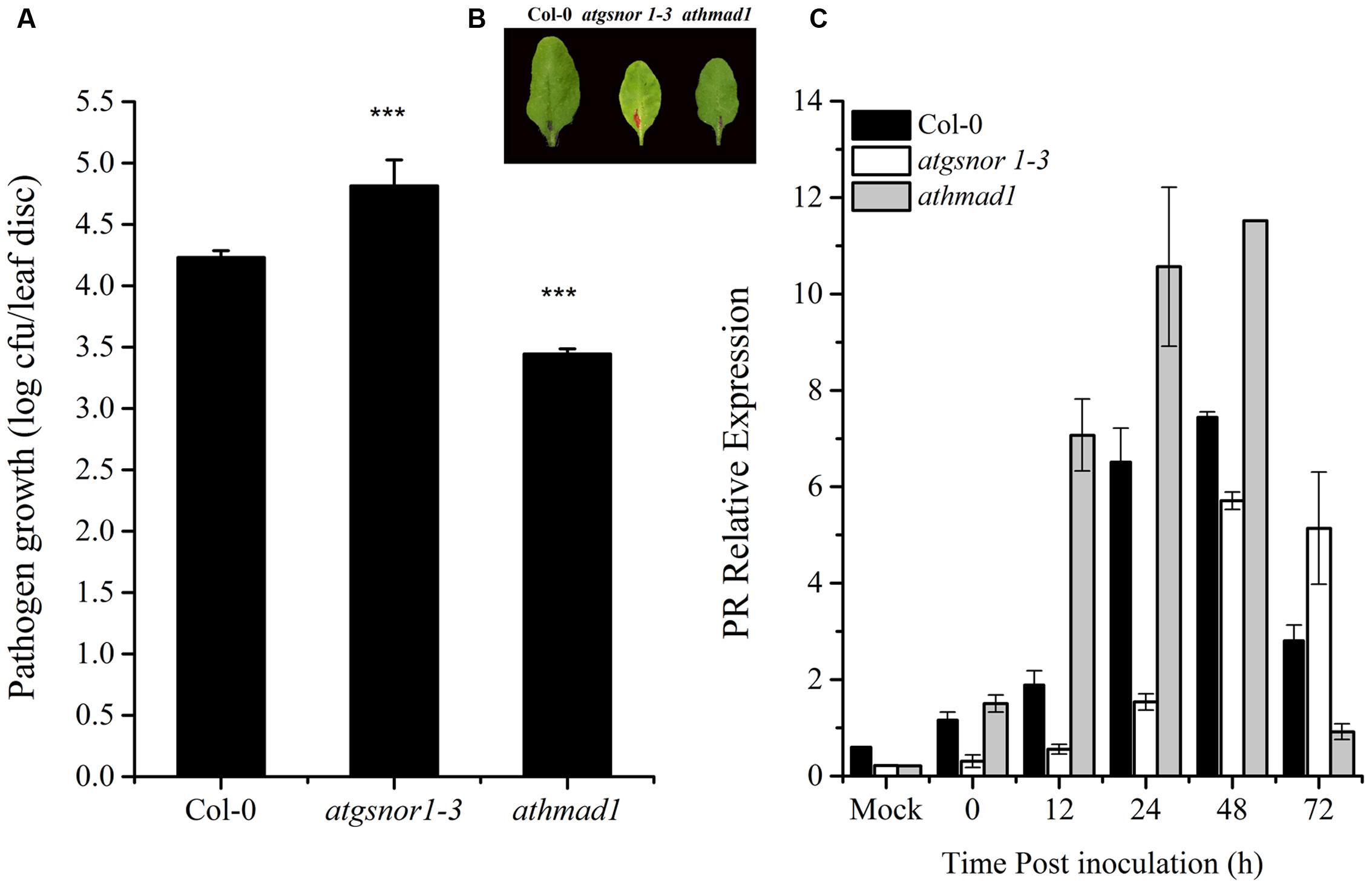
FIGURE 4. AtHMAD1 negatively regulates the basal defense system. Pathogen growth from the inoculated leaves with virulent Pst DC3000 at 5 × 105 colony forming units (CFU) mL-1after 2 days post inoculation (DPI; A). Development of symptoms in wild type (WT) and transgenic lines following infiltration with virulent Pst DC3000 (B). Transcript accumulation of the PR gene (SA-dependent pathway) in the indicated Arabidopsis genotypes in response to pressure infiltration of Pst DC3000 using qPCR (C). The expression values were normalized using the constitutively expressed actin. All data points represent the mean of four replicates. Error bars represent ± SE (n = 4) with asterisks indicate highly significant differences (99% confidence level) from WT plants. The experiment was repeated three times with similar results (Student’s t-test, P < 0.0000265).
We also determined whether induction of PR genes would have an effect on the expression of PDF1.2 in athmad1 plants following inoculation with virulent Pst DC3000. PDF1.2 is a marker gene for JA pathway. Literature reports suggested a negative interaction between SA and JA pathway (Penninckx et al., 1996; Niki et al., 1998). Therefore, we sought to determine interaction between SA and JA pathway in athmad1 mutants. Expression of PDF1.2 in athmad1 plants was observed to be significantly lower than that of WT plants (Supplementary Figure S4).
AtHMAD1 also Negatively Regulates R-Gene Mediated Resistance
To determine if AtHMAD1 function is required for R-gene mediated resistance, we infiltrated plants with Pst DC3000 (avrB). Our results indicated early induction and high transcript accumulation of the PR gene over time compared to WT (Figure 5). We also examined whether AtHMAD1 is involved in the JA defense pathway, by studying the expression level of the JA-dependent pathway gene PDF1.2. In contrast, PDF1.2 expression was significantly lower compared to WT (Supplementary Figure S5), indicating an antagonistic relationship between PR and PDF1.2 during plant defense responses.
