- 1Institute of Disease Control and Prevention, Academy of Military Medical Sciences, Beijing, China
- 2National Institutes for Food and Drug Control, Beijing, China
Bordetella pertussis is an important human respiratory pathogen. Here, we describe a loop-mediated isothermal amplification (LAMP) method for the rapid detection of B. pertussis in clinical samples based on a visual test. The LAMP assay detected the BP485 target sequence within 60 min with a detection limit of 1.3 pg/μl, a 10-fold increase in sensitivity compared with conventional PCR. All 31 non-pertussis respiratory pathogens tested were negative for LAMP detection, indicating the high specificity of the primers for B. pertussis. To evaluate the application of the LAMP assay to clinical diagnosis, of 105 sputum and nasopharyngeal samples collected from the patients with suspected respiratory infections in China, a total of 12 B. pertussis isolates were identified from 33 positive samples detected by LAMP-based surveillance targeting BP485. Strikingly, a 4.5 months old baby and her mother were found to be infected with B. pertussis at the same time. All isolates belonged to different B. pertussis multilocus sequence typing groups with different alleles of the virulence-related genes including four alleles of ptxA, six of prn, four of tcfA, two of fim2, and three of fim3. The diversity of B. pertussis carrying toxin genes in clinical strains indicates a rapid and continuing evolution of B. pertussis. This combined with its high prevalence will make it difficult to control. In conclusion, we have developed a visual detection LAMP assay, which could be a useful tool for rapid B. pertussis detection, especially in situations where resources are poor and in point-of-care tests.
Introduction
Bordetella pertussis, a Gram-negative bacterium first reported in 1906 (1), is an important human-specific respiratory pathogen that causes whooping cough (pertussis) in humans. Although the morbidity and mortality of B. pertussis have been dramatically reduced since the introduction of Pw and Pa vaccines into the routine immunization schedule of infants, pertussis is considered as the most prevalent vaccine-preventable disease (2). It was revealed that neonates are now commonly infected by adolescents and adults, before transmitting whooping cough further to infants and other groups. This change in transmission pattern from “child to child” to “adult to child” may be the major cause for the rising incidence of pertussis, especially in developed countries with high vaccination coverage (3–7). In China, pertussis is a reportable infectious disease, and although the number of reported cases has been decreasing, pertussis remains endemic. A diagnosis is made on the basis of clinical symptoms, not using diagnostic tests such as culture, PCR, and serologic analysis. Therefore, the reported low incidence may, in fact, be due to substantial underreporting caused by misdiagnosis. However, timely and accurate diagnosis is necessary for controlling the disease.
Various diagnostic tests have been established, such as culture (8), PCR (9, 10), real-time PCR (11, 12), and enzyme-linked immunosorbent assay (13). Methods based on molecular biology or immunology have become universal and are increasingly replacing culture-based methods, as they are more rapid and sensitive. However, all of the above methods are time-consuming and require specialized equipment.
Loop-mediated isothermal amplification (LAMP) is a simple and cost-effective nucleic acid amplification method, which can be applied to rapidly detect B. pertussis. A LAMP assay can be completed within 1 h at a constant temperature using simple instruments. Further advantages include its high sensitivity and specificity. LAMP has previously been used to detect B. pertussis (14–17). Previously described PCR and LAMP assays have typically used IS481, which is present in more than 200 copies in the B. pertussis genome, as the target sequence (18, 19). However, because B. holmesii and some B. bronchiseptica strains contain the same IS481 elements, these assays lacked specificity (20, 21). In 2008, Probert et al. compared the genomes of B. pertussis, B. bronchiseptica, and B. parapertussis, and tested the BP283 and BP485 sequences for the specific detection of B. pertussis using real-time PCR. They reported an improvement in specificity over assays targeting only IS481 (22). Our study is the first to use BP485 as the target sequence in a LAMP assay. The purpose of the present study was to develop a rapid and simple test for B. pertussis detection, based on LAMP technology, which requires only basic equipment and allows immediate interpretation of results by visual inspection.
Materials and Methods
Bacterial Isolates, Identification, and MLST Typing
Thirty-four bacterial strains were used for specificity analysis, which are listed in Table 1. Three B. pertussis vaccine strains were tested, including strain 18530, which originated from United States, strain P3s10, which has been used to produce Pw vaccines since the early 1960s, and strain CS, which has been used to produce Pa vaccines since 1995. Paired sputum and nasopharyngeal swabs were collected from 105 hospitalized patients using rayon-tipped swabs (Copan Diagnostics; Corona, CA, USA) between the spring of 2010 and spring of 2014. The patients (age range, 14 days to 35 years; median, 1.33 years; 52 females, 50 males) came from rural areas of China and had been treated for coughs and pneumonia for more than 7 days. Ten pairs of sputum samples and nasopharyngeal swabs from healthy people were also collected as controls. One swab or sputum sample was immediately placed in Regan-Lowe transport medium for culture and the other was stored at −70°C in an empty sterile tube for bacterial nucleic acid isolation.
