- 1 Department of Neurosurgery, Yale University School of Medicine, New Haven, CT, USA
- 2 Child Study Center, Yale University School of Medicine, New Haven, CT, USA
Regulation of maternal behavior in the immediate postpartum period involves neural circuits in reward and homeostasis systems responding to cues from the newborn. Our aim was to assess one specific regulatory mechanism: the role that olfaction plays in the onset and modulation of parenting behavior. We focused on changes in gene expression in olfactory brain regions, examining nine genes found in previous knockout studies to be necessary for maternal behavior. Using a quantitative PCR (qPCR)-based approach, we assessed changes in gene expression in response to exposure to pups in 11 microdissected olfactory brain regions. Over the first postpartum days, all nine genes were detected in all 11 regions (at differing levels) and their expression changed in response to pup exposure. As a general trend, five genes (Dbh, Esr1, FosB, Foxb1, and Oxtr) were found to decrease their expression in most of the olfactory regions examined, while two genes (Mest and Prlr) were found to increase expression. Nos1 and Peg3 levels remained relatively stable except in the accessory olfactory bulb (AOB), where greater than fourfold increases in expression were observed. The largest magnitude expression changes in this study were found in the AOB, which mediates a variety of olfactory cues that elicit stereotypic behaviors such as mating and aggression as well as some non-pheromone odors. Previous analyses of null mice for the nine genes assessed here have rarely examined olfactory function. Our data suggest that there may be olfactory effects in these null mice which contribute to the observed maternal behavioral phenotypes. Collectively, these data support the hypothesis that olfactory processing is an important sensory regulator of maternal behavior.
Introduction
Regulation of maternal behavior in the immediate postpartum period involves neural circuits in reward and homeostasis systems responding to newborn sensory cues from the somatosensory, visual, auditory, and olfactory systems. Olfactory regulation of maternal behavior is less well understood than other sensory modalities, especially visual and auditory.
The few studies examining olfactory cues and early maternal behavior have studied maternal sensitivity to olfactory cues as a means of identifying key characteristics of their own offspring. Mothers are reliably able to identify their own child’s odor (Porter et al., 1983; Kaitz et al., 1987) with up to about 90% accuracy in as little as 10 min after birth (Kaitz et al., 1987). New mothers find baby-related odors have greater hedonic value (Fleming et al., 1993). Moreover, first-time mothers, with higher levels of circulating cortisol, are better able to identify their own infant’s odor (Fleming et al., 1997). Consistent with this small literature are studies in animal models where many aspects of social behavior, such as gender identification, control of mating and aggression responses, and pup recognition are mediated by olfaction. We have elected to perform our study in mice as rodents have been the mainstay of biomedical research and the insights provided into the workings of mammalian systems have proved to be applicable to human biological systems (Shively and Clarkson, 2009).
In many animals a functioning olfactory system is crucial for maternal behavior (Levy et al., 2004; Levy and Keller, 2009). For example, largely anosmic mice that lack the second messenger adenylyl cyclase type 3 (AC3) have been shown to have impaired maternal behavior as they fail to retrieve pups, do not construct well-defined nests, and do not exhibit maternal aggression (Wang and Storm, 2011). Likewise when vomeronasal (VNO)-specific signal transduction cascade components are mutated (e.g., mice that lack the TrpC2 channel) deficits are observed in maternal behaviors such as aggression and nest building, indicating that the VNO pathway is also involved in their regulation (Kimchi et al., 2007; Hasen and Gammie, 2009, 2011).
Many genes have been identified from mouse genetic studies which are necessary for the expression of one or more aspects of maternal behavior. Deletion of the following nine genes, dopamine beta-hydroxylase (Dbh; Thomas and Palmiter, 1997), prolactin receptor (Prlr; Ormandy et al., 1997; Lucas et al., 1998), nitric oxide synthase 1 (Nos1; Gammie and Nelson, 1999), oxytocin receptor (Oxtr; Takayanagi et al., 2005), estrogen receptor alpha (Esr1; Ogawa et al., 1996, 1998), forkhead box B1 (Foxb1; Wehr et al., 1997), mesoderm-specific transcript (Mest; Lefebvre et al., 1998), paternally expressed gene 3 (Peg3; Li et al., 1999), and FBJ murine osteosarcoma viral oncogene homolog B (FosB; Brown et al., 1996) have a variety of effects on different aspects of maternal behavior, from nest building to licking and grooming. In the majority of these studies olfaction was not evaluated, or if it was there were issues with experimental design (e.g., odors were tested which were either not behaviorally relevant or only activated one of the two olfactory systems were assessed, mice were not tested during the postpartum period; reviewed in Leckman and Herman, 2002). Interestingly, each of these nine genes are expressed in olfactory regions at ages when mice are sexually mature [postnatal day 28 (P28) or P56; Allen Brain Atlas Resources (Internet). Seattle, WA, USA: Allen Institute for Brain Science. ©2009. Available from: http://www.brain-map.org]. As maternal behavior has long been known to depend upon detection of olfactory cues from the pup (Noirot, 1969), we reasoned that aspects of the observed maternal behavioral phenotypes in null mice may have olfactory component.