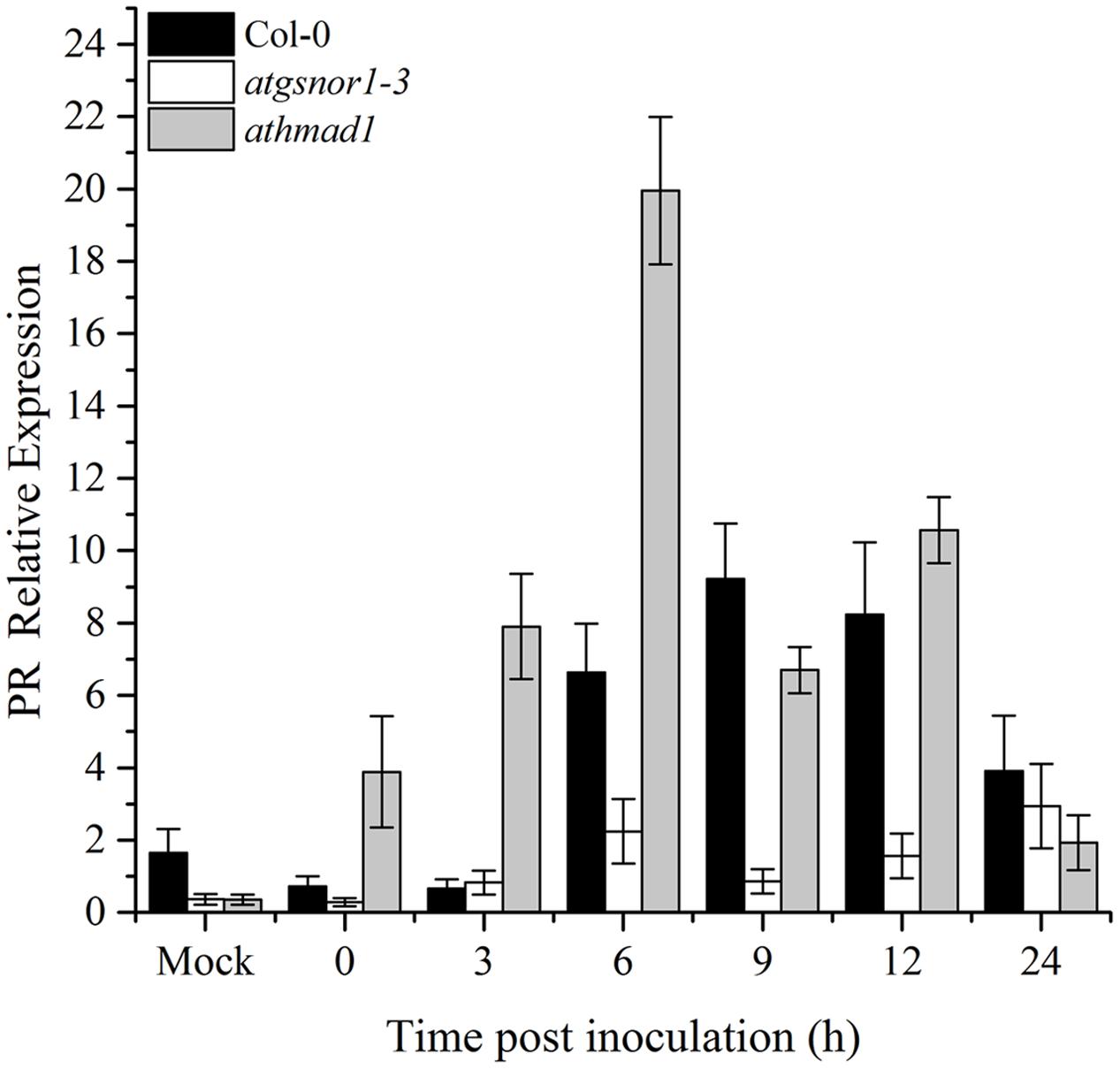
FIGURE 5. Role of AtHMAD1 in R-gene mediated resistance. Early induction and high transcript accumulation of the PR gene in athmad1 plants in response to Pst DC3000 (avrB) inoculation at 5 × 105 colony forming units (CFU) mL-1 in the indicated Arabidopsis genotypes. All data points represent the mean of three replicates. All the data was analyzed statistically using analysis of variance (P < 0.05) followed by Standard Error. Error bars represent ± SE (n = 3). The experiment was repeated three times with similar results.
Pathogen-Triggered Hypersensitive Response (HR) Is Induced in athmad1 Plants
Effector-triggered immunity ultimately culminates in the hyper sensitive response (HR), that is, the intentional execution of plant cells at the site of infection. To identify the possible role of AtHMAD1 in HR induction, we challenged plants with avirulent Pst DC3000 (avrB). To visualize cell death in vivo, the inoculated leaves were stained with lactophenol trypan blue. An increase in trypan blue staining was observed which was used as a method to detect and quantify cell death. The athmad1 plants showed an increased HR after 12 and 24 h of inoculation compared to WT plants (Figure 6B,), that was also confirmed by quantifying the cell death (Figure 6C) indicating that AtHMAD1 negatively regulates HR. For further confirmation, we quantified cell death induced electrolyte leakage. The athmad1 plants showed higher levels of electrolyte leakage over time, followed by the atgsnor1-3 plants with significant difference (P < 0.05) at 2 and 6 hpi compared to WT (Figure 6A).
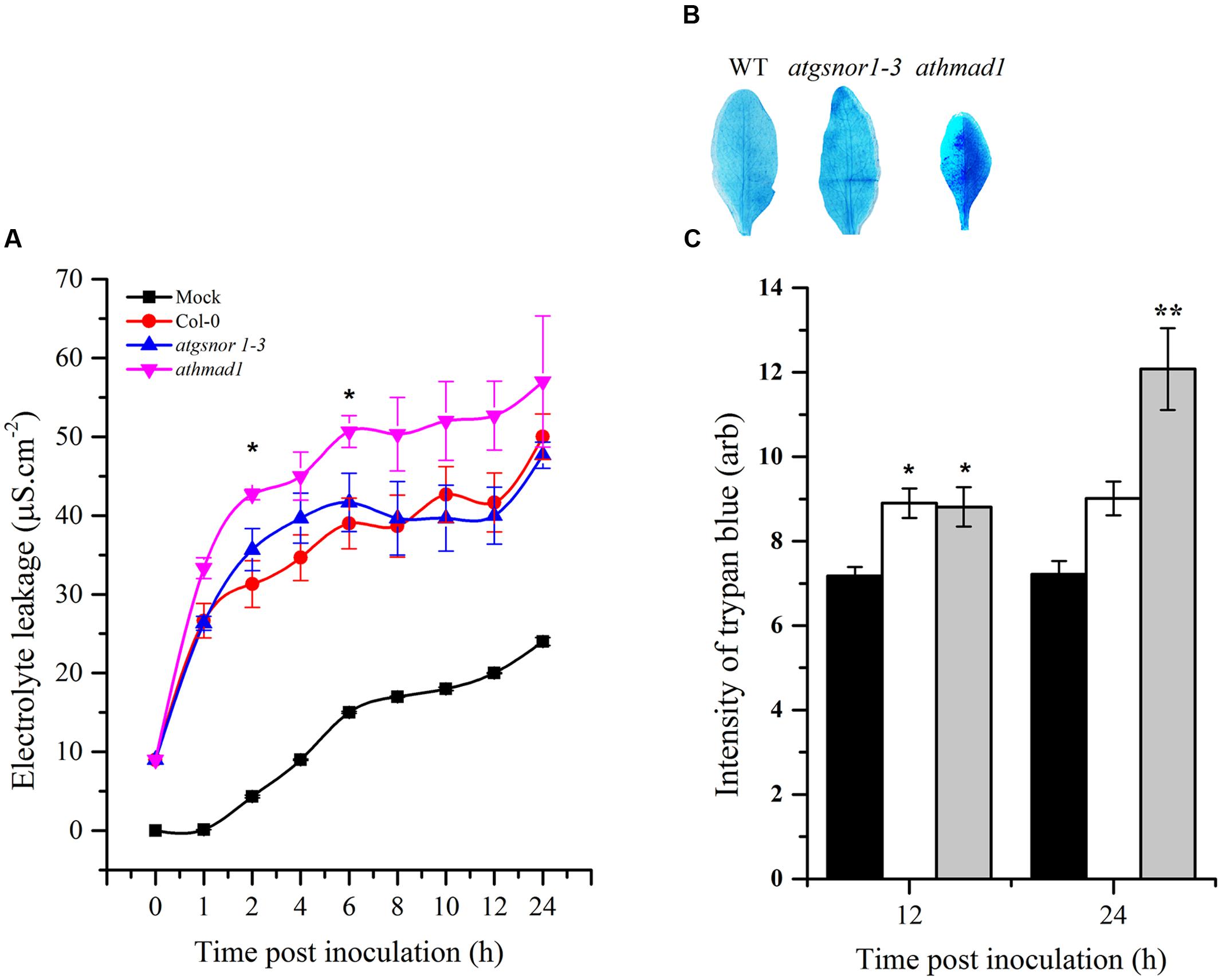
FIGURE 6. Induced HR response of athmad1 plants following attempted Pst DC3000 (avrB) infection. (A) Electrolyte leakage; (B) Visualization of cell death by trypan blue staining; and (C) Its quantification using the Luminosity tool of Photoshop by fixing the values for the rectangular marquee tool. Values represent the means of three random replicates. The error bars represent ± SE (n = 3), and asterisks represent significant differences compared to WT plants, (Student’s t-test with a 95% confidence level).