Bordetella pertussis strains were cultured on Bordet–Gengou agar supplemented with 15% defibrinated sheep blood for 4–5 days at 37°C. Species identification was carried out using the Phoenix Automated Microbiology System (BD Diagnostic Systems) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Genomic DNA was extracted from all samples using TIANamp Bacteria DNA Kit (TIANGEN Co., Ltd., Beijing, China). The sequences of 16S ribosomal DNA (rDNA) and BP485 were validated by PCR-based sequencing and showed 100% identity with those previously reported.
Seven housekeeper genes including adk, fumC, glyA, tyrB, icd, pepA, and pgm were detected by PCR. The allele number for each gene was assigned on the basis of the information in the B. pertussis MLST database1. Moreover, the strains were screened for the presence of 5 virulence-related genes (prn, ptxA, tcfA, fim2, and fim3) by PCR as previously reported (23).
Primer Design for LAMP
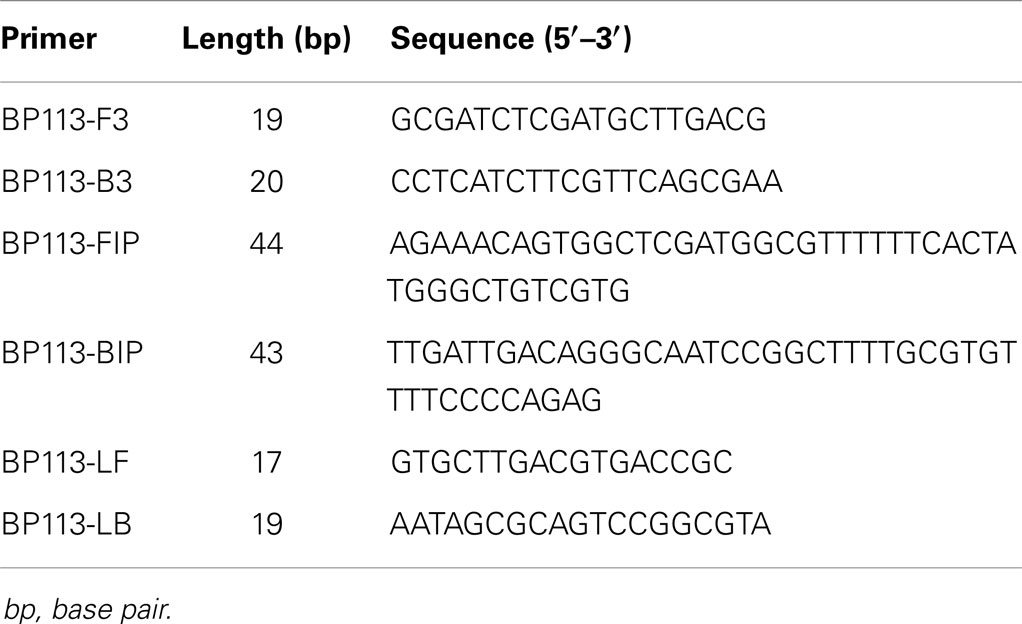
The sequence of BP485 (Accession number: CP002695.1) was downloaded from NCBI GenBank database and further analyzed by Primer Explorer Version 42. Five primer sets were designed and synthesized commercially (Sangon Biotech Co., Ltd., Shanghai, China).
LAMP Assay
Loop-mediated isothermal amplification assays were carried out using the Loopamp DNA Amplification kit (Eiken Chemical Co., Ltd., Tochigi, Japan). Briefly, the LAMP amplification system in a final volume of 25 μl contained 12.5 μl reaction mixture, 1 μl Bst DNA polymerase, 2 μl template for real-time turbidimeter, and another 1 μl calcein/Mn2+ solution for visual detection. Primers were used at a concentration of 40 pmol for FIP and BIP, 20 pmol for LB and LF, and 5 pmol for F3 and B3. The reaction mix was overlaid with a sealing agent mainly comprised wax to prevent cross contamination of samples by aerosol. During the amplification, the protectant melted and became liquid without disturbing the reaction, avoiding volatilization of amplified products after reaction, and the protectant solidified as the temperature in the tubes decreased (Patent: ZL201210371448.5 in China).
Amplification was monitored using two methods, real-time changes in turbidity or visual detection of a color change. Pyrophosphate ions are released during LAMP amplification that with the addition of magnesium ions (Mg2+) in the reaction buffer form a white magnesium pyrophosphate precipitate (24), which can be monitored every 6 s at 650 nm using a real-time turbidimeter, and the data are transformed into turbidity curves. For visual detection, 1 μl of calcein/Mn2+ fluorescent detection reagent was added to the reaction. LAMP amplification results in a green fluorescent emission as a result of magnesium ions forming a complex with calcein. The color change from orange to green when samples are positive is visible to the naked eye (25). Each experiment was performed at least three times.