As a first measure to test this hypothesis we took the approach of determining whether gene expression changed in wild type olfactory brain regions during the immediate postpartum period. We took this approach because the information obtained would allow one to design appropriate olfactory behavioral tests to test null mice for olfactory deficits. Using a qPCR approach (which provides a quantitative measure of a gene transcription), we compared gene expression of the nine genes in maternal olfactory brain regions at postpartum day 0 (PPD0) and postpartum day 1 (PPD1) relative to expression levels in virgin mice that had never been exposed to pups. We found that, as a general trend, Dbh, Esr1, Foxb1, FosB, and Oxtr decreased in expression in most regions after exposure to pups, while Nos1, Mest, Peg3, and Prlr increased in most regions during the postpartum period. Moreover, largest changes were seen in the accessory olfactory pathway which is known to mediate some olfactory cues (pheromones) which elicit stereotypic behaviors, as well as some non-pheromone odors too. Collectively these data support the hypothesis that olfactory cues regulate maternal behavior during the postpartum period. Moreover, these data highlight the necessity of designing appropriate olfactory behavioral tests which activate specific olfactory pathways and also performing these tests during the initial postpartum period which we see these changes in gene expression.
Materials and Methods
Animals
Virgin (12 weeks old) CD-1 (Charles River) and dams with litters on PPD0 and PPD1 were sacrificed with CO2 and brains dissected (n = 3). Note, the nulliparous mice for each group were purchased, shipped and housed together in the same room, and tissue collected for all groups with 60 min. Specifically, control mice were age matched, to nulliparous-females delivered pregnant 17 days which gave birth in our animal room. Also note dams were not removed from litters until immediately before sacrifice, to minimize stress response and possible changes in gene expression. Brains were dissected in ice-cold PBS and microdissected regions (see below) were collected in tubes on dry ice. Animal protocols were reviewed and approved by the Yale Animal Care and Use Committee.
Microdissection and RNA Extraction
Eleven regions were microdissected for RNA extraction. These olfactory brain regions were selected to encompass olfactory processing centers in both the main and accessory pathways. From the main olfactory pathway we collected samples from the main olfactory bulb (MOB), olfactory tubercle (OT), anterior piriform cortex (aPC), anterior and posterior lateral amygdala (aLA; pLA), entorhinal cortex (EC), anterior/dorsal hippocampus (aHC), and posterior/ventral hippocampus (pHC). Samples collected from the accessory olfactory pathway were from the accessory olfactory bulb (AOB) and anterior and posterior medial amygdala (aMA; pMA). Figure 1 details the major components of the main (red) and accessory (blue) olfactory pathways. Dissected regions are highlighted in gray. Tertiary projections are indicated in purple and centrifugal projections back into the OB and AOB are indicated in green.
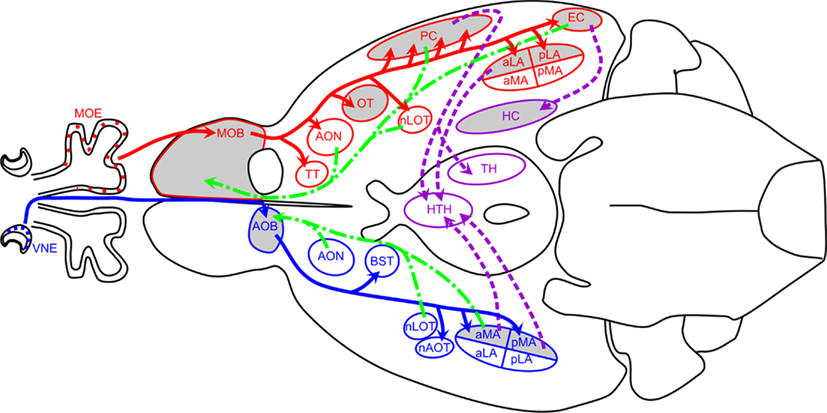
Figure 1. Schematic of mouse olfactory circuits. Main olfactory pathway is shown in red, accessory olfactory pathway in blue, back projections in green, and intracortical projections in purple. Regions highlighted in gray were analyzed in this study. Adapted in part from (Dulac and Wagner, 2006; Baum and Kelliher, 2009; de Castro, 2009). Abbreviations: aLA, anterior lateral amygdala; aMA, anterior medial amygdala; AOB, accessory olfactory bulb; AON, anterior olfactory nucleus; AOT, accessory olfactory tract; BST, bed nucleus of the stria terminalis; EC, entorhinal cortex; HC, hippocampus; HTH, hypothalamus; LOT, lateral olfactory tract; MOB, main olfactory bulb; MOE, main olfactory epithelium; nAOT, nucleus of the accessory olfactory tract; nLOT, nucleus of the lateral olfactory tract; OT, olfactory tubercle; PC, the piriform cortex; pLA, posterior lateral amygdala; pMA, posterior medial amygdala; TH, thalamus; TT, tenia tecta; VMH, ventromedial hypothalamus; VNE, vomeronasal epithelium.
Brains were sliced into 1 mm thick slices using a Rodent Brain Matrix (mouse 30 g coronal; Electro Microscopy Sciences, Hatfield, PA, USA). Specific regions were manually dissected from slices, with the aid of a mouse brain atlas and identifiable landmarks. Samples were stored in Eppendorf tubes on dry ice. As the majority of samples were small, and therefore yielded low amounts of RNA, samples were pooled. This was unfortunate, as it precluded statistical analyses and limited us to observing trends in gene expression. RNA was extracted using RNeasy Lipid Tissue Mini Kit (Qiagen, Germantown, MD, USA) following manufacturer’s instructions.
cDNA was produced in triplicate from each sample (to minimize error and ensure accurate replication of RNA pool) using iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. Optimally 500 ng of total RNA was transcribed, but in some small samples this was reduced to 100–200 ng. For any given brain region, cDNA samples were transcribed from equal amounts of RNA in all three samples (V, PPD0, and PPD1) that were going to be compared to facilitate downstream qPCR analyses.
QPCR Protocol
Primers
We designed the following primers for qPCR using PerlPrimer v1.1.19 software (Marshall, 2004). Primers (see Table 1) were designed to span an intron–exon boundary (to exclude genomic amplification) and to amplify a product between 100 and 150 bp. Each primer pair amplified a single band when visualized on an agarose gel.
