- Department of Plant Physiology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
The phloem-feeding whitefly Bemisia tabaci is a serious pest to a broad range of host plants, including many economically important crops such as tomato. These insects serve as a vector for various devastating plant viruses. It is known that whiteflies are capable of manipulating host-defense responses, potentially mediated by effector molecules in the whitefly saliva. We hypothesized that, beside putative effector proteins, small RNAs (sRNA) are delivered by B. tabaci into the phloem, where they may play a role in manipulating host plant defenses. There is already evidence to suggest that sRNAs can mediate the host-pathogen dialogue. It has been shown that Botrytis cinerea, the causal agent of gray mold disease, takes advantage of the plant sRNA machinery to selectively silence host genes involved in defense signaling. Here we identified sRNAs originating from B. tabaci in the phloem of tomato plants on which they are feeding. sRNAs were isolated and sequenced from tomato phloem of whitefly-infested and control plants as well as from the nymphs themselves, control leaflets, and from the infested leaflets. Using stem-loop RT-PCR, three whitefly sRNAs have been verified to be present in whitefly-infested leaflets that were also present in the whitefly-infested phloem sample. Our results show that whitefly sRNAs are indeed present in tomato tissues upon feeding, and they appear to be mobile in the phloem. Their role in the host-insect interaction can now be investigated.
Introduction
Bemisia tabaci (Hemiptera), commonly known as whitefly, is a polyphagous insect that is a threat for many crops across the globe. These insects can reduce crop yield in a number of ways; (1) through transmission of yield-limiting plant viruses (Navas-Castillo et al., 2011); (2) via honeydew excrement, which results in growth of sooty molds leading to a reduction of photosynthesis (Walling, 2008) or the release of the glycoside of salicylic acid (VanDoorn et al., 2015) or; (3) ingestion of phloem sap thereby depleting plants of photosynthetic compounds (Buntin et al., 1993).
Plants can defend themselves against herbivores and pathogens in various ways e.g., via physical barriers, volatile or non-volatile compounds, and through induction of defense responses controlled by various phytohormones (Walling, 2008; Kant et al., 2015). Trichomes can act both as physical barriers and as metabolite production facilities. Trichomes on the leaves will hinder small herbivores in their movement and finding suitable feeding places (Simmons and Gurr, 2005). In addition, trichomes can produce specialized metabolites such as repellent volatiles or exudates that can be toxic or that trap herbivores (Simmons and Gurr, 2005; Walling, 2008; Bleeker et al., 2009). The phytohormones involved in herbivore-defense responses are predominantly jasmonic acid (JA) and salicylic acid (SA). SA can antagonize the JA-mediated signaling responses (Koornneef and Pieterse, 2008). Adult whiteflies feeding on tomato induce the SA-response thereby suppressing the JA-response (Shi et al., 2014). During the feeding of whitefly nymphs on Arabidopsis, transcript levels of SA-induced genes became higher while JA-related transcript levels decreased (Kempema et al., 2007; Zarate et al., 2007).
After hatching from the egg, whitefly nymphs are mobile and will select the site where they will feed and develop into an adult while being immobile. Feeding is initiated by insertion of a specialized mouthpiece (stylet) through the leaf surface toward the phloem sieve elements in a mostly intercellular fashion (Pollard, 1955; Jiang et al., 1999; Jiang and Walker, 2003). This insertion is facilitated by the excretion of gel-like saliva, in a similar way as an aphid, and other stylet- and phloem-feeding insects (Jiang et al., 1999; Moreno et al., 2011). After the stylet enters the sieve element, watery saliva is excreted and ingestion of phloem sap starts (Jiang et al., 1999; Jiang and Walker, 2003). Plants try to close the opening made by the stylet by depositing callose and proteins (Kempema et al., 2007) and phloem-feeding insects try to counteract this (Will et al., 2007).
There is evidence that herbivore saliva contains factors that can manipulate plant defenses (Will et al., 2013; Sharma et al., 2014; Su et al., 2015; Peng et al., 2016; Villarroel et al., 2016). For hemipterans most knowledge stems from work with aphids: several salivary proteins (effectors) have been identified that affect aphid reproductive rate (Bos et al., 2010; Pitino and Hogenhout, 2013). The aphid salivary proteins C002, Mp1, and Mp2 increase fecundity, while Mp10 and Mp42 reduce aphid fecundity (Bos et al., 2010; Pitino and Hogenhout, 2013). The production of effector proteins by aphids seems to be analogous to that of plant pathogens to establish disease. Such plant pathogens can interfere with the defense response of their host by secreting effectors that interact with host proteins and modulate these to their benefit.
Besides effector-protein interactions, small non-coding RNAs (sRNAs) between 21 and 24 nucleotides long (nts) have been shown to mediate interactions between hosts and pathogens (Knip et al., 2014; Baulcombe, 2015). Regarding plants, one of the best-studied examples is the Botrytis cinerea infection of Arabidopsis and tomato (Weiberg et al., 2013). After fungal infection, 73 sRNAs from Botrytis were found in infected leaves (Weiberg et al., 2013). These Botrytis sRNAs take advantage of the plant's own silencing machinery to mediate their action (i.e., targeting ARGONAUTE 1). Another example comes from the green peach aphid (Myzus persicae) that displays reduced fecundity on Arabidopsis mutants affected in their miRNA biogenesis pathway (i.e., Dicer-like1 dcl1 and Argonaute1 ago1; Kettles et al., 2013). These results indicate that sRNA pathways are not only involved in plant resistance against a phloem-feeding insect, but also suggest that aphids produce sRNAs that can influence plant-defense responses (Kettles et al., 2013).
Here we show that whiteflies transfer sRNAs to the host plant they are feeding from. To detect whitefly-specific sRNAs within the phloem of tomato plants small RNA sequencing was utilized. These sRNAs are detected in isolated phloem sap indicating they are mobile. The presence of three whitefly sRNAs in tomato was confirmed by means of stem-loop RT-PCR. Our findings are, to our knowledge, the first confirmation of the transfer of insect sRNA to phloem.
Methods and Materials
Whitefly Rearing and Tomato Infestation
Whiteflies (B. tabaci biotype B) were reared in a climatised chamber (Snijders, Tilburg; 28°C, 16 h light 150 μE m−2 s−1, RH 75%) as previously described (Bleeker et al., 2011), on a diet of cucumber plants (Cucumis sativus, Ventura, RijkZwaan). Two weeks after sowing, 5 tomato plants (Solanum lycopersicum, cultivar Moneymaker) were placed in a netted insect dome (60 × 60 × 90 cm) and infested with ±200 adult whiteflies (greenhouse 22–25°C, 16/8 h photoperiod at 500 μE m−2 s−1). Aiming for a consistent treatment with the different instar stages present, whiteflies (±100) were added 3 times per week until week 4 after sowing. In week 6 after sowing the samples for small RNA sequencing were collected.
Phloem, Nymph, and Leaf Collection for sRNA-seq
For phloem collection plants were kept in the greenhouse under standard greenhouse conditions (22–25°C, 16/8 h photoperiod at 500 μE m−2 s−1). Phloem sap was collected from control and whitefly-infested leaflets using the “EDTA” method (King and Zeevaart, 1974) during the light-period (see also Figure S1 and Tetyuk et al., 2013). Adult whiteflies were removed from treated leaflets by aspiration. Leaflets with a high density of nymphs were excised and the petioles were carefully submerged in phloem collection buffer (5 mM EDTA, 5 mM phosphate buffer pH 6.8). The petioles of 3–6 leaflets were then cut once more while submerged in buffer and placed in a 2-ml Eppendorf vial containing phloem-collection buffer to bleed for 30 min under high humidity. After this the leaflets were transferred to collection tubes with fresh phloem-collection buffer supplemented with protease inhibitor (1 Complete Protease Inhibitor Tablet (Roche) 100 ml−1 water) and phloem samples were collected for 6 h under high humidity before being snap frozen in liquid nitrogen. Nymphs (1st, 2nd, and 3rd instar) were collected from a total of 4 infested leaflet using an insect pin, pooled, and transferred to 100% acetone. For the infested sample (LW) in addition to nymphs, eggs were removed as accurate as possible as well, after which the leaflet samples were separately snap frozen in liquid nitrogen (n = 4). Untreated control leaflets (LC) were harvested in exactly the same way and at the same time point (n = 4). For an additional control, leaflets with the eggs remaining were included (LE, n = 4). For this adult whiteflies were placed on leaflets for 24 h after which the adults were removed.