PCR Assay
PCRs were setup using 12.5 μl of PCR MasterMix reagents (Tiangen Biotech Co., Ltd., Beijing, China), 1 μM BP-113F3 and BP-113B3 primers, and 2 μl of DNA template in a final volume of 25 μl. PCR was performed using the following cycling conditions: initial PCR activation, 95°C for 5 min; amplification, 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; final extension, 72°C for 7 min. PCR products were visualized on a 1% agarose gel stained with ethidium bromide. Images were documented with a Gel Doc EQ imaging system (Bio-Rad).
Results
Optimization of LAMP Assay
BP485 was targeted for detection of B. pertussis by a LAMP assay for the first time. Five different primer sets were initially tested, four of which resulted in successful amplification as shown in Figure 1. The BP-113 primer set amplified the target sequence within the shortest time and was therefore chosen as the optimal primer set (Table 2).
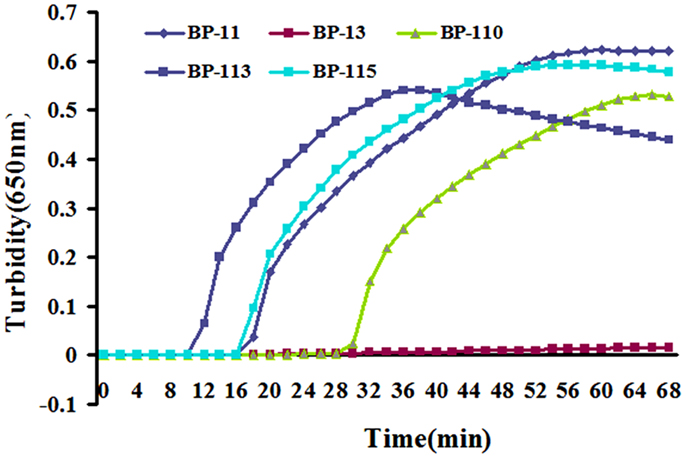
Figure 1. Five designed primer sets were tested by real-time turbidimeter at 650 nm every 6 s. BP-113 group possess optimal efficiency and was chosen to be final primers for LAMP assay.
Reaction temperatures from 60 to 67°C were compared for optimal amplification. Amplification efficiency was highest at 64°C (Figure 2) and was therefore used for later experiments.
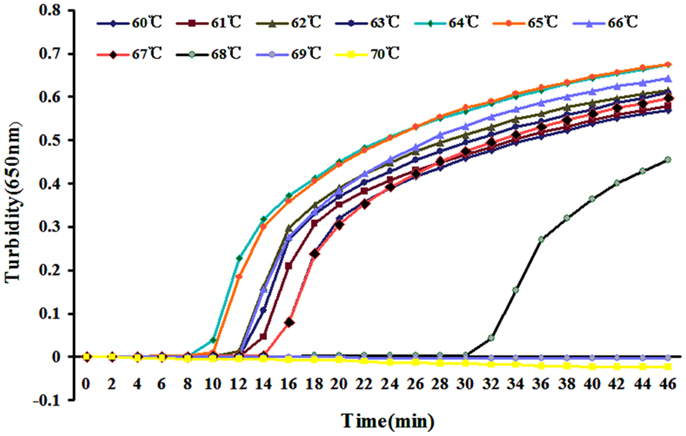
Figure 2. Different temperatures for LAMP reaction were tested using real-time turbidimeter at 650 nm.
Specificity of LAMP Assay
To evaluate the specificity of LAMP detection for B. pertussis, genomic DNA was extracted from B. pertussis ATCC18530, B. pertussis ATCC58003, B. pertussis ATCC53894, and 31 other pathogenic respiratory bacteria and tested using real-time turbidity or visual detection of color change as readouts. Both methods of analysis positively identified the B. pertussis isolates. All other strains, including the blank control, tested negative, indicating that the LAMP assay was specific to B. pertussis (Figure 3).