Quantitative PCR
Quantitative PCR was performed using iQ SYBR Green supermix (Bio-Rad) in a 20 μl reaction volume, using 0.6 μl iScript cDNA reaction and 500 nM of primers, following manufacturer’s instructions. Master mixes were made for each primer and cDNAs were added independently. Using a Chromo4 Real-Time PCR machine (Bio-Rad) run settings were according to recommended protocol. Briefly, an enzyme activation step for 10 min at 95°C, then 45 cycles of 15 s denaturation step at 95°C, 45 s annealing at 55°C, and 30 s extension at 72°C, and fluorescence data collection at the end of the denaturation step. Melt-curve analysis followed each run with ramping from 55 to 95°C, with fluorescence data collection in 0.5°C increments. Each sample, comprising cDNA from three animals, was run in triplicate.
Data Analysis
The quantity of PCR product was determined on a cycle-by-cycle basis by monitoring the fluorescence values in each sample. Opticon Monitor 3.1.32 MJ software (Bio-Rad) normalized for background fluorescence and permitted setting a threshold at which fluorescence data were analyzed. This threshold was chosen at a level during the exponential phase where the reactions had entered a constant rate of amplification. The same threshold, with a value of 0.2, was used for all runs in this experiment. This yielded a CT value for each sample, the cycle number at which the fluorescence reached threshold, and this was taken as a measure of the abundance of cDNA target present. Standard curve analyses were performed for each primer set to determine reaction efficiencies over a range of cDNA concentrations.
Results were analyzed with the delta delta CT method (ΔΔCT) which is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments (Schmittgen and Livak, 2008). Briefly, CT data for each gene of interest was normalized to CT values for the housekeeping gene GAPDH. This yielded a ΔCT value, or the number of cycles between GAPDH and the gene of interest. The ΔCT values of the two experimental groups, PPD0 and PPD1, were than normalized to the Virgin control sample, giving a ΔΔCT value. This was converted into expression fold change using the 2−ΔΔCT (Schmittgen and Livak, 2008), with Virgin set to 1.0. Values under 1 therefore indicate downregulation, while values above 1 indicate upregulation. PCR efficiency was calculated from standard curves and was accounted for when calculating ΔΔCT. The values across the three plates were then averaged, and variance measured. Graphs were made using GraphPad Prism software.
Results
Baseline gene expression for each of the nine genes studied was established in virgin mice (age and weight matched at 12 weeks; Figure 2). Surprisingly, apart from Prlr (Figure 2), individual genes had fairly stable levels of expression between regions, as indicated by similar ΔCT value. All data points are fairly close except for the Prlr, which had two levels of expression: very low expression in the OB, AOB, OT, and aPC (ΔCT values between ∼14 and 17) with the remaining regions having higher expression (ΔCT values between ∼8 and 11), an average 64-fold (26) difference in expression. Note the error bars in Figure 2 show SD, highlighting the minimal variability observed.
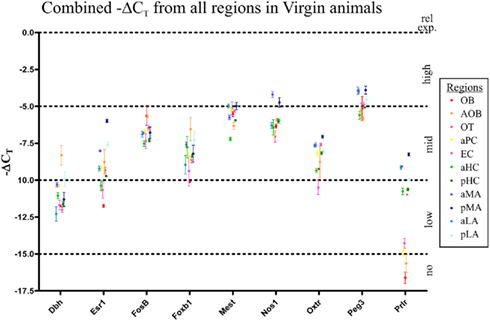
Figure 2. Relative gene expression levels in virgin mice of the nine genes of interest in the 11 olfactory brain regions. Gene expression was normalized against housekeeping gene expression (Gapdh) to determine the ΔCT value (i.e., PCR cycle at which gene was detected above threshold). The earlier a gene is detected, the lower its ΔCT value and the higher the level of expression. To graph this, we plot the −ΔCT value, for easier interpretation: physically higher points on the graph represent higher gene expression. Gene expression from all 11 gene regions examined were plotted (−ΔCT ± SD) to compare expression between genes. All nine genes were found in all 11 regions, although relative levels of expression differed. Interestingly, only Prlr showed a largely dichotomy in expression between regions, with seven the regions (pMA, aMA, pLA, aLA, aHC, pHC, and EC) having significantly higher expression than the other four regions (OB, AOB, aPC, and OT). All other genes generally had between two-fold and eight-fold variation in expression between regions (i.e., ΔCT values within two to four cycles).
To examine changes in gene expression in the postpartum period we elected to look at PPD0 to see if exposure to pups elicited changes in gene expression in olfactory regions, as well as at PPD1 to see if expression remained stable or showed further changes after continued exposure to pups. To compare expression, we normalized expression in the virgin animal to 1 and determined whether expression in PPD0 and PPD1 regions differed from 1. This approach did not allow us to compare expression between genes, but there was no biological reason to assume expression of one gene was linked to another, other than a common link to maternal regulation. Moreover, because we had to pool samples due to limited amounts of RNA, these data could not be statically analyzed and hence we were limited to observing trends in gene expression.
Main Olfactory Pathway
In most mammals, olfactory information is processed through two distinct, parallel, yet non-overlapping pathways: the main olfactory system, which processes information from small volatile odorants, and the accessory olfactory system, which process information from aqueous-soluble odorants that are actively pumped in from the nasal cavity (Dulac and Wagner, 2006). In addition to segregation of the primary inputs, the projections to the central nervous system (CNS) are also segregated and the pathways only converge after many synapses (Figure 1).