Total and Small RNA Isolation
Total RNA from phloem samples was isolated using concentrated TRIzol reagent (Life Technologies). The leaf samples (for each treatment four replicates pooled) and one nymph sample were ground in liquid nitrogen. Total RNA was isolated using the E.Z.N.A.® MicroElute RNA Clean Up Kit (Omega Bio-Tek). Briefly, TRIzol Reagent (Life Technologies) and chloroform was added according to the manufacturer's instructions. After centrifugation, the RNA-containing aqueous phase was collected, mixed with 1.5 volume of 100% ethanol and applied to a MicroElute spin column (Omega Bio-Tek). The column was washed according to the manufacturers's instructions: once with RWT buffer (Qiagen), once with RPE washing buffer (Qiagen) and finally with 80% ethanol. The RNA concentration was measured on a NanoDrop ND-2000 (Thermo Scientific) and RNA integrity was examined using the 2200 TapeStation System with Agilent RNA ScreenTapes (Agilent Technologies).
Total RNA was spiked with ERCCs spike-in mix 1 (Life Technologies) as well as a synthetic spike-in set for Size Range Quality Control (SRQC) together with an External Reference for Data Normalization (ERDN; Locati et al., 2015). Both phloem samples from the control and whitefly-infested plants were not spiked. The total RNA was divided in a large and a small fraction. The large RNA fraction was bound to a mirVana™ spin column (mirVana™ miRNA Isolation Kit, Life Technologies) according to the manufacturer's instructions. Small RNAs (<200 nts) were purified from the flow-through by adding ethanol to a final concentration of 65% (v/v) and bound to an E.Z.N.A.® MicroElute spin column. The column was washed once with RWT buffer, once with RPE buffer and once with 80% ethanol (Qiagen). The concentration and integrity of small RNA was examined as described above.
Next-Generation Sequencing
Bar-coded small RNA libraries of the 6 different samples were generated according to the manufacturer's protocols using the Ion Total RNA-Seq Kit v2 and the Ion Xpress™ RNA-Seq bar-coding kit (Life Technologies). The size distribution and yield of the bar-coded libraries were assessed using the 2200 TapeStation System with Agilent D1K ScreenTapes (Agilent Technologies). Sequencing templates were prepared on the Ion Chef™ System using the Ion PI Hi-Q Chef Kit (Life Technologies). Sequencing was performed on an Ion Proton™ System using Ion PI v3 chips (Life Technologies) according to the manufacturer's instructions.
Bioinformatic Analyses
Bioinformatic analyses were done using the Snakemake workflow management tool (Köster and Rahmann, 2012) to generate bioinformatic pipelines. Software used was Bowtie2 v2.1.0 (Langmead and Salzberg, 2012), Samtools v1.2 (Li et al., 2009), Python v3.3.3, Python package Pandas 0.14.1, and Biopython 1.64 (Cock et al., 2009), STAR v2.4.0 (Dobin et al., 2013), R v3.2.1 (R Core Team, 2016). All sequences <18 and >40 nts were removed. Contaminating sequences were removed by alignment to plant virus databases (Adams and Antoniw, 2006), other types of RNA (rRNA, tRNA, snoRNA, degraded messenger RNA, mitochondrial RNA) using the RFAM 12.0 database for tomatoes (excluding microRNA; Nawrocki et al., 2015) and the publically available tomato transcriptome [ITAG2.3, solgenomics.net Tomato Genome Consortium, 2012]. sRNAs were normalized for comparisons and expressed as RPKM (Reads Per Kilobase per Million mapped reads; Table S1).
The online psRNATarget tool (Dai and Zhao, 2011) was used to retrieve mRNA targets of selected miRNAs (using the Solanum lycopersicum ITAG2.4 cDNA reference) with default parameters: a seed region length of 20 nts to score complementarity between target and miRNA and a target accessibility (maximum energy required to open the mRNA secondary structure around the target site) of 25.
Stem-Loop RT-PCR of Small RNAs
Stem-loop RT-PCR of small RNAs was conducted to confirm findings of the sRNA-seq. For this analyses total RNA was isolated from four biological replicates of leaflet samples using the method described above and primers for the specific small RNAs were designed (Varkonyi-Gasic et al., 2007; Kramer, 2011; Table S2). A total of 100 ng RNA per sample was used for a reverse transcriptase reaction (RevertAid H Minus reverse Transcriptase, Thermo fisher) in a total volume of 20 μL with small RNA specific RT-primers (Table S2). A pulse reverse-transcriptase reaction (RT) was used (Varkonyi-Gasic et al., 2007). For the stem-loop RT-PCRs, 2 μL of RT reaction was used as template in a total volume of 50 μL for 35 cycles with an annealing temperature of 58°C. PCR products were cloned with CloneJET PCR cloning kit (Thermo scientific) and sequenced to validate amplicon specificity.
Results
Small RNA Sequencing: Detection of Small RNAs in Phloem and Leaves
Phloem samples were collected from uninfested tomato leaflets (Phloem control, PC) and compared to phloem samples from tomato leaflets infested with whitefly nymphs and eggs (Phloem whitefly, PW, Figure 1). Whitefly nymphs (WN) were separately collected from leaflets of the same plants used to obtain phloem sap exudates (Figure 1). After sRNA sequencing we obtained 31,231,948 sRNAs in the WN sample and 5,370,176 and 7,321,768 sRNAs in the PC and PW samples respectively (see also Table S1). To determine which sRNAs originated from whiteflies and were present in phloem, a bioinformatic pipeline was designed (Figure 1). For this analysis, sequences shorter than 18 nucleotides (nts) and longer than 40 nts were removed. Next, we removed sequences that aligned to plant viruses (Adams and Antoniw, 2006), other types of RNA (rRNA, tRNA, snoRNA, degraded messenger RNA, mitochondrial RNA) using the RFAM 12.0 database for tomatoes (excluding microRNA; Nawrocki et al., 2015) and the publically available tomato transcriptome (ITAG2.3, Tomato Genome Consortium, 2012). To eliminate sequences present in PW but unrelated to whitefly infestation, PC sRNAs were excluded from the PW sRNAs (PW–PC, Figure 1). Of these PW-PC sRNAs a final 144,646 non-redundant sequences overlapped with WN sequence (Figure 1, Table S1) and were regarded as putatively transferred from the insect into the phloem.