athmad1 Plants Showed High Induction of SAR Associated Genes
A number of SAR associated signals have been identified so far. These include SA and its methylated derivative MeSA and azelaic acid (Park et al., 2007; Shah and Zeier, 2013; Wang et al., 2014; Wendehenne et al., 2014). To determine the role of AtHMAD1 in SAR activation, we studied the induction of PR and G3Pdh genes over time in systemic leaves following Pst DC3000 (avrB) inoculation. High induction of PR gene was observed in systemic leaves of athmad1 plants at earlier time points (0, 3, and 6 hpi) compared to WT plants and atgnsor 1-3 (Figure 7A). Furthermore, after 24 h of inoculation, high expression levels of G3Pdh (the detail description about this gene is given in Discussion section) was observed in systemic leaves of athmad1 plants in comparison to those of WT plants (Figure 7B). Whereas transcripts of PDF1.2 were significantly reduced in athmad1 plants compared to WT plants (Supplementary Figure S6), indicating that AtHMAD1 negatively regulates SAR.
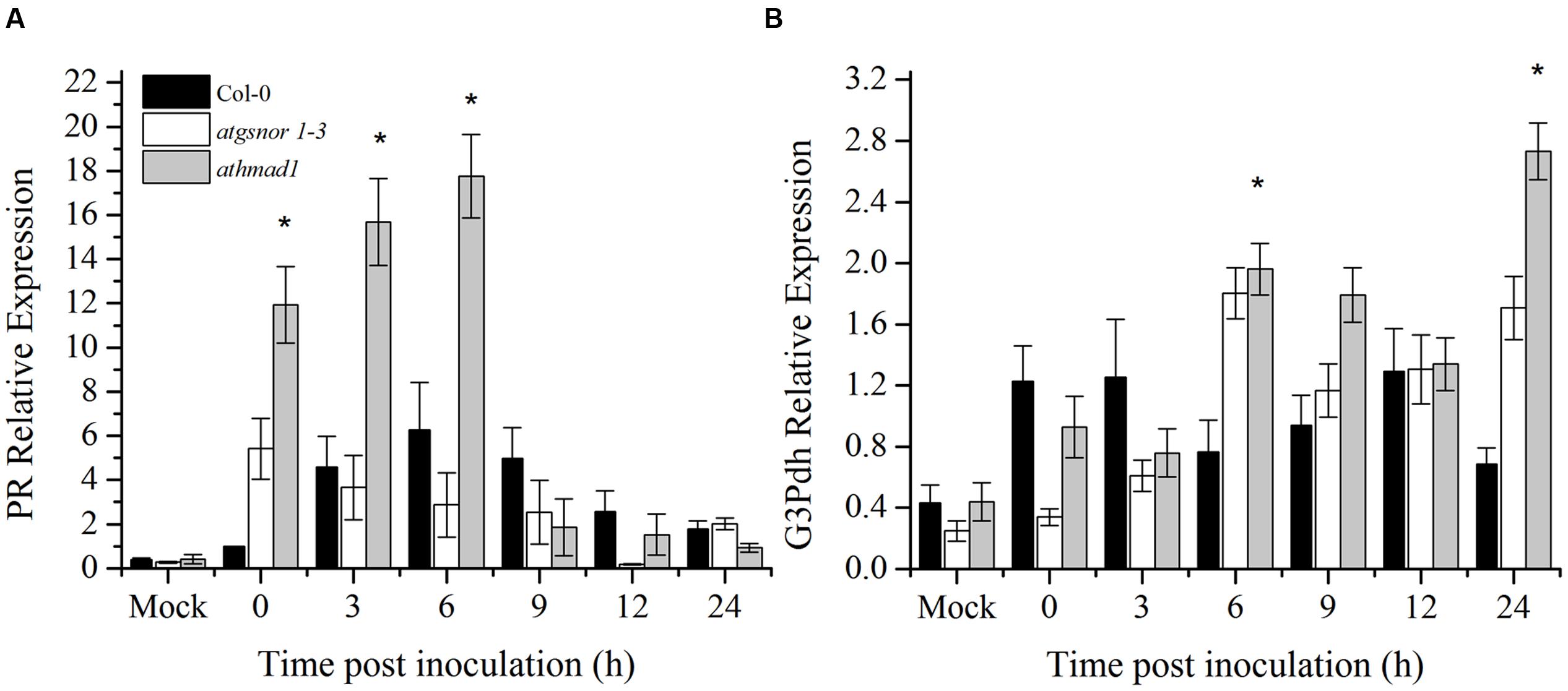
FIGURE 7. Transcript accumulation of genes involved in SAR. Expression level of (A) the PR gene and (B) G3Pdh (Glycerol-3-phosphate dehydrogenase) following attempted Pst DC3000 (avrB) infiltration at 5 × 105 CFU. Leaf samples were collected from systemic leaves at various intervals over 24 hpi. 0 hpi represents samples collected immediately after pathogen inoculation. The experiment was repeated three times with similar results. All the data was analyzed statistically using analysis of variance (P < 0.05) followed by Standard Error. Error bars represent ± SE (n = 3). Asterisks represent significant differences compared to WT plants, (Student’s t-test with a 95% confidence level).
Exogenous Application of SA Induces PR Gene, but Does Not Induce SAR-Associated Genes in athmad1 Plants
An early induction and high expression of the PR gene was observed in athmad1 plants compared to WT and atgsnor1-3 plants, following the application of SA spray (Figure 8A). On the other hand, G3Pdh expression was not induced with exogenous application of SA in athmad1 plants compared to WT (Figure 8B), indicating that AtHMAD1 may interact with SAR upstream of G3Pdh. Another reason for this might be that the NO and G3Pdh pathway is distinct from the SA pathway, as described by El-Shetehy et al. (2015), who reported that exogenous SA is unable to restore SAR in ROS- or NO-deficient mutants.
