- Donald Danforth Plant Science Center, St Louis, MO, USA
Millet is a common name for a group of polyphyletic, small-seeded cereal crops that include pearl, finger and foxtail millet. Millet species are an important source of calories for many societies, often in developing countries. Compared to major cereal crops such as rice and maize, millets are generally better adapted to dry and hot environments. Despite their food security value, the genetic architecture of agronomically important traits in millets, including both morphological traits and climate resilience remains poorly studied. These complex traits have been challenging to dissect in large part because of the lack of sufficient genetic tools and resources. In this article, we review the phylogenetic relationship among various millet species and discuss the value of a genetic model system for millet research. We propose that a broader adoption of green foxtail (Setaria viridis) as a model system for millets could greatly accelerate the pace of gene discovery in the millets, and summarize available and emerging resources in S. viridis and its domesticated relative S. italica. These resources have value in forward genetics, reverse genetics and high throughput phenotyping. We describe methods and strategies to best utilize these resources to facilitate the genetic dissection of complex traits. We envision that coupling cutting-edge technologies and the use of S. viridis for gene discovery will accelerate genetic research in millets in general. This will enable strategies and provide opportunities to increase productivity, especially in the semi-arid tropics of Asia and Africa where millets are staple food crops.
Introduction
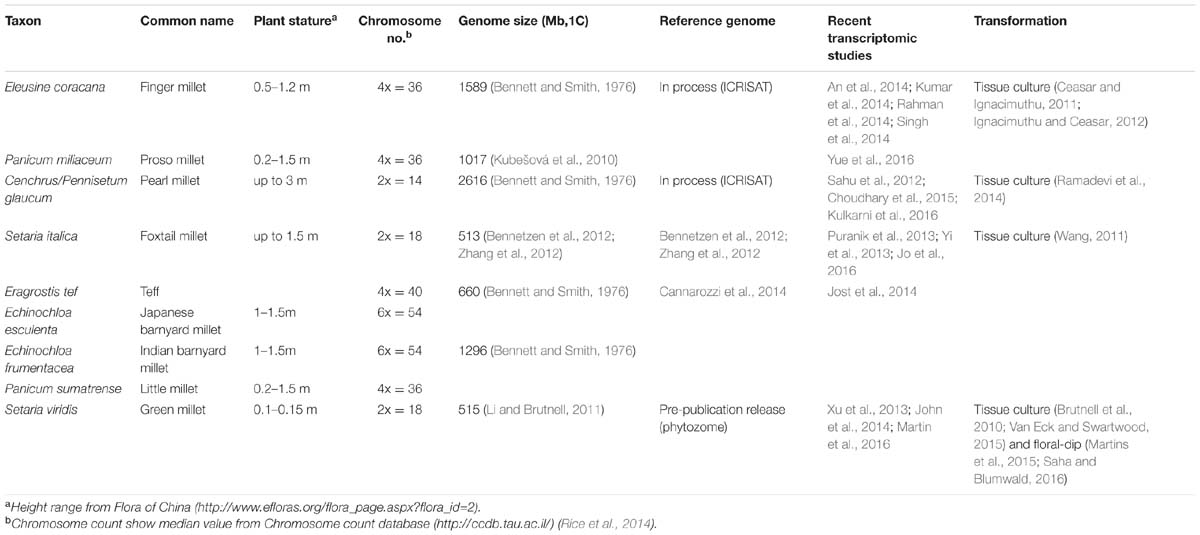
Although less prominent than major crops such as rice, maize, and wheat, the polyphyletic millets are important food sources worldwide. Generally, millets are some of the most well-adapted crops to drought, heat, and low nutrient input conditions (Dwivedi et al., 2011; Goron and Raizada, 2015; Saha et al., 2016). Given the increasing global population and decreasing arable lands, the stress tolerant millets are ideal candidates for crop production in climates that are not suitable for major crops. This is especially important for millet-growing developing countries in Asia and Africa. However, common features of millets, including complex polyploid genomes, large plant stature, and long generation times (Table 1) hinder both breeding and genetic research (Goron and Raizada, 2015; Saha et al., 2016).
In this review, we discuss the recent development of several genetic and genomic resources in the model grass Setaria viridis (green foxtail) and its domesticated relative S. italica (foxtail millet). We provide several use cases that demonstrate the value of these resources and their potential to provide new opportunities for breeding and research in millets. S. viridis was originally developed as a genetic model for bioenergy feedstocks and panicoid food crops like switchgrass, sorghum, and maize (Doust et al., 2009; Li and Brutnell, 2011; Diao et al., 2014; Brutnell, 2015; Brutnell et al., 2015; Muthamilarasan and Prasad, 2015), and as a model for C4 photosynthesis (Brutnell et al., 2010, 2015; Huang and Brutnell, 2016). S. viridis, like all millet species, is a member of the PACMAD clade of grasses (Figure 1). Previous work in genome organization (Benabdelmouna et al., 2001) and diversity (Huang et al., 2014) shows S. viridis is most closely related to and interfertile with foxtail millet. Genetic resources are largely shared between foxtail millet and S. viridis, but we emphasize on S. viridis in this review because of its nature as an ideal lab organism. Similar to the dicot model Arabidopsis thaliana, S. viridis has a short life span (6∼8 weeks under greenhouse conditions), small plant stature (less than 30 cm at maturity) and small diploid genome (∼500 Mb).