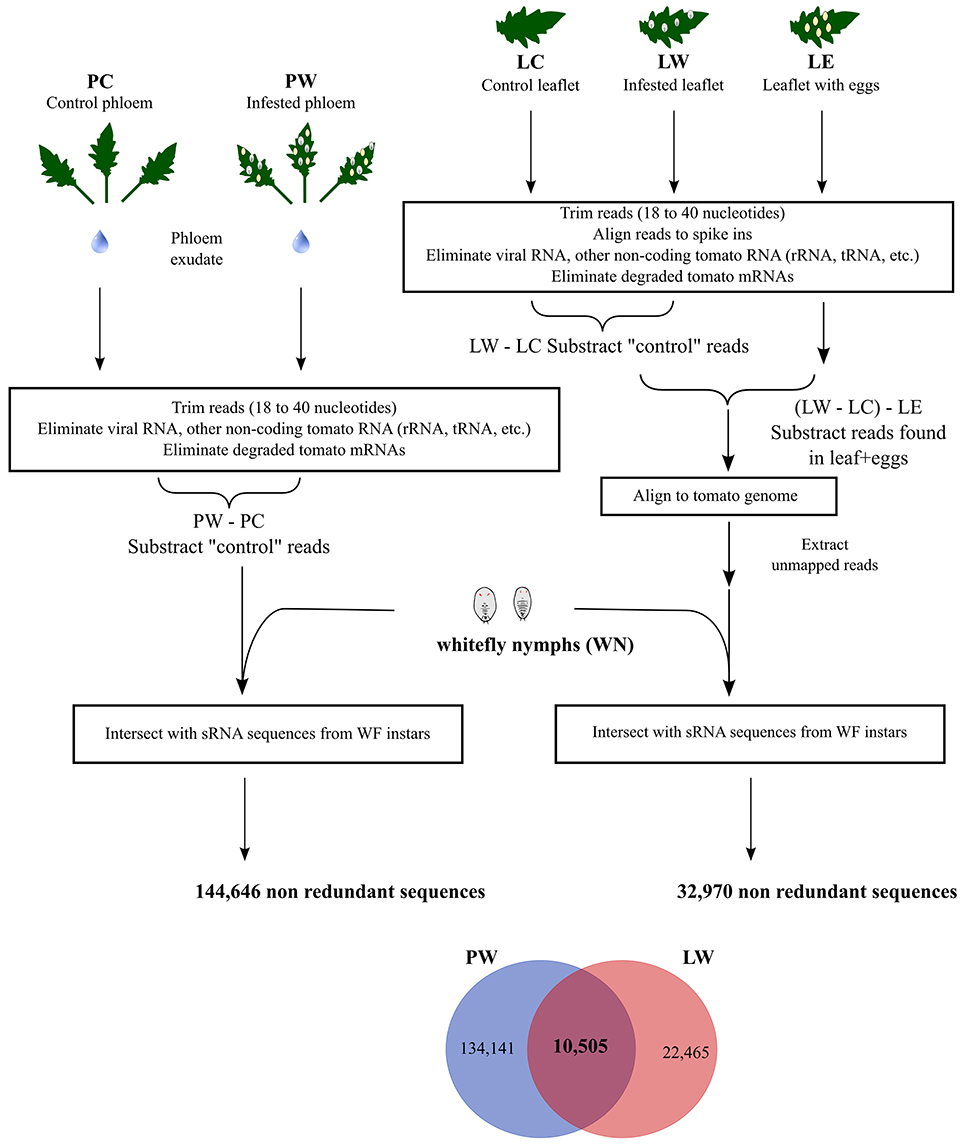
Figure 1. Bioinformatic pipeline. Small RNAs (sRNAs) were isolated from tomato-phloem exudates from either control leaflets (PC) or leaflets infested with whitefly nymphs (PW), and from control tomato leaflets (LC), whitefly nymphs (WN), leaflets first infested with whiteflies but with adults and nymphs subsequently removed (LW) and tomato leaflets with only eggs (LE). A bioinformatic workflow (see main text for details) was implemented yielding 10,505 non-redundant small RNAs present in feeding whitefly nymphs plus identified in both infested tomato phloem and leaflets.
In order to further validate whitefly-specific sRNAs present in tomato, we additionally sequenced sRNAs isolated from whole tomato leaflets (Figure 1, Table S1). We obtained 36,793,380 sRNAs from uninfested tomato leaflets (Leaf Control, LC), 33,780,469 sRNAs from infested leaflets after removal of nymphs (Leaf whitefly, LW) and 32,730,583 sRNAs from leaflets with only eggs (Leaf Eggs, LE). LC sRNAs were subtracted from the LW sRNAs (LW–LC, Figure 1). Next, to correct for any whitefly-specific egg sRNA that could have been left on the leaf surface of LW, the LE sequences were removed (Figure 1). The remaining whitefly nymph sRNAs were subsequently aligned against the tomato genome and the unmapped sequences were aligned with the tomato-fed WN sequences to find nymph-specific sRNAs. By doing so, we ended up with 32,970 non-redundant sequences (Figure 1). Finally, we searched (qualitatively) for sRNAs that would be both present amongst the 144,464 sRNAs coming from PW sample and the 32,970 sRNAs coming from infested LW sample, and found in WN nymph sample. This resulted in 10,505 non-redundant (Figure 1, Table S1) putative whitefly sRNAs found in both phloem exudate and leaflets containing phloem of whitefly-infested plants.
Length Distribution
The sRNA-length distributions from the different libraries ranging from 18 to 40 nts are shown in Figure 2. The sRNA length distributions of the three leaf samples (LC, LW, and LE) were very comparable with an expected major peak at 24 nts (35–40% of all sequences) and a minor peak at 21 nts (Figure 2). Compared to tomato leaf samples, both phloem samples exhibited a slightly different length distribution with a peak at 23–24 nts (~18–25% of all sequences). However, the sRNA length distribution of the whitefly nymphs (WN) was distinctly different from the other samples with two major peaks; one at 22 nts (~12% of all sequences) and one at 29–30 nts (~15–20% of all sequences; Figure 2). The 29–30 nt sequences, apparent in the nymph sample, appeared to be a well-defined peak in the phloem sample from the nymph-infested leaflet (PW), compared to the control phloem (PC).
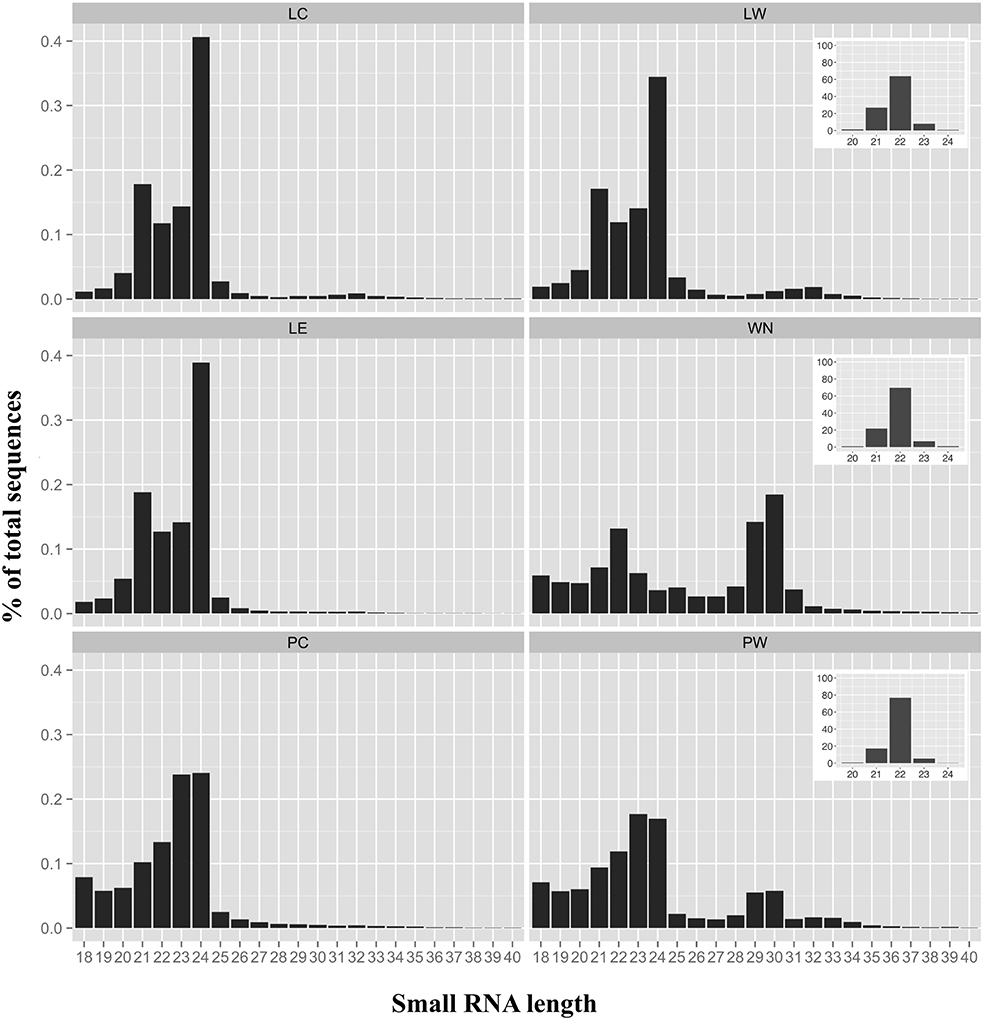
Figure 2. Size distribution of sRNAs. Size distributions and percentages of small RNAs (sRNAs) between 18 and 40 nucleotides are indicated for each sample. LC, control leaflets (not infested with whitefly nymphs); LW, leaflets first infested with whiteflies but with adults and nymphs subsequently removed; LE, leaflets with whitefly eggs; WN, whitefly nymphs; PC, phloem control (not infested); PW, phloem infested with whitefly nymphs. Insets for LW, WN, and PW represent the size distributions and percentages of the sRNAs matching previously described whitefly miRNAs (Guo et al., 2013).