In the main olfactory system, the olfactory sensory neurons synapse with mitral cells in the MOB. Mitral cells in turn project to multiple regions of paleocortex, including the anterior olfactory nucleus (AON), the OT, the piriform cortex (PC; considered to be the primary olfactory cortex), the lateral amygdala (LA) and EC (Lledo et al., 2005). These connections are indicated in Figure 1 in red. Information is further relayed to the hippocampus as well as the thalamus, which in turn projects to the orbitofrontal cortex. These intracortical connections are indicated in purple in Figure 1. It is here that information is integrated with both the accessory olfactory system and other sensory systems.
At PPD0 and PPD1, few genes were observed to show greater than two-fold changes in expression in the OB (Figure 3A). Dbh and Foxb1 both showed approximately a two-fold decrease in expression, while the Oxtr was found to have ∼1.5-fold decrease in expression only at PPD1. Prlr showed approximately a two-fold increase in expression while both Mest and Nos1 were found to have ∼1.5-fold increase in expression. In the OT, however, more genes were observed to change expression in the immediate postpartum period (Figure 3B). Again, Dbh and Foxb1 both showed approximately a two-fold decrease in expression at both PPD0 and PPD1. Oxtr was found to have a ∼2.5-fold decrease in expression only at PPD0, while FosB had a ∼1.5-fold decrease in expression only at PPD1 followed by a nearly two-fold decrease at PPD1. Prlr increased expression ∼1.5 to 2-fold and Mest increased expression ∼two-fold at PPD1. Changes of larger magnitude were observed in the piriform cortex (aPC; Figure 3C), with Prlr increasing expression ∼three-fold at both PPD0 and PPD1 while Oxtr, FosB, Foxb1 decreased expression ∼ three-fold.
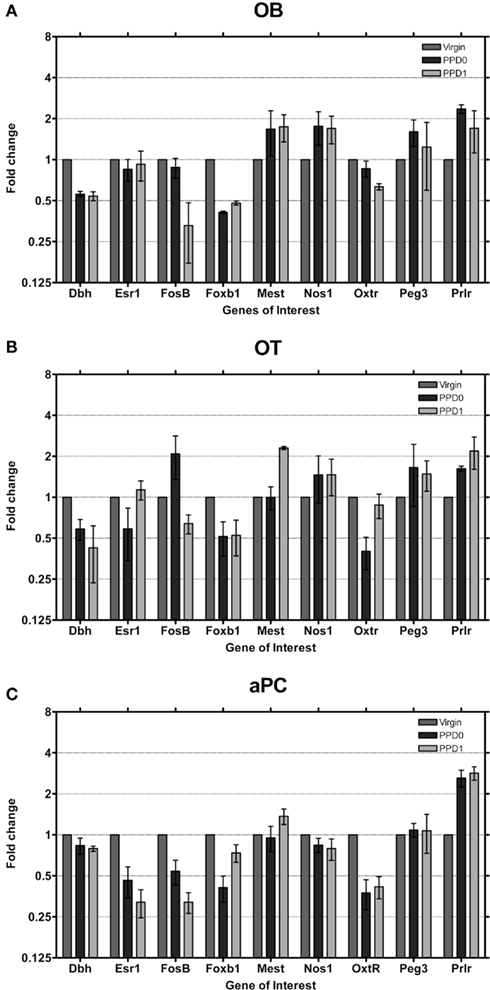
Figure 3. Changes in fold expression (determined by CT method) at PPD0 and PPD1 in the (A) OB, (B) OT, and (C) aPC. Mean ± SD are plotted on a log 2 scale, such that fold increases and decreases are represented linearly.
In the lateral amygdala (aLA and pLA; Figures 4A,B) similar changes in expression were seen, with decreases in Dbh and Foxb1 continuing to be a common trend in all main olfactory system areas. The pLA had the most pronounced decreases in expression of these two genes observed in the main olfactory system, to levels we would consider close to the “unexpressed” range (i.e., detected more than 15 cycles after the detection of the housekeeping gene). Mest was found to increase in both aLA and pLA at PPD1 by two-fold. The Prlr was found to also increase expression two-fold in the pLA at PPD1.
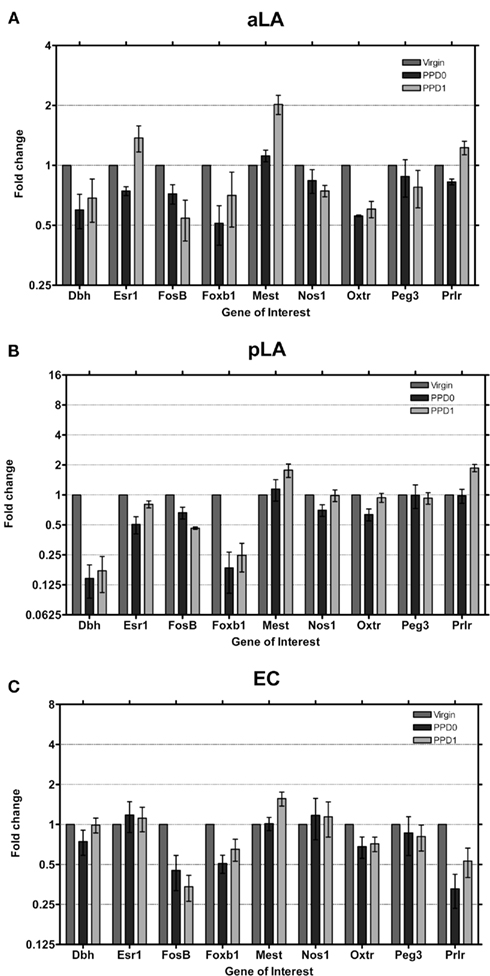
Figure 4. Changes in fold expression (determined by CT method) at PPD0 and PPD1 in the (A) aLA, (B) pLA, and (C) EC. Mean ± SD are plotted on a log 2 scale, such that fold increases and decreases are represented linearly.
