- Department of Brain and Cognitive Sciences, Picower Institute for Learning and Memory, Massachusetts Institute of Technology, Cambridge, MA, USA
MicroRNAs (miRNAs) are small non-coding RNAs conserved throughout evolution whose perceived importance for brain development and maturation is increasingly being understood. Although a plethora of new discoveries have provided novel insights into miRNA-mediated molecular mechanisms that influence brain plasticity, their relevance for neuropsychiatric diseases with known deficits in synaptic plasticity, such as schizophrenia and autism, has not been adequately explored. In this review we discuss the intersection between current and old knowledge on the role of miRNAs in brain plasticity and function with a focus in the potential involvement of brain expressed miRNAs in the pathophysiology of neuropsychiatric disorders.
Introduction
When Victor Ambros (Lee et al., 1993) and Gary Ruvkun (Wightman et al., 1993) first discovered that a small non-coding RNA lin-4 was able to repress the translation of lin -14, a gene important for developmental timing of C. elegans, the scientific community accepted the findings as an “oddity” of worm biology. Since then a plethora of studies have solidified the notion that these small non-coding RNAs, which are named microRNAs (miRNAs), are evolutionary conserved, are responsible for regulating the expression of the majority of protein coding genes, and play a critical role in diverse cellular functions.
The biogenesis of miRNAs starts when they are transcribed from intergenic or intronic genomic regions into primary miRNA precursor molecules, known as pri -miRNAs (reviewed in Bartel, 2004; Krol et al., 2010a). Pri -miRNAs are cleaved inside the nucleus by the components of the microprocessor complex, Drosha and Dgcr8, to generate hairpin structured RNA hairpins called pre-miRNAs (Bartel, 2004; Krol et al., 2010a). Pre-miRNAs are exported into the cytoplasm and undergo further cleavage by RNaseIII enzyme Dicer, thereby forming a complementary duplex of two miRNA strands (Bartel, 2004; Krol et al., 2010a). The miRNA duplex is unwound and one of the strands, the mature miRNA, is incorporated into a large miRNA-induced silencing complex (miRISC), which serves to detect and bind into complementary sequences inside messenger RNAs (mRNAs). As a result of an efficient binding of the mature miRNA to its target, which is usually in the 3′ untranslated region (3′ UTR) of the mRNA, translational inhibition and/or mRNA cleavage occur with mechanisms that are still not fully understood (Bartel, 2004; Pillai et al., 2007; Krol et al., 2010a). Through this miRNA-mediated control of gene expression, miRNAs are known to not only restrict the expression of their target genes in certain cell types or during specific developmental periods, but also to “fine tune” the levels of co-expressed targets so that a desired biological response can occur.
The mammalian brain is characterized by a notable abundance of miRNAs. Targeted deletion studies of the miRNA processing enzyme Dicer in different cellular populations in the mouse brain have provided strong evidence for the significance of miRNAs in the development and maturation of both neuronal and glial cells, with consequent abnormalities in brain morphology and connectivity (Cuellar et al., 2008; Davis et al., 2008; Shin et al., 2009; Dugas et al., 2010; Konopka et al., 2010; Tao et al., 2011). However, the fact that Dicer can process other types of small non-coding RNAs does not allow us to exclude miRNA-independent mechanisms behind these observed phenotypes. In addition, deletion of another miRNA processing enzyme Dgcr8 in neurons, results in alterations in dendritic branching, excitatory synaptic transmission, and short term plasticity (Stark et al., 2008; Fenelon et al., 2011; Schofield et al., 2011).
miRNAs in Brain Maturation and Function
A large number of studies have identified miRNAs that are important for brain development and neuronal differentiation, which have been described in previous reviews (Li and Jin, 2010; Bian and Sun, 2011). However the focus of this review is on miRNAs that have been proven to influence brain maturation and plasticity, mechanisms that are known to be perturbed in psychiatric diseases (Table 1). Dendritic spines are protrusions at branches of a neuron’s dendritic tree that form the post-synaptic end of a synapse. Their structural properties are known to reflect the degree of brain maturation and their dynamics are an important component of brain plasticity. Moreover, alterations in dendritic spine density have been reported in numerous neuropsychiatric disorders, including schizophrenia and autism spectrum disorders (ASD; Penzes et al., 2001). Interestingly, a loss of dendritic spines has been reported to occur in schizophrenia (Glantz and Lewis, 2000; Kolomeets et al., 2005; Sweet et al., 2009), whereas an increase in the number of spines has been proposed for ASD (Hutsler and Zhang, 2000). In both diseases, however, the changes in dendritic spine density are limited to specific brain regions and cortical layers (Glantz and Lewis, 2000; Hutsler and Zhang, 2000; Kolomeets et al., 2005; Sweet et al., 2009).
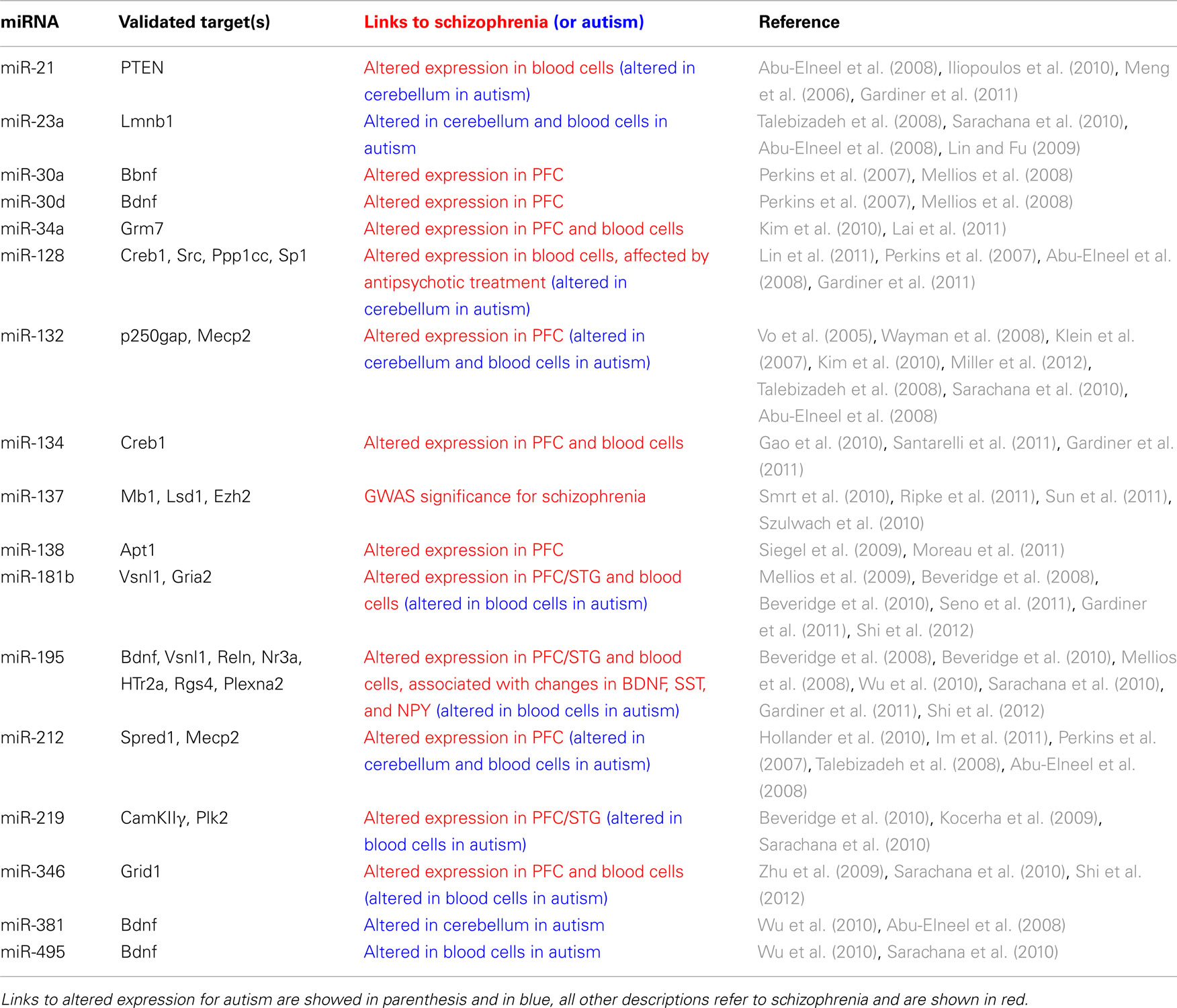
Table 1. MicroRNAs involved in brain plasticity and maturation with links to schizophrenia and autism spectrum disorders.