Next, the sRNAs from whitefly-infested samples LW, WN, and PW were compared to 185 whitefly miRNA sequences found in two different biotypes of B. tabaci (Guo et al., 2013). All previously described whitefly miRNAs had a length comprised between 20 and 24 nts (Guo et al., 2013). Accordingly, 150 sRNAs of our WN sample could be exactly matched to the whitefly miRNAs published earlier (Table S3) and had a length predominantly centered around 22 nts, a feature characteristic of insect miRNAs (Figure 2, insets). The most abundant miRNAs found in the WN sample were miR-276a, miR-317 and miR-14 that appear to be conserved as well in other insects e.g., Bombyx mori, Apis mellifera, Drosophila melanogaster (Yin et al., 2016; Table S3).
Confirmation of Small RNAs in Leaf Samples
A sensitive stem-loop RT-PCR (Varkonyi-Gasic et al., 2007) was used to confirm the presence or absence of whitefly sRNAs in four biological replicates of whitefly-infested tomato leaflets (LW), non-infested leaflets (LC), leaflets with eggs (LE), and the B. tabaci nymph sample (WN). To verify there was no sRNA from whitefly nymph contaminating the LW sRNAs other than those transferred by the whitefly, a 29-nt sRNA (# 29691) was amplified as this sRNA proved particularly abundant in the WN sample. Figure 3A shows that sRNA #29691 was indeed specific to the nymph sample and was absent in the LW, LC, or LE samples, indicating that there is no whitefly nymph contamination in the LW sample after the infestation and that nymphs had been successfully removed. Sample quality was further checked using a known B. tabaci specific sRNA (Bta_miR2A; Guo et al., 2013) and a known tomato miR172 that is conserved among land plants (Taylor et al., 2014). Accordingly, the plant-specific miR172 was detected in all leaf samples (Figure 3B) while the whitefly Bta_miR2A was detected in the nymph samples of B. tabaci reared on tomato and in the leaf samples containing eggs (LW and LE; Figure 3C).
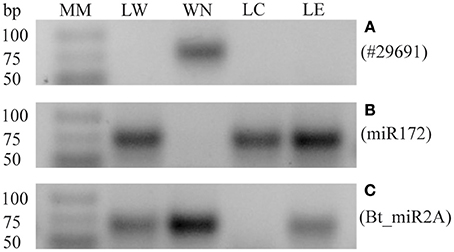
Figure 3. Quality check of leaflet samples by stem-loop RT-PCRs. (A) Expression of nymph-specific sRNA #29691 in: leaflet first infested with whiteflies but with adults and nymphs subsequently removed (LW), whitefly nymphs (WN), control leaflet (LC), or leaflet with eggs (LE), showing a specific band in WN only. (B) Validation of the expression of tomato specific sRNA Sly_miR172 in the different samples. Expression is detected in LW, LC, and LE, but not in the WN sample. (C) Expression of the known whitefly-specific miRNA Bta_miR2A is also detected in LE. Similar results were obtained in four biological replicates. Molecular Mass (MM), GeneRuler Ultra Low Range DNA ladder (Thermo Scientific).
Whitefly Small RNAs Transferred to Tomato
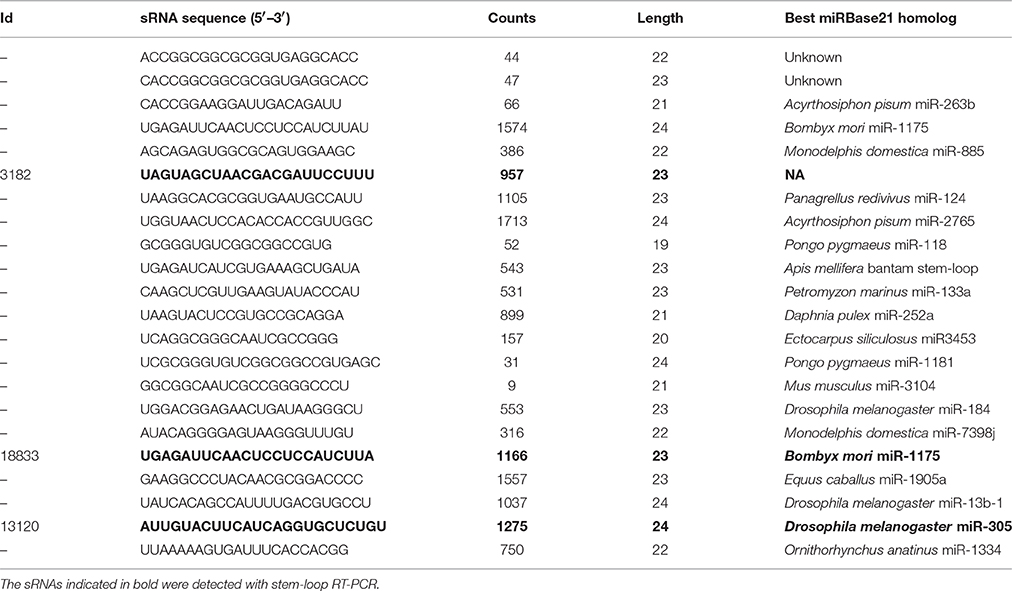
sRNA sequencing of the phloem samples identified the presence of mobile whitefly-originating sRNAs. We selected three whitefly candidate sRNAs from the final list (Table 1) for validation using stem-loop PCR on leaflets. The criteria for selecting these specific candidates from the sRNAseq data were (1) a length between 23 and 24 nt (Figure 2), (2) present among highest counts in nymphs, (3) presence in whitefly-infested leaflets (LW), in phloem from leaflets infested with whiteflies (PW) and in the B. tabaci nymph (WN) sample, while absent in the control leaflet (LC), absent in leaflets with only eggs (LE), and absent in the phloem control sample, and finally (4) preferably matching an insect-like or an unknown small RNA in the miRBase. From the three selected sRNAs, sRNA #13120, and #18833 were annotated as insect miR305 and miR1175-3p, respectively, using the miRBase (Kozomara and Griffiths-Jones, 2014). sRNAs #13120 (Figure 4A) and #18833 (Figure 4B) were present in nymph and were found back in three out of four LW samples while being absent in all LC and LE samples. sRNA #3182 did not provide a match in the miRBase but could be amplified in nymph and all infested leaf samples, however it was found in one out of four control samples. Overall, whitefly sRNAs could be detected within the leaflet samples (Figure 4C) on which nymph feeding took place. Since two out of three candidate small RNAs were found exclusively in the infested samples PW and LW, we conclude that whiteflies transfer small RNAs to the phloem, which then have the potential to move.