In EC, the last region we studied in the main olfactory system (Figure 4C), in contrast to all the other studied areas, Dbh expression was not changed in the early postpartum period. Foxb1 was downregulated as observed in the other regions. Both the Oxtr and Prlr were found to decrease, which is surprising for Prlr as it was already expressed at very low levels to start with (Figure 2). Mest was observed to increase ∼1.5-fold, similar to the increase seen in the LA and OT.
Even though the LA and EC do not receive exclusive innervations from the main olfactory system, the changes in gene expression observed are consistent with changes observed in the OB, OT, and aPC (i.e., decreases in Dbh, Foxb1, and Oxtr). This suggests that system wide changes are maintained along the pathway, and may underlie some aspects of maternal behavior.
Accessory Olfactory System
Sensory information detected in the VNO epithelium is processed in the accessory olfactory system. VNO sensory neurons synapse on AOB mitral cells, which project to four nuclei of the limbic system: the bed nucleus of the stria terminalis (BST), the nucleus of the accessory olfactory tract (nAOT), the aMA and the pMA (Dulac and Wagner, 2006). Projections from medial amygdala connect to hypothalamus, where they converge in some hypothalamic areas with main olfactory information.
The most striking changes in gene expression observed were in the AOB (Figure 5A) where seven genes exhibited moderate (i.e., greater than two-fold change in expression) or strong changes (i.e., greater than four-fold change in expression). Dbh and Foxb1 in particular downregulate eight-fold, a dramatic change sustained over both days. It is important to note that this downregulation brings expression into the “unexpressed” range. Esr1 also downregulates strongly, to nearly one-fourth of baseline, on both days. Interestingly, in the AOB, all three of these genes have high baseline levels to begin with (Figure 2), relative to olfactory bulb and other primarily olfactory structures.
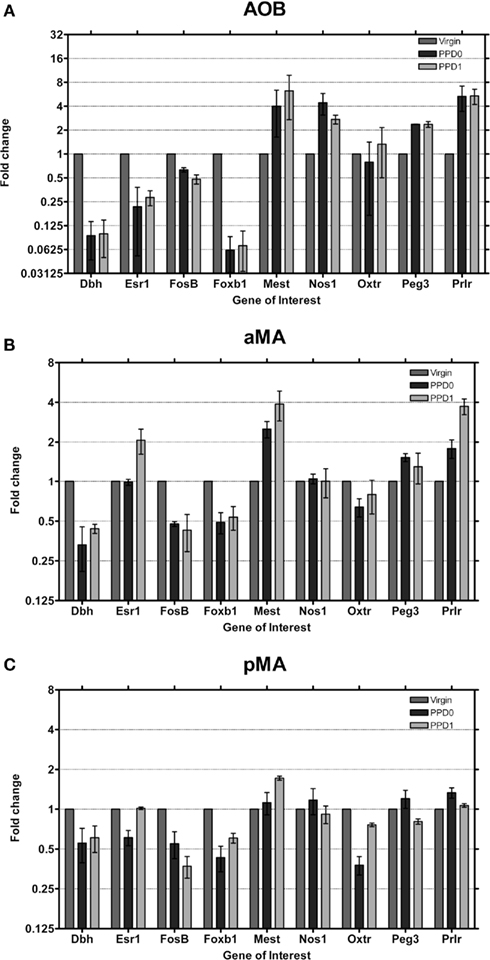
Figure 5. Changes in fold expression (determined by CT method) at PPD0 and PPD1 in the (A) AOB, (B) aMA, and (C) pMA. Mean ± SD are plotted on a log 2 scale, such that fold increases and decreases are represented linearly.
Concurrently, Mest1, Nos1, Peg3, and Prlr upregulate more than two-fold (Prlr increased ∼six-fold). As mentioned above, however, initial Prlr levels were extremely low in AOB, so while a six-fold increase in Prlr levels in the AOB may have some effect, it may not be comparable to other, similarly large fold changes. FosB shows a more modest change, with an ∼two-fold decrease in expression. Therefore we show that in the AOB, a region known to be important in mediating a variety of social odor cues, eight out of nine of maternally relevant genes display large magnitude changes in expression in the early postpartum period.
In both the aMA and pMA (Figures 5B,C) we saw smaller effects than in the AOB; however, changes largely mirrored those seen in the AOB. Dbh, FosB, and Foxb1 downregulated their expression while Mest and Prlr increased expression. In contrast to the AOB, Esr1 showed a two-fold increase at PPD1 in the aMA.
Hippocampus
The olfactory systems provide substantial and direct input to the hippocampus (Staubli et al., 1995). EC densely innervates granule cells in the dentate gyrus, as well as connecting to hippocampal pyramidal cells in CA1 and CA3 (Staubli et al., 1995; Canto et al., 2008). aHC was analyzed independently to pHC, as these regions are known to regulate different information (Woollett and Maguire, 2009).
We found the single largest gene expression change in the aHC (Figure 6A). Prlr was observed to undergo a ∼16-fold increase in expression at PPD1, after an initial decrease in expression at PPD0. We did not observe changes in Prlr expression in pHC (Figure 6B), suggesting that this is specific to the aHC. Dbh and FosB were observed to decrease in both regions, while Mest was found to increase on PPD1 in both regions. Foxb1 expression decreased only in aHC at PPD0, again consistent with the functional dichotomy of these regions.