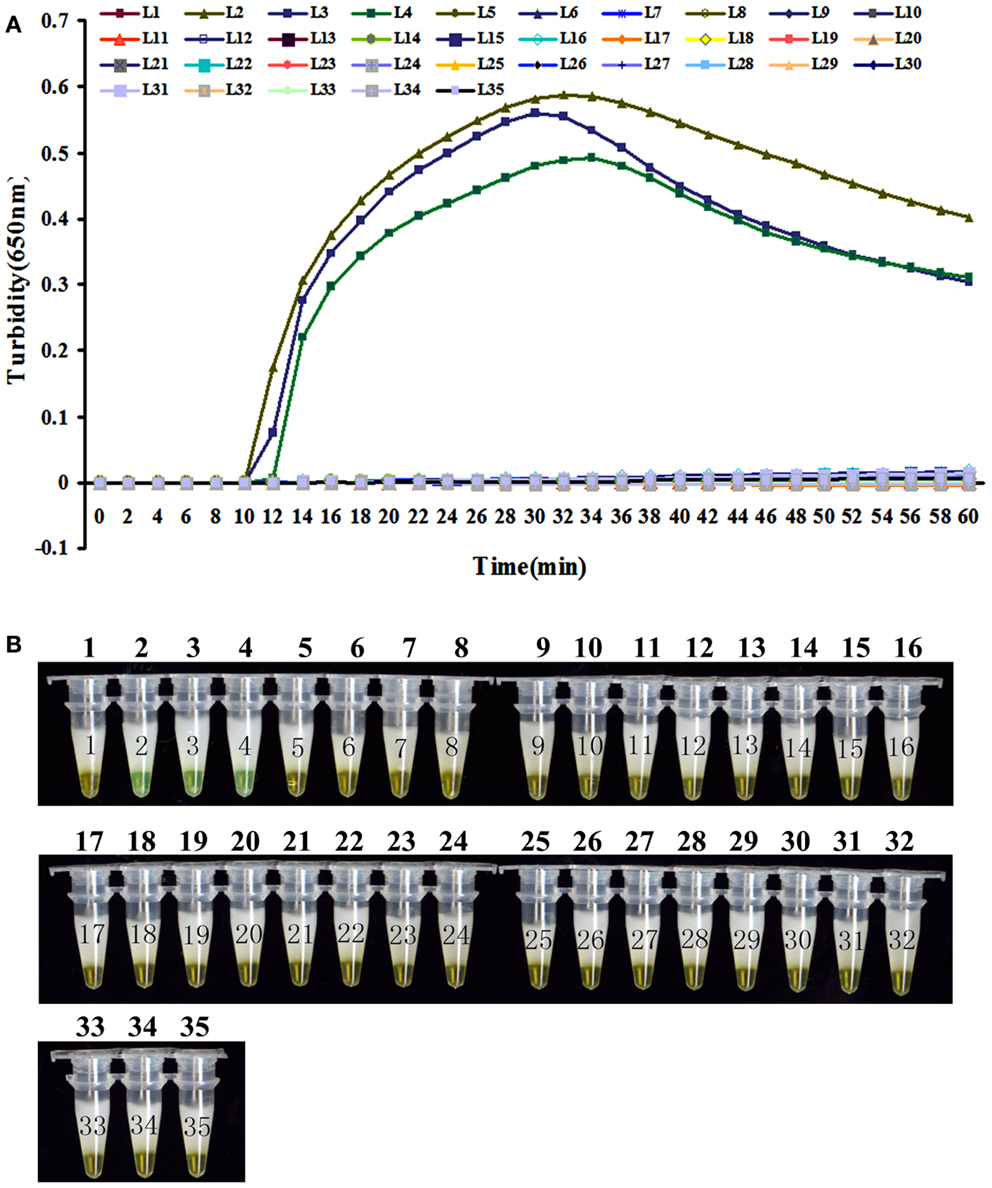
Figure 3. The specificity detection of LAMP assay by real-time turbidimeter (A) or addition calcein to the reaction tube (B). Amplification was performed at 64°C for 60 min. 1, NC; 2, B. pertussis ATCC 18530; 3, B. pertussis CMCC 58003; 4, B. pertussis ATCC 53894; 5, B. parapertussis CMCC 58302; 6, B. parapertussis ATCC 15237; 7, B. parapertussis ATCC BAA-587; 8, B. parapertussis ATCC 53893; 9, B. bronchiseptica ATCC BAA-588; 10, B. bronchiseptica CMCC 58401; 11, B. bronchiseptica ATCC 4617; 12, B. holmesii ATCC 51541; 13, B. avium ATCC 35086; 14, B. hinzii ATCC 51730; 15, B. petrii ATCC BAA-461; 16, Corynebacterium diphtheriae CMCC 38001; 17, Haemophilus influenzae CMCC 58534; 18, Betahaemolytic streptococcus group A CMCC 32213; 19, Streptococcus pneumonia CMCC 31001; 20, Neisseria meningitides group B CMCC 29022; 21, Neisseria meningitides group C CMCC29026; 22, Neisseria meningitides group Y CMCC 29028; 23, Neisseria meningitides group A CMCC 29202; 24, Mycobacterium tuberculosis 4368; 25, Neisseria meningitides NM29019; 26, Streptococcus pneumonia SP112-07; 27, Legionella pneumophila LP9135; 28, Haemophilus influenzae M5216; 29, Klebsiella pneumonia 46117; 30, Vibrio parahaemolyticus 5474; 31, Salmonella enteritidis 50326-1; 32, Salmonella paratyphi A 86423; 33, Shigella flexneri 4536; 34, Shigella sonnei 2531; 35, EIEC 44825.