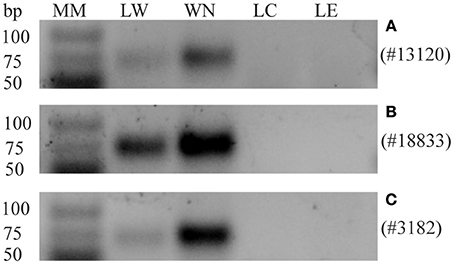
Figure 4. Detection of candidate transferred whitefly sRNAs in the nymph and leaflet samples by stem-loop RT-PCRs. Expression of sRNAs in: leaflet first infested with whiteflies but with adults and nymphs subsequently removed (LW), whitefly nymphs (WN), control leaflet (LC) or leaflet with eggs (LE). (A) sRNA #13120, a specific band is detected in LW and WN but not in LC and LE. (B) #18833, a specific band is detected in LW and WN but not in LC and LE. Similar results for #13120 and #18833 were obtained in four biological replicates. (C) #3182, a specific band is detected in LW and WN but not in LC and LE in three out of four replicates. One biological replicate showed also a band in LC and LE. Molecular Mass (MM), GeneRuler Ultra Low Range DNA ladder (Thermo Scientific).
Prediction of Whitefly sRNAs Targets in Tomato
To get insight into putative roles of these three whitefly sRNAs, the online psRNAtarget tool was used (Dai and Zhao, 2011) to predict putative tomato mRNA targets. By doing so, putative targets for the sRNAs #13120 and #18833 were found. sRNA #13120 is predicted to target four different exocyst complex proteins of which two are expressed in tomato leaves (Table 2). Another interesting putative targets of sRNA #18833 are three closely related xylanase inhibitors (Table 2). They are moderately similar (between 56 and 58% identity at the amino acid level) to a previously described tomato xyloglucan-specific endoglucanase inhibitor (Qin et al., 2003).
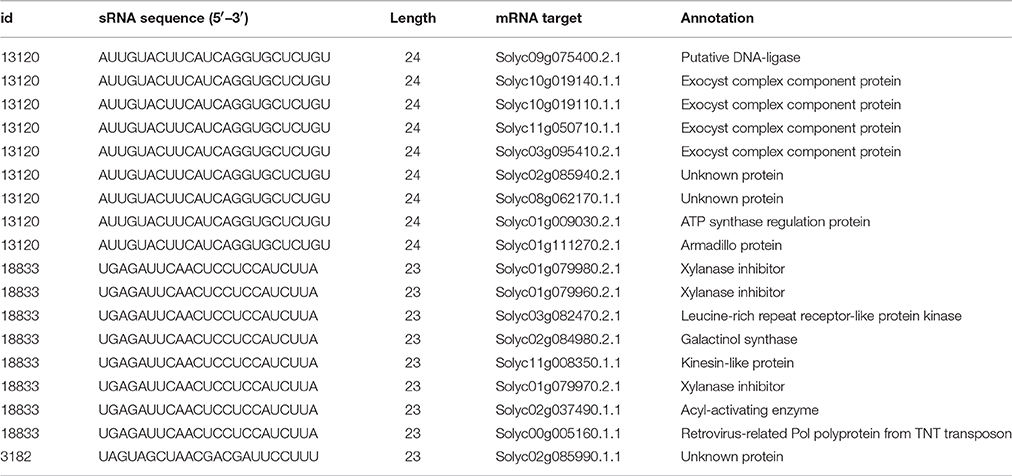
Table 2. List of predicted mRNA targets from the selected whitefly small RNA found in tomato tissues.
Discussion
Cross-Kingdom Interactions Mediated by sRNAs
Besides ingesting plant sap, phloem-feeding insects such as aphids manipulate plant defenses by secreting protein effectors that can improve host colonization and reproductive fitness (Louis and Shah, 2013). However, the precise molecular mode of action of aphid effectors remains elusive. Along with an array of notorious plant-viruses (Rosen et al., 2015), whiteflies theoretically could also transfer effector proteins into the phloem while feeding. Here we show that B. tabaci also appears to transfer sRNAs into the tomato phloem. Transfer of sRNAs from whiteflies could putatively be involved in transcriptional or post-transcriptional gene silencing inside the host and form an additional way for the insect to manipulate host defenses. There is increasing experimental evidence that sRNAs can mediate cross-kingdom interactions between plant and microorganisms (Knip et al., 2014), with perhaps the most convincing examples being those of Botrytis-Arabidopsis and Botrytis-tomato (Weiberg et al., 2013). One of the major difficulties in the field is to establish with certainty that a specific sRNA has been transferred by the “invader organism” into the host rather than being produced by the attacked host. Genome availability of B. cinerea, tomato and Arabidopsis thaliana was a major advantage that led to the discovery that sRNA can promote fungal pathogenicity (Weiberg et al., 2013). We did not have a draft or complete B. tabaci genome sequence to our disposal and this work therefore relied on a bioinformatic pipeline to search for whitefly sRNAs present in the phloem of whitefly-infested tomato (Figure 1). To further verify the sequences found in the tomato phloem, the sRNAs present only in the PW phloem were compared to the sRNAs of the whitefly nymph (WN) sample. Additionally, they were cross-referenced to sRNAs from whitefly-infested leaflets (LW) after elimination of non-infected leaves (LC) and leaflet with eggs (LE), in case not all eggs were removed from the LW leaflet surface. sRNAs were subsequently aligned to the tomato genome to remove tomato sRNAs especially from repetitive regions (Figure 1), leaving us with potentially 10,505 whitefly-specific sRNAs present in plant tissue. The final LW sample (nymph sRNAs in LW–LC–LE) contains less phloem than phloem exudate itself (nymph sRNAs in PW–PC) as the starting material contains many other tomato cell types that have been consequently filtered out. The remaining 22,465 sequences that do not match the phloem sequences can contain e.g., whitefly sRNAs from phloem companion cells. The phloem exudate samples are more concentrated and thus contain more (134,141) putative whitefly sRNAs than the leaflet samples.
A great diversity of sRNAs has been found in the phloem sap of several species including pumpkin, cucumber, lupin, and Arabidopsis, in the absence of major pathogen infection or pest infestation (Yoo et al., 2004). These phloem sRNAs typically had a length between 18 and 25 nts with a major peak at 23 nts (Yoo et al., 2004), which is consistent with the sRNA length distribution found in our non-infested control samples (LC and PC, Figure 2). In leaves, a major peak at 24 nts is very common (see, Itaya et al., 2008 for an example in tomato), which has long been associated with transcriptional gene silencing especially of repetitive sequences e.g., transposons (Borges and Martienssen, 2015). In both phloem and leaflet samples, we found conserved miRNAs such as miR156 and miR172 known to act in concert to regulate flowering time (Spanudakis and Jackson, 2014) and miR159 previously identified in cucurbit phloem (Yoo et al., 2004). Finding such miRNAs among the most abundant phloem sRNAs is consistent with previous studies (Yoo et al., 2004; Rodriguez-Medina et al., 2011; Bhogale et al., 2014). We also observed Solanaceae-specific miR482 and miR6022 among the most abundant miRNAs in the non-infested phloem PC and LC sRNAs (Table S4). The collection of phloem was performed after an initial “bleed” period of 30 min to limit sample contamination by other types of cellular content. Nymphs were feeding on the leaflet at 2–3 cm distance from the petiole phloem collection site indicating mobile whitefly sRNAs in the plant phloem. It was estimated that contamination of phloem exudates by companion cell breakage composed around 2% of the exudate (Atkins et al., 2011). Thus, it is likely that the identified sRNAs in the phloem originate from the sieve elements although some contamination from neighboring cells cannot be completely ruled out.