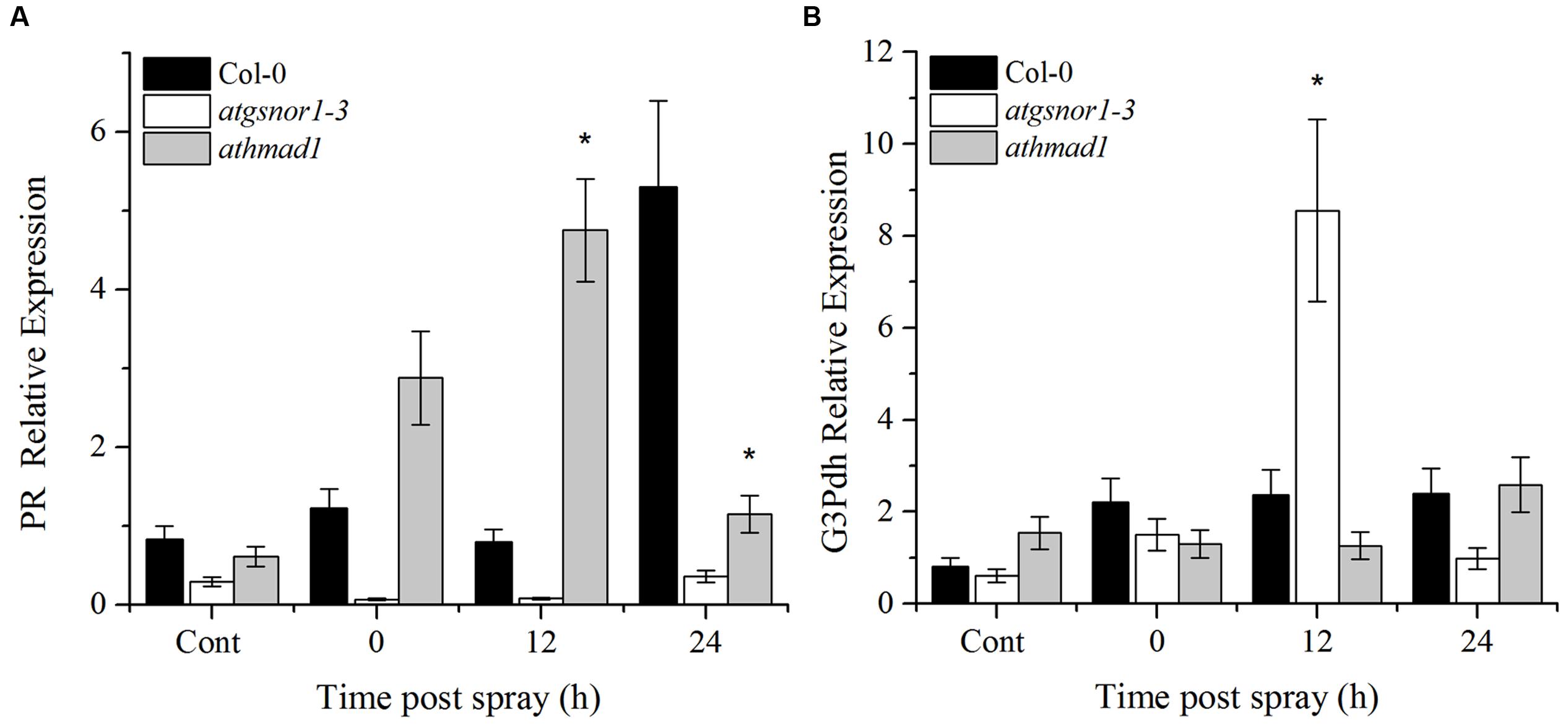
FIGURE 8. Transcriptional analysis of exogenous SA application. (A) PR gene expression; and (B) G3Pdh (Glycerol-3-phosphate dehydrogenase) expression in response to 1 mM SA using RT-qPCR. The corresponding values were calculated relative to those of the constitutively expressed Actin gene. All the data was analyzed statistically using analysis of variance (P < 0.05) followed by Standard Error Error bars represent ± SE (n = 3). Asterisks represent significant differences compared to WT plants, (Student’s t-test with a 95% confidence level).
Pathogen-Triggered SNO Accumulation Was Reduced in athmad1 Plants Following the Application of SA Spray
The major route of NO signaling in plants is considered a direct one, involving S-nitrosylation to form SNOs. We observed a gradual increase in SNO accumulation in athmad1 leaves after 6, 12, and 24 h of inoculation with the pathogen, and this difference was most significant after 24 h (Figure 9A). We then sprayed plants with 1 mM SA to examine the interaction between AtHMAD1 and SA during SNO accumulation in plants. SNO content was significantly reduced at 12 and 24 h following the application of SA spray in athmad1 plants, compared to WT control plants (Figure 9B). Published reports suggest that SA positively regulates S-nitrosoglutathione reductase (GSNOR) (Feechan et al., 2005). Thus, a reduction in SNO contents following exogenous application of SA might be due to SA-mediated GSNOR induction that reduced the elevated NO levels within the cell.
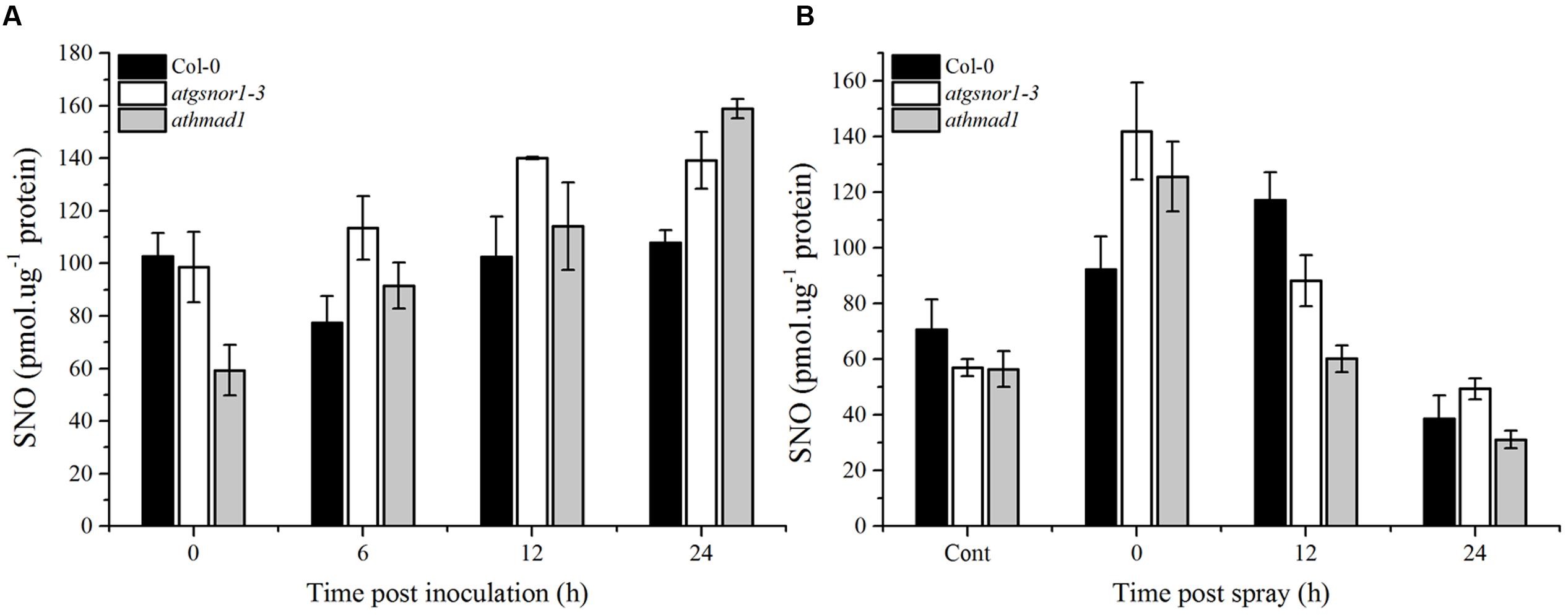
FIGURE 9. Accumulation of S-nitrosothiol (SNO) in response to (A) Pst DC3000 (avrB) inoculation and (B) exogenous SA application. The indicated genotypes were infiltrated with Pst DC3000 (avrB) or 1mM SA. control plants were sprayed with distilled water. The total level of cellular SNO was determined over time using NO analyzer (NOA-280i, Sievers, USA). Error bars represent ± SE (n = 3).