The first demonstration that miRNAs are localized in the synapse and can regulate dendritic spine structure came when miR-134 was found to be expressed in hippocampal dendritic spines and shown to be capable of reducing spine width by targeting spine growth-promoting kinase Limk1 (Schratt et al., 2006). Since then, a number of miRNAs such as miR-132 (Vo et al., 2005; Wayman et al., 2008; Edbauer et al., 2010; Impey et al., 2010), miR-125b (Edbauer et al., 2010), miR-138 (Siegel et al., 2009), and miR-137 (Smrt et al., 2010) have been shown to regulate dendritic spine structure and morphology. In the case of miR-138 it reduces spine size by targeting depalmitoylation enzyme acyl-protein thioesterase 1 (Apt1; Siegel et al., 2009), whereas miR-125b reduces spine width potentially by targeting NMDA receptor 2a (NR2a; Edbauer et al., 2010). A negative impact on spine maturation was also shown for miR-137, with the proposed mechanism of action being its effect on ubiquitin ligase Mind Bomb 1 (Mb1; Siegel et al., 2009). Neuronal activity and cAMP response binding protein (CREB) signaling was found to regulate the expression of miR-132, which has been shown to increase dendritic spine density and size and promote mature spine morphology by targeting spine inhibitor GTPase p250GAP (Vo et al., 2005; Wayman et al., 2008; Impey et al., 2010). In newborn hippocampal neurons miR-132 plays an additional role by influencing dendritic branching and synaptic integration (Magill et al., 2010; Luikart et al., 2011). Notably, growth factors such as brain-derived neurotrophic factor (BDNF) are known to interfere with miRNA expression and function. For example, BDNF can induce the expression of miR-132 and co-transcribed miR-212 (Remenyi et al., 2010) and inhibit the actions of miR-134 (Schratt et al., 2006), both in an activity-dependent manner.
Despite the fact that most of the initial discoveries pertaining to the role of miRNAs in brain maturation were based in neuronal cultures or slices, recent studies have employed new methods for in vivo manipulation of miRNA levels in mouse brain, and have provided important insights into the interplay between miRNA-mediated structural effects and their impact in brain plasticity and function. The most well studied miRNA family is that of miR-132/miR-212, two miRNAs of the same family that share sequence similarities, and are expressed from the same genomic location. Both miR-132 and miR-212 have been demonstrated to respond to cocaine treatment and in vivo manipulation of miR-212 expression was found to modulate cocaine-induced plasticity and cocaine seeking behavior by indirectly promoting CREB signaling (Hollander et al., 2010). A follow-up study revealed that miR-132/212 were involved into a complex activity-dependent loop with BDNF and Methyl-CPG-binding protein (Mecp2), that resulted in the observed effects in cocaine plasticity (Im et al., 2011). A similar interplay between Mecp2 and miR-132 expression, that involves BDNF, has been previously shown to occur in neuronal cultures (Klein et al., 2007).
Furthermore, two independent studies have recently shown that miR-132 is an experience-dependent miRNA in mouse visual cortex, which regulates dendritic spine morphology in vivo, and whose balanced expression is needed to maintain visual cortex plasticity (Mellios et al., 2011; Tognini et al., 2011). In the first study miR-212 was included together with miR-132 in the subset of miRNAs that showed significant changes in their expression following visual deprivation, an established paradigm for studying cortical plasticity (Mellios et al., 2011). In vivo inhibition of miR-132 function in led to increased levels of spine inhibitor p250GAP and reduced dendritic spine density and immature morphology (Mellios et al., 2011). In the second study, on the other hand, in vivo upregulation of miR-132 expression promoted spine maturation (Tognini et al., 2011). Notably, both of these manipulations abrogated ocular dominance plasticity, the ability of neurons of the visual cortex to adjust their synaptic weights in response to altered visual drive following visual deprivation during a specific sensitive period of plasticity known as the critical period. These finding suggest that balanced levels of miR-132 are required for brain plasticity, and that too much or too little miR-132 expression can result in precocious or delayed cortical maturation, respectively. The effect of miR-132 in experience-dependent cortical maturation is very likely to not be limited to the visual cortex, since multiple studies have shown that miR-132 expression is induced in other brain regions in an experience-dependent manner (Nudelman et al., 2010).
Furthermore, a number of recent studies have shown that a subset of brain expressed miRNAs can affect learning and memory. Specifically, miR-134 expression in the hippocampus of Sirtuin 1 (Sirt1) knockout mice, which display memory deficits during fear conditioning and aberrant long term potentiation (LTP), was shown to be increased, and in vivo inhibition of miR-134 expression in hippocampus, was able to rescue both the memory and plasticity impairments (Gao et al., 2010). The novel mechanism proposed by this study was that SIRT1 normally inhibits miR-134 transcription, and miR-134 targets CREB, which in turn increases BDNF expression (Gao et al., 2010). Two more studies described CREB-targeting miRNAs as being important for plasticity and memory, with miR-124a shown to block serotonin-induced long term plasticity in Aplysia (Rajasethupathy et al., 2009), and miR-128b proposed to regulate the formation of fear extinction memory in mice (Lin et al., 2011). The latter study also demonstrated that miR-128b, which is an intronic miRNA, targets its host gene, regulator of calmodulin signaling (Src, or Rpp21), along with a number of other plasticity-related genes such as Reelin (Reln), protein phosphatase 1c gamma (Ppp1cc), and trans-acting transcription factor 1 (Sp1). Of note, miR-124 is an example of a brain enriched miRNA that acts in a temporal and cell-specific manner, since it can also affect neuronal development (Cheng et al., 2009), differentiation, including neurite branching (Makeyev et al., 2007; Visvanathan et al., 2007; Yu et al., 2008), and is able in parallel to influence the activation of brain microglia (Ponomarev et al., 2011), in which it is also abundantly expressed (see also Figure 1). Finally, transgenic mice overexpressing miR-132 were also shown to have impairments in novel object recognition memory (Hansen et al., 2010). Surprisingly, the conditional and transient deletion of Dicer1 in adult mouse forebrain caused an unexpected improvement in learning and memory (Konopka et al., 2010).