Sensitivity of the LAMP Assay Versus PCR for B. pertussis
To compare the detection limit of LAMP using either real-time turbidity measurements or color change with traditional PCR, 10-fold serial dilutions of genomic DNA extracted from B. pertussis ATCC18530 (130 ng/μl to 1.3 fg/μl) were tested. The results were shown in Figure 4. The detection limits of real-time turbidity and visual detection were both 1.3 pg/μl, which was 10-fold more sensitive than traditional PCR assay.
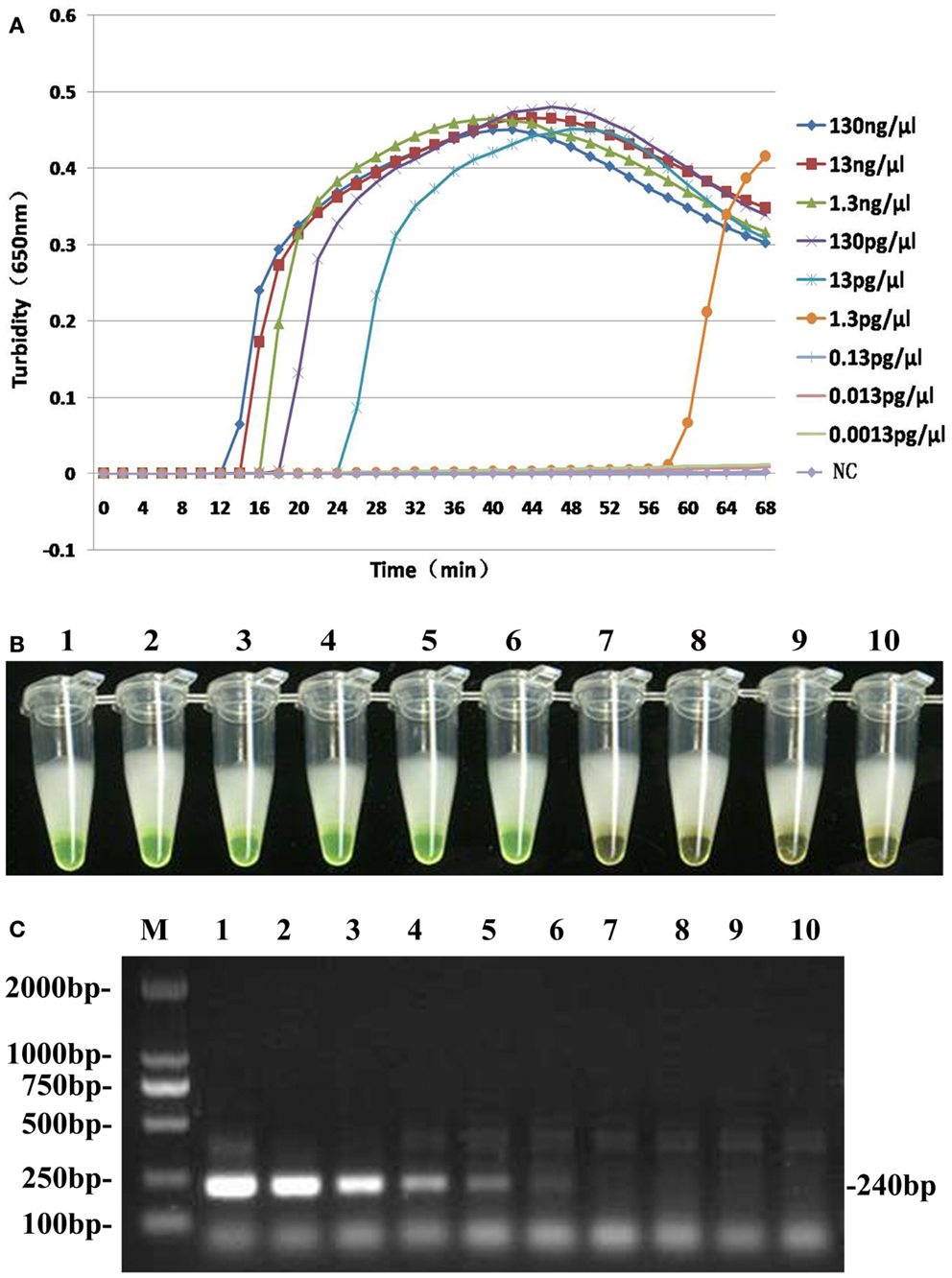
Figure 4. Comparative sensitivities of both LAMP assay (A,B) and traditional PCR (C). Lane M: DL2000 Marker; Tubes 1–9 and lanes 1–9: 10-fold serial dilution pure genomic DNA extracted from B. pertussis (130 ng/μl to 0.0013 pg/μl); Tube 10 and lane 10: ddH2O as negative control.