Discussion
Heavy metal associated domain containing proteins have been shown to play a pivotal role in plant growth and stress perception (Williams et al., 2000; de Abreu-Neto et al., 2013). HMA domain containing genes not only serve as metal ion transporters for different essential functions and/or detoxification, but are also important reserviors of essential metal ions that are required for growth and development (Williams et al., 2000; Clemens, 2001; Grotz and Guerinot, 2006). By using a variety of metal-associated loss of function mutants and/or over-expressor transgenic genotypes, a number of published functional genomic studies have shown the importance of metal biology in plant growth and development in the model plant Arabidopsis (Dykema et al., 1999; Barth et al., 2009; Tehseen et al., 2010), as well as various cultivated crops such as soybean, tobacco (Dykema et al., 1999), and rice (de Abreu-Neto et al., 2013). Both ROIs and RNIs regulates metal ion homeostasis in plants (Burniston and Wilson, 2008). Heavy metal-associated domain containing genes are metallochaperones that have the HMA domain Cys-XX-Cys metal ion binding motif within the first loop of a ferredoxin like structural fold (βαββαβ) that is mainly responsible for the delivery of metal ions within the cell (Robinson and Winge, 2010).
Under normal conditions, plants produce basal levels of NO that usually increase in response to biotic and/or abiotic stresses. By utilizing high throughput transcriptomic analysis, we identified a list of 14 genes that showed differential expression in response to 1 mM CysNO (Supplementary Table S2). The use of spatiotemporally inducible promoters is considered important for increased resistance to plant pathogens. The expression patterns and regulation strategies of only a few plant promoters induced in response to phyto-pathogens have been characterized so far (Gurr and Rushton, 2005). Furthermore, besides the core promoter area, other auxiliary transcription regulatory elements like enhancers, silencers, and insulators can be found at 5′ upstream and 3′ downstream of the promoter region and introns (Cawley et al., 2004). The promoter analysis for cis-regulatory elements in AtHMA domain containing genes indicated the presence of various important motifs and elements that regulate gene expression under various environmental conditions and during pathogen ingress (Figure 2). For instance, the presence of ABRE that is involved in ABA responses; ERE, that are ethylene responsive elements; and EIRE, involved in maximal elicitor-mediated activation, suggests the possible role of AtHMA domain containing genes in drought tolerance, growth, and defense against various pathogens (Figure 2).
Structure prediction and domain binding analysis of AtHMAD1 suggest its possible role in metal binding and/or transport, thus regulating various physiological and molecular processes under biotic and abiotic stress. AtHMAD1 showed high PM for Cu, an indication of a true binding interaction (Supplementary Figures S1A-D). Interestingly, Cu is the co-factor for important enzyme nitrite reductase that catalyzes the formation of NO through a reversible reaction with ferrocytochrome-C in the presence of two hydrogen protons thus controlling the cellular RNS levels. In addition, it is also a co-factor for other important enzymes like galactose oxidase, 2-furoyl-CoA dehydrogenase, nitrous oxide reductase, NO reductase, cytochrome-c oxidase, tyrosinase, etc7. These results suggests the putative regulatory role of AtHMAD1 in the regulation of enzyme function during important cellular processes, particularly under stressful conditions.
Gene interaction studies of AtHMAD1 (AT1G51090) yielded interesting information about its association with other genes (Supplementary Figure S2). Its strong association with Gibberellin 2-Oxidase 6 (GA2ox6:AT1G02400) predicts the importance of this gene in plant growth and development. We observed reduced hypocotyl emergence, CDF, and shoot growth (Figures 3A–C) under conditions of oxidative and nitrosative stress in athmad1 plants compared to WT. This reduction could be attributed to excess accumulation of ROS and RNIs, in response to changes in the redox environment as described by García-Mata and Lamattina (2001), Uchida et al. (2002), and Kopyra and Gwóźdź (2003). Altogether, these results imply that the AtHMAD1 gene product negatively regulates CDF and shoot growth.
The athmad1 loss of function mutant showed greater resistance to the virulent pathogen Pst DC3000 compared to WT plants (Figure 4). On the other hand, the susceptible mutant atgsnor1-3 showed significantly higher bacterial growth. This is because of the already established fact that S-nitrosoglutathione reductase (GSNOR) is required for both basal and R-gene mediated resistance (Feechan et al., 2005). Studies reveal that cell death occurs mainly because of ROIs and RNIs that are produced by NADPH oxidase and nitrosative bursts, respectively (Delledonne et al., 1998; Yun et al., 2011). Early induction and high transcript accumulation of the PR gene in response to Pst DC3000 in the athmad1 mutant compared to WT suggests that AtHMAD1 protein regulates plant defense through the SA-dependent pathway.
Plant defense starts with the recognition of PAMPs by plant pattern recognition receptors (PRRs), resulting in PAMP-triggered immunity (PTI). Some pathogens have evolved to evade PTI by deploying certain effector molecules into the host system, thereby leading to effector-triggered susceptibility (Jones and Dangl, 2006). However, different effector molecules are recognized by plant disease resistance (R) genes encoded by the nucleotide binding site leucine rich repeat (NBS-LRR) genes that recognize these effector molecules (Jones and Dangl, 2006). Published reports suggest that following pathogenic infection, SA accumulation is responsible for the induction of PR genes (Durrant and Dong, 2004). ETI culminates with HR. Therefore, to investigate the possible role of AtHMAD1 in ETI, we also examined the HR following inoculation with Pst DC3000 (avrB). The athmad1 plants displayed enhanced pathogen-triggered cell death and early induction of PR gene expression compared to WT plants (Figures 5 and 6B,C). Earlier reports suggest that R gene-mediated resistance is conferred because of interaction between avr and R-gene products that serves to limit the growth of virulent pathogens inside the host plant (Glazebrook et al., 1996; Parker et al., 1996). A similar study has been conducted in rice showing that avirulent strain of X. oryzae pv. oryzae which show sensitivity to Cu, release effector molecules that induces rice susceptibility gene XA13. This further triggers Cu removal from xylem vessels by inducing its ability to bind with COPT1 and COPT5 which are copper transporter proteins thus facilitating the spread of X. oryzae. The study has been validated through reverse genetics studies and it was suggested that recessive allele of XA13 confers enhanced resistance to X. oryzae pv oryzae infection (Freund et al., 2008). Our results suggested reduced transcript accumulation of PDF1.2 in athmad1 plants compared to WT plants after attempted inoculation of both Pst DC3000 and Pst DC3000 (avrB). This is consistent with the findings of Niki et al. (1998) who described the antagonistic effects of SA and JA during regulation of defense pathways. It was also reported that PDF1.2 showed high levels of expression in the SA-deficient mutant NahG (Penninckx et al., 1996). Therefore, low levels of transcript accumulation of PDF1.2 in samples showing increased PR gene expression indicates negative interaction between the two immune activators. The notion that AtHMAD1 modulates the SA-dependent pathway can also be supported by the levels of SNO observed in athamd1. Our results suggests that after Pst DC300 (avrB) inoculation, a gradual increase in SNO levels was observed in athamd1 plants compared to WT (Figure 9A), however, after SA spray the SNO levels decreased over time in athmad1 compared to WT (Figure 9B). Earlier reports suggested that high levels of SNO were observed in nox-1 and atgsnor1-3 plants that blunted the SA-dependent pathway (Feechan et al., 2005; Yun et al., 2016). In contrast, atgsnor1-1 and atgsnor1-2 plants showed increased SA levels and reduced basal SNO levels (Feechan et al., 2005). Cellular NO levels can be regulated via the NO donor, GSNO that can be reduced to GSSH (Glutathione disulfide) and NH3 in the presence of GSNOR. Thus, a reduction in SNO contents following exogenous application of SA might be due to SA-mediated GSNOR induction that reduced the elevated NO levels within the cell.