The presence of whitefly nymphs on tomato leaflets caused 29–30 nts sRNAs to appear in the phloem of infested plants (Figure 2). These longer sRNAs were indeed also observed in the whitefly nymphs (Figure 2). A similar sRNA length distribution has previously been found in adult whiteflies (Guo et al., 2013; Wang et al., 2016) and similar sized sRNAs (between 26 and 30 nts) have been reported for other insects e.g., cotton-melon aphid (Aphis gossypii) or the brown planthopper (Nilaparvata lugens; Chen et al., 2012; Sattar et al., 2012). These longer sRNAs are assumed to be Piwi-interacting RNAs (piRNAs) and are known to be a large class of non-coding RNAs in animals, specifically linked to genome stability in germ-line cells and silence transposons (Vagin et al., 2006). In Drosophila (D. melanogaster), for example, piRNAs are produced in a Dicer-independent manner from transposon-rich genomic clusters and specifically silence transposon expression in the germline (Iwasaki et al., 2015). Sattar et al. (2012) found that sRNAs with a length between 26 and 27 were overrepresented in the cotton-melon aphid, A. gossyppii, when infesting melon plants containing the Vat aphid resistance gene. Similar to B. tabaci, there is no annotated genome for A. gossypii but in this case the authors could make use of an A. pisum transposon database to show that around 50% of these 26–27 sRNAs actually derived from transposons. Another ~5% matched from the primary endosymbionts in aphids, Buchnera aphodicola (Sattar et al., 2012). In this study, we cannot completely rule out that the 29–30 nts sRNA originate from tomato. Nevertheless, when trying to align the five most abundant 29 or 30 nts sRNA to the tomato Heinz genome sequence (Figure 1), no full-length alignments were found. Assuming these particular sRNAs are indeed piRNAs involved in insect germline development, it remains elusive as to if and how the enrichment in the phloem of whitefly-infested leaves (Figure 2) is biologically relevant.
Insect Salivary Small RNAs Transferred into Host
Our bioinformatic pipeline identified whitefly sRNAs in tomato phloem of leaflets where nymphs were feeding. These sRNAs most likely found their way into the phloem via the whitefly saliva. During feeding whiteflies salivate into the phloem after which they ingests phloem sap (Figure S1; Pollard, 1955; Jiang et al., 1999; Jiang and Walker, 2003). Since whitefly nymphs are immobile and feed for long periods of time, one can expect to find components of whitefly saliva in the phloem. Salivary glands of phloem feeding insects like aphids and whitefly have been subjected to RNA sequencing and proteomics to obtain insight in the transcriptome and proteome (Carolan et al., 2011; Su et al., 2012; Rao et al., 2013). Also, aphid saliva has been collected and used for proteomics studies (Rao et al., 2013; Chaudhary et al., 2015). In the current study, phloem from whitefly-infested tomato was used, as obtaining salivary glands from adult whiteflies, though feasible (Ghanim et al., 2001; Su et al., 2012), proved too challenging in nymphs. In addition, the saliva composition of insects is not necessarily the same as the composition of the salivary gland, which includes cell membranes and ducts (Rao et al., 2013). Moreover, in order to collect sufficient saliva, it requires the culturing of large amounts of adult whiteflies for a prolonged period on an artificial diet (Su et al., 2015; VanDoorn et al., 2015), which was technically not feasible. Also, it has been reported that the composition of insect saliva differs when feeding on artificial diet and on different plant species (Habibi et al., 2001; Cooper et al., 2010). Finally, as nymphs are immobile while feeding for long periods of time it might increase the chances of actually identifying whitefly sRNAs in phloem.
To our knowledge, miRNAs have so far only been identified in the saliva of mosquito (Aedes aegypti; Maharaj et al., 2015). Interestingly, miRNAs closely related (one nucleotide difference) to our #13120 (Bta_miR305-pGtoU) and #18833 (Bta_miR1175-3p+A) were also found in the saliva of A. aegypti, particularly after sucrose feeding (Maharaj et al., 2015). Whether saliva secreted miRNAs are conserved among fluid-feeding insects remains to be seen. Since sRNAs of prokaryotes are generally bigger than 100 nt (Gottesman and Storz, 2011) and our cut-off for analysis was <40 nt, it is unlikely that the miRNAs presented here originate from symbionts present in the whitefly. Nevertheless, we aligned the 10,505 sRNAs to the genome of Rickettsia sp. Strain MEAM1 (Genbank AJWD00000000.2), and found no matches.
A possible source of contamination for the sRNA sequencing of leaflet samples could be part of nymphs still attached to the leaflet. However, in Figure 3A it was shown that nymph tissue was removed from leaflets or at least below the level of detection. The bands visible in the LW samples (Figures 4A–C) are therefore very unlikely to originate from nymph tissue still present on the leaflets. The plant specific sRNA miR172 was only found in leaf samples and not in the whitefly nymphs (Figure 3B) though this could have been possible since sRNAs have been found previously ingested by aphids (Sattar et al., 2012). Similarly, Bta_miR2A was detected only in the nymph and whitefly-infested samples (Figure 3C), showing that the miR2A of the eggs on leaflet (LE) samples was even detectable by stem-loop RT-PCR.
The three sRNAs investigated here were very likely transferred from whitefly into tomato. All three candidates were identified in whitefly-infested material and in the nymphs themselves. sRNA #13120 (Bta_miR305-pGtoU) (Figure 4A) and #18833 (Bta_miR1175-3p+A) (Figure 4B) have been detected in LW samples but not in the LC or LE sample. Family members of two of these candidates have been previously identified in whitefly (Guo et al., 2013). For the third candidate (#3182), no similarity was found with previously identified whitefly sRNAs or with other sequences in the miRBase. This sRNA has been detected in all of the replicates of whitefly-infested leaf samples and is very abundant in the nymph sample but could be detected once out of 4 in control leaflets. Despite the fact that it cannot be completely ruled out, it is highly unlikely that #3182 derives from tomato. Sly-miR172, a very abundant tomato miRNAs present in our phloem sample could not be detected in our nymph sample while #3182 was found in the insect in a relative high level.
Small RNA as Effectors?
Pathogens and insects are known to transfer proteins into host plant cells in order to suppress host immunity (Dangl et al., 2013; Will et al., 2013; Su et al., 2015). In fact, it has been postulated that phloem-feeding insects employ a suit of proteins that are passed from the saliva into the phloem during feeding which could act as effector proteins that suppress plant defenses. Besides proteins, non-protein salivary factors can act as an effector (Su et al., 2015). Here we describe, for the first time, the transfer of putative salivary non-coding sRNAs from whitefly and postulate that they might target tomato host proteins. Small RNAs could facilitate the interaction between organisms by improving the attackers chance of survival (Weiberg et al., 2013; Knip et al., 2014) or improve fecundity (Sattar et al., 2012). In plants, the high base complementarity between the sRNA and the target mRNA has been successfully used to predict post-transcriptional regulations by sRNAs (Ding et al., 2012). Further validation and characterization of these mRNA targets is currently under investigation.
Author Contributions
PvK and MG contributed equally to this paper and shared first authors. MG analyzed the RNAseq data and PvK did the wet-lab experiments. PB conceived the project and supervised MG. RS supervised PvK and contributed to discussions. All four authors wrote and carefully read and approved the final manuscript.