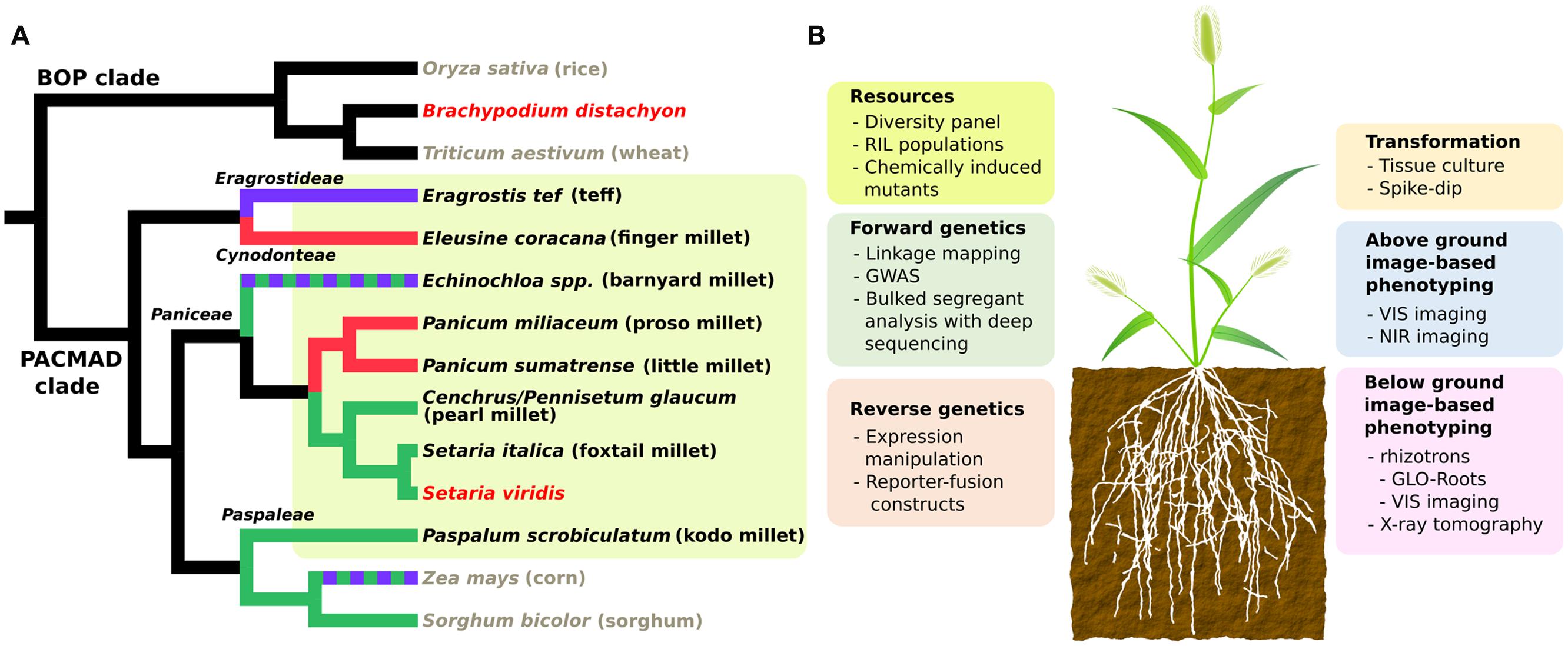
FIGURE 1. (A) Cladogram showing phylogenetic relationships and photosynthetic subtypes of millets and other Poaceae species. Black, gray and red taxa names represent millets, major crops and model grasses, respectively. Millet lineages are also highlighted in yellow. Green, red, purple, and black branch colors represent three subtypes of C4 (NADP-malic enzyme, NAD-malic enzyme and phosphoenolpyruvate phosphatase) and C3 photosynthesis respectively. Dashed colors represents mixed subtypes of C4. Tree topology is extracted from Grass Phylogeny Working Group II (2011). (B) Summary chart of available genetic resources and technologies for Setaria viridis. RIL, recombinant inbred line; GWAS, genome wide association study; VIS, visual; NIR, near infra-red.
Phylogeny and Photosynthetic Subtypes of Millets
Despite the common small grain nature, millets include grasses from a broad range of phylogenetic clades. We compared the phylogenetic relationship among eight small-seed cereal crops along with other major crops and model species in the Poaceae family based on a previous study (Grass Phylogeny Working Group II, 2011). In this phylogeny, “millet” refers to species from at least four distinct tribes of PACMAD grasses: Paniceae, Paspaleae, Cynodonteae, and Eragrostideae (Figure 1A). This polyphyletic nature is also reflected by independent domestications of various millets in different areas of the world (Dwivedi et al., 2011; Goron and Raizada, 2015). Five out of eight species belong to tribe Paniceae, including three major species: pearl millet (Cenchrus/Pennisetum glaucum), foxtail millet and proso millet (Panicum milliacum), along with the model grass S. viridis (Figure 1A). Close phylogenetic relatedness generally implies shared genetic mechanism behind complex traits. That is, the more closely related two species are the easier it is to translate genetic discoveries between them. Therefore, compared to other grass models and major crops (Figure 1A), S. viridis is the most suitable model for most millets from a phylogenetic perspective.
A key feature shared by all millets is C4 photosynthesis, regardless of their separate domestication history. Most C4 plants, including all the C4 grasses utilize specialized bundle sheath and mesophyll cells (Kranz anatomy) to concentrate CO2 in the vicinity of ribulose bisphosphate carboxylase/oxygenase. This machinery reduces photorespiration and increases water use efficiency in C4 plants (Rawson et al., 1977), especially under drought and heat stress. C4 plants also have a better nitrogen use efficiency, namely they require less nitrogen input to achieve similar photosynthetic rates as C3 plants (Sage et al., 1987; Sage and Pearcy, 1987a,b). These features of C4 correspond nicely with, and likely contribute to the climatic resilience and low soil nutrient demands of millets. Thus, dissecting the genetic basis of C4 is an important route to understand the mechanism underlying climatic resilience in millets.
Setaria viridis promises to greatly accelerate the pace of discovery in dissecting C4 photosynthesis in grasses (Brutnell et al., 2010; Huang and Brutnell, 2016). While genetic screens for C4 related mutants in S. viridis are currently ongoing, comparative genomics has already provided new insights. For example, Huang et al. (2016) searched for signals of adaptive evolution in two independently evolved C4 lineages, Setaria and the maize-sorghum clade to identify a candidate gene list for C4. The results also indicated a potential for “cross species engineering” of C4 transporters. John et al. (2014) showed an 87% correlation between the bundle sheath/mesophyll expression specificity between S. viridis and maize, indicating phylogenetically conserved genetic modules controlling C4 development. These findings can be generalized to understand C4 in other millets. Downstream of candidate gene identification, S. viridis as a transformable C4 model system also plays a key role in functional characterizations (Martins et al., 2015; Van Eck and Swartwood, 2015; Huang and Brutnell, 2016; Saha and Blumwald, 2016).