Systemic acquired resistance is another important plant defense system induced by primary infection. It provides broad spectrum disease resistance and protects other parts of the plant from secondary infections (El-Shetehy et al., 2015). Earlier reports have shown that NO and ROS are required for activation of SAR (Alvarez et al., 1998; Song and Goodman, 2001). The accumulated evidence suggests that a number of SAR signals act to confer resistance in distant tissues to subsequent infections (El-Shetehy et al., 2015). Azelaic acid (AzA) and Glycerol 3 phosphate (G3P) are important SAR signaling molecules (Yu et al., 1999, 2013). Exogenous application of AzA increases the expression of G3P biosynthesis genes, GLY1 and GLI1 that encodes G3P dehydrogenase and glycerol kinase, respectively. Reports suggested that AzA and G3P-induced signaling was connected somehow in the SAR pathway. Recently, a study by Yu et al. (2013) suggested that SAR inducer AzA conferred SAR in wild type but not in plants defective in the G3P biosynthesis genes GLI1 and G3Pdh (Chanda et al., 2011). Furthermore, G3P levels were also significantly increased after AzA application which was consistent with increased G3Pdh expression suggesting that AzA-mediated SAR require G3Pdh gene (Yu et al., 2013). Therefore, we also studied the AtHMAD1 role in SAR signaling by studying the expression of marker genes (PR and G3Pdh) (Wang et al., 2014) both in local and systemic leaves. High induction of PR and G3Pdh genes was observed in systemic leaves of athmad1 plants overtime compared to WT suggesting negative role of ATHMAD1 in SAR.
Together all these results suggest that AtHMAD1 gene product negatively regulates multiple modes of disease resistance such as basal defense, R-gene mediated resistance, hypertensive cell death response, and systemic acquired disease resistance. Furthermore, it negatively regulates shoot growth while positively regulates root growth under nitrosative stress condition. This suggests a regulatory role for AtHMAD1 in NO biology.
Author Contributions
QI, AH, B-WY designed the experiments QI, NF, AH, B-GM, AS executed the experiments S-UL, K-MK, AS analyzed the data, QI, AH, B-WY wrote the manuscript.
Funding
This work was supported by a grant from the Next-Generation Biogreen 21 Program (SSAC, Grant No: PJ01110201), Rural Development Administration, Republic of Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01712/full#supplementary-material
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer IBR and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Footnotes
- ^ http://www.arabidopsis.org/
- ^ http://string-db.org/
- ^ http://geneontology.org/
- ^ http://arabidopsis.info/
- ^ www.photoshop.com
- ^ http://geneontology.org/
- ^ http://enzyme.expasy.org/
References
Alvarez, M. E., Pennell, R. I., Meijer, P. J., Ishikawa, A., Dixon, R. A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. doi: 10.1016/S0092-8674(00)81405-1
Apostol, I., Heinstein, P. F., and Low, P. S. (1989). Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: role in defense and signal transduction. Plant Physiol. 90, 109–116. doi: 10.1104/pp.90.1.109
Arasimowicz, M., and Floryszak-Wieczorek, J. (2007). Nitric oxide as a bioactive signalling molecule in plant stress responses. Plant Sci. 172, 876–887. doi: 10.1016/j.plantsci.2007.02.005
Barth, O., Vogt, S., Uhlemann, R., Zschiesche, W., and Humbeck, K. (2009). Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol. Biol. 69, 213–226. doi: 10.1007/s11103-008-9419-0
Beligni, M. V., and Lamattina, L. (1999). Nitric oxide counteracts cytotoxic processes mediated by reactive oxygen species in plant tissues. Planta 208, 337–344. doi: 10.1007/s004250050567
Burniston, M. T., and Wilson, D. J. (2008). Radioiodine ablation outcomes after imaging with I-123 or I-131: Is no news good news? J. Nucl. Med. 49, 166–166. doi: 10.2967/jnumed.107.047076
Caarls, L., Pieterse, C. M., and Van Wees, S. C. (2015). How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6:170. doi: 10.3389/fpls.2015.00170
Cawley, S., Bekiranov, S., Ng, H. H., Kapranov, P., Sekinger, E. A., Kampa, D., et al. (2004). Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116, 499–509. doi: 10.1016/S0092-8674(04)00127-8
Chanda, B., Xia, Y., Mandal, M. K., Yu, K. S., Sekine, K. T., Gao, Q. M., et al. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43, 421–427. doi: 10.1038/ng.798
Cho, D. H., Nakamura, T., Fang, J. G., Cieplak, P., Godzik, A., Gu, Z., et al. (2009). S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science 324, 102–105. doi: 10.1126/science.1171091
Clarke, A., Desikan, R., Hurst, R. D., Hancock, J. T., and Neill, S. J. (2000). NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 24, 667–677. doi: 10.1046/j.1365-313x.2000.00911.x
Clemens, S. (2001). Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212, 475–486. doi: 10.1007/s004250000458
Culotta, E., and Koshland, D. E. (1992). No news is good-news. Science 258, 1862–1865. doi: 10.1126/science.1361684
de Abreu-Neto, J. B., Turchetto-Zolet, A. C., de Oliveira, L. F. V., Zanettini, M. H. B., and Margis-Pinheiro, M. (2013). Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 280, 1604–1616. doi: 10.1111/febs.12159
Dellagi, A., Brisset, M. N., Paulin, J. P., and Expert, D. (1998). Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant Microbe Interact. 11, 734–742. doi: 10.1094/MPMI.1998.11.8.734
Delledonne, M., Xia, Y., Dixon, R. A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. doi: 10.1038/29087
Dupont, C. L., Butcher, A., Valas, R. E., Bourne, P. E., and Caetano-Anolles, G. (2010). History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl. Acad. Sci. U.S.A. 107, 10567–10572. doi: 10.1073/pnas.0912491107
Durner, J., Wendehenne, D., and Klessig, D. F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 95, 10328–10333. doi: 10.1073/pnas.95.17.10328
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Dykema, P. E., Sipes, P. R., Marie, A., Biermann, B. J., Crowell, D. N., and Randall, S. K. (1999). A new class of proteins capable of binding transition metals. Plant Mol. Biol. 41, 139–150. doi: 10.1023/A:1006367609556
El-Shetehy, M., Wang, C. X., Shine, M. B., Yu, K. S., Kachroo, A., and Kachroo, P. (2015). Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. 10:e998544. doi: 10.1080/15592324.2014.998544
Espunya, M. C., De Michele, R., Gómez-Cadenas, A., and Martínez, M. C. (2012). S-Nitrosoglutathione is a component of wound- and salicylic acid-induced systemic responses in Arabidopsis thaliana. J. Exp. Bot. 63, 3219–3227. doi: 10.1093/jxb/ers043
Feechan, A., Kwon, E., Yun, B.-W., Wang, Y., Pallas, J. A., and Loake, G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102, 8054–8059. doi: 10.1073/pnas.0501456102
Foster, M. W., Liu, L. M., Zeng, M., Hess, D. T., and Stamler, J. S. (2009). A genetic analysis of nitrosative stress. Biochemistry 48, 792–799. doi: 10.1021/bi801813n
Fourcroy, P., Vansuyt, G., Kushnir, S., Inze, D., and Briat, J. F. (2004). Iron-regulated expression of a cytosolic ascorbate peroxidase encoded by the APX1 gene in Arabidopsis seedlings. Plant Physiol. 134, 605–613. doi: 10.1104/pp.103.029876
Freund, C., Schmalz, H. G., Sticht, J., and Kuhne, R. (2008). Proline-rich sequence recognition domains (PRD): ligands, function and inhibition. Handb. Exp. Pharmacol. 186, 407–429. doi: 10.1007/978-3-540-72843-6_17
Fukuda, Y. (1997). Interaction of tobacco nuclear protein with an elicitor-responsive element in the promoter of a basic class I chitinase gene. Plant Mol. Biol. 34, 81–87. doi: 10.1023/A:1005737128339
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001. doi: 10.1126/science.1175550
Gao, Q. M., Venugopal, S., Navarre, D., and Kachroo, A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476. doi: 10.1104/pp.110.166876
García-Mata, C., and Lamattina, L. (2001). Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 126, 1196–1204. doi: 10.1104/pp.126.3.1196
Glazebrook, J., Rogers, E. E., and Ausubel, F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982.