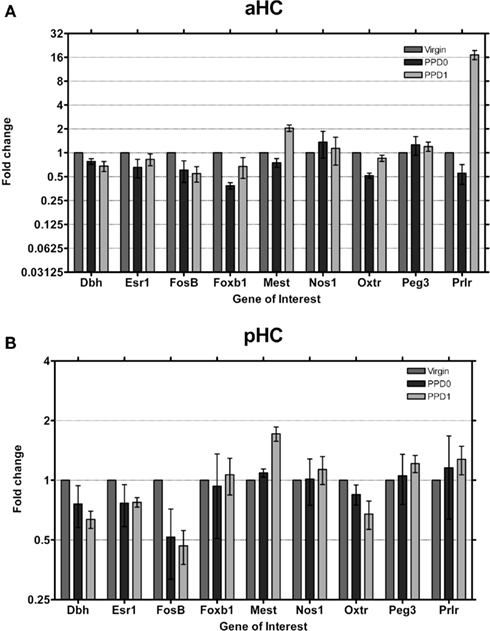
Figure 6. Changes in fold expression (determined by CT method) at PPD0 and PPD1 in the (A) aHC and (B) pHC. Mean ± SD are plotted on a log 2 scale, such that fold increases and decreases are represented linearly.
Discussion
Our results show that the expression of nine known genes regulating early maternal behavior is differentially modulated in olfactory brain regions in the early postpartum period. Interestingly, we did not see large changes in most genes between PPD0 and PPD1 indicating that increased exposure to pups did not elicit further widespread changes in gene expression. We find the largest changes in the AOB, the first component of the accessory system which mediates a variety of olfactory cues including many known to elicit stereotypic behaviors. Our data support the hypothesis that behavioral phenotypes observed in mice which lack expression of these genes may have an olfactory component. Moreover, our data provide a basis to design appropriate olfactory behavioral tests to assess olfaction in these null mice. This is important because if a gene shows changes in expression in the accessory olfactory system, it would be important to use an odor which is detected in the VNO to assess olfactory function. To date many studies have lacked this important information, and have concluded that olfaction is “intact” after testing with one or two odorants which may not reveal the existing deficit. Similarly, tests of olfactory function have not been performed in this immediate postpartum period which, given the dynamic changes in gene expression we demonstrate, may also fail to reveal olfactory deficits. Collectively our data demonstrate how variable and responsive gene expression is during this period in the maternal olfactory brain, and highlight the importance of testing olfactory function in maternal behavioral paradigms in the immediate postpartum period (PPD0 and PPD1).
Olfactory cues have well established roles in eliciting maternal behaviors in rodents. In rodents, as in most mammals, maternal behaviors emerge at or close to parturition. These behaviors include nest building, nursing, pup retrieval, and aggression. Anosmic mice show impaired maternal behavior, with the majority of females eating their progeny (Gandelman et al., 1971a, b; Vandenbergh, 1973; Seegal and Denenberg, 1974). Experience mitigates these effects, as olfactory bulbectomy in multiparous mice does not result in cannibalism (Seegal and Denenberg, 1974). Likewise, mice which lack AC3, a component of the olfactory signal transduction cascade in the main OE, are largely anosmic and have impaired maternal behavior, although they do not cannibalize their young (Wang and Storm, 2011). In contrast, removal of the VNO does not result in cannibalism (Lepri et al., 1985). This apparent lack of dependence on VNO signaling for maternal behavior may be a reflection, however, of recent evidence showing that that some pheromones initially thought to be solely processed by the VNO, are also detected in the MOE in mice (Mandiyan et al., 2005; Liberles and Buck, 2006; Wang et al., 2006). While they do not display cannibalism, mice with surgically removed VNOs do display impaired maternal behavior, which is mirrored in mice which lack Trp2C, a component of the signal transduction cascade in VNO neurons (Kimchi et al., 2007).
In contrast to the studies above, which looked at the role of sensory innervation in olfactory circuits, we were interested in seeing whether known maternally regulated genes had effects in olfactory cortical circuits. From expression databases (i.e., Alan Brain Atlas) we knew that all nine genes were expressed in olfactory regions and were present in a spatiotemporal window to be modulated in the postpartum period. Our data (summarized in Figure 7) indicates that these genes do undergo regulation of expression in the postpartum period. We observed some general trends in the expression changes: Five genes (Dbh, Esr1, Foxb1, FosB, and Oxtr) were found to decrease their expression, while Mest and Prlr increased their expression. Nos1 and Peg3 were largely stable across regions, apart from a region-specific increase in the AOB.

Figure 7. Summary diagram of trends in gene expression changes at PPD0 and PPD1. Using mean fold expression, we plotted changes in expression in a matrix of region vs. gene. This matrix highlights that the largest magnitude changes (both increase and decrease in expression) were found in the accessory olfactory pathway (i.e., AOB and aMA). It also highlights that many genes tended to follow trends in expression in the immediate postpartum period, generally showing either a decrease or an increase in expression in most regions, rather than having variable changes. Note, genes were determined to have no change in expression if the mean fell between 0.9 and 1.1 (i.e., observed changes were less than 10% of virgin controls).
In light of these findings, it is interesting to note that analyses of most lines of null mice for these genes either did not include an assessment of olfaction, or only checked to see if the mice were anosmic (reviewed in (Leckman and Herman, 2002). The exception was the analyses of Dbh−/− mice (Brennan et al., 1990; Thomas and Palmiter, 1997) which also checked the accessory olfactory system by assessing whether pregnancy block was intact. In no case were salient maternal odors such as pup urine tested. Wang and Storm (2011) used a pup/odor preference test to assess olfaction in AC3−/− mice which would also be useful for testing lines of null mice with maternal phenotypes. This test involves placing pups (anesthetized to eliminate ultrasonic vocalizations) in one opaque chamber and assessing how much time a dam spends exploring/sniffing the empty vs. pup containing chamber. Using this paradigm to test olfaction in null mice may more accurately tease out the contribution that olfaction plays in the observed phenotype.