Advances of Forward Genetics in Setaria and Other Millets
Classical forward genetic approaches such as linkage and association mapping have been widely applied in most millet species (Table 1). However, the lack of high density marker maps is a major limiting factor for the resolution of these applications. Although many quantitative trait loci (QTLs) have been identified for various agronomic traits such as plant height, flowering time, lodging, and drought tolerance (Mauro-Herrera et al., 2013; Parvathaneni et al., 2013; Sato et al., 2013; Babu et al., 2014; Qie et al., 2014; Mauro-Herrera and Doust, 2016; Rajput et al., 2016), the QTL intervals are often large (>1 Mb) and difficult to fine map. A partial solution is to generate high density linkage maps using technologies like genotyping by sequencing (Moumouni et al., 2015; Fang et al., 2016; Rajput et al., 2016), but the ultimate solution is to build high-quality reference genomes. To date, foxtail millet remains the only millet that has a chromosomal scale genome assembly (Bennetzen et al., 2012; Zhang et al., 2012), while Eragrostis tef has a draft genome (Cannarozzi et al., 2014), and the genome sequencing of finger millet and pearl millets are still ongoing (Table 1). Complete genome sequencing not only enables high density maps (Fang et al., 2016), but also large scale genome wide association studies (GWAS; Jia et al., 2013). Recently, a pre-publication release of an S. viridis genome de novo assembly became available through phytozome1. A panel of accessions in S. viridis with a greater genetic diversity than foxtail millet was also assembled for ongoing GWAS (Huang et al., 2014).
Molecular markers are often shared across multiple grass species, further enabling the use of a model species to accelerate gene discovery. For example, Rajput et al. (2014) showed 62% of a total of 339 microsatellite markers are shared between switchgrass and proso millet. One important application of reference genomes is to assist marker development and inform the selection of candidate genes (Parvathaneni et al., 2013). With a closer phylogenetic relationship, more shared synteny and no complicated duplication history, S. viridis is generally a better reference than sorghum or maize for both purposes. For example, Hu et al. (2015) examined a diverse panel of pearl millet and showed that shared markers and size of syntenic regions between Setaria and pearl millet is more than double of those between sorghum and pearl millet. In addition, S. viridis allelic variation can be directly introgressed into foxtail millet through interspecific crosses. Such crosses result in dense molecular markers and additional phenotypic variations, thus greatly facilitating genetic mapping of traits such as flowering time, tillering, and drought tolerance (Mauro-Herrera et al., 2013; Qie et al., 2014; Mauro-Herrera and Doust, 2016).
The short life cycle and small genome of Setaria makes it an ideal fit for bulked segregant analysis (BSA). BSA was originally developed for rapid gene mapping in F2 generations (Michelmore et al., 1991). When coupled with deep sequencing technologies, BSA can be conducted faster and without prior knowledge of markers (Takagi et al., 2015). Empirically, the expense of this approach correlates with genome size, and the time to discovery largely depends on the generation time, so this approach is most suitable for model systems. Using this method, Li et al. (2016) mapped a yellow–green leaf mutation in foxtail millet to a chlorophyll biosynthesis related gene SiYGL1. Masumoto et al. (2016) mapped a branching panicle mutation, a yield related trait in foxtail millet, to a candidate gene NEKODE1. In chemically induced mutants of S. viridis, BSA can be expected to define causative mutations to a one to few gene interval within two generations (<7 months). This approach will greatly facilitate genetic dissection of traits such as seed size, inflorescence architecture, flowering time, and climatic resilience (Brutnell, 2015; Brutnell et al., 2015).
Setaria viridis as a Model System to Dissect Gene Function in Millets
Reverse genetics is a powerful tool that enables gene validation and characterization from transcriptomic datasets and/or forward genetics. In light of recent advances in plant biotechnology, reverse genetics is becoming a faster and cheaper routine. There are several important features for a model species to have successful reverse genetic applications: (1) Plant transformation is often the most limiting step for most species and therefore it should not be recalcitrant to Agrobacterium-mediated transformation (Gelvin, 2003; Ceasar and Ignacimuthu, 2009; Plaza-Wüthrich and Sonia, 2012; Tadele and Plaza-Wüthrich, 2013). (2) Controlled crosses and prolific seed production are also essential for rapid genetic analyses (Li and Brutnell, 2011; Brutnell, 2015). (3) Short life cycle and plant size is highly advantageous to conduct experiments in controlled environments, and to reduce costs (Brutnell et al., 2010). (4) Transcriptomic and genomic information facilitates the selection of candidate genes and inference of potential function based on orthology and/or synteny compared to its relatives (Huang et al., 2016; Huang and Brutnell, 2016). Unfortunately the majority of features are not inherent to most millet species, except in Setaria. To date, the techniques and methods of reverse genetics in millets are still very limited, thus a genetic model for millets is greatly needed (Goron and Raizada, 2015).
In recent years, remarkable technical advances were made in the development of resources and techniques for conducting reverse genetics in S. viridis. Its inbreeding nature and the ability to perform crosses (Jiang et al., 2013) not only facilitates the generation of homozygous offspring carrying the allele of interest but also enables controlled outcrosses to different populations (i.e., for complementation assays). Agrobacterium tumefaciens-mediated gene transfer in S. viridis has been successfully developed and first generation events can be produced within 15 weeks (Brutnell et al., 2010; Van Eck and Swartwood, 2015). Alternatively, floral-dip protocols are being developed and would accelerate immensely the pace of gene discovery by reducing the time of callus generation (Martins et al., 2015; Saha and Blumwald, 2016). Together with the rise of genome editing technology using CRISPR/Cas9, model species like S. viridis hold the key to accelerate reverse genetic discoveries in C4 grasses. It is now possible to generate biallelic mutations and begin downstream gene function characterizations within 1 year, a timeframe which is nearly impossible to match in most crop species. More subtle gene expression manipulations are also possible using modified versions of Cas9 (dCas9) and adding an activator and/or repressor motif to enhance or repress gene expression (Piatek et al., 2015; Zhang et al., 2015). These features and technological advancements in S. viridis are especially important for timely characterizations of candidate genes underlying complex traits, including the development of Kranz anatomy and stress tolerance.