Funding
This work was supported by funding from the European Union (H2020, DURETO project #655656) to MG and the NWO VIDI grant 12988 awarded to PB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HJ and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The authors would like to thank Suzanne Hoogstrate for careful reading of the manuscript. We are grateful to Selina van Leeuwen, Wim Ensink, and Marina van Olst, Wim de Leeuw, and Han Rauwerda of the “RNA Biology and Applied Bioinformatics' group (SILS, University of Amsterdam) for sRNAs sequencing and bioinformatics support and thank Ludek Tikovsky, Harold Lemereis, and Fernando Inchauste Callahuara for taking care of the tomato plants. This work was supported by funding from the European Union (H2020, DURETO project #655656) and by the Dutch Technology Foundation STW, part of the Netherlands Organization for Scientific Research (NWO, VIDI project #12988).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01759/full#supplementary-material
References
Adams, M. J., and Antoniw, J. F. (2006). DPVweb: a comprehensive database of plant and fungal virus genes and genomes. Nucleic Acids Res. 34, D382–D385. doi: 10.1093/nar/gkj023
Atkins, C. A., Smith, P. M., and Rodriguez-Medina, C. (2011). Macromolecules in phloem exudates–a review. Protoplasma 248, 165–172. doi: 10.1007/s00709-010-0236-3
Baulcombe, D. C. (2015). VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol. 26, 141–146. doi: 10.1016/j.pbi.2015.06.007
Bhogale, S., Mahajan, A. S., Natarajan, B., Rajabhoj, M., Thulasiram, H. V., and Banerjee, A. K. (2014). MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. Andigena. Plant Physiol. 164, 1011–1027. doi: 10.1104/pp.113.230714
Bleeker, P. M., Diergaarde, P. J., Ament, K., Guerra, J., Weidner, M., Schuetz, S., et al. (2009). The role of specific tomato volatiles in tomato-whitefly interaction. Plant Physiol. 151, 925–935. doi: 10.1104/pp.109.142661
Bleeker, P. M., Diergaarde, P. J., Ament, K., Schütz, S., Johne, B., Dijkink, J., et al. (2011). Tomato-produced 7-epizingiberene and R-curcumene act as repellents to whiteflies. Phytochemistry 72, 68–73. doi: 10.1016/j.phytochem.2010.10.014
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Bos, J. I., Prince, D., Pitino, M., Maffei, M. E., Win, J., and Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6:e1001216. doi: 10.1371/journal.pgen.1001216
Buntin, D. G., Gilbertz, D. A., and Oetting, R. D. (1993). Chlorophyll loss and gas exchange in tomato leaves after feeding injury by Bemisia tabaci (Homoptera: Aleyrodidae). J. Econ. Entomol. 86, 517–522. doi: 10.1093/jee/86.2.517
Carolan, J. C., Caragea, D., Reardon, K. T., Mutti, N. S., Dittmer, N., Pappan, K., et al. (2011). Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): a dual transcriptomic/proteomic approach. J. Proteome Res. 10, 1505–1518. doi: 10.1021/pr100881q
Chaudhary, R., Atamian, H. S., Shen, Z., Briggs, S. P., and Kaloshian, I. (2015). Potato aphid salivary proteome: enhanced salivation using resorcinol and identification of aphid phosphoproteins. J. Proteome Res. 14, 1762–1778. doi: 10.1021/pr501128k
Chen, Q., Lu, L., Hua, H., Zhou, F., Lu, L., and Lin, Y. (2012). Characterization and comparative analysis of small RNAs in three small RNA libraries of the brown planthopper (Nilaparvata lugens). PLoS ONE 7:e32860. doi: 10.1371/journal.pone.0032860
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Cooper, W. R., Dillwith, J. W., and Puterka, G. J. (2010). Salivary proteins of Russian Wheat Aphid (Hemiptera: Aphididae). Environ. Entomol. 39, 223–231. doi: 10.1603/EN09079
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Ding, J., Zhou, S., and Guan, J. (2012). Finding microRNA targets in plants: current status and perspectives. Genomics Proteomics Bioinformatics 10, 264–275. doi: 10.1016/j.gpb.2012.09.003
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Ghanim, M., Rosell, R. C., Campbell, L. R., Czosnek, H., Brown, J. K., and Ullman, D. E. (2001). Digestive, salivary, and reproductive organs of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) B type. J. Morphol. 248, 22–40. doi: 10.1002/jmor.1018
Gottesman, S., and Storz, G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Guo, Q., Tao, Y. L., and Chu, D. (2013). Characterization and comparative profiling of miRNAs in invasive Bemisia tabaci (Gennadius) B and Q. PLoS ONE 8:e59884. doi: 10.1371/journal.pone.0059884
Habibi, J., Backus, E. A., Coudron, T. A., and Brandt, S. L. (2001). Effect of different host substrates on hemipteran salivary protein profiles*. Entomol. Exp. Appl. 98, 369–375. doi: 10.1046/j.1570-7458.2001.00794.x
Itaya, A., Bundschuh, R., Archual, A. J., Joung, J. G., Fei, Z., Dai, X., et al. (2008). Small RNAs in tomato fruit and leaf development. Biochim. Biophys. Acta 1779, 99–107. doi: 10.1016/j.bbagrm.2007.09.003
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-Interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433. doi: 10.1146/annurev-biochem-060614-034258
Jiang, Y. X., Lei, H., Collar, J. L., Martin, B., Muniz, M., and Fereres, A. (1999). Probing and feeding behavior of two distinct biotypes of Bemisia tabaci (Homoptera: Aleyrodidae) on tomato plants. J. Econ. Entomol. 92, 357–366. doi: 10.1093/jee/92.2.357
Jiang, Y. X., and Walker, G. P. (2003). Electrical penetration graphs of the nymphal stage of Bemisia argentifolii. Entomol. Exp. Appl. 109, 101–111. doi: 10.1046/j.1570-7458.2003.00093.x
Kant, M. R., Jonckheere, W., Knegt, B., Lemos, F., Liu, J., Schimmel, B. C., et al. (2015). Mechanisms and ecological consequences of plant defence induction and suppression in herbivore communities. Ann. Bot. 115, 1015–1051. doi: 10.1093/aob/mcv054
Kempema, L. A., Cui, X., Holzer, F. M., and Walling, L. L. (2007). Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 143, 849–865. doi: 10.1104/pp.106.090662
Kettles, G. J., Drurey, C., Schoonbeek, H. J., Maule, A. J., and Hogenhout, S. A. (2013). Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol. 198, 1178–1190. doi: 10.1111/nph.12218
King, R. W., and Zeevaart, J. A. (1974). Enhancement of Phloem exudation from cut petioles by chelating agents. Plant Physiol. 53, 96–103. doi: 10.1104/pp.53.1.96
Knip, M., Constantin, M. E., and Thordal-Christensen, H. (2014). Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 10:e1004602. doi: 10.1371/journal.pgen.1004602
Koornneef, A., and Pieterse, C. M. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Köster, J., and Rahmann, S. (2012). Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. doi: 10.1093/bioinformatics/bts480
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Kramer, M. F. (2011). Stem-loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. Chapter 15: Unit 15.10. doi: 10.1002/0471142727.mb1510s95
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Locati, M. D., Terpstra, I., de Leeuw, W. C., Kuzak, M., Rauwerda, H., Ensink, W. A., et al. (2015). Improving small RNA-seq by using a synthetic spike-in set for size-range quality control together with a set for data normalization. Nucleic Acids Res. 43, e89. doi: 10.1093/nar/gkv303