The largest changes in gene expression we observed were in the accessory olfactory system, specifically in the AOB and aMA. In the main olfactory system the changes were predominantly observed downstream from the OB, in the OT, aPC, and pLA. The further downstream one gets, the less olfactory specific a region becomes. There is sensory integration in the amygdala from multiple sources, yet it is interesting to note how the changes largely mirror changes upstream. For example, Dbh was observed to decrease in all regions examined, apart from EC. Functionally this would result in a decrease in epinephrine/norepinephrine synthesized in these areas and most likely a concomitant increase in dopamine. This argues for continuity in gene expression in olfactory circuits and suggests that these changes may be behaviorally relevant.
It is perhaps not surprising that we saw the largest magnitude changes in the accessory olfactory system. This pathway is responsible for mediating many innate behaviors such as gender recognition, mating behavior, and aggression, as well as female estrus cycle (reviewed in (Dulac and Wagner, 2006). In addition to pheromone sensing, this pathway can also processes a variety of odors that are not pheromones (Trinh and Storm, 2003; Ben-Shaul et al., 2010). This circuit, in contrast to the main olfactory pathway, bypasses cortical areas and directly projects to the limbic system. This short synaptic distance from sensory input to effector targets in the hypothalamus (see Figure 1) suggest that this pathway mediates pre-programmed responses (Dulac and Wagner, 2006). The large magnitude changes we detected in the immediate postpartum period were relatively stable from PPD0 to PPD1 (Figure 7) which supports the model of genetically pre-programmed maternal behavioral responses rather than adaptive, experienced-based responses proposed by Dulac and Wagner (2006).
In summary our data show that known maternally regulated genes also have regulated gene expression in the olfactory system during the immediate postpartum period. This pattern of regulation appears to be synchronized across the main and accessory olfactory circuits, with the largest magnitude changes seen in the accessory pathway. Our data also highlight the importance of evaluating olfaction using salient odors in the immediate postpartum period when assessing maternal behavior deficits.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was support in part by NIH DC007600 (Helen B. Treloar) and NIH P50 DA022446 and R01 DA026437 (Linda C. Mayes). This publication was also made possible by CTSA Grant Number UL1 RR024139 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
References
Baum, M. J., and Kelliher, K. R. (2009). Complementary roles of the main and accessory olfactory systems in mammalian mate recognition. Annu. Rev. Physiol. 71, 141–160.
Ben-Shaul, Y., Katz, L. C., Mooney, R., and Dulac, C. (2010). In vivo vomeronasal stimulation reveals sensory encoding of conspecific and allospecific cues by the mouse accessory olfactory bulb. Proc. Natl. Acad. Sci. U.S.A. 107, 5172–5177.
Brennan, P., Kaba, H., and Keverne, E. B. (1990). Olfactory recognition: a simple memory system. Science 250, 1223–1226.
Brown, J. R., Ye, H., Bronson, R. T., Dikkes, P., and Greenberg, M. E. (1996). A defect in nurturing in mice lacking the immediate early gene fosB. Cell 86, 297–309.
Canto, C. B., Wouterlood, F. G., and Witter, M. P. (2008). What does the anatomical organization of the entorhinal cortex tell us? Neural Plast. 2008, 381243.
de Castro, F. (2009). Wiring olfaction: the cellular and molecular mechanisms that guide the development of synaptic connections from the nose to the cortex. Front. Neurosci. 3:52.
Dulac, C., and Wagner, S. (2006). Genetic analysis of brain circuits underlying pheromone signaling. Annu. Rev. Genet. 40, 449–467.
Fleming, A. S., Corter, C., Franks, P., Surbey, M., Schneider, B., and Steiner, M. (1993). Postpartum factors related to mother’s attraction to newborn infant odors. Dev. Psychobiol. 26, 115–132.
Fleming, A. S., Steiner, M., and Corter, C. (1997). Cortisol, hedonics, and maternal responsiveness in human mothers. Horm. Behav. 32, 85–98.
Gammie, S. C., and Nelson, R. J. (1999). Maternal aggression is reduced in neuronal nitric oxide synthase-deficient mice. J. Neurosci. 19, 8027–8035.
Gandelman, R., Zarrow, M. X., and Denenberg, V. H. (1971a). Stimulus control of cannibalism and maternal behavior in anosmic mice. Physiol. Behav. 7, 583–586.
Gandelman, R., Zarrow, M. X., Denenberg, V. H., and Myers, M. (1971b). Olfactory bulb removal eliminates maternal behavior in the mouse. Science 171, 210–211.
Hasen, N. S., and Gammie, S. C. (2009). Trpc2 gene impacts on maternal aggression, accessory olfactory bulb anatomy and brain activity. Genes Brain Behav. 8, 639–649.
Hasen, N. S., and Gammie, S. C. (2011). Trpc2-deficient lactating mice exhibit altered brain and behavioral responses to bedding stimuli. Behav. Brain Res. 217, 347–353.
Kaitz, M., Good, A., Rokem, A. M., and Eidelman, A. I. (1987). Mothers’ recognition of their newborns by olfactory cues. Dev. Psychobiol. 20, 587–591.
Kimchi, T., Xu, J., and Dulac, C. (2007). A functional circuit underlying male sexual behaviour in the female mouse brain. Nature 448, 1009–1014.
Leckman, J. F., and Herman, A. E. (2002). Maternal behavior and developmental psychopathology. Biol. Psychiatry 51, 27–43.