Stress tolerance is probably the most explored trait in millets (Charu Lata, 2015; Tadele, 2016). In foxtail millet, several studies have reported on candidate genes regulating drought stress. For example, overexpression of SiLEA14, a homolog of the Late embryogenesis abundant (LEA) proteins showed increased salt/drought tolerance and improved growth in foxtail millet (Wang et al., 2014). One important component of abiotic stress responses are Dehydration-Responsive Element Binding (DREB) transcription factors (Li et al., 2014). An abscisic acid (ABA)-responsive DREB-binding protein gene, cloned from foxtail millet (SiARDP), was shown to mediate a response that increases tolerance to drought and high salinity stress (Li et al., 2014). Similarly, Lata et al. (2011) identified a DREB2-like gene (SiDREB2) that is associated with dehydration tolerance and developed an allele-specific marker for tolerant accessions. Technical advances in Setaria can also be useful for other millet species for the purposes of functional complementation of orthologous genes. Two recent studies found a NAC and a bZIP transcription factor from finger millet can enhance abiotic tolerance in rice and tobacco, respectively (Babitha et al., 2015; Rahman et al., 2016). As reverse genetic tools advance in S. viridis, the pace of gene discovery will also accelerate, enabling the identification of candidate genes that can be introduced into other grasses to confer enhanced abiotic stress tolerance. It will also facilitate the testing of candidate gene function as genes isolated from related millet species can be introduced into S. viridis and phenotypes rapidly characterized.
High-Throughput Phenotyping as a Critical Tool to Advance Millet Research
With the rapid development of genetic tools in Setaria, it is critical to have advanced phenotyping techniques to maximize the value of these resources. Automated high-throughput hardware platforms and corresponding software packages are transforming the field of plant-based phenotyping (Yang et al., 2013; Fahlgren et al., 2015b; Rahaman et al., 2015). Here we highlight phenotyping platforms and software packages that have been utilized for Setaria and millet research.
Above ground architectural traits such as plant height, biomass and leaf area are important traits for plant breeding (Duvick, 2005). To obtain this information in a high-throughput manner, images are acquired from plants by scanner-based systems or conveyer belt systems under controlled (Fahlgren et al., 2015a; Neilson et al., 2015) or field environments (Vadez et al., 2015). One advantage of these platforms is they allow measurements in a time-dependent manner. For example, Fahlgren et al. (2015a) studied drought responses in Setaria using a conveyer belt-based platform. Through image analysis, the authors found that S. viridis grows faster and earlier than foxtail millet though they have similar biomass at later time points. S. viridis was also found to respond faster to water limitations than foxtail millet. In parallel to 2D images, 3D images can be generated using scanner-based systems. For example, Vadez et al. (2015) used 3D scanning to characterize variations in leaf areas between breeding populations in pearl millet.
Physiological traits can also be measured using specialized imaging systems. For example, using near infra-red (NIR) imaging, Fahlgren et al. (2015a) found strong water content differences between Setaria treated with and without water limitation. In addition, fluorescence imaging efficiently measures photosynthesis rate in 2D leaves (Attaran et al., 2014; Cruz et al., 2016), but it is still challenging to measure 3D plants due to confounding height effects (Fahlgren et al., 2015a). Spectroscopy imaging can also be used to examine stress responses (Fahlgren et al., 2015b; Rahaman et al., 2015), but so far this technology has not been utilized in millet research.
Below ground traits contribute greatly to crop performance, but are challenging to image. Therefore, methods for obtaining root images is critical. Rhizotrons are root visualizing systems which hold a thin volume of soil or nutrient substrates between two plastic sheets (Neufeld et al., 1989; Rellán-Álvarez et al., 2015; Passot et al., 2016). This system has been utilized in pearl millets to measure root growth rates (Passot et al., 2016). In S. viridis, transgenic lines with a constitutively expressed luciferase reporter provides an imaging system with a cleaner background, known as Growth and Luminescence Observatory for Roots (GLO-Roots; Rellán-Álvarez et al., 2015; Sebastian et al., 2016). Using GLO-Roots, Sebastian et al. (2016) found suppression of crown root growth as a key phenotypic response under water-limiting conditions. To capture 3D structures of root tissues, X-ray tomography has also been utilized in pearl millet, though the system operates at lower throughput (Passot et al., 2016).
As phenotyping systems rapidly develop, it is important to have software packages that can efficiently extract biologically meaningful information from images. Though software such as ImageJ is available (Schneider et al., 2012; Lobet et al., 2013), a new generation of high-throughput, customizable and open-source software is much needed (Fahlgren et al., 2015a; Knecht et al., 2016; Singh et al., 2016). Among them, PlantCV is the first package that has pipelines optimized specifically for Setaria (Fahlgren et al., 2015a). Importantly, the small size and rapid growth of S. viridis will facilitate its use in both controlled and field-based phenotyping platforms where access to such facilities is often rate limiting.
Conclusion
Since Setaria was initially proposed as a model system for the panicoid grasses (Doust et al., 2009; Brutnell et al., 2010), genetic resources in Setaria have been rapidly accumulating. The outstanding model system features of Setaria greatly accelerated gene discovery using both classical mapping approaches and new approaches such as BSA coupled with deep sequencing. Availability of transformation techniques along with gene editing technology has also allowed S. viridis to be an ideal platform for molecular characterization of gene function. In the meantime, high-throughput phenotyping in Setaria has broadened millet research into new dimensions, such as discovery of novel time-dependent traits in plant architecture and physiology.
It is important to note that the use of S. viridis is not a substitute for millet research. Rather, S. viridis is positioned to become the model for hypothesis testing and genome engineering in order to increase the pace of yield gains and trait enhancements in millets. Usages of this model include but are not limited to, translating mapped genes and QTLs from Setaria to other millets, validating candidate genes from other millets in S. viridis, and adopting well-established high-throughput phenotyping strategies in Setaria to other millets. Finally, fundamental understandings of important complex traits such as C4 photosynthesis and stress tolerance in Setaria will greatly benefit studies of these commonly shared features in all millets, and create new opportunities to accelerate millet breeding and genetic engineering.