Grant, J. J., and Loake, G. J. (2000). Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 124, 21–30. doi: 10.1104/pp.124.1.21
Greenberg, J. T., and Yao, N. (2004). The role and regulation of programmed cell death in plant–pathogen interactions. Cell Microbiol. 6, 201–211. doi: 10.1111/j.1462-5822.2004.00361.x
Grotz, N., and Guerinot, M. L. (2006). Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta Mol. Cell Res. 1763, 595–608. doi: 10.1016/j.bbamcr.2006.05.014
Gurr, S. J., and Rushton, P. J. (2005). Engineering plants with increased disease resistance: How are we going to express it? Trends Biotechnol. 23, 283–290. doi: 10.1016/j.tibtech.2005.04.009
Haas, C. E., Rodionov, D. A., Kropat, J., Malasarn, D., Merchant, S. S., and de Crecy-Lagard, V. (2009). A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. doi: 10.1186/1471-2164-10-470
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Huffman, D. L., and O’Halloran, T. V. (2001). Function, structure, and mechanism of intracellular copper trafficking proteins. Annu. Rev. Biochem. 70, 677–701. doi: 10.1146/annurev.biochem.70.1.677
Hussain, A., Mun, B. G., Imran, Q. M., Lee, S. U., Adamu, T. A., Shahid, M., et al. (2016). Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Frontiers Plant Sci. 7:975. doi: 10.3389/fpls.2016.00975
Imran, Q. M., Kamran, M., Rehman, S. U., Ghafoor, A., Falak, N., Kim, K. M., et al. (2016). GA mediated OsZAT-12 expression improves salt resistance of rice. Int. J. Agric. Biol. 18, 330–336. doi: 10.17957/IJAB/15.0090
Jabs, T., Dietrich, R. A., and Dangl, J. L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273, 1853–1856. doi: 10.1126/science.273.5283.1853
Jaffrey, S. R., Erdjument-Bromage, H., Ferris, C. D., Tempst, P., and Snyder, S. H. (2001). Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3, 193–197. doi: 10.1038/35055104
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kallberg, M., Wang, H., Wang, S., Peng, J., Wang, Z., Lu, H., et al. (2012). Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522. doi: 10.1038/nprot.2012.085
Kim, J. S., Mizoi, J., Yoshida, T., Fujita, Y., Nakajima, J., Ohori, T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52, 2136–2146. doi: 10.1093/pcp/pcr143
Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. doi: 10.2307/3869093
Kopyra, M., and Gwóźdź, E. A. (2003). Nitric oxide stimulates seed germination and counteracts the inhibitory effect of heavy metals and salinity on root growth of Lupinus luteus. Plant Physiol. Biochem. 41, 1011–1017. doi: 10.1016/j.plaphy.2003.09.003
Latson, L. (2011). No news is good news, but. Catheter. Cardiovasc. Interv. 78, 583–583. doi: 10.1002/ccd.23348
Leister, R. T., Ausubel, F. M., and Katagiri, F. (1996). Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl. Acad. Sci. U.S.A. 93, 15497–15502. doi: 10.1073/pnas.93.26.15497
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
López, M. A., Bannenberg, G., and Castresana, C. (2008). Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr. Opin. Plant Biol. 11, 420–427. doi: 10.1016/j.pbi.2008.05.002
MauchMani, B., and Slusarenko, A. J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212. doi: 10.1105/tpc.8.2.203
Medina-Rivera, A., Defrance, M., Sand, O., Herrmann, C., Castro-Mondragon, J. A., Delerce, J., et al. (2015). RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 43, W50–W56. doi: 10.1093/nar/gkv362
Mi, H. Y., Muruganujan, A., Casagrande, J. T., and Thomas, P. D. (2013). Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566. doi: 10.1038/nprot.2013.092
Mukumoto, F., Hirose, S., Imaseki, H., and Yamazaki, K. (1993). DNA-sequence requirement of a tata element-binding protein from Arabidopsis for transcription in-vitro. Plant Mol. Biol. 23, 995–1003. doi: 10.1007/Bf00021814
Nakao, M., Nakamura, R., Kita, K., Inukai, R., and Ishikawa, A. (2011). Non-host resistance to penetration and hyphal growth of Magnaporthe oryzae in Arabidopsis. Sci. Rep. 1:171. doi: 10.1038/Srep00171
Niki, T., Mitsuhara, I., Seo, S., Ohtsubo, N., and Ohashi, Y. (1998). Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 39, 500–507. doi: 10.1093/oxfordjournals.pcp.a029397
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Park, S. W., Kaimoyo, E., Kumar, D., Mosher, S., and Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
Parker, J. E., Holub, E. B., Frost, L. N., Falk, A., Gunn, N. D., and Daniels, M. J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. doi: 10.1105/tpc.8.11.2033
Pastore, A., Piemonte, F., Locatelli, M., Lo Russo, A., Gaeta, L. M., Tozzi, G., et al. (2001). Determination of blood total, reduced, and oxidized glutathione in pediatric subjects. Clin. Chem. 47, 1467–1469.