Lefebvre, L., Viville, S., Barton, S. C., Ishino, F., Keverne, E. B., and Surani, M. A. (1998). Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet. 20, 163–169.
Lepri, J. J., Wysocki, C. J., and Vandenbergh, J. G. (1985). Mouse vomeronasal organ: effects on chemosignal production and maternal behavior. Physiol. Behav. 35, 809–814.
Levy, F., and Keller, M. (2009). Olfactory mediation of maternal behavior in selected mammalian species. Behav. Brain Res. 200, 336–345.
Levy, F., Keller, M., and Poindron, P. (2004). Olfactory regulation of maternal behavior in mammals. Horm. Behav. 46, 284–302.
Li, L., Keverne, E. B., Aparicio, S. A., Ishino, F., Barton, S. C., and Surani, M. A. (1999). Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284, 330–333.
Liberles, S. D., and Buck, L. B. (2006). A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650.
Lledo, P. M., Gheusi, G., and Vincent, J. D. (2005). Information processing in the mammalian olfactory system. Physiol. Rev. 85, 281–317.
Lucas, B. K., Ormandy, C. J., Binart, N., Bridges, R. S., and Kelly, P. A. (1998). Null mutation of the prolactin receptor gene produces a defect in maternal behavior. Endocrinology 139, 4102–4107.
Mandiyan, V. S., Coats, J. K., and Shah, N. M. (2005). Deficits in sexual and aggressive behaviors in Cnga2 mutant mice. Nat. Neurosci. 8, 1660–1662.
Marshall, O. J. (2004). PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20, 2471–2472.
Noirot, E. (1969). Changes in responsiveness to young in the adult mouse. V. Priming. Anim. Behav. 17, 542–546.
Ogawa, S., Eng, V., Taylor, J., Lubahn, D. B., Korach, K. S., and Pfaff, D. W. (1998). Roles of estrogen receptor-alpha gene expression in reproduction-related behaviors in female mice. Endocrinology 139, 5070–5081.
Ogawa, S., Taylor, J. A., Lubahn, D. B., Korach, K. S., and Pfaff, D. W. (1996). Reversal of sex roles in genetic female mice by disruption of estrogen receptor gene. Neuroendocrinology 64, 467–470.
Ormandy, C. J., Camus, A., Barra, J., Damotte, D., Lucas, B., Buteau, H., Edery, M., Brousse, N., Babinet, C., Binart, N., and Kelly, P. A. (1997). Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 11, 167–178.
Porter, R. H., Cernoch, J. M., and McLaughlin, F. J. (1983). Maternal recognition of neonates through olfactory cues. Physiol. Behav. 30, 151–154.
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108.
Seegal, R. F., and Denenberg, V. H. (1974). Maternal experience prevents pup-killing in mice induced by peripheral anosmia. Physiol. Behav. 13, 339–341.
Shively, C. A., and Clarkson, T. B. (2009). The unique value of primate models in translational research. Nonhuman primate models of women’s health: introduction and overview. Am. J. Primatol. 71, 715–721.
Staubli, U., Le, T. T., and Lynch, G. (1995). Variants of olfactory memory and their dependencies on the hippocampal formation. J. Neurosci. 15, 1162–1171.
Takayanagi, Y., Yoshida, M., Bielsky, I. F., Ross, H. E., Kawamata, M., Onaka, T., Yanagisawa, T., Kimura, T., Matzuk, M. M., Young, L. J., and Nishimori, K. (2005). Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 102, 16096–16101.
Thomas, S. A., and Palmiter, R. D. (1997). Impaired maternal behavior in mice lacking norepinephrine and epinephrine. Cell 91, 583–592.
Trinh, K., and Storm, D. R. (2003). Vomeronasal organ detects odorants in absence of signaling through main olfactory epithelium. Nat. Neurosci. 6, 519–525.
Vandenbergh, J. G. (1973). Effects of central and peripheral anosmia on reproduction of female mice. Physiol. Behav. 10, 257–261.
Wang, Z., Balet Sindreu, C., Li, V., Nudelman, A., Chan, G. C., and Storm, D. R. (2006). Pheromone detection in male mice depends on signaling through the type 3 adenylyl cyclase in the main olfactory epithelium. J. Neurosci. 26, 7375–7379.
Wang, Z., and Storm, D. R. (2011). Maternal behavior is impaired in female mice lacking type 3 adenylyl cyclase. Neuropsychopharmacology, 36, 772–781.
Wehr, R., Mansouri, A., de Maeyer, T., and Gruss, P. (1997). Fkh5-deficient mice show dysgenesis in the caudal midbrain and hypothalamic mammillary body. Development 124, 4447–4456.
Keywords: olfactory bulb, accessory olfactory bulb, olfactory tubercle, piriform cortex, entorhinal cortex, amygdala, hippocampus
Citation: Canavan SV, Mayes LC and Treloar HB (2011) Changes in maternal gene expression in olfactory circuits in the immediate postpartum period. Front. Psychiatry 2:40. doi: 10.3389/fpsyt.2011.00040
Received: 28 February 2011; Accepted: 15 June 2011;
Published online: 01 July 2011.
Edited by:
Joan Irene Morrell, Rutgers, The State University of New Jersey, USAReviewed by:
Stephen Gammie, University of Wisconsin – Madison, USALévy Frédéric, Institut National de la Recherche Agronomique, France
Copyright: © 2011 Canavan, Mayes and Treloar. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Helen B. Treloar, Department of Neurosurgery, Yale University School of Medicine, P.O. Box 208082, 333 Cedar Street, New Haven, CT 06520-8082, USA. e-mail: helen.treloar@yale.edu