Author Contributions
PH and TB conceived of the manuscript. PH, CS, CC, YC, and TB wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work is supported by US Department of Energy (DE-SC0008769) and National Science Foundation (1546882) to PH, CC, YC, and TB and a US Department of Agriculture – National Institute of Food and Agriculture Postdoctoral Fellowship (2014-67012-22269) to CS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors thank R. Parvathaneni for helpful comments on the manuscript.
Footnotes
References
An, J., Shen, X., Ma, Q., Yang, C., Liu, S., and Chen, Y. (2014). Transcriptome profiling to discover putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.). PLoS ONE 9:e99940. doi: 10.1371/journal.pone.0099940
Attaran, E., Major, I. T., Cruz, J. A., Rosa, B. A., Koo, A. J. K., Chen, J., et al. (2014). Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 165, 1302–1314. doi: 10.1104/pp.114.239004
Babitha, K. C., Ramu, S. V., Nataraja, K. N., Sheshshayee, M. S., and Udayakumar, M. (2015). EcbZIP60, a basic leucine zipper transcription factor from Eleusine coracana L. improves abiotic stress tolerance in tobacco by activating unfolded protein response pathway. Mol. Breed. 35:181. doi: 10.1007/s11032-015-0374-6
Babu, B. K., Kalyana Babu, B., Agrawal, P. K., Dinesh, P., and Anil, K. (2014). Comparative genomics and association mapping approaches for opaque2 modifier genes in finger millet accessions using genic, genomic and candidate gene-based simple sequence repeat markers. Mol. Breed. 34, 1261–1279. doi: 10.1007/s11032-014-0115-2
Benabdelmouna, A., Shi, Y., Abirached-Darmency, M., and Darmency, H. (2001). Genomic in situ hybridization (GISH) discriminates between the A and the B genomes in diploid and tetraploid Setaria species. Genome 44, 685–690. doi: 10.1139/gen-44-4-685
Bennett, M. D., and Smith, J. B. (1976). Nuclear dna amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 274, 227–274. doi: 10.1098/rstb.1976.0044
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. doi: 10.1038/nbt.2196
Brutnell, T. P. (2015). Model grasses hold key to crop improvement. Nat. Plants 1:15062. doi: 10.1038/nplants.2015.62
Brutnell, T. P., Bennetzen, J. L., and Vogel, J. P. (2015). Brachypodium distachyon and Setaria viridis: model genetic systems for the grasses. Annu. Rev. Plant Biol. 66, 465–485. doi: 10.1146/annurev-arplant-042811-105528
Brutnell, T. P., Wang, L., Swartwood, K., Goldschmidt, A., Jackson, D., Zhu, X.-G., et al. (2010). Setaria viridis: a model for C4 photosynthesis. Plant Cell 22, 2537–2544. doi: 10.1105/tpc.110.075309
Cannarozzi, G., Plaza-Wüthrich, S., Esfeld, K., Larti, S., Wilson, Y. S., Girma, D., et al. (2014). Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genomics 15:581. doi: 10.1186/1471-2164-15-581
Ceasar, S. A., and Ignacimuthu, S. (2009). Genetic engineering of millets: current status and future prospects. Biotechnol. Lett. 31, 779–788. doi: 10.1007/s10529-009-9933-4
Ceasar, S. A., and Ignacimuthu, S. (2011). Agrobacterium-mediated transformation of finger millet (Eleusine coracana (L.) Gaertn.) using shoot apex explants. Plant Cell Rep. 30, 1759–1770. doi: 10.1007/s00299-011-1084-0
Charu Lata, M. (2015). Advances in omics for enhancing abiotic stress tolerance in millets. Proc. Indian Natl. Sci. Acad. 81, 397–417. doi: 10.16943/ptinsa/2015/v81i2/48095
Choudhary, M., Jayanand, and Padaria, J. C. (2015). Transcriptional profiling in pearl millet (Pennisetum glaucum L.R. Br.) for identification of differentially expressed drought responsive genes. Physiol. Mol. Biol. Plants 21, 187–196. doi: 10.1007/s12298-015-0287-1
Cruz, J. A., Savage, L. J., Zegarac, R., Hall, C. C., Satoh-Cruz, M., Davis, G. A., et al. (2016). Dynamic environmental photosynthetic imaging reveals emergent phenotypes. Cell Syst. 2, 365–377. doi: 10.1016/j.cels.2016.06.001
Diao, X., Schnable, J., Bennetzen, J. L., and Li, J. (2014). Initiation of Setaria as a model plant. Front. Agric. Sci. Eng. 1:16. doi: 10.15302/J-FASE-2014011
Doust, A. N., Kellogg, E. A., Devos, K. M., and Bennetzen, J. L. (2009). Foxtail millet: a sequence-driven grass model system. Plant Physiol. 149, 137–141. doi: 10.1104/pp.108.129627
Duvick, D. N. (2005). The contribution of breeding to yield advances in maize (Zea mays L). Adv. Agron. 86, 83–145.
Dwivedi, S., Sangam, D., Hari, U., Senapathy, S., Charles, H., Kenji, F., et al. (2011). “Millets: genetic and genomic resources,” in Plant Breeding Reviews, ed. J. Janick (Hoboken, NJ: Wiley-Blackwell), 247–375.