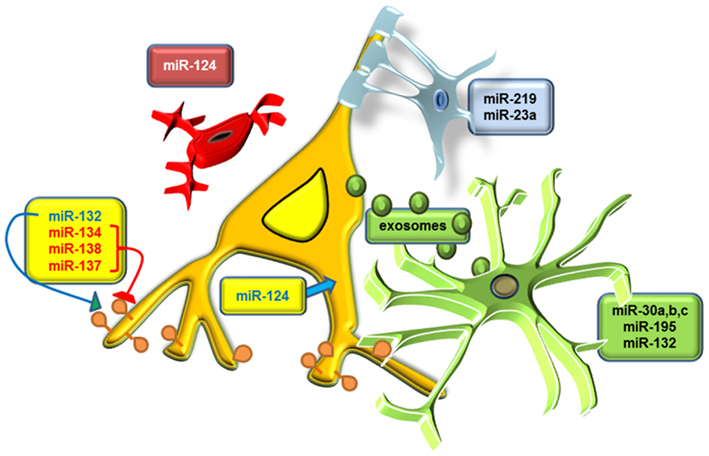
Figure 1. Cell-specific expression of miRNAs regulates neuronal and glial functions. Drawing showing examples of miRNAs enriched in different cellular populations in the central nervous system. Aneuron is shown in yellow–orange with miR-132 activating and miR-134, miR-137, and miR-138 inhibiting spine growth. Expression of miR-132 is present (albeit in lower levels) in astrocytes (shown in green), which also express high levels of miR-195 and miR-30a, miR-30b, and miR-30c. Secretion of microvesicles that are known to contain miRNAs (exosomes) by astrocytes is also depicted. Expression of miR-124 is shown in both neurons, where it promotes neurite outgrowth, and microglia (shown in red), where it suppresses their activation. Oligodendrocytes (shown in light blue) express high levels of miR-219 and miR-23a, which regulate their differentiation.
miRNAs in Schizophrenia
The first psychiatric disease to be linked to miRNA expression and function was Tourette’s syndrome, following the report that mutations in the 3′UTR of SLITRK1 were affecting the targeting by miR-189 in a limited subset of patients (Abelson et al., 2005). However, a plethora of postmortem studies using subjects diagnosed with schizophrenia provided the strongest support for aberrant miRNA expression in psychiatric diseases (Perkins et al., 2007; Beveridge et al., 2008, 2010; Mellios et al., 2009, 2010; Zhu et al., 2009; Kim et al., 2010; Moreau et al., 2011; Santarelli et al., 2011; Miller et al., 2012). Among the reported dysregulated miRNAs are those described above as being important regulators of brain maturation and plasticity (Table 1). The known limitations of postmortem studies, however, and differences in miRNA quantification techniques, have not allowed for a comprehensive description of which miRNAs are reliably affected in schizophrenia, and more importantly which might be important for the pathogenesis of this highly heterogeneous psychiatric disorder. In addition to individual miRNA expression the possibility that miRNA processing might be perturbed as a whole in a subset of subjects of schizophrenia has been proposed (Beveridge et al., 2010; Santarelli et al., 2011): although debatable, the idea deserves to be further investigated, especially in light of the fact that patients suffering from DiGeorge 22q11,2 deletion syndrome, which harbor microdeletions that affect miRNA processing gene Dgcr8, display a 30-fold increase in the prevalence of schizophrenia and schizoaffective disorder (Lindsay et al., 1995; Murphy et al., 1999; Stark et al., 2008).
A subset of miRNA altered in the brains of subjects diagnosed with schizophrenia, such as miR-132/212 (Perkins et al., 2007; Kim et al., 2010; Miller et al., 2012), miR-219 (Beveridge et al., 2010), and miR-195 (Beveridge et al., 2008, 2010) deserves special attention. In the case of miR-132 and miR-212, their prementioned links to activity-dependent synaptic plasticity, and neuronal maturation, are of interest given the known abnormalities in synaptic plasticity and connectivity observed in schizophrenia. The fact that NMDA inhibition is reported to affect the stability of mature miR-132 in neuronal cultures (Krol et al., 2010b) is of additional importance since NMDA hypofunction is hypothesized to be linked to the core of schizophrenia pathophysiology. On a similar note, miR-219, whose expression is downregulated in response to acute pharmacological blockade of NMDA receptors with diclozapine and in NMDA receptor 1 (Nr1) knockout mice, can target calcium/calmodulin-dependent protein kinase IIγ subunit (CamKIIγ; Kocerha et al., 2009). However, both miR-219 and CamKIIγ levels are increased in the prefrontal cortex (PFC) and superior temporal gyrus (STG) of patients with schizophrenia (Beveridge et al., 2010), suggesting an additional level of complexity. Another notable similarity between miR-219 and miR-132 is that they were both shown to influence genes important for circadian clock entrainment (Cheng et al., 2007), which is of potential relevance to schizophrenia given the observed deficits in circadian synchronization (Lamont et al., 2010).
Another miRNA that deserves notice is miR-195 which was found to be increased in the brain of subjects with schizophrenia (Beveridge et al., 2010), and which has been proven to regulate numerous schizophrenia-related genes, such as Bdnf (Mellios et al., 2008), Reln (Beveridge et al., 2010), Visinin-like 1 (Vsnl1; Beveridge et al., 2010), 5-hydroxytryptamine (serotonin) receptor 2a (Htr2a; Beveridge et al., 2010), and glutamate receptor, ionotropic, N-methyl-D-aspartate 3A (Grin3; Beveridge et al., 2010). The presence of two GABAergic expressed genes (Reln, Vsnl1) in the above list of validated miR-195 targets, and the fact that BDNF is known to influence GABAergic gene expression and function, position miR-195 as an intriguing candidate for schizophrenia research given the known deficits in GABAergic gene expression and inhibitory neuron function in schizophrenia (Akbarian et al., 1995; Guidotti et al., 2000; Costa et al., 2004; Hashimoto et al., 2008). The findings that disease-related changes in miR-195 are inversely correlated to those of BDNF protein levels, which in turn are associated with the observed changes in somatostatin (Sst) and neuropeptide Y (Npy) mRNA levels in human PFC of subjects with schizophrenia (Mellios et al., 2009), adds value to the above hypothesis. Last, but not least, an in silico study of schizophrenia-related miRNAs and transcription factors, depicted miR-195 as the core component of the predicted schizophrenia regulatory gene networks (Guo et al., 2010).
Despite the known neuronal-specific targets of schizophrenia-related miRNAs it is tempting to speculate that disease-related changes in the expression of a subset of miRNAs in schizophrenia might not be limited to neurons but could be also prevalent in glial cell populations. For example, miR-219 expression is abundantly expressed in oligodendrocytes and regulates their maturation (Dugas et al., 2010), and miR-132, miR-195, and many members of the miR-30 family are also expressed in astrocytes (Moser and Fritzler, 2010; Mor et al., 2011; Numakawa et al., 2011; Figure 1). In addition, the findings of the existence of microvesicles secreted by astrocytes called exosomes that contain miRNAs and could be taken up by neurons introduce a novel unexplored mechanism between cell to cell communication in the brain (Faure et al., 2006; Smalheiser, 2007; Valadi et al., 2007; Figure 1). Differential expression of miRNAs within different neuronal populations was recently discovered in mouse neocortex, with the example of miR-34a, a miRNA altered in the PFC of subjects diagnosed with schizophrenia (Kim et al., 2010), being particularly enriched in parvalbumin expressing interneurons (He et al., 2012). Future studies using similar cell-specific methods for miRNA detection, including exosome profiling, are needed to elucidate the role of non-neuronal miRNAs in schizophrenia.