Penninckx, I. A. M. A., Eggermont, K., Terras, F. R. G., Thomma, B. P. H. J., DeSamblanx, G. W., Buchala, A., et al. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. doi: 10.1105/tpc.8.12.2309
Pompella, A., Visvikis, A., Paolicchi, A., De Tata, V., and Casini, A. F. (2003). The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 66, 1499–1503. doi: 10.1016/S0006-2952(03)00504-5
Rizhsky, L., Liang, H., Shuman, J., Shulaev, V., Davletova, S., and Mittler, R. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. doi: 10.1104/pp.103.033431
Robinson, N. J., and Winge, D. R. (2010). Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562. doi: 10.1146/annurev-biochem-030409-143539
Rubino, J. T., and Franz, K. J. (2012). Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J. Inorg. Biochem. 107, 129–143. doi: 10.1016/j.jinorgbio.2011.11.024
Rushton, P. J., and Somssich, I. E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1, 311–315. doi: 10.1016/1369-5266(88)80052-9
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H. Y., and Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. doi: 10.1105/tpc.8.10.1809
Satoh, S., Kimura, T., Toda, M., Maekawa, M., Ono, S., Narita, H., et al. (1997). Involvement of L-type-like amino acid transporters in S-nitrosocysteine-stimulated noradrenaline release in the rat hippocampus. J. Neurochem. 69, 2197–2205. doi: 10.1046/j.1471-4159.1997.69052197.x
Shah, C. M., Bell, S. E., Locke, I. C., Chowdrey, H. S., and Gordge, M. P. (2007). Interactions between cell surface protein disulphide isomerase and S-nitrosoglutathione during nitric oxide delivery. Nitric Oxide 16, 135–142. doi: 10.1016/j.niox.2006.08.001
Shah, J., and Zeier, J. (2013). Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 4:30. doi: 10.3389/Fpls.2013.00030
Sharma, A., Hussain, A., Mun, B. G., Imran, Q. M., Falak, N., Lee, S. U., et al. (2016). Comprehensive analysis of plant rapid alkalization factor (RALF) genes. Plant Physiol. Biochem. 106, 82–90. doi: 10.1016/j.plaphy.2016.03.037
Shi, S., Wang, G., Wang, Y., Zhang, L., and Zhang, L. (2005). Protective effect of nitric oxide against oxidative stress under ultraviolet-B radiation. Nitric Oxide 13, 1–9. doi: 10.1016/j.niox.2005.04.006
Shirsat, A., Wilford, N., Croy, R., and Boulter, D. (1989). Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol. Gen. Genet. 215, 326–331. doi: 10.1007/BF00339737
Singh, D., and Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6:895. doi: 10.3389/Fpls.2015.00895
Song, F. M., and Goodman, R. M. (2001). Activity of nitric oxide is dependent on, but is partially required for function of, salicylic acid in the signaling pathway in tobacco systemic acquired resistance. Mol. Plant Microbe Interact. 14, 1458–1462. doi: 10.1094/Mpmi.2001.14.12.1458
Stamler, J., Singel, D., and Loscalzo, J. (1992). Biochemistry of nitric oxide and its redox-activated forms. Science 258, 1898–1902. doi: 10.1126/science.1281928
Sudhakar, C., Lakshmi, A., and Giridarakumar, S. (2001). Changes in the antioxidant enzyme efficacy in two high yielding genotypes of mulberry (Morus alba L.) under NaCl salinity. Plant Sci. 161, 613–619. doi: 10.1016/S0168-9452(01)00450-2
Tada, Y. (2009). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins (August, pg 952, 2008). Science 325, 1072–1072. doi: 10.1126/science.1156970
Tehseen, M., Cairns, N., Sherson, S., and Cobbett, C. S. (2010). Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2, 556–564. doi: 10.1039/c003484c
Tuteja, N., Chandra, M., Tuteja, R., and Misra, M. K. (2004). Nitric oxide as a unique bioactive signaling messenger in physiology and pathophysiology. J. Biomed. Biotechnol. 2004, 227–237. doi: 10.1155/S1110724304402034
Uchida, A., Jagendorf, A. T., Hibino, T., Takabe, T., and Takabe, T. (2002). Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 163, 515–523. doi: 10.1016/S0168-9452(02)00159-0
Uehara, T., Nakamura, T., Yao, D. D., Shi, Z. Q., Gu, Z. Z., Ma, Y. L., et al. (2006). S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517. doi: 10.1038/nature04782
Valderrama, R., Corpas, F. J., Carreras, A., Fernández-Ocaña, A., Chaki, M., Luque, F., et al. (2007). Nitrosative stress in plants. FEBS Lett. 581, 453–461. doi: 10.1016/j.febslet.2007.01.006
van Dam, N. M. (2009). How plants cope with biotic interactions. Plant Biol. 11, 1–5. doi: 10.1111/j.1438-8677.2008.00179.x
Wang, C. X., El-Shetehy, M., Shine, M. B., Yu, K. S., Navarre, D., Wendehenne, D., et al. (2014). Free radicals mediate systemic acquired resistance. Cell Rep. 7, 348–355. doi: 10.1016/j.celrep.2014.03.032
Wendehenne, D., Gao, Q. M., Kachroo, A., and Kachroo, P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20, 127–134. doi: 10.1016/j.pbi.2014.05.012
Wernimont, A. K., Huffman, D. L., Lamb, A. L., O’Halloran, T. V., and Rosenzweig, A. C. (2000). Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat. Struct. Mol. Biol. 7, 766–771. doi: 10.1038/78999
Williams, L. E., Pittman, J. K., and Hall, J. L. (2000). Emerging mechanisms for heavy metal transport in plants. Biochim. Biophys. Acta Biomembr. 1465, 104–126. doi: 10.1016/S0005-2736(00)00133-4
Wittkopp, P. J., and Kalay, G. (2012). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Xiong, L., Schumaker, K. S., and Zhu, J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl. 1), S165–S183. doi: 10.1105/tpc.000596
Yu, D. Q., Cen, C., Li, B. J., and Fu, J. R. (1999). Systemic acquired disease resistance and signal transduction in plant. Acta Bot. Sin. 41, 115–124.
Yu, K. S., Soares, J. M., Mandal, M. K., Wang, C. X., Chanda, B., Gifford, A. N., et al. (2013). A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 3, 1266–1278. doi: 10.1016/j.celrep.2013.03.030
Yun, B.-W., Feechan, A., Yin, M., Saidi, N. B. B., Le Bihan, T., Yu, M., et al. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. doi: 10.1038/nature10427
Yun, B. W., Skelly, M. J., Yin, M., Yu, M., Mun, B. G., Lee, S. U., et al. (2016). Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol. 211, 516–526. doi: 10.1111/nph.13903
Zai, A., Rudd, M. A., Scribner, A. W., and Loscalzo, J. (1999). Cell-surface protein disulfide isomerase catalyzes transnitrosation and regulates intracellular transfer of nitric oxide. J. Clin. Investig. 103, 393–399. doi: 10.1172/Jci4890
Zhao, L., Zhang, F., Guo, J., Yang, Y., Li, B., and Zhang, L. (2004). Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 134, 849–857. doi: 10.1104/pp.103.030023
Zhu, J., Li, S., Marshall, Z. M., and Whorton, A. R. (2008). A cystine-cysteine shuttle mediated by xCT facilitates cellular responses to S-nitrosoalbumin. Am. J. Physiol. Cell Physiol. 294, C1012–C1020. doi: 10.1152/ajpcell.00411.2007
Keywords: R-gene mediated resistance, basal defense, HR response, nitrosative stress, RNA-seq profile
Citation: Imran QM, Falak N, Hussain A, Mun B-G, Sharma A, Lee S-U, Kim K-M and Yun B-W (2016) Nitric Oxide Responsive Heavy Metal-Associated Gene AtHMAD1 Contributes to Development and Disease Resistance in Arabidopsis thaliana. Front. Plant Sci. 7:1712. doi: 10.3389/fpls.2016.01712
Received: 22 September 2016; Accepted: 31 October 2016;
Published: 21 November 2016.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Robin K. Cameron, McMaster University, CanadaInes Ben Rejeb, University of Neuchâtel, Switzerland
Copyright © 2016 Imran, Falak, Hussain, Mun, Sharma, Lee, Kim and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Wook Yun, bwyun@knu.ac.kr
†These authors have contributed equally to this work.
 Q. Muhammad Imran
Q. Muhammad Imran Noreen Falak
Noreen Falak Adil Hussain
Adil Hussain Bong-Gyu Mun
Bong-Gyu Mun Arti Sharma1
Arti Sharma1 Sang-Uk Lee
Sang-Uk Lee Kyung-Min Kim
Kyung-Min Kim Byung-Wook Yun
Byung-Wook Yun