Fahlgren, N., Feldman, M., Gehan, M. A., Wilson, M. S., Shyu, C., Bryant, D. W., et al. (2015a). A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol. Plant 8, 1520–1535. doi: 10.1016/j.molp.2015.06.005
Fahlgren, N., Noah, F., Gehan, M. A., and Ivan, B. (2015b). Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 24, 93–99. doi: 10.1016/j.pbi.2015.02.006
Fang, X., Dong, K., Wang, X., Liu, T., He, J., Ren, R., et al. (2016). A high density genetic map and QTL for agronomic and yield traits in Foxtail millet [Setaria italica (L.) P. Beauv]. BMC Genomics 17:336. doi: 10.1186/s12864-016-2628-z
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Goron, T. L., and Raizada, M. N. (2015). Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front. Plant Sci. 6:157. doi: 10.3389/fpls.2015.00157
Grass Phylogeny Working Group II (2011). New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 193, 304–312. doi: 10.1111/j.1469-8137.2011.03972.x
Hu, Z., Mbacké, B., Perumal, R., Guèye, M. C., Sy, O., Bouchet, S., et al. (2015). Population genomics of pearl millet (Pennisetum glaucum (L.) R. Br.): comparative analysis of global accessions and Senegalese landraces. BMC Genomics 16:1048. doi: 10.1186/s12864-015-2255-0
Huang, P., and Brutnell, T. P. (2016). A synthesis of transcriptomic surveys to dissect the genetic basis of C4 photosynthesis. Curr. Opin. Plant Biol. 31, 91–99. doi: 10.1016/j.pbi.2016.03.014
Huang, P., Feldman, M., Schroder, S., Bahri, B. A., Diao, X., Zhi, H., et al. (2014). Population genetics of Setaria viridis, a new model system. Mol. Ecol. 23, 4912–4925. doi: 10.1111/mec.12907
Huang, P., Pu, H., Studer, A. J., Schnable, J. C., Kellogg, E. A., and Brutnell, T. P. (2016). Cross species selection scans identify components of C4 photosynthesis in the grasses. J. Exp. Bot. doi: 10.1093/jxb/erw256 [Epub ahead of print].
Ignacimuthu, S., and Ceasar, S. A. (2012). Development of transgenic finger millet (Eleusine coracana (L.) Gaertn.) resistant to leaf blast disease. J. Biosci. 37, 135–147. doi: 10.1007/s12038-011-9178-y
Jia, G., Huang, X., Zhi, H., Zhao, Y., Zhao, Q., Li, W., et al. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957–961. doi: 10.1038/ng.2673
Jiang, H., Barbier, H., and Brutnell, T. (2013). Methods for performing crosses in Setaria viridis, a new model system for the grasses. J. Vis. Exp. 80:e50527. doi: 10.3791/50527
Jo, Y., Lian, S., Cho, J. K., Choi, H., Kim, S.-M., Kim, S.-L., et al. (2016). De novo transcriptome assembly of Setatria italica variety taejin. Genom Data 8, 121–122. doi: 10.1016/j.gdata.2016.05.001
John, C. R., Smith-Unna, R. D., Woodfield, H., Covshoff, S., and Hibberd, J. M. (2014). Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165, 62–75. doi: 10.1104/pp.114.238667
Jost, M., Esfeld, K., Burian, A., Cannarozzi, G., Chanyalew, S., Kuhlemeier, C., et al. (2014). Semi-dwarfism and lodging tolerance in tef (Eragrostis tef) is linked to a mutation in the -Tubulin 1 gene. J. Exp. Bot. 66, 933–944. doi: 10.1093/jxb/eru452
Knecht, A. C., Campbell, M. T., Caprez, A., Swanson, D. R., and Walia, H. (2016). Image harvest: an open-source platform for high-throughput plant image processing and analysis. J. Exp. Bot. 67, 3587–3599. doi: 10.1093/jxb/erw176
Kubešová, M., Moravcová, L., Suda, J., Jarošík, V., and Pyšek, P. (2010). Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia 82, 81–96.
Kulkarni, K. S., Zala, H. N., Bosamia, T. C., Shukla, Y. M., Sushil, K., Fougat, R. S., et al. (2016). De novo transcriptome sequencing to dissect candidate genes associated with pearl millet-downy mildew (Sclerospora graminicola Sacc.) interaction. Front. Plant Sci. 7:847. doi: 10.3389/fpls.2016.00847
Kumar, A., Anil, K., Gaur, V. S., Anshita, G., and Gupta, A. K. (2014). De novo assembly and characterization of developing spikes transcriptome of finger millet (Eleusine coracana): a minor crop having nutraceutical properties. Plant Mol. Biol. Rep. 33, 905–922. doi: 10.1007/s11105-014-0802-5
Lata, C., Bhutty, S., Bahadur, R. P., Majee, M., and Prasad, M. (2011). Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. J. Exp. Bot. 62, 3387–3401. doi: 10.1093/jxb/err016
Li, C., Yue, J., Wu, X., Xu, C., and Yu, J. (2014). An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 65, 5415–5427. doi: 10.1093/jxb/eru302
Li, P., and Brutnell, T. P. (2011). Setaria viridis and Setaria italica, model genetic systems for the panicoid grasses. J. Exp. Bot. 62, 3031–3037. doi: 10.1093/jxb/err096
Li, W., Tang, S., Zhang, S., Shan, J., Tang, C., Chen, Q., et al. (2016). Gene mapping and functional analysis of the novel leaf color gene SiYGL1 in foxtail millet [Setaria italica (L.) P. Beauv]. Physiol. Plant. 157, 24–37. doi: 10.1111/ppl.12405
Lobet, G., Draye, X., and Périlleux, C. (2013). An online database for plant image analysis software tools. Plant Methods 9:38. doi: 10.1186/1746-4811-9-38
Martin, A. P., Palmer, W. M., Christopher, B., Christin, A., Lunn, J. E., Furbank, R. T., et al. (2016). A developing Setaria viridis internode: an experimental system for the study of biomass generation in a C4 model species. Biotechnol. Biofuels 9:45. doi: 10.1186/s13068-016-0457-6
Martins, P. K., Nakayama, T. J., Ribeiro, A. P., da Cunha, B. A. D. B., Nepomuceno, A. L., Harmon, F. G., et al. (2015). Setaria viridis floral-dip: a simple and rapid Agrobacterium-mediated transformation method. Biotechnol. Rep. 6, 61–63. doi: 10.1016/j.btre.2015.02.006