Dissemination of B. pertussis in Clinical
A total of 105 clinical sputum samples and nasopharyngeal swabs were collected for LAMP-based surveillance of BP485 from patients with suspected respiratory infections in China during 2010–2014. Ten pairs of sputum samples and nasopharyngeal swabs from healthy people were collected as controls. All clinical samples were analyzed by LAMP and PCR simultaneously. As shown in Table 3, of the 105 clinical samples, LAMP detected 33 positive samples while 23 were detected by PCR. Then, B. pertussis was successfully cultured from 12 samples, all of which had been positively identified by LAMP. The healthy control samples all tested negative in each of the assays.
The sequence analysis of the BP485 gene from the B. pertussis isolates confirmed conservation with the nucleotide sequences of reported genes. Multilocus sequence typing (MLST) analysis of B. pertussis showed that five of the seven housekeeping genes belonged to a single genotype, similar to reported global genotypes. However, the adk and tyrB housekeeping genes had two different genotypes belonging to type 1 and type 3, respectively.
To further characterize the virulence-related genes of the 12 isolated strains, the alleles of ptxA, prn, tcfA, fim2, and fim3 were analyzed. All isolates had virulence-related genes belonging to different genotypes with all four ptxA alleles, six prn alleles, four tcfA alleles, two fim2 alleles (fim2-1 and fim2-2), and three fim3 alleles (fim3-1, fim3-2, and fim3-4) present across the samples. Our data show that B. pertussis is widespread in respiratory infections, and the diverse genotypic features indicate on-going evolution.
Discussion
Pertussis, also known as whooping cough, is an acute respiratory infectious disease caused by B. pertussis, and can cause severe disease, particularly in infants. Although vaccines are widely used in most countries, the morbidity of pertussis is rising, even in developed countries with high vaccination coverage. Remarkably, many adolescents and adults become infected despite having been vaccinated as infants as a result of waning antibody titers, which can result in further transmission to children. Unfortunately, conventional wisdom holds that pertussis is a pediatric disease, leading to substantial under-diagnosis in adolescents and adults. Pertussis is highly infectious, and therefore rapid and simple methods to aid diagnosis would be advantageous in controlling disease spread.
Several diagnostic techniques have been established for detecting B. pertussis, e.g., culture, serological assays, and, most commonly, PCR-based methods. These approaches are either time-consuming or costly and frequently require skilled technicians and precision instruments. They are therefore not suitable for situations where resources are limited and fast results are required such as for point-of-care assessments. Since its introduction in 2000, a nucleic amplification method known as LAMP (26) has been widely used in various fields, including clinical diagnosis, food safety, livestock breeding, and even determination of sex (27, 28). A number of reports indicated that LAMP had a better sensitivity and specificity for detecting pathogens than other commonly used methods. Here, we described a LAMP method for detecting BP485 of B. pertussis by color change.
Although many target genes, such as IS481, have been used to diagnose pertussis, we chose BP485, which, unlike IS481, is able to distinguish B. pertussis from B. bronchiseptica and B. parapertussis (22). Specificity evaluation was performed here using 3 B. pertussis strains and 31 homologous species by both real-time turbidimeter and visual color change methods. All B. pertussis strains tested positive and all 31 non-pertussis respiratory tract pathogens, including B. bronchiseptica and B. parapertussis, tested negative. These results demonstrated that LAMP targeting BP485 had a high specificity for the detection of B. pertussis and that using turbidity or color change as readouts gave comparable results. The sensitivity of LAMP to detect BP485 was compared with traditional PCR using 10-fold serial dilution of genomic DNA extracted from B. pertussis. The detection limit of the LAMP assay used here was 1.3 pg/μl, 10-fold higher than conventional PCR, in agreement with previous reports. Furthermore, LAMP can be completed in one hour using a simple thermostat, whereas a PCR run takes approximately 3 h and requires a relatively complicated thermal cycler. LAMP is therefore a more rapid and simple technique making it better suited to field use.
Two readouts were used for the LAMP assay to detect amplification products, real-time turbidity and visual observation of color change. Detection by color change was as sensitive and specific as real-time turbidity measurements. As this color change is visible to the naked eye, it would further simplify the assay. It is worth noting that the use of a wax sealant over the reaction mixture was essential to minimize cross contamination by aerosol, which frequently resulted in false positives.