Louis, J., and Shah, J. (2013). Arabidopsis thaliana-Myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Front. Plant Sci. 4:213. doi: 10.3389/fpls.2013.00213
Maharaj, P. D., Widen, S. G., Huang, J., Wood, T. G., and Thangamani, S. (2015). Discovery of mosquito saliva microRNAs during CHIKV infection. PLoS Negl. Trop. Dis. 9:e0003386. doi: 10.1371/journal.pntd.0003386
Moreno, A., Garzo, E., Fernandez-Mata, G., Kassem, M., Aranda, M. A., and Fereres, A. (2011). Aphids secrete watery saliva into plant tissues from the onset of stylet penetration. Entomol. Exp. Appl. 139, 145–153. doi: 10.1111/j.1570-7458.2011.01117.x
Navas-Castillo, J., Fiallo-Olivé, E., and Sanchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Nawrocki, E. P., Burge, S. W., Bateman, A., Daub, J., Eberhardt, R. Y., Eddy, S. R., et al. (2015). Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137. doi: 10.1093/nar/gku1063
Peng, H. C., Mantelin, S., Hicks, G. R., Takken, F. L., and Kaloshian, I. (2016). The conformation of the plasma membrane-localized SlSERK1 - Mi-1.2 complex is altered by a potato aphid derived effector. Plant Physiol. 171, 2211–2222. doi: 10.1104/pp.16.00295
Pitino, M., and Hogenhout, S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe Interact. 26, 130–139. doi: 10.1094/MPMI-07-12-0172-FI
Pollard, D. G. (1955). FEEDING HABITS OF THE COTTON WHITEFLY, BEMISIA TAB AC I GENN. (HOMOPTERA: ALEYRODIDAE). Ann. Appl. Biol. 43, 664–671. doi: 10.1111/j.1744-7348.1955.tb02510.x
Qin, Q., Bergmann, C. W., Rose, J. K., Saladie, M., Kolli, V. S., Albersheim, P., et al. (2003). Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J. 34, 327–338. doi: 10.1046/j.1365-313X.2003.01726.x
Rao, S. A., Carolan, J. C., and Wilkinson, T. L. (2013). Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS ONE 8:e57413. doi: 10.1371/journal.pone.0057413
Rodriguez-Medina, C., Atkins, C. A., Mann, A. J., Jordan, M. E., and Smith, P. M. (2011). Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.). BMC Plant Biol. 11:36. doi: 10.1186/1471-2229-11-36
Rosen, R., Kanakala, S., Kliot, A., Cathrin Pakkianathan, B., Farich, B. A., Santana-Magal, N., et al. (2015). Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 15, 1–8. doi: 10.1016/j.coviro.2015.06.008
Sattar, S., Addo-Quaye, C., Song, Y., Anstead, J. A., Sunkar, R., and Thompson, G. A. (2012). Expression of small RNA in Aphis gossypii and its potential role in the resistance interaction with melon. PLoS ONE 7:e48579. doi: 10.1371/journal.pone.0048579
Sharma, A., Khan, A. N., Subrahmanyam, S., Raman, A., Taylor, G. S., and Fletcher, M. J. (2014). Salivary proteins of plant-feeding hemipteroids - implication in phytophagy. Bull. Entomol. Res. 104, 117–136. doi: 10.1017/S0007485313000618
Shi, X., Pan, H., Xie, W., Jiao, X., Fang, Y., Chen, G., et al. (2014). Three-way interactions between the tomato plant, tomato yellow leaf curl virus, and Bemisia tabaci (Hemiptera: Aleyrodidae) facilitate virus spread. J. Econ. Entomol. 107, 920–926. doi: 10.1603/EC13476
Simmons, A. T., and Gurr, G. M. (2005). Trichomes of Lycopersicon species and their hybrids: effects on pests and natural enemies. Agric. For. Entomol. 7, 265–276. doi: 10.1111/j.1461-9555.2005.00271.x
Spanudakis, E., and Jackson, S. (2014). The role of microRNAs in the control of flowering time. J. Exp. Bot. 65, 365–380. doi: 10.1093/jxb/ert453
Su, Q., Oliver, K. M., Xie, W., Wu, Q., Wang, S., and Zhang, Y. (2015). The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Funct. Ecol. 29, 1007–1018. doi: 10.1111/1365-2435.12405
Su, Y. L., Li, J. M., Li, M., Luan, J. B., Ye, X. D., Wang, X. W., et al. (2012). Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS ONE 7:e39303. doi: 10.1371/journal.pone.0039303
Taylor, R. S., Tarver, J. E., Hiscock, S. J., and Donoghue, P. C. (2014). Evolutionary history of plant microRNAs. Trends Plant Sci. 19, 175–182. doi: 10.1016/j.tplants.2013.11.008
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Tetyuk, O., Benning, U. F., and Hoffmann-Benning, S. (2013). Collection and analysis of Arabidopsis phloem exudates using the EDTA-facilitated Method. J. Vis. Exp. e51111. doi: 10.3791/51111
Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. doi: 10.1126/science.1129333
VanDoorn, A., de Vries, M., Kant, M. R., and Schuurink, R. C. (2015). Whiteflies glycosylate salicylic acid and secrete the conjugate via their honeydew. J. Chem. Ecol. 41, 52–58. doi: 10.1007/s10886-014-0543-9
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F., and Hellens, R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12–12. doi: 10.1186/1746-4811-3-12
Villarroel, C. A., Jonckheere, W., Alba, J. M., Glas, J. J., Dermauw, W., Haring, M. A., et al. (2016). Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 86, 119–131. doi: 10.1111/tpj.13152
Walling, L. L. (2008). Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866. doi: 10.1104/pp.107.113142
Wang, B., Wang, L., Chen, F., Yang, X., Ding, M., Zhang, Z., et al. (2016). MicroRNA profiling of the whitefly Bemisia tabaci Middle East-Aisa Minor I following the acquisition of Tomato yellow leaf curl China virus. Virol. J. 13, 20. doi: 10.1186/s12985-016-0469-7
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Will, T., Furch, A. C. U., and Zimmermann, M. R. (2013). How phloem-feeding insects face the challenge of phloem-located defenses. Front. Plant Sci. 4:336. doi: 10.3389/fpls.2013.00336
Will, T., Tjallingii, W. F., Thonnessen, A., and van Bel, A. J. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. U.S.A. 104, 10536–10541. doi: 10.1073/pnas.0703535104
Yin, C., Shen, G., Guo, D., Wang, S., Ma, X., Xiao, H., et al. (2016). InsectBase: a resource for insect genomes and transcriptomes. Nucleic Acids Res. 44, D801–D807. doi: 10.1093/nar/gkv1204
Yoo, B. C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y. M., et al. (2004). A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000. doi: 10.1105/tpc.104.023614
Keywords: small-RNA, whitefly, RNAseq, phloem, tomato
Citation: van Kleeff PJM, Galland M, Schuurink RC and Bleeker PM (2016) Small RNAs from Bemisia tabaci Are Transferred to Solanum lycopersicum Phloem during Feeding. Front. Plant Sci. 7:1759. doi: 10.3389/fpls.2016.01759
Received: 30 May 2016; Accepted: 08 November 2016;
Published: 24 November 2016.
Edited by:
Linda Walling, University of California, Riverside, USAReviewed by:
Hailing Jin, University of California, Riverside, USASaskia A. Hogenhout, John Innes Centre (BBSRC), UK
Raffaella Balestrini, National Research Council, Italy
Shai Morin, Hebrew University of Jerusalem, Israel
Copyright © 2016 van Kleeff, Galland, Schuurink and Bleeker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra M. Bleeker, p.m.bleeker@uva.nl
†These authors have contributed equally to this work.
 Paula J. M. van Kleeff
Paula J. M. van Kleeff Marc Galland
Marc Galland Robert C. Schuurink
Robert C. Schuurink Petra M. Bleeker
Petra M. Bleeker