Masumoto, H., Hisato, M., Hiroki, T., Yohei, M., Ryohei, T., and Kenji, F. (2016). Genetic analysis of NEKODE1 gene involved in panicle branching of foxtail millet, Setaria italica (L.) P. Beauv., and mapping by using QTL-seq. Mol. Breed. 36:59. doi: 10.1007/s11032-016-0481-z
Mauro-Herrera, M., and Doust, A. N. (2016). Development and genetic control of plant architecture and biomass in the panicoid grass. Setaria. PLoS ONE 11:e0151346. doi: 10.1371/journal.pone.0151346
Mauro-Herrera, M., Wang, X., Barbier, H., Brutnell, T. P., Devos, K. M., and Doust, A. N. (2013). Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3 3, 283–295. doi: 10.1534/g3.112.005207
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Moumouni, K. H., Kountche, B. A., Jean, M., Hash, C. T., Vigouroux, Y., Haussmann, B. I. G., et al. (2015). Construction of a genetic map for pearl millet, Pennisetum glaucum (L.) R. Br., using a genotyping-by-sequencing (GBS) approach. Mol. Breed. 35:5. doi: 10.1007/s11032-015-0212-x
Muthamilarasan, M., and Prasad, M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. doi: 10.1007/s00122-014-2399-3
Neilson, E. H., Edwards, A. M., Blomstedt, C. K., Berger, B., Moller, B. L., and Gleadow, R. M. (2015). Utilization of a high-throughput shoot imaging system to examine the dynamic phenotypic responses of a C4 cereal crop plant to nitrogen and water deficiency over time. J. Exp. Bot. 66, 1817–1832. doi: 10.1093/jxb/eru526
Neufeld, H. S., Durall, D. M., Rich, P. M., and Tingey, D. T. (1989). A rootbox for quantitative observations on intact entire root systems. Plant Soil 117, 295–298. doi: 10.1007/BF02220725
Parvathaneni, R. K., Jakkula, V., Padi, F. K., Faure, S., Nagarajappa, N., Pontaroli, A. C., et al. (2013). Fine-mapping and identification of a candidate gene underlying the d2 dwarfing phenotype in pearl millet. Cenchrus americanus (L.) Morrone. G3 (Bethesda) 3, 563–572. doi: 10.1534/g3.113.005587
Passot, S., Sixtine, P., Fatoumata, G., Daniel, M., Mikaël, L., Soazig, G., et al. (2016). Characterization of pearl millet root architecture and anatomy reveals three types of lateral roots. Front. Plant Sci. 7:829. doi: 10.3389/fpls.2016.00829
Piatek, A., Ali, Z., Baazim, H., Li, L., Abulfaraj, A., Al-Shareef, S., et al. (2015). RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 13, 578–589. doi: 10.1111/pbi.12284
Plaza-Wüthrich, S., and Sonia, Z. (2012). Millet improvement through regeneration and transformation. Biotechnol. Mol. Biol. Rev. 7, 48–61. doi: 10.5897/bmbr12.001
Puranik, S., Swati, P., Sahu, P. P., Mandal, S. N., B, V. S., Parida, S. K., et al. (2013). Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 8:e64594. doi: 10.1371/journal.pone.0064594
Qie, L., Jia, G., Zhang, W., Schnable, J., Shang, Z., Li, W., et al. (2014). Mapping of quantitative trait locus (QTLs) that contribute to germination and early seedling drought tolerance in the interspecific cross Setaria italica × Setaria viridis. PLoS ONE 9:e101868. doi: 10.1371/journal.pone.0101868
Rahaman, M. M., Chen, D., Gillani, Z., Klukas, C., and Chen, M. (2015). Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front. Plant Sci. 6:619. doi: 10.3389/fpls.2015.00619
Rahman, H., Jagadeeshselvam, N., Valarmathi, R., Sachin, B., Sasikala, R., Senthil, N., et al. (2014). Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol. Biol. 85, 485–503. doi: 10.1007/s11103-014-0199-4
Rahman, H., Ramanathan, V., Nallathambi, J., Duraialagaraja, S., and Muthurajan, R. (2016). Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol. 16(Suppl. 1):35. doi: 10.1186/s12896-016-0261-1
Rajput, S. G., Santra, D. K., and James, S. (2016). Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). Mol. Breed. 36:37. doi: 10.1007/s11032-016-0460-4
Rajput, S. G., Tammy, P.-H., and Santra, D. K. (2014). Development and characterization of SSR markers in proso millet based on switchgrass genomics. Am. J. Plant Sci. 5, 175–186. doi: 10.4236/ajps.2014.51023
Ramadevi, R., Rao, K. V., and Reddy, V. D. (2014). Agrobacterium tumefaciens-mediated genetic transformation and production of stable transgenic pearl millet (Pennisetum glaucum [L.] R. Br.). Vitro Cell. Dev. Biol. Plant 50, 392–400. doi: 10.1007/s11627-013-9592-y
Rawson, H. M., Begg, J. E., and Woodward, R. G. (1977). The effect of atmospheric humidity on photosynthesis, transpiration and water use efficiency of leaves of several plant species. Planta 134, 5–10. doi: 10.1007/BF00390086
Rellán-Álvarez, R., Lobet, G., Lindner, H., Pradier, P.-L., Sebastian, J., Yee, M.-C., et al. (2015). GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. Elife 4:e07597. doi: 10.7554/eLife.07597
Rice, A., Anna, R., Lior, G., Shiran, A., Moshe, E., Kopelman, N. M., et al. (2014). The Chromosome Counts Database (CCDB) - a community resource of plant chromosome numbers. New Phytol. 206, 19–26. doi: 10.1111/nph.13191
Sage, R. F., and Pearcy, R. W. (1987a). The nitrogen use efficiency of C3 and C4 plants: II. Leaf nitrogen effects on the gas exchange characteristics of Chenopodium album (L.) and Amaranthus retroflexus (L.). Plant Physiol. 84, 959–963.
Sage, R. F., and Pearcy, R. W. (1987b). The nitrogen use efficiency of C3 and C4 plants: I. Leaf nitrogen, growth, and biomass partitioning in Chenopodium album (L.) and Amaranthus retroflexus (L.). Plant Physiol. 84, 954–958.
Sage, R. F., Pearcy, R. W., and Seemann, J. R. (1987). The nitrogen use efficiency of C3 and C4 plants : III. Leaf nitrogen effects on the activity of carboxylating enzymes in Chenopodium album (L.) and Amaranthus retroflexus (L.). Plant Physiol. 85, 355–359.