Application of the assay to samples taken from hospital admissions of cough and pneumonia indicated that B. pertussis was prevalent in the community with over 30% of samples testing positive. The cases included a 35-year-old female patient from rural China who was admitted for treatment in the city hospital after suffering with cough and pneumonia for 14 days. Her baby had contracted whooping cough from her as a result of close contact. This was an example of increasing numbers of “adult to child” transmissions, which may be a major factor in the rising incidence of pertussis.
In conclusion, the visual LAMP assay developed here for detecting B. pertussis allows for positive samples to be identified by the naked eye immediately after amplification is completed. The visual method is as sensitive and specific as real-time turbidity measurements and is more sensitive than PCR. It is a rapid, simple, and low-cost method to diagnose B. pertussis, making it particularly useful for point-of-care testing where time and resources are limited.
Author Contributions
JY and SZ helped conceive project and designed experiments. WL, YX, and HL performed and wrote the manuscript. XZ, LL, YZ, MZ, XW, XW, and SH designed and executed experiments. DD and LH helped to edit the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Christian Riedel for advice on the preparation of the manuscript and Fengjing Li for technical assistance and helpful discussions. Funding: This work was supported by a grant from the National Natural Science Foundation of China (31370093) to JY, Mega-projects of Science and Technology Research of China (Grant 2011ZX10004-001 and 2011ZX10004-205), and a grant from the National High Technology Research and Development Program of China (863 Program; grant no. SS2014AA022210).
Footnotes
References
2. Warfel JM, Beren J, Merkel TJ. Airborne transmission of Bordetella pertussis. J Infect Dis (2012) 206:902–6. doi: 10.1093/infdis/jis443
3. Güris D, Strebel PM, Bardenheier B, Brennan M, Tachdjian R, Finch E, et al. Changing epidemiology of pertussis in the United States: increasing reported incidence among adolescents and adults, 1990-1996. Clin Infect Dis (1999) 28:1230–7. doi:10.1086/514776
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Wendelboe AM, Njamkepo E, Bourillon A, Floret DD, Gaudelus J, Gerber M, et al. Transmission of Bordetella pertussis to young infants. Pediatr Infect Dis J (2007) 26:293–9. doi:10.1097/01.inf.0000258699.64164.6d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Lee GM, Lett S, Schauer S, LeBaron C, Murphy TV, Rusinak D, et al. Societal costs and morbidity of pertussis in adolescents and adults. Clin Infect Dis (2004) 39:1572–80. doi:10.1086/425006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Baughman AL, Cortese MM, Slade BA. Incidence of Bordetella pertussis infection in adolescents and adults. Clin Infect Dis (2007) 44:149–50. doi:10.1086/510081
7. Mitka M. Age range widens for pertussis vaccine: boosters advised for adolescents and adults. JAMA (2006) 295:871–2. doi:10.1001/jama.295.8.871
8. Van den Bossche D, De Bel A, De Smet D, Heylen O, Vekens E, Vandoorslaer K, et al. Prevalence of Bordetella holmesii and Bordetella bronchiseptica in respiratory tract samples from Belgian patients with pertussis-like symptoms by sensitive culture method and mass spectrometry. Acta Clin Belg (2013) 68:341–8. doi:10.2143/ACB.3341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Herwegh S, Carnoy C, Wallet F, Loïez C, Courcol RJ. Development and use of an internal positive control for detection of Bordetella pertussis by PCR. J Clin Microbiol (2005) 43:2462–4. doi:10.1128/JCM.43.5.2462-2464.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Holberg-Petersen M, Jenum PA, Mannsåker T, Melby KK. Comparison of PCR with culture applied on nasopharyngeal and throat swab specimens for the detection of Bordetella pertussis. Scand J Infect Dis (2011) 43:221–4. doi:10.3109/00365548.2010.538855
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Gao F, Mahoney JC, Daly ER, Lamothe W, Tullo D, Bean C. Evaluation of a multitarget real-time PCR assay for detection of Bordetella species during a pertussis outbreak in New Hampshire in 2011. J Clin Microbiol (2014) 52:302–6. doi:10.1128/JCM.01656-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Litt DJ, Jauneikaite E, Tchipeva D, Harrison TG, Fry NK. Direct molecular typing of Bordetella pertussis from clinical specimens submitted for diagnostic quantitative (real-time) PCR. J Med Microbiol (2012) 61:1662–8. doi:10.1099/jmm.0.049585-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Riffelmann M, Thiel K, Schmetz J, Wirsing von Koenig CH. Performance of commercial enzyme-linked immunosorbent assays for detection of antibodies to Bordetella pertussis. J Clin Microbiol (2010) 48:4459–63. doi:10.1128/JCM.01371-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Kamachi K, Toyoizumi-Ajisaka H, Toda K, Soeung SC, Sarath S, Nareth Y, et al. Development and evaluation of a loop-mediated isothermal amplification method for rapid diagnosis of Bordetella pertussis infection. J Clin Microbiol (2006) 44:1899–902. doi:10.1128/JCM.44.5.1899-1902.