In addition to the alterations in miRNA expression in schizophrenia a growing number of genetic studies has provided intriguing links between miRNAs and schizophrenia. Initially, single nucleotide polymorphisms (SNPs) within miR-206 and miR-198 were shown to be nominally associated with schizophrenia (Hansen et al., 2007). Furthermore, another SNP in pre-miR-30e was strongly linked to the disease (Xu et al., 2010). Notably, the expression of miR-30e is known to be altered in the PFC of subjects with schizophrenia, together with other members of the miR-30 family (Perkins et al., 2007), including miR-30a and miR-30d, which have been proven to target BDNF (Mellios et al., 2009; Wu et al., 2010). In addition, another member of the miR-30 family, miR-30b, was shown to be predominantly affected in female subjects with schizophrenia, and to be influenced by a disease-related SNP in estrogen receptor alpha (Mellios et al., 2010). However, the most robust genetic evidence came from a recent meta-analysis of genome-wide association studies (GWAS) for schizophrenia that reported that a SNP within intronic miR-137 displayed the most significant association with the disease (Ripke et al., 2011). This impressive finding was accompanied by the observation that four more significant loci contained miR-137 targets (Ripke et al., 2011). Other than the pre-mentioned reported effect of miR-137 in dendritic spine maturation (Smrt et al., 2010), it is of interest that Mecp2 inhibits miR-137 expression, which in turn can target other chromatin modifying genes, such as Lysine (K)-specific demethylase 1a (KDM1A, also known as LSD1; Sun et al., 2011), and Histone-lysine N-methyl-transferase, Enhancer of zeste (Drosophila) homolog 2 (EZH2; Szulwach et al., 2010). Notably, a recent finding demonstrated that miR-137-mediated regulation of LSD1 can also regulate neuronal differentiation and migration by affecting neural stem cell renewal (Sun et al., 2011). Furthermore, the interplay between miR-137 and genes important for chromatin remodeling is of importance given the hypothesized role of epigenetics in schizophrenia (Akbarian, 2010), and especially since a large number of other schizophrenia-related miRNAs, such as miR-132/212, miR-195 are known to be sensitive to epigenetic regulation (Li et al., 2011; Tognini et al., 2011; Zhang et al., 2011). Interestingly, recent work has uncovered that the deficits in miR-132 in schizophrenia are accompanied by an increase in miR-132 target DNA methyl-transferase alpha (DNMT3a; Miller et al., 2012), thus creating an epigenetically regulated double negative feedback loop. Further work is warranted to determine whether epigenetic regulation of miRNA expression could play an important role in the pathophysiology of the disease.
miRNAs in Autism Spectrum Disorders
Unlike the large number of studies examining the role of miRNAs in schizophrenia, little is known about the potential importance of miRNAs for ASD, and especially for the non-syndromic ASD subtypes. So far two studies have determined the expression of miRNAs in lymphoblastoid cell cultures of ASD patients (Abu-Elneel et al., 2008; Talebizadeh et al., 2008; Sarachana et al., 2010; Seno et al., 2011), and one study has identified dysregulated miRNAs in the cerebellum of ASD patients (Abu-Elneel et al., 2008). Among the most promising results (Table 1) is the finding of altered expression of miR-132/212, the two miRNAs of the same family described above to be dysregulated in schizophrenia (Perkins et al., 2007; Kim et al., 2010; Miller et al., 2012) and to be important for the control of experience-dependent cortical plasticity (Mellios et al., 2011; Tognini et al., 2011). Notably, miR-132 effects on synaptic structure are modulated by FMRP (Edbauer et al., 2010), the product of the Fmr1 gene responsible for Fragile-X Mental Retardation, the leading cause of mental retardation in adults and a subtype of syndromic ASD (D’Hulst and Kooy, 2009). Intriguingly, miR-132 is known to be induced by endotoxins and regulate the immune response (Taganov et al., 2006; Shaked et al., 2009; Lagos et al., 2010), as well as to be affected by cytomegalovirus infection (Wang et al., 2008), which are known to contribute to autism pathogenesis (Stubbs et al., 1984; Onore et al., 2012). In addition, miR-132 levels are reduced in Rett Syndrome, another subtype of syndromic ASD, caused by mutations in Mecp2 (Klein et al., 2007; Wu et al., 2010). As mentioned above, Mecp2 can increase the expression of miR-132 through its effect on BDNF, and decrease miR-137 by binding to its promoter and inducing transcriptional inhibition (Klein et al., 2007; Wu et al., 2010). However, the effect of Mecp2 on miRNAs is far more complex, since Mecp2 is known to regulate a large number of miRNAs in the mouse brain, and influence a plethora of miRNA-mediated functions with potential importance for brain disease (Wu et al., 2010; de Leon-Guerrero et al., 2011).
Furthermore, miR-195, miR-219, miR-346, miR-181b, and miR-128b, all known to be altered in schizophrenia (Beveridge et al., 2008, 2010; Zhu et al., 2009), exhibit changes in ASD samples as well (Abu-Elneel et al., 2008; Talebizadeh et al., 2008; Sarachana et al., 2010; Seno et al., 2011), strengthening the argument for the existence of common molecular pathways behind the pathophysiology of both neuropsychiatric diseases. Interestingly, and as mentioned above, miR-195 is known to target BDNF (Mellios et al., 2009) and miR-128b to inhibit CREB (Lin et al., 2011), whereas miR-132/212 are induced by BDNF and positively regulate CREB signaling (Hollander et al., 2010; Remenyi et al., 2010), which, in conjunction with two more BDNF regulating miRNAs (miR-495, miR-381; Wu et al., 2010) altered in autism (Talebizadeh et al., 2008; Sarachana et al., 2010), allows us to speculate that activity-dependent miRNAs could be affected in ASD. Such abnormalities in activity-dependent BDNF/CREB-related miRNAs are expected to have an impact on synaptic connectivity and maturation, which are known to be perturbed in ASD. As in the case of schizophrenia, the cell-specificity of ASD-related alterations in miRNA expression warrants further investigation, since differentially expressed miRNAs are known to be also expressed in astrocytes and oligodendrocytes (Dugas et al., 2010; Moser and Fritzler, 2010; Numakawa et al., 2011; Mor et al., 2011; Lin and Fu, 2009. In the case of miR-23a, for example, which together with miR-132 is altered in three out of four ASD studies (Abu-Elneel et al., 2008; Talebizadeh et al., 2008; Sarachana et al., 2010), there is strong evidence suggesting that it is an oligodendrocyte enriched miRNA that regulates their maturation by inhibiting lamin B1 (Lin and Fu, 2009; see also Figure 1). Another pathway that is emerging to be regulated by ASD altered miRNAs and that is also affected by BDNF signaling, is that of phosphoinositide 3-kinase (PI3K)/protein kinase B (PKB also known as Akt), with the best example being the reported changes in miR-21, a miRNA known to target phosphatase and tensin homolog (PTEN; Meng et al., 2006; Iliopoulos et al., 2010), which is responsible for inactivating Akt. Given that patients with PTEN mutations represent a substantial proportion of syndromic ASD (Varga et al., 2009), further research is warranted to elucidate the importance of miR-21 for ASD pathogenesis.
Conclusion
The increasing number of studies uncovering novel miRNA-mediated mechanisms that influence brain maturation and plasticity, in conjunction with the parallel findings for the potential involvement of such miRNAs in schizophrenia and autism, elevates the importance of miRNAs for brain function and dysfunction to a new level. It is tempting to speculate that a subset of the reported dysregulated in neuropsychiatric disorders miRNAs might turn out to be integral for shedding light on the elusive pathophysiology or pathogenesis of these complex and enigmatic disorders. Moreover, given the fact that both autism and schizophrenia pathogenesis appears to be influenced by mechanisms related to neurodevelopment, priority should be given to ASD or schizophrenia altered miRNAs with specific developmental patterns. For example, the experience-dependent miR-132/212 miRNA family holds great promise for schizophrenia, since its expression in mouse PFC peaks around adolescence (Miller et al., 2012), which coincides with the age of onset of schizophrenia. In theory, if a miRNA is indeed regulating genes and pathways that could trigger the onset of the disease, it is expected that its dysregulation will have the biggest impact at the time point when its expression reaches the maximum, because this is when its targets will be most robustly affected. Moreover, in the case where alterations in specific miRNAs are only an epiphenomenon of the disease, they could be ideal biomarker candidates, given the increased stability and durability of miRNAs, even in harsh handling conditions (Chen et al., 2008). Indeed, recent studies are exploring this possibility and have already reported some aberrantly expressed miRNAs in blood cells that could be associated with schizophrenia symptomatology, including miRNAs known to be altered in the brains of schizophrenia subjects as well, or affected by antipsychotic treatment (Gardiner et al., 2011; Lai et al., 2011; Woelk et al., 2011; Shi et al., 2012; Table 1). Ideally, the use of blood expressed miRNAs as biomarkers can be expanded to monitor the progress and response to therapeutic treatment of a disease, such as in the case of miR-134 and bipolar disorder (Rong et al., 2011). Lastly, the emerging technologies of induced neurons (Ambasudhan et al., 2011), or induced pluripotent stem cell-derived (Pedrosa et al., 2011) neurons, from patient fibroblasts might be an alternative for uncovering novel miRNA biomarkers, as well as for discovering new treatment options.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abelson, J. F., Kwan, K. Y., O’Roak, B. J., Baek, D. Y., Stillman, A. A., Morgan, T. M., Mathews, C. A., Pauls, D. L., Rasin, M. R., Gunel, M., Davis, N. R., Ercan-Sencicek, A. G., Guez, D. H., Spertus, J. A., Leckman, J. F., Dure, L. S. IV, Kurlan, R., Singer, H. S., Gilbert, D. L., Farhi, A., Louvi, A., Lifton, R. P., Sestan, N., and State, M. W. (2005). Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 310, 317–320.
Abu-Elneel, K., Liu, T., Gazzaniga, F. S., Nishimura, Y., Wall, D. P., Geschwind, D. H., Lao, K., and Kosik, K. S. (2008). Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 9, 153–161.
Akbarian, S., Kim, J. J., Potkin, S. G., Hagman, J. O., Tafazzoli, A., Bunney, W. E. Jr., and Jones, E. G. (1995). Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of Schizophrenics. Arch. Gen. Psychiatry 52, 258–266.
Ambasudhan, R., Talantova, M., Coleman, R., Yuan, X., Zhu, S., Lipton, S. A., and Ding, S. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118.
Beveridge, N. J., Gardiner, E., Carroll, A. P., Tooney, P. A., and Cairns, M. J. (2010). Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 15, 1176–1189.
Beveridge, N. J., Tooney, P. A., Carroll, A. P., Gardiner, E., Bowden, N., Scott, R. J., Tran, N., Dedova, I., and Cairns, M. J. (2008). Dysregulation of miRNA 181b in the temporal cortex in Schizophrenia. Hum. Mol. Genet. 17, 1156–1168.
Bian, S., and Sun, T. (2011). Functions of noncoding RNAs in neural development and neurological diseases. Mol. Neurobiol. 44, 359–373.
Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., Guo, J., Zhang, Y., Chen, J., Guo, X., Li, Q., Li, X., Wang, W., Zhang, Y., Wang, J., Jiang, X., Xiang, Y., Xu, C., Zheng, P., Zhang, J., Li, R., Zhang, H., Shang, X., Gong, T., Ning, G., Wang, J., Zen, K., Zhang, J., and Zhang, C.-Y. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006.
Cheng, H. Y., Papp, J. W., Varlamova, O., Dziema, H., Russell, B., Curfman, J. P., Nakazawa, T., Shimizu, K., Okamura, H., Impey, S., and Obrietan, K. (2007). microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829.
Cheng, L. C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408.
Costa, E., Davis, J. M., Dong, E., Grayson, D. R., Guidotti, A., Tremolizzo, L., and Veldic, M. (2004). A GABAergic cortical deficit dominates Schizophrenia pathophysiology. Crit. Rev. Neurobiol. 16, 1–23.
Cuellar, T. L., Davis, T. H., Nelson, P. T., Loeb, G. B., Harfe, B. D., Ullian, E., and McManus, M. T. (2008). Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 105, 5614–5619.
Davis, T. H., Cuellar, T. L., Koch, S. M., Barker, A. J., Harfe, B. D., McManus, M. T., and Ullian, E. M. (2008). Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322–4330.
de Leon-Guerrero, S. D., Pedraza-Alva, G., and Perez-Martinez, L. (2011). In sickness and in health: the role of methyl-CpG binding protein 2 in the central nervous system. Eur. J. Neurosci. 33, 1563–1574.
D’Hulst, C., and Kooy, R. F. (2009). Fragile X syndrome: from molecular genetics to therapy. J. Med. Genet. 46, 577–584.
Dugas, J. C., Cuellar, T. L., Scholze, A., Ason, B., Ibrahim, A., Emery, B., Zamanian, J. L., Foo, L. C., McManus, M. T., and Barres, B. A. (2010). Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611.
Edbauer, D., Neilson, J. R., Foster, K. A., Wang, C. F., Seeburg, D. P., Batterton, M. N., Tada, T., Dolan, B. M., Sharp, P. A., and Sheng, M. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384.
Faure, J., Lachenal, G., Court, M., Hirrlinger, J., Chatellard-Causse, C., Blot, B., Grange, J., Schoehn, G., Goldberg, Y., Boyer, V., Kirchhoff, F., Raposo, G., Garin, J., and Sadoul, R. (2006). Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 31, 642–648.
Fenelon, K., Mukai, J., Xu, B., Hsu, P. K., Drew, L. J., Karayiorgou, M., Fischbach, G. D., Macdermott, A. B., and Gogos, J. A. (2011). Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. U.S.A. 108, 4447–4452.
Gao, J., Wang, W. Y., Mao, Y. W., Graff, J., Guan, J. S., Pan, L., Mak, G., Kim, D., Su, S. C., and Tsai, L. H. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109.
Gardiner, E., Beveridge, N. J., Wu, J. Q., Carr, V., Scott, R. J., Tooney, P. A., and Cairns, M. J. (2011). Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol. Psychiatry.
Glantz, L. A., and Lewis, D. A. (2000). Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry 57, 65–73.
Guidotti, A., Auta, J., Davis, J. M., Di-Giorgi-Gerevini, V., Dwivedi, Y., Grayson, D. R., Impagnatiello, F., Pandey, G., Pesold, C., Sharma, R., Uzunov, D., and Costa, E. (2000). Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch. Gen. Psychiatry 57, 1061–1069.
Guo, A. Y., Sun, J., Jia, P., and Zhao, Z. (2010). A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst. Biol. 4, 10. doi:10.1186/1752-0509-4-10
Hansen, K. F., Sakamoto, K., Wayman, G. A., Impey, S., and Obrietan, K. (2010). Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 5, e15497. doi:10.1371/journal.pone.0015497
Hansen, T., Olsen, L., Lindow, M., Jakobsen, K. D., Ullum, H., Jonsson, E., Andreassen, O. A., Djurovic, S., Melle, I., Agartz, I., Hall, H., Timm, S., and Wang, A. G. Werge, T. (2007). Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE 2, e873. doi:10.1371/journal.pone.0000873
Hashimoto, T., Arion, D., Unger, T., Maldonado-Aviles, J. G., Morris, H. M., Volk, D. W., Mirnics, K., and Lewis, D. A. (2008). Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with Schizophrenia. Mol. Psychiatry 13, 147–161.
He, M., Liu, Y., Wang, X., Zhang, M. Q., Hannon, G. J., and Huang, Z. J. (2012). Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73, 35–48.
Hollander, J. A., Im, H. I., Amelio, A. L., Kocerha, J., Bali, P., Lu, Q., Willoughby, D., Wahlestedt, C., Conkright, M. D., and Kenny, P. J. (2010). Striatal microRNA controls cocaine intake through CREB signalling. Nature 466, 197–202.
Hutsler, J. J., and Zhang, H. (2000). Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 1309, 83–94.
Iliopoulos, D., Jaeger, S. A., Hirsch, H. A., Bulyk, M. L., and Struhl, K. (2010). STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 39, 493–506.
Im, H. I., Hollander, J. A., Bali, P., and Kenny, P. J. (2011). MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 13, 1120–1127.
Impey, S., Davare, M., Lasiek, A., Fortin, D., Ando, H., Varlamova, O., Obrietan, K., Soderling, T. R., Goodman, R. H., and Wayman, G. A. (2010). An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell. Neurosci. 43, 146–156.
Kim, A. H., Reimers, M., Maher, B., Williamson, V., McMichael, O., McClay, J. L., van den Oord, E. J., Riley, B. P., Kendler, K. S., and Vladimirov, V. I. (2010). MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr. Res. 124, 183–191.
Klein, M. E., Lioy, D. T., Ma, L., Impey, S., Mandel, G., and Goodman, R. H. (2007). Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10, 1513–1514.
Kocerha, J., Faghihi, M. A., Lopez-Toledano, M. A., Huang, J., Ramsey, A. J., Caron, M. G., Sales, N., Willoughby, D., Elmen, J., Hansen, H. F., Orum, H., Sakari, K., Kenny, P. J., and Wahlestedta, C. (2009). MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. U.S.A. 106, 3507–3512.
Kolomeets, N. S., Orlovskaya, D. D., Rachmanova, V. I., and Uranova, N. A. (2005). Ultrastructural alterations in hippocampal mossy fiber synapses in Schizophrenia: a postmortem morphometric study. Synapse 57, 47–55.
Konopka, W., Kiryk, A., Novak, M., Herwerth, M., Parkitna, J. R., Wawrzyniak, M., Kowarsch, A., Michaluk, P., Dzwonek, J., Arnsperger, T., Wilczynski, G., Merkenschlager, M., Theis, FJ., Köhr, G., Kaczmarek, L., and Schütz, G. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842.
Krol, J., Loedige, I., and Filipowicz, W. (2010a). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610.
Krol, J., Busskamp, V., Markiewicz, I., Stadler, M. B., Ribi, S., Richter, J., Duebel, J., Bicker, S., Fehling, H. J., Schubeler, D., Oertner, T. G., Schratt, G., Bibel, M., Roska, B., and Filipowicz, W. (2010b). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631.
Lagos, D., Pollara, G., Henderson, S., Gratrix, F., Fabani, M., Milne, R. S., Gotch, F., and Boshoff, C. (2010). miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12, 513–519.
Lai, C. Y., Yu, S. L., Hsieh, M. H., Chen, C. H., Chen, H. Y., Wen, C. C., Huang, Y. H., Hsiao, P. C., Hsiao, C. K., Liu, C. M., Yang, P.-C., Hwu, H.-G., and Chen, W. J. (2011). MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE 6, e21635. doi:10.1371/journal.pone.0021635
Lamont, E. W., Coutu, D. L., Cermakian, N., and Boivin, D. B. (2010). Circadian rhythms and clock genes in psychotic disorders. Isr. J. Psychiatry Relat. Sci. 47, 27–35.
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854.
Li, D., Zhao, Y., Liu, C., Chen, X., Qi, Y., Jiang, Y., Zou, C., Zhang, X., Liu, S., Wang, X., Zhao, D., Sun, Q., Zeng, Z., Dress, A., Lin, M. C., Kung, H. F., Rui, H., Liu, L. Z., Mao, F., Jiang, B. H., and Lai, L. (2011). Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin. Cancer Res. 17, 1722–1730.
Li, X., and Jin, P. (2010). Roles of small regulatory RNAs in determining neuronal identity. Nat. Rev. Neurosci. 11, 329–338.
Lin, Q., Wei, W., Coelho, C. M., Li, X., Baker-Andresen, D., Dudley, K., Ratnu, V. S., Boskovic, Z., Kobor, M. S., Sun, Y. E., and Bredy, T. W. (2011). The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat. Neurosci. 14, 1115–1117.
Lin, S. T., and Fu, Y. H. (2009). miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model. Mech. 2, 178–188.
Lindsay, E. A., Morris, M. A., Gos, A., Nestadt, G., Wolyniec, P. S., Lasseter, V. K., Shprintzen, R., Antonarakis, S. E., Baldini, A., and Pulver, A. E. (1995). Schizophrenia and chromosomal deletions within 22q11.2. Am. J. Hum. Genet. 56, 1502–1503.
Luikart, B. W., Bensen, A. L., Washburn, E. K., Perederiy, J. V., Su, K. G., Li, Y., Kernie, S. G., Parada, L. F., and Westbrook, G. L. (2011). miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS ONE 6, e19077. doi:10.1371/journal.pone.0019077
Magill, S. T., Cambronne, X. A., Luikart, B. W., Lioy, D. T., Leighton, B. H., Westbrook, G. L., Mandel, G., and Goodman, R. H. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 20382–20387.
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448.
Mellios, N., Galdzicka, M., Ginns, E., Baker, S. P., Rogaev, E., Xu, J., and Akbarian, S. (2010). Gender-specific reduction of estrogen-sensitive small RNA, miR-30b, in subjects with Schizophrenia. Schizophr. Bull. doi: 10.1093/schbul/sbq091
Mellios, N., Huang, H. S., Baker, S. P., Galdzicka, M., Ginns, E., and Akbarian, S. (2009). Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with Schizophrenia. Biol. Psychiatry 65, 1006–1014.
Mellios, N., Huang, H. S., Grigorenko, A., Rogaev, E., and Akbarian, S. (2008). A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum. Mol. Genet. 17, 3030–3042.
Mellios, N., Sugihara, H., Castro, J., Banerjee, A., Le, C., Kumar, A., Crawford, B., Strathmann, J., Tropea, D., Levine, S. S., Edbauer, D., and Sur, M. (2011). miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat. Neurosci. 14, 1240–1242.
Meng, F., Henson, R., Lang, M., Wehbe, H., Maheshwari, S., Mendell, J. T., Jiang, J., Schmittgen, T. D., and Patel, T. (2006). Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130, 2113–2129.
Miller, B. H., Zeier, Z., Xi, L., Lanz, T. A., Deng, S., Strathmann, J., Willoughby, D., Kenny, P. J., Elsworth, J. D., Lawrence, M. S., Roth, R. H., Edbauere, D., Kleimand, R. J., and Wahlestedt, C. (2012). MicroRNA-132 dysregulation in Schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl. Acad. Sci. U.S.A. 109, 3125–3130.
Mor, E., Cabilly, Y., Goldshmit, Y., Zalts, H., Modai, S., Edry, L., Elroy-Stein, O., and Shomron, N. (2011). Species-specific microRNA roles elucidated following astrocyte activation. Nucleic Acids Res. 39, 3710–3723.
Moreau, M. P., Bruse, S. E., David-Rus, R., Buyske, S., and Brzustowicz, L. M. (2011). Altered microRNA expression profiles in postmortem brain samples from individuals with Schizophrenia and bipolar disorder. Biol. Psychiatry 69, 188–193.
Moser, J. J., and Fritzler, M. J. (2010). The microRNA and messengerRNA profile of the RNA-induced silencing complex in human primary astrocyte and astrocytoma cells. PLoS ONE 5, e13445. doi:10.1371/journal.pone.0013445
Murphy, K. C., Jones, L. A., and Owen, M. J. (1999). High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry 56, 940–945.
Nudelman, A. S., DiRocco, D. P., Lambert, T. J., Garelick, M. G., Le, J., Nathanson, N. M., and Storm, D. R. (2010). Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20, 492–498.
Numakawa, T., Yamamoto, N., Chiba, S., Richards, M., Ooshima, Y., Kishi, S., Hashido, K., Adachi, N., and Kunugi, H. (2011). Growth factors stimulate expression of neuronal and glial miR-132. Neurosci. Lett. 505, 242–247.
Onore, C., Careaga, M., and Ashwood, P. (2012). The role of immune dysfunction in the pathophysiology of autism. Brain Behav. Immun. 26, 383–392.
Pedrosa, E., Sandler, V., Shah, A., Carroll, R., Chang, C., Rockowitz, S., Guo, X., Zheng, D., and Lachman, H. M. (2011). Development of patient-specific neurons in schizophrenia using induced pluripotent stem cells. J. Neurogenet. 25, 88–103.
Penzes, P., Cahill, M. E., Jones, K. A., VanLeeuwen, J. E., and Woolfrey, K. M. (2001). Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 3, 285–293.
Perkins, D. O., Jeffries, C. D., Jarskog, L. F., Thomson, J. M., Woods, K., Newman, M. A., Parker, J. S., Jin, J., and Hammond, S. M. (2007). microRNA expression in the prefrontal cortex of individuals with Schizophrenia and schizoaffective disorder. Genome Biol. 8, R27.
Pillai, R. S., Bhattacharyya, S. N., and Filipowicz, W. (2007). Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 17, 118–126.
Ponomarev, E. D., Veremeyko, T., Barteneva, N., Krichevsky, A. M., and Weiner, H. L. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat. Med. 17, 64–70.
Rajasethupathy, P., Fiumara, F., Sheridan, R., Betel, D., Puthanveettil, S. V., Russo, J. J., Sander, C., Tuschl, T., and Kandel, E. (2009). Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63, 803–817.
Remenyi, J., Hunter, C. J., Cole, C., Ando, H., Impey, S., Monk, C. E., Martin, K. J., Barton, G. J., Hutvagner, G., and Arthur, J. S. (2010). Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem. J. 428, 281–291.
Ripke, S., Sanders, A. R., Kendler, K. S., Levinson, D. F., Sklar, P., Holmans, P. A., Lin, D. Y., Duan, J., Ophoff, R. A., Andreassen, O. A., Scolnick, E., Cichon, S., St Clair, D., Corvin, A., Gurling, H., Werge, T., Rujescu, D., Blackwood, D. H., Pato, C. N., Malhotra, A. K., Purcell, S., Dudbridge, F., Neale, B. M., Rossin, L., Visscher, P. M., Posthuma, D., Ruderfer, D. M., Fanous, A., Stefansson, H., Steinberg, S., Mowry, B. J., Golimbet, V., De Hert, M., Jönsson, E. G., Bitter, I., Pietiläinen, O. P., Collier, D. A., Tosato, S., Agartz, I., Albus, M., Alexander, M., Amdur, R. L., Amin, F., Bass, N., Bergen, S. E., Black, D. W., Børglum, A. D., Brown, M. A., Bruggeman, R., Buccola, N. G., Byerley, W. F., Cahn, W., Cantor, R. M., Carr, V. J., Catts, S. V., Choudhury, K., Cloninger, C. R., Cormican, P., Craddock, N., Danoy, P. A., Datta, S., de Haan, L., Demontis, D., Dikeos, D., Djurovic, S., Donnelly, P., Donohoe, G., Duong, L., Dwyer, S., Fink-Jensen, A., Freedman, R., Freimer, N. B., Friedl, M., Georgieva, L., Giegling, I., Gill, M., Glenthøj, B., Godard, S., Hamshere, M., Hansen, M., Hansen, T., Hartmann, A. M., Henskens, F. A., Hougaard, D. M., Hultman, C. M., Ingason, A., Jablensky, A. V., Jakobsen, K. D., Jay, M., Jürgens, G., Kahn, R. S., Keller, M. C., Kenis, G., Kenny, E., Kim, Y., Kirov, G. K., Konnerth, H., Konte, B., Krabbendam, L., Krasucki, R., Lasseter, V. K., Laurent, C., Lawrence, J., Lencz, T., Lerer, F. B., Liang, K. Y., Lichtenstein, P., Lieberman, J. A., Linszen, D. H., Lönnqvist, J., Loughland, C. M., Maclean, A. W., Maher, B. S., Maier, W., Mallet, J., Malloy, P., Mattheisen, M., Mattingsdal, M., McGhee, K. A., McGrath, J. J., McIntosh, A., McLean, D. E., McQuillin, A., Melle, I., Michie, P. T., Milanova, V., Morris, D. W., Mors, O., Mortensen, P. B., Moskvina, V., Muglia, P., Myin-Germeys, I., Nertney, D. A., Nestadt, G., Nielsen, J., Nikolov, I., Nordentoft, M., Norton, N., Nöthen, M. M., O’Dushlaine, C. T., Olincy, A., Olsen, L., O’Neill, F. A., Orntoft, T. F., Owen, M. J., Pantelis, C., Papadimitriou, G., Pato, M. T., Peltonen, L., Petursson, H., Pickard, B., Pimm, J., Pulver, A. E., Puri, V., Quested, D., Quinn, E. M., Rasmussen, H. B., Réthelyi, J. M., . Ribble, R., Rietschel, M., Riley, B. P., Ruggeri, M., Schall, U., Schulze, T. G., Schwab, S. G., Scott, R. J., Shi, J., Sigurdsson, E., Silverman, J. M., Spencer, C. C., Stefansson, K., Strange, A., Strengman, E., Stroup, T. S., Suvisaari, J., Terenius, L., Thirumalai, S., Thygesen, J. H., Timm, S., Toncheva, D., van den Oord, E., van Os, J., van Winkel, R., Veldink, J., Walsh, D., Wang, A. G., Wiersma, D., Wildenauer, D. B., Williams, H. J., Williams, N. M., Wormley, B., Zammit, S., Sullivan, P. F., O’Donovan, M. C., Daly, M. J., Gejman, P. V., and Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. (2011). Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976.
Rong, H., Liu, T. B., Yang, K. J., Yang, H. C., Wu, D. H., Liao, C. P., Hong, F., Yang, H. Z., Wan, F., Ye, X. Y., Xu, D., Zhang, X., Chao, C. A., and Shen, Q. J. (2011). MicroRNA-134 plasma levels before and after treatment for bipolar mania. J. Psychiatr. Res. 45, 92–95.
Santarelli, D. M., Beveridge, N. J., Tooney, P. A., and Cairns, M. J. (2011). Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry 69, 180–187.
Sarachana, T., Zhou, R., Chen, G., Manji, H. K., and Hu, V. W. (2010). Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2, 23.
Schofield, C. M., Hsu, R., Barker, A. J., Gertz, C. C., Blelloch, R., and Ullian, E. M. (2011). Monoallelic deletion of the microRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev. 6, 11.
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., and Greenberg, M. E. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289.
Seno, M., Hu, P., Gwadry, F. G., Pinto, D., Marshall, C. R., Casallo, G., and Scherer, S. W. (2011). Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 1380, 85–97.
Shaked, I., Meerson, A., Wolf, Y., Avni, R., Greenberg, D., Gilboa-Geffen, A., and Soreq, H. (2009). MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973.
Shi, W., Du, J., Qi, Y., Liang, G., Wang, T., Li, S., Xie, S., Zeshan, B., and Xiao, Z. (2012). Aberrant expression of serum miRNAs in schizophrenia. J. Psychiatr. Res. 46, 198–204.
Shin, D., Shin, J. Y., McManus, M. T., Ptacek, L. J., and Fu, Y. H. (2009). Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann. Neurol. 66, 843–857.
Siegel, G., Obernosterer, G., Fiore, R., Oehmen, M., Bicker, S., Christensen, M., Khudayberdiev, S., Leuschner, P. F., Busch, C. J., Kane, C., Hübel, K., Dekker, F., Hedberg, C., Rengarajan, B., Drepper, C., Waldmann, H., Kauppinen, S., Greenberg, M. E., Draguhn, A., Rehmsmeier, M., Martinez, J., and Schratt, G. M. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716.
Smalheiser, N. R. (2007). Exosomal transfer of proteins and RNAs at synapses in the nervous system? Biol. Direct 2, 35.
Smrt, R. D., Szulwach, K. E., Pfeiffer, R. L., Li, X., Guo, W., Pathania, M., Teng, Z. Q., Luo, Y., Peng, J., Bordey, A., Jin, P., and Zhao, X. (2010). MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070.
Stark, K. L., Xu, B., Bagchi, A., Lai, W. S., Liu, H., Hsu, R., Wan, X., Pavlidis, P., Mills, A. A., Karayiorgou, M., and Gogos, J. A. (2008). Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 40, 751–760.
Stubbs, E. G., Ash, E., and Williams, C. P. (1984). Autism and congenital cytomegalovirus. J. Autism Dev. Disord. 14, 183–189.
Sun, G., Ye, P., Murai, K., Lang, M. F., Li, S., Zhang, H., Li, W., Fu, C., Yin, J., Wang, A., Ma, X., and Shi, Y. (2011). miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2, 529.
Sweet, R. A., Henteleff, R. A., Zhang, W., Sampson, A. R., and Lewis, D. A. (2009). Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology 34, 374–389.
Szulwach, K. E., Li, X., Smrt, R. D., Li, Y., Luo, Y., Lin, L., Santistevan, N. J., Li, W., Zhao, X., and Jin, P. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141.
Taganov, K. D., Boldin, M. P., Chang, K. J., and Baltimore, D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486.
Talebizadeh, Z., Butler, M. G., and Theodoro, M. F. (2008). Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res. 1, 240–250.
Tao, J., Wu, H., Lin, Q., Wei, W., Lu, X. H., Cantle, J. P., Ao, Y., Olsen, R. W., Yang, X. W., Mody, I., Sofroniew, M. V., and Sun, Y. E. (2011). Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J. Neurosci. 31, 8306–8319.
Tognini, P., Putignano, E., Coatti, A., and Pizzorusso, T. (2011). Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat. Neurosci. 14, 1237–1239.
Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J. J., and Lotvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659.
Varga, E. A., Pastore, M., Prior, T., Herman, G. E., and McBride, K. L. (2009). The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet. Med. 11, 111–117.
Visvanathan, J., Lee, S., Lee, B., Lee, J. W., and Lee, S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749.
Vo, N., Klein, M. E., Varlamova, O., Keller, D. M., Yamamoto, T., Goodman, R. H., and Impey, S. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431.
Wang, F. Z., Weber, F., Croce, C., Liu, C. G., Liao, X., and Pellett, P. E. (2008). Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 82, 9065–9074.
Wayman, G. A., Davare, M., Ando, H., Fortin, D., Varlamova, O., Cheng, H. Y., Marks, D., Obrietan, K., Soderling, T. R., Goodman, R. H., and Impey, S. (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 105, 9093–9098.
Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862.
Woelk, C. H., Singhania, A., Perez-Santiago, J., Glatt, S. J., and Tsuang, M. T. (2011). The utility of gene expression in blood cells for diagnosing neuropsychiatric disorders. Int. Rev. Neurobiol. 101, 41–63.
Wu, H., Tao, J., Chen, P. J., Shahab, A., Ge, W., Hart, R. P., Ruan, X., Ruan, Y., and Sun, Y. E. (2010). Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 107, 18161–18166.
Xu, Y., Li, F., Zhang, B., Zhang, K., Zhang, F., Huang, X., Sun, N., Ren, Y., Sui, M., and Liu, P. (2010). MicroRNAs and target site screening reveals a pre-microRNA-30e variant associated with schizophrenia. Schizophr. Res. 119, 219–227.
Yu, J. Y., Chung, K. H., Deo, M., Thompson, R. C., and Turner, D. L. (2008). MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp. Cell Res. 314, 2618–2633.
Zhang, S., Hao, J., Xie, F., Hu, X., Liu, C., Tong, J., Zhou, J., Wu, J., and Shao, C. (2011). Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis 32, 1183–1189.
Keywords: microRNA, schizophrenia, autism, synaptic plasticity, neuropsychiatric disorder
Citation: Mellios N and Sur M (2012) The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front. Psychiatry 3:39. doi: 10.3389/fpsyt.2012.00039
Received: 15 December 2011; Accepted: 11 April 2012;
Published online: 25 April 2012.
Edited by:
Daniela Tropea, Trinity College Dublin, IrelandReviewed by:
Daniela Tropea, Trinity College Dublin, IrelandClaudia Bagni, Catholic University of Leuven Medical School, Belgium
Copyright: © 2012 Mellios and Sur. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Nikolaos Mellios, Picower Institute for Learning and Memory, Massachusetts Institute of technology, 43 Vassar Street, Cambridge, MA 02139, USA. e-mail: nmellios@mit.edu