Saha, D., Dipnarayan, S., Channabyre Gowda, M. V., Lalit, A., Manjusha, V., and Bansal, K. C. (2016). Genetic and genomic resources of small millets. CRC Crit. Rev. Plant Sci. 35, 56–79. doi: 10.1080/07352689.2016.1147907
Saha, P., and Blumwald, E. (2016). Spike-dip transformation of Setaria viridis. Plant J. 86, 89–101. doi: 10.1111/tpj.13148
Sahu, P. P., Gupta, S., Malaviya, D. R., Roy, A. K., Kaushal, P., and Prasad, M. (2012). Transcriptome analysis of differentially expressed genes during embryo sac development in apomeiotic non-parthenogenetic interspecific hybrid of Pennisetum glaucum. Mol. Biotechnol. 51, 262–271. doi: 10.1007/s12033-011-9464-9
Sato, K., Kei, S., Yohei, M., Ken, N., and Kenji, F. (2013). Construction of a foxtail millet linkage map and mapping of spikelet-tipped bristles 1(stb1) by using transposon display markers and simple sequence repeat markers with genome sequence information. Mol. Breed. 31, 675–684. doi: 10.1007/s11032-012-9825-5
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Sebastian, J., Yee, M.-C., Goudinho Viana, W., Rellán-Álvarez, R., Feldman, M., Priest, H. D., et al. (2016). Grasses suppress shoot-borne roots to conserve water during drought. Proc. Natl. Acad. Sci. U.S.A. 113, 8861–8866. doi: 10.1073/pnas.1604021113
Singh, A., Ganapathysubramanian, B., Singh, A. K., and Sarkar, S. (2016). Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 21, 110–124. doi: 10.1016/j.tplants.2015.10.015
Singh, U. M., Chandra, M., Shankhdhar, S. C., and Kumar, A. (2014). Transcriptome wide identification and validation of calcium sensor gene family in the developing spikes of finger millet genotypes for elucidating its role in grain calcium accumulation. PLoS ONE 9:e103963. doi: 10.1371/journal.pone.0103963
Tadele, Z. (2016). “Drought adaptation in millets,” in Abiotic and Biotic Stress in Plants - Recent Advances and Future Perspectives, eds A. K. Shanker and C. Shanker (Rijeka: InTech), 639–662. doi: 10.5772/61929
Tadele, Z., and Plaza-Wüthrich, S. (2013). Regeneration and Transformation of Millets. Lakewood, NJ: LAP Lambert Academic Publishing.
Takagi, H., Tamiru, M., Abe, A., Yoshida, K., Uemura, A., Yaegashi, H., et al. (2015). MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 33, 445–449.
Vadez, V., Kholová, J., Hummel, G., Zhokhavets, U., Gupta, S. K., and Hash, C. T. (2015). LeasyScan: a novel concept combining 3D imaging and lysimetry for high-throughput phenotyping of traits controlling plant water budget. J. Exp. Bot. 66, 5581–5593. doi: 10.1093/jxb/erv251
Van Eck, J., and Swartwood, K. (2015). Setaria viridis. Methods Mol. Biol. 1223, 57–67. doi: 10.1007/978-1-4939-1695-5_5
Wang, M., Li, P., Li, C., Pan, Y., Jiang, X., Zhu, D., et al. (2014). SiLEA14, a novel atypical LEA protein, confers abiotic stress resistance in foxtail millet. BMC Plant Biol. 14:290. doi: 10.1186/s12870-014-0290-7
Wang, M.-Z. (2011). Culturing of immature inflorescences and Agrobacterium-mediated transformation of foxtail millet (Setaria italica). Afr. J. Biotechnol. 10, 16466-16479. doi: 10.5897/ajb10.2330
Xu, J., Jiajia, X., Yuanyuan, L., Xiuling, M., Jianfeng, D., Kai, W., et al. (2013). Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research. Plant Mol. Biol. 83, 77–87. doi: 10.1007/s11103-013-0025-4
Yang, W., Duan, L., Chen, G., Xiong, L., and Liu, Q. (2013). Plant phenomics and high-throughput phenotyping: accelerating rice functional genomics using multidisciplinary technologies. Curr. Opin. Plant Biol. 16, 180–187. doi: 10.1016/j.pbi.2013.03.005
Yi, F., Xie, S., Liu, Y., Qi, X., and Yu, J. (2013). Genome-wide characterization of microRNA in foxtail millet (Setaria italica). BMC Plant Biol. 13:212. doi: 10.1186/1471-2229-13-212
Yue, H., Hong, Y., Le, W., Hui, L., Wenjie, Y., Xianghong, D., et al. (2016). De novo assembly and characterization of the transcriptome of broomcorn millet (Panicum miliaceum L.) for gene discovery and marker development. Front. Plant Sci. 7:1083. doi: 10.3389/fpls.2016.01083
Zhang, F., Puchta, H., and Thomson, J. G. (2015). Advances in New Technology for Targeted Modification of Plant Genomes. Berlin: Springer.
Keywords: Setaria viridis, foxtail millet, bulked segregant analysis, stress tolerance, high-throughput phenotyping, model grass, C4 photosynthesis
Citation: Huang P, Shyu C, Coelho CP, Cao Y and Brutnell TP (2016) Setaria viridis as a Model System to Advance Millet Genetics and Genomics. Front. Plant Sci. 7:1781. doi: 10.3389/fpls.2016.01781
Received: 22 September 2016; Accepted: 11 November 2016;
Published: 28 November 2016.
Edited by:
Manoj Prasad, National Institute of Plant Genome Research, IndiaReviewed by:
Kevin Murphy, Washington State University, USAChandra Bhan Yadav, University of Milan, Italy
Copyright © 2016 Huang, Shyu, Coelho, Cao and Brutnell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pu Huang, phuang@danforthcenter.org
 Pu Huang
Pu Huang Christine Shyu
Christine Shyu Carla P. Coelho
Carla P. Coelho Yingying Cao
Yingying Cao Thomas P. Brutnell
Thomas P. Brutnell