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Loeffelholz M. Towards improved accuracy of Bordetella pertussis nucleic acid amplification tests. J Clin Microbiol (2012) 50:2186–90. doi:10.1128/JCM.00612-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Nakamura A, Sakano T, Nakayama T, Shimoda H, Okada Y, Hanayama R, et al. Neonatal pertussis presenting as acute bronchiolitis: direct detection of the Bordetella pertussis genome using loop-mediated isothermal amplification. Eur J Pediatr (2009) 168:347–9. doi:10.1007/s00431-008-0744-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Nakamura Y, Kamachi K, Toyoizumi-Ajisaka H, Otsuka N, Saito R, Tsuruoka J, et al. Marked difference between adults and children in Bordetella pertussis DNA load in nasopharyngeal swabs. Clin Microbiol Infect (2011) 17:365–70. doi:10.1111/j.1469-0691.2010.03255.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Nat Genet (2003) 35:32–40. doi:10.1038/ng1227
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Register KB, Sanden GN. Prevalence and sequence variants of IS481 in Bordetella bronchiseptica: implication for IS481-based detection of Bordetella pertussis. J Clin Microbiol (2006) 44:4577–83. doi:10.1128/JCM.01295-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Reischl U, Lehn N, Sanden GN, Loeffelholz MJ. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J Clin Microbiol (2001) 39:1963–6. doi:10.1128/JCM.39.5.1963-1966.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Riffelmann M, Wirsing von Konig CH, Caro V, Guiso N. Nucleic acid amplification test for diagnosis of Bordetella infections. J Clin Microbiol (2005) 43:4925–9. doi:10.1128/JCM.43.10.4925-4929.2005
22. Probert WS, Ely J, Schrader K, Atwell J, Nossoff A, Kwan S. Identification and evaluation of new target sequences for specific detection of Bordetella pertussis by real-time PCR. J Clin Microbiol (2008) 46:3228–31. doi:10.1128/JCM.00386-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Mooi FR, Hallander H, Wirsing von König CH, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis (2000) 19:174–81. doi:10.1007/s100960050455
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun (2001) 289:150–4. doi:10.1006/bbrc.2001.5921
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc (2008) 3:877–82. doi:10.1038/nprot.2008.57
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res (2000) 28:E63. doi:10.1093/nar/28.12.e63
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Hirayama H, Kageyama S, Moriyasu S, Sawai K, Minamihashi A. Embryo sexing and sex chromosomal chimerism analysis by loop-mediated isothermal amplification in cattle and water buffaloes. J Reprod Dev (2013) 59:321–6. doi:10.1262/jrd.2013-028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Yang L, Li J, Bi Y, Xu L, Liu W. Development and application of a reverse transcription loop-mediated isothermal amplification method for rapid detection of duck hepatitis A virus type 1. Virus Genes (2012) 45:585–9. doi:10.1007/s11262-012-0798-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: BP485, B. pertussis, LAMP, sensitivity, specificity, rapid diagnosis, prevalence
Citation: Liu W, Xu Y, Dong D, Li H, Zhao X, Li L, Zhang Y, Wei X, Wang X, Huang S, Zeng M, Huang L, Zhang S and Yuan J (2015) Survey and rapid detection of Bordetella pertussis in clinical samples targeting the BP485 in China. Front. Public Health 3:39. doi: 10.3389/fpubh.2015.00039
Received: 28 November 2014; Accepted: 11 February 2015;
Published online: 05 March 2015.
Edited by:
Paulo Marcos Zech Coelho, Fundação Oswaldo Cruz – FIOCRUZ, BrazilReviewed by:
Li Xu, Cornell University, USAMalgorzata Anna Mikaszewska-Sokolewicz, The Medical University of Warsaw, Poland
Copyright: © 2015 Liu, Xu, Dong, Li, Zhao, Li, Zhang, Wei, Wang, Huang, Zeng, Huang, Zhang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Yuan and Liuyu Huang, Institute of Disease Control and Prevention, Academy of Military Medical Sciences, No.20 Dongda Street, Fengtai District, Beijing 100071, China e-mail: yuanjing6216@163.com; huangliuyuly@163.com;
Shumin Zhang, National Institutes for Food and Drug Control, No.2 Tiantan Xili, Beijing 100050, China e-mail: zhangsm@nifdc.org.cn
†Wei Liu, Yinghua Xu and Derong Dong have contributed equally to this work.
 Wei Liu
Wei Liu Yinghua Xu2†
Yinghua Xu2† Derong Dong
Derong Dong Huan Li
Huan Li Xiangna Zhao
Xiangna Zhao Xiao Wei
Xiao Wei Xuesong Wang
Xuesong Wang Liuyu Huang
Liuyu Huang Jing Yuan
Jing